Abstract
Vaginal microbiota-host interactions are linked to preterm birth (PTB), which continues to be the primary cause of global childhood mortality. Due to population size, the majority of PTB occurs in Asia, yet there have been few studies of the pregnancy vaginal microbiota in Asian populations. Here, we characterized the vaginal microbiome of 2689 pregnant Chinese women using metataxonomics and in a subset (n = 819), the relationship between vaginal microbiota composition, sialidase activity and leukocyte presence and pregnancy outcomes. Vaginal microbiota were most frequently dominated by Lactobacillus crispatus or L. iners, with the latter associated with vaginal leukocyte presence. Women with high sialidase activity were enriched for bacterial vaginosis-associated genera including Gardnerella, Atopobium and Prevotella. Vaginal microbiota composition, high sialidase activity and/or leukocyte presence was not associated with PTB risk suggesting underlying differences in the vaginal microbiota and/or host immune responses of Chinese women, possibly accounting for low PTB rates in this population.
Subject terms: Microbial communities, Bacteria, Anatomy
Introduction
There is substantial evidence implicating the pregnancy vaginal microbiota in shaping maternal and neonatal health outcomes1–3. The dominance of the vaginal niche by commensal Lactobacillus species is often considered “optimal” due to their ability to prevent pathogen colonization through competitive exclusion, in part achieved through the production of antimicrobial compounds and production of lactic acid4,5. Recent studies have highlighted L. crispatus dominance as being protective against preterm birth (PTB)2,6–9 and neonatal sepsis following preterm prelabour rupture of membranes1. By contrast, colonization by L.iners7,10–12 or Lactobacillus species depleted, high diversity compositions are associated with an increased risk of PTB2,7,9,10,13–15. Despite the majority of PTBs (60%) occurring in Asia16, molecular-based characterization of the vaginal microbiota in pregnancy and its relationship with PTB has largely been restricted to Northern American and European populations. Moreover, ethnicity is now recognized as a potential confounder of the relationship between the vaginal microbiome and PTB, particularly between Caucasian and women of African-descent in North American or European populations2,9,10,17.
The Amsel criteria18 and Nugent scoring system19 are commonly used to diagnose Bacterial Vaginosis (BV), a condition characterized by a loss of vaginal lactobacilli and overgrowth of anaerobes, which is associated with a two-fold increased risk of PTB20–23. The Amsel criteria requires microscopy and is subject to potential interobserver bias24, whereas Nugent scoring is limited by the requirement for laboratory access. Enzymatic-based assays for rapid BV diagnosis may offer an objective, point-of-care alternative to BV diagnosis in clinical settings, including during pregnancy25,26. These assays often work by measuring microbial sialidase (neuraminidase), produced by BV-associated bacteria such as Gardnerella vaginalis, which removes sialic acid from sialoglycoconjugates including those on the surface of vaginal epithelial cells, providing a nutrient source and exposing glycan-binding sites for bacterial adhesion. Additionally, sialidase is thought to mediate biofilm formation and the establishment of sub-optimal vaginal microbiota compositions27–29. High vaginal sialidase levels have previously been associated with an increased risk of PTB30 and with failure of cervical cerclage31, a procedure used to reinforce the cervical opening in women at risk of preterm delivery due to cervical shortening. Sialidase-producing taxa associated with BV have also been implicated in chorioamnionitis, a risk factor for PTB that is characterized by inflammation of the fetal membranes32. Although BV is not classified as an inflammatory syndrome, the disease has been associated with the presence of vaginal leukocytes, which have been purported to offer predictive value in identifying upper reproductive tract infections33–35 and PTB36–38. Quantification of leukocyte counts using vaginal wet mount microscopy could therefore represent an easily accessible and cost-effective method to determine cervicovaginal inflammation39.
In this study, we characterized the bacterial component of the vaginal microbiome in 2689 Chinese women sampled at mid-pregnancy and in a subset (n = 819), explored the relationship between vaginal microbiota composition, sialidase activity, and leukocyte presence with risk of PTB.
Results
Study population
A total of 2796 women, with a median (IQR) maternal age of 29 years (26–32 years), met the study inclusion criteria and were recruited between November 2015 to December 2018. The median (IQR) time of sampling was 16+4 weeks+days gestation (range 15+6–17+6) after exclusion of women with samples collected in the third trimester. Of these, 1397 delivered at the same hospital and maternal and neonatal outcomes data were obtained. The median (IQR) gestation at delivery was 39+3 (38+5–40+1) weeks+days. The PTB (<37 weeks) rate in the cohort was 5.44% (76/1397) (Supplementary Data 1). The remaining 1356 women delivered elsewhere and due to data protection, pregnancy outcome data were not available. There was no significant difference in the gestation of sampling between women with and without available outcome data (Mann-Whitney Test, p > 0.05; Supplementary Fig. 1).
The pregnancy vaginal microbiota of Chinese women
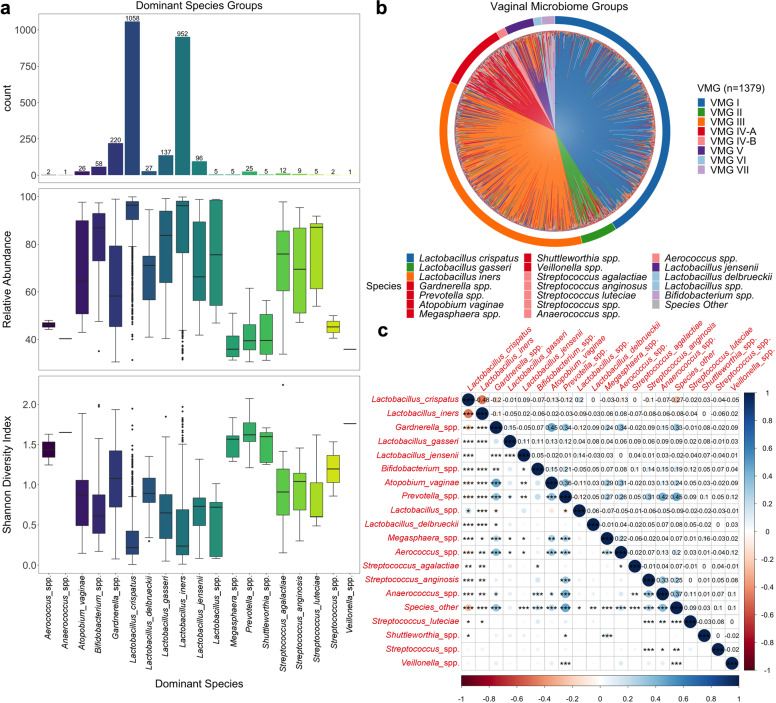
The 2796 sequenced vaginal samples generated a total of 58,582,840 reads with a mean read count of 20,952 per sample. Of these samples, 2689 passed library size and microbiome classification criteria (Supplementary Fig. 1). Only those samples collected in the first and second trimester were included in subsequent analyses (2646/2689, 98.4%) After removal of kit and reagent contaminants, a total of 82 taxa were detected and included in subsequent analyses. Vaginal microbiota profiles were classified into 19 groups based upon the dominant (>30% relative abundance) taxa observed within each sample2. At the genus level, the majority of samples were dominated by Lactobacillus (2275/2646, 85.98%), which were predominately L. crispatus (1058/2646, 39.98%) or L. iners (952/2646, 35.98%) dominated (Fig. 1a). The median relative abundance of L. crispatus and L. iners in these samples was 96.37% and 96.19% respectively and the alpha diversity as measured by the Shannon Index was similar between the two groups (median, 0.22 and 0.24) (Supplementary Table 1). Conversely, microbiota dominated by BV-associated species including Gardnerella spp. (220/2646, 8.31%), Prevotella spp. (25/2646, 0.94%) and Atopobium spp. (26/2646, 0.98%) had lower median relative abundances (<70%) and higher alpha diversity (median Shannon Index >0.85) compared to samples classified as L. crispatus or L. iners dominated (median Shannon Index <0.25, Kruskall-Wallis Test, p < 0.001) (Fig. 1a and Supplementary Table 1). The same relationships between alpha diversity and dominant vaginal taxa profiles were observed when read count data was normalized using trimmed mean of M values normalization (Supplementary Fig. 2). When these dominant taxa profiles were grouped into Vaginal Microbiome Groups (VMG) consistent with previously reported “community state types”40 or “vagitypes”2, the most prevalent was VMG I (L. crispatus dominated, 1058/2646, 39.98%), followed by VMG III (L. iners dominated, 952/2646, 35.98%), VMG IV-A (BV-associated taxa, 282/2646, 10.66%), VMG II (L. gasseri dominated, 137/2646, 5.18%), VMG V (L. jensenii dominated, 96/2646, 3.63%) and IV-B (Non-BV taxa, 31/2646, 1.17%). Groups dominated by other Lactobacillus species or Bifidobacterium species were classified into VMG VI (32/2646, 1.21%) and VII (58/2646, 2.19%) (Fig. 1b and Supplementary Table 2), respectively. The proportions of VMGs in women with and without available pregnancy outcome data (Supplementary Fig. 3) were similar (Fisher’s Exact Test, p = 0.24). Additional analyses also showed that vaginal microbiota compositional characteristics and their relationship with clinical outcomes were not impacted by the type of extraction kits used (see Supplementary Fig. 4).
