Summary
Background
There is a critical need to better understand the mechanisms that drive local cell invasion and metastasis to develop new therapeutics targeting metastatic disease. Bioelectricity is an important mediator of cellular processes and changes in the resting membrane potential (RMP) are associated with increased cancer cell invasion. However, whether the RMP can be used to target invading cancer cells is unknown.
Methods
We employed both genetic and pharmacological manipulation of potassium channel activity and characterized the effects on breast cancer cell migration and invasion in vitro, and metastasis in an animal model of breast cancer.
Findings
Our data demonstrate that altering the RMP of triple-negative breast cancer (TNBC) cells by manipulating potassium channel expression increases in vitro invasion, in vivo tumour growth and metastasis, and is accompanied by changes in gene expression associated with cell adhesion.
Interpretation
We describe a novel mechanism for RMP-mediated cell migration involving cadherin-11 and the MAPK pathway. Importantly, we identify a new strategy to target metastatic TNBC in vivo by repurposing an FDA-approved potassium channel blocker. Our results demonstrate that bioelectricity regulates cancer cell invasion and metastasis which could lead to a new class of therapeutics for patients with metastatic disease.
Funding
This work was supported by the National Institutes of Health (R00-CA207866 to M.J.O.), Tufts University (Start-up funds from the School of Engineering to M.J.O., Tufts Collaborates Award to M.J.O. and M.L.), Allen Discovery centre program (Paul G. Allen Frontiers Group (12,171) to M.L.), and Breast Cancer Alliance Young Investigator Grant to M.J.O, Laidlaw Scholar funding to D.S. M.L. also gratefully acknowledges support of the Barton Family Foundation.
Keywords: Migration, Ion channel, Drug therapy, Cell invasion, Cell adhesion
Research in context.
Evidence before this study
While it is widely known in the field that bioelectric properties of cancer cells differ from their normal counterparts, we performed a literature search of primary peer-reviewed papers to gather evidence of the impact of potassium ion channel activity on cancer cell invasion and metastasis. Most studies of potassium channels have focused on its role in regulating tumour cell proliferation and cell cycle progression. We explored prior work using PubMed with the following terms: “potassium channel cancer,” “membrane potential cancer,” and “ion channels metastasis.” After determining that potassium channels were indeed known to influence cancer cell migration and that the link to resting membrane potential was largely unknown, we next searched for the clinical relevance of potassium channels and breast cancer. For this, we searched patient mRNA data using cBioPortal, an open-access online cancer database. From this search we determined that overexpression of potassium channels was indeed associated with triple-negative breast cancer.
Added value of this study
Most studies on the role of ions and ion channels focus on individual channels which are upregulated in cancer. Here, we instead focused on the whole cell changes in the bioelectric properties that all ion channels regulate. This removes the burden on finding the most important ion channel regulating these cellular properties and instead focuses on identifying strategies to target the cellular state. Furthermore, previous studies do not investigate the effect of potassium channel overexpression, which we see in triple-negative breast cancer patient tumours. Lastly, there are no published studies dissecting how whole-cell changes in bioelectric properties, which is a defining features of tumour cells, regulates gene expression.
Implications of all the available evidence
Our research demonstrates that alteration of breast cancer cells through overexpression of K+ channels leads to enhanced cell invasion, tumour growth, and metastasis. We also identify a novel hyperpolarization-driven mechanism of cell migration mediated by cadherin-11 and MAPK signalling. These data suggest that potassium channels are important in driving breast cancer invasion and metastasis, and, coupled with patient data showing potassium channels are upregulated in TNBC tumours, are a viable target for cancer therapies. Our data supports this implication by identifying a novel FDA-approved drug, amiodarone, that can decrease breast cancer cell migration in vitro and metastasis in vivo by targeting the bioelectric state of the cell. Approving new drugs for cancer treatment is expensive (up to 1 billion dollars) and lengthy (at least 10 years). Overall, 310 FDA-approved drugs have been implicated in regulating cancer behaviors, but to be prescribed by clinicians, these drugs would need to undergo additional small scale clinical trials. For this to occur, additional scientific evidence to support a certain class of drugs is necessary to help prioritize what drugs are likely to prove most beneficial to patients. Our data suggest that amiodarone may be a good candidate for further study.
Alt-text: Unlabelled box
Introduction
Despite identifying pro-metastatic cues and signalling pathways that drive invasion, a cellular process that supports tumour growth, dissemination, colonization, and ultimately metastasis, there are no clinically available treatments that directly target invading cells in cancer. As a result, metastasis remains the leading cause of death in cancer patients. The triple-negative breast cancer (TNBC) subtype accounts for approximately 15% of all breast cancer cases, and is associated with a poorer 5-year prognosis, increased likelihood of metastasis and shorter overall survival than the other subtypes.1,2 Several clinical trials are ongoing to identify new agents to treat TNBC, however current strategies benefit only a subset of patients and have limited efficacy. Therefore, there is an urgent need for more effective treatments. Targeting bioelectricity-mediated signalling pathways of tumour cells is an emerging strategy to treat cancer.3, 4, 5, 6, 7
All cells generate and receive bioelectric signals through the flow of ions through transmembrane ion channel, transporters, and pumps, controlling the resting membrane potential (RMP). Cellular RMP, the membrane potential at which the net ionic currents across the plasma membrane are zero, is maintained at a negative value.8 The RMP is dependant on the concentration gradient of the most abundant ions (K+, Na+, Ca2+, and Cl−), membrane permeability to these ions, and ion channel expression and activity.8 As result of changes in these factors, the RMP of cells can either become more negative (hyperpolarized) or more positive (depolarized). The bioelectric properties of cancer cells differ vastly from their normal counterparts: the RMP is more depolarized, and they have dysregulated ion channel expression and activity.9, 10, 11 Previous work has demonstrated that Na+ and K+ ion channel activity can regulate migration, invasion, and metastasis in breast cancer.12, 13, 14, 15 K+ channels, which selectively conduct K+ ions across the membrane down their electrochemical gradient, generally dominate the ion conductance at the resting state of normal cells, causing the RMP to be more positive than the equilibrium potential for K+.8,16,17 Patients with apocrine breast cysts, which have a higher risk of developing into cancer, have an elevated K+ concentration in the cyst fluid18 suggesting a potential correlation between K+ ions and cancer. Indeed, K+ channels are known to drive cancer cell migration via calcium-dependant14,19,20 and independent mechanisms.21, 22 For example, knockdown of the Ca2+-activated potassium channel SK3 in breast cancer cells, which regulates K+ currents in these cells, reduced migration in a transwell assay in vitro.14 A voltage-gated potassium channel, Kv10.1 has also been implicated in driving hyperpolarization of MDA-MB-231 breast cancer cells through Orai1-mediated Ca2+ entry.71 In contrast, Kv11.1 regulates breast cancer cell migration via effects on the epithelial-to-mesenchymal transition, stemness and β-catenin signalling.21 The ether-a-go-go potassium channel EAG2 is enriched at the trailing edge of migrating medulloblastoma cells and regulates cell volume dynamics to facilitate cell motility independent of Ca2+ signalling.92 Despite these observations, it remains unknown whether K+-driven cancer cell migration requires changes in the RMP and whether manipulating the RMP via K+ channel activity can be used to inhibit migrating cells. Ion channels are the second-most common target for existing pharmaceuticals and there are many available FDA-approved drugs targeting K+ channels for non-cancer indications that could be repurposed for cancer therapy.23,24 Understanding the role of the RMP and K+ channel activity in cell migration and cancer metastasis could lead to new treatment strategies for TNBC.
Here, we investigate the effect of manipulating the RMP through K+ channel-driven mechanisms on TNBC cell invasion and metastasis. We find, unexpectedly, that hyperpolarizing the RMP through overexpression of K+ ion channels increases breast cancer cell migration, invasion, tumour growth and metastasis. For the first time, we characterize gene expression changes driven by hyperpolarization in TNBC cells, which lead to upregulation of pathways involved in cell adhesion, in particular the expression of cadherin-11. We demonstrate that cadherin-11-mediated MAPK signalling is involved in hyperpolarization-driven cell migration and invasion. Lastly, reversing the RMP state by blocking endogenous K+channels in TNBC cells with the FDA-approved drug amiodarone decreases in vitro migration and metastasis in vivo. Our results demonstrate that targeting K+ channel-mediated changes in RMP may provide a new strategy for treating metastatic disease.
Materials and methods
Ethics
Tufts (Animal Assurance #A-3115–01) is registered with the United States Department of Agriculture and is committed to comply with the Guide for the Care and Use of Laboratory Animals 8th edition (NIH Publication No. 85–23), the provisions of the Animal Welfare Act, and all applicable federal and state laws and regulations. The Oudin lab animal protocol #M2020–31 entitled “Investigating the role of the microenvironment in cancer metastasis” was approved on April 27th, 2020 by the Tufts University-Tufts Medical centre Institutional Animal Care and Use Committee.
Reagents
Reagents were purchased from Fisher Scientific (Hampton, NH) or Sigma (St. Louis, MO) unless otherwise specified. All reagents were validated by the manufacturer and/or have been previously cited in the literature. Antibodies used were: anti-α-Tubulin (T9026; Sigma, St. Louis, MO), anti-pFAK397 (3283; Cell Signalling Technology, Danvers, MA), anti-pPaxillin118 (44–722 G; ThermoFisher Scientific, Waltham, MA), anti-cadherin-11 (71–7600; ThermoFisher Scientific, Waltham, MA), anti–phospho-p44/42 ERK (Thr202/Tyr204; 4370; Cell Signalling Technology, Danvers, MA), anti-p44/42 ERK (9102; Cell Signalling Technology, Danvers, MA), anti-Ki67 (ab15580; Abcam, Cambridge, UK), AlexaFluor 488 anti-rabbit secondary (A-11,008; ThermoFisher Scientific, Waltham, MA), and AlexaFluor 647 anti-rabbit secondary (A-21,244; ThermoFisher Scientific, Waltham, MA). ECM substrates used were: Collagen I (CB-40,236; Fisher Scientific, Hampton, NH). Inhibitors used were: Amiodarone hydrochloride (40–955–0; Tocris Bioscience, Minneapolis, MN), Carvedilol (C3993, Sigma, St. Louis, MO), Imipramine hydrochloride (I7379; Sigma, St. Louis, MO), and Thioridazine hydrochloride (T9025, Sigma, St. Louis, MO).
Cell lines
The MDA-MB-231 (HTB-126), MDA-MB-468 (HTB-132), BT-20 (HTB-19) breast cancer cell lines, the HEK293T cell line, and MCFA10 breast epithelial cell line (CRL-10,317) were obtained from ATCC (Manassa, VA) and cultured in DMEM with 10% FBS (SH30071.03; Cytiva; Marlborough, MA) and penicillin-streptomycin glutamine (10,378,016; ThermoFisher Scientific; Waltham, MA). Cell lines were previously validated by the manufacturer using routine STR profiling and purchased new at the start of the study. SUM-159 cells and LM2 cells were gifts from the lab of Prof. Frank Gertler at MIT. LM2 cells were cultured in DMEM with 10% FBS (SH30071.03; Cytiva; Marlborough, MA) and penicillin-streptomycin glutamine (10,378,016; ThermoFisher Scientific; Waltham, MA) and SUM-159 cells were cultured in F-12 (ThermoFisher Scientific, 11,765,062) with 5% FBS, 1% PSG, 5 µg/ml insulin (ThermoFisher Scientific, 12,585,014), 1 µg/ml hydrocortisone (H0888, Sigma, St. Louis, MO), 20 ng/ml EGF (ThermoFisher Scientific, PHG0311). Cells were passaged regularly when they reached approximately 70% confluency and used between p5 and p15 for all experiments. Cells were routinely checked for the presence of mycoplasma by a PCR based method using a Universal Mycoplasma Detection Kit (30–1012 K; ATCC, Manassas, VA). Only mycoplasma negative cells were used in this study.
Plasmids and lentiviral infection
Viruses were produced as described previously.25 For infections, 1 µg of constructs expressing human ion channel coding proteins RFP, Kv1.5-T2A-his-EGFP, or KCNJ2(Kir2.1)-Y242F-EGFP were used. For each construct, HEK293T cells at a confluency of 80% were used to transfect for virus production. Constructs were mixed with 0.5 µg helper (encoding gag/pol) and 0.1 µg encoding coat protein (pMD2) along with 6 µL TransIT-Lenti transfection reagent (MIR6603; Mirus Bio; Madison, WI) in 200 uL Opti-MEM (31,985,062; ThermoFisher Scientific; Waltham, MA). Culture media of HEK293T cells was replaced with Opti-MEM and 200µL of transfection mixture and cells were kept in transfection media overnight. Transfection media was replaced with DMEM containing 1% Pen/Strep the following day. Two days post-transfection media containing virus was collected and stored at −80 °C. Collection was repeated on the following day and the total virus collected was combined. To transduce cells, MDA-MB-231 or MDA-MB-468 cells were seeded in six-well plates at 2 × 105 cells per well in 2 mL DMEM containing 10 μg/mL polybrene and virus. The plates were centrifuged at 1500 r.p.m., 4 °C for 1 h and incubated at 37 °C. Medium was changed after 16 h. Infected cells were sorted by FACS and gating on eGFP-derived green fluorescence.
