Abstract
The G-quadruplexes (G4s) are a class of DNA secondary structures with guanine rich DNA sequences that can fold into four stranded non-canonical structures. At the genomic level, their pivotal role is well established in DNA replication, telomerase functions, constitution of topologically associating domains, and the regulation of gene expression. Genome instability mediated by altered G4 formation and assembly has been associated with multiple disorders including cancers and neurodegenerative disorders. Multiple tools have also been developed to predict the potential G4 regions in genomes and the whole genome G4 maps are also being derived through sequencing approaches. Enrichment of G4s in the cis-regulatory elements of genes associated with tumorigenesis has accelerated the quest for identification of G4-DNA binding ligands (G4DBLs) that can selectively bind and regulate the expression of such specific genes. In this context, the analysis of G4DBL responsive transcriptome in diverse cancer cell lines is inevitable for assessment of the specificity of novel G4DBLs. Towards this, we assembled the transcripts differentially regulated by different G4DBLs and have also identified a core set of genes regulated in diverse cancer cell lines in response to 3 or more of these ligands. With the mode of action of G4DBLs towards topology shifts, folding, or disruption of G4 structure being currently visualized, we believe that this dataset will serve as a platform for assembly of G4DBL responsive transcriptome for comparative analysis of G4DBLs in multiple cancer cells based on the expression of specific cis-regulatory G4 associated genes in the future.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12079-021-00637-z.
Keywords: PhenDC3, TMPyP4, 360A, APTO-253, AQ1, Centriole assembly
Introduction
Guanine-rich DNA sequences which folds into four stranded non-canonical structure are called G-quadruplex DNA (G4 DNA) structures. G4s consists of several G-tetrad layers that comprise four planar guanines linked through Hoogsteen hydrogen bonding and exhibits an extremely high stability under physiological conditions with monovalent metal cations such as Na + and K + (Bacolla and Wells 2009). They adopt various alternative conformations based on the sequence motifs and its interactions with different proteins into parallel, hybrid and anti-parallel polymorphic G4 topologies (Gellert et al. 1962; Ma et al. 2020). The structural regulatory role of G4s involve both DNA and RNA and the roles played by the RNA G-quadruplexes (RNA G4s) are also gaining more attention in recent years.
The endogenous G4-DNA structures have been widely studied in the telomeric regions (Henderson et al. 1987; Granotier et al. 2005; Bryan 2020). Human G4-DNA maps using G4-seq have identified over 700,000 G4-DNA structures including over 450,000 that were not predicted by computational tools (Chambers et al. 2015). Formation of G-quadruplexes within the genome can be triggered by biological events such as replication, transcription, and repair processes that involves temporal dissociation of DNA strands (Johnson 2020). With the identification of a large number of G-quadruplex-protein interactions, the role of G4 DNA in chromatin remodeling is well perceived for comparative epigenetic analysis and epigenetic therapy potential (Sengupta et al. 2019; Reina and Cavalieri 2020; Varizhuk et al. 2019).
