Abstract
To investigate the role of insulin receptor substrate 1 (IRS-1) and IRS-2, the two ubiquitously expressed IRS proteins, in adipocyte differentiation, we established embryonic fibroblast cells with four different genotypes, i.e., wild-type, IRS-1 deficient (IRS-1−/−), IRS-2 deficient (IRS-2−/−), and IRS-1 IRS-2 double deficient (IRS-1−/− IRS-2−/−), from mouse embryos of the corresponding genotypes. The abilities of IRS-1−/− cells and IRS-2−/− cells to differentiate into adipocytes are approximately 60 and 15%, respectively, lower than that of wild-type cells, at day 8 after induction and, surprisingly, IRS-1−/− IRS-2−/− cells have no ability to differentiate into adipocytes. The expression of CCAAT/enhancer binding protein α (C/EBPα) and peroxisome proliferator-activated receptor γ (PPARγ) is severely decreased in IRS-1−/− IRS-2−/− cells at both the mRNA and the protein level, and the mRNAs of lipoprotein lipase and adipocyte fatty acid binding protein are severely decreased in IRS-1−/− IRS-2−/− cells. Phosphatidylinositol 3-kinase (PI 3-kinase) activity that increases during adipocyte differentiation is almost completely abolished in IRS-1−/− IRS-2−/− cells. Treatment of wild-type cells with a PI 3-kinase inhibitor, LY294002, markedly decreases the expression of C/EBPα and PPARγ, a result which is associated with a complete block of adipocyte differentiation. Moreover, histologic analysis of IRS-1−/− IRS-2−/− double-knockout mice 8 h after birth reveals severe reduction in white adipose tissue mass. Our results suggest that IRS-1 and IRS-2 play a crucial role in the upregulation of the C/EBPα and PPARγ expression and adipocyte differentiation.
Recently there has been a dramatic increase in the prevalence of obesity attributable to excessive white adipose tissue both in Western countries and in Japan. Because adipocytes play a critical role in energy balance, understanding the molecular mechanisms of adipocyte differentiation may provide clues to strategies for the prevention and treatment of obesity.
The mechanisms of adipocyte differentiation have been extensively studied in preadipocyte culture systems. Characterization of the regulatory regions of adipose-specific genes has led to the identification of key transcription factors in the complex transcriptional cascade that occurs during adipocyte differentiation (36); these factors include peroxisome proliferator-activated receptor γ (PPARγ) (19, 44), CCAAT/enhancer binding protein (C/EBP) (12, 23, 32, 46, 52, 56), and adipocyte differentiation and determination factor 1 (ADD1)-sterol regulatory element binding protein 1c (SREBP1c) (17, 18, 36, 45).
PPARγ is induced before the transcriptional activation of most adipocyte-specific genes, and expression of PPARγ has been shown to be sufficient to induce growth arrest and to initiate adipogenesis in exponentially growing fibroblast cell lines, thus demonstrating its critical role in the regulation of adipocyte differentiation (2, 15, 43). In addition, PPARγ-deficient cells fail to differentiate into adipocytes, indicating that PPARγ plays a pivotal role in adipocyte differentiation (21, 29). Most of the PPARγ target genes in adipose tissue including the genes encoding phosphoenolpyruvate carboxykinase (41), lipoprotein lipase (LPL) (33) and adipocyte fatty acid binding protein (A-FABP or aP2) (42) are directly implicated in lipogenic pathways.
C/EBPα is the most highly expressed member of C/EBP family in adipose tissue and in liver and has been implicated in the maintenance of the terminally differentiated adipocyte phenotype (4, 9, 25, 56). C/EBPα is induced relatively late during adipogenesis in culture, after the induction of PPARγ but before the induction of many of the enzymes and proteins characteristic of fully differentiated cells (53), and it transcriptionally activates a large number of adipocyte-specific genes (8). C/EBPα-null mice fail to develop white adipose tissue (48), and C/EBPα-deficient cells fail to differentiate into adipocytes (51). C/EBPβ (1, 7, 9, 10, 56) and C/EBPδ (4, 16, 49) are induced very early and have been shown to play a crucial role in initiating the differentiation of preadipocytes by activating the expression of PPARγ (39, 50, 52, 56). An in vitro study of adipocyte differentiation in C/EBPβ−/− C/EBPδ−/− embryonic fibroblast (EF) cells shows that the expression of both PPARγ and C/EBPα is severely reduced (39).
In addition, there is a large amount of literature describing extracellular factors that influence adipogenic potentials. These include insulin-like growth factor 1 (IGF-1) and insulin. However, little is known about the signal transduction pathways by which these hormones regulate the expression of these transcription factors and promote adipogenesis.
IRS-1 and IRS-2 are the two most ubiquitously expressed members of the IRS family of proteins, which can bind signaling proteins with Src-homology-2 domains (SH2 proteins), such as p85 regulatory subunit of phosphatidylinositol 3-kinase (PI 3-kinase), subsequent to the activation of receptors for insulin, IGF-1, and several cytokines (26, 28, 37). IRS-1 plays an important role in the metabolic actions of insulin and IGF-1 mainly in skeletal muscle and adipose tissue, and IRS-2 plays an important role in the metabolic actions of these hormones in the liver (3, 55). The roles of IRS-1 and IRS-2 in adipocyte differentiation, on the other hand, have not been reported.
To investigate the role of IRS-1 and IRS-2 in adipocyte differentiation, we intercrossed mice heterozygous for each of two null alleles (Irs1+/− [38] and Irs2+/− [22]) and generated primary EF cells that were wild type, IRS-1 deficient (IRS-1−/−), IRS-2 deficient (IRS-2−/−), or IRS-1 IRS-2 double deficient (IRS-1−/− IRS-2−/−).
We show here that IRS-1 IRS-2 double-deficient cells have no ability to differentiate into adipocytes. We also show that the simultaneous lack of both IRS-1 and IRS-2 dramatically decreases the expression of C/EBPα and PPARγ at both the mRNA and the protein levels during adipocyte differentiation. Thus, our data provide the first direct evidence for the essential role of IRS-1 and IRS-2 in the expression of adipose-specific transcriptional factors, such as the C/EBP family and PPARγ, and adipocyte differentiation.
MATERIALS AND METHODS
Materials.
The probe for Northern blot and RNase protection assay to 36B4 was a generous gift from Naoya Yahagi (University of Tokyo) (34). The polyclonal antibodies to IRS-1, phosphotyrosine, PPARγ, and C/EBPα were from Santa Cruz Biotechnology, Inc. Polyclonal antibodies to phospho-mitogen-activated protein kinase (MAPK) and MAPK were from New England Biolabs, Inc. The medium and penicillin-streptomycin solution were purchased from Gibco, Inc. Fetal bovine serum (FBS) was from JRH Biosciences. The nitrocellulose paper used for the immunoblots was from Schleicher & Schuell, Inc. The 3-isobutyl-1-methylxanthine (IBMX) and human recombinant insulin were purchased from Sigma Chemical Co. Dexamethasone (DEX) was purchased from Wako Pure Chemical Industries, Ltd. L-type Wako TG-H (S-R1 and S-R2) for triglyceride measurement was purchased from Wako Pure Chemical Industries, Ltd. BCA Protein Assay Reagent (Pierce) was used for the protein assay.
Preparation of wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− EFs.
