Abstract
Background
Deoxyribonucleic acid from invasive, non-invasive and 9th week embryo can be a resource for the determination of fetal sex using highly sensitive and specific multiplex PCR.
Methods
A total of 402 DNA samples were used to test the newly developed novel multiplex PCR including male specific (3 genes: SRY, DAZ2 and TSPY1) Y-biomarkers and internal control, ACTB. The study isolated cffDNA (Cell-free fetal DNA; n = 73) from mother’s plasma, serum and urine, fetal DNA from 9th week embryo and cord blood, and fetal DNA from CD71+ve nucleated red blood cells (fNRBC; n = 73). Paternal and maternal DNA from buccal cells (n = 20) and blood (n = 232) used for male and female confirmation.
Results
The study observed that SRY alone cannot be a suitable Y-biomarker. Confirmation from any two Y-biomarkers is mandatory for male fetus identification. Direct sequencing of the gel eluted multiplex and single amplicons confirmed the specific sequences. Presence of two out of 3 Y-biomarkers OR single Y-biomarker with >1,000,000 intensity is considered positive for male. The multiplex PCR is suitable for determining sex from all source of fetal DNA including highly degraded cffDNA and can detect the sex using 0.5ng DNA. Individual marker-based real-time qPCR followed by combined melt curve analysis showed distinguished melt curve peaks for the markers.
Conclusion
The multiplex PCR achieved 100% accuracy on fetal DNA from fNRBC for early determinations (<13 weeks) of gender. The developed novel and simple multiplex PCR and individual qPCR can be adopted in all types of laboratories for determining human fetal gender using fetal DNA from fNRBC. Early identification of gender can support to prepare for possible X-linked analysis, reduce anxiety in mother, strengthen a bond between mother and fetus, and effective decision making. Non-invasive source of fetal DNA from fNRBC preferred for identifying gender to reduce the risk of invasive procedures in early (8–13 weeks) pregnancy.
Keywords: cffDNA, male PCR, invasive, non-invasive, Y biomarkers, fetal gender, fNRBC
Introduction
Determination of fetal gender in early pregnancy is highly essential to identify pregnancies at risk of X-linked inherited disorders. Over 100 X-linked inherited diseases have been discovered in humans so far, including muscular dystrophy, fragile X syndrome and hemophilia.1 Fetal gender determination tests were developed to help in the management or presentation of X-linked inherited disorders. Early fetal gender determination is accurately achieved by invasive procedures either by chorionic villus sampling in the first trimester or amniocentesis early in the second trimester.2 However, these procedures are associated with risk of pregnancy loss and are expensive which makes these protocols less acceptable to some patients.3 Therefore, many patients know fetal gender using 2D ultrasound which hold accuracy 100% as from 20 weeks of gestation.4 Yet, the high accuracy of sonography can be only achieved in late second trimester. The accuracy of ultrasound in early pregnancy is affected by the gestational age and it is operator dependent which makes the accuracy around 70% at 11 weeks of gestation.5 Therefore, it is pivotal to explore less expensive and non-invasive diagnostic methods with high accuracy rate that can be done early in the first trimester.
Fetal DNA is more reliable and can be extracted and used early in pregnancy.6 Fetal DNA can be obtained by non-invasive methods from maternal peripheral blood as cell free fetal DNA (cffDNA) or fetal DNA that circulates freely in the maternal blood,7 or by invasive procedures such as chronic villus and amniocentesis sampling.8 Fetal nucleated red blood cells (fNRBCs) are the cells circulating in peripheral blood of pregnant mother. FNRBC contains whole genome of the fetus may serve for source for sex determination.9 The availability and concentration of fetal DNA is important to have prominent results. In non-invasive methods low concentration DNA can be obtained.10 This raises the need for specific, sensitive and optimal technique for fetal gender determination. Placental extracellular vesicles in pregnant mother is a relatively new field that is growing rapidly, observed as a novel mode of communication from the fetus to its pregnant mother and can influence the maternal cell multitude.11 Exosome DNA (exoDNA) or vesicular fetal DNA is also an emerging option for determining fetal gender.12–15
Several studies have targeted the Y chromosome specific biomarkers and genes. Different Y chromosome sequences have been used for gender determination. Mainly, single-copy sex determination region Y (SRY) is well-reported.16 However, SRY is a single copy thus, not sensitive enough for fetal gender determination. A combination of multiple copies of sequences in Y chromosome such as Deleted in Azoospermia 2 gene (DAZ2) and Testis Specific Protein Y-linked 1 gene (TSPY1) also known as (DYS14) will be more sensitive and specific for gender determination. The multiplex single tube PCR assay with three Y- biomarkers presented here is sensitive and specific and can detect male fetal DNA from any source of samples using conventional multiplex PCR method.