Fig. 1. Vaginal microbiome composition and structure.
a Bar chart displaying number of samples within each dominant species group. Box plots show relative abundance of species and Shannon diversity index of each dominant species group with bounds of the box representing the first and third quartiles, the center line representing the median and whiskers as min-to-max values. b Microbiome profiles of women with outcome data classified by vaginal microbiome groups (VMG). Color of species denotes the VMG group classification with VMG I (L. crispatus dominated, blue), II (L. gasseri dominated, green), III (L. iners dominated, orange), IV-A (bacterial vaginosis associated, red), IV-B (non-BV taxa, pink), V (L. jensenii dominated, purple), VI (Other Lactobacillus, light blue), and VII (Bifidobacterium dominated, light purple). c Correlation plot for dominant species showing correlation strength (circles), relationship (positive, blue; negative, red) and significance (***, p < 0.001; **, p < 0.01; *, p < 0.05).
Relative abundance of L. crispatus was negatively correlated with most vaginal taxa including L. iners (r = −0.48, p < 0.001). L. iners was also negatively correlated to most other vaginal taxa except Megasphaera spp. (r = 0.06, p < 0.05) and Aerococcus spp. (r = 0.08, p < 0.01), where weak positive correlations were observed. Several BV-associated taxa were positively correlated with each other including Gardnerella spp., with Atopobium vaginae (r = 0.45, p < 0.001), Prevotella spp. (r = 0.34, p < 0.001), Aerococcus spp. (r = 0.34, p < 0.001) and Other Species (r = 0.33, p < 0.001); Atopobium vaginae with Prevotella spp. (r = 0.36, p < 0.001), Megasphaera spp. (r = 0.29, p < 0.01), and Aerococcus spp. (r = 0.31, p < 0.001); and Prevotella spp. with the non-BV taxa Streptococcus anginosus (r = 0.31, p < 0.001) and Anaerococcus spp. (r = 0.42, p < 0.001), as well as Other Species (r = 0.45, p < 0.001) (Fig. 1c).
Sialidase activity and vaginal microbiota structure in pregnancy
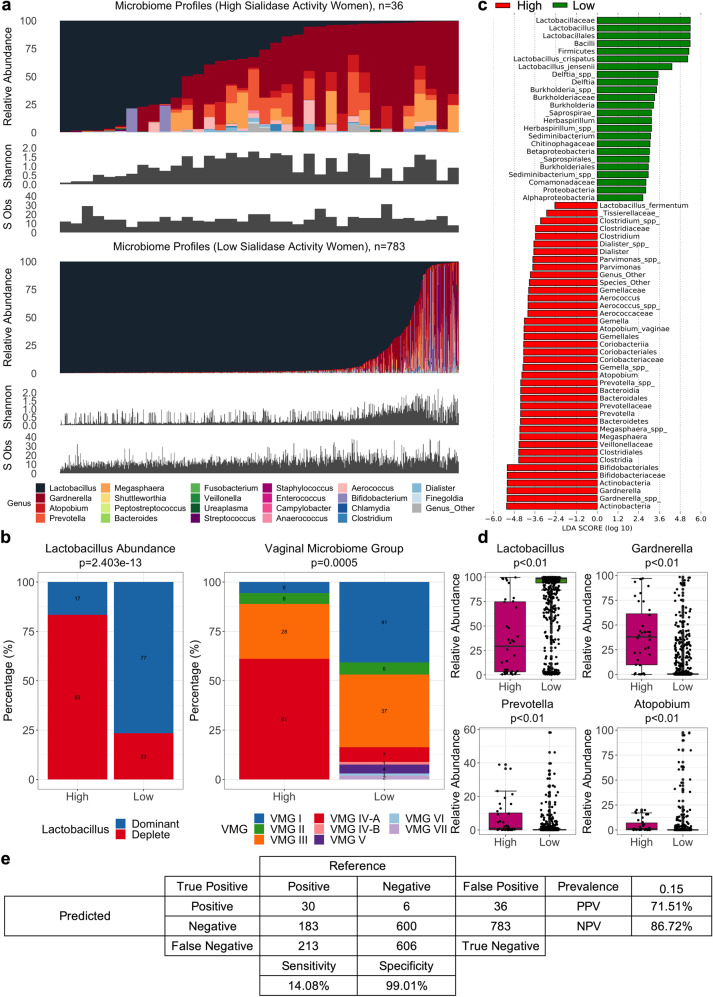
High vaginal sialidase activity was strongly associated with increased bacterial alpha diversity (n = 36; median (IQR) Shannon index 0.93 (0.54–1.55)) compared to women with low vaginal sialidase activity (n = 783; median (IQR) Shannon index 0.32 (0.16–0.77), Mann-Whitney Test, p < 0.001) (Fig. 2a). Women with high sialidase activity had significantly higher prevalence of Lactobacillus species depleted microbiota (30/36, 83.33% versus 183/783, 23.37%; Fisher’s Exact Test, p = 2.403e-13) and VMG IV-A (22/36, 61.11% vs. 58/783, 7.40%; Fisher’s Exact Test, p = 0.0005) (Fig. 2b, Supplementary Fig. 5). VMG I (2/36, 5.56% versus 319/783, 40.74%) and II (2/36, 5.56% versus 48/783, 6.13%) were rarely observed in women with high sialidase activity whereas prevalence of VMG III was similar in women with high and low sialidase activity (10/36, 27.78% vs. 288/783, 36.78%). Samples classified as VMG IV-B (11/783, 1.40%), V (35/783, 4.47%), VI (9/783, 1.15%), and VII (15/783, 1.92%) were only associated with low sialidase activity. Differential abundance analysis using LDA and LefSe of samples with high and low sialidase activity confirmed L. crispatus and L. jensenii as being enriched in low sialidase activity samples whereas high activity was characterized by enrichment with BV-associated genera including Gardnerella, Atopobium, Prevotella, and Megasphaera (Fig. 2c). In these women, the higher total relative abundance of BV-type taxa (Gardnerella spp., Atopobium vaginae, Prevotella spp. and Megasphaera spp.) was observed compared to women with low sialidase activity (Fig. 2d and Supplementary Table 3). Using metataxonomics-defined Lactobacillus depleted (positive) and Lactobacillus dominated (negative) as the reference test, sialidase activity in this cohort was found to have low sensitivity (14.08%), high specificity (99.01%), moderate positive predictive value (PPV, 71.51%), and high negative predictive value (NPV, 86.72%) for Lactobacillus spp. depleted high diversity vaginal microbiota compositions (Fig. 2e).
