Abstract
The flexibility and specificity of ubiquitin-dependent proteolysis are mediated, in part, by the E3 ubiquitin ligases. One class of E3 enzymes, SKp1/cullin/F-box protein (SCF), derives its specificity from F-box proteins, a heterogeneous family of adapters for target protein recognition. Grr1, the F-box component of SCFGrr1, mediates the interaction with phosphorylated forms of the G1 cyclins Cln1 and Cln2. We show that binding of Cln2 by SCFGrr1 was dependent upon its leucine-rich repeat (LRR) domain and its carboxy terminus. Our structural model for the Grr1 LRR predicted a high density of positive charge on the concave surface of the characteristic horseshoe structure. We hypothesized that specific basic residues on the predicted concave surface are important for recognition of phosphorylated Cln2. We show that point mutations that converted the basic residues on the concave surface but not those on the convex surface to neutral or acidic residues interfered with the capacity of Grr1 to bind to Cln2. The same mutations resulted in the stabilization of Cln2 and Gic2 and also in a spectrum of phenotypes characteristic of inactivation of GRR1, including hyperpolarization and enhancement of pseudohyphal growth. It was surprising that the same residues were not important for the role of Grr1 in nutrient-regulated transcription of HXT1 or AGP1. We concluded that the cationic nature of the concave surface of the Grr1 LRR is critical for the recognition of phosphorylated targets of SCFGrr1 but that other properties of Grr1 are required for its other functions.
Ubiquitin-mediated proteolysis is now widely recognized as an important regulatory process in eukaryotes. Its usefulness in biological regulation is due to the tremendous flexibility and specificity of the ubiquitination machinery. That machinery comprises an elaborate assemblage of components that includes the E3 ubiquitin ligases. Two classes of E3 complexes, the anaphase promoting complex and the Skp1/cullin/F-box protein (SCF) complex have come under intense scrutiny due largely to their involvement in regulation of the cell division cycle. Both factors are now understood to comprise modular sets of adapters that confer specificity on relatively nonspecific E2 ubiquitin-conjugating enzymes. Furthermore, both classes of regulatory complexes are highly conserved among eukaryotes (reviewed in reference 14). One key to understanding the regulation conferred by these factors is to understand the basis for substrate discrimination.
SCF complexes comprised of Skp1, Cdc53/cullin, Rbx1/Roc1, and an F-box protein associate with ubiquitin-conjugating enzymes (Ubcs) to mediate the interaction of SCF with specific substrates (21, 24, 48, 50). SCF comprises a family of protein complexes, each with a different F-box protein component. The nature of the F-box protein determines the specificity of the interaction between Ubc-SCF and its targets. Related complexes have been demonstrated in yeast, plant, and animal cells. Although 11 F-box proteins have been identified in the budding yeast genome most have yet to be assigned to a specific function. Three (Cdc4, Grr1, and Met30) have been shown clearly to associate with SCF (7, 42). SCF complexes containing each of those proteins have been shown to interact with a distinct group of ubiquitination targets. For example, SCFCdc4 is specifically required for the ubiquitination of the CDK inhibitors Sic1 (9, 49) and Far1 (13) and the replication protein Cdc6 (8). SCFGrr1 targets the G1 cyclins Cln1 and Cln2 (2, 42) and the putative Cdc42 effector Gic2 (17) for ubiquitination, and SCFMet30 has been shown to be involved in recognition of both the morphogenesis checkpoint kinase Swe1 (19) and transcription factor Met4 (18, 45). Involvement of F-box proteins as specificity factors for ubiquitination is not restricted to yeasts but appears to be a common theme among eukaryotes. Among the best-characterized mammalian F-box proteins are β-TRCP (reviewed in reference 22) and Skp2 (6, 33, 36, 52), which play roles in the recognition of IκBα and of p27 and E2F1, respectively. Thus, the E2-SCF ubiquitinating complexes comprise an evolutionarily conserved system that is widely exploited in the regulation of diverse biological processes.
Although there does not seem to be any consistent feature in the proteins targeted by SCF for ubiquitination, many of the targets that have been studied to date are phosphorylated. In some cases those proteins have been shown to be phosphorylated and to depend on phosphorylation for proteolysis. In several cases, the target proteins were shown to bind to SCF only in their phosphorylated state. Phosphorylation-dependent binding, ubiquitination, and degradation of Cln1 and Cln2 by SCFGrr1 (30, 42, 49, 55), of Sic1 by SCFCdc4 (9, 49), and of IκBα by SCFβTcRP (reviewed in reference 22) have been demonstrated using a combination of in vivo mutational analysis and in vitro reconstituted ubiquitination systems. In each case, targeting of the proteins for ubiquitination has been shown to depend on a specific F-box protein and the phosphorylation state of the protein. Thus, ubiquitination of specific targets not only depends on the identity of that target but also on its state.
In addition to their involvement in targeting proteins for ubiquitin-mediated proteolysis, there are a number of examples of the involvement of F-box proteins in transcriptional regulation. Although in some cases SCF-dependent ubiquitination and even proteolysis may be involved, in most cases the mechanism has yet to be established. Met30 has been shown to be important for repression of MET gene expression (51), and Grr1 is required for induction of the expression of hexose and amino acid permeases encoded by the HXT genes (reviewed in reference 38) and AGP1 (15), respectively. Although Met30 clearly regulates MET gene expression via SCF-dependent ubiquitination of the transcription factor Met4, ubiquitin-mediated proteolysis appears to be unimportant for that regulation (18; but see reference 45). The role of Grr1 in nutrient-regulated transcription in yeasts is even less clear. Induction of the HXT1 gene in response to glucose is defective in grr1Δ mutants. That defect is suppressed by inactivation of the transcriptional repressor, Rgt1 (39–41, 53). This has led to the hypothesis that Grr1 inactivates Rgt1 or a coregulator in response to glucose (10, 31), perhaps via ubiquitin-mediated proteolysis. Grr1 has also been shown to be required for transcriptional induction of the amino acid permease gene AGP1 in the presence of exogenous amino acids. However, because the relevant transcription factors have not been identified, neither the targets nor the precise role of Grr1 in that process has been established.
These and other observations clearly define a role for F-box proteins in determining the specificity of SCF-target interactions. However, the basis for that specificity and the nature of the protein-protein interaction sites remain to be elucidated. This class of proteins is characterized by the presence of an F box. Although their other features are less conserved, many contain motifs recognized as protein-protein interaction domains conserved throughout biological systems. The leucine-rich repeat (LRR) is one such motif (reviewed in reference 26). An LRR domain is comprised of multiple LRRs, each consisting of a beta strand and an alpha helix separated by a variable region, which all fold into a horseshoe structure that forms a parallel beta sheet on the concave surface with helices on the convex surface. These domains are found in a variety of proteins of disparate function, including the F-box proteins Grr1 and Skp2.
We investigated the LRR domain of Grr1 as a potential site for target recognition. Grr1 contains 12 complete LRRs and 1 partial LRR belonging to the LRR cysteine-containing subfamily. We investigated whether the LRR domain of Grr1 interacts with its substrates and characterized the basis of the specificity of Grr1 for phosphorylated substrates. As was previously shown (23, 31), we found that the LRR region is essential for the functioning of Grr1 and its ability to bind target proteins. Molecular modeling of the Grr1 LRR revealed an unusually high density of cationic charges on the concave surface of the horseshoe. Based on that model, we showed that those positively charged residues are important for binding phosphorylated G1 cyclin. In contrast, the same residues were shown not to be important for assembly of the SCFGrr1 complex. The inability of Grr1 to bind phosphorylated targets resulted in their stabilization and in phenotypes consistent with inactivation of SCFGrr1. However, the same mutations had no effect on some of the other Grr1-dependent functions. We concluded that the positively charged surface of the LRR is critical for the recognition of at least one class of phosphorylated SCFGrr1 targets.
MATERIALS AND METHODS
Yeast strains and culture.
Yeast strains used are listed in Table 1. All strains were isogenic with 15 Daub, W303a, or Σ1278b, as indicated. Culture conditions and medium were as indicated and were prepared by standard methods.
