Abstract
Background & Aims
Hypoxia and hypoxia-inducible factors (HIFs) are involved in chronic liver disease progression. We previously showed that hepatocyte HIF-2α activation contributed significantly to nonalcoholic fatty liver disease progression in experimental animals and human patients. In this study, using an appropriate genetic murine model, we mechanistically investigated the involvement of hepatocyte HIF-2α in experimental nonalcoholic steatohepatitis (NASH)-related carcinogenesis.
Methods
The role of HIF-2α was investigated by morphologic, cellular, and molecular biology approaches in the following: (1) mice carrying hepatocyte-specific deletion of HIF-2α (HIF-2α-/- mice) undergoing a NASH-related protocol of hepatocarcinogenesis; (2) HepG2 cells stably transfected to overexpress HIF-2α; and (3) liver specimens from NASH patients with hepatocellular carcinoma.
Results
Mice carrying hepatocyte-specific deletion of HIF-2α (hHIF-2α-/-) showed a significant decrease in the volume and number of liver tumors compared with wild-type littermates. These effects did not involve HIF-1α changes and were associated with a decrease of cell proliferation markers proliferating cell nuclear antigen and Ki67. In both human and rodent nonalcoholic fatty liver disease–related tumors, HIF-2α levels were strictly associated with hepatocyte production of SerpinB3, a mediator previously shown to stimulate liver cancer cell proliferation through the Hippo/Yes-associated protein (YAP)/c-Myc pathway. Consistently, we observed positive correlations between the transcripts of HIF-2α, YAP, and c-Myc in individual hepatocellular carcinoma tumor masses, while HIF-2α deletion down-modulated c-Myc and YAP expression without affecting extracellular signal-regulated kinase 1/2, c-Jun N-terminal kinase, and AKT-dependent signaling. In vitro data confirmed that HIF-2α overexpression induced HepG2 cell proliferation through YAP-mediated mechanisms.
Conclusions
These results indicate that the activation of HIF-2α in hepatocytes has a critical role in liver carcinogenesis during NASH progression, suggesting that HIF-2α–blocking agents may serve as novel putative therapeutic tools.
Keywords: HIF-2α, NAFLD, NASH, Hepatocellular Carcinoma
Abbreviations used in this paper: α-SMA, α-smooth muscle actin; CCN, cyclin; CDAA, choline-deficient L-amino acid–defined; CLD, chronic liver disease; DEN, diethylnitrosamine; ERK, extracellular signal-regulated kinase; HCC, hepatocellular carcinoma; HIF, hypoxia-inducible factor; IHC, immunohistochemistry; JNK, c-Jun N-terminal kinase; mRNA, messenger RNA; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PCNA, proliferating cell nuclear antigen; Q-PCR, quantitative real-time polymerase chain reaction; SB3, SerpinB3; siRNA, small interfering RNA; WT, wild-type; YAP, Yes-associated protein
Graphical abstract
Summary.
Hypoxia-inducible factor 2α (HIF-2α) is up-regulated in nonalcoholic fatty liver disease progression. Experiments performed in mice carrying hepatocyte-specific deletion of HIF-2α provide mechanistic evidence, re-inforced by analyses on cancer cells and human samples, that HIF-2α is critical for nonalcoholic steatohepatitis–related liver carcinogenesis.
