Abstract
Background:
High-risk human papillomaviruses (HR HPV) cause nearly all cervical cancers and, in the USA, the majority of head and neck cancers (HNSCCs). NFX1-123 is overexpressed in cervical cancers, and NFX1-123 partners with the HR HPV type 16 E6 oncoprotein to affect multiple growth, differentiation, and immune response genes. However, neither the expression of NFX1-123, nor the levels of these genes, have been investigated in HPV positive (HPV+) or negative (HPV−) HNSCCs.
Methods:
The Cancer Genome Atlas Splicing Variants Database and HNSCC cell lines were used to quantify expression of NFX1-123 and cellular genes increased in cervical cancers.
Results:
NFX1-123 was increased in HPV+ HNSCCs compared to HPV− HNSCCs. LCE1B, KRT16, SPRR2G, and FBN2 were highly expressed in HNSCCs compared to normal tissues. Notch1 and CCNB1IP1 had greater expression in HPV+ HNSCCs compared to HPV− HNSCCs.
Conclusion:
NFX1-123, and a subset of its known targets, were increased in HPV+ HNSCCs.
Keywords: Head and neck squamous cell carcinoma, oropharyngeal cancer, human papillomavirus, NFX1-123, Notch1
INTRODUCTION
Head and Neck Squamous cell carcinomas (HNSCC) account for 4.8% of all cancers with similar percentage of cancer death worldwide 1. The majority of HNSCCs arise from squamous epithelial cells lining the oral cavity, pharynx, larynx and nasal cavity. There are several etiological factors including alcohol and tobacco that contribute to HNSCC, but about 25% of HNSCCs globally are caused by high-risk (HR) human papillomaviruses (HPVs) 2,3, and this percentage is on the rise 4. HR HPVs cause cancers at other anatomic sites – including cervical, vulvar, vaginal, penile, and anal cancers 5. In developed countries, non-HPV associated (HPV−) HNSCC cases have decreased by 50%; however HPV associated (HPV+) HNSCCs have increased by 225% 4,6–8, and this shift may be due in part to changes in sexual behaviors 9–12. As rates of HPV+ HNSCCs increase, understanding the common and unique mechanisms driving HPV associated cancers at these anatomic sites is paramount. It may lead to new ways to screen, diagnosis, and treat patients with HPV+ HNSCCs.
HR HPVs universally express two viral oncogenes, E6 and E7, in their cancers. These oncogenes themselves have no enzymatic function, but they partner with cellular proteins to collaboratively dysregulate pathways in infected host cells in order to drive growth, longevity, and immune evasion. We and others have identified that HR HPV type 16 E6 (16E6) binds to two splice variants of the NFX1 gene, NFX1-91 and NFX1-123 13–15. NFX1-123 is highly expressed in cervical cancer cell lines 16 and in primary cervical cancers 17,18. When overexpressed in primary keratinocytes with 16E6, NFX1-123 decreases innate immune pathway genes, augments cellular growth, increases telomerase activity, upregulates Notch1 levels and its canonical and non-canonical pathways, and protects against cellular arrest 15,16,19–21. NFX1-123 also functions directly to augment JNK signaling in keratinocytes, increasing cellular differentiation markers and late HPV gene expression that is triggered by cellular differentiation, all while supporting continued cellular growth 22. A recent review on NFX1 and its isoforms NFX1-123 and NFX1-91 highlighted the functional significance of NFX1-123 in human diseases and cancer 23. We have recently confirmed that many genes upregulated by NFX1-123 and 16E6 in primary human foreskin keratinocytes (HFKs) were also overexpressed in cervical cancers when compared to the normal cervix 17. This indicates that NFX1-123 plays a functional role in cervical cancer, and it also suggests that NFX1-123 may be important in HPV+ cancers at other anatomic sites.
Although there are FDA-approved vaccines to prevent HPV infections, and protection again HPV-associated head and neck cancers was recently added as a clinical indication for the nonavalent vaccine, most males and females have not received this vaccine and remain at-risk for oropharyngeal HPV infections and HPV+ HNSCCs 24. The standard treatment options for HPV+ HNSCCs are surgery, radiotherapy or chemotherapy. In the case of metastatic disease, the treatment options are adjuvant chemoradiotherapy combined with radiotherapy 25,26. Disease-free survival has been reported as equivalent for any of these therapeutic strategies in non-metastatic HPV+ HNSCC 25,27,28, and prognosis is better for HPV+ than HPV− HNSCC patients. Unfortunately, all of the available treatment strategies, while highly effective for survival, lead to significant morbidities for patients. There is an urgent need to develop alternative, non-inferior treatment strategies – ones that leverage functional biomarkers that are dually associated with prognosis and may be targeted with novel therapeutic agents 29.
Recognizing that NFX1-123 is highly expressed in cervical cancers when compared to normal cervical tissue, and that it has a functional role in gene expression and pathways required for the HR HPV life cycle and cervical cancer progression, we wanted to investigate if this also was seen in HNSCCs broadly, or if there was a specific association with HPV+ HNSCCs. Our results revealed that NFX1-123, but no other splice variant of the NFX1 gene, was specifically highly expressed in HPV+ HNSCCs. Of the 25 genes previously published as highly expressed in HFKs with 16E6 and overexpressed NFX1-123 21, 14 of which were also highly expressed in cervical cancer 17, four had greater expression in HNSCCs, and two had greater expression specifically in HPV+ HNSCCs.
MATERIALS AND METHODS
NFX1-123 and NFX1-91 mRNA expression analysis:
To identify the NFX1-123 splice variant expression in HNSCC, The Cancer Genome Atlas Data (TCGA) splice variant database TSVdb was utilized 30. NFX1 splice variants were searched in HNSCC and exon usage values were downloaded. All six splice variants available in the TSVdb were downloaded, and the known expressed NFX1 isoforms were identified through sequence verification in the UCSC Genome Browser (https://genome.ucsc.edu)
There were 6 alternative splice variants of NFX1 reported in TSVdb, namely:
| NFX1 splice variant | Exons | Protein size aa | |
|---|---|---|---|
| isoform_uc011lnw | NFX1 transcript Variant 3 | 16 | 832 |
| isoform_uc003zso | NFX1 transcript Variant 3 | 16 | 833 |
| isoform_uc003zsp | NFX1 transcript Variant 2 | 21 | 1024 |
| isoform_uc010mjr | NFX1 transcript Variant 2 | 21 | 1025 |
| isoform_uc003zsq | NFX1 transcript Variant 1 | 24 | 1120 |
| isoform_uc003zsr | NFX1 transcript Variant 1 | 24 | 1121 |
Using the UCSC Genome Browser, the longer isoform of NFX1 expressed in epithelial cells, NFX1-123 (isoform_uc003zsq), was identified with 1120 amino acids. The transcript of NFX1-123 (including UTRs) started from position: hg19ch9:33,290,418-33,371,155; size 80,738; total exon count: 24 strand: +. The coding region of NFX1-123 started from position: hg19ch9:33,290,571-33,369,976; size 79,406; coding exon count: 24.