Various biological functions of G4-DNA may be derived from their sequence, stability, location in the genome, local environments, and combinations of these factors. G-quadruplex-stabilizing ligands have been used to study and understand such G-quadruplex functions (Law et al. 2010). The abundance of G4s in the cis-regulatory/promoter regions (Huppert et al. 2008; Verma et al. 2009) has accelerated the quest for G4-binding ligands (G4DBLs) that can selectively bind and stabilize G4s to suppress the expression of specific genes, especially, the genes associated with tumorigenesis. G4 DNA binding ligands are reported to block the progression of DNA polymerase and sometimes causes severe DNA damage, such as strand breakage (Schiavone et al. 2016; De Magis et al. 2019). Multiple G-quadruplex ligands (BRACO19, Pyridostatin, Phen-DC3, TMPyP4, L2H2-6OTD, and L1H1-7OTD) have been used for biochemical and biophysical studies on G-quadruplexes and their mode of action towards topology shifts, folding or disruption of G4 structure are being visualized (Jonchhe et al. 2018; O'Hagan et al. 2019). In particular, ligands with a specificity towards certain G-quadruplexes over others are desirable for the detailed investigation of the features and functions of individual G-quadruplexes in the genome (Monchaud and Teulade-Fichou 2008; Asamitsu et al. 2019). Based on the number of contiguous G-tracts and the residues between them, G4 sequences can adopt various topologies such as parallel, anti-parallel and hybrid type conformations (Burge et al. 2006). These structures are studied with the help of X-ray crystallography, NMR spectroscopy and Fluorescence imaging in combination with circular dichroism spectroscopy. These are dependent on custom labelled oligonucleotides as native bases for efficient analysis (Maleki et al. 2017). Most of these chemical probes may not be efficient in differentiating subtle differences in conformation of different G4 structures and thereby small molecule targeted inhibition of gene regulations may not be accurate due to the lack of conformational specificity. In this context, the ligands that has the potential to derive spatio-temporal control over the DNA structures are desirable (O'Hagan et al. 2019). Amidst the challenges associated with their identification for potential clinical use, the analysis of the specific-G4DBL responsive transcriptome in cancer cells is inevitable for the assessment of specificity of novel G4DBLs. Hence, we assembled the publicly available transcriptome datasets in response to diverse G4DBLs and have derived the core set of G4DBL-responsive genes that can serve as a platform for comparative analysis to screen and evaluate new G4DNA binding ligands.
Materials and methods
Screening and annotation of genes regulated by G4-DNA binding ligands
Research articles extracted by the PubMed search using the term (“G-quadruplex”) were screened for RNA-sequencing or microarray datasets in response to any G4-DNA binding ligands. The names of specific G4-DNA binding ligands obtained from these articles were also used for further search to obtain studies pertaining to their responsive transcriptome. The inclusion criteria for the genes differentially regulated by G4DBLs compared to the unstimulated or control were defined to an absolute fold change value of 1.5 with a p-value < 0.05. The annotation was restricted to the human cell line/ systems.
Assembly of G4DBL responsive genes and the extraction of core responsive genes
For uniform annotation and comparative analysis, the protein coding genes were first mapped to their corresponding Gene Symbol and Entrez Gene ID. Considering datasets from each of the studies as distinct and non-synchronous in their experimental contexts, we have also accommodated the study-specific information such as concentration of the ligands, time period of stimulation, control or reference used (such as whether unstimulated or DMSO was used a control), name of the cell line and the cell type that were stimulated, experiment type (whether, microarray or RNA-Seq), and also the PubMed ID reference to the research article.
The differentially regulated genes in two or more cancer cell models that were unidirectional in expression (either up-regulated or down-regulated) across all the time points in temporal datasets in response to three or more G4DBLs were categorized as core responsive genes.
Visualization of G4 sequences in genes
The core responsive genes were extracted from the annotated datasets as mentioned above. Human G4 whole genome map derived by G4-Seq (involving polymerase stop assay coupled with Illumina next-generation sequencing) using the G4 stabilizing ligand ‘Pyridostatin’ in primary human B lymphocytes (NA18507) was used to map the G4-DNA sequences in these core responsive genes (Chambers et al. 2015). The integrative genome browser (IGV) was used for visualization (Robinson et al. 2011).
Gene set enrichment analysis
The enrichment of differentially regulated genes for their involvement in biological processes and signaling pathways based on their pre-assembled database in the Cytoscape BINGO and gProfiler web server, respectively (Raudvere et al. 2019; Maere et al. 2005). A p-value < 0.05 was considered as a cut-off for selection of biological processes.