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Source |
|---|---|---|
| CWY231 | 15Daub | 44 |
| YHY209 | 15Daub his2::GAL1-CLN2-FLAG::HIS2 | S. Lanker |
| K699 | W303 | K. Nasmyth |
| YHY565 | W303 GRR1-6His::KanMX2 | This study |
| YHY284 | W303 grr1::LEU2 | This study |
| YHY597 | W303 grr1ΔL-6His::KanMX2 | This study |
| YHY599 | W303 grr1ΔC-6His::KanMX2 | This study |
| YHY668 | W303 grr1-B4Q-6His::KanMX2 | This study |
| YHY602 | W303 grr1-B4E-6His::KanMX2 | This study |
| YHY654 | W303 grr1-R485Q-6His::KanMX2 | This study |
| YHY600 | W303 grr1-R680Q-6His::KanMX2 | This study |
| YHY670 | W303 grr1-R680E-6His::KanMX2 | This study |
| YHY643 | W303 grr1-K498E-6His::KanMX2 | This study |
| YHY604 | W303 grr1-K498E R550E-6His::KanMX2 | This study |
| YHY671 | W303 grr1-R680Q R709Q-6His::KanMX2 | This study |
| YHY672 | W303 grr1-R680E R709E-6His::KanMX2 | This study |
| YHY285 | W303 leu2::GAL1-CLN2::LEU2 | This study |
| YHY595 | W303 leu2::GAL1-CLN2::LEU2 grr1ΔL-6His::KanMX2 | This study |
| YHY669 | W303 leu2::GAL1-CLN2::LEU2 grr1-B4Q-6His::KanMX2 | This study |
| YHY603 | W303 leu2::GAL1-CLN2::LEU2 grr1-B4E-6His::KanMX2 | This study |
| L5366 | MATa/α ura3-52/ura3-52 | 28 |
| RLVsc504 | MATa/α ura3-52/ura3-52 leu2::hisG/leu2::hisG his3::hisG/HIS3; grr1::LEU2/grr1::LEU2 | This study |
| RLVsc485 | MATa/α ura3-52/ura3-52 leu2::hisG/LEU2 his3::hisG/HIS3; grr1ΔL::KanMX2/grr1ΔL::KanMX2 | This study |
| RLVsc486 | MATa/α ura3-52/ura3-52 leu2::hisG/LEU2 his3::hisG/HIS3; grr1ΔC::KanMX2/grr1ΔC::KanMX2 | This study |
| RLVsc492 | MATa/α ura3-52/ura3-52 leu2::hisG/LEU2 his3::hisG/HIS3 grr1-B4E::KanMX2/grr1-B4E::KanMX2 | This study |
| HLY876 | MATa/α ura3-52/ura3-52 leu2::hisG/leu2::hisG; trp1::hisG/trp1::hisG cln1::ura3::TRP1/cln1::ura3::TRP1; cln2::LEU2/cln2::LEU2 | 32 |
| HLY1540 | MATa/α ura3-52/ura3-52 leu2::hisG/leu2::hisG; his3::hisG/his3::hisG grr1::LEU2/grr1::LEU2 cln1::ura3::HIS3/cln1::ura3::HIS3 cln2::LEU2/cln2::LEU2 | 32 |
Pseudohyphal growth was evaluated on synthetic low-ammonia dextrose plates (SLAD) which contained 50 μM ammonium sulfate, 6.8 g of yeast nitrogen base per liter without amino acids or ammonium sulfate, 2% dextrose, and 2% washed agar (12). Agar was washed five times as a 2% (wt/vol) suspension with deionized water for 30 min per wash. After the final wash, the agar was sterilized by autoclaving at 4% (wt/vol) in deionized water and diluting to a 2% (wt/vol) final concentration with 2× filter-sterilized liquid media. Yeast extract, peptone, and yeast nitrogen base were from Difco Laboratories, and agar was from Angus. Other reagents were obtained from Sigma Chemical Co.
Plasmids.
Plasmids used in this study are listed in Table 2. Full-length GRR1 was cloned by adding an NcoI-NdeI fragment encompassing the first 840 nucleotides generated by PCR into pADH1-grr1ΔN (31). All pADH1-derived plasmids contained a single hemagglutinin (HA) epitope at the amino-terminal end of the GRR1 open reading frame. The grr1 point mutants were constructed by site-directed mutagenesis with the pALTER mutagenesis system (Promega). Fragments encompassing the mutated site or sites were subcloned into pADH1-GRR1 for use in interaction studies and into the pKAN-6His vector (modified from pKHA3) (Kanr) (35; S. B. Haase, M. Wolff, and S. I. Reed, unpublished results) for targeted integration into the GRR1 locus. grr1ΔL was constructed by deleting the EcoRV-StuI fragment encoding amino acids 447 to 754 from the open reading frame, whereas grr1ΔC was constructed by adding the first 840 nucleotides of the open reading frame of GRR1 to a grr1ΔNC construct that was generated by PCR (nucleotides 840 to 2700, amino acids 280 to 900). grr1ΔNCF was generated by PCR and included nucleotides 1171 to 2700, which encode amino acids 391 to 900. All DNA fragments generated by PCR were sequenced to confirm fidelity prior to use.
TABLE 2.
Plasmids used in this study
| Plasmid | Parent and insert | Source |
|---|---|---|
| YCplac33 | YCplac33 | 11 |
| pYH210 | pADH1-GRR1-HA | This study |
| pBM3425 | pADH1-grr1ΔN-HA | 31 |
| pYH460 | pADH1-grr1ΔC-HA | This study |
| pYH475 | pADH1-grr1ΔL-HA | This study |
| pYH233 | pADH1-grr1ΔNCF-HA | This study |
| pYH 340 | pADH1-grr1-K498Q-HA | This study |
| pYH 336 | pADH1-grr1-R550Q-HA | This study |
| pYH 353 | pADH1-grr1-R680Q-HA | This study |
| pYH 350 | pADH1-grr1-R709Q-HA | This study |
| pYH 374 | pADH1-grr1-K498Q R550Q-HA | This study |
| pYH 338 | pADH1-grr1-R680Q R709Q-HA | This study |
| pYH 499 | pADH1-grr1-B4Q-HA | This study |
| pYH 426 | pADH1-grr1-R485Q-HA | This study |
| pYH 357 | pADH1-grr1-K498E-HA | This study |
| pYH 379 | pADH1-grr1-R550E-HA | This study |
| pYH 345 | pADH1-grr1-R680E-HA | This study |
| pYH 352 | pADH1-grr1-R709E-HA | This study |
| pYH 341 | pADH1-grr1-K498E R550E-HA | This study |
| pYH 339 | pADH1-grr1-R680E R709E-HA | This study |
| pYH 380 | pADH1-grr1-B4E-HA | This study |
| pYH 428 | pADH1-grr1-R485E-HA | This study |
| pYH 378 | pADH1-grr1-Y655A-HA | This study |
| pYH 430 | pADH1-grr1-K622E-HA | This study |
| pPK102 | YCp-CUP1p-SKP1-GST | 19 |
| PMJ383 | YCp-GAL1-GIC2-GFP | 17 |
| pYH226 | YCp-CDC53-6Xmyc | This study |
Antibodies.
The 12CA5 anti-HA monoclonal antibody was derived from ascites fluid provided by I. Wilson, The Scripps Research Institute, La Jolla, Calif. The antibody was conjugated to protein A-Sepharose for use in immunoprecipitation experiments. Affinity-purified polyclonal anti-Cln2 (57) and anti-Cdc28 (56) antibodies were prepared as previously described. Anti-6His (Quiagen), anti-myc (Santa Cruz Biotechnology) and anti-HA (Babco) were obtained commercially. Polyclonal anti-glutathione S-transferase (anti-GST) was a generous gift from S. Reed (The Scripps Research Institute).
Preparation of yeast cell extracts.
Exponentially growing yeast cells were pelleted, washed with cold 1× phosphate-buffered saline, and resuspended in yeast lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40, 10% glycerol) containing 50 mM sodium fluoride, protease inhibitors (0.4 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml, and 1 μg of aprotinin per ml), and phosphatase inhibitors (0.1 mM sodium orthovanadate, 5 mM EDTA, 5 mM EGTA, and 10 mM sodium pyrophosphate). Cells were lysed with glass beads by using seven cycles of vortexing for 1 min followed by a 1-min incubation on ice. Extracts were collected after centrifugation for 15 min at 14,000 rpm in an Eppendorf S417R microcentrifuge. The protein concentrations of the lysate were determined by the Bio-Rad protein assay (Bio-Rad Laboratories).
Immunoprecipitation and immunoblotting.
Immunoprecipitation was carried out by incubating 750 to 1,000 μl of yeast extract containing 0.5 to 1.0 mg of total yeast protein with anti-HA (12CA5) antibody conjugated on protein A-Sepharose beads (Sigma) for 1 h at 4°C with gentle rocking. The beads were then washed three times with 1 ml of yeast lysis buffer, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and resolved by SDS-PAGE. For immunoblotting, proteins were transferred to Imobilon membranes (Millipore Corp.) by semidry transfer at 200 mA for 2 to 6 h. The proteins were detected using Super Signal (Pierce). Dilutions of the antibodies for Western blots were 1:1,000 for anti-Cln2 and anti-6His antibodies (Quiagen), 1:2,000 for anti-HA (Babco) and anti-myc antibodies (Santa Cruz Biotechnology), 1:5,000 for anti-GST antibody, and 1:10,000 for anti-Cdc28 antibody.
Determination of the stability of Cln2 and Gic2.
For determination of Cln2 and Gic2-green fluorescent protein (GFP) half-life by promoter shut-off, cell strains were grown in 2% raffinose until optical density at 600 nm (OD600) was 0.5 to 0.8. Galactose was added to a concentration of 2% to induce expression driven by the GAL1 promoter for 45 min in the case of CLN2 and for 3 h in the case of GIC2. Glucose was added to a concentration of 2% to repress the expression. Samples were taken before the induction and at the indicated time following repression by glucose. Cln2 abundance was assayed by immunoblotting with anti-Cln2 antibody (57), and Gic2-GFP abundance was assayed using an anti-GFP antibody (Boehringer-Mannheim).
Structural modeling of the LRR of Grr1.
Coordinates used for three repeats of the LRR from Grr1 were those from the previous model of Kajava (20). Those repeats contain three β-strands covered on one face by two α-helices. Side chains of the three repeats were replaced by those of the Grr1 molecule using XFIT (34), and their positions were energy minimized using X-PLOR 3.8 (5) with the CHARMM22 all-atoms force field (4). To assemble the 12 repeats of Grr1, we superimposed the Grr1 repeats on the crystal structure of the RNase inhibitor (27) using InsightII (MSI, Inc., San Diego, Calif.). Side chains were replaced by XFIT and subsequently energy minimized in X-PLOR. Finally, two insertions required in the 6th and 12th repeats and a deletion in the 11th repeat were carried out in TURBO-FRODO (46). Insertions increased the length of both C-terminal helices by almost a turn (three residues). This model was optimized by several rounds of refinement using TURBO-FRODO and X-PLOR.