The IRS-1 IRS-2 double-heterozygous mice (IRS-1+/− IRS-2+/− mice) were generated by intercrosses of IRS-1-deficient mice (38) and IRS-2-deficient mice (22). To obtain embryos at 13.5 days past coitus, conceptuses that were obtained by fertilization in vitro of ova from IRS-1+/− IRS-2+/− female mice with sperm from IRS-1+/− IRS-2+/− male mice were implanted into pseudopregnant foster mothers, as previously described (21). The foster mothers were sacrificed at 13.5 days past coitus, the embryos were dissected from the uterus, and the extra-embryonic membranes and viscera were removed. The embryos were cut into small pieces with scissors and soaked for 30 min in 4 ml of 0.25% trypsin-EDTA at room temperature with shaking, and then inactivated with α-modified Eagle medium (αMEM) supplemented with 10% heat-inactivated FBS, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. The cells were then suspended by pipetting and were plated on two 10-cm dishes per embryo. After 48 h, adherent cells were trypsinized, counted, and replated at a density of 1.2 × 105 cells/cm2 in αMEM with 10% heat-inactivated FBS, 50 U of penicillin per ml, and 50 μg of streptomycin per ml to induce adipocyte differentiation.
Induction of adipocyte differentiation.
The passage number of the EF cells used in these studies was within two passages. Induction of adipocyte differentiation was performed as previously described (39). In brief, cells were cultured on 24- or 6-well or 10-cm plastic dishes and propagated to confluence. Two days later, the medium was replaced with standard differentiation induction medium containing 0.5 mM IBMX, 1 μM DEX, 5 μg of insulin per ml, 10% FBS, 50 U of penicillin per ml, and 50 μg of streptomycin per ml, and the medium was renewed every other day.
Oil-Red O staining and triglyceride-protein assay.
Oil-Red O staining solution (0.5% Oil-Red O in isopropyl alcohol solution-distilled water [60:40]) was filtered through the Whatman no. 1 filter paper and, after the cells were washed with phosphate-buffered saline (PBS), they were stained with the filtered staining solution for 30 min at 37°C and then washed with distilled water three times.
For the triglyceride-protein assay, the cells were washed with PBS twice, 0.8 ml of homogenizing buffer (150 mM sodium chloride, 10 mM Tris-HCl [pH 8.0], 0.1% Triton X-100) was added to each well of a 24-well plate, and the adherent cells were homogenized with Polytron. The homogenate was filtered with Samprep (0.2 μm [pore size]; Millipore), and the concentration of triglyceride in the filtered homogenate was measured by using L-type Wako TG-H (S-R1 and S-R2), and a triolein diluted with the homogenizing buffer was used as the standard. A 125-μl volume of the homogenate, 50 μl of S-R1, and 25 μl of S-R2 were used. The protein concentration of the same homogenate solution was measured with a bicinchoninic acid (BCA) assay kit.
Retrovirus-mediated gene transfer.
To rescue adipocyte differentiation of EF cells from IRS1−/− IRS2−/−, the PPARγ2 expression vector for retrovirus-mediated gene transfer was constructed by ligating the BstXI fragment from the pBabe-mPPARγ2-puro (kindly provided by Bruce M. Spiegelman) into the BstXI site of pMX-puro (27). Mouse EF cells were infected with equal titers of each recombinant virus as described previously (44), with some modification.
PI 3-kinase assay.
PI 3-kinase activity in EF cells was determined in immunoprecipitates with antibodies to phosphotyrosine as described previously (55), with some modification. In brief, EF cells were lysed with lysis buffer (0.25 M sucrose; 5 mM EDTA; 5 mM EGTA; 20 mM Tris-HCl, pH 7.4; 0.2 mM Na3VO4; 10 mM NaF; 10 mM sodium PPi, 1 mM phenylmethylsulfonyl fluoride; 10 μg of leupeptin per ml; 10 μg of aprotinin per ml) containing 1% NP-40. The lysates were cleared by centrifugation at 12,000 rpm in a microcentrifuge at 4°C, and the protein content of the supernatant was determined. Then, 50 μg of the cellular protein was subjected to immunoprecipitation with monoclonal antiphosphotyrosine antibody (PY20; Santa Cruz) by using protein G-Sepharose, and the buffer was changed to PI 3-kinase assay buffer (100 mM sodium chloride; 25 mM Tris-HCl, pH 7.4; 0.5 mM EGTA). The PI 3-kinase activity was then measured as described previously (55).
RNA preparation, Northern blot analysis and RNase protection assay.
Total RNA was prepared from EF cells with Trizol (Gibco-BRL) according to the manufacturer's instructions.
Northern blot analysis was performed by a standard protocol. A 6-μg sample of total RNA was electrophoresed through denaturing formaldehyde-agarose (1%) gel and then transferred to a Hybond N+ nylon membrane (Amersham). cRNA probes for C/EBPβ, C/EBPδ, C/EBPα, PPARγ, LPL, and 36B4 were labeled by in vitro transcription with Strip-EZ RNA (Ambion) and [α-32P]UTP (NEN Life Science Products, Boston, Mass.). The cDNA probe for aP2 was labeled by random priming with the Megaprime DNA Labeling System (Amersham) and [α-32P]dCTP (NEN Life Science Products). A probe for SREBP1c that does not detect SREBP1a was designed and constructed as described previously (35).
The RNase protection assay to measure SREBP1c mRNA and PPARγ was performed with RPA III (Ambion). A 10-μg sample of total RNA was hybridized with the cRNA probe, which was prepared with MAXIscript (Ambion) and [α-32P]UTP (NEN Life Science Products).
Immunoblot analysis.
EF cells were lysed in a lysis buffer containing 1% NP-40, and insoluble materials were removed by centrifugation. Proteins in the supernatants were assayed with a BCA protein assay kit (Pierce) and then subjected to immunoblotting analysis.
The tyrosine-phosphorylated proteins associated with PI 3-kinase were analyzed by immunoprecipitating equal amounts of protein (100 μg) with anti-PI 3-kinase p85 rabbit antiserum (anti-p85PAN; Upstate 06-195) and immunoblot analysis with the antiphosphotyrosine, PY20-horseradish peroxidase (HRP) conjugate antibody, or 4G10-HRP conjugate antibody.
MAPK and phospho-MAPK were analyzed by electrophoresing 10 μg of protein from the cell lysates via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% polyacrylamide gel and immunoblotting with PhosphoPlus p44/p42 MAPK (Thr202-Tyr204) Antibody Kit (New England Biolabs, Inc.) according to the manufacturer's instructions.
PPARγ and C/EBPα were analyzed by electrophoresing 6 μg of protein from the cell lysates by SDS-PAGE on a 12% polyacrylamide gel, followed by immunoblotting with PPARγ (H-100) rabbit polyclonal immunoglobulin G (IgG) (1:500) and C/EBPα (14AA) rabbit polyclonal IgG (1:500).
Histologic analysis.
To generate IRS-1−/− IRS-2−/− mice, conceptuses that were obtained by in vitro fertilization of ova from IRS-1+/− IRS-2+/− female mice and sperm from IRS-1−/− IRS-2+/− male mice were implanted into pseudopregnant foster mothers, as previously described (21). At 8 h after birth, neonates were fixed in 10% formaldehyde in PBS. Genomic DNA was prepared from the tail of each neonate, and the genotype was determined by PCR analysis. Neonate was cut into 10-μm sections, and the sections were mounted on silanized slides. The adipose tissue was stained with hematoxylin and eosin. All sections were examined by light microscopy.
RESULTS
Adipocyte differentiation of IRS-1−/− cells is impaired.
