Abstract
The pancreas consists of several specialized cell types that display a remarkable ability to alter cellular identity in injury, regeneration, and repair. The abundant cellular plasticity within the pancreas appears to be exploited in tumorigenesis, with metaplastic, dedifferentiation, and transdifferentiation processes central to the development of pancreatic intraepithelial neoplasia and intraductal papillary neoplasms, precursor lesions to pancreatic ductal adenocarcinoma. In the face of shifting cellular identity, the cell of origin of pancreatic cancer has been difficult to elucidate. However, with the extensive utilization of in vivo lineage-traced mouse models coupled with insights from human samples, it has emerged that the acinar cell is most efficiently able to give rise to both intraductal papillary neoplasms and pancreatic intraepithelial neoplasia but that acinar and ductal cells can undergo malignant transformation to pancreatic ductal adenocarcinoma. In this review, we discuss the cellular reprogramming that takes place in both the normal and malignant pancreas and evaluate the current state of evidence that implicate both the acinar and ductal cell as context-dependent origins of this deadly disease.
Keywords: acinar cells, ductal cells, cell-of-origin, pancreatic cancer, IPMN, PanIN, lineage-tracing, mouse models
Abbreviations used in this paper: ADM, acinar-ductal metaplasia; CAC, centroacinar cell; IPMN, intraductal papillary mucinous neoplasm; PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma
Despite recent advances in cancer treatment, overall survival from pancreatic cancer remains dismal with few efficacious therapeutic options. Nearly 80% of pancreatic cancer patients present with unresectable or metastatic disease, and those few who undergo complete resection can expect a still dismal median survival of ~20 months.1 Overall, the survival at 5 years is approximately 8%, and owing to increasing incidence, pancreatic cancer is projected to become the second-leading cause of cancer-related death in the United States.2
Pancreatic ductal adenocarcinoma (PDAC) consists of several hallmark features—an extensive desmoplastic reaction, a hypoxic local tumor microenvironment, a relative restriction of immunologic surveillance, and a pervasive resistance to conventional chemotherapy.3 On average, PDAC lesions harbor 60–70 mutations and display considerable tumor heterogeneity with variation noted among several identifiable subclones.4 Genomic assessments have revealed that the KRAS gene is near-universally mutated, occurring in approximately 90% of cases.5 Three tumor suppressors are as well often mutated—CDKN2A (~50%), TP53 (60%–70%), and SMAD4 (40%–50%), with SMAD4 typically associated with increased metastatic burden.6
Notably, well-differentiated PDAC carries the morphological appearance of abundant ductal structures with wide expression of the ductal marker cytokeratin 19.7 Thus, by histology alone, the putative cell of origin was long thought to be the ductal cell. However, the discovery and characterization of precursor neoplastic lesions in the pancreas that are associated with PDAC, either pancreatic intraepithelial neoplasia (PanIN) or intraductal papillary mucinous neoplasms (IPMNs), have shed new light on the beginnings of PDAC. In this review, we assess the current state of evidence regarding the beginnings of these pancreatic premalignant lesions and of malignant PDAC, and offer our perspectives on the uncertainties that remain.
Lessons from the Endocrine Pancreas
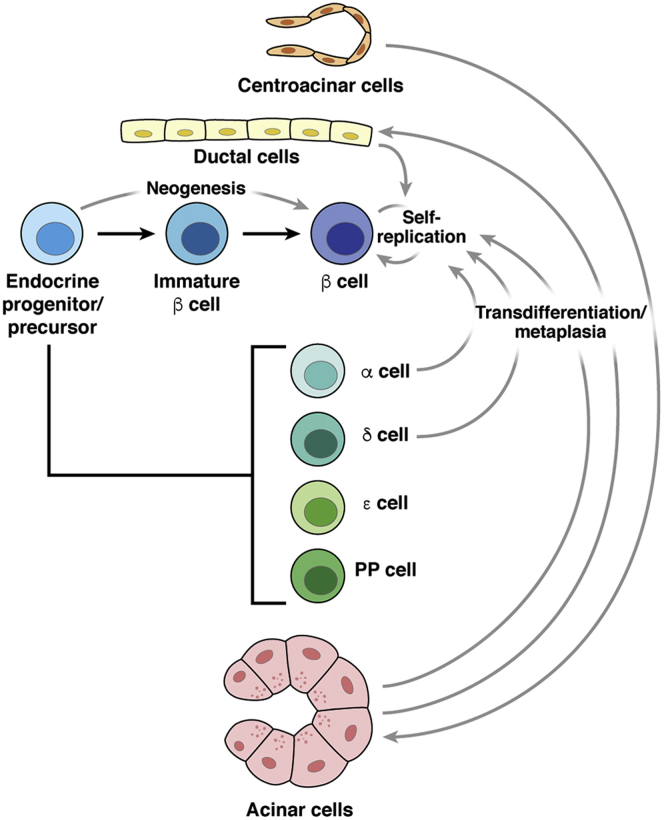
Much of the ambiguity regarding the potential cellular origins of pancreatic cancer rests in our newfound understanding of plasticity among terminally differentiated cells of the pancreas, particularly in the endocrine pancreas (as summarized in Figure 1). In initial diabetes-related studies, experimental models of rodent injury, including pancreatectomy, appear to induce the self-replication of β cells, rather than differentiation of precursor cells or transdifferentiation from another compartment.8 This mechanism of self-replication appears essential to homeostasis in the setting of pregnancy, obesity, or induced insulin resistance. However, the response to near-complete ablation of the β cell population within the endocrine pancreas using a diphtheria toxin–based system is quite different. Lineage-tracing of terminally differentiated cells revealed that glucagon-producing α cells, rather than a latent progenitor population, are largely responsible for restoration of β cell populations.9 Interestingly, in young mice, somatostatin-producing δ cells (but not α cells) undergo spontaneous conversion into β cells, proceeding through a dedifferentiated intermediate.10 This α-to-β cell conversion appears to be inducible by the administration of gamma aminobutyric acid, and arises not from a terminally differentiated α cell, but a precursor cell lining the duct.11 The concept of persistent endogenous progenitors in the ductal lining is supported by earlier work suggesting that Ngn3+ hormone negative cells are expanded with pancreatic duct ligation, differentiate into β cells, and can be purified from the adult pancreas.12 Together, these key findings demonstrate that β cell repopulation can occur through a multitude of routes—transdifferentiation of other cell types, self-replication, or neogenesis from a pool of endocrine progenitor cells—depending on the timing and nature of the physiologic insult.13,14 These studies in the endocrine pancreas make clear that plasticity is an abundant feature of the normal pancreas, and that in disease, one cellular compartment may be repopulated by another.
Figure 1.
Cellular plasticity within the normal pancreas. Pancreatic progenitors arise from definitive endoderm; ‘Tip’ and ‘trunk’ compartment specialization follows formation of microlumens within the pancreatic bud and gives rise to major cell types as indicated. These cell types can be repopulated by transdifferentiation, self-replication, and neogenesis in the setting of injury or insult.
Plasticity Within the Exocrine Compartment
Multiple lines of evidence also support abundant cellular plasticity within the exocrine compartment in response to pancreatic injury (see Figure 1). Acute pancreatitis, induced by either pancreatic duct ligation or by the exogenous administration of the cholecystokinin analog caerulein, features release of digestive enzymes from the zymogen granules of acinar cells. In response to this injury, acinar cells lose polarity and flatten somewhat, taking on a cuboidal-columnar morphology and resembling ductal precursors in the embryonic pancreas. Despite not being completely flat and elongated as a typical ductal cell, this process has been termed acinar-ductal metaplasia (ADM), largely owing to the acquisition of a mucinous cytoplasm and surface expression of cytokeratin 19, both of which are characteristic of ductal cells.15 In addition to ADM, the caerulein-treated mouse pancreas features infiltration by inflammatory cells, expansion of the interstitium, and loss of acinar cell area. In their acquisition of a “duct-like” phenotype, acinar cells specifically lose key features of their cell identity, including transcription factors essential to acinar cell fate (Ptf1a, Mist1).15,16 In mouse and human models, acinar cells undergoing ductal metaplasia frequently will simultaneously express acinar-specific enzymes (amylase, trypsin, elastases, carboxypeptidases, etc.) in addition to CK19 and the HMG-box transcription factor SOX9 that are found in ductal cells.15,17 At best, then, so-called ADM could be characterized as incomplete given the persistence of acinar-specific gene expression, with several of the key transcription factors and marker proteins summarized in Table 1.
Table 1.