RT-PCR.
Grr1 mutant strains were grown in 2% galactose in rich medium to mid-log phase. Glucose was added to half of the culture to a 4% concentration, and cells were collected 90 min after induction of glucose. For AGP1 reverse transcriptase PCR (RT-PCR), only the glucose-induced cells were used.
Total yeast cell RNA was isolated using the RNeasy minikit (Qiagen) as recommended by the manufacturer. The first strand of cDNA was synthesized from 3 μg of total RNA by using SuperScript II reverse transcriptase (RT) (Life Technologies) with oligo(dT)12–18. A PCR program with 94°C for 3 min, followed by 20 cycles of 1 min at 94°C, 1 min at 54°C, and 2 min at 72°C was performed to analyze the level of AGP1, HXT1, and ACT1 in each cDNA preparation. A PCR from equal amounts of RNA (without RT) was also performed to check DNA contamination, if any. Amplified products were analyzed by gel electrophoresis. Different amounts of cDNA were tested for PCR to make sure that 20 cycles of amplification quantitatively represented the level in each of the cDNA samples. The results shown in Fig. 8 below were PCR amplified from one-fortieth of each cDNA preparation.
FIG. 8.
Nutrient-regulated transcription is not affected by mutations in LRR of Grr1. (A) The capacity of wild-type cells or cells carrying the indicated grr1 mutation to induce HXT1 gene expression was evaluated by analysis of the HXT1 mRNA. Total RNA was isolated from strains carrying the indicated GRR1 allele either prior to (G, galactose) or following 90 min of induction with 4% glucose (D, dextrose). The HXT1 mRNA level in each strain was determined using RT-PCR. PCRs for ACT1 were carried out using an equal amount of RNA to ensure equal loading. Samples were performed without reverse transcriptase (-RT) to evaluate the extent of DNA contamination in each RNA preparation. (B) The expression of AGP1 in wild-type cells and cells carrying the indicated grr1 allele under inducing conditions in rich medium. AGP1 expression was analyzed (left panel) by RT-PCR, and (right panel) Northern hybridization. AGP1 and ACT1 transcript levels were determined by RT-PCR (as described for panel A) and Northern hybridization using RNA isolated from glucose-grown cells.
The primers used were as follows: AGP1, 5′-CGTCGTCGAAGTCTCTATACG-3′ and 5′-GGTCCGTTCCTCAAACGTTCCC-3′; HXT1, 5′-GGAATCTGGTCGTTCAAAGGCC-3′ and 5′-GGTTGGTCATCATGCATTAGG-3′; and ACT1, 5′-GAAGCTCAATCCAAGAGAGG-3′ and 5′-GAGGAGCAATGATCTTGACC-3′.
Filamentous assay conditions.
To evaluate pseudohyphal development, cells were pregrown for 2 days on synthetic minimal (SD) medium at 30°C and then transferred to SLAD. To avoid disturbing the agar surface and the colony density-dependent inhibition of pseudohypha formation (58), single unbudded cells were carefully placed 1 cm from each other by using the needle of a dissecting microscope. Between 20 and 100 colonies, each derived from a single unbudded cell, were analyzed for each strain. Pseudohyphal growth was evaluated by multiple criteria. First, colony and cell morphologies were monitored after 1 day of growth on SLAD plates (an assay of the extent of early morphological differentiation). Next, the cell and colony morphologies were evaluated after 5 days of growth on SLAD before (total growth) and after mechanically washing the noninvasive cells from the plate surface (invasive growth).
Microscopy and imaging.
Cells from suspension cultures were imaged with differential interference contrast (DIC) optics on a Nikon Eclipse E800 microscope using IPLAB Spectrum software and a Photmometrix Quantix charge-coupled device camera. Images were processed for publication with Adobe Photoshop software.
Microcolonies and colonies growing on plates were imaged from below through the agar and plastic petri dish, by using a Nikon Labophot microscope. Pixera VCS image acquisition software and a Pixera charge-coupled device camera were used to capture images at a resolution of 1,280 by 1,024 pixels. Images were processed for publication with Adobe Photoshop software.
RESULTS
Protein-protein interaction domain containing LRR and additional domains in the C terminus are required for Cln2 binding.
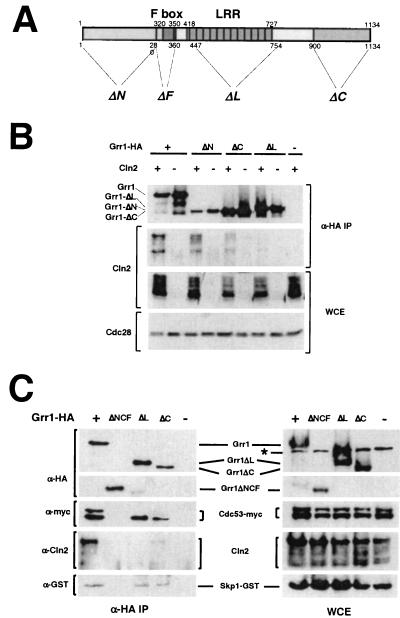
The interaction between the E2-E3 complex Cdc34-SCF and its substrates depends on both the nature of the F-box protein component of the complex and the identity and phosphorylation state of the substrate. The interaction of Grr1 with phosphorylated G1 cyclins Cln1 and Cln2 can be demonstrated in vivo (23, 55) (Fig. 1B) as well as in vitro (49, 50). Those complexes also contain SCF components including Cdc53 and Skp1 (1, 9, 42, 49, 55) (Fig. 1C). We have evaluated the importance of various Grr1 domains (Fig. 1A) for the interaction between Grr1 and Cln2 by analyzing the abundance of Cln2 present in immune complexes prepared from extracts of yeast cells expressing wild-type and mutant forms of epitope-tagged Grr1 (Grr1-HA) from the ADH1 promoter. In agreement with previous analyses of these same deletion mutants with the yeast two-hybrid assay (31), we found that deletion of the amino-terminal 280 amino acids of Grr1 (Grr1ΔN) caused only a modest defect in the capacity to specifically interact with Cln2 protein in vivo (Fig. 1C). In contrast, a more dramatic effect was caused by deletion of either the C-terminal 234 amino acids (Grr1ΔC) or the 308-amino-acid domain that comprises the majority of the LRRs (Grr1ΔL). Deletion of the carboxy terminus results in a dramatic reduction in binding to Cln2 compared to that observed with wild-type Grr1. However, the small amount of Cln2 protein that forms a complex with Grr1ΔC, like the protein binding to wild-type Grr1, is primarily in the phosphorylated form. Finally, Grr1 lacking the predicted LRR region was completely deficient in Cln2 binding (Fig. 1B), consistent with a previous report (23). Based on this analysis, it appears that both the LRR and, to a somewhat lesser extent, the carboxy-terminal sequences of Grr1 are important in the interaction of Grr1 with its substrates.
FIG. 1.
(A) Domain structure of Grr1 and positions of deletion mutations. Domains are indicated across the top of the diagram, and positions of the deletion mutations are indicated across the bottom of the diagram. Residue numbers are indicated for the end points of the deletions. (B) Comparison of Cln2 binding to full-length and domain deletion mutations of Grr1. Anti-HA immunoprecipitates were prepared using extracts (WCE) from 15Daub strains expressing epitope-tagged CLN2 from the GAL1 promoter and wild-type or deletion mutation forms of GRR1-HA from the ADH1 promoter. Components of the extracts and immune complexes were evaluated using either anti-HA (12CA5), anti-Cln2, or anti-Cdc28 antibodies. (C) Interaction of Grr1 derivatives with Cln2 and the SCF components, Skp1 and Cdc53. Anti-HA immunoprecipitates were prepared using extracts (WCE) from 15Daub strains carrying CDC53-myc, SKP1-GST expressed from the CUP1 promoter, GAL-CLN2-FLAG, and wild-type or deletion mutation forms of GRR1-HA from the ADH1 promoter. Components of the extracts and immune complexes were visualized using either anti-HA (12CA5), anti-Cln2, anti-myc, or anti-GST antibodies.
The finding that the LRR is essential for Cln2 binding suggested that it might also be sufficient for Cln2 binding. We therefore prepared a construct having just the region encoding the LRR and an HA tag under the ADH1 promoter. We then evaluated the ability of this construct to bind to Cln2 in vivo by coimmunoprecipitation. Although the isolated LRR was stable when expressed in yeasts and accumulated to a level comparable to that of full-length Grr1, it was not capable of binding to Cln2 (data not shown).
There is some evidence that assembly of an SCF complex is required for the interaction of F-box proteins with their targets (49). Thus, we evaluated whether the failure of some of these mutant proteins to interact with phosphorylated Cln2 was a consequence of their failure to form a productive SCF complex. The HA-tagged Grr1 constructs were introduced into cells expressing Cdc53-myc and GST-Skp1, and anti-HA immune complexes were analyzed for the presence of the appropriate tagged protein. Both Cdc53 and Skp1 were present in the immune complexes containing Grr1ΔN (data not shown), Grr1ΔC, Grr1ΔL, and wild-type Grr1, whereas Grr1ΔNCF failed to interact with either Cdc53 or Skp1 (Fig. 1C) (31). Despite their ability to interact with SCF components, both Grr1ΔL and Grr1ΔC differed from the wild type and Grr1ΔN in a couple of ways. First, they exhibited a modest but reproducible reduction in the amount of Skp1 that was bound (Fig. 1C, lanes ΔL and ΔC). This was more apparent in the case of Grr1ΔL, since this form was more abundant in the immune complexes. A more striking observation was the failure of Grr1ΔL and Grr1ΔC to associate with the modified, low-mobility form of Cdc53. The lower mobility form of Cdc53, which was present in the whole-cell extracts (Fig. 1C, lanes ΔL and ΔC), has been shown to be covalently modified with the ubiquitin-like polypeptide Rub1 (29, 55). Although SCFCdc4 stability appears to be affected by inactivation of RUB1 (29), neither the relevance of the specific defect of Grr1 binding to Rub1-derivatized Cdc53 nor the importance of Rub1 derivatization for the integrity or function of the SCFGrr1 complex has been clearly established.
