Abstract
Lipid droplets (LDs) are the main organelles for lipid storage, and their surfaces contain unique proteins with diverse functions, including those that facilitate the deposition and mobilization of LD lipids. Among organelles, LDs have an unusual structure with an organic, hydrophobic oil phase covered by a phospholipid monolayer. The unique properties of LD monolayer surfaces require proteins to localize to LDs by distinct mechanisms. Here we review the two pathways known to mediate direct LD protein localization: the CYTOLD pathway mediates protein targeting from the cytosol to LDs, and the ERTOLD pathway functions in protein targeting from the endoplasmic reticulum to LDs. We describe the emerging principles for each targeting pathway in animal cells and highlight open questions in the field.
Keywords: Lipid droplets, protein targeting, amphipathic helix, hairpin motif, phospholipid monolayer, bilayer membranes
Lipid Droplets as dynamic cellular organelles
Lipid droplets (LDs) are ubiquitous organelles, found in eukaryotic and some prokaryotic cells, that are hubs of cellular lipid metabolism and act primarily to store neutral lipids. They consist of a core of triglycerides (TGs) or sterol esters (SEs) that is bounded by a monolayer of phospholipids. Through the action of lipases acting at the LD surface, neutral lipid components are released to generate membrane lipids or metabolic fuel. Other functions of LDs include storing proteins, preventing the toxic accumulation of lipids, providing reservoirs of lipid-soluble drugs and vitamins, and serving as cellular hubs for the replication of some viruses [1–5]. These broad functions of LDs underlie the increasing identification of LD functional roles in different biological processes, including for instance energy metabolism and roles in innate immunity [6].
LD proteins number in the tens to hundreds and vary among species and cell types (see Box 1). Many are important enzymes involved in cellular lipid metabolism (see below). Increasingly, variations in LD proteins have been recognized as risk factors for metabolic and other human diseases. For example, variations in PNPLA3 or HSB17B13, which encode metabolic enzymes that localize to LDs, modify the risk for human non-alcoholic fatty liver disease (NAFLD) (see Glossary) and/or non-alcoholic steatohepatitis (NASH) [7, 8].
TEXT BOX 1: Methods and criteria to identify lipid droplet (LD) proteins.
LD proteomes have been characterized by a variety of techniques in many different cell types and in different species [14, 46, 47, 97, 98]. However, the interpretation of many of these studies is complicated by the copurification of contaminants as LD proteins. This can be due to the unspecific binding of non-LD proteins to the LD oil phase during cell fractionation and to the close association between LDs and other organelles, such as the ER and mitochondria. Additionally, microscopy studies that claim the LD localization of proteins based on their overexpression should be approached with caution. This is because overexpression can saturate a protein’s normal localization sites within the cell, leading to their aberrant binding to the LD oil-water interface.
Ideally, the classification of a protein as a bona fide LD protein should be supported by multiple lines of evidence. These might include assessing the LD localization of the endogenous protein in cells by fluorescence microscopy, identifying it in LD cellular fractions by biochemistry methods, and when possible, performing super-resolution or electron microscopy to show that the protein directly binds to the LD surface [15, 46, 47, 60, 99, 100].
How proteins recognize and localize to LD surfaces has emerged as a fascinating and important aspect of cell biology with biomedical relevance. Here, focusing on animal cells, we review some of the major LD proteins and the principles of how they target to this organelle.
LD proteins and their functions
The discovery of specific LD proteins helped to define LDs as cellular organelles. In particular, the identification of perilipins as LD proteins suggested these are regulated organelles harboring specific proteins [9]. Perilipins regulate lipolysis in response to extracellular signals [10]. Analogously, in plants, oleosins decorate LDs, particularly in seeds, where they likely perform similar structural and regulatory functions [11, 12].
The majority of bona fide LD proteins are involved in lipid metabolism. Examples of such proteins include enzymes that act in neutral lipid synthesis, such as fatty acyl-CoA synthetases ACSL3 and ACSL5 [13, 14], glycerol-3-phosphate acyltransferase 4 (GPAT4) [15], or the yeast squalene monooxygenase Erg1, which is involved in sterol synthesis [16]. In Drosophila cells, targeting neutral lipid synthesis enzymes (e.g., GPAT4) to the LD surface enables LD-localized TG synthesis, which is catalyzed in an endoplasmic reticulum (ER)-LD compartment by diacylglycerol acyltransferase 2 (DGAT2), and thus the expansion of initial LDs [15, 17]. Although little is known about the relative activities of many lipid metabolic enzymes at the LD surface compared with the ER, in yeast, the DGAT2 orthologue Dga1p exhibits different activities in the corresponding subcellular fractions [18].
Enzymes of phospholipid synthesis and transport also localize to LDs. These include the rate-limiting enzyme controlling phosphatidylcholine (PC) synthesis, CTP:phosphocholine cytidylyltransferase (CCT; [19]), and enzymes of the Lands cycle that remodel phospholipid acyl chains (LPCATs; [20]), which are found on the LDs of some cell types [20, 21]. In the case of CCT, LD binding occurs in response to PC deficiency on the growing LD surface and activates the enzyme to increase PC synthesis to meet demands [19, 22, 23]. In humans, mutations in the LD- and membrane-binding motif of the CCT isoform encoded by PCYT1A are associated with lipodystrophy [24, 25]. Several lipid transfer proteins, such as PC transfer protein [26] and ORP5 [27], have also been localized to LDs, where they might be involved in the transfer of specific phospholipids or sterols to or from the LD monolayer.
LD proteins also include lipases that degrade neutral lipids, including the major TG lipase adipose TG lipase (ATGL/PNPLA2; [28]), its activator comparative gene identification-58 (CGI-58), and the major diacylglycerol (DAG) lipase hormone-sensitive lipase (HSL; [29]). These proteins act in concert to degrade TG to fatty acids (FAs) and glycerol, a process best understood in adipocytes (reviewed in [30, 31]). Deficiency of ATGL or CGI-58 results in human neutral lipid storage disease [32] characterized by the accumulation of fat in various tissues of the body.
Another putative lipase, PNPLA3, is a LD protein with direct connections to human metabolic disease. A common variant of PNPLA3 (I148M) is a major risk factor for the development of NAFLD/NASH [7, 33]. PNPLA3 has been suggested to transfer fatty acyl chains between TGs and phospholipids [34]. The PNPLA3 (I148M) variant appears to be catalytically compromised and accumulates on the surface of LDs [35], likely due to a reduction in ubiquitylation and protein degradation [36]. A current model posits that the accumulation of PNPLA3 (I148M) on LD surfaces results in a defect in TG mobilization from LDs, perhaps via diminished ATGL activity by sequestering its activator CGI-58, leading to excessive LD accumulation and ultimately NAFLD/NASH [37]. Reducing the amount of PNPLA3 (I148M) on the surface of LDs might thus provide a therapeutic avenue to treat these metabolic conditions [38].
Similarly, genetic studies revealed that loss-of-function alleles for HSD17B13, encoding the LD protein 17-β-hydroxysteroid-dehydrogenase 13 (HSD17B13), is associated with protection from NASH in humans [8]. Although the enzymatic function of HSD17B13 is poorly defined, its N-terminal domain has similarity to perilipin proteins and is required for its LD targeting and activity [39]. How HSD17B13 promotes NASH is unknown. The protective allele of HSD17B13 is associated with changes in PC levels in liver [40].
Unexpectedly, some transcription factors were found to be LD proteins [41, 42]. These include the transcription factor Max-like protein X (MLX) and MLX family members (MLXIP, MLXIPL/ChREBP; [43]), which are master regulators of glucose and lipid metabolism [44]. The binding of these transcription factors to LDs provides a potential additional layer of regulation of their activities in gene expression, where studies in cells have shown that LD binding acts in a feedback mechanism to downregulate their transcriptional activity [41]. A current model suggests that glucose metabolites activate these transcription factors, upregulating de novo lipogenesis and lipid storage in LDs, among other responses. This triggers binding of these transcription factors to LDs in the cytosol, thereby attenuating the transcriptional response. Conversely, the LD protein perilipin 5 appears to promote expression of PGC-1α/SIRT target genes, including mitochondrial proteins [42], further suggesting a relationship between LD metabolism and metabolic gene expression.
