Abstract
Understanding how a regulatory protein occupies its sites in vivo is central to understanding gene regulation. Using the yeast Gal4 protein as a model for such studies, we have found 239 potential Gal4 binding sites in the yeast genome, 186 of which are in open reading frames (ORFs). This raises the questions of whether these sites are occupied by Gal4 and, if so, to what effect. We have analyzed the Saccharomyces cerevisiae ACC1 gene (encoding acetyl-coenzyme A carboxylase), which has three Gal4 binding sites in its ORF. The plasmid titration assay has demonstrated that Gal4 occupies these sites in the context of an active ACC1 gene. We also find that the expression of the ACC1 is reduced fourfold in galactose medium and that this reduction is dependent on the Gal4 binding sites, suggesting that Gal4 bound to the ORF sites affects transcription of ACC1. Interestingly, removal of the Gal4 binding sites has no obvious effect on the growth in galactose under laboratory conditions. In addition, though the sequence of the ACC1 gene is highly conserved among yeast species, these Gal4 binding sites are not present in the Kluyveromyces lactis ACC1 gene. We suggest that the occurrence of these sites may not be related to galactose regulation and a manifestation of the “noise” in the occurrence of Gal4 binding sites.
Yeast cells grown in galactose medium express a set of genes (GAL genes) in order to utilize galactose (15). The activation of the GAL genes is effected by a powerful transcription activator, Gal4, which recognizes one or several upstream activating sequences (UASs) in the promoters of Gal4-regulated genes through its N-terminal DNA binding domain (DBD). Understanding how a regulatory protein such as Gal4 occupies its sites in vivo and the consequences of occupancy is central to understanding gene regulation. We have previously identified 239 putative Gal4 binding sites in the yeast genome using a computational approach (Q. Li and S. A. Johnston, submitted for publication). Among them, 29 novel Gal4 binding sites in the promoters have been intensively studied and new genes regulated by Gal4 have been identified. It is notable that 186 Gal4 binding sites are located in the open reading frames (ORFs) of genes, but there are only about 60 Gal4 dimers per yeast cell (E. H. Xu, Q. Li, A. Vonica, and S. A. Johnston, unpublished data). This raises the interesting questions of whether these binding sites in ORFs compete with the promoter sites for occupancy by Gal4 protein and whether occupancy confers regulation. Here we examine the specific question of whether Gal4 protein occupies sites in the ACC1 gene and, if so, what the consequences are. The results of this analysis may have implications for interpreting genome-wide expression patterns.
Gal4 binding to its putative sites in the ORFs has not been investigated. UASGAL sites in promoters tend to be nucleosome free (25, 36). It is quite possible that Gal4 sites in ORFs are nucleosome covered and that this may block Gal4 occupancy. However, it has been noted that, at least in vitro, Gal4 protein can effectively compete with nucleosomes for binding UASGAL sites (38, 39). Gal4 may also affect nucleosome stability through recruitment of remodeling complexes (4, 31, 40).
Even if Gal4p does occupy sites in ORFs, it may or may not affect transcription. There have been several reports of other putative regulatory protein binding sites in the ORFs of genes in yeast. Multiple downstream elements in the yeast transposons Ty1 and Ty2 have been shown to either activate or repress transcription (7). The LPD1 gene, which encodes the lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase, is also regulated by elements in the ORFs. Two downstream activation sites and two downstream repressor sites activate and repress the transcription of LPD1 genes, respectively (30). Similar downstream regulatory elements have been observed in the SRP1 (serine-rich protein 1) (6) and PGK (phosphoglycerate kinase) genes (20). In addition to these natural ORF sites, when three Leu3p binding sites were introduced into a LacZ reporter its transcription was down-regulated threefold (17).
We are interested in constructing a global picture of the occupancy of Gal4 protein on the yeast genome. Since a large portion of the putative Gal4p binding sites are in ORFs, it is important to make an assessment of the potential of these sites to contribute to Gal4p binding. In this report we focus on analysis of the UASGAL sites in the ACC1 gene because it is the only ORF in the yeast genome with more than one binding site. We found that the three UASGAL sites in ACC1 are occupied by Gal4p in vivo and that this leads to a novel form of regulation. Disruption of these sites does relieve the ACC1 regulation by Gal4p but has no effect on growth on galactose. These observations suggest that the occurrence of these sites might be a by-product of the random distribution of the sites in the genome and has implications for interpretation of genome-wide expression analysis.
MATERIALS AND METHODS
Strains, media, and genetic methods.
Strain W303a (a ade2-1 leu2-3,112 ura3-1 his3-115 trp1-1 can1-100) was used as a parental strain throughout this study. Strain YJ0Z (a Δgal4 Δgal80 ura3-52 leu2-3,112 his3 ade1 trp1 MEL1) has an integrated LacZ reporter gene controlled by the GAL1 promoter. Strain SC18 (a Δgal80 ura3-52 leu2-3,112 his3 ade1 trp1 MEL1) was used in the plasmid titration assay. To generate yeast strain SC785 with the two Gal4 binding sites removed from the ACC1 ORF, the plasmid YIP352-ACC1(ΔS2&S3) was cut with SpeI and integrated into the ACC1 locus of W303a to create a duplication. The integrated strain was then grown in yeast extract-peptone-dextrose and plated on 5′-fluoroorotic acid. Surviving colonies were picked. ACC1 DNA covering these two sites was obtained by PCR and sequenced (Oligo184 [TCACTTGGAAGTTCAACTGC] and Oligo185 [CGACTTGTAATCTTGGGTTC]). Yeast cells were grown in complete synthetic media with the addition of 2% glucose, raffinose, or galactose unless described otherwise. Yeast transformation was performed using the Saccharomyces cerevisiae EasyComp transformation kit from Invitrogen.
