Abstract
Purpose
Agonists of the stimulator of interferon genes (STING) play a key role in activating the STING pathway by promoting the production of cytokines. In this study, we investigated the antitumor effects and activation of the systemic immune response of treatment with DMXAA (5,6-dimethylxanthenone-4-acetic acid), a STING agonist, in EML4-ALK lung cancer and CT26 colon cancer.
Materials and Methods
The abscopal effects of DMXAA in the treatment of metastatic skin nodules were assessed. EML4-ALK lung cancer and CT26 colon cancer models were used to evaluate these effects after DMXAA treatment. To evaluate the expression of macrophages and T cells, we sacrificed the tumor-bearing mice after DMXAA treatment and obtained the formalin-fixed paraffin-embedded (FFPE) tissue and tumor cells. Immunohistochemistry and flow cytometry were performed to analyze the expression of each FFPE and tumor cell.
Results
We observed that highly infiltrating immune cells downstream of the STING pathway had increased levels of chemokines after DMXAA treatment. In addition, the levels of CD80 and CD86 in antigen-presenting cells were significantly increased after STING activation. Furthermore, innate immune activation altered the systemic T cell-mediated immune responses, induced proliferation of macrophages, inhibited tumor growth, and increased numbers of cytotoxic memory T cells. Tumor-specific lymphocytes also increased in number after treatment with DMXAA.
Conclusion
The abscopal effect of DMXAA treatment on the skin strongly reduced the spread of EML4-ALK lung cancer and CT26 colon cancer through the STING pathway and induced the presentation of antigens.
Keywords: STING agonist, skin metastasis, lung cancer, T cell-mediated immune response, macrophages
INTRODUCTION
The development of immune checkpoint inhibitors (ICIs) has revolutionized the treatment of multiple solid cancer types, and ICIs have emerged as an effective treatment option even in the first-line setting. Immune checkpoint proteins, such as PD-1 or CTLA-4, have emerged as promising targets of immunotherapy and have been approved for use in the treatment of certain cancer types.1 Tumor cells can evade immunosurveillance and progress through different mechanisms, including activation of immune checkpoint pathways that suppress the antitumor immune responses. Mechanistically, ICIs reinvigorate the antitumor immune responses by interrupting the coinhibitory signaling pathways and promote immune-mediated elimination of tumor cells.2 However, ICI treatments have shown a low response rate and a limited proportion of responders. Therefore, the development of different treatment strategies that target the innate immune responses is urgently needed.3
Several previous studies have shown that the activation of the stimulator of interferon genes (STING) pathway through the administration of a STING agonist triggers an immune response that leads to an increase in the production of immune cells within the tumor and is associated with tumor size reduction.4,5,6 Furthermore, the activation of the STING pathway in tumor-resident host antigen-presenting cells (APCs) is required for the induction of CD8+ T-cell response against tumor-derived antigens in vivo.7,8 In addition, the intratumoral injection of a STING agonist can induce tumor regression and generate systemic immune responses that inhibit distant metastases and long-lived immunologic memory.9 In the basal state, STING exists in the endoplasmic reticulum (ER) as an adapter transmembrane protein with its C-terminal domain residing in the cytosol. However, when cytosolic DNA is detected from viruses, bacteria, or parasites, the STING protein undergoes conformational changes and is transported from the ER to the Golgi body to the perinuclear endosome through a mechanism that seemingly requires the components of the autophagosome. Consequently, STING recruits TANK-binding kinase 1 (TBK1), which phosphorylates STING and enables it to be more accessible for binding interferon (IFN) regulatory factor 3 (IRF3). TBK1 then phosphorylates IRF3, which translocates to the nucleus to promote the transcription of IFN-β and other innate immune genes.
These mechanisms occur in APCs in the tumor microenvironment, which in turn drives the effective processing of antigens by the CD8a+/CD103+ dendritic cells (DCs) and subsequent presentation of antigenic peptides on major histocompatibility complex (MHC) class I molecules to cytotoxic CD8+ T cells. This process is known as cross-priming.10 Significantly, the activation of the STING pathway induces a spontaneous antitumor CD8+ T cell response related to the expression of type I IFN genes. In addition, cyclic dinucleotides (CDNs) (such as GMP and c-di-AMP)-mediated STING pathway has been shown to boost antitumor immune response and substantially inhibit tumor growth. Therefore, many STING agonists have been developed and are currently undergoing clinical trials.11,12
EML4-ALK is a distinct molecular subset of lung cancer that does not respond to checkpoint inhibitors.13 The tumor immune microenvironment showed a lack of immunogenicity both before and after treatment with ALK TKI. For these socalled “cold tumors,” a different treatment strategy other than checkpoint inhibitors is required. In this study, we developed a soft tissue metastasis model of EML4-ALK transgenic mouse to test the abscopal effect of STING agonist. Soft tissue metastasis of lung cancer is associated with poor prognosis, and urgent medical intervention is required.
In this study, we investigated a mouse STING agonist, DMXAA (5,6-dimethylxanthenone-4-acetic acid), which can induce innate immune responses using the syngeneic tumor model of colon cancer with peritoneal metastases. In particular, macrophages are usually divided into two types: M1 and M2. M1 macrophages are generally stimulated by IFN-γ and/or LPS and provide an antitumor phenotype with pro-inflammatory cytokines such as tumor necrosis factor (TNF)-α; interleukin (IL)-6, and IL-12. M2 macrophages are involved in tissue repair and immunosuppressive functions and polarized by IL-4, IL-13, and other factors.14 Therefore, memory cytotoxic T cells and tumor-specific lymphocytes were increased and, for the first time, showed profound antitumor responses in peritoneal seeding lesions after treatment with DMXAA. We noted that treatment with DMXAA showed systemic activation of macrophage activity and increased the antigen presentation.
MATERIALS AND METHODS
Experimental animals
All animal experiments were conducted in accordance with the ethical standards of the Institutional Animal Care and Use Committee of Avison Bio Medical Research Center in Yonsei University (IACUC number, 2016-0093). Six-week-old female BALB/c mice were purchased from Orient Bio (Seongnam, South Korea). According to the SPF guidelines, the mice were maintained under pathogen-free conditions (at room temperature with 40%–60% humidity). EML4-ALK mice were generated as previously reported.15
Magnetic resonance imaging
Mice were anesthetized with isoflurane delivered in 100% oxygen. To minimize the motion effects, respiratory gating was performed, and magnetic resonance signal was synchronized with the respiratory cycles. The magnetic resonance imaging (MRI) protocols were optimized at 9.4 Tesla 94120 (Bruker Biospec, Billerica, MA, USA) to assess for pulmonary parenchyma. An MRI was performed every week, and two independent operators interpreted the MRI scan images. Progressive disease was defined as >20% increase in tumor burden from baseline. Partial response was defined as >30% decrease in tumor burden from baseline. Tumor burden changes between -30% to +20% were defined as stable disease. The overall survival was measured when the mice died or developed a moribund condition.
Cell lines
The CT26 (CRL-2638) murine colon carcinoma cell line was purchased from the ATCC (Manassas, VA, USA). CT26 cells with integrated Enhanced Green Fluorescent protein (EGFP) and luciferase genes were established using the lentiviral vector (LPP-HLUC-LV201-025, GeneCopoeia; Rockville, MD, USA). The cells were cultured in a 75-cm2 culture plate with RPMI 1640 (HyClone; Logan, UT, USA), and transfection was performed when the cell density reached 30%–40% confluency. After 24 hours of culture, the culture medium was replaced with new RPMI 1640 medium. Puromycin (2 µg/mL) was added for colony selection. Transfection was assessed by conducting a polymerase chain reaction (PCR) using an EGFP protein and a luciferase targeting primer in an in vivo optical imaging system (IVIS, PerkinElmer; Waltham, MA, USA). Mycoplasma was tested for using a PCR-based detection kit (data not shown). All experiments were performed within 20 passages. Carboxyfluorescein succinimidyl ester (CFSE) labeling of CT26 cells was performed using the CellTrace CFSE Cell Proliferation Kit (Thermo Fisher; Waltham, MA, USA), according to the manufacturer’s instructions. Briefly, the cells were stained with CFSE, incubated at 37℃ for 10 min, and then washed with a phosphate-buffered saline two times. In order to obtain the cell lysate, the labeled cells were subjected to five freeze/thaw cycles. The lysate was filtered using a 0.45-µm syringe filter and stored in a deep freezer before use.