Nonalcoholic fatty liver disease (NAFLD) is emerging as the most common cause of chronic liver disease (CLD) worldwide, with a global prevalence of 25% in the general population and even higher among obese individuals and/or patients affected by type II diabetes mellitus.1, 2, 3 Approximately 20%–30% of NAFLD patients can develop nonalcoholic steatohepatitis (NASH), which is characterized by hepatocyte injury and lobular inflammation, and can progress to fibrosis, cirrhosis, and liver failure.1,2 NAFLD patients also show a steadily increasing trend to develop hepatocellular carcinoma (HCC),4, 5, 6 the most common primary liver cancer (70%–90%), representing the fourth leading cause of cancer mortality worldwide, and with a minority of patients surviving at 5 years from diagnosis, despite treatment. Moreover, NAFLD-associated HCC also can arise in the noncirrhotic liver,4, 5, 6 a worrisome issue considering the high prevalence of NAFLD in the general population and the lack of validated therapy for this disease.1,2
In recent years, increasing evidence has shown that hepatic hypoxia is involved in CLD progression and in HCC development by sustaining angiogenesis, fibrogenesis, and, possibly, inflammatory and autophagy responses.7, 8, 9 HCC is considered as a hypoxic tumor, with a reported median oxygen tension lower than 1.0%.10,11 The cellular response to hypoxia mainly relies on heterodimeric transcriptional hypoxia-inducible factors (HIFs). These factors consist of an oxygen-sensitive α-subunit (HIF-1α or HIF-2α) and a constitutive β-subunit (HIF-1β).12,13 Although in the liver HIF-1α and HIF-2α can modulate common transcriptional programs, they often up-regulate distinct and nonoverlapping responses.12,13 Studies in HCCs with different etiologies and HCC cell lines have indicated that HIF-1α activation may contribute to tumor development by stimulating cell proliferation, metabolic changes, angiogenesis, invasion, and metastasis.8,10,11 Furthermore, HIF-1α overexpression is associated with a poor prognosis and HCC resistance to therapy.10,14 Conversely, the contribution of HIF-2α to HCC development is less well characterized in relation to conflicting results concerning its impact on liver carcinogenesis, particularly on cell survival and proliferation,15, 16, 17, 18, 19 and also as a consequence of using nonmechanistic/genetic in vivo experimental approaches.20 Furthermore, it has been suggested that the knockdown of HIF-1α enhances the expression of HIF-2α and vice versa.19 In the setting of NAFLD, it has been shown that HIF-2α, but not HIF-1α, can up-regulate genes involved in fatty acid synthesis/uptake and lipid storage, while it down-regulates those involved in fatty acid catabolism.21,22 In a previous study we showed that hepatocyte-specific HIF-2α deletion resulted in a significant decrease of fatty liver, parenchymal injury, lobular inflammation in NAFLD, and ameliorated disease progression toward fibrosis.23 More recently, a study performed on a limited number of NASH patients carrying HCC proposed that HIF-2α expression may be