Materials and Methods
Ethics Statement
The Institutional Review Board at Imam Abdulrahman Bin Faisal University approved the study. IRB approval number: IRB-2017-13-137 dated 07 June 2017 and extended on 14 Dec 2020.
Primers Designed
The primers were designed (Primer sequences are in the patent disclosure at US Patent office; Sequences will be available on request) for ACTB, SRY, DAZ2 and TSPY1 with 541, 769, 269, and 329–356 bp, respectively. Primers were manually strengthened for high accuracy, specificity and narrow annealing temperature difference and confirmed for its suitability using primer BLAST. The primers were synthesised from Integrated DNA Technologies, Inc. (Leuven, Belgium).
Collection of Samples
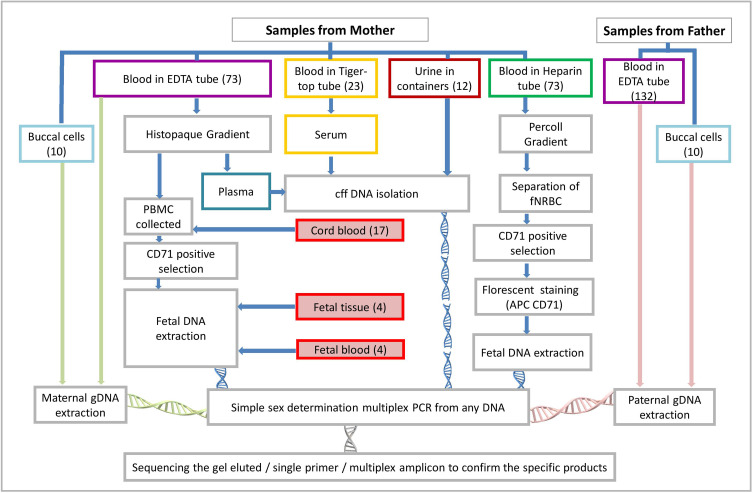
All the samples (n=402) used in the study were collected at King Fahd Hospital of the University, at Imam Abdulrahman Bin Faisal University. Detailed sources of cffDNA and fetal DNA are presented in Figure 1. Total of 402 DNA samples used in the study: miscarried fetus at 9th week of pregnancy (n=4) and in utero fetus ≤13 weeks (n=24) [cffDNA: 8 weeks = 2, 9 weeks = 2, 10 weeks = 2, 11 weeks = 2, 12 weeks = 3, 13 weeks = 1; fetal DNA from fNRBCs: 8 weeks = 2, 9 weeks = 2, 10 weeks = 2, 11 weeks = 2, 12 weeks = 3, 13 weeks = 1]. In utero fetus >13 weeks (n = 122) [cffDNA = 61; fetal DNA from fNRBCs = 61]. Furthermore, adult gDNA from buccal cells (n = 20; 10♀, 10♂) and blood samples (n = 232; 100♀, 132♂) were also used to authenticate the single tube novel multiplex PCR. Maternal blood (3 to 5 mL) and cord blood (3 to 5 mL) samples were collected in lithium heparin or EDTA vacutainer from women at delivery. Samples were processed 8–10 hours of collection for cffDNA extraction. Serum (n=23) samples were collected in tubes without clot activator or gel, and urine (1 to 2 oz) (n=12) samples collected in sterile containers. Urine samples were centrifuged at 3000 rpm for 10 min in 20–24 °C twice to remove cells then stored at −20 °C for cffDNA extraction. Tissue (n=4) (separated cautiously from maternal tissue to avoid contamination) samples from the miscarriage at the 9th week of pregnancy and blood samples were collected from the fetus. Miscarriage sample was collected in RNAprotect Cell Reagent (Qiagen, Hilden, Germany). Except four miscarried fetus all the other samples are in utero fetus. All the sample were collected as per the standard protocols after receiving signed informed consent.
Figure 1.
Flow chart of the sample collection and source of DNA for identifying fetal sex.
Abbreviations: PBMC, peripheral blood mononuclear cell; gDNA, genomic deoxyribonucleic acid; cff DNA, cell-free fetal deoxyribonucleic acid; PCR, polymerase chain reaction; fNRBC, fetal nucleated red blood cell.