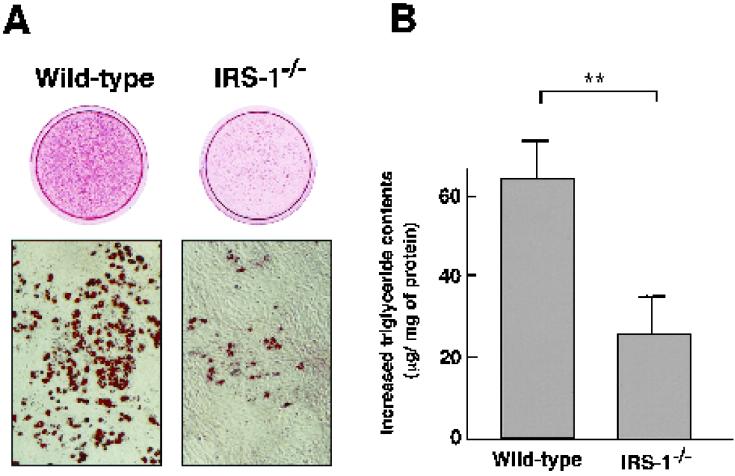
Wild-type and IRS-1−/− cells were induced to differentiation into adipocytes. Oil-Red O staining was performed 8 days after induction of adipocyte differentiation (Fig. 1A), and the increase in intracellular triglyceride content was measured (Fig. 1B). The ability of IRS-1−/− cells to differentiate into adipocytes was found to be approximately 60% lower than that of wild-type cells. These results were reproduced in six independent IRS-1−/− cells with six independent wild-type cells as controls.
FIG. 1.
Ability of wild-type and IRS-1−/− cells to differentiate into adipocytes. (A) Oil-Red O staining for fat accumulation in wild-type and IRS-1−/− cells at day 8 after induction. Cells were grown to confluence and then induced to differentiation by exposure to IBMX, DEX, and insulin, as described in Materials and Methods. (B) Increase in intracellular triglyceride content from day 0 to day 8 after induction. The assays were performed as described in Materials and Methods. The data represent the means ± the standard errors of the means from four experiments. Wild-type, n = 6; IRS-1−/−, n = 6; ∗∗, P < 0.01.
PI 3-kinase activation during adipocyte differentiation in IRS-1−/− cells was decreased.
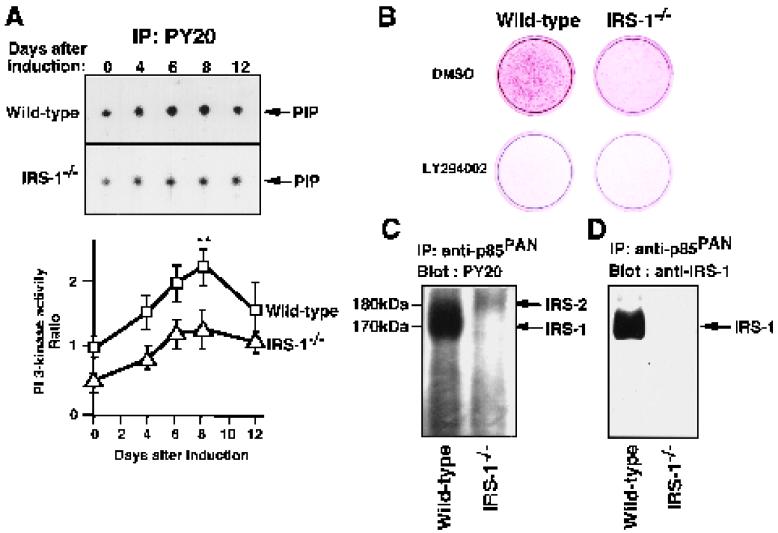
PI 3-kinase activities during adipocyte differentiation were measured in immunoprecipitates with antibody to phosphotyrosine from lysates of wild-type cells and IRS-1−/− cells (Fig. 2A). PI 3-kinase activity increased during adipocyte differentiation and reached a maximum at 8 days after induction, a result consistent with observations in adipocyte cell lines 3T3-L1 and 3T3-F442A (31). The increase in PI 3-kinase activity of in IRS-1−/− cells during adipocyte differentiation was approximately 50% less than that in the wild-type cells from 0 to 8 days after induction.
FIG. 2.
Role of PI 3-kinase in adipocyte differentiation of wild-type and IRS-1−/− cells. (A) PI 3-kinase activities during adipocyte differentiation. The antiphosphotyrosine antibody (PY20) immunoprecipitates from 50 μg of protein of cellular lysates of wild-type and IRS-1−/− cells at the day indicated after induction into adipocytes were subjected to a PI 3-kinase assay. The autoradiograms of the spots corresponding to PIP are shown in the upper panel. The radioactivity in the spots was measured, and the results are shown in the lower panel, expressed as the ratios of the values for the respective cells to those for wild-type cells at day 0 after induction. The data represent the means ± the standard errors of the means from three independent experiments. ∗∗, P < 0.01. (B) Effect of the PI 3-kinase inhibitor LY294002 on adipocyte differentiation of wild-type and IRS-1−/− cells. LY294002 (20 μM) was added to adipocyte differentiation medium, and the medium was changed at 36-h intervals. As a control, 0.1% dimethyl sulfoxide was added to the medium, and the medium was changed at the same intervals. At day 8 after induction, the intracellular fat accumulation was assessed by staining with Oil-Red O. (C) Tyrosine-phosphorylated proteins associated with p85 subunit of PI 3-kinase. Total lysates (100 μg of protein each) of cells at day 8 after induction were immunoprecipitated with anti-p85PAN (antibody to the p85 subunit of PI 3-kinase), and the immunoprecipitates were subjected to SDS-PAGE followed by immunoblotting with PY20 conjugated to HRP. (D) Association of IRS-1 with p85 subunit of PI 3-kinase. Total lysates (100 μg of protein each) of cells at day 8 after induction were immunoprecipitated with anti-p85PAN, and the immunoprecipitates were subjected to SDS-PAGE, followed by immunoblotting with anti-IRS-1 antibody.
To address the importance of the increase in PI 3-kinase activity during adipocyte differentiation, we investigated the effect of a synthetic PI 3-kinase inhibitor, LY294002, on the ability of wild-type and IRS-1−/− cells to differentiate into adipocytes. LY294002 (20 μM) was shown to completely block adipocyte differentiation of both wild-type and IRS-1−/− cells (Fig. 2B), strongly suggesting that PI 3-kinase activity was required for adipocyte differentiation.
We next studied the tyrosine-phosphorylated proteins associated with the p85 subunit of PI 3-kinase during adipocyte differentiation and found that a 170-kDa protein in wild-type cells, but a 180-kDa protein in IRS-1−/− cells, was the major tyrosine phosphorylated protein associated with the p85 subunit of PI 3-kinase (Fig. 2C). The 170-kDa protein was confirmed to be IRS-1 because this protein was detected with anti-IRS-1 antobody in wild-type cells and was not detected in IRS-1−/− cells (Fig. 2D). The 180-kDa protein was strongly suggested to be IRS-2, because this protein had a molecular mass 10 kDa larger than IRS-1 and was detected with antiphosphotyrosine antibody in IRS-1−/− cells, but not in IRS-2−/− cells or IRS-1−/− IRS-2−/− cells (Fig. 3A).
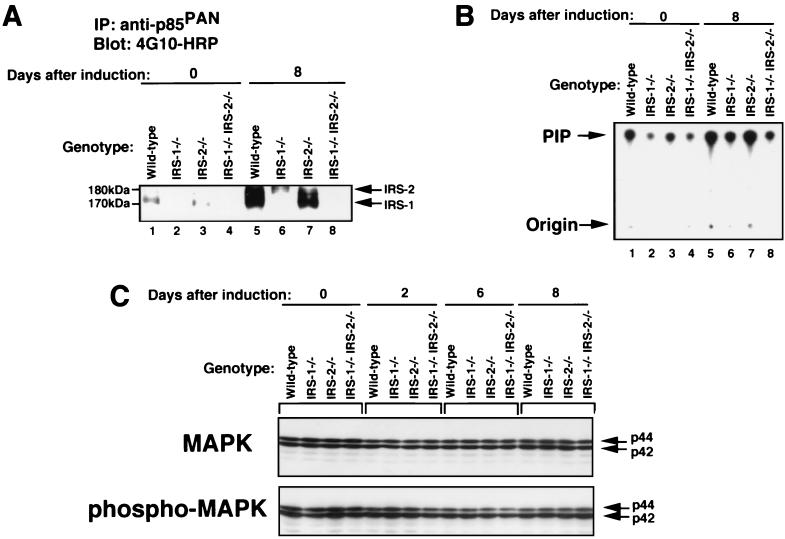
FIG. 3.