Summary of Key Transcription Factors and Markers of Exocrine and Endocrine Cells
| Key Protein | Family | Function | Onset of Expression | Expression in |
|||
|---|---|---|---|---|---|---|---|
| Adult Pancreas | ADM | PanIN | PDAC | ||||
| Amylase | Secretory zymogen | Carbohydrate digestion | e13.5 | Acinar | + | – | – |
| Cpa1 | Secretory zymogen | Cleaves C-terminal branched-chain amino acids from dietary proteins | e10.5 | Acinar | + | – | – |
| Dclk1 | Doublecortin | Microtubule-polymerizing activity | No embryonic study | Ductal, islets, and acinar (minority of cells) | + | ++ | ++ |
| Elastase | Chymotrypsin-like | Carbohydrate digestion | e12.5 | Acinar | + | – | – |
| GATA4 | Zinc-finger transcription factors | Regulators of the expansion and maintenance of MPCs | e9.5-e10.5 | Acinar and B cell | +++ | +++ | ++ |
| GATA6 | Zinc-finger transcription factors | Regulators of the expansion and maintenance of MPCs | e9.5-e10.5 | Endocrine and exocrine | + | + | – |
| Hnf1β | Homeodomain-containing family TF | Require for pancreatic precursors generation | e8.5 | Ductal and centroacinar | + | + | + (variable) |
| Hnf6 | ONECUT class of cut homeodomain proteins | Essential regulator of Pdx1; endocrine cell differentiation | e8.5 | Ductal | + | – | – |
| Krt19 | Type I keratin | Intermediate filament protein key to cellular structure | e13.5 | Ductal and centroacinar | +++ | ++++ | +++ |
| Mist1 | Basic helix-loop-helix TF | Acinus polarity, cell-cell junctions, and the processing of zymogen granules | e12.5 | Acinar | – | – | – |
| Ngn3 | Basic helix-loop-helix TF | Initiates endocrine developmental program | e9.5-e10.5 | Low expression in islet | + | – | – |
| Nr5a2 | Nuclear hormone receptor family | MPC formation; normal acinar differentiation and function | e7.5 | Acinar | + | – | – |
| Pdx1 | TF of ParaHox gene cluster | Determine fate specification of pancreatic MPCs | e8.5-9.0 | B cell and low level exocrine cells | ++ | ++ | ++ |
| Ptf1a | Basic helix-loop-helix TF | Acinar enzyme gene activator; determine fate specification of pancreatic MPCs | e9.5 | Acinar | + (low) | + (low) | – |
| Sox9 | Member of the SRY/HMG box (Sox) family | Proliferation, survival, and maintenance of pancreatic progenitors | e9.5 | Ductal and centroacinar | +++ | ++++ | ++ |
ADM, acinar-ductal metaplasia; MPC, mesodermal precursor cell; PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma; TF, transcription factor.
Further complicating matters is that ADM cannot be viewed as a “pure” transdifferentiation event (ie, direct conversion from acinar to ductal), as it is accompanied by features suggestive of a dedifferentiated phenotype (ie, reversion within the pancreatic lineage). Extensive immunohistochemistry of the pancreas following caerulein administration has shown that the pancreatic progenitor marker Pdx1, Notch, and β-catenin are all reactivated.18,19 Notably, differentiated ductal cells do not display abundant Notch signaling or β-catenin expression, reaffirming that the acquired acinar cell fate is not entirely ductal in nature. In vitro experimentation has demonstrated the activation of the progenitor marker Nestin over the course of acinar cell identity loss.20 More recent work has shown that the expression of pluripotency genes Oct4, Sox2, Klf4, and Myc are sufficient to induce dedifferentiation of the acinar cell and potentiate ADM formation.21 Klf4 overexpression alone also potentiates loss of acinar cell identity and is required for ADM formation.22 In elegant studies by Real and colleagues, haploinsufficiency of the acinar-defining transcription factor Nr5a2 is capable of driving a low-level inflammatory state that destabilizes terminally differentiated acinar cells.23 Together, these findings suggest that acinar cells readily acquire features of both ductal and progenitor cells in response to pancreatic injury, a process that has been recently described as “paligenosis.”24
Within the exocrine compartment, ductal cells can also alter their identity, as has been observed in mice expressing the transgenic diphtheria toxin receptor under the control of the Pdx1 promoter. Diphtheria toxin administration in these mice ablates the acinar and endocrine compartments, which is then repopulated by cells of ductal origin adopting the pancreatic progenitor program.25 Despite this, many of the in vivo studies utilizing nonspecific pancreatic injury, however, fail to identify significant injury to or plasticity among ductal cells—a feature that has been attributed to the elevated expression of the antiapoptotic protein Bcl-2—suggesting that the plasticity within the exocrine compartment is most evident in acinar cells.
Loss of acinar cell identity in response to caerulein-induced pancreatitis is reversible, with the exocrine pancreas undergoing “redifferentiation” to regain its normal architecture and function. Regeneration of the acinar compartment is dependent on the reactivation of several developmental pathways, including β-catenin signaling,26 Hedgehog signaling,27 and the Notch pathway.28,29 In addition to developmental signals, bona fide regeneration of the acinar compartment also relies on preservation of acinar-specific transcription factors, including Nr5a230 and Ptf1a.31 In support of redifferentiation as the means of regeneration, the enforced expression of pluripotency genes tends to lead to permanent loss of acinar cell identity, rather than to induction of a transient “facultative progenitor” state.21,22 Regeneration of the acinar compartment also does not occur in Kras-driven metaplasia, likely owing to a similar inability to undergo this process of redifferentiation as Kras inactivation allows for reversion of both ADM and PanIN.32 Regeneration of the acinar cell thus appears to be dependent on the ability to reactivate normal developmental pathways to acquire determinants of the differentiated acinar cell type.
Acinar cell plasticity can be exploited to derive other cell types. Inactivation of c-Myc within the pancreas has been shown via lineage tracing to induce the transdifferentiation of acinar cells into adipocytes.33 This transdifferentiation was also evident during caerulein-induce injury alone, suggesting that acinar cell fate is permissive of even mesenchymal transdifferentiation. In addition, acinar cells can be used to derive endocrine-derived cells following induction of Pdx1, Ngn3, and Mafa—all key transcription factors in β cell development.34 In other work, suppression of Ptf1a activity is also permissive of an acinar-to-endocrine conversion.35 Lineage-tracing reveals that acinar cells specifically can undergo a transdifferentiation to an endocrine phenotype following pancreatic duct ligation with or without streptozotocin.36
Further plasticity in the exocrine pancreas exists among centroacinar cells (CACs) and specific niche populations. Sox9–positive CACs can differentiate into acinar cells,37 and can do so in response to pancreatic injury (Figure 1).38 Similarly, the DCLK1+ subpopulation of acinar cells are expanded with pancreatic injury and are necessary for regeneration following caerulein-mediated injury.39 Therefore, acinar cells, representing over 90% of the pancreas mass, are the most abundant source of cells highly amenable to a change in identity in noncancer contexts; it should serve as no surprise then that these cells could give rise to cancer in the appropriate contexts.
Premalignant Lesions of the Pancreas and Their Origins
Invariably, PDAC does not appear to arise de novo, but instead from progressive dysplasia in precursor lesions derived from the neoplastic transformation of normal cells; the origins of these lesions have thus long thought to be central to understanding the beginnings of PDAC.
