Abstract
Trichomoniasis is a common and widespread sexually-transmitted infection, caused by the protozoan parasite Trichomonas vaginalis. T. vaginalis lacks the biosynthetic pathways for purines and pyrimidines, making nucleoside metabolism a drug target. Here we report the first comprehensive investigation into purine and pyrimidine uptake by T. vaginalis. Multiple carriers were identified and characterised with regard to substrate selectivity and affinity. For nucleobases, a high affinity adenine transporter, a possible guanine transporter and a low affinity uracil transporter were found. Nucleoside transporters included two high affinity adenosine/guanosine/uridine/cytidine transporters distinguished by different affinities to inosine, a lower affinity adenosine transporter, and a thymidine transporter. Nine Equilibrative Nucleoside Transporter (ENT) genes were identified in the T. vaginalis genome. All were expressed equally in metronidazole-resistant and -sensitive strains. Only TvagENT2 was significantly upregulated in the presence of extracellular purines; expression was not affected by co-culture with human cervical epithelial cells. All TvagENTs were cloned and separately expressed in Trypanosoma brucei. We identified the main broad specificity nucleoside carrier, with high affinity for uridine and cytidine as well as purine nucleosides including inosine, as TvagENT3. The in-depth characterisation of purine and pyrimidine transporters provides a critical foundation for the development of new anti-trichomonal nucleoside analogues.
Graphical Abstract
The sexually-transmitted disease trichomoniasis is caused by the parasite Trichomonas vaginalis, which is unable to make its own purine and pyrimidine nucleotides for DNA and RNA synthesis. This makes nucleotide metabolism a potential drug target. Antimetabolites and inhibitors will need to enter through purine/pyrimidine transporters. This paper describes these carriers in T. vaginalis cells in detail, identifies nine nucleoside transporter genes and characterises one in detail after cloning into a different cell type.
Plain Language Summary
New drugs against the sexually transmitted infection trichomoniasis are needed. The Trichomonas vaginalis parasite has a genetic weakness in that it cannot make the nucleoside building blocks for its own DNA and RNA and must take these up from its environment using transport proteins. Here we report an in-depth study of the nucleoside transporters of T. vaginalis, studying them as a complex whole in the parasite. In addition, we cloned each of the transporters individually for expression in a different cell type, allowing the separate characterisation of one transporter at a time. We present a model of a minimum of six transporters in the parasite and identify the gene encoding the most efficient one via the expression strategy. We also report that the parasite seems to use all of the nucleoside transporters at the same time, regardless of growth conditions.
Introduction
Trichomoniasis can be said to be a neglected and underestimated sexually transmitted infection (STI), even though several hundred million people world-wide are infected annually (WHO, 2012). Although symptoms are usually mild or even absent (Edwards et al., 2016), the potential sequelae are severe. These include adverse pregnancy outcomes (Silver et al., 2014) and increased transmission of viral infections as a result of damage to the epithelial layers of the reproductive tracts, including HIV (Kissinger and Adamski, 2013), HSV-2 (Gottlieb et al., 2004; Kissinger, 2015) and HPV (Feng et al., 2018; Raffone et al., 2021), increasing the incidence of AIDS, genital herpes and cervical neoplasia, respectively. Vertical transmission during birth has also been documented (Peters et al., 2021). Although the infection is particularly ignored in men, as a result of its often-asymptomatic nature, here too are severe long-term risks, including diminished fertility, urethritis, prostatitis and, again, higher risk of HIV infection (Van Gerwen et al., 2021).
The infection is routinely treated with the relatively cheap nitro-imidazole drug metronidazole or, in a minority of cases, its more recent derivative tinidazole (Kissinger, 2015). However, reports of clinical resistance to these drugs have increased. Although the level of resistance is generally mild-to-moderate (Marques-Silva et al., 2021) the adverse effects of the drugs, including neurologic maladies, nausea, metallic taste, and hypersensitivity (Muzny et al., 2020), are such that dosage cannot easily be increased. The need for alternative anti-trichomonal therapies when nitroimidazoles are ineffective or not tolerated is therefore undisputed. Among the most promising targets in trichomonads is their nucleotide metabolism, as they lack the ability to synthesise either purines (Heyworth et al., 1982) or pyrimidines (Wang et al., 1984; Heyworth et al., 1984) de novo. This makes the parasites vulnerable to inhibitors of key enzymes of the nucleoside salvage pathways and to subversive substrates. Nucleoside analogues with strong antitrichomonal activity have been identified and include formycin A (Munagala and Wang, 2003), adenine arabinoside, 2′-F,2′-deoxyadenosine, and 2′-F,2′-deoxyarabinoadenosine (Shokar et al., 2012). Very recently, we reported on a range of 7-substituted,7-deazaadenosine analogues with mid-nanomolar activity against T. vaginalis in vitro and one compound, 7-(4-Cl-phenyl),7-deazaadenosine, was shown to be efficacious in a murine model of vaginal trichomonad infection (Natto et al., 2021).
T. vaginalis must salvage extracellular purines and pyrimidines, and expresses 5’-ecto-nucleotidases and NTPDases to hydrolyse nucleotides to their corresponding nucleosides (Menezes et al., 2016; Tasca et al., 2003), which are then internalised by transporters. Most nucleoside antimetabolites rely on those same transporters and thus their substrate selectivity is a key determinant as to which analogues will be efficiently taken up (Campagnaro and De Koning, 2020). In Trypanosoma brucei, for instance, sensitivity to tubercidin (7-deazaadenosine) and cordycepin (3’-deoxyadenosine) depends on the expression of the TbAT1 aminopurine transporter (Geiser et al., 2005), while sensitivity to 7-Br,3’-deoxytubercidin does not (Hulpia et al., 2019). Similarly, sensitivity to tubercidin and formycin B in Leishmania donovani depends on the NT1 and NT2 nucleoside transporters, respectively (Galazka et al., 2006; Vasudevan et al., 2001), and sensitivity to adenine arabinoside (AraA) in Toxoplasma gondii on the TgAT1 adenosine transporter (Chiang et al., 1999). However, nucleoside and nucleobase transport have been poorly studied in T. vaginalis. A single report from 1988 describes two nucleoside transport activities, one that transports all nucleosides and one selective for adenosine, guanosine and uridine (Harris et al., 1988); neither was inhibited by nucleobases, although adenine and guanine have both been shown to be incorporated into the T. vaginalis nucleotide pool (Munagala and Wang, 2003). Yet, the genome of T. vaginalis contains nine genes of the Equilibrative Nucleoside Transporter (ENT) family (TrichDB.org), to which, to date, all protozoan nucleoside and nucleobase transporters have been attributed (De Koning et al., 2005; Campagnaro and De Koning, 2020), although there are some indications that there may be some protozoan nucleobase transport activities that are not encoded by ENT genes (De Koning, 2007; Campagnaro et al., 2018a). We therefore performed a comprehensive examination of nucleoside and nucleobase transport in T. vaginalis trophozoites with the objective to begin the process of assigning specific transport activities to individual genes, as well assessing their relative levels of expression and their regulation in the presence and absence of substrate and feeder cells. Expression of the individual TvagENT genes in a Trypanosoma brucei cell line allowed us to identify the main high affinity, broad specificity nucleoside transporter, which turned out to be encoded by TvagENT3.
Results
Nucleoside transport in T. vaginalis trophozoites
For all permeants (i.e, the substrate for which permeation is being measured, as opposed to inhibitors that are only potential permeants), time course experiments were first undertaken to establish (1) whether uptake could be discerned at a certain radiolabel concentration, and (2) that uptake is linear and through zero over a given period. If these conditions are not met, the parameters determined might be of the rate-limiting step, which could be a metabolic enzyme rather than the transporter for this radiolabel. In all cases, care was taken to use a very low starting permeant concentration so as to obtain the most accurate Km and Ki values and Hill slopes, and avoid (partial) saturation of very high affinity transporters as much as possible. This has the added benefit of extending the linear range of uptake as the low rate of permeant entry at those concentrations will not easily saturate the downstream metabolic reactions. For some permeants the lower concentration limit was determined by detectability of low rates of uptake. Higher radiolabel concentrations were used when specifically probing the existence of lower-affinity transporters and in these cases, again, linearity of uptake was first established. Dose-response experiments used incubation times near the middle of the established linear uptake period. Linearity was queried using the Prism runs test for deviation of linearity. Uptake was considered significant if the slope was significantly non-zero (F-test, Prism) and saturability by high levels of unlabelled permeant was tested with the function for significant difference between linear regression lines (F-test). All transporter data presented in figures are single experiments with data points representing the mean and SEM of triplicate determinations (unless otherwise indicated), and are representative of multiple similar repeats. For the transport experiments with T. vaginalis trophozoites, close attention was paid to the Hill slope of inhibition experiments, as a Hill slope above −1 (usually between −1 and −0.5) will indicate the uptake of permeants by more than one transporter with non-identical Km for the inhibitor. In a complex cellular system with multiple transporters of overlapping substrate selectivity this is an important parameter and a Hill slope that is consistently >-1 permits the plotting of the inhibitor data to a bi-phasic inhibitor model.
1. Adenosine uptake.
Submicromolar concentrations of [3H]-adenosine were rapidly taken up by T. vaginalis trophozoites. Figure 1A shows that transport of 0.25 µM [3H]-adenosine was linear over 60 s with a rate of 1.15 pmol(107 cells)−1s−1. The uptake was >99% inhibited by 1 mM unlabelled adenosine, and the remainder was not significantly different from zero (P=0.35, F-test). The uptake was therefore saturable and most likely carrier-mediated. The inset in Figure 1A is a technical control showing that stopping the uptake with 1 mL of 1 mM ice-cold adenosine did stop all uptake, as previously reported for other cell types.
Figure 1.
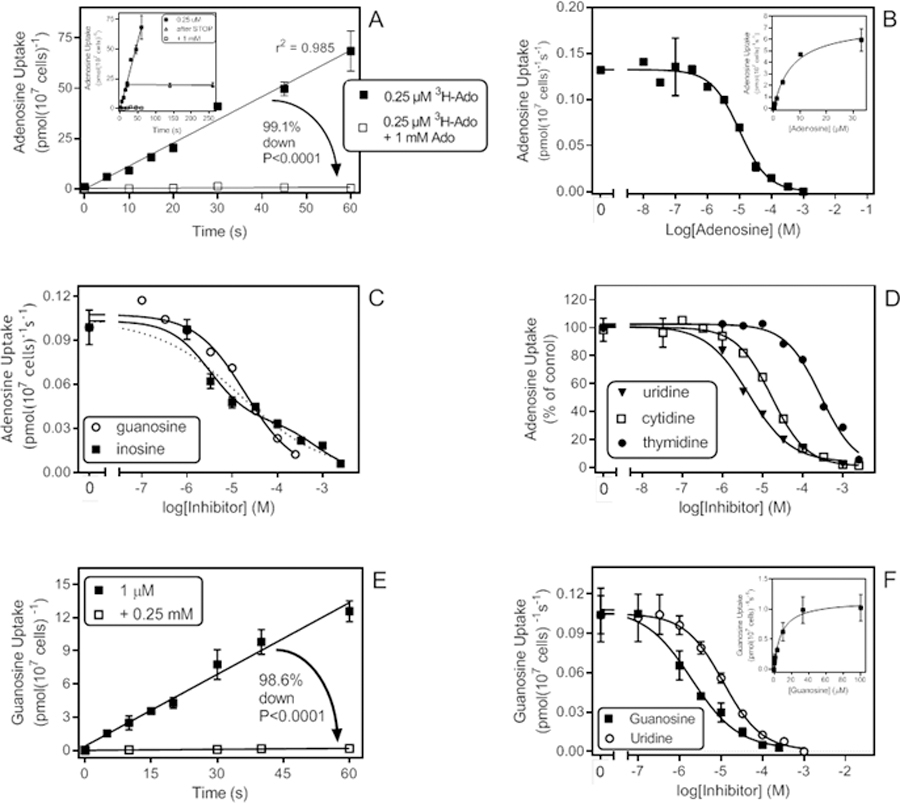
Adenosine and guanosine uptake in T. vaginalis trophozoites. A. Transport of 0.25 µM [3H]-adenosine in the presence or absence of 1 mM unlabelled adenosine. In the presence of unlabelled adenosine uptake over 60 s was not significantly different from zero (0.0091 ± 0.0085 pmol(107 cells)−1s−1), P=0.35, F-test). In contrast, uninhibited uptake was highly significant (1.15 ± 0.06 pmol(107 cells)−1s−1), P<0001). Inset: Same data, but also showing two extra data points representing samples that were incubated for 20 s with the [3H]-adenosine but left uncentrifuged for an additional 120 or 240 s after the addition of ice-cold 1 mM adenosine to stop the uptake. Uptake over the additional 240 s was not significantly different from zero (−0.0021, pmol(107 cells)−1s−1), P=0.62). B. Dose-dependent inhibition of 0.15 µM [3H]-adenosine transport over 30s, with unlabelled adenosine, plotted to a sigmoid curve, Hill slope −0.974. Inset: Conversion of the same data to a Michaelis-Menten saturation plot. C. Dose-dependent inhibition of 0.15 µM [3H]-adenosine transport over 30s, with unlabelled inosine, plotted to a biphasic curve for two-site competition, or with guanosine, plotted to a one-site inhibition curve with variable slope. D. As C, showing monophasic inhibition of adenosine transport with three pyrimidine nucleosides. Hill slopes were −0.99, −0.89 and −0.82 for cytidine, thymidine and uridine, respectively. E. Transport of 1 µM [3H]-guanosine with a rate of 0.22 ± 0.01 pmol(107 cells)−1s−1, significantly different from zero (P<0.0001) and not significantly non-linear (P=0.71). In the presence of 0.25 mM unlabelled guanosine, the rate was reduced to 0.0030 ± 0.0004 pmol(107 cells)−1s−1 (r2=0.96; significantly different from zero (P=0.004), not significantly non-linear (P=0.90). F. Transport of 0.5 µM [3H]-guanosine in the presence of various concentrations of unlabelled guanosine (Hill slope = −0.77) and uridine (Hill slope = −0.94). Inset: Conversion of the guanosine inhibition data to a Michaelis-Menten saturation plot. In all transport graphs symbols/error bars represent the mean and SEM of triplicate determinations of single representative experiments.
