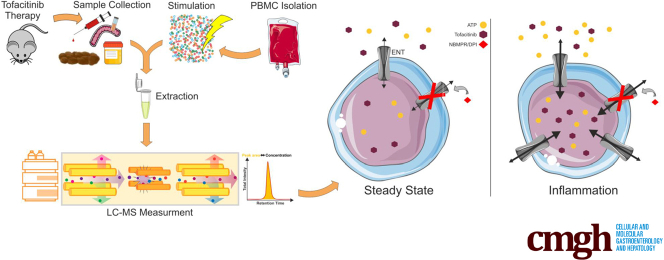
Abstract
Objective
By interfering with multiple cytokines, human Janus kinase inhibitors (JAKis) are of growing importance in the treatment of malignant and inflammatory conditions. Although tofacitinib has demonstrated efficacy as the first-in-class JAKi in ulcerative colitis many aspects concerning its mode of action and pharmacokinetics remain unresolved.
Design
We studied tofacitinib’s impact on various primary human innate and adaptive immune cells. In-depth in vivo studies were performed in dextran sodium sulfate–induced colitis in mice. Immune populations were characterized by flow cytometry and critical transcription factors and effector cytokines were analyzed. Pharmacokinetics of tofacitinib was studied by liquid chromatography–tandem mass spectrometry.
Results
Tofacitinib inhibited proliferation in CD4+ and CD8+ T cells along with Th1 and Th17 differentiation, while Th2 and regulatory T cell lineages were largely unaffected. Monocytes and macrophages were directed toward an anti-inflammatory phenotype and cytokine production was suppressed in intestinal epithelial cells. These findings were largely reproducible in murine cells of the inflamed mucosa in dextran sulfate sodium colitis. Short-term treatment with tofacitinib had little impact on the mouse microbiota. Strikingly, the degree of inflammation and circulating tofacitinib levels showed a strong positive correlation. Finally, we identified inflammation-induced equilibrative nucleoside transporters as regulators of tofacitinib uptake into leukocytes.
Conclusions
We provide a detailed analysis of the cell-specific immune-suppressive effects of the JAKis tofacitinib on innate and adaptive immunity and reveal that intestinal inflammation critically impacts tofacitinib’s pharmacokinetics in mice. Furthermore, we describe an unappreciated mechanism—namely induction of equilibrative nucleoside transporters—enhancing baseline cellular uptake that can be inhibited pharmaceutically.
Keywords: pharmacokinetics, inflammatory bowel disease, mucosal inflammation, JAK inhibitor, tofacitinib
Abbreviations used in this paper: 3D, 3-dimensional; ASV, amplicon sequence variant; ATP, adenosine triphosphate; CD, Crohn’s disease; CFSE, carboxyfluorescein succinimidyl ester; DI, division index; DSS, dextran sulfate sodium; ELISA, enzyme-linked immunosorbent assay; ENT, equilibrative nucleoside transporter; FSC, forward scatter; JAK, Janus kinase; JAKi, Janus kinase inhibitor; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IFNγ, interferon γ; IHC, immunohistochemistry; IL, interleukin; LC-MS/MS, liquid chromatography–tandem mass spectrometry; LPS, lipopolysaccharide; M-CSF, macrophage colony-stimulating factor; mRNA, messenger RNA; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PHA, phytohemagglutinin; PI, proliferation index; STAT, signal transducers and activator of transcription; Th, T helper; TNF-α, tumor necrosis factor α; UC, ulcerative colitis
Graphical abstract
Summary.
We present graduated effects on innate and adaptive immune cells involved in inflammatory bowel disease with preference for T cell pathways. Moreover, we identified equilibrative nucleoside transporters as molecular basis for cellular uptake of tofacitinib—upregulated during inflammation and accessible by pharmaceuticals.
Crohn’s disease (CD) and ulcerative colitis (UC) represent the 2 major forms of chronic inflammatory bowel diseases (IBDs).1,2 They are characterized by sustained or recurrent inflammation of the intestinal mucosa. Persons affected experience diarrhea, rectal blood, and abdominal pain, being detrimental to their quality of life. Over time repeated flares of mucosal inflammation may result in irreversible damage of the macro- and microstructural integrity of the gastrointestinal tract, providing the basis for complications such as fistulas, strictures, and colitis-associated carcinogenesis.3,4
Concerning the therapeutic options in IBD the last years have seen a widening of the therapeutic armamentarium—specifically for moderate to severe disease courses. Monoclonal antibodies against tumor necrosis factor α (TNF-α), α4β7 integrin, and the p40 subunit of interleukin (IL)-12 and IL-23 have proven to be effective in a proportion of patients.5 However, up to 30% of IBD patients do not respond to such therapies at all, and of those who initially benefit, many will eventually lose response to treatment or develop intolerance.6 A vast body of research has been dedicated to the unravelling of the underlying mechanisms. The observation that antibody trough concentrations correlate positively with clinical efficacy gave rise to seek a reason in the pharmacokinetics. However, a marked increase of the induction dosing of adalimumab failed to demonstrate superiority,7 suggesting that the reasons for low trough levels are more complex including antigen-dependent and immunologic factors. The mechanism of action of therapeutic antibodies goes beyond the simple neutralization of the target structures and involve modulatory effects on innate immune cells.8,9 Thus, after 20 years, many questions regarding aspects of pharmacokinetics and mechanisms of therapeutic antibodies remain unanswered.
Cytokine networks in the inflamed mucosa are sophisticated and subjected to multiple layers of regulation by microbial, genetic and immunological factors. Thus, targeting a single cytokine in patients with IBD may induce alternative compensatory or redundant pathways. An novel approach to optimize response rates in IBD is the use of multicytokine blockers, which inhibit shared cytokine signalling pathways.10 Janus kinase (JAK)–signal transducers and activator of transcription (STAT) (JAK-STAT) pathway operates downstream of over 50 cytokines and growth factors. Loss or gain of function mutations in humans and mice result in detrimental immunological phenotypes.11 The human JAK-STAT family comprises 4 JAKs and 7 STATs. Mechanistically, cytokine receptor–ligand engagement activates receptor-associated JAKs that phosphorylate specific tyrosine residues on their receptors. These serve as a docking site for STATs that are in turn also phosphorylated by adjacent JAKs. Phosphorylated STATs dimerize and translocate into the nucleus to regulate gene transcription.11 Thus, interfering with JAK-STAT signalling opens the possibility to block or modulate the activity of several cytokines simultaneously.
This has prompted the development of orally available, small-molecule JAK inhibitors (JAKis) for the treatment of a wide range of inflammatory conditions. In 2018, tofacitinib was the first JAKi to be approved to treat moderately to severely active UC.12 Tofacitinib is a reversible competitive inhibitor that binds to the adenosine triphosphate (ATP) binding site in the catalytic cleft of the kinase domains of JAK1 and JAK3. As an oral, small-molecule JAKi, tofacitinib is rapidly absorbed, with a bioavailability of 93%, reaching peak plasma concentration within 1 hour, and with a terminal half-life of approximately 3 hours.13
However, regarding the mechanistic underpinnings of tofacitinib in intestinal inflammation there are many open questions, eg, the unexpected failure of tofacitinib to demonstrate efficacy in CD.14 A possible explanation may be that tofacitinib engages cell type–specific effects capturing cell populations and related cytokines relevant for UC but not relevant for CD. Furthermore, there is a substantial lack of information regarding tofacitinib’s pharmacokinetics during inflammation both on a systemic level and on a cellular level. This prompted us to examine cell type–specific effects of tofacitinib in primary human cells of the innate and adaptive immune system and to reproduce such findings in vivo, in a mouse model of experimental colitis, in consideration of a detailed systemic, tissue- and cell-specific pharmacokinetic workup.
Results
Tofacitinib Potently Suppresses Peripheral Blood T Cell Proliferation
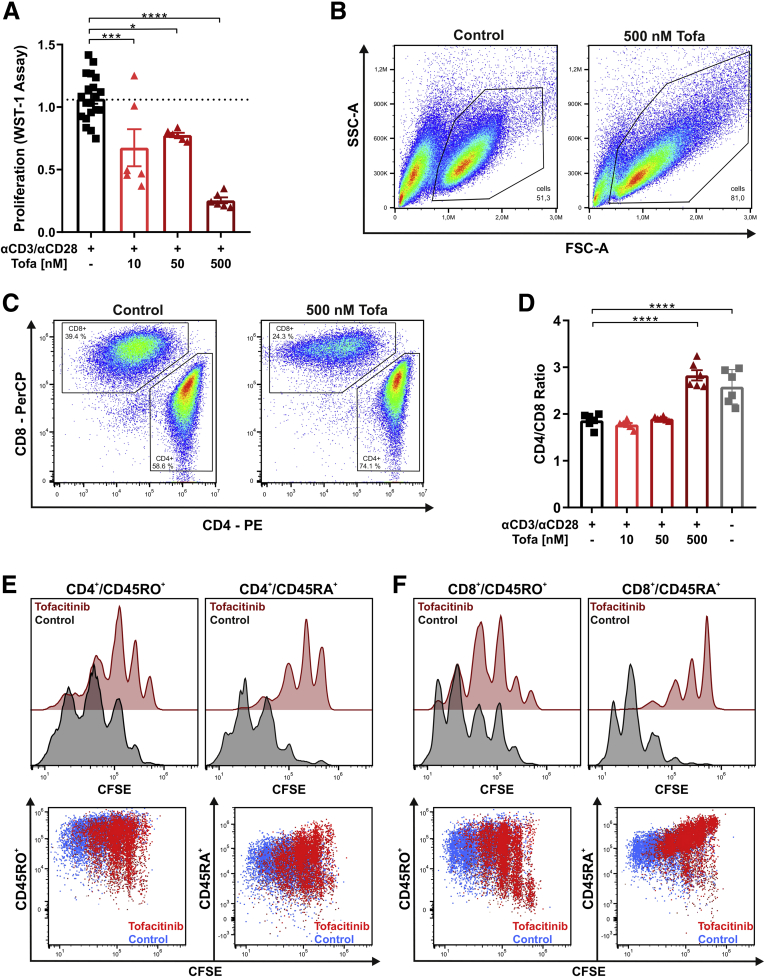
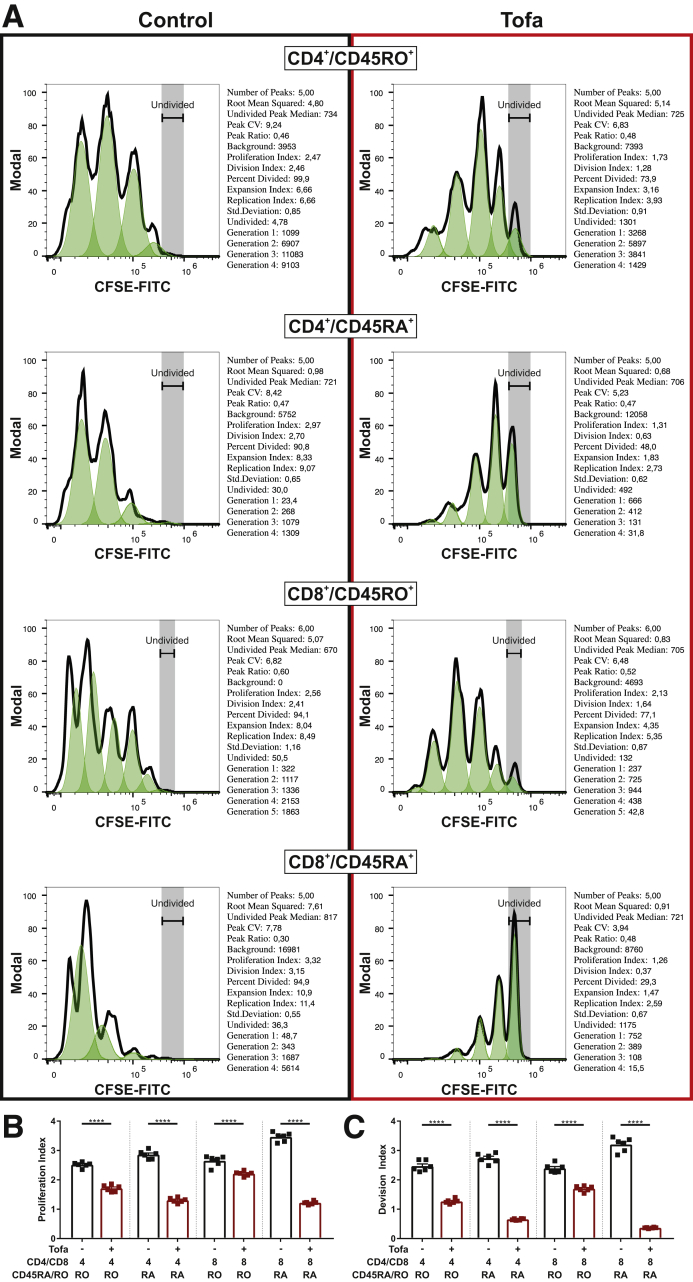
Given the close linkage of T cell proliferation with the IL-2 receptor–JAK1–JAK3 axis,15 we first studied effects of tofacitinib on proliferation of CD3+ peripheral T cell isolated from human peripheral blood. Cells were restimulated with CD3/CD28 with or without increasing concentrations of tofacitinib. Effects of tofacitinib on T cell proliferation were determined by WST-1 assay and flow cytometry. Treatment with tofacitinib dose-dependently decreased cellular proliferation as indicated by a reduced conversion of WST-1 to formazan by mitochondrial dehydrogenases (Figure 1A) and proliferation assessed in flow cytometric carboxyfluorescein succinimidyl ester (CFSE) experiments (Figure1E and F and Figure 2). The flow cytometry gating strategy is outlined in Figure 3. Remarkably, the concentrations of tofacitinib used did not induce excessive cell death (Figure 4). As for the WST-1 assay tofacitinib resulted in a reduction in both CD4+ and CD8+ positive T cells (Figure 1C), resulting in a steady-state CD4/CD8 ratio at a tofacitinib concentration of 500 nM (Figure 1D). To decipher the proliferation inhibitory potential of tofacitinib on naïve and effector memory T cells, CFSE assays were performed in combination with CD4 and CD8 along with CD45RA (naïve) and CD45RO (effector/memory) lineage markers. Proliferation index (PI) and division index (DI), both commonly used indices to evaluate proliferation, were strongly reduced in CD4+/CD45RO+ (PI: 2.47 to 1.73; DI: 2.46 to 1.28) and CD4+/CD45RA+ (PI: 2.97 to 1.31; DI: 2.70 to 0.63) (Figure 1E), as well as in CD8+/CD45RO+ (PI: 2.56 to 2.13; DI: 2.41 to 1.64) and CD8+/CD45RA+ (PI: 3.32 to 1.26; DI: 3.15 to 0.37) (Figure 1F), indicative of a particularly strong effect on the naïve CD8+ cytotoxic T cell subset.