Pancreatic Intraepithelial Neoplasia
First described by Brat et al40 in the analysis of lesions associated with human PDAC, PanIN represents flat or papillary epithelial lesions that develop progressive dysplasia. PanINs are graded based on the level of atypia as either PanIN-1, -2, or -3, and are almost always associated with KRAS mutation41,42 and, with greater atypia, associated tumor-suppressor loss.43 Indeed, the majority of human PDAC is found in association with PanIN,44 suggesting that PDAC occurs either as a transformation of underlying PanIN or requires a concomitant PanIN lesion. Interestingly, recent sequencing of human PanINs not associated with PDAC suggests that tumor suppressor inactivation is infrequent among isolated PanIN.45
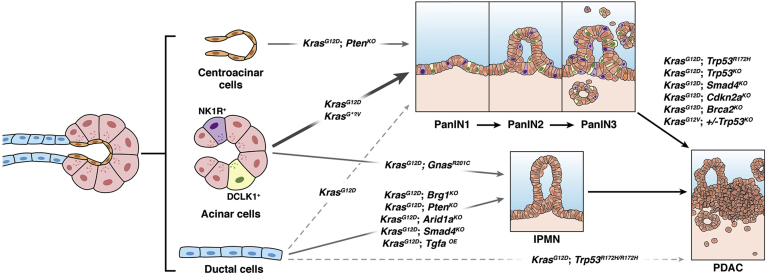
Mounting evidence clearly demonstrates that acinar cells are the likely cell of origin for PanINs.46, 47, 48 In initial studies, the expression of a Myc transgene under the control of the Ela (elastase) promoter was found to drive loss of elastase expression and of acinar cell differentiation, as well as the accumulation of duct-like structures.49 It was not until the subsequent development of Cre-lox technology that the ability to direct conditional expression of driver oncogenes allowed for further mouse models of PanIN. In one early study, direction of mutated Kras to progenitor cells of the pancreas clearly recapitulated the PanIN histology in mice, suggesting that the rodent model parallels the histological features of malignant precursors to human PDAC.50 Usage of the cytokeratin 19 promoter to drive mutant Kras expression in ductal cells, however, failed to induce neoplasia and showed only a lymphocytic infiltration in the pancreas.51 In seminal work from Guerra et al,52 the expression of KrasG12V was restricted to acinar and CACs using the Elastase promoter and coupled to a tetracycline-regulated (tetO) system. In this system, PanINs developed by 2–3 months of age in the absence of doxycycline and progressed to PDAC by 10–12 months of age.52 In contemporaneous work, however, targeting of KrasG12D to Nestin-expressing cells was shown to generate PanIN with the same frequency as pan-pancreatic drivers such as p48 or Pdx1,53 suggesting a potential progenitor origin for PanIN. As mentioned previously, Nestin upregulation is a hallmark finding of acinar cell dedifferentiation, such that these findings also do not exclude acinar cells as the source of the “progenitor” population.
Additional support for acinar cells as the source of PanIN has arisen from the use of additional promoters specific to the acinar cell population, as illustrated in Figure 2. In an early study, cloning of KrasG12D in-frame in to the locus of Mist1—a basic helix-loop-helix transcription factor expressed primarily in acinar cells—generated acinar-ductal metaplasia and PanIN.54 Usage of a tamoxifen-inducible Elastase-Cre (ElaCreERT) in conjunction with the LSL-KrasG12D-allele has also been shown to generate PanIN from acinar cells.55 Importantly, this study also utilized a Cre-dependent Notch1 gain-of-function transgene containing green fluorescent protein to label acinar cells and were able to show that PanIN lesions were derived from green fluorescent protein–expressing cells. Habbe et al56 took these studies one step further by using both ElaCreERT and Mist1CreERT2 alleles coupled with KrasG12D to show that PanINs arise spontaneously alongside ADM lesions and even “biphenotypic” cells displaying features of both acinar and ductal differentiation. Critically, Cre recombinase induction in the adult using these drivers is specific to the acinar cell population and excludes CACs. Together, these efforts provide strong evidence that acinar cells readily generate PanINs, both with and without superimposed pancreatitis.
Figure 2.
Cell-of-origin of pancreatic neoplasia and adenocarcinoma in mouse models. The predominant cell types of the exocrine compartment of the pancreas – acinar, centroacinar, and ductal cells – give rise to pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasms (IPMNs) in the mouse in the genetic contexts indicated above. Subsequent pancreatic ductal adenocarcinoma (PDAC) usually requires tumor suppressor loss in addition to oncogene activation.
Despite these studies, the capacity of ductal cells to generate PanINs was not completely evaluated until a few years ago. In key work from Kopp et al,57 this question was answered elegantly using Ptf1aCreER and Sox9CreER alleles to drive mutant Kras expression in acinar and ductal and CACs, respectively. The authors observed mutant Kras could not generate significant PanIN derived from Sox9-expressing ductal and CACs despite the fact that PanIN lesions express Sox9. While other reports suggest that ductal cells can give rise to PanIN,58 acinar cells are far more efficient at undergoing loss of terminally differentiated identity to express specific ductal markers (including Sox9) that are hallmarks of ADM and PanIN, suggesting that Sox9 (while essential to ADM and PanIN development)59,60 does not confess cell of origin. Additional evidence showing that inactivation of mutant Kras in early and established PanIN restores acinar cell area and acinar-specific transcriptional programs also highlights the acinar cell as the likely source of PanIN.32 Data from humans do support this, as ADM found in association with PanIN frequently does contain the same KRAS mutations, suggesting that coexistent ADM/PanIN arise from the same preneoplastic acinar cell.61 Furthermore, ADM but not ductal cells are typically found in proximity to neoplastic precursor lesions, again supporting an acinar origin for PanIN.62
Intraductal Papillary Mucinous Neoplasms
Unlike PanINs, IPMNs are macroscopic lesions that are characterized by cystic appearance radiographically, intraluminal mucin, and papillary growth. Insofar as IPMNs resemble large ducts and can involve the main pancreatic duct, these precursor lesions are thought to arise from ductal cells more proximally in the arborization of the pancreatic ductal tree. Indeed, ductal cell populations are heterogeneous and consist of 2 groups—those connecting to the acini and those forming the terminal duct—with spatial localization impacting gene expression and morphology.63,64 In humans, IPMN appears to be associated with 15% of pancreatic cancer, but the vast majority of clinically identified IPMNs do not progress to malignancy despite bearing many of the same oncogenic drivers (activating KRAS mutations in particular) To what degree IPMNs and PanINs are related remains unknown, but the former also features frequent mutations to GNAS that are quite rare in “conventional” PanIN-associated PDAC,65 suggesting distinct origins. Moreover, intestinal-type IPMNs tend to proceed to histologically distinct colloid carcinoma that is associated with more favorable outcome.66 Tubular variant adenocarcinomas arising from gastric and pancreatobiliary histologic subtype IPMNs, however, are indistinguishable from classical PanIN-derived PDAC—whether this reflects convergent mechanisms of tumorigenesis between PanIN and IPMN or if there is concomitant PanIN that progresses to PDAC is unknown.
Mouse models have shed light on the origins of IPMN, with ductal cells historically considered the likely precursor. Early studies utilizing KrasG12D as the oncogene coupled with transgenic overexpression of Tgfa generated large cystic and papillary structures mimicking IPMN (in addition to PanIN); these were seen using p48-Cre mice but not when using Elastase-CreER mice—suggesting that adult acinar cells do not readily give rise to IPMN.67 The ability of ductal cells to specifically serve as cells of origin for IPMNs has been observed with mutant Kras activation coupled with loss of Brg1, the catalytic subunit of SWI/SNF chromatin remodeling complexes. Loss of Brg1 in acinar cells using the Ptf1a-CreER driver abrogates the development of PanIN, whereas loss in ductal cells (using Hnf1b-CreERT2 mice) potentiates the development of rare Kras-driven IPMN-like lesions.68 Both Brg1 deletion and loss of Arid1a (another SWI/SNF subunit) in ductal cells has been shown to drive dedifferentiation of these cells in vitro.69,70 Similarly, development of macroscopic, cystic intraductal papillary lesions has been demonstrated with loss of the tumor suppressor phosphatase Pten or Liver kinase B1 (Lkb1) directed specifically to ductal cells using the Sox9-CreERT2 allele.71,72 Together, these findings certainly demonstrate the competency of ductal cells to give rise to IPMN-like lesions in mice, as summarized in Figure 2.
The aforementioned mouse models invariably have utilized mutant Kras as the driver oncogene for IPMN development. However, approximately 60% of IPMNs feature mutations to GNAS, which encodes a protein that functions downstream of several G-protein coupled receptors to activate adenylyl cyclase to generate cyclic adenosine monophosphate.65,73,74 Coupling of one of the frequent Gnas mutations (R201H) to KrasG12D in the pancreas using Ptf1a-Cre generated cystic tumors consisting of marked dilated ducts lined with papillary dysplastic epithelia, resembling IPMN.75 Interestingly, a more recent study coupled another hotspot Gnas mutation (GnasR201C) to KrasG12D to generate IPMNs but discovered that the same phenotypes were observed even with usage of the Ptf1a-CreER allele to direct recombination events solely in adult acinar cells.76 This study calls into question the notion that acinar cells generate PanIN and ductal cells generate IPMN, and suggests that neoplastic fate may be as dependent on the specific driver mutation as it is on the cell type in which the mutation arises.
Perspectives on the Origins of PDAC
Beginning with the initial mouse models using oncogenic Kras in Pdx1+ pancreatic progenitors,50 there have been a multitude of studies describing specific murine allelic configurations to generate PDAC.77, 78, 79, 80, 81, 82, 83, 84, 85 However, it is only with the advent of in vivo lineage tracing that specificity regarding the ability of a particular cell type to yield invasive disease has been addressed (and is summarized in Figure 2 and Table 2). In the following, we address the state of the evidence regarding each potential cellular precursor of PDAC.