Voltage sensitive and calcium dyes
Changes in RMP were measured using DiBAC (ThermoFisher Scientific, Waltham, MA) as described previously.26 MDA-MB-231 or MDA-MB-468 cells, cultured in black clear-bottomed 96-well plates, were rinsed once with 200 μL of Fluorobrite™ medium (ThermoFisher Scientific, Waltham, MA) and solutions containing 1 μM DiBAC were added to cells and incubated in a cell incubator for 15 min to ensure dye distribution across the membrane. Intracellular calcium was quantified using X-Rhod-1 (ThermoFisher Scientific, Waltham, MA). X-Rhod-1 was reconstituted with DMSO to make a 5 μM stock solution. Cultured cells were rinsed once with 200 μL of Fluorobrite™ medium and X-Rhod-1 diluted in Fluorobrite™ medium at a concentration of 1:1000 was added to cells, incubated in a cell incubator for 60 min, and rinsed in Fluorobrite™ medium. Assays were carried out at 37 °C and 5% CO2 concentration in an incubation chamber fitted to a Keyence BZX700 fluorescence microscope. Changes in DiBAC fluorescence were measured within an hour following incubation to ensure that the dye signal reached a steady state at excitation and emission wavelengths of 488 and 520 nm, respectively. Three regions of interest per well and three wells per condition were imaged. DiBAC or X-Rhod-1 mean pixel intensity excluding background values was measured using ImageJ.
Electrophysiology
Patch clamp experiment was conducted in the whole cell configuration at 37±1 C. Cells were passaged and 1.5 × 105 cells were seeded in an uncoated 2 cm plastic dish 24 h prior to the experiment. On the day of the experiment cells were superfused with an extracellular solution containing (in mM): NaCl 140, KCl 5.4, CaCl2 1.8, MgCl2 1, HEPES-NaOH 10, Glucose 5.5, pH = 7.4. 1 mM of BaCl2 and 10 mM of 4-AP (4-Aminopyridine) were added to the extracellular solution to dissect Kir2.1-mediated and Kv1.5-mediated currents, respectively. Either RFP- or GFP-positive cells were identified and used for the analysis. Borosilicate glass pipettes had a resistance of 5–7 MΩ when filled with an intracellular-like solution containing (in mM): 130 K-Gluconate, 10 NaCl, 5 EGTA-KOH, 2 MgCl2, 2 CaCl2, 2 ATP (Na-salt), 5 creatine phosphate, 0.1 GTP, 10 HEPES-KOH; pH 7.2. Resting membrane potential (RMP) recordings were carried out in the I/0 configuration; after the stabilization of the signal for more than 10 s, the extracellular solutions containing either treatments (BaCl2 and 4-AP) were exchanged until stabilization of the RMP. Kir2.1-mediated current was recorded in the voltage clamp configuration with a ramp protocol from −100 mV to 100 mV (duration of 100 ms) preceded by a 20 ms long step at −100 mV from a holding potential of 0 mV and analysed as the Barium-sensitive current. Kv1.5-mediated current was recorded in the voltage clamp configuration with voltage test steps from a holding potential of 0 mV in the range −60/80 mV (ΔV = +10 mV; duration of 1.75 s each step) and analysed as 4-AP-sensitive current. Currents were normalized to cell capacitance. Density current-voltage plots were then reconstructed for both conductances; for the Kv1.5-mediated current, steady state current was used. Cell capacitance compensation was applied during current recordings and liquid junction potential correction was applied as previously reported.27
In vitro drug studies
MDA-MB-231 and MDA-MB-468 cells were seeded at 5 × 103 cells/well on uncoated 96 well polystyrene plates allowed to adhere for 24 h. Cells were then treated with various concentrations (0.001, 0.01, 0.1, 1, 5, 10, 50, 100, 500, and 1000 µM) of Amiodarone hydrochloride (40–955–0; Tocris Bioscience, Minneapolis, MN), Carvedilol (C3993,Sigma, St. Louis, MO), Imipramine hydrochloride (I7379; Sigma, St. Louis, MO), and Thioridazine hydrochloride (T9025, Sigma, St. Louis, MO) reconstituted in DMSO according to the manufacturer's instructions, or 1% DMSO and incubated for 3 days. After establishing IC50 values, 5 and 10 µM was chosen for downstream assays.
Immunocytochemistry
Cells were plated on 96 well polystyrene dishes (MatTek, Ashland, MA) and allowed to adhere for 24 h. Cells were then fixed for 20 min in 4% paraformaldehyde, then permeabilized with 0.2% TritonX-100, blocked with 5% BSA and incubated with primary antibodies overnight at 4 °C. Cells were stained using DAPI (D1306; ThermoFisher Scientific, Waltham, MA) and phalloidin (A12390; ThermoFisher Scientific, Waltham, MA) and simultaneously incubated with fluorescently labelled secondary antibodies at room temperature for one hour. Imaging was performed using a Keyence BZ-X710 microscope (Keyence, Elmwood park, NJ) and mean fluorescent intensity was quantified using ImageJ.
Western blot
Standard procedures were used for protein electrophoresis and Western blotting. Protein lysates were collected from 231-Control, 231-Kv1.5, 231-Kir2.1 with and without CDH11 siRNA treatment and separated by SDS–polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, blocked with 5% nonfat dry milk solution, and incubated in primary antibody overnight at 4 °C. Proteins were detected using horseradish peroxidase–conjugated secondary antibodies. Imaging was performed using a ChemiDoc MP imaging system (12,003,154; Bio-Rad, Hercules, CA).
Cell viability assay
Cells were seeded on uncoated polystyrene plates allowed to adhere for 24 h. Medium was then changed to medium containing vehicle (DMSO) or ion channel drug and incubated for 48 h. PrestoBlue™ Cell Viability Reagent (A13261; Invitrogen, Carlsbad, CA) was added to each well according to the manufacturer's instructions and incubated for 25 min at 37 °C. Fluorescence was then read on a plate reader at 562 nm. Background was corrected to control wells containing only cell culture media (no cells). A dose-response curve was generated for each drug and the IC50 was calculated. Data are the result of at least three independent experiments with three technical replicates per experiment.
2D migration assay
For 2D migration, cells were plated on uncoated plastic dishes (MatTek, Ashland, MA) and allowed to adhere for a minimum of one hour. Cells were imaged overnight with images acquired every 10 min for 16 h in an environmentally controlled chamber within the Keyence BZ-X710 microscope (Keyence, Elmwood park, NJ). Cells were then tracked using VW-9000 Video Editing/Analysis Software (Keyence, Elmwood Park, NJ) and both cell speed and distance migrated were calculated using a custom MATLAB script vR2020a (MathWorks, Natick, MA). Data are the result of at least three independent experiments with three fields of view per experiment and an average of eight cells tracked per field of view.
3D single cell invasion assay
The day of the experiment, single cell suspensions were formed by mixing cells within a collagen I matrix with collagen I protein, 10 mM NaOH, 7.5% 10x DMEM and 50% 1x DMEM, added to a 24 well glass bottom culture plate, and incubated for one hour at 37°C. Cells were imaged overnight with images acquired every 10 min for 16 h in an environmentally controlled chamber within the Keyence BZ-X710 microscope (Keyence, Elmwood park, NJ). Cells were then tracked using VW-9000 Video Editing/Analysis Software (Keyence, Elmwood Park, NJ) and both cell speed and distance migrated were calculated using a custom MATLAB script vR2020a (MathWorks, Natick, MA). Data are the result of at least three independent experiments with minimum six fields of view per experiment and an average of eight cells tracked per field of view.
3D invasion assay
Cells were seeded in low-attachment plates in media, followed by centrifugation to form spheroids. Spheroids were grown for three days after which matrix was added to each well, which includes collagen I protein, 10 mM NaOH, 7.5% 10x DMEM and 50% 1x DMEM. The spheroids in matrix were then spun down and a further 50μL of media added to each well. Following another seven days of growth (three days for amiodarone treatment studies), spheroids were imaged as a Z-stack using a Keyence BZ-X710 microscope (Keyence, Elmwood Park, NJ) and Z-projection images analysed using ImageJ. Data are the result of at least three independent experiments with three technical replicates per experiment.
Cell adhesion assay
Cells were plated on glass-bottomed dishes (MatTek, Ashland, MA) coated with 20 μg/ml collagen I and allowed to adhere for 24 h. Cells were then fixed for 20 min in 4% paraformaldehyde, then permeabilized with 0.2% TritonX-100, blocked with 5% BSA and incubated with primary antibodies overnight at 4 °C. Cells were stained using DAPI (D1306; ThermoFisher Scientific, Waltham, MA) and phalloidin (A12390; ThermoFisher Scientific, Waltham, MA) and simultaneously incubated with fluorescently labelled secondary antibodies at room temperature for one hour. Imaging was performed using a Keyence BZ-X710 microscope (Keyence, Elmwood park, NJ) and CellProfiler v3.1.8 (Carpenter et al.) was used for imaging analysis using a custom pipeline to quantify cell area and shape parameters based on the phalloidin channel.
RNA sequencing and analysis
RNA was extracted from three separate biological replicates of MDA-MB-231-Control, 231-Kv1.5, and 231-Kir2.1 cells using the RNA MiniPrep Kit (R1057; Zymo Research; Irvine, CA). Samples were confirmed to have sufficient quality with Agilent Bioanalyzer before undergoing library preparation by Tufts University Genomics Core using the TruSeq stranded mRNA kit (20,020,594; Illumina; San Diego, CA). Samples were run on a HiSeq2500 platform at a depth of 50 million single-end 50 base pair reads to allow for transcript quantification.28 Quality control of raw read was performed using FastQC29 and adaptor trimming and read filtering was done using Trimmomatic30 with standard settings. Reads were mapped to the reference human genome (assembly GRCh38) with STAR.31 Differential gene expression analysis was performed using EdgeR in R,32,33 genes with cpm less than 1 were filtered, and a fold change threshold of 1.5 was used. Pathway enrichment analysis was performed using Gen Set Enrichment Analysis (GSEA).34,35 Raw reads and processed data files were uploaded to the Gene Expression Omnibus (GEO) under accession code GSE171150.
CDH11 knockdown
Knockdown of CDH11 was conducted as described previously.36 MDA-MB-231 cells were passaged and incubated at 1.0 × 105 cells per well in a six well polystyrene coated plate for 24 h. Cells were transfected with On-Target™ plus Human CDH11 siRNA-Smartpool (L-013,493–00–0005, Dharmacon, Lafayette, CO, US) that contains 4 siRNA mix. CDH11 #1: 5′-GUGAGAACAUCAUUACUUA-3′. CDH11 #2: 5′-GGACAUGGGUGGACACAUAUG-3′. CDH11 #3: 5′-GGAAAUAGCGCCAAGUUAG-3. CDH11 #4: 5′-CCUUAUGACUCCAUUCAAA-3′ using Lipofectamine RNAiMAX transfection reagent (13,778,030; ThermoFisher Scientific, Waltham, MA) in serum-free DMEM. Silencer™ Negative Control #1 siRNA (AM4611; ThermoFisher Scientific, Waltham, MA) was used as a negative control for transfection. The final concentration of siRNA was 80 nM. The siRNA-transfected cells were incubated for 48 h and used for functional assays.
Animal experiments
Animal procedures were carried out in full accordance with established standards set forth in the Guide for the Care and Use of Laboratory Animals, 8th edition (NIH Publication No. 85–23). For xenograft tumours, 2 × 106 MDA-MB-231 cells in PBS and 20% collagen I were injected into the fourth right mammary fat pad of 6-wk-old female NOD-SCID-γ (NSG) mice (005,557; JAX, Bar harbor, ME). For each experiment animals were categorized based on tumour size and assigned into experimental groups such that each group had the same distribution of starting tumour size. For the channel overexpressing lines, 10 mice each were injected with either MDA-MB-231 control, Kir2.1, or Kv1.5 cells and tumour growth was monitored for seven weeks. For the amiodarone experiment MDA-MB-231 GFP cells were injected into 20 animals and 0.05 mg/kg amiodarone or DMSO control was administered i.p. beginning at five weeks post-injection every day for two weeks, followed by injection of 0.5 mg/kg amiodarone or DMSO control for an additional 10 days. During the drug injection period, tumour growth was measured biweekly and body weight weekly. Mice were euthanized by CO2 and tumours and lungs were excised once tumours reached a volume of 1.5cm3 or animal wellbeing endpoints were met. Animals that met wellbeing endpoints before the end of the study were excluded from analysis.