Results
Assembly of G4-DNA binding ligand induced differential transcriptome of cancer cell lines
A number of studies had been carried out to identify global or specific targets for G4DBLs (Izbicka et al. 1999; Pennarun et al. 2005; Hwang et al. 2019; Zorzan et al. 2016; Ahmed et al. 2020; Gray et al. 2019; Local et al. 2018) in human cell systems. A literature survey was carried out to identify published research articles that have undertaken global transcriptomic analysis to study the targets or effects of diverse G4DBLs in cancer cell lines. Following a defined criterion, we compiled the transcripts differentially regulated in response to 7 distinct G4DBLs reported in published research articles (Table 1). These G4DBLs were namely TMPyP4 (CID: 135,398,505), TMPyP4-PT (Zheng et al. 2016), 360A (CID: 11,430,774), AQ1 (CID: 135,567,364), CMO3 (Marchetti et al. 2018), PhenDC3 (CID: 44,449,504), and APTO-253 (CID: 11,960,271) (Kim et al. 2021). Together, following the inclusion criteria for considering differentially expressed genes, we assembled 25,228 temporal differential gene regulation events in response to these 7 G4DBLs across their diverse concentration and stimulation time points in six different human cancer cell line systems (Supplementary Table 1). There were 12,667 unique genes identified across these datasets that could be considered as the responsive gene set together for all these G4DBLs.
Table 1.
An overview of the assembled transcriptomic datasets
| G4DBL | Concentration of G4DBL | Time point | Differential expression type | No: of differentially regulated genes | Cell line_ Cell type | Experiment type | Reference |
|---|---|---|---|---|---|---|---|
| 360A | 10 μM | 48 h | Down | 215 | HeLa-S3 Cervix Carcinoma Cell line | Microarray | Halder et al. 2012, BMC Res Notes |
| Up | 312 | ||||||
| APTO-253 | 500 nM | 24 h | Down | 416 | MV4-11 Acute Myeloid Leukemia Cells line | RNA-Seq | Local et al. 2018, Mol Cancer Ther |
| 1277 | |||||||
| AQ1 | 2 mM | 12 h | Up | 3322 | HMC1.2 Mast Cell Leukemia Cell line | Zorzan et al. 2018, Sci Rep | |
| 2117 | |||||||
| 1 mM | Down | 2293 | |||||
| Up | 1349 | ||||||
| CMO3 | 400 nM | 6 h | Down | 832 | PANC1 Pancreatic Ductal Adenocarcinoma Cell line | Marchetti et al. 2018, J Med Chem | |
| Up | 644 | ||||||
| Down | 1636 | MIAPACA2 Pancreatic Ductal Adenocarcinoma Cell line | |||||
| Up | 1638 | ||||||
| 24 h | Down | 1567 | PANC1 Pancreatic Ductal Adenocarcinoma Cell line | ||||
| Up | 1462 | MIAPACA2 Pancreatic Ductal Adenocarcinoma Cell line | |||||
| Down | 1694 | ||||||
| Up | 1860 | ||||||
| PhenDC3 | 10 μM | 48 h | Down | 970 | HeLa-S3 Cervix Carcinoma Cell line | Microarray | Halder et al. 2012, BMC Res Notes |
| Up | 1242 | ||||||
| TMPyP4 | 100 μM | 24 h | Down | 9 | Verma et al. 2008, J Med Chem | ||
| 59 | |||||||
| 48 h | 67 | ||||||
| 0.5 μM | 43 | A549 Lung Cancer Cell line | RNA-Seq | Zheng et al. 2016, Sci Rep | |||
| Up | 56 | ||||||
| 2 μM | Down | 40 | |||||
| Up | 59 | ||||||
| TMPyP4-PT | 0.5 μM | Down | 18 | ||||
| Up | 81 |
Core responsive transcripts of multiple G4-DNA binding ligands
We next sought to extract a set of core genes for the analysis of their regulatory potential by multiple G4DBLs. Towards this, we filtered genes that were differentially regulated by 3 or more G4DBLs in our annotated datasets. A large number of genes differentially regulated across multiple G4DBL concentration and time points of stimulation in multiple cell line datasets showed oscillatory patterns of expression. Hence, we filtered out 49 genes that were up-regulated and 71 genes that were down-regulated consistently in a unidirectional manner across different time points of G4DBL stimulation (Supplementary Fig. 1). Considering that these genes were also regulated in 2 or more distinct cell lines/types, they are henceforth referred to as core responsive genes of the seven distinct G4DBLs from our annotated datasets.
