Abstract
Aims and Background:
Identifying fibrosis in non-alcoholic fatty liver disease (NAFLD) is essential to predict liver-related outcomes and inform treatment decisions. A protein-based signature of fibrosis could serve as a valuable, non-invasive diagnostic tool. This study sought to identify circulating proteins associated with fibrosis in NAFLD.
Methods:
We used aptamer-based proteomics to measure 4783 proteins in two cohorts (Cohort A and B). Targeted, quantitative assays coupling aptamer-based protein pull down and mass spectrometry (SPMS) validated the profiling results in a bariatric and NAFLD cohort (Cohort C and D, respectively). Generalized linear modelling-logistic regression assessed the candidate proteins to classify fibrosis.
Results:
From the multiplex profiling, 16 proteins differed significantly by fibrosis in cohorts A (n=62) and B (n=98). Quantitative and robust SPMS assays were developed for 8 proteins and validated in Cohorts C (n=71) and D (n=84). The protein A disintegrin and metalloproteinase with thrombospondin motifs like 2 (ADAMTSL2) accurately distinguished NAFL/NASH with fibrosis stage 0–1 (F0–1) from at-risk NASH with fibrosis stage 2–4 with an AUROC of 0.83 and 0.86 in Cohorts C and D, respectively, and from NASH with significant fibrosis (F2–3) with an AUROC of 0.80 and 0.83 in Cohorts C and D, respectively. An 8-protein panel distinguished NAFL/NASH F0–1 from at-risk NASH (AUROC 0.90 and 0.87 in Cohort C and D, respectively) and NASH F2–3 (AUROC 0.89 and 0.83 in Cohorts C and D, respectively). The 8-protein panel and ADAMTSL2 protein had superior performance to the NAFLD fibrosis score and Fibrosis-4 score.
Conclusion:
The ADAMTSL2 protein and an 8-protein soluble biomarker panel are highly associated with at-risk NASH and significant fibrosis with superior performance to standard of care fibrosis scores.
Keywords: proteomics, ADAMTSL2, non-alcoholic fatty liver disease, non-alcoholic steatohepatitis, fibrosis, biomarker
Graphical Abstact
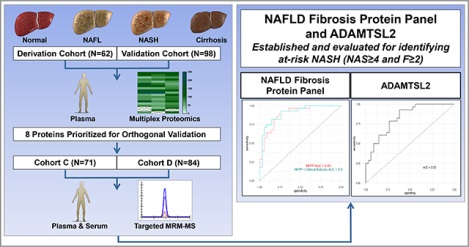
Lay Summary:
Non-alcoholic fatty liver disease (NAFLD) is one of the most common causes of liver disease worldwide. Diagnosing NAFLD and identifying fibrosis (scarring of the liver) currently requires a liver biopsy. Our study identified novel proteins found in blood which may identify fibrosis without the need for a liver biopsy.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is an accelerating cause of liver disease worldwide, impacting 25% of adults.[1] Non-alcoholic fatty liver (NAFL) is defined by excess steatosis in the liver and can progress to non-alcoholic steatohepatitis (NASH), characterized by steatosis, lobular inflammation, hepatocyte ballooning and, frequently, hepatic fibrosis and/or cirrhosis.[2] NASH can result in decompensated liver disease, hepatocellular carcinoma and increased liver-related, cardiovascular, and all-cause mortality.[3, 4]
The identification of individuals with NASH who are at the highest risk of progression to end-stage liver disease is essential for risk stratification and management. Fibrosis stage is the most prognostic histological feature of NAFLD, with higher stage associated with increased risk of death and liver transplantation.[5] In addition, a NAFLD activity score (NAS) ≥4 in the setting of fibrosis stage 2 or higher (F≥2) is considered at-risk NASH, with a high risk of disease progression.[2] Histologic analysis of liver biopsy is the gold standard for diagnosis but its invasive nature, sampling variability and potential for complications precludes use as a tool for identifying at-risk NASH.[6] While predictive scores including the NAFLD fibrosis score (NFS) and Fibrosis-4 (FIB-4) score have been developed to stage fibrosis in NAFLD, performance varies by population, indeterminant results are often reported and insight into the pathogenesis of fibrosis is not provided. Direct markers of fibrosis, such as N-terminal type III collagen propeptide (Pro-C3), the Enhanced Liver Fibrosis (ELF) score, and the NIS4 have been used to predict fibrosis and/or at-risk NASH but are limited by low positive predictive values or have only been evaluated in at-risk NASH and not significant fibrosis subsets alone. [2, 7, 8]
Human plasma and serum are important biological sources for biomarker and pathway discovery due to the abundance of circulating proteins involved in complex biological processes.[9] While the large number and range of protein concentrations can make assessment challenging, advances in large-scale proteomic profiling allow for the rapid detection and relative quantification of thousands of proteins from a single sample. One such multiplexed affinity assay (SomaScan®) uses modified single-stranded DNA aptamers to specifically bind target proteins in a high-throughput assay format with capture and microarray readouts.[10, 11] This method has been successfully utilized in discovering biomarkers including those associated with cardiovascular and neurologic disorders.[12, 13]
The present study applied the aptamer-based profiling platform to plasma and serum from adults with histologically-defined NAFLD for the identification of proteins associated with fibrosis and the development of a disease panel for the non-invasive identification of adults with at-risk NASH.
Materials and Methods
Study Design
Design, Setting and Participants
We conducted a nested case-control study drawing from three unique, prospectively enrolled cohorts of adults with NAFLD; Cohort A, (Discovery Cohort) from the Massachusetts General Hospital (MGH) Study of Brain Natriuretic Peptide (BNP) in Bariatric Surgery, Cohort B (Discovery Cohort) and Cohort C (Validation Cohort), both from the MGH NAFLD Cohort Registry and Cohort D (Validation Cohort) from the Beth Israel Deaconess Medical Center (BIDMC) NAFLD Clinic Cohort.
Cohort A (Discovery Cohort) included men and women age ≥18 years who underwent Roux-en-Y gastric bypass including standard of care liver biopsies at the time of bariatric surgery. Plasma was collected at baseline, 1–2 months and 6 months post-operatively.[14] Subjects with pre-existing cardiac disease, chronic kidney disease and uncontrolled hypertension were excluded.
The MGH NAFLD Cohort Registry includes more than 2,000 adults with NAFLD, diagnosed by imaging or liver biopsy and individuals who have undergone bariatric surgery with standard of care intra-operative liver biopsies as described previously.[15, 16]. Subjects are followed longitudinally with collection of plasma, serum and, in a subset, liver tissue.
The NAFLD Clinic Cohort at Beth Israel Deaconess Medical Center includes 205 adults with a histological diagnosis of NAFLD enrolled between December 2009 and 2016 and followed longitudinally.
For Cohorts B, C and D, subjects were selected using the following criteria: (1) men and women age ≥18 years; (2) hepatitis C antibody and hepatitis B surface antigen negative and (3) contemporaneous liver biopsy and plasma samples (within 6 months for MGH cohort in individuals with a clinical biopsy or 6 months before bariatric surgery in those with a biopsy during surgery, 3 months for the BIDMC Registry). Subjects were excluded for (1) alcohol use >20g daily for women or >30 g daily for men, (2) decompensated cirrhosis, (3) chronic use of methotrexate, amiodarone, corticosteroids or tamoxifen; or (4) other causes of chronic liver disease.