LDs can also act as protein storage depots. For example, LDs temporarily sequester a subset of histones in Drosophila embryos (e.g., H2A and H2B) through a LD adaptor protein, Jabba [1, 45]. This allows for the storage and proper balance of the massive amounts of histones needed during rapid embryonic nuclear divisions, and it prevents excess histones from interfering with normal embryonic development [1].
Also found on LDs are the enzymes of dolichol synthesis (e.g., Rer2; [46]) and other enzymes involved in the proximal steps of the pathway generating glycans on dolichol for N-linked glycosylation in the ER (e.g., ALG14; [46, 47]). The significance and function of the LD localization of these proteins are unknown, although dolichols stored as dolichol esters are likely found in LDs [48]. Similarly, UBX domain-containing protein 8 (UBXD8), a protein of the ubiquitin-proteasome system, is consistently identified as a LD protein [49, 50]. In the ER, UBXD8 functions in ER-associated protein degradation (ERAD) by recruiting the large-protein-extraction ATPase VCP/p97 [51]. It also has been reported to be a sensor of unsaturated FAs, regulating their incorporation into neutral lipids [52]. The function of LD-localized UBXD8 is unclear.
A number of other proteins localize to the interface between LDs and other organelles: Mdm1 localizes to LD-vacuole contact sites in yeast [53], and Snazarus localizes to LDs near the plasma membrane in Drosophila larvae lipid deposits [54]. Little is known about the LD targeting mechanisms of these proteins, and they are reviewed elsewhere [55].
Some proteins of important intracellular pathogens also localize to LDs. For instance, flaviviruses, such as hepatitis C, Dengue, and Zika virus, use the surfaces of LDs as part of their replication site in cells [5]. The hepatitis C core protein localizes to LDs, and its overexpression is sufficient to cause steatosis in mice [56]. Notably, viperin, an innate immune response protein, also localizes to LDs [57], suggesting that these organelles serve as battlegrounds during viral infection. Further, a number of proteins involved in innate immunity in response to bacterial infections have been found in LD fractions of cells in which LDs are induced by lipopolysaccharide [6]. Thus, an important function of LDs in immune cells is being increasingly recognized and is reviewed elsewhere [58].
Principles of protein targeting to LDs
The formation of LDs from the ER [59, 60] results in the generation of an organic phase in the cytosol, effectively converting cells into emulsions. The LD oil phase is covered by a monolayer of phospholipids that makes up the interface with the aqueous cytosol and acts as a surfactant to lower surface tension (Figure 1, left). As a consequence, LD proteins must recognize and localize to an interface that has unusual biophysical properties among cellular organelles. How proteins interact with this phase boundary is governed by the principles of emulsion physics [61].
Figure 1. Unique features of the lipid droplet (LD) surface influence protein targeting.
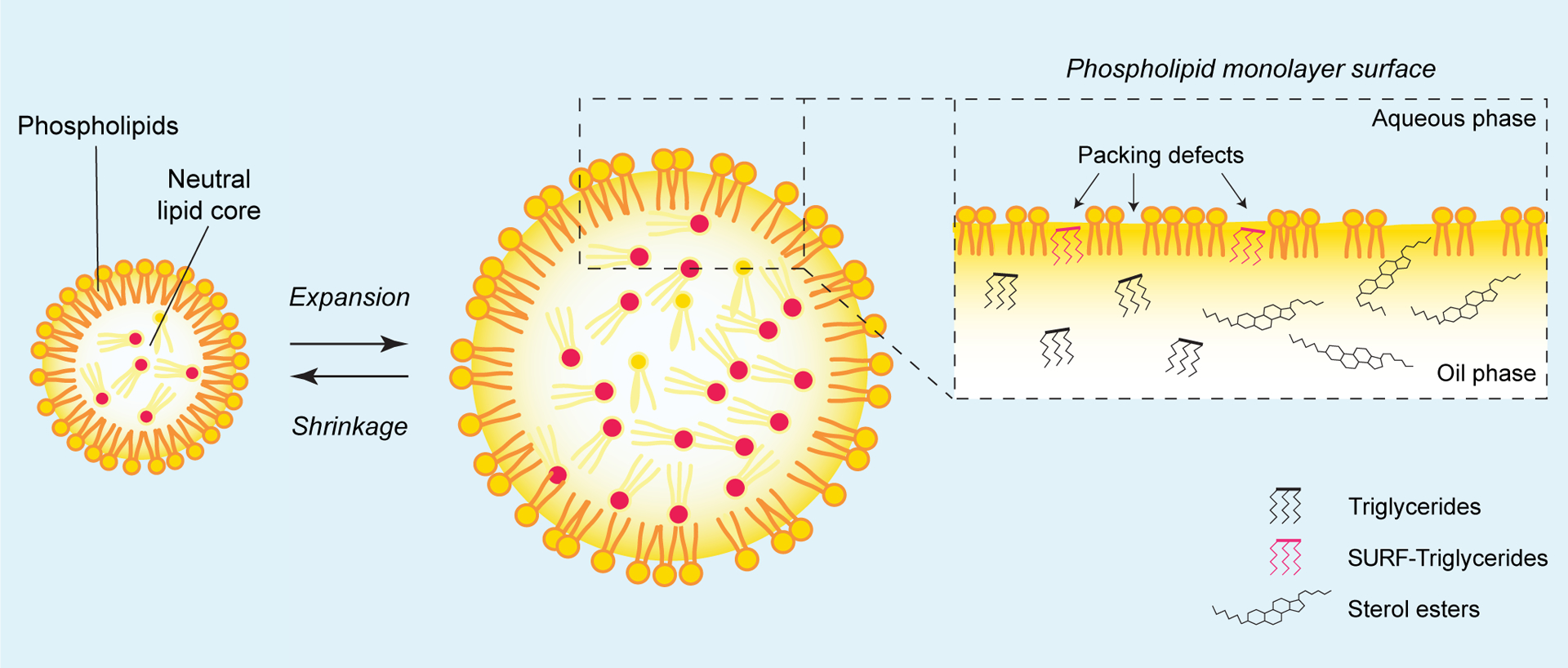
Left: LDs possess a unique structure consisting of a core of neutral lipids [e.g., triglycerides (TGs) or sterol esters] surrounded by a monolayer of phospholipids. Middle: Due to their dynamic character, LDs undergo constant expansion and shrinkage, which can affect the phospholipid packing density of the LD surface. Inset: Properties, such as phospholipid packing defects, limited surface area, and the neutral lipids that underlie the phospholipid monolayer are all features that affect the mechanisms of LD protein targeting. For instance, LDs can be rich in TGs or sterol esters, which can influence their surface properties and the proteins that target to LDs. In the case of TG-rich LDs, some TG molecules can be located at the LD surface, at sites where phospholipid packing defects exist, and align themselves with the phospholipid monolayer plane. These are known as surface-oriented TG molecules (SURF-TG, magenta). SURF-TG behave as structural components of the LD monolayer, while also altering its surface tension to promote the LD targeting of specific proteins.
Studies of protein targeting to LDs have yielded a number of important insights. First, LD protein targeting entails mechanisms that differ markedly from those for targeting proteins to bilayer membrane-bound organelles, such as the ER, mitochondria, or peroxisomes. For example, there appears to be no dedicated protein machinery on LDs that mediates the direct insertion of proteins into the LD surface, nor have specific lipids (such as different phosphoinositides employed by other organelles) been identified as markers for directing proteins to the LD surface.