Fetal Cells Isolation
Maternal and cord blood samples were subjected to density gradient centrifugation using histopaque −1077. Centrifugation was performed at 1430 rpm for 45 min. The plasma was collected in 1.5 mL tubes and cell layer between plasma and histopaque was recovered then washed with PBS. For positive selection of anti-CD71, cells were incubated for 30 min at 4–8 °C with a 1:10 CD71 MicroBeads, human (Miltenyi Biotec, Bergisch Gladbach, Germany). Magnetic selection was achieved using the mini-MACS system (Miltenyi Biotec). Double gradient isolation method was also applied for the separation of fNRBCs.17
DNA Extraction
DNA from fetal blood was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to manufacturer instructions. DNA from fetal tissue was extracted using Puregene Cell and Tissue kit (Qiagen, Germany) according to manufacturer protocol. cffDNA from maternal plasma, serum and urine were extracted using the QiAamp MinElute ccfDNA mini Kit (Qiagen, Germany). Concentrations of extracted DNA was estimated using nanodrop (Nanodrop 8000, Thermo Scientific, DE, USA) and ensured the quantity >0.5 ng/µL and quality A260/A280 ratio of 1.8±0.2. Approximately 5% of the samples were quantified using Qubit 2.0 fluorometer (Qubit™ dsDNA HS Assay Kits; Life Technologies, CA, USA).
Gender Specific Multiplex PCR
Individual PCR for the amplicons of ACTB (541 bp), SRY (769 bp), DAZ2 (269 bp) and TSPY1 (329–356 bp) were completed before designing the multiplex recipe. The PCR amplification reactions were set a total volume of 25 µL containing: Absolute master mix 12.5 µL (MOLEQULE-ON, Auckland, New Zealand), Primer 1 µL (10 nM) SRYaF, Primer 1 µL (10 nM) SRYaR, Primer 1 µL (10 nM) ActinF, Primer 1 µL (10 nM) ActinR, Primer 1 µL (10 nM) DYS14F, Primer 1 µL (10 nM) DYS14R, Primer 1 µL (10 nM) DAZ2aF, Primer 1 µL (10 nM) DAZ2aR, Template DNA 25 ng and Dis H2O to 25 µL. The PCR thermal cycling for the multiplex: Step 1: 95 °C for 10 minutes, Step 2: 95 °C for 1 minute, Step 3: 68.3 °C for 1 minute, Step 4: 72 °C for 1 minute, Step 5: Go to step 2, 35 cycles, Step 6: 72 °C for 5 minutes, Step 7: Store at 4 °C. Amplicons were visualized using 2% agarose electrophoresis at 100 volt for one hour.
Real-Time PCR (qPCR) Amplification
The same primers were used individually in Real-Time PCR (qPCR) amplification, to validate the specificity of these primers in qPCR. Samples from different maternal sources and male and female samples as positive and negative controls were analyzed through qPCR assay using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA) with 5 to 10 ng of DNA and performed in duplicates. The reactions were carried out on a 7500 Fast Real-Time PCR system (Applied Biosystems, USA), using the 7500 Software v2.0.6 (Applied Biosystems, Life Technologies). Amplification method: 50 °C/2 minute; 95 °C/10 minutes; 45 amplification cycles including a denaturation step at 95 °C/15 seconds, annealing at 60 °C/1 minute and a extension at 72 °C/1 minute. Melting Curve stage were performed as follow: 95 °C/15 seconds, 60 °C/1 minute to 95/1 minute with 0.5% increase.
Results
The designed primer pairs for ACTB (541 bp), SRY (769 bp), DAZ2 (269 bp) and TSPY1 (329–356 bp) genes were tested for the specific application of the respective gene in the male and no non-specific amplification in female at the specific temperature (Supplementary File 1). All the four pairs of primers were amplified individually to confirm the amplicons free from non-specific amplicons in female (n=110; buccal cells=10; blood=100) and male (n=142; buccal cells=10; blood=132) samples with gDNA (Supplementary File 1). The temperature gradient multiplex PCR was done to confirm the annealing temperature (68.3°C) with specific amplicons and without non-specific amplicons. Amplicons of concentration dependent multiplex PCR confirms the stable specific products in various concentration of DNA. Multiplex PCR based amplification from various samples like paternal DNA from father’s blood, maternal DNA from mother’s blood, paternal DNA from father’s buccal cells, maternal DNA from mother’s buccal cells, cffDNA from maternal urine, cffDNA from maternal serum, cffDNA from maternal plasma, cffDNA from cord blood, fetal DNA from fetal blood, fetal DNA from fetal tissue, and fetal DNA from fetal fNRBCs confirms the possibility and specificity (Figure 2). Hundred percentage accuracy was achieved on identifying the gender in the adult gDNA [♀n=110; buccal cells=10; blood=100 and ♂ n=142; buccal cells=10; blood=132).