Fig. 2. Comparison between women in sialidase sub-cohort.
a Microbiome composition, diversity (Shannon Index) and richness (species observed) for all women with high (n = 36) and low (n = 783) sialidase activity. b Proportion of Lactobacillus abundance groups (Lactobacillus dominant and Lactobacillus deplete) and vaginal microbiome groups (VMG) for women with high or low sialidase activity. Statistical significance based on Fisher’s Exact Test. c LDA showing effect size of differentially abundant taxa associated with microbiota of women with high (red) or low (green) sialidase activity. d Comparison of relative abundance between main BV-associated bacteria identified as significantly different (Mann-Whitney Test, p < 0.05) between women with high and low sialidase activity (see Supplementary Table 3). The bounds of the box represent the first and third quartiles, center line represents the median, and whiskers show min-to-max values. e Confusion matrix with Lactobacillus abundance (dominant as positive/deplete as negative) as the reference test and sialidase result (High/Low) as the predicted test. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated.
Leukocyte presence and vaginal microbiota composition during pregnancy
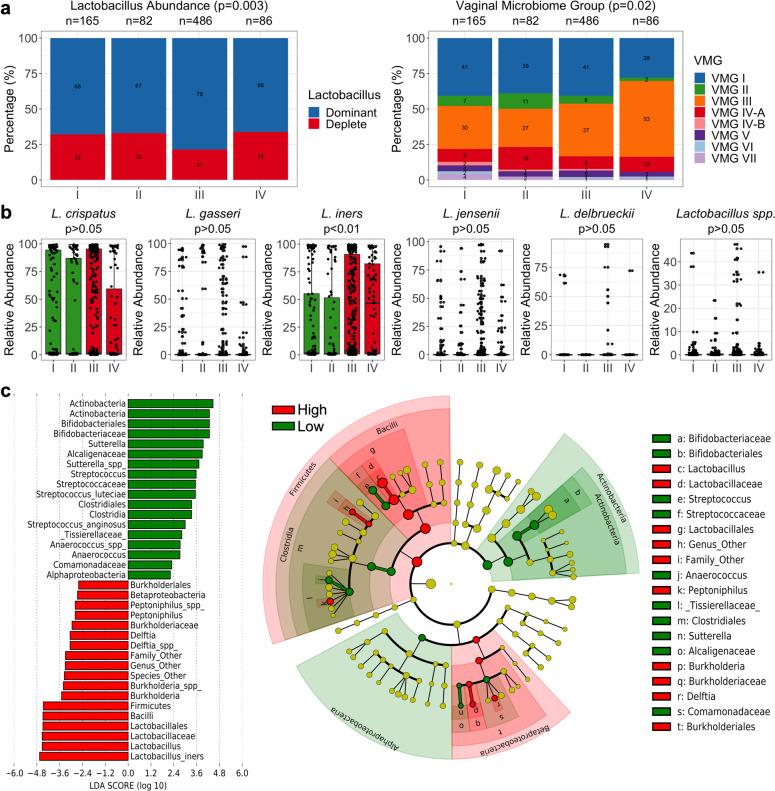
Women with leukocyte grade III wet mounts were associated with a small but significantly higher proportion of Lactobacillus dominated microbiomes (382/486, 78.60%) compared to women with leukocyte grades I (112/165, 67.88%), II (55/82, 67.07%), and IV (57/86, 66.28%; Fisher’s Exact Test, p = 0.003). There were also higher proportions of VMG III in women with leukocyte grades III (180/486, 37.03%) and IV (46/86, 53.49%) compared to women with grades I (50/165, 30.30%) and II (22/82, 26.83%; Fisher’s Exact Test, p = 0.02). Consistent with this, relative abundance levels of L. iners were highest in women with leukocyte grades III and IV (Kruskall-Wallis Test, p < 0.01) (Fig. 3b). Similar results were observed when using LDA and LefSe analysis to identify differential taxa between leukocyte low (grades I-II) and leukocyte high (grades III-IV) wet mounts with L. iners having the strongest effect size of the differentially abundant features identified (Fig. 3c).
Fig. 3. Vaginal microbiome composition and leukocyte presence.
a Stacked bar showing Lactobacillus abundance groups and vaginal microbiome groups (VMG) of women with leukocyte grades of I (0-5 counts/Hp), II (5-15 counts/Hp), III (15-30 counts/Hp) and IV (>30 counts/Hp). b Boxplots showing relative abundance of L. crispatus, L. gasseri, L. iners, L. jenseni, L. delbruecki, and Lactobacillus spp. in women with leukocyte low (grades I and II, green) and high (grades III and IV, red). The bounds of the box represent the first and third quartiles, center line represents the median, and whiskers show min-to-max values. c LDA of differentially abundant taxa and cladogram displaying bacterial clades and nodes identified as differentialy abundance from LDA analysis in vaginal microbiota of women with leukocyte low (grades I-II, green) and high (grades III-IV, red). Statistical significance of categorical variables based on Fisher’s Exact Test and continuous variables based on Kruskal-Wallis Test.
Vaginal microbiota, sialidase activity, and leukocyte presence are not associated with risk of PTB and chorioamnionitis in pregnant Chinese women
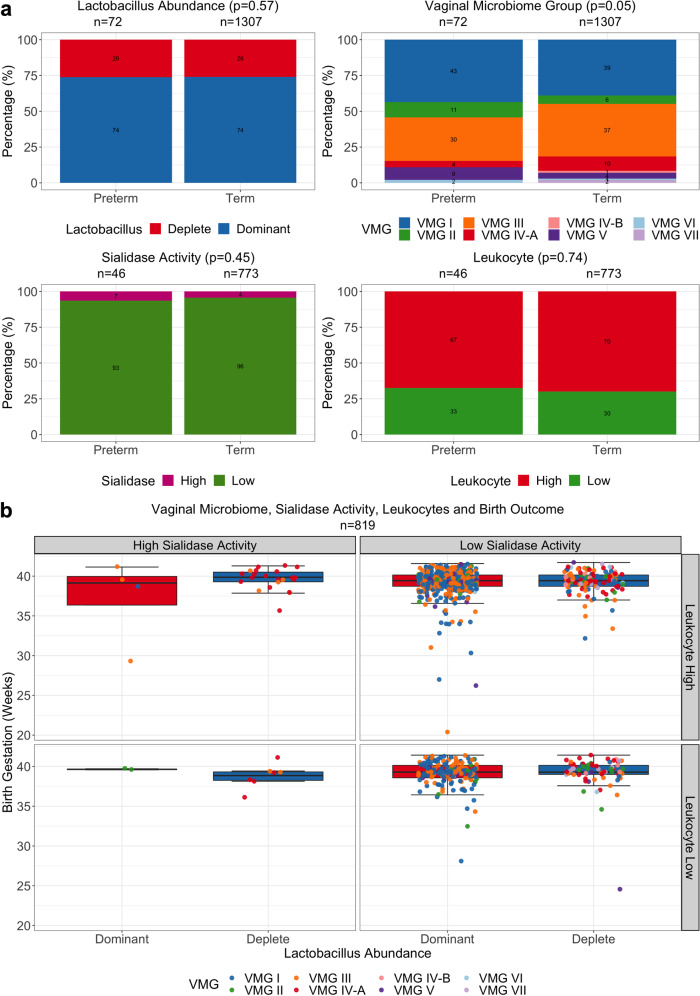
In women with available outcome data (n = 1379), the proportion of Lactobacillus dominated or depleted vaginal microbiota (p = 0.57; Fisher’s Exact test) and VMGs (p = 0.05; Fisher’s Exact test) were similar between women delivering preterm or term, including when preterm births were divided into early (<34 weeks gestation) and late (≥34 weeks gestation)(Supplementary Fig. 6). Likewise, proportions of high or low sialidase activity (p = 0.45; Fisher’s Exact test) and leukocyte high/low (p = 0.74; Fisher’s Exact test) were comparable between women subsequently experiencing preterm or term deliveries (Fig. 4a). Apart from women who had Lactobacillus dominated microbiomes, high sialidase activity, and leukocyte low which had insufficient sample size (n = 2), no significant difference was observed in birth gestation based on Lactobacillus abundance, VMG prevalence, sialidase activity, and leukocyte wet mount results (Kruskal-Wallis Test, p > 0.05) (Fig. 4b and Supplementary Table 4). Logistic regression results similarly showed vaginal microbiome Lactobacillus composition (p = 0.83), sialidase activity (p = 0.45) and leukocyte high or low (p = 0.70) did not significantly contribute to birth outcome (Supplementary Table 5). Finally, no relationships were observed between vaginal microbiota composition, sialidase activity and/or leukocyte presence, and chorioamnionitis (Supplementary Fig. 7).