Diagnosis of Liver Disease
For Cohort A, pathology reports from liver biopsies underwent independent review by two clinicians. Normal liver histology (NLH) was defined by the absence of steatosis, hepatocyte ballooning, lobular inflammation and fibrosis. NAFL was defined by steatosis score ≥1, ballooning score of 0, lobular inflammation score of 0–1 and fibrosis stage 0, or no report of lobular inflammation, ballooning or fibrosis. NASH was defined by steatosis score ≥1, ballooning score ≥1 and lobular inflammation score ≥1. Fibrosis was staged using the modified Brunt criteria.[17] When specific scores were not available, reports of the presence of ballooning and lobular inflammation were used.
For Cohorts B, C and D, all biopsies were reviewed by a hepatopathologist in a blinded manner using the NASH Clinical Research Network scoring criteria.[17] Steatosis, NASH and fibrosis were defined as above. Subjects were classified as NLH, NAFL, NASH F0, NASH F1, NASH F2, NASH F3 and NASH cirrhosis. Subjects with at-risk NASH (NAS ≥4 and F≥2) or NASH with significant fibrosis (F2–3) were grouped for comparisons to subjects with NAFL/NASH F0–1. Two subjects in validation Cohort C had fibrosis stage ≥2 but NAS3 (with 1 in each component).
Sample Collection
Fasting plasma samples (Cohorts A-C) or serum samples (Cohort D) were collected for measurement with the multiplex assay. Blood was collected in EDTA-treated tubes for plasma or serum separating tubes (SST) for serum, centrifuged at 1.9 g for 15 minutes within 120 minutes of collection, and the supernatant aliquoted and frozen at −80°C.
Proteomic profiling assay
The SomaScan proteomic profiling platform utilizes SOMAmers® (Slow-Off rate Modified Aptamers) that bind to target proteins with high affinity and specificity.[18] The expanded assay includes 5,034 SOMAmers, of which 4,783 measure human proteins from 4,137 distinct human genes with femtomole (fM) limits of detection over a wide range of protein levels in plasma or serum (>8 logs of concentration). The platform exhibits median limits of detection and quantification (LOD/LOQ) of 40 fM and 100 fM, respectively and ~5% coefficients of variation for median intra- and inter- assay variability.[19] A hybridization array to capture SOMAmers quantitatively determines the protein present by converting the assay signal (relative fluorescence units, RFUs) to analyte relative abundance.[12, 13] Assays were performed by SomaLogic in collaboration with Novartis according to the protocol described by our group and others. [20–23].
Validation using SOMAmer-Pulldown Mass Spectrometry
SOMAmers are well-suited for use in multiplexed protein enrichment strategies from serum or plasma coupled with quantitation by mass spectrometry.[20,21] Proteins are affinity enriched, eluted and digested prior to targeted multiple reaction monitoring (MRM) mass spectrometry. Accurate quantitation is achieved by the use of stable isotope-labeled peptide external calibration curves. We refer to this technique as SOMAmer-pulldown mass spectrometry (SPMS) which allows for both specific and sensitive orthogonal validation while achieving lower limits of protein detection in the nanogram per milliliter range. Performance characteristics of this method were assessed (see Supplementary Methods) and analytical validation established using a subset of Cohort A (31 subjects). Protein level changes by SPMS were directionally consistent to the relative protein abundance as determined by SomaScan. All SPMS was performed by Novartis.
SomaScan Microarray Data Analysis
Open source microarray analysis software from the R/Bioconductor consortium was used to analyze the microarray data (http://www.bioconductor.org/). arrayQualityMetrics was used for microarray technical quality assessment, and data was background corrected and normalized by SomaLogic using spike in controls and calibrator samples.[24, 25] Differentially measured protein levels were calculated using a moderated t-statistic in limma applying a simple regression model comparing patients grouped by disease stage as assessed by histology. The p-values were adjusted using Benjamini-Hochberg to control for the false discovery rate (FDR). Proteins were considered significantly changed if they had a false discovery rate (FDR) adjusted p-value <0.05 and a fold change greater than 20%.
Analysis from SOMAmer-pull down mass spectrometry
Two analytical approaches, generalized linear modelling-logistic regression (GLM-LR) and random forest classification, were used to assess the ability of a single protein (A disintegrin and metalloproteinase with thrombospondin motifs like 2; ADAMTSL2) or an 8-protein signature to classify fibrotic NASH. Multivariate models were trained using Cohorts C and D, comparing NAFL/NASH F0–1 to either at-risk NASH or NASH F2–3. Random forest classifier for at-risk NASH trained on Cohort D was tested on Cohort C. Machine learning methods were provided through the R/Bioconductor framework using the caret library (package version 6.0–86).[26] Model performance was evaluated using the area under the receiver operating characteristic curve (AUROC) using pROC (package version_1.17.0.1).[27] Accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the models was determined using standard definitions.[28–30] Continuous and categorical variables were assessed using the student’s t-test and Fisher’s exact test, respectively, across the key comparisons between cohorts and incorporated into the models.
The FIB-4 score and NFS were used as comparative fibrosis risk scores in Cohort D, with metadata available to calculate results in 81 subjects. The presence of advanced fibrosis was defined as >2.67 (≥36 years) for FIB-4 and >0.67 (≥36 years) for the NFS. The absence of advanced fibrosis was defined as <1.3 (36–65 years) or <2.0 (≥65 years) for FIB-4 and <−1.455 (36–65 years) or <0.12 (≥65 years) for the NFS. [31]
All studies were approved by the MGH and BIDMC institutional review boards and all participants provided written informed consent.
Results
Baseline Characteristics
Overall, 315 subjects from three bariatric surgery cohorts (Cohort A n=62 subjects, Cohort B n=98, Cohort C n=71) and one NAFLD cohort (Cohort D n=84) were included in the study (Table 1). Mean age ranged from 46 to 50 years, 45–75% of subjects were women, and diabetes mellitus was present in 32%−58% of subjects. Lower mean BMI (p<0.0001), lower prevalence of diabetes (p<0.01) and higher mean ALT and AST levels (p<0.0001 and p<0.0005, respectively) were observed when comparing validation Cohort D to Cohort C.
Table 1.
