Abstract
Over 90% of the COVID-19 patients manifest mild/moderate symptoms or are asymptomatic. Although comorbidities and dysregulation of immune response have been implicated in severe COVID-19, the host factors that associate with asymptomatic or mild infections have not been characterized. We have collected serial samples from 23 hospitalized COVID-19 patients with mild symptoms and measured the kinetics of SARS-CoV-2 viral load in respiratory samples and markers of inflammation in serum samples. We monitored seroconversion during the acute phase of illness and quantitated the amount of total IgG against the receptor-binding domain of SARS-CoV-2 and estimated the virus neutralization potential of these antibodies. Viral load decreased by day 8 in all the patients but the detection of viral RNA in saliva samples did not correlate well with viral RNA detection in nasopharyngeal/oropharyngeal swab samples. 25% of the virus-positive patients had no detectable neutralizing antibodies in the serum and in other cases, the efficiency of antibodies to neutralize SARS-CoV-2 B1.1.7 strain was lower as compared to the circulating virus isolate. Decrease in viral load coincided with increase in neutralizing antibodies and interferon levels in serum. Most patients showed no increase in inflammatory cytokines such as IL-1β or IL-6, however, elevated levels of IL-7 and other inflammatory mediators such as TNF-α and IL-8 was observed. These data suggest that most mild infections are associated with absence of inflammation coupled with an active innate immune response, T-cell activation and neutralizing antibodies.
Keywords: SARS-CoV-2, COVID-19, Antibodies, PRNT, Cytokines, Inflammation
1. Introduction
Coronavirus disease of 2019 (COVID-19) has infected close to 150 million people worldwide leading to over 3 million deaths (as on 28th April 2021). Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a positive-sense RNA virus from the family Coronaviridae, is the causative agent of COVID-19. The RNA genome of SARS-CoV-2 ranges from 26 to 32 kb in length and similar to other coronaviruses that have jumped species to infect humans, SARS-CoV-2 is believed to have been in circulation in animals such as bats for several decades before infecting humans [1]. The genome of SARS-CoV-2 shares about 96% identity with that of bat coronavirus and is about 80% identical to the SARS-CoV-1 [2]. There are close to 300 vaccines under development for COVID-19 and few of the vaccines have already been approved for human use [3]. Efforts to repurpose drugs and to manage the cytokine storm have identified a number of drugs that have been recommended for use in COVID-19 [4].
Most of the patients with COVID-19 exhibit mild or no symptoms and severe clinical symptoms are observed in patients in the older age group (>65 years) or patients with underlying comorbidities such as hypertension and diabetes [5]. Impaired immune response and cytokine storm has been implicated in clinical manifestations observed in severe disease [6, 7, 8]. Increased viral load has been shown to correlate with disease severity [6, 9] and some of the antivirals such as remdesivir which inhibit the viral RNA-dependent-RNA-polymerase of SARS-CoV-1, Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and SARS-CoV-2 has been shown to reduce time to recovery and mortality in COVID-19 patients [10]. As is the case with RNA viruses, coronaviruses accumulate mutations and evolve at a rate similar to other RNA viruses [11, 12]. Variations in the viral genome and emergence of novel clades has been implicated in increased fitness and transmission [13] and recent emergence of SARS-CoV-2 variants which escape neutralization have been linked to increased transmission, severe disease and higher mortality [14, 15, 16, 17]. In this study, we monitored the kinetics of viral load in respiratory samples and saliva and markers of inflammation in the serum of hospitalized COVID-19 patients who displayed mild/moderate symptoms and recovered from illness. Our results suggest a clear inverse correlation between neutralizing antibody titers, interferon response with viral load and identifies inflammatory mediators in the serum of COVID-19 patients with mild/moderate symptoms. We also show that the serum samples from patients who were infected during early phases of the pandemic last year showed reduced neutralization capacity against B.1.1.7 SARS-CoV-2 variant.