Fig. 4. Association between the vaginal microbiome, sialidase activity and leukocyte presence with birth outcome.
a Stacked bar plots showing proportion of Lactobacillus abundance (deplete, red; or dominant, blue), vaginal microbiome groups (VMG), sialidase activity (high, pink; or low, red) and leukocyte low (grades I-II, green) or high (grades III-IV, red) in preterm and term groups. Statistical significance based on Fisher’s Exact Test. b Boxplots showing birth gestation in weeks grouped by Lactobacillus abundance, sialidase activity, and leukocyte presence or absence with dot points colored by VMG. The bounds of the box represent the first and third quartiles, center line represents the median, and whiskers show min-to-max values. No significant difference between boxplots with n ≥ 3 based on Kruskal-Wallis Test, p > 0.05.
Discussion
In this large cross-sectional study of 2646 pregnant Chinese women, the vaginal microbiota in mid-pregnancy was characterized by Lactobacillus spp. dominance and low diversity. This pattern is consistent with previous descriptions of the healthy mid-pregnancy vaginal microbiome in Caucasian, African and Asian women from European and Northern American populations6,10,13,17,41. Similar results have also been reported in small cohort studies of Chinese and Indian women42,43. One of these studies examined the vaginal microbiota of 113 Chinese women and reported L. crispatus dominance in 45.1% and L. iners dominance in 31.9% of women sampled at a mean gestation of 17 weeks42. In our cohort, L. crispatus (40%) and L. iners (36%) dominated vaginal microbiota were also the most frequently observed profiles. In previous work, we have observed that the most prevalent vaginal microbiota community compositions in European Caucasian women are those dominated by L. crispatus (50%) followed by L. iners (25%), whereas the inverse has been reported for Asian and Black women (L. iners, 48% and 50% v L. crispatus, 26% and 20%, respectively)6. Higher frequency of L. iners dominated vaginal microbiota has also been reported in Indian43, Karen and Burman44, Hispanic and African American pregnant women9,10,17. Our results therefore provide further evidence that ethnic and/or geographical differences are a major source of variation in the underlying structure and composition of the vaginal microbiome during pregnancy.
Correlation analyses in our patient cohort showed that L. crispatus was negatively correlated with almost all other vaginal taxa, highlighting its well-described exclusionary behavior in the vaginal niche that appears to be broadly consistent across different ethnic groups6,13,17,45. However, we also observed a strong negative correlation between L. iners and most other vaginal taxa. This was surprising given that L.iners has been shown to co-occur with various BV-taxa in other ethnic groups including Caucasian and African American women9,46. The highly negative correlation observed between L. iners and L. crispatus might be attributable to overlapping ecological functions in the vagina5,47. Compared to L. crispatus, L. iners has a substantially smaller genome, thought to be indicative of a symbiotic and/or parasitic role in the vaginal niche5,47,48. Similar to G. vaginalis, L. iners can produce pore-forming toxins such as inerolysin and vaginolysin, which can lead to lysis of host cells and release of carbon sources, particularly during times of nutrient scarcity5,47,48. This behavior may explain the observed positive relationship between L.iners and leukocyte presence in our study cohort. Consistent with these findings, a recent study of 83 healthy Chinese pregnant women reported a positive association between leukocyte esterase concentrations and increased L. iners levels determined using quantitative real-time PCR49. These women were also reported to have increased presence of white blood cells and other non-Lactobacillus morphotypes observed under microscopy. In vitro experiments using human vaginal epithelial cells have also demonstrated that L. iners stimulates increased pro-inflammatory cytokine production compared to L. crispatus50,51. Despite these findings, we did not observe any relationship between L. iners dominance or any vaginal microbiota profile and subsequent risk of PTB in this cohort. This is in contrast to previous studies of predominantly Caucasian women by ourselves and others that have reported a relationship between L.iners and increased risk of PTB or cervical shortening, which is clinically used as a marker of PTB risk6,7,12,52. Our data are consistent with earlier reports in Hispanic and African American women indicating that L. iners dominance is not a risk factor for PTB in these ethnic groups.
In our sub-cohort analysis, high vaginal sialidase activity was strongly associated with Lactobacillus depleted microbiota enriched with BV-associated taxa including Gardnerella, Prevotella, Atopobium, and Megasphaera species. This finding is consistent with previous studies in both pregnant and nonpregnant women where the production of sialidase by BV-associated taxa, such as Gardnerella spp. and Prevotella spp., is thought to be important for pathogenesis27,53,54. In this context, desialylation of glycolipids and/or glycoproteins (e.g. immunoglobulins, cytokines, cellular receptors, mucins, and antimicrobial molecules) can decrease the ability of host defence responses to recognize and bind to microbes whilst increasing bacterial adherence, invasion, and tissue breakdown27,30,54,55. Interestingly, high sialidase activity was observed in some Lactobacillus-dominated vaginal microbiota. These often contained low relative aundance of BV-associated taxa indicating that perhaps even low levels of sialidase-producing bacteria may be sufficient to produce high sialidase activity. It is also possible that sialidase was produced by other microorganisms including viruses and fungi that were not assessed in our study56,57. In contrast to these findings, a proportion of samples (23%) harboring Lactobacillus spp. depleted vaginal microbiota enriched with BV-associated taxa did not have high sialidase activity. This was supported by our findings indicating sialidase activity has high specificity (99%) but low sensitivity (14%) in predicting high-diversity, Lactobacillus spp. depleted vaginal microbiota. Low sialidase activity in these samples could be due to several factors: 1) the production of sialidase by BV-taxa, including Gardnerella spp., is strain-dependent;27,30,31 2) binding of sialidase to the BV test kit substrate may be influenced by vaginal pH;26,58 3) the amount of sialidase produced by BV-taxa maybe insufficient to reach the detection limit of the BV test kit; and/or 4) the bacterial load of BV-taxa, which was not quantified in our study, may be inadequate to produce sufficient sialidase for detection by the BV test kit. Overall, our findings suggest that high sialidase activity is significantly associated with BV-taxa but cannot be used as an accurate proxy for high-diversity, Lactobacillus spp. depleted vaginal microbiota in pregnant women.