Demographics and NAFLD liver histological grading of discovery and validation cohorts†
| Variable | SomaScan | SOMAmer-Pulldown Multiplex MRM Mass Spectrometry | ||
|---|---|---|---|---|
| Cohort | Discovery Cohort A (n=62) | Discovery Cohort B (n=98) | Validation Cohort C (n=71) | Validation Cohort D (n=84) |
| Age, years (SD) | 46 (11) | 48 (13) | 48 (12) | 50 (13) |
| Sex, female – yes (%) | 47 (75) | 69 (70) | 47 (66) | 38 (45)* |
| Diabetes mellitus – yes (%) | 29 (46) | 45 (46) | 41 (58) | 27 (32)* |
| BMI (kg/m2) | 47 (7) | 45 (6) | 46 (7) | 34 (7)*** |
| ALT (U/L) | 44 (37) | 27 (15) | 50 (42) | 74 (40)*** |
| AST (U/L) | 32 (24) | 26 (10) | 35 (28) | 50 (24)** |
| Alkaline phosphatase (U/L) | 91 (22) | 81 (24) | 88 (33) | 80 (27) |
| Total Bilirubin (mg/dL) | 0.5 (0.2) | 0.5 (0.3) | 0.5 (0.2) | 0.7 (0.8) |
| Glucose (mg/dL) | 113 (38) | 105 (31) | 128 (60) | 113 (40) |
| LDL (mg/dL) | 102 (32) | 98 (32) | 101 (36) | 107 (39) |
| HDL (mg/dL) | 52 (13) | 44 (12)* | 45 (16) | 46 (12) |
| Triglycerides (mg/dL) | 172 (100) | 127 (69)* | 157 (80) | 189 (97) |
| Liver Histology – subjects (%) | ||||
| Normal | 4 (6) | 19 (19) | 10 (14) | NA |
| NAFL | 31 (50) | 30 (31) | 8 (11) | 12 (14) |
| NASH Fibrosis Stage 0 | 13 (21) | 9 (9) | 10 (14) | 19 (23) |
| NASH Fibrosis Stage 1 | 5 (8) | 23 (23) | 10 (14) | 12 (14) |
| NASH Fibrosis Stage 2 | 5 (8) | 10 (10) | 18 (25) | 21 (25) |
| NASH Fibrosis Stage 3 | 3 (5) | 5 (5) | 10 (14) | 12 (14) |
| NASH Fibrosis Stage 4 | 1 (2) | 2 (2) | 5 (7) | 8 (10) |
Data reported as mean (standard deviation) unless otherwise noted. Significant differences between Cohort A/B and Cohort C/D for demographic data reported using one-way ANOVA for continuous variables and Fisher’s Exact test for categorical variables.
p<0.01,
p<0.0005,
p<0.0001
NAFL: non-alcoholic fatty liver; NASH: non-alcoholic steatohepatitis; ALT: alanine aminotransferase; AST: aspartate aminotransferase; LDL: low-density lipoprotein; HDL: high-density lipoprotein
Proteomic Profiles in Discovery Cohorts and Selection of Candidates for Validation
A total of 234 and 304 proteins were identified by SomaScan in Cohorts A and B, respectively, which differed between NLH and any of the NAFLD phenotypes (adjusted p<0.05). (Figure 1) Twenty-four proteins were significantly different in at-risk NASH subjects (adjusted p<0.05). ADAMTSL2 was included given its biological plausibility, directional consistency and conserved effect size in both cohorts (Cohort A p=0.01, Cohort B p=0.08 both adjusted for the 5,034 multiple comparisons). The proteins were refined to 16 candidates that demonstrated stepwise directionally consistent relationships between phenotypes and statistically significant protein values between at-risk NASH compared to NAFL/NASH F0–1 or NLH. These 16 candidates were selected for validation and quantification by SPMS in two additional cohorts (C and D). Accurate and robust SPMS assays were established for the following 8 proteins, referred to as the NAFLD Fibrosis Protein Panel (NFPP): ACY1, ADAMTSL2, ADH4, ALDOC, ASL, ENPP7, FBP1 and FTCD. SPMS assays could not be established for eight proteins (NFASC, CHST9, COLEC11, POR, FAH, SELE, THBS2 and TREM2) due to challenges in reproducibility, stability and/or sensitivity. Protein levels as determined by SOMAscan in the two discovery cohorts and by SPMS in the two validation cohorts are presented in Supplementary Figures 1 and 2. Statistical results across NAFLD phenotypes in the two discovery cohorts are presented in Supplementary Table 1. Biological function and tissue expression for NFPP proteins is provided in Supplementary Table 2.
Figure 1:
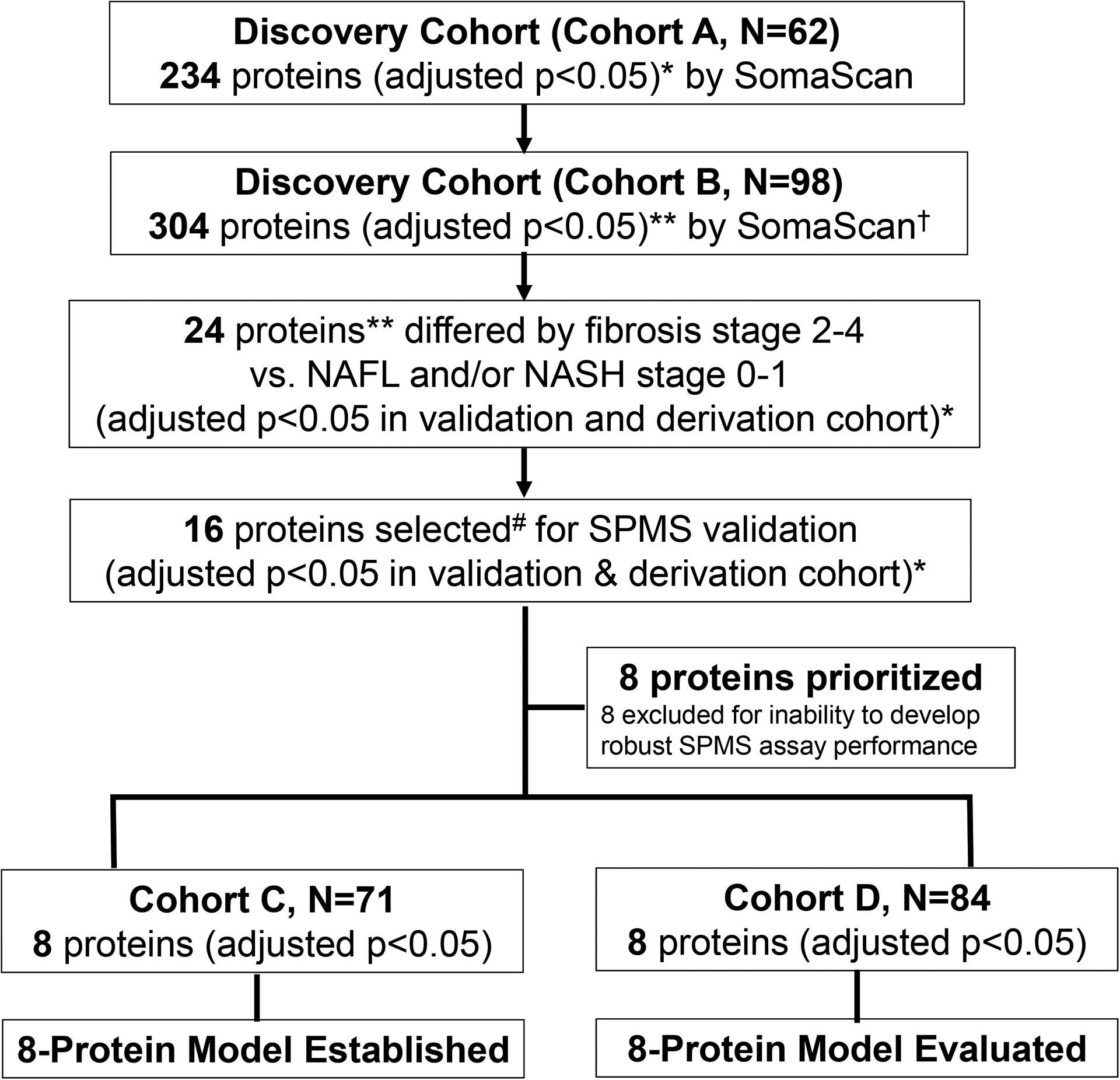
Flow Diagram of Protein Selection for NAFLD Fibrosis Protein Panel.