2. Results
2.1. Kinetics of viral load in NP/OP and saliva samples in mild/asymptomatic COVID-19 patients
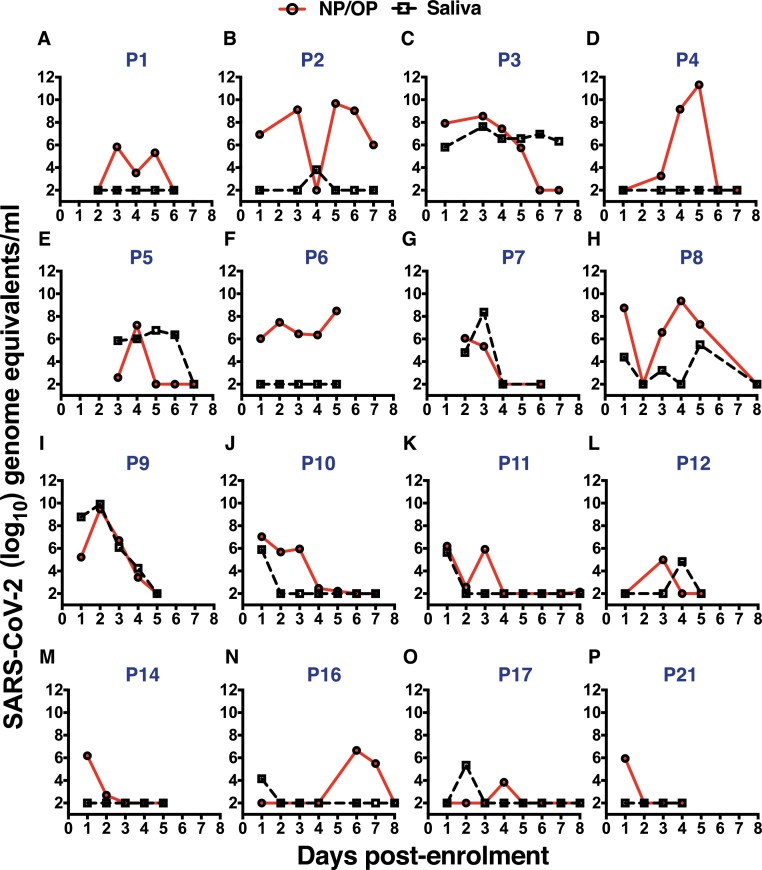
The median age of the patients was 35 (26–45; 25th and 75th percentile) and the mean duration of hospitalization was 12.4 ± 4.9 days (Mean ± SD). Out of the 23 patients 14 were male and 9 were female. We measured daily viral load in the NP/OP samples and saliva samples by RT-PCR from patients depending on the consent for sampling. We collected saliva samples early in the morning for estimation of viral load in saliva and compared with the NP/OP sample. Viral load in NP/OP swab samples peaked between day 2 to 5 in almost all patients, and in over half of the patients we observed a biphasic increase in viral RNA levels (Fig. 1 ). The peak viral RNA levels, as measured by the copy numbers of the N gene, ranged from 106 to 1012 copies per ml of virus transport medium. Surprisingly, 7 of the 23 samples were RT-PCR negative in subsequent serial sampling (Supplementary Figure S1) suggesting a false-positive RT-PCR test or other unknown reasons for negative RT-PCR results. In the remaining 16 patients viral RNA levels peaked between 3 and 5 days and was below the level of detection by day 8 (Fig. 1). Saliva has been proposed as an alternate sample for SARS-CoV-2 diagnosis and previous reports have shown good correlation between NP/OP samples and saliva for detecting viral RNA. Therefore, we collected saliva samples from all 23 samples to verify the suitability of saliva as a diagnostic specimen. Detection of viral RNA in saliva was inconsistent and no viral RNA was detected in 5 of the 16 saliva samples with true-positive NP/OP RT-PCR (Fig. 1A, 1D, 1F, 1M and 1P). Saliva samples from false-positive NP/OP RT-PCR samples were negative (Supplementary Figure S1). Incidentally, previous reports have also reported 13–15% lower positivity rates in saliva samples as compared to NP/OP samples [18, 19] suggesting a need for establishing standardized sampling protocols, transport and isolation procedures for making saliva as a reliable alternative to NP/OP samples.