Table 2.
Development of Premalignant Lesions and PDAC From Varying Cells of Origin
| Mutation | Embryonic Expression |
Acinar Expression |
Ductal Expression |
|||
|---|---|---|---|---|---|---|
| Pathology | Cre Driver | Pathology | Cre Driver | Pathology | Cre Driver | |
| KrasG12D | PanIN/PDAC | Ptf1a-Cre32,50 Pdx1-Cre50 |
PanIN/late PDAC | Ptf1a-CreER57,86 | No lesions or cancer | Sox9-CreER57,86 |
| Early PanIN | CK19-CreERT258 | |||||
| Sporadic PanIN | Mist1-CreER56,87 Elastase-CreERT255,56 Dclk1-CreERT39 |
PDAC | Sox9-CreERT2 with PDL88 | |||
| PanIN | Elastase-CreERT2 with caerulein59 Mist1-CreER with caerulein87 Dclk1-CreERT with caerulein39 |
PDAC | CK19-CreERT2 with PDL88 | |||
| No lesions or cancer | Elastase-CreERTM with PDL88 | |||||
| sporadic PanIN | Pdx1-IRES-Cre21 Nestin-Cre53 |
|||||
| KrasG12V | PanIN/PDAC | Elas-tTA; tetO-Cre52 | No lesions or cancer | Elas-tTA; tetO-Cre52 | ||
| PanIN/PDAC | Elas-tTA; tetO-Cre with caerulein52 | |||||
| KrasG12D; p53R172H | PanIN/PDAC | Pdx1-Cre77 | PanIN/PDAC | Ptf1a-CreER86 Mist1-CreER89 |
No lesions or cancer | Hnf1b-CreERT289 |
| KrasG12D; p53flox/+ | PanIN/PDAC | Ptf1a-CreER86 | ||||
| KrasG12D; p53flox/flox | PanIN/PDAC | Pdx1-Cre80 | PanIN/PDAC | Ptf1a-CreER90,86 Elastase-CreERT2 with caerulein91 |
PDAC | Sox9-CreER90,86 |
| Nonmucinous tubular lesions/PDAC | CK19-CreER91 | |||||
| KrasG12D; p53flox/- or flox/R270H or flox/R172H | PanIN/PDAC | Ptf1a-CreER86 | PDAC | Sox9-CreER86 | ||
| KrasG12D; p53R172H/R172H | PDAC | Hnf1b-CreERT289 | ||||
| KrasG12V; p53+/– | PanIN/PDAC | Elas-tTA; tetO-Cre52 | ||||
| KrasG12D; p16–/– | PanIN/PDAC | Pdx1-Cre80,78 | ||||
| KrasG12D; p16/Arfflox/flox or –/– | PanIN/PDAC | Pdx1-Cre80,78 | ||||
| KrasG12D; Smad4flox/flox | IPMN/PDAC | Pdx1-Cre79 | ||||
| KrasG12D; Tgfbr2flox/flox | PanIN/PDAC | Ptf1a-Cre92 | ||||
| KrasG12V; p16/Arflox/+ | PanIN | Elas-tTA; tetO-Cre93 | ||||
| KrasG12V; p16/Arflox/lox | PanIN/PDAC | Elas-tTA; tetO-Cre93 | ||||
| KrasG12D; Lkb1lox/lox | PanIN/PDAC | Pdx1-Cre84 | IPMN/premature death | Sox9-CreERT272 | ||
| KrasG12D; Brg1f/f | IPMN/PDAC | Ptf1a-Cre68,70 | No lesions or cancer | Ptf1a–CreER68 | Few IPMN | Hnf1b-CreERT268 |
| KrasG12D; Arid1af/+ | PanIN | Ptf1a-Cre70 | PanIN | Ptf1a-CreERT2 with caerulein70 | No lesions or cancer | Hnf1b-CreERT270 |
| KrasG12D; Arid1af/f | PanIN and IPMN/PDAC | Ptf1a-Cre70 | PanIN | Ptf1a-CreERT2 with caerulein70 | Few IPMN | Hnf1b-CreERT270 |
| KrasG12D; Fbw7F/F | PanIN/PDAC | Elastase-CreERT2 with caerulein91 | Nonmucinous tubular lesions/PDAC | CK19-CreER91 | ||
| GNASR201H | Early IPMN | Ptf1a-Cre75 | ||||
| KrasG12D; GNASR201H | IPMN/cystic tumor | Ptf1a-Cre75 | ||||
| KrasG12D; GNASR201C | IPMN | Ptf1a-Cre76 | IPMN | Ptf1a-CreER76 | ||
| KrasG12D; GNASR201C; p53LoxP/+ | IPMN/PDAC | Ptf1a-CreER76 | ||||
| Ptenflox/flox | PanIN and papillary adenocarcinoma/PDAC | Pdx1-Cre94 | No lesions or cancer | Elastase-CreERTM94 | IPMN/PDAC—spontaneous Kras mutation | Sox9-CreERT271 |
| Ptenflox/flox; p53–/– | Papillary adenocarcinomas | Pdx1-Cre94 | ||||
| KrasG12D; Ptenflox/flox | IPMN/PDAC | Sox9-CreERT271 | ||||
Note: Blank cells indicate models that have not been evaluated.
ADM, acinar-ductal metaplasia; IPMN, intraductal papillary mucinous neoplasm; PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma; PDL, pancreatic duct ligation.
Acinar Cells
Given the evidence in the mouse model outlined previously to show that acinar cells give rise to PanIN, it would appear evident that these cells likely serve as the cell of origin for PDAC. Indeed, several lineage-tracing mouse models that incorporate mutant p53 with acinar-directed Kras activation largely support this possibility, with mice developing invasive PDAC.93,95 Usage of the more mature acinar-specific driver Mist1-CreERT2 coupled to KrasG12D and mutant p53 also leads to invasive PDAC and relatively rapid demise among mice that mirror early nonselective Pdx1-Cre and p48-Cre models.89,87
However, several critical caveats remain. First, the bulk of evidence is derived from cell type–specific mouse models and therefore addresses only the competence of murine cell types to give rise to a histology similar to that seen in associated with human PDAC. In all of the available genetic mouse models, oncogenic Kras is directed to all cells of the corresponding lineage; this does not completely mimic what is likely a sporadic event in humans, as acinar cells adjacent to PanINs largely bear wild-type KRAS.96 It is therefore quite possible that the phenotypes derived from widespread Kras activation in acinar cells, which represent 95% of pancreatic exocrine cells, might be quite different from the human scenario wherein presumably only few cells acquire oncogenic mutations. On the other hand, many of the mouse models do employ tamoxifen- or doxycycline-inducible oncogenic Kras, with efficiencies of recombination far less than the total acinar cell population. In addition, oncogenic Kras is necessary but not sufficient for the development of neoplasia (as injury/inflammation is frequently necessary), such that widespread Kras activation in these models should not necessarily be viewed as a limitation of these studies.
Second, acinar cells are a highly heterogeneous population of cells that may feature variable differentiation states. Single-cell analysis of acinar cells has a identified a progenitor-like subpopulation with long-term self-renewal capacity that expresses the progenitor transcript STMN1.97 STMN1+ populations are expanded with pancreatic injury, suggesting a potential role in regeneration. Therefore, the possibility remains that it is not acinar cells as a whole that represent the cell of origin, but rather is a subpopulation of acinar cells whose behavior and function are quite different from the bona fide acinar population. On the other hand, expansion of STMN1+ populations could simply reflect that this is another gene whose expression is induced in injury, and not necessarily outgrowth of an acinar subset.
Third, many of the mouse models also employ caerulein, which appears to selectively injure acinar cell populations. Intriguingly, mouse models that employ pancreatic duct ligation as an alternative means of inducing pancreatic inflammation show different exocrine cell predispositions to tumorigenesis. In this model, acinar cells are surprisingly recalcitrant to tumorigenesis undergoing apoptosis after ADM, suggesting that the specific mode of acinar cell injury may impact the likelihood of tumor information.88
Regardless, the enormous plasticity and heterogeneity of acinar cells can thus be seen as either one reason it is the likely cell of origin for PanIN-associated PDAC, or a potential confounding variable in studies targeting genetic events to this diverse cell population. The previous caveats notwithstanding, the preponderance of evidence shows that acinar cells are highly capable of generating PanIN and IPMN in response to oncogenic Kras and co-occurring mutant Kras/Gnas, respectively, and that precursor lesions derived from acinar cells can progress to invasive disease. The transcriptional and epigenetic determinants of acinar cell plasticity, particularly in the context of oncogenic stress, thus appear to be worthwhile avenues for further investigation.