Figure 1.
Tofacitinib suppresses T cell proliferation. (A) Proliferation of pan T cells stimulated with αCD3/αCD28 in the absence or presence of the indicated tofacitinib concentrations, measured by WST-1 assay. (B) Cellular appearance of αCD3/αCD28-stimulated pan T cells with (right) or without (left) tofacitinib in the FSC and side scatter (SSC) channels. (C, D) Representative flow cytometry plots showing CD4+ to CD8+ ratios (C) in pan T cells treated with tofacitinib, which was summarized in a graph that includes (D) unstimulated control subjects. (E, F) Proliferation analysis of tofacitinib-treated (red) or control-treated (black) pan T cells showing CFSE staining patterns of CD4+/CD45RO+ and CD4+/CD45RA+ cells as well as CD8+/CD45RO+ and CD8+CD45RA+ cells. Representative flow cytometric (FC) plots are depicted below the respective histograms. Differences were analyzed by ordinary 1-way analysis of variance followed by Bonferroni’s multiple comparisons test. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Tofa, tofacitinib; WST, water soluble tetrazolium salt.
Figure 2.
Statistics for proliferation data. (A) Statistical data from the CFSE proliferation experiments of different T cell subsets, with vehicle-treated cells on the left and tofacitinib treated cells on the right. (B) Statistical summary of the proliferation index displayed as a graph. (C) Statistical summary of the division index displayed as a graph. All bars represent means and symbols represent individual values. Differences between vehicle-treated and tofacitinib-treated cells were calculated by multiple t tests. ∗∗∗∗P < .0001.
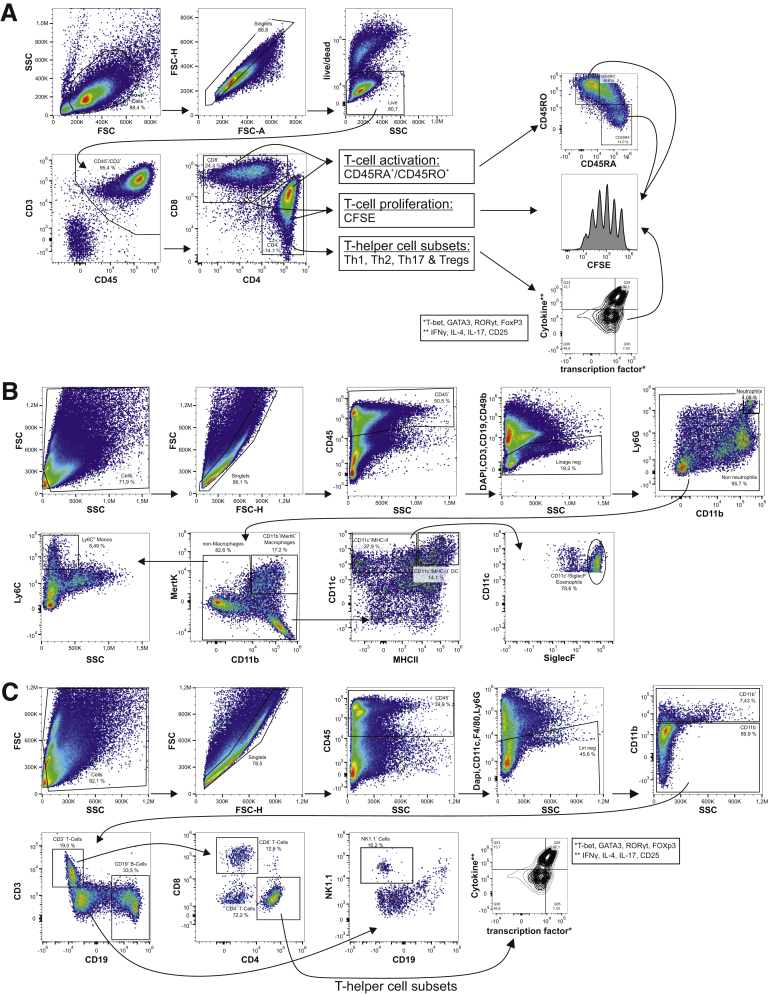
Figure 3.
Gating strategies for flow cytometric analysis. (A) Representative gating strategy for human T cell experiments. (B) Representative gating strategy for murine lamina propria infiltrating innate immune cells. (C) Representative gating strategy for murine lamina propria infiltrating adaptive immune cells.
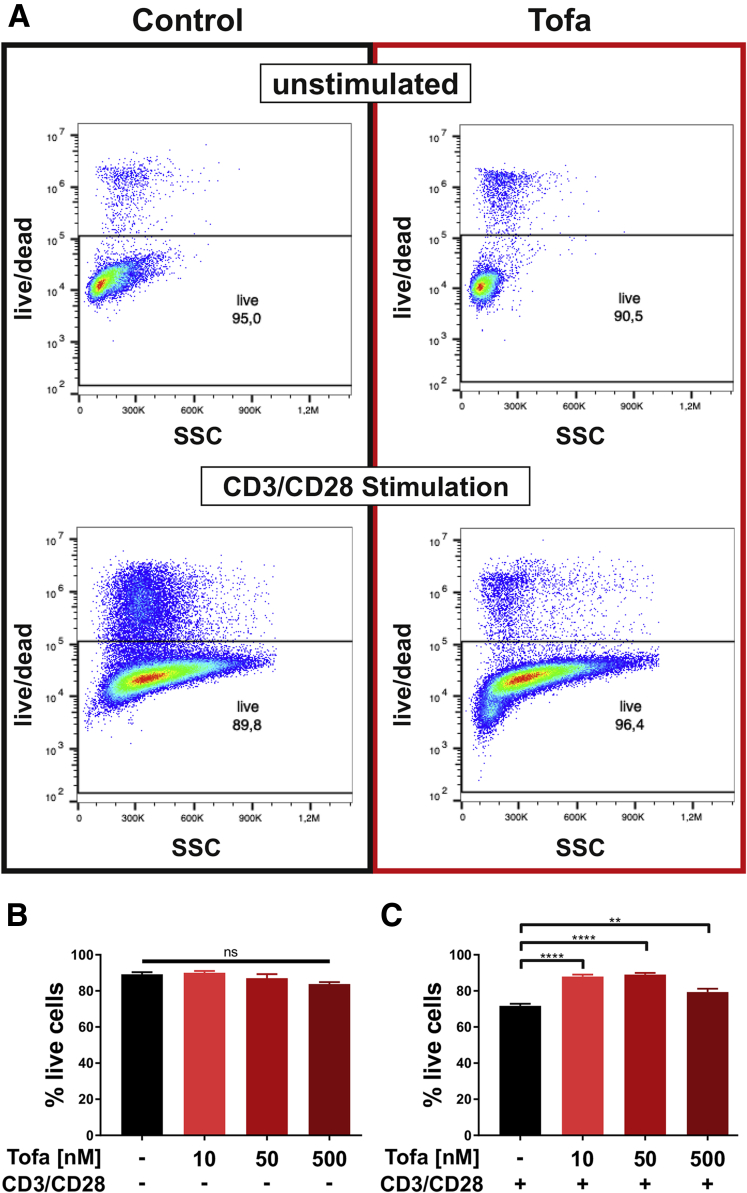
Figure 4.