Construction of plasmids. (i) pMEL-β-gal(ACC1-S2&S3).
Briefly, a 0.75-kb fragment containing the MEL1 promoter was fused to lacZ, and the natural UASMEL in the promoter was deleted and replaced with a unique XhoI site. The reporter gene was inserted into PRS316 (29). The 158-bp PCR product containing sites 2 and 3 from ACC1 was digested with XhoI and ligated into the pMEL-β-gal plasmid (Oligo176 [AAACTCGAGCGGTAGAGACTGTTCCGTTC] and Oligo177 [AAACTCGAGCGGCAAACTAGCCA]).
YEP351-ACC1 and PRS316-ACC1.
An approximately 9-kb SacI genomic fragment containing a functional ACC1 gene was obtained from S. Kohlwein and inserted into the multiple cloning sites of YEP351 and PRS316.
YIP352-ACC1 (ΔS2&S3).
A 5.4-kb BamHI fragment which contains the 5′ half of a functional ACC1 gene was excised from PRS316-ACC1 and cloned into YIP352. Site 2 and site 3 were replaced by making four point mutations in the crucial CGG and CCG triplets. Site-directed mutagenesis was performed using the QuickChange kit from Stratagene and confirmed by sequencing.
Northern and Southern blotting.
Yeast cells were grown to mid-log phase in 100 ml of synthetic medium plus either raffinose or galactose to extract total RNA. Cells were harvested and resuspended in 1 ml of RNA extraction buffer (100 mM Tris HCl [pH 7.5], 100 mM LiCl, 10 mM iodoacetate, 1 mM EDTA, 1% sodium dodecyl sulfate). Then 1 ml of phenol-chloroform and 1 ml of glass beads were added, and the solution was vortexed at maximum speed for 60 s. The liquid phase was transferred to new tubes and centrifuged at 14,500 × g for 10 min. Phenol-chloroform extraction was repeated three times. The supernatant was transferred to new tubes, and 1/10 volume of 3 M sodium acetate (pH 5.0) and 2.5 volumes of cold 100% ethanol were added. The solution was chilled at −80°C for at least 5 min and centrifuged at 3,600 × g for 10 min. The pellet was washed with 5 ml of 80% cold ethanol and dried in the air. Ten micrograms of total RNA was loaded in each lane of the 1.2% agarose gel (1× MEA buffer [20 mM morpholineethanesulfonic acid, 1 mM EDTA, 5 mM sodium acetate], 5% formaldehyde, 0.1% diethyl pyrocarbonate [DEPC]). The gel was run at 2.5 V/cm for approximately 2 h and then washed with DEPC-treated water for 30 min. The RNA was then transferred to a nylon transfer membrane (MAGNA, 0.45 μm; MSI) using the Posi-blotter from Stratagene.
Southern blots were performed using the standard protocol (28). One microgram of yeast genomic DNA was digested with the appropriate restriction endonucleases. Radioactive DNA probes were synthesized using a random primer labeling kit (Gibco BRL). The probed membrane was exposed to a PhosphorImager screen (Molecular Dynamics) overnight, and radioactive bands were analyzed with ImageQuant software.
Growth competition assay.
Wild-type and mutant yeast cells were mixed together at approximately a 1:1 ratio. The mixed culture was then diluted about 1,000-fold in the desired medium and incubated at 30°C until it reached the same optical density as before the dilution. This allowed the mixed culture to go through 10 doublings. The culture was then diluted and grown for another 10 doublings. The final culture was collected, and the genomic DNA was prepared and digested with HpaII for Southern blotting. A 306-bp ACC1 fragment was used as a probe. The relative fitness of the mutant strain was calculated by the number of mutant yeast cells divided by the number of wild-type yeast cells in the culture after a certain number of doublings, with the fitness of the wild-type yeast strain set at 1.00.
Assays for α-Gal and β-Gal.
A 10-ml yeast culture was grown to an optical density at 600 nm of 0.4 to 0.6. Cell extracts were prepared, and the α-galactosidase (α-Gal) and β-Gal assays were performed as previously described (16). The protein concentrations were determined using the Coomassie blue G250 assay (Pierce). One β-Gal unit is defined as 1 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per ml per min. One α-Gal unit is defined as 1 nmol of p-nitrophenyl α-d-galactopyranoside hydrolyzed per min per ml.
Plasmid titration assay.
The ability of Gal4 binding sites to bind Gal4 protein was determined by the binding site's ability to decrease MEL1 expression. Sc18 cells were transformed with plasmid containing Gal4 binding sites and control plasmid. A 10-ml yeast culture was grown using the appropriate selection medium to an optical density at 600 nm of 0.6 to 0.8. Cell extracts were prepared for the α-Gal assay, and at the same time, total DNA was purified from the remaining extract. For plasmid YEP351, the copy number was determined using a quantitative Southern blot. The total DNA was digested with EcoRI and PstI. A 1.2-kb EcoRI fragment from YEP351 containing most of LEU2 was used as a probe. The genomic DNA produced a 2.2-kb band which is easily distinguished from the 1.2-kb band from YEP351. The copy number was calculated from the relative intensities of the two bands.
DNA sequencing.
The ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase was used for sequencing. One hundred nanograms of PCR product or 1 μg of plasmid DNA was used as the template. The sample was loaded on an ABI 310 genetic analyzer.