A series of dose-response inhibition experiments with unlabelled adenosine (Fig. 1B) allowed the determination of Km as 6.2 ± 0.6 µM (n=5) and the Vmax as 9.5 ± 2.4 pmol(107 cells)−1s−1 (n=4) (Table 1). These dose-response experiments were performed at 0.15 µM of [3H]-adenosine to ensure that the label concentration was far below the Km. The resulting curves were apparently monophasic; however, the average Hill slope was −0.90 ± 0.04, with values ranging from −1.01 to −0.76 (n=5), leaving open the possibility of more than one transporter with very similar but non-identical Km values. Indeed, the Hill slope for guanosine inhibition of [3H]-adenosine uptake was even more indicative of multiple carriers (−0.68 ± 014, n=3). If taken as monophasic (plotted to a sigmoid equation with variable slope), the apparent guanosine Ki,app would be 12.2 ± 2.4 µM (Table 1; Fig. 1C). The biphasic inhibition of adenosine uptake was clearest when using inosine as inhibitor (Figure 1C) and here an equation for two binding sites was in all cases the significantly better fit, yielding a high affinity Ki,app of 5.2 ± 1.2 µM and a low affinity interaction Ki,app of 347 ± 127 µM (n=3). Thus, the combined inhibitor evidence strongly suggests that two similar high-affinity adenosine transporters are expressed in T. vaginalis, with similar affinity for adenosine and guanosine as well as uridine (Ki,app = 3.7 ± 1.0 µM; Hill slope −0.97 ± 0.10; n = 3) and cytidine (Ki,app = 17.9 ± 4.4 µM; Hill slope −0.73 ± 0.13; n=3) (Figure 1D). These two adenosine transporters both displayed much lower affinity for thymidine (Ki,app = 274 ± 45 µM; n=3) and the nucleobases hypoxanthine (Ki,app = 185 ± 37 µM; n=2) and adenine (Ki,app > 1 mM).
Table 1.
Kinetic parameters and inhibitor profile of T. vaginalis nucleoside transporters (Average and SEM, µM).
| Adenosine (HA) |
Guanosine (HA) |
Inosine (LA) |
Uridine (HA) |
Cytidine (HA) |
Thymidine (HA) |
Adenosine (LA) |
Cytidine (LA) |
Thymidine (LA) |
|
|---|---|---|---|---|---|---|---|---|---|
| Km,app | 6.2 ± 0.6 | 9.0 ± 0.6 | 196 ± 18 | 5.5 ± 1.8 | 7.2 ± 1.8 | 13.2 ± 0.8 | 59 ± 6 | 348 ± 18 | 470 ± 104 |
| Vmax 1 | 9.5 ± 2.4 | 0.91 ± 0.25 | 2.6 ± 0.4 | 1.5 ± 0.5 | 0.83 ± 0.06 | 6.7 ± 1.8 | 31 ± 2 | 6.7 ± 1.8 | 9.4 ± 2.5 |
| Vmax/Km | 1.5 | 0.10 | 0.013 | 0.27 | 0.12 | 0.05 | 0.52 | 0.019 | 0.020 |
| Ki,app | |||||||||
| Adenosine | 4.1 ± 0.9 | 6.4 ± 1.7 | 8.2 ± 0.6 | 2.2 ± 0.3 | 39 ± 8 | 357 ± 32 | 73 ± 6.9 | ||
| Inosine (HA) | 5.2 ± 1.2 | 14.1 ± 5.7 | 3.8 ± 0.9 | ND | 78 ± 15 3 | ND | ND | ND | |
| Inosine (LA) | 348 ± 127 | ND | 231 ± 23 | ND | N/A | ND | ND | ND | |
| Guanosine | 12.2 ± 2.4 | 7.4 ± 2.9 | ND | 16.7 ±2.6 | 36 ± 7 | >250 | ND | ND | |
| Uridine | 3.7 ± 1.0 | 7.9 ± 1.3 | 3.6 ± 1.1 | 2.0 ± 0.5 | 12 ± 1 | 376 ± 63 | ND | ND | |
| Thymidine | 274 ± 45 | 206 ± 62 | ND | ND | ND | 557 ± 135 | ND | ||
| Cytidine | 17.9 ± 4.4 | 19 ± 0.9 | ND | 15.6 ± 2.3 | ~15 2 | 116 ± 5 | ~250 2 | ||
| Adenine | ~1000 | ND | ND | ND | ND | >1000 | >1000 | ND | ND |
| Hypoxanthine | >500 | ND | ND | ND | ND | >1000 | >1000 | ND | ND |
| Uracil | ND | ND | ND | ND | ND | >1000 | >10000 | ND | ND |
HA, high affinity; LA, low affinity; ND, not determined; N/A, not applicable.
Units for Vmax are pmol(107 cells)−1s−1.
Estimated rough average from three separate experiments yielding biphasic plots.
Based on monophasic plots. This transport activity has only one Ki,app for inosine.
2. Guanosine uptake.
The above findings suggest the expression of two similar adenosine transporters with, judging by the Hill slopes, slightly different affinities for adenosine, cytidine and guanosine. This should be mirrored when using [3H]-guanosine as radiolabel, unless there is a separate adenosine-insensitive guanosine transporter as well. Uptake of 1 µM [3H]-guanosine was linear for at least 60 s and almost completely saturable (98.6% reduced) by 250 µM guanosine (Figure 1E). Sigmoid inhibition plots showed that the Hill slopes for nucleoside inhibitors were indeed indicative of two transporters with somewhat different affinity: −0.67 ± 0.03 (adenosine), −0.78 ± 0.08 (guanosine) and −0.72 ± 0.06 (cytidine), respectively. Using the EC50 values from plotting to a monophasic sigmoid curve with variable slope apparent Ki,app values of 4.1 ± 0.9 µM and 18.6 ± 0.9 µM were obtained for adenosine and cytidine, respectively, and a Km,app of 9.0 ± 0.6 µM was calculated for guanosine (n = 3 for all) (Figure 1F). It is acknowledged that most of these Ki,app values are highly likely to be composites of at least two separate transport activities. Also consistent with the [3H]-adenosine transport data was that uridine again displayed a Hill slope close to −1 (−0.91 ± 0.02) and high affinity (Ki,app 7.8 ± 1.3 µM) while affinity for thymidine was much lower (Ki,app 206 ± 62 µM). Finally, the Hill slope for inosine (−0.75 ± 0.07) was again indicative of at least two separate transport activities, with Ki,app 6.3 ± 2.6 and 146 ± 26 µM, respectively. It can be tentatively concluded on the evidence that guanosine is taken up by the same transporters as adenosine, at least at low permeant concentrations.
3. Uridine uptake.
Uptake of 1 μM [3H]-uridine was linear for at least 240 s (r2 =0.97; not significantly non-linear (runs test, P=0.14)) and fully saturable with 1 mM unlabelled uridine (not significantly different from zero (runs test, P=0.15) (Fig. 2A). Subsequent inhibition plots were performed with an incubation time of 30 s. Unlabelled uridine dose dependently (0.1 – 2500 µM) inhibited uptake of [3H]-uridine, with an average Hill slope of −1.04 ± 0.05 (n=3), yielding an average Km,app of 5.49 ± 1.80 µM and Vmax of 1.49 ± 0.51 pmol(107 cells)−1s−1 (n=3) (Table 1). Inhibition by adenosine and cytidine yielded Ki,app values of 8.16 ± 0.59 µM and 15.6 ± 2.3 µM (n=3), respectively, with Hill slopes ≅ 0.90 (Fig. 2B,C). Inosine inhibition was again clearly biphasic (Fig. 2C), with a Hill slope of −0.51 ± 0.02 (n=3) and plotting to a 2-site sigmoidal curve yielded Ki,app values of 3.80 ± 0.90 µM and 231 ± 23 µM.
Figure 2.
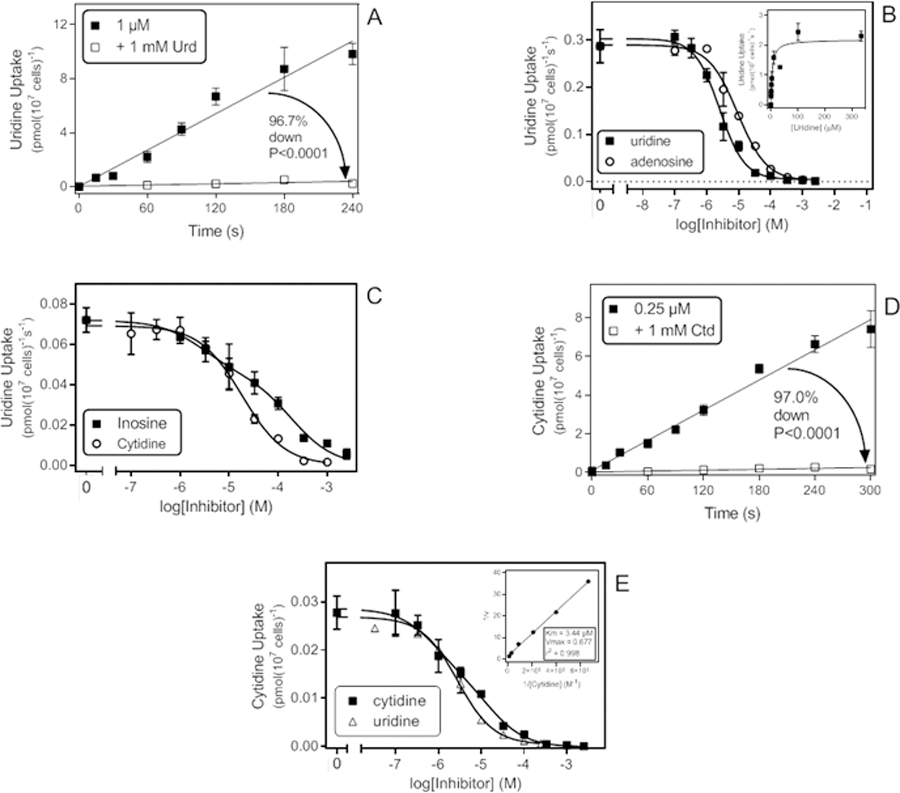
Transport of pyrimidine nucleosides by T. vaginalis trophozoites. A. Time course of 1 µM [3H]-Uridine uptake. The rate was 0.045 ± 0.003 pmol(107 cells)−1s−1 (r2 =0.97; not significantly non-linear (runs test, P=0.14)) which was significantly different from zero (P<0.0001, F-test) but in the presence of 1 mM uridine the rate (0.0015 ± 0.0008 pmol(107 cells)−1s−1 (r2 =0.55 )) was not significantly different from zero (P=0.15) as well not significantly non-linear (P=0.50). The two slopes were highly significantly different P<0.0001). The rates were obtained by linear regression. B. Transport of 0.5 µM [3H]-Uridine over 30 s: inhibition plot with unlabelled uridine and adenosine using sigmoid curves with variable slopes. Inset: Conversion of the uridine inhibition data to a Michaelis-Menten saturation plot. C. Transport of [3H]-Uridine (0.5 µM, 30 s), inhibition by cytidine and inosine plotted to a sigmoid dose-response curve with variable slope and a two-site competition curve, respectively. D. Time course of transport of 0.25 µM [3H]-cytidine. In the absence of inhibitor, the rate was 0.026 ± 0.001 pmol(107 cells)−1s−1 (r2 = 0.99; not significantly non-linear P=0.50, significantly different from zero P<0.001); in the presence of 1 mM unlabelled cytidine the rate was 0.00079 ± 0.00024 pmol(107 cells)−1s−1 (r2 =0.73; not significantly non-linear, P=0.30; significantly different from zero, P = 0.029). E. Inhibition of 0.15 µM [3H]-cytidine transport over 30 s by unlabelled cytidine and uridine, plotted to a sigmoid inhibition curve with variable. Inset: double-reciprocal Lineweaver-Burke plot from the inhibition data.
4. Cytidine uptake.
Uptake of 0.25 µM [3H]-cytidine was linear for at least 5 min (r2 = 0.99), and saturable with 1 mM unlabelled cytidine although the inhibition was not complete (97.0% and significantly non-zero by F-test, P= 0.029), indicating the presence of at least one low(er)-affinity carrier for cytidine (Fig. 2D). Indeed, inhibition profiles, using just 0.15 µM [3H]-cytidine, consistently showed a mixed high affinity and low affinity component, with an average Hill slope of −0.74 ± 0.06 (n=4). The higher affinity component was dominant at the chosen radiolabel concentration and resolved to a Km,app of 7.19 ± 1.77 µM (n=4) (Fig. 2E) but no consistent estimate for the lower affinity component could be obtained. Dose-response experiments yielded Ki,app values of 1.98 ± 0.50 µM for uridine (n=3; Hill slope −0.88 ± 0.09; Fig. 2E) and 2.18 ± 0.29 for adenosine (n=4; Hill slope −0.71 ± 0.06).
5. Thymidine uptake.
Uptake of 1 µM thymidine was linear for at least 180 s (r2 = 0.98) with a rate of 0.139 ± 0.008 pmol(107 cells)−1s−1. This was strongly (90.6%, P<0.0001, F-test) but incompletely inhibited by 1 mM unlabelled thymidine (slope significantly non-zero, P=0.020; Fig. 3A). At the lower concentration of 0.125 µM, the averaged rate was determined as 0.0045 ± 0.0012 pmol(107 cells)−1s−1 (n=4; slope non-zero, P = 0.0002) and was likewise only partially saturated by 1 mM thymidine (Fig. 3B), consistent with a mixed higher and lower affinity uptake of thymidine. At the lower permeant concentration of 125 nM [3H]-thymidine, the higher affinity component was highly dominant and dose-dependent thymidine uptake was near-monophasic (Hill slope = −0.88 ± 0.01, n=3), yielding a Km,app of 13.2 ± 0.8 µM and a Vmax of 0.69 ± 0.37 pmol(107 cells)−1s−1 (n=3) (Fig. 3C). This transporter appeared not to discriminate between thymidine and uridine (Fig. 3C), which displayed a Ki,app of 12.2 ± 1.2 µM with a Hill slope of −0.76 ± 0.12 (n=3), but had lower affinity for purine nucleosides, with moderate affinity for adenosine, guanosine and inosine (Ki,app of 39.5 ± 8.1 µM (n=4), 36.5 ± 7.3 µM (n=4) and 77.7 ± 15.1 µM (n=3), respectively (Table 1). The three purine nucleosides all displayed near-monophasic inhibition with Hill slopes consistently near −1. In contrast, inhibition by cytidine appeared to be biphasic (Fig. 3D), although the two individual Ki values could not be determined separately from the available data, as large 95% confidence intervals (95%-CI) were returned in each case. From a series of three experiments, they could be estimated as being approximately 15 µM and 250 µM. None of the nucleobases, adenine, hypoxanthine and uracil, inhibited transport of 125 nM [3H]-thymidine by more than 50% at 1 mM (n=3).
Figure 3.