Elevated sialidase activity has previously been associated with spontaneous PTB and late miscarriage in a North American study of 1806 pregnant women when measured at 12 weeks gestation30, and with early PTB and low birth weight in a mid-trimester study of 579 Danish women59. Additionally, in a recent study of 85 Chinese women, sialidase activity was associated with subsequent cervical cerclage failure, a risk factor for PTB31. However, in our study, no significant association was found between elevated sialidase activity and PTB. Our findings are similar to a previous nested case-control study of 126 pregnant women, where elevated sialidase activity measured at mid-pregnancy was not associated with PTB risk53. The observed differences between studies could be due to different methods for measuring sialidase activity where a single cut-off was used rather than a quantitative measurement. An association between vaginal leukocytes and preterm labor or risk factors of PTB including reproductive tract infections, histologic chorioamnionitis and inflammatory cytokines have also previously been reported33,34,39,60–62. Here, we classified leukocyte presence based on vaginal wet mount microscopy, which has been shown to be useful for identification of cervicovaginal inflammation in pregnant women39,63. Logistic regression analyses indicated that neither leukocyte presence, sialidase activity nor vaginal microbiota contributed to preterm risk in our study cohort. The rate of PTB in our study population was 5.4% which is slightly lower than the background rate in China (7.2%)64, but comparable to published preterm birth rates for the local population in Guangzhou (4.15% and 6.0%)65–67. These rates are considerably less than the global average of ~10%68. Our findings therefore highlight differences in the vaginal microbiota and/or host immune response as a potential contributing factor to reduced incidence of PTB in Chinese women.
There are several limitations to our study that should be considered when interpreting the results. Birth outcome data was unavailable for 48% of the women in our cohort. These women did not return to the recruitment site of Nanfang Hospital and due to data protection considerations, were unable to be contacted for follow-up. Although vaginal microbiota composition in these women was comparable to those with available outcome data, non-observed outcomes may represent a potential source of bias. Further, the pregnancy vaginal microbiota can be impacted by a variety of factors including pregnancy history, smoking status, genetics and hygiene habits21,69,70. This information was not available for our patient cohort and cross-sectional sampling of the vaginal microbiota did not permit us to assess the potential role of vaginal microbiota dynamics in shaping clinical outcomes17,71,72. Our study was also limited to the assessment of the bacterial component of the vaginal microbiota via 16S rRNA gene sequencing, which does not provide sufficient resolution to account for strain-level variations associated with potential function of microbial communities73,74. Future studies could apply function-based profiling approaches to enable a deeper insight into potential relationships between predicted function of microbiome communities within different ethnic groups and preterm birth risk phenotype.
In conclusion, our study provides new insight into the vaginal microbial composition and structure of Chinese pregnant women. Although sialidase activity was predictive of high diversity vaginal microbiota compositions, neither were associated with increased risk of PTB. Lactobacillus spp. depleted vaginal microbiomes with high sialidase activity and leukocyte presence are not associated with higher risk of PTB in Chinese women. Our results provide further evidence that ethnicity is an important determinant of microbiota-host interactions during pregnancy and highlight the need for further investigations into the mechanisms underpinning these relationships.
Methods
Study design
This prospective study was reviewed and approved ([2013] EC (100)) by the Ethical Committee of Nanfang Hospital, Southern Medical University, Guangzhou, China. Written, informed consent was obtained from all participants. Pregnant women at their first prenatal visit were recruited from the outpatient clinic of Nanfang Hospital of Southern Medical University in Guangzhou, China, from January 2015 to December 2018. All women were recruited consecutively, with no disruption to recruitment during this period. Women who received antibiotics, prebiotics or probiotics within 30 days prior to vaginal swab collection and/or who had sexual activity within 48 h of sample collection were excluded. Metadata collected included maternal age, mode of labor, gestation at delivery in days (calculated using the last menstrual period and/or ultrasound data) and chorioamnionitis (Supplementary Data 1).
Sample collection and processing
Vaginal samples were collected using a sterile swab that was inserted into the vaginal posterior fornix, before being gently rotated 360° for approximately 20 rotations prior to removal. All swabs were placed immediately on ice and were then stored at −20 °C within 1 h of collection before being transferred to −80 °C within 24 h for long-term storage. For DNA extraction, swabs were immersed and vigorously mixed in 500 μL of sterile water before the solution was transferred to a clean 2 mL centrifuge tube. The sample was then vortexed for 5 sec before being centrifuged at 13,800 g for 10 min. The supernatant was removed, and the pellet retained for DNA extraction. For PCR negative controls (n = 82), 2 μL of diethylpyrocarbonate (DEPC) water was used as non-template control. For DNA extraction kit negative controls (n = 21), 500 μL of sterile water was used instead of test samples for the DNA extraction process.
DNA extraction and 16S rRNA (V4) amplicon sequencing
DNA was extracted from vaginal swabs manually using the BioTeke bacterial DNA extraction kit (BioTeke Corporation, Cat #DP7001) per manufacturer’s instructions (n = 1176) or via an automated protocol using the Bioeasy bacterial DNA extraction kit (Shenzhen Bioeasy Biotechnology Co. Ltd., Cat #YRMBN7001) on a Thermo ScientificTM KingFisherTM Flex Purification System (n = 1620). The V4 region of bacterial 16S rRNA was amplified using barcoded V4F 5’ GTGCCAGCMGCCGCGGTAA 3’ forward and V4R 5’ CTACCNGGGTATCTAAT 3’ reverse primers. The PCR condition included an initial denaturation step at 94 °C for 5 min, 30 cycles of 94 °C for 30 sec, 52 °C for 30 sec, 72 °C for 45 sec and a final extension step at 72 °C for 5 min. Each 20 μl reaction volume consisted of 10 μl of AceQqPCR SYBR Green Master Mix (Vazyme, Nanjing, China), 0.4 μL prime V4F, 0.4 μL primer V4R, 0.4 μL ROX Reference Dye 2, 2 μL template DNA, 8.8 μL sterilized distilled water. Equimolar amplicon suspensions were combined and subjected to paired-end 101 bp sequencing on an Illumina HiSeq sequencer at the Beijing Genomics Institute (BGI; Beijing, China).
Sialidase enzyme activity detection
A total of 848 women had an additional vaginal swab collected for detection of sialidase enzyme activity. Sialidase enzyme activity was measured as per manufacturer’s instructions using a single-enzyme BV kit based on the colorimetric method (Zhuhai DL Biotech. Co. Ltd). Briefly, each swab sample was immersed into a BV test bottle solution containing 5-bromo-4-chloro-3-indolyl-α-D-N-acetylneuraminic acid (BCIN) as a substrate75. The BCIN substrate hydrolyzes and reacts with the added 1 – 2 drops of BV chromogenic solution containing hydroxide and potassium acetate, resulting in a color reaction that was measured using the BV-10 analyser (Zhuhai DL Biotech. Co. Ltd). The resulting sialidase activity is defined based on enzyme unit (U), where one U is the amount of sialidase required to catalyze 1 nmol of BCIN per minute in a ml. The amount of sialidase activity corresponded to color changes in the BV test bottle with ≥7.8U/ml for a positive result (blue or green) and 0 – 7.8U/ml for a negative result (yellow). The samples were defined as having high or low sialidase activity based on ≥7.8U/ml or 0 – 7.8U/ml, respectively26.
Leukocyte wet mount
Preparation of leukocyte wet mounts was performed by agitating vaginal swabs in 1 mL of saline solution and a single drop of the fluid was then placed on a glass slide76,77. A cover slip was applied on the droplet and the slides were then inspected under 400x maginification. The average white blood cell count (non-clumped) in 10 nonadjacent fields of magnificant were determined. Leukocyte counts were categorized into grades I (0–5 counts/Hp), II (5–10 counts/Hp), III (10–15 counts/Hp) or IV (>30 counts/Hp). Grades I and II were further classified as leukocyte low (<15 counts/Hp) and grades III-IV as high (>15 counts/Hp)63.