Protein purification.
The pGEX-CS-Gal4(1–147) expression plasmid was transformed into Escherichia coli strain BL21(pLysS). GST-CS-Gal4(1–147) was purified as previously described (5, 33). Tobacco etch virus protease was added to cleave Gal4(1–147) from the glutathione S-transferase (GST) beads. The digest reaction mixture was incubated for 1 to 2 h at 30°C with gentle shaking. The supernatant was collected for further biochemical studies.
Gel shift.
For the assay of Gal4 binding to ACC1, two PCR oligonucleotides, AAACTCGAGCGGTAGAGACTGTTCCGTTC (Acc1.a) and AAACTCGAGCGGCAGAGACATAACCG (Acc1.b), were synthesized according to the sequence of ACC1. Two XhoI sites were created for subsequent cloning purposes. The 158-bp PCR product which contains sites 2 and 3 was labeled with [γ-32P]ATP (Amersham) with T4 kinase (Promega). Gal4 DBD (positions 1 to 147) and labeled oligonucleotides were mixed in binding buffer (25 mM Tris [pH 7.5], 50 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM spermidine, 10 μg of bovine serum albumin per ml, 50 μg of salmon sperm DNA/ml, and complete protease inhibitor). The binding reaction mixture was incubated for 20 min at 4°C. The mixtures were loaded on 6% native polyacrylamide gel. The gel was dried in vacuum and finally exposed to a PhosphorImager screen for 30 min. In the competition assays, the consensus UASGAL was formed by annealing c1 (TCGAGCGGAGGACTGTCCTCCGG) and c2 (TCGACCGGAGGACAGTCCTCCGC). The mutated Gal4 binding site was formed by annealing m1 (TCGAGCGAAGGACTGTCCTCCAG) and m2 (TCGACTGGAGGACAGTCCTTCGC).
Degenerate PCR.
A BLAST search was run using the coding sequence of ACC1 against the Candida albicans DNA database (http://candida.stanford.edu) to determine the conserved residues of ACC1. Based on this comparison, two degenerate PCR primers were synthesized from the highly conserved region of ACC1 (ACC1.d1 [TGGTCNGGTACNGGTGTNGAY] and ACC1.d2 [CNACTTGNAATCTTGGGTTC]). A 552-bp PCR product was obtained from the Kluyveromyces lactis genomic DNA. Sequencing was performed using both primers, and the sequence was aligned with the ACC1 sequence of S. cerevisiae.
RESULTS
Gal4 binds to its sites in the ACC1 gene in vitro.
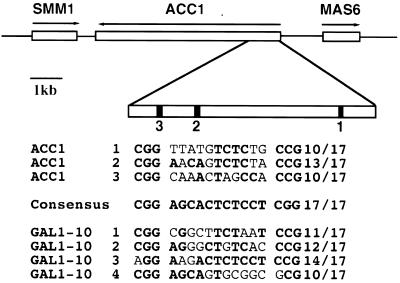
By sequence analysis, we found three potential Gal4 binding sites in the ORF of the ACC1 gene, which encodes acetyl-coenzyme A carboxylase. The ACC1 sites are present in a cluster, resembling those of GAL1-10 genes (Fig. 1). Site 2 and site 3 are separated by only 150 bp. Site 2 is the closest to the consensus UASGAL, with 13 matches out of 17 bases, suggesting that it is a strong site. Since the strength of a UASGAL generally corresponds to its similarity to the consensus UASGAL (35), the predicted strengths of these three sites in the ORF are comparable to those of the UASGAL sequences present in well-documented GAL genes, such as GAL1-10 genes (Fig. 1).
FIG. 1.
There are three Gal4 binding sites in the ORF of the ACC1 gene. The arrows indicate the direction of transcription. The ORF of ACC1 is 6.7 kb. In the 1-kb 5′ region of the ACC1 ORF there are three Gal4 binding sites. Sites 2 and 3 are closer to each other. The Gal4 binding sites in GAL1-10 are also listed. The sequences matching the consensus UASGAL are in boldface type. The fraction listed is the total number of matches out of the 17 bases. The consensus sequence was generalized from all previously identified GAL genes (Li and Johnston, submitted).
We performed a gel shift assay to determine if these predicted sites actually bind Gal4 protein. A 150-bp DNA fragment containing sites 2 and 3 was obtained by PCR and labeled as a probe in a gel shift experiment. A purified Gal4 DBD (amino acids 1 to 147) binds the probe very well (Fig. 2A). Complex C1 is formed by a Gal4 DBD binding one site, and complex C2 is formed when both sites are occupied by Gal4 (Fig. 2A, lanes 2 to 6). Interestingly, a third complex C3 is observed when the Gal4 level is increased (Fig. 2A, lane 6). We suggest that this complex is formed by the occupancy of a third weak UASGAL-like site (CAGCCTTTTCCATCTCG; boldface type indicates conserved positions) between site 2 and site 3 by Gal4 at a higher concentration. Based on these types of experiments, the estimated Kd of Gal4 binding to one site is about 1 nM while the Kd of a Gal4 DBD binding to a consensus sequence is ∼0.1 nM in large (>50-bp) DNA fragments (K. Melcher and S. A. Johnston, unpublished data). The specific binding of Gal4 to sites 2 and 3 is supported by the results of competition experiments with cold consensus UASGAL (Fig. 2B, lanes 3 to 5) and a mutated Gal4 binding site (lanes 6 to 8). Gal4 DNA complexes were completely disrupted at a 10-fold excess of consensus UASGAL over the labeled probe. The mutated Gal4 binding site has two point mutations in the CGG and CCG triplets.