* P value adjusted for FDR multiple hypothesis testing
** ADAMTSL2 adjusted p=0.08 in Cohort B
# 16 proteins selected by effect size, consistent trends and directionality
† SomaScan measures relative abundance of proteins, SPMS provides precise quantification of protein levels.
Performance of ADAMTSL2 to Distinguish NASH with Significant Fibrosis (F2–3) and At-Risk NASH
Of the eight proteins evaluated, ADAMTSL2 was the strongest individual protein classifier of fibrosis. ADAMTSL2 accurately classified NASH F2–3 or at-risk NASH relative to NAFL/NASH F0–1 with an AUROC of 0.80 (p=2.3×10−3) or 0.83 (p=1.3×10−3) in Cohort C and 0.83 (p=2.8×10−4) or 0.86 (p=7.1×10−7) in Cohort D, respectively. (Figure 2) For at-risk NASH subjects, sensitivity of 70% and 68%, specificity of 79% and 86%, PPV of 79% and 82%, and NPV of 69% and 74% were obtained in Cohorts C and D, respectively. The full performance characteristics of ADAMTSL2 to classify significant fibrosis and at-risk NASH are provided in Table 2.
Figure 2. ROC curves of ADAMTSL2 for the identification of NASH F2–3 and at-risk NASH.
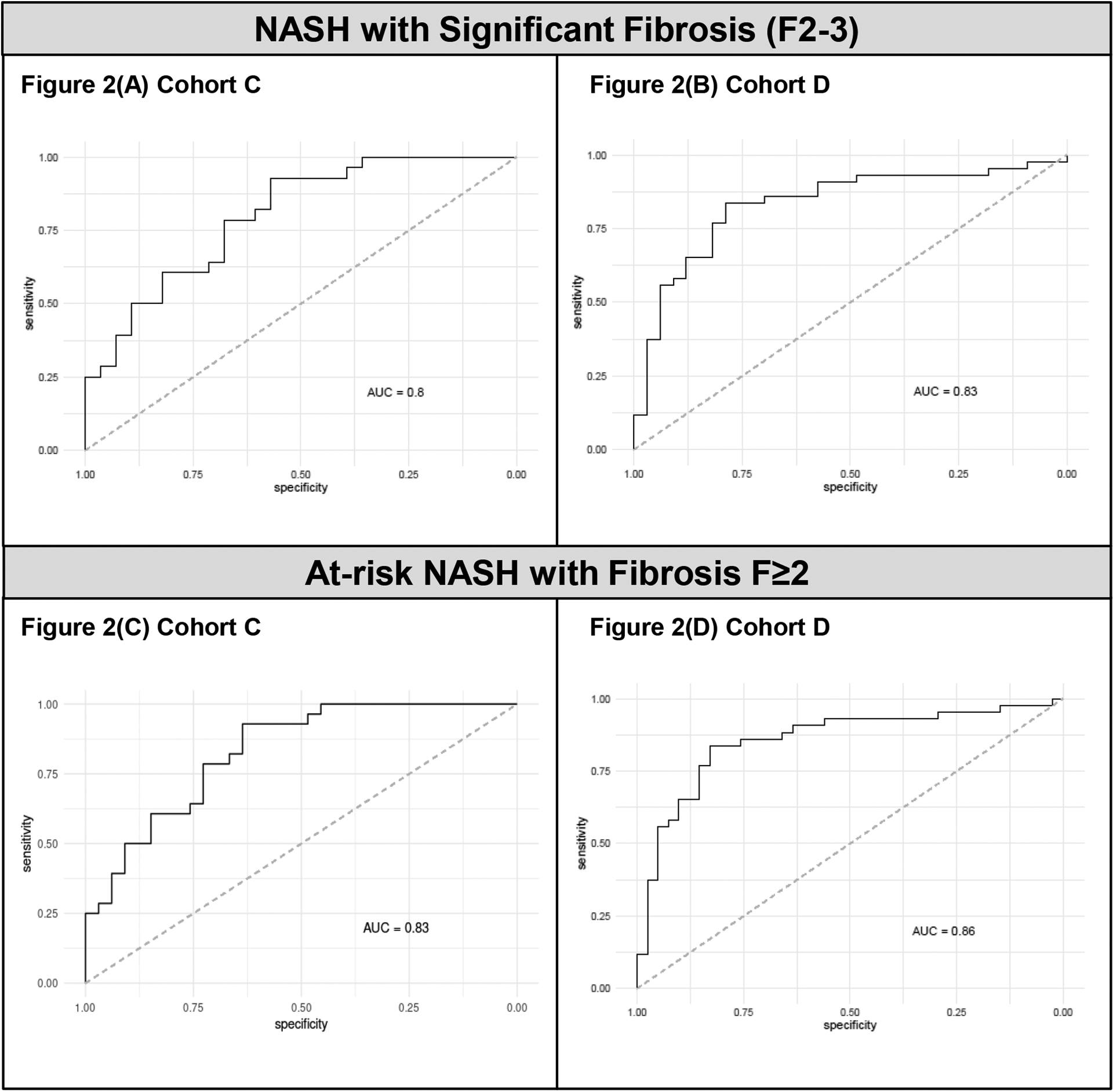
(A) In Cohort C, ADAMTSL2 AUROC = 0.80 (95%CI 0.56–0.81) for NASH F2–3. (B) In Cohort D, ADAMTSL2 AUROC = 0.83 (95%CI 0.65–0.85) for NASH F2–3. (C) In Cohort C, ADAMTSL2 AUROC = 0.83 (95%CI 0.61–0.84) for at-risk NASH. (D) In Cohort D, ADAMTSL2 AUROC = 0.86 (95%CI 0.67–0.86) for at-risk NASH.
Table 2.