Ductal Cells
With recent data, the capacity of ductal cells to function as the cell of origin for PDAC has become increasingly unclear. Certainly, the capacity of these cells to undergo a cell identity change outside of cancer contexts appears limited, particularly as compared with acinar cells. Accordingly, it should be noted that while ductal cells may not effectively produce PanIN, there are multiple contexts in which these cells can give direct rise to PDAC. Specifically, Bailey et al89 demonstrated the ability of ductal cells (selected using the Hnf1b-CreERT2) allele to generate PDAC in the setting of 2 copies of mutant p53 but without associated PanIN lesions. In a similar study employing deletion of p53 using a floxed allele, a duct-specific driver of Cre recombinase (Sox9-CreER) combined with KrasG12D was observed to generate PDAC more rapidly than with an acinar-specific driver (Ptf1a-CreER).90 Further work by Flowers et al86 employing a comprehensive panel of genetically engineered mice largely recapitulate these findings, namely that p53 mutation or deletion is required for PDAC development from pancreatic ductal cells (in the absence of superimposed injury). Interestingly, duct- and acinar-origin tumors appear molecularly distinct, and map closely to basal and squamous and classical and progenitor transcriptional subtypes, respectively.86 In all of these aforementioned studies, there is a paucity of low-grade PanIN that accompanies ductal-driven PDAC, reinforcing the notion that ductal cells are not competent to generate PanIN but may give rise to PDAC. These findings thus raise the distinct possibility that in humans a greater proportion of PDAC may in fact arise from ductal cells, and that coincident PanIN found in patients might arise from separate mutational events in distinct acinar cells. However, genetic sequencing of PanINs and associated PDAC suggest more shared than distinct drivers, indicating that independent coevolution of acinar-derived PanIN and ductal-derived PDAC is likely infrequent.87 On the other hand, Ferreira et al91 also demonstrated using Fbw7 deletion that ductal-driven PDAC arises independent of PanIN and that, importantly, orthotopic transplantation induces the development of bystander PanIN, suggesting that not all PanIN lesions are precursors of PDAC.
As mentioned previously, there are multiple genetic mouse models in which the development of IPMN can be driven specifically from a ductal origin. It should be noted, however, that the available models clearly demonstrating ductal-derived IPMNs utilize additional genetic events to either transforming growth factor β signaling pathway components (Tgfa, Smad4), chromatin remodelers (Brg1, Arid1a), to Pten or to Lkb1, all of which are infrequently mutated in either premalignant or invasive IPMN. The findings by Bardeesy et al76 that co-occurring Gnas/Kras mutations—which actually recapitulate the driver mutations found in human disease—are associated with a nonductal origin of IPMN are therefore highly compelling. While specific mouse models can generate ductal-derived IPMN, it remains unclear if ductal cells are frequent cells of origin in the human setting. Nevertheless, the frequency of basal and squamous subtype PDAC, which might be more likely to be ductal-derived, and the emergence of mouse models employing duct-specific inactivation of p53 or utilizing alternative forms of pancreatic injury (eg, pancreatic duct ligation) demonstrate that ductal-driven tumorigenesis is quite possible from common driver mutations but not with an apparent low-grade IPMN or PanIN precursor.
Centroacinar Cells
Several studies do point to CACs, which are present at the junction between terminal acini and ductal cells, as potential precursors to PDAC. Stanger et al were able to identify an expansion and proliferation of these cells in Pten-deficient mice, and a development of ductal metaplasia and malignant transformation in Pdx1-Cre but not Elastase-CreERTM mice.94 Notably, Sox9-Cre KrasG12D mice, wherein mutant Kras is expressed in both ductal and centroacinar cells, do not demonstrate robust PanIN. Perhaps these 2 contradictory findings can be reconciled by the fact that the driver mutations are different, and that Pten deletion is a sensitivity specific to the ductal–CAC compartment.71,94 As a putative contribution of CACs appears restricted to only rare models of pancreatic neoplasia, and utilizing Pten deficiency that is typically not observed in human disease, there appears to be little evidence to implicate CACs as the cell of origin of PDAC.
Cancer Stem Cells and Niche Populations
As to the role of niche populations within the exocrine compartment, there are increasing data to point to specific neuroendocrine cells (among acinar cells) responsive to neuronal input and important to PanIN progression98 such that ablation of sensory neurons impacts the development of PanIN and PDAC.99, 100, 101 Cancer stem cells have also been proposed to exist in the pancreas, as defined by enhanced tumorigenicity of the CD44+ CD24+ ESA+ or CD133+ CXCR4+ populations of epithelial cells.102,103 Largely, these cancer stem cells have been shown to have enhanced clonogenic growth in vitro and heightened metastatic potential in vivo, and also bear substantial overlap with the niche population of so-called tuft cells in the pancreas that express Dclk1 and acetylated tubulin (AcTub) on their cell surface.104 AcTubHI Dclk1+ cells bear the same “stem cell” markers (CD133, CD24, ESA, etc.) and have been shown to be important to PanIN development and progression.104,92 Direction of oncogenic Kras to the Dclk1+ population appears as efficient at initiating PanIN as usage of the Mist1 promoter, and deletion of Dclk1 diminishes PanIN formation substantially.92 Importantly, lineage tracing suggests the vast majority of Dclk1+ cells, which represent a rare population (0.1%-0.5% of all cells) in the normal pancreas belong to the acinar lineage, such that these data reinforce the notion that acinar cells are the cell of origin for pancreatic neoplasia and cancer. Whether these are subsets of acinar cells from which premalignant lesions and PDAC exclusively arise remains a subject for further investigation. In addition, there are no data to our knowledge that demonstrate that these cancer stem cells are a fixed population of acinar cells and not a stochastically acquired phenotype among acinar cells exhibiting transcriptional plasticity.
Conclusions
As the usage of genetically engineered mouse models of PanIN, IPMN, and PDAC arising from both precursor lesions has evolved, so too has the current state of knowledge regarding the cell of origin of pancreatic cancer. The body of literature yields a complex, perhaps muddled, picture that should be no surprise given the abundant plasticity within the normal endocrine and exocrine pancreas. In keeping with the observations from pancreatic injury, acinar cells are easily reprogrammed to give rise to neoplastic PanIN and IPMN lesions. Though human PDAC appears to arise largely in the context of PanIN and IPMN, genetically engineered mice can seemingly bypass premalignant lesions, particularly in the context of PDAC that is generated from the ductal lineage. Overall, pancreatic cancer can be seen as arising from 3 routes—from underlying PanIN, from IPMN, and perhaps from bypassing either of these intermediates. The data suggest that PanIN is most easily generated in the mouse through the expression of oncogenic Kras in acinar cells; IPMN, by contrast, can variably arise from acinar cells (in the context of mutant Kras and Gnas) or ductal cells (in the context of chromatin remodeling or transforming growth factor β signaling mutations). PDAC that arises in the background of a normal pancreas is observed using ductal drivers and specific modes of injury or tumor suppressor loss. The specific roles of CACs, as well as that of niche populations within the acinar and ductal compartments that bear heightened tumor-initiating potential, remain yet to be fully elucidated. Regardless, the multitude of models have made clear that driver mutations may function differently depending on the affected population. In the end, there may not be a cell of origin per se of pancreatic cancer, but rather there may be susceptibilities that are dependent on the intersection between cell type, niche population, mode of injury, and the precise mutational events that destabilize the normal cell.