Viability data and statistics for pan T cells. (A) Representative fluorescence-activated cell sorting plots for live/dead staining of unstimulated (top) and αCD3/αCD28 stimulated (bottom) pan T cells under control (left) and tofacitinib (right) treatment. (B) Summarized graph for the percent live cells for the unstimulated condition. (C) Summarized graph for the percent live cells for the αCD3/αCD28 stimulated condition. All bars represent means and error bar the SD. Differences between treatment conditions were calculated using 1-way analysis of variance followed by Tukey’s multiple comparison test. ns = not significant; ∗∗∗∗ P < .0001; ∗∗ P < .01
Impact of Tofacitinib on Peripheral Blood T Helper Cell Functions
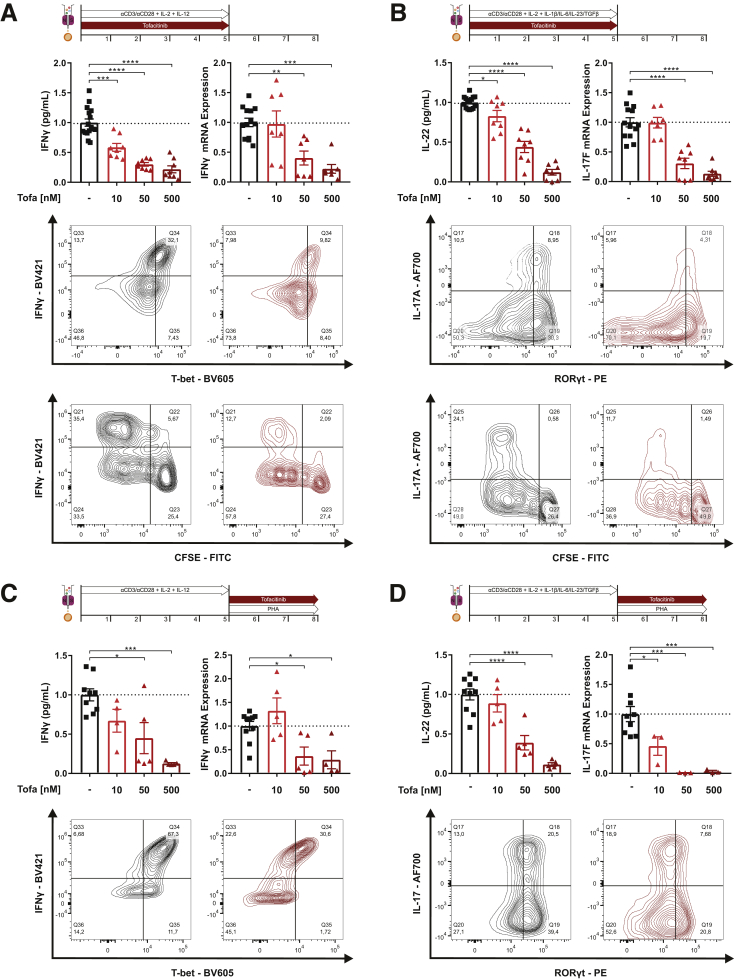
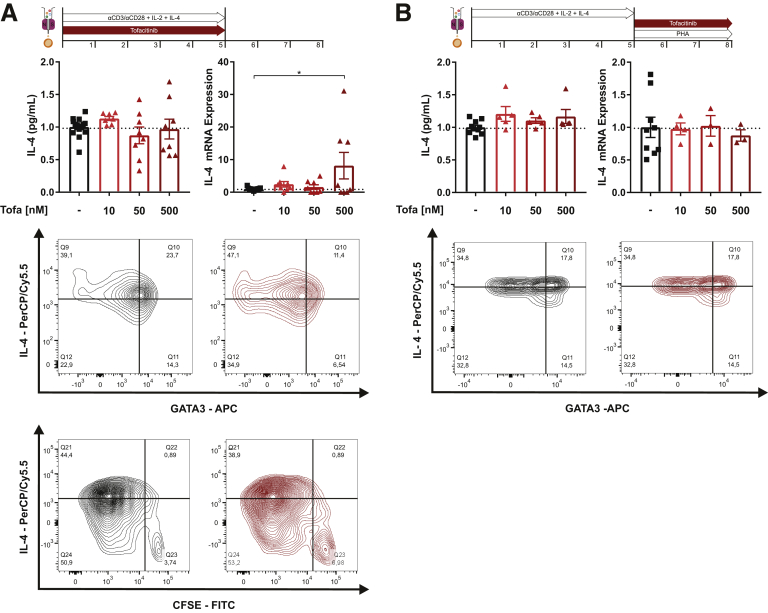
CD4+ positive helper T cells orchestrate immune reactions in IBD.16 Thus, we examined the impact of JAK1 and JAK3 modulation by tofacitinib on Th differentiation and effector functions of T cells isolated from human peripheral blood. As outlined in Figure 5A, we first determined the impact of tofacitinib on naïve CD4+ Th1 differentiation. Compared with untreated control cells, tofacitinib resulted in a dose-dependent up to 5-fold reduction on interferon γ (IFNγ) production both on the protein and the messenger RNA (mRNA) level. This was paralleled by a tofacitinib-induced suppression of T-bet, the key transcriptional regulator of Th1 lineage commitment.17 Despite tofacitinib’s antiproliferative capacity the observed effects were not primarily explained by a reduced T helper (Th) cell proliferation. To determine whether tofacitinib affects Th17 differentiation, naïve Th cells were differentiated under Th17 polarizing conditions, as depicted in Figure 5B. As for the Th1 conditions, tofacitinib strongly and dose-dependently suppressed Th17 differentiation as for IL17A, IL-17F, and IL-22 production. Tofacitinib reduced the intracellular immunopositivity for RORγt, the lineage defining transcription factor of Th17 cells,18 again without completely interfering with T cell proliferation. Noteworthy, in Th2 polarizing conditions, tofacitinib lacked an effect on IL-4 production, and the number of GATA3-positive cells was decreased in the presence of 500 nM tofacitinib (Figure 6). To determine tofacitinib’s potential to interfere with effector functions of differentiated Th cells, we differentiated naïve Th cells under the previously mentioned conditions and restimulated with phytohemagglutinin (PHA) in presence or absence of this JAK1/3 inhibitor. As outlined in Figure 5C, tofacitinib strongly suppressed IFNγ production, particularly at concentration of 50 nM and 500 nM. Similarly, tofacitinib strongly suppressed Th17 effector cytokines in PHA-restimulated Th17 cells (Figure 5D).
Figure 5.
Tofacitinib suppresses the differentiation and activation of human Th1 and Th17 cells. (A) Analysis of tofacitinib’s effect on Th1 differentiation showing IFNγ production by ELISA and quantitative polymerase chain reaction (qPCR) (top), flow cytometric (FC) analysis of CD4+/IFNγ+/T-bet+ cells (middle) and analysis of proliferation (CFSE) in combination with IFNγ production (bottom). (B) Analysis of tofacitinib’s impact on Th17 differentiation studying IL-22 release by ELISA and IL-17F production by qPCR (top), FC analysis of CD4+/IL-17A+/RORγt+ cells (middle), and analysis of proliferation (CFSE) in combination with IL-17 production (bottom). (C) Analysis of tofacitinib’s effect on differentiated Th1 cells showing IFNγ production by ELISA and qPCR (top) and fluorescence-activated cell sorting analysis of CD4+/IFNγ+/T-bet+ cells (bottom). (D) Analysis of tofacitinib’s effect on differentiated Th17 cells by measuring IL-22 release by ELISA and IL-17F production by qPCR (top) and FC analysis of CD4+/IL-17A+/RORγt+ cells (bottom). Differences were analyzed by ordinary 1-way analysis of variance followed by Bonferroni’s multiple comparisons test and were normalized to the levels observed in untreated control subjects. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. AF, Alexa Fluor; BV, Brilliant Violet; Tofa, tofacitinib.
Figure 6.
Tofacitinib’s effect on differentiation and activation of Th2 cells. (A) Production of the Th2 effector cytokine IL-4 was studied by ELISA and quantitative polymerase chain reaction (top), flow cytometric analysis of CD4+/IL-4+/GATA3+ cells (middle) and analysis of proliferation (CFSE) in combination with IL-4 production (bottom). (B) Analysis of tofacitinib’s impact on differentiated Th2 cells by IL-4 ELISA and quantitative polymerase chain reaction (top) and fluorescence-activated cell sorting analysis of CD4+/IL-4+/GATA3+ cells (bottom). qPCR data are shown as relative mRNA expression. ILTofa, tofacitinib.
Tofacitinib Suppresses Immune Function of Innate Immune Cells
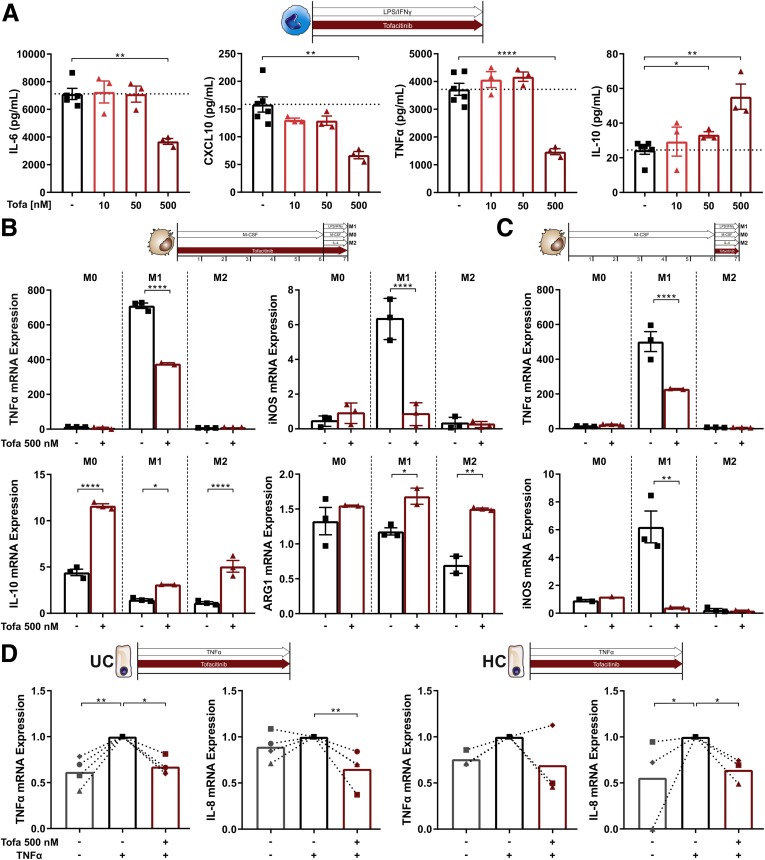
Recently, cells of the innate immune system have been brought back into the limelight of intestinal immunity.19 To determine a potential impact on innate immune cells, we first studied the effect of tofacitinib on LPS/IFNγ stimulated CD14+ monocytes isolated from human peripheral blood (Figure 7A). Tofacitinib exerted anti-inflammatory effects on monocytes—suppression of the proinflammatory cytokines IL-6, CXCL10 (IP-10), TNF-α, and induction of the anti-inflammatory cytokine IL-10—at a tofacitinib concentration of 500 nM. To determine the effect of tofacitinib on macrophage differentiation, CD14+ monocytes isolated from peripheral blood were grown in the presence of macrophage colony-stimulating factor (M-CSF) with or without tofacitinib for 6 days and then subjected to M0, M1, or M2 polarizing conditions. As shown in Figure 7B, tofacitinib suppressed M1 transcripts including TNF-α and inducible nitric oxide synthase 2, which were strongly induced by LPS/IFNγ. Under M0 and M2 conditions tofacitinib skewed macrophage polarization toward M2 indicated by induction of ARG1 (arginase 1) and/or IL-10. When monocytes were differentiated without tofacitinib for 6 days (Figure 7C) and restimulated under the aforementioned conditions in the presence of tofacitinib, again M1 cytokine production was markedly suppressed. The aforementioned M2-promoting properties were not observed under these conditions. Intestinal epithelial cells (IECs), one of the evolutionary most ancient innate immune cell populations, orchestrate the mutualistic relationship between the host and the intestinal microenvironment.20 To study the effects of tofacitinib on IECs, colonic organoids were prepared form tissue biopsies of patients with ulcerative colitis and healthy control subjecs. Intestinal crypts were isolated from the biopsies and cultured into 3-dimensional (3D) organoids with Matrigel and specific growth media. Because intestinal organoids form a closed 3D structure, they were reseeded as monolayers to allow stimulation of the apical side of the cells. Monolayers were analyzed unstimulated, with TNF-α stimulation as well as with TNF-α stimulation and tofacitinib treatment (Figure 7D). IECs from UC patients demonstrated an increased baseline proinflammatory tone, as indicated by elevated baseline IL-8 expression and a disposition to react to TNF-α stimulation. Both in IECs from UC patients and healthy control subjects excess TNF-α and IL-8 production was significantly suppressed by tofacitinib (Figure 7D).