Functional analysis of core responsive genes of G4-DNA binding ligands
Based on multiple studies that have analysed the effect of G4-DNA binders, it could be presumed that binding of the G4DBLs especially in the G-quadruplex structures of promoters may mostly lead to the negative regulation of expression of those genes. For example, Chelerythrine, a G4DBL identified to interact with the G4 structures on promoter regions of VEGEF, BCL2 and KRAS is reported to decrease the expression of these genes (Jana et al. 2017). Hoechst 33,258, a well-known ligand that binds to the promoter region of the human MYC gene is also reported to decrease its expression (Maiti et al. 2003). Hence, we carried out enrichment analysis of 49 up-regulated and 71 down-regulated genes separately to identify the potential biological process perturbed by them.
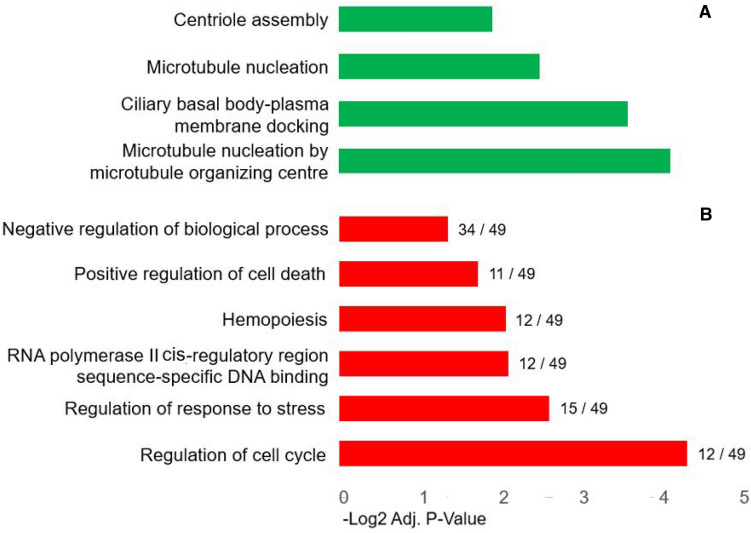
Within the set of 71 core genes that were down-regulated, a set of 8 genes (centromere protein J, CENPJ; centromere protein I, CENPI; centrosomal protein 192 kDa, CEP192; centrosomal protein 72 kDa, CEP72; centrosomal protein 250 kDa, CEN250; tubulin polymerization promoting protein, TPPP; tubulin gamma 1, TUBG1; RAB3A interacting protein, RAB3IP; Coiled-Coil Domain Containing 78, CCDC78) were enriched for centriole assembly, microtubule nucleation and ciliary basal body-plasma membrane docking (Fig. 1). Interestingly, treatment with another G4DBL, HXDV, have also been shown to result in defective chromosome alignment and spindle fibre assembly in HEK293 cells (Tsai et al. 2009).
Fig. 1.
Top selected biological processes perturbed by (a) up-regulated (49) and (b) down-regulated (71) core response genes
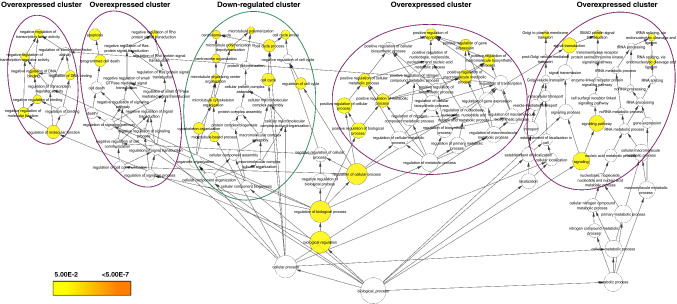
Further, we presumed that the genes up-regulated consistently at multiple time points may mostly orientate towards the cell response to suppression of gene expression as a result of G4DBLs binding to specific G-quadruplex structures in the promoter or other regions in the gene body. In this context, the 49 core genes upregulated were enriched for the RNA polymerase II cis-region sequence-specific DNA binding, response to stress, hemopoiesis, regulation of cell cycle and positive regulation of cell death. A biological process enrichment of these 120 genes together using Cytoscape BINGO analysis tool segregating these process is represented in Fig. 2. Together, leading to the positive regulation of apoptotic processes, it also implies that the genes involved in the centrosome assembly could be selectively targeted for anti-cancer therapy.