The shorter isoform of NFX1 expressed in epithelial cells, NFX1-91 (isoform_uc003zso), was also identified with 833 amino acids. The transcript of NFX1-91 (including UTRs) started from position: hg19 chr9:33,290,418-33,348,721; Size: 58,304; total exon count: 16 Strand: +. The coding region of NFX1-91 started from position: hg19 chr9:33,290,571-33,347,738; Size: 57,168; coding exon count: 16.
The expression values of all NFX1 isoforms were presented as log2RSEM. From the TCGA source data, HPV positive and negative cases were marked in the TSVdb data, and expression comparison of NFX1-123 between HPV negative and positive tumors as well as between tumor adjacent normal tissue and primary tumors was completed. The NFX1 isoforms data from 567 head and neck samples, which included 520 HNSCC primary tumors, 44 normal solid tissue samples, and 2 metastatic tumors, was downloaded and divided into their appropriate groups. For differential analysis of NFX1-123 expression, we compared its expression between normal solid tissue samples and primary tumors. Within in the group of primary tumors, we compared expression between HPV negative and HPV positive primary tumors, and the anatomic locations of the HPV negative HNSCCs. Specifically, primary tumors from the mouth floor, buccal mucosa, oral tongue, larynx, OPC (HPV negative and HPV positive) were separated and expression of NFX1-123 and NFX1-91 was quantified.
NFX1-123 regulated genes mRNA expression analysis:
The Cancer Genome Atlas Data (TCGA) data of UCSC Xenabrowser (https://xenabrowser.net) was used to obtain normalized mRNA gene expression data in HNSCC samples from the Genomic Data Commons (GDC) TCGA 31. In the GDC TCGA, gene expression from 520 HNSCCs and 44 adjacent normal solid tissues were quantified and compared. Of the 25 genes previously identified by our laboratory as upregulated two fold or more in HFKs with co-expressed 16E6 and overexpressed NFX1-123 splice variant 21, 19 of them (CCNB1IP1, TGM1, SLPI, KRT16, SPRR2G, LCE1B, FBN2, RPS29, PPL, RAB7B, CEBPD, ALDH3B2, LCE2B, LOR, BNIPL, RAET1G, FOXA2, IMPA2, and Notch1), whose expression levels in cervical cancer we recently published 17, had available expression data across these tissue types for analysis. Normal and HNSCC primary tumors as well as HPV positive and negative primary tumors were compared for gene expression analyses.
Cell Culture:
HPV negative HNSCC cell lines (HN4, SCC61 and SCC1) and HPV 16 positive HNSCC cell lines (SCC47, SCC104 and SCC152) were a kind gift of Dr. Ian Morgan, and they were authenticated by confirmation of HPV gene expression. The HPV negative HNSCC cell line FaDu and the HPV 16 positive HNSCC cell line SCC090 were purchased from ATCC. The anatomical sites of these cell lines are as listed: HN4 - Larynx; SCC61 - Tongue; SCC1 - Mouth Floor; FaDu - Hypopharynx; SCC47 - Tongue; SCC104 - Mouth Floor; SCC152 - Hypopharynx; SCC90 - Tongue. All cell lines were cultured in DMEM with 10% FBS and penicillin and streptomycin. All cell culture studies were approved by the Indiana University IBC.
Western blot analysis:
Forty micrograms of whole cell protein extracts from HPV negative and positive HNSCC cells were separated by gel electrophoresis in 4-20% precast Mini-Protein TGX gels and transferred using the Transfer-Blot Turbo PVDF transfer pack and the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). Protein detection was performed with the following primary antibodies: NFX1-123 (Rabbit, Novus Biologicals, Centennial, CO, USA, cat# NBP1-49933, 1:1000 dilution; or Rabbit polyclonal generated, gift from Dr. Ann Roman, 1:1000 dilution); p53 (Mouse monoclonal DO-1, Millipore Sigma, St. Louis, MO, USA, cat# OP43, 1:1000 dilution); p16 (Rabbit monoclonal, Abcam, Cambridge, MA, USA, cat# ab81278, 1:1000 dilution), KRT16 (Rabbit monoclonal, Abcam, Cambridge, MA, USA, cat# ab76416, 1:500 dilution), FBN2 (Rabbit polyclonal, Proteintech, Rosemont, IL, USA, cat# 20252-1-AP, 1:500 dilution), Notch1 ( Rat monoclonal 5B5, Cell Signaling Technology, Danvers, MA, USA, cat# 3447, 1:500 dilution), CCNB1IP1 ( Rabbit polyclonal, Invitrogen-ThermoFisher Scientific, Waltham, MA, USA, cat# PA5-68386, 1:500 dilution) and GAPDH (Mouse monoclonal, Invitrogen-ThermoFisher Scientific, Waltham, MA, USA, cat# AM4300, 1:100,000 dilution). The following secondary antibodies HRP conjugated were used at a 1:5000 dilution: Rabbit (Santa Cruz Biotechnology, Dallas, TX, USA), Mouse (Cell Signaling Technology, Danvers, MA, USA) or Rat (Cell Signaling Technology, Danvers, MA, USA). Membranes were blocked for one hour at room temperature in 2% ECL prime western blot blocking agent (Amersham GE Healthcare, Pittsburgh, PA, USA), and incubated in primary antibodies diluted in 2% ECL prime western blot blocking agent at 4°C overnight. Blots were washed with PBS-T and incubated with their respective HRP conjugated secondary antibodies in 2% ECL blocking agent for one hour at room temperature. The blots were washed with PBS-T and developed with ECL Prime Western Blot detection reagent, and images were captured with the ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA).
Quantification of western blot relative protein expression was determined using ImageJ (NIH, 1.47v, Washington, D.C., USA). For each cell line, band intensity was normalized to its loading control GAPDH, and then fold differences in protein expression was determined relative to the expression level of the protein of interest in HPV negative cells.
Quantitative PCR:
cDNA synthesis: HNSCC cells were collected at three separate time points to assure biologic reproducibility, then lysed with 1ml TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA was isolated as per the manufacturer’s instructions. The RNA quantity and purity were determined by Nanodrop spectrophotometer. Following the manufacturer’s instructions, cDNA was synthesized from 2 micrograms of RNA with SuperScript IV VILO Master Mix plus ezDNase enzyme kit (ThermoFisher Scientific, Waltham, MA, USA).