PI 3-kinase and MAPK activities during adipocyte differentiation in wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells. (A) Tyrosine-phosphorylated proteins associated with p85 subunit of PI 3-kinase in cells with four different genotypes, wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells. Total lysates (100 μg of protein each) of cells at day 0 and day 8 after induction were immunoprecipitated with anti-p85PAN (antibody to the p85 subunit of PI 3-kinase), and the immunoprecipitates were subjected to SDS-PAGE, followed by immunoblotting with antiphosphotyrosine antibody (4G10) conjugated to HRP. (B) PI 3-kinase activity associated with tyrosine phosphorylated proteins in wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells. The immunoprecipitates with PY20 from cell lysates (50 μg of protein) were subjected to the PI 3-kinase assay. The autoradiogram of the thin-layer chromatograph is shown. (C) Levels of MAPK protein (upper panel) and phosphorylation of MAPK (lower panel) during adipocyte differentiation. Total lysates (10 μg of protein each) of cells with the four different genotypes were subjected to immunoblotting as described in Materials and Methods using antibody specific to either MAPK or phospho-MAPK.
Since this residual PI 3-kinase activity associated with IRS-2 appeared to partially rescue the ability of IRS1−/− cells to differentiate into adipocytes, we then investigated the ability of IRS-2−/− cells and IRS-1−/− IRS-2−/− cells to differentiate into adipocytes.
PI 3-kinase and MAPK activities during adipocyte differentiation in wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells.
We performed immunoblotting analysis to identify the tyrosine-phosphorylated proteins associated with the p85 subunit of PI 3-kinase during adipocyte differentiation (Fig. 3A). At day 0 after induction, tyrosine-phosphorylated 170-kDa proteins, which were recognized by anti-IRS-1 antibody (data not shown), were weakly detected in wild-type and IRS-2−/− cells (Fig. 3A; lanes 1 and 3). At 8 days after induction, tyrosine phosphorylation of IRS-1 associated with PI 3-kinase in wild-type and IRS-2−/− cells was markedly increased (Fig. 3A, lanes 5 and 7). In IRS-1−/− cells, tyrosine phosphorylation of the 180-kDa protein associated with the p85 subunit of PI 3-kinase was induced (Fig. 3A, lane 6). This protein was shown to be IRS-2 because it was not detected in IRS-2−/− cells and IRS-1−/− IRS-2−/− cells (Fig. 3A, lanes 7 and 8). In IRS-1−/− IRS-2−/− cells, tyrosine-phosphorylated proteins associated with the p85 subunit of PI 3-kinase were not detected in the 170- to 180-kDa range (Fig. 3A, lane 8).
The PI 3-kinase activities which were immunoprecipitated with PY20 during adipocyte differentiation were measured in lysates from each of the four genotypes (Fig. 3B). IRS-1−/− cells had 50% less PI 3-kinase activity than the wild-type cells, whereas the PI 3-kinase activity of IRS-2−/− cells was similar to that of wild-type cells. As we predicted, the increase in PI 3-kinase activity during adipocyte differentiation was largely abolished in IRS-1−/− IRS-2−/− cells. These findings indicated that a major portion of PI 3-kinase activity which increased during adipocyte differentiation was associated with IRS-1 and IRS-2.
A second important pathway activated through IRS-1 and IRS-2 is the MAPK cascade. The amount and phosphorylation of MAPK protein was very similar among the cells of each of the four genotypes and remained constant during adipocyte differentiation (Fig. 3C).
IRS-1−/− IRS-2−/− cells failed to differentiate into adipocytes.
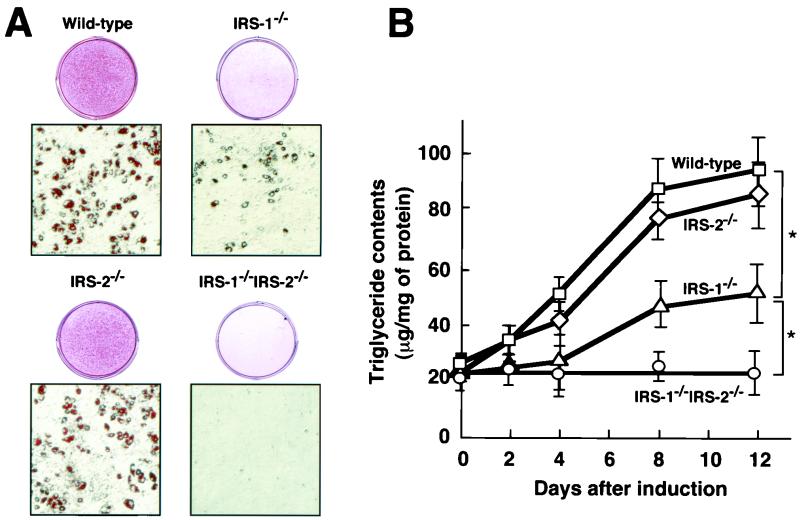
The EF cells with each of the four genotypes were induced to differentiation into adipocytes. Oil-Red O staining was performed 8 days after induction to adipocyte differentiation (Fig. 4A). The ability of IRS-1−/− cells to differentiate into adipocytes was again approximately 60% lower than that of wild-type cells. The ability of IRS-2−/− cells to differentiate into adipocytes was approximately 15% lower than that of wild-type cells (Fig. 4B). Surprisingly, IRS-1−/− IRS-2−/− cells were completely unable to differentiate into adipocytes (Fig. 4).
FIG. 4.
Adipocyte differentiation of EF cells with the four different genotypes, i.e., wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells. (A) Oil-Red O staining for fat accumulation in cells with the four different genotypes. Wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells were grown to confluence, exposed to IBMX, DEX, and insulin to induce differentiation as described in Materials and Methods and then stained with Oil-Red O at day 8 day after induction. (B) Intracellular triglyceride content during adipocyte differentiation of wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells. The data represent the means ± the standard errors of the means from the analysis of wild type (n = 3), IRS-1−/− (n = 4), IRS-2−/− (n = 4), and IRS-1−/− IRS-2−/− (n = 3) cells. ∗, P < 0.05.
Expression of transcriptional factors for adipocyte differentiation and adipogenic markers.