Chemicals
DMXAA (Vadimezan, CAS no. 117570-53-3) was purchased from Selleck Chemicals (Houston, TX, USA). DMXAA was diluted with Hanks Balanced Salt Solution (HBSS) for in vivo experiments. For in vitro experiments, DMXAA was diluted with total media [10% fetal bovine serum (FBS) and 1% penicillin and streptomycin] and filtered using a 0.45-µm syringe filter.
Bone marrow macrophage and peritoneal macrophage cultivation
Peritoneal macrophages (PMs) and bone marrow macrophages (BMMs)16 were harvested from BALB/c mice according to the protocols that were described previously.17 The purity of macrophages was assessed by conducting a flow cytometry analysis (BD Biosciences; San Jose, CA, USA) using the surface marker F4/80. The 90%-pure macrophages were used for the ex vivo experiments.
Flow cytometry assay
The macrophages were harvested in Flow Cytometry Staining Buffer (FACS Buffer) (3% bovine serum albumin, 1 mM EDTA, and 0.025% NaN3). The cells were initially blocked with anti-mouse CD16/CD32 monoclonal antibodies (Fc-Blocker, BioLegend; San Diego, CA, USA) for 10 min at 4℃ and stained for surface markers using PE-Cy5-labeled anti-mouse CD80 (clone 16-10A1, BioLegend), APC/Cy7-labeled anti-mouse CD86 (clone GL-1, BioLegend), and PE-labeled anti-mouse H2 (clone M1/42, BioLegend) and incubated in the dark for 30 min at 4℃. The cells were washed, resuspended in 200 µL of FACS buffer, and fixed with a 4% paraformaldehyde. The splenocytes, mesenteric lymph node (MLN) cells, and axillary lymph node (LN) cells were harvested in FACS buffer. The cells were initially blocked with anti-mouse CD16/CD32 monoclonal antibodies (Fc-Blocker, BioLegend) for 10 min at 4℃ and stained for surface markers using APC/Cy7-labeled anti-mouse CD3 (clone 17A2, BioLegend), PE-labeled anti-mouse CD4 (clone GK1.5 BioLegend), PE-Cy5-labeled anti-mouse CD8a (clone 53-6.7 BioLegend), APC-labeled anti-mouse CD69 (clone H1.2F3, BioLegend), PerCP/Cy 5.5-labeled anti-mouse CD44 (clone IM7, BioLegend), FITC-labeled anti-mouse CD19 (clone 6D5, Bio-Legend), and PE-labeled anti-mouse CD11c (clone N418, Bio-Legend) and incubated in the dark for 30 min at 4℃. The cells were washed, resuspended in 200 µL of FACS buffer, and fixed with 4% paraformaldehyde. The data were acquired using a BD LSRFortessa flow cytometer (Becton Dickinson; San Jose, CA, USA) and analyzed using the FlowJo Software (Tree Star; Ashland, OR, USA).
Immunohistochemistry
The numbers of tumor-infiltrating CD3+ T cells were measured by immunohistochemistry (IHC). The fluorescence image is a re-implemented mixed image of the 3, 3′-diaminobenzidine result obtained through a multispectral image analysis, and the yellow (upper) and red (lower) colors indicate the CD3+ cells. All tumor tissue was analyzed, and 15 to 40 fields were analyzed per sample. The results were calculated by quantifying the number of total cells and the number of CD3+ cells per mm2. The whole slide scan and cell segmentation were performed in order to quantify the IHC results, and the degree of CD3-positive total T cell infiltration was measured using the Vectra Polaris and InForm software. IHC was performed on the automatic staining machine, LEICA BOND RX. Digital images of IHC slides were obtained using a whole slide scanner. Image deconvolution was performed using the InForm software. The slides were stained with CD3e (CD3-12, Cell Signaling Technology; Beverly, MA, USA) and F4/80 (D2S9R, Cell Signaling Technology).
Enzyme-linked immunosorbent assay
To assess cytokine production, the culture medium-supernatant of PMs was collected. Bronchoalveolar lavage (BAL) fluid was harvested as previously described.18 The concentrations of IFN-β (cat no. # 439407, BioLegend) and IL-12p40 (Cat No. # 431604 BioLegend) were measured using an enzyme-linked immunosorbent assay (ELISA) kit (BioLegend).
Enzyme-linked immune absorbent spot
The BALB/c mice were treated in three groups: control, DMXAA treatment group, and DMXAA and anti-IFN-β treatment group. CT26 GFP/luciferase cells (5×106 cells) were injected into the right thighs of mice in Groups 2 and 3 before drug treatment. One week later, 1×105 CT26 GFP/luciferase-expressing cells were injected into all groups intraperitoneally. Next, DMXAA 500 µg was administered to Groups 2 and 3 every 3 days, and anti-IFN-β 600 µg (Leinco Technologies Inc.; St Louis, MO, USA) was administered to Group 3 every 3 days. Antigen-specific T-cell responses were measured via IFN-β enzyme-linked immune absorbent spot (ELISpot). Briefly, ELISpot 96-well plates (BD Biosciences) were coated with anti-mouse IFN-γ capture antibodies and incubated at 4℃ overnight (BD Biosciences). The next day, the plates were blocked for 2 hours with cell culture media containing 10% FBS and 1% penicillin-streptomycin. Approximately 1×105 LN cells from the mice were added to each well and stimulated with 500 µg CT26 GFP/luciferase antigen for 24 hours at 37℃ in 5% CO2. After 24 hours of stimulation, the cells were washed with distilled water and incubated for 2 hours at room temperature with biotinylated anti-mouse IFN-γ antibodies (BD Biosciences). The plates were washed, and streptavidin–alkaline phosphatase was added (BD Biosciences). Then, the plates were incubated for 2 hours at room temperature. The plates were washed, and 3-amino-9-ethylcarbazole (Sigma; St. Louis, MO, USA) and N,N-dimethylformamide (Sigma) were added to each well. The plates were then rinsed with distilled water and dried at room temperature for 24 hours. The spots detected on the ELISpot assay were counted using an automated ELISpot reader (ELHR03; Strasberg, Germany).
Measurement of nitric oxide
The macrophages were incubated in 6-well plates for 24 hours. The cells were treated with 50 µg/mL of DMXAA in a time-dependent manner. The concentration of nitrite in the culture supernatant was measured using the Griess reaction assay technique: 100 µL of supernatant and 100 µL of Griess reagent (Sigma) were mixed, followed by spectrophotometric measurement at 540 nm after 15 min using a VersaMax Microplate Reader (Molecular Devices; Union City, CA, USA). The nitrite concentration was determined by comparing it with a standard curve of sodium nitrite (Sigma) in the medium.
Immunofluorescence staining
Macrophages (5×104 cells) were seeded directly in an 8-well chamber slide (Lab-Tek; Waltham, MA, USA) and treated with DMXAA in a time-dependent manner. The macrophages were then fixed with 4% paraformaldehyde overnight at 4℃. Immunofluorescence staining was performed using the PerkinElmer antibody diluent/Block kit protocol (PerkinElmer) according to the manufacturer’s protocol. The cell nuclei were stained with DAPI (cat no. #4083). The pIRF3 (cat no. #3033) and NF-κB p65 (cat no. #8242) antibodies were purchased from Cell Signaling Technology.