Performance Characteristics of ADAMTSL2 and the NAFLD Fibrosis Protein Panel (NFPP) by Generalized Linear Modeling Logistic Regression (GLM-LR)
| Comparison | Classifier/Model | Cohort | AUROC (95% CI) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|
| NAFL/NASH F0–1 vs. NASH F2–3 | ADAMTSL2 | C | 0.80 (0.56–0.81) | 61% | 79% | 74% | 67% |
| D | 0.83 (0.65–0.85) | 58% | 91% | 83% | 74% | ||
| NFPP | C | 0.89 (0.70–0.91) | 71% | 93% | 91% | 76% | |
| D | 0.83 (0.65–0.85) | 64% | 86% | 78% | 76% | ||
| NFPP + clinical features | C | 0.90 (0.72–0.92) | 79% | 89% | 88% | 81% | |
| D | 0.87 (0.73–0.91) | 70% | 93% | 88% | 80% | ||
| NAFL/NASH F0–1 vs. at-risk NASH | ADAMTSL2 | C | 0.83 (0.61–0.84) | 70% | 79% | 79% | 69% |
| D | 0.86 (0.67–0.86) | 68% | 86% | 82% | 74% | ||
| NFPP | C | 0.90 (0.72–0.92) | 82% | 86% | 87% | 80% | |
| D | 0.87 (0.68–0.87) | 73% | 84% | 81% | 77% | ||
| NFPP + clinical features | C | 0.90 (0.68–0.89) | 79% | 82% | 84% | 77% | |
| D | 0.86 (0.68–0.87) | 73% | 84% | 81% | 77% |
Performance of the NFPP to Distinguish NASH with Significant Fibrosis (F2–3) and At-Risk NASH
The NFPP classified NASH F2–3 or at-risk NASH with an AUROC of 0.89 (p=6.2×10−7) or 0.90 (p=1.2 × 10−6) in Cohort C and 0.83 (p=2.7×10−4) or 0.87 (p=2.1 × 10−7) in Cohort D, respectively.(Figure 3) For at-risk NASH subjects, sensitivity of 82% and 73%, specificity of 86% and 84%, PPV of 87% and 81%, and NPV of 80% and 77% were obtained in Cohorts C and D, respectively. Full performance characteristics are noted in Table 2. Comparable results were observed when applying random forest classification models (Supplementary Table 3).
Figure 3. ROC curves of the NAFLD Fibrosis Protein Panel (NFPP) for the identification of NASH F2–3 and at-risk NASH.
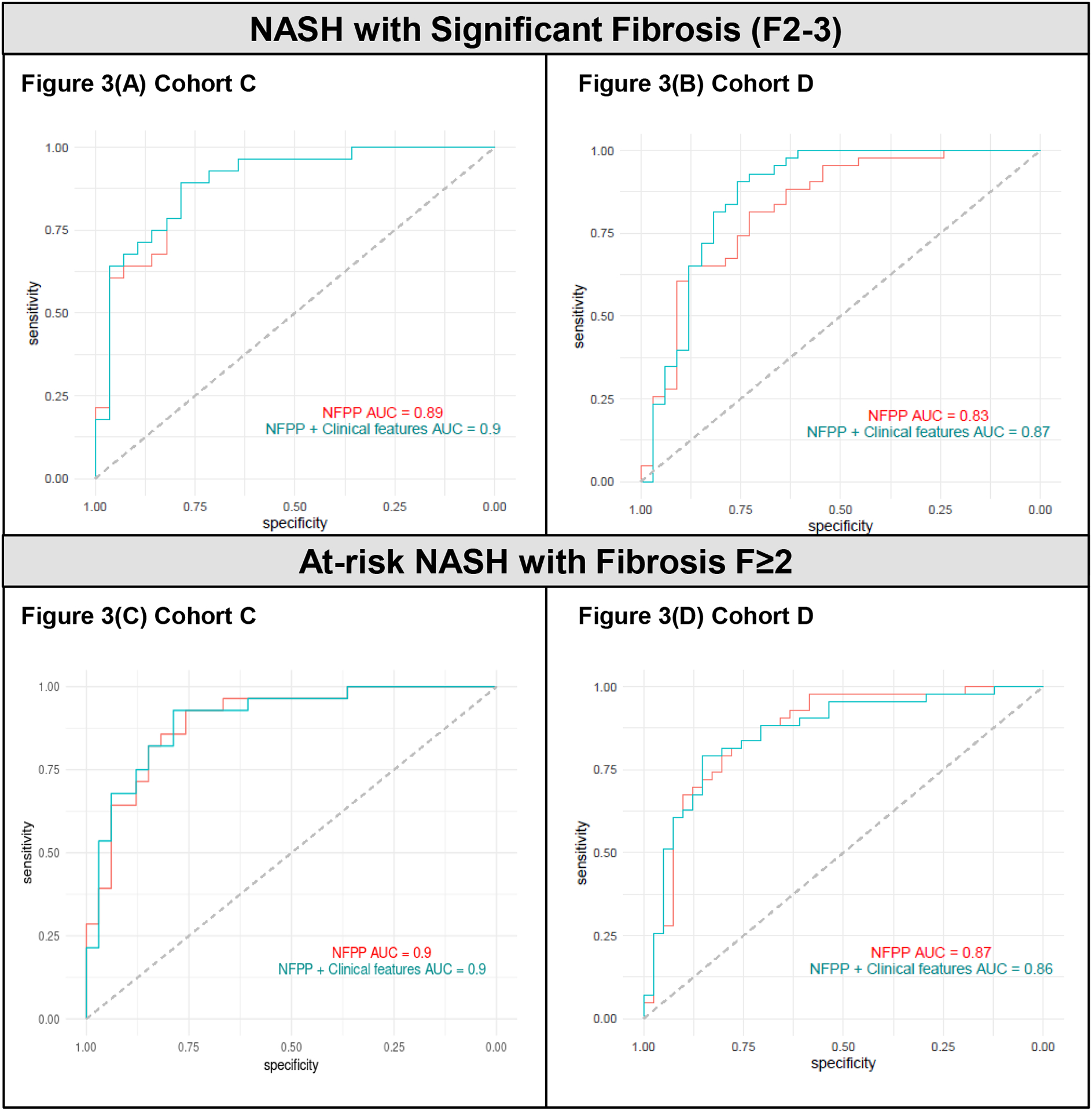
(A) In Cohort C, NFPP AUROC = 0.89 (95%CI 0.70–0.91) and NFPP plus Clinical Features AUROC = 0.90 (95%CI 0.72–0.92) for NASH F2–3. (B) In Cohort D, NFPP AUROC = 0.83 (95%CI 0.65–0.85) and NFPP plus Clinical Features AUROC = 0.87 (95%CI 0.71–0.90) for NASH F2–3. (C) In Cohort C, NFPP AUROC = 0.90 (95%CI 0.72–0.92) and NFPP plus Clinical Features AUROC = 0.90 (95%CI 0.68–0.89) for at-risk NASH. (D) In Cohort D, NFPP AUROC = 0.87 (95%CI 0.68–0.87) and NFPP plus Clinical Features AUROC = 0.86 (95%CI 0.68–0.87) for at-risk NASH. Clinical features include age, BMI, gender, and diabetes status.