Fig. 1.
SARS-CoV-2 RNA detection in serial samples of hospitalized patients.(A-P) Total RNA was isolated from virus transport medium containing nasopharyngeal/oropharyngeal swabs or from saliva samples collected on indicated days from hospitalized COVID-19 patients. SARS-CoV-2 N gene was detected by quantitative RT-PCR. Respective patient numbers are indicated.
2.2. Kinetics of antibody generation in mild/asymptomatic COVID-19 patients
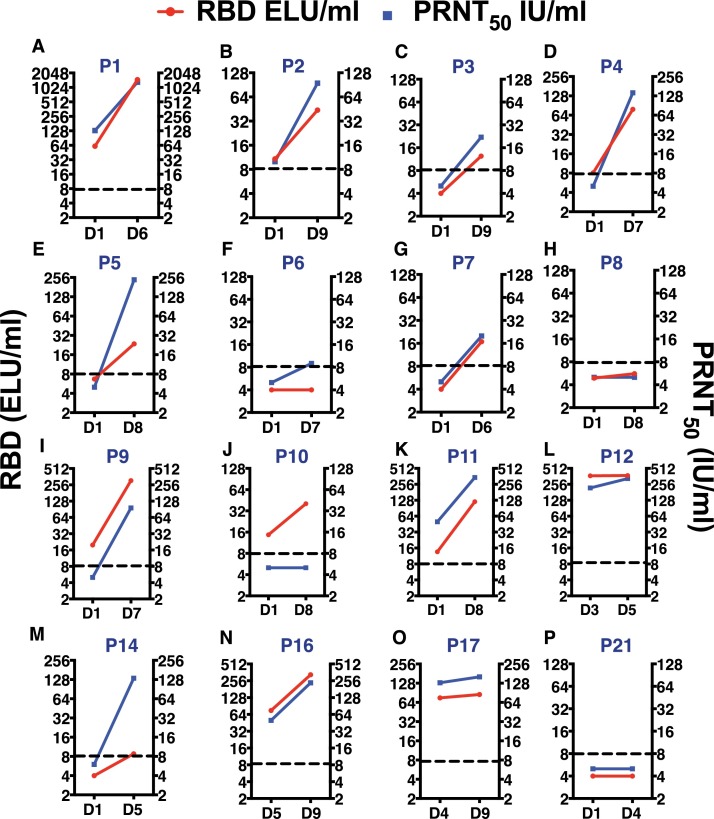
We next looked for the presence of SARS-CoV-2 antibodies (total IgG) in paired serum samples collected at the time of enrolment, and last sample collected between day 4 to day 9 post-admission. 21 of the 23 patients consented for providing blood samples for at least two time points. Antibodies to RBD-domain of SARS-CoV-2 Spike protein was measured by an in-house quantitative ELISA [20]. None of the patients who were negative by RT-PCR subsequent to hospitalization showed any presence of anti-RBD IgGs (Supplementary Figure S1) further reiterating that these patients were likely to be false positives in RT-PCR. Median RBD antibody titers in virus positive patients on the last sample collected was 42.15 ELU/ml (9.7 - 259.6; 25th and 75th percentile respectively). 7 of the 16 patients had antibodies against the RBD in the first sample collected after hospitalization suggesting that these patients had been infected for at least a week or more (Fig. 2 A, 2I-L, 2 N and 2O) and these patients had relatively lower levels of viral RNA (Fig. 1A, 1I-L, 1 N and 1O) compared to samples where the antibody levels were below the level of detection clearly suggesting virus neutralization by these antibodies. Interestingly, 3 of the 16 patients who were RT-PCR positive in serial sampling (106–108 copy numbers/ml at peak) did not have detectable levels of IgGs in their serum samples (Fig. 2F, 2H and 2P) and two of these patients had a relatively prolonged RT-PCR positivity (Fig. 1F and 1H). It is possible that these patients were in the early stages of illness and yet to generate antibodies.