FIG. 2.
Gel shift assay of sites 2 and 3. A 0.1-pmol portion of radiolabeled PCR fragment containing sites 2 and 3 from the ACC1 ORF was used. C1, C2, and C3 represent one, two, and three Gal4 DBD dimers bound to the probe, respectively. (A) Lanes 1 to 6, 0, 0.0005, 0.0024, 0.012, 0.06, and 0.3 pmol of purified Gal4DBD(1–147). (B) For lanes 2 to 8, 0.15 pmol of Gal4DBD(1–147) was used in binding reactions. For competition, 0.01, 0.1, and 1.0 pmol of cold consensus UASGAL were added (lanes 3 to 5). Then 0.01, 0.1, and 1.0 pmol of cold mutated Gal4 binding sites were added to (lanes 6 to 8).
The Gal4 binding sites in the ORF of ACC1 activate transcription when inserted into a promoter.
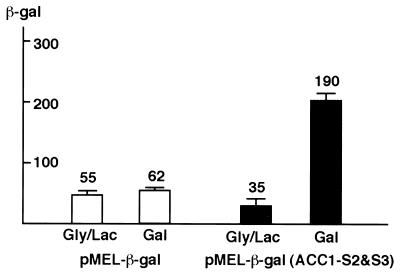
We then tested if the Gal4 binding sites in the ACC1 ORF can replace an authentic UAS and activate transcription. We used a reporter construct (pMEL-β-gal) which fuses a β-Gal reporter gene to a MEL1 promoter. The single, natural UASGAL in MEL1 promoter was eliminated and replaced with a cloning site. MEL1 encodes α-Gal and is a member of the GAL regulon. The 150-bp PCR fragment containing sites 2 and 3 was inserted into the MEL1 promoter. As shown in Fig. 3, sites 2 and 3 can induce transcription sixfold in the presence of galactose. The wild-type MEL1 gene is activated 50-fold in galactose. This experiment demonstrates that these sites can function like a natural UASGAL when placed in the promoters and that there are no intrinsic difference between the Gal4 binding sites in the ACC1 ORF and those previously found in the promoters of GAL genes. The activation level of sites 2 and 3 is lower than expected, given that the ACC1 sites have a Kd comparable to that of the MEL1 UASGAL (35). The lower activation might be due to effects of the sequence between the two sites.
FIG. 3.
The Gal4 binding sites in the ORF can function as UASGAL. Sites 2 and 3 were cloned into a reporter construct, pMEL-β-gal, in which the UASGAL of MEL1 gene was removed and replaced with a cloning site. W303a cells transformed with the pMEL-β-gal and pMel-ACC1(S2&S3)-β-gal were grown in glycerol-lactic acid and galactose. β-Gal activity was measured as described in the text.
Gal4 binds its sites in the ACC1 ORF in vivo.
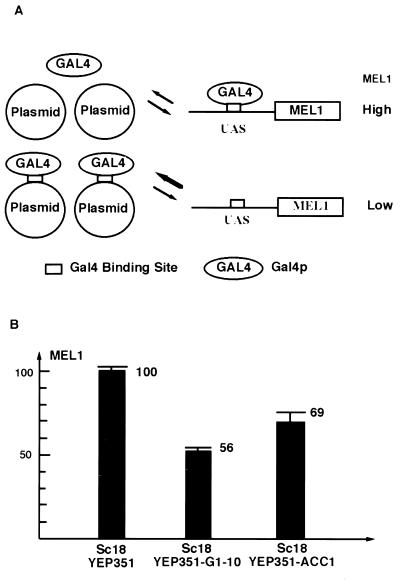
We have developed a plasmid titration assay to address occupancy of DNA by transcription activators in vivo (34; Xu et al., unpublished). Expression of MEL1 is Gal4 dependent and activated in galactose (23). Gal4 activates the transcription of MEL1 through a single UASGAL in its promoter (19), which is not fully occupied under normal conditions as evidenced by increased level of MEL1 expression when Gal4 is overexpressed. When exogenous Gal4 binding sites are introduced into yeast cells, the MEL1 expression level decreases as a result of the titration of Gal4 protein. Therefore, the relative occupancy of Gal4 on the sites of the plasmid is reflected by the relative decrease in the expression level of MEL1 (Fig. 4A).
FIG. 4.
Gal4 occupies the sites in ACC1 ORF. (A) The principle of the plasmid titration assay. There is one single weak UASGAL in the promoter of MEL1 (α-Gal gene). The expression of MEL1 reflects the available Gal4. When Gal4 binding sites are introduced by a multicopy plasmid, a decreased level of Gal4 will result in a lower level of MEL1. (B) Sc18 cells transformed with Yep351, YEP351-Gal1-10 promoter, and YEP351-ACC1. There are four Gal4 binding sites in the promoter of GAL1-10. The average copy number was determined by Southern blot.
As a positive control, the four Gal4 binding sites from the GAL1-10 promoter in a multicopy YEP351 vector were transformed into yeast cells. This plasmid reduced the MEL1-encoded α-Gal activity to 56% of its original level through titration of Gal4 protein (Fig. 4B). There are four Gal4 binding sites in the GAL1-10 promoter, two of which are strong sites (Fig. 1). The copy number of GAL1-10 promoter determined by Southern blotting was approximately 19 per cell. To investigate whether the Gal4 binding sites in the ACC1 ORF can cause a similar effect, we cloned a functional genomic fragment of ACC1 into the YEP351 vector. The ACC1 plasmid was then transformed into yeast cells. In these cells, the MEL1 expression level was reduced to 69% of its original level (Fig. 4B). There were approximately 10 copies of the ACC1 gene per cell. Though it is possible that multiple copies of ACC1 reduce MEL1 expression through other mechanisms, since the observed titration effect for ACC1 is as expected for a DNA containing three UASGAL sequences, we think it likely that the effect on MEL1 is through titration of Gal4 by the ACC1 fragment.