Quantitative real-time PCR (qRT-PCR): qRT-PCR was used to determine relative mRNA expression of NFX1-123, LCE1B, KRT16, SPRR2G, FBN2 and Notch1. Gene expression was determined using either specific PCR primers or TaqMan probes. Primers were used for the following mRNAs: NFX1-123 (Forward 5’-CCACAGCTTCCCTCCCA-3’ and Reverse 5’-CCTGGACGTCAAAATAGTCAA-3’); Notch1 (Forward 5’-GCCGAACCAATACAACCCTCTGC-3’ and Reverse 5’-GGTAGCTCATCATCTGGGACAGG-3’); Keratin 16 (Forward 5’-TCGAGGACCTGAGGAACAAG-3’ and Reverse 5’-GGGCCAGTTCATGCTCATAC-3’); and 36B4 (Forward 5’-TGCCAGTGTCTGTCTGCAGA-3’ and Reverse 5’-ACAAAGGCAGATGGATCAGC-3’). TaqMan probes were used for the following mRNAs: LCE1B (Hs00866755_s1); SPRR2G (Hs00972901_s1); FBN2 (Hs00266592_m1); and GAPDH (Hs02786624_g1). qRT-PCR was performed using the QuantStudio 3 Real-Time PCR System (Applied Biosystems-ThermoFisher Scientific, Waltham, MA, USA). PowerUp SYBR Green Master Mix was used for assays with primers, and TaqMan Fast Advanced Master Mix was used for assays with TaqMan probes (Applied Biosystems, Waltham, MA, USA). Each biologic sample was amplified by qPCR in triplicate for technical replicates, and values were normalized to 36B4 for primer assays or GAPDH for TaqMan probe assays. A relative standard curve was generated and used to determine expression values. The expression of each gene was compared between HPV negative (n=4) and HPV positive (n=4) cell lines. Data presented are of one biologic sample for each cell line; they are representative of all experiments conducted.
Statistical analysis:
Gene expression data was analyzed for statistical significance using Mann-Whitney test and comparison ranks, and results were considered significant if the p-value was <0.05. qRT-PCR gene expression data was analyzed for statistical significance using student’s t-tests, and results were considered significant if the p-value was <0.05. All exact p-values are shown (GraphPad Prism 8.0.2 Software, San Diego, CA, USA).
RESULTS
NFX1-123, but not NFX1-91, was increased in human papillomavirus positive head and neck squamous cell carcinomas
NFX1-123 was previously identified as a protein partner of 16E6, and its expression is increased in cervical cancer cell lines and primary cervical cancers 16–18 . There are three splice variants of the NFX1 gene in NCBI, and two are expressed at the protein level in epithelial cells and keratinocytes. Those two splice variants, NFX1-123 and NFX1-91, have been the focus of studies in cervical cancers, but have not been studied in HNSCCs. NFX1-123 and NFX1-91 have different, and often opposing, functional roles in collaborative gene regulation with 16E6, as well as different subcellular localization. As such, their levels of expression may be differentially regulated during cancer development and progression, in concert with or separate from the presence of HR HPV. To parse the expression of these two NFX1 splice variants in HNSCCs, we utilized The Cancer Genome Atlas Splice Variant database (TSVdb).
For NFX1-123, its mRNA expression was not quantifiably different in HNSCC primary tumors when compared to adjacent normal solid tissue (Fig. 1A). However, when analyzing within HNSCCs, NFX1-123 had greater expression in HPV+ HNSCCs when compared to HPV− HNSCCs (Fig, 1B). These findings revealed that, like cervical cancers that are nearly universally HPV+, HPV+ HNSCCs also had higher expression of NFX1-123 mRNA.
Figure 1. NFX1-123 increased in HPV positive HNSCC.
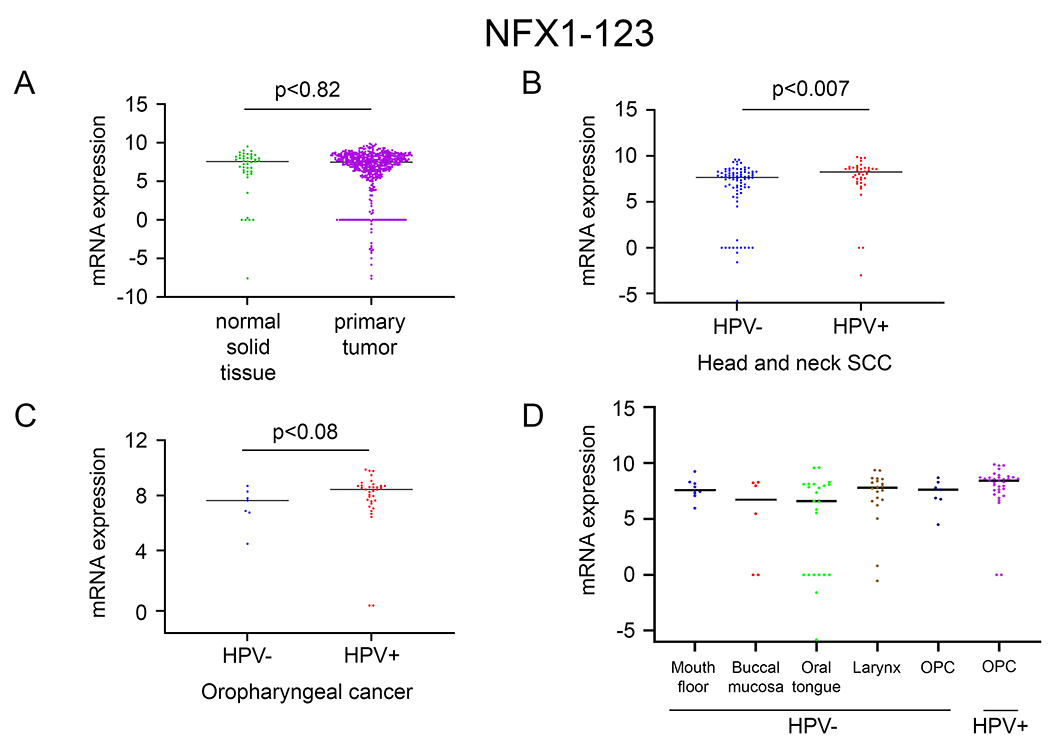
Using the TSVdb, NFX1-123 expression was quantified in: A. primary tumors of HNSCC (n=520) compared to adjacent normal solid tissue (n=44); B. HPV positive HNSCC (n=39) compared to HPV negative HNSCC (n=80); C. HPV positive oropharyngeal cancer (OPC) (n=34) compared to HPV negative OPC (n=7). D. NFX1-123 mRNA expression in HPV positive OPCs compared to HPV negative OPCs and at different anatomical cancer sites (mouth floor, buccal mucosa, oral tongue, larynx).
