Abstract
Inosine monophosphate dehydrogenase (IMPDH) is a key regulatory enzyme in the de novo synthesis of the purine base guanine. Dominant mutations in human IMPDH1 cause photoreceptor degeneration for reasons that are unknown. Here, we sought to provide some foundational information on Impdh1a in the zebrafish retina. We found that in zebrafish, gene subfunctionalization due to ancestral duplication resulted in a predominant retinal variant expressed exclusively in rod and cone photoreceptors. This variant is structurally and functionally similar to the human IMPDH1 retinal variant and shares a reduced sensitivity to GTP-mediated inhibition. We also demonstrated that Impdh1a forms prominent protein filaments in vitro and in vivo in both rod and cone photoreceptor cell bodies, synapses, and to a lesser degree, in outer segments. These filaments changed length and cellular distribution throughout the day consistent with diurnal changes in both mRNA and protein levels. The loss of Impdh1a resulted in a substantial reduction of guanine levels, although cellular morphology and cGMP levels remained normal. Our findings demonstrate a significant role for IMPDH1 in photoreceptor guanine production and provide fundamental new information on the details of this protein in the zebrafish retina.
Keywords: Zebrafish, photoreceptors, purine metabolism, inosine monophosphate dehydrogenase, IMPDH, metabolic filaments, photoreceptor metabolism
Abbreviations: IHC, immunohistochemistry; IMPDH, inosine monophosphate dehydrogenase; IS, inner segment; TEM, transmission electron microscopy; tv, transcript variant
Inosine monophosphate dehydrogenase (IMPDH) is a universally conserved enzyme that catalyzes the rate-limiting step in de novo GTP synthesis, converting the precursor inosine-5′-monophosphate (IMP) into xanthine-5′-monophosphate through a covalent intermediate, reducing NAD+ in the process. IMP is the first stable purine nucleotide in the pathway, and it is the precursor of both ATP and GTP. IMPDH plays a critical role in balancing flux between adenine and guanine nucleotide synthesis. Downstream guanine nucleotide products and ATP are allosteric effectors of Impdh (1, 2, 3, 4, 5). Impdh forms filaments containing stacked octamers (6, 7, 8). Recent findings show that human IMPDH2 and the IMPDH1 human retinal variants form filaments that maintain a protein conformation that resists GTP inhibition (3, 9).
Vertebrates have two IMPDH isoforms, IMPDH1 and IMPDH2. IMPDH2 is abundant in proliferating cells. IMPDH1 is widely expressed at low levels in most tissues (10). In contrast, IMPDH1 expression in photoreceptors is robust (11), which is likely because of the critical importance of both 3′-5′ cGMP and ATP in photoreceptors. cGMP is a signaling molecule that is generated from GTP in the dark and is hydrolyzed to 5′-GMP in the light. cGMP levels in photoreceptor outer segments regulate opening and closing of cation channels to control the membrane potential that drives signaling in response to light (12). In addition, energy consumption by photoreceptor neurons is uniquely high. Na+/K+ efflux in the inner segment compensates for Na+ influx to maintain membrane potential. This consumes up to 108 ATP per second in a mouse rod photoreceptor (13).
In humans, at least nine different mutations in IMPDH1 are linked to photoreceptor death, in the diseases autosomal dominant retinitis pigmentosa and Leber’s congenital amaurosis (14). Disease symptoms, which range from infancy in the most severe cases to late middle age, vary depending on the mutation. The molecular mechanisms causing IMPDH1-related photoreceptor degeneration are unknown, although many hypotheses have been proposed (15, 16, 17, 18, 19, 20, 21). Recent work has focused on dissecting the structure and function of the primary variant expressed in retina. In humans, this variant has extensions at both the N- and C-termini compared with the canonical enzyme, and a prevailing hypothesis from in vitro analysis of retinopathy mutations is that altered activity of the retinal variant could explain the cause of degeneration. However, although some retinopathy mutations show defects in allosteric regulation and filament formation, these effects are identical in the canonical form of the enzyme (9).
A recent study examining the role of endogenous IMPDH1 in mouse retina indicated a role for the enzyme in cGMP synthesis during prolonged bright light. This response correlated with enzyme phosphorylation and filament formation (22). Mutations that disrupt normal cGMP metabolism in photoreceptors cause cell degeneration and blindness (23). Thus, one hypothesis is that IMPDH1 mutations cause degeneration by elevating cGMP. Indeed, one class of retinopathy mutations reduces enzyme inhibition by GTP, which could result in elevated cellular cGMP (9). However, a second class of mutations has no obvious structural or functional phenotype (9). Disease models to study IMPDH1 retinopathy mutations would provide the ability to dissect the causes of disease in vivo.
Zebrafish are widely used to study human mutations causing retinal degeneration (24, 25, 26, 27, 28, 29, 30, 31). In this study, we provide fundamental information on Impdh1 in the zebrafish retina. We determine the primary splice variant present in the retina and show its exclusive expression and ability to form filaments in rods and cones. We demonstrate biochemical and structural similarity between the zebrafish and human retinal variant of IMPDH1, uncover dynamic changes in photoreceptor filaments throughout the day, and use metabolomics to elucidate an important role for Impdh1 in purine nucleotide homeostasis in photoreceptors. Our study provides the foundation for using zebrafish to determine Impdh1 function in normal and diseased retina.
Results
IMPDH expression in zebrafish retina
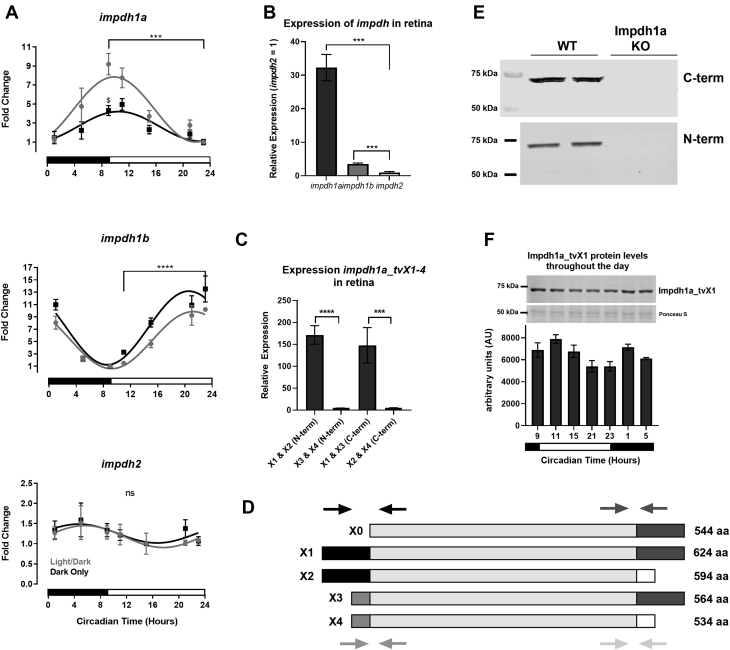
Using primers that recognize all reported major splice variants for impdh1a, 1b, and 2 (Table 1), we found that peak expression of impdh1a and impdh1b transcript levels in the zebrafish retina occur at different times during the day; impdh1a shows highest expression at light onset (9 AM) and impdh1b peaks at dark onset (11 PM). To test whether expression patterns might be circadian, we also analyzed expression in fish retina in darkness after a 24 h dark incubation. In general, the changes in expression were maintained in constant darkness, however, there was a light-dependent increase in the expression for impdh1a, which we did not detect for impdh1b. imdph2 expression remained constant over a 24-h period (Fig. 1A). Using the maximal mRNA levels for each gene (1a, 1b, and 2), we found that impdh1a is the most abundant impdh transcript in the zebrafish retina, with more than 9-fold the level of imdph1b and more than 32-fold the level of imdph2 (Fig. 1B).
Table 1.