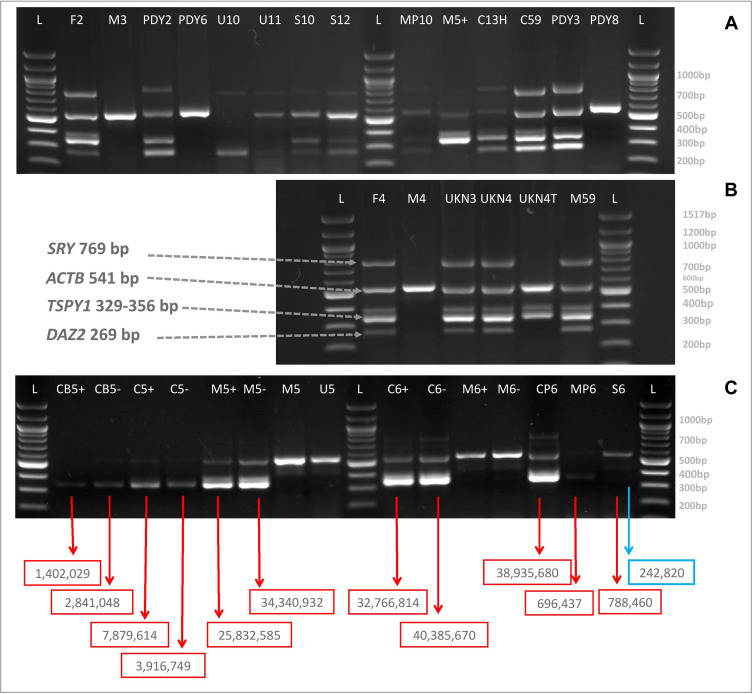
Figure 2.
(A and B) Representatives of the multiplex amplicons of DNA from various resources. (C) Intensity of amplicons of single Y marker. Red box indicates the acceptable intensities. Blue box indicates the intensity less than acceptable intensities.
Abbreviations: L, 100 bp ladder; F, paternal DNA from father blood; M, maternal DNA from mother blood; PDY3, paternal DNA from father’s buccal cells; PDY8, maternal DNA from mother’s buccal cells; U, cffDNA from maternal urine; S, cffDNA from maternal serum; MP, cffDNA from maternal plasma; C, fetal DNA of fetal cell isolated from cord blood; UKN3 and UKN4, fetal DNA from fetal blood; UKN4 TISSUE, fetal DNA from fetal tissue; M59, fetal DNA of fetal cell isolated from maternal blood; C59, fetal DNA of fetal cell isolated from cord blood; M5+ and C13H, fetal DNA from CD71+ve cells from cord blood.
Presence of two amplicons out of 3 Y markers shall be considered positive for male. Some of the amplicons were observed very faint and not able to decide for their presence. In order to confirm the presence of amplicons and to avoid human intervention on deciding the presence or absence of amplicon, the intensity of amplicons of single Y-biomarker were calculated using Image Lab (Bio-Rad, USA) (Figure 2). Single Y-biomarker amplicon with >1,000,000 intensity is considered to be positive for male. cffDNA from urine (n=2; >13 weeks of pregnancy) positive for two Y-biomarkers, while they are negative for ACTB gene. Fetal DNA (n=1; <13 weeks of pregnancy) from NRBC’s were also positive for two Y-biomarkers, while they are negative for ACTB.
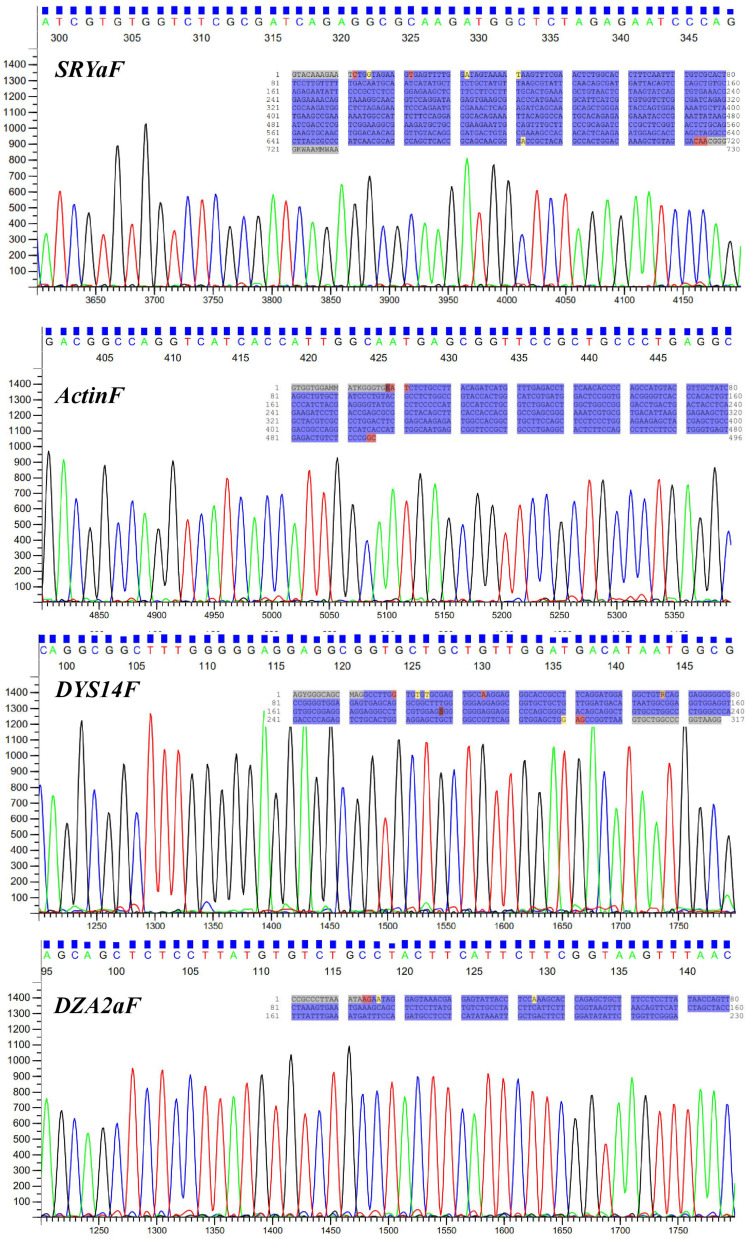