Fig. 2.
Combined Gene Ontology biological process enrichment analysis of core response genes of 7 G4DBLs
Visualization of G-quadruplex potential in G4-whole genome map
The G-quadruplex structure formation potential of the 71 core down-regulated genes and 49 up-regulated genes were visualized in the G4-Seq derived human G4 whole genome map by Chambers et al. (2015). Evidently, based on this a Pyridostatin treated ChIP-Seq data derived from primary human B lymphocytes, there were no enrichment of the G-quadruplex regions in the promoters for either up- or down- regulated gene set (Supplementary Fig. 2). Hence, it could be considered that most of these genes have a high propensity towards specific multiple G-quadruplex folding topologies and that the RNA G-quadruplex binding of the G4DBLs may also contribute to the regulation of their gene expression.
Discussion
Based on a whole genome screening for potential G4s, around 55.5% of the genes are estimated to contain a potential G4 motif within ± 1 kb region centered at their transcription start sites (Verma et al. 2008). One of the impending challenges associated with defining the expression of the genes regulated by G4DBLs is the intrinsic formation of a plethora of G4 topologies. This is also evident from the scarce number of genes that are down-regulated in response to each of these ligands compared to the expected number of cis-regulatory G-quadruplex harbored regions in genes. Ligands are being developed that can switch G4 topology, direct the selective covalent modification of nucleic acid structures, or respond to external stimuli to permit spatiotemporal control of their activity. An evaluation of such selective G4DBLs demands the analysis of gene expression profiles for their specificity. Towards this, we have derived a set of core genes (71 down-regulated and 49 up-regulated) that are transcriptionally regulated by 3 or more G4DBLs in multiple cancer cell line systems. With over 12,000 genes assembled as responsive to 7 distinct G4DBLs, our resource would also serve as a platform for investigation of the G-quadruplex topologies in these genes based on the specific G4DBL binding properties. To our knowledge, this is the first resource of G4DBL responsive transcriptome. We also propose to consider the RNA-level G4-quadruplex formation potential of these genes for the evaluation of specific G4DBL target effects in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Amjesh Revikumar is a recipient of National Post-Doctoral Fellowship (PDF/2017/001652) from the Science and Engineering Research Board, Department of Science and Technology (DST), Government of India. Rajesh Raju is a recipient of the Young Scientist Award (YSS/2014/000607) from the Science and Engineering Research Board, Department of Science and Technology (DST), Government of India. Anjana Aravind is a recipient of the Junior Research Fellowship from the Council of Scientific & Industrial Research (CSIR), Government of India. Akhina Palollathil is a recipient of Junior Research Fellowship from the University Grants Commission (UGC), Government of India.
Abbreviations
- G4
G-quadruplex
- G4DBLs
G4-DNA binding ligands
- TMPyP4
5,10,15,20–Tetrakis(N-methyl-4-pyridyl)porphyrin
- TMPyP4-PT
Metallated analogues of TMPyP4 with Zn(II), Pt(II)
- 360A
2-N,6-N-Bis(1-methylquinolin-1-ium-3-yl)pyridine-2,6-dicarboxamide
- AQ1
4-[(7-Chloroquinolin-4-yl)amino]-2-(diethylaminomethyl)phenol
- CMO3
Phenanthroline-1,3,6,8 (2H,7H)-tetraone
- PhenDC3
N2,N9-Bis(1-Methylquinolin-3-Yl)-1,10-Phenanthroline-2,9-Dicarboxamide
- APTO-253
L2-(5-fluoro-2-methyl-1H-indol-3-yl)-1H-imidazo[4,5f][1,10]phenanthroline;hydrochloride
Funding
Not applicable.