increased in NAFLD-related HCC vs HCC of a different etiology.20 In the present study, by using mice carrying hepatocyte conditional deletion of HIF-2α and additional in vitro approaches, we provide mechanistic and unequivocal evidence that HIF-2α plays a critical role in the development of NASH-related carcinogenesis by promoting liver cancer cell proliferation. Finally, we also provide confirming evidence that HIF-2α expression is up-regulated in a high percentage of human patients carrying NASH-related HCC.
Results
Hepatocyte-Specific Deletion of HIF-2α Reduces the Development of NAFLD-Associated HCC (hepatocellular carcinoma)
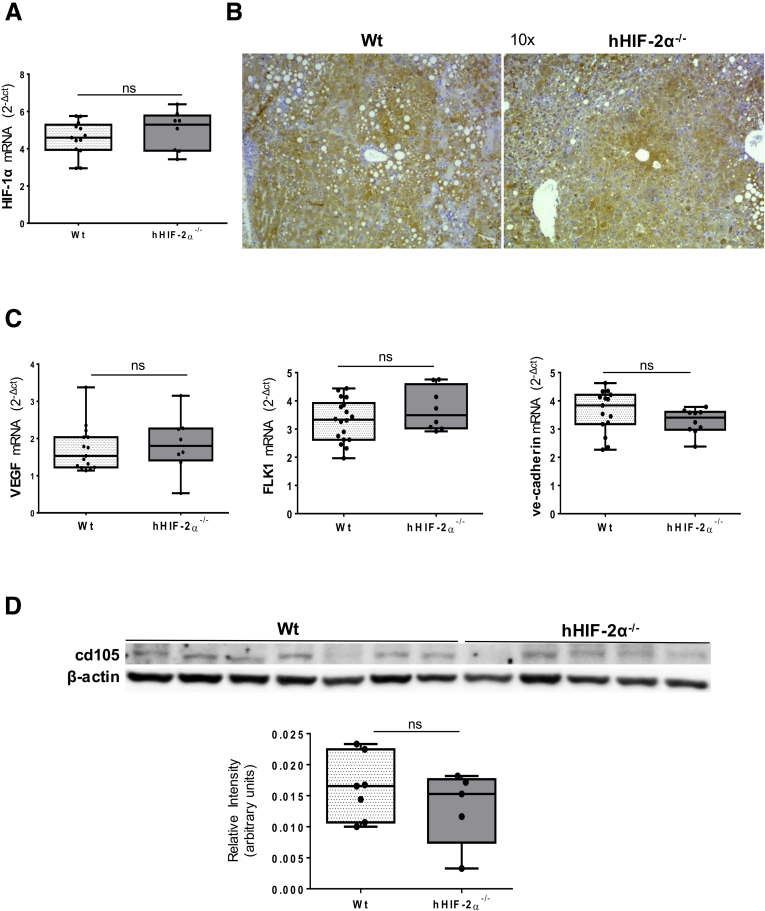
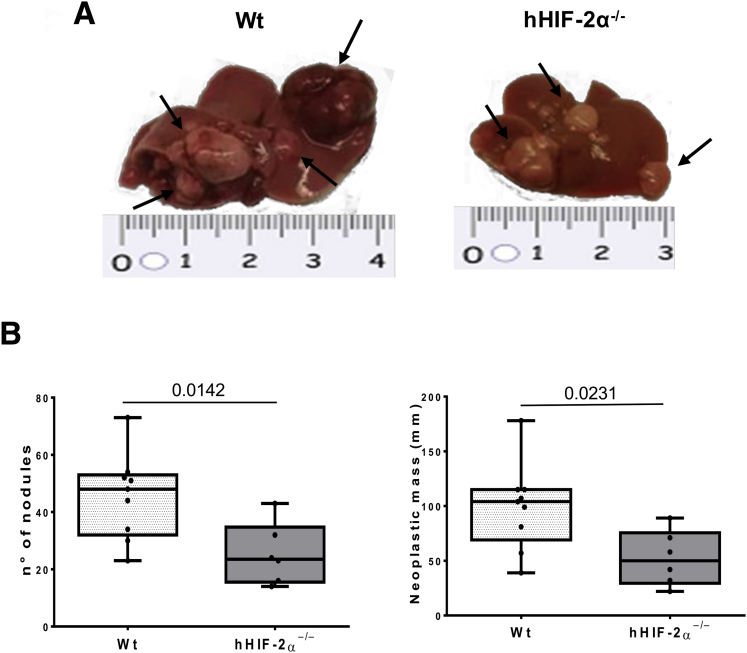
To mechanistically investigate the role of HIF-2α in the development of NAFLD-related primary liver cancer we used mice carrying a hepatocyte-specific HIF-2α deletion (hHIF-2α–/– mice) already used in a previous study that unequivocally outlined the relevant role of HIF-2α in either human or murine NAFLD progression.23 In the present study, these mice and related control littermates were submitted to an established murine model of NAFLD-associated hepatocarcinogenesis based on a single injection of diethylnitrosamine (DEN) at 2 weeks of age and the subsequent induction of steatohepatitis by the administration of a choline-deficient L-amino acid–defined (CDAA) diet for 25 weeks (Figure 1A).24 The mouse HCC arising in wild-type (WT) mice (characterized by nuclear atypia, pleomorphism, and increased mitotic activity, resembling human Edmonson–Steiner G1/G2 grading) showed diffuse parenchymal cell fat accumulation (Figure 1B and C) consistent with the features of the steatohepatitic HCC often detected among NAFLD patients.25 Although tumor cell morphology was not appreciably modified, H&E staining showed reduced fatty infiltration in liver tumors from mice lacking HIF-2α (Figure 1B and C).The analysis of HIF-2α protein levels in individual mouse HCCs from WT mice exposed to DEN/CDAA treatment showed that HIF-2α expression was up-regulated in cancer cells compared with healthy livers of WT mice fed with the choline-sufficient control diet (Figure 2A). hHIF-2α–/– mice submitted to the DEN/CDAA protocol developed mouse HCCs with a HIF-2α messenger RNA (mRNA) and protein content greatly lower than those from WT mice (Figure 2B and C), as expected. Similarly, neoplastic cells showed a reduced expression of HIF-2α–dependent genes such as CXCR4 and EPO (Figure 2D and E). On the other hand, the transcripts and protein levels of HIF-1α as well as the transcript levels of HIF-1α–related pro-angiogenic factors such as vascular endothelial growth factor-A, vascular endothelial growth factor-2, and vascular endothelial cadherin, were not significantly different between liver tumors from hHIF-2α–/– and WT mice (Figure 3A–C). Moreover, no significant change was observed concerning protein levels of CD105 (endoglin), a pro-angiogenic factor, between tumors from hHIF-2α–/– and WT mice (Figure 3D). These data overall suggest that hepatocyte-specific HIF-2α