Oropharyngeal cancers are the most common anatomic location for HPV+ HNSCCs, and indeed this subgroup of HPV+ HNSCCs (HPV+ OPCs) expressed higher levels of NFX1-123 when compared to HPV− OPCs (Fig. 1C). Although this difference was not statistically significant, the trend matched the general HPV+ HNSCC data. NFX1-123 expression in HNSCC primary tumors across different anatomical sites (mouth floor, buccal mucosa, tongue, larynx and oropharynx) matched this finding; greater NFX1-123 expression in HPV+ OPCs when compared to all other HPV− primary HNSCC tumors (Fig. 1D).
In contrast to NFX1-123, the shorter splice variant of NFX1, NFX1-91, was downregulated in HNSCC primary tumors when compared to adjacent normal solid tissue (Fig. 2A). Additionally, neither NFX1-91, nor any other isoform of NFX1 (data not shown), was changed in HPV+ HNSCCs when compared to HPV− HNSCCs (Fig. 2B). In HPV+ and HPV− OPCs, there was also no significant difference in NFX1-91 mRNA expression (Fig. 2C), and this held true across HNSCCs from different anatomical sites (Fig. 2D).
Figure 2. NFX1-91 was not significantly increased in HNSCC or HPV positive HNSCC.
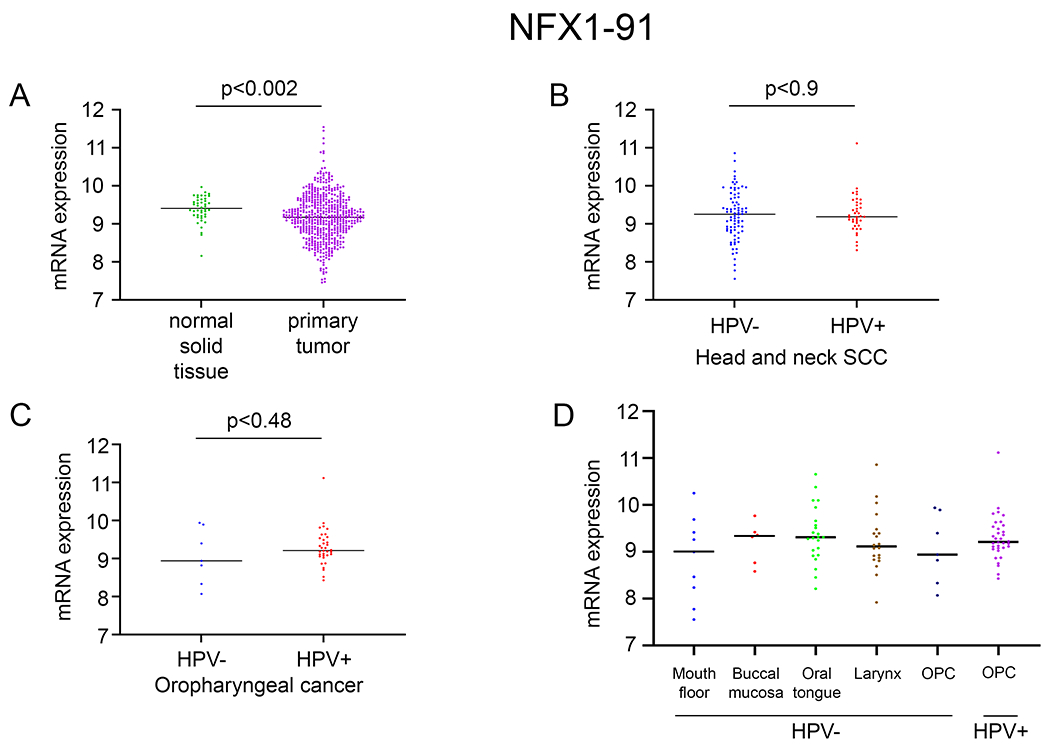
Using the TSVdb, NFX1-91 expression was quantified in: A. primary tumors of HNSCC (n=520) compared to adjacent normal solid tissue (n=44); B. HPV positive HNSCC (n=39) compared to HPV negative HNSCC (n=80); C. and HPV positive oropharyngeal cancer (OPC) (n=34) compared to HPV negative OPC (n=7). D. NFX1-91 mRNA expression in HPV positive OPCs compared to HPV negative OPCs and at different anatomical cancer sites (mouth floor, buccal mucosa, oral tongue, larynx).
Because NFX1-123 had greater expression levels in HPV+ HNSCCs in the TCGA database, we wanted to determine if NFX1-123 mRNA and protein levels were greater in HPV+ HNSCC cell lines compared to HPV− HNSCC cell lines (Fig. 3). Four HPV 16+ and four HPV− HNSCC cell lines were collected for total RNA and protein analysis. Although it is recognized that these head and neck cancer cell lines originated from different anatomic sites, we found that, as a collection, NFX1-123 mRNA was significantly higher in HPV+ HNSCC cell lines when compared to HPV− HNSCC cell lines (Fig. 3A). The same was seen in NFX1-123 protein expression (Fig. 3B), and ImageJ densitometry analysis of whole cell protein western blots for NFX1-123 confirmed this (Fig. 3B). Therefore, greater NFX1-123 mRNA and protein correlated with the presence HPV 16 in HNSCC cell lines and correlated with NFX1-123 mRNA levels in HPV+ HNSCCs in the TCGA database.
Figure 3. High expression of NFX1-123 in HPV positive OPC HNSCC cell lines.