Fig. 2.
Quantitation of RBD antibodies and PRNT50 titers. (A-P) Serum samples collected on the day-of-admission (DOA) and day-of-discharge (DOD) from the hospital were used to estimate antibodies against the RBD of SARS-CoV-2 spike protein by quantitaive ELISA. The same sample was also used to determine the PRNT50 titers using wild type virus neutralization assay. Data was expressed as international units as the reference reagent was calibrated against the WHO international reference reagent.
2.3. Assessing neutralizing capacity of antibodies
We next assessed whether the antibodies detected in these patients were capable of neutralizing the wild type virus isolated from one of the patients. We performed plaque reduction neutralization titre (PRNT) assay on the same paired samples that were used for RBD ELISA (Fig. 2). None of the virus-negative and RBD-negative samples had any detectable neutralizing antibodies (Supplementary Figure S1). All but one sample (Fig. 2J) which was positive for RBD antibody was capable of neutralizing SARS-CoV-2 in the PRNT assay (Fig. 2A-E, 2 G, 2I, 2 K and 2M-O) showing a strong correlation between RBD antibodies, virus neutralization and recovery from illness in concordance with previous reports [21]. Median PRNT50 titers in virus-positive patients on the last sample collected was 115 IU/ml (11.8 - 235.8; 25th and 75th percentile respectively). The three virus-positive, RBD-negative samples were also negative for virus neutralization in the PRNT assay (Fig. 2F, 2H and 2P). Nevertheless, these patients recovered from illness and resolved the viral load indicating a role for mucosal immune responses and T-cell responses in resolving the infection. Recent emergence of SARS-CoV-2 variants which escape neutralization by pre-existing antibodies from natural infection or vaccination is a major concern to tackle the ongoing pandemic [16, 17]. We next determined whether the neutralizing antibodies from COVID-19 patients collected from this study in May 2020 were capable of neutralizing the B.1.1.7 variant. We determined the PRNT50 titers for the Indian and B.1.1.7 isolates and found that the antibodies from previous infection had a significantly lower PRNT50 values for B.1.1.7 variant (Median titers of 143 vs 76) (Supplementary Figure S2).
2.4. Kinetics of inflammatory mediators in mild/asymptomatic COVID-19 patients
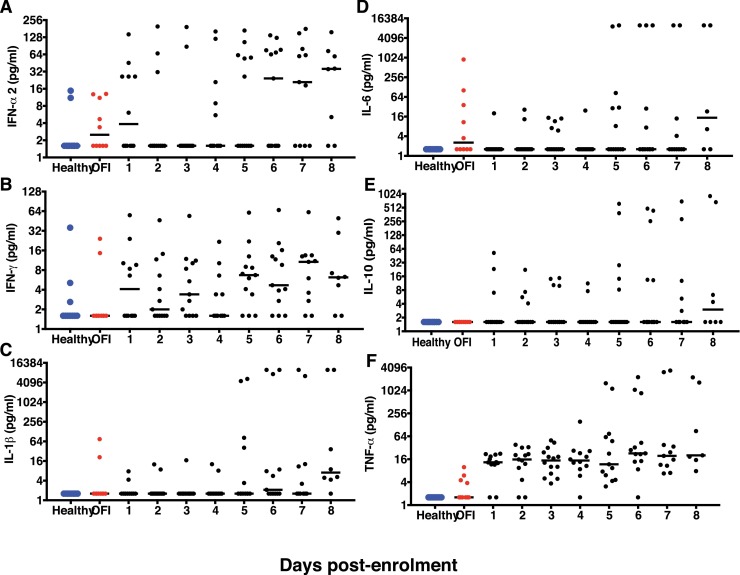
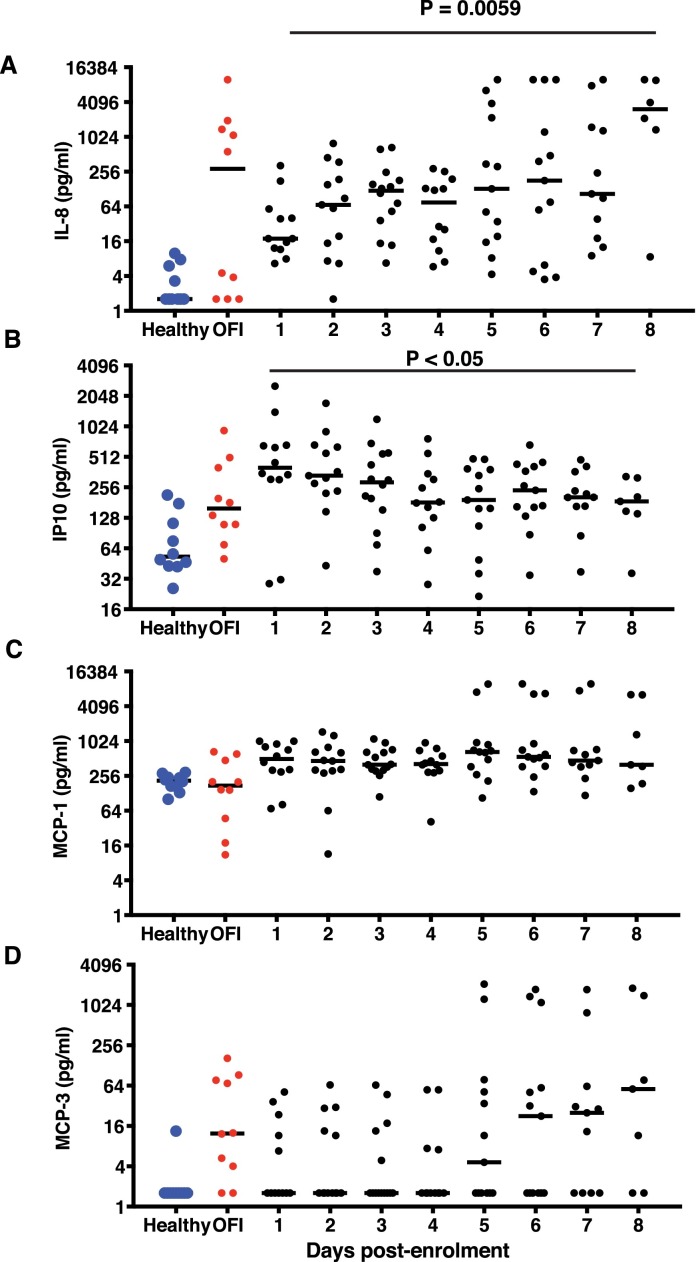
As previous reports implicate both virus and host factors in severe COVID-19 disease [6, 22], we were interested in estimating the levels of some of the inflammatory mediators along with interferons in the serum samples collected daily from our cohort of patients to understand the correlates of recovery from disease. We measured serum inflammatory mediators in three categories of age-matched samples: Healthy (n = 10), other febrile