Chorioamnionitis diagnosis
Chorioamnionitis was diagnosed using a combination of clinical symptoms and histological assessment. Clinical symptoms included the presence of high maternal fever (≥39 °C), maternal tachycardia (>100 beats/min) and malodorous amniotic fluid. Histological chorioamnionitis was diagnosed based on the presence of inflammatory cells in the chorionic plate and/or the chorioamniotic membranes consistent with acute inflammation and the presence of neutrophils in the wall of the umbilical vessels and Wharton’s jelly78,79. Staging and grading of chorioamnionitis was performed following the guidelines of the Amsterdam Placental Workshop Group Consensus Statement80.
Bioinformatics and statistical analyses
The raw sequencing reads were processed using the DADA2 package (v1.6.0) in R (v3.4.3)81. The raw paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcodes and primer sequences. The reads were then processed using the DADA2 package (v1.6.0) in R (v3.4.3) according to the following steps: quality controls, dereplication, error rate calculations, sequences denoising, paired-end reads merging, amplicon sequence variant (ASV) table constructing and removal of chimeras. The sequences in the ASV table were annotated using the RDP Classifier82 with the GreenGenes database83. The sequences assigned to Lactobacillus were further classified into species level using UCLUST84 with the customized Lactobacillus database (V4 region extracted from representative sequences of Lactobacillus species from RDP database).
All downstream bioinformatic and statistical analyses were performed in R (v4.0.0)85 unless otherwise stated. Low abundance taxa accounting for less than 0.1% within-sample relative abundance and present in less than 1% of samples were filtered using phyloseq (v1.32.0)86 and genefilter (v1.70.0)87 packages. This left a total of 167 ASVs representing 94 unique taxa. Extraction kit and reagent negative controls (n = 103) were used to identify potential contaminants using the decontam package (v1.9.0) with a prevalence threshold of 0.1 and 0.5 respectively88. A total of 12 contaminants were assigned as Pseudomonas veronii, Bifidobacterium adolescentis, Klebsiella spp., Enhydrobacter spp., Chryseobacterium spp., Pedobacter spp., Bradyrhizobium spp., Methylobacterium spp., Escherichia coli, Acinetobacter johnsonii and Sneathia spp., and Species_Other. Samples with a library size of <2000 reads were excluded from the analysis (n = 90) (Supplementary Fig. 1).
Each vaginal bacterial profile in the cohort was initially classified on the basis of the highest relative abundance of the dominant species taxon within the sample2,89. Dominant species with <30% abundance were classified as “Other” (n = 16) and a single sample dominated by Mesorhizobium spp. was classified as “Outlier”. These samples were excluded from subsequent analyses. A total of 19 dominant (>30% relative abundance) species groups were then classified into VMG consisting of VMG I (L. crispatus); II (L. gasseri); III (L. iners); IV-A (BV-associated species; Atopobium vaginae, Gardnerella spp., Megasphaera spp., Prevotella spp., Shuttleworthia spp., Veillonella spp.)40,69; IV-B (Non-BV taxa; Aerococcus spp., Anaerococcus spp., Streptococcus agalactiae, Streptococcus anginosus, Streptococcus luteciae, Streptococcus spp.)90; V (L. jensenii); VI (Other Lactobacillus; L. delbrueckii, Lactobacillus spp.) and VII (Bifidobacterium spp.). Analyses were also performed on samples classified into Lactobacillus species dominated and depleted groups based on Lactobacillus abundance of greater or lower than 90%91.
Data were visualized using ggplot2 package (v3.3.1)92. Statistical difference for continuous variables between groups were determined using either Mann-Whitney Test with Bonferroni p-adjusted values or Kruskal-Wallis Test with pairwise comparison between groups and Bonferroni p-adjusted values. Association between categorical groups were determined using Fisher’s Exact Test. For all statistical tests, significance was based on p < 0.05 unless otherwise stated. Spearman’s correlation plots were generated from relative abundance using corrplot package (v0.84) with ward.D2 agglomerative method93. The Shannon diversity index and richness (number of species observed) were computed using the vegan package (v2.5–6)94, the former calculated for both raw and normalized read counts using the trimmed mean of M-values normalization method in the edgeR package (v3.30.3)95. LefSe analysis was performed using per-sample normalization to sum of values to 1 M with a Linear Discriminant Analysis (LDA) threshold of 2.0 for discriminative features and an alpha of 0.05 for factorial Kruskal-Wallis test among classes and alpha 0.05 for pairwise Wilcoxon test between subclasses96. The caret package (v6.0–86)97 was used to compute confusion matrices with a prevalence of 0.15 and perform multiple logistic regression using gestation at delivery (days) as a binary outcome (Preterm, <259 days and Term, ≥259 days) for the response variable and Lactobacillus abundance, sialidase activity and leukocyte wet mount results as explanatory variables.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank all patients who have participated in this study. This work was funded by National Natural Science Foundation of China (NSFC81925062, 82002201), National Key R&D Program of China (2019YFA0802300) and the March of Dimes European Prematurity Research Centre at Imperial College London.
Author contributions
S.N. and M.C. contributed equally to this study. S.N., M.C., D.A.M., and H.Z. conceived and designed the study. Clinical sampling and coordination of metadata were performed by M.C., X.W., Z.D.Z., H.S., Y.H., and Y.W. Experiments were performed by M.C., Z.Y., and W.Q. Data processing and analysis were performed S.N., M.C., and D.A.M. S.N. and M.C. prepared all figures and tables. S.N. and D.A.M. wrote the first draft of the manuscript. All authors critically reviewed, read, and approved the final manuscript.
Data availability
Raw data files for the sequence data used in this study are publicly available through the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession numbers (PRJNA706523).
Code availability
Key analysis code and processed datasets are publicly available the GitHub repository; https://github.com/sherrianne/NgChen_2021_ChinaPregnancyCohort.
Competing interests
The authors declare that there are no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sherrianne Ng, Muxuan Chen.
Contributor Information
David A. MacIntyre, Email: d.macintyre@imperial.ac.uk
Hongwei Zhou, Email: biodegradation@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-021-00261-0.