Figure 7.
Tofacitinib skews innate immune cells toward a more regulatory phenotype. (A) Quantification of proinflammatory (IL-6, TNF-α, C-X-C motif ligand 10 [CXCL10]) and anti-inflammatory (IL-10) cytokines produced by stimulated CD14+ monocytes with or without tofacitinib. (B, C) Analysis of TNF-α and inducible nitric oxide synthase (iNOS) (M1) and IL-10 and arginase 1(ARG1) (M2) mRNA expression of CD14+ monocytes differentiated toward M0, M1 or M2 macrophages with or without tofacitinib (B) from day 0 or (D) from day 6. Induction of proinflammatory cytokines (TNF-α, IL-8) was assayed in intestinal organoid monolayers from UC patients (UC, left) or healthy control subjects (HC, right). Unstimulated organoids are shown in gray, TNF-α–stimulated untreated organoids are shown in black and TNF-α–stimulated organoids treated with tofacitinib are shown in red. Differences in A–C were analyzed by ordinary 1-way analysis of variance followed by Bonferroni’s multiple comparisons test and for D with a Kruskal-Wallis test, quantitative polymerase chain reaction data are shown as relative mRNA expression. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. HC, healthy control subjects; Tofa, tofacitinib.
Tofacitinib Ameliorates Inflammation in the DSS Model of Experimental Colitis
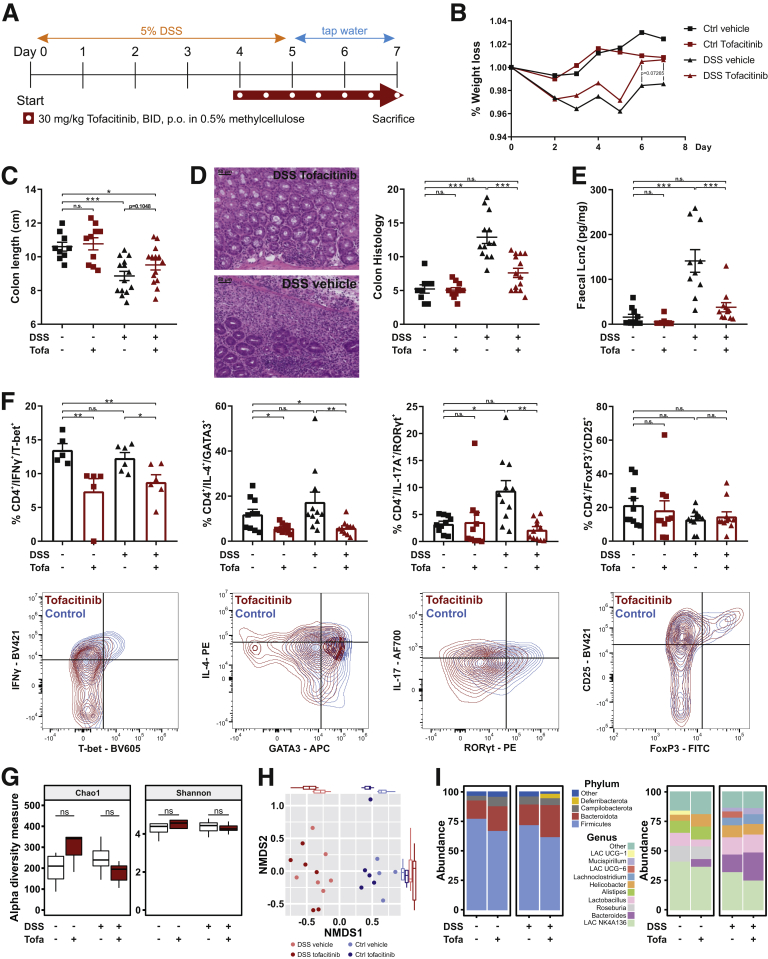
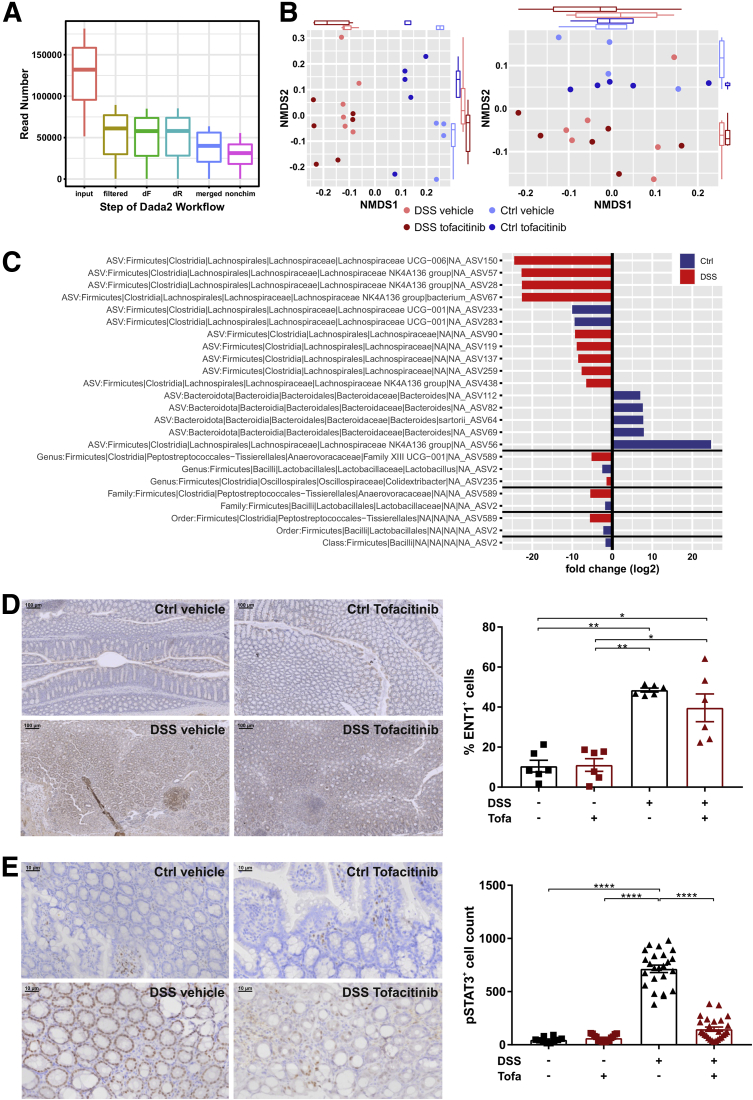
To verify the in vitro human findings within the complex environment of a living organism’s colon, we next performed a DSS colitis model in female BALB/c mice that received 30 mg/kg tofacitinib twice a day in a therapeutic setting starting from day 4. The experimental outline is illustrated in Figure 8A. As control subjects served rodents without DSS again with or without tofacitinib. Within DSS-treated animals tofacitinib-treated mice tended to have a lesser weight loss and to recover quicker than vector-treated animals (Figure 8B). Accordingly, the difference between the colon lengths—DSS-induced shortening of the colon is considered a hallmark of this model21—was not statistically different (Figure 8C). However, treatment with tofacitinib resulted in a significant reduction in the histologic severity of colitis which was further corroborated by a strong reduction of the fecal inflammatory biomarker lipocalin 2,22 that both returned to near control levels (Figure 8D and E). Additionally, we could show on mouse tissue sections that pSTAT3 is significantly increased upon DSS exposure, which is almost reduced to baseline levels under tofacitinib therapy (Figure 9E). To confirm tofacitinib’s effects on the Th cell compartment, we isolated colonic lamina propria cells and studied alterations in the distributions of Th1, Th2, Th17, and regulatory T cells by flow cytometry. As summarized in Figure 8F, treatment with DSS did not result in an expansion of CD4+/IFNγ+/T-bet+ Th1 and CD4+/IL4+/GATA3+ Th2 cells. Nonetheless, tofacitinib significantly reduced the proportion of Th1 and Th2 cells both in the steady state and after DSS exposure. Interestingly, DSS treatment resulted in a significant increase in CD4+/IL17A+/RORγt+ T cells. This expansion was completely blocked by tofacitinib which did not affect the Th17 pool in the steady state (Figure 8F). Tofacitinib had no effect on CD4+/CD25+/FoxP3+ regulatory T cells (Figure 8F). Another interesting aspect is related to a potential reciprocal influence between the xenobiotic nucleoside tofacitinib and the gut microbiome. This aspect is supported by recent data showing that certain nucleoside analogues are metabolized by microbial enzymes,23 and secondly dietary nucleoside are capable of altering gut microbial compositions.24 Expectedly, we observed strong DSS-induced alterations in the composition of the colonic microbiota. However, tofacitinib—during the short period of 3.5 days in our experimental setup—did not affect alpha or beta diversity neither in the steady state nor after DSS exposure based in 16S ribosomal DNA signatures (Figure 8G–I, Figure 9A–D).
Figure 8.
Tofacitinib ameliorates DSS colitis. (A) Experimental outline used for the DSS colitis model. (B) Time-resolved weight changes during the DSS model. (C) Assessment of colon lengths in control and DSS-treated animals with or without tofacitinib. (D) Representative histology (left) histologic scoring (right) in DSS- or water-treated animals receiving tofacitinib or vector control. The scale bars indicate 50 μm. (E) Quantification of fecal Lcn2 by ELISA. (F) Flow cytometry based analysis of Th cell subpopulations in the inflamed gut of the experimental animals with or without tofacitinib treatment. (G–I) Microbiome analysis including (G) Chao1 and Shannon indices as α-diversity measures, (H) β-diversity using Bray-Curtis dissimilarity, and (I) differences in the taxonomic compositions, of tofacitinib- and vector-treated animals with or without DSS exposure. In B, Differences were calculated by multiple t tests. For C–E, 1-way analysis of variance followed by Bonferroni’s multiple comparisons test and for F, a Kruskal-Wallis test was used. ∗P < .05; ∗∗P < .01;∗∗∗P < .001; ∗∗∗∗P < .0001. BID, twice per day; Ctrl, Control; DSSLAC, Lachnospiraceae; Lcn, lipocalin; NMDS, nonmetric multidimensional scaling; Tofa, tofacitinib.
Figure 9.
Microbiome studies and IHC. (A) Procession of the input reads throughout the dada2 workflow of the microbiome analysis. (C) Analysis of β-diversity using unweighted (left) or weighted Unifrac analysis. (C) Depiction of all significant log2 fold changes in taxonomic abundance from different taxonomic levels. (D) Representative IHC stainings of ENT1 on mouse tissue section of DSS- and vehicle-treated animals in the presence or absence of tofacitinib (left) with the quantification displayed as graph (right). (E) Representative IHC stainings of pSTAT3 on mouse tissue section of DSS- and vehicle-treated animals in the presence or absence of tofacitinib (left) with the quantification displayed as graph (right). dF, denoised forward; dR, denoised reverse; NMDS, nonmetric multidimensional scaling.