illness (OFI) (n = 10), and virus-positive COVID-19 patients (n = 16). The NP/OP RT-PCR false-positive samples were excluded from cytokine analysis. Healthy and OFI samples were collected at one time point only. Each COVID-19 patient contributed at least two serial samples. Multiplex luminex assay was performed to measure IFN-α2, IFN-γ, IL-1β, IL6, IL-7, IL-8, IL-10, IL12p40, IL12p70, Monocyte Chemotactic Protein 1(MCP-1), MCP-3, TNF-α and gamma interferon inducible protein −10 (IP-10) in serial samples. Less than half of the virus-positive patients showed enhanced IFN-α2 after day 5 of enrolment (Fig. 3 A). Similar trend was observed for IFN-γ where a median of 8–16 pg/ml was detected in less than half of the samples beyond day 5 (Fig. 3B). Increased IFN-α2 and IFN-γ at later stages of infection coincided with recovery and clearance of virus by day 8 of enrolment. Other inflammatory markers such as IL-1β, IL-6 and IL-10 (Fig. 3C-E) did not show any significant increase in these patients indicating a clear absence of inflammation. Although all the samples showed a moderate increase in the levels of TNF-α, it was not statistically significant (Fig. 3F) which is in agreement with previous reports (23). We next looked at the cytokines that influence T-cell functions namely the pro-inflammatory IL-12p40 homodimer and IL-12p70, the active IL-12 heterodimer secreted by dendritic cells and macrophages that promotes Th1 response and leads to IFN-γ and TNF-α production. Both IL-12p40 and IL-12p70 levels did not show any significant increase (Supplementary Figure S3B and S3C). IL-7, a cytokine produced by epithelial cells and DCs and promotes biogenesis and proliferation of lymphocytes was increased in samples collected on day 6–8 suggesting a cue for activation of T-cell responses. Of the four chemokines analysed in this study, only IL-8 and IP-10 showed a significant increase (Fig. 4 A and 4B) in almost all of the samples relative to OFI controls indicating a critical role for these chemoattractants in recruiting innate immune cells. Interestingly, the levels of the pro-inflammatory chemokine MCP-1 did not show any variation, and MCP-3 levels were very heterogenous during the course of infection in any of the patients, suggesting lack of severe inflammatory response (Fig. 4C and 4D).
Fig. 3.
Quantitation of serum interferons and inflammatory markers. (A-F) Levels of indicated analytes in serum samples collected serially from hospitalizedpatients was estimated by multiplex luminex bead assays. Bar represents the median values. Healthy and other febrile illness samples were from a single time-point.
Fig. 4.
Quantitation of serum chemokines. (A-D) Levels of indicated analytes in serum samples collected serially from hospitalized patients was estimated by multiplex luminex bead assays. Bar represents the median values. Healthy and other febrile illness samples were from a single time-point. P values were determined by ANOVA.