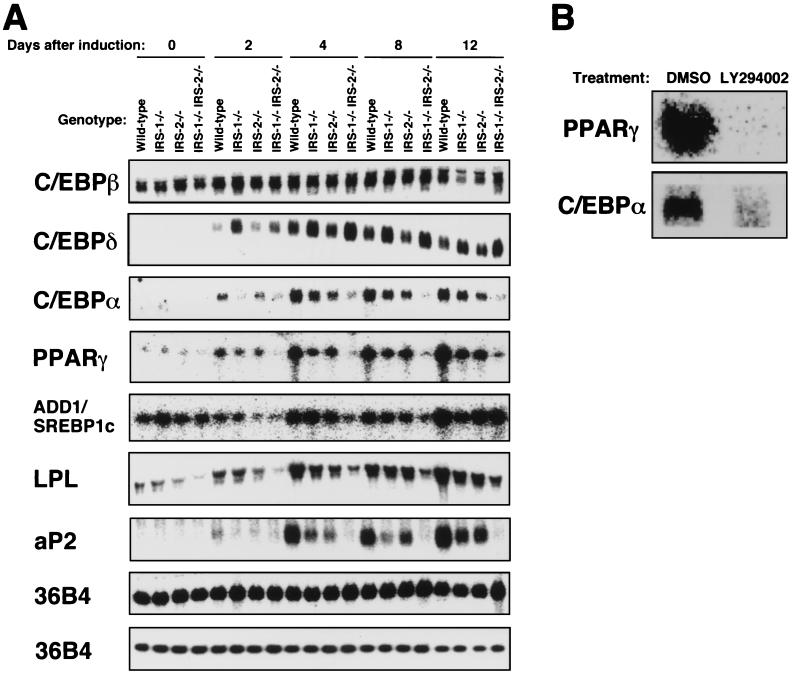
To identify the molecular mechanism for the defective adipocyte differentiation in IRS-1−/− IRS-2−/− cells, we performed Northern blot analysis to investigate gene expression of transcriptional factors for adipocyte differentiation and adipogenic markers (Fig. 5A). The levels of C/EBPβ expression in IRS-1−/− and IRS-1−/− IRS-2−/− cells were comparable to those of wild-type cells. Somewhat surprisingly, C/EBPδ expression in IRS-1−/− and IRS-1−/− IRS-2−/− cells was greater than that in wild-type cells. By contrast, the expression of C/EBPα in IRS-1−/− IRS-2−/− cells was markedly reduced. The expression of PPARγ was also severely decreased in IRS-1−/− IRS-2−/− cells. Although the expression of SREBP1c (ADD1) was significantly reduced 4 and 8 days after induction in IRS-1−/− IRS-2−/− cells, it caught up with that of wild-type cells by 12 days after induction. The expression of LPL and aP2 mRNAs was severely decreased but not abolished in IRS-1−/− IRS-2−/− cells.
FIG. 5.
(A) Northern blotting analysis and RNase protection assay of transcriptional factors for adipocyte differentiation and adipogenic markers in wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells during adipocyte differentiation. Total RNAs were extracted from adipocytes at days 0, 2, 4, 8, and 12 after induction, and a 6-μg sample of the total RNA was subjected to electrophoresis and hybridization with probes consisting of 32P-labeled cRNAs encoding C/EBPβ, C/EBPδ, C/EBPα, and 36B4 (upper panel) and with probes consisting of 32P-labeled cDNAs encoding aP2 and LPL. In an RNase protection assay, 10 μg of the total RNA was hybridized with cRNA encoding PPARγ, SREBP1c, and 36B4 (lower panel). 36B4 encodes acidic ribosomal phosphoprotein P0 and was used as a loading control. (B) Effect of the PI 3-kinase inhibitor LY294002 on expression of the mRNAs of PPARγ and C/EBPα in wild-type cells. LY294002 (20 μM) was added to adipocyte differentiation medium, and the medium was changed at 36-h intervals. As a control, 0.1% dimethyl sulfoxide was added to the medium, and the medium was changed at the same intervals. At day 8 after induction, total RNAs were extracted from wild-type cells, and a 6-μg sample of total RNA was electrophoresed and subjected to Northern blotting with PPARγ and C/EBPα.
To further study the role of PI 3-kinase in adipocyte differentiation in EF cells, the mRNAs of PPARγ and C/EBPα in wild-type cells were examined by Northern blotting. Expressions of both PPARγ and C/EBPα at 8 days after induction were severely decreased in wild-type cells treated with 20 μM LY294002 (Fig. 5B).
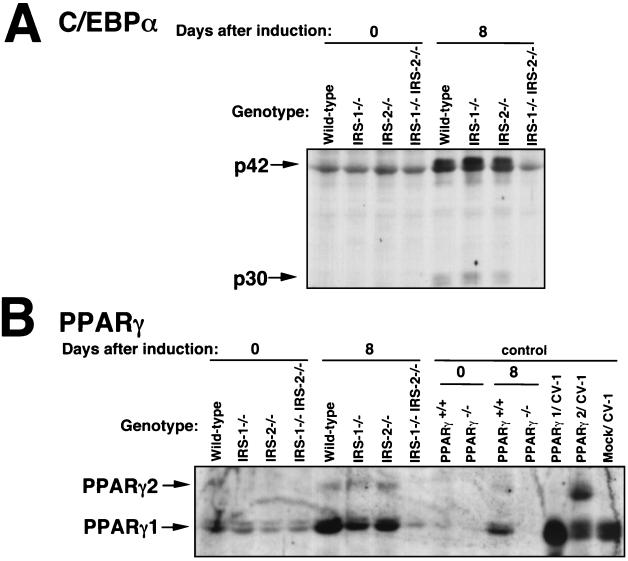
Expression of PPARγ and C/EBPα was markedly reduced on the protein level in IRS-1−/− IRS-2−/− cells.
Immunoblotting analysis was performed to assess the expression of PPARγ and C/EBPα on the protein level. The expression of C/EBPα was markedly reduced on the protein level in IRS-1−/− IRS-2−/− cells at 8 days after induction (Fig. 6A), and the PPARγ protein level in IRS-1−/− IRS-2−/− cells was also markedly reduced (Fig. 6B). These results were consistent with those obtained by Northern blotting analysis of the IRS-1−/− IRS-2−/− cells.
FIG. 6.
Immunoblot analysis of C/EBPα (A) and PPARγ (B) in wild-type, IRS-1−/−, IRS-2−/−, and IRS-1−/− IRS-2−/− cells during adipocyte differentiation. Cells were grown to confluence and then exposed to IBMX, DEX, and insulin to induce differentiation. At days 0 and 8 after induction, the cells were lysed, and the lysates (6 μg of protein each) were subjected to SDS-PAGE, followed by immunoblotting with anti-C/EBPα antibody (A) and anti-PPARγ antibody (B) as described in Materials and Methods. In an immunoblot analysis of PPARγ(B), PPARγ-deficient cells described in a previous report (21) and the CV-1 cells in which PPARγ1 or PPARγ2 was overexpressed were also used as control samples.
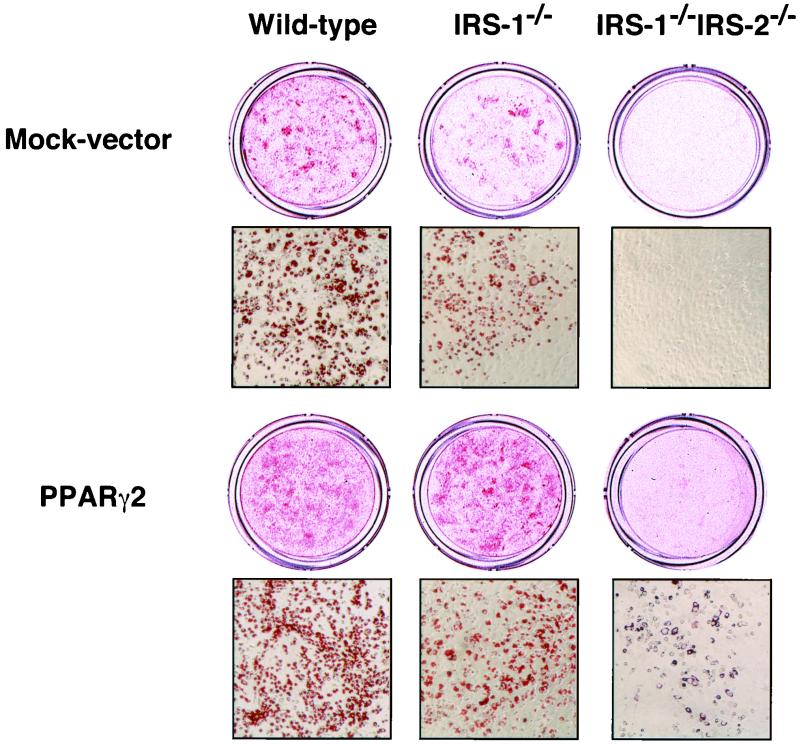
Retroviral expression of PPARγ2 in IRS-1−/− IRS-2−/− cells partially rescued the lack of adipocyte differentiation.