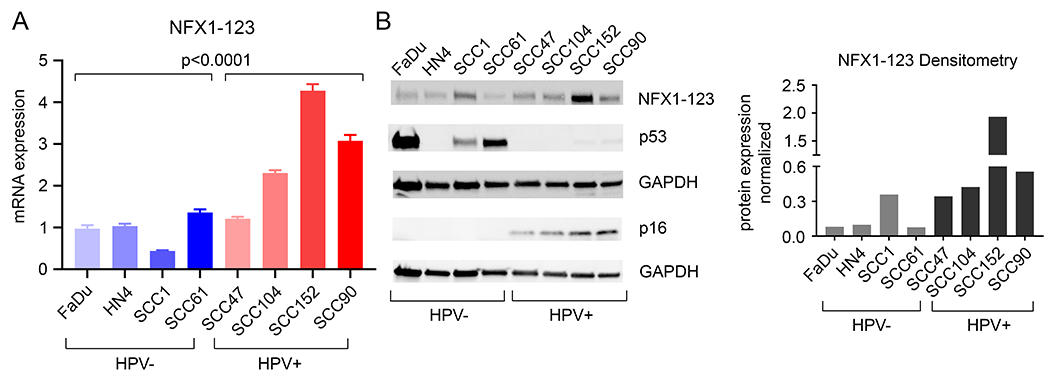
A. Quantitative PCR analysis of NFX1-123 was performed to determine the mRNA expression and was normalized to 36B4 as an internal control. Differential NFX1-123 expression between HPV negative and positive HNSCC cell lines was determined. B. Western blot analysis of NFX1-123 in HPV negative and positive HNSCC cell lines. HPV positivity was confirmed with p16 expression. p53, a protein degraded by HPV16 E6, was not detected in HPV positive cells. HN4 HPV negative cells do not express p53 due to a splice site mutation in these cells. GAPDH was used as a loading control. ImageJ analysis of NFX1-123 expression, normalized to GAPDH, is shown.
A subset of genes highly expressed in keratinocytes with overexpressed NFX1-123 and 16E6 as well as in primary cervical cancers were also increased in head and neck squamous cell carcinomas
Utilizing primary keratinocytes as the biologic background, we previously published that NFX1-123, in cooperation with 16E6, upregulated 25 different genes that are involved in the regulation of cell growth, longevity, and differentiation 21. Recently we studied the expression of 19 of these and confirmed the overexpression of 14 in primary cervical cancers when compared to the normal cervix 17. Based on these findings, we wanted to examine two things. First, we wanted to determine if these 19 genes were also modulated in HNSCC primary cancers when compared to normal solid tissues. Second, we wanted to determine how expression patterns of these genes in primary HNSCCs related to HPV16+ or HPV− HNSCC cell lines.
First, when compared to adjacent normal solid tissue, HNSCCs as a broad category had greater expression of four of these 19 genes: late cornified envelope 1B (LCE1B); keratin 16 (KRT16); small proline rich protein 2G (SPRR2G); and fibrillin 2 (FBN2) (Fig. 4A). Second, we wanted to investigate the expression patterns of these genes in HPV+ and HPV− HNSCC cell lines. We recognize that HNSCCs are more diverse in origin and anatomic location than cervical cancers, so as a corollary, we looked to see if there was an association with differential gene expression not only due to HPV status but also due to anatomic site. We evaluated the mRNA and protein expression of these genes by qPCR and western blot analysis, respectively. We found that three of these genes (LCE1B, KRT16 and SPRR2G) had high mRNA levels in HPV+ HNSCC cell lines compared to HPV− HNSCC cell lines (Fig. 4B). Two genes were quantified at the protein level (K16 and FBN2), and there was more heterogeneity across cell lines, likely reflecting their variation in origin (Fig 4C). Protein expression of Keratin 16 was very high in one HPV+ HNSCC cell line (SCC104, mouth floor), matching its mRNA expression. Both protein isoforms of FBN2 were moderately increased in HPV+ HNSCC cell lines, though an increase in mRNA was not observed in these cell lines. These results indicated that HPV+ status in HNSCC cell lines as a group correlated with higher mRNA levels of three genes that had previously been identified as also increased in cervical cancers and in keratinocytes co-expressing 16E6 and overexpressed NFX1-12317,21. These commonalities represent a subset of genes and pathways that are increased in HPV+ cancers.
Figure 4. Genes with greater expression in cervical cancers also increased in HNSCCs compared to adjacent normal solid tissues.
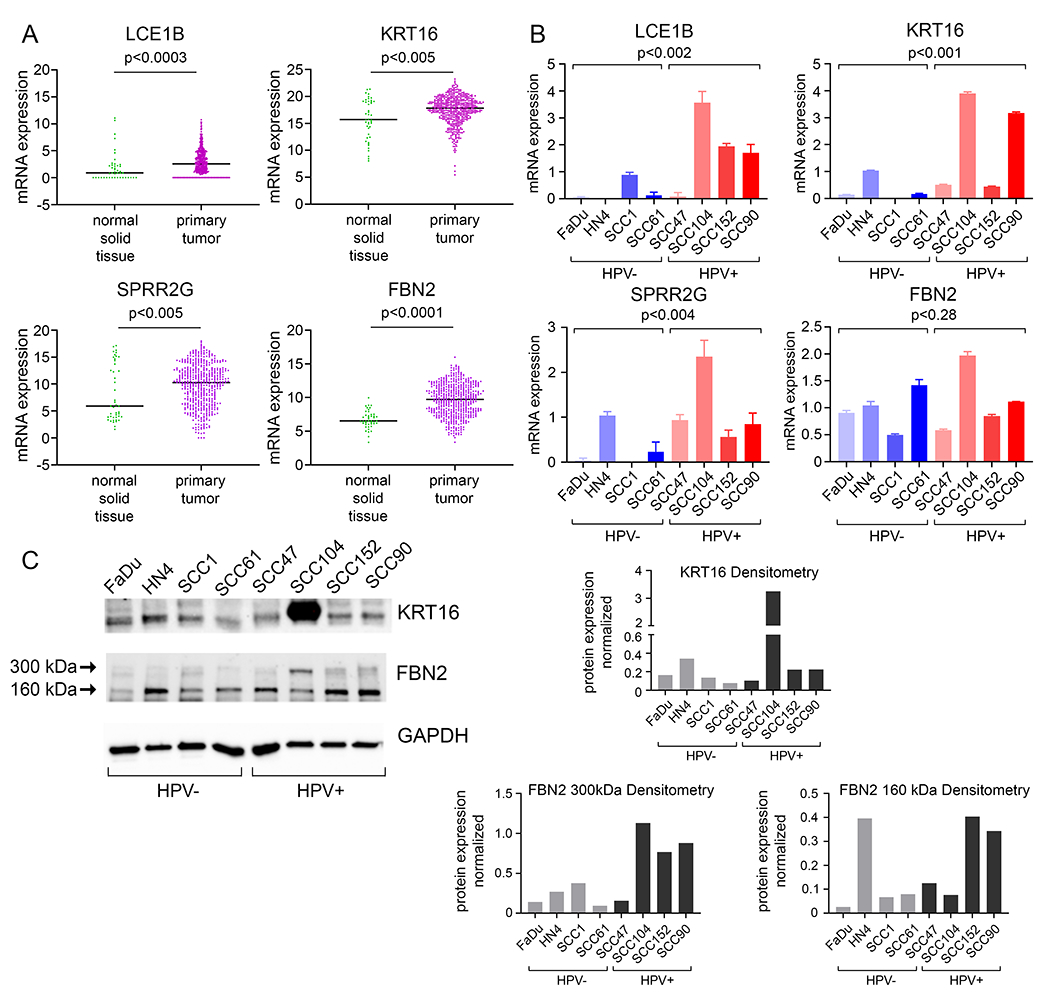
A. mRNA expression of LCE1B, KRT16, SPRR2G and FBN2 in primary tumors of HNSCC (n=520) compared to adjacent normal solid tissue (n=44). B. Quantitative PCR analysis of LCE1B, KRT16, SPRR2G and FBN2 in HPV negative and positive HNSCC cell lines. Differential gene expression between HPV negative and positive HNSCC cell lines were determined and was normalized to GAPDH (LCE1B, SPRR2G, FBN2) or 36B4 (KRT16) as an internal control. C. Western blot analysis of KRT16 and FBN2 in HPV negative and positive HNSCC cell lines. Two isoforms of FBN2 (300kDa and 160kDa) were detected. GAPDH was used as a loading control. ImageJ analysis, normalized to GAPDH, of KRT16 and the two isoforms of FBN2 is shown.