Second, although the phospholipid monolayer covering LDs has a composition similar to the ER membrane [62, 63], its biophysical properties are unusual (Figure 1, inset). Similar to bilayer membranes, the monolayer surface includes phospholipid packing defects, which are sites where the phospholipid tails are dynamically exposed to water in the cytosol. Molecular modeling suggests that packing defects are more frequent, larger, and more persistent on the LD surface than in bilayer membranes [64, 65]. Such modelling studies also indicate that LD packing defects are chemically distinct: the TG acyl chains are intercalated into the phospholipid tail region, and in some instances, entire TG molecules are integrated so that their glycerol backbones align with the phospholipids and act as monolayer components [65]. These surface-oriented TG molecules (SURF-TG) (Figure 1, inset) are unique to LD packing defects and slightly increase LD surface tension relative to bilayer membranes. Additionally, the types of exposed neutral lipids may vary (i.e., sterol esters or TGs) and influence the surface properties of LDs, resulting in different proteins binding to specific types of LDs [41, 66].
Third, the LD surface changes dynamically during the LD lifecycle in ways that may affect protein targeting (Figure 1, left and middle). For example, during LD expansion, the LD surface stretches and the integration of new SURF-TG molecules into the monolayer decreases the induced surface tension, enabling new proteins to target altered surface properties [67] (see details below). Similarly, when LDs shrink, weakly bound proteins become displaced due to molecular crowding [68] and competition for the fewer available packing defects. This provides a mechanism to modulate the protein composition of LDs, based on the biophysical properties of the LD surface and the protein binding to the interface.
Given these unique features of the LD monolayer surface, how do proteins detect and localize to LDs? At least two major and distinct protein targeting pathways appear to operate in cells. In one, which we term CYTOLD (for “cytosol to LD”) targeting, proteins are initially translated in the cytosol and subsequently bind to the LD surface. In the second, ERTOLD (for “ER to LD”) targeting, proteins are initially inserted into the ER membrane and then relocalize from there to the LD surface.
CYTOLD targeting pathway
The CYTOLD targeting pathway (Figure 2) begins with the translation of proteins in the cytosol. Examples of proteins that use CYTOLD targeting are listed in Table 1. Many CYTOLD-targeted proteins contain amino acid sequences with the potential to form amphipathic helices. Some sequences are folded regions of a protein, but others are unstructured in the cytosol until they associate with a bilayer membrane or LD monolayer, which facilitates their folding into amphipathic helices with hydrophobic and polar residues on opposing faces (Figure 2, left and middle). Perilipins, for instance, are CYTOLD-targeted proteins that contain many of these sequences, with each corresponding to an 11-amino acid repeat [69]. Perilipin 3 (PLIN3) appears to bind LDs early after their formation [60, 70]. These amphipathic sequences might be shielded from aggregation in the cytosol by binding chaperones. The cytosolic pool of perilipin 1 (PLIN1) is rapidly degraded by the ubiquitin-proteasome system if the protein is not bound to LDs. Thus, the LD pool of PLIN1 seems to be protected from degradation [71].
Figure 2. CYTOLD pathway for lipid droplet (LD) protein targeting.
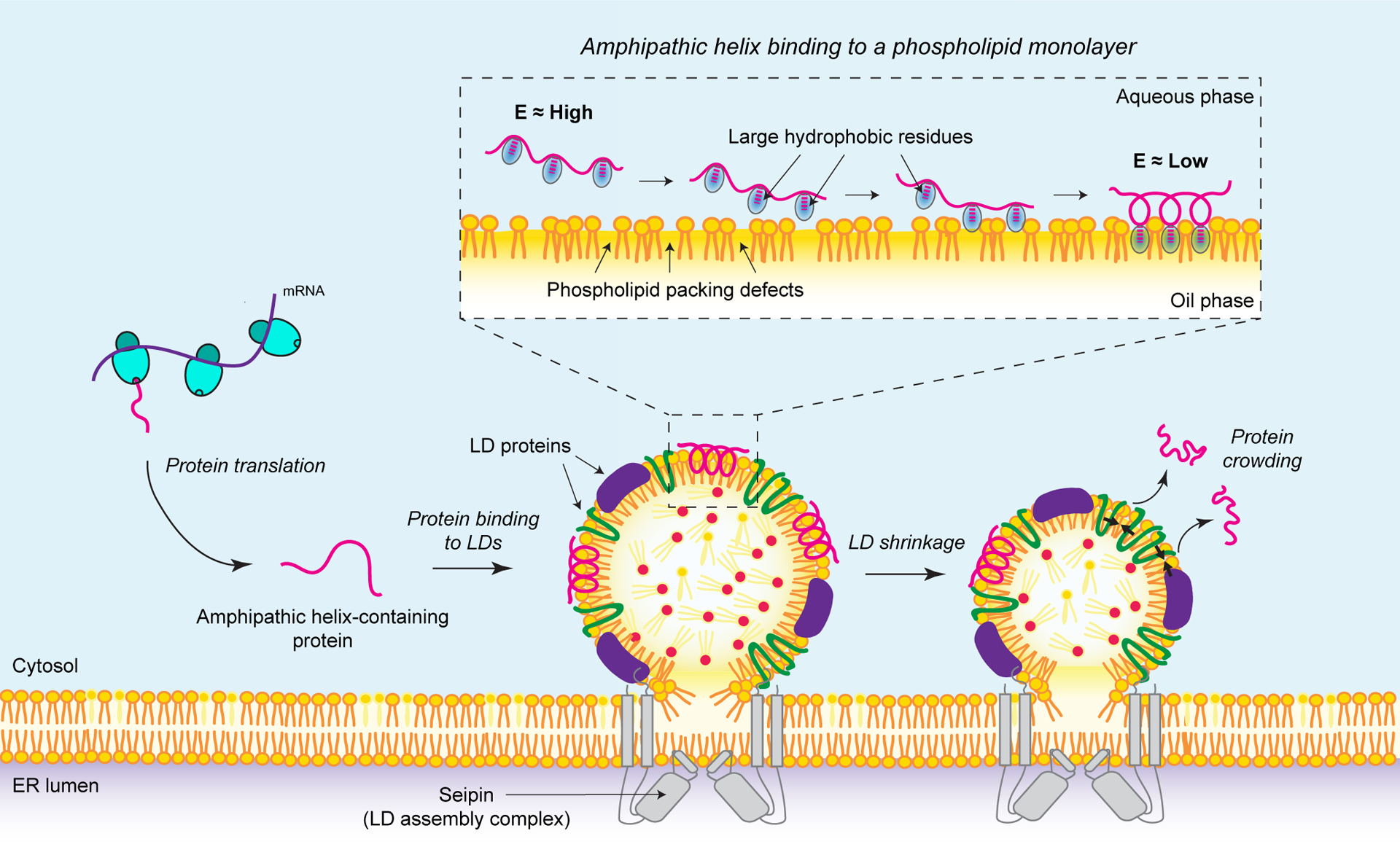
Proteins that access and bind the surface of LDs from the cytosol via the “cytosol to LD” (CYTOLD) pathway are soluble and often characterized by containing one or more amphipathic helix (AH) motifs for LD binding. Left: A model for the CYTOLD pathway starts with the synthesis of proteins in the cytosol. Middle: Once fully released from the protein synthesis machinery, AH-containing proteins are thought to be unfolded in the cytosol and interact with packing defects at the surface of LDs (which form within the endoplasmic reticulum (ER) membrane at assembly complexes containing seipin). Inset: The AH motifs of these proteins mediate their binding to the LD monolayer surface through a thermodynamic mechanism: i) first, the unfolded AH motif is in a high free energy (E) state in the cytosol; ii) large hydrophobic residues in its sequence bind to phospholipid packing defects on the LD surface, anchoring the protein to it; iii) as more large hydrophobic residues bind to packing defects, the AH motif folds on the surface of LDs. This renders the protein in a lower free energy state and stabilizes its binding to the surface of LDs. Right: As the metabolic needs of the cell change and neutral lipids are mobilized, LDs may shrink in size leading to an increase in protein density at their surfaces. As a consequence, weakly bound proteins, such as AH-containing proteins, may be displaced from the LD surface due to protein crowding, releasing them back into the cytosol.