3. Discussion
Of all the coronavirus epidemics/pandemics of recent past, COVID-19 has had the greatest impact on global health and economy. Although most of the individuals are asymptomatic or with mild symptoms, the disease affects the elderly and people with co-morbidities the most [5], indicating a critical role for innate immune response in limiting the viral load and spread. Most asymptomatic or individuals with mild symptoms harbor high viral loads in their respiratory tract and contribute to disease transmission [24]. Previous report suggested reduced interferon response in severe patients as compared to mild/moderate cases 8–12 days after the onset of symptoms [6] and viral genetics was not found to correlate with severe disease [25]. We focussed our efforts to monitor the kinetics of viral load, detect and quantitate neutralizing antibodies and inflammatory mediators in the serum samples of hospitalized COVID-19 patients who displayed mild to no symptoms but were RT-PCR positive for SARS-CoV-2 during screening at the hospital or during contact tracing. Most of the patients were enrolled within 48 h of testing positive by RT-PCR and are likely to be within 7–10 days of exposure to the virus. Despite the large variation in viral load (about 4 orders of magnitude), the clinical features and disease outcome did not vary as all patients were able to clear the virus and recover from the disease. This suggests that immune response in younger adults is capable of clearing the infection and age and imbalance in immune response could contribute to severe disease in older individuals and patients with co-morbidities. We observed that decrease in viral loads coincided with increase in IFN responses and neutralizing antibodies. Although our study does not have relative comparison between samples from different severities, previous reports have shown that low serum interferons preceded respiratory failure in critical COVID-19 cases [6] whereas we observed an increase in both IFN-α and IFN-γ in most of the patients who recovered and were discharged in the present study clearly indicating a critical role for interferon responses in recovery from COVID-19. Our results show that there was moderate to no induction of pro-inflammatory cytokines or chemokines such as IL-6 and TNF-α in any of the patients. IL-8, the primary chemoattractant for neutrophil recruitment has been shown to play an important role in acute respiratory distress syndrome (ARDS) and in recruitment of neutrophils to the site of injury in lungs in sepsis [26]. We speculate that generation of high levels of IL-8 may lead to ARDS observed in severe COVID-19 patients. Therefore, we hypothesize that regulation of IL-8 signaling and neutrophil activity could be an important determinant of clinical outcomes in COVID-19 patients.