Individual PCR amplicons and gel-eluted amplicons of multiplex PCR were confirmed for the specific gene products using direct sequencing (Figure 3). The gel-eluted amplicons of SRY, ACTB, DAZ2 and TSPY1 genes from multiplex PCR were confirmed for the specific sequence and absence of background noise. For early gender determinations ≤13 weeks of pregnancy and the >13 weeks of pregnancy, the multiplex PCR obtained 100% accuracy on fetal DNA from fNRBC. However, the cffDNA from plasma (n=36), serum (n=23) and urine (n=12) showed 83.56% of accuracy on determining the gender. Among the plasma, serum and urine, the study observed little high accuracy on cffDNA from plasma (Table 1).
Figure 3.
Electropherograms of gel-eluted amplicons of multiplex PCR. Electropherograms generated from the forward primers of the multiplex PCR amplicons. Inside blue highlighted sequence indicates the complete sequences of the amplicons.
Table 1.
Distribution of Samples with Number of Y-Biomarker
| Weeks of Pregnancy | Source of DNA | Number of Sample | Failed to Detect | 1Y-Biomarker | ≥2Y-Biomarker | Female |
|---|---|---|---|---|---|---|
| ≤13 weeks | cffDNA | 12 | 6 | 2 | 0 | 4 |
| Fetal DNA from fNRBCs | 12 | 0 | 1 | 7 | 4 | |
| >13 weeks | cffDNA | 61 | 6 | 23 | 26 | 6 |
| Fetal DNA from fNRBCs | 61 | 0 | 25 | 31 | 5 |
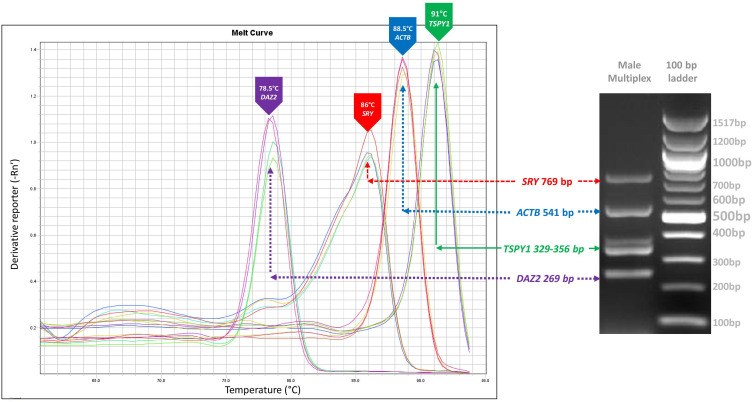
qPCR results showed high specificity for the primers to detect male fetal DNA in the tested samples (Figure 4). The melt curve analysis for individual markers (Y-biomakers and housekeeping) revealed that the derivative of fluorescence vs temperature shows distinguished melt curve peaks for DAZ2, SRY, ACTB and TSPY1 genes at 78.5 °C, 86 °C, 88.5 °C, and 91 °C, respectively. Combined analysis melt curve analysis for individual markers with any one Y-biomakers along with housekeeping gene considered.