Table 1.
Examples of CYTOLD-targeted proteins
| Protein name | Function | References | Cell types (examples) |
|---|---|---|---|
| Perilipins (1–5) | Coat the surface of LDs early during their biogenesis, preventing their breakdown by lipases and regulating their metabolism | [9, 42, 69, 73] | 3T3-L1 adipocytes, mouse Leydig tumour cells (MLTC-1), HeLa, COS-7, Drosophila S2, S. cerevisiae (yeast) |
| Comparative gene identification-58 (CGI-58/AB5HD) | Co-activator of the lipase ATGL/PNPLA2 | [101, 102] | 3T3-L1 adipocytes |
| CTP:phosphocholine cytidylyltransferase (CCT) | Catalyzes the key rate-limiting step in the CDP-choline pathway for phosphatidylcholine biosynthesis | [19] | Raw267.2 and bone marrow-derived macrophages (BMDMs), Drosophila S2 |
| Max-like protein X (MLX) and members of the MLX family of transcription factors | Activate the transcription of glycolytic genes in a glucose-responsive manner | [41] | THP-1 macrophages, mouse primary hepatocytes, HEPG2, U2OS, COS-7, SUM159 |
Much of our current understanding of the CYTOLD targeting pathway comes from the analysis of model peptides (e.g., peptides derived from perilipins or CCT) for which biochemical, biophysical, and molecular dynamics simulation studies have been performed [19, 64–66, 69, 72, 73]. Experimental studies of CCT indicated that multiple large hydrophobic residues (e.g., leucines, phenylalanines, or tryptophans) were required to bind LDs; complementary simulations demonstrated initial association of these residues with phospholipid packing defects [64] (Figure 2, inset). Subsequent simulations showed that a single tryptophan is critical for the initial association of the peptide with the glycerol moiety in SURF-TG packing defects, and that three additional phenylalanines require neighboring defects to be present in expanding LDs [67]. Large tryptophan residues also appear to be required for the binding of CGI-58 to LDs [74]. The resulting model of large hydrophobic residues binding to LD surface packing defects predicts that parameters that affect the abundance of packing defects on LDs (e.g., phospholipid composition, protein abundance, or the underlying oil expansion or shrinkage) impact the LD binding of amphipathic helical proteins [64, 65, 67].
Once LD binding is initiated, proteins that are unstructured in the cytosol, such as CCT, fold into an α-helix [75, 76], stabilized by hydrophobic interactions with the acyl chains of phospholipids and TGs at the LD surface and polar/charged interactions with phospholipid head groups and water. Thus, CYTOLD targeting is often energetically coupled to protein folding at the surface of LDs, which leads to very slow unbinding rates for strongly associated proteins, such as perilipins.
The principles of CYTOLD targeting may play a large role in governing the protein composition of LDs. Studies have shown that the amphipathic helical regions of many cellular proteins can bind to LD monolayer surfaces [64, 68], thus prompting the question of how specificity is maintained for genuine LD proteins. One feature that may contribute to determining specificity is the competition between proteins for available LD surface. For instance, increasing protein density at the LD surface, which occurs during lipolysis, leads to macromolecular crowding and the displacement of more weakly bound proteins [68] (Figure 2, right). Similarly, competition for packing defects may limit LD adsorption and binding of bona fide LD proteins and it may be a key factor governing LD protein composition.
Another mechanism that potentially regulates the dynamic changes in LD protein composition is the post-translational modification of LD proteins. CCT is known to undergo phosphorylation [77], which may affect its localization. Further, high fat diet-induced hepatic lipid accumulation in mice causes changes in the phosphoproteome that include phosphorylation events correlating with the localization of proteins to LDs [78].
ERTOLD targeting pathway
Examples of proteins that target LDs via the ERTOLD pathway (Figure 3) are shown in Table 2. ERTOLD-targeted proteins commonly contain hydrophobic, membrane-embedded sequences, often disrupted in the middle by prolines, which suggests they adopt a hairpin conformation in the ER membrane [79]. Proteins that completely span the ER bilayer and have luminal loops or domains will not target to LDs [15], as embedding of the luminal hydrophilic segments in neutral lipids would be energetically unfavorable.
Figure 3. ERTOLD pathway for lipid droplet (LD) protein targeting.
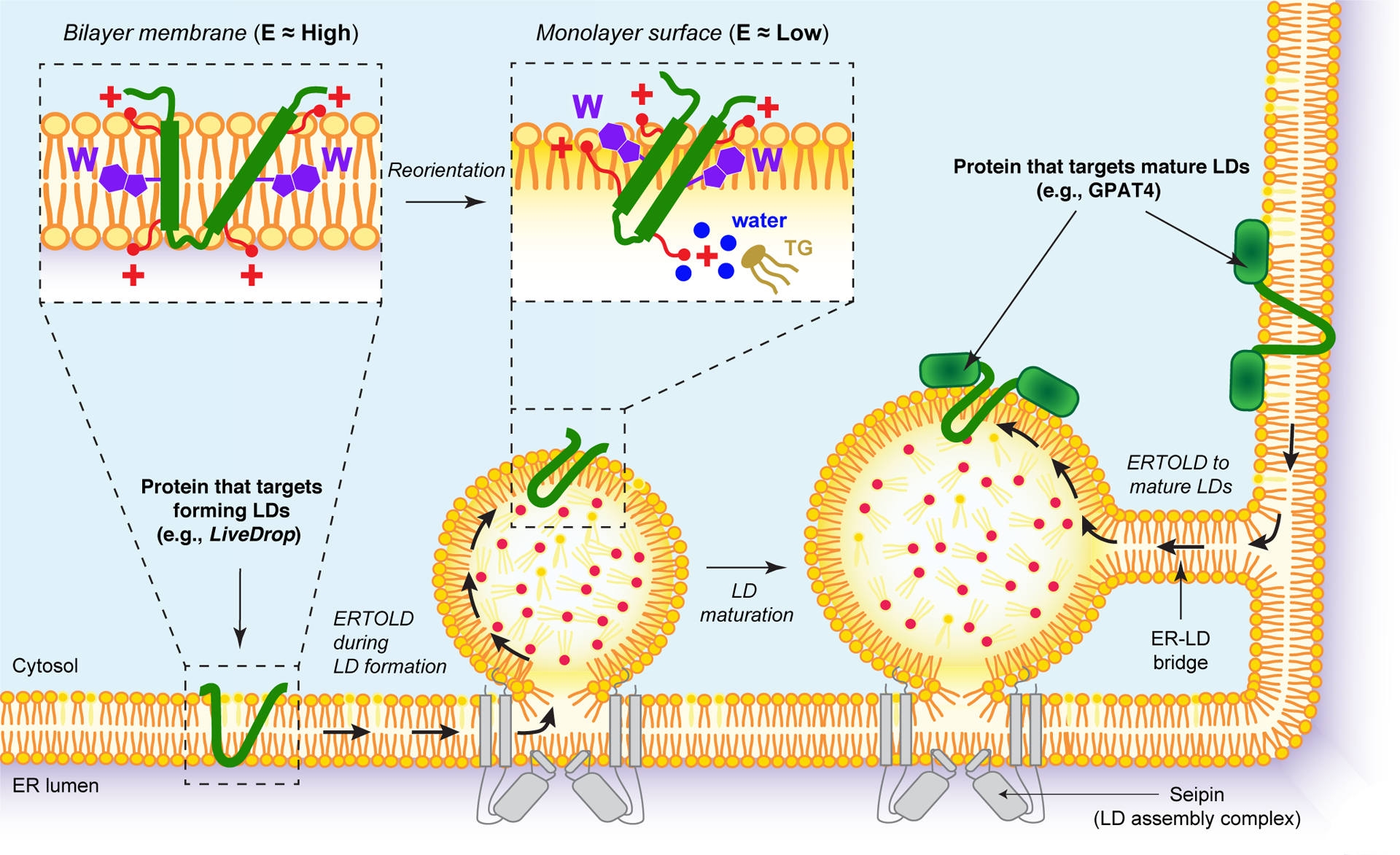
Proteins that access and bind the surface of LDs from the endoplasmic reticulum (ER) via the “ER to LD” (ERTOLD) pathway usually contain hydrophobic, membrane-embedded motifs that are predicted to acquire a hairpin conformation in bilayer membranes. Left: A model for the ERTOLD pathway starts with proteins being synthesized and inserted into the ER membrane. Middle: During LD formation at ER sites defined by a LD assembly complex (which includes seipin), some hairpin-containing proteins, such as LiveDrop, relocalize to nascent LDs. Insets: Sequence features of these hairpin motifs mediate their LD targeting and accumulation: i) in the ER bilayer membrane, specific sequence features of the hairpin motif (i.e., positively charged residues, red, “+”) anchor the protein to the cytosolic and luminal leaflets of the membrane; ii) this positions large hydrophobic residues, such as tryptophans (purple, “W”), in an energetically unfavorable position within the middle of the hydrophobic plane of the ER membrane; iii) upon LD localization, some positively charged residues are released and reorient towards the monolayer interface (simulations suggest that others may remain embedded below the LD surface, interacting with water molecules that access the LD core); iv) this also allows the reorientation of tryptophans towards the phospholipid packing defects, lowering the free energy (E) state of the protein and further stabilizing the LD surface. Right: At later stages, specific hairpin-containing proteins, such as GPAT4, can be recruited to mature LDs from the ER, presumably via membrane bridges, to mediate LD-localized triglycerides (TG) synthesis.