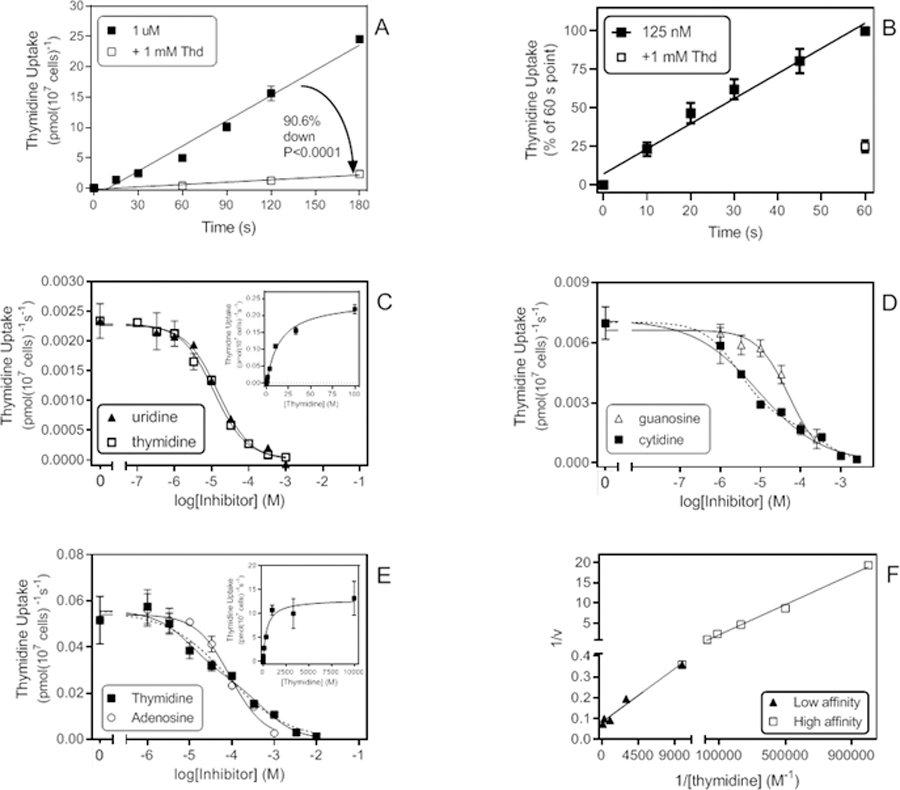
Thymidine uptake in trophozoites of T. vaginalis. A. Time course of 1 µM [3H]-thymidine, displaying a rate of 0.139 ± 0.008 pmol(107 cells)−1s−1 (r2 =0.98; not significantly non-linear, P = 0.20; significantly different from zero, P < 0.0001). In the presence of 1 mM thymidine, the rate was reduced by 90.6% to 0.013 ± 0.0018 pmol(107 cells)−1s−1 (r2 =0.98; not significantly non-linear, P = 0.67; significantly different from zero, P = 0.02). B. Time course of 0.125 µM [3H]-thymidine. Data are presented as the average of 4 experiments over 60 s. Rate was 0.0045 ± 0.0012 pmol(107 cells)−1s−1 (n=4), inhibited 75% by 1 mM unlabelled thymidine. C. Dose-dependent inhibition of 0.125 µM [3H]-thymidine uptake by thymidine and uridine. Thymidine: Hill slope = −0.89, EC50 = 11.7 µM, r2 = 0.996; Uridine: Hill slope = −0.95, EC50 = 15.3 µM, r2 = 0.993. Inset: conversion to Michaelis-Menten saturation plot, Km = 15.1 µM, Vmax = 0.245 pmol(107 cells)−1s−1 for this experiment. Incubation time was 30 s. D. Dose-dependent inhibition of 0.125 µM [3H]-thymidine transport by guanosine and cytidine (Hill slopes of −1.09 and −0.54, respectively). EC50 (guo) = 51.2 µM in this experiment (r2 0.98). 2-transporter plot for cytidine (dashed line) yielded EC50 values of 2.86 µM and 322 µM (r2 0.99). E. Inhibition plots with 1 µM [3H]-thymidine as radiotracer. Hill slopes for thymidine and adenosine were −0.61 and −1.09, respectively. When plotted to a 2-site equation (dashed line), the thymidine inhibition curve yielded EC50 values of 10.7 µM and 529 µM (r2 = 0.99); adenosine EC50 = 89.8 µM in this experiment. Inset: conversion to saturation curve, yielding Km = 394 µM, Vmax = 12.9 pmol(107 cells)−1s−1 (r2 = 0.98).F. Lineweaver-Burk double reciprocal plot from the thymidine dose-response data in C. Low affinity component: Km 340 µM, Vmax 12.0 pmol(107 cells)−1s−1, r2 0.98; Higher affinity Km 23.6 µM, Vmax 2.2 pmol(107 cells)−1s−1, r2 0.99.
At 1 µM [3H]-thymidine, transport was more clearly a mixture of the higher and lower affinity thymidine flux (Fig. 3E), reflected in an average Hill slope of −0.69 ± 0.08 (n=3). For the lower affinity component, a Michaelis-Menten curve could now be obtained and this yielded a Km,app of 470 ± 104 µM (Inset of Fig. 3E). A double reciprocal Lineweaver-Burk plot (Fig. 3F) yielded highly similar estimates as well as estimates for the higher affinity Km,app, which averaged ~20 µM over 3 experiments. This low affinity thymidine flux was sensitive to inhibition by adenosine (Fig. 3E) with Ki,app 73.0 ± 6.9 µM, which indicated that the same transporter could potentially also function as a low affinity adenosine transporter.
6. Low affinity adenosine transport.
The presence of a low affinity adenosine transport capability was suspected not just on the basis of inhibition on the low affinity thymidine flux but on the Hill slope for adenosine inhibition of multiple permeants being above −1, including the inhibition curves for 150 nM [3H]-adenosine (section 1). We probed the presence of this low-affinity carrier using a concentration of 20 µM [3H]-adenosine to fully saturate the high affinity adenosine uptake. The Km,app was determined as 59.4 ± 6.1 µM and Vmax as 31 ± 2 pmol(107 cells)−1s−1 (n=3; Fig. 4A), not statistically different from the Ki value for adenosine on the low affinity thymidine transport activity (P = 0.29, Student’s unpaired t-test). Similarly, the Ki,app for thymidine, at 557 ± 135 µM (n=4; Fig. 4B) was not significantly different from the thymidine low affinity Km,app (P = 0.66), an indication that this may be the same transporter. Apparent Ki values for uridine (Fig. 4B) and cytidine were 376 ± 63 µM and 116 ± 5 µM, respectively (n=3). No inhibition above 50% was observed at the highest tested concentrations of guanosine (250 µM, n=2), adenine (1 mM, n=3), hypoxanthine (1 mM, n=3) or uracil (10 mM, n=2). We conclude that this is a high capacity transporter with modest affinity for adenosine and low affinity for the pyrimidine nucleosides.
Figure 4.
Low affinity adenosine transport. A. Inhibition of transport of 20 µM [3H]-adenosine by unlabelled adenosine and cytidine; incubation time was 30 s. Cytidine: EC50 = 290 µM, Hill slope −1.06, r2 = 0.96; Adenosine: EC50 = 60.5 µM, Hill slope −0.83, r2 = 0.98 for this experiment. Inset: Conversion of adenosine inhibition data to Michaelis-Menten plot, Km,app = 63.5 µM, r2 = 0.99. B. Inhibition of 20 µM [3H]-adenosine by unlabelled thymidine and uridine. Thymidine: EC50 = 817 µM, Hill slope −1.39, r2 = 0.99; Uridine: EC50 = 519 µM, Hill slope −1.21, r2 = 0.97.
7. Inosine transport.
Inosine uptake could be measured at 1 µM at a rate of 0.0081 pmol(107 cells)−1s−1, which was significantly (P < 0.0001, F-test) but incompletely inhibited by 1 mM unlabelled inosine (88%, significantly non-zero uptake, P = 0.0018; Fig. 5A). Dose-response experiments yielded a Km,app of 196 ± 18 µM, with a Hill slope −0.95 ± 15, consistent with a single transporter and with the approximately 90% inhibition by 1 mM adenosine in the time course experiment; the Vmax was 2.25 ± 0.37 pmol(107 cells)−1s−1 (n=3) (Fig. 5B). Inhibition by adenosine was monophasic (Hill slope −0.93 ± 0.04) and produced an apparent Ki of 6.36 ± 1.69 µM (n=3; Fig. 5B). Ki,app values for guanosine and uridine were 7.41 ± 2.92 µM and 3.60 ± 1.14 µM, respectively (n=3), both with Hill slopes of approximately −1. This inhibition pattern is very close to that of the high affinity adenosine transport described above. However, the high-affinity adenosine transport split in two parts, with high and with low affinity for inosine (Fig. 1C). The uptake described in this section clearly delineates the transport component with the lower inosine affinity. We did not observe any high affinity inosine transport, the rate of which may have been insufficient to be clearly observed over the high capacity lower affinity uptake activity.
Figure 5.
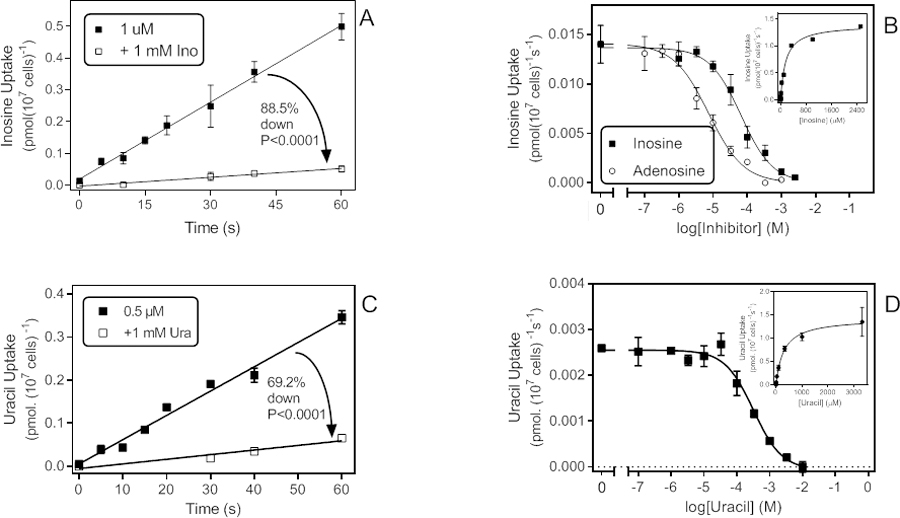
Low affinity uptake of some purine and pyrimidine substrates. A. Time course of 1 µM [3H]-inosine transport in the presence and absence of 1 mM unlabelled inosine. Rate was 0.0081 ± 0.0002 pmol(107 cells)−1s−1 in the absence of unlabelled permeant (r2 = 0.99; not significantly non-linear P = 0.97, significantly different from zero P < 0.0001); in the presence of 1 mM unlabelled inosine the rate was 0.00092 ± 0.00009 pmol(107 cells)−1s−1 (r2 =0.97; not significantly non-linear, P=0.90; significantly different from zero, P = 0.0018). The two slopes were significantly different (F-test, P<0001). B. Dose-dependent inhibition of 1 µM [3H]-inosine uptake by unlabelled inosine or adenosine. Inosine: EC50 = 67.1 µM, Hill slope −0.93, r2 = 0.99; Adenosine: EC50 = 74.8 µM, Hill slope −0.85, r2 = 0.99. Inset: conversion of the inosine inhibition data to a Michaelis-Menten saturation curve, Km,app = 157 µM, r2 = 0.99. C. Time course for the uptake of 0.5 µM [3H]-uracil, with rate 0.0056 ± 0.0003 pmol(107 cells)−1s−1 (r2 =0.98; not significantly non-linear, P=0.89; significantly different from zero, P < 0.0001). In the presence of 1 mM unlabelled uracil, the rate was reduced by 81% to 0.0011 ± 0.0002 pmol(107 cells)−1s−1 (r2 =0.94; not significantly non-linear, P=0.67; significantly different from zero, P < 0.031). D. Dose-dependent uptake of 0.5 µM [3H]-uracil over 20 s; EC50 = 247 µM, r2 0.97 in this experiment. Inset: conversion to Michaelis-Menten saturation plot, Km = 298 µM, Vmax = 1.43 pmol(107 cells)−1s−1, r2 = 0.997.
8. Uracil transport.
We next probed whether T. vaginalis is able to salvage nucleobases from its environment, starting with uracil. Uptake of 0.5 µM [3H]-uracil was linear for at least 60 s and partially inhibited by 1 mM unlabelled uracil (69.2%. P<0.0001) (Fig. 5C). The rate in the presence of 1 mM uracil was significantly different from zero (P<0.031), apparently indicating low affinity uptake. Accordingly, dose-dependent inhibition of 0.5 µM [3H]-uracil uptake over 20 s yielded mono-phasic curves (Hill slope −1.00 ± 0.09, n=3) and a Km,app of 257 ± 58 µM, Vmax 1.58 ± 0.08 pmol(107 cells)−1s−1 (Fig. 5D). Other pyrimidine nucleobases were similarly able to inhibit this transporter, with Ki,app for cytosine, thymine and 5F-uracil at 334 ± 73 µM, 57 ± 8 µM and 265 ± 57, respectively (all n=3). In contrast, pyrimidine nucleosides (uridine, thymidine, up to 10 mM) and purine nucleosides (adenosine, inosine, up to 1 mM) were unable to inhibit uracil transport, even at high concentrations. We conclude that this is a low affinity pyrimidine nucleobase transporter. However, in preliminary experiments saturable uptake of cytosine and thymine over 120 s was not significant at 0.25 µM and 0.5 µM, respectively.
9. Transport of purine nucleobases.
Transport of [3H]-guanine was measurable at 2 µM (as lower concentration yielded insufficient signal), producing a rate of 0.017 ± 0.002 pmol(107 cells)−1s−1 (Fig. 6A). The slope of the linear regression line was significantly non-zero (P = 0.0035) and not significantly non-linear (P = 0.50; r2 = 0.96) but limits to guanine solubility did not allow stringent testing of saturability or a dose-dependent inhibition with unlabelled guanine.
Figure 6.