A recent report suggested immunosuppression as a major cause for COVID-19 disease rather than cytokine storm and administering IL-7 ex vivo enhanced the capacity of T-cells to produce IFN-γ [23]. IL-7 has also been shown to play a key role in T-cell-mediated response to influenza virus infection [27]. We also observed, at later stages of infection, a trend showing increase in IL-7 and IL-12p70 suggesting activation of T cells. These observations envisage an important role for IL-7 in T-cell-mediated response to COVID-19 infections.
4. Methods
4.1. Clinical samples
23 patients who were positive for COVID-19 by RT-PCR were enrolled into the study after obtaining informed consent. Details of sample collection and diagnosis is described in supplementary information.
4.2. Quantitative RT-PCR
Quantitation of SARS-CoV-2 viral RNA levels by reverse transcription polymerase chain reaction (RT-PCR) as described earlier [28]. Detailed methodology is described in supplementary information.
4.3. Multiplex cytokine/chemokine assays
25 μl of serum was used for in magnetic bead-based multiplex chemokine and cytokine luminex assay as per the manufacturer's instructions (Merck-Millipore). Assays were performed in duplicates along with quality controls as described previously [29]. Further details is provided in supplementary information.
4.4. Quantitative ELISA
Recombinant spike protein receptor binding domain (RBD) ELISA was performed as described earlier [20] with minor modifications. Please see supplementary information for a detailed description of the process.
4.5. Plaque reduction neutralization titre assay
Plaque-purified Indian isolate [30] and B1.1.7 isolate were used in the study for neutralization assays (NCBI accession ID: MW422884 and MW881790). Detailed description of the method is provided in the supplementary information.
5. Data analysis
Data was analysed and final graphs were prepared using GraphPad Prism (Version 7.0e) software. All experiments were performed with two or more replicates and graphs have been prepared representing data from at least two independent experiments with n ≥ 6 unless otherwise indicated. Statistical significance was estimated by one-way ANOVA using Dunnett's test was applied for multiple comparisons. All COVID-19 samples were compared with OFI samples. For paired t-test, P values were obtained by Wilcoxon matched-pairs signed rank test.
Funding information
This work was supported by funding to GRM by the Biotechnology Industry Research Assistance Council (BIRAC) (BT/NBM0099/02/18). GRM also acknowledges the funding support provided by the Department of Biotechnology (DBT) through IndCEPI
Mission and Translational Research Program. RP acknowledges funding support from CSIR (MLP-2005), and Fondation Botnar (CLP-0031). The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Author contributions
AA, SG, NAK, HS, SP and RP performed experiments and analyzed the data. NV, AD and AKP coordinated the study at clinical site and contributed reagents. GRM conceived the study, designed the experiments, and analyzed data. AA, SG and GRM wrote the manuscript. All authors have reviewed and approved the final version of the manuscript.
Ethical approval
The study was approved by the Institutional ethics committees for human research at ESIC Hospital and Medical College (No. 134/A/11/16/Academics/MC/2016/134) and THSTI (THS 1.8.1/ (93)). Informed consent was obtained from all the participants.