Gal4 represses the transcription of ACC1.
In the case of the artificial addition of Leu3 binding sites to an ORF, the observed repression was proposed to be due to the DNA binding of Leu3 (17). Gal4 can exist in two states when bound to DNA. In noninducing medium (raffinose), Gal4 is bound to DNA but transcriptionally inactive due to Gal80 repression. In inducing medium (galactose), Gal4 is bound to its site and capable of activating transcription. We first examined whether these two states had any effect on the transcription of the ACC1 gene. RNA was extracted from the W303 strain grown on either galactose or raffinose and probed for ACC1 transcript levels. Surprisingly, the ACC1 level was approximately fourfold lower when Gal4p was in the inducing (galactose) state (Fig. 5, lanes 1 and 2).
FIG. 5.
Gal4 represses the transcription of ACC1. (A) Wild-type (WT) ACC1 mRNA levels in raffinose (lane 1) and galactose (lane 2) and mutant ACC1 mRNA levels in raffinose (lane 3) and galactose (lane 4). In the mutant ACC1, two Gal4 binding sites, sites 2 and 3, were eliminated without changing the codons.
If the repression on galactose of ACC1 transcription is directly due to Gal4p binding of the ACC1 ORF, disruption of the Gal4 binding sites should relieve this repression. The CGG triplets in the Gal4 binding sites (CGGN5TN5CCG) are crucial for Gal4 binding, as a single mutation in the triplet reduces the binding of Gal4 500-fold and completely compromises the ability of a UASGAL to activate transcription in vivo (35). We inactivated sites 2 and 3 in ACC1 by introducing two silent point mutations into each site. The 5.4-kb 5′ half of ACC1 carrying these mutations was reconstructed and cloned into YIP352, an integration vector. The plasmid was linearized by cutting in the middle of the ACC1 DNA fragment and integrated into the endogenous locus of ACC1 to create a partial duplication of ACC1. Finally, the duplicated region was excised by selection against the URA3 marker. This strain has a single ACC1 allele with the sites 2 and 3 eliminated by silent mutations in the CGG sites. As is evident by comparing lanes 3 and 4 in Fig. 5, eliminating the Gal4 binding sites relieves the galactose repression of ACC1. This demonstrates that the down-regulation of ACC1 in galactose medium is dependent on these Gal4 binding sites.
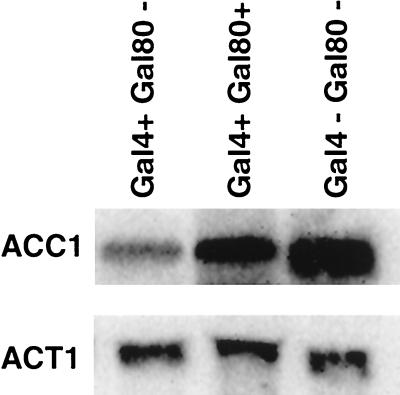
To further investigate the mechanism of the repression, we determined the relative levels of ACC1 gene in gal4 and gal80 deletion backgrounds (Fig. 6). In agreement with the proposal of Gal4-mediated regulation of ACC1, the repression is abolished when both GAL4 and GAL80 are deleted (Fig. 6, lane 3). When GAL80 alone is deleted, the ACC1 transcription is repressed even in raffinose (Fig. 6, lane 1). These data suggest that Gal4 binding itself does not repress ACC1 transcription but that the activation domain must also be accessible, either through induction by galactose or deletion of GAL80. This is the first example of Gal4 acting as a negative regulator.
FIG. 6.
Active Gal4 is required to repress AccI mRNA. The ACC1 mRNA levels were measured by Northern blot. The sizes of ACC1 mRNA and ACT1 mRNA are 7.2 and 1.2 kb, respectively. ACT1 was used as a control for quantification of ACC1 mRNA levels.
The repression of ACC1 by Gal4 does not affect the growth of yeast S. cerevisiae in galactose.
It is generally assumed that activator binding and regulations are physiologically relevant (26). The ACC1 gene is essential and is regulated by the level of phospholipids. One possibility is that the Gal4 binding sites in ACC1 reflect a linkage of the two regulatory systems and that ACC1 regulation plays an undiscovered role in galactose growth. To test for this possibility we used a very sensitive mixed culture growth competition assay to see if the growth rate in galactose medium is altered when sites 2 and 3 are eliminated from ACC1 (Fig. 7). Briefly, wild-type and mutant yeast cells were mixed together at a 1:1 ratio, the mixed culture was grown for a certain number of doublings, and the ratio of wild-type to mutant forms was determined. The site 2 and 3 mutations disrupt a natural HpaII site (CCGG is changed to CAGG). This creates a restriction fragment length polymorphism convenient for discriminating between the wild-type and mutant strains. After 10, 20, and 30 doublings, the ratio of wild-type and mutant remained unchanged in both raffinose and galactose medium (Fig. 7, lanes 2 to 8). This protocol should readily detect effects of 1% or more on fitness. Therefore the elimination of repression of ACC1 transcription by Gal4 does not detectably affect growth in galactose.