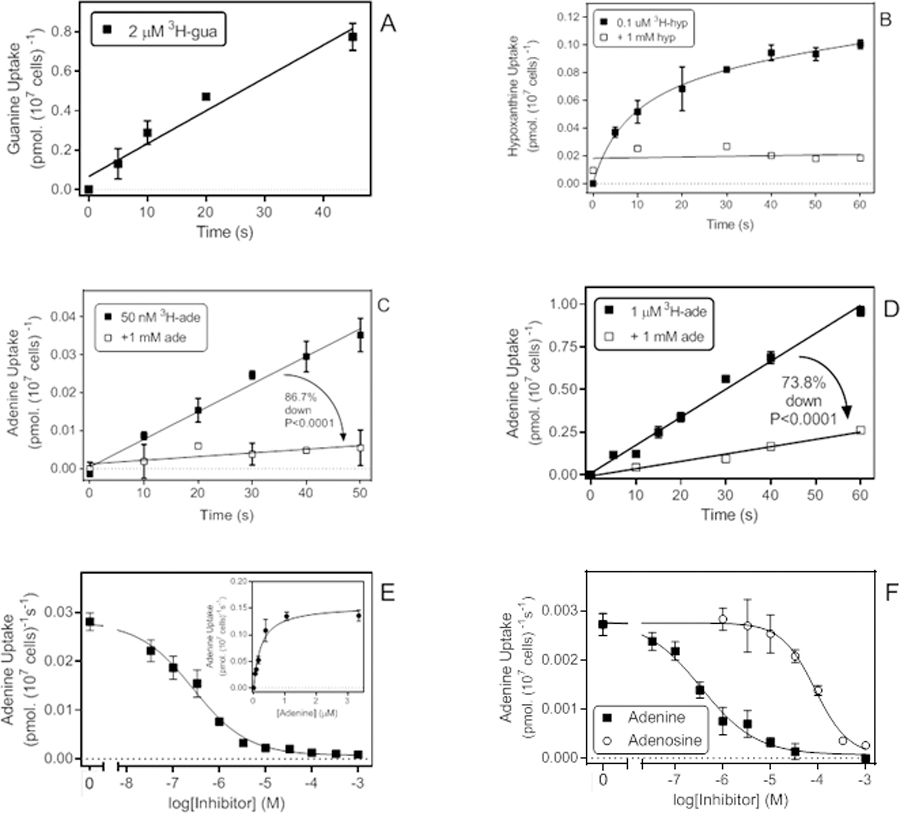
Transport of purine nucleobases. A. Uptake of 2 µM [3H]-guanine with a rate of 0.0167 ± 0.0020 pmol(107 cells)−1s−1 (r2 =0.96; not significantly non-linear, P = 0.50; significantly different from zero, P = 0.0035). B. Uptake of 0.1 µM [3H]-hypoxanthine was non-linear and instead plotted to a curve for saturation binding. In the presence of 1 mM unlabelled hypoxanthine linear regression was applied and yielded a slope that was not significantly different from zero (P=0.74). C. Transport of 50 nM [3H]-adenine in the presence and absence of 1 mM unlabelled adenine, yielding rates of 0.00073 and 0.000097 pmol(107 cells)−1s−1, respectively (P < 0.0001). In the absence of 1 mM adenine the rate was significantly non-zero (P < 0.0001) but in the presence of saturating adenine it was not (P = 0.064). D. Transport of 1 µM [3H]-adenine in the presence and absence of 1 mM unlabelled adenine, yielding rates of 0.0164 and 0.00429 pmol(107 cells)−1s−1, respectively (P < 0.0001). Both rates were significantly non-zero (P < 0.0001 and P = 0.0016, respectively), the regression lines not significantly non-linear, and r2 values were 0.989 and 0.976, respectively. E. Inhibition of [3H]-adenine uptake by unlabelled adenine. EC50 was 0.27 µM for this experiment, Hill slope was −0.74, r2 = 0.992. Inset: conversion to a Michaelis-Menten saturation plot, yielding a Km,app of 0.23 µM and Vmax of 0.155 pmol(107 cells)−1s−1, r2 = 0.979. F. inhibition of 50 nM [3H]-adenine transport by unlabelled adenine (EC50 = 0.35 µM, Hill slope −0.75, r2 = 0.987) and adenosine (EC50 = 85.9 µM, Hill slope −1.23, r2 = 0.994). When plotted to a two transporter model the adenine inhibition curve exhibited a high affinity EC50 of 0.23 µM (81% of flux) and a lower affinity component of 17 µM, but the latter had a large degree of uncertainty.
Uptake of 0.1 µM [3H]-hypoxanthine was low and non-linear over 60 s (Fig. 6B), which seems to indicate that hypoxanthine is not (or very slowly) metabolised by T. vaginalis, as the unmetabolized hypoxanthine equilibrates across the plasma membrane. At 1 µM [3H]-hypoxanthine no specific uptake was observed over 300 s, as the rate was not significantly different in the presence or absence of 1 mM unlabelled hypoxanthine (P = 0.73; data not shown) and we conclude that, at least in culture, T. vaginalis takes up at most very little hypoxanthine.
In contrast, we observed significant uptake of [3H]-adenine, both at 50 nM (0.00073 pmol(107 cells)−1s−1; non-zero P<0.0001) (Fig. 6C) and 1 µM (0.016 pmol(107 cells)−1s−1; non-zero P<0.0001) (Fig. 6D). At both label concentrations, saturation by 1 mM unlabelled adenine was highly significant (P<0.0001): uptake of 50 nM [3H]-adenine was 86.7% inhibited by 1 mM unlabelled adenine (not significantly non-zero, P = 0.064), whereas uptake of 1 µM [3H]-adenine was inhibited by 73.8% (significantly non-zero, P = 0.018). The significant uptake at 50 nM pointing to a high affinity transporter and the incomplete inhibition with 1 mM adenine pointing to a low affinity transporter, a dose-dependent inhibition experiment was performed with 50 nM of [3H]-adenine, which favours uptake through the high-affinity system, yielding a Km, app of 0.41 ± 0.097 µM and Vmax of 0.091 ± 0.027 pmol(107 cells)−1s−1 (n=4) (Fig. 6E). The adenine transporter displayed only low affinity for adenosine (Ki,app 155 ± 48 µM, n=3) (Fig. 6F).
10. Summary of purine and pyrimidine transport by T. vaginalis trophozoites.
Our data show that T. vaginalis trophozoites grown in suspension culture express multiple distinct transporters for salvage of nucleosides and nucleobases, with overlapping substrate specificities. It appears that most natural purines and pyrimidine nucleosides can be taken up by multiple transporters (Table 1). This is an important conclusion but these observations make construction of an unambiguous model for purine and pyrimidine salvage based on analysis of whole-organism cellular uptake studies challenging. Equally, it is difficult to construct a full model of nutrient salvage by an organism through a reductive approach of characterising single transporters through heterologous expression alone - clearly, the two approaches are complementary in order to arrive at a full understanding of purine/pyrimidine salvage. Nonetheless, the cells express at least two high affinity, broad specificity transporters for purine and pyrimidine nucleoside, one of which has high affinity for inosine, one low affinity. Neither has significant affinity for nucleobases. Of the potential substrates, adenosine appears to be taken up more efficiently than the others, as expressed by Vmaz/Km,app (Table 1) and uridine appears to be the best pyrimidine substrate. Thymidine was salvaged relatively poorly, and through separate higher and lower affinity transporters, which may make T. vaginalis vulnerable to antifolates. We also found low affinity uptake of cytidine but are unable to say, based on the current data, whether this is a separate transport activity or the same carrier that is responsible for the low affinity thymidine uptake. Separate nucleobase transporters were also found, including a high affinity and a low affinity adenine carrier as well as likely uptake of guanine, although this remains largely uncharacterised. The only pyrimidine nucleobase uptake we observed was low affinity, low capacity uracil uptake.
ENT genes of T. vaginalis
In order to gain further insights into purine and pyrimidine salvage in T. vaginalis, we next probed its genome for genes of the Equilibrative Nucleoside Transporter (ENT) family, which to date is the only gene family linked to nucleoside/nucleobase transport in protozoa (Campagnaro and De Koning, 2020). BLAST and keyword searches of TrichDB identified nine putative ENT family members, which we designated TvagENT1–9, with 1011 – 3377 bp and 9 – 11 predicted trans-membrane domains (TMDs) (Table 2). Their sequences and a multiple alignment of the 9 genes is shown in the Supplemental materials (Fig. S1). The alignment shows that most of the variation in length is from high variability in the size of the N-terminal domain, most pronounced for TvagENT2, which lacks 135 N-terminal amino acids relative to TvagENT1, including all of TMD1 and part of TMD2, posing the question whether this transporter can be fully functional; it is the only TvagENT with less than 10 TMDs. From a phylogenetic tree (Fig. 7) it can be seen that all TvagENTs are more similar to each other than to other protozoan or human ENT genes, and that the TvagENTs essentially split into two clades, with one consisting of TvagENT1, 2, 3, 6 and 8, and the other of TvagENT4, 5, 7 and 9.
Table 2.
ENT family genes in Trichomonas vaginalis.
| Name | Accession Code | Size (bp / aa) | TM domains |
|---|---|---|---|
| TvagENT1 | TVAG_166380 | 1377 / 458 | 11 |
| TvagENT2 | TVAG_192810 | 1011 / 336 | 9 |
| TvagENT3 | TVAG_271560 | 1275 / 424 | 10 |
| TvagENT4 | TVAG_441760 | 1203 / 400 | 10 |
| TvagENT5 | TVAG_483030 | 1200 / 399 | 11 |
| TvagENT6 | TVAG_053320 | 1266 / 421 | 10 |
| TvagENT7 | TVAG_101510 | 1206 / 401 | 11 |
| TvagENT8 | TVAG_271570 | 1287 / 428 | 10 |
| TvagENT9 | TVAG_341290 | 1227 / 408 | 10 |
Source: TrichDB.org
Figure 7.
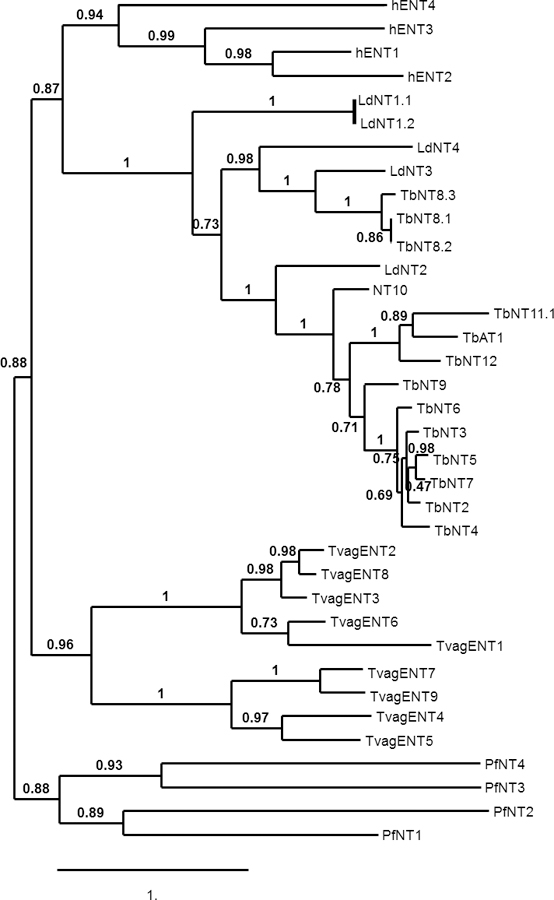
Phylogenetic tree of human and protozoan ENT family genes including T. vaginalis. Multiple alignment (MUSCLE), phylogeny (PhyML) and tree rendering (TreeDyn) were performed using phylogeny.fr (http://www.phylogeny.fr/index.cgi). Accession codes for all genes are listed in the Supplemental materials). The Scale bar indicates the number of substitutions of a given site.
Expression levels of TvagENTs in trophozoites
The relative level of expression of the TvagENTs in trophozoites was determined using qRT-PCR, standardised to the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH), in a panel of three metronidazole-sensitive and three -resistant T. vaginalis isolates. The expression pattern of the TvagENTs was very similar in all 6 isolates, with the highest expression consistently found for TvagENT5 (Fig. 8). An exception was TvagENT9, which was expressed at a low level in isolates G3 and S1469 but robustly in all other strains (Fig. 8). To explore whether substrate availability had an effect on TvagENT expression, we assessed expression in the presence of 100 µM of adenosine, hypoxanthine or inosine. For TvagENT3–9, no significant effects on expression were observed but, interestingly, expression of TvagENT1 was reduced under these conditions and expression of TvagENT2 increased, although the latter trend only reached significance for incubation with inosine (Fig. 9A). We also tested whether the presence of (and feeding on) HeLa cell layers would change the level of ENT expression, but no significant changes were observed (Fig. 9B). Together, these results indicate that the TvagENT expression is largely constitutive regardless of high substrate availability or the presence of human feeder cells.
Figure 8.
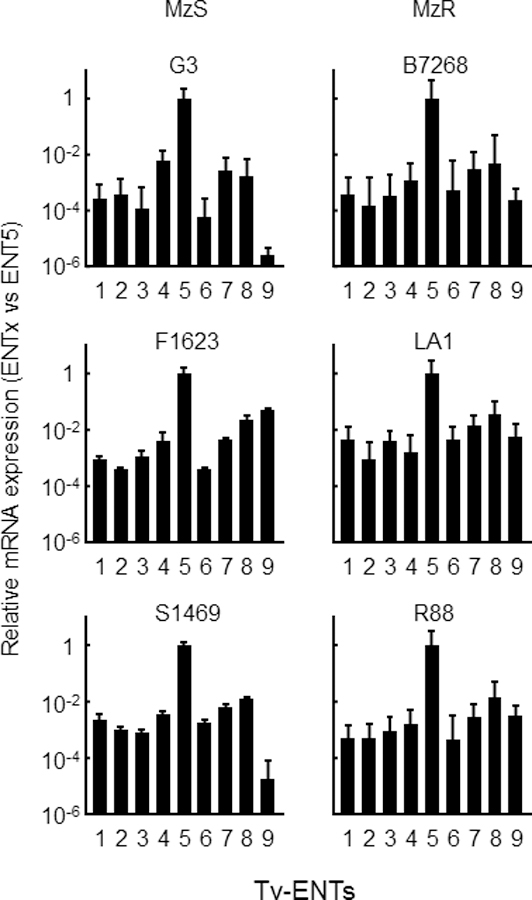
Expression patterns of ENT genes in diverse T. vaginalis strains. RNA was extracted from the indicated T. vaginalis strains, three of which are sensitive to metronidazole (MzS) and the other resistant to metronidazole (MzR), grown at log phase. Expression levels of the indicated TvagENTs were analyzed by quantitative PCR Raw Ct values were standardized by GAPDH, and all values are expressed relative to Tv ENT5. Bars represent means and SEM of 3–4 independent experiments.
Figure 9.