Two genes highly expressed in keratinocytes with 16E6 and overexpressed NFX1-123 as well as in primary cervical cancers were also increased in human papillomavirus positive head and neck squamous cell carcinomas
Returning to head and neck cancers in the TCGA database, we wanted next to determine whether genes shown to be highly expressed in primary cervical cancers were also differentially expressed within HNSCCs based on HPV status. We found significant upregulation of Notch1 and Cyclin B1 interacting protein 1 (CCNB1IP1) mRNA in HPV+ HNSCCs when compared to HPV− HNSCCs (Fig. 5A). When analyzing anatomic sites for HNSCCs, Notch1 remained highly expressed in HPV+ OPCs (Fig. 5B).
Figure 5. Genes with greater expression in cervical cancers were also increased in HPV positive HNSCCs as well as HPV positive Oropharyngeal cancers (OPC).
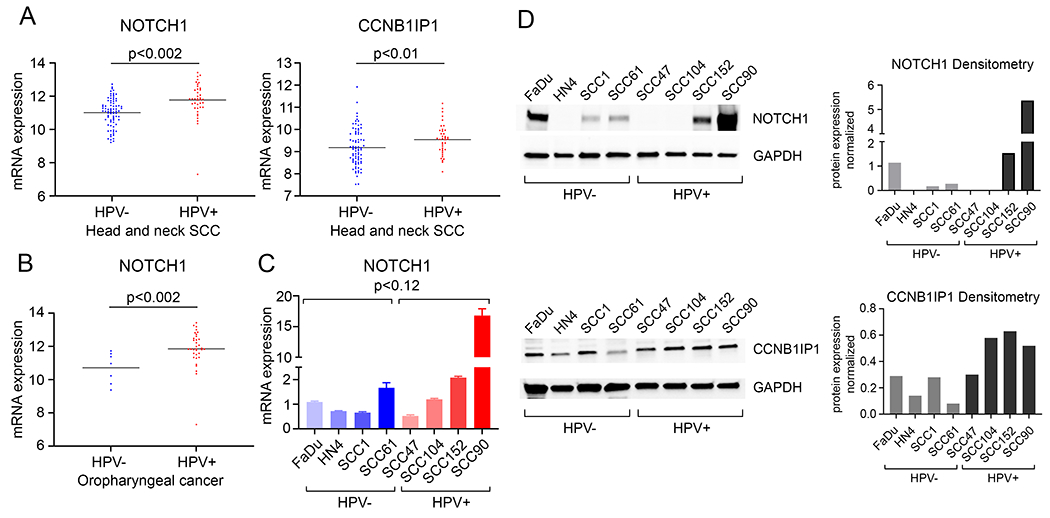
A. mRNA expression of Notch1 and CCNB1IP1 in HPV positive HNSCC (n=39) compared to HPV negative HNSCC (n=80). B. mRNA expression of Notch1 in HPV positive oropharyngeal cancer (OPC) (n= 34) compared to HPV negative OPC (n=7). C. Quantitative PCR analysis of Notch1 in HPV negative and positive HNSCC cell lines was determined and normalized to 36B4 as an internal control. D. Western blot analysis of Notch1 (upper) and CCNB1IP1 (lower) in HPV negative and positive HNSCC cell lines. GAPDH was used as a loading control. ImageJ analysis, normalized to GAPDH, of Notch1 and CCNB1IP1 is shown.
Again, we quantified expression of Notch1 and CCNB1IP1 protein, and Notch1 mRNA, in HPV+ and HPV− HNSCC cell lines to confirm these differences. Notch1 mRNA levels were variable, with a broader range of expression in HPV+ HNSCC cell lines from various anatomic sites. While there was very high expression of Notch1 in SCC90, a HPV+ HNSCC cell line, as a group, these data did not reach significance when compared to HPV− HNSCC cell lines (Fig. 5C). Notch1 protein levels were high in two of the four HPV+ HNSCC cell lines, matching the relative increases in Notch1 mRNA in these cell lines, and CCNB1IP protein was moderately increased as well (Fig. 5D). Taken as a whole, these findings highlight the similarities and differences in cervical and head and neck cancers, the heterogeneity of the origins of HNSCCs, and the correlations between NFX1-123 expression and downstream target genes in the presence of HR HPV.
DISCUSSION
For the first time, we found NFX1-123, a functional protein partner of 16E6, was highly expressed in primary HPV+ head and neck cancers and in HPV 16+ head and neck cancer cell lines (Figs. 1 and 3). This increase in expression was specific to the NFX1-123 isoform of NFX1, as NFX1-91 was equivalently expressed at the mRNA level in HNSCCs regardless of HPV status (Fig. 2). These data mirror our recent findings of NFX1-123 overexpression in cervical cancer cell lines and in primary cervical cancers, which are HPV positive in 98% of cases 16–18.
Previous studies in primary keratinocytes to identify genes that were collaboratively regulated by NFX1-123 and the viral oncogene 16E6 found 25 genes with two fold or more increased mRNA expression 21. These genes are involved in cellular differentiation, and they complemented findings we have published on the short and long-term augmented growth of keratinocytes and cervical cancer cell lines due to NFX1-123, 16E6, and telomerase activity 15,16,18,21. We confirmed that more than half of the genes were also significantly overexpressed in cervical cancers 17; however, different anatomic sites for HPV positive cancers may require dysregulation of different genes and pathways in order to establish precancerous lesions and frank cancers. Additionally, cancers at these anatomic sites that are HPV negative may dysregulate shared or unique genes and pathways.