Primers
| Target/NCBI reference sequence | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Gene amplification | ||
| impdh1a/NM_001002177.1 | ACAGAGAACAGATTGGTGGATCCATGGCTGATTACCTGATCAGCGG | GAGTGCGGCCGCTTAGCTGCTGTGTTTAGTGACGGC |
| impdh1b/NM_001014369.2 | ACAGAGAACAGATTGGTGGATCCATGGCAGACTATCTGATAAGCGGAGGAA | GAGTGCGGCCGCTTAGTAAAGACGCTTCTCGAAGGAGTGAAGTC |
| impdh2/NM_201464.1 | ACAGAGAACAGATTGGTGGATCCATGGCGGACTATTTAATCAGCGG | CTCGAGTGCGGCCGCTTAAAACAGGCGCTTCTCATAGCTGT |
| impdh1a_tvX1/XM_005159007.4 impdh1a_tvX2/XM_005159008.4 | TTTGGGGAAATCAGTCCAAC | GAAGCCCGAACAAAACAAAA |
| impdh1a_tvX3/XM_009293441.2 | GAAGCCCGAACAAAACAAAA | GGAGACCCTTCATCCACAGC |
| impdh1a_tvX4/MZ851785 | GGAGACCCTTCATCCACAGC | GTTGAGAAATGAGCGCCTCT |
| Genotyping | ||
| impdh1aˆsa23234 | GACGCCACTCATTTCCTCTC | CTTTCCGGACCTCGTTAGC |
| rhoˆfl9 | CCCTTCGAAGAGGAAGAGG | TCGAGAGTGTCTGGAAGGAGA |
| qPCR | ||
| impdh1a (all variants) | CATTGCAGGAATTCAACACG | GAACTCCACCCTCAACCTGA |
| impdh1b (all variants) Set 1 | AGCAAACTAGTCGGCATCGT | GGCAGGAGCGACTACAAGAT |
| impdh1b (all variants) Set 2 | ATCTGATAAGCGGAGGAACG | CCAGGCAAGATCAGGAAGTC |
| impdh2 (all variants) | TCTGGAGACGGCCTTACCTA | ATAGGCGACGAGATCAGTGG |
| impdh1a_tvX1 & X2 (N-term) | AATGGCCCGTTTACTCACAG | GTAATCAGCCATGCTCTCTGG |
| impdh1a_tvX3 & X4 (N-term) | GGCGAATATTCAGCAGAAGG | ATCACCGATGGCAAAGAGTT |
| impdh1a_tvX1 & X3 (C-term) | CATTGCAGGAATTCAACACG | TAAATGGCAAAAAAGAATATGA |
| impdh1a_tvX2 & X4 (C-term) | CATTGCAGGAATTCAACACG | ATAACCGTTTCTCGTAAGAAT |
| es1 NM_131039.1 | TCACAACACCCGTGAACCTT | GGTCTCCCACATGAAGGTCG |
| gnat1 NM_131868.2 | AGTGCATACGACATGGTGCT | GATGTGGTGGCGAAGTAACG |
Figure 1.
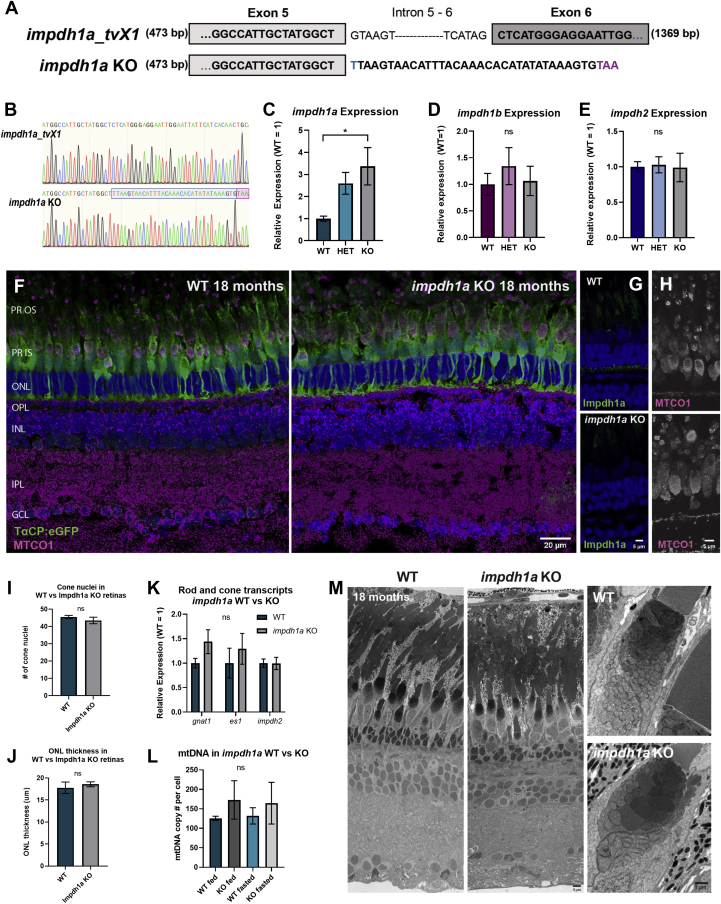
Impdh1a_tv1a is the predominant variant in the retina.A, relative expression levels of all transcript variants for impdh1a, 1b, and 2 throughout the day. For each graph, minimum expression = 1: impdh1a timepoints measured relative to 23:00, impdh1b relative to 9:00, and impdh2 relative to 15:00. The black and white bars indicate lights on (9:00) and lights out (23:00) for animals under normal light/dark cycle, $ denotes p ≤ 0.05 normal light/dark compared with dark only at 9:00. B, comparison of relative maximum expression levels of impdh1a (9:00), 1b (23:00), and 2 (5:00) transcripts. C, comparison of relative expression levels for all impdh1a transcripts detected in the retina at 9:00. N = 6 animals, the samples were in triplicate. D, schematic of impdh1a transcripts X1 to X4 and 544 (X0, NM_001002177.1); primer location for (C) indicated by arrows for both C-terminal and N-terminal reactions. See Table 1 for primer/transcript information. E, Western blot of Impdh1a protein in WT and impdh1a23234 zebrafish retinas using antibodies that recognize either X1 and X3 (C-terminal) or X1 and X2 (N-terminal) N = 2. F, quantification of Impdh1a_tvX1 protein levels throughout the day using the C-terminal antibody, and inset shows representative Western blot with Ponceau S used as a loading control. Trend p =0.04, N = 2 animals per time point. N = 3 animals per time/data point for all other panels except where noted. impdh, inosine monophosphate dehydrogenase; tv, transcript variant. ns p > 0.05, ∗ p ≤ 0.05 , ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001.
We then used primer combinations of four imdph1a splice variants listed in NCBI to determine which transcript is primarily expressed (see Table 1). We found that the predominant transcript variant (tv) is X1, and we identified a new transcript which we designate as X4 (Fig. 1, C and D). Using unique N-terminal and C-terminal primers to distinguish the predicted variants, we found that X1 is expressed ∼150 times more than the X2 to 4. To further confirm our findings, we developed two antibodies that detect the C-terminus of X1, X3, and impdh1a (544) and the N-terminus of X1 and X2. These antibodies are specific and verified that only the X1 (624 aa) variant is expressed at significant levels in the zebrafish retina (Fig. 1E). No protein was detected in the fish strain impdh1asa23234, which contains a splice site mutation in impdh1a that introduces a premature stop codon (32). The diurnal change in mRNA expression was partially recapitulated in protein levels on Western blots (Fig. 1F).
The zebrafish retinal variant is structurally and functionally conserved with human IMPDH1
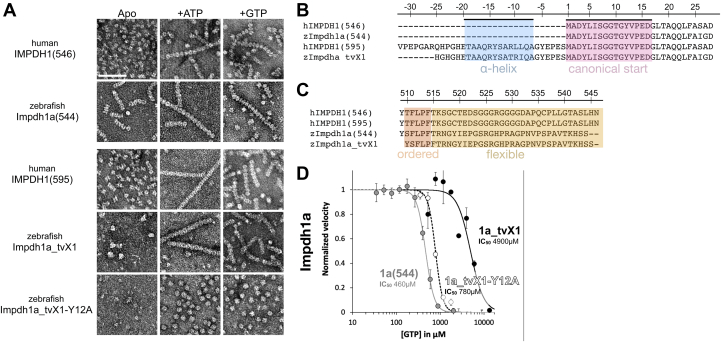
Structural features and filament assembly properties are broadly conserved between zebrafish Impdh1a (544) and human IMPDH1 (9) (Fig. 2A). Negative stain electron microscopy reveals that, like human IMPDH1, addition of ATP or GTP stimulates filament assembly of Impdh1a (544) and Impdh1a_tvX1. Both ATP and GTP drive assembly of filaments made of stacked octamers (6, 33, 34). Previous studies have shown that IMPDH octamers are in an extended conformation in the active state, and that GTP binding induces a compressed and inactive conformation (3, 9). Similarly, both zebrafish Impdh1a (544) and Impdh1a_tvX1 filaments are in the active/extended state with a rise 111 Å and 107 Å, respectively, when bound to ATP or the inhibited/compressed state with a rise of 86 Å and 100 Å, respectively, when bound to GTP. At this resolution, the zebrafish protein was very similar to the human retinal variants. The designed nonassembly point mutation Y12A breaks assembly of both IMPDH1a variants (9, 35, 36).
Figure 2.
Structural and functional conservation between human and zebrafish IMPDH1.A, negative stain EM of purified human retinal variants IMPDH1(546) & IMPDH1(595) and zebrafish Impdh1a (544) & Impdh1a_tvX1. The scale bar represents 100 nm. B and C, sequence alignment of human retinal variants IMPDH1(546) and IMPDH1(595), zebrafish Impdh1a (544) NM_001002177.1, and zebrafish retinal variant Impdh1a_tvX1 (XM_005159007.4). B, conserved N-terminal alpha-helix in blue and gene beginning in pink. C, residues in orange (ordered) are resolved in cryo-EM structures of the human protein (9), remainder of C-terminus in yellow is flexible and not resolved in structures (9). D, GTP-inhibition curves of zebrafish Impdh1a (544) (solid gray line), Impdh1a_tvX1 (solid black line), and nonassembly Y12A protein (dashed black line). Each data point represents a triplicate and the error bars are standard deviation. impdh, inosine monophosphate dehydrogenase; tv, transcript variant.