Figure 4.
qPCR melt curve analysis of 3 Y markers (SRY, DAZ2 and TSPY1) and housekeeping (ACTB) genes.
Discussion
The ability to determine the fetus’s gender early in pregnancy was an enormous leap in prenatal diagnosis, specifically in the prevention or management of X-linked inherited disorders. The easiest method to determine fetal gender is massive parallel sequencing (MPS).18 However, it will be expensive to subject all pregnancies with risk of X-linked inherited disorders to MPS. The developed single tube multiplex PCR in the study is more cost effective. The use of SRY, TSPY1 and DAZ are well studied19–21 and showed high sensitivity for fetal gender determination, mostly through Real-Time PCR. However, good concentration of DNA was needed. A major advantage of the present study is that it can detect low concentration of DNA (< 5 ng/µL). The present study included DNA from all the possible samples either from the mother or fetus, both in invasive and non-invasive samples. The study isolated the cffDNA (Cell-free fetal DNA) from mother’s plasma, serum and urine, fetal DNA from 9th week embryo and cord blood, fetal DNA from the nucleated red blood cells (positive for CD71). Paternal and maternal DNA from buccal cells were used as the positive DNA source for male and female, respectively. Presence of two out of 3 Y-biomarkers OR single Y-biomarker with >1,000,000 intensity is considered positive for male. The fetal gender was confirmed by ultrasound after 20 weeks of pregnancy or at birth. The multiplex PCR is an appropriate methodology for determining sex from all source of fetal DNA including highly degraded cffDNA, however accuracy is not very high.
According to our study single non-invasive maternal cffDNA sample is not sufficient to determine fetal gender with highest accuracy. From different periods of pregnancy male DNA was detected in only 61 out of 73. This results comes in contrary with the scientific literature that reports higher sensitivity in early pregnancy (87.5–99.1%) and it increases as the pregnancy progress (97–100%).20,22 This is due to the use of less volume of sample. Present study used only <1 mL of plasma or serum is being used to extract cffDNA due to the less availability of blood samples. If more volume of blood samples, then we can have higher accuracy due to the availability of high cffDNA. However, we are not able to get more blood due to restriction in the IRB approval. In a study conducted by Kazachkova et al, the failure to detect Y chromosome biomarkers in cffDNA is influenced by gestational age coupled with low cffDNA concentration in maternal samples.19 In addition, type of PCR used is proposed by Costa et al and Levi et al as an influence on detecting the Y chromosome.23,24
Fetal DNA from fNRBC are the most suitable for early determination of fetal gender as it shows 100% accuracy in early pregnancy samples. Two studies used different techniques to enrich fNRBC: Byeon et al, used double enrichment process using RBC hyperaggregation and WBC depletion and Zhang et al, employ microfluidic chip.25,26 Both studies reported full accuracy in detected Y chromosome in the enrichment fNRBC as like the present study using double enrichment process.17
The multiplex PCR correctly identified the gender of the fetuses in mothers carrying male fetus (n=67) and female (n=6) fetus using fetal DNA from fNRBC. The gender was confirmed either by ultrasound after 20 weeks of pregnancy or at birth. The multiplex PCR has 100% accuracy identifying fetal gender from all adult sample.
The gender of four fetal tissue samples was determined using multiplex PCR. One of the fetal tissue reported with homozygous ASIC5Saudi mutation.27 As the fetal tissue samples were collected from aborted 9-week fetuses therefore clinical confirmation of the gender of the fetus could not be achieved. Early prenatal diagnosis of gender and identification of disease associated mutations can provide effective decision making in the mother among high-consanguinity population to decide within the allowed time.27,28 The eight pairs of primers designed in the study is highly specific, sensitive, unique and well matched for the simultaneous detection of the male specific Y-biomarkers, SRY, DAZ2 and TSPY1 genes and the internal control, ACTB gene for both conventional PCR and real-time qPCR. The specificity of the primers were confirmed through sequencing. This can lower expenses, increase accuracy due to 3Y biomarkers, minimize the risk for invasive sampling,16 simplify the process because of the single tube, and can be adoptable for the low income regions and laboratories with minimal facilities.
Conclusion
The gender of adult gDNA was correctly identified with 100% accuracy. The Y chromosome based markers using the presently developed novel multiplex PCR can be a reliable methodology for the determination of fetal sex using invasive and non-invasive source of fetal DNA, with 100% accuracy on fetal DNA from fNRBC, which cannot be done in the first trimester by means of ultrasonography due to the incomplete external genitalia. The shortest possible time is sufficient to determine using single tube multiplex PCR with 3 Y-biomarkers or individual real-time qPCR. The study confirms the gender determination procedure using fNRBC from male-bearing and female-bearing pregnancies with full accuracy. The study confirms the application of non-invasive and invasive source of fetal DNA for the early identification of gender, which can help to reduce depression and anxiety in mother, can strengthen a bond between mother and fetus, prepare for possible X-linked analysis, effective decision making, and can reduce the risk of invasive procedures.