Table 2.
Examples of ERTOLD-targeted proteins
| Protein name | Function | References | Cell types (examples) |
|---|---|---|---|
| Glycerol-3-phosphate acyltransferase 4 (GPAT4) | Catalyzes the conversion of glycerol-3-phosphate and long-chain acyl-CoA to lysophosphatidic acid (LPA), which is the first and rate-limiting step of the de novo TG synthesis pathway | [15] | Drosophila S2, COS-7 |
| Acyl-CoA synthetase long Chain family member 3 (ACSL3) and 5 (ACSL5) | Catalyze the esterification of FAs with CoA in order to chemically “activate” them for further TG synthesis | [13, 14] | COS-7, HepG2, mouse enterocytes |
| Adipose TG lipase (ATGL) or PNPLA2 | Catalyzes the initial step of TG hydrolysis in adipocyte and non-adipocyte LDs | [28] | HeLa |
| Oleosins | Coat the surface of oil bodies (i.e., LDs from plant cells) to stabilize them, especially in seeds that undergo desiccation | [11] | Maize seeds |
| UDP-N-acetylglucosaminyltransferase subunit (ALG14) | Catalyzes the second step of the dolichol-linked oligosaccharide pathway | [46, 47, 103] | Drosophila S2, HeLa, S. cerevisiae (yeast) |
| Acyl-CoA:lysophosphatidylcholine acyltransferase (LPCAT) | Catalyzes the conversion of lysophosphatidylcholine (LPC) into phosphatidylcholine (PC), as part of the phospholipid remodeling pathways | [20, 47, 104] | Drosophila S2, human A431 |
| Thioredoxin reductase-like selenoprotein or SelT-like protein | Probably has thioredoxin reductase-like oxidoreductase activity | [47] | Drosophila S2 |
| 1-acylglycerol-3-phosphate O-acyltransferase 3 (AGPAT3) | Catalyzes the conversion of LPA into phosphatidic acid (PA), as part of the de novo biosynthesis pathways of diacyl phospholipids and TGs | [47] | Drosophila S2 |
| Multiple C2 and transmembrane domain-containing protein 2 (MCTP2) | Defines ER domains that serve as formation sites for preperoxisomal vesicles and LDs | [47, 105] | Drosophila S2, COS-7, S. cerevisiae (yeast) |
| UBX domain-containing protein 8 (UBXD8) | Recruits the ATPase p97/VCP to facilitate the extraction or dislocation of ERAD substrates from the ER membrane | [49] | Huh7, HeLa |
| Spastin | When localized to the surface of LDs, it tethers LDs to peroxisomes and supports the traffic of FAs between both organelles | [100] | HeLa |
| Fatty acid transport protein (FATP) | Mediates the vectorial acylation of FAs, which drives their movement across membranes and results in the production of acyl-CoA | [68] | Drosophila S2 |
| Lipid droplet-associated hydrolase (LDAH) | Serine hydrolase | [106] | Drosophila S2, HeLa |
| Associated with LD protein 1 (ALDI) | Putative methyltransferase | [107] | COS-7, rat hepatocytes |
| Methyltransferase-like protein 7A (METTL7A) or AAM-B | Putative methyltransferase | [108] | HeLa, COS-7, S. cerevisiae (yeast) |
| Ancient ubiquitous protein 1 (AUP1) | Involved in ERAD and, when localized to LDs, it binds the E2 conjugase Ube2g2 which links LDs to the ubiquitination machinery | [109, 110] | COS-7, human Huh7, human A431, dog MDCK |
| Caveolin-1 (Cav1) and caveolin-2 (Cav2) | Associate with each other to form a complex that targets membrane lipids and drives the formation of caveolae | [111] | Human A431, Fisher rat thyroid (FRT), baby hamster kidney (BHK) |
| NAD(P)H steroid dehydrogenase-like (NSDHL) | Sequentially removes two C-4 methyl groups from the cholesterol precursor lanosterol during cholesterol biosynthesis | [112, 113] | HeLa, COS-7, Chinese hamster ovary (CHO) |
| Lipid droplet assembly factor 1 (LDAF1) or promethin | Interacts with seipin in the ER to form the LD assembly complex (LDAC), which defines the sites of LD formation | [60, 114] | SUM159 breast cancer cells, MCF7 breast cancer cells |
The biogenesis of ERTOLD-targeted proteins begins with their translation and insertion into the ER membrane. The initial ER insertion step may occur through the canonical translocon-mediated pathway. UBXD8 and possibly other ER proteins use an alternative, post-translational pathway requiring the cytosolic protein Pex19 and the ER protein Pex3, which are best known for their roles in peroxisome biogenesis [80].
Once in the ER, proteins following the ERTOLD pathway appear to target the LD surface by moving from the ER bilayer membrane to the LD monolayer surface via ER-LD membrane connections [15, 81] (Figure 3, left). Early studies with fluorescence recovery after photobleaching in the yeast Saccharomyces cerevisiae suggested that yeast LDs are always physically connected to the ER, allowing proteins to travel to the LD surface [82]. In yeast, fly, and mammalian cells, physical connections between LDs and the ER appear to be maintained at sites of LD formation via LD assembly complexes (LDAC) that contain seipin [60, 81, 83–87]. In Drosophila cells, a few ER-localized proteins (e.g., the membrane-embedded region of GPAT4 (known as LiveDrop; [79]), LDAF-1 [60], or ACSL3 [13]) have been shown to target LDs during their formation. Many other ERTOLD-targeted proteins, including full-length GPAT4, do not localize to LDs during their biogenesis but instead target mature LDs well after their formation [15] (Figure 3, right). What determines which proteins localize to LDs during their formation or at later stages is unclear.