Histology
Fixation, processing and staining of tissue sections from tumours and lungs was carried out as previously described.37 Tumours and lungs dissected from NSG mice were fixed in 10% paraformaldehyde and embedded in paraffin. Tissue was sectioned using a microtome at 10 µm thickness. For H&E staining: standard procedures were followed, including deparaffinizing, hydration, staining with Hematoxylin (GHS280; SIGMA, St. Louis, MO) and counterstaining with Eosin (HT110180; SIGMA, St. Louis, MO). Lung metastases were quantified by three independent observers blinded to treatment using ImageJ.
Statistical analysis
GraphPad Prism v8.4.3 was used for generation of graphs and statistical analysis. For data with normal distribution: for comparison between two groups, an unpaired two-tailed Student's t-test was used and a p-value of ≤0.05 considered significant, and for comparison between multiple groups a one-way ANOVA with Dunnett's multiple testing correction was used with a corrected p-value of ≤0.05 considered significant. Patch clamp data were analysed with Clampfit10 (Axon) and Origin Pro 9. Linear fitting was performed reporting the adjusted R2 value. Data represent mean ± S.E.M. unless otherwise stated. Sample sizes used to calculate statistical significance were at least three biological replicates. Sample size was determined by power analysis (Power and Precision 4) and previous experiments done in our lab. We determined that 10 animals per group will be adequate for 94% power to detect a difference in burden (two-tailed alpha of 0.5).
Role of funding source
The funder of the study has no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all data in the study and accept responsibility to submit for publication.
Results
TNBC cell RMP is controlled by K+ ions
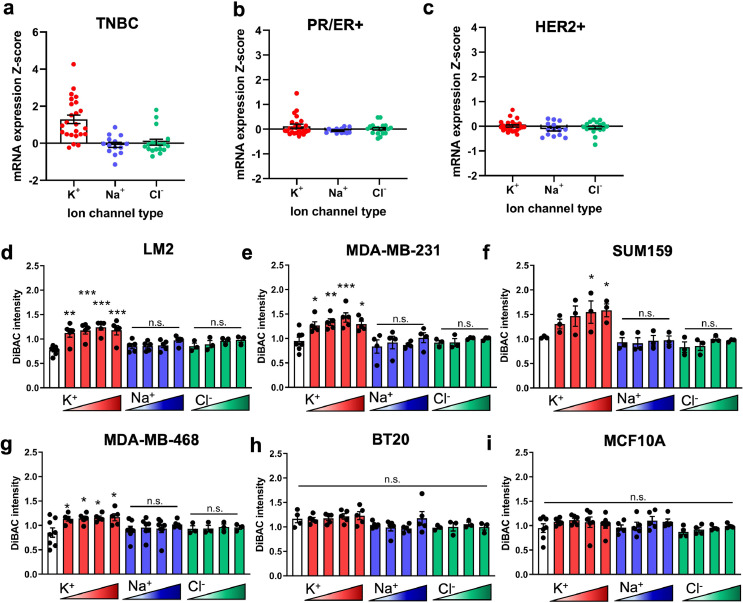
Given that the cellular RMP is determined by ion channel expression and activity, we first aimed to determine the ion channel expression profile of human breast cancers. Analysis of the TCGA dataset using cBioPortal23,38 for mRNA expression of ion channel types revealed that K+channel encoding genes are largely upregulated in invasive ductal carcinoma TNBC patient samples compared to unaltered diploid samples, whereas Na+and Cl−channel genes were both upregulated and downregulated (Fig. 1a-c). Additionally, we found larger z-score values for K+ channel expression in TNBC patients (Fig. 1a) compared to ER/PR+ (Fig. 1b) or HER2+ (Fig. 1c). Although it is known that K+ channel activity regulates the RMP of normal cell types, this has not been previously established for TNBC cells. Therefore to determine which ion species are dominant in setting the RMP of TNBC cells, we incubated human breast cancer cell lines (LM2, MDA-MB-231, SUM159, MDA-MB-468, and BT20) and immortalized normal breast epithelial MCF10A cells in a series of extracellular solutions that are balanced osmotically and differ only in the concentration of one ion26 (Figure S1). Rather than targeting a single ion channel, this approach allowed us to distinguish the cumulative effect on RMP of an ion species, by measuring cellular RMP using DiBAC, a voltage-sensitive membrane dye.26,39 We observed that incubating cells with increasing extracellular concentration of K+ (ranging from 5.4 mM to 135.4 mM) results in significant depolarization of the cell population in metastatic lines LM2, MDA-MB-231, SUM159, and MDA-MB-468 (Fig. 1d-g) but had no effect on the poorly metastatic BT20 or healthy epithelial MCF10A line (Fig. 1h-i) compared to physiological K+ concentration (white bars). Incubation in Na+ (ranging from 28 mM to 140 mM) or Cl− solutions (ranging from 30.2 mM to 151 mM) had no effect on the RMP for any lines. Interestingly the increase in RMP was also associated with the reported metastatic potential of the cell lines;40 in the presence of increased extracellular K+ concentration, lines with a higher metastatic potential had a relatively greater depolarization than low metastatic potential or healthy MCF10As. These data show that K+ channels are upregulated in human breast cancer tumours and suggest that TNBC cell RMP is regulated by the activity of K+channels rather than that of Na+ or Cl−.
Fig. 1.
The RMP of metastatic TNBC cells is driven by K+ions. Expression distribution mRNA Z-scores for ion channels in invasive ductal breast carcinoma relative to unaltered diploid expression from TCGA dataset categorized by (a) TNBC, (b) oestrogen and progesterone receptor (ER/PR+) and (c) human epidermal growth factor receptor 2 (HER2+) subtypes. The RMP of five TNBC lines, (d) LM2 (**p = 0.0004, ***p<0.0001), (e) MDA-MB-231 (*p = 0.0415, **p = 0.0084, ***p = 0.0007), (f) SUM159 (*p = 0.0489, 0.0331), (g) MDA-MB-468 (*p = 0.0387, 0.0317, 0.0217, 0.0153), and (h) BT20, and one healthy epithelial breast cell line, (i) MCF10A, when incubated in ionic solutions of increasing concentration of a single ion, K+, Na+, or Cl− and compared to physiological concentrations (white bar). Data are shown as mean ± S.E.M, n = 3–7 biological replicates. Significance was determined using a one-way ANOVA with Dunnett's post hoc test to compare each set of ionic solutions to physiological concentration.
K+ channel-driven RMP hyperpolarization increases TNBC cell invasion
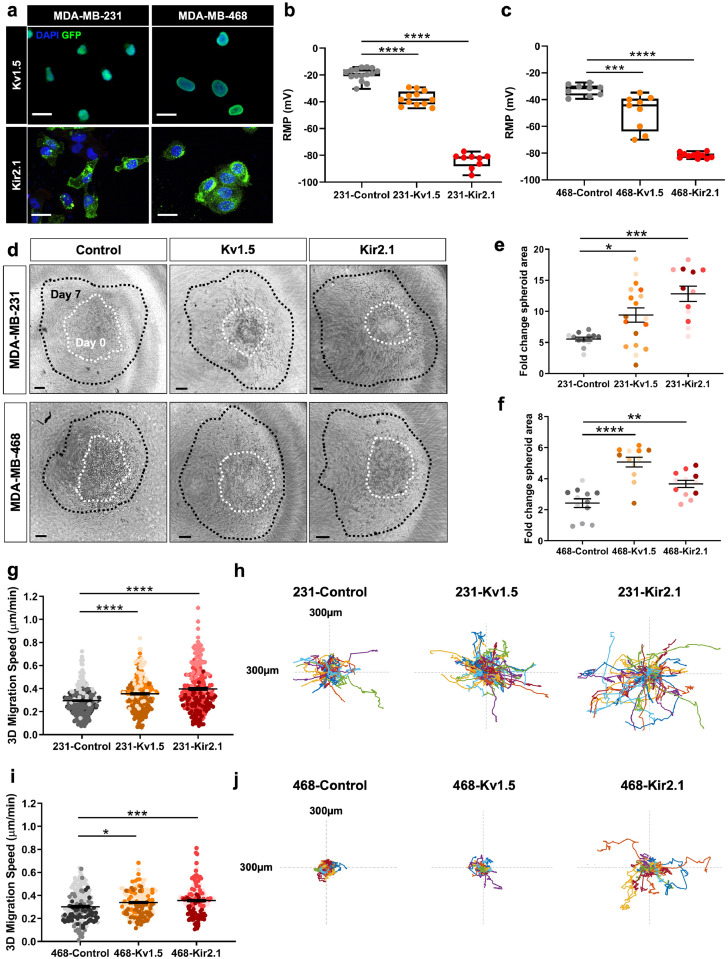
To study the effect of K+conductance and resulting change in the RMP on breast cancer cell metastatic potential, we engineered MDA-MB-231 and MDA-MB-468 cell lines to stably express two different types of GFP-tagged K+ channels: Kv1.5, a voltage-gated channel (231-Kv1.5 and 468-Kv1.5), and Kir2.1, a constitutively open, inwardly-rectifying channel (231-Kir2.1 and 468-Kir2.1) (Fig. 2a). These channels were chosen as a tool to hyperpolarize the RMP, as has been previously demonstrated25,41, 42, 43 and compared against their respective negative control expressing a fluorophore alone (231-Control and 468-Control). The observed nuclear localization of GFP in the Kv1.5 expressing cells (Fig. 2a) is due to the Thosea asigna virus 2A (T2A) sequence linking Kv1.5 and histone GFP in the Kv1.5 construct, which during translation initiates ribosomal skipping and as a result the histone GFP is cleaved and shuttled to the nucleus.44 Expression of either channel resulted in a significantly hyperpolarized RMP in both cell lines relative to wildtype MDA-MB-231 cells, with a change in voltage of approximately −18 mV for Kv1.5 and −40 mV for Kir2.1 (Fig. 2b-c). We confirmed these cell lines did have a significantly more negative RMP relative to MCF10A healthy breast epithelial cells, which have an average RMP of −27.1 mV compared to an average RMP of −19.5 mV for wildtype MDA-MB-231 cells (Figure S2a). The difference in RMP produced with K+ channel overexpression is within a range similar to that produced with the use of pharmacological agents.45,46 We confirmed that this change in RMP was accompanied by an increase in outward K+current for both MDA-MB-231 and MDA-MB-468 derived lines (Figure S2b, d), although the amount of current measured varied considerably between the two lines, with both 231-Kv1.5 and -Kir2.1 lines generating a higher K+ current than their MDA-MB-468 counterparts. We also confirmed that pharmacological blocking of the channels with an inhibitor specific to each channel type resulted in a reversal of the observed hyperpolarization (Figure S2c, e).
Fig. 2.
Overexpression of K+ channels induces RMP hyperpolarization and increases 3D cell invasion. (a) Expression of GFP-tagged Kv1.5 and Kir2.1 channel constructs by lentiviral transduction into MDA-MB-231 and MDA-MB-468 breast cancer cell lines visualized with GFP. Scale bar = 10 µm. Electrophysiology measurement of the RMP in K+ channel overexpressing lines compared to negative controls in (b) MDA-MB-231 (****p<0.0001) and (c) MDA-MB-468 (***p = 0.0002, ****p<0.0001) cells. (d) Representative images at day 7 (black) overlayed with outlines of spheroid area at day 0 (white). Scale bar = 100 µm. The fold change in spheroid area in (e) MDA-MB-231 (*p = 0.0246, ***p = 0.0001) (n = 190 cells/group for statistical analysis) and (f) MDA-MB-468 (**p = 0.0066, ****p<0.0001) (n = 190 cells/group for statistical analysis) lines. Quantification of 3D single cell invasion and representative rose plots in (g-h) MDA-MB-231 (****p<0.0001) (n = 348 cells/group for statistical analysis) and (i-j) MDA-MB-468 (*p = 0.0141, ***p = 0.0001) (n = 121 cells/group for statistical analysis) lines. Data are pooled from three or more biological replicates and shown as mean ± S.E.M. Different colour shades within group represent samples from different replicates. Significance was determined using a one-way ANOVA with Dunnett's multiple comparisons test.