We concluded that although the elimination of the LRR of Grr1 affected the nature of the SCFGrr1 complexes formed, it did not inhibit the capacity to form those complexes. Nevertheless, the complexes that did form failed to bind to phosphorylated Cln2. Elimination of the carboxy-terminal domain of Grr1 had a similar but less dramatic effect on the capacity of Grr1 to bind to phosphorylated Cln2. Subsequent analysis demonstrated that mutations in the LRR domain, which had no effect on the binding of Grr1 to Rub1-modified Cdc53, were also defective in binding to phosphorylated Cln2 (see below). Therefore, we consider it likely that the defect in the LRR domain of Grr1ΔL is sufficient to explain the inability of that protein to bind to Cln2.
Predicted structure of LRR of Grr1 reveals a potential binding site for phosphorylated substrates.
The apparent importance of the LRR for the interaction between Grr1 and phosphorylated Cln2 as well as the impact of LRR deletion on formation of SCFGrr1 prompted us to analyze this motif in greater detail. Consistent with the observations described above, LRRs in a number of proteins have been shown to be important for protein-protein interactions. Structural models for a number of the subfamilies of LRRs have been proposed based on the conservation of the motif between members of those subfamilies (20, 26) and the crystal structure of the porcine ribonuclease inhibitor (RI) (25–27). The basic repeat units of a number of distinguishable classes of LRRs have been defined. The Grr1 LRRs fall into the cysteine-containing LRR family (20). Using that repeat, we derived an alignment for the 12 LRRs of Grr1 (Fig. 2A). This repeat structure differed somewhat from previously suggested repeat alignments for Grr1, but it was most consistent with the repeat unit derived from structural modeling by Kajava (20) for the cysteine-containing family.
FIG. 2.
(A) Alignment of LRRs. The LRRs of Grr1 used to construct the structural model are shown. Residues indicated in green are those that are generally conserved between repeats. The residues that have been altered in the mutations presented in this study are depicted in red letters. Structural properties (β-sheet by the arrow; α-helix by the helix) of the repeats are indicated by the diagram at the top. (B) Space-filling model. Structural prediction for the LRR of Grr1. A tube diagram is overlaid on a space-filling model derived for the LRR domain comprised of the residues presented in panel A. Amino acid side chains are shown only for those residues that have been altered by mutation. Red and blue surfaces of the space-filling model represent negatively and positively charged regions of the surface of that domain, respectively.
A structure for the Grr1 LRR was predicted based on the X-ray crystal structure of the porcine RI complex (27) and the modeling of Kajava (20) for cysteine-rich LRRs. Several properties of the predicted structure (Fig. 2B) are immediately apparent. First, the 12 LRRs form a horseshoe-like structure with a concave surface formed by the parallel packing of the β-sheets and a convex surface formed by the α-helices. Strikingly, the concave surface contains a high density of basic residues, whereas the convex surface has a random distribution of charged residues. The electrostatic surface potential of the LRR was calculated at the solvent-accessible surface (1.4 Å away from the molecular surface), using a physiological ionic strength (150 mM), and mapped back onto the molecular surface. This process was designed to reveal the electrostatic potential actually experienced by atoms that come into contact with the surface of the LRR. The positively charged electrostatic potential was enhanced by the depth of the concave region of LRR (reduced solvent-accessible area) and by a relatively sparse distribution of negatively charged residues (Fig. 2A). Together, these properties resulted in this strong positive electrostatic potential.
Although the general shape predicted for the Grr1 LRR was a conserved characteristic of LRR structures, the basic character of the concave surface was not conserved. This property of the Grr1 LRR immediately suggested a model in which the positive surface potential of the concave surface is involved in the interaction with the negatively charged phosphate residues of the phosphorylated target. This idea becomes even more attractive when one realizes that the interaction between ribonuclease and the LRR of the RI involves a substantial contribution of its concave surface.
Basic residues localized on the putative binding surface but not the opposite surface affect binding of SCFGrr1 to Cln2.
The structure of the Grr1 LRR derived from computational modeling indicates an abundance of basic residues on the concave surface. In contrast, a relatively random distribution of charged residues was observed on the convex surface predicted by the model. This is particularly striking when considered in the context of the role of Grr1 as an adapter for recognition of phosphorylated targets by SCF. At least in the case of Cln2, those phosphorylated residues appear to be clustered (30; S. Lanker, unpublished data). Based on the predicted structure, we hypothesized that either the basic character of the concave surface or the specific distribution of basic residues on that surface determines the capacity of SCFGrr1 to recognize phosphorylated Cln2.
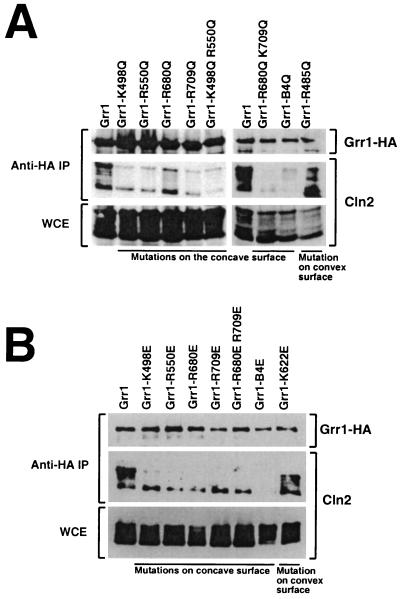
To examine the role of the positively charged residues on the concave surface of the Grr1 LRR, we undertook site-directed mutagenesis of several of these residues, choosing four basic residues, K498 (LRR 4), R550 (LRR 6), R680 (LRR 11), and R709 (LRR 12). These residues were predicted to project prominently into the pocket formed by the concave surface of the LRR region and therefore were expected to make a substantial contribution to the basic character of that surface. Mutations changing each of those residues to either neutral glutamine (Q) or acidic glutamate (E) were introduced into Grr1 alone or in combination. In addition to these four basic residues, mutations in two additional residues were also constructed (Fig. 2). Glutamine (Q) or glutamate (E) was introduced in place of the R485 (LRR 3) and K622 (LRR 8) basic residues, which were predicted by our model to reside on the convex surface of the LRR and therefore to be unimportant for binding phosphorylated substrates.
Each of the mutant grr1 genes was tagged with an HA epitope, placed under control of the constitutive ADH1 promoter and introduced into an otherwise wild-type yeast strain. The mutant forms of Grr1-HA were all expressed and accumulated to a level that was comparable to that of similarly expressed wild-type Grr1-HA (Fig. 3 and data not shown). The capacity of the mutated Grr1-HA proteins to bind phosphorylated Cln2 was evaluated by coimmunoprecipitation from extracts of cells expressing Cln2 from the inducible GAL1 promoter. All of the mutant proteins that had lesions predicted to lie on the concave surface of the LRR were compromised in their capacity to bind Cln2 relative to wild-type Grr1, whereas those proteins involving residues on the convex surface were largely unaffected (Fig. 3 and data not shown). Nevertheless, there was some variability in the Cln2 binding efficiency observed between the mutant Grr1 proteins. First, binding of Cln2 to those Grr1 mutant proteins with glutamate (E) in place of the basic residue (Fig. 3B) was more severely affected in every case than binding to those with glutamine (Q) in the same position (Fig. 3A). This is consistent with our model, since the presence of like charges on both Grr1 and the phosphorylated substrate is expected to be repulsive. Next, some of the single-residue mutants were less severely affected than others. For example, the R680Q mutant retained approximately half of the wild-type level of Cln2 binding (Fig. 3A). It is interesting that, according to our computational model, R680 was predicted to participate in a salt bridge with D657 (data not shown) and may therefore be less available for interactions with a charged ligand. Mutants having more than one mutated residue were, in general, more severely affected than those carrying single-site mutations. In fact, the mutant in which all four basic residues were replaced with glutamate (Grr1-B4E) was completely defective in the ability to bind Cln2. This included the capacity to bind the highest mobility form of Cln2, which is presumably unmodified. In contrast, using the other mutants that had glutamate in place of the basic residues, we observed binding of the highest-mobility form of Cln2. We are uncertain whether this form actually binds or is formed during preparation of the samples, but it was clear that, whatever the source, it was without consequence in terms of Cln2 instability (see below). Finally, consistent with our prediction, mutations altering basic residues on the convex surface (R485 and K622) had little or no effect on Cln2 binding relative to wild-type Grr1 regardless of whether the replacement was with glutamine or glutamate (Fig. 3 and data not shown).
FIG. 3.
Analysis of Cln2 binding by Grr1 with alterations in the basic residues of the LRR. (A) Cln2 binding by Grr1 with basic residues of the LRR replaced by glutamine (Q). Anti-HA immunoprecipitates were prepared from extracts (WCE) of strains expressing wild-type or mutant GRR1-HA expressed from the ADH1 promoter and CLN2 expressed from the GAL1 promoter. Immunoblots were probed with the anti-HA or anti-Cln2 as indicated. Grr1-B4Q represents the protein with all four basic residues on the concave surface of the LRR converted to glutamine. (B) Interaction of Grr1 point mutants with glutamate replacement of basic residues with Cln2. This experiment was similar to that shown in panel A, except that the basic residues in the LRR of Grr1 were replaced by glutamate (E) instead of glutamine. Grr1-B4E represents the protein with all four basic residues on the concave surface of the LRR converted to glutamate.