Real-time PCR
RNA was isolated from PMs using a total RNA isolation kit (Qiagen; Valencia, CA, USA), and cDNA was produced using an oligo-dT premix (Elpis Biotech; Daejeon, South Korea) and 1 µg RNA according to the manufacturer’s protocol. Real-time RT-PCR was performed in a final volume of 20 µL containing SYBR Green PCR Master Mix (Enzynomics; Daejeon, South Korea), poly-A cDNA as the PCR template, and 20 µM of primers. The PCR reaction was performed on the QuantStudio 5 Real-Time PCR System (Applied Biosystems; Foster City, CA, USA) using the following cycling parameters: 60℃ for 10 min, 95℃ for 10 min, 60 cycles of 95℃ for 15 sec, and 60℃ for 40 sec, and a dissociation stage of 95℃ for 15 sec, 60℃ for 1 min, 95℃ for 15 sec, and 60℃ for 15 sec. The primers were used to amplify a 100–120-bp fragment that corresponds to the following gene targets: IFN-β: 5′-CAG CTC CAA GAA AGG ACG AAC-3′ (forward), 5′-GGC AGT GTA ACT CTT CTG CAT-3′ (reverse); CCL2: 5′-TTA AAA ACC TGG ATC GGA ACC AA-3′ (forward), 5′-GCA TTA GCT TCA GAT TTA CGG GT-3′ (reverse); CCL3: 5′-TTC TCT GTA CCA TGA CAC TCT GC-3′ (forward), 5′-CGT GGA ATC TTC CGG CTG TAG-3′ (reverse); CCL4: 5′-TTC CTG CTG TTT CTC TTA CAC CT-3′ (forward), 5′-CTG TCT GCC TCT TTT GGT CAG-3′ (reverse); CCL5: 5′-GCT GCT TTG CCT ACC TCT CC-3′ (forward), 5′-TCG AGT GAC AAA CAC GAC TGC-3′ (reverse); CCL7: 5′-GCT GCT TTC AGC ATC CAA GTG-3′ (forward), 5′-CCA GGG ACA CCG ACT ACT G-3′ (reverse); CCL12: 5′-ATT TCC ACA CTT CTA TGC CTC CT-3′ (forward), 5′-ATC CAG TAT GGT CCT GAA GAT CA-3′ (reverse); CXCL10: 5′-CCA AGT GCT GCC GTC ATT TTC-3′ (forward), 5′-GGC TCG CAG GGA TGA TTT CAA-3′ (reverse); and ′-actin: 5′-GGC TGT ATT CCC CTC CAT CG-3′ (forward), 5′-CCA GTT GGT AAC AAT GCC ATG T-3′ (reverse).
T lymphocyte proliferation assay with 5-bromo-2′-deoxyuridine assay
The control and 5-bromo-2′-deoxyuridine (BrdU) groups were injected with 1.5 mg/mL (200 µL) of BrdU (BioLegend) intraperitoneally. After 15 hours, the control group was injected with 1.2 mg/mL (200 µL) of BrdU and 200 of µL HBSS, while the BrdU group was injected with 1.2 mg/mL (200 µL) of BrdU intraperitoneally and 500 µg of DMXAA intramuscularly. After 24 hours, the mice were sacrificed and the spleen, LNs, and MLNs were collected. The cells were analyzed by flow cytometry.
Antigen presentation assay
The macrophages were incubated in 6-well plates for 24 hours. The cells were treated with or without CFSE-labeled CT26 lysate or treated with 50 µg/mL of DMXAA in a time-dependent manner. Then, the cells were harvested and stained with FACS antibodies.
Chemokine array
The supernatant was stored at -80℃ until use. Chemokine array analysis was performed according to the manufacturer’s protocol (RayBiotech, Inc.; Norcross, GA, USA). Briefly, the membranes were blocked with a blocking buffer for 30 min. The supernatant was incubated with membranes coated with 96 anti-mouse chemokine antibody cocktails overnight. The membranes were incubated with biotinylated detection antibodies and a streptavidin-peroxidase conjugate overnight. Finally, the membranes were developed, and chemokine proteins were quantitated using the Image Quant LAS4000 software (Fujifilm; Tokyo, Japan), and the expression level of each chemokine was analyzed using the ImageJ software (NIH; Bethesda, MD, USA).
Measurement of survival rate
The BALB/c mice were organized into three groups: control group, DMXAA treatment group, DMXAA and anti-IFN-β treatment group. The CT26 GFP/luciferase cells (5×106 cells) were injected in the right thighs of mice in Groups 2 and 3. One week later, 1×105 CT26 GFP/luciferase cells were injected intraperitoneally in all treatment groups. After that, 500 µg of DMXAA was administered in Groups 2 and 3 every 3 days, while 600 µg of anti-IFN-β (Leinco Technologies Inc.) was administered in Group 3 every 3 days. The mice were injected with 2 mg of endotoxin-free luciferase substrate, luciferin (VivoGlo luciferin, Promega; Madison, WI, USA), and scanned using an IVIS (PerkinElmer).
In vivo luciferase measurement
Using an IVIS, the luciferase activity was monitored in living animals. The animals were injected intraperitoneally with luciferin (100 µL, 10 mg/mL). After 15 min, the animals were placed in a dark chamber and exposed to isoflurane anesthesia. The photon images were obtained, and the photon signal was quantified using the Living Image software (PerkinElmer).
Statistical analysis
All data were analyzed for the mean and standard deviation. Data analysis was performed using the Prism 6.0 software (GraphPad Software, Inc.; San Diego, CA, USA), and significant differences were evaluated using the paired-samples t-test and one-way analysis of variance. A p-value of <0.05 was considered statistically significant.
RESULTS
DMXAA treatment reduced tumor growth in an EML-ALK NSCLC mouse model
We generated an EML4-ALK transgenic mouse model to mimic EML4-ALK-mediated skin metastasis as previously described.15 Previous studies included transgenic mice with inducible expression of two common human EGFR mutants found in human lung cancer. Human EML4-ALK fusion proteins are more important in tumor proliferation and survival than in oncogenic mutation in vivo. Our conditional model for this fusion gene was used to evaluate the efficacy of ALK inhibitors and to discover the mechanisms of acquired resistance. It was also used to measure the proliferation of tumors that mimics human conditions after development of acquired resistance. In particular, an in vivo model is very useful for exploring the best treatment strategy utilizing a novel drug that failed to inhibit the growth of ALK armamentarium.
We observed that neither anti-PD-1 nor anti-PD-L1 was effective in the EML4-ALK tumor model (Supplementary Fig. 1A, only online), showing tumor progression within 4 weeks of treatment. Skin metastasis eventually occurred. However, when 500 µg of DMXAA was intratumorally injected in the group with a metastatic skin nodule, we observed a significant decrease in the tumor size (Fig. 1A and Supplementary Fig. 1B, only online) compared to that in the control group (n=4 per group).
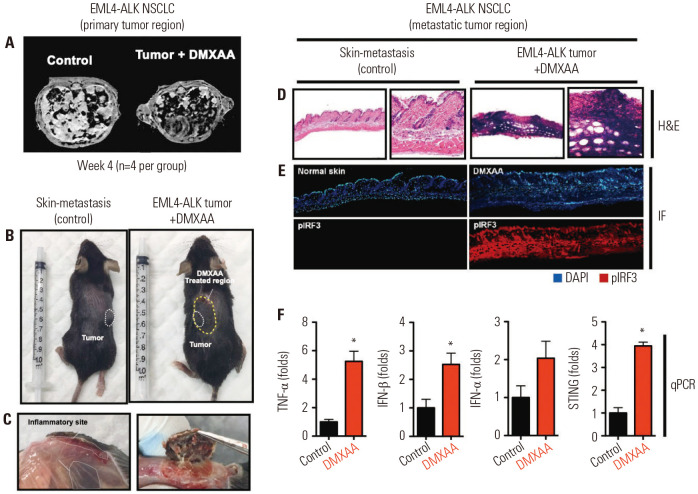
Fig. 1. DMXAA reduced tumor growth and induces strong antitumor response in EML-ALK NSCLC model. (A) MRI images of the primary tumor site of the control and DMXAA (500 µg)-treated tumor-bearing mice. (B) Skin nodule in the EML4-ALK mice. White dotted line indicates the metastatic site, while yellow dotted line indicates the DMXAA-treated region. (C) Skin inflammation observed in the control and DMXAA-treated sites. (D) H&E staining, (E) DAPI (upper) and pIFR3 expression (lower) images of normal skin (control) and DMXAA-treated site. (F) Expression levels of TNF-α, IFN-β, IFN-α, and STING. The results are expressed as mean±SD. *p<0.05. TNF, tumor necrosis factor; IL, interleukin; IFN, interferon; STING, stimulator of interferon genes; H&E, hematoxylin and eosin; qPCR, real-time PCR.