Clinical features were evaluated as potential confounders to NASH fibrosis classification using the NFPP. In Cohort C, there were no statistically significant differences by fibrosis group. In Cohort D, the at-risk NASH group had higher mean age (54 ± 12 years vs. 47 ± 13 years, p=0.013) and diabetes prevalence (54% vs. 12%, p<0.0001) than those with NAFL/NASH F0–1. Gender (66% vs. 45% female, p<0.01) and BMI (46 kg/m2 vs. 34 kg/m2, p<0.001) differed across Cohort C and D, respectively. Including these four clinical features in the model resulted in low or minimal impact to performance. (Figure 3 and Table 2)
Performance of ADAMTSL2 and the NFPP Compared to NFS and FIB-4 to Distinguish At-Risk NASH
In Cohort D, the NFS demonstrated an AUROC of 0.70 (p=0.009) for at-risk NASH classification. Addition of ADAMTSL2 or the NFPP improved performance of the NFS, increasing AUROC to 0.86 (p=4.7×10−6) or 0.89 (p=1.8×10−9), respectively. (Figure 4A) Sensitivity improved from 55% for NFS alone to 70% and 80% with the addition of ADAMTSL2 or the NFPP, respectively. Specificity improved from 73% for NFS alone to 80% and 85% with the addition of ADAMTSL2 or the NFPP, respectively. Similarly, PPV and NPV improved from 67% and 63%, respectively, for NFS alone to 78% and 73% with the addition of ADAMTSL2 and to 84% and 81% with the addition of the NFPP, respectively. (Supplementary Table 4)
Figure 4. ROC curves of the NAFLD Fibrosis Score (NFS) and Fibrosis-4 (FIB-4) score with the addition of ADAMTSL2 and the NAFLD Fibrosis Protein Panel (NFPP) for the identification of at-risk NASH in Cohort D.
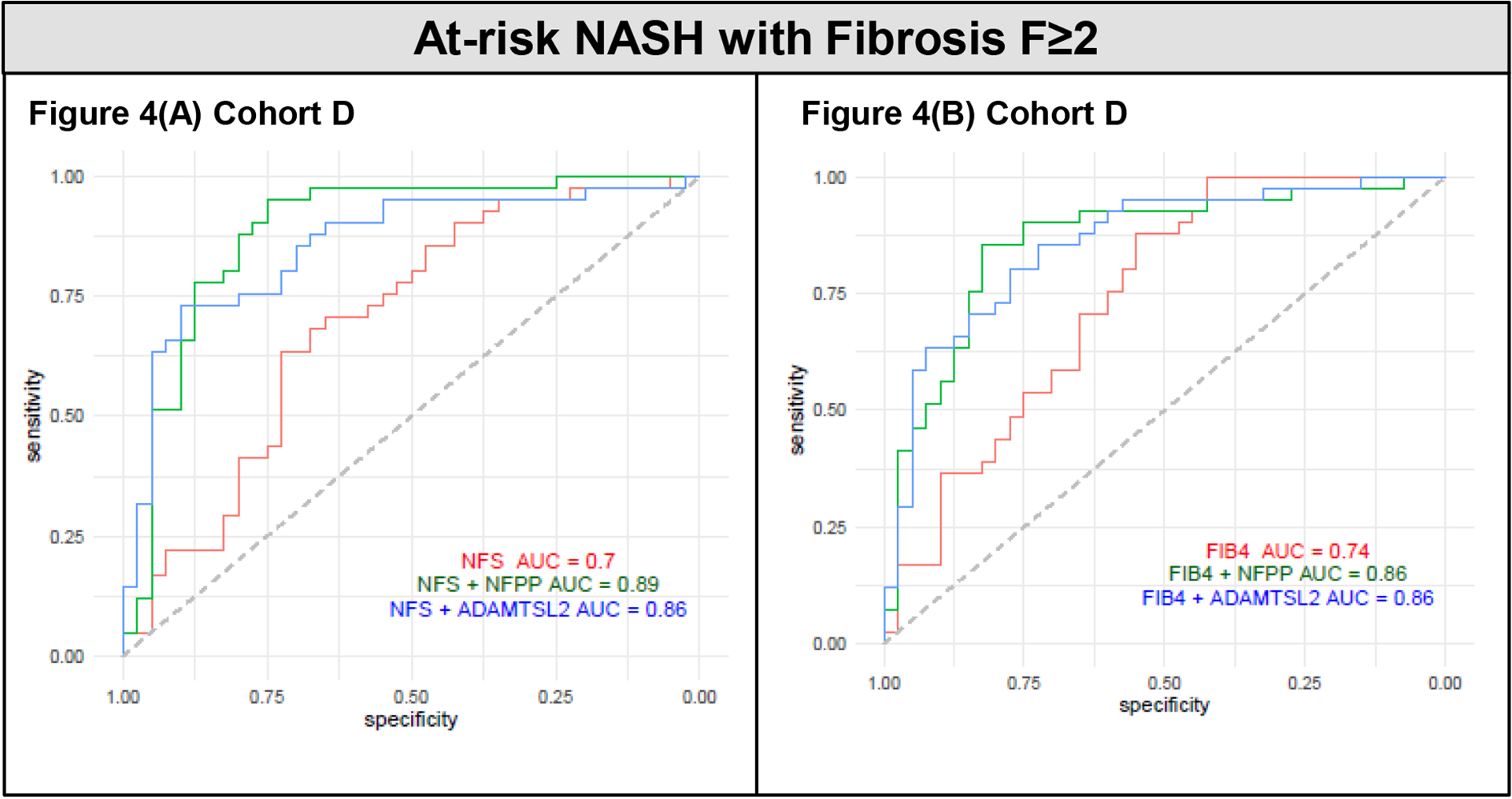
(A) NFS AUROC = 0.70 (95% CI 0.53–0.75), NFS plus ADAMTSL2 AUROC = 0.86 (95% CI 0.64–0.84), and NFS plus NFPP AUROC = 0.89 (95% CI 0.73–0.90). (B) FIB-4 AUROC = 0.74 (95% CI 0.60–0.81), FIB-4 plus ADAMTSL2 AUROC = 0.86 (95% CI 0.69–0.87), and FIB-4 plus NFPP AUROC = 0.86 (95% CI 0.73–0.90).
In Cohort D, the FIB-4 score had an AUROC of 0.74 (p=9.8 × 10−5) for identifying at-risk NASH. Addition of ADAMTSL2 or the NFPP improved performance of the FIB-4 score, increasing AUROC to 0.86 (p=1.3×10−7) or 0.86 (p=1.8×10−9), respectively. (Figure 4B) While sensitivity was 55% for both the NFS and FIB-4, other assay metrics were higher (specificity 88%, PPV 81% and NPV 67%) for FIB-4. Addition of ADAMTSL2 or the NFPP improved performance of the FIB-4 score, with sensitivity of 72% and 75%, specificity of 85% and 90%, PPV of 83% and 88%, and NPV of 76% and 79%, respectively. (Supplementary Table 4)
Performance characteristics for NASH F2–3 subjects are provided in Supplementary Table 4.
Discussion
Using the SomaScan method, we identified circulating protein biomarkers associated with fibrotic NASH in two histologically-defined cohorts and validated the findings in two additional cohorts using a rigorous, specific, and quantitative complementary technique (SOMAmer-pulldown mass spectrometry; SPMS). We identified a single protein (ADAMTSL2) and developed an 8-protein panel (the NAFLD Fibrosis Panel; NFPP) that accurately classified at-risk NASH (NAS ≥4, F≥2) and NASH with significant fibrosis (F2–3).