deletion does not result in major changes in angiogenic response. Interestingly, both the number and the size of mouse HCCs that developed in hHIF-2α–/– mice were reduced by 45% and 48%, respectively, compared with those developed in the liver of WT mice (Figure 4A and B). In addition, the extracellular matrix of mouse HCCs originating in hHIF-2α–/– mice also had a significant lower prevalence of α-smooth muscle actin (α-SMA)–positive myofibroblasts compared with that of the tumors arising in WT control mice (Figure 5A). This was accompanied by a significant reduction in the mRNA levels of matrix metalloprotease 9 and α-SMA (Figure 5B and C), and by a trend for decreased collagen Sirius Red staining (Figure 5D), despite that differences did not reach statistical significance. Altogether, these data suggest that the hepatocyte-specific deletion of HIF-2α might affect tumor growth and impact on the formation of extracellular matrix within the tumor, making the microenvironment less favorable for tumor progression. On the other hand, parameters related to inflammatory response were found to be decreased significantly in the tumors detected in hHIF-2α–/– mice vs those in WT mice, including macrophage infiltration, detected by immunohistochemistry (IHC) (Figure 6A) and transcript levels for F4/80 (Figure 6B), as well as transcript levels for programmed death-ligand 1 and Interferon Regulatory Factor 4 (Figure 6C and D). These results once again suggest that in hHIF-2α–/– mice the tumor microenvironment might be less favorable for tumor progression.
Figure 1.
Experimental NAFLD/NASH-related HCC: the DEN–CDAA murine model. (A) Graphic representation of the rodent model of NAFLD-associated hepatocarcinogenesis based on a single injection of DEN at 2 weeks of age and the subsequent induction of steatohepatitis by the administration of a CDAA diet for 25 weeks. (B and C) H&E staining was performed on paraffin-embedded HCC tumor masses from WT mice (n = 9) or from hHIF-2α–/– (n = 6). Original magnification is indicated. Alb, albumin.
Figure 2.
Validation of the DEN–CDAA hepatocyte-specific deletion of HIF-2α murine model. (A) Western blot analysis of HIF-2α performed in healthy liver of 8 WT mice fed with the choline-sufficient control diet (WT CSAA) or in HCC tumor masses from 5 WT mice treated with the DEN–CDAA protocol (WT DEN-CDAA). HIF-2α expression analyzed by (B) Q-PCR or (C) Western blot analysis in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/–. Q-PCR analysis of (D) CXCR4 and (E) EPO transcripts performed in WT or in hHIF-2α–/–. The mRNA values are expressed as fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentile) comprise 80% of the values. (B, D, and E) Statistical differences were assessed by the Student t test or Mann–Whitney test for nonparametric values. For the Western blot analysis, Bio-Rad (Hercules, CA) Quantity One software was used to perform the densitometric analysis. Equal loading was evaluated by reprobing membranes for Vinculin or β-actin. (A and C) Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Figure 3.