Next, we investigated the impact of K+ channel-driven hyperpolarization on 3D cell invasion. Spheroids made from control and K+channel-expressing cells were encapsulated in collagen I matrices and grown for seven days. The change in spheroid area was quantified as a measurement of cell invasion into the surrounding matrix, a model which most accurately reflects the physical properties of tumours.47 We found that overexpression of Kv1.5 and Kir2.1 significantly increased 3D cell invasion in both cell lines when compared to the negative control (Fig. 2d-f). 3D invasion was also tested using a single cell assay where cells were dispersed in a collagen I matrix and cell speed was monitored over 16 h. In both MDA-MB-231 and MDA-MB-468 lines, overexpression of Kv1.5 and Kir2.1 caused a significant increase in 3D single cell migration speed (Fig. 2g-j). In addition, we evaluated the effects of channel expression on 2D migration, demonstrating an increase in cell speed in the 231-Kv1.5 and 231-Kir2.1 lines as compared to the control (Figure S3a-c) that is reduced with the application of 4-aminopyridine (4-AP) or barium chloride, blockers specific for Kv1.5 and Kir2.1 respectively (Figure S3d-e). There was no significant change in 2D migration in channel-overexpressing MDA-MB-468 lines (Figure S3f-g). Lastly, cell proliferation was measured by quantification of Ki67 expression in cells. In the MDA-MB-231 line, Kv1.5 overexpression had no effect on proliferation, whereas in the MDA-MB-231-Kir2.1 and MDA-MB-468 lines proliferation was decreased compared to the negative controls (Figure S3h-i). Taken together, these data demonstrate that K+ channel-driven RMP hyperpolarization increases TNBC cell migration and invasion.
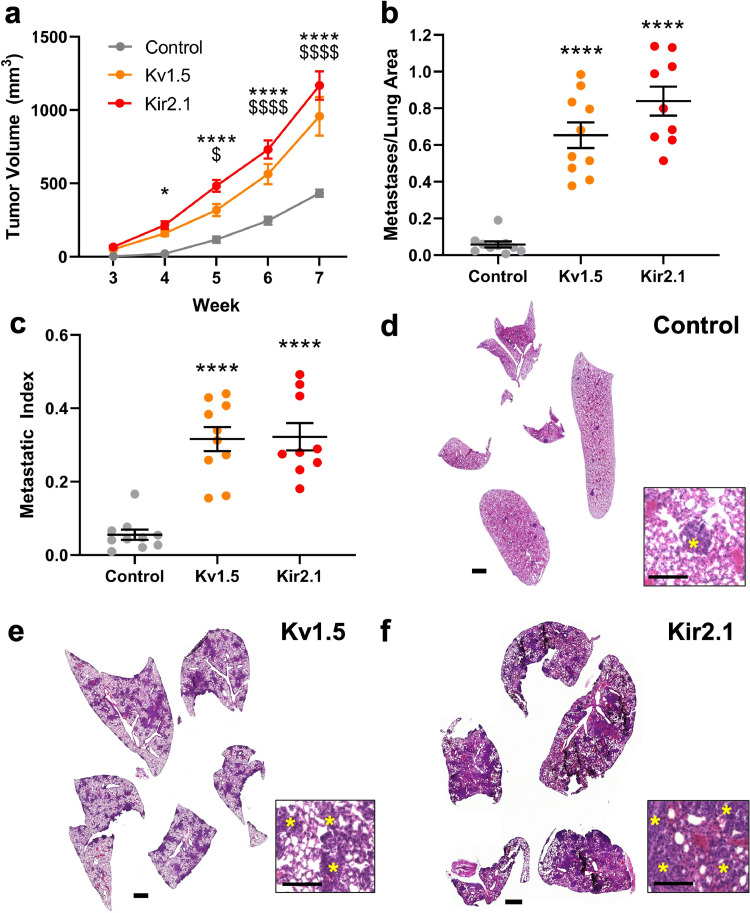
We next investigated the effect of K+ channel-driven RMP hyperpolarization on in vivo tumour growth and metastasis in a mouse xenograft model. NOD-SCID-γ mice were injected with 231-Kv1.5, 231-Kir2.1, or 231-Control lines in the fourth mammary fat pad and tumour growth was tracked over time. Beginning at four weeks post-injection to the terminal time point, tumour volume was significantly greater in mice in the 231-Kv1.5 and 231-Kir2.1 group compared to the 231-Control group (Fig. 3a). Animals were sacrificed at seven weeks and metastasis to the lungs was quantified using H&E staining and expressed as the number of metastases relative to total lung area (Fig. 3b) and the number of metastases relative to final tumour volume (metastatic index, Fig. 3c). Our data show that both the 231-Kv1.5 and 231-Kir2.1 group had significantly more metastases present than the 231-Control group (Fig. 3d-f). Combined, these data demonstrate that K+ channel-driven RMP hyperpolarization enhances TNBC cell migration and invasion in vitro, and tumour growth and metastasis in vivo.
Fig. 3.
K+ channel-driven RMP hyperpolarization increases TNBC tumour growth and metastasis. (a) Tumour volume measured over time in 231-Kv1.5 and 231-Kir2.1 vol compared to the 231-Control group. *p = 0.0191, ****p<0.0001 for 231-Kir2.1 compared to 231-Control; $p = 0.0153, $$$$p<0.0001 for 231-Kv1.5 compared to 231-Control. (b) The number of metastases per lung and (c) the metastatic index was significantly increased in the 231-Kv1.5 and Kir2.1 group compared to 231-Control (****p<0.0001). Significance was determined using a one-way ANOVA with Dunnett's multiple comparisons test. Representative lung tissue sections stained with H&E for (d) 231-Control, (e) 231-Kv1.5, and (f) 231-Kir2.1 group. Scale bar = 1 mm. Inset scale bar =100 µm. Yellow asterisks indicate metastases. Data are shown as mean ± S.E.M. N = 9–10 animals/group.
K+ channel-driven RMP hyperpolarization drives upregulation of genes associated with cell adhesion and MAPK signalling
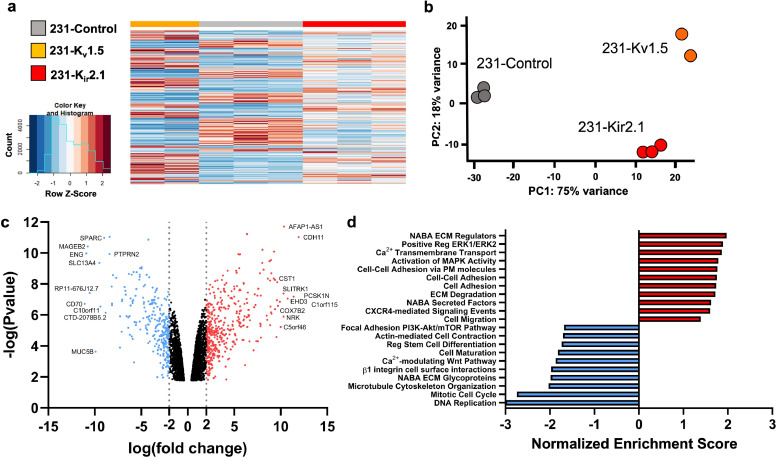
After determining that overexpression of K+ channels and hyperpolarization result in profound phenotypic changes in TNBC cell lines, we wanted to determine the mechanism by which this contributes to cell migration. K+ channels have been shown to drive cell migration by regulating Ca2+ levels in cells,17 so we first measured intracellular Ca2+ levels in our cells using X-Rhodhamine, a calcium-sensitive fluorescent dye. We found no significant changes in intracellular Ca2+ levels in the K+-overexpressing cells in both MDA-MB-231 and MDA-MB-468 cells (Fig S4a,b). Further, we found no significant changes in Ca2+ levels in cells treated with the different K+ solutions described in Fig. 1 which also altered the membrane potential (Fig S4c,d). These data suggest that K+-induced hyperpolarization in TNBC cells drives migration via mechanisms other than Ca2+ signalling. To determine whether changes in gene expression could explain these different phenotypes, we performed RNA sequencing on the 231-Kv1.5, 231-Kir2.1 and 231-Control lines. Differential expression analysis revealed a significant number of genes to be differentially expressed in channel overexpressing lines compared to the control (Fig. 4a). Principal component (PC) analysis determined that 75% of the overall variance was found in PC1 between groups separating control and K+ channel-expressing cells, indicating that the change in cellular RMP has significant effects on gene expression (Fig. 4b). To focus on genes that were commonly dysregulated in both 231-Kv1.5 and 231-Kir2.1 lines we combined differentially-expressed genes from both and analysed the average fold change in their expression compared to the 231-Control group, generating a list of significantly upregulated and downregulated genes (Fig. 4c). We next analysed enrichment for functional pathways and generated normalized enrichment scores for differentially expressed genes shared by both 231-Kv1.5 and 231-Kir2.1 lines, using a false discovery rate (FDR) threshold of 0.1 and significance of p<0.05. Relevant positively enriched pathways include ECM regulators, ERK1/2 regulation, MAPK activity, cell adhesion and cell migration (Fig. 4d). Together, these results demonstrate that K+ channel-driven RMP hyperpolarization drives significant changes in gene expression, upregulation of pathways involved in cell adhesion.
Fig. 4.
K+ channel-driven RMP hyperpolarization upregulates genes associated with cell adhesion. (a) Heat map clustering of differentially expressed genes in 231-Kv1.5 and 231-Kir2.1 compared to 231-Control. (b) Principal component (PC) analysis showing clustering and variance captured for cell lines. (c) Volcano plot demonstrating log(fold change) and log(p value) of upregulated and downregulated gene expression in the combined group of 231-Kv1.5 and 231-Kir2.1. The top 10 genes with highest fold change are labelled. (d) Normalized enrichment score for a subset of functional pathways upregulated and downregulated in the combined 231-Kv1.5/231-Kir2.1 group compared to 231-Control. N = 3 for 231-Control, 231-Kir2.1; n = 2 for 231-Kv1.5.
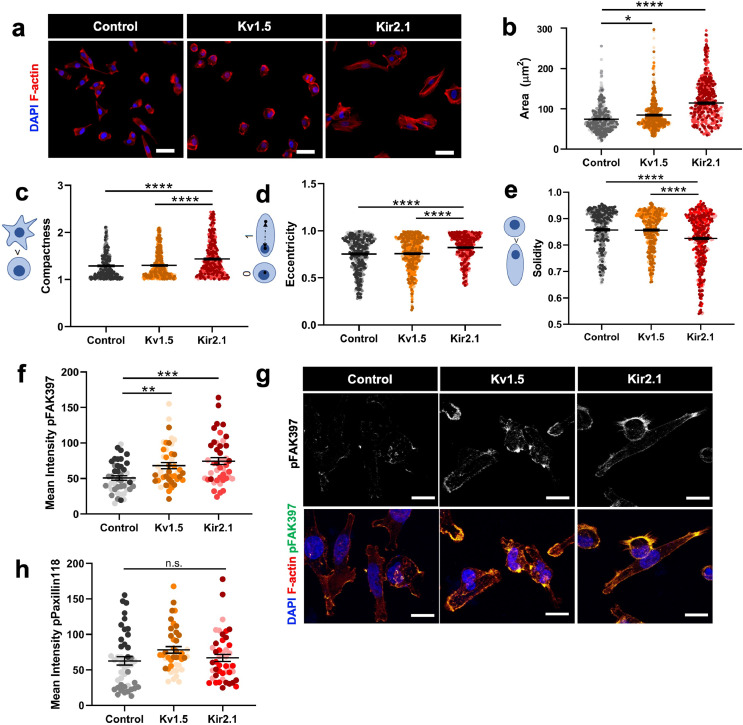
The observed upregulation of genes involved in cell adhesion suggests that overexpression of K+ channels may alter the adhesion and morphology of TNBC cells, which our lab and others have demonstrated is highly predictive of 3D invasive behaviour driving metastasis.48,49 We characterized cell area and morphology of MDA-MB-231 K+ channel-overexpressing lines cultured on collagen I-coated glass plates. Quantification of cell area using F-actin staining (Fig. 5a) revealed that 231-Kv1.5 and 231-Kir2.1 cells have a significantly larger area than 231-Control cells (Fig. 5b). Cell compactness, the ratio of the area of a cell compared to a circle of the same perimeter, and eccentricity, the ratio of the minor to major cell axis, were found to be increased in 231-Kir2.1 cells compared to 231-Control and 231-Kv1.5 cells (Fig. 5c-d). Cell solidity, the ratio of the area of a cell to a convex hull of the cell, was significantly decreased (Fig. 5e). These morphological features indicate that 231-Kir2.1 cells are on average more elongated, have more protrusions, and greater boundary irregularity compared to 231-Control and 231-Kv1.5 cells. An important step in cell migration and invasion is the formation of focal adhesions between the intracellular actin cytoskeleton and the ECM. To test if K+ channel-driven hyperpolarization is linked to focal adhesion formation, we investigated levels of phosphorylated focal adhesion kinase protein (pFAK397), a regulator of adhesion turnover, and phosphorylated paxillin (pPax118), a focal adhesion adaptor protein phosphorylated by FAK, in our channel overexpressing cell lines. We found that 231-Kv1.5 and 231-Kir2.1 cells have significantly higher levels of pFAK when compared to 231-Control cells (Fig. 5f-g), but did not detect a difference in pPax118 expression (Fig. 5h). Combined, our results suggest that K+ channel-driven RMP hyperpolarization is associated with changes in cell shape and focal adhesion signalling.