Human IMPDH1 retinal variants include N- and C-terminal extensions that play significant roles in modulating GTP-feedback sensitivity and structure (9). Zebrafish Impdh1a (544) includes the C-terminal extension but not the N-terminal extension, whereas Impdh1a_tvX1 has both (Fig. 2, B and C). Critical amino acids in both termini of the human retinal variants are conserved in the zebrafish proteins (Fig. 2, B and C). Human IMPDH1 forms polymorphic filaments, whereas the N-terminal helix in the retinal spice variant IMPDH1(595) controls filament assembly and only permits one filament architecture that resists GTP inhibition (9). This N-terminal helix is conserved in the zebrafish Impdh1a_tvX1, and we hypothesize the helix plays the same role in controlling assembly of the zebrafish protein. In humans, the C-terminal extension decreases sensitivity to GTP, but the mechanism of action is still unknown. The C-terminal end is conserved between human and zebrafish, which suggests a conserved function.
We found that zebrafish Impdh1a_tvX1 resists GTP inhibition in a similar fashion to the human retinal variants (9) (Fig. 2D). Zebrafish Impdh1a has an IC50 for GTP of 460 μM, whereas Impdh1a_tvX1 is more than tenfold less sensitive to GTP, with an IC50 of 4900 μM. When the nonassembly mutation Y12A is introduced to Impdh1a_tvX1, the IC50 for GTP drops to near Impdh1a levels (780 μM). This pattern is similar to the human IMPDH1 retinal variant, in which the effect of the N-terminal extension on decreasing GTP sensitivity is dependent on filament assembly (9).
Impdh1a_tvX1 is expressed exclusively in rods and cones and forms filaments
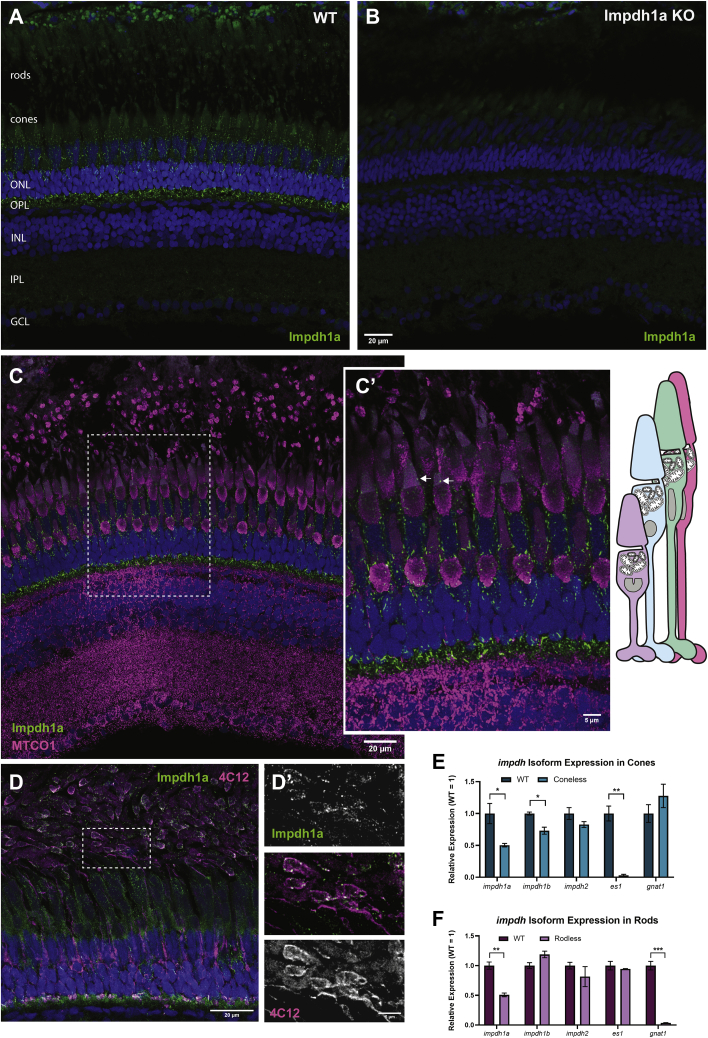
Using our antibody to the C-terminus of Impdh1a_tvX1, we next analyzed the cellular distribution of protein in the retina. We used immunohistochemistry (IHC) and high-resolution confocal imaging to localize Impdh1a_tvX1 in WT zebrafish retinas. As in Western blots, our antibody was highly specific and labeled WT retinas (Fig. 3A) but not retinas from a fish strain with a splice site mutation, impdh1asa23234 and consequently no IMPDH1a protein (Fig. 3B; also see Fig. 1E; (32)).
Figure 3.
Impdh1a_tvX1 is expressed exclusively in rods and cones.A, representative IHC images showing Impdh1a staining of WT and (B) KO adult zebrafish retina at 11:00 AM using the C-terminal antibody. C, Impdh1a (green) and MTCO1 (magenta) immunostaining of WT-pigmented retina. The nuclei are blue. C′, magnified section of image in (C) showing Impdh1a filament localization in cones. The arrows indicate where Impdh1a filaments appear to form in the OS. Note the tiering of photoreceptors normal in adult zebrafish retina. D, representative Impdh1a staining (green) and 4C12 (magenta) of bleached retina showing the localization of Impdh1a with known zebrafish rod marker. The nuclei are stained blue. D′, magnified section of (D) showing overlap of Impdh1a with 4C12 (rod) staining. E, qPCR quantification of impdh1a, 1b, and 2 transcripts in coneless retina and (F) qPCR analysis of relative impdh1a, 1b, and 2 expression in rodless retina. es1 and gnat1 are cone and rod specific genes, respectively. N = 3 animals and the error bars are standard error. IHC, immunohistochemistry; impdh, inosine monophosphate dehydrogenase. ns p > 0.05, ∗ p ≤ 0.05 , ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001.
In WT retinas, Impdh1a_tvX1 is expressed only in rods and cones (Fig. 3, C and D). In both rods and cones, filaments are easily detected in the cell body and synapse and in outer segments of cones although at much lower levels (arrow pointing to outer segment filament, Fig. 3C'). The retina shown in Figure 3, D and D′ is bleached, so filaments in rod cell bodies (co-stained with rod marker 4C12 (magenta), and outlined with dotted white box) are visible and not hidden by melanin within the retinal pigment epithelium.
To quantify the relative expression levels of impdh1a_tvX1 in rods and cones, we used zebrafish mutants that lack cones (37) or rods due to degeneration (38). Analysis by qPCR at a single time point determined that ∼50% of the retina’s impdh1a transcript and ∼27% of the retina’s impdh1b transcript is expressed in cones (Fig. 3E). As expected, the remaining ∼50% impdh1a expression is in rods (Fig. 3F). An insignificant amount of impdh2 is expressed in either rods or cones. Although the relative abundance in each cell type may change throughout the day, we conclude that Impdh1a_tvX1 is robustly expressed in both rod and cone photoreceptors, consistent with our ability to easily detect filaments in both cell types.
Impdh1a filaments are dynamic
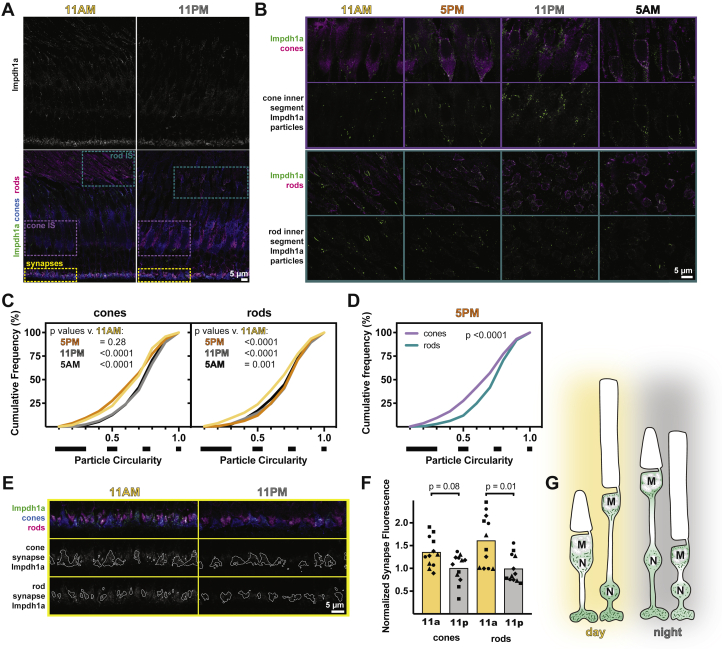
The retinal variant of human IMPDH1 and zebrafish Impdh1 (Impdh1a_tvX1) stabilizes a filament conformation that resists allosteric inhibition by GTP (Fig. 2 and (9)). This suggests that filaments may dynamically localize to regions of high GTP demand. To analyze this, we quantified filament length throughout the inner segment (IS) portion of the cell at four different times of the day, 11 AM, 5 PM, 11 PM, and 5 AM, in both rods and cones. For the synapse, we quantified the overall change in fluorescence intensity at these timepoints because the high concentration of Impdh1a_tvX1 in this region of the cell made it difficult to resolve filaments even with image deconvolution. We distinguished rods from cones using the 4C12 antibody (rods) and a transgenic strain expressing GFP specifically in cone photoreceptors (Fig. 4A and Table 2).