Non-invasive testing (NITs) to identify fibrosis and risk stratify patients with NAFLD is a growing field. Currently, several algorithmic fibrosis scores are used in clinical practice. The NFS and FIB4 scores combine clinical variables and laboratory results to predict the presence of fibrosis stage 3–4 with AUROCs of 0.82–0.88 and 0.765. [32] [33] Subsequent evaluation resulted in AUROCs ranging from 0.66–0.71 for the NFS and 0.73–0.76 for FIB-4, depending on population prevalence of advanced fibrosis.[34, 35] The NIS4 panel, incorporating miR-34a-5p, alpha-2-macroglobulin, YKL-40 and hemoglobin A1C, was recently developed to assess at-risk NASH and demonstrated AUROCs of 0.76–0.83.[2] Pro-C3, a marker of type III collagen formation, distinguishes advanced fibrosis with AUROC of 0.73–0.78 and when used as part of the ADAPT score with age, diabetes and platelet count, identified advanced fibrosis with an AUROC of 0.86.[7, 8] In a recent meta-analysis, the ELF score, composed of tissue inhibitor of metalloproteinases 1 (TIMP-1), hyaluronic acid (HA) and amino-terminal propeptide of type III procollagen (PIIINP), was highly sensitive for excluding fibrosis but exhibited low PPV and sensitivity for detecting advanced and significant fibrosis except in high prevalence settings (>50%).[36] In the present study, both the NFPP and ADAMTSL2 showed comparable performance in the at-risk population (AUROC 0.83–0.90) and warrants direct comparison to emerging non-invasive biomarkers. The NFPP and ADAMTSL2 maintain their predictive ability in NASH with significant fibrosis (F2–3) (AUROC 0.80–0.89). Evaluation of biomarkers to identify at-risk NASH in the absence of cirrhosis ensures biomarker performance is not driven solely by large differences seen in patients with severe disease.
The rule-in approach for NAFLD seeks to accurately predict which patients with NASH are at risk of progressive disease. Both the NFPP and ADAMTSL2 exhibit high PPV for classification of at-risk NASH, 87% and 81% for the NFPP and 79% and 82% for ADAMTSL2 in Cohorts C and D, respectively. When compared to existing clinical scores in Cohort D, the NFPP and ADAMTSL2 show superior (NFS PPV 67%) or comparable (FIB-4 PPV 81%) performance. Addition of the NFPP or ADAMTSL2 improved performance of the NFS (PPV 84% and 78%, respectively) and FIB-4 score (PPV 88% and 83%, respectively). Notably, the results suggest addition of a single soluble biomarker to existing clinical scores could extend their performance into F2–4 populations. Thus, for ruling in at-risk NASH, both the NFPP and ADAMTSL2 were superior to the commonly used NFS and FIB-4 score and improved their performance.
The findings in this report strongly support ADAMTSL2 as a novel circulating classifier of NASH fibrosis. ADAMTSL2 was previously identified as a plasma marker of NASH fibrosis in a preclinical model [37] and included as part of a gene expression signature associated with NAFLD progression derived from a meta-analysis of published human studies.[38] Existing data suggests a role for ADAMTSL2 in extracellular matrix (ECM) biology. ADAMTSL2 interacts with latent TGF-β-binding protein-1 (LTBP1), involved in TGF-β tissue specific sequestration in the ECM, as well as with fibrillin-1 and fibrillin-2 (FBN1, FBN2), proteins involved in the modulation of microfibril formation.[39, 40] ADAMTSL2 also interacts with lysyl oxidases (LOX) which are involved in microfibril formation, ECM-associated signaling and hepatic fibrosis progression.[41] Potential interplay between LOX and ADAMTSL2 is intriguing given the known role of LOX in liver fibrosis and interest as targets for pharmacological therapy for NASH. Thus, the identification and validation of ADAMTSL2 may provide insight into NASH fibrosis pathophysiology.
The present study has several notable strengths including the use of 3 separate cohorts with distinct populations, those with obesity undergoing bariatric surgery who generally have a lower prevalence of NAFLD fibrosis and those followed at a NAFLD Specialty Clinic where rates of fibrosis are higher than the general population. The SomaScan assay allows for a relatively unbiased approach to detect thousands of proteins at low circulating levels with high precision.[12] This was complemented by the use of SPMS to allow orthogonal validation of SOMAmer selectivity, precise quantification of identified proteins and validation in distinct cohorts. Further, results were consistent across serum and plasma samples collected from different centers and of different age.
Our study is cross-sectional in nature and thus cannot distinguish whether the proteins identified play a role in fibrosis progression or result from it. Future studies must test whether these proteins associate with increased risk for progression of fibrotic disease in longitudinal cohorts of NAFLD and other liver conditions not explored here. Sample selection bias is possible for the bariatric surgery cohorts based on the availability of pre-operative serum/plasma, however similar baseline characteristics across all cohorts suggest the measured samples are representative of the larger repository. Eight proteins could not be robustly assessed using the SPMS approach which may have strengthened our classifier score. Heterogeneity of protein levels precluded a focus on distinguishing fibrosis stage 1, reflecting the significant variability of early-stage disease as evidenced by high levels of variations in markers of fibrogenesis in other work.[42] Our study did not examine the performance of all emerging fibrosis biomarkers, including Pro-C3 or the ELF score. Comparisons within the same cohort are warranted to directly compare the performance characteristics of these biomarkers to ADAMTSL2 and the NFPP. Finally, it is important to acknowledge that the findings of any biomarker study are influenced by the chosen platform and our findings were confined to the SomaScan and SPMS platforms.
The present study utilized a proteomics-based assay to identify eight circulating biomarkers that accurately identify at-risk patients in need of liver biopsy and aggressive management and may be useful to better understand the pathophysiology of fibrosis development in NAFLD. In particular, ADAMTSL2 warrants further investigation as a non-invasive biomarker of significant and advanced fibrosis. The ability to non-invasively identify patients with the highest risk of disease progression could ultimately contribute to better risk stratification, appropriate disease management and consideration for clinical trial enrollment.
Supplementary Material
Using an aptamer-based profiling platform we identified an individual and panel of circulating proteins that non-invasively identify significant fibrosis at-risk NASH in 316 adults with NAFLD.
An 8-protein panel can distinguish NAFL/NASH fibrosis stage 0–1 from fibrosis stage 2–4 with an AUROC 0.87–0.89.
The ADAMTSL2 protein alone can distinguish NAFL/NASH fibrosis stage 0–1 from fibrosis stage 2–4 with an AUROC 0.83–0.86.
Both the 8-protein panel and ADAMTSL2 showed superior performance to the NAFLD Fibrosis Score and Fibrosis-4 Score.
Financial support statement:
This work was supported by the National Institute of Health through R01 DK114144 (KEC), R01HL132320 (REG and TJW), R01HL133870 (REG)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: RP, JL, SMR, JJ, NF, CU & LLJ are employees and stockholders of Novartis. KEC serves on the scientific advisory board for Novo Nordisk and BMS and has received grant funding from Boehringer-Ingelheim, BMS and Novartis. All other authors have no conflicts of interest to declare.
Data availability statement:
Data are available on request due to privacy/ethical restrictions. SomaScan data are available through a data use agreement with Novartis Institutes for BioMedical Research (lori.jennings@novartis.com).