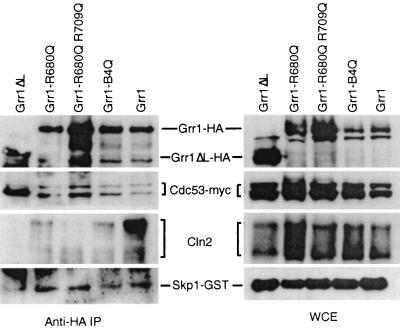
Since both the grr1ΔLRR and grr1ΔC mutations resulted in some alterations in the nature and extent of binding to SCF components (Fig. 1), it was conceivable that the point mutants in this region might be similarly affected. Therefore, we evaluated the capacity of these mutant proteins to assemble into SCF complexes. The constructs carrying both glutamine and glutamate replacements of basic residues were introduced into cells carrying Myc epitope-tagged Cdc53 and GST-Skp1, and their capacity to bind Grr1 was evaluated by coimmunoprecipitation (Fig. 4). All of the Grr1 point mutants that were tested bound to both Cdc53 and Skp1 at levels comparable to that of wild-type Grr1. Unlike the Grr1ΔL protein, which is completely deficient in the LRR, the point mutants bound to both the unmodified and the Rub1-modified forms of Cdc53 efficiently (Fig. 4). Thus, the capacity of Grr1 to interact with modified Cdc53 is independent of the presence of basic residues in the LRR and is not strictly associated with the inability of Grr1 to interact with Cln2. Together, these observations support our hypothesis that charged interactions on the concave surface of the LRR but not those on the convex surface are required for the interaction between Grr1 and Cln2.
FIG. 4.
Interaction of Grr1 point mutant proteins with Skp1, Cdc53, and Cln2. Anti-HA immunoprecipitates were prepared from strains expressing the wild type or the indicated mutation of GRR1-HA from the ADH1 promoter along with SKP1-GST expressed from the CUP1 promoter, CDC53-myc, and GAL-CLN2, as described in the legend to Fig. 1C.
grr1 mutants deficient in Cln2 binding are also defective in proteolysis of Cln2.
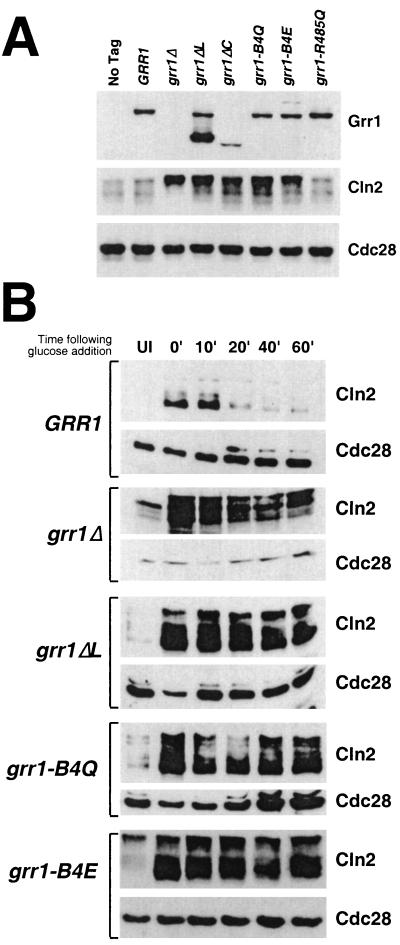
To obviate problems associated with the use of overexpressed proteins, single copies of each of the deletion mutant constructs as well as several of the point mutant constructs were introduced into cells under control of the native GRR1 promoter by targeted integration. Each of the point mutant products accumulated to levels equivalent to or modestly higher than that of the wild-type protein. The grr1ΔL product accumulated to a level that is approximately two- to threefold higher than that of the wild-type protein when expressed from that promoter (Fig. 5A). A similar increase relative to the wild-type protein was observed when grr1ΔL was expressed in multiple copies from the ADH1 promoter (Fig. 1C). In contrast, the Grr1ΔC protein accumulated to approximately one-half the level of the wild-type Grr1 (Fig. 5A; data not shown). It is likely that the altered accumulation of these proteins is a consequence of their altered stability, which is known to be dependent on SCF function (59). However, we have not evaluated the stability of these Grr1 derivatives.
FIG. 5.
(A) Accumulation of Grr1-6His and Cln2 proteins in various point and deletion mutations of GRR1. Twenty-five micrograms of whole-cell extract from asynchronously growing cells expressing the wild type or the indicated GRR1-6His form from its endogenous promoter were analyzed by immunoblotting using anti-His, anti-Cln2, and anti-Cdc28 antibodies, respectively. (B) Stability of Cln2 in cells having point and deletion mutations in Grr1. Cells carrying CLN2 under the inducible GAL1 promoter and wild-type GRR1 or its derivatives (grr1Δ, grr1ΔL, grr1-B4Q, and grr1-B4E) under its native promoter were pregrown under noninducing conditions (2% raffinose). GAL1-CLN2 was expressed for 45 min by addition of 2% galactose and then repressed by addition of 2% glucose. Extracts prepared from samples taken at the times indicated were analyzed by immunoblotting using anti-Cln2 and anti-Cdc28 antibodies. UI, uninduced.
An effect of the grr1 mutations on Cln2 stability was immediately suggested by the abundance of the Cln2 protein in the various grr1 mutant strains. Each of the strains shown to be deficient in Cln2 binding also had an elevated steady-state level of Cln2 (Fig. 5A). In contrast, the grr1-R485Q mutant, the product of which we showed binds to Cln2 efficiently, accumulates Cln2 protein to the same extent as wild-type cells.
We analyzed the stability of Cln2 in strains expressing the various deletion and point mutations. Single copies of the mutations were introduced under their own promoter into a strain carrying an integrated copy of CLN2 expressed from the GAL1 promoter in addition to the wild-type CLN2 gene. The abundance of Cln2 was then evaluated over a time course following repression of CLN2 expression by the addition of glucose (Fig. 5B). Whereas the half-life of Cln2 in cells expressing wild-type Grr1 was approximately 10 min, each of the strains expressing mutant Grr1 exhibited an extended Cln2 half-life. As previously demonstrated, introduction of either the grr1Δ or the grr1ΔL mutation resulted in a dramatic stabilization of Cln2 (Fig. 5B). Strikingly, replacement of all four basic residues on the concave surface was also associated with a dramatically lengthened half-life for Cln2 that was consistent with the effect of those mutations on Cln2 binding. Both the grr1-B4Q and the grr1-B4E mutants also caused a similar increase in the stability of Cln2 without the associated defect in SCF formation (Fig. 4 and 5B). The various single-site point mutants that were analyzed resulted in more modest increases in Cln2 stability than either Grr1ΔLRR or one of the quadruple-point mutant proteins (data not shown). Finally, the Grr1ΔC mutant, which exhibited a partial defect in Cln2 binding, resulted in a less dramatic stabilization of Cln2 (data not shown). Because Grr1ΔC is reduced in abundance relative to wild-type Grr1, it was difficult to distinguish the effect of reduced binding to Cln2 from the effect of reduced abundance of the Grr1ΔC protein. Nevertheless, its effect is less severe than that of Grr1ΔL, as well as those of most of the point mutants (Fig. 5A and data not shown). We conclude that the stability of Cln2 in strains expressing single copies of the various grr1 mutations was largely consistent with the capacity of the mutants to bind to Cln2.
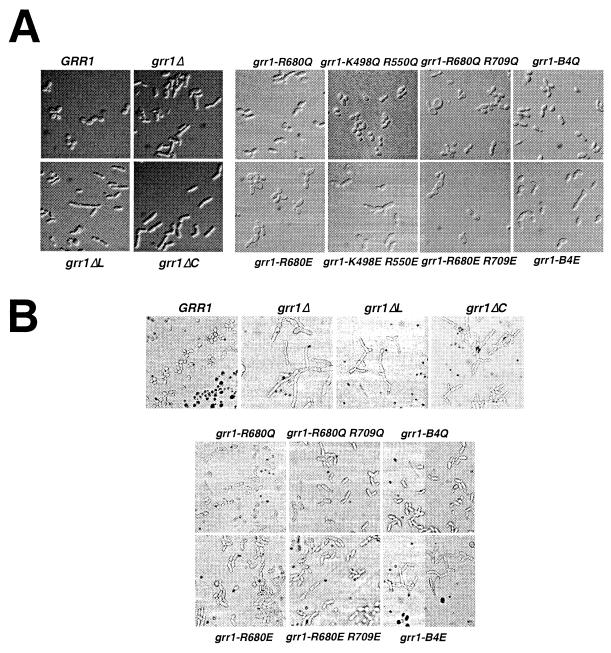
Morphological consequences of grr1 mutations.