Figure 4.
Impdh1a localization in photoreceptors is dynamic.A, representative IHC images of zebrafish outer retina at 11 AM and 11 PM stained with antibodies against Impdh1a (top white and bottom green) and colabeled (bottom panels) with transgenic cone-targeted GFP (blue) and rods (4C12, magenta). B, detailed IHC images of photoreceptor inner segments throughout the day. Impdh1a (green) counterstained with cone or rod markers (magenta). C, cumulative frequency distributions of particle circularity ratios in rods and cones throughout the day. N = 4 animals, three experiments each. D, comparison of particle circularity ratios in cones and rods at 5 PM. E, detailed IHC images of photoreceptor synapses at 11 AM and 11 PM. Top, Impdh1a (green) counterstained with cone (blue) or rod (magenta) markers. Middle and bottom, Impdh1a signal overlapping with cone or rod synapse masks, respectively. F, quantification of mean synapse fluorescence in cones and rods at 11 AM and 11 PM, normalized to 11 PM. Each symbol is a different animal, three experiments for each animal. G, hypothesized model for Impdh1a dynamics throughout the day. In daytime, longer Impdh1a filaments occupy the inner segments, surrounding mitochondrial clusters (M) and nuclei (N). More subtle changes occur at synapses, with slightly reduced Impdh1a immunoreactivity at night. IHC, immunohistochemistry; impdh, inosine monophosphate dehydrogenase.
Table 2.
Antibodies/dyes
| Name | Target | Species | Clonality | Manufacturer | Catalog # | IHC dilution | WB dilution |
|---|---|---|---|---|---|---|---|
| Impdh1aC | Impdh1a C-terminus | Rabbit | in-house | N/A | 1:5000 | 1:500 | |
| Impdh1aN | Impdh1a N-terminus | Rabbit | in-house | N/A | N/A | 1:500 | |
| 4C12 | Unknown rod epitope | Mouse | mAb | Fadool lab | gift | 1:200 | 1:200 |
| MTCO1 | complex IV mitochondria | Mouse | mAb | Abcam | ab14705 | 1:1000 | 1:1000 |
| GFP | Transgenic GFP | Chicken | pAb | Abcam | ab13970 | 1:5000 | N/A |
| HOESCHT | nuclei | Invitrogen | H3569 | 5 μM | N/A |
We detected Impdh1a_tvX1 in the IS of both rods and cones at all timepoints (Fig. 4B). In cones, Impdh1a_tvX1 aggregates were more elongated (filamentous) during the day and more rounded (circular) at night (Fig. 4C, left). In contrast, rod ISs had smaller changes in filament shape throughout the day (Fig. 4C, right), with the most elongated filaments at 5 PM. However, even at 5 PM, the filaments in rods tended to have higher circularity than in cones, indicating an overall increase in filament length in cone ISs compared with rod ISs (Fig. 4D). At the synapse, the overall fluorescence intensity of Impdh1a_tvX1 decreased at night compared with daytime in both rods and cones (Fig. 4, E and F). These results are summarized in a schematic in Figure 4G.
Loss of Impdh1a significantly alters nucleotide homeostasis but not cellular structure in retina
Given the abundance and selectivity of Impdh1a_tvX1 in zebrafish photoreceptors, we hypothesized an important role for this enzyme. To determine this, we analyzed the essential splice site mutant Impdh1asa23234 (32), which lacks the Impdh1a protein both in Western blots and in IHC (see Figs. 1E and 3B). Thus, the sa23234 allele generates a gene knock-out (impdh1a KO). We sequenced the Impdh1asa23234 transcript and found, as predicted, that an intron was retained leading to a premature stop codon (Fig. 5, A and B). Further, consistent with previous studies demonstrating negative feedback regulation of IMPDH transcription (39), we found a significant upregulation of Impdh1asa23234 transcript in homozygotes (Fig. 5C). In contrast, impdh1b and impdh2 expression were not substantially altered by the loss of Impdh1a protein (Fig. 5, D and E).
Figure 5.
Loss of impdh1a does not alter the expression of impdh1b or impdh2 and does not cause photoreceptor degeneration.A, schematic of impdh1a KO mutation, a G > T mutation at essential splice site between exon 5 and 6 resulting in a premature stop codon 11 amino acids after exon 5. B, primary sequence of impdh1a_tvX1 compared with impdh1a KO. Blue marks start of retained intron, purple marks premature stop codon in impdh1a KO. qPCR analysis of (C) impdh1a, (D) impdh1b, and (E) impdh2 transcripts at 11:00 AM in WT, HET, and impdh1a KO retinas. N = 6 for WT and heterozygous animals and N = 5 for KO animals. F, representative IHC images of 18 month old impdh1a WT and KO retinas with mitochondria (MTCO1) stained magenta. Cone photoreceptors express eGFP under the cone transducin promotor (TaCP:eGFP) and are stained with eGFP antibody to visualize cones (green). The nuclei are stained in blue. The images are maximum intensity projections of 20 stacks 0.3 μm per step. G, control demonstrating lack of Impdh1a protein at 18 months in impdh1a KO retinas. H, mitochondrial (MTCO1) staining of impdh1a WT and KO retinas showing normal mitochondrial localization and morphology in the KO retina at 18 months. I, cone nuclei quantification and (J) outer nuclear layer thickness (rod nuclei) comparing impdh1a WT and KO zebrafish retinas at 18 months. K, rod- (gnat1) and cone- (es1) specific transcripts in WT compared with KO. impdh2 transcript, which is not found in rod or cones, used as a control. N = 5 animals for WT and N = 3 animals for KO. L, whole retina mtDNA copy number after feeding or after 18 to 24 h fasting; N = 3 for all the samples. M, representative TEM images of 18 month old WT and KO retinas showing high resolution morphology of retinal layers (left and middle panel) and mitochondria (right) in 18 months impdh1a WT and KO retinas. impdh, inosine monophosphate dehydrogenase; tv, transcript variant. ns p > 0.05, ∗ p ≤ 0.05.
The retinal layers were intact and photoreceptor size and organization appeared normal in the KO compared with WT at 18 months (Fig. 5, F and M, left and middle panels). As a control, we verified that Impdh1a protein remained absent in the KO at this age (Fig. 5G). Cone photoreceptor number and outer nuclear layer (rod layer) thickness were not significantly different between WT and KO (Fig. 5, I and J). This is consistent with near normal expression of a single rod and cone-specific transcript in KO compared with WT retina (Fig. 5K).
Because mitochondrial DNA replication continues in differentiated cells independent of nuclear DNA replication, we carefully analyzed photoreceptor mitochondrial morphology in IHC and transmission electron microscopy (TEM) in the KO compared with WT. Overall photoreceptor mitochondrial distribution, morphology, and cristae looked normal (Fig. 5, H and M, right). We also did not detect changes in mitochondrial DNA copy number using qPCR in either fed or fasted animals (Fig. 5L).
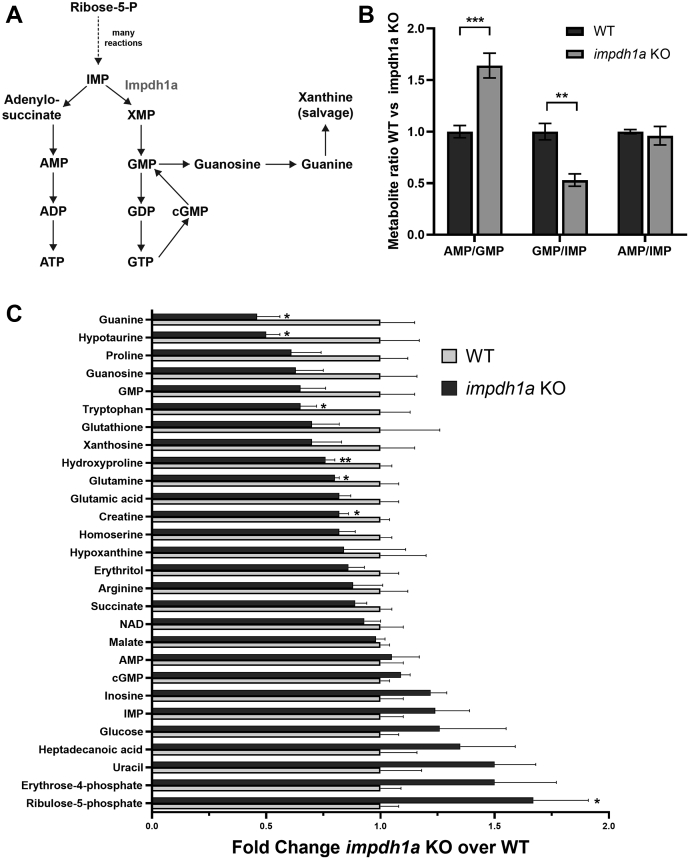
To determine whether there was an imbalance in purine nucleotide synthesis pathways (Fig. 6A), we compared total metabolite levels in retinas from impdh1a KO with WT (Fig. 6, B and C). We found a striking imbalance in purine nucleotides consistent with the predominant expression of this rate limiting enzyme. Guanine levels were reduced by more than 2-fold, but remarkably the steady state cGMP levels under ambient illumination were not affected. Increases in ribulose 5-phosphate also suggest that nucleotide synthesis pathways are disrupted because of the loss of impdh1a.