Tofacitinib Pharmacokinetics Is Influenced by Inflammation
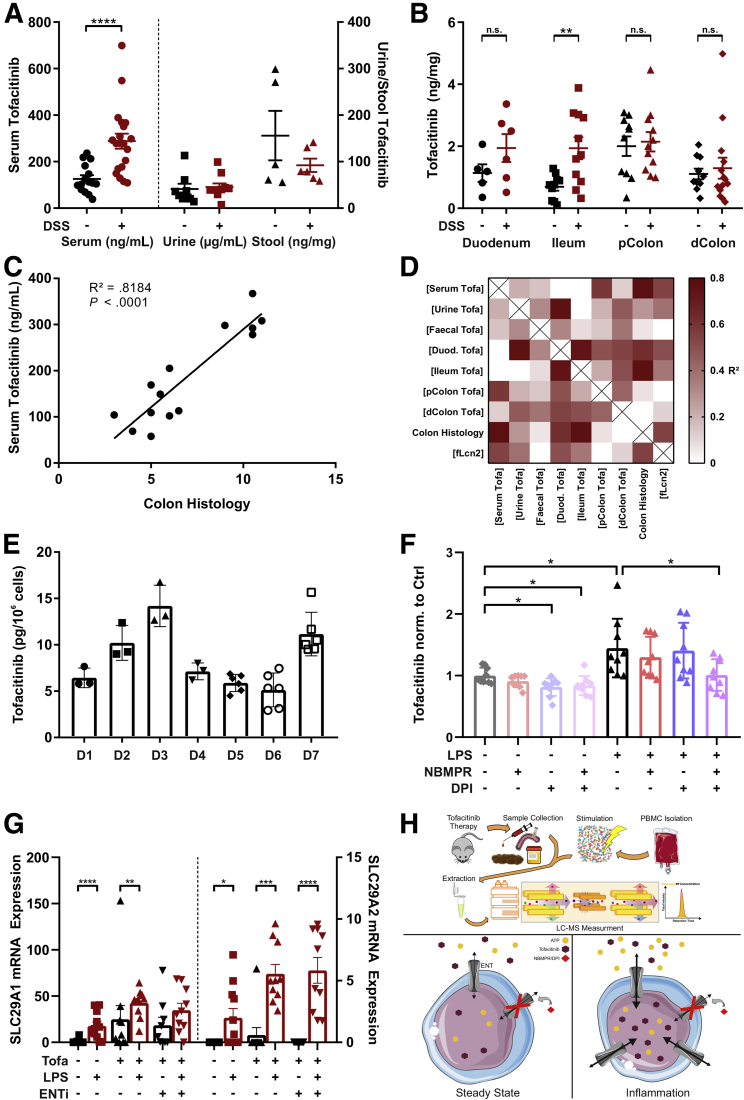
So far, little is known about a potential impact of systemic or local inflammation on the pharmacokinetics of tofacitinib, particularly in the context of intestinal inflammation. Thus, in a first step, we determined tofacitinib concentrations in mouse sera by liquid chromatography–tandem mass spectrometry (LC-MS/MS). The experimental setup is outlined in Figure 10H. Interestingly, we observed elevated circulating concentrations of tofacitinib in DSS-treated animals compared with water-treated control subjects (Figure 10A). To test if this effect could be explained by a reduced tofacitinib excretion, tofacitinib was next measured in urine and stool samples. As shown in Figure 10A, tofacitinib levels were comparable in both the urine and the stool of DSS- vs control-treated animals. To assess whether an increased tofacitinib uptake could explain elevated serum concentrations, we determined tissue concentrations in duodenal, ileal, proximal, and distal colonic scrapings. Tissue concentrations of tofacitinib were largely comparable along the gastrointestinal axis with an exception in the ileum where mucosal tofacitinib concentrations were higher in DSS-treated than in water-treated rodents (Figure 10B). As drug metabolism is known to be susceptible to perturbation by mediators of acute and chronic inflammation, we correlated serum tofacitinib concentrations with colonic histologic disease severity. This resulted in an extraordinarily strong correlation (R2 = 0.818, P < .0001) suggesting an association with intestinal inflammation (Figure 10C). Figure 10D highlights additional correlations between various tofacitinib concentrations and inflammatory parameters, namely histologic disease activity and fecal lipocalin 2. Numerous compounds have been shown to be preferentially taken up by inflamed cells and tissues. To study tofacitinib uptake by white blood cells, we first assessed the steady state uptakes of peripheral blood mononuclear cell (PBMCs) of 7 different donors. As demonstrated in Figure 10E, we found relevant fluctuations in steady-state uptakes between the different donors. From a structural perspective tofacitinib represents an ATP analogue. Thus, we hypothesized that tofacitinib may utilize equilibrative nucleoside transporters (ENTs) to facilitate its cellular entrance.25 As shown in Figure 10F, tofacitinib uptake was indeed significantly increased after LPS stimulation, and this effect was abrogated by incubation with a combination of ENT inhibitors. In support of such a mechanism we show that the expression of ENT1 and ENT2 were induced by LPS irrespectively of the presence of ENT inhibitors. Additionally, we could also show an increase in ENT1 expression with immunohistochemistry (IHC) staining on mouse tissue in the DSS group (Figure 9D). Figure 10H, lower panel, graphically summarizes the latter findings.
Figure 10.
Systemic and cellular pharmacokinetics of tofacitinib. (A, B) Quantification of tofacitinib (A) in serum, urine, and stool samples and (B) in the mucosa of various intestinal sections analyzed by LC-MS/MS analysis. (C) Correlation of serum tofacitinib levels with colonic histologic inflammation. (D) Correlation matrix of various tofacitinib concentrations along with colonic histology and fecal Lcn2 levels. (E) Steady-state tofacitinib uptake into PBMCs from healthy volunteers. (F) Comparison of tofacitinib uptake at baseline and after stimulation with LPS in the presence or absence of the nucleoside transport inhibitors NBMPR and dipyridamole. (G) Analysis of the mRNA expression of the equilibrative nucleoside transporters (ENTs) 1 and 2 with or without stimulation. (H) The experimental setup of the LC-MS/MS studies (top). A proposed mechanism regulating tofacitinib cellular uptake (bottom). Differences in A and B were calculated using a 1-way analysis of variance followed by Bonferroni’s multiple comparisons test. Correlations in C and D were calculated using the Pearson correlation coefficient. For F, a Kruskal-Wallis test was performed, and for G, a Mann-Whitney test between unstimulated and stimulated samples was used. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. D, donor; dColon, distal Colon; DPI, dipyridamole; Duod, duodenum; fLcn2; fecal lipocalin 2; NBMPR, nitrobenzylthioinosine; pColon, proximal Colon.
Discussion
Herein we describe distinct effects of the JAK1/JAK3 inhibitor tofacitinib on cells of the innate and adaptive immune system. Specifically, tofacitinib interfered with the proliferation of naïve and effector or memory cytotoxic and helper T cells. Tofacitinib strongly suppressed Th1 and Th17 differentiation and effector function while showing only a minor effect on Th2 and Treg biology. Furthermore, at higher concentrations, tofacitinib impacted on primary cells of the innate immune system including monocytes, macrophages, and human intestinal epithelial organoids. Tofacitinib skewed monocyte and macrophage function toward a regulatory M2 phenotype. Most in vitro effects were reproducible in an experimental model of colitis and a detailed pharmacokinetic workup revealed a relevant impact of intestinal inflammation on systemic and tissue pharmacokinetic of tofacitinib. Finally, we identified ENTs—which are upregulated in activated immune cells—as promotors of cellular tofacitinib uptake, a mechanism pharmacologically accessible by specific inhibitors.
Historically, CD and UC have been characterized as prototypic Th1- and Th2-mediated diseases.26 Based on genetic and molecular studies, today, immune-mediated diseases range on a continuum from autoimmunity to autoinflammation with IBD categorized as a polygenic autoinflammatory disorder.27 As a JAK1 and JAK3 inhibitor, tofacitinib is likely to affect both innate and adaptive immune signaling. Lethality observed with JAK1 genetic ablation and marked defects in T cell development observed in JAK3 knockout animals underscores the importance of these receptor kinases.11 In line with previously published data,28 we observed a strong suppression of primary Th1 and Th17 responses. However, in our hands, the effects on Th2 and particularly regulatory T cells responses were less pronounced. Moreover, we demonstrated that besides its effect on the differentiation of naïve CD4+ T cells, tofacitinib strongly suppresses effector functions of differentiated effector Th cells. An impact of tofacitinib on VZV-specific CD4+ T cells has been suggested to contribute to the risk for Herpes zoster virus infections,29 which could be further aggravated by suppression of specific cytotoxic T cells as supported by our data.
Monocytes and macrophages are increasingly recognized as important gatekeepers of intestinal immune homeostasis.30 Polarization of macrophages toward M2 emerges as a relevant anti-inflammatory mechanism in experimental models and in humans.8,9,31 Indeed, several studies reported tofacitinib-induced monocyte and macrophage polarization toward a regulatory phenotype.32,33 We complement this information by showing that tofacitinib inhibits M1 and promotes M2 macrophage differentiation, yet in differentiated macrophages, tofacitinib suppresses effector cytokines in LPS-stimulated M1, but not in M0 or M2 macrophages. Another important innate gut immune cell, namely the IEC, orchestrates the first line of defense against luminal contents.34 By generating colonic organoids from UC patients and healthy control subjects, we demonstrated that tofacitinib suppresses TNF-α-induced production of proinflammatory cytokines rendering IECs an additional tofacitinib cellular target. To reproduce these findings within the context of a living organism DSS-treated wildtype animals received tofacitinib twice a day in a therapeutic manner. In contrast to previously published data,33 tofacitinib significantly ameliorated DSS colitis with respect to histologic disease activity and the fecal biomarker lipocalin 2. In contrast to other models of experimental colitis, T and B cells are not required for DSS colitis.31 However, DSS induced a Th17 response that was entirely suppressed by tofacitinib. Furthermore, tofacitinib suppressed steady-state activity of Th1 and Th2 cells but did not affect regulatory T cells.
From a structural perspective tofacitinib represents a pyrrolo-pyrimidine mimicking the nucleotide ATP. As it has been demonstrated that dietary nucleosides may shape gut microbial composition,24 and microbial enzymes even contribute to drug pharmacokinetics of certain nucleoside analogs,23 we studied the impact of tofacitinib on gut microbial compositions in the steady state and during intestinal inflammation. Arguably, owing to the short treatment duration, we could not observe significant effects on alpha- or beta-diversity measures. On a taxonomic level, several amplicon sequence variants were significantly affected warranting further studies on long-term effects of tofacitinib on the microbiome and vice versa.
Given the enormous body of data on the pharmacokinetics of therapeutic antibodies in IBD, the lack of data regarding the pharmacokinetic properties of tofacitinib during intestinal inflammation is surprising.13 Thus, we set up an LC-MS/MS based strategy to study systemic and tissue concentrations of tofacitinib. We observed increased tofacitinib serum concentrations in animals treated with DSS. Given the fact that monoclonal antibodies are known to be lost into the feces during intestinal inflammation,35 this finding was counterintuitive. Strikingly, serum concentrations strongly correlated with the severity of intestinal inflammation. As tofacitinib concentrations were comparable in the urine and the feces between DSS and vector treated animals, we hypothesized that the observed increase may originate from the interference of inflammatory mediators with tofacitinib metabolizing enzymes, which has to be substantiated in future studies.36 The sensitivity of our diagnostic set-up further enabled us to study mechanisms associated with the cellular uptake of tofacitinib. Already in the steady state, we observed a marked variation of spontaneous tofacitinib uptake into leukocytes. This steady-state uptake was significantly enhanced after stimulation with LPS. Based on the structural relationship with ATP, we hypothesized that tofacitinib may utilize the same adenosine cell membrane transporters.25 Strikingly, LPS stimulation induced a significant upregulation of ENT1 and 2, and this upregulation was blocked by specific ENT inhibitors highlighting an involvement of this transporter system in the cellular delivery of tofacitinib and may even propose some preference for activated immune cells.