The ERTOLD targeting of proteins to mature, established LDs appears to happen via ER-LD membrane bridges that form after LD biogenesis. In Drosophila cells, where the LD targeting of GPAT4 has been well characterized, ER-LD membrane bridges can be visualized by electron tomography [15]. Fluorescence recovery after photobleaching experiments suggest that these membrane bridges are used by GPAT4 to rapidly relocalize from the ER to the LD surface [15].
How ER-LD membrane bridges are established and maintained remains a mystery. Genome-wide screens searching for LD phenotypes identified members of the Arf1/COPI membrane trafficking complexes as required for normal LD morphology and the LD targeting of some ER proteins [88, 89]. The Arf1/COPI machinery is best known for its function in the formation of coated vesicles that mediate retrograde vesicular trafficking in the secretory pathway [90]. How Arf1/COPI participates mechanistically in ER-LD bridge formation in cells is unknown, but it might be by modulating the properties of the LD surface. Specifically, in vitro studies have shown that Arf1/COPI proteins can mediate the budding of nano-LDs from LD surfaces [91], thereby removing phospholipids and increasing the surface tension of LDs, possibly rendering them in a fusion-competent state that allows them to connect to the ER [92].
Many ERTOLD-targeted proteins not only localize to LDs but also accumulate there. This accumulation requires an energy difference that prevents proteins from equilibrating between the ER and LDs. Otherwise, a high proportion of most proteins would stay in the massively more abundant ER. How then do some ERTOLD-targeted proteins accumulate at LDs? Although this question has not been examined in the context of full-length proteins, recent studies of short, membrane-embedded hairpin motifs, performed in cells and using molecular simulations, have shed light on the mechanism of ERTOLD targeting and LD accumulation (Figure 3, insets). Studies of the membrane-embedded domains of GPAT4 and ALG14, which likely adopt a hairpin conformation in the ER membrane, suggest that a specific combination and distribution of sequence features are important for their accumulation at LD surfaces. Each of these hairpin motifs possess positively charged residues within their hydrophobic segments, which appear to anchor them to the luminal leaflet of the ER bilayer membrane. This forces some residues into regions of the bilayer membrane that are energetically less favorable. For example, tryptophans, which are most stable in the phospholipid glycerol region where they satisfy both hydrophobic interactions and hydrogen bonding, are positioned in the middle of the phospholipid tails region [79]. Upon relocalization to the LD surface, this strain appears to be relieved as tryptophans and positively charged residues reorient towards the interface and surface of the LD phospholipid monolayer, resulting in an energetically more favorable conformation of the hairpin motifs (Figure 3, insets). Thus, the current model posits that the free energy difference for a hairpin motif to be in the ER and at a LD surface provides the driving force for its LD accumulation [79].
This thermodynamic model does not rule out additional factors that might contribute to the selective accumulation of ER proteins on LDs. For instance, differential oligomerization states in the different compartments may contribute. Also, some ERTOLD-targeted proteins may be degraded faster in the ER than on the LD surface, potentially contributing to the apparent accumulation of such proteins on LDs [93]. However, this mechanism does not appear to be sufficient to explain the LD accumulation of the LiveDrop sequence from GPAT4 [79].
Are there other pathways for protein localization to LDs?
Whether pathways other than CYTOLD and ERTOLD exist for LD protein targeting remains to be determined. For instance, one could imagine that lipid anchors, such as palmitate, myristate, or prenyl groups, found covalently attached to some proteins, could mediate targeting to LDs. Indeed, proteomic studies routinely identify a large number of lipidated Rab proteins on LDs, and some of them are associated with LD phenotypes when depleted from specific cell types [94–96]. However, microscopy evidence showing endogenous Rab proteins localizing specifically to LDs remains sparse. Therefore, at least some Rab proteins seem to partition to LDs nonspecifically during the biochemical purification of LDs.
Concluding remarks
Since the discovery of perilipins as specific LD proteins, new LD proteins have been identified, and the molecular understanding of how these proteins target to LDs via the CYTOLD and ERTOLD pathways has grown enormously. These studies have yielded fascinating cell biology insights. Perhaps more importantly, the associations between many LD proteins (e.g., ATGL, PNPLA3, HSD17B13, and CCT) and human metabolic diseases highlight the need to better understand the principles of LD protein targeting. Yet many important and compelling aspects of LD protein targeting remain mysterious (see Outstanding Questions). We anticipate that additional studies employing new proteomic, microscopy, structural biology, and biophysical approaches will rapidly lead to further insights into the mechanisms of LD protein targeting and its functional consequences.
Outstanding questions.
What is the composition of the lipid droplet (LD) proteome in different cell types (e.g., brown adipocytes, cardiac myocytes, neutrophils)?
What are the functional consequences of having different populations of LDs in the same cell?
How does the LD proteome change with, and influence, the LD lifecycle under different cellular conditions?
Are there other LD targeting pathways besides CYTOLD and ERTOLD?
What factors control the specificity of the LD protein composition?
Will the emergence of sequence-specific features for proteins that target to LDs enable computational predictions of LD localization?
Are chaperones involved in the initial steps of the CYTOLD pathway?
How are the ER-LD membrane bridges used for late ERTOLD targeting established? What factors control this process?
How do the core neutral lipids affect the targeting of proteins to LDs?
How does the phospholipid composition of the LD monolayer surface affects LD protein targeting?
How are proteins selectively removed from LDs? Can ERTOLD-targeted proteins migrate back to the ER for ERAD removal?
What machinery mediates selective autophagy (lipophagy) of LDs?
Are LDs important for detoxifying the ER from specific proteins or lipids?
How does the LD targeting of specific proteins facilitate the replication of some viruses?
How does aberrant LD protein targeting leads to metabolic diseases?
Highlights.
Lipid droplet (LD) organelles contain specific proteins, including metabolic enzymes, that differ among cell types.
Proteins targeting the monolayer surface of lipid droplets do not follow the same principles as proteins targeting bilayer membranes.
The targeting of proteins to lipid droplet surfaces is governed by the biophysical features of their unique oil-water interface.
LD protein targeting occurs via two major pathways, CYTOLD and ERTOLD targeting.
Elucidating the principles driving these protein targeting pathways has implications for further understanding physiology and metabolic diseases.
Acknowledgments
We thank the many members of the Farese & Walther laboratory and our many colleagues who have contributed to our understanding of this topic, and Gary Howard for editorial assistance. Our work on LD cell biology is supported by NIH grants R01GM124348 (to R.V.F) and R01GM097194 (to T.C.W). T.C.W is an investigator of the Howard Hughes Medical Institute.
Glossary
- Dolichol
membrane lipid that corresponds to isoprenoid polymers, also known as polyisoprenoid alcohols. They are synthesized on the cytosolic side of the ER membrane and exist as a mixture of species of different chain lengths. During N-linked protein glycosylation, they function as lipid carriers of the glycan molecules destined to be conjugated to proteins.
- LD assembly complex (LDAC)
protein complex of ~600 kDa that forms in the ER membrane by oligomers of seipin and accessory proteins such as LDAF1 (Ldb16 and Ldo protein in yeast). LDACs define the sites of LD formation in the ER membrane and are thought to promote the nucleation and phase separation of TG lenses.
- Lipid anchor
lipids covalently attached to proteins and also integrated into membranes, allowing the stable interaction of proteins with membranes. There are three main types of lipid anchors: isoprene polymers (e.g., farnesyl, geranylgeranyl), FAs (e.g., saturated myristic acid and palmitic acid), and glycosylphosphatidylinositol (GPI).
- Lipodystrophy
conditions in which patients are unable to produce and maintain healthy fat tissue, resulting in metabolic complications (e.g., fatty liver, insulin resistance). They are caused by preventing the differentiation, survival, or functionality of adipocytes. Lipodystrophies can be classified based on their origin (i.e., congenital vs acquired) or on the extent the fat tissue is compromised (i.e., generalized vs. partial).