Hepatocyte-specific deletion of HIF-2α does not affect HIF-1α expression. Liver expression of HIF-1α evaluated by (A) Q-PCR and (B) immunohistochemical analysis in HCCs from 9 WT or from 6 hHIF-2α–/–. Original magnification is indicated. (C) Gene expression of vascular endothelial growth factor (VEGF), FLK1, and vascular endothelialcadherin evaluated by Q-PCR in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice (C). The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentile) comprise 80% of the values. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. (D) WB analysis of cd105 protein levels in HCCs from 7 WT mice or from 5 hHIF-2α–/– mice. For the Western blot analysis, Bio-Rad Quantity One software was used to perform the densitometric analysis. Equal loading was evaluated by reprobing membranes for β-actin. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Figure 4.
Hepatocyte-specific deletion of HIF-2α significantly affects the development of experimental liver tumors. (A and B) Reduction in number and neoplastic mass measured in HCC tumors (indicated by arrows) from 9 WT mice or 6 hHIF-2α–/–. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentile) comprise 80% of the values. (B) Statistical differences were assessed by the Student t test.
Figure 5.
Hepatocyte-specific deletion of HIF-2α significantly affects liver fibrosis. Liver fibrosis was evaluated morphologically in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice, by (A) IHC analysis of α-SMA and by (D) Sirius Red staining. ImageJ software analysis was performed to evaluate the amount of fibrosis. Data in graphs are expressed as means ± SEM. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. (A and D) Original magnification is indicated. Q-PCR analysis of (B) α-SMA and (C) matrix metalloprotease 9 transcripts performed in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice. The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentile) comprise 80% of the values. (B and C) Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Figure 6.
Hepatocyte-specific deletion of HIF-2α significantly affects the inflammatory response. (A) IHC analysis of F4/80 performed on paraffin-embedded HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice (A). ImageJ software analysis was performed to evaluate the amount of F4-/80-positive areas. Data in graphs are expressed as means ± SEM. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. Original magnification is indicated. (B–D) Q-PCR analysis of (B) F4/80, (C) programmed death-ligand 1, (D) IRF-4 transcripts performed in HCCs from 9 WT mice or from 6 hHIF-2α–/– mice. The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentile, whereas horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Hepatocyte-Specific HIF-2α Deletion Affects HCC Proliferative Capacity
On the basis of the relevant reduction of both the number and the size of tumor masses that developed in hHIF-2α–/– mice, we next investigated whether this effect might be related to a modulation in the proliferative capacity of cancer cells. Indeed, data in the literature indicate that in nonliver tumors HIF-2α has a greater oncogenic capacity than HIF-1α, and is able to promote tumor proliferation, stemness, and radioresistance and chemoresistance.16,19,26, 27, 28 The transcriptional analysis of the specimens obtained from tumor and peritumoral tissue of WT mice showed that the transcripts of the cell proliferation markers proliferating cell nuclear antigen (PCNA) and nuclear antigen Ki67 were increased significantly in the tumor masses (Figure 7A and C) and correlated positively with HIF-2α (Figure 7B and D). Consistently, the lack of HIF-2α resulted in decreased PCNA and Ki67 expression in mouse HCCs, without affecting the expression of these markers in peritumoral areas (Figure 7A and C). IHC analysis confirmed a selective reduction in PCNA staining (Figure 8A), as well as a significant decrease in transcript levels for cyclin E1 (CCNE1) and cyclin E2 (CCNE2) in the tumors from hHIF-2α–/– mice (Figure 8B). At the protein level, PCNA reduction was accompanied by a concomitant increase in the cellular content of the cell-cycle inhibitors p53 and p21 (Figure 9A–C). Previous studies have shown that HIF-2α can positively regulate c-Myc expression, which, in turn, directly stimulates PCNA production in HCC cells under hypoxic conditions, thus promoting cancer cell proliferation and HCC resistance to the tyrosine kinase inhibitor sorafenib.29,30 Accordingly, we observed that c-Myc mRNA levels in mouse HCCs from WT mice correlated positively (r = 0.72; P = .008; 95% CI, 0.25–0.92) with those of HIF-2α (Figure 9D). In line with the reduction in cell proliferation, c-Myc mRNA and protein levels were decreased significantly in the tumors that developed in hHIF-2α–/– mice (Figure 9E and F). Such an effect apparently was unrelated to the modulation of signal pathways involving extracellular signal-regulated kinase (ERK)1/2, c-Jun N-terminal kinase (JNK), and AKT (Figure 10A–C).