Fig. 5.
K+ channel-driven RMP hyperpolarization alters cell morphology and focal adhesion signalling. (a) Representative images of overexpressing cell lines stained with phalloidin for F-actin. Scale bar = 10 µm. (b) Quantification of F-actin-based cell area (*p = 0.0202, ****p<0.0001) (n = 498 cells/group for statistical analysis). Quantification of c) compactness, (d) eccentricity, and (e) solidity (****p<0.0001) (n = 498 cells/group for statistical analysis). (f) Quantification of pFAK397 expression in overexpressing lines compared to 231-Control (**p = 0.0065, ***p = 0.0002) (n = 47 cells/group for statistical analysis). (g) Representative images of pFAK397 expression. Scale bar = 10 µm. (h) Quantification of pPax118 in overexpressing lines compared to 231-Control (n = 47 cells/group for statistical analysis). Data are pooled from three or more biological replicates and shown as mean ± S.E.M. Different colour shades within group represent samples from different replicates. Significance was determined using a one-way ANOVA with Dunnett's multiple comparisons test.
Cadherin-11 is necessary for K+ channel-driven cell migration
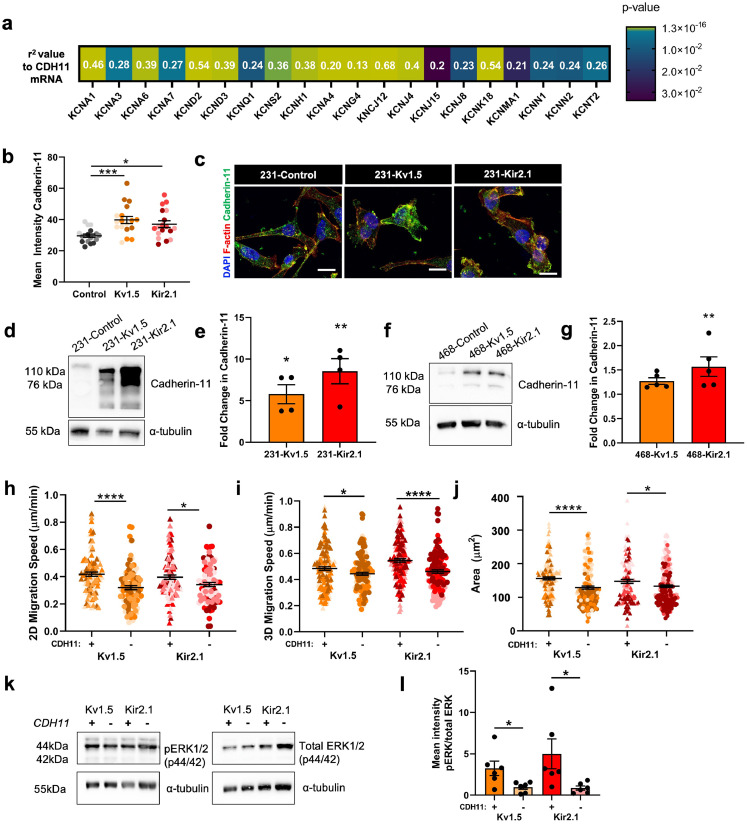
Cadherin-11 is a member of the cadherin super-family which mediates homophilic cell-cell adhesion in a calcium-dependant manner by interacting with the cytoplasmatic catenins, α, β, and γ, and has been shown to drive metastasis in epithelial cancers.50, 51, 52 Specifically in breast cancer, cadherin-11 has been associated with cell migration, EMT, and enhanced metastasis.36,53, 54, 55 Mining of the publicly-available patient TCGA dataset using cBioPortal38 revealed that CDH11 mRNA levels are positively correlated with mRNA levels of 20 of 52 total K+ channels in TNBC patients (Fig. 6a, Figure S5a-b). Due to this, as well as its significant 11-fold genetic upregulation in the 231-Kv1.5 and 231-Kir2.1 cell lines, and its known involvement in metastasis, we chose to investigate the role of cadherin-11 in the K+ channel-driven effects observed. To determine if the increased expression of the CDH11 gene seen in our RNAseq results (Fig. 4c) corresponds to increased cadherin-11 protein expression, we quantified cadherin-11 in K+-overexpressing 231 cell lines. Compared to 231-Control cells, 231-Kv1.5 and 231-Kir2.1 expressed significantly higher levels of cadherin-11 as determined by immunocytochemistry (Fig. 6b-c). Significantly higher levels of cadherin-11 were also detected using western blot in both MDA-MB-231 Kv1.5 and Kir2.1 (Fig. 6d-e) and MDA-MB-468 Kir2.1 (Fig. 6f-g) lines.
Fig. 6.
Cadherin-11 mediates the pro-migratory effects of K+ channel-driven hyperpolarization. (a) Heatmap demonstrating significant correlation of CDH11 mRNA expression with K+channel genes in TNBC patients from TCGA dataset. Pearson's coefficient values for each gene are listed and colour scale indicates degree of statistical significance. (b) Quantification of cadherin-11 expression in 231 cell lines (*p = 0.0168, ****p<0.0001) (n = 18 images for statistical analysis). (c) Representative images of cadherin-11 expression (green) colocalized with F-actin (red). Scale bar = 10 µm. Representative western blot and quantification of cadherin-11 expression in (d-e) 231 (*p = 0.0231, **p = 0.0016) (n = 4 replicates for statistical analysis) and (f-g) 468 (**p = 0.0064) (n = 5 replicates for statistical analysis) cell lines. (h) 2D migration speed following CDH11 knockdown by siRNA relative to control siRNA (*p = 0.0143, ****p<0.0001) (n = 96 cells/group for statistical analysis). (i) 3D migration speed following CDH11 knockdown by siRNA relative to control siRNA (*p = 0.0290, ****p<0.0001) (n = 120 cells/group for statistical analysis). (j) Quantification of F-actin-based cell area following CDH11 knockdown by siRNA relative to control siRNA (*p = 0.0363, ****p<0.0001) (n = 131 cells/group for statistical analysis). (k) Representative western blot and (l) quantification of the ratio of pERK1/2 to total ERK1/2 with and without CDH11 knockdown (*p = 0.032 for Kv1.5, *p = 0.0471 for Kir2.1). Data are pooled from three or more biological replicates and shown as mean ± S.E.M. Different colour shades within group represent samples from different replicates. Significance was determined using a one-way ANOVA with Dunnett's multiple comparisons test.
We next induced knockdown of CDH11 with siRNA which has been used previously in breast cancer cells56 and measured the effects on cell migration and adhesion in the MDA-MB-231 K+ channel-overexpressing lines. An 80–90% reduction in CDH11 expression was achieved with siRNA knockdown after 48 h which was verified with cadherin-11 immunostaining (Figure S5c) and western blot (Figure S5d-e). This CDH11 knockdown resulted in a significant decrease in 2D cell migration speed (Fig. 6h), 3D cell single cell invasion (Fig. 6i), as well as a decrease in cell area (Fig. 6j) in both 231-Kv1.5 and 231-Kir2.1 cell lines. Cadherin-11 has been previously linked to ERK phosphorylation,52,57 and ERK in turn can promote cell migration by phosphorylating FAK and Paxillin to drive focal adhesion formation.58 Furthermore, MAPK signalling pathway-related genes were enriched in our RNAseq dataset (Fig. 4d). Consistent with this, we observed that knockdown of CDH11 results in a significant decrease in the ratio of pERK1/2 to total ERK1/2 (Fig. 6k-l). Taken together, these results suggest that the increase in 2D migration and 3D invasion of hyperpolarized cells is driven by cadherin-11 and MAPK signalling.
Pharmacological blockage of K+ channels decreases migration and metastasis
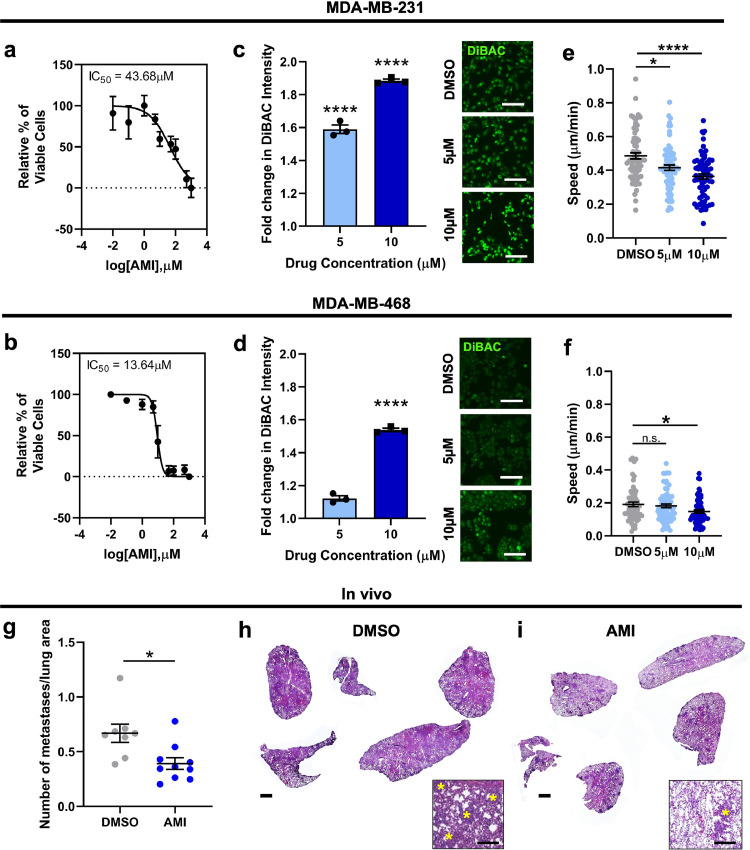
Our results using K+ channel overexpressing breast cancer lines suggest that K+ channel activity and the subsequent hyperpolarization of the RMP can increase cell invasion and metastasis. To determine the feasibility of targeting the RMP of breast cancer cells to treat metastatic disease, we next investigated the effect of K+ channel blockers on cell invasion and metastasis. We mined the online repurposed drug database ReDO23,59 and selected four clinically available drugs that are known to block K+ channels and have been associated with anti-cancer effects in other tumour types:59 amiodarone, carvedilol, imipramine, and thioridazine. An initial in vitro screen was performed with MDA-MB-231 and MDA-MB-468 cell lines, with all drugs inducing significant tumour cell death with IC50s in the low micromolar range (Fig. 7a-b; Figure S6a-c). Next, we measured the change in RMP elicited by each drug using DiBAC at two doses in the range of the IC50 and determined that all drugs tested caused depolarization of the RMP in MDA-MB-231 and MDA-MB-468 cells (Fig. 7c-d; Figure S6d-e). Due to the relatively large depolarization produced by amiodarone, we moved forward with this drug for subsequent experiments. Amiodarone is a class III anti-arrhythmic drug known to block voltage-gated and inward rectifier K+ channels60, 61, 62 and has been shown to improve survival of hepatocellular carcinoma patients.63 Treatment of MDA-MB-231 and MDA-MB-468 cells with amiodarone at 5 and 10 µM resulted in a significant reduction in 2D cell migration for MDA-MB-231 cells at both concentrations (Fig. 7e) and for MDA-MB-468 cells at 10 µM (Fig. 7f). Amiodarone treatment also significantly decreased 2D migration but not viability of the MDA-MB-231 K+ channel overexpressing Kv1.5 and Kir2.1 lines (Figure S7). Interestingly, we observed a reduction in cadherin-11 expression in MDA-MB-231 cells with amiodarone treatment of approximately 70% and 50% with 5 µM and 10 µM respectively (Figure S8a-b) as well as a reduction in total ERK1/2 levels that was statistically significant with administration of 10 µM (Figure S8c-d). Amiodarone treatment also reduced cadherin-11 expression in MDA-MB-468 cells at 10 µM (Figure S8e-f) but had no significant effect on total ERK1/2 levels (Figure S8g-h). To determine if the decrease in 2D migration is associated with changes in Ca2+ concentration, we quantified intracellular Ca2+ in MDA-MB-231 cells treated with amiodarone, finding that there was no significant difference between groups (Figure S8i). Lastly, we saw a decrease in proliferation of MDA-MB-231 and MDA-MB-468 cells treated with 10 µM amiodarone as measured by Ki67 expression (Figure S8j-l).