References
- 1.Brown RG, et al. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med. 2018;16:9. doi: 10.1186/s12916-017-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fettweis JM, et al. The vaginal microbiome and preterm birth. Nat. Med. 2019;25:1012–1021. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kindinger LM, et al. Relationship between vaginal microbial dysbiosis, inflammation, and pregnancy outcomes in cervical cerclage. Sci. Transl. Med. 2016;8:350ra102. doi: 10.1126/scitranslmed.aag1026. [DOI] [PubMed] [Google Scholar]
- 4.Amabebe E, Anumba DOC. The vaginal microenvironment: the physiologic role of Lactobacilli. Front Med (Lausanne) 2018;5:181. doi: 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.France MT, Mendes-Soares H, Forney LJ, Schloss PD. Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina. Appl. Environ. Microbiol. 2016;82:7063–7073. doi: 10.1128/AEM.02385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindinger LM, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5:6. doi: 10.1186/s40168-016-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne, M. S. et al. A specific bacterial DNA signature in the vagina of Australian women in midpregnancy predicts high risk of spontaneous preterm birth (the Predict1000 study). Am J Obstet Gynecol, 10.1016/j.ajog.2020.08.034 (2020). [DOI] [PubMed]
- 8.Stafford GP, et al. Spontaneous Preterm Birth Is Associated with Differential Expression of Vaginal Metabolites by Lactobacilli-Dominated Microflora. Front Physiol. 2017;8:615. doi: 10.3389/fphys.2017.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan BJ, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl Acad. Sci. USA. 2017;114:9966–9971. doi: 10.1073/pnas.1705899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elovitz MA, et al. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat. Commun. 2019;10:1305. doi: 10.1038/s41467-019-09285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kindinger LM, et al. The effect of gestational age and cervical length measurements in the prediction of spontaneous preterm birth in twin pregnancies: an individual patient level meta-analysis. BJOG. 2016;123:877–884. doi: 10.1111/1471-0528.13575. [DOI] [PubMed] [Google Scholar]
- 12.Petricevic L, et al. Characterisation of the vaginal Lactobacillus microbiota associated with preterm delivery. Sci. Rep. 2014;4:5136. doi: 10.1038/srep05136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown RG, et al. Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes. Transl. Res. 2019;207:30–43. doi: 10.1016/j.trsl.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiulio DB, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl Acad. Sci. USA. 2015;112:11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freitas AC, Bocking A, Hill JE, Money DM, Group VR. Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome. 2018;6:117. doi: 10.1186/s40168-018-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blencowe H, et al. Born too soon: the global epidemiology of 15 million preterm births. Reprod. Health. 2013;10(Suppl 1):S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano MG, et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019;25:1001–1011. doi: 10.1038/s41591-019-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amsel R, et al. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 1983;74:14–22. doi: 10.1016/0002-9343(83)91112-9. [DOI] [PubMed] [Google Scholar]
- 19.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donders GG, et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG: Int. J. Obstet. Gynaecol. 2009;116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- 21.Guaschino S, De Seta F, Piccoli M, Maso G, Alberico S. Aetiology of preterm labour: bacterial vaginosis. BJOG. 2006;113(Suppl 3):46–51. doi: 10.1111/j.1471-0528.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- 22.Leitich H, et al. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am. J. Obstet. Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 23.Hillier SL, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N. Engl. J. Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 24.Colonna, C. & Steelman, M. in StatPearls (2021).
- 25.Madhivanan P, et al. Performance of BVBlue rapid test in detecting bacterial vaginosis among women in Mysore, India. Infect. Dis. Obstet. Gynecol. 2014;2014:908313. doi: 10.1155/2014/908313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myziuk L, Romanowski B, Johnson SC. BVBlue test for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 2003;41:1925–1928. doi: 10.1128/JCM.41.5.1925-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briselden AM, Moncla BJ, Stevens CE, Hillier SL. Sialidases (Neuraminidases) in bacterial vaginosis and bacterial vaginosis-associated microflora. J. Clin. Microbiol. 1992;30:663–666. doi: 10.1128/jcm.30.3.663-666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy L, et al. The presence of the putative Gardnerella vaginalis sialidase A gene in vaginal specimens is associated with bacterial vaginosis biofilm. PLoS One. 2017;12:e0172522. doi: 10.1371/journal.pone.0172522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung HS, Ehlers MM, Lombaard H, Redelinghuys MJ, Kock MM. Etiology of bacterial vaginosis and polymicrobial biofilm formation. Crit. Rev. Microbiol. 2017;43:651–667. doi: 10.1080/1040841X.2017.1291579. [DOI] [PubMed] [Google Scholar]
- 30.Cauci S, Culhane JF. High sialidase levels increase preterm birth risk among women who are bacterial vaginosis-positive in early gestation. Am. J. Obstet. Gynecol. 2011;204:142 e141–142 e149. doi: 10.1016/j.ajog.2010.08.061. [DOI] [PubMed] [Google Scholar]
- 31.Fang J, Chen L, Chen Z, Jiang X, Pan M. Association of the vaginal microbiota with pregnancy outcomes in Chinese women after cervical cerclage. Reprod. Biomed. Online. 2020;41:698–706. doi: 10.1016/j.rbmo.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N. Engl. J. Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 33.Yudin MH, et al. Vaginal polymorphonuclear leukocytes and bacterial vaginosis as markers for histologic endometritis among women without symptoms of pelvic inflammatory disease. Am. J. Obstet. Gynecol. 2003;188:318–323. doi: 10.1067/mob.2003.105. [DOI] [PubMed] [Google Scholar]
- 34.Geisler WM, Yu S, Venglarik M, Schwebke JR. Vaginal leucocyte counts in women with bacterial vaginosis: relation to vaginal and cervical infections. Sex. Transm. Infect. 2004;80:401–405. doi: 10.1136/sti.2003.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakakha M. Leukorrhea and bacterial vaginosis as in-office predictors of cervical infection in high-risk women. Obstet. Gynecol. 2002;100:808–812. doi: 10.1016/s0029-7844(02)02147-6. [DOI] [PubMed] [Google Scholar]
- 36.Ramsey PS, et al. Use of vaginal polymorphonuclear to epithelial cell ratios for the prediction of preterm birth. Obstet. Gynecol. 2005;105:139–144. doi: 10.1097/01.AOG.0000148269.36622.0a. [DOI] [PubMed] [Google Scholar]
- 37.Verstraelen H, et al. Modified classification of Gram-stained vaginal smears to predict spontaneous preterm birth: a prospective cohort study. Am. J. Obstet. Gynecol. 2007;196:528 e521–526. doi: 10.1016/j.ajog.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Simhan HN, Caritis SN, Krohn MA, Hillier SL. Elevated vaginal pH and neutrophils are associated strongly with early spontaneous preterm birth. Am. J. Obstet. Gynecol. 2003;189:1150–1154. doi: 10.1067/s0002-9378(03)00582-9. [DOI] [PubMed] [Google Scholar]
- 39.Nenadić DB, Pavlović MD. Cervical fluid cytokines in pregnant women: Relation to vaginal wet mount findings and polymorphonuclear leukocyte counts. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008;140:165–170. doi: 10.1016/j.ejogrb.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Gajer, P. et al. Temporal Dynamics of the Human Vaginal Microbiota. Sci. Transl. Med.4, 132ra52 (2013). [DOI] [PMC free article] [PubMed]
- 41.Stout MJ, et al. Early pregnancy vaginal microbiome trends and preterm birth. Am. J. Obstet. Gynecol. 2017;217:356 e351–356 e318. doi: 10.1016/j.ajog.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y, Huang Y, Zhang Z, Yu F, Zheng Y. Exploring profile and potential influencers of vaginal microbiome among asymptomatic pregnant Chinese women. PeerJ. 2019;7:e8172. doi: 10.7717/peerj.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta O, et al. Vaginal microbiome of pregnant Indian women: insights into the genome of dominant Lactobacillus species. Micro. Ecol. 2020;80:487–499. doi: 10.1007/s00248-020-01501-0. [DOI] [PubMed] [Google Scholar]
- 44.Kumar M, et al. Vaginal microbiota and cytokine levels predict preterm delivery in Asian women. Front Cell Infect. Microbiol. 2021;11:639665. doi: 10.3389/fcimb.2021.639665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verstraelen, H. et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiology9, 116 (2009). [DOI] [PMC free article] [PubMed]