Acknowledgments
The authors thank the Dean, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia for her continuous support and encouragement. Authors thank Mr. Ranilo. M. Tumbaga, Mr. Horace T. Pacifico, and Mrs. Jee Entusiasamo Aquino for their assistance. This study was supported by The Deanship of Scientific Research, Imam Abdulrahman Bin Faisal University (To Dr J Francis Borgio, Grant No: 2017-100-IRMC). The authors thank the Institute for Research and Medical Consultations (IRMC) for instrumentation and facilities.
Data Sharing Statement
All data will be available on reasonable request from the corresponding author.
Ethics Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board (IRB) at Imam Abdulrahman Bin Faisal University. IRB approval number: IRB-2017-13-137 dated 07 June 2017 and extended on 14 Dec 2020.
Disclosure
Dr J Francis Borgio report a patent US Patent office pending to Not yet filed (Multiplex PCR Methodology for identifying sex in early pregnancy samples). The authors declare no other conflicts of interest in this work.
References
- 1.Germain DP. General aspects of X-linked diseases. In: Mehta A, Beck M, Sunder-Plassmann G, editors. Fabry Disease: Perspectives from 5 Years of FOS. ISBN 978-1-903539-03-3. Oxford:Oxford PharmaGenesis; 2006. [PubMed] [Google Scholar]
- 2.Colmant C, Morin-Surroca M, Fuchs F, Fernandez H, Senat M-V. Non-invasive prenatal testing for fetal sex determination: is ultrasound still relevant? Eur J Obstet Gynecol Reprod Biol. 2013;171:197–204. doi: 10.1016/j.ejogrb.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 3.Bakker M, Birnie E, Pajkrt E, Bilardo CM, Snijders RJM. Low uptake of the combined test in the Netherlands - which factors contribute?. Prenat Diagn. 2012;32:1305–1312. doi: 10.1002/pd.4001 [DOI] [PubMed] [Google Scholar]
- 4.Gelaw SM, Bisrat H. The role of ultrasound in determining fetal sex. Ethiop J Health Dev. 2012;25:6. [Google Scholar]
- 5.Manzanares S, Benítez A, Naveiro-Fuentes M, López-Criado MS, Sánchez-Gila M. Accuracy of fetal sex determination on ultrasound examination in the first trimester of pregnancy: fetal sex determination in first trimester. J Clin Ultrasound. 2016;44:272–277. doi: 10.1002/jcu.22320 [DOI] [PubMed] [Google Scholar]
- 6.Rijnders RJP, Van Der Luijt RB, Peters EDJ, et al. Earliest gestational age for fetal sexing in cell-free maternal plasma. Prenat Diagn. 2003;23:1042–1044. doi: 10.1002/pd.750 [DOI] [PubMed] [Google Scholar]
- 7.Drury S, Hill M, Chitty LS, One C. Cell-free fetal DNA testing for prenatal diagnosis. In: Makowski GS, editor. Advances in Clinical Chemistry. Vol. 76. ISBN 0065-2423. Elsevier; 2016:1–35. [DOI] [PubMed] [Google Scholar]
- 8.Sillence KA, Roberts LA, Hollands HJ, et al. Fetal sex and RHD genotyping with digital PCR demonstrates greater sensitivity than real-time PCR. Clin Chem. 2015;61:1399–1407. doi: 10.1373/clinchem.2015.239137 [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Cheng L, Sun Y, et al. Enhanced isolation of fetal nucleated red blood cells by enythrocyte-leukocyte hybrid membrane-coated magnetic nanoparticles for noninvasive pregnant diagnostics. Anal Chem. 2021;93:1033–1042. doi: 10.1021/acs.analchem.0c03933 [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhu Z, Gao Y, et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod Sci. 2015;22:1429–1435. doi: 10.1177/1933719115584445 [DOI] [PubMed] [Google Scholar]
- 11.Tong M, Chamley LW. Placental extracellular vesicles and feto-maternal communication. Cold Spring Harb Perspect Med. 2015;5:a023028. doi: 10.1101/cshperspect.a023028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Repiská G, Konečná B, Shelke GV, Lässer C, Vlková BI, Minárik G. Is the DNA of placental origin packaged in exosomes isolated from plasma and serum of pregnant women? Clin Chem Lab Med. 2018;56:e150–e153. doi: 10.1515/cclm-2017-0560 [DOI] [PubMed] [Google Scholar]