We have analyzed the morphological phenotypes associated with the various grr1 mutants (Fig. 6). The mutants have been analyzed both when growing in rich liquid medium (yeast extract-peptone-dextrose [YEPD] liquid; Fig. 6A) or on rich solid medium (YEPD plate; Fig. 6B). Consistent with their effect on Cln2 stability, both the grr1ΔL and grr1ΔC mutants caused cells to become elongated and to bud in a unipolar fashion. This was particularly apparent in cells growing on solid medium (Fig. 6B). The grr1 point mutants exhibited a gradation of severity that was roughly consistent with their effect on Cln2 proteolysis. Mutations resulting in neutralization of basic residues on the predicted convex surface of the LRR had no phenotypic effect, whereas those neutralizing basic residues on the predicted concave surface had a pronounced effect on cell polarity, bud site selection, and, to a lesser extent, abscission (Fig. 6 and data not shown). Whereas single- and double-point mutations had a relatively modest effect on cell morphology, the effect of the quadruple-point mutations (grr1-B4Q and grr1-B4E) on polarized growth and bud site selection became more pronounced. Finally, replacement with an acidic residue generally resulted in more severe phenotypes than replacement with a neutral residue at the same position. Nevertheless, whereas grr1-B4E mutations are similar to the grr1Δ mutations in the severity of their effects on morphological phenotypes, they have less severe effects on growth rate (data not shown).
FIG. 6.
(A) Morphological phenotypes of point and deletion mutations of GRR1 growing in rich liquid medium. Cells grown to late log phase were analyzed by DIC microscopy and digital imaging. (B) Morphological phenotypes of point and deletion mutations of GRR1 growing on rich solid medium. Cells were grown on rich glucose medium (YEPD) for 4 h prior to photomicrography.
Despite the general increase in severity of the phenotype that was observed with an increasing number of mutations, there are subtle distinctions between the morphological phenotypes of the four different basic residue mutants (Fig. 6 and data not shown). For example, mutations affecting residue R680 result in cell enlargement rather than hyperpolarization. This is very noticeable when comparing the neutral and acidic replacements of R680 and R709 to the same replacements of K498 and R550. However, the single R680E mutant also exhibits significant enlargement. The source of the cell enlargement is unknown, but it is unlikely to be a consequence of hyperaccumulation of Cln2. Other, more subtle phenotypic differences are apparent on closer examination but have not been further evaluated.
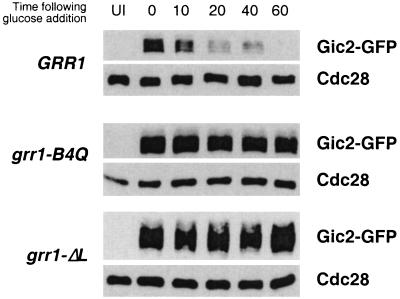
Mutations in GRR1 affecting Cln2 binding and stability also result in stabilization of Gic2, a target for SCFGrr1-dependent proteolysis.
The fact that mutations on the putative concave surface of the Grr1 LRR affected the capacity of SCFGrr1 to bind to Cln2 and resulted in its stabilization suggested that the same mutations would also result in the stabilization of other SCFGrr1 targets. The stability of the putative Cdc42 effectors Gic1 and Gic2 has been shown to be phosphorylation dependent and to be mediated by SCFGrr1 (17). To determine whether the basic residues on the concave surface of the Grr1 LRR were required for the degradation of the Gic2 protein, we analyzed the stability of Gic2-GFP expressed from the repressible GAL1 promoter in cells having the grr1-B4Q allele at the chromosomal locus as the only source of Grr1. The Gic2-GFP fusion protein was rapidly degraded when the GAL1 promoter was repressed by the addition of glucose (Fig. 7). However, the same protein was dramatically stabilized in grr1ΔL mutants, as well as in cells carrying grr1-B4Q (Fig. 7) and grr1-B4E (not shown) mutations. Thus, mutations that either eliminate the LRR domain of Grr1 or neutralize the charge on its concave surface result in stabilization of the Gic2 protein, presumably due to the failure of Grr1 to bind to Gic2 and promote its ubiquitination. We conclude that the concave surface of the Grr1 LRR, by virtue of its basic nature, participates in binding to phosphorylated targets of the SCFGrr1 ubiquitin ligase.
FIG. 7.
Stability of Gic2 in cells having point and deletion mutations in Grr1. Cells carrying GIC2-GFP under the inducible GAL1 promoter and wild-type GRR1 or its derivatives (grr1ΔL and grr1-B4Q) under its native promoter were pregrown under noninducing conditions (2% raffinose). GAL1-GIC2-GFP was expressed for 3 h by addition of 2% galactose and then repressed by addition of 2% glucose. Extracts prepared from samples taken at the times indicated were analyzed by immunoblotting using anti-GFP and anti-Cdc28 antibodies. UI, uninduced.
Mutations in GRR1 have differential effects on Cln2 stability and other established functions of Grr1.
Grr1 has been implicated in a number of cellular functions in addition to proteolysis of G1 cyclins and Gic1/2. Genetic analysis has implicated GRR1 as a regulator of the expression of several nutrient responsive genes. Of those, the best characterized are the systems governing expression of the HXT genes, which encode a family of hexose permeases in response to glucose (reviewed in reference 38) and which govern the expression of the broad-specificity amino acid permease gene AGP1 in response to amino acids (15). We have investigated the effect of specific grr1 mutations on these functions (Fig. 8). A summary of the properties of a representative set of mutants is presented in Table 3.
TABLE 3.
Properties of representative set of grr1 mutants
| Mutant | Cln2 bindinga | Cln2 instability | Polarized growth | Induction of HXT1 | Expression of AGP1 | Pseudohyphal growth |
|---|---|---|---|---|---|---|
| GRR1 | ++++ | ++++ | − | ++++ | ++++ | ++ |
| grr1Δ | − | − | ++++ | − | +/− | ++++ |
| grr1ΔL | − | − | ++++ | ++ | +++ | ++++ |
| grr1ΔC | − | + | ++ | ++ | +++ | ++++ |
| grr1-B4Q | −/+ | +/− | ++ | +++ | ++++ | ND |
| grr1-B4E | − | +/− | +++ | +++ | +++ | ++++ |
| grr1-R485Q | ++++ | ND | − | ++++ | ++++ | ND |
Cln2 binding was tested in 15Daub strains expressing GRR1 from the ADH1 promoter and CLN2 expressed from the GAL1 promoter. All other data was obtained using W303 strains, except for the pseudohyphal growth assays, which were performed using strains of the Σ1278b background. The extent to which each phenotype is manifested ranges from − (none detectable) to ++++ (easily detectable). ND, not determined.
The four yeast hexose transporters encoded by HXT1–4 are each controlled somewhat differently by the availability of glucose. HXT1 is specifically induced in the presence of high levels of glucose (4%) and is repressed in the presence of most other carbon sources. We have evaluated expression of HXT1 in cells growing in either glucose or galactose by monitoring the abundance of HXT1 mRNA by RT-PCR using HXT1-specific primers (Fig. 8A). This approach obviates the problem of cross-hybridization of probes with transcripts of the other highly conserved HXT genes. As previously described, the strong induction of HXT1 expression observed in wild-type cells grown in glucose-containing medium is lost when cells are deficient in GRR1. Surprisingly, both grr1ΔL, and grr1ΔC, as well as the various point mutations of GRR1, retain the capacity to induce HXT1 expression in response to glucose. Thus, despite their dramatic effect on both Cln2 proteolysis and cellular morphology, the deletion mutations and mutations in basic residues of the LRR remained responsive to induction by glucose. Furthermore, grr1-R485Q, the neutral replacement on the predicted convex surface of the LRR, had no noticeable effect on glucose regulation of HXT1.
A similar analysis was performed to evaluate the effect of grr1 mutations on aromatic amino acid transport. Induction of the expression of the amino acid permease gene AGP1 has been shown to depend on GRR1 (15). We have performed two distinct but related assays. First, AGP1 transcript levels were monitored by RT-PCR and by Northern analysis (Fig. 8B). As previously reported, AGP1 expression in the presence of extracellular amino acids was significantly reduced in grr1Δ mutants relative to wild-type cells. However, as observed for HXT1 induction by glucose, each of the internal deletion mutations and the point mutations replacing basic residues on the concave surface of the LRR resulted in little or no effect on the accumulation of AGP1 transcripts. Since the grr1Δ mutant exhibited residual RT-PCR signal, we confirmed the result using Northern analysis. Again, although the grr1Δ mutant expressed a detectable amount of AGP1 transcript, that level was consistently lower than the transcript level expressed by grr1ΔL and grr1ΔC mutant or wild-type cells. Thus, the effect of accumulation of the AGP1 transcript in the various grr1 mutants growing under inducing conditions was similar to that of the accumulation of the HXT1 transcript in the same mutants growing in the presence of glucose.
Together the results of these analyses suggest that the role of Grr1 in the regulation of these transcriptional induction pathways involves recognition mechanisms that are distinct from those involved in the recognition of Cln2. It remains possible that the ability of the grr1ΔL, grr1ΔC, and point mutants to mediate the transcriptional responses but not Cln2 proteolysis results from quantitative rather than qualitative differences in Grr1 and grr1Δ functions rather than from differences in the ability to interact with specific targets. This consideration is especially relevant when considering grr1ΔC. However, because the effect of grr1ΔL on Cln2 binding, proteolysis, and various other cellular functions is similar to that of grr1Δ, it seems doubtful that its activity in this regard is significantly higher. Yet, like the point mutations, grr1ΔL has little effect on these transcriptional responses. Since the targets of Grr1 involved in either of these processes are unknown, their capacity to interact with Grr1 cannot be evaluated. Although it is generally assumed that these processes involve ubiquitination, that has not been established.