Fig. 7.
Amiodarone treatment depolarizes TNBC RMP resulting in decreased cell migration. (a) Dose response curves of cell viability in MDA-MB-231 and (b) MDA-MB-468 cells cultured with varying concentrations of amiodarone (AMI). RMP of (c) MDA-MB-231 and (d) MDA-MB-468 cells measured by fold change in DiBAC intensity after amiodarone treatment at IC50 concentrations (****p<0.0001) and representative DiBAC images. Scale bar = 60 µm. Cell migration speed following treatment with amiodarone in (e) MDA-MB-231 (*p = 0.0167, ****p<0.0001) and (f) MDA-MB-468 (*p = 0.0267) cells (n = 72 cells/group for statistical analysis). Data are pooled from three or more biological replicates and shown as mean ± S.E.M. Significance was determined using a one-way ANOVA with Dunnett's multiple comparisons test. (g) Number of MDA-MB-231 derived metastases per area of lung after daily i.p. injections of amiodarone at 0.05 mg/kg (14 days) followed by at 0.5 mg/kg (10 days) or DMSO for a total of 24 days (*p = 0.01) (n = 8 animals DMSO, n = 10 animals AMI for statistical analysis). Representative lung tissue sections stained with H&E from animals treated with (h) DMSO or (i) amiodarone. Scale bar = 1 mm. Inset scale bar = 150 µm. Yellow asterisks indicate metastases. Data shown as mean ± S.E.M. Significance was determined using a Student's t-test. N = 8–10 animals per group.
Next, we investigated the effect of blocking K+ channels with amiodarone on in vivo tumour growth and metastasis in a mouse xenograft model. NOD-SCID-γ mice were injected with MDA-MB-231 cells in the 4th mammary fat pad and tumour growth was tracked over time. When tumours reached an average volume of 200mm3 mice received daily i.p. injections of 0.05 mg/kg amiodarone or DMSO for 14 days, followed by 0.5 mg/kg amiodarone or DMSO for 10 days. Quantification of the metastases in the lungs at sacrifice revealed that animals treated with amiodarone had significantly fewer metastases than vehicle-treated animals (Fig. 7g-i). We observed a trend towards a lower tumour volume in animals treated with amiodarone compared to the DMSO control, but it was not statistically significant (Figure S6f). Overall, our results demonstrate that depolarization of TNBC cells via amiodarone treatment leads to decreased migration and a reduction in the number of lung metastases in vivo.
Discussion
Here we show that hyperpolarization of TNBC cells through overexpression of K+ channels leads to enhanced cell invasion, tumour growth, and metastasis. We provide the first characterization of hyperpolarization-induced changes in gene expression in cancer, which included upregulation of genes associated with cell adhesion and MAPK signalling. We also identify a novel hyperpolarization-driven mechanism of cell migration mediated by cadherin-11 and MAPK signalling. These effects were observed with the overexpression of two different types of K+ channels whose activity is modulated by different stimuli, suggesting that the effects seen are a result of the RMP hyperpolarization and not a channel-specific phenomenon. Lastly, we demonstrate that the bioelectric state of breast cancer cells can be targeted pharmacologically using the K+ channel blocker amiodarone to reduce breast cancer cell migration and lung metastasis in a mouse model of TNBC.
Our study demonstrates that hyperpolarization of breast tumour cells increases invasion and metastasis and that depolarization of cells can be used to reduce metastatic burden. Previously it has been reported that breast cancer cells are less negative than their normal counterparts, and that this depolarization is therefore associated with tumour progression. However, this finding arises largely from observations of individual cells in vitro experiments, and has not been well studied in patient samples. One study found that the membrane potential of 9 infiltrating ductal carcinomas was more depolarized than benign breast tissue, however this is a small sample size and the cancer subtype of each sample is not known.64 Additionally, a caveat with this method is that as soon as the tissue is extracted and placed on a dish, the bioelectric properties will be altered. In another study, the surface impedance of the skin of women with cancerous masses was found to be more electropositive relative to the skin of women with benign masses.65 While this is not a measure of cellular RMP, the authors do suggest that increase impedance is caused by an increase in K+ channel activity, which is in line with our findings. Further, it is well known that transient hyperpolarization is needed for progression from G1 phase to the S phase of the cell cycle and for replication to begin.66, 67, 68 Tumors proliferate at a much high rate than healthy tissues, suggesting that more cells are likely to be transiently hyperpolarized in a tumour compared to healthy tissue. The overall bioelectrical properties of a tumour are challenging to quantify due to 1) heterogeneity within tumors, 2) the fact that it is not a function of single cells' RMP but of many cells' bioelectrical state and their connections via gap junctions, and 3) the lack of available methods to quantify this in vivo. However, there is ample evidence from these published studies to support the concept that tumorigenesis may in fact be associated with hyperpolarization, which is in line with our findings that potassium channels are upregulated in TNBC and that hyperpolarization increases tumour progression and metastasis.
We further show that K+ channels modulate tumour cell invasion via changes in the cellular RMP that impact cell adhesion and MAPK signalling pathways. Ion channels have been previously shown to regulate tumour cell migration. Indeed, blocking sodium channels inhibits cell migration and invasion in MDA-MB-231 and MCF7 breast cancer cells,9,69,70 however whether these effects are RMP-dependant is not discussed. Several potassium channels that are known to impact K+ currents have also been linked to migration. Knockdown of the Ca2+-activated potassium channel SK3 in breast cancer cells, which regulates K+ currents in these cells, reduced migration in a transwell assay in vitro.14 A voltage-gated potassium channel, Kv10.1 has also been implicated in driving hyperpolarization of MDA-MB-231 breast cancer cells through Orai1-mediated Ca2+ entry, and silencing Kv10.1 results in cell depolarization, reduced Ca2+ influx, and reduced cell migration without change in cell proliferation.71 In contrast, activating the voltage-gated K+ channel Kv11.1 has been shown to inhibit metastasis and EMT in a mouse breast cancer model,21 however effects of Kv11.1 on cell migration and metastasis have not been shown to be dependant on changes in the RMP. In contrast, our results suggest that hyperpolarization via K+ channel expression leads to increased breast cancer cell invasion.71 In line with our results are other reports that hyperpolarization can drive migration in cancer and other cell types.14,71,72 For example, blocking K+ channel KCa2.3 activity in hyperpolarized MDA-MB-435 melanoma cancer cells inhibits 3D Matrigel invasion71 and 2D migration,73 respectively. Furthermore, hyperpolarization has also been linked to a redistribution of F-actin towards the plasma membrane and an increase in adherens junction stability in bovine corneal endothelial cells,74 consistent with our observed upregulation of cell adhesion-associated genes. Our results also support a model of hyperpolarization-driven transcription of CDH11 and increased cadherin-11 levels in TNBC cells. Cadherin-11 has been previously shown to drive cell invasion and metastasis by two main pathways. First, it regulates TNBC cell migration by increasing β-catenin nuclear localization and the transcription of Wnt targets Met, c-Myc, c-Jun, MMP7, Sox2, CD44, KLF4.36 However, the mRNA levels of these targets were unchanged in our dataset with the MDA-MB-231 or MDA-MB-468. Secondly, Cadherin-11 promotes paxillin phosphorylation, and adhesion formation,52 as well as Rac-driven actin dynamics,58 leading to downstream MAPK signalling and cell motility. We also observed that in vitro amiodarone treatment significantly reduced cadherin-11 levels in MDA-MB-231 cells, which supports our findings of a correlation between RMP, cadherin-11, and cell migration. Although further work will be needed to fully elucidate the mechanisms involved, overall, our results suggest that control of cancer cell migration and invasion by the RMP may be more complex than previously appreciated.
In addition to demonstrating that TNBC RMP is an important regulator of cell migration, invasion, and metastasis, we also show that it can be used to target metastasis by depolarizing MDA-MB-231 cells with low-dose amiodarone. Although we did see an anti-proliferative effect of amiodarone in vitro at the higher concentration in MDA-MB-231 and MDA-MB-468 cells, in xenograft mice there was a significant reduction in lung metastases with no difference in tumour volume, suggesting that at the dosage used the anti-proliferative effect of amiodarone was minimal. It would be interesting to investigate whether higher doses of amiodarone would have effects on tumour size, in addition to effects on metastasis. It is worth noting that in addition to blocking outward potassium channels, amiodarone is reported to block inward sodium and calcium currents in cardiac cells under certain conditions75, 76, 77 therefore we cannot fully attribute the observed depolarization of MDA-MB-231 cells with amiodarone to K+ channel blocking alone. Nevertheless, we show that the depolarization produced by amiodarone treatment results in reduced TNBC cell migration and metastasis, demonstrating that the cellular RMP is a targetable feature of TNBC cells that can be exploited to treat metastasis. Importantly, our results demonstrate that the anti-cancer effects of amiodarone can be achieved with lower doses than the equivalent standard therapeutic levels used for cardiac indications,78,79 avoiding previously reported off-target toxicity.80,81 Ion channel blockers such as amiodarone present a rich resource for drug repurposing due to their ubiquity and known safety profiles.5,82,83 Interestingly, amiodarone has also been shown to enhance the toxicity of standard of care chemotherapy drugs for TNBC such as doxorubicin, albeit in other tumour types.84, 85, 86 Amiodarone decreased the resistance of cancer cells to doxorubicin in vitro87 and increased efficacy of doxorubicin in a phase II clinical trial,88 both in hepatocellular carcinoma. In prostate cancer cell culture, liposomes loaded with amiodarone and doxorubicin have enhanced toxicity compared to doxorubicin alone.86 Given that chemotherapeutic drugs impact cell proliferation, and that transient hyperpolarization is needed for progression through the cell cycle,89, 90, 91 it is possible that disruption of the cellular RMP by blocking K+ channels with amiodarone also could enhance the effect of chemotherapeutic drugs.
In conclusion, we demonstrate that TNBC cell hyperpolarization drives migration, invasion and metastasis which can be targeted by depolarizing the RMP pharmacologically. This hyperpolarization is associated with striking changes in gene expression including upregulation of cell adhesion and MAPK pathways involving cadherin-11 signalling. Our results reveal the importance of RMP for pro-migratory signalling pathways in cancer and identify a new therapeutic target for breast cancer metastasis.
CRediT authorship contribution statement
Samantha L. Payne: Project administration, Conceptualization, Visualization, Methodology, Formal analysis, Writing – original draft, Validation. Priyanka Ram: Methodology, Software, Formal analysis. Deepti H. Srinivasan: Methodology, Supervision. Thanh T. Le: Formal analysis. Michael Levin: Project administration, Conceptualization, Visualization. Madeleine J. Oudin: Project administration, Conceptualization, Visualization, Writing – original draft, Validation.
Declaration of interests
ML receives funding from Augmanity Inc. for a separate project. There are no other conflicts.
Acknowledgments
Acknowledgements
We thank Mattia Bonzanni for performing electrophysiology experiments, and David Kaplan for support. We also thank members of the Oudin lab for manuscript review. We thank Steven Kwok and the staff at the Tufts University School of Medicine Laser Cytometry core facility for performing FACS. (S10 OD016196–01). We also thank the Tufts University Core Facility Genomics for performing RNA sequencing (NIH 1S10OD016196–01), the Tufts University Animal Histology Core for processing tissue samples and acknowledge NIH Research Infrastructure grant S10 OD021624 for the microscopy core.
This work was supported by the National Institutes of Health (R00-CA207866 to M.J.O.), Tufts University (Start-up funds from the School of Engineering to M.J.O., Tufts Collaborates Award to M.J.O. and M.L.), Allen Discovery centre program (Paul G. Allen Frontiers Group (12171) to M.L.), and Breast Cancer Alliance Young Investigator Grant to M.J.O, Laidlaw Scholar funding to D.S. M.L. also gratefully acknowledges support of the Barton Family Foundation.
Data sharing statement
All RNA-seq raw data generated for the present study, along with counts matrices and metadata for each sample, are publicly available in GEO under accession code GSE171150. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103767.