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. Epub 2015/12/29. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SA, Ratziu V, Boursier J, Francque S, Bedossa P, Majd Z, et al. A blood-based biomarker panel (NIS4) for non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis: a prospective derivation and global validation study. Lancet Gastroenterol Hepatol. 2020;5(11):970–85. Epub 2020/08/09. doi: 10.1016/S2468-1253(20)30252-1. [DOI] [PubMed] [Google Scholar]
- 3.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67(1):123–33. Epub 2017/08/13. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henson JB, Simon TG, Kaplan A, Osganian S, Masia R, Corey KE. Advanced fibrosis is associated with incident cardiovascular disease in patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2020. Epub 2020/02/12. doi: 10.1111/apt.15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angulo P, Kleiner DE, Dam-Larsen S, Adams LA, Bjornsson ES, Charatcharoenwitthaya P, et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2015;149(2):389–97 e10. Epub 2015/05/04. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, et al. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128(7):1898–906. Epub 2005/06/09. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 7.Boyle MTD, Schattenberg JM, Ratziu V, Bugianessi E, Petta S, Oliveira CP, Govaere O, Younes R, McPherson S, Bedossa P, Nielsen MJ, Karsdal M, Leeming D, Kendrick S, Anstee QM. Performance of the PRO-C3 collagen neo-epitope biomarker in non-alcoholic fatty liver disease. JHEP Reports. 2019;1(3):188–98. doi: 10.1016/j.jhepr.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels SJ, Leeming DJ, Eslam M, Hashem AM, Nielsen MJ, Krag A, et al. ADAPT: An Algorithm Incorporating PRO-C3 Accurately Identifies Patients With NAFLD and Advanced Fibrosis. Hepatology. 2019;69(3):1075–86. Epub 2018/07/18. doi: 10.1002/hep.30163. [DOI] [PubMed] [Google Scholar]
- 9.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56(2):177–85. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 10.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol. 2012;29(5):543–9. doi: 10.1016/j.nbt.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, et al. Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease. Circulation. 2016;134(4):270–85. Epub 2016/07/23. doi: 10.1161/CIRCULATIONAHA.116.021803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattlecker M, Kiddle SJ, Newhouse S, Proitsi P, Nelson S, Williams S, et al. Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement. 2014;10(6):724–34. doi: 10.1016/j.jalz.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Chen-Tournoux A, Khan AM, Baggish AL, Castro VM, Semigran MJ, McCabe EL, et al. Effect of weight loss after weight loss surgery on plasma N-terminal pro-B-type natriuretic peptide levels. Am J Cardiol. 2010;106(10):1450–5. Epub 2010/11/10. doi: 10.1016/j.amjcard.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimberly WT, O’Sullivan JF, Nath AK, Keyes M, Shi X, Larson MG, et al. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight. 2017;2(9). Epub 2017/05/05. doi: 10.1172/jci.insight.92989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Sullivan JF, Morningstar JE, Yang Q, Zheng B, Gao Y, Jeanfavre S, et al. Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J Clin Invest. 2017;127(12):4394–402. Epub 2017/10/31. doi: 10.1172/JCI95995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21. Epub 2005/05/26. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 18.Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O’Connell D, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A. 2012;109(49):19971–6. Epub 2012/11/10. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, et al. Assessment of Variability in the SOMAscan Assay. Sci Rep. 2017;7(1):14248. Epub 2017/10/29. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob J, Ngo D, Finkel N, Pitts R, Gleim S, Benson MD, et al. Application of Large-Scale Aptamer-Based Proteomic Profiling to Planned Myocardial Infarctions. Circulation. 2018;137(12):1270–7. doi: doi: 10.1161/CIRCULATIONAHA.117.029443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–73. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–7. Epub 2019/12/04. doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hathout Y, Brody E, Clemens PR, Cripe L, DeLisle RK, Furlong P, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2015;112(23):7153–8. Epub 2015/06/04. doi: 10.1073/pnas.1507719112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25(3):415–6. Epub 2008/12/25. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. http://somalogic.com/wp-content/uploads/2017/06/SSM-071-Rev-0-Technical-Note-SOMAscan-Data-Standardization.pdf.
- 26.Kuhn M. Building predictive models in R using the caret package. Journal of Statistical Software. 2008;28(5). [Google Scholar]
- 27.Robin XTN, Hainard A, Tiberti N,Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(77). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman DG, Bland JM. Diagnostic tests 2: Predictive values. BMJ. 1994;309(6947):102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ. 1994;308(6943):1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007;31(4):306–15. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- 31.McPherson S, Hardy T, Dufour J-F, Petta S, Romero-Gomez M, Allison M, et al. Age as a Confounding Factor for the Accurate Non-Invasive Diagnosis of Advanced NAFLD Fibrosis. Am J Gastroenterol. 2017;112(5):740–51. Epub 2016/10/11. doi: 10.1038/ajg.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45(4):846–54. Epub 2007/03/30. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 33.Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–25. Epub 2006/05/27. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 34.Wong VW, Adams LA, de Ledinghen V, Wong GL, Sookoian S. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat Rev Gastroenterol Hepatol. 2018;15(8):461–78. Epub 2018/05/31. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 35.Karsdal MA, Daniels SJ, Holm Nielsen S, Bager C, Rasmussen DGK, Loomba R, et al. Collagen biology and non-invasive biomarkers of liver fibrosis. Liver Int. 2020;40(4):736–50. Epub 20200219. doi: 10.1111/liv.14390. [DOI] [PubMed] [Google Scholar]
- 36.Vali Y, Lee J, Boursier J, Spijker R, Loffler J, Verheij J, et al. Enhanced liver fibrosis test for the non-invasive diagnosis of fibrosis in patients with NAFLD: A systematic review and meta-analysis. J Hepatol. 2020;73(2):252–62. Epub 2020/04/11. doi: 10.1016/j.jhep.2020.03.036. [DOI] [PubMed] [Google Scholar]
- 37.Fernando H, Wiktorowicz JE, Soman KV, Kaphalia BS, Khan MF, Shakeel Ansari GA. Liver proteomics in progressive alcoholic steatosis. Toxicol Appl Pharmacol. 2013;266(3):470–80. Epub 2012/12/04. doi: 10.1016/j.taap.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryaboshapkina M, Hammar M. Human hepatic gene expression signature of non-alcoholic fatty liver disease progression, a meta-analysis. Sci Rep. 2017;7(1):12361. Epub 2017/09/29. doi: 10.1038/s41598-017-10930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40(9):1119–23. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubmacher D, Apte SS. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 2015;47:34–43. Epub 2015/05/11. doi: 10.1016/j.matbio.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aviram R, Zaffryar-Eilot S, Hubmacher D, Grunwald H, Maki JM, Myllyharju J, et al. Interactions between lysyl oxidases and ADAMTS proteins suggest a novel crosstalk between two extracellular matrix families. Matrix Biol. 2019;75–76:114–25. Epub 2018/05/15. doi: 10.1016/j.matbio.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y, Oseini A, Gagnon R, Charles ED, Sidik K, Vincent R, et al. An Evaluation of the Collagen Fragments Related to Fibrogenesis and Fibrolysis in Nonalcoholic Steatohepatitis. Sci Rep. 2018;8(1):12414. Epub 2018/08/19. doi: 10.1038/s41598-018-30457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request due to privacy/ethical restrictions. SomaScan data are available through a data use agreement with Novartis Institutes for BioMedical Research (lori.jennings@novartis.com).