Figure 6.
Steady state guanine levels are dramatically reduced in impdh1a KO retinas.A, a schematic of purine nucleotide synthesis pathway. B, the ratio of metabolites directly connected to Impdh enzyme activity. C, quantification from LC/MS/MS analysis of a subset of relevant metabolites and metabolites at 11:00 AM with significant changes between WT and impdh1a KO. The experiment was repeated twice, total N = 5 or 6 animals, and the error bars are standard error. impdh, inosine monophosphate dehydrogenase. ns p > 0.05, ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001.
Discussion
The main findings from our study are the following: (1) Impdh1a_tvX1 is the most abundant impdh transcript in zebrafish retina, (2) key structural and biochemical features are conserved between the zebrafish and human predominant retinal isoforms, (3) in retina, Impdh1a_tvX1 is only present in rods and cones where it forms abundant and dynamic filaments, and (4) loss of Impdh1a function significantly alters nucleotide balance in retina but does not cause degeneration. Zebrafish are firmly established as a powerful genetic model with highly conserved photoreceptor physiology (40, 41). Our new and significant findings presented here provide the foundation to genetically dissect the cause of IMPDH1-induced retinopathy using zebrafish.
IMPDH transcripts
Based on previous work (8, 36, 42), we predicted that zebrafish photoreceptors primarily express Impdh1a, however, none of these studies determined which transcript variant predominates. We sought to identify the specific transcript expressed because tissue and cell-type specific functions often depend on the expressed isoform (43, 44). Gene duplications, which are common in zebrafish, provide gene specialization (subfunctionalization) (45). Our findings presented in Figures 1 and 3 show that for the study of Impdh1 in the retina, the zebrafish offers excellent cell class specificity of gene expression; we detect a predominant single transcript variant (X1) of impdh1a that is expressed specifically in rods and cones. In contrast, the impdh1b gene is more broadly expressed throughout the zebrafish (36) and at lower levels in the retina. We also confirm that in the retina, the Impdh1 paralogs show alternating circadian patterns of expression with nearly tenfold daily changes, whereas Impdh2 does not (8). Although protein levels showed a similar trend, the changes are smaller than for mRNA consistent with the complicated temporal and spatial relationships between protein and mRNA levels (46).
Structural features of IMPDH
We have demonstrated that key structural and functional properties of IMPDH1 splice variants are conserved between the well-characterized human enzyme and zebrafish. Human IMPDH1 assembles polymorphic filaments upon binding the allosteric effectors ATP and GTP (9, 46). These structural features are conserved in zebrafish Impdh1a and Imph1a_tvX1, and the point mutation Y12A disrupts assembly in both species, suggesting that the assembly mechanism is also conserved (Fig. 2A). In humans, IMPDH1 retinal splice variants have specific structural features in the N- and C-terminal splice extensions that reduce sensitivity to GTP inhibition. These sequences are conserved in zebrafish, with the C-terminal extension part of Impdh1a (544) and the N-terminal extension added in the splice variant Impdh1a_tvX1 (Fig. 2, B and C). In humans, the effect of the N-terminal extension in reducing GTP sensitivity is dependent on filament assembly, as it functions to bias assembly into a hyperactive filament conformation (9). To test conservation of this property, we introduced the nonassembly Y12A mutation in Impdh1a_tvX1 and found that this increased sensitivity to GTP to the level of Impdh1a, which lacks the N-terminal extension, indicating that the effect of the N-terminal extension is dependent on polymerization of the zebrafish enzyme (Fig. 2D). Thus, the regulation of Impdh1a_tvX1 by polymerization and N- and C-terminal extensions mirrors their effects in human IMPDH1, bolstering the case for the zebrafish retina as a relevant and appropriate model to study the role of IMPDH1 in the retina.
IMPDH filaments
Impdh1a_tvX1 forms robust filaments in both rods and cones (Figs. 3 and 4). This is consistent with recent findings in mice and zebrafish (22, 36, 47) although Impdh filaments, as measured by the appearance of aggregates, were only detected in mouse photoreceptors after exposure to bright light and then only in outer segments and not elsewhere in the cell (22). This increase in filaments within the outer segment correlated with elevated cGMP synthesis in the whole retina and led Plana-Bonamaisó et al. (22) to propose that filaments in photoreceptor outer segments are required for continued cGMP production during phototransduction. Recent work with the human retinal variant and our work presented here on the zebrafish retinal variant (Fig. 2) show that filaments favor a conformation that resists GTP inhibition, which would be predicted to increase guanine synthesis (9).
Here, we detailed filament dynamics during normal day-night light cycles (Fig. 4). Our results show that under normal lighting conditions, few filaments were detected in outer segments. In contrast, the filaments are abundant in synapses and throughout the cell body, around both the nucleus and mitochondria in both rods and cones. These results suggest an important role for nucleotide synthesis throughout the cell, consistent with the role of guanine nucleotides in many cellular processes and the robust vesicular trafficking characteristic of photoreceptors. In Xenopus laevis, 80 million rhodopsin molecules are transported/day from the inner to outer segment (48), and synaptic vesicle release in photoreceptors is graded thus requiring large numbers of vesicles (49). However, the recycling of guanine nucleotides that occurs in these processes and in phototransduction might not require significant new synthesis of guanine. Filament formation could also provide a scaffold to organize signaling molecules (50). Increased filaments in cones during the day could support more guanine synthesis and is consistent with the extended photopic range of cones compared with rods (51). Reduced filament signals at night at the synapse could reflect the known disassembly of the ribbon that occurs in zebrafish cone pedicles (52).
Metabolic role of IMPDH
The overall morphology of the Impdh1a KO retina using IHC and TEM appeared largely normal (Fig. 5). We demonstrate that this is not because of increased transcription of either impdh1b or impdh2. This suggested that either impdh1a is not responsible for significant guanine production in photoreceptors or that although guanine production is perturbed in the KO, the photoreceptors are able to regulate the key signaling molecule associated with cell degeneration, cGMP. To analyze the importance of impdh1a contribution to guanine production, we conducted metabolomic analyses comparing WT and impdh1a KO retinas. We detected large changes in guanine in KO retinas compared with WT, consistent with a significant role for Impdh1a_tvX1 in de novo purine synthesis in photoreceptors, but notably cGMP levels were normal (Fig. 6). We also found no change in mtDNA copy number due to the reduced guanine levels in the KO, even when guanine was potentially removed as a food source in fasted animals. Previous work in mice found that the loss of Impdh1 also did not cause significant loss of photoreceptors at least through 10 months, although ERGs revealed a reduction in the dark-adapted a-wave responses starting at 5 months (53). A careful physiological analysis done at different ages under different illumination conditions may also reveal subtle changes in the photoresponse in the zebrafish KO. The increase in ribulose-5-phosphate may reflect a change in nucleotide metabolism from the pentose phosphate pathway. The key enzymes such as phosphoribosylpyrophosphate synthetase and amidophosphoribosyltransferase are inhibited by both guanine and adenine containing nucleotides (54). Future experiments analyzing metabolic flux with different fuels will dissect the regulation of purine synthesis in photoreceptors.
Conclusion
Our study provides a detailed evaluation of the main IMPDH1 transcript/protein in zebrafish retina. We have determined that the zebrafish protein shares structural and functional similarity with the human protein expressed in the retina. This foundational study provides essential new information required to exploit zebrafish for dissecting purine metabolism in normal and diseased photoreceptors.
Experimental procedures
Zebrafish maintenance and retina collection
Zebrafish were maintained and used in accordance with the guidelines of experimental protocols approved by the IACUC of the University of Washington in Seattle (protocol number: 3113-02). The strains used were AB, AB/roya9 (55), the transgenic strain Tg(gnat2:EGFP) (56) and impdh1asa23234, which was obtained from the Zebrafish International Research Center. For experiments with impdh1asa23234, controls were sibling WT fish. impdh1asa23234 fish were identified by genotyping (Table 1). Coneless and rodless strains were Tg(gnat2:mcu-2A-RFP)w248Tg (37) and rhofl9 (38), respectively. To ensure that all cones had degenerated Tg(gnat2:mcu-2A-RFP)w248Tg, fish were between 12 and 18 months old. Males and females were used in equal numbers for all experiments, and no differences due to sex were noted. Fish ages were between 4 and 12 months old unless stated otherwise.