Four genes, which had higher expression in keratinocytes with 16E6 co-expression and overexpressed NFX1-123, were significantly overexpressed in primary HNSCCs when compared to adjacent normal tissues: LCE1B, KRT16, SPRR2G, and FBN2 (Fig. 4A) 21. The smaller overlap in shared genes, when compared to HNSCCs, is not surprising. This has also been seen in DNA damage repair pathways genes that are commonly co-opted in HPV+ cervical cancers; in HNSCCs, only a subset of these genes also had differential expression 32. This may reflect the heterogeneity of HNSCCs, or the differences in the anatomic sites themselves. Indeed, our studies in HPV+ and HPV− HNSCCs demonstrated the variation of gene expression that can be seen in these cancers. As such, further work in this area is needed.
Upon further review of the four genes with greater expression in HNSCC tumors (Fig. 4), it is interesting to note that FBN2 was increased in keratinocytes with 16E6 and overexpressed NFX1-123 21 but was not statistically significantly overexpressed in cervical cancers 17. FBN2 is a constituent of larger extracellular glycoproteins that multimerize to form microfibrils, and it is an important structure in the extracellular matrix by directly influencing tumor angiogenesis. It is also known that FBN2 influences the sequestration of TGF-β by microfibrils, permitting higher concentrations of active TGF-β in the tumor microenvironment 33,34. This may be of greater importance in head and neck cancers, although there is heterogeneity in the HNSCC cell lines we analyzed (Fig. 4). LCE1B and SPRR2G are both proteins involved in late epithelial differentiation 21,35, and KRT16 is a protein activated during wound healing and can function as an immune regulator 36. These epithelial-specific genes are representative of the anatomic tissues and sites from which these cancers, and specifically HPV+ cancers, originate. As such, the dysregulation of differentiation, wound healing, and extracellular matrix genes and pathways is logical. It is an important caveat to note that our studies of LCE1B and SPRR2G expression in HNSCC cell lines were of mRNA but not protein; their protein expression is typically difficult to quantify in whole cell extracts as they are insoluble.
One gene that we found to be increased in primary cervical cancers 17, and now in HPV+ HNSCCs, is Notch1 (Fig. 5). Notch1 is a master cell fate regulator, and Notch1 and its downstream pathway is known to be important in cervical cancer development and progression 37–40 as well as in HPV+ head and neck cancers 37,41. Studies have identified that Notch1 expression is modified in HPV+ head and neck cancers, through either overexpression or by mutation41,42, and we have confirmed that Notch1 was increased in a subset of HPV+ HNSCCs (Fig. 5). Because the expression of Notch1 was not linked categorically to the HPV status of a HNSCC cell line in our studies, this highlights that the regulation of Notch1 may be more heterogeneous in these cancer types. There are several reasons that could underlie this variation across HNSCC cell lines, and even within HPV+ HNSCC cell lines, when compared to cervical cancer cell lines. One that may be most salient is the varied anatomic sites where HNSCCs can develop. These differences in cell origin are noted in our findings, and future studies are warranted that focus on the mucosal epithelial cell types that become cancers as well as the differences in the tumor microenvironment that augment or blunt gene expression in head and neck sub-anatomic sites.
A second gene, cyclin B interacting protein (CCNB1IP1), also had significantly greater mRNA expression in HPV+ HNSCCs when compared to HPV− HNSCCs (Fig. 5A), and that mirrors our findings in primary cervical cancers and in keratinocytes overexpressing NFX1-123 and co-expressing 16E6 17,21. Although its protein expression only moderately increased in HPV+ HNSCC cell lines (Fig. 4D), and mRNA expression was not determined, its role in cells is important. CCNB1IP1 is an E3 ubiquitin ligase that functions in cell cycle progression 43,44; the maintenance of cell cycle progression during HPV cancer oncogenesis is fundamental, and it is known that this is co-opted by HR HPV oncogenes E6 and E7.
Although our primary interest was to determine the differential expression of NFX1-123 and its regulated genes in the presence or absence of HPV in head and neck cancers, we also recognized that these genes could function as future targets for screening and diagnosis and could identify cellular pathways for novel treatment interventions. It is well established that HPV+ HNSCCs have a better prognosis and respond better to the treatments when compared to HPV− HNSCCs. It is interesting to consider ways in which clinicians could identify patients who merit different, or decelerated, treatments based on known prognostic, functional biomarkers. NFX1-123 has multiple protein motifs that define its function in cells. Those include a PAM2 motif, with which it binds cytoplasmic poly(A) binding proteins and regulates the mRNA of Notch1 and the catalytic subunit of telomerase, hTERT 15,19,21. NFX1-123 also includes a PHD/RING domain that has E3 ubiquitin ligase functions 14,45,46. We published that NFX1-123 bound the deubiquitinase interacting protein USP9X (also known as USP/FAFX) 15, and this interaction was recently confirmed 13. Interestingly, the expression of USP9X is increased by 16E6 47 and USP9X expression is also increased in cancers 48. These findings, as a whole, indicate the multilayered regulation of mRNA stability and protein degradation that could be orchestrated by NFX1-123 and 16E6. The functional consequences of these mechanisms of action in HPV-associated cancers are still being studied, but there are unique and common gene targets that may be leveraged in novel screening and treatment protocols.