To evaluate the local response to DMXAA, we dissected the metastatic skin nodule in the control and DMXAA-treated groups (Fig. 1B) and observed the control and DMXAA-treated sites for the presence of inflammation (Fig. 1C). The spleen was also examined. The spleen from a tumor-bearing mouse was bigger than that from the control. The spleen from a tumor-bearing mouse treated with DMXAA was the same as or larger than that from a tumor-bearing mouse with no treatment (Supplementary Fig. 2, only online). Next, hematoxylin and eosin (H&E) staining was performed, and DAPI staining and pIRF3 expression were observed under an immunofluorescence microscope. We observed a dense infiltration of immune cells on H&E staining, with overexpression of DMXAA and pIRF3 on immunofluorescence microscopy (Fig. 1D and E). The DMXAA-treated skin exhibited a downstream signaling activation, IRF-3 phosphorylation, and production of IFN-β and other types of cytokines. IFN-β is a subfamily of type 1 IFN. It is one of main cytokines induced by the STING pathway, by either the exogenous CDNs produced by bacterial infection or through the binding of a structurally distinct endogenous CDN produced by a host cyclic GMP-AMP synthetase in response to sensing cytosolic double-stranded DNA-like virus infection.19,20 The levels of TNF-α, IFN-β, and STING expression were significantly upregulated in DMXAA-treated tumors than in controls, as shown on quantitative PCR assay (Fig. 1F).
In particular, the levels of TNF-α, IFN-β, and STING were significantly increased in the DMXAA-treated group than in the control group (*p<0.05). In addition, the level of IFN-α increased in the DMXAA-treated group compared to the control group, but this result was not significant. This finding suggests that STING activation may induce a type 1 IFN-mediated immune response.
DMXAA treatment induced strong antitumor immune response in the EML4-ALK NSCLC mouse model
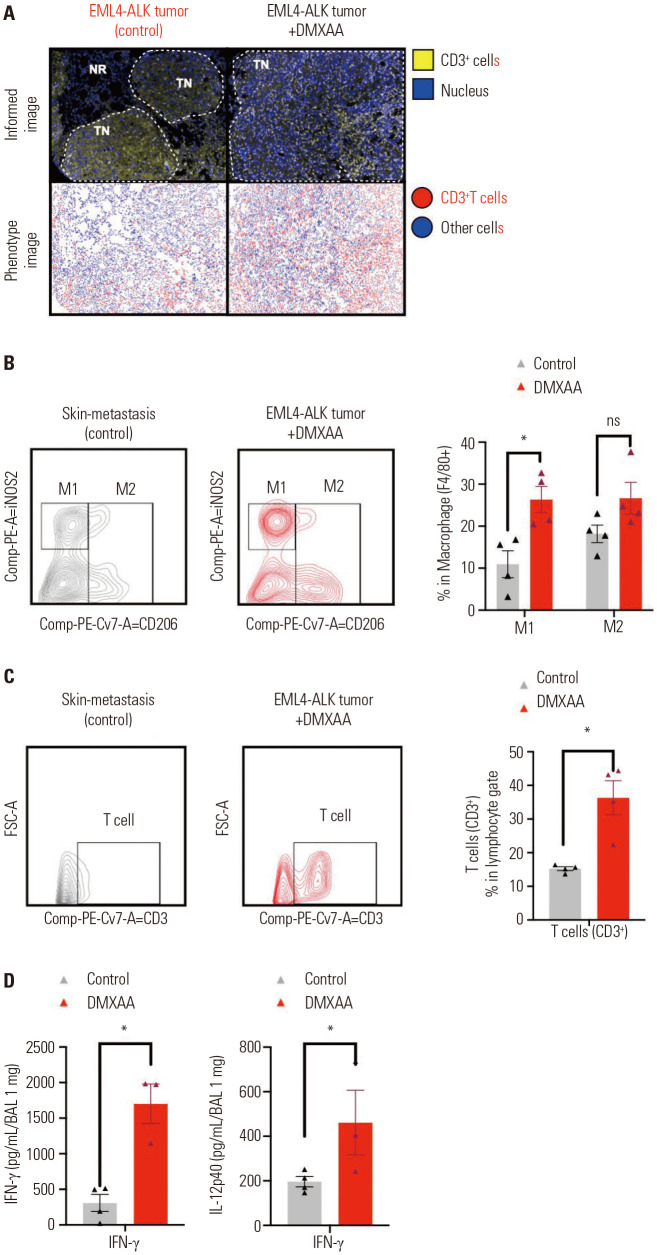
Previous results had shown that STING mediated the secretion of cytokines. We aimed to investigate the changes in the lung tumor microenvironment and determine whether these cytokines are able to recruit immune cells, which is induced by treatment with DMXAA. We sacrificed the mice and collected the tumor tissue for multiplex IHC of immune cells as described in the Methods section. We observed an increase in the infiltration of CD3+ T cells in the EML4-ALK tumor treated with DMXAA (Fig. 2A). Flow cytometry analyses of the infiltrated macrophages and T cells were performed, and the results showed that DMXAA treatment induced the expression of M1 macrophage phenotype (F4/80+CD206-) (Fig. 2B) and CD3+ T cells (Fig. 2C) in tumors. The expression levels of M1 macrophages and CD3+ T cells were significantly increased in the DMXAA-treated group than in the control group (*p<0.05); the expression of M2 macrophages (F4/80+CD206+) was also increased in the DMXAA-treated group than in the control group. The gating strategy used to identify the macrophages and T cells in the tumor tissues is described in Supplementary Fig. 3 (only online). In addition, the IFN-γ and IL-12p40 concentrations in BAL fluid were analyzed by conducting an ELISA test, which showed significant upregulation in the DMXAA-treated group (Fig. 2D) (*p<0.05). Taken together, these results suggest that STING stimulation induces the secretion of type 1 IFN and results in the recruitment of T cells and M1 macrophages in the tumor.
Fig. 2. DMXAA treatment enhanced the infiltration of CD3+ T cells and macrophages in the tumor. (A) Representative IHC and phenotype images of the control and DMXAA-treated tumor-bearing mice. T cells are marked as yellow (upper) or red (lower). (B) Flow cytometry analysis of the macrophages and (C) T cells. The gating strategy is shown in the left panel; the numbers of M1, M2, and T cells of treatment group were higher than those in the control group. The percentages of M1, M2, and T cells are presented in a bar graph (right panel). (D) The IFN-γ and IL-12p40 concentrations in BAL fluid analyzed using an ELISA assay. The results are expressed as mean±SD. *p<0.05. ns, not significant; IFN, interferon; IL, interleukin; IHC, immunohistochemistry; BAL; Bronchoalveolar lavage; ELISA, enzyme-linked immunosorbent assay.
Association between DMXAA-induced antitumor immune response and IFN gene signatures
The recruitment of cytokines and immune cells were induced by treatment with DMXAA in the EML4-ALK model (Figs. 1 and 2). The STING-mediated immune response is initiated by cytosolic self-DNA or microbial DNA species recognized by Toll-like receptors or RIG-1-like receptors.21 In the tumor microenvironment, the STING-triggered innate signaling is similar to the host defense against pathogen invasion. The STING pathway stimulates the release of innate immune cells that reside in the skin, which then triggers an adaptive immune response. We evaluated this mechanism using a syngeneic mouse model of CT26 colon cancer, which is reported to be a non-immunogenic tumor.
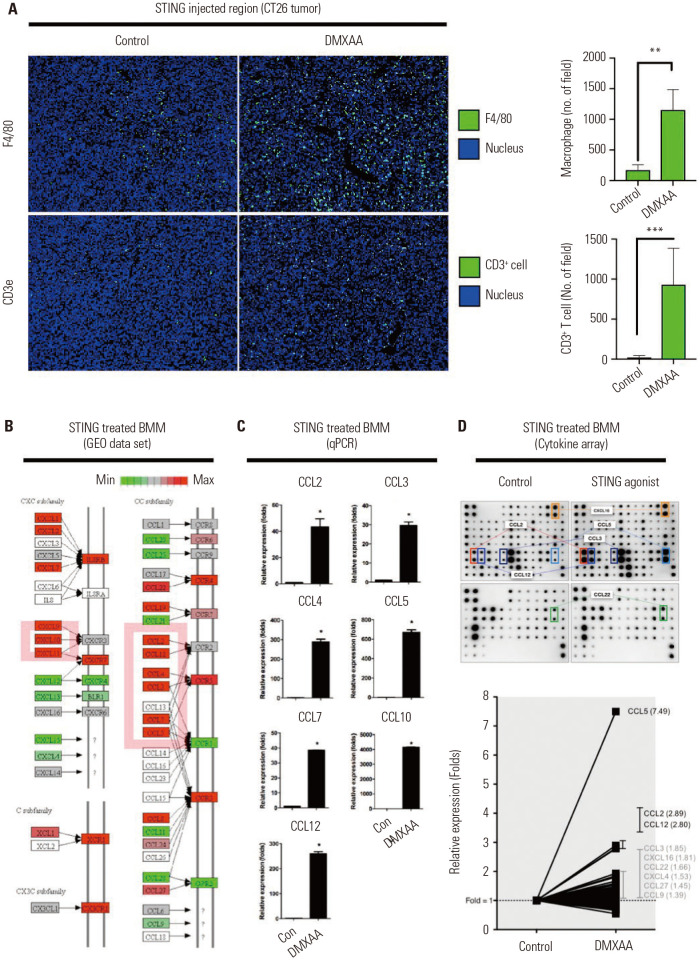
To confirm whether the recruitment of macrophages occurs after treatment with DMXAA (Fig. 2B), we performed a multiplex IHC analysis of the STING injection lesion using F4/80 and CD3e antibodies, which are unique markers of murine macrophages and T cells. We found that the levels of F4/80+ (**p<0.01) and CD3+ T cells (***p<0.001) were significantly upregulated per unit field (Fig. 3A).