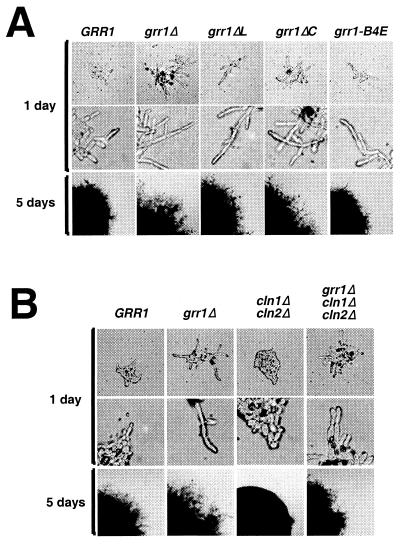
In addition to its involvement in nutrient-regulated gene expression, GRR1 has recently been implicated as a potential transducer of nutrient signals during filamentous differentiation (3, 32). Analysis of the effect of grr1 mutations on the capacity of cells to undergo filamentous differentiation yielded a strikingly different result from that of their effect on nutrient-regulated transcription. Assays of pseudohyphal differentiation of diploid strains were carried out in the Σ1278b background, which undergoes pseudohyphal growth on nitrogen-deficient SLAD plates. Wild-type and GRR1-deficient strains were analyzed along with grr1ΔLRR, grr1ΔC, and grr1-B4E mutants (Fig. 9A). All four strains carrying grr1 mutations were found to be hyperinducible for filamentous differentiation, acquiring a highly elongated morphology and becoming hyperinvasive relative to the wild-type strain. Although the various mutants were indistinguishable in terms of their cell morphology, the grr1-B4E mutant did not appear to be as hyperinvasive as the other mutants. Thus, unlike the effect of these same mutations on the glucose induction of HXT1 or AGP1, their effect on pseudohyphal growth was consistent with the effect of the same mutations on Cln2 binding and stability. We found the elongated cell phenotype in liquid medium caused by the grr1Δ mutation to be largely suppressed by the inactivation of CLN1 and CLN2, whereas pseudohyphal differentiation appeared to be only modestly affected (Fig. 9B). Both the extent of cell elongation and invasion of the solid substrate are only modestly reduced in the grr1Δ cln1Δ cln2Δ strain relative to the grr1Δ strain. Both phenotypes were strongly enhanced relative to the cln1Δ cln2Δ strain. The analysis of the grr1 deletion and point mutations yielded similar results (data not shown). We concluded that, although the enhancement of pseudohyphal growth caused by inactivation of GRR1 correlates with the stabilization of G1 cyclins, it is not dependent on CLN1 and CLN2 and must therefore be a consequence of the altered regulation of another Grr1 target. The nature of that target remains to be determined. A similar finding was made in a previous study by Loeb et al. (32), but their interpretation was somewhat different.
FIG. 9.
Pseudohyphal differentiation is enhanced by mutations in the LRR of GRR1. (A) Σ1278b strains carrying either grr1Δ, grr1ΔL, grr1ΔC, or grr1-B4E mutations were grown on SLAD plates for 5 days at 30°C. Photomicrographs were taken after 1 day and 5 days of growth. (B) Effect of inactivation of CLN1 and CLN2 on pseudohyphal differentiation in grr1Δ strains. Σ1278b strains carrying the indicated mutations were treated as described for panel A.
DISCUSSION
SCF formation and Cln2 binding by grr1 mutants.
Grr1 plays a central role in the regulation of SCF-dependent ubiquitination of a number of proteins including Cln2. SCF binding and Cln2 ubiquitination are dependent on phosphorylation of Cln2. We have shown here that the LRR is important for binding phosphorylated Cln2 and for its subsequent proteolysis. Furthermore, we showed that basic residues, predicted by our three-dimensional model to lie on the concave surface of the Grr1 LRR, are critical for the interaction between Cln2 and Grr1. In contrast, basic residues on the convex surface are not important for that interaction nor do they affect Cln2 proteolysis. These results reveal the domain of Grr1 required for the Grr1-Cln2 interaction as well as defining the probable binding surface as predicted by our model. We suggest that electrostatic interactions between basic residues on the concave surface of the LRR and the phosphoryl groups on Cln2 provide the binding energy for this interaction. Similar electrostatic interactions are characteristic of the contacts observed by the X-ray crystal structure of RI and RNase A (25) on which our structural model for the Grr1 LRR was based. Although the charged residues of RI are not as clearly concentrated on the concave surface of the LRR, about a third of the contacts between the LRR of RI and the RNase A molecule are between acidic and basic residues. In that case, primarily acidic residues are contributed by the LRR whereas the binding partner contributes predominantly basic residues to the interaction (27). The importance of electrostatic interactions was further supported by our demonstration that Cln2 phosphorylation is required for recognition by SCFGrr1 (30, 55). Thus, Grr1 serves as target-specific adapter for ubiquitination by providing a specific binding pocket for interaction with the target.
Several lines of evidence suggest that the interaction between Grr1 and Cln2 is dependent upon properties in addition to the basic character of the LRR. First, although the LRR domain of Grr1 is stable when expressed in yeasts, the isolated domain does not interact stably with Cln2 (data not shown). Next, the carboxy terminus of Grr1 is important for productive interactions since a Grr1 mutant lacking the region of the carboxy terminus adjacent to the LRR fails to bind stably to Cln2. In contrast, Grr1 lacking the 280-amino-acid amino terminus adjacent to the F box appears to bind Cln2 relatively efficiently. Although elimination of the C terminus does not appear to be as detrimental as elimination of the LRR, it does result in a substantial decrease in Cln2 binding and stabilization. Support for a role of the carboxy-terminal domain of Grr1 is provided by the recently reported structure of a cocomplex between human Skp1 and Skp2, an LRR-containing F box protein involved in recognition of the CDK inhibitor p27 (47). In that complex, the C terminus of Skp2 wraps around the concave face of the LRR and interacts with the F box domain, thereby positioning it to interact with substrate bound to Skp2 via the LRR. However, a role for the LRR of Skp2 in binding substrates has yet to be established. This, together with our results, suggests that the carboxy-terminal domains of these proteins participate in binding by facilitating interactions between substrates and the LRR.
All of the Grr1 proteins with alterations in basic residues were effective in forming SCF complexes, despite their defect in binding Cln2. Both the LRR-deficient and the carboxy-terminally truncated mutants of Grr1 failed to bind to the lower mobility form of Cdc53, which is modified by the ubiquitin-like protein Rub1. There is no established role for Rub1 modification in SCF function in budding yeast cells, and its elimination results in no detectable phenotypic consequences. In contrast, the homolog NEDD8 is essential for viability in fission yeasts (37) and is required for degradation of some SCF targets in animal cells (43). Since both forms of Cdc53 are present at wild-type levels in the Grr1ΔL-expressing cells, it appears either that Grr1ΔL and Grr1ΔC fail to form such complexes or that the complexes, once formed, are more unstable than those formed with unmodified Cdc53. The latter would be consistent with the suggestion that Rub1 is involved in regulating cullin abundance (29).
Differences between requirements for the Grr1-Cln2 interaction and the regulation of other putative targets of Grr1.
The role of Grr1 in SCF-dependent ubiquitination extends beyond Cln2 to other protein targets including the Cdc42 interactor Gic2 (17). We show that basic residues on the concave surface of the LRR are required for the instability of Gic2. In addition, Grr1 also plays a role in several systems of nutrient-regulated transcription. The direct targets of Grr1 in those systems are unknown. In fact, it has not been established in either of these cases that the role of Grr1 is in the context of protein ubiquitination. We have shown here that, although disruption of Grr1 results in failure to induce either the HXT1 or AGP1 transcripts, the other deletion mutations or point mutations we have constructed result in little or no defect in the induction of those genes. In contrast, all of those mutations interfere with the ubiquitin-dependent degradation of Cln2. Thus, we consider it likely that the protein domains of Grr1 involved in recognition of phosphorylated Cln2 are unimportant for recognition of the targets required for the regulation of these transcription systems. Alternatively, the capacity of these mutant proteins to function in those pathways may be a reflection of differences in the efficiency of the interactions required for transcriptional activation and G1 cyclin proteolysis. The differences observed in the requirements for recognition of the relevant targets in these transcriptional pathways may reflect differences in the basis for recognition. For example, recognition of those targets may not involve protein phosphorylation. Resolution of these issues awaits the identification of the relevant targets of Grr1 in those pathways.
What is the role of Grr1 in these transcriptional regulatory systems? Our understanding of both systems is largely derived from genetic studies. Induction of HXT1 gene expression by glucose has been shown to involve components of the SCF system in addition to Grr1, including Cdc53, Skp1, and, at least in our experiments, Cdc34 (H.-C. Chang and C. Wittenberg, unpublished results; for a different result, see reference 31). This is consistent with a role for Grr1 in proteolysis of a negative regulator of transcription (10; reviewed in reference 38). However, evidence that Rgt1 is regulated at the level of ubiquitination or proteolysis is lacking. Recently reports regarding the role of SCFMet30 in transcriptional regulation of MET gene expression have come to conflicting conclusions. Experimental support has been presented for the involvement of ubiquitin-mediated proteolysis of the transcription factor Met4 (45). In contrast, we have recently shown that, although the transcriptional regulator Met4 is ubiquitinated under conditions that inhibit MET gene expression, proteolysis is unnecessary to inactivate MET gene transcription (18). Instead ubiquitination interferes with the function of Met4 as a transcriptional activator, presumably by interfering with its ability to bind a coactivator. That system provides an attractive model for regulation of Rgt1, especially because it has been proposed to act as both a positive and a negative regulator in the presence of different carbon sources (39). For instance, in the absence of glucose, Rgtl may be present in a nonubiquitinated form and act as a repressor via recruitment of the corepressors Ssn6 and Tup1. Conversely, in the presence of glucose it might become ubiquitinated in a Grr1-dependent manner, disrupting the interaction with its corepressors, thereby converting it to a transcriptional activator. Alternatively, ubiquitination may regulate the activity of the corepressors or other regulators via either proteolytic or nonproteolytic mechanisms. Because the central elements for transcriptional regulation of AGP1 are currently unknown, it is difficult to predict the role of Grrl in that regulation.