FIG. 7.
The Gal4 regulation of ACC1 has no effect on growth. The wild-type (WT) and mutant (S2,3−) yeast cells were mixed in a 1:1 ratio and grown in raffinose (Raf) or galactose (Gal) for 10 (lanes 2 and 6), 20 (lanes 3 and 7), or 30 (lanes 5 and 8) generations. Southern blots were performed using a ACC1 probe, and the genomic DNA was digested with HpaII. In the site 2 and 3 mutant, one HpaII site was destroyed.
The repression of ACC1 by Gal4 is not conserved in K. lactis
K. lactis is closely related to S. cerevisiae, and the galactose system is highly conserved between the two organisms. The K. lactis equivalent of the Gal4 protein, Lac9, is able to recognize identical binding sequences (2, 27). ACC1 is highly conserved between many eukaryotic species. If the ACC1 UASGAL sites play a role in galactose growth, they might also be present in the K. lactis ACC1 gene. Therefore, we cloned the relevant part of the K. lactis ACC1 gene to determine if these sites are also present. In order to locate the regions in ACC1 which are likely to be conserved in K. lactis, we aligned the ACC1 sequence from the yeast C. albicans with that from S. cerevisiae. Degenerate oligonucleotides were made, and a PCR fragment of ACC1 was obtained from K. lactis genomic DNA and sequenced. The sequence of K. lactis ACC1 in this region is highly similar to the counterpart in S. cerevisiae, with 76% identity at the DNA level. However, none of the Gal4 (Lac9) sites are present in the K. lactis ACC1 gene. In the K. lactis ACC1 gene, the same amino acids are encoded but the Gal4 binding sites are not conserved (Fig. 8). This comparison between the two yeast strains indicates that the regulation of ACC1 by Gal4 is not conserved, at least in K. lactis.
FIG. 8.
Alignment of ACC1 DNA sequences from K. lactis (KL) and S. cerevisiae (SC). The Gal4 binding sites are underlined and in bold type.
DISCUSSION
In this study, we investigated the Gal4 binding sites in the ORF of ACC1, showing that Gal4 can bind to these sites both in vitro and in vivo. In galactose, Gal4 represses the transcription of the ACC1 gene when bound to the ORF. This repression requires both Gal4 binding and the induction of Gal4 or deletion of the Gal80 negative regulator. The absence of these UASGAL sites eliminates the regulation but does not affect the growth potential in galactose medium, and these sites are not present in K. lactis ACC1, indicating that these sites may not have a critical or conserved biological role.
The Gal4 binding sites in the ACC1 gene are occupied.
In order to activate the transcription of GAL genes, Gal4 must occupy its sites in the promoters, and the transcription output depends on the relative occupancy of Gal4 (Xu et al., unpublished). Theoretically, Gal4's occupancy is affected by the concentration of Gal4 protein, the number of specific binding sites, the amount of nonspecific genomic DNA, and the binding affinity of Gal4 to specific and nonspecific DNA. Gal4 is expressed at a low level, about 60 dimers per cell, and it is known that high levels of a potent transcription activator like Gal4 can be toxic to the cell (1, 9). How are these few Gal4 dimers able to locate the UASGALs in the yeast genome? Besides the UASGAL in the promoters of GAL genes, we have found ∼300 potential Gal4 binding sites in the genome, ∼80% of which are in the coding regions.
We found that the ACC1 gene on a multicopy plasmid can decrease the transcription of MEL1 to a level similar to that observed with the GAL1-10 promoter. These results suggest that Gal4 occupies the sites in the ORF of ACC1 quite well. This also suggests that if nucleosomes are present on the UASGAL in ACC1, they do not affect the accessibility of Gal4 binding. It has been shown that Gal4p can compete nucleosomes for binding a UASGAL (4, 18) and that this process may be aided by various chromatin-remodeling complexes like the SWI-SNF complex (22). Though the ACC1 gene was placed in a yeast expression plasmid in the titration assay, it behaved identically with respect to regulation. It is well documented that plasmid DNA is packed with nucleosomes when transformed into yeast nuclei (13).
Besides nucleosomes, Gal4 binding sites in an ORF would encounter the polymerase II (Pol II) complex. Gal4p may be displaced from its sites by Pol II transcription. Furthermore, in the transcribed region of ACC1, the DNA double strands have to be separated from each other to allow the transcription to occur. Since Gal4 does not bind to single-stranded DNA, Gal4 would have to dissociate from its sites at the moment when the Pol II passes. Because transcription is a very rapid process and the initiation frequency is relatively low even for a highly expressed gene (14), Gal4 may not be forced off its sites in an ORF for long. Gal4 binding in the ACC1 ORF is presumably a dynamic process.
It is also notable that the Gal4 binding sites in the promoters and ORFs have different contexts that may be important for their occupancy by Gal4. In the promoters, there are potential sites for other general DNA binding transcription factors such as TATA binding protein. There is evidence that transcription activators bind cooperatively with them (3, 12, 34). Since this context is absent in the ORFs, Gal4 binding to UASs in ORFs could be weakened. However, our data imply that the Gal4 binding sites in the ACC1 ORF do not require a promoter context for binding. Three Gal4 binding sites in the ACC1 ORF in a high-copy-number plasmid decreased the MEL1 expression to a level comparable to that of four Gal4 binding sites in the GAL1-10 promoter, indicating that Gal4 binds to these ORF sites equally well even if they are not located in a promoter. It is notable that Gal4 binding sites in the ACC1 ORF are in a cluster and they are strong sites. In contrast, the other Gal4 binding sites in the ORFs are isolated and most of them are predicted to have weak affinity.