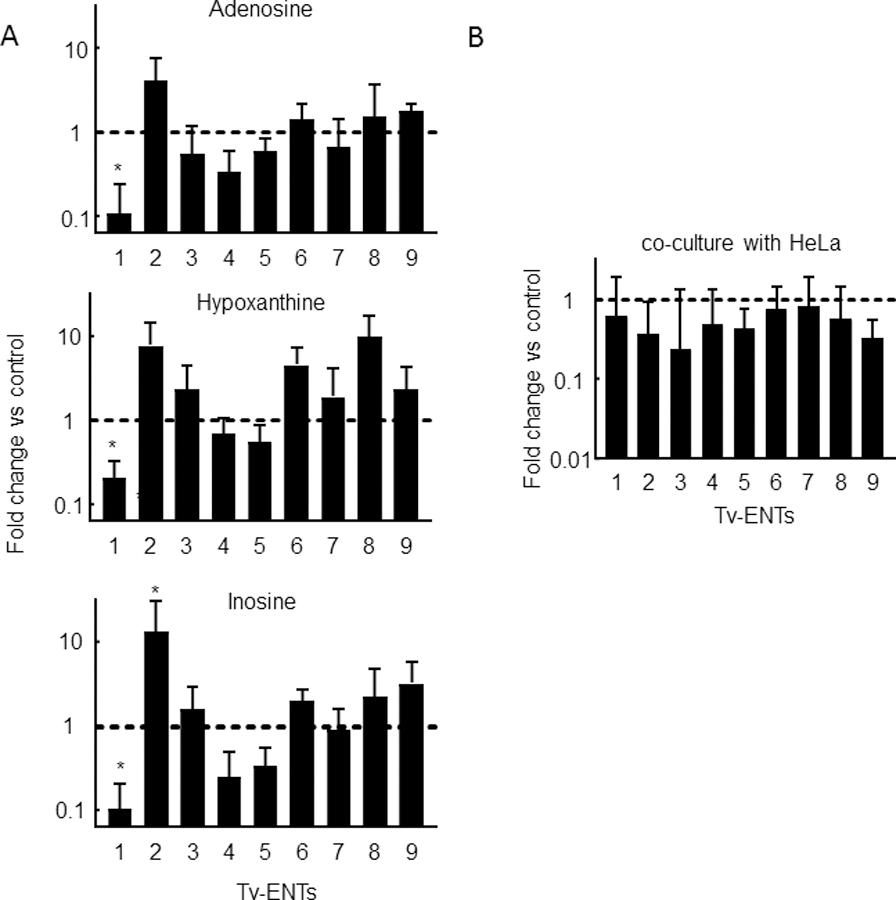
Impact of culture conditions on T. vaginalis ENT expression patterns. A. T. vaginalis G3 trophozoites were incubated with or without (control) the indicated nucleosides (100 µM) overnight. RNA was extracted and analyzed by quantitative PCR for ENT expression. B. T. vaginalis G3 were incubated with (experimental) or without (control) HeLa cells in 6 well plates with 25% TYM and 75 % DMEM for 6 hours. RNA was extracted and analyzed by qPCR. Ct values were standardized by GAPDH and expression levels were normalized to controls. Bars represent the means and SEM of 3–4 independent experiments; *p<0.05 vs control condition by ANOVA and Dunnett’s post-hoc.
Heterologous expression of TvagENTs in Trypanosoma brucei brucei TbAT1-KO
To start assigning gene identities to the observed nucleoside and nucleobase transport activities of T. vaginalis trophozoites, each individual gene was to be expressed in a T. b. brucei strain, TbAT1-KO, in which we had successfully characterised nucleoside transporters from several other protozoan species including Trypanosoma cruzi (Campagnaro et al., 2018b), Trypanosoma congolense (Munday et al., 2013), Leishmania spp (Alzahrani et al., 2017) and Toxoplasma gondii (Campagnaro et al., manuscript in preparation).
As T. b. brucei is able to synthesise its own pyrimidine nucleotides (Ali et al., 2013a), bloodstream forms are known to have a very limited ability to take up any pyrimidines (Gudin et al., 2006; Ali et al., 2013a,b) except uracil, which is efficiently taken up by the U3 transporter (Ali et al., 2003a). However, the P1 transporter sub-family, consisting of at least 8 distinct ENT-family genes (Sanchez et al., 2002; Al-Salabi et al., 2007; Campagnaro and De Koning, 2020), is also capable of some thymidine uptake (Ali et al., 2013b) with affinity of 44 µM (De Koning and Jarvis, 1999), although P1-type transporters are principally high affinity, broad specificity purine nucleoside carriers (De Koning et al., 1998; Campagnaro and De Koning, 2020). For these reasons the TbAT1-KO strain is particularly useful for the expression and characterisation of transporters of uridine and, particularly, cytidine, which was fortuitous as these are high affinity substrates of the main broad specificity nucleoside transporter(s) of T. vaginalis (Table 1).
The nucleotide sequences of the nine TvagENTs were codon-optimised for T. brucei codon usage bias using the Codon Adaptative Index described by Rashmi and Swati (2013). The thus optimised sequences were custom synthesised and cloned into the pHD1336 expression vector (Biebinger et al., 1997; Munday et al., 2015) for expression in the T. b. brucei strain TbAT1-KO, from which the aminopurine transporter TbAT1 had been deleted by homologous recombination (Matovu et al., 2003). Plasmid DNA was then used to transfect bloodstream forms of T. b. brucei TbAT1-KO followed by selection on blasticidin, and cloned out by limiting dilution. The presence and integrity of the constructs was verified by Sanger sequencing. For each TvagENT, three clones were screened by qRT-PCR for expression. The highest-expressing clone was selected for each gene of interest, to be used in nucleoside transport experiments.
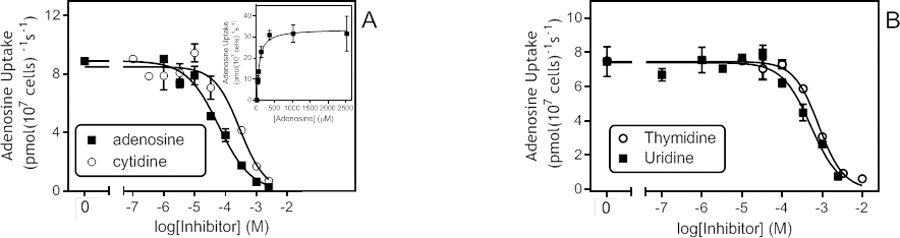
All TvagENT-expressing T. b. brucei lines were tested for their ability to take up 0.5 µM [3H]-cytidine over 10 min. Fig. 10A shows the uptake rates calculated from the linear regression lines. Although several TvagENTs exhibited significant cytidine uptake above control (ENT2 (P = 0.029), ENT5 (P = 0.0084), ENT6 (P = 0.0024) and ENT9 (P = 0.015)), uptake by TvagENT3 was by far the highest. Uptake of cytidine by TvagENT3 was linear (non-zero uptake P = 0.0009; r2 = 0.95; not significant non-linear, P = 0.70) (Fig. 10B). It was inhibited completely by 400 µM of adenosine, uridine or inosine (slope not significantly non-zero, P >0.2) but only incompletely by thymidine (62.6%, slope significantly different from zero, P = 0.0074) (Fig. 10C). The cytidine Km,app for this transporter was determined at 10.2 ± 2.6 µM (n=4, Fig. 10D). The same graph also shows monophasic inhibition curves for inosine with Ki,app 3.3 ± 0.07 µM (Hill slope −1.08 ± 0.02, n=3) and for guanosine with Ki,app 2.9 ± 0.14 µM (Hill slope −1.33 ± 0.09, n=3), i.e. no discrimination between the two oxopurine nucleosides (Fig. 10D). The inhibition profile of TvagENT3 was consistent with it encoding the inosine-sensitive, broad selectivity transporter observed in T. vaginalis, with high affinity for adenosine (Ki = 0.87 ± 0.15 µM, Hill slope −1.04 ± 0.07; n=5), somewhat lower affinity for uridine (Ki = 11.3 ± 1.8 µM, Hill slope −0.98 ± 0.08; n=4) (Fig. 10E) and low affinity for thymidine (Ki = 286 ± 12 µM, Hill slope −1.17 ± 0.15; n=4) (Fig. 10F). We also tested 2’F,2’-deoxyadenosine on this transporter, as it is an extraordinarily potent inhibitor of P1-mediated adenosine transport in T. brucei bloodstream forms (Ki = 63 ± 9 nM; Ranjbarian et al., 2017) and could be used to specifically inhibit this uptake if it displayed low affinity for TvagENT3. However, 2’F, 2’-deoxyadenosine also displayed high affinity for the T. vaginalis transporter (Ki = 1.11 ± 0.38 µM, Hill slope −0.97± 0.15; n=4) (Fig. 10F).
Figure 10.
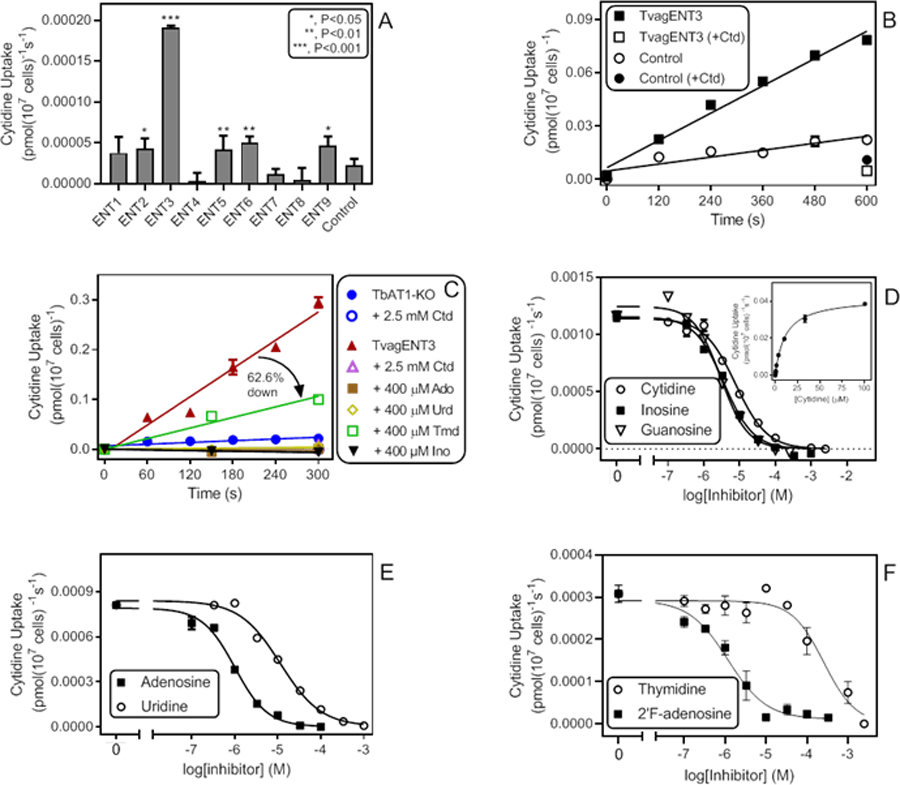
Cytidine uptake in T. brucei expressing TvagENT3. A. Uptake of 0.5 µM [3H]-cytidine by T. brucei TbAT1-KO cells expressing one of the indicated TvagENT genes, as calculated from linear regression of uptake over 600 s with time points at 0, 180, 360 and 600 s. Bars represent average and SEM of the calculated slope. B. Uptake of 0.5 µM [3H]-cytidine by T. brucei TbAT1-KO cells expressing TvagENT3 over 600 s (r2 0.982, significantly non-zero P = 0.0001, not significantly non-linear P = 0.40). Control cells were T. brucei TbAT1-KO transfected with the empty vector r2 0.83, significantly non-zero P = 0.012, not significantly non-linear P = 0.90). [3H]-Cytidine uptake by either strain was strongly inhibited by the addition of 1 mM unlabelled cytidine (+Ctd). C. Transport of 0.5 µM [3H]-cytidine by T. brucei TbAT1-KO cells expressing TvagENT3. Control cells were T. brucei TbAT1-KO transfected with the empty vector. Transport of the uninhibited uptake (closed red triangles; r2 = 0.968, significantly different from zero P = 0.0004) was partly inhibited by 400 µM thymidine (62.6 %, significantly different from zero P = 0.0074) and completely inhibited by uridine, adenosine and inosine (not significantly different from zero, P > 0.15). D. Inhibition by cytidine, inosine and guanosine (Hill slopes −0.99, −1.07 and-1.15, respectively) of transport of 0.5 µM [3H]-cytidine over ** s. Inset: conversion of the cytidine inhibition data to a Michaelis-Menten saturation plot with Km 10.9 µM and Vmax 0.042 pmol(107 cells)−1s−1, r2 = 0.998. E. Inhibition by adenosine and uridine (Hill slopes −1.1 and −0.93, respectively), F. Inhibition by thymidine and 2’F-adenosine, with Hill slopes of −1.1 and-0.92, respectively.
As expected, a screen using a low concentration of [3H]-thymidine was not particularly revealing, due to the relatively high background, although cells expressing TvagENT3 again displayed the highest rate, closely followed by those expressing TvagENT6 (P >0.05; Fig. 11A). Uptake of 0.25 µM [3H]-uridine suffered from similar issues but highlighted TvagENT6 as the most efficient uridine transporter (Fig. 11B). The high background rate in control cells was caused by uridine uptake through a uracil transporter (Alzahrani et al., 2017). Thus, the addition of 250 µM of uracil to the assay buffer reduced background uridine uptake by ~85% (Fig. 11C) and allowed the determination of Km and Ki values (Fig. 11D). The uridine Km,app for TvagENT6 was 169 ± 39 µM with a Vmax of 0.092 ± 0.021 pmol(107 cells)−1s−1 (n=5). Ki,app values for adenosine and inosine were 47.2 ± 6.4 µM (n=4) and 40.0 ± 9.5 µM (n=5), respectively (Fig. 11D). The transporter was not significantly inhibited by adenine up to 1 mM (n = 2). From this partial characterisation, it appears that TvagENT6 encodes a transporter with moderate affinity for purine nucleosides, lower affinity for uridine, and probably some limited capacity for thymidine uptake.
Figure 11.