Appendix. Supplementary materials
References
- 1.Scott L.C., Mobley L.R., Kuo T.M., Il'yasova D. Update on triple-negative breast cancer disparities for the United States: a population-based study from the United States Cancer Statistics database, 2010 through 2014. Cancer. 2019;125(19):3412–3417. doi: 10.1002/cncr.32207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent R., Hanna W.M., Trudeau M., Rawlinson E., Sun P., Narod S.A. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115(2):423–428. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang J., Lu Z., Wu C., Li Y., Kong Y., Zhou R., et al. Evaluation of the anticancer and anti-metastasis effects of novel synthetic sodium channel blockers in prostate cancer cells in vitro and in vivo. Prostate. 2019;79(1):62–72. doi: 10.1002/pros.23711. [DOI] [PubMed] [Google Scholar]
- 4.Yang M., Kozminski D.J., Wold L.A., Modak R., Calhoun J.D., Isom L.L., et al. Therapeutic potential for phenytoin: targeting Nav1.5 sodium channels to reduce migration and invasion in metastatic breast cancer. Breast Cancer Res Treat. 2012;134(2):603–615. doi: 10.1007/s10549-012-2102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuszynski J.M., Tilli T., Levin M. Ion channel and neurotransmitter modulators as electroceutical approaches to the control of cancer. Curr Pharm Des. 2017;23:4827–4841. doi: 10.2174/1381612823666170530105837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernet B.T., Adams D.S., Lobikin M., Levin M. Use of genetically encoded, light-gated ion translocators to control tumorigenesis. Oncotarget. 2016;7(15) doi: 10.18632/oncotarget.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernet B.T., Levin M. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in a Xenopus model. Dis Model Mech. 2013;6(3):595–607. doi: 10.1242/dmm.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright S.H. Generation of resting membrane potential. Adv Physiol Educ. 2004;28(4):139–142. doi: 10.1152/advan.00029.2004. [DOI] [PubMed] [Google Scholar]
- 9.Fraser S.P., Diss J.K.J., Chioni A.M., Mycielska M.E., Pan H., Yamaci R.F., et al. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11(15):5381–5389. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 10.Al Ahmad M., Al Natour Z., Mustafa F., Rizvi T.A. Electrical characterization of normal and cancer cells. IEEE Access. 2018;6:25979–25986. [Google Scholar]
- 11.Djamgoz M.B.A., Fraser S.P., Brackenbury W.J. In vivo evidence for voltage-gated sodium channel expression in carcinomas and potentiation of metastasis. Cancers (Basel) [Internet] 2019 Oct 28;11(11):1675. doi: 10.3390/cancers11111675. https://www.mdpi.com/2072-6694/11/11/1675 cited 2019 Dec 4] Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roger S., Rollin J., Barascu A., Besson P., Raynal P.-.I., Iochmann S., et al. Voltage-gated sodium channels potentiate the invasive capacities of human non-small-cell lung cancer cell lines. Int J Biochem Cell Biol [Internet] 2007 Jan 1;39(4):774–786. doi: 10.1016/j.biocel.2006.12.007. https://www.sciencedirect.com/science/article/pii/S1357272507000180 cited 2019 May 4]Available from. [DOI] [PubMed] [Google Scholar]
- 13.Pardo-Pastor C., Rubio-Moscardo F., Vogel-González M., Serra S.A., Afthinos A., Mrkonjic S., et al. Piezo2 channel regulates RhoA and actin cytoskeleton to promote cell mechanobiological responses. Proc Natl Acad Sci U S A [Internet]. 2018 Feb 20;115(8):1925–1930. doi: 10.1073/pnas.1718177115. http://www.ncbi.nlm.nih.gov/pubmed/29432180 cited 2019 Aug 5] Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potier M., Joulin V., Roger S., Besson P., Jourdan M.-.L., LeGuennec J.-.Y., et al. Identification of SK3 channel as a new mediator of breast cancer cell migration. Mol Cancer Ther [Internet] 2006 Nov 1;5(11):2946–2953. doi: 10.1158/1535-7163.MCT-06-0194. http://www.ncbi.nlm.nih.gov/pubmed/17121942 cited 2019 May 5] Available from. [DOI] [PubMed] [Google Scholar]
- 15.Gradek F., Lopez-Charcas O., Chadet S., Poisson L., Ouldamer L., Goupille C., et al. Sodium channel nav1.5 controls epithelial-to-mesenchymal transition and invasiveness in breast cancer cells through its regulation by the salt-inducible kinase-1. Sci Rep [Internet] 2019;9(1):18652. doi: 10.1038/s41598-019-55197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X., Jan L.Y. Targeting potassium channels in cancer. J Cell Biol. 2014;206(2):151–162. doi: 10.1083/jcb.201404136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne S., Levin M., Oudin M. Bioelectric control of metastasis in solid tumors. Bioelectricty. 2019;1(3) doi: 10.1089/bioe.2019.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Şişman A.R., Sis B., Canda T., Önvural B. Electrolytes and trace elements in human breast cyst fluid. Biol Trace Elem Res. 2009;128(1):18–30. doi: 10.1007/s12011-008-8253-1. [DOI] [PubMed] [Google Scholar]
- 19.Chantome A., Potier-Cartereau M., Clarysse L., Fromont G., Marionneau-Lambot S., Gueguinou M., et al. Pivotal role of the lipid raft SK3-Orai1 complex in human cancer cell migration and bone metastases. Cancer Res [Internet] 2013 Aug 1;73(15):4852–4861. doi: 10.1158/0008-5472.CAN-12-4572. http://www.ncbi.nlm.nih.gov/pubmed/23774210 cited 2019 May 4] Available from. [DOI] [PubMed] [Google Scholar]
- 20.Schwab A., Fabian A., Hanley P.J., Stock C. Role of ion channels and transporters in cell migration. Physiol Rev [Internet] 2012 Oct;92(4):1865–1913. doi: 10.1152/physrev.00018.2011. http://www.physiology.org/doi/10.1152/physrev.00018.2011 cited 2019 May 3] Available from. [DOI] [PubMed] [Google Scholar]
- 21.Breuer E.K., Fukushiro-Lopes D., Dalheim A., Burnette M., Zartman J., Kaja S., et al. Potassium channel activity controls breast cancer metastasis by affecting β-catenin signaling. Cell Death Dis. 2019 Mar 1;10(3) doi: 10.1038/s41419-019-1429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.d'Alessandro G., Catalano M., Sciaccaluga M., Chece G., Cipriani R., Rosito M., et al. KCa3. 1 channels are involved in the infiltrative behavior of glioblastoma in vivo. Cell Death Dis. 2013;4(8):e773. doi: 10.1038/cddis.2013.279. –e773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantziarka P., Verbaanderd C., Sukhatme V., Rica Capistrano I., Crispino S., Gyawali B., et al. Redo_DB: the repurposing drugs in oncology database. Ecancermedicalscience. 2018;12:1–19. doi: 10.3332/ecancer.2018.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prevarskaya N., Skryma R., Shuba Y. Ion channels in cancer: are cancer hallmarks oncochannelopathies? Physiol Rev. 2018;98(2):559–621. doi: 10.1152/physrev.00044.2016. [DOI] [PubMed] [Google Scholar]
- 25.Sousounis K., Erdogan B., Levin M., Whited J.L. Precise control of ion channel and gap junction expression is required for patterning of the regenerating axolotl limb. Int J Dev Biol. 2020;64:485–494. doi: 10.1387/ijdb.200114jw. (10-11–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonzanni M., Payne S.L., Adelfio M., Kaplan D.L., Levin M., Oudin M.J. Defined extracellular ionic solutions to study and manipulate the cellular resting membrane potential. Biol Open. 2020;9(bio048553) doi: 10.1242/bio.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neher E. 6]Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol. 1992;207:123–131. doi: 10.1016/0076-6879(92)07008-c. [DOI] [PubMed] [Google Scholar]
- 28.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., et al. A survey of best practices for RNA-seq data analysis. Genome Biol [Internet] 2016 Dec 26;17(1):13. doi: 10.1186/s13059-016-0881-8. http://genomebiology.com/2016/17/1/13 cited 2019 Mar 18] Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. Babraham Bioinformatics, Babraham Institute, Cambridge; United Kingdom: 2010. FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics [Internet] 2013 Jan 1;29(1):15–21. doi: 10.1093/bioinformatics/bts635. https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/bts635 cited 2019 Mar 18] Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varet H., Brillet-Guéguen L., Coppée J.-.Y., Dillies M.-.A. SARTools: a DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE. 2016;11(6) doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha V.K., Lindgren C.M., Eriksson K.-.F., Subramanian A., Sihag S., Lehar J., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet [Internet] 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci [Internet] 2005 Oct 25;102(43) doi: 10.1073/pnas.0506580102. http://www.pnas.org/content/102/43/15545.abstract 15545 LP –15550. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satriyo P.B., Bamodu O.A., Chen J.-.H., Aryandono T., Haryana S.M., Yeh C.-.T., et al. Cadherin 11 inhibition downregulates β-catenin, deactivates the canonical WNT signalling pathway and suppresses the cancer stem cell-like phenotype of triple negative breast cancer. J Clin Med. 2019;8(2):148. doi: 10.3390/jcm8020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roussos E.T., Balsamo M., Alford S.K., Wyckoff J.B., Gligorijevic B., Wang Y., et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011;124(13):2120–2131. doi: 10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. AACR; 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams D.S., Levin M. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4 (3) and CC2-DMPE. Cold Spring Harb Protoc. 2012;2012(4) doi: 10.1101/pdb.prot067702. pdb-prot067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yankaskas C.L., Thompson K.N., Paul C.D., Vitolo M.I., Mistriotis P., Mahendra A., et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat Biomed Eng. 2019;3(6):452–465. doi: 10.1038/s41551-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMillen P., Novak R., Levin M. Toward decoding bioelectric events in xenopus embryogenesis: new methodology for tracking interplay between calcium and resting potentials in vivo. J Mol Biol. 2020;432(2):605–620. doi: 10.1016/j.jmb.2019.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams D.S., Uzel S.G.M., Akagi J., Wlodkowic D., Andreeva V., Yelick P.C., et al. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen–Tawil Syndrome. J Physiol. 2016;594(12):3245–3270. doi: 10.1113/JP271930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pai V.P., Lemire J.M., Paré J.-.F., Lin G., Chen Y., Levin M. Endogenous gradients of resting potential instructively pattern embryonic neural tissue via notch signaling and regulation of proliferation. J Neurosci. 2015;35(10):4366–4385. doi: 10.1523/JNEUROSCI.1877-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szymczak-Workman A.L., Vignali K.M., Vignali D.A.A. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb Protoc. 2012;2012(2) doi: 10.1101/pdb.ip067876. pdb-ip067876. [DOI] [PubMed] [Google Scholar]
- 45.Roger S., Besson P., Le Guennec J.-.Y. Involvement of a novel fast inward sodium current in the invasion capacity of a breast cancer cell line. Biochim Biophys Acta - Biomembr [Internet] 2003 Oct 13;1616(2):107–111. doi: 10.1016/j.bbamem.2003.07.001. https://www.sciencedirect.com/science/article/pii/S0005273603002347 cited 2019 May 4] Available from. [DOI] [PubMed] [Google Scholar]
- 46.Laniado M.E., Fraser S.P., Djamgoz M.B.A. Voltage-gated K+ channel activity in human prostate cancer cell lines of markedly different metastatic potential: distinguishing characteristics of PC-3 and LNCaP cells. Prostate. 2001;46(4):262–274. doi: 10.1002/1097-0045(20010301)46:4<262::aid-pros1032>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 47.Wu P.-.H., Gambhir S.S., Hale C.M., Chen W.-.C., Wirtz D., Smith B.R. Particle tracking microrheology of cancer cells in living subjects. Mater Today [Internet] 2020;39:98–109. https://www.sciencedirect.com/science/article/pii/S1369702120300985 Available from. [Google Scholar]