- Lipolysis
the catabolism or degradation of the TGs stored in LDs. It occurs in three sequential steps, mediated by the enzymes ATGL, HSL, and monoglyceride lipase (MGL), and produces FAs and glycerol.
- Non-alcoholic fatty liver disease (NAFLD)
liver condition characterized by the excessive accumulation of fat in the liver (i.e., hepatic steatosis) that is caused by metabolic disease, often with insulin resistance, rather than excessive alcohol intake, medication, or hereditary disorders. Specifically, it’s defined by the accumulation of large and heterogeneously sized LDs, consisting mainly of TGs, in more than 5% of hepatocytes.
- Non-alcoholic steatohepatitis (NASH)
condition that results from the progression of NAFLD to more serious conditions, such as hepatocyte inflammation and liver tissue fibrosis. Chronic liver fibrosis and damage can lead to cirrhosis and liver failure.
- Oleosins
proteins of around 15–26 kDa that are characteristic of LDs from plant cells, particularly, of seed LDs. They play a structural role as they coat the surface of LDs, providing stability and preventing them from coalescing with other LDs. Oleosins contain a long, hydrophobic hairpin motif (72 amino acids) which is involved in its targeting to LDs from the ER.
- Seipin
protein encoded by the Berardinelli-Seip congenital lipodystrophy type 2 (BSCL2) gene, which can harbor mutations causing lipodystrophy. It forms oligomers in the ER membrane and it is involved in defining the sites of LD formation (as part of the LDAC) and in stabilizing the ER-LD connections.
References
- 1.Li Z et al. (2014) Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol 24 (13), 1485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chitraju C et al. (2017) Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab 26 (2), 407–418.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbrugge SE et al. (2016) Multifactorial resistance to aminopeptidase inhibitor prodrug CHR2863 in myeloid leukemia cells: down-regulation of carboxylesterase 1, drug sequestration in lipid droplets and pro-survival activation ERK/Akt/mTOR. Oncotarget 7 (5), 5240–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spicher L and Kessler F (2015) Unexpected roles of plastoglobules (plastid lipid droplets) in vitamin K1 and E metabolism. Curr Opin Plant Biol 25, 123–9. [DOI] [PubMed] [Google Scholar]
- 5.Cloherty APM et al. (2020) Hijacking of Lipid Droplets by Hepatitis C, Dengue and Zika Viruses-From Viral Protein Moonlighting to Extracellular Release. Int J Mol Sci 21 (21), 7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch M et al. (2020) Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370 (6514), eaay8085. [DOI] [PubMed] [Google Scholar]
- 7.Romeo S et al. (2008) Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40 (12), 1461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abul-Husn NS et al. (2018) A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med 378 (12), 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg AS et al. (1991) Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem 266 (17), 11341–6. [PubMed] [Google Scholar]
- 10.Sztalryd C and Brasaemle DL (2017) The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim Biophys Acta Mol Cell Biol Lipids 1862 (10 Pt B), 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu RD and Huang AH (1990) Oleosin KD 18 on the surface of oil bodies in maize. Genomic and cDNA sequences and the deduced protein structure. J Biol Chem 265 (4), 2238–43. [PubMed] [Google Scholar]
- 12.Tzen JT and Huang AH (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117 (2), 327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poppelreuther M et al. (2012) The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res 53 (5), 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Aquila T et al. (2015) Characterization of the proteome of cytoplasmic lipid droplets in mouse enterocytes after a dietary fat challenge. PLoS One 10 (5), e0126823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilfling F et al. (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24 (4), 384–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leber R et al. (1998) Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell 9 (2), 375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFie PJ et al. (2018) Diacylglycerol acyltransferase-2 contains a c-terminal sequence that interacts with lipid droplets. Biochim Biophys Acta Mol Cell Biol Lipids 1863 (9), 1068–1081. [DOI] [PubMed] [Google Scholar]
- 18.Gao Q et al. (2017) Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J Cell Biol 216 (10), 3199–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krahmer N et al. (2011) Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab 14 (4), 504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moessinger C et al. (2011) Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem 286 (24), 21330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouchoux J et al. (2011) The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol Cell 103 (11), 499–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang HK et al. (2013) The membrane-binding domain of an amphitropic enzyme suppresses catalysis by contact with an amphipathic helix flanking its active site. J Mol Biol 425 (9), 1546–64. [DOI] [PubMed] [Google Scholar]
- 23.Lee J et al. (2014) Structural basis for autoinhibition of CTP:phosphocholine cytidylyltransferase (CCT), the regulatory enzyme in phosphatidylcholine synthesis, by its membrane-binding amphipathic helix. J Biol Chem 289 (3), 1742–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornell RB et al. (2019) Disease-linked mutations in the phosphatidylcholine regulatory enzyme CCTα impair enzymatic activity and fold stability. J Biol Chem 294 (5), 1490–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne F et al. (2014) Mutations disrupting the Kennedy phosphatidylcholine pathway in humans with congenital lipodystrophy and fatty liver disease. Proc Natl Acad Sci U S A 111 (24), 8901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan SA et al. (2015) Quantitative analysis of the murine lipid droplet-associated proteome during diet-induced hepatic steatosis. J Lipid Res 56 (12), 2260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du X et al. (2020) ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J Cell Biol 219 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova E et al. (2006) ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep 7 (1), 106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egan JJ et al. (1992) Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci U S A 89 (18), 8537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zechner R et al. (2017) Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol 18 (11), 671–684. [DOI] [PubMed] [Google Scholar]
- 31.Yang A and Mottillo EP (2020) Adipocyte lipolysis: from molecular mechanisms of regulation to disease and therapeutics. Biochem J 477 (5), 985–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radner FP et al. (2010) Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J Biol Chem 285 (10), 7300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smagris E et al. (2015) Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61 (1), 108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsche MA et al. (2018) Patatin-like phospholipase domain–containing protein 3 promotes transfer of essential fatty acids from triglycerides to phospholipids in hepatic lipid droplets. J Biol Chem 293 (18), 6958–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.BasuRay S et al. (2019) Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc Natl Acad Sci U S A 116 (19), 9521–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.BasuRay S et al. (2017) The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 66 (4), 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y et al. (2019) PNPLA3, CGI-58, and Inhibition of Hepatic Triglyceride Hydrolysis in Mice. Hepatology 69 (6), 2427–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz BE et al. (2020) Discovery and Targeting of the Signaling Controls of PNPLA3 to Effectively Reduce Transcription, Expression, and Function in Pre-Clinical NAFLD/NASH Settings. Cells 9 (10), 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y et al. (2020) Characterization of essential domains in HSD17B13 for cellular localization and enzymatic activity. J Lipid Res 61 (11), 1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luukkonen PK et al. (2020) Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. JCI Insight 5 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mejhert N et al. (2020) Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression. Mol Cell 77 (6), 1251–1264.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallardo-Montejano VI et al. (2016) Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat Commun 7, 12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billin AN and Ayer DE (2006) The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol 302, 255–78. [DOI] [PubMed] [Google Scholar]
- 44.Filhoulaud G et al. (2013) Novel insights into ChREBP regulation and function. Trends Endocrinol Metab 24 (5), 257–68. [DOI] [PubMed] [Google Scholar]
- 45.Li Z et al. (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol 22 (22), 2104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Currie E et al. (2014) High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res 55 (7), 1465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krahmer N et al. (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 12 (5), 1115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valtersson C et al. (1985) The influence of dolichol, dolichol esters, and dolichyl phosphate on phospholipid polymorphism and fluidity in model membranes. J Biol Chem 260 (5), 2742–51. [PubMed] [Google Scholar]