In summary, we present a detailed analysis of the cell-specific effects of the JAKi tofacitinib on innate and adaptive immunity. We identify intestinal inflammation as a decisive modulator of the systemic pharmacokinetics of tofacitinib in mice, which needs to be studied and confirmed in humans. Finally, we decipher an important membrane transport mechanism that regulates cellular uptake of tofacitinib into activated immune cells, suggesting a model that explains a preferred uptake of tofacitinib into activated immune cells and a potential starting point to interfere with and channel such an uptake.
Materials And Methods
Human T Cell Experiments
PBMCs were isolated from healthy volunteer whole blood samples using Lymphoprep (Axis Shield, Dundee, United Kingdom) according to the manufacturer’s instructions. Pan T cells were isolated using a Pan T Cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany), stimulated with αCD3/αCD28 for 3 days in the presence of tofacitinib (10–500 nM) or dimethyl sulfoxide and the proliferation was measured using a WST-1 assay. For flow cytometric analyses, pan T cells were stained with CFSE prior to stimulation with αCD3/αCD28 for 5 days in the presence of tofacitinib (10–500 nM) or dimethyl sulfoxide. Human naïve CD4+ T cells were isolated from PMBCs using a naïve CD4+ isolation kit (Miltenyi Biotec). For cytokine and RNA measurements the cells were stimulated with αCD3/αCD28 and IL-12 (Th1), IL-4 (Th2) or IL-6, IL-1β, TGF-β, and IL-23 (Th17) for 5 days. Tofacitinib or dimethyl sulfoxide was either added at the start of the experiment or after the differentiation (day 5) and cells were restimulated with PHA. For flow cytometric analysis naïve CD4+ T cells were stained with CFSE prior to differentiation as mentioned above.
Human Monocyte and Macrophage Experiments
PBMCs were isolated from healthy volunteer whole blood samples using Lymphoprep (Axis Shield) according to the manufacturer’s protocol. Monocytes were isolated using a CD14+ MACS positive selection kit (Miltenyi Biotec). For cytokine measurements monocytes were plated and stimulated with 100 ng/mL LPS and 50 ng/mL IFNγ for 24 hours in the presence of tofacitinib (10–500 nM) or DMSO. For macrophage differentiation experiments, monocytes were differentiated with 25 ng/mL M-CSF for 6 days and then polarized overnight to M0 macrophages in the absence of cytokines, M1 macrophages with 100 ng/mL LPS and 50 ng/mL IFNγ, or M2 macrophages with 20 ng/mL IL-4. Tofacitinib or dimethyl sulfoxide treatment was added to the cells either on day 0 or on day 6 of the experiments.
Human Intestinal Organoids
Human organoids were cultured from IEC isolates of biopsy specimens retrieved from endoscopy of UC patients and healthy control subjects using IntestiCult Organoid Growth Medium (STEMCELL Technologies, Vancouver, Canada) and a protocol adapted from the manufacturer’s instructions. Briefly, biopsies were flushed with ice-cold phosphate-buffered saline (PBS) and minced into small pieces. Tissue was then transferred to Gentle Cell Dissociation Reagent (STEMCELL Technologies) and incubated at 4°C on a rocking platform for 30 minutes. After centrifugation, crypts were transferred to 1 mL of ice-cold 1% bovine serum albumin/Dulbecco’s modified Eagle medium. Crypts were dissolved by gentle mixing and passed through a 70-μm cell strainer. Crypts (n = 500) per well were seeded in 50 μL Matrigel (BD, Franklin Lakes, NJ; 356231) on a prewarmed 24-well plate and allowed to solidify for 10 minutes at 37°C, after which 500 μL IntestiCult Growth Medium supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Biochrome, Cambridge, United Kingdom, 0257F) was added. Medium was exchanged 3 times per week and organoids passaged with a split ratio of 1:6 every 7–14 days. For monolayers, organoids (24-well plate) were harvested, disrupted, trypsinized, washed, and seeded in a 96 well plate, precoated with 5% Matrigel. When the organoid monolayers had reached a confluence of >80%, the medium was changed to Dulbecco’s modified Eagle medium/F12 and after 1 hour cells are stimulated with 50 ng/mL TNF-α for 6 hours in the presence of Tofacitinib or dimethyl sulfoxide.
Flow Cytometric Analysis of Human T Cells
The respective cells were stained with CFSE prior to differentiation or stimulation. At the end of the experiment cells were stimulated with PHA for 4 hours in the presence of monensin (BD GolgiStop; BD, Franklin Lakes, NJ) and brefeldin A (BD GolgiPlug). Cells and OneComp eBeads (eBioscience) were stained with respective antibodies and isotype control subjects according to standard procedures for combined intracellular/surface stainings. Typically, 100,000–600,000 cells were analyzed using a CytoFLEX S (Beckman Coulter, Brea, CA) and FlowJo Software version 10.6 (FlowJo, Ashland, OR). Antibodies are outlined in Table 1 (human) and Table 2 (mouse).
Table 1.
Human Antibodies Used for Flow Cytometry
| Specificity | Fluorophore | Clone | Purpose | Company |
|---|---|---|---|---|
| CD3 | APC-Cy7 | UCHT1 | T cells | BioLegend |
| CD4 | PE | SK3 | T helper cells | BioLegend |
| CD8 | PerCP | HIT8a | Cytotoxic T cells | BioLegend |
| CD45 | BV785 | 2D1 | Leukocytes | BioLegend |
| CD45RA | PE-Cy7 | HI100 | naïve/resting T cells | BioLegend |
| CD45RO | APC | UCHC1 | activated/memory T cells | BioLegend |
| CD45RO | PE-Dazzle 595 | UCHC1 | activated/memory T cells | BioLegend |
| CFSE | FITC | N/A | Proliferation | BioLegend |
| GATA3 | APC | 16E10A2 | TF Th2 cells | BioLegend |
| IFNγ | BV421 | 4S.B3 | Th1 Cytokine | BioLegend |
| IL-4 | PerCp-Cy5.5 | MP4-25D2 | Th2 Cytokine | BioLegend |
| IL-17A | AF700 | BL168 | Th17 Cytokine | BioLegend |
| Live/Dead | Violet 510 | N/A | Viability | Tonbo Bioscience |
| RORγt | PE | AF-KJS-9 | TF Th17 cells | eBioscience |
| T-bet | BV 605 | 4B10 | TF Th1 cells | BioLegend |
Table 2.
Mouse Antibodies Used for Flow Cytometry
| Specificity | Fluorophore | Clone | Purpose | Company |
|---|---|---|---|---|
| CD3 | eFluor450 | 17A2 | T cells | eBioscience |
| CD3 | superbright600 | 145-2C11 | T cells | eBioscience |
| CD4 | AF700 | RM4-5 | T helper cells | BioLegend |
| CD4 | APC-eFluor780 | GK1.5 | T helper cells | eBioscience |
| CD8 | FITC | 53-6.7 | Cytotoxic T cells | BD Bioscience |
| CD8 | PB | 558106 | 53-6.7 | BD Bioscience |
| CD11b | APC-Cy7 | M1/70 | Macrophages | eBioscience |
| CD11b | PerCP | M1/70 | Macrophages | eBioscience |
| CD11c | PE | N418 | Dendritic cells | eBioscience |
| CD11c | eFluor450 | N418 | Dendritic cells | eBioscience |
| CD19 | eFluor450 | eBio1D3 | B cells | eBioscience |
| CD19 | PE-Cy7 | 6D5 | B cells | eBioscience |
| CD25 | PE-Cy7 | PC61 | regulatory T cells | BioLegend |
| CD45 | FITC | 104 | Leukocytes | eBioscience |
| CD45 | APC | 30-F11 | Leukocytes | BioLegend |
| CD45 | APC-Cy7 | 30-F11 | Leukocytes | BioLegend |
| CD49b | eFluor450 | DX5 | NK cells | eBioscience |
| F4/80 | PE-eF610 | BM8 | Macrophages | eBioscience |
| F4/80 | eFluor450 | BM8 | Macrophages | eBioscience |
| FoxP3 | FITC | FJK-16s | TF Tregs | eBioscience |
| GATA3 | AF647 | 16E10A23 | TF Th2 cells | BioLegend |
| GR1 | eFluor450 | RB6-8C5 | Myeloid Cells | eBioscience |
| IFNγ | PE-Cy7 | XMG1.2 | Th1 Cytokine | BD Bioscience |
| IL-4 | PE | 11B11 | Th2 Cytokine | BioLegend |
| IL-17A | AF488 | TC11-18H10 | Th17 Cytokine | BD Bioscience |
| Live/Dead | DAPI | N/A | Viability | BioLegend |
| Live/Dead | Violet 510 | N/A | Viability | Tonbo Bioscience |
| Ly6C | PerCP-Cy5.5 | HK1.4 | Monocytes | eBioscience |
| Ly6G | superbright600 | RB6-8C5 | Neutrophils | eBioscience |
| MertK | PE-CY7 | DS5MMER | Macrophages | eBioscience |
| MHCII | AF700 | M5/114.15.2 | APCs | eBioscience |
| NK1.1 | PE | PK136 | NK cells | BioLegend |
| RORγt | APC | AFKJS-9 | TF Th17 cells | eBioscience |
| SiglecF (CD170) | eF660 | 1RNM44N | Eosinophils | eBioscience |
| T-bet | PE | eBio4B10 | TF Th1cells | eBioscience |
| TNF-α | PerCP-Cy5.5 | MP6-XT22 | Th1 Cytokine | BioLegend |
Preparation of Mouse Intestinal Single Cells for Flow Cytometry
Mice were sacrificed and colons harvested, flushed with ice-cold ,PBS and cut longitudinally. Tissue was minced followed by a 90-minute digestion period at 37°C on a shaking platform in RPMI + 0.5% bovine serum albumin containing 128 U/mL collagenase IV (C1889; Sigma, St Louis, MO) and 10 U/mL DNAse II (Sigma D8764). After straining and washing, the intestinal cells were spun down, split in 2 parts, and resuspended in complete medium and stimulated with PHA for 4 hours (adaptive panel) or resuspended in FACS Buffer (innate Panel). Cells and OneComp eBeads (eBioscience, San Diego, CA) were stained with respective antibodies and isotype control subjects according to standard procedures for combined intracellular/surface stainings. Typically, 100,000–600,000 cells were analyzed using a CytoFLEX S (Beckman Coulter) and FlowJo Software version 10.6.
Fluorescence-Activated Cell Sorting Gating Strategy
The gating strategy for analyzing differentiation and proliferation of T cells and for the mucosal cellular infiltrate is outlined in Figure 3. Briefly, cells were gated using forward scatter (FSC)/side scatter characteristics. Singlets were selected by comparing FSC height and FSC area. Neutrophils were identified as CD45+, Lin1− (Lin1=CD3, CD19, CD49b, DAPI), and Ly6G+ cells. Macrophages were identified by CD45+, Lin1−, Ly6G−, CD11b+, and MertK+. Monocytes were characterized by CD45+, Lin1−, Ly6G−, and Ly6Chi. Dendritic cells were characterized by CD45+, Lin1−, Ly6G−, CD11c+, and MHCII+. Eosinophils were characterized by CD45+, Lin1−, Ly6G−, CD11c+, and SiglecF+. Th cells were identified by Lin2− (Lin2=CD11c, F4/80, Ly6G, DAPI), CD3+, CD19−, CD4+. Cytotoxic T cells were identified by Lin2−, CD3+, CD19−, CD8+. B cells were defined as Lin2−, CD3–, and CD19+. NK cells were identified by Lin2−, CD3−, CD19+, and NK1.1+.