Supplementary information
This article includes supplementary figures and figure legends.
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.The authors declare the following financial interests/personal relationships which may be considered as potential competing interests
Acknowledgements
We thank Amresh Kumar Singh for technical support and all members of CCV lab and bioassay lab for their technical help and critical inputs. We thank all the patients who consented to participate in the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.105060.
Appendix. Supplementary materials
References
- 1.Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tregoning J.S., Brown E.S., Cheeseman H.M., Flight K.E., Higham S.L., Lemm N.M., et al. Vaccines for COVID-19. Clin. Exp. Immunol. 2020 doi: 10.1111/cei.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heimfarth L., Serafini M.R., Martins-Filho P.R., Quintans J.S.S., LJ Quintans-Junior. Drug repurposing and cytokine management in response to COVID-19: a review. Int. Immunopharmacol. 2020;88 doi: 10.1016/j.intimp.2020.106947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajgain K.T., Badal S., Bajgain B.B., Santana M.J. Prevalence of comorbidities among individuals with COVID-19: a rapid review of current literature. Am. J. Infect. Control. 2021;49(2):238–246. doi: 10.1016/j.ajic.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han H., Ma Q., Li C., Liu R., Zhao L., Wang W., et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020;9(1) doi: 10.1080/22221751.2020.1770129. 1123-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5) doi: 10.1172/JCI137244. 2620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020;8(9) doi: 10.1016/S2213-2600(20)30354-4. e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., et al. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n579. n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. medRxiv. 2021 doi: 10.1038/s41586-021-03426-1. 2021.02.01.21250959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. bioRxiv. 2021 doi: 10.1038/s41586-021-03398-2. 2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- 17.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021 doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 18.To K.K.-.W., Tsang O.T.-.Y., Leung W.-.S., Tam A.R., Wu T.-.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhuri S., Thiruvengadam R., Chattopadhyay S., Mehdi F., Kshetrapal P., Shrivastava T., et al. Comparative evaluation of SARS-CoV-2 IgG assays in India. J. Clin. Virol. 2020;131 doi: 10.1016/j.jcv.2020.104609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476–488. doi: 10.1016/j.cell.2020.12.015. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remy K.E., Mazer M., Striker D.A., Ellebedy A.H., Walton A.H., Unsinger J., et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson M.A., Quandelacy T.M., Kada S., Prasad P.V., Steele M., Brooks J.T., et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4(1) doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 26.Groeneveld A.B.J., editor A Central Role of Interleukin-8 in the Pathogenesis of ARDS1998; Berlin, Heidelberg: Springer Berlin Heidelberg.
- 27.Plumb A.W., Patton D.T., Seo J.H., Loveday E.K., Jean F., Ziegler S.F., et al. Interleukin-7, but not thymic stromal lymphopoietin, plays a key role in the T cell response to influenza A virus. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0050199. e50199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharaj A., Das S.J., Sharanabasava P., Lodha R., Kabra S.K., Sharma T.K., et al. Visual Detection of SARS-CoV-2 RNA by Conventional PCR-Induced Generation of DNAzyme Sensor. Front. Mol. Biosci. 2020;7(444) doi: 10.3389/fmolb.2020.586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singla M., Kar M., Sethi T., Kabra S.K., Lodha R., Chandele A., et al. Immune Response to Dengue Virus Infection in Pediatric Patients in New Delhi, India–Association of Viremia, inflammatory mediators and monocytes with disease severity. PLoS Negl. Trop. Dis. 2016;10(3) doi: 10.1371/journal.pntd.0004497. e0004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anantharaj A., Gujjar S., Kumar S., Verma N., Wangchuk J., Khan N.A., et al. Kinetics of viral load, immunological mediators and characterization of a SARS-CoV-2 isolate in mild COVID-19 patients during acute phase of infection. medRxiv. 2020 2020.11.05.20226621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.