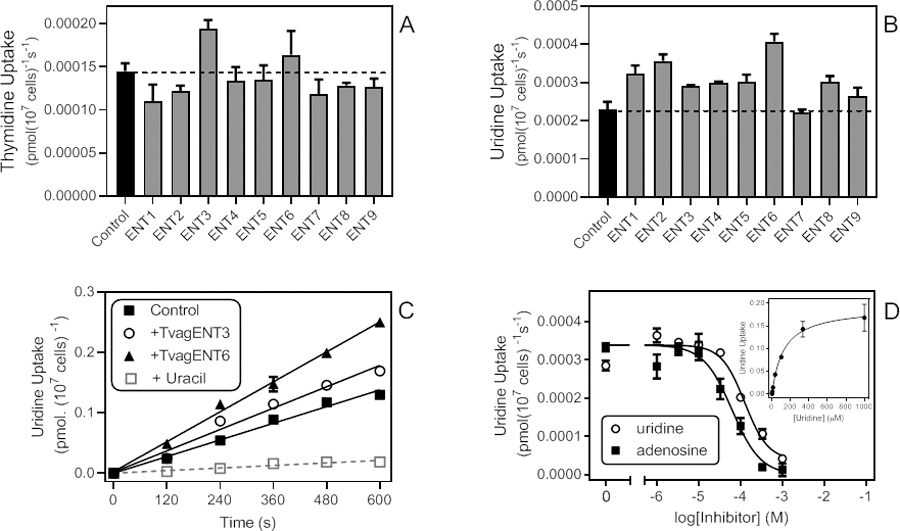
Uptake of thymidine and uridine in T. brucei expressing TvagENTs. A. Uptake rates of 0.25 µM [3H]-thymidine, determined by linear regression of a time course over 300 s, for all nine TvagENTs. Bars show average and SEM of the slope. The dashed line indicates the level of uptake in control cells with empty vector pHD1336. B. Uptake rates of 0.25 µM [3H]-uridine; linear regression of a time course over 600 s, for all nine TvagENTs (measured at 0, 2, 4, 6, 8 and 10 min). C. Separate time course of 0.25 µM [3H]-uridine uptake by three cell lines, highlighting the high background uptake in the control (empty vector) cells. This was reduced by 84.7% in the presence of 250 µM uracil (P < 0.0001). Uptake by cells expressing TvagENT3 and TvagENT6 was significantly higher than empty vector control (P = 0.0332 and P<0.0001, respectively). D. Dose-response curves for inhibition of uptake of 0.5 µM [3H]-uridine over 240 s, by adenosine (r2 = 0.972) and uridine (r2 = 0.972). Inset: conversion of the uridine inhibition data to a Michaelis-Menten saturation curve, showing Km,app = 125.4 µM for this experiment.
Discussion
In this study we have attempted to map out the nucleoside and nucleobase transport activities of T. vaginalis. The effort is complicated as trophozoites express multiple such transporters, with overlapping substrate selectivities and a wide range of substrate affinities. This makes it hard to characterise single carriers by kinetic analysis of whole-cell transport assays only. Yet, the whole cell analysis helped to establish which purine and pyrimidines are salvaged by the parasite and with what affinities and rates. The efficiency of uptake, defined as Vmax/Km, was highest for high-affinity adenosine transport, at 1.5, compared to 0.10 for high affinity guanosine uptake, making adenosine the preferred purine substrate. Uptake of adenine was relatively weak with low Vmax, with an efficiency of 0.22. For pyrimidines the highest affinity and efficiency was observed for uridine followed by cytidine and then thymidine. Although uracil uptake is robust in kinetoplastid parasites (De Koning and Jarvis, 1998; Papageorgiou et al., 2005; Alzahrani et al., 2017) and Toxoplasma gondii (Natto and De Koning, unpublished), as well as other microbes including Saccharomyces cerevisiae, Aspergillus nidulans and E. coli (De Koning and Diallinas, 2000; Campagnaro and De Koning, 2020), uptake of uracil (efficiency 0.0061) and other pyrimidine nucleobases was marginal.
The observations of efficient uptake of purine nucleosides rather than nucleobases are largely consistent with observations and models from before the T. vaginalis genome was published (Carlton et al., 2007). Hypoxanthine has been reported to not or barely be incorporated into the T. vaginalis nucleotide pool (Heyworth et al., 1982; Munagala and Wang, 2003) and indeed we found little or no uptake of this nucleobase. However, adenine and guanine could be salvaged and incorporated into nucleotides (Munugala and Wang, 2003). Miller and Lindstead (1983) reported that they were unable to detect any phosphoribosyltransferase activity with any of the purine nucleobases, only with uracil, but the same authors did find that the purine nucleobases, including hypoxanthine and xanthine, could be converted by cell-free extracts of T. vaginalis to nucleosides using purine nucleoside phosphorylase (PNP). This enzyme has been isolated from extracts (Miller and Miller, 1985) and cloned (Munagala and Wang, 2002), and the structure has been elucidated (Rinaldo-Matthis et al., 2007). For the pyrimidine nucleobase, a uracil phosphoribosyltransferase activity was reported in T. vaginalis extracts by Miller and Lindstead (1983) but not by Wang and Cheng (1984), who attributed the incorporation of uracil to a uridine phosphorylase. We were unable to identify a candidate gene for uracil phosphoribosyltransferase in the T. vaginalis genome but did identify a candidate uridine phosphorylase (XP_001323814.1; TrichDB).
We observed very little overlap between nucleoside transporters and nucleobase transporters in T. vaginalis, as is generally the case for protozoan purine transporters (De Koning et al., 2005; Campagnaro and De Koning, 2020), although there are some notable exceptions like the TbAT1 adenosine/adenine transporter of T. b. brucei (De Koning and Jarvis, 1999), the UUT1 uridine-uracil transporter of Leishmania major (Alzahrani et al., 2017) and the Plasmodium falciparum NT1 carrier that transports both hypoxanthine and adenosine (Quashie et al., 2008). The strict separation of nucleoside and nucleobase transport activities in T. vaginalis appears to fit well with the organism’s overall preference for nucleosides over nucleobases. Indeed, it is possible that the nucleobase carriers have more of a sensory/regulatory role than one of providing significant amounts of nutrients, judging by their very low Vmax and/or efficiency, and the observation that the presence of hypoxanthine influenced the expression of some of the TvagENTs, especially TvagENT1.
The most active nucleoside transporter in T. vaginalis trophozoites appears to most efficiently transport adenosine, in keeping with adenosine in the form of ATP being most likely the most abundant purine available to it for salvage and T. vaginalis expressing a number of apyrases and ecto-phosphohydrolases to convert extracellular nucleotides to the corresponding nucleosides (de Jesus et al., 2006; de Aguiar Matos et al., 2001). The same transporter also displayed high affinity for uridine, guanosine and cytidine (Table 1). Our analysis strongly suggests there are at least two such transporters expressed in trophozoites, one with high affinity for inosine, here provisionally designated NT-a until the gene ID is definitively known, and one with low affinity, NT-b. In addition, we found a lower affinity adenosine > cytidine > uridine > thymidine transporter (NT-c), and a thymidine transporter with similar affinity for uridine and moderate affinity for adenosine and guanosine (NT-d). Together, our analysis suggests a minimum of 4 nucleoside transporters and up to three nucleobase transporters in T. vaginalis (NT-e for adenine, NT-f for uracil, NT-g for guanine) (Fig. 12). However, the characterisation of guanine transport has been incomplete, without Km or inhibition profile and we cannot exclude the possibility that it overlaps with the NT-e adenine transporter activity or one of the nucleoside transport activities.
Figure 12.
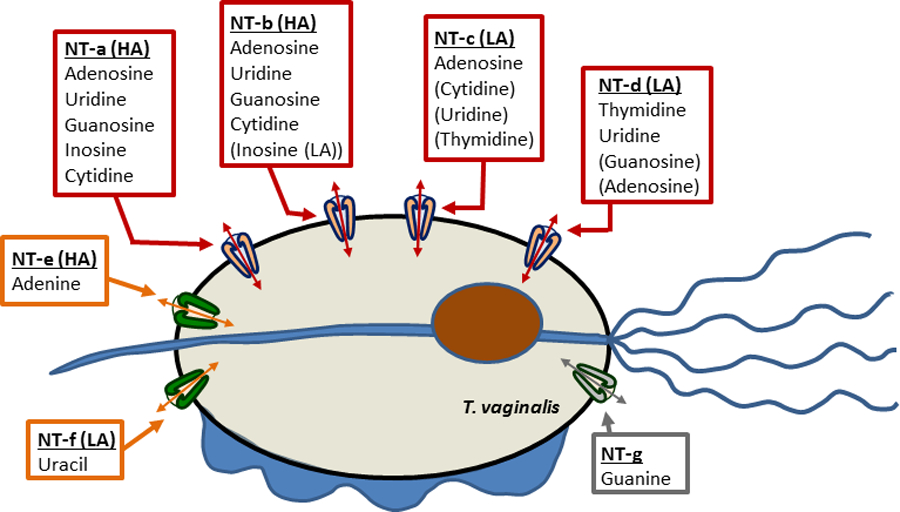
Diagram of nucleoside (red boxes and arrows) and nucleobase (orange boxes and arrows) transporters in Trichomonas vaginalis. HA, high affinity; LA, (relatively) low affinity. Substrates indicated in brackets are of substantial lower affinity than those without brackets. Arrows indicate bi-directional traffic (equilibrative) but it is not yet known whether (some of) the transporters are active transporters, which would imply mono-directionality. The guanine uptake may be mediated by a separate transporter, which would be designated NT-g. However, at this juncture the possibility that the guanine uptake is mediated by one of the other transporters rather than a separate gene product cannot be excluded.
The low affinity [3H]-inosine transport is most likely mediated by the NT-b activity as the Ki values for adenosine, guanosine and uridine are all very close to those obtained with [3H]-adenosine. Similarly, high affinity uptake of uridine and cytidine appears to be mediated jointly by NT-a and NT-b. The low affinity [3H]-thymidine uptake could well be mediated by NT-c, considering the adenosine Ki value is highly similar to the Km for NT-c and the NT-c Ki for thymidine is similar to the Km for low affinity thymidine uptake.
Although our comprehensive transporter characterizations give a well-supported view of nucleoside and nucleobase uptake in T. vaginalis, the data cannot resolve how many and which ENT-family (or other gene family) genes are involved in the various transport processes. As a complement to the functional transport studies in trophozoites, we therefore identified the TvagENT genes from the TrichDB database, had them synthesised in the Trypanosoma brucei codon preference and introduced them individually in the TbAT1-KO strain of T. b. brucei. The new strains were screened for the transport of pyrimidine nucleosides and uptake of cytidine was the most effective tool, identifying TvagENT3 as the carrier that mediated its uptake most strongly, at conditions designed to identify the high affinity flux. The inhibition profile aligned quite closely to that of NT-a, with high affinity for inosine, adenosine, guanosine and uridine but not thymidine or nucleobases. TvagENT3 is functionally related to the broad specificity T. brucei P1-type transporters (De Koning and Jarvis, 1999), although the relatively high affinity for cytidine may be unique amongst protozoan transporters characterised to date, and like P1, its broad specificity/high affinity should be valuable in targeting cytotoxic nucleoside analogues to the T. vaginalis interior (Geiser et al., 2005; Hulpia et al., 2019; Ranjbarian et al., 2017). Indeed, we have very recently reported the identification of a series of strongly antitrichomonal nucleoside analogues (Natto et al., 2021) and other nucleoside analogues with activity against this parasite have been reported (Munagala and Wang, 2003; Shokar et al., 2012).
After TvagENT3, TvagENT6 displayed the most significant cytidine uptake as well as the most robust uridine uptake, and we used uptake of [3H]-uridine in the presence of 250 µM uracil for a partial characterisation of this carrier. In this case, the characterisation indicated alignment with the NT-c activity observed in trophozoites, as the affinities for adenosine and uridine, in particular, are very similar. Definitive assignment any gene ID to a specific transport activity in T. vaginalis trophozoites, however, will require further studies, such as gene deletions or the identification of sufficiently specific inhibitors.
In this work we present a model of multiple high and low-affinity, broad specificity nucleobase transporters and separate nucleobase transporters in T. vaginalis. Only a single study of purine or pyrimidine transport in this species has previously been reported (Harris et al., 1988). That study did not address nucleobase uptake but did describe expression of at least two high affinity, broad specificity nucleoside transporters with overlapping substrate specificity. Indeed, their Km values for adenosine (3.9 µM), guanosine (13.9 µM) and uridine (2.5 µM) are remarkably similar to those we report here for the NT-a activity (6.2, 12.2 and 3.7 µM, respectively using [3H]-adenosine as substrate). In addition, we identified three additional nucleoside transport activities and three nucleobase transport activities in trophozoites and identified the genes encoding two of the main nucleoside transporters through heterologous expression. This study thus presents the most complete model yet of nucleoside/nucleobase transport in a trichomonad, or indeed any protozoan that is auxotrophic for both purines and pyrimidines. We find that the main transporters of T. vaginalis are able to take up purine nucleosides and pyrimidine nucleosides with similar affinity. In keeping with most protozoa no pyrimidine-specific nucleoside transporters were identified (the sole documented exception is the T. cruzi NT2 transporter (Campagnaro et al., 2018b)), although NT-d did display higher affinity for uridine and thymidine than for adenosine and guanosine. Mixed purine-pyrimidine nucleoside transporters have been described in Leishmania (Vasudevan et al., 1998) and Toxoplasma gondii (De Koning et al., 2003), whereas purine-specific nucleoside transporters are the norm in T. brucei (De Koning and Jarvis, 1997, 1999) and Plasmodium falciparum (Quashie et al., 2008). A uracil transporter, NT-f, was also identified and uracil-specific transporters were previously reported in L. major (Papageorgiou et al., 2005) and T. brucei (De Koning and Jarvis, 1998). The one new feature of the T. vaginalis transporters, relative to those of other protozoa, is the high affinity for cytidine by NT-a and NT-b.
The characterisation of the T. vaginalis purine and pyrimidine transporters provides a critical foundation for the design of novel trichomonacidal agents that target the nucleoside metabolism in the parasite.
4. Experimental Procedures
4.1. Parasite strains and cell cultures
The following T. vaginalis strains were used: Metronidazole-sensitive G3 (ATCC PRA-98), F1623 (Brown et al., 1999) and S1469 R88 (kindly provided by Dr. Evan Secor, US Centers for Disease Control and Prevention); metronidazole-resistant B7268 (Bradic et al., 2017), LA1 (Goldman et al., 2009) and R88 (kindly provided by Dr. Secor). Trophozoites were grown in vitro, as described, at 37 °C in modified Diamond’s media (MDM) with 10% heat-inactivated horse serum (HIHS; Gibco Life Technologies), with the pH adjusted to 6.3−6.4 (Natto et al., 2015). The MDM composition was 20 g/L trypticase peptone (BD Biosciences, Sparks, USA), 10 g/L yeast extract (Formedium Ltd., UK), 5 g/L maltose monohydrate (Sigma, U.K.), 1 g/L L-ascorbic acid (Sigma), 1 g/L KCl, 1 g/L KHCO3, 1 g/L KH2PO4, 0.5 g/L K2HPO4, and 0.1 g/L FeSO4·2H2O (Natto et al., 2012). Culture flasks were completely filled and tightly capped to maintain anaerobic conditions. Cell density was determined using a haemocytometer (Camlab, Cambridge, U.K.) and phase-contrast microscopy at 40× magnification, or a Beckman Coulter Z2 particle counter.
Bloodstream T. brucei of strain TbAT1-KO, which lacks the P2/TbAT1 aminopurine transporter and the HAPT pentamidine transporter (Matovu et al., 2003), were grown in complete HMI-11 medium (Hirumi and Hirumi, 1989) complemented with 10% Fetal Bovine Serum at 37 °C and 5% CO2, exactly as described (Gudin et al., 2006).