Data availability
The 25,228 temporal differential gene regulation events in response to 7 G4DBLs (with their diverse concentrations and stimulation time points) in six different human cancer cell lines is provided in the ‘.xlsx’ format (Supplementary table 1).
Declarations
Conflict of interest
Authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amjesh Revikumar, Email: amjesh@gmail.com.
Rajesh Raju, Email: rajeshraju@yenepoya.edu.in.
References
- Ahmed AA, Marchetti C, Ohnmacht SA, Neidle S. A G-quadruplex-binding compound shows potent activity in human gemcitabine-resistant pancreatic cancer cells. Sci Rep. 2020;10(1):12192. doi: 10.1038/s41598-020-68944-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamitsu S, Bando T, Sugiyama H. Ligand design to acquire specificity to intended G-quadruplex structures. Chemistry. 2019;25(2):417–430. doi: 10.1002/chem.201802691. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog. 2009;48(4):273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- Bryan TM. G-quadruplexes at telomeres: Friend or foe? Molecules. 2020;25(16):3683. doi: 10.3390/molecules25163686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucl Acids Res. 2006;34(19):5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33(8):877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, Capranico G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci USA. 2019;116(3):816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, Mailliet P, Dutrillaux B, Boussin FD. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucl Acids Res. 2005;33(13):4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LT, Puig Lombardi E, Verga D, Nicolas A, Teulade-Fichou MP, Londono-Vallejo A, Maizels N. G-quadruplexes sequester free heme in living cells. Cell Chem Biol. 2019;26(12):1681–1691 e5. doi: 10.1016/j.chembiol.2019.10.003. [DOI] [PubMed] [Google Scholar]
- Halder R, Riou JF, Teulade-Fichou MP, Frickey T, Hartig JS. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. BMC Res Notes. 2012;5(1):1. [Google Scholar]
- Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51(6):899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucl Acids Res. 2008;36(19):6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IP, Mailliet P, Hossard V, Riou JF, Bugaut A, Roger L. Investigating the effect of mono- and dimeric 360A G-quadruplex ligands on telomere stability by single telomere length analysis (STELA) Molecules. 2019;24(3):577. doi: 10.3390/molecules24030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59(3):639–644. [PubMed] [Google Scholar]
- Jana J, Mondal S, Bhattacharjee P, Sengupta P, Roychowdhury T, Saha P, Kundu P, Chatterjee S. Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-Quadruplex structures at their promoter regions. Sci Rep. 2017;7:40706. doi: 10.1038/srep40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB. Fundamentals of G-quadruplex biology. Annu Rep Med Chem. 2020;54:3–44. doi: 10.1016/bs.armc.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonchhe S, Pandey S, Emura T, Hidaka K, Hossain MA, Shrestha P, Sugiyama H, Endo M, Mao H. Decreased water activity in nanoconfinement contributes to the folding of G-quadruplex and i-motif structures. Proc Natl Acad Sci U S A. 2018;115(38):9539–9544. doi: 10.1073/pnas.1805939115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. PubChem in 2021: new data content and improved web interfaces. Nucl Acids Res. 2021;49(D1):D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143(3):367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Local A, Zhang H, Benbatoul KD, Folger P, Sheng X, Tsai CY, Howell SB, Rice WG. APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute Myeloid Leukemia cells. Mol Cancer Ther. 2018;17(6):1177–1186. doi: 10.1158/1535-7163.MCT-17-1209. [DOI] [PubMed] [Google Scholar]