If the lack of PPARγ expression was responsible for the observed lack of lipid accumulation in IRS-1−/− IRS-2−/− cells, it was thought that forced expression of PPARγ might rescue adipocyte differentiation. To determine whether it would, we performed retrovirus-mediated PPARγ gene transfer into IRS-1−/− IRS-2−/− cells. The results of Oil-Red O staining showed that forced expression of PPARγ2 in IRS-1−/− cells completely rescued the impaired adipocyte differentiation. Forced expression of PPARγ2 in IRS-1−/− IRS-2−/− cells, however, rescued it only in part, not to the same level as in mock-vector-transferred wild-type cells (Fig. 7), suggesting that IRS-1 and IRS-2 are required for full adipocyte differentiation in addition to their roles to induce PPARγ2.
FIG. 7.
Retrovirus-mediated reexpression of PPARγ improved the differentiation capacity of IRS-1−/− IRS-2−/− cells. Wild-type, IRS-1−/−, or IRS-1−/− IRS-2−/− cells were transfected with pMX-mPPARγ2-puro and mock vector by retrovirus-mediated gene transfer as described in Materials and Methods. At day 8 of differentiation, cells were stained with Oil-Red O to assess the intracellular fat accumulations.
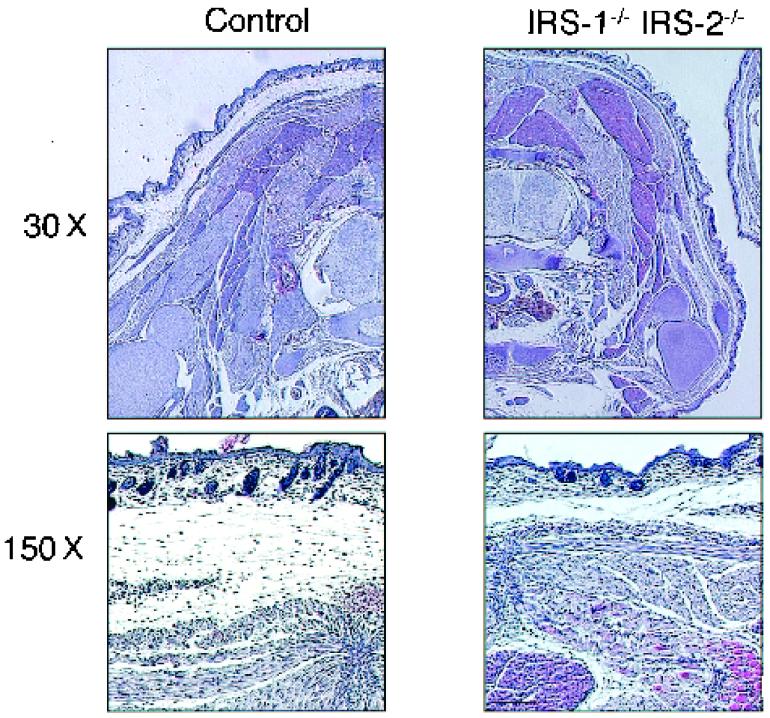
WAT mass of IRS-1−/− IRS-2−/− double-knockout mice was markedly reduced.
To address whether the in vitro effects observed in this study are translated into effects on adiposity in vivo, we tried to produce IRS-1−/− IRS-2−/− double-knockout mice. Interestingly, they were carried to term (R. Suzuki et al., unpublished data). Because white adipose tissue (WAT) appears at birth during mouse development, mice at the 8-h postnatal point were examined histologically (Fig. 8). At 8 h after birth, control animals had abundant subcutaneous WAT. In contrast, WAT mass was dramatically reduced, but not abrogated, in the newborn IRS-1−/− IRS-2−/− double-knockout mice. Surprisingly, brown adipose tissue (BAT) mass in IRS-1−/− IRS-2−/− double-knockout mice was almost the same as that in control mice.
FIG. 8.
Histological analyses of IRS-1−/− IRS-2−/− double-knockout mice showed severe reduction in WAT but not in BAT. Transverse sections at the level of the neck were made from ∼8-h control (left) and IRS-1−/− IRS-2−/− (right) mice. The sections were subjected to hematoxylin and eosin staining. Magnifications: ×30 (top) and ×150 (bottom).
DISCUSSION
Adipocyte differentiation is an important aspect of energy balance. Although cascades and networks of transcriptional factors during adipocyte differentiation have been investigated rather thoroughly, the signaling mechanism by extracellular hormones and growth factors regulating the cascades and networks of adipose-specific transcriptional factors remains unclear (30). Insulin and IGF-1 have been reported to play important roles in adipocyte differentiation (5, 13, 14). In this study, we investigated the role of IRS-1 and IRS-2, the two major common substrates for insulin and IGF-1 receptor tyrosine kinases, in adipocyte differentiation. Using a gene ablation strategy, we found that the abilities of IRS-1−/− cells and IRS-2−/− cells to differentiate into adipocytes were reduced by 60 and 15%, respectively, compared to that of wild-type cells. These results indicate that even though both IRS-1 and IRS-2 are required for full capacity of adipocyte differentiation, IRS-1 plays a more role in adipocyte differentiation. Moreover, it is possible that IRS-2 and IRS-1 may have rescued the phenotype of IRS-1−/− and IRS-2−/− cells, respectively, in adipocyte differentiation. Therefore, to fully understand the role of IRS-1 and IRS-2 in adipocyte differentiation, we established IRS-1 IRS-2 double-deficient EF cells. Our results clearly show that IRS-1−/− IRS-2−/− cells have no ability to differentiate into adipocytes; providing the first direct evidence that IRS-1 and IRS-2 are absolutely essential to adipocyte differentiation.
Since IRS-1 and IRS-2 are important mediators of insulin and IGF-1 actions, these data may imply that the absence of IRS-1 and IRS-2 impairs hormonal activation of adipogenesis. It should be noted, however, that this study does not address which tyrosine kinase(s) phosphorylate IRS-1 and IRS-2 during adipocyte differentiation. Therefore, it is possible that tyrosine phosphorylation of IRS-1 and IRS-2 by putative upstream tyrosine kinase(s) other than insulin and IGF-1 receptor tyrosine kinases plays a crucial role in in vitro adipocyte differentiation or in vivo adipogenesis. Data from Chaika et al. (6) suggest that the insulin receptor tyrosine kinase was still capable of stimulating adipocyte differentiation even when mutated such that it could no longer phosphorylate IRS proteins.
The present study strongly suggests the important role of tyrosine-phosphorylated IRS-1 and IRS-2 and PI 3-kinase activation by three lines of experimental results: (i) adipocyte differentiation capacity parallels PI 3-kinase activity, which increases during adipocyte differentiation, among the four genotypes; (ii) PI 3-kinase inhibitor completely blocks adipocyte differentiation; and (iii) phosphorylation of MAP kinase is unaltered in any of the four genotypes. Consistent with our results, several studies (31, 40, 47, 54) have reported that PI 3-kinase activity plays a role in adipocyte differentiation by using dominant-negative p85, wortmannin, or LY294002, inhibitors of PI 3-kinase. Moreover, Kohn et al. and Magun et al. have demonstrated the role of activation of PI 3-kinase and one of its downstream mediators, Akt, to induce adipocyte differentiation by constitutive active form of Akt Ser/Thr kinase (20, 24). Taken together with these reports, the present study strongly suggests that PI 3-kinase activity activated through induction of tyrosine phosphorylation of IRS-1 and IRS-2 is essential to adipocyte differentiation.