Fig. 3. Profiling gene expression on DMXAA-stimulated macrophages. (A) Representative IHC images of vehicle-treated (control) and DMXAA-treated CT26 tumor-bearing mice (left). Tumor-infiltrated macrophages and CD3+ T cells analyzed per unit field (right). (B) Differential gene expression for chemokine pathway from KEGG. The pathway showed significantly regulated genes, which are highlighted in red. (C) Peritoneal macrophages treated with 50 µg/mL of DMXAA for 24 hours. The mRNA levels of these chemokines in macrophages were detected by real-time PCR. (D) Supernatants were collected within 24 hours after treatment, and a chemokine array containing 96 cytokines/chemokines in duplicate was analyzed (upper). The relative expression of chemokines with fold change (lower). The results are expressed as mean±SD. *p<0.05; **p<0.01; ***p<0.001. STING, stimulator of interferon genes; BMM, bone marrow macrophages; GEO, Gene Expression Omnibus; qPCR, real-time PCR; IHC, immunohistochemistry; KEGG, Kyoto Encyclo-pedia of Genes and Genomes; PCR, polymerase chain reaction.
DMXAA treatment triggered the macrophages to produce chemokines and cytokines in order to induce an innate immune response and an adaptive immune response, such as T cell recruitment. In order to pre-screen the expression of chemokines and cytokines, the Gene Expression Omnibus (GEO) data set (GSE7194) of DMXAA-treated PMs were obtained from the NCBI database and analyzed using the Gene Set Enrichment Analysis (GSEA) and Differentially Expressed Gene (DEG) methods. The IFN signaling was the major pathway in the DMXAA-treated group (Supplementary Fig. 4, only online). The DMXAA-treated macrophages uniquely expressed CCL3, CCL4, CCL5 CCL12, CXCL9, CCL2, CXCL11, CXCL10, and CCL7 transcripts compared to the control (Fig. 3B). The expression of these chemokines could be correlated with the actual mRNA and protein levels. To validate our findings, the total mRNA from DMXAA-treated macrophages were extracted and analyzed using the qPCR method. The mRNA levels of CCL2, CCL3, CCL4, CCL5, CCL7, CXCL10, and CCL12 were significantly greater in the DMXAA-treated macrophages (Fig. 3C). These results were strongly correlated with the results of the analysis of the GEO data set. In addition, the following chemokines from DMXAA-treated macrophages were expressed at higher levels on the supernatant samples on the cytokine array: CCL2, CCL3, CCL5, CCL12, CXCL16, and CCL12 (Fig. 3D). CCL2, CCL5, and CCL12 were universally increased on the microarray, qPCR, and protein array. Altogether, these data suggest that DMXAA-induced chemokines may induce innate immune responses and DC recruitment.
DMXAA enhanced antigen presentation by activating the co-stimulatory receptors and MHC pathway
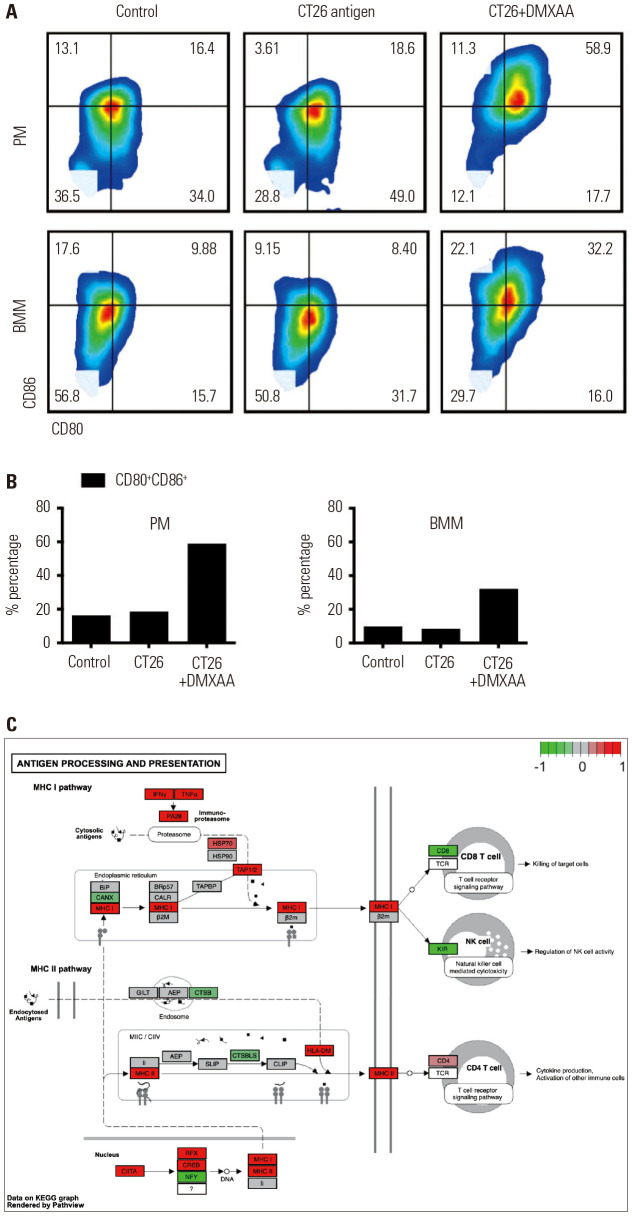
As shown in Fig. 4C, the upregulation of CD80 and CD86 surface proteins is important for binding the CD28 transmembrane domain of T cells, which could potentiate the antigen presentation ability. We hypothesized that, based on previous observations, stimulation of the STING pathway would also increase the antigen presentation. As expected, after treatment with DMXAA, the CD80 and CD86 levels in both peritoneal and BMMS were increased compared to those in the control group (Fig. 4A). In PMs, the CD86+CD80+ value in the control group was 16.4%, which was similar to that in the CT26 antigen-treated group (18.6%); by contrast, the group treated with a combination of CD26 antigen and DMXAA showed a CD86+CD80+ value of 58.9%. In BMMs, the CD86+CD80+ value in the control group was 9.8%, which was similar to that in the CT26 antigen-treated group (8.4%); on the contrary, the group treated with a combination of CD26 antigen and STING agonist showed a CD86+CD80+ value of 32.2%. The upregulated expression of CD80 and CD86 occurred within 24 hours after DMXAA treatment (Fig. 4B). Treatment with DMXAA stimulated the macrophages to produce chemokines and cytokines in order to induce an innate immune response and trigger an adapt immune response, such as T cell recruitment. In order to pre-screen the chemokine and cytokine expression, the GEO data set (GSE7194) of DMXAA-treated PMs were obtained from the NCBI database and analyzed using the GSEA and DEG methods. IFN signaling was the major pathway in DMXAA-treated group (Fig. 4C).
Fig. 4. Reinforcement of tumor antigen-presenting activity via CD80/CD86 after DMXAA treatment. (A) Transient induction of CD80 and CD86 in control (vehicle-treated group), CT26 antigen-treated, and CT26 antigen+DMXAA treated macrophages isolated from the peritoneum and bone marrow. (B) The CD80 and CD86 expression levels on the surface increase in a time-dependent manner. The percentages of CD80 and CD86 double-positive cells increased from 24.4% at 0 h to 66.3% at 24 hours of DMXAA treatment in the PMs (left) and BMMs (right). No changes were observed in the macrophages treated with only a CT26 antigen. (C) DMXAA-related alteration of gene expression associated with antigen processing and presentation in macrophages. The entire MHC I and MHC II signaling pathways are presented. The molecules with higher or lower gene expression compared to the control are highlighted in red and green, respectively. PM, peritoneal macrophages; BMM, bone marrow macrophages; MHC, major histocompatibility complex.