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Adrien Grimont is supported by the Prevent Cancer Foundation Fellowship. Rohit Chandwani is supported by the AACR-Pancreatic Cancer Action Network Pathway to Leadership Award, American Surgical Association Foundation Fellowship Award. Steven D. Leach is supported by the Ring Family Foundation and by the National Cancer Institute R01CA204228.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Moyer M.T., Gaffney R.R. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2139–2141. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue C.A., Fu B., Yachida S., Luo M., Abe H., Henderson C.M., Vilardell F., Wang Z., Keller J.W., Banerjee P., Herman J.M., Cameron J.L., Yeo C.J., Halushka M.K., Eshleman J.R., Raben M., Klein A.P., Hruban R.H., Hidalgo M., Laheru D. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feig C., Gopinathan A., Neesse A., Chan D.S., Cook N., Tuveson D.A. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hezel A.F., Kimmelman A.C., Stanger B.Z., Bardeesy N., Depinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 7.Cao D., Maitra A., Saavedra J.A., Klimstra D.S., Adsay N.V., Hruban R.H. Expression of novel markers of pancreatic ductal adenocarcinoma in pancreatic nonductal neoplasms: additional evidence of different genetic pathways. Mod Pathol. 2005;18:752–761. doi: 10.1038/modpathol.3800363. [DOI] [PubMed] [Google Scholar]
- 8.Dor Y., Brown J., Martinez O.I., Melton D.A. Adult pancreatic beta-cells are formedby self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 9.Thorel F., Népote V., Avril I., Kohno K., Desgraz R., Chera S., Herrera P.L. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J.N., Gu G., Furuyama K., Thorel F., Gribble F.M., Reimann F., Herrera P.L. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Othman N., Vieira A., Courtney M., Record F., Gjernes E., Avolio F., Hadzic B., Druelle N., Napolitano T., Navarro-Sanz S., Silvano S., Al-Hasani K., Pfeifer A., Lacas-Gervais S., Leuckx G., Marroquí L., Thévenet J., Madsen O.D., Eizirik D.L., Heimberg H., Kerr-Conte J., Pattou F., Mansouri A., Collombat P. Long-term GABA administration induces alpha cell-mediated beta-like cell neogenesis. Cell. 2017;168:73–85.e11. doi: 10.1016/j.cell.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., D'Hoker J., Stangé G., Bonné S., De Leu N., Xiao X., Van de Casteele M., Mellitzer G., Ling Z., Pipeleers D., Bouwens L., Scharfmann R., Gradwohl G., Heimberg H. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q., Melton D.A. Pancreas regeneration. Nature. 2018;557:351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguayo-Mazzucato C., Bonner-Weir S. Pancreatic β cell regeneration as a possible therapy for diabetes. Cell Metab. 2018;27:57–67. doi: 10.1016/j.cmet.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L., Shi G., Schmidt C.M., Hruban R.H., Konieczny S.F. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziv O., Glaser B., Dor Y. The plastic pancreas. Dev Cell. 2013;26:3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Jensen J.N., Cameron E., Garay M.V., Starkey T.W., Gianani R., Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto Y., Maitra A., Ghosh B., Zechner U., Argani P., Iacobuzio-Donahue C.A., Sriuranpong V., Iso T., Meszoely I.M., Wolfe M.S., Hruban R.H., Ball D.W., Schmid R.M., Leach S.D. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 20.Means A.L., Meszoely I.M., Suzuki K., Miyamoto Y., Rustgi A.K., Coffey R.J., Jr., Wright C.V., Stoffers D.A., Leach S.D. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 21.Shibata H., Komura S., Yamada Y., Sankoda N., Tanaka A., Ukai T., Kabata M., Sakurai S., Kuze B., Woltjen K., Haga H., Ito Y., Kawaguchi Y., Yamamoto T., Yamada Y. In vivo reprogramming drives Kras-induced cancer development. Nat Commun. 2018;9:2081. doi: 10.1038/s41467-018-04449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei D., Wang L., Yan Y., Jia Z., Gagea M., Li Z., Zuo X., Kong X., Huang S., Xie K. KLF4 is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell. 2016;29:324–338. doi: 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobo I., Martinelli P., Flández M., Bakiri L., Zhang M., Carrillo-de-Santa-Pau E., Jia J., Sánchez-Arévalo Lobo V.J., Megías D., Felipe I., Del Pozo N., Millán I., Thommesen L., Bruland T., Olson S.H., Smith J., Schoonjans K., Bamlet W.R., Petersen G.M., Malats N., Amundadottir L.T., Wagner E.F., Real F.X. Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature. 2018;554:533–537. doi: 10.1038/nature25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willet S.G., Lewis M.A., Miao Z.F., Liu D., Radyk M.D., Cunningham R.L., Burclaff J., Sibbel G., Lo H.G., Blanc V., Davidson N.O., Wang Z.N., Mills J.C. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 2018;37:e98311. doi: 10.15252/embj.201798311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Criscimanna A., Speicher J.A., Houshmand G., Shiota C., Prasadan K., Ji B., Logsdon C.D., Gittes G.K., Esni F. Duct cells contribute to regeneration of endocrine and acinar cells following pancreatic damage in adult mice. Gastroenterology. 2011;141:1451–1462. doi: 10.1053/j.gastro.2011.07.003. 1462.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris J.P., 4th, Cano D.A., Sekine S., Wang S.C., Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fendrich V., Esni F., Garay M.V., Feldmann G., Habbe N., Jensen J.N., Dor Y., Stoffers D., Jensen J., Leach S.D., Maitra A. Hedgehog signaling is required for effective regeneration of exocrine pancreas. Gastroenterology. 2008;135:621–631. doi: 10.1053/j.gastro.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Y., Büchler P., Gazdhar A., Giese N., Reber H.A., Hines O.J., Giese T., Büchler M.W., Friess H. Pancreatic regeneration in chronic pancreatitis requires activation of the notch signaling pathway. J Gastrointest Surg. 2006;10:1230–1241. doi: 10.1016/j.gassur.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Siveke J.T., Lubeseder-Martellato C., Lee M., Mazur P.K., Nakhai H., Radtke F., Schmid R.M. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134:544–555. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 30.von Figura G., Morris J.P., 4th, Wright C.V., Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2014;63:656–664. doi: 10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang C.Q., Hale M.A., Azevedo-Pouly A.C., Elsässer H.P., Deering T.G., Willet S.G., Pan F.C., Magnuson M.A., Wright C.V., Swift G.H., MacDonald R.J. Transcriptional maintenance of pancreatic acinar identity, differentiation, and homeostasis by PTF1A. Mol Cell Biol. 2016;36:3033–3047. doi: 10.1128/MCB.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galbán S., Galbán C.J., Rakshit S., Flannagan K.S., Adsay N.V., Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonal C., Thorel F., Ait-Lounis A., Reith W., Trumpp A., Herrera P.L. Pancreatic inactivation of c-Myc decreases acinar mass and transdifferentiates acinar cells into adipocytes in mice. Gastroenterology. 2009;136:309–319.e9. doi: 10.1053/j.gastro.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hesselson D., Anderson R.M., Stainier D.Y. Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Curr Biol. 2011;21:712–717. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan F.C., Bankaitis E.D., Boyer D., Xu X., Van de Casteele M., Magnuson M.A., Heimberg H., Wright C.V. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., Hosokawa S., Elbahrawy A., Soeda T., Koizumi M., Masui T., Kawaguchi M., Takaori K., Doi R., Nishi E., Kakinoki R., Deng J.M., Behringer R.R., Nakamura T., Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 38.Kopp J.L., Dubois C.L., Schaffer A.E., Hao E., Shih H.P., Seymour P.A., Ma J., Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westphalen C.B., Takemoto Y., Tanaka T., Macchini M., Jiang Z., Renz B.W., Chen X., Ormanns S., Nagar K., Tailor Y., May R., Cho Y., Asfaha S., Worthley D.L., Hayakawa Y., Urbanska A.M., Quante M., Reichert M., Broyde J., Subramaniam P.S., Remotti H., Su G.H., Rustgi A.K., Friedman R.A., Honig B., Califano A., Houchen C.W., Olive K.P., Wang T.C. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell. 2016;18:441–455. doi: 10.1016/j.stem.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brat D.J., Lillemoe K.D., Yeo C.J., Warfield P.B., Hruban R.H. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am J Surg Pathol. 1998;22:163–169. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 41.Klimstra D.S., Longnecker D.S. K-ras mutations in pancreatic ductal proliferative lesions. Am J Pathol. 1994;145:1547–1550. [PMC free article] [PubMed] [Google Scholar]