To further clarify the mechanism by which IRS-1 and IRS-2 fulfill their role in adipocyte differentiation, we investigated the expression of transcriptional factors for adipocyte differentiation and adipogenic markers. The results show that the mRNA expression of C/EBPα and PPARγ is severely decreased in IRS-1−/− IRS-2−/− cells, although the expression of C/EBPβ is unaltered and the expression of C/EBPδ is even slightly increased. It therefore seems possible that IRS-1 and IRS-2 upregulate the expression of C/EBPα and PPARγ, but the molecular link between IRS-1, IRS-2, and PI 3-kinase and the upregulation of these transcription factors is unclear and requires further investigation. The expression of LPL and aP2, whose promoters have a PPAR response element (PPRE), is parallel to the expression of PPARγ. Although the expression of SREBP1c is at least in part dependent on the expression of PPARγ, as previously reported (21), SREBP1c has been reported to be also regulated by many other factors (11). At day 12 after induction the expression of SREBP1c in IRS-1−/− IRS-2−/− cells is induced to almost the same amount as that in wild-type cells. It indicates that upstream signaling pathway(s) other than IRS-1 and IRS-2 may be involved in the regulation of the expression of SREBP1c. Nevertheless, normal expression of SREBP1c at day 12 is not sufficient to induce adipocyte differentiation in IRS-1−/− IRS-2−/− cells.
To further study the role of PI 3-kinase in adipocyte differentiation, we examined whether inhibition of PI 3-kinase during adipocyte differentiation affects the mRNA expressions of PPARγ and C/EBPα in wild-type cells. As shown in Fig. 5B, mRNAs of both PPARγ and C/EBPα decrease significantly in wild-type cells treated with 20 μM LY294002. It strongly suggests that the inhibition of PI 3-kinase blocks adipogenesis due to severe reduction in expression of both PPARγ and C/EBPα mRNAs. Our present data appear to differ from those of a previous report (31) that the inhibition of PI 3-kinase in 3T3-F442A cells by expressing dominant-negative p85 does not affect the expression of the transcription factor PPARγ at the mRNA level using insulin and 5,8,11,14-eicosatetraynoic acid (ETYA) as inducers for adipocyte differentiation (31). The reasons for the apparent discrepancy between our data and those of Sakaue et al. (31) are unknown at present but could be related to differences in methods such as cell type (primary EF cells versus 3T3-F442A) or inducers for differentiation (IBMX, DEX, and insulin versus ETYA and insulin) or means to inhibit PI 3-kinase (LY294002 versus dominant-negative p85). On the other hand, our present result that PI 3-kinase inhibition using LY294002 decreases PPARγ mRNA is consistent with the study by Xia and Serrero (54), in which they demonstrated treatment of 3T3-L1 adipocytes with LY294002 inhibited PPARγ expression and adipocyte differentiation induced by IBMX, DEX, and insulin. Identification of signals downstream of IRS-1 and IRS-2 mediating the upregulation of PPARγ and C/EBPα mRNA expression is an important subject of future research.
Restoration of PPARγ to IRS-1−/− IRS-2−/− cells by retrovirus-mediated gene transfer partially rescues the defective adipocyte differentiation. This is consistent with the hypothesis that one of the roles of IRS-1 and IRS-2 in adipocyte differentiation is upregulation of mRNA expression of PPARγ. On the other hand, the fact that PPARγ is unable to completely rescue the defective adipocyte differentiation suggests that IRS-1 and IRS-2 may stimulate adipocyte differentiation via distinct mechanism(s) in addition to induction of PPARγ.
Histological analyses of IRS-1−/− IRS-2−/− double-knockout mice show severe reduction in WAT but not in BAT. These data may provide the evidence that the essential role of IRS-1 and IRS-2 in adipocyte differentiation in vitro can be extended to that in development of WAT in vivo. It should be noted, however, that WAT mass is not completely abrogated in IRS-1−/− IRS-2−/− double-knockout mice. These differences in adipogenesis between in vitro and in vivo have been previously observed in the report of Tanaka et al. on C/EBPβ and C/EBPδ knockout mice (39). In in vitro differentiation experiments, the only differentiation stimuli are MIX, DEX, INS and FBS; however, in vivo, there are many other adipogenic factors such as growth hormone, IGF-1, and prostaglandins. With the restricted number of stimuli in vitro, IRS-1 and IRS-2 is absolutely required for the expression of PPARγ and C/EBPα and for adipocyte differentiation. However, the lack of adipogenesis may be partially rescued with additional stimuli in vivo which are not absolutely dependent upon the presence of IRS-1 and IRS-2.
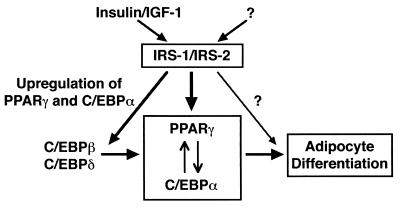
In conclusion, this study is the first to report that IRS-1 and IRS-2 play an essential role in adipocyte differentiation and adipogenesis. Moreover, we propose that IRS-1 and IRS-2 play crucial roles in adipocyte differentiation through the upregulation of mRNA expression of PPARγ and C/EBPα subsequent to induction of C/EBPβ and C/EBPδ (Fig. 9).
FIG. 9.
Schematic model of the roles of IRS-1 and IRS-2 in adipogenesis. IRS-1 and IRS-2 play a crucial role in the upregulation of the expression of C/EBPα and PPARγ and adipocyte differentiation.
ACKNOWLEDGMENTS
We thank B. M. Spiegelman and N. Yahagi for providing PPARγ2 expression vector and 36B4 cDNA probe, respectively, and E. Yoshida-Nagata and H. Chiyonobu for excellent technical assistance.
This work was supported by a grant-in-aid for creative basic research (10NP0201) from the Ministry of Education, Science, Sports, and Culture of Japan (to T. Kadowaki) and by health science research grants (Research on Human Genome and Gene Therapy) from the Ministry of Health and Welfare (to T. Kadowaki).