Enzyme-Linked Immunosorbent Assay
For the analysis of cytokine production in cell culture supernatants the following commercially available enzyme-linked immunosorbent assay (ELISA) kits have been used according to the manufacturers’ instructions: Human IL-22 DuoSet ELISA (DY782), Human TNFα DuoSet ELISA (DY210), Human IL-6 DuoSet ELISA (DY206), Human IL-10 DuoSet ELISA (DY217B), and Human CXCL10/IP-10 DuoSet ELISA (DY266) from R&D Systems (Minneapolis, MN) and BD OptEIA Set Human IFN-γ (555142) and BD OptEIA Set Human IL-4 (555194) from BD Biosciences. For the FLCN2 ELISA the stool was diluted in PBS at a concentration of 50 mg/mL, homogenized by vortexing for 5 minutes, and centrifuged for 10 minutes at 3000 g, and the supernatant was stored at –20°C until analyzed. FLCN2 was assayed using the Mouse Lipocalin-2/NGAL DuoSet ELISA (DY1857; R&D Systems). The samples were diluted as required and results calculated from the standard curve.
Gene Expression Analysis
Cells were homogenized by vigorous pipetting using Tri Reagent (Invitrogen, Waltham, MA). RNA extraction was carried out according to the manufacturer’s instructions. Total RNA concentration was quantified at 260 nm right after isolation, using a Nanodrop 1000 (Peqlab, Erlangen, Germany). RNA was converted to complementary DNA using an M-MLV reverse transcriptase in combination with hexamer primers (Thermo Fisher Scientific, Waltham, MA). The complementary DNA sequences were amplified by polymerase chain reaction using gene-specific primers in combination with SYBR-green chemistry. For denaturation, primer annealing and elongation, the following polymerase chain reaction conditions were chosen: 95°C for 2 minutes followed by 45 cycles of 95°C for 30 seconds, Tm for 30 seconds, and 72°C for 30 seconds. Tm was adjusted for each primer pair. Data were analyzed with Stratagene’s MxPro software (Stratagene, San Diego, CA). Relative expressions were calculated using the delta Ct method, using actin-beta as a housekeeping gene. The primers used are outlined in Table 3.
Table 3.
Sequences of Quantitative Polymerase Chain Reaction Primer Pairs
| Primer Name | Forward Sequence | Reverse Sequence | Tm |
|---|---|---|---|
| Actin-β | TGGAAGAGTGCCTCAGGGC | GAAGAGCTACGAGCTGCCTGA | 60°C |
| ARG1 | TGGACAGACTAGGAATTGGCA | CCAGTCCGTCAACATCAAAACT | 56°C |
| IFNγ | GTGGCCCGGATGTGAGAAG | GGAGCCCTTGTCGGATGATG | 62°C |
| IL-4 | TCGGTAACTGACTTGAATGTCCA | TCCTTTTTCGCTTCCCTGTTTT | 60°C |
| IL-8 | TGTCACTGCAAATCGACACCT | TCTGCTCTGTGAGGCTGTTC | 60°C |
| IL-10 | ACTGAGAGTGATTGAGAGTGGAC | AACCCTCTGCACCCAGTTTTC | 60°C |
| IL-17F | CTGCACCAGACCATGCTTCA | TCTTCAGAAATGTCAGCGCGT | 60°C |
| iNOS | GGGAGAACCTGAAGACCCTCA | TGCTCTTGTTTTCACAGGGAAG | 60°C |
| SLC29A1 (ENT1) | ATGGCCCTGTGCCTTAGTAGT | AGCTTTGCATTCATGGTCTTGA | 60°C |
| SLC29A2 (ENT2) | GAGGAGCTGGTCAACATCAAC | GCTCCATACCATGCTGCCA | 60°C |
| TNF-α | ATGACAGTGAAGACCCTGCAT | TTGGGGATTTTCCGAGCTGC | 60°C |
Animal Studies and Colitis Induction
BALB/c mice were bred and maintained under specific pathogen-free conditions at the animal facility of the Medical University Innsbruck. All experiments were approved by the Austrian Ministry of Science and Research (BMBWF-66,011/0097-V/3b/2019). The DSS colitis model was performed as previously described.31 Briefly, acute colitis was induced in 8-week-old female BALB/c mice with 5% DSS (MP Biomedicals, Solon, OH, MW 36,000–50,000) ad libitum for 5 consecutive days, followed by a tap water period until the end of experiments. Control mice received tap water during the entire study period. Mice were orally gavaged 30 mg/kg bodyweight tofacitinib (MedChemExpress, Monmouth Junction, NJ, HY-40354A) or vehicle control (0.5% methylcellulose; Sigma, St Louis, MO, M0387) twice daily in a therapeutic manner starting on day 4 until the end of experiments. Disease activity and body weight was monitored on a daily basis. At the end of the experiment, mice were euthanized, stool and blood samples were collected, and intestinal tissue was removed and flushed with ice-cold PBS. For mucosal scrapings, intestinal tissue was opened longitudinally, scraped with glass slides, and immediately snap-frozen into liquid nitrogen. For IHC, colons were fixed in 10% buffered formalin and further transferred into an automatic tissue processor. For flow cytometric analysis 2 cm of colon were collected and stored in RPMI medium until further analysis.
Histological Procedures and Scoring
A total of 5 μm formalin-fixed, paraffin-embedded tissue sections were stained with hematoxylin and eosin and Slides were scanned on an IntelliSite Ultra-Fast Scanner (Philips digital pathology; Philips Healthcare, Best, the Netherlands), examined with a CaseViewer software module (3DHISTECH, Budapest, Hungary) in a blinded fashion. Histology scores were assessed using a semi-quantitative score as previously described.31
IHC Analysis
A total of 5 μm formalin-fixed, paraffin-embedded tissue sections were processed for IHC according to the manufacturer’s protocol. Briefly, tissue sections were deparaffinized and rehydrated and subsequently underwent antigen unmasking with a citrate buffer for 30 minutes in a steamer and 20 minutes at room temperature. Next, we performed peroxidase blocking and protein blocking steps and incubated the slides with the primary antibody for 1 hour at room temperature. Following a 30-minute incubation with the second antibody, the slides are developed with the substrate and counterstained with hematoxylin. Finally, sections are dehydrated, mounted in Eukitt, and scanned on an IntelliSite Ultra-Fast Scanner (Philips digital pathology). The appropriate scans are then analyzed with a CaseViewer software module (3DHISTECH), and positive cells are quantified with a script written in octave.
Microbiome Screening (16S Sequencing)
Amplicon-based sequencing was performed on an Illumina Miseq device with V3 chemistry using the 8F-YM/517R primer pair to amplify the V1-V3 regions of the bacterial 16S rRNA fragment (5′-AGAGTTTGATYMTGGCTCAG-3′ and 5′-ATTACCGCGGCTGCTGG-3′; expected amplicon size 510 bp). Sequences were demultiplexed and further processed using DADA2 according to the published workflow.37 Reads were trimmed at a length of 295 and 285 for the F- and R-read respectively after removing primers. A maximum rate of expected errors of 2 and 4 were accepted for the F- and R-reads, respectively. Error rates were learned and absolute sequence variants inferred as demonstrated in the DADA2 tutorial (https://benjjneb.github.io/dada2/tutorial.htmL). F- and R-Reads were then merged and sequence table was generated which included 17829 unique sequences. Chimera were removed using the removeBimeraDenovo command in DADA2. The overall chimera rate was 22%, resulting in a total number of 2652 nonchimeric sequences. Taxonomic information including the species level was assigned using the pretrained classifier based on the SILVa v138 database.38 A phylogenetic tree was generated using the DECIPHER39 and phangorn40 R packages. Prior to statistical analysis, amplicon sequence variants (ASVs) classified as mitochondria or chloroplasts as well as singleton ASVs were removed. Metagenomics analysis was performed using phyloseq,41 assessing measures of α-diversity (Shannon and Chao1 indices) and β-diversity (Bray-Curtis dissimilarity, Weighted/Unweighted Unifrac). Diversity measures were compared between tofacitinib- and vehicle-treated groups separately for the DSS-exposed and the control mice using Student’s t test (for α-diversity) or Permanova (for β-diversity; implemented in the vegan package).42 Abundance of ASVs was compared statistically using a negative binomial model implemented in the R package DESeq2.43,44 In silico prediction of metagenome function was carried out using PICRUSt245 on the ASV reference sequences. Bioinformatics analysis was done in R v4.0.2 (R Foundation for Statistical Computing, Vienna, Austria), using the following general purpose packages: tidyverse v1.3,46 Biostrings v2.56.0,47 rstatix v0.6.0,48 ggpubr v0.4.0,49 cowplot v1.0.0,50 and ggsci v2.9.51
Uptake Experiments
PBMCs were isolated from healthy volunteer whole blood samples using Lymphoprep (Axis Shield) according to the manufacturer’s protocol. The cells were plated in 12-well plates (15 × 106 cells/well). On the next day cells were treated with 100 nM tofacitinib for 2 hours. After tofacitinib treatment the cells were immediately put on ice. The cells are washed with ice-cold PBS, scraped, and collected. The cells were centrifuged for 10 minutes at 4°C and 300 g and the pellet washed twice with ice-cold PBS. Cell numbers were quantified by a LUNA automated cell counter (Logos Biosystems) and the cell pellet was snap frozen in liquid nitrogen for later analyses. For the uptake inhibitor experiments, PBMCs were isolated as described above and plated in 12-well plates (15 × 106 cells/well). On the next day, cells were pretreated with LPS or a vector control to be then exposed to 100 nM tofacitinib or vector for 2 hours in the presence or absence of 1 μM NBMPR, 10 μM dipyridamole, or a combination. The cells were then harvested and processed as described previously.
Liquid Chromatography-Tandem Mass Spectrometry
Tofacitinib was quantified by LC-MS/MS. After the addition of an isotope labeled internal standard 13C3-tofacitinib (Clearsynth CS-O-10298), preanalytical workup included “dilute-and-shoot” (urine samples), protein precipitation (serum samples), and chemical and mechanical lysis (tissue and cell samples). Samples were analyzed on an LC 1100 series HPLC system (Agilent, Waldbronn, Germany) hyphenated to a QTRAP 4000 (Sciex, Herndon, VA) mass spectrometer by monitoring specific precursor-to-fragment ion transitions of tofacitinib and the internal standard. Quantification was accomplished with linear calibration models generated from analyzing donated reference samples.
Reagents
The following reagents were used for cell stimulation, differentiation or treatment:αCD3 (10 μg/mL; BioLegend, San Diego, CA, 300438), αCD8 (2 μg/mL; BioLegend, 302934), dipyridamole (10 μM, R&D, 0691), IFNγ (20 ng/mL; PeproTech, Rocky Hill, NJ, 300-02), IL-1β (10 ng/mL; PeproTech, 200-01B), IL-2 (10 ng/mL; PeproTech, 200-02), IL-4 (20-25 ng/mL; PeproTech, 200-04), IL-6 (10 ng/mL; PeproTech, 200-06), IL-12 (10 ng/mL; PeproTech, 200-12), IL-23 (10 ng/mL; PeproTech, 200-23), ionomycin (10 μg/mL; Sigma, I0634), LPS (20 ng/mL to 1 μg/mL, Invivogen, tlrl-3pelps), M-CSF (10-25 ng/mL; PeproTech, 300-25), NBMPR (1 μM; R&D, 2924), PHA (4 μg/mL; Sigma, L1686), PMA (1 μg/mL; Sigma, P1585), transforming growth factor β (1 ng/mL; Miltenyi, 130-095-067), TNF-α (50 ng/mL; PeproTech, 300-01A), tofacitinib citrate (Tofa, 10-500 nM; MedChemExpress, HY-40354A), SLC29A1/ENT1 antibody (LSBio, #LS-B3385), and pSTAT3 antibody (Cell Signaling Technology, Danvers, MA; 9145).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8.1 software package (GraphPad Software, San Diego, CA), Excel 2016 (Microsoft, Redmond, WA), and FlowJo software version 10.6. Differences between multiple groups were analyzed using 1-way analysis of variance with Bonferroni’s post tests for multiple comparisons of parametric data, or Kruskal-Wallis tests combined with Dunn post tests for nonparametric data. Descriptive results are expressed using mean ± SD. Correlation was calculated with Spearman’s correlation for nonparametric samples.