DMXAA treatment promoted tumor-reactive T cell activation in vivo
To evaluate whether DMXAA could induce the expression of tumor-reactive T cells by stimulating APCs in vivo, we injected 500 µg of DMXAA in the experimental mice and collected the spleen, LN, and MLN after 24 hours. We aimed to investigate whether the STING pathway was essential in the tumor-reactive T cell activation. We used anti-IFN-β to inhibit the STING pathway and examined the induction of tumor-reactive T cells. The DMXAA group and DMXAA+anti-IFN-β group were injected with 5×106 CT26 GFP/luciferase cells in the right flanks. A week later, the control, DMXAA, and DMXAA+anti-IFN-β (n=6) mice were inoculated with 1×105 CT26 GFP/luciferase cells by intraperitoneal injection. Then, they were administered either an HBSS (control group) or 500 µg of DMXAA. The DMXAA+anti-IFN-β group was injected additionally with 600 µg of anti-IFN-β intraperitoneally every 3 days (Supplementary Fig. 5, only online).
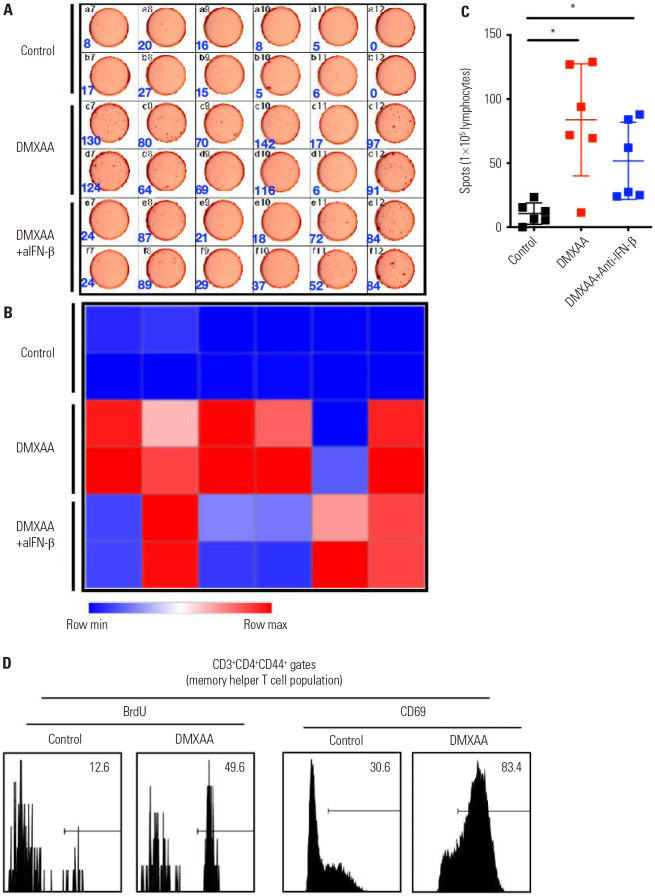
IFN-γ ELISpot assay was performed in the collected lymphocytes from MLN to measure the antigen-specific T cells (Fig. 5A). Interestingly, the IFN-β-depleted group showed partially abrogated tumor-specific T cells (Fig. 5B). The differences in the results were verified using a dot plot graph. The levels of tumor-specific T cells was significantly increased in the DMXAA-treated group than in the control group (*p<0.05). Moreover, the levels of tumor-specific T cells significantly increased in the DMXAA+anti-IFN-β group compared to the control group (*p<0.05). However, the levels of tumor-specific T cells significantly decreased in the DMXAA+anti-IFN-β group compared to the DMXAA-treated group (Fig. 5C). Moreover, we confirmed that BrdU is a proliferation marker and CD69 is an early activation marker of T cells in the DMXAA-treated group and control group. The proportion of BrdU-positive population was 49.6% in the DMXAA-treated group, while that in the control group was only 12.6%. In addition, the CD69-positive population was 83.4% in DMXAA-treated group, while that in the control group was only 30.6% (Fig. 5D). These data demonstrate that the STING pathway is essential for activating the helper and cytotoxic T cells, especially the tumor-specific T cells, which play an important role in eliminating tumors with IFN-β.
Fig. 5. DMXAA promotes strong T cell activation, especially tumor-reactive T cells, in vivo. (A and B) The DMXAA and DMXAA+anti-IFN-β groups of BALB/c mice were inoculated with 5×106 CT26 GFP/luciferase cells in the right flank (n=6). One week later, the control (vehicle treated group), DMXAA, and DMXAA+anti-IFN-β groups were injected with 1×105 CT26 GFP/luciferase cells intraperitoneally. They received an intratumoral injection of either an HBSS or 500 µg of DMXAA or an intraperitoneal injection of 600 µg of anti-IFN-β every 3 days. The frequency of tumor-specific IFN-γ producing T cells was assessed by ELISpot. (C) The cells were analyzed using ImageJ software and visualized using Prism 9. (D) Memory T cell populations were analyzed with BrdU and CD69. The results are expressed as mean±SD. *p<0.05. IFN, interferon; HBSS, Hanks Balanced Salt Solution; ELISpot, enzyme-linked immune absorbent spot.
DMXAA induced an abscopal effect in vivo
Type 1 IFN plays an important role in anticancer immune response both locally and systemically. We observed that STING stimulation activates the expression of adaptive immune cells.
Tumor-specific T cells were attracted by type I IFN cytokines in the primary and metastatic sites. We hypothesized that this activation occurs systemically and is able to preserve the abscopal site of metastasis.22 To confirm the role of type 1 IFN, the experimental mice were organized into three groups (control, DMXAA, and DMXAA+anti-IFN-β groups). The individual mice were injected with 5×106 CT26 GFP/luciferase cells into the right flanks. A week later, the control (n=6), DMXAA, and DMXAA+anti-IFN-β groups (n=10) were inoculated with 1×105 CT26 GFP/luciferase cells intraperitoneally.
The mice were intratumorally injected with an HBSS (control group) or 500 µg of DMXAA (DMXAA and DMXAA+anti-IFN-β groups), while anti-IFN-β was administered in the DMXAA+anti-IFN-β group. Secondary tumor growth was measured using an IVIS (Fig. 6A). DMXAA treatment resulted in the suppression of tumor growth in the abdominal cavity of the DMXAA-treated group at day 3. However, the injection of neutralizing IFN-β modestly reduced the antitumor effects of DMXAA at day 3. Western blot analysis was performed using the CT26 cells to confirm the expression of IFN-β. The CT26 cells were treated with 50 µg/mL of DMXAA and measured at each indicated time point, as shown in Supplementary Fig. 6 (only online). The expression of pIRF3 did not increase until 30 min after DMXAA treatment, but it eventually increased 60 min after DMXAA treatment. Therefore, in order to measure the expression levels of IFN-β based on the expression of phosphorylated IRF3, we treated PM and CT26 cells with 50 µg/mL of DMXAA in a time-dependent manner. ELISA was performed using the supernatant. Our results showed that the expression levels of IFN-β significantly increased in PMs for 6 hours, but it eventually decreased thereafter. In contrast, IFN-β was not expressed in the CT26 cells. This finding suggested that the IFN-β is secreted by PMs and not by tumors (Supplementary Fig. 6, only online).