- 48.Baskaran J.P., Weldy A., Guarin J., Munoz G., Shpilker P.H., Kotlik M., et al. Cell shape, and not 2D migration, predicts extracellular matrix-driven 3D cell invasion in breast cancer. APL Bioeng. 2020;4(2):26105. doi: 10.1063/1.5143779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu P.-.H., Phillip J.M., Khatau S.B., Chen W.-.C., Stirman J., Rosseel S., et al. Evolution of cellular morpho-phenotypes in cancer metastasis. Sci Rep. 2015;5(1):1–10. doi: 10.1038/srep18437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews J.L., Kim A.C., Hens J.R. The role and function of cadherins in the mammary gland. Breast Cancer Res. 2012;14(1):1–10. doi: 10.1186/bcr3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Guo Z., Chen H., Dong Z., Pan Z.K., Ding H., et al. HOXC8-dependent cadherin 11 expression facilitates breast cancer cell migration through Trio and Rac. Genes Cancer. 2011;2(9):880–888. doi: 10.1177/1947601911433129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langhe R.P., Gudzenko T., Bachmann M., Becker S.F., Gonnermann C., Winter C., et al. Cadherin-11 localizes to focal adhesions and promotes cell–substrate adhesion. Nat Commun. 2016;7(1):1–10. doi: 10.1038/ncomms10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pohlodek K., Tan Y.Y., Singer C.F., Gschwantler‑Kaulich D. Cadherin-11 expression is upregulated in invasive human breast cancer. Oncol Lett. 2016;12(6):4393–4398. doi: 10.3892/ol.2016.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pishvaian M.J., Feltes C.M., Thompson P., Bussemakers M.J., Schalken J.A., Byers S.W. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59(4):947–952. [PubMed] [Google Scholar]
- 55.Chen J.-.H., Huang W.-.C., Bamodu O.A., Chang P.M.-.H., Chao T.-.Y., Huang T.-.H. Monospecific antibody targeting of CDH11 inhibits epithelial-to-mesenchymal transition and represses cancer stem cell-like phenotype by up-regulating miR-335 in metastatic breast cancer, in vitro and in vivo. BMC Cancer. 2019;19(1):1–13. doi: 10.1186/s12885-019-5811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanagisawa M., Anastasiadis P.Z. p120 catenin is essential for mesenchymal cadherin–mediated regulation of cell motility and invasiveness. J Cell Biol. 2006;174(7):1087–1096. doi: 10.1083/jcb.200605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madarampalli B., Watts G.F.M., Panipinto P.M., Nguygen H.N., Brenner M.B., Noss E.H. Interactions between cadherin-11 and platelet-derived growth factor receptor-alpha signaling link cell adhesion and proliferation. Biochim Biophys Acta - Mol Basis Dis [Internet] 2019;1865(6):1516–1524. doi: 10.1016/j.bbadis.2019.03.001. https://www.sciencedirect.com/science/article/pii/S0925443919300754 Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Gao Y., Imsland F., Gu X., Feng C., Liu R., et al. The crest phenotype in chicken is associated with ectopic expression of HOXC8 in cranial skin. PLoS ONE. 2012;7(4):e34012. doi: 10.1371/journal.pone.0034012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pantziarka P., Bouche G., Meheus L., Sukhatme V., Sukhatme V.P., Vikas P. The repurposing drugs in oncology (ReDO) project. Ecancermedicalscience [Internet] 2014;8:442. doi: 10.3332/ecancer.2014.442. http://www.ncbi.nlm.nih.gov/pubmed/25075216 cited 2019 Mar 21] Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato R., Koumi S., Singer D.H., Hisatome I., Jia H., Eager S., et al. Amiodarone blocks the inward rectifier potassium channel in isolated guinea pig ventricular cells. J Pharmacol Exp Ther. 1994;269(3):1213–1219. [PubMed] [Google Scholar]
- 61.Zhang Y., Colenso C.K., El Harchi A., Cheng H., Witchel H.J., Dempsey C.E., et al. Interactions between amiodarone and the hERG potassium channel pore determined with mutagenesis and in silico docking. Biochem Pharmacol [Internet] 2016;113:24–35. doi: 10.1016/j.bcp.2016.05.013. https://www.sciencedirect.com/science/article/pii/S000629521630106X Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H., Kim H.S., Kim H.W., Shin S.E., Jung W.-.K., Ha K.-.S., et al. The class III anti-arrhythmic agent, amiodarone, inhibits voltage-dependent K+ channels in rabbit coronary arterial smooth muscle cells. Naunyn Schmiedebergs Arch Pharmacol [Internet] 2016;389(7):713–721. doi: 10.1007/s00210-016-1232-8. [DOI] [PubMed] [Google Scholar]
- 63.Huang S.-.T., Hsu W.-.F., Huang H.-.S., Yen J.-.H., Lin M.-.C., Peng C.-.Y., et al. Improved survival in hepatocellular carcinoma patients with cardiac arrhythmia by amiodarone treatment through autophagy. Int J Mol Sci. 2019;20(16):3978. doi: 10.3390/ijms20163978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marino A.A., Iliev I.G., Schwalke M.A., Gonzalez E., Marler K.C., Flanagan C.A. Association between cell membrane potential and breast cancer. Tumor Biol. 1994;15(2):82–89. doi: 10.1159/000217878. [DOI] [PubMed] [Google Scholar]
- 65.Marmo A.A., Morris D.M., Schwalke M.A., Iliev I.G., Rogers S. Electrical potential measurements in human breast cancer and benign lesions. Tumor Biol. 1994;15(3):147–152. doi: 10.1159/000217885. [DOI] [PubMed] [Google Scholar]
- 66.Blackiston D.J., McLaughlin K.A., Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8(21):3527–3536. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.WSMZWKCCD A.G. Evidence for an early G1 ionic event necessary for cell cycle progression and survival in the MCF-7 human breast carcinoma cell line. J Cell Physiol. 1998;176(3):456–464. doi: 10.1002/(SICI)1097-4652(199809)176:3<456::AID-JCP2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 68.Urrego D., Tomczak A.P., Zahed F., Stühmer W., Pardo L.A. Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc B Biol Sci. 2014;369(1638) doi: 10.1098/rstb.2013.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang M., James A.D., Suman R., Kasprowicz R., Nelson M., O'Toole P.J., et al. Voltage-dependent activation of Rac1 by Nav1. 5 channels promotes cell migration. J Cell Physiol. 2020;235(4):3950–3972. doi: 10.1002/jcp.29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo Q., Wu T., Wu W., Chen G., Luo X., Jiang L., et al. The functional role of voltage-gated sodium channel Nav1.5 in metastatic breast cancer [Internet]. Vol. 11. Front Pharmacol. 2020:1111. doi: 10.3389/fphar.2020.01111. https://www.frontiersin.org/article/10.3389/fphar.2020.01111 Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chantome A., Girault A., Potier M., Collin C., Vaudin P., Pagès J.-.C., et al. KCa2.3 channel-dependent hyperpolarization increases melanoma cell motility. Exp Cell Res [Internet] 2009 Dec 10;315(20):3620–3630. doi: 10.1016/j.yexcr.2009.07.021. https://www.sciencedirect.com/science/article/pii/S0014482709003334 [cited 2019 May 5] Available from. [DOI] [PubMed] [Google Scholar]
- 72.Rao J.N., Platoshyn O., Li L., Guo X., Golovina V.A., Yuan J.X.-.J., et al. Activation of K+ channels and increased migration of differentiated intestinal epithelial cells after wounding. Am J Physiol Physiol. 2002;282(4):C885–C898. doi: 10.1152/ajpcell.00361.2001. [DOI] [PubMed] [Google Scholar]
- 73.Mayorga-Flores M., Chantôme A., Melchor-Meneses C.M., Domingo I., Titaux-Delgado G.A., Galindo-Murillo R., et al. Novel blocker of onco SK3 channels derived from scorpion toxin tamapin and active against migration of cancer cells. ACS Med Chem Lett. 2020;11(8):1627–1633. doi: 10.1021/acsmedchemlett.0c00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nin V., Hernández J.A., Chifflet S. Hyperpolarization of the plasma membrane potential provokes reorganization of the actin cytoskeleton and increases the stability of adherens junctions in bovine corneal endothelial cells in culture. Cell Motil Cytoskeleton. 2009;66(12):1087–1099. doi: 10.1002/cm.20416. [DOI] [PubMed] [Google Scholar]
- 75.Varró A., Virág L., Papp J.G. Comparison of the chronic and acute effects of amiodarone on the calcium and potassium currents in rabbit isolated cardiac myocytes. Br J Pharmacol [Internet] 1996 Mar 1;117(6):1181–1186. doi: 10.1111/j.1476-5381.1996.tb16713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lalevée N., Barrère-lemaire S., Gautier P., Nargeot J., Richard S. Effects of amiodarone and dronedarone on voltage-dependent sodium current in human cardiomyocytes. J Cardiovasc Electrophysiol [Internet] 2003 Aug 1;14(8):885–890. doi: 10.1046/j.1540-8167.2003.03064.x. [DOI] [PubMed] [Google Scholar]
- 77.Banach M., Popławska M., Borowicz-Reutt K.K. Amiodarone, a multi-channel blocker, enhances anticonvulsive effect of carbamazepine in the mouse maximal electroshock model. Epilepsy Res [Internet] 2018;140:105–110. doi: 10.1016/j.eplepsyres.2018.01.003. https://www.sciencedirect.com/science/article/pii/S0920121117304588 Available from. [DOI] [PubMed] [Google Scholar]
- 78.Steinberg E., Fluksman A., Zemmour C., Tischenko K., Karsch-Bluman A., Brill-Karniely Y., et al. Low dose amiodarone reduces tumor growth and angiogenesis. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-75142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamilton D., Nandkeolyar S., Lan H., Desai P., Evans J., Hauschild C., et al. Amiodarone: a comprehensive guide for clinicians. Am J Cardiovasc Drugs. 2020:1–10. doi: 10.1007/s40256-020-00401-5. [DOI] [PubMed] [Google Scholar]
- 80.Basaria S., Cooper D.S. Amiodarone and the thyroid. Am J Med. 2005;118(7):706–714. doi: 10.1016/j.amjmed.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 81.Martino E., Bartalena L., Bogazzi F., Braverman L.E. The effects of amiodarone on the thyroid. Endocr Rev. 2001;22(2):240–254. doi: 10.1210/edrv.22.2.0427. [DOI] [PubMed] [Google Scholar]
- 82.Kale V.P., Amin S.G., Pandey M.K. Targeting ion channels for cancer therapy by repurposing the approved drugs. Biochim Biophys Acta (BBA)-Biomembranes. 2015;1848(10):2747–2755. doi: 10.1016/j.bbamem.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 83.Leanza L., Manago A., Zoratti M., Gulbins E., Szabo I. Pharmacological targeting of ion channels for cancer therapy: in vivo evidences. Biochim Biophys Acta (BBA)-Molecular Cell Res. 2016;1863(6):1385–1397. doi: 10.1016/j.bbamcr.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 84.Chauffert B., Martin M., Hammann A., Michel M.F., Martin F. Amiodarone-induced enhancement of doxorubicin and 4′-deoxydoxorubicin cytotoxicity to rat colon cancer cells in vitro and in vivo. Cancer Res. 1986;46(2):825–830. [PubMed] [Google Scholar]
- 85.van der Graaf W.T.A., de Vries E.G.E., Timmer-Bosscha H., Meersma G.J., Mesander G., Vellenga E., et al. Effects of amiodarone, cyclosporin A, and PSC 833 on the cytotoxicity of mitoxantrone, doxorubicin, and vincristine in non-P-glycoprotein human small cell lung cancer cell lines. Cancer Res. 1994;54(20):5368–5373. [PubMed] [Google Scholar]
- 86.Theodossiou T.A., Galanou M.C., Paleos C.M. Novel amiodarone− doxorubicin cocktail liposomes enhance doxorubicin retention and cytotoxicity in du145 human prostate carcinoma cells. J Med Chem. 2008;51(19):6067–6074. doi: 10.1021/jm800493j. [DOI] [PubMed] [Google Scholar]
- 87.Favoulet P., Cercueil J.P., Faure P., Osmak L., Isambert N., Beltramo J.L., et al. Increased cytotoxicity and stability of lipiodol-pirarubicin emulsion compared to classical doxorubicin-lipiodol: potential advantage for chemoembolization of unresectable hepatocellular carcinoma. Anticanc Drugs. 2001;12(10):801–806. doi: 10.1097/00001813-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 88.Boulin M., Ciboulet A., Guiu B., Maillard E., Bonnetain F., Minello A., et al. Randomised controlled trial of lipiodol transarterial chemoembolisation with or without amiodarone for unresectable hepatocellular carcinoma. Dig Liver Dis. 2011;43(11):905–911. doi: 10.1016/j.dld.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 89.Strobl J.S., Cather C., Wang S., Melkoumian Z., Woodfork K.A., Davidson A.G., et al. Evidence for an early G1 ionic event necessary for cell cycle progression and survival in the MCF-7 human breast carcinoma cell line. J Cell Physiol. 2002;176(3):456–464. doi: 10.1002/(SICI)1097-4652(199809)176:3<456::AID-JCP2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 90.Wonderlin W.F., Woodfork K.A., Strobl J.S. Changes in membrane potential during the progression of MCF-7 human mammary tumor cells through the cell cycle. J Cell Physiol. 1995;165(1):177–185. doi: 10.1002/jcp.1041650121. [DOI] [PubMed] [Google Scholar]
- 91.Zhu L., Yu X., Xing S., Jin F., Yang W.-.J. Involvement of AMP-activated protein kinase (AMPK) in regulation of cell membrane potential in a gastric cancer cell Line. Sci Rep [Internet] 2018;8(1):6028. doi: 10.1038/s41598-018-24460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang X., He Y., Dubuc A.M., et al. EAG2 potassium channel with evolutionarily conserved function as a brain tumor target. Nat Neurosci. 2015;18:1236–1246. doi: 10.1038/nn.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.