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altiok S, Xu M, Spiegelman B M. PPARγ induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruning J C, Winnay J, Cheatham B, Kahn C R. Differential signaling by insulin receptor substrate 1 (IRS-1) and IRS-2 in IRS-1-deficient cells. Mol Cell Biol. 1997;17:1513–1521. doi: 10.1128/mcb.17.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3–L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 5.Catalioto R M, Gailard D, Ailhaud G, Negrel R. Terminal differentiation of mouse preadipocyte cells: the mitogenic-adipogenic role of growth hormone is mediated by the protein kinase C signalling pathway. Growth Factors. 1992;6:255–264. doi: 10.3109/08977199209026932. [DOI] [PubMed] [Google Scholar]
- 6.Chaika O V, Chaika N, Volle D J, Wilden P A, Pirrucello S J, Lewis R E. CSF-1 receptor/insulin receptor chimera permits CSF-1-dependent differentiation of 3T3–L1 preadipocytes. J Biol Chem. 1997;272:11968–11974. doi: 10.1074/jbc.272.18.11968. [DOI] [PubMed] [Google Scholar]
- 7.Chang C J, Chen T T, Lei H Y, Chen D S, Lee S C. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol Cell Biol. 1990;10:6642–6653. doi: 10.1128/mcb.10.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christy R J, Kaestner K H, Geiman D E, Lane M D. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3–L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3–L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 10.Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990;4:1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- 11.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim J B, Ferre P, Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freytag S O, Paielli D L, Gilbert J D. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 13.Green H, Morikawa M, Nixon T. A dual effector theory of growth-hormone action. Differentiation. 1985;29:195–198. doi: 10.1111/j.1432-0436.1985.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 14.Guller S, Corin R E, Yuan-Wu K, Sonenberg M. Up-regulation of vinculin expression in 3T3 preadipose cells by growth hormone. Endocrinology. 1991;129:527–533. doi: 10.1210/endo-129-1-527. [DOI] [PubMed] [Google Scholar]
- 15.Hu E, Tontonoz P, Spiegelman B M. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc Natl Acad Sci USA. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama R, Sasai Y, Nakanishi S. Molecular characterization of transcription factors that bind to the cAMP responsive region of the substance P precursor gene. cDNA cloning of a novel C/EBP-related factor. J Biol Chem. 1991;266:15525–15531. [PubMed] [Google Scholar]
- 17.Kim J B, Spiegelman B M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 18.Kim J B, Wright H M, Wright M, Spiegelman B M. ADD1/SREBP1 activates PPARγ through the production of endogenous ligand. Proc Natl Acad Sci USA. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohn A D, Summers S A, Birnbaum M J, Roth R A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3–L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 21.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Nagai R, Tobe K, Kimura S, Kadowaki T. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 22.Kubota N, Tobe K, Terauchi Y, Eto K, Yamauchi T, Suzuki R, Tsubamoto Y, Komeda K, Nakano R, Miki H, Satoh S, Sekihara H, Sciacchitano S, Lesniak M, Akanuma Y, Aizawa S, Nagai R, Kimura S, Taylor S, Kadowaki T. Disruption of insulin receptor substrate-2 causes type 2 diabetes due to liver insulin resistance and lack of compensatory β-cell hyperplasia. Diabetes. 2000;49:1800–1889. doi: 10.2337/diabetes.49.11.1880. [DOI] [PubMed] [Google Scholar]
- 23.Lin F T, Lane M D. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3–L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magun R, Burgering B M, Coffer P J, Pardasani D, Lin Y, Chabot J, Sorisky A. Expression of a constitutively activated form of protein kinase B (c-Akt) in 3T3–L1 preadipose cells causes spontaneous differentiation. Endocrinology. 1996;137:3590–3593. doi: 10.1210/endo.137.8.8754791. [DOI] [PubMed] [Google Scholar]
- 25.Mandrup S, Lane M D. Regulating adipogenesis. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 26.Myers M G, Jr, Sun X J, Cheatham B, Jachna B R, Glasheen E M, Backer J M, White M F. IRS-1 is a common element in insulin and insulin-like growth factor-I signaling to the phosphatidylinositol 3′-kinase. Endocrinology. 1993;132:1421–1430. doi: 10.1210/endo.132.4.8384986. [DOI] [PubMed] [Google Scholar]
- 27.Onishi M, Nosaka T, Misawa K, Mui A L, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridderstrale M, Degerman E, Tornqvist H. Growth hormone stimulates the tyrosine phosphorylation of the insulin receptor substrate-1 and its association with phosphatidylinositol 3-kinase in primary adipocytes. J Biol Chem. 1995;270:3471–3474. doi: 10.1074/jbc.270.8.3471. [DOI] [PubMed] [Google Scholar]
- 29.Rosen E D, Sarraf P, Troy A E, Bradwin G, Moore K, Milstone D S, Spiegelman B M, Mortensen R M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 30.Rosen E D, Walkey C J, Puigserver P, Spiegelman B M. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 31.Sakaue H, Ogawa W, Matsumoto M, Kuroda S, Takata M, Sugimoto T, Spiegelman B M, Kasuga M. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J Biol Chem. 1998;273:28945–28952. doi: 10.1074/jbc.273.44.28945. [DOI] [PubMed] [Google Scholar]
- 32.Samuelsson L, Stromberg K, Vikman K, Bjursell G, Enerback S. The CCAAT/enhancer binding protein and its role in adipocyte differentiation: evidence for direct involvement in terminal adipocyte development. EMBO J. 1991;10:3787–3793. doi: 10.1002/j.1460-2075.1991.tb04948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoonjans K, Peinado-Onsurbe J, Lefebvre A M, Heyman R A, Briggs M, Deeb S, Staels B, Auwerx J. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 34.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty A H, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, Gotoda T, Ishibashi S, Yamada N. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 35.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Investig. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiegelman B M, Hu E, Kim J B, Brun R. PPAR gamma and the control of adipogenesis. Biochimie. 1997;79:111–112. doi: 10.1016/s0300-9084(97)81500-3. [DOI] [PubMed] [Google Scholar]
- 37.Sun X J, Wang L M, Zhang Y, Yenush L, Myers M G, Jr, Glasheen E, Lane W S, Pierce J H, White M F. Role of IRS-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 38.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomiyama K, Nakata H, Sasa H, Arimura S, Nishio E, Watanabe Y. Wortmannin, a specific phosphatidylinositol 3-kinase inhibitor, inhibits adipocytic differentiation of 3T3–L1 cells. Biochem Biophys Res Commun. 1995;212:263–269. doi: 10.1006/bbrc.1995.1965. [DOI] [PubMed] [Google Scholar]
- 41.Tontonoz P, Hu E, Devine J, Beale E G, Spiegelman B M. PPAR gamma 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1995;15:351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. mPPARγ 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 43.Tontonoz P, Hu E, Spiegelman B M. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr Opin Genet Dev. 1995;5:571–576. doi: 10.1016/0959-437x(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 44.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPARγ 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 45.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umek R M, Friedman A D, McKnight S L. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 47.Usui I, Haruta T, Iwata M, Takano A, Uno T, Kawahara J, Ueno E, Sasaoka T, Kobayashi M. Retinoblastoma protein phosphorylation via PI 3-kinase and mTOR pathway regulates adipocyte differentiation. Biochem Biophys Res Commun. 2000;275:115–120. doi: 10.1006/bbrc.2000.3201. [DOI] [PubMed] [Google Scholar]
- 48.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Impaired energy homeostasis in C/EBPα knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 49.Williams S C, Cantwell C A, Johnson P F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z, Bucher N L, Farmer S R. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol Cell Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z, Rosen E D, Brun R, Hauser S, Adelmant G, Troy A E, McKeon C, Darlington G J, Spiegelman B M. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 52.Wu Z, Xie Y, Bucher N L, Farmer S R. Conditional ectopic expression of C/EBPβ in NIH-3T3 cells induces PPARγ and stimulates adipogenesis. Genes Dev. 1995;9:2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Xie Y, Morrison R F, Bucher N L, Farmer S R. PPARγ induces the insulin-dependent glucose transporter GLUT4 in the absence of C/EBPα during the conversion of 3T3 fibroblasts into adipocytes. J Clin Investig. 1998;101:22–32. doi: 10.1172/JCI1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia X, Serrero G. Inhibition of adipose differentiation by phosphatidylinositol 3-kinase inhibitors. J Cell Physiol. 1999;178:9–16. doi: 10.1002/(SICI)1097-4652(199901)178:1<9::AID-JCP2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 55.Yamauchi T, Tobe K, Tamemoto H, Ueki K, Kaburagi Y, Yamamoto-Honda R, Takahashi Y, Yoshizawa F, Aizawa S, Akanuma Y, Sonenberg N, Yazaki Y, Kadowaki T. Insulin signalling and insulin actions in the muscles and livers of insulin-resistant, insulin receptor substrate 1-deficient mice. Mol Cell Biol. 1996;16:3074–3084. doi: 10.1128/mcb.16.6.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh W C, Cao Z, Classon M, McKnight S L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]