Given these considerations, we feel the UASGAL sites in ORFs will have little contribution to Gal4 binding on a genomic scale. The ACC1 ORF is unique in that it has three relatively strong sites. This may facilitate cooperative binding of Gal4 protein. We note that this cooperativity is not evident in vitro (Fig. 2 and unpublished data), but Gal4 without its activation domain does not bind cooperatively (W. V. Ding and S. A. Johnston, unpublished data). Single sites are almost all weaker sites. Weak sites (for example, the MEL1 UAS) are occupied by Gal4, but this is in the context of a promoter where cooperative interaction with other transcription factors can come into play (34).
Gal4 represses ACC1 expression by a novel mechanism.
Leu3, another acidic activator, regulates the gene involved in leucine biosynthesis and responds to the levels of the metabolic intermediate α-isopropylmalate, which serves as a sensor of leucine availability (8). When three Leu3 binding sites were artificially introduced within the RPL16-lacZ reporter, downstream of the start of transcription, the expression of the reporter gene was inhibited threefold (17). Leu3 and Gal4 belong to a super class of zinc finger DNA binding transactivators. They recognize sequences with a similar motif (32). In the case of ACC1, the Gal4 binding sites naturally exist in the downstream region of an ORF.
Unlike higher eukaryotes, placement of a UAS from the CYC1 gene in a downstream intron cannot activate transcription (11). This is consistent with the data presented here. The inhibition of transcription we observed for the ACC1 gene and possibly that in the case of LEU3 could be for at least three possible reasons.
First, Gal4 protein binding itself may present a “roadblock” for RNA polymerase. If the roadblock model is correct, binding of the Gal4-Gal80 complex to the ACC1 UASGAL in noninducing medium would be expected to repress. However, ACC1 transcription is repressed only in galactose, and the repression is relieved in noninducing raffinose medium. We feel that this argues against a simple roadblock model.
Second, bound Gal4 may remodel the chromatin structure so that it is unfavorable for the transcription of ACC1. Activators such as Gal4 are known to recruit chromatin-remodeling machines such as SWI-SNF and SAGA complexes (10, 21). In vitro, SWI-SNF can alter nucleosome structure in a manner that facilitates the binding of transcription factors (37). SWI-SNF affects the nuclease sensitivity pattern of chromatin in some promoters and may facilitate binding of other transcription factors. It is possible that Gal4 binding downstream of the promoter may also recruit the chromatin remodeling machines and alter the chromatin structure in a way that interferes with transcription. However, such modification of chromatin structure would have a negative effect when it occurs downstream of the promoter.
Third, bound Gal4 may interfere with Ino2 and Ino4, the requisite activators of ACC1, or other transcription factors on the ACC1 promoter. For example, the Gal4p activation domain could compete for transcription factors required by Ino2 and Ino4. The repression may be connected to Gal4's function of activating transcription. It is generally believed that the transcription machinery must be recruited to the promoter by an activator, AD, for transcription activation (24). Upon binding to the promoter, a transcription activator can increase the recruitment by binding direct targets in the transcription machinery, which leads to transcriptional activation. Since there are a limited number of general transcriptional factors, many of these targets must be shared by different transcription activators. Thus, the presence of a powerful activator such as Gal4 could compete with the Ino2-Ino4 activator complex in the vicinity for some common targets. As a result, the activation of the ACC1 gene by Ino2-Ino4 could be reduced (Fig. 9). We called this phenomenon “local squelching,” which is similar to the “squelching” caused by overexpression of a strong transcriptional activator. For example, overexpression of Gal4 inhibits the expression of a gene which is normally activated by Gcn4 (9). Analogous to this, a similar squelching effect of Ino2-Ino4 could be caused by proximally bound Gal4, which creates a high “local concentration” of its activation domain. This type of squelching presumably would be against promoter-bound rather than free elements.
FIG. 9.
A proposed mechanism for Gal4 repressing of ACC1. The Ino2-Ino4 heterodimer activates transcription of ACC1 by binding to the inositol-choline response element (ICRE) and recruiting the general transcription factors. When Gal4 binds in the ORF of ACC1, Gal4 titrates the general transcription factors and competes with Ino2-Ino4. This local squelching effect results in the repression of ACC1 transcription.
Biological role of the UASGAL in the ACC1 gene.
It is generally assumed that binding and regulation by a regulatory protein imply that the target gene is part of a physiologically relevant pathway (26). Probably the most important aspect of the work we report here is its implication relative to this assumption. Is the regulation of ACC1 by Gal4 a manifestation of an undiscovered metabolic link between galactose metabolism and long-chain fatty acids synthesis, or does it reflect “noise” in regulation due to the random distribution of Gal4 sites? We feel that our data support the latter interpretation—that is, there may be no positive selection to maintain these sites. This conclusion is based on the following considerations. First, the total number of Gal4 binding sites in the yeast genome is almost exactly the number predicted to occur at random, and the number of sites in ORFs is what would be predicted to occur at random (Li and Johnston, submitted). So there does not appear to be any strong selection to limit binding sites to promoters. Second, we did not observe any adverse effect of eliminating the Gal4 binding sites in the ORF of ACC1. Neither growth on rich glucose medium (data not shown) nor growth on galactose was affected by eliminating the Gal4 regulation of the ACC1. In order not to overlook a small growth potential of the wild-type over the mutant, we used a growth competition assay which is very sensitive and can easily pick up a growth difference of 1%. However, we cannot exclude the possibility that this regulation is meaningful for S. cerevisiae under conditions in nature. Third, in a related yeast, K. lactis, we find that the ACC1 gene encodes the identical amino acid sequence in the corresponding regions where Gal4 binding sites are located but that the Gal4 (Lac9) binding sites are not present. The GAL system regulation in these two species is very similar. Therefore, if the Gal4 regulation of ACC1 in S. cerevisiae is biologically relevant, it has not been conserved between the two species.