Human cervix epithelial HeLa (ATCC CCL-2) cells were grown in DMEM medium containing 10 % FBS and 5% CO2. For co-culture of T. vaginalis with HeLa cells, subconfluent HeLa monolayers were exposed to doses of 106 G3 trophozoites at a 1:1 ratio in 75 % DMEM and 25 % MDM under 5 % CO2 atmosphere at 37°C for 6 hours. Total cellular RNA (attached trophozoites and HeLa cell) was harvested from the co-cultures for subsequent analysis. HeLa cells alone and trophozoites alone were used as controls.
4.2. Transport assays
Transport assays with T. vaginalis trophozoites and T. brucei bloodstream forms were performed identically and essentially as described previously for T. brucei (Wallace et al., 2002) and Leishmania promastigotes (Alzahrani et al., 2017). Briefly, cells of either species were grown in larger culture flasks (75 cm2 U-shaped Canted Neck Culture Flask with Vent Cap (Corning) for T. brucei, 25 mL glass Universals, completely filled with medium for T. vaginalis), harvested and washed twice by centrifugation (10 min, 1000 × g, room temperature) into an assay buffer (AB (Campagnaro et al., 2018a)) and resuspended at 108 cells/mL. Aliquots of 100 µL (i.e. 107 cells) were incubated for predetermined times with 100 µL of a solution containing [3H]-labelled nucleoside or nucleobase plus a competitive inhibitor, depending on the experiment, each at 2× the final concentration, on top of an oil layer (7:1 v/v mix of di-n-butylphthalate (DBH) and light mineral oil (Sigma)) in a microfuge tube. To stop the uptake of radiolabel, 1 mL of an ice-cold solution of unlabelled permeant, usually at 1 mM or 2.5 mM, was added, followed by immediate centrifugation of the cells through the oil layer (90 s, 13,000 × g). The tubes were flash-frozen in liquid nitrogen and the tips of the tubes, containing the cell pellet cut off into scintillation tubes. Cell were lysed with 2% SDS for at least 1 h under agitation on a rocking platform, and after scintillation fluid (Scintlogic U, Lablogic) was added, the vials were shaken and left to stand overnight in the dark. Radioactivity was determined using a Hidex 300SL scintillation counter and background radiation (from vials to which no radiolabel was added) was subtracted. Radiolabel associated with the cell pellet but not internalised was also subtracted, and defined as radioactivity in the pellet after incubation of the same number of cells in the presence of saturating levels of non-radiolabelled permeant. All assay points were determined in triplicate in each experiment. Results were plotted and analysed with GraphPad Prism (versions 8 and 9), using the inbuilt statistics packages to determine linearity, difference between two lines, goodness of fit, and significances from zero slope etc. Time courses were commonly plotted by linear regression and inhibition experiments used incubation times well within the linear phase of uptake. Dose-response inhibition experiments for the determination of 50% inhibition concentrations (IC50) were plotted to a 4-parameter sigmoid curve with variable slope and Km values through a Michaelis-Menten saturation plot (V0 = Vmax × [S] / (Km + [S]) in which [S] is the substrate concentration) or double reciprocal Lineweaver-Burk plot. Plots typically contained 6–8 points over the relevant range plus a zero-inhibitor control, all in triplicate. Ki values were calculated from IC50s using the Cheng-Prusoff equation Ki = IC50 / [1 + ([S] + Km) (Cheng and Prusoff, 1973). In all cases, care was taken to use a very low permeant concentration so as to obtain the most accurate Km and Ki values and Hill slopes, and avoid saturation of very high affinity transporters. This has the added benefit of extending the linear range of uptake as the low rate of permeant entry at those concentrations will not easily saturate the downstream metabolic reactions. The following radiolabels were used: [5-3H]-cytidine from American Radiolabeled Chemicals (ARC), 20 Ci/mmol; [3H]-uridine from ARC, 60 Ci/mmol; [methyl-3H]-thymidine from ARC, 71.7 Ci/mmol; [3H]-adenosine from Amersham, 27.0 Ci/mmol); [3H]-inosine from Moravek, 7.9 Ci/mmol; [3H]-guanosine from Moravek, 12.9 Ci/mmol; [3H]guanine from Moravek, 8.4 Ci/mmol; [3H]-adenine from Moravek, 27 Ci/mmol; [3H]-hypoxanthine from Amersham, 19 Ci/mmol; [3H]-uracil from Perkin-Elmer 30.3 Ci/mmol; [3H]-cytosine from Moravek, 25.6 Ci/mmol; [3H]-thymine from Moravek, 56.3 Ci/mmol.
4.3. RNA extraction and real-time RT-PCR of T. vaginalis
Total cellular RNA was extracted from lysates made with TRIzol (Invitrogen, 1 ml per 107 cells or confluent 10 cm2 flask) and purified with DirectZol kit (Zymo) according to the manufacturer’s protocol. RNA was quantified by absorbance at 260 nm. RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), and quantitative RT-PCR (qRT-PCR) analysis was done with PerfeCTa SYBR Green FastMix (Quantabio) in a Step-one real-time PCR machine (Applied Biosystems) using the primer pairs listed in Supplement table. Relative changes in target mRNA levels were calculated by the 2ΔΔCt method (Dann et al., 2015), with GAPDH mRNA as the reference standard and normalized to ENT5 mRNA levels. Primers for GAPDH: forward 5′-GCCGCAAGCTCTATCCAAAG-3’; reverse primer 5′-CGGCCACCGATTGACTTAAC-3’.
4.4. Expression of TvagENTs in T. brucei
The full-length coding sequences of 9 TvagENT genes (Table 2) were retrieved from TrichDB.org by BLASTP and keyword searches. The nucleotide sequences were optimised for expression in T. brucei using a codon optimisation algorithm at https://www.idtdna.com/CodonOpt (Rashmi and Swati, 2013). The thus optimised sequences were synthesised by BaseClear BV, except for TvagENT8, which was synthesised by Genewiz and all delivered in the vector pUC57-Amp. Each gene was amplified by PCR using primers that introduced 5’-HindIII and 3’-BamH1 restriction sites (Supplemental Table 2); the same enzymes were used to digest pHD1336 (Biebinger et al., 1997) and the PCR products were ligated in using T4 DNA ligase (NEB). The constructs were used to transform E. coli XL1-blue competent cells (Agilent), which were then grown on ampicillin agar plates. Colonies were screened by PCR using the vector specific primer HDK528 (CTCTAGAGGATCCTATGCGTGACTGAGTGAGCC) and the relevant reverse primer for the inserted gene (Supplemental Table 2) and, if yielding the correct size of amplicon, further verified by Sanger sequencing (Source BioScience, Nottingham, UK). Plasmid DNA from selected colonies was linearised by digestion with NotI before transfection into T. brucei strain TbAT1-KO (Matovu et al., 2003) using an Amaxa Nucleofector, program X-001 (Burkard et al., 2011). Transfectants were grown and cloned out, by limiting dilution, in complete HMI-11/FBS media containing 5 µg ml−1 blasticidin S. Correct integration of the expression cassettes was analysed by PCR.
4.5. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR) of TvagENTs expressed in T. brucei
Cells were harvested from 11 mL flasks, each containing ~3 x 106 cells/mL for one clonal line expressing a TvagENT in T. brucei TbAT1-KO, by centrifugation at 2,000 RPM for 10 min. The supernatant was discarded and RNA was isolated using the NucleoSpin® RNA isolation kit in accordance with the manufacturer’s instructions. RNA was eluted off the column with 15 µL RNAase-free water and the concentration determined using a Nanodrop spectrophotometer.
cDNA was produced using the Precision nanoScript2 Reverse Transcription kit (Primer Design). Primers were annealed to 9 μL of RNA sample using 0.5 μL oligo-dT primers and 0.5 μL Random nonamer primers for 5 minutes at 65 °C and subsequently cooled on ice for 5 min. The extension master mix consisted of 5 μL nanoScript2 4x buffer, 1 μL 10 mM dNTP, 2.5 μL RNAse free water and 1 μL nanoScript2 enzyme. This was added to 10 µL RNA in water, briefly vortexed, vortexed and incubated at room temperature for 5 min before extending at 42 °C and a heat denaturation step at 75 °C for 10 min.
Quantitative PCR. qPCR primers (Supplemental Table 1) were designed using Primer Express 3000 software, selecting for primers with >50% GC content and the forward and reverse being of similar length. Primers were diluted in RNAse free water to 100 µM stock concentration and stored at −20 °C. The forward and reverse primers were then combined at a concentration of 3 μM each. 15 μL of master mix, consisting of 10 μL PrecisionPLUS, 1 μL primer mix and 4 μL RNase free water, was added to wells in a 96-well plate. cDNA was diluted to 4 ng/μL using RNAse free water and 5 μL of cDNA from each clone was added to the master mix. Plates were designed to have three repeats for the TvagENT and the GPI8 control gene, and four repeats for the empty vector control as well as four no cDNA controls for both the TvagENT and the GPI8 primers. The qPCR was run on a 7500 Real Time PCR system for 40 cycles in accordance with the PrecisionFAST Mastermix protocol and analysed using SDS software.
4.6. Data analysis
All values for IC50, Km and Vmax are presented as means ± SEM of at least three independent determinations in triplicate but individual plots shown as Figures are always single experiments with data points being the means ± SEM of the triplicates. Hill slopes were calculated by plotting dose-response inhibition data to an equation for a sigmoid plot with variable slope (4 parameter) in GraphPad Prism. Rates of transport in timecourses were calculated by linear regression in Prism and the in-build statistical analysis were used to determine deviation from linearity, significance of deviation from zero slope (flat line), and significance of difference in slope between two lines within the same experiment.
For mRNA measurements, experiments were repeated at least three times, fold changes were expressed as mean and S.E.M. of log10-transformed expression values. Significance was evaluated by ANOVA with Dunnett’s post-hoc test using Prism (Graphpad software).
Supplementary Material
Table 3.
Percent amino acid identity among T. vaginalis ENT proteins
| TvagENT1 | TvagENT2 | TvagENT3 | TvagENT4 | TvagENT5 | TvagENT6 | TvagENT7 | TvagENT8 | TvagENT9 | |
|---|---|---|---|---|---|---|---|---|---|
| TvagENT1 | 100 | 42.2 | 39.6 | 28.4 | 26.8 | 49.9 | 24.2 | 41.2 | 22.2 |
| TvagENT2 | 42.2 | 100 | 83.2 | 22.4 | 26.6 | 56.6 | 29.2 | 86.4 | 24.8 |
| TvagENT3 | 39.6 | 83.2 | 100 | 24.0 | 25.9 | 54.3 | 28.4 | 77.7 | 25.1 |
| TvagENT4 | 28.4 | 22.4 | 24.0 | 100 | 53.7 | 25.2 | 38.6 | 23.5 | 42.0 |
| TvagENT5 | 26.8 | 26.6 | 25.9 | 53.7 | 100 | 26.7 | 44.8 | 26.8 | 42.6 |
| TvagENT6 | 49.9 | 56.6 | 54.3 | 25.2 | 26.3 | 100 | 26.2 | 52.2 | 24.7 |
| TvagENT7 | 24.2 | 29.2 | 28.4 | 38.6 | 44.8 | 26.2 | 100 | 26.7 | 72.8 |
| TvagENT8 | 41.2 | 86.4 | 77.7 | 23.5 | 26.8 | 52.2 | 26.7 | 100 | 23.9 |
| TvagENT9 | 22.2 | 24.8 | 25.1 | 42.0 | 42.6 | 24.7 | 72.8 | 23.9 | 100 |
Acknowledgements
The authors thank Professor Jeremy Mottram (University of York, UK) and Dr. Evan Secor for gifts of T. vaginalis strains. This research was supported by a personal fellowship to MJN from the Saudi Arabian Ministry of Education, a studentship to TAA from the Saudi Arabian Ministry of Education, MIA by a studentship from the Libyan government, NBQ by a Getfund studentship from the Ghanaian government, a project grant from the Medical Research Council (84733), and National Institutes of Health grants DK120515 and AI158612.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest regarding the publication of this paper.
Data availability statement
Sequences used are those of the publicly available database trichDB. All other relevant data are presented in the main text and Supporting Information sections of the paper.