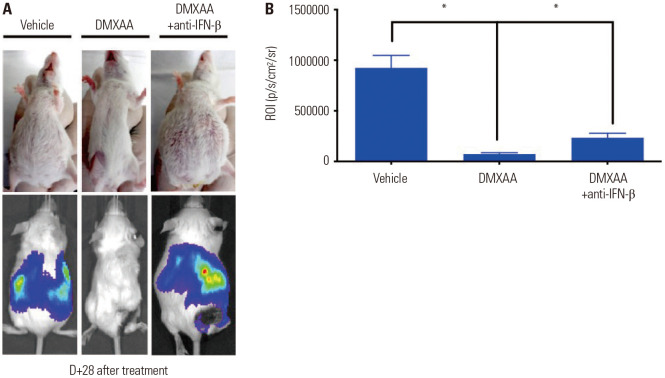
Fig. 6. IFN-β played an important role in the DMXAA-induced antitumor response in vivo. The DMXAA and DMXAA+anti-IFN-β groups of BALB/c mice were inoculated with 5×106 CT26 GFP/luciferase cells in the right flank (n=10). One week later, the control (vehicle treated group), DMXAA, and DMXAA+ anti-IFN-β groups were injected with 1×105 CT26 GFP/luciferase cells intraperitoneally. They received an intratumoral injection of HBSS or 500 µg of DMXAA or an intraperitoneal injection of 600 µg of anti-IFN-β every 3 days. (A) In vivo imaging of CT26 GFP/luciferase tumor in BALB/c mice at day 3 after drug treatment. (B) Region of interest of panel A. The results are expressed as mean±SD. *p<0.05. IFN, interferon; HBSS, Hanks Balanced Salt Solution.
After 28 days of drug treatment, the DMXAA-treated group showed absence of tumor in the abdomen (Fig. 6A). The quantitative analyses results are shown in Fig. 6B; DMXAA treatment resulted in a profound tumor remission, whereas DMXAA+anti-IFN-β treatment did not result in the profound inhibition of tumors. The ROI of the DMXAA-treated group was significantly lower than that of the control and DMXAA+anti-IFN-β groups (*p<0.05) (Fig. 6B). Taken together, treatment with DMXAA can induce an antitumor effect by promoting an abscopal effect, while treatment with anti-IFN-β partially suppresses the antitumor effects of DMXAA.
DISCUSSION
We observed that the innate immune responses were activated after treatment with the STING agonist, DMXAA. The quick innate immune responses were initiated within 1 hour via the translocation of pIRF3 and pp63 into the nucleus of the macrophages. The innate genes were recovered within 12 hours. The swift responses reinforced the presentation of antigens.
Macrophage is a major APC found in the skin. Our results showed the enhancement of antigen presentation after treatment with DMXAA. Cancer cells primarily contain neoantigens, but their levels are not sufficient to stimulate the antigen presentation due to the decreased immune potential. The transient treatment of STING agonist on macrophages increased the expression of CD80 and CD86. In addition, type 1 IFNs induced the activation and development of DCs. CD14+ monocytes circulating in the blood developed into mature DCs after treatment with type 1 IFNs. The mature DC highly expresses class 1 MHC molecules as well as CD40, CD80, and CD86.23 The type 1 IFNs enhance antigen recognition and engraftment.23 The production of tumor-targeting T cells was increased in the DMXAA+CT26 antigen-treated group.
The anticancer effect can be due to the connections between the initiation of innate immune responses with type 1 IFNs and the induction of antigen presentation of APC. The DMXAA-treated macrophages express chemokines and cytokines and were previously used in the microarray assay. The GEO data set was re-analyzed using GSEA and DEG. The gene set was upregulated in the IFN-related pathways [Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome]. Among them, the levels of chemokines were markedly increased (log-FC >2, p<0.05). The in silico data were analyzed by mRNA and cytokine array. As predicted, the chemokines with the highest expression levels were CCL5 (RANTES), CCL2 (MCP1), and CCL12 (MCP5). The expression of CCL5 and CCL2 promoted the migration of mature or immature DCs in a dose-dependent manner.24 The murine DCs expresses Ccr2 (ligands: CCL2, CCL5, and CCL12) and Ccr1 (ligand: CCL5).24 The migration of DC could be related to the expression of the three chemokines. In this article, we defined the DC recruitment and activation of antigen presentation. These reactions correlated with T cell activation and memory Th1 cell proliferation. The increase in and activation of T cells were induced by the production of tumor-killing T cells.
These results could explain the anticancer effect with abscopal treatment. Our results showed a wide range of involvement in the cancer-immune cycle. The cancer-immune cycle was initially described by Chen and Mellman,9 Iwasaki, et al.25 The cycle involves cancer characterization, expression of neoantigens and MHC class-antigen binding, and T cell events with cancer killing. The cancer-immune cycle is beneficial in understanding the immuno-oncology drug treatment. All seven steps are important for cancer eradication. PD-1, PD-L1, and CTLA4 are well-known targets of cancer treatment. The tumor-infiltrating T cells are generally abundant. However, the induction of innate immune responses via the STING pathway is widely involved in antigen presentation as well as T cell activation and proliferation. Our data showed the connection between innate immune responses and adaptive immune responses for cancer therapy.
Taken together, our results suggest the abscopal effect of DMXAA treatment in peritoneal metastatic colon cancer. The effect of STING agonist was widely elaborated in the cancer-immune cycle: antigen presentation, T cell activation, and memory Th1 cell proliferation. An important type 1 IFN-β was partially involved in the improvement of antigen presentation and anticancer effect. This suggests that the other immune elements play similar roles as that of IFN-β. However, significant barriers to the STING pathway were observed, which have been addressed in recent clinical studies. DMXAA was administered intratumorally. This limits the use of DMXAA to accessible tumors. In addition, the effect of systemic administration of STING agonists remains unknown, as the overactivation of STING is associated with a wide range of autoimmune conditions. In order to overcome the limitations of the STING agonist, further studies should be conducted to elucidate the unexplained mechanism of the STING pathway and to enhance the tumor-specific immune response.
ACKNOWLEDGEMENTS
This work was supported by the Research Foundation of Yonsei University (no. 6-2017-0104) and the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2019R1A2C4069993).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Chun-Bong Synn, Dong Kwon Kim, Chang Gon Kim, Min Hee Hong, Sun Min Lim, and Kyoung-Ho Pyo.
- Data curation: Chun-Bong Synn, Dong Kwon Kim, Mi Ran Yun, Sun Min Lim, and Kyoung-Ho Pyo.
- Formal analysis: Chun-Bong Synn, Dong Kwon Kim, Youngseon Byeon, Sun Min Lim, and Kyoung-Ho Pyo.
- Funding acquisition: Sun Min Lim and Kyoung-Ho Pyo.
- Investigation: Chun-Bong Synn, Dong Kwon Kim, Mi Ran Yun, Ji Min Lee, Wongeun Lee, Seul Lee, You-Won Lee, Doo Jae Lee, Hyun-Woo Kim, Chang Gon Kim, Min Hee Hong, Jung Dong Park, Sun Min Lim, and Kyoung-Ho Pyo.
- Methodology: Chun-Bong Synn, Dong Kwon Kim, Jae Hwan Kim, Young Seob Kim, Eun Ji Lee, You-Won Lee, Chang Gon Kim, Min Hee Hong, Sun Min Lim, and Kyoung-Ho Pyo.
- Project administration: Chun-Bong Synn, Sun Min Lim, and Kyoung-Ho Pyo.
- Resources: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Software: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Supervision: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Validation: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Visualization: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Writing—original draft: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Writing—review & editing: Chun-Bong Synn, Dong Kwon Kim, Sun Min Lim, and Kyoung-Ho Pyo.
- Approval of final manuscript: all authors.
SUPPLEMENTARY MATERIALS
DMXAA induces tumor reduction in EML4-ALK mice. (A) MRI images of control and anti-PD-1-treated tumor-bearing mice showing no response to anti-PD-1 treatment. (B) Percentage of tumor reduction in DMXAA-treated mice compared to that in tumor-bearing mice. The results are expressed as mean±SD. *p<0.05.
Spleen images from EML4-ALK NSCLC mice. Spleen size comparison between control, tumor-bearing, and DMXAA-treated tumor-bearing mice.
Immune cell infiltration assay by flow cytometry. Gating method for the identification of macrophages (A) and T cells (B) after treatment with DMXAA on tumor-bearing mice in Fig. 2.
Schematic design of GSEA and DEG using GEO data set. GEO, Gene Expression Omnibus; STING, stimulator of interferon genes; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclo-pedia of Genes and Genomes; DEG; Differentially Expressed Gene; FWER, family-wise error rate.