- 49.Suzuki M et al. (2012) Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell 23 (5), 800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zehmer JK et al. (2009) Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci 122 (Pt 20), 3694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye Y et al. (2017) A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front Mol Biosci 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JN et al. (2010) Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc Natl Acad Sci U S A 107 (50), 21424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hariri H et al. (2019) Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J Cell Biol 218 (4), 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ugrankar R et al. (2019) Drosophila Snazarus Regulates a Lipid Droplet Population at Plasma Membrane-Droplet Contacts in Adipocytes. Dev Cell 50 (5), 557–572.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiam AR and Dugail I (2019) Lipid droplet-membrane contact sites - from protein binding to function. J Cell Sci 132 (12), jcs230169. [DOI] [PubMed] [Google Scholar]
- 56.Moriya K et al. (1997) Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol 78 (Pt 7), 1527–31. [DOI] [PubMed] [Google Scholar]
- 57.Hinson ER and Cresswell P (2009) The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A 106 (48), 20452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monson EA et al. (2021) Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walther TC et al. (2017) Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung J et al. (2019) LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev Cell 51 (5), 551–563.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thiam AR et al. (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14 (12), 775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartz R et al. (2007) Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res 48 (4), 837–47. [DOI] [PubMed] [Google Scholar]
- 63.Tauchi-Sato K et al. (2002) The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem 277 (46), 44507–12. [DOI] [PubMed] [Google Scholar]
- 64.Prévost C et al. (2018) Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev Cell 44 (1), 73–86.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S and Swanson JMJ (2020) The Surface and Hydration Properties of Lipid Droplets. Biophys J 119 (10), 1958–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chorlay A and Thiam AR (2020) Neutral lipids regulate amphipathic helix affinity for model lipid droplets. J Cell Biol 219 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S et al. (2021) Stressed Lipid Droplets: How Neutral Lipids Relieve Surface Tension and Membrane Expansion Drives Protein Association. J Phys Chem B 125 (21), 5572–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kory N et al. (2015) Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev Cell 34 (3), 351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowe ER et al. (2016) Conserved Amphipathic Helices Mediate Lipid Droplet Targeting of Perilipins 1–3. J Biol Chem 291 (13), 6664–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hänisch J et al. (2006) Eukaryotic lipid body proteins in oleogenous actinomycetes and their targeting to intracellular triacylglycerol inclusions: Impact on models of lipid body biogenesis. Appl Environ Microbiol 72 (10), 6743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu G et al. (2006) Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta 1761 (1), 83–90. [DOI] [PubMed] [Google Scholar]
- 72.Ajjaji D et al. (2019) Dual binding motifs underpin the hierarchical association of perilipins1–3 with lipid droplets. Mol Biol Cell 30 (5), 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Čopič A et al. (2018) A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat Commun 9 (1), 1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boeszoermenyi A et al. (2015) Structure of a CGI-58 motif provides the molecular basis of lipid droplet anchoring. J Biol Chem 290 (44), 26361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taneva S et al. (2003) Lipid-induced conformational switch in the membrane binding domain of CTP:phosphocholine cytidylyltransferase: a circular dichroism study. Biochemistry 42 (40), 11768–76. [DOI] [PubMed] [Google Scholar]
- 76.Dunne SJ et al. (1996) Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase. Biochemistry 35 (37), 11975–84. [DOI] [PubMed] [Google Scholar]
- 77.Yue L et al. (2020) Differential dephosphorylation of CTP:phosphocholine cytidylyltransferase upon translocation to nuclear membranes and lipid droplets. Mol Biol Cell 31 (10), 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krahmer N et al. (2018) Organellar Proteomics and Phospho-Proteomics Reveal Subcellular Reorganization in Diet-Induced Hepatic Steatosis. Dev Cell 47 (2), 205–221 e7. [DOI] [PubMed] [Google Scholar]
- 79.Olarte MJ et al. (2020) Determinants of Endoplasmic Reticulum-to-Lipid Droplet Protein Targeting. Dev Cell 54 (4), 471–487.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schrul B and Kopito RR (2016) Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat Cell Biol 18 (7), 740–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salo VT et al. (2016) Seipin regulates ER-lipid droplet contacts and cargo delivery. Embo j 35 (24), 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacquier N et al. (2011) Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 124 (Pt 14), 2424–37. [DOI] [PubMed] [Google Scholar]
- 83.Wang H et al. (2016) Seipin is required for converting nascent to mature lipid droplets. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salo VT et al. (2019) Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev Cell 50 (4), 478–493 e9. [DOI] [PubMed] [Google Scholar]
- 85.Choudhary V et al. (2020) Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J Cell Biol 219 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grippa A et al. (2015) The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J Cell Biol 211 (4), 829–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szymanski KM et al. (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A 104 (52), 20890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo Y et al. (2008) Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453 (7195), 657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beller M et al. (2008) COPI complex is a regulator of lipid homeostasis. PLoS Biol 6 (11), e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nickel W et al. (2002) Vesicular transport: the core machinery of COPI recruitment and budding. J Cell Sci 115 (Pt 16), 3235–40. [DOI] [PubMed] [Google Scholar]
- 91.Thiam AR et al. (2013) COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc Natl Acad Sci U S A 110 (33), 13244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilfling F et al. (2014) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3, e01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruggiano A et al. (2016) Spatial control of lipid droplet proteins by the ERAD ubiquitin ligase Doa10. Embo j 35 (15), 1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bekbulat F et al. (2020) RAB18 Loss Interferes With Lipid Droplet Catabolism and Provokes Autophagy Network Adaptations. J Mol Biol 432 (4), 1216–1234. [DOI] [PubMed] [Google Scholar]
- 95.Xu D et al. (2018) Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J Cell Biol 217 (3), 975–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jayson CBK et al. (2018) Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol Biol Cell 29 (17), 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bersuker K et al. (2018) A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev Cell 44 (1), 97–112.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brasaemle DL et al. (2004) Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem 279 (45), 46835–42. [DOI] [PubMed] [Google Scholar]
- 99.Gemmink A et al. (2018) Super-resolution microscopy localizes perilipin 5 at lipid droplet-mitochondria interaction sites and at lipid droplets juxtaposing to perilipin 2. Biochim Biophys Acta Mol Cell Biol Lipids 1863 (11), 1423–1432. [DOI] [PubMed] [Google Scholar]
- 100.Chang CL et al. (2019) Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J Cell Biol 218 (8), 2583–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Subramanian V et al. (2004) Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem 279 (40), 42062–71. [DOI] [PubMed] [Google Scholar]
- 102.Yamaguchi T et al. (2004) CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem 279 (29), 30490–7. [DOI] [PubMed] [Google Scholar]
- 103.Saka HA et al. (2015) Chlamydia trachomatis Infection Leads to Defined Alterations to the Lipid Droplet Proteome in Epithelial Cells. PLoS One 10 (4), e0124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moessinger C et al. (2014) Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joshi AS et al. (2018) Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat Commun 9 (1), 2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kory N et al. (2017) Mice lacking lipid droplet-associated hydrolase, a gene linked to human prostate cancer, have normal cholesterol ester metabolism. J Lipid Res 58 (1), 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turro S et al. (2006) Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic 7 (9), 1254–69. [DOI] [PubMed] [Google Scholar]
- 108.Zehmer JK et al. (2008) Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci 121 (11), 1852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stevanovic A and Thiele C (2013) Monotopic topology is required for lipid droplet targeting of ancient ubiquitous protein 1. J Lipid Res 54 (2), 503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Spandl J et al. (2011) Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem 286 (7), 5599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pol A et al. (2004) Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol Biol Cell 15 (1), 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ohashi M et al. (2003) Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J Biol Chem 278 (38), 36819–29. [DOI] [PubMed] [Google Scholar]
- 113.Caldas H and Herman GE (2003) NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum Mol Genet 12 (22), 2981–91. [DOI] [PubMed] [Google Scholar]
- 114.Castro IG et al. (2019) Promethin Is a Conserved Seipin Partner Protein. Cells 8 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]