Acknowledgements:
This study was supported by NIH R01 CA 172742 to R.A.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. International journal of cancer. 2017;141(4):664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndiaye C, Mena M, Alemany L, et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 2014;15(12):1319–1331. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhai L, Tumban E. Gardasil-9: A global survey of projected efficacy. Antiviral Res. 2016;130:101–109. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza G, Westra WH, Wang SJ, et al. Differences in the Prevalence of Human Papillomavirus (HPV) in Head and Neck Squamous Cell Cancers by Sex, Race, Anatomic Tumor Site, and HPV Detection Method. JAMA Oncol. 2017;3(2):169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–1435. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi AK, Graubard BI, Broutian T, et al. NHANES 2009-2012 Findings: Association of Sexual Behaviors with Higher Prevalence of Oral Oncogenic Human Papillomavirus Infections in U.S. Men. Cancer Res. 2015;75(12):2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Souza G, Agrawal Y, Halpern J, Bodison S, Gillison ML. Oral sexual behaviors associated with prevalent oral human papillomavirus infection. J Infect Dis. 2009;199(9):1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza G, Wentz A, Kluz N, et al. Sex Differences in Risk Factors and Natural History of Oral Human Papillomavirus Infection. J Infect Dis. 2016;213(12):1893–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnelle C, Whiteman DC, Porceddu SV, Panizza BJ, Antonsson A. Past sexual behaviors and risks of oropharyngeal squamous cell carcinoma: a case-case comparison. Int J Cancer. 2017;140(5):1027–1034. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Lu D, Gao J, et al. Identification of a USP9X Substrate NFX1-123 by SILAC-Based Quantitative Proteomics. Journal of proteome research. 2019. [DOI] [PubMed] [Google Scholar]
- 14.Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18(18):2269–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzenellenbogen RA, Egelkrout EM, Vliet-Gregg P, Gewin LC, Gafken PR, Galloway DA. NFX1-123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. J Virol. 2007;81(8):3786–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA. Human papillomavirus 16E6 and NFX1-123 potentiate Notch signaling and differentiation without activating cellular arrest. Virology. 2015;478:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chintala S, Levan J, Robinson K, Quist K, Katzenellenbogen RA. Genes Regulated by HPV 16 E6 and High Expression of NFX1-123 in Cervical Cancers. Onco Targets Ther. 2020;13:6143–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vliet-Gregg PA, Robinson KL, Levan J, Matsumoto LR, Katzenellenbogen RA. NFX1-123 is highly expressed in cervical cancer and increases growth and telomerase activity in HPV 16E6 expressing cells. Cancer Lett. 2019;449:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J Virol. 2009;83(13):6446–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levan J, Vliet-Gregg PA, Robinson KL, Katzenellenbogen RA. Human papillomavirus type 16 E6 and NFX1-123 mislocalize immune signaling proteins and downregulate immune gene expression in keratinocytes. PloS one. 2017;12(11):e0187514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA. NFX1-123 and human papillomavirus 16E6 increase Notch expression in keratinocytes. J Virol. 2013;87(24):13741–13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levan J, Vliet-Gregg PA, Robinson KL, Matsumoto LR, Katzenellenbogen RA. HPV type 16 E6 and NFX1-123 augment JNK signaling to mediate keratinocyte differentiation and L1 expression. Virology. 2019;531:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chintala S, Katzenellenbogen RA. NFX1, Its Isoforms and Roles in Biology, Disease and Cancer. Biology (Basel). 2021;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumban E A Current Update on Human Papillomavirus-Associated Head and Neck Cancers. Viruses. 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinstein AJ, Shay SG, Chang E, Lewis MS, Wang MB. Treatment outcomes in veterans with HPV-positive head and neck cancer. Am J Otolaryngol. 2017;38(2):188–192. [DOI] [PubMed] [Google Scholar]
- 26.Lee LA, Huang CG, Liao CT, et al. Human papillomavirus-16 infection in advanced oral cavity cancer patients is related to an increased risk of distant metastases and poor survival. PLoS One. 2012;7(7):e40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar CR, Smith RV, Garg MK, et al. Human papillomavirus-associated head and neck squamous cell carcinoma survival: a comparison by tumor site and initial treatment. Head Neck Pathol. 2014;8(1):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whang SN, Filippova M, Duerksen-Hughes P. Recent Progress in Therapeutic Treatments and Screening Strategies for the Prevention and Treatment of HPV-Associated Head and Neck Cancer. Viruses. 2015;7(9):5040–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsahafi E, Begg K, Amelio I, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10(8):540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun W, Duan T, Ye P, et al. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics. 2018;19(1):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kono T, Hoover P, Poropatich K, et al. Activation of DNA damage repair factors in HPV positive oropharyngeal cancers. Virology. 2020;547:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Loon K, Yemelyanenko-Lyalenko J, Margadant C, Griffioen AW, Huijbers EJM. Role of fibrillin-2 in the control of TGF-beta activation in tumor angiogenesis and connective tissue disorders. Biochim Biophys Acta Rev Cancer. 2020;1873(2):188354. [DOI] [PubMed] [Google Scholar]
- 34.Gopal S, Veracini L, Grall D, et al. Fibronectin-guided migration of carcinoma collectives. Nat Commun. 2017;8:14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koh LF, Ng BK, Bertrand J, Thierry F. Transcriptional control of late differentiation in human keratinocytes by TAp63 and Notch. Exp Dermatol. 2015;24(10):754–760. [DOI] [PubMed] [Google Scholar]
- 36.Lessard JC, Pina-Paz S, Rotty JD, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A. 2013;110(48):19537–19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Zhang R, Zhou S, Ji Y. Overexpression of Notch1 is associated with the progression of cervical cancer. Oncol Lett. 2015;9(6):2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues C, Joy LR, Sachithanandan SP, Krishna S. Notch signalling in cervical cancer. Exp Cell Res. 2019;385(2):111682. [DOI] [PubMed] [Google Scholar]
- 39.Rong C, Feng Y, Ye Z. Notch is a critical regulator in cervical cancer by regulating Numb splicing. Oncol Lett. 2017;13(4):2465–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khelil M, Griffin H, Bleeker MCG, et al. Delta-like ligand-Notch1 signalling is selectively modulated by HPV16 E6 to promote squamous cell proliferation and correlates with cervical cancer prognosis. Cancer Res. 2021. [DOI] [PubMed] [Google Scholar]
- 41.Fukusumi T, Califano JA. The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma. J Dent Res. 2018;97(6):645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaka AS, Nowacki NB, Kumar B, et al. Notch1 Overexpression Correlates to Improved Survival in Cancer of the Oropharynx. Otolaryngol Head Neck Surg. 2017;156(4):652–659. [DOI] [PubMed] [Google Scholar]
- 43.Confalonieri S, Quarto M, Goisis G, et al. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene. 2009;28(33):2959–2968. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida W, Tomikawa J, Inaki M, et al. An insulator element located at the cyclin B1 interacting protein 1 gene locus is highly conserved among mammalian species. PLoS One. 2015;10(6):e0131204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13(1):7–12. [DOI] [PubMed] [Google Scholar]
- 46.Jackson PK, Eldridge AG, Freed E, et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10(10):429–439. [DOI] [PubMed] [Google Scholar]
- 47.Rolen U, Kobzeva V, Gasparjan N, et al. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45(4):260–269. [DOI] [PubMed] [Google Scholar]
- 48.Lu Q, Zhang FL, Lu DY, Shao ZM, Li DQ. USP9X stabilizes BRCA1 and confers resistance to DNA-damaging agents in human cancer cells. Cancer Med. 2019;8(15):6730–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]