Footnotes
Conflicts of interest A.R.M. is receiving research support from AbbVie and Takeda under the framework of the Christian Doppler Research Society. He has received further consultation fees and/or speaker honoraria from AbbVie, Merck Sharp & Dohme, Takeda, Janssen-Cilag, Amgen, Sandoz and Pfizer. H.T. has received speaker honoraria from AbbVie, Janssen-Cilag, Merck Sharp & Dohme and Takeda. The remaining authors disclose no conflicts.
Funding This work was supported by the Christian Doppler Research Association (to A.R.M) and we gratefully acknowledge the support by the Austrian Federal Ministry of Science, Research, and Economy and the National Foundation for Research, Technology, and Development.
References
- 1.Crohn B.B., Ginsberg L., Oppenheimer G.D. Regional ileitis: a clinicaland pathological entity. JAMA. 1932;99:1323–1329. [Google Scholar]
- 2.Cameron H.C., Rippmann C. Statistics of ulcerative colitis from the London hospitals, provided to form a basis for the above discussion: Guy's Hospital. Proc R Soc Med. 1909;2:100–106. [PMC free article] [PubMed] [Google Scholar]
- 3.Roda G., Ng S.C., Kotze P.G., Argollo M., Panaccione R., Spinelli A., Kaser A., Peyrin-Biroulet L., Danese S. Crohn’s disease. Nat Rev Dis Prim. 2020;6:1–19. doi: 10.1038/s41572-020-0156-2. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T., Siegmund B., Le Berre C., Wei S.C., Ferrante M., Shen B., Bernstein C.N., Danese S., Peyrin-Biroulet L., Hibi T. Ulcerative colitis. Nat Rev Dis Prim. 2020;6:74. doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- 5.Lamb C.A., Kennedy N.A., Raine T., Hendy P.A., Smith P.J., Limdi J.K., Hayee B.H., Lomer M.C., Parkes G.C., Selinger C. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut. 2019;68:s1–s106. doi: 10.1136/gutjnl-2019-318484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melmed G.Y., Targan S.R. Future biologic targets for IBD: potentials and pitfalls. Nat Rev Gastroenterol Hepatol. 2010;7:110–117. doi: 10.1038/nrgastro.2009.218. [DOI] [PubMed] [Google Scholar]
- 7.Colombel J., Panés J., D’Haens G., Schreiber S., Panaccione R., Peyrin-Biroulet L., Loftus E., Jr., Danese S., Louis E., Armuzzi A. OP01 Higher vs. standard adalimumab maintenance regimens in patients with moderately to severely active ulcerative colitis: Results from the SERENE-UC maintenance study. J Crohn's Colitis. 2020;14:S001. [Google Scholar]
- 8.Vos A.C.W., Wildenberg M.E., Duijvestein M., Verhaar A.P., van den Brink G.R., Hommes D.W. Anti–tumor necrosis factor-α antibodies induce regulatory macrophages in an Fc region-dependent manner. Gastroenterology. 2011;140:221–230. doi: 10.1053/j.gastro.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Zeissig S., Rosati E., Dowds C.M., Aden K., Bethge J., Schulte B., Pan W.H., Mishra N., Zuhayra M., Marx M. Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease. Gut. 2019;68:25–39. doi: 10.1136/gutjnl-2018-316023. [DOI] [PubMed] [Google Scholar]
- 10.Neurath M.F. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 11.O'Shea J.J., Schwartz D.M., Villarino A.V., Gadina M., McInnes I.B., Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandborn W.J., Su C., Sands B.E., D’Haens G.R., Vermeire S., Schreiber S., Danese S., Feagan B.G., Reinisch W., Niezychowski W. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2017;376:1723–1736. doi: 10.1056/NEJMoa1606910. [DOI] [PubMed] [Google Scholar]
- 13.Dowty M.E., Lin J., Ryder T.F., Wang W., Walker G.S., Vaz A., Chan G.L., Krishnaswami S., Prakash C. The pharmacokinetics, metabolism, and clearance mechanisms of tofacitinib, a janus kinase inhibitor, in humans. Drug Metab Dispos. 2014;42:759–773. doi: 10.1124/dmd.113.054940. [DOI] [PubMed] [Google Scholar]
- 14.Panés J., Sandborn W.J., Schreiber S., Sands B.E., Vermeire S., D'Haens G., Panaccione R., Higgins P.D., Colombel J.-F., Feagan B.G. Tofacitinib for induction and maintenance therapy of Crohn's disease: results of 2 phase IIb randomised placebo-controlled trials. Gut. 2017;66:1049–1059. doi: 10.1136/gutjnl-2016-312735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki T., Kawahara A., Fujii H., Nakagawa Y., Minami Y., Liu Z.-J., Oishi I., Silvennoinen O., Witthuhn B.A., Ihle J.N. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 16.Neurath M.F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970–979. doi: 10.1038/s41590-019-0415-0. [DOI] [PubMed] [Google Scholar]
- 17.Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov, McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. (2006). The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 19.Corridoni D., Chapman T., Ambrose T., Simmons A. Emerging mechanisms of innate immunity and their translational potential in inflammatory bowel disease. Frontiers in medicine. 2018;5:32. doi: 10.3389/fmed.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaser A., Zeissig S., Blumberg R.S. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okayasu I., Hatakeyama S., Yamada M., Ohkusa T., Inagaki Y., Nakaya R. (1990). A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 22.Zollner A., Schmiderer A., Reider S.J., Oberhuber G., Pfister A., Texler B., Watschinger C., Koch R., Effenberger M., Raine T., Tilg H., Moschen A.R. Fecal biomarkers in inflammatory bowel diseases: calprotectin versus lipocalin-2–a comparative study. J Crohn's Colitis. 2021;15:43–54. doi: 10.1093/ecco-jcc/jjaa124. [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363 doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doo E.-H., Chassard C., Schwab C., Lacroix C. Effect of dietary nucleosides and yeast extracts on composition and metabolic activity of infant gut microbiota in PolyFermS colonic fermentation models. FEMS Microbiol Ecol. 2017;93:fix088. doi: 10.1093/femsec/fix088. [DOI] [PubMed] [Google Scholar]
- 25.Pastor-Anglada M., Pérez-Torras S. Who is who in adenosine transport. Front Pharmacol. 2018;9:627. doi: 10.3389/fphar.2018.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand S. Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut. 2009;58:1152–1167. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 27.McGonagle D., McDermott M.F. (2006). A proposed classification of the immunological diseases. PLoS Med. 2006;3:e297. doi: 10.1371/journal.pmed.0030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghoreschi K., Jesson M.I., Li X., Lee J.L., Ghosh S., Alsup J.W., Warner J.D., Tanaka M., Steward-Tharp S.M., Gadina M. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550) J Immunol. 2011;186:4234–4243. doi: 10.4049/jimmunol.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almanzar G., Kienle F., Schmalzing M., Maas A., Tony H.-P., Prelog M. Tofacitinib modulates the VZV-specific CD4+ T cell immune response in vitro in lymphocytes of patients with rheumatoid arthritis. Rheumatology. 2019;58:2051–2060. doi: 10.1093/rheumatology/kez175. [DOI] [PubMed] [Google Scholar]
- 30.Na Y.R., Stakenborg M., Seok S.H., Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16:531–543. doi: 10.1038/s41575-019-0172-4. [DOI] [PubMed] [Google Scholar]
- 31.Gerner R.R., Klepsch V., Macheiner S., Arnhard K., Adolph T.E., Grander C., Wieser V., Pfister A., Moser P., Hermann-Kleiter N. NAD metabolism fuels human and mouse intestinal inflammation. Gut. 2017;67:1813–1823. doi: 10.1136/gutjnl-2017-314241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordes F., Lenker E., Spille L.J., Weinhage T., Bettenworth D., Kessel C., Schmidt H.H., Foell D., Varga G. Tofacitinib reprograms human monocytes of IBD patients and healthy control subjects toward a more regulatory phenotype. Inflamm Bowel Dis. 2020;26:391–406. doi: 10.1093/ibd/izz213. [DOI] [PubMed] [Google Scholar]
- 33.De Vries L., Duarte J., De Krijger M., Welting O., Van Hamersveld P., Van Leeuwen-Hilbers F., Moerland P., Jongejan A., D’Haens G., De Jonge W. A JAK1 selective kinase inhibitor and tofacitinib affect macrophage activation and function. Inflamm Bowel Dis. 2019;25:647–660. doi: 10.1093/ibd/izy364. [DOI] [PubMed] [Google Scholar]
- 34.Martini E., Krug S.M., Siegmund B., Neurath M.F., Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandse J.F., van den Brink G.R., Wildenberg M.E., van der Kleij D., Rispens T., Jansen J.M., Mathôt R.A., Ponsioen C.Y., Löwenberg M., D’Haens G.R. Loss of infliximab into feces is associated with lack of response to therapy in patients with severe ulcerative colitis. Gastroenterology. 2015;149:350–355.e2. doi: 10.1053/j.gastro.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Mallick P., Taneja G., Moorthy B., Ghose R. Regulation of drug-metabolizing enzymes in infectious and inflammatory disease: implications for biologics–small molecule drug interactions. Expert Opin Drug Metab Toxicol. 2017;13:605–616. doi: 10.1080/17425255.2017.1292251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaren MR. Silva SSU taxonomic training data formatted for DADA2, Silva version 138 [dataset]. Available at: https://zenodo.org/record/3986799#.YVGwGmYzYeU. Accessed October 9, 2020.
- 39.Wright ES. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R Journal 206;8:352-359.
- 40.Schliep K.P. Phangorn: phylogenetic analysis in R. Bioinformatics. 2010;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. Vegan: community ecology package, Version 2.4-6. https://cran.r-project.org/web/packages/vegan/index.htmL Available at:
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickham H., Averick M., Bryan J., Chang W., McGowan L.D.A., François R., Grolemund G., Hayes A., Henry L., Hester J. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 47.Pagès H., Aboyoun P., Gentleman R., DebRoy S. Biostrings: Efficient manipulation of biological strings. R package version 2.56.0. https://bioconductor.org/packages/release/bioc/htmL/Biostrings.htmL Available at:
- 48.Kassambara A. Rstatix: pipe-friendly framework for basic statistical tests, Version 0.6.0. https://cran.r-project.org/web/packages/rstatix/index.htmL Available at: Accessed October 9, 2020.
- 49.Kassambara A. Ggpubr: 'Ggplot2' based publication ready plots, Version 0.4.0. https://cran.r-project.org/web/packages/ggpubr/index.htmL Available at:
- 50.Wilke C. OCowplot: streamLined plot theme and plot annotations for 'ggplot2', Version 1.0.0. Accessed October 9, 2020.
- 51.Xiao N. Scientific journal and sci-fi themed color palettes for'ggplot2', Version 2.9. https://cran.r-project.org/web/packages/ggsci/vignettes/ggsci.htmL Available at: Accessed October 9, 2020.