DMXAA activated the STING pathway in the peritoneal macrophage. Peritoneal macrophages were treated with 50 µg/mL of DMXAA for the indicated time points. (A) Schematic diagram of IFN-β expression. (B) The macrophages were stimulated with 50 µg/mL of DMXAA for the indicated time points. The amounts of pIRF3, total IRF3, pp65, total p65, total I-B-a, and β-actin were measured by western blot analysis. (C and D) The macrophages were cultured on coated chamber slides with total media (5×103 cells/chamber). The macrophages were stimulated with 50 µg/mL of DMXAA for the indicated time points and stained with specific antibodies against pIRF3, NF-κB, and DAPI. (E) The expression of STING-related genes (IFN-β, IFN-α, and TNF-α) on macrophages (upper) was measured at the indicated time points (0–24 hours). The amount of IFN-β was measured by ELISA, while the NO-related gene, NOS2, was measured by real-time PCR; the nitric oxide level was also measured (lower). (F) IFN signaling gene levels and IFN-α/β signaling gene levels were analyzed with Reactome. The results are expressed as mean±SD. *p<0.05. IFN, interferon; STING, stimulator of interferon genes; TNF, tumor necrosis factor; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; PCR, polymerase chain reaction.
DMXAA stimulated pIRF signal in the STING pathway but expressed no IFN-β on CT26. (A) The pIRF and NF-κB levels in DMXAA-treated CT26 cells were measured by western blot analysis. The CT26 cells were treated with 50 µg/mL of DMXAA at the indicated time points. (B) To determine the expression of IFN-β, peritoneal macrophages and CT26 cells were treated with 50 µg/mL of DMXAA at the indicated time points. Then, a culture supernatant is harvested, and ELISA was performed. The results are expressed as mean±SD. *p<0.05. IFN, interferon; PM, peritoneal macrophages; STING, stimulator of interferon genes; ELISA, enzyme-linked immunosorbent assay.
References
- 1.Abuodeh Y, Venkat P, Kim S. Systematic review of case reports on the abscopal effect. Curr Probl Cancer. 2016;40:25–37. doi: 10.1016/j.currproblcancer.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Adams S. Toll-like receptor agonists in cancer therapy. Immunotherapy. 2009;1:949–964. doi: 10.2217/imt.09.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baaten BJ, Li CR, Bradley LM. Multifaceted regulation of T cells by CD44. Commun Integr Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du H, Xu T, Cui M. cGAS-STING signaling in cancer immunity and immunotherapy. Biomed Pharmacother. 2021;133:110972. doi: 10.1016/j.biopha.2020.110972. [DOI] [PubMed] [Google Scholar]
- 5.Jiang M, Chen P, Wang L, Li W, Chen B, Liu Y, et al. cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol. 2020;13:81. doi: 10.1186/s13045-020-00916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity. 2010;32:104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Cotter SE, Dunn GP, Collins KM, Sahni D, Zukotynski KA, Hansen JL, et al. Abscopal effect in a patient with metastatic Merkel cell carcinoma following radiation therapy: potential role of induced antitumor immunity. Arch Dermatol. 2011;147:870–872. doi: 10.1001/archdermatol.2011.176. [DOI] [PubMed] [Google Scholar]
- 11.Daei Farshchi Adli A, Jahanban-Esfahlan R, Seidi K, Samandari-Rad S, Zarghami N. An overview on Vadimezan (DMXAA): the vascular disrupting agent. Chem Biol Drug Des. 2018;91:996–1006. doi: 10.1111/cbdd.13166. [DOI] [PubMed] [Google Scholar]
- 12.Motedayen Aval L, Pease JE, Sharma R, Pinato DJ. Challenges and opportunities in the clinical development of STING agonists for cancer immunotherapy. J Clin Med. 2020;9:3323. doi: 10.3390/jcm9103323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyo KH, Lim SM, Park CW, Jo HN, Kim JH, Yun MR, et al. Comprehensive analyses of immunodynamics and immunoreactivity in response to treatment in ALK-positive non-small-cell lung cancer. J Immunother Cancer. 2020;8:e000970. doi: 10.1136/jitc-2020-000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohkuri T, Kosaka A, Nagato T, Kobayashi H. Effects of STING stimulation on macrophages: STING agonists polarize into “classically” or “alternatively” activated macrophages? Hum Vaccin Immunother. 2018;14:285–287. doi: 10.1080/21645515.2017.1395995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pyo KH, Lim SM, Kim HR, Sung YH, Yun MR, Kim SM, et al. Establishment of a conditional transgenic mouse model recapitulating EML4-ALK–positive human non–small cell lung cancer. J Thorac Oncol. 2017;12:491–500. doi: 10.1016/j.jtho.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DG, Martinelli R, Besra GS, Illarionov PA, Szatmari I, Brazda P, et al. Identification and characterization of a novel anti-inflammatory lipid isolated from Mycobacterium vaccae, a soil-derived bacterium with immunoregulatory and stress resilience properties. Psychopharmacology (Berl) 2019;236:1653–1670. doi: 10.1007/s00213-019-05253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun F, Xiao G, Qu Z. Murine bronchoalveolar lavage. Bio Protoc. 2017;7:e2287. doi: 10.21769/BioProtoc.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamell EF, Wolchok JD, Gnjatic S, Lee NY, Brownell I. The abscopal effect associated with a systemic anti-melanoma immune response. Int J Radiat Oncol Biol Phys. 2013;85:293–295. doi: 10.1016/j.ijrobp.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn J, Barber GN. STING signaling and host defense against microbial infection. Exp Mol Med. 2019;51:1–10. doi: 10.1038/s12276-019-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vatner RE, Cooper BT, Vanpouille-Box C, Demaria S, Formenti SC. Combinations of immunotherapy and radiation in cancer therapy. Front Oncol. 2014;4:325. doi: 10.3389/fonc.2014.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DMXAA induces tumor reduction in EML4-ALK mice. (A) MRI images of control and anti-PD-1-treated tumor-bearing mice showing no response to anti-PD-1 treatment. (B) Percentage of tumor reduction in DMXAA-treated mice compared to that in tumor-bearing mice. The results are expressed as mean±SD. *p<0.05.
Spleen images from EML4-ALK NSCLC mice. Spleen size comparison between control, tumor-bearing, and DMXAA-treated tumor-bearing mice.
Immune cell infiltration assay by flow cytometry. Gating method for the identification of macrophages (A) and T cells (B) after treatment with DMXAA on tumor-bearing mice in Fig. 2.
Schematic design of GSEA and DEG using GEO data set. GEO, Gene Expression Omnibus; STING, stimulator of interferon genes; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclo-pedia of Genes and Genomes; DEG; Differentially Expressed Gene; FWER, family-wise error rate.
DMXAA activated the STING pathway in the peritoneal macrophage. Peritoneal macrophages were treated with 50 µg/mL of DMXAA for the indicated time points. (A) Schematic diagram of IFN-β expression. (B) The macrophages were stimulated with 50 µg/mL of DMXAA for the indicated time points. The amounts of pIRF3, total IRF3, pp65, total p65, total I-B-a, and β-actin were measured by western blot analysis. (C and D) The macrophages were cultured on coated chamber slides with total media (5×103 cells/chamber). The macrophages were stimulated with 50 µg/mL of DMXAA for the indicated time points and stained with specific antibodies against pIRF3, NF-κB, and DAPI. (E) The expression of STING-related genes (IFN-β, IFN-α, and TNF-α) on macrophages (upper) was measured at the indicated time points (0–24 hours). The amount of IFN-β was measured by ELISA, while the NO-related gene, NOS2, was measured by real-time PCR; the nitric oxide level was also measured (lower). (F) IFN signaling gene levels and IFN-α/β signaling gene levels were analyzed with Reactome. The results are expressed as mean±SD. *p<0.05. IFN, interferon; STING, stimulator of interferon genes; TNF, tumor necrosis factor; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; PCR, polymerase chain reaction.
DMXAA stimulated pIRF signal in the STING pathway but expressed no IFN-β on CT26. (A) The pIRF and NF-κB levels in DMXAA-treated CT26 cells were measured by western blot analysis. The CT26 cells were treated with 50 µg/mL of DMXAA at the indicated time points. (B) To determine the expression of IFN-β, peritoneal macrophages and CT26 cells were treated with 50 µg/mL of DMXAA at the indicated time points. Then, a culture supernatant is harvested, and ELISA was performed. The results are expressed as mean±SD. *p<0.05. IFN, interferon; PM, peritoneal macrophages; STING, stimulator of interferon genes; ELISA, enzyme-linked immunosorbent assay.