It is generally thought that gene regulation networks are biologically relevant, in that there is positive selection pressure to maintain them. However, we propose that at least in the case of Gal4 regulation of ACC1, the regulation is not under selection. The three sites appear to have occurred by chance and to exert regulation on ACC1 with little or no effect on galactose growth or, apparently, selection against the regulation. Considering the large number of Gal4 binding sites found in ORFs, it is not improbable to have multiple Gal4 binding sites present in a single ORF by chance. Since there are many more DNA binding transcriptional activators like Gal4 in the yeast genome, the case of ACC1 may not be unique. There may be multiple binding sites in the ORFs for other activators. Those sites may regulate transcription by a similar mechanism, and this proposed mechanism of repression may be common. In contrast to the apparent situation with ACC1, some of these ORF regulatory sites may reflect biologically relevant regulation. However, particularly with the advent of the global analysis of gene expression, the ACC1 example illustrates the point that some portion of coregulation may be the product of noise in the random distribution of binding sites.
ACKNOWLEDGMENTS
We thank S.A.J. lab members for helpful discussions and S. Kohlwein for providing us a plasmid carrying ACC1. We are grateful to Fernando Gonzalez for reading the manuscript and to Liping Sun for providing consensus UASGAL. John Shelton helped us with some figures.
This work was supported by grants to S.A.J. from NIH (40070).
REFERENCES
- 1.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 2.Breunig K D, Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987;7:4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Farmer G, Zhu H, Prywes R, Prives C. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- 4.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 5.Ding W V, Johnston S A. The DNA binding and activation domains of Gal4p are sufficient for conveying its regulatory signals. Mol Cell Biol. 1997;17:2538–2549. doi: 10.1128/mcb.17.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fantino E, Marguet D, Lauquin G J. Downstream activating sequence within the coding region of a yeast gene: specific binding in vitro of RAP1. protein. Mol Gen Genet. 1992;236:65–75. doi: 10.1007/BF00279644. [DOI] [PubMed] [Google Scholar]
- 7.Farabaugh P, Liao X B, Belcourt M, Zhao H, Kapakos J, Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friden P, Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988;8:2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 10.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 11.Guarente L, Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box.”. Proc Natl Acad Sci USA. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horikoshi M, Hai T, Lin Y S, Green M R, Roeder R G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao C L, Carbon J. Characterization of a yeast replication origin (ars2) and construction of stable minichromosomes containing cloned yeast centromere DNA (CEN3) Gene. 1981;15:157–166. doi: 10.1016/0378-1119(81)90125-6. [DOI] [PubMed] [Google Scholar]
- 14.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Broach J R, Jones E W, editors. The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 16.Johnston S A, Hopper J E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkpatrick C R, Schimmel P. Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol Cell Biol. 1995;15:4021–4030. doi: 10.1128/mcb.15.8.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 19.Liljestrom P L. The nucleotide sequence of the yeast MEL1 gene. Nucleic Acids Res. 1985;13:7257–7268. doi: 10.1093/nar/13.20.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellor J, Dobson M J, Kingsman A J, Kingsman S M. A transcriptional activator is located in the coding region of the yeast PGK gene. Nucleic Acids Res. 1987;15:6243–6259. doi: 10.1093/nar/15.15.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 23.Post-Beittenmiller A M, Hamilton R W, Hopper J E. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1238–1245. doi: 10.1128/mcb.4.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 25.Reagan S M, Majors J E. The chromatin structure of the GAL1 promoter forms independently of Reb1p in Saccharomyces cerevisiae. Mol Gen Genet. 1998;259:142–149. doi: 10.1007/s004380050799. [DOI] [PubMed] [Google Scholar]
- 26.Ren B, Robert F, Wyrick J J, Aparicio O, Jennings E G, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert T L, Wilson C J, Bell S P, Young R A. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 27.Salmeron J M, Jr, Johnston S A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986;14:7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinclair D A, Kornfeld G D, Dawes I W. Yeast intragenic transcriptional control: activation and repression sites within the coding region of the Saccharomyces cerevisiae LPD1 gene. Mol Cell Biol. 1994;14:214–225. doi: 10.1128/mcb.14.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor I C, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 32.Todd R B, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 33.Van Hoy M, Leuther K K, Kodadek T, Johnston S A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 34.Vashee S, Kodadek T. The activation domain of GAL4 protein mediates cooperative promoter binding with general transcription factors in vivo. Proc Natl Acad Sci USA. 1995;92:10683–10687. doi: 10.1073/pnas.92.23.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vashee S, Xu H, Johnston S A, Kodadek T. How do “Zn2 cys6” proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J Biol Chem. 1993;268:24699–24706. [PubMed] [Google Scholar]
- 36.Weiss E, Ruhlmann C, Oudet P. Transcriptionally active SV40 minichromosomes are restriction enzyme sensitive and contain a nucleosome-free origin region. Nucleic Acids Res. 1986;14:2045–2058. doi: 10.1093/nar/14.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 38.Workman J L, Kingston R E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 39.Workman J L, Taylor I C, Kingston R E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- 40.Yudkovsky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]