Fish were maintained in the University of Washington ISCRM aquatics facility at 27.5 °C on a 14-h/10-h light/dark cycle, with broad-spectrum white light and daily feedings around 10 AM, 12 PM, and 5 PM 18 to 24 h before experimental timepoints, the fish siblings were fasted to minimize effects of feeding on Impdh distribution and nucleotide metabolism. The fish were euthanized in an ice bath after cervical dislocation and enucleation; for dark timepoints, euthanasia and dissections were performed under infrared light.
Immunohistochemistry
Eyes from transgenic adult zebrafish in the AB or AB/roya9 background were placed into fixative (4% paraformaldehyde in PBS, pH 7.3) at room temperature (rt), pierced across the cornea with a needle, and the vitreous cavity was flushed with fixative. After incubation for 1 to 2 h at rt, the eyes were stored for up to 48 h at 4 °C, rinsed in PBS, and then cryoprotected in a 20% sucrose gradient overnight. For experiments measuring rod photoreceptors, the eyes were bleached with 10% H2O2 in PBS overnight at 37 °C before sucrose gradient to remove pigmentation of the retinal pigment epithelium. The following day, the anterior halves of the eye were dissected away, and eyecups were embedded and frozen in OCT cryomolds. The eyecups were cryosectioned at 14 μm; for time point analyses, the sections from all animals were arranged on slides for parallel staining and analysis. The sections were washed in PBS, then blocked for 30 min in blocking buffer (2% normal donkey serum, 2 mg/ml bovine serum albumin, and 0.3% Triton X-100 in PBS). Primary antibodies diluted in the blocking buffer were then applied to cryosections overnight at 4 °C. The antibodies and stains are reported in Table 2. Secondary antibodies conjugated to AlexaFluor dyes were diluted at 1:2000 in blocking buffer and incubated on sections for 1 h in darkness at RT.
IHC imaging and analysis
Imaging was performed on a Leica SP8 confocal microscope with a 20× or 63× oil objective and 12 bit depth using Leica LAS-X software (RRID:SCR_013673). For quantitative imaging with the 63× objective, Z-stack images of photoreceptor regions 100 to 200 μm from the optic nerve were acquired every 0.3 μm at 5000 × 625 pixel resolution for synapses and 4096 × 2048 pixel resolution for inner segments. The Z-stacks from each animal and region were blindly analyzed using Image J software (RRID:SCR_002285). For all, the cone and rod Impdh1a signals were isolated by creating masks with either cone Tg(gnat2:eGFP) (RRID:ZDB-GENO-070829-1, (55)) or the rod marker 4C12 (ZDB-ATB-090506-2). Three technical replicates of staining and imaging were performed for each animal. Representative images were postprocessed with Leica Lightning deconvolution; brightness and contrast were adjusted equally to ease visualization.
For inner segments
Three micrometer Z depth (ten frames) was projected at maximum intensity, then cone and rod Impdh1a signals were isolated. The binary Impdh1a images were created to include pixels with intensities greater than half-maximum, a Gaussian blur (radius 2) was applied, and Impdh1a particles larger than 20 pixels2 were selected. Particle dimensions were measured, and circularity was calculated using the following equation: Circularity ratio = Minor axis/Major axis. Cumulative frequency distributions of particle circularity were compared using Kolmogorov–Smirnov tests.
For synapses
Cone and rod Impdh1a signals were isolated from a single frame where GFP, 4C12, and Impdh1a were clearly visualized. Mean Impdh1a fluorescence intensity was measured for rods and cones separately. Statistical analyses were performed using ANOVA with Kruskal–Wallis multiple comparisons.
Data analysis
Data were processed using Microsoft Excel (RRID:SCR_016137); statistical tests were performed using GraphPad Prism (RRID:SCR_002798).
For measurements of cone nuclei and outer nuclear layer thickness
Retinas from 18-month-old WT and impdh1a KO zebrafish were dissected, sectioned, and stained with 5 μM HOESCHT to visualize nuclei. For each retina, the outer nuclear layer and cone nuclei were measured from 185 μm2 fields in triplicate under 63× magnification. The stack for each field was generated from maximum intensity projection of 20 stacks, 0.3 μM apart (6 μM). The cone nuclei were manually counted, and outer nuclear layer (rod nuclear layer) thickness was quantified using Image J.
Antibodies and Western analysis
The company Pacific Immunology (https://www.pacificimmunology.com) generated polyclonal antibodies in rabbits from the following N and C terminal sequences (N terminus ERYVDGDREGYQIDYRRIVGD-Cys and C terminus Cys-TRNGYIEPGSRGHPRAGPNVPSPAVTKHSS). The antibodies were provided affinity purified. Western blots were conducted, as previously described (57). After Western blot transfer, the membranes were stained with Ponceau S (Sigma #P7170), washed with water, and imaged as a loading control.
Transmission electron microscopy
TEM preparation and imaging was done, as previously described (37).
Metabolomics
Fish were fasted for 18 to 24 h. The retinas were rapidly dissected from eyes, rinsed in Krebs–Ringer bicarbonate buffer (98.5 mM NaCl, 4.9 mM KCl, 1.2 mM KH2PO4, 1.2 mM MGSO4–7H2O, 20 mM Hepes, 2.6 mM CaCl–2H2O, and 25.9 mM NaHCO3) and then immediately frozen. The retina samples were homogenized in 140 μl cold 80% methanol (methanol:water (80:20 V/V)) using a microtube homogenizer. The samples were stored on dry ice for 30 min, then centrifuged at 15,000 RPM for 15 min. The supernatant was dried by the gel pump (Savant), and then the dried extract was reconstituted with 100 μl of mobile phase (a mixture of A:B at 30:70 in V/V) for targeted metabolomics, as reported (58, 59). The metabolite extracts were analyzed by a Shimadzu LC Nexera X2 UHPLC coupled with a QTRAP 5500 LC-MS/MS (AB Sciex). An ACQUITY UPLC BEH Amide analytic column (2.1 × 50 mm, 1.7 μm) (Waters Corp) was used for chromatographic separation. The mobile phase was (A) water with 10 mM ammonium acetate (pH 8.9) and (B) acetonitrile/water (95/5) with 10 mM ammonium acetate (pH 8.2) (All solvents were LC–MS Optima grade from Fisher Scientific). The total run time was 11 min with a flow rate of 0.5 ml/min and an injection volume of 5 μl. The gradient elution was 95 to 61% B in 6 min, 61 to 44% B at 8 min, 61 to 27% B at 8.2 min, and 27 to 95% B at 9 min. The column was equilibrated with 95% B at the end of each run. The source and collision gas was N2. The ion source conditions in positive and negative mode were as follows: curtain gas (CUR) = 25 psi, collision gas (CAD) = high, ion spray voltage (IS) = 3800/−3800 V, temperature (TEM) = 500 °C, ion source gas 1 (GS1) = 50 psi, and ion source gas 2 (GS2) = 40 psi. Each metabolite was tuned with standards for optimal transitions. D4-nicotinamide (Cambridge Isotope Laboratories) was used as an internal standard. The extracted MRM peaks were integrated using MultiQuant 3.0.3 software (AB Sciex).
qRT-PCR and identification of transcripts
Extraction of RNA, cDNA synthesis, and qRT-PCR amplification were conducted, as previously described using TATA-box binding protein as a reference gene (37, 60). The primers are listed in Table 1. For impdh1b amplification, primer set 1 was used for Figures 1, A and B and 5C, and primer set 2 was used for Figures 3, F and G and 5C. Mitochondrial DNA quantification was performed, as described (61).
Protein purification
Purified Impdh protein was prepared, as described previously (35). BL21(DE3) E. coli transformed with a pSMT3-Kan vector expressing N-terminal 6xHis-SMT3/SUMO-tagged Impdh were cultured in LB at 37 °C until reaching an A600 of 0.9 then induced with 1 mM IPTG for 4 h at 30 °C and pelleted. The remainder of the purification was performed at 4 °C. The pellets were resuspended in lysis buffer (50 mM KPO4, 300 mM KCl, 10 mM imidazole, and 800 mM urea, pH 8) and lysed with an Emulsiflex-05 homogenizer. The lysate was cleared by centrifugation and SUMO-tagged IMPDH chromatographically purified with HisTrap FF columns (GE Healthcare Life Sciences) and an Äkta Start chromatography system. After on-column washing with lysis buffer and elution (50 mM KPO4, 300 mM KCl, and 500 mM imidazole, pH 8), the peak fractions were treated with 1 mg ULP1 protease (62) per 100 mg IMPDH for 1 h at 4 °C, after the addition of DTT to 1 mM and urea to 800 mM. Protein was then concentrated using a 30,000 MWCO Amicon filter and subjected to size-exclusion chromatography using Äkta Pure system and a Superose 6 column preequilibrated in filtration buffer (20 mM Hepes, 100 mM KCl, 800 mM urea, and 1 mM DTT, pH 8). The peak fractions were concentrated using a 10,000 MWCO Amicon filter, then flash-frozen in liquid nitrogen and stored at −80 °C.