- 47.Witkin, S. S., Moron, A. F., Linhares, I. M. & Forney, L. J. Influence of Lactobacillus crispatus, Lactobacillus iners and Gardnerella vaginalis on bacterial vaginal composition in pregnant women. Arch Gynecol Obstet, 10.1007/s00404-021-05978-z (2021). [DOI] [PubMed]
- 48.Petrova MI, Reid G, Vaneechoutte M, Lebeer S. Lactobacillus iners: friend or foe? Trends Microbiol. 2017;25:182–191. doi: 10.1016/j.tim.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Zheng N, et al. Lactobacillus iners is associated with vaginal dysbiosis in healthy pregnant women: a preliminary Study. Biomed. Res Int. 2019;2019:6079734. doi: 10.1155/2019/6079734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anahtar MelisN, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doerflinger SY, Throop AL, Herbst-Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J. Infect. Dis. 2014;209:1989–1999. doi: 10.1093/infdis/jiu004. [DOI] [PubMed] [Google Scholar]
- 52.Witkin, S. S. et al. Vaginal Biomarkers That Predict Cervical Length and Dominant Bacteria in the Vaginal Microbiomes of Pregnant Women. mBio10, 02242–19 (2019). [DOI] [PMC free article] [PubMed]
- 53.Andrews WW, et al. The Preterm Prediction Study: Failure of midtrimester cervical sialidase level elevation to predict subsequent spontaneous preterm birth. Am. J. Obstet. Gynecol. 1999;180:1151–1154. doi: 10.1016/s0002-9378(99)70609-5. [DOI] [PubMed] [Google Scholar]
- 54.Cauci S, et al. Immunoglobulin A response against Gardnerella vaginalis hemolysin and sialidase activity in bacterial vaginosis. Am. J. Obstet. Gynecol. 1998;178:511–515. doi: 10.1016/s0002-9378(98)70430-2. [DOI] [PubMed] [Google Scholar]
- 55.Cauci S, et al. Determination of immunoglobulin A against Gardnerella vaginalis hemolysin, sialidase, and prolidase activities in vaginal fluid: implications for adverse pregnancy outcomes. J. Clin. Microbiol. 2003;41:435–438. doi: 10.1128/JCM.41.1.435-438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwerdtfeger SM, Melzig MF. Sialidases in biological systems. Pharmazie. 2010;65:551–561. doi: 10.1002/chin.201047267. [DOI] [PubMed] [Google Scholar]
- 57.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 58.Agarwal, K. & Lewis, A. L. Vaginal sialoglycan foraging by Gardnerella vaginalis: Mucus barriers as a meal for unwelcome guests? Glycobiology, 10.1093/glycob/cwab024 (2021). [DOI] [PMC free article] [PubMed]
- 59.Cauci S, McGregor J, Thorsen P, Grove J, Guaschino S. Combination of vaginal pH with vaginal sialidase and prolidase activities for prediction of low birth weight and preterm birth. Am. J. Obstet. Gynecol. 2005;192:489–496. doi: 10.1016/j.ajog.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 60.Matsubara S, Yamada T, Minakami H, Sato I. Stimulated polymorphonuclear leukocytes in vaginal secretions from patients with preterm labor. Gynecol. Obstet. Invest. 1998;45:35–40. doi: 10.1159/000009921. [DOI] [PubMed] [Google Scholar]
- 61.Yamada T, et al. Relation between viability of vaginal polymorphonuclear leukocytes and presence of histologic chorioamnionitis. Acta Obstet. Gynecol. Scand. 2000;79:818–823. [PubMed] [Google Scholar]
- 62.Yamada T, Minakami H, Matsubara S, Yatsuda T, Sato I. Changes in polymorphonuclear leukocytes in the vagina of patients with preterm labor. Gynecol. Obstet. Invest. 1998;45:32–34. doi: 10.1159/000009920. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, et al. Computerized detection of leukocytes in microscopic leukorrhea images. Med Phys. 2017;44:4620–4629. doi: 10.1002/mp.12381. [DOI] [PubMed] [Google Scholar]
- 64.Guo T, et al. The association between ambient temperature and the risk of preterm birth in China. Sci. Total Environ. 2018;613-614:439–446. doi: 10.1016/j.scitotenv.2017.09.104. [DOI] [PubMed] [Google Scholar]
- 65.He JR, et al. Ambient Temperature and the Risk of Preterm Birth in Guangzhou, China (2001-2011) Environ. Health Perspect. 2016;124:1100–1106. doi: 10.1289/ehp.1509778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu J, et al. Increasing trends in incidence of preterm birth among 2.5 million newborns in Guangzhou, China, 2001 to 2016: an age-period-cohort analysis. BMC Public Health. 2020;20:1653. doi: 10.1186/s12889-020-09739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miao H, et al. Adverse birth outcomes in Guangdong province, China, 2014-2017: a spatiotemporal analysis of 2.9 million births. BMJ Open. 2019;9:e030629. doi: 10.1136/bmjopen-2019-030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chawanpaiboon S, et al. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob. Health. 2019;7:e37–e46. doi: 10.1016/S2214-109X(18)30451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Onderdonk AB, Delaney ML, Fichorova RN. The human microbiome during bacterial vaginosis. Clin. Microbiol Rev. 2016;29:223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turovskiy Y, Sutyak Noll K, Chikindas ML. The aetiology of bacterial vaginosis. J. Appl Microbiol. 2011;110:1105–1128. doi: 10.1111/j.1365-2672.2011.04977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacIntyre DA, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015;5:8988. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romero R, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2:4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Douglas GM, et al. Overcoming barriers to early disease intervention. Nat. Biotechnol. 2020;38:669–688. doi: 10.1038/s41587-020-0550-z. [DOI] [PubMed] [Google Scholar]
- 74.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiggins R, et al. Use of 5-bromo-4-chloro-3-indolyl-alpha-D-N-acetylneuraminic acid in a novel spot test To identify sialidase activity in vaginal swabs from women with bacterial vaginosis. J. Clin. Microbiol. 2000;38:3096–3097. doi: 10.1128/jcm.38.8.3096-3097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kottmel A, Petersen EE. Vaginal wet mount. J. Sex. Med. 2013;10:2616–2619. doi: 10.1111/jsm.12339. [DOI] [PubMed] [Google Scholar]
- 77.Galinanes S, et al. Vaginal inflammatory status in pregnant women with normal and pathogenic microbiota in lower genital tract. ISRN Obstet. Gynecol. 2011;2011:835926. doi: 10.5402/2011/835926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tita AT, Andrews WW. Diagnosis and management of clinical chorioamnionitis. Clin. Perinatol. 2010;37:339–354. doi: 10.1016/j.clp.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoon BH, et al. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 2001;185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 80.Khong TY, et al. Sampling and Definitions of Placental Lesions: Amsterdam Placental Workshop Group Consensus Statement. Arch. Pathol. Lab Med. 2016;140:698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- 81.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 85.R Development Core Team, R. R: A Language and Environment for Statistical Computing. (2011).
- 86.McMurdie, P. J. & Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE8, 0061217 (2013). [DOI] [PMC free article] [PubMed]
- 87.genefilter: methods for filtering genes from high-throughput experiments (2020).
- 88.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brooks JP, et al. Changes in vaginal community state types reflect major shifts in the microbiome. Micro. Ecol. Health Dis. 2017;28:1303265. doi: 10.1080/16512235.2017.1303265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van de Wijgert, J. H. H. M. et al. Pathobionts in the Vaginal Microbiota: Individual Participant Data Meta-Analysis of Three Sequencing Studies. Front. Cell. Infect. Microbiol.10, 00129 (2020). [DOI] [PMC free article] [PubMed]
- 91.Fu, M. et al. Alterations in vaginal microbiota and associated metabolome in women with recurrent implantation failure. mBio11, 03242-19 (2020). [DOI] [PMC free article] [PubMed]
- 92.Wickham, H. ggplot2: Elegant graphics for data analysis. (Springer Verlag, 2009).
- 93.corrplot: Visualization of a correlation matrix v. 0.84 (2017).
- 94.vegan: Community ecology package v. 2.5-6 (2019).
- 95.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biology12, r60 (2011). [DOI] [PMC free article] [PubMed]
- 97.Kuhn, M. Building predictive models in R using the caret package. J. Stat. Software28, i05 (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data files for the sequence data used in this study are publicly available through the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession numbers (PRJNA706523).
Key analysis code and processed datasets are publicly available the GitHub repository; https://github.com/sherrianne/NgChen_2021_ChinaPregnancyCohort.