- 42.Biankin A.V., Kench J.G., Dijkman F.P., Biankin S.A., Henshall S.M. Molecular pathogenesis of precursor lesions of pancreatic ductal adenocarcinoma. Pathology. 2003;35:14–24. [PubMed] [Google Scholar]
- 43.Moskaluk C.A., Hruban R.H., Kern S.E. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 44.Hruban R.H., Adsay N.V., Albores-Saavedra J., Compton C., Garrett E.S., Goodman S.N., Kern S.E., Klimstra D.S., Klöppel G., Longnecker D.S., Lüttges J., Offerhaus G.J. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 45.Hosoda W., Chianchiano P., Griffin J.F., Pittman M.E., Brosens L.A., Noë M., Yu J., Shindo K., Suenaga M., Rezaee N., Yonescu R., Ning Y., Albores-Saavedra J., Yoshizawa N., Harada K., Yoshizawa A., Hanada K., Yonehara S., Shimizu M., Uehara T., Samra J.S., Gill A.J., Wolfgang C.L., Goggins M.G., Hruban R.H., Wood L.D. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J Pathol. 2017;242:16–23. doi: 10.1002/path.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner M., Lührs H., Klöppel G., Adler G., Schmid R.M. Malignant transformation of duct-like cells originating from acini in transforming growth factor transgenic mice. Gastroenterology. 1998;115:1254–1262. doi: 10.1016/s0016-5085(98)70098-8. [DOI] [PubMed] [Google Scholar]
- 47.Wagner M., Greten F.R., Weber C.K., Koschnick S., Mattfeldt T., Deppert W., Kern H., Adler G., Schmid R.M. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid R.M. Acinar-to-ductal metaplasia in pancreatic cancer development. J Clin Invest. 2002;109:1403–1404. doi: 10.1172/JCI15889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandgren E.P., Quaife C.J., Paulovich A.G., Palmiter R.D., Brinster R.L. Pancreatic tumor pathogenesis reflects the causative genetic lesion. Proc Natl Acad Sci U S A. 1991;88:93–97. doi: 10.1073/pnas.88.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 51.Brembeck F.H., Schreiber F.S., Deramaudt T.B., Craig L., Rhoades B., Swain G., Grippo P., Stoffers D.A., Silberg D.G., Rustgi A.K. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 2003;63:2005–2009. [PubMed] [Google Scholar]
- 52.Guerra C., Schuhmacher A.J., Cañamero M., Grippo P.J., Verdaguer L., Pérez-Gallego L., Dubus P., Sandgren E.P., Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Carrière C., Seeley E.S., Goetze T., Longnecker D.S., Korc M. The Nestin progenitor lineage is the compartment of origin for pancreatic intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2007;104:4437–4442. doi: 10.1073/pnas.0701117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuveson D.A., Zhu L., Gopinathan A., Willis N.A., Kachatrian L., Grochow R., Pin C.L., Mitin N.Y., Taparowsky E.J., Gimotty P.A., Hruban R.H., Jacks T., Konieczny S.F. Mist1-KrasG12D knock-in mice develop mixed differentiation metastatic exocrine pancreatic carcinoma and hepatocellular carcinoma. Cancer Res. 2006;66:242–247. doi: 10.1158/0008-5472.CAN-05-2305. [DOI] [PubMed] [Google Scholar]
- 55.De La O J.P., Emerson L.L., Goodman J.L., Froebe S.C., Illum B.E., Curtis A.B., Murtaugh L.C. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habbe N., Shi G., Meguid R.A., Fendrich V., Esni F., Chen H., Feldmann G., Stoffers D.A., Konieczny S.F., Leach S.D., Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopp J.L., von Figura G., Mayes E., Liu F.F., Dubois C.L., Morris J.P., 4th, Pan F.C., Akiyama H., Wright C.V., Jensen K., Hebrok M., Sander M. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ray K.C., Bell K.M., Yan J., Gu G., Chung C.H., Washington M.K., Means A.L. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One. 2011;6:e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimont A., Pinho A.V., Cowley M.J., Augereau C., Mawson A., Giry-Laterrière M., Van den Steen G., Waddell N., Pajic M., Sempoux C., Wu J., Grimmond S.M., Biankin A.V., Lemaigre F.P., Rooman I., Jacquemin P. SOX9 regulates ERBB signalling in pancreatic cancer development. Gut. 2015;64:1790–1799. doi: 10.1136/gutjnl-2014-307075. [DOI] [PubMed] [Google Scholar]
- 60.Prévot P.P., Simion A., Grimont A., Colletti M., Khalaileh A., Van den Steen G., Sempoux C., Xu X., Roelants V., Hald J., Bertrand L., Heimberg H., Konieczny S.F., Dor Y., Lemaigre F.P., Jacquemin P. Role of the ductal transcription factors HNF6 and Sox9 in pancreatic acinar-to-ductal metaplasia. Gut. 2012;61:1723–1732. doi: 10.1136/gutjnl-2011-300266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi C., Hong S.M., Lim P., Kamiyama H., Khan M., Anders R.A., Goggins M., Hruban R.H., Eshleman J.R. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol Cancer Res. 2009;7:230–236. doi: 10.1158/1541-7786.MCR-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basturk O., Hong S.M., Wood L.D., Adsay N.V., Albores-Saavedra J., Biankin A.V., Brosens L.A., Fukushima N., Goggins M., Hruban R.H., Kato Y., Klimstra D.S., Klöppel G., Krasinskas A., Longnecker D.S., Matthaei H., Offerhaus G.J., Shimizu M., Takaori K., Terris B., Yachida S., Esposito I., Furukawa T. Baltimore Consensus Meeting. A revised classification system and recommendations from the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol. 2015;39:1730–1741. doi: 10.1097/PAS.0000000000000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rovira M., Scott S.G., Liss A.S., Jensen J., Thayer S.P., Leach S.D. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M., Melton D.A., Yanai I. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu J., Matthaei H., Maitra A., Dal Molin M., Wood L.D., Eshleman J.R., Goggins M., Canto M.I., Schulick R.D., Edil B.H., Wolfgang C.L., Klein A.P., Diaz L.A., Jr., Allen P.J., Schmidt C.M., Kinzler K.W., Papadopoulos N., Hruban R.H., Vogelstein B. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mino-Kenudson M., Fernández-del Castillo C., Baba Y., Valsangkar N.P., Liss A.S., Hsu M., Correa-Gallego C., Ingkakul T., Perez Johnston R., Turner B.G., Androutsopoulos V., Deshpande V., McGrath D., Sahani D.V., Brugge W.R., Ogino S., Pitman M.B., Warshaw A.L., Thayer S.P. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut. 2011;60:1712–1720. doi: 10.1136/gut.2010.232272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siveke J.T., Einwächter H., Sipos B., Lubeseder-Martellato C., Klöppel G., Schmid R.M. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12:266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 68.von Figura G., Fukuda A., Roy N., Liku M.E., Morris Iv J.P., Kim G.E., Russ H.A., Firpo M.A., Mulvihill S.J., Dawson D.W., Ferrer J., Mueller W.F., Busch A., Hertel K.J., Hebrok M. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roy N., Malik S., Villanueva K.E., Urano A., Lu X., Von Figura G., Seeley E.S., Dawson D.W., Collisson E.A., Hebrok M. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–671. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimura Y., Fukuda A., Ogawa S., Maruno T., Takada Y., Tsuda M., Hiramatsu Y., Araki O., Nagao M., Yoshikawa T., Ikuta K., Yoshioka T., Wang Z., Akiyama H., Wright C.V., Takaori K., Uemoto S., Chiba T., Seno H. ARID1A maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2018;155:194–209. doi: 10.1053/j.gastro.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 71.Kopp J.L., Dubois C.L., Schaeffer D.F., Samani A., Taghizadeh F., Cowan R.W., Rhim A.D., Stiles B.L., Valasek M., Sander M. Loss of Pten and activation of Kras synergistically induce formation of intraductal papillary mucinous neoplasia from pancreatic ductal cells in mice. Gastroenterology. 2018;154:1509–1523.e5. doi: 10.1053/j.gastro.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collet L., Ghurburrun E., Meyers N., Assi M., Pirlot B., Leclercq I.A., Couvelard A., Komuta M., Cros J., Demetter P., Lemaigre F.P., Borbath I., Jacquemin P. Kras and Lkb1 mutations synergistically induce intraductal papillary mucinous neoplasm derived from pancreatic duct cells. Gut. 2020;69:704–714. doi: 10.1136/gutjnl-2018-318059. [DOI] [PubMed] [Google Scholar]
- 73.Hosoda W., Sasaki E., Murakami Y., Yamao K., Shimizu Y., Yatabe Y. GNAS mutation is a frequent event in pancreatic intraductal papillary mucinous neoplasms and associated adenocarcinomas. Virchows Arch. 2015;466:665–674. doi: 10.1007/s00428-015-1751-6. [DOI] [PubMed] [Google Scholar]