IMPDH activity assays
Protein aliquots were diluted in the activity buffer (20 mM Hepes, 100 mM KCl, and 1 mM DTT, pH 7.0). The reactions (100 μl total) were performed with 1 μM protein, 1 mM ATP, 1 mM IMP, 300 μM NAD+, and varying GTP in 96 well UV transparent plates (Corning model 3635). NADH production was measured by optical absorbance (340 nm) in real-time using a Varioskan Lux microplate reader (Thermo Scientific) at 25 °C, 1 measurement/min, for 15 min; the absorbance was correlated with NADH concentration using a standard curve. Each data point was performed in triplicate, and averages are graphed with standard deviation error bars.
Negatively stained electron microscopy
Protein preparations were applied to glow-discharged continuous carbon EM grids and negatively stained with 2% uranyl formate. The grids were imaged by transmission electron microscopy using a FEI Morgagni at 100 kV acceleration voltage and a Gatan Orius CCD. Micrographs were collected at a nominal 22,000× magnification (pixel size 3.9 Å).
Data availability
Raw data used to generate graphs are presented in supporting Excel files 1 through 6.
Supporting information
This article contains supporting information (Supplemental Data Figures 1-6).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Stanley Kim for maintaining the SLU fish facility.
Author contributions
W. M. C., A. L. B., J. M. K., and S. E. B. conceptualization; W. M. C., A. L. B., M. M. G., D. C. B., J. M. K., and S. E. B. formal analysis; W. M. C., A. L. B., M. M. G., D. C. B., Y. W., Z. S. C., and J. D. investigation; S. E. B. writing–original draft; W. M. C., A. L. B., M. M. G., D. C. B., Y. W., and J. M. K. writing–review and editing; W. M. C., A. L. B., and M. M. G. visualization; W. M. C., J. M. K., and S. E. B. supervision; J. M. K. and S. E. B. project administration; A. L. B., J. M. K., and S. E. B. funding acquisition.
Funding and additional information
The research presented in this article was supported by NIH grants EY031546 (J. M. K. and S. E. B.), EY030732 (A. L. B.), EY031324 (J. D.), and EY001730 (UW Vision Core). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Enrique De La Cruz
Footnotes
Present address for Daniel C. Brock: Neurobiology, Neurodegeneration and Repair Laboratory, National Eye Institute, National Institutes of Health, MSC0610, 6 Center Drive, Bethesda, Maryland 20892, USA.
Supporting information
References
- 1.Buey R.M., Fernandez-Justel D., Marcos-Alcalde I., Winter G., Gomez-Puertas P., de Pereda J.M., Luis Revuelta J. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 2017;7:2648. doi: 10.1038/s41598-017-02805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buey R.M., Ledesma-Amaro R., Velazquez-Campoy A., Balsera M., Chagoyen M., de Pereda J.M., Revuelta J.L. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 2015;6:8923. doi: 10.1038/ncomms9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson M.C., Kollman J.M. Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. Elife. 2020;9 doi: 10.7554/eLife.53243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labesse G., Alexandre T., Gelin M., Haouz A., Munier-Lehmann H. Crystallographic studies of two variants of Pseudomonas aeruginosa IMPDH with impaired allosteric regulation. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1890–1899. doi: 10.1107/S1399004715013115. [DOI] [PubMed] [Google Scholar]
- 5.Labesse G., Alexandre T., Vaupre L., Salard-Arnaud I., Him J.L., Raynal B., Bron P., Munier-Lehmann H. MgATP regulates allostery and fiber formation in IMPDHs. Structure. 2013;21:975–985. doi: 10.1016/j.str.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Calise S.J., Carcamo W.C., Krueger C., Yin J.D., Purich D.L., Chan E.K. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell. Mol. Life Sci. 2014;71:2963–2973. doi: 10.1007/s00018-014-1567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carcamo W.C., Calise S.J., von Mühlen C.A., Satoh M., Chan E.K. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int. Rev. Cell Mol. Biol. 2014;308:35–74. doi: 10.1016/B978-0-12-800097-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Li G., Gorling B., Luy B., Du J., Yan J. Integrative analysis of circadian transcriptome and metabolic network reveals the role of de novo purine synthesis in circadian control of cell cycle. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrell A.L., Nie C., Said M., Simonet J.C., Fernández-Justel D., Johnson M.C., Quispe J., Buey R.M., Peterson J.R., Kollman J.M. IMPDH1 retinal variants control filament architecture to tune allosteric regulation. bioRxiv. 2021 doi: 10.1101/2021.08.03.454821. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senda M., Natsumeda Y. Tissue-differential expression of two distinct genes for human IMP dehydrogenase (E.C.1.1.1.205) Life Sci. 1994;54:1917–1926. doi: 10.1016/0024-3205(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 11.Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M., Bäckström A., Danielsson F., Fagerberg L., Fall J., Gatto L., et al. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 12.Yarfitz S., Hurley J.B. Transduction mechanisms of vertebrate and invertebrate photoreceptors. J. Biol. Chem. 1994;269:14329–14332. [PubMed] [Google Scholar]
- 13.Okawa H., Sampath A.P., Laughlin S.B., Fain G.L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008;18:1917–1921. doi: 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowne S.J., Sullivan L.S., Mortimer S.E., Hedstrom L., Zhu J., Spellicy C.J., Gire A.I., Hughbanks-Wheaton D., Birch D.G., Lewis R.A., Heckenlively J.R., Daiger S.P. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2006;47:34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozhevnikova E.N., van der Knaap J.A., Pindyurin A.V., Ozgur Z., van Ijcken W.F., Moshkin Y.M., Verrijzer C.P. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol. Cell. 2012;47:133–139. doi: 10.1016/j.molcel.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 16.McGrew D.A., Hedstrom L. Towards a pathological mechanism for IMPDH1-linked retinitis pigmentosa. Adv. Exp. Med. Biol. 2012;723:539–545. doi: 10.1007/978-1-4614-0631-0_68. [DOI] [PubMed] [Google Scholar]
- 17.McLean J.E., Hamaguchi N., Belenky P., Mortimer S.E., Stanton M., Hedstrom L. Inosine 5'-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem. J. 2004;379:243–251. doi: 10.1042/BJ20031585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortimer S.E., Hedstrom L. Autosomal dominant retinitis pigmentosa mutations in inosine 5'-monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem. J. 2005;390:41–47. doi: 10.1042/BJ20042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimer S.E., Xu D., McGrew D., Hamaguchi N., Lim H.C., Bowne S.J., Daiger S.P., Hedstrom L. IMP dehydrogenase type 1 associates with polyribosomes translating rhodopsin mRNA. J. Biol. Chem. 2008;283:36354–36360. doi: 10.1074/jbc.M806143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam L.C., Kiang A.S., Campbell M., Keaney J., Farrar G.J., Humphries M.M., Kenna P.F., Humphries P. Prevention of autosomal dominant retinitis pigmentosa by systemic drug therapy targeting heat shock protein 90 (Hsp90) Hum. Mol. Genet. 2010;19:4421–4436. doi: 10.1093/hmg/ddq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X.T., Mion B., Aherne A., Engel P.C. Molecular recruitment as a basis for negative dominant inheritance? Propagation of misfolding in oligomers of IMPDH1, the mutated enzyme in the RP10 form of retinitis pigmentosa. Biochim. Biophys. Acta. 2011;1812:1472–1476. doi: 10.1016/j.bbadis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Plana-Bonamaisó A., López-Begines S., Fernández-Justel D., Junza A., Soler-Tapia A., Andilla J., Loza-Alvarez P., Rosa J.L., Miralles E., Casals I., Yanes O., de la Villa P., Buey R.M., Méndez A. Post-translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. Elife. 2020;9 doi: 10.7554/eLife.56418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iribarne M., Masai I. Neurotoxicity of cGMP in the vertebrate retina: From the initial research on rd mutant mice to zebrafish genetic approaches. J. Neurogenet. 2017;31:88–101. doi: 10.1080/01677063.2017.1358268. [DOI] [PubMed] [Google Scholar]
- 24.Baye L.M., Patrinostro X., Swaminathan S., Beck J.S., Zhang Y., Stone E.M., Sheffield V.C., Slusarski D.C. The N-terminal region of centrosomal protein 290 (CEP290) restores vision in a zebrafish model of human blindness. Hum. Mol. Genet. 2011;20:1467–1477. doi: 10.1093/hmg/ddr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessieur E.M., Fogerty J., Gaivin R.J., Song P., Perkins B.D. The ciliopathy gene ahi1 is required for zebrafish cone photoreceptor outer segment morphogenesis and survival. Invest. Ophthalmol. Vis. Sci. 2017;58:448–460. doi: 10.1167/iovs.16-20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link B.A., Collery R.F. Zebrafish models of retinal disease. Annu. Rev. Vis. Sci. 2015;1:125–153. doi: 10.1146/annurev-vision-082114-035717. [DOI] [PubMed] [Google Scholar]