References
- Ali JAM, Tagoe D, Munday JC, Donachie A, Morrison LJ & De Koning HP (2013a) Pyrimidine biosynthesis is not an essential function for Trypanosoma brucei bloodstream forms. PLoS One, 8, e58034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali JAM, Creek DJ, Burgess K, Allison HC, Field MC, Mäser P & De Koning HP (2013b) Pyrimidine salvage in Trypanosoma brucei bloodstream forms and the trypanocidal action of halogenated pyrimidines. Molecular Pharmacology, 83, 439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Salabi MI, Wallace LJM, Lüscher A, Mäser P, Candlish D, Rodenko B et al. (2007) Molecular interactions underlying the unusually high affinity of a novel Trypanosoma brucei nucleoside transporter. Molecular Pharmacology, 71, 921–929. [DOI] [PubMed] [Google Scholar]
- Alzahrani KJH, Ali JAM, Eze AA, Looi WL, Tagoe DNA, Creek DJ et al. (2017) Functional and genetic evidence that nucleoside transport is highly conserved in Leishmania species: implications for pyrimidine-based chemotherapy. International Journal for Parasitology Drugs and Drug Resistance, 7, 206–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebinger S, Wirtz LE, Lorenz P & Clayton C (1997) Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Molecular and Biochemical Parasitology, 85, 99–112. [DOI] [PubMed] [Google Scholar]
- Bradic M, Warring SD, Tooley GE, Scheid P, Secor WE, Land KM et al. (2017) Genetic indicators of drug resistance in the highly repetitive genome of Trichomonas vaginalis. Genome Biology and Evolution, 9, 1658–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Upcroft JA, Dodd HN, Chen N & Upcroft P (1999) Alternative 2-keto acid oxidoreductase activities in Trichomonas vaginalis. Molecular and Biochemical Parasitology, 98, 203–214. [DOI] [PubMed] [Google Scholar]
- Burkard G, Fragoso C & Roditi I (2007) Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Molecular and Biochemical Parasitology, 153, 220–223. [DOI] [PubMed] [Google Scholar]
- Campagnaro GD, Alzahrani KJH, Munday JC & De Koning HP (2018a) Trypanosoma brucei bloodstream forms express highly specific and separate transporters for adenine and hypoxanthine; evidence for a new protozoan purine transporter family? Molecular and Biochemical Parasitology, 220, 46–56. [DOI] [PubMed] [Google Scholar]
- Campagnaro GD, de Freitas Nascimento J, Girard RBM, Silber AM & De Koning HP (2018b) Cloning and characterisation of the Equilibrative Nucleoside Transporter family of Trypanosoma cruzi: ultra-high affinity and selectivity to survive in the intracellular niche. Biochimica et Biophysica Acta General Subjects, 1862, 2750–2763. [DOI] [PubMed] [Google Scholar]
- Campagnaro GD & De Koning HP (2020) Purine and pyrimidine transporters of pathogenic protozoa - conduits for therapeutic agents. Medicinal Research Reviews, 40, 1679–1714. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science, 315, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y-C & Prusoff WH (1973) Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- Chiang CW, Carter N, Sullivan WJ Jr, Donald RG, Roos DS, Naguib FN et al. (1999) The adenosine transporter of Toxoplasma gondii. Identification by insertional mutagenesis, cloning, and recombinant expression. Journal of Biological Chemistry, 274, 35255–35261. [DOI] [PubMed] [Google Scholar]
- Dann SM, Manthey CF, Le C, Miyamoto Y, Gima L, Abrahim A et al. (2015) L-17A promotes protective IgA responses and expression of other potential effectors against the lumen-dwelling enteric parasite Giardia. Experimental Parasitology, 156, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Matos JA, Pires Borges F, Tasca T, Reis Bogo M, de Carli GA, Fauth MG et al. (2001) Characterisation of an ATP diphosphohydrolase (Apyrase, EC 3.6.1.5) activity in Trichomonas vaginalis. International Journal for Parasitology, 31, 770–775. [DOI] [PubMed] [Google Scholar]
- De Jesus JB, Ferreira MA, Cuervo P, Britto C, e Silva-Filho FC & Meyer-Fernandes JR (2006) Iron modulates ecto-phosphohydrolase activities in pathogenic trichomonads. Parasitology International, 55, 285–290. [DOI] [PubMed] [Google Scholar]
- De Koning HP, Al-Salabi MI, Cohen A, Coombs GH & Wastling JM (2003) Identification and characterisation of high affinity purine nucleoside and nucleobase transporters in Toxoplasma gondii. International Journal for Parasitology, 33, 821–831. [DOI] [PubMed] [Google Scholar]
- De Koning HP, Bridges DJ & Burchmore R (2005) Purine transporters of protozoa: from biology to therapy. FEMS Microbiology Reviews, 29, 987–1020. [DOI] [PubMed] [Google Scholar]
- De Koning H & Diallinas G (2000) Nucleobase transporters (review). Molecular Membrane Biology, 17, 75–94. [DOI] [PubMed] [Google Scholar]
- De Koning HP (2007) Pyrimidine transporters of protozoa – A class apart? Trends in Parasitology, 23, 190 [Google Scholar]
- De Koning HP, Watson CJ & Jarvis SM (1998) Characterisation of a nucleoside/proton symporter in procyclic Trypanosoma brucei brucei. Journal of Biological Chemistry, 273, 9486–9494. [DOI] [PubMed] [Google Scholar]
- De Koning HP & Jarvis SM (1997) Purine nucleobase transport in bloodstream forms of Trypanosoma brucei brucei is mediated by two novel transporters. Molecular and Biochemical Parasitology, 89, 245–258. [DOI] [PubMed] [Google Scholar]
- De Koning HP & Jarvis SM (1998) A highly selective, high affinity transporter for uracil in Trypanosoma brucei brucei; evidence for proton-dependent transport. Biochemistry and Cell Biology, 76, 853–858. [DOI] [PubMed] [Google Scholar]
- De Koning HP & Jarvis SM (1999) Adenosine transporters in bloodstream forms of T. b. brucei: Substrate recognition motifs and affinity for trypanocidal drugs. Molecular Pharmacology, 56, 1162–1170. [DOI] [PubMed] [Google Scholar]
- Edwards T, Burke P, Smalley H & Hobbs G (2016) Trichomonas vaginalis: Clinical relevance, pathogenicity and diagnosis. Critical Reviews in Microbiology, 42, 406–417. [DOI] [PubMed] [Google Scholar]
- Feng R-M, Wang MZ, Smith JS, Dong L, Chen F, Pan Q-J et al. (2018) Risk of high-risk human papillomavirus infection and cervical precancerous lesions with past or current trichomonas infection: a pooled analysis of 25,054 women in rural China. Journal of Clinical Virology, 99–100, 84–90. [DOI] [PubMed] [Google Scholar]
- Galazka J, Carter NS, Bekhouche S, Arastu-Kapur S & Ullman B (2006) Point mutations within the LdNT2 nucleoside transporter gene from Leishmania donovani confer drug resistance and transport deficiency. International Journal of Biochemistry and Cell Biology, 38, 1221–1229. [DOI] [PubMed] [Google Scholar]
- Geiser F, Lüscher A, De Koning HP, Seebeck T & Mäser P (2005) Molecular pharmacology of adenosine transport in Trypanosoma brucei: P1/P2 revisited. Molecular Pharmacology, 68, 589–595. [DOI] [PubMed] [Google Scholar]
- Goldman LM, Upcroft JA, Workowski K & Rapkin A (2009) Treatment of metronidazole-resistant Trichomonas vaginalis. Sexual Health, 6, 345–347. [DOI] [PubMed] [Google Scholar]
- Gottlieb SL, Douglas JM jr., Foster M, Schmid DS, Newman DR, Baron AE et al. (2004) Incidence of Herpes Simplex Virus Type 2 infection in 5 sexually transmitted disease (STD) clinics and the effect of HIV/STD risk-reduction counseling. Journal of Infectious Diseases, 190, 1059–1067. [DOI] [PubMed] [Google Scholar]
- Gudin S, Quashie NB, Candlish D, Al-Salabi MI, Jarvis SM, Ranford-Cartwright LC & De Koning HP (2006) Trypanosoma brucei: A survey of pyrimidine transport activities. Experimental Parasitology, 114, 103–108. [DOI] [PubMed] [Google Scholar]
- Harris DI, Beechey RB, Linstead D & Barrett J (1988) Nucleoside uptake by Trichomonas vaginalis. Molecular and Biochemical Parasitology, 29, 105–116. [DOI] [PubMed] [Google Scholar]
- Heyworth PG, Gutteridge WE & Ginger CD (1982) Purine metabolism in Trichomonas vaginalis. FEBS Letters, 141, 106–110. [DOI] [PubMed] [Google Scholar]
- Heyworth PG, Gutteridge WE & Ginger CD Pyrimidine metabolism in Trichomonas vaginalis. FEBS Letters, 176, 55–60. [DOI] [PubMed] [Google Scholar]
- Hirumi H & Hirumi K (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. Journal of Parasitology, 75, 985–989. [PubMed] [Google Scholar]
- Hulpia F, Mabille D, Campagnaro GD, Schumann G, Maes L, Roditi I et al. (2019) Combining tubercidin and cordycepin scaffolds results in highly active candidates to treat late-stage sleeping sickness. Nature Communications, 10, 5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger P (2015) Epidemiology and treatment of trichomoniasis. Current Infectious Disease Reports, 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger P & Adamski A (2013) Trichomoniasis and HIV interactions: a review. Sexually Transmitted Infections, 89, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Silva M, Lisboa C, Gomes N & Rodrigues AG (2021) Trichomonas vaginalis and growing concern over drug resistance. A systematic review. Journal of the European Academy of Dermatology and Venereology, 35, 2007–2021. [DOI] [PubMed] [Google Scholar]
- Matovu E, Stewart M, Geiser F, Brun R, Mäser P, Wallace LJM et al. (2003) The mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryotic Cell, 2, 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes CB, Durgante J, de Oliveira RR, Dos Santos VH, Rodrigues LF, Garcia SC et al. (2016) Trichomonas vaginalis NTPDase and ecto-5’-nucleotidase hydrolyze guanine nucleotides and increase extracellular guanosine levels under serum restriction. Molecular and Biochemical Parasitology, 207, 10–18. [DOI] [PubMed] [Google Scholar]
- Miller RL & Lindstead D (1983) Purine and pyrimidine metabolising activities in Trichomonas vaginalis extracts. Molecular and Biochemical Parasitology, 7, 41–51. [DOI] [PubMed] [Google Scholar]
- Miller RL & Miller WH (1985) Purine salvage enzymes in Trichomonas vaginalis. Pediatric Research, 19, 776. [DOI] [PubMed] [Google Scholar]
- Munagala NR & Wang CC (2003) Adenosine is the primary precursor of all purine nucleotides in Trichomonas vaginalis. Molecular and Biochemical Parasitology, 127, 143–149. [DOI] [PubMed] [Google Scholar]
- Munagala N & Wang CC (2002) The purine nucleoside phosphorylase from Trichomonas vaginalis is a homologue of the bacterial enzyme. Biochemistry, 41, 10382–10389. [DOI] [PubMed] [Google Scholar]
- Munday JC, Rojas López KE, Eze AA, Delespaux V, Van Den Abbeele J, Rowan T et al. (2013) Functional expression of TcoAT1 reveals it to be a P1-type nucleoside transporter with no capacity for diminazene uptake. International Journal for Parasitology Drugs and Drug Resistance, 3, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday JC, Tagoe DNA, Eze AA, Krezdorn JA, Rojas López KE et al. (2015) Functional analysis of drug resistance-associated mutations in the Trypanosoma brucei adenosine transporter 1 (TbAT1) and the proposal of a structural model for the protein. Molecular Microbiology, 96, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzny CA, Van Gerwen OT & Kissinger P (2020) Updates in Trichomonas treatment including persistent infection and 5-nitroimidazole hypersensitivity. Current Opinion in Infectious Disease, 33, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natto MJ, Eze AA & De Koning HP (2015) Protocols for the routine screening of drug sensitivity in the human parasite Trichomonas vaginalis. Methods in Molecular Biology, 1263, 103–110. [DOI] [PubMed] [Google Scholar]
- Natto MJ, Hulpia F, Kalkman ER, Baillie S, Alhejeli A, AlSiari T, et al. (2021) Deazapurine nucleoside analogues for the treatment of Trichomonas vaginalis. ACS Infectious Diseases, 7, 1752–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natto MJ, Savioli F, Quashie NB, Dardonville C, Rodenko B & De Koning HP (2012) Validation of novel fluorescence assays for the routine screening of drug susceptibilities of Trichomonas vaginalis. Journal of Antimicrobial Chemotherapy, 67, 933–943. [DOI] [PubMed] [Google Scholar]
- Papageorgiou IG, Yakob L, Al-Salabi MI, Diallinas G, Soteriadou K & De Koning HP (2005) Identification of the first pyrimidine nucleobase transporter in Leishmania: similarities with the Trypanosoma brucei U1 transporter and antileishmanial activity of uracil analogues. Parasitology, 130, 275–283. [DOI] [PubMed] [Google Scholar]
- Peters RP, Feucht UD, De Vos L, Ngwepe P, McIntyre JA, Klausner JD & Medina-Marino A (2021) Mother-to-child transmission of Chlamydia trachomatis, Neisseria gonorrhoeae, and Trichomonas vaginalis in HIV-infected pregnant women in South Africa. International Journal of STD & AIDS, 32, 799–805. [DOI] [PubMed] [Google Scholar]
- Quashie NB, Dorin-Semblat D, Bray PG, Biagini GA, Doerig C, Ranford-Cartwright LC & De Koning HP (2008) A comprehensive model of purine uptake by the malaria parasite Plasmodium falciparum: Identification of four purine transport activities in intraerythrocytic parasites. Biochemical Journal, 411, 287–295. [DOI] [PubMed] [Google Scholar]
- Raffone A, Travaglino A, Angelino A, Esposito R, Orlandi G, Toscano P, et al. (2021) Gardnerella vaginalis and Trichomonas vaginalis infections as risk factors for persistence and progression of low-grade precancerous cervical lesions in HIV-1 positive women. Pathology Research and Practice, 219, 153349. [DOI] [PubMed] [Google Scholar]
- Ranjbarian F, Vodnala M, Alzahrani KJH, Ebiloma GU, De Koning HP & Hofer A (2017) 9-(2-Deoxy-2-fluoro-ß-D-arabinofuranosyl) is a potent antitrypanosomal adenosine analogue that circumvents transport-related drug resistance. Antimicrobial Agents and Chemotherapy, 61, e02719–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashmi M & Swati D (2013) Comparative genomics of trypanosomatid pathogens using codon usage bias. Bioinformation, 9, 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo-Matthis A, Wing C, Ghanem M, Deng H, Wu P, Gupta A et al. (2007) Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues. Biochemistry, 46, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MA, Tryon R, Green J, Boor I & Landfear SM (2002) Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct biochemical functions. Journal of Biological Chemistry, 277, 21499–21504. [DOI] [PubMed] [Google Scholar]
- Shokar A, Au A, An SH, Tong E, Garza G, Zayas J et al. (2012) S-Adenosylhomocysteine hydrolase of the protozoan parasite Trichomonas vaginalis: potent inhibitory activity of 9-(2-deoxy-2-fluoro-β,D-arabinofuranosyl)-adenine. Bioorganic and Medicinal Chemistry Letters, 22, 4203–4205. [DOI] [PubMed] [Google Scholar]
- Silver BJ, Guy RJ, Kaldor JM, Jamil MS & Rumbold AR (2014) Trichomonas vaginalis as a cause of perinatal morbidity: A systematic review and meta-analysis. Sexually Transmitted Diseases, 41, 369–376. [DOI] [PubMed] [Google Scholar]
- Tasca T, Bonan CD, Carli GA, Battastini AM & Sarkis JJ (2003) Characterization of an ecto-5’-nucleotidase (EC 3.1.3.5) activity in intact cells of Trichomonas vaginalis. Experimental Parasitology, 105, 167–173. [DOI] [PubMed] [Google Scholar]
- Van Gerwen OT, Camino AF, Sharma J, Kissinger PJ & Muzny CA (2021) Epidemiology, natural history, diagnosis, and treatment of Trichomonas vaginalis in men. Clinical Infectious Diseases, 73, 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan G, Carter NS, Drew ME, Beverley SM, Sanchez MA, Seyfang A et al. (1998) Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proceedings of the National Academy of Sciences USA, 95, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan G, Ullman B & Landfear SM (2001) Point mutations in a nucleoside transporter gene from Leishmania donovani confer drug resistance and alter substrate selectivity. Proceedings of the National Academy of Sciences USA, 98, 6092–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace LJ, Candlish D & De Koning HP (2002) Different substrate recognition motifs of human and trypanosome nucleobase transporters: Selective uptake of purine antimetabolites. Journal of Biological Chemistry, 277, 26149–26156. [DOI] [PubMed] [Google Scholar]
- Wang CC & Cheng H-W (1984) Salvage of pyrimidine nucleosides by Trichomonas vaginalis. Molecular and Biochemical Parasitology, 10, 171–184. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2012). Global incidence and prevalence of selected curable sexually transmitted infections – 2008. ISBN 978 92 4 150383 9 [Available from: https://www.who.int/reproductivehealth/publications/rtis/stisestimates/en/]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences used are those of the publicly available database trichDB. All other relevant data are presented in the main text and Supporting Information sections of the paper.