- Ma Y, Iida K, Nagasawa K. Topologies of G-quadruplex: biological functions and regulation by ligands. Biochem Biophys Res Commun. 2020;531(1):3–17. doi: 10.1016/j.bbrc.2019.12.103. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Maiti S, Chaudhury NK, Chowdhury S. Hoechst 33258 binds to G-quadruplex in the promoter region of human c-myc. Biochem Biophys Res Commun. 2003;310(2):505–512. doi: 10.1016/j.bbrc.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Maleki P, Ma Y, Iida K, Nagasawa K, Balci H. A single molecule study of a fluorescently labeled telomestatin derivative and G-quadruplex interactions. Nucl Acids Res. 2017;45(1):288–295. doi: 10.1093/nar/gkw1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti C, Zyner KG, Ohnmacht SA, Robson M, Haider SM, Morton JP, Marsico G, Vo T, Laughlin-Toth S, Ahmed AA, Di Vita G, Pazitna I, Gunaratnam M, Besser RJ, Andrade ACG, Diocou S, Pike JA, Tannahill D, Pedley RB, Evans TRJ, Wilson WD, Balasubramanian S, Neidle S. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J Med Chem. 2018;61(6):2500–2517. doi: 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex ligands. Org Biomol Chem. 2008;6(4):627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- O'Hagan MP, Morales JC, Galan MC. Binding and beyond: What else can G-Quadruplex ligands do? Eur J Org Chem. 2019;31–32:4995–5017. [Google Scholar]
- Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, Boussin FD. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24(18):2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucl Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina C, Cavalieri V. Epigenetic modulation of chromatin states and gene expression by G-quadruplex structures. Int J Mol Sci. 2020;21(11):4172. doi: 10.3390/ijms21114172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavone D, Jozwiakowski SK, Romanello M, Guilbaud G, Guilliam TA, Bailey LJ, Sale JE, Doherty AJ. PrimPol Is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol Cell. 2016;61(1):161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Ganguly A, Chowdhury S. Promise of G-Quadruplex structure binding ligands as epigenetic modifiers with anti-cancer effects. Molecules. 2019;24(3):582. doi: 10.3390/molecules24030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Qi H, Lin CP, Lin RK, Kerrigan JE, Rzuczek SG, LaVoie EJ, Rice JE, Pilch DS, Lyu YL, Liu LF. A G-quadruplex stabilizer induces M-phase cell cycle arrest. J Biol Chem. 2009;284(34):22535–22543. doi: 10.1074/jbc.M109.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varizhuk A, Isaakova E, Pozmogova G. DNA G-quadruplexes (G4s) modulate epigenetic (Re)programming and chromatin remodeling: transient genomic G4s assist in the establishment and maintenance of epigenetic marks, while persistent G4s may erase epigenetic marks. BioEssays. 2019;41(9):e1900091. doi: 10.1002/bies.201900091. [DOI] [PubMed] [Google Scholar]
- Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51(18):5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucl Acids Res. 2009;37(13):4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XH, Nie X, Liu HY, Fang YM, Zhao Y, Xia LX. TMPyP4 promotes cancer cell migration at low doses, but induces cell death at high doses. Sci Rep. 2016;6:26592. doi: 10.1038/srep26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzan E, Da Ros S, Musetti C, Shahidian LZ, Coelho NF, Bonsembiante F, Letard S, Gelain ME, Palumbo M, Dubreuil P, Giantin M, Sissi C, Dacasto M. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget. 2016;7(16):21658–21675. doi: 10.18632/oncotarget.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzan E, Elgendy R, Giantin M, Dacasto M, Bonsembiante F, Sissi C. Wholetranscriptome profiling of canine and human in vitro models exposed to a G-Quadruplex binding small molecule. Sci Rep. 2018;8(1):1. doi: 10.1038/s41598-018-35516-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 25,228 temporal differential gene regulation events in response to 7 G4DBLs (with their diverse concentrations and stimulation time points) in six different human cancer cell lines is provided in the ‘.xlsx’ format (Supplementary table 1).