- 27.Liu F., Qin Y., Yu S., Soares D.C., Yang L., Weng J., Li C., Gao M., Lu Z., Hu X., Liu X., Jiang T., Liu J.Y., Shu X., Tang Z., et al. Pathogenic mutations in retinitis pigmentosa 2 predominantly result in loss of RP2 protein stability in humans and zebrafish. J. Biol. Chem. 2017;292:6225–6239. doi: 10.1074/jbc.M116.760314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minegishi Y., Sheng X., Yoshitake K., Sergeev Y., Iejima D., Shibagaki Y., Monma N., Ikeo K., Furuno M., Zhuang W., Liu Y., Rong W., Hattori S., Iwata T. CCT2 mutations evoke leber congenital amaurosis due to chaperone complex instability. Sci. Rep. 2016;6:33742. doi: 10.1038/srep33742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiwaki Y., Komori A., Sagara H., Suzuki E., Manabe T., Hosoya T., Nojima Y., Wada H., Tanaka H., Okamoto H., Masai I. Mutation of cGMP phosphodiesterase 6alpha'-subunit gene causes progressive degeneration of cone photoreceptors in zebrafish. Mech. Dev. 2008;125:932–946. doi: 10.1016/j.mod.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Raghupathy R.K., Zhang X., Liu F., Alhasani R.H., Biswas L., Akhtar S., Pan L., Moens C.B., Li W., Liu M., Kennedy B.N., Shu X. Rpgrip1 is required for rod outer segment development and ciliary protein trafficking in zebrafish. Sci. Rep. 2017;7:16881. doi: 10.1038/s41598-017-12838-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stearns G., Evangelista M., Fadool J.M., Brockerhoff S.E. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J. Neurosci. 2007;27:13866–13874. doi: 10.1523/JNEUROSCI.3136-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kettleborough R.N.W., Busch-Nentwich E.M., Harvey S.A., Dooley C.M., de Bruijn E., van Eeden F., Sealy I., White R.J., Herd C., Nijman I.J., Fényes F., Mehroke S., Scahill C., Gibbons R., Wali N., et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carcamo W.C., Satoh M., Kasahara H., Terada N., Hamazaki T., Chan J.Y., Yao B., Tamayo S., Covini G., von Muhlen C.A., Chan E.K. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Y., Gu J., Makhov A.M., Griffith J.D., Mitchell B.S. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J. Biol. Chem. 2006;281:206–212. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- 35.Anthony S.A., Burrell A.L., Johnson M.C., Duong-Ly K.C., Kuo Y.M., Simonet J.C., Michener P., Andrews A., Kollman J.M., Peterson J.R. Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell. 2017;28:2600–2608. doi: 10.1091/mbc.E17-04-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keppeke G.D., Chang C.-C., Antos C.L., Peng M., Sung L.-Y., Andrade L.E.C., Liu J.-L. IMPDH forms the cytoophidium in zebrafish. Dev. Biol. 2021;478:89–101. doi: 10.1016/j.ydbio.2021.05.017. [DOI] [PubMed] [Google Scholar]
- 37.Hutto R.A., Bisbach C.M., Abbas F., Brock D.C., Cleghorn W.M., Parker E.D., Bauer B.H., Ge W., Vinberg F., Hurley J.B., Brockerhoff S.E. Increasing Ca(2+) in photoreceptor mitochondria alters metabolites, accelerates photoresponse recovery, and reveals adaptations to mitochondrial stress. Cell Death Differ. 2020;27:1067–1085. doi: 10.1038/s41418-019-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zelinka C.P., Sotolongo-Lopez M., Fadool J.M. Targeted disruption of the endogenous zebrafish rhodopsin locus as models of rapid rod photoreceptor degeneration. Mol. Vis. 2018;24:587–602. [PMC free article] [PubMed] [Google Scholar]
- 39.Glesne D.A., Collart F.R., Huberman E. Regulation of IMP dehydrogenase gene expression by its end products, guanine nucleotides. Mol. Cell. Biol. 1991;11:5417–5425. doi: 10.1128/mcb.11.11.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoon M., Okawa H., Della Santina L., Wong R.O. Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res. 2014;42:44–84. doi: 10.1016/j.preteyeres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J.S., Kefalov V.J. The cone-specific visual cycle. Prog. Retin. Eye Res. 2011;30:115–128. doi: 10.1016/j.preteyeres.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higdon C.W., Mitra R.D., Johnson S.L. Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao X., Sinha S., Belcastro M., Woodard C., Ramamurthy V., Stoilov P., Sokolov M. Splice isoforms of phosducin-like protein control the expression of heterotrimeric G proteins. J. Biol. Chem. 2013;288:25760–25768. doi: 10.1074/jbc.M113.486258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy D., Cieply B., Carstens R., Ramamurthy V., Stoilov P. The musashi 1 controls the splicing of photoreceptor-specific exons in the vertebrate retina. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Fernández-Justel D., Núñez R., Martín-Benito J., Jimeno D., González-López A., Soriano E.M., Revuelta J.L., Buey R.M. A nucleotide-dependent conformational switch controls the polymerization of human IMP dehydrogenases to modulate their catalytic activity. J. Mol. Biol. 2019;431:956–969. doi: 10.1016/j.jmb.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 48.Papermaster D.S., Schneider B.G., DeFoe D., Besharse J.C. Biosynthesis and vectorial transport of opsin on vesicles in retinal rod photoreceptors. J. Histochem. Cytochem. 1986;34:5–16. doi: 10.1177/34.1.2934469. [DOI] [PubMed] [Google Scholar]
- 49.Sterling P., Matthews G. Structure and function of ribbon synapses. Trends Neurosci. 2005;28:20–29. doi: 10.1016/j.tins.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Simonet J.C., Burrell A.L., Kollman J.M., Peterson J.R. Freedom of assembly: Metabolic enzymes come together. Mol. Biol. Cell. 2020;31:1201–1205. doi: 10.1091/mbc.E18-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingram N.T., Sampath A.P., Fain G.L. Why are rods more sensitive than cones? J. Physiol. 2016;594:5415–5426. doi: 10.1113/JP272556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emran F., Dowling J.E. Larval zebrafish turn off their photoreceptors at night. Commun. Integr. Biol. 2010;3:430–432. doi: 10.4161/cib.3.5.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aherne A., Kennan A., Kenna P.F., McNally N., Lloyd D.G., Alberts I.L., Kiang A.S., Humphries M.M., Ayuso C., Engel P.C., Gu J.J., Mitchell B.S., Farrar G.J., Humphries P. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum. Mol. Genet. 2004;13:641–650. doi: 10.1093/hmg/ddh061. [DOI] [PubMed] [Google Scholar]
- 54.Becker M.A., Kim M. Regulation of purine synthesis de novo in human fibroblasts by purine nucleotides and phosphoribosylpyrophosphate. J. Biol. Chem. 1987;262:14531–14537. [PubMed] [Google Scholar]
- 55.D'Agati G., Beltre R., Sessa A., Burger A., Zhou Y., Mosimann C., White R.M. A defect in the mitochondrial protein Mpv17 underlies the transparent casper zebrafish. Dev. Biol. 2017;430:11–17. doi: 10.1016/j.ydbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennedy B.N., Alvarez Y., Brockerhoff S.E., Stearns G.W., Sapetto-Rebow B., Taylor M.R., Hurley J.B. Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Invest. Ophthalmol. Vis. Sci. 2007;48:522–529. doi: 10.1167/iovs.06-0975. [DOI] [PubMed] [Google Scholar]
- 57.Bisbach C.M., Hutto R.A., Poria D., Cleghorn W.M., Abbas F., Vinberg F., Kefalov V.J., Hurley J.B., Brockerhoff S.E. Mitochondrial Calcium Uniporter (MCU) deficiency reveals an alternate path for Ca(2+) uptake in photoreceptor mitochondria. Sci. Rep. 2020;10:16041. doi: 10.1038/s41598-020-72708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grenell A., Wang Y., Yam M., Swarup A., Dilan T.L., Hauer A., Linton J.D., Philp N.J., Gregor E., Zhu S., Shi Q., Murphy J., Guan T., Lohner D., Kolandaivelu S., et al. Loss of MPC1 reprograms retinal metabolism to impair visual function. Proc. Natl. Acad. Sci. U. S. A. 2019;116:3530–3535. doi: 10.1073/pnas.1812941116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B., Zhang T., Liu W., Wang Y., Xu R., Zeng S., Zhang R., Zhu S., Gillies M.C., Zhu L., Du J. Metabolic features of mouse and human retinas: Rods versus cones, macula versus periphery, retina versus RPE. iScience. 2020;23:101672. doi: 10.1016/j.isci.2020.101672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giarmarco M.M., Brock D.C., Robbings B.M., Cleghorn W.M., Tsantilas K.A., Kuch K.C., Ge W., Rutter K.M., Parker E.D., Hurley J.B., Brockerhoff S.E. Daily mitochondrial dynamics in cone photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28816–28827. doi: 10.1073/pnas.2007827117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Artuso L., Romano A., Verri T., Domenichini A., Argenton F., Santorelli F.M., Petruzzella V. Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio) Biochim. Biophys. Acta. 2012;1817:1002–1011. doi: 10.1016/j.bbabio.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 62.Mossessova E., Lima C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data used to generate graphs are presented in supporting Excel files 1 through 6.