- 13.Konečná B, Tóthová Ľ, Repiská G. Exosomes-associated DNA-new marker in pregnancy complications? Int J Mol Sci. 2019;20:E2890. doi: 10.3390/ijms20122890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaşa B, Şahin O, Öcüt E, Seven M, Sözer S. Assessment of fetal rhesus D and gender with cell-free DNA and exosomes from maternal blood. Reprod Sci. 2021;28:562–569. doi: 10.1007/s43032-020-00321-4 [DOI] [PubMed] [Google Scholar]
- 15.Czernek L, Düchler M. Exosomes as messengers between mother and fetus in pregnancy. Int J Mol Sci. 2020;21:E4264. doi: 10.3390/ijms21124264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill M, Finning K, Martin P, et al. Non-invasive prenatal determination of fetal sex: translating research into clinical practice. Clin Genet. 2011;80:68–75. doi: 10.1111/j.1399-0004.2010.01533.x [DOI] [PubMed] [Google Scholar]
- 17.Nemescu D, Constantinescu D, Gorduza V, Carauleanu A, Caba L, Navolan DB. Comparison between paramagnetic and CD71 magnetic activated cell sorting of fetal nucleated red blood cells from the maternal blood. J Clin Lab Anal. 2020;34. doi: 10.1002/jcla.23420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda Y, Kato T, Kato A, et al. Potentially effective method for fetal gender determination by noninvasive prenatal testing for X‐linked disease. Congenit Anom. 2019;59:88–92. doi: 10.1111/cga.12302 [DOI] [PubMed] [Google Scholar]
- 19.Kazachkova N, Gontar J, Verlinsky O, Ilyin I. Successful early fetal sex determination using cell-free fetal DNA isolated from maternal capillary blood: a pilot study. Eur J Obstet Gynecol Reprod Biol X. 2019;3:100038. doi: 10.1016/j.eurox.2019.100038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boggian FCTS, Pinto ALC, Silvestre MA, Jaime JC, Silva KSF, Silva CTX. Research article noninvasive pre-natal diagnosis of sex by maternal cell-free plasma fetal DNA analysis. Genet Mol Res. 2020;19. doi: 10.4238/gmr18559 [DOI] [Google Scholar]
- 21.Khorram Khorshid HR, Zargari M, Sadeghi MR, Edallatkhah H, Shahhosseiny MH, Kamali K. Early fetal gender determination using real-time PCR analysis of cell-free fetal DNA during 6th–10th weeks of gestation. Acta Med Iran. 2013;51:209–214. [PubMed] [Google Scholar]
- 22.Casanova J, Jacob C, Milot H, Cacia S. Accurate fetal sex determination from maternal blood at 8 weeks gestation. IPCB. 2019;5:135–137. doi: 10.15406/ipcb.2019.05.00164 [DOI] [Google Scholar]
- 23.Costa JM, Benachi A, Gautier E, Jouannic JM, Ernault P, Dumez Y. First-trimester fetal sex determination in maternal serum using real-time PCR. Prenat Diagn. 2001;21:1070–1074. doi: 10.1002/pd.219 [DOI] [PubMed] [Google Scholar]
- 24.Levi JE, Wendel S, Takaoka DT. Determinação pré-natal do sexo fetal por meio da análise de dna no plasma materno. Rev Bras Ginecol Obstet. 2003;25:687–690. doi: 10.1590/S0100-72032003000900011 [DOI] [Google Scholar]
- 25.Byeon Y, Ki C-S, Han K-H. Isolation of nucleated red blood cells in maternal blood for non-invasive prenatal diagnosis. Biomed Microdevices. 2015;17:118. doi: 10.1007/s10544-015-0021-3 [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Yang Y, Li X, et al. Frequency-enhanced transferrin receptor antibody-labelled microfluidic chip (FETAL-Chip) enables efficient enrichment of circulating nucleated red blood cells for non-invasive prenatal diagnosis. Lab Chip. 2018;18:2749–2756. doi: 10.1039/c8lc00650d [DOI] [PubMed] [Google Scholar]
- 27.Al Qahtani NH, AbdulAzeez S, Almandil NB, et al. Whole-genome sequencing reveals exonic variation of ASIC5 gene results in recurrent pregnancy loss. Front Med. 2021;8:699672. doi: 10.3389/fmed.2021.699672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.AbdulAzeez S, Al Qahtani NH, Almandil NB, et al. Genetic disorder prenatal diagnosis and pregnancy termination practices among high consanguinity population, Saudi Arabia. Sci Rep. 2019;9:17248. doi: 10.1038/s41598-019-53655-8 [DOI] [PMC free article] [PubMed] [Google Scholar]