The relationship between the hyperpolarized phenotype, nutrient sensing, and pseudohyphal morphogenesis is also unclear. We expected, based on the relationship between Grr1 and Cln2 stability and on the capacity of Cln2, when overexpressed, to induce hyperpolarization, that the hyperinvasive phenotype of grr1Δ mutants was a consequence of the hyperaccumulation of G1 cyclins. In fact, evidence supporting that hypothesis has been reported (39). However, we have found that the inactivation of Cln1 and Cln2 has little effect on the capacity of grr1Δ mutations to induce pseudohyphal differentiation in terms of either morphological differentiation or the capacity to invade agar. Furthermore, the induction of pseudohyphal growth is at least partially independent of the established signaling pathways since invasive growth can be induced in grr1Δ mutants lacking both TEC1 and FLO8 (R. LaValle and C. Wittenberg, unpublished data). We conclude that Grr1 targets one or more elements of a TEC1/FLO8 independent pathway to suppress filamentation. The involvement of Grr1 in transcriptional regulation of nutrient permeases and the importance of nutrient signaling in the induction of pseudohyphal differentiation are intriguing in this regard. We have not yet analyzed the importance of amino acid or glucose permeases in the pseudohyphal growth signal or analyzed the potential targets of Grr1 in that response.
LRR as recognition domain for phosphorylated proteins.
The capacity of proteins to interact specifically with each other has long been recognized as a fundamental property of living systems and is the basis for the assembly of the macromolecular complexes central to many biological processes. Regulated protein-protein interaction is a critical component of many biological regulatory mechanisms. Protein phosphorylation is one of the most widely studied and perhaps most common mechanisms governing regulated protein recognition. Despite that fact, the basis for recognition is only poorly understood. This may be a consequence of the wide array of solutions to this specific recognition problem.
LRRs are thought to be involved in protein-protein interaction. Deletion analysis of a number of proteins including Grr1 has confirmed that supposition. Although this property of LRR domains appears to be conserved, the nature and roles of the proteins that they bind are varied. Structural studies of LRRs, although relatively limited, suggest that it is the concave surface that is important in the interaction with other proteins. There also appears to be growing support for the importance of electrostatic interactions in the recognition process. The interaction demonstrated between RI and RNase A (25), as well as our findings concerning Grr1, are consistent with both of those proposals. In addition, structural modeling of the LRR of the acid-labile subunit of the insulin-like growth factor binding protein (IGFBP-3) complex reveals that, like in the LRR of Grr1, a highly charged patch is predicted on the concave surface (16). However, unlike Grr1, that patch is predicted to be acidic rather than basic. Whereas the ring structure of that molecule has been confirmed by electron microscopic analysis, the importance of the acidic patch in its interaction with insulin-like growth factor–insulin-like growth factor binding protein 3 (IGF-IGFBP-3) complex has not been established. It is not clear whether such electrostatic interactions are universally involved in LRR protein interactions. Our mutational analysis of Grr1 indicated that the basic character of the concave surface of the LRR is unimportant for regulation of transcriptional activation of HXT1 and AGP1. The properties of Grr1 important for that regulation remain to be established.
Protein phosphorylation figures centrally in the targeting of proteins for SCF-dependent ubiquitination. This has been established in a wide variety of regulatory systems in eukaryotes. However, not all of those targeting interactions involve LRR-containing F-box proteins. In addition, F-box proteins with WD40 repeats are also prevalent and in at least some cases are involved in targeting events that require phosphorylation. Their role in phosphorylation-dependent ubiquitination of the mammalian NF-κB inhibitor IκB by SCFβTRCP (22) and of the yeast CDK inhibitor Sic1 by SCFCdc4 (9, 49, 54) has been well established. Both of those systems rely on interactions that occur via the WD40 repeats of their respective F-box proteins, βTRCP/E3RSIκB and Cdc4. Phosphorylation is implicated in a number of other SCF-dependent ubiquitination events that involve LRR- and WD40 repeat-containing F-box proteins. This study has established one mechanism by which such interactions can be implemented. However, the specifics of the interactions utilized among this large class of protein recognition events will require further mutagenesis and/or structural studies. Despite their involvement in phosphorylation-dependent interactions, LRR and WD40 repeat motifs in F-box proteins, as in their wider context, are not restricted to such interactions nor are interactions involving phosphorylation restricted to those involving LRR and WD40 motifs.
ACKNOWLEDGMENTS
We thank Frank Li, Mark Johnston, Hao-ping Liu, and Matthias Peter for providing plasmids and yeast strains and Andre Kajava for providing coordinates derived from his modeling of cysteine-rich leucine rich repeats. We also thank the TSRI cell cycle group, including members of the laboratories of C. McGowan, S. Reed, P. Russell, and C. Wittenberg, for helpful comments and discussion during the course of this work.
Y.H. was supported by a fellowship from the Lymphoma and Leukemia Society (formerly the Leukemia Society of America). R.L.V. is supported by an AIDS fellowship from the National AIDS Program, Istituto Superiore di Sanita, Rome, Italy. This work was supported by U.S. Public Health Service grants GM59759 to S.L. and GM43487 to C.W.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 3.Blacketer M J, Madaule P, Myers A M. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimalization, and molecular dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 5.Brunger A T. X-PLOR manual, version 3.1. New Haven, Conn: Yale University; 1992. [Google Scholar]
- 6.Carrano A C, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies R J. SCF and cullin/ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 8.Drury L S, Perkins G, Diffley J F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 10.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geitz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 12.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 15.Iraqui I, Vissers S, Bernard F, de Craene J O, Boles E, Urrestarazu A, Andre B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janosi J B, Ramsland P A, Mott M R, Firth S M, Baxter R C, Delhanty P J. The acid-labile subunit of the serum insulin-like growth factor-binding protein complexes. Structural determination by molecular modeling and electron microscopy. J Biol Chem. 1999;274:23328–23332. doi: 10.1074/jbc.274.33.23328. [DOI] [PubMed] [Google Scholar]
- 17.Jaquenoud M, Gulli M P, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser P, Flick K, Wittenberg C, Reed S I. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser P, Sia R A, Bardes E G, Lew D J, Reed S I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajava A V. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 21.Kamura T, Conrad M N, Yan Q, Conaway R C, Conaway J W. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 23.Kishi T, Yamao F. An essential function of Grr1 for the degradation of Cln2 is to act as a binding core that links Cln2 to Skp1. J Cell Sci. 1998;111:3655–3661. doi: 10.1242/jcs.111.24.3655. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa K, Skowyra D, Elledge S J, Harper J W, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 25.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 26.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 28.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin, Cln2, induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 31.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeb J D, Kerentseva T A, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCF, SKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 34.McRee D E. StalView: a visual protein crystallographic software system for X11/XView. J Mol Graph. 1992;10:44–47. [Google Scholar]
- 35.Mondesert G, Clarke D J, Reed S I. Identification of genes controlling growth polarity in the budding yeast Saccharomyces cerevisiae: a possible role of N-glycosylation and involvement of the exocyst complex. Genetics. 1997;147:421–434. doi: 10.1093/genetics/147.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama K, Nagahama H, Minamishima Y A, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, Kitagawa M, Hatakeyama S. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osaka F, Saeki M, Katayama S, Aida N, Toh E A, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozcan S, Schulte F, Freidel K, Weber A, Ciriacy M. Glucose uptake and metabolism in grr1/cat80 mutants of Saccharomyces cerevisiae. Eur J Biochem. 1994;224:605–611. doi: 10.1111/j.1432-1033.1994.00605.x. [DOI] [PubMed] [Google Scholar]
- 42.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Read M A, Brownell J E, Gladysheva T B, Hottelet M, Parent L A, Coggins M B, Pierce J W, Podust V N, Luo R S, Chau V, Palombella V J. Nedd8 modification of cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson H E, Wittenberg C, Cross F R, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 45.Rouillon A, Barbey R, Patton E E, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roussel A, Cambillau C. TURBO-FRODO: Silicon Graphics geometry partners directory. Mountain View, Calif: SGI; 1995. pp. 77–78. [Google Scholar]
- 47.Schulman B A, Carrano A C, Jeffrey P D, Bowen Z, Kinnucan E R, Finnin M S, Elledge S J, Harper J W, Pagano M, Pavletich N P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 48.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 50.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 51.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsvetkov L M, Yeh K H, Lee S J, Sun H, Zhang H. p27Kip1 ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 53.Vallier L G, Coons D, Bisson L F, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma R, Feldman R M, Deshaies R J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willems A R, Lanker S, Patton E E, Craig K L, Nasson T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 56.Wittenberg C, Richardson S L, Reed S I. Subcellular localization of a protein kinase required for cell cycle initiation in Saccharomyces cerevisiae: evidence for an association between the CDC28 gene product and the insoluble cytoplasmic matrix. J Cell Biol. 1987;105:1527–1538. doi: 10.1083/jcb.105.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 58.Wright R M, Repine T, Repine J E. Reversible pseudohyphal growth in haploid Saccharomyces cerevisiae is an aerobic process. Curr Genet. 1993;23:388–391. doi: 10.1007/BF00312623. [DOI] [PubMed] [Google Scholar]
- 59.Zhou P, Howley P M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]