- 74.Tan M.C., Basturk O., Brannon A.R., Bhanot U., Scott S.N., Bouvier N., LaFemina J., Jarnagin W.R., Berger M.F., Klimstra D., Allen P.J. GNAS and KRAS mutations define separate progression pathways in intraductal papillary mucinous neoplasm-associated carcinoma. J Am Coll Surg. 2015;220:845–854.e1. doi: 10.1016/j.jamcollsurg.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taki K., Ohmuraya M., Tanji E., Komatsu H., Hashimoto D., Semba K., Araki K., Kawaguchi Y., Baba H., Furukawa T. GNAS(R201H) and Kras(G12D) cooperate to promote murine pancreatic tumorigenesis recapitulating human intraductal papillary mucinous neoplasm. Oncogene. 2016;35:2407–2412. doi: 10.1038/onc.2015.294. [DOI] [PubMed] [Google Scholar]
- 76.Patra K.C., Kato Y., Mizukami Y., Widholz S., Boukhali M., Revenco I., Grossman E.A., Ji F., Sadreyev R.I., Liss A.S., Screaton R.A., Sakamoto K., Ryan D.P., Mino-Kenudson M., Castillo C.F., Nomura D.K., Haas W., Bardeesy N. Mutant GNAS drives pancreatic tumourigenesis by inducing PKA-mediated SIK suppression and reprogramming lipid metabolism. Nat Cell Biol. 2018;20:811–822. doi: 10.1038/s41556-018-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., Tuveson D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 78.Aguirre A.J., Bardeesy N., Sinha M., Lopez L., Tuveson D.A., Horner J., Redston M.S., DePinho R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bardeesy N., Cheng K.H., Berger J.H., Chu G.C., Pahler J., Olson P., Hezel A.F., Horner J., Lauwers G.Y., Hanahan D., DePinho R.A. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006:203130–203146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bardeesy N., Aguirre A.J., Chu G.C., Cheng K.H., Lopez L.V., Hezel A.F., Feng B., Brennan C., Weissleder R., Mahmood U., Hanahan D., Redston M.S., Chin L., Depinho R.A. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy A.L., Morton J.P., Manoharan I., Nelson D.M., Jamieson N.B., Pawlikowski J.S., McBryan T., Doyle B., McKay C., Oien K.A., Enders G.H., Zhang R., Sansom O.J., Adams P.D. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol Cell. 2011;42:36–49. doi: 10.1016/j.molcel.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skoulidis F., Cassidy L.D., Pisupati V., Jonasson J.G., Bjarnason H., Eyfjord J.E., Karreth F.A., Lim M., Barber L.M., Clatworthy S.A., Davies S.E., Olive K.P., Tuveson D.A., Venkitaraman A.R. Germline Brca2 heterozygosity promotes Kras(G12D)–driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 83.Hanlon L., Avila J.L., Demarest R.M., Troutman S., Allen M., Ratti F., Rustgi A.K., Stanger B.Z., Radtke F., Adsay V., Long F., Capobianco A.J., Kissil J.L. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010;70:4280–4286. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morton J.P., Jamieson N.B., Karim S.A., Athineos D., Ridgway R.A., Nixon C., McKay C.J., Carter R., Brunton V.G., Frame M.C., Ashworth A., Oien K.A., Evans T.R., Sansom O.J. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139:586–597.e1–6. doi: 10.1053/j.gastro.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Russell R., Perkhofer L., Liebau S., Lin Q., Lechel A., Feld F.M., Hessmann E., Gaedcke J., Güthle M., Zenke M., Hartmann D., von Figura G., Weissinger S.E., Rudolph K.L., Möller P., Lennerz J.K., Seufferlein T., Wagner M., Kleger A. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat Commun. 2015;6:7677. doi: 10.1038/ncomms8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flowers B.M., Xu H., Mulligan A.S., Hanson K.J., Seoane J.A., Vogel H., Curtis C., Wood L.D., Attardi L.D. Cell of origin influences pancreatic cancer subtype. Cancer Discov. 2021;11:660–677. doi: 10.1158/2159-8290.CD-20-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McAllister F., Bailey J.M., Alsina J., Nirschl C.J., Sharma R., Fan H., Rattigan Y., Roeser J.C., Lankapalli R.H., Zhang H., Jaffee E.M., Drake C.G., Housseau F., Maitra A., Kolls J.K., Sears C.L., Pardoll D.M., Leach S.D. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 2014;25:621–637. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi C., Pan F.C., Kim J.N., Washington M.K., Padmanabhan C., Meyer C.T., Kopp J.L., Sander M., Gannon M., Beauchamp R.D., Wright C.V., Means A.L. Differential cell susceptibilities to KrasG12D in the setting of obstructive chronic pancreatitis. Cell Mol Gastroenterol Hepatol. 2019;8:579–594. doi: 10.1016/j.jcmgh.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bailey J.M., Hendley A.M., Lafaro K.J., Pruski M.A., Jones N.C., Alsina J., Younes M., Maitra A., McAllister F., Iacobuzio-Donahue C.A., Leach S.D. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–4288. doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- 90.Lee A.Y.L., Dubois C.L., Sarai K., Zarei S., Schaeffer D.F., Sander M., Kopp J.L. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut. 2019;68:487–498. doi: 10.1136/gutjnl-2017-314426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferreira R.M.M., Sancho R., Messal H.A., Nye E., Spencer-Dene B., Stone R.K., Stamp G., Rosewell I., Quaglia A., Behrens A. Duct- and acinar-derived pancreatic ductal adenocarcinomas show distinct tumor progression and marker expression. Cell Rep. 2017;21:966–978. doi: 10.1016/j.celrep.2017.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ijichi H., Chytil A., Gorska A.E., Aakre M.E., Fujitani Y., Fujitani S., Wright C.V., Moses H.L. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guerra C., Collado M., Navas C., Schuhmacher A.J., Hernández-Porras I., Cañamero M., Rodriguez-Justo M., Serrano M., Barbacid M. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stanger B.Z., Stiles B., Lauwers G.Y., Bardeesy N., Mendoza M., Wang Y., Greenwood A., Cheng K.H., McLaughlin M., Brown D., Depinho R.A., Wu H., Melton D.A., Dor Y. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 95.Ji B., Tsou L., Wang H., Gaiser S., Chang D.Z., Daniluk J., Bi Y., Grote T., Longnecker D.S., Logsdon C.D. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–1082.e6. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wollny D., Zhao S., Everlien I., Lun X., Brunken J., Brüne D., Ziebell F., Tabansky I., Weichert W., Marciniak-Czochra A., Martin-Villalba A. Single-cell analysis uncovers clonal acinar cell heterogeneity in the adult pancreas. Dev Cell. 2016;39:289–301. doi: 10.1016/j.devcel.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 97.Makohon-Moore A.P., Matsukuma K., Zhang M., Reiter J.G., Gerold J.M., Jiao Y., Sikkema L., Attiyeh M.A., Yachida S., Sandone C., Hruban R.H., Klimstra D.S., Papadopoulos N., Nowak M.A., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature. 2018;561:201–205. doi: 10.1038/s41586-018-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha S., Fu Y.Y., Grimont A., Ketcham M., Lafaro K., Saglimbeni J.A., Askan G., Bailey J.M., Melchor J.P., Zhong Y., Joo M.G., Grbovic-Huezo O., Yang I.H., Basturk O., Baker L., Park Y., Kurtz R.C., Tuveson D., Leach S.D., Pasricha P.J. PanIN neuroendocrine cells promote tumorigenesis via neuronal cross-talk. Cancer Res. 2017;77:1868–1879. doi: 10.1158/0008-5472.CAN-16-0899-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saloman J.L., Albers K.M., Li D., Hartman D.J., Crawford H.C., Muha E.A., Rhim A.D., Davis B.M. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A. 2016;113:3078–3083. doi: 10.1073/pnas.1512603113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., Wicha M., Clarke M.F., Simeone D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 101.Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 102.Renz B.W., Takahashi R., Tanaka T., Macchini M., Hayakawa Y., Dantes Z., Maurer H.C., Chen X., Jiang Z., Westphalen C.B., Ilmer M., Valenti G., Mohanta S.K., Habenicht A.J.R., Middelhoff M., Chu T., Nagar K., Tailor Y., Casadei R., Di Marco M., Kleespies A., Friedman R.A., Remotti H., Reichert M., Worthley D.L., Neumann J., Werner J., Iuga A.C., Olive K.P., Wang T.C. β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 2018;33:75–90.e7. doi: 10.1016/j.ccell.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Renz B.W., Tanaka T., Sunagawa M., Takahashi R., Jiang Z., Macchini M., Dantes Z., Valenti G., White R.A., Middelhoff M.A., Ilmer M., Oberstein P.E., Angele M.K., Deng H., Hayakawa Y., Westphalen C.B., Werner J., Remotti H., Reichert M., Tailor Y.H., Nagar K., Friedman R.A., Iuga A.C., Olive K.P., Wang T.C. Cholinergic signaling via muscarinic receptors directly and indirectly suppresses pancreatic tumorigenesis and cancer stemness. Cancer Discov. 2018;8:1458–1473. doi: 10.1158/2159-8290.CD-18-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bailey J.M., Alsina J., Rasheed Z.A., McAllister F.M., Fu Y.Y., Plentz R., Zhang H., Pasricha P.J., Bardeesy N., Matsui W., Maitra A., Leach S.D. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146:245–256. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]