Figure 7.
HIF-2α expression positively correlates with markers of HCC proliferative capacity. (A and C) qPCR analysis of (A) PCNA and (C) Ki67 transcripts performed in peritumoral tissue or HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice. The mRNA values are expressed as fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. Statistical differences were assessed by a 1-way analysis of variance test with the Tukey correction for multiple comparisons or the Student t test. (B and D) Relationship between HIF-2α and (B) PCNA or (D) Ki67 mRNA in HCCs from 9 WT mice. The values represent the relative mRNA content. The correlation analysis was performed with the Pearson r test.
Figure 8.
Hepatocyte-specific HIF-2α deletion impact on HCC proliferative capacity. (A) IHC analysis of PCNA performed on paraffin-embedded HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice (A). ImageJ software analysis was performed to evaluate the number of PCNA-positive nuclei per microscopic field. Data in graphs are expressed as means ± SEM. Statistical differences were assessed by the Student t test. Original magnification is indicated. (B) qPCR analysis of CCNE1 and CCNE2 transcripts performed in HCCs from 9 WT mice or from 6 hHIF-2α–/– mice. The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Figure 9.
Hepatocyte-specific HIF-2α deletion impact on HCC proliferative capacity. (A–C and F) WB analysis for (A) PCNA, (B) p21, (C) p53, and (F) c-Myc performed in HCCs from 6 WT mice or from 5 hHIF-2α–/– mice. Bio-Rad Quantity One software was used to perform the densitometric analysis (data are expressed as the fold change relative to the normalized WT expression). Equal loading was evaluated by reprobing membranes for β-actin. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. (D) Relationship between HIF-2α and c-Myc mRNA in HCCs from 9 WT mice. The values represent the relative mRNA content. The correlation analysis was performed with the Pearson r test. (E) Q-PCR analysis of the c-MYC transcript performed in HCCs from 9 WT mice or from 6 hHIF-2α–/– mice. The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
Figure 10.
Hepatocyte-specific HIF-2α deletion affects HCC proliferative capacity without the involvement of the ERK, JNK, and AKT signal pathways. Western blot analysis of (A) P-AKT, (B) phospho-ERK (extracellular regulated kinase), and (C) phospho-JNK (c-Jun-aminoterminal kinase) in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice. Bio-Rad Quantity One software was used to perform the densitometric analysis. Equal loading was evaluated by reprobing membranes for the relative nonphosphorylated protein AKT, ERK, JNK, and β-actin or vinculin. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
HIF-2α Overexpression Supports HepG2 Cell Growth In Vitro
To investigate the role of HIF-2α in HCC growth we performed cell culture experiments using HepG2 cells stably transfected to overexpress HIF-2α (H/2α cells), as well as related control cells transfected with the empty pCMV6 vector (H/V6 cells). Figure 11A shows that in H/2α cells HIF-2α protein levels increased in a time-dependent manner along with transcripts of HIF-2α target genes such as the chemokine receptor CXCR4 and EPO (Figure 11B). Furthermore, bromodeoxyuridine incorporation assay, cell count, and crystal violet assay indicated that H/2α cells had a more proliferative phenotype compared with H/V6 cells (Figure 11C–E). This was confirmed further by flow cytometry analysis of the cell cycle, which outlined a significant shift toward the S phase in the H/2α cells (Figure 11F). These changes were accompanied by increased expression of PCNA and the c-Myc oncogene in H/2α cells compared with control H/V6 cells and by a parallel reduction in the levels of cell-cycle inhibitor p53 and p21 (Figure 11A).
Figure 11.
HIF-2α overexpression supports HepG2 cell growth in vitro. (A) Western blot analysis of HIF-2α, c-Myc, PCNA, p53, and p21 levels performed on HepG2 stably transfected to overexpress HIF-2α (H/2α) or in control HepG2 cells transfected with empty vector (H/V6) at different time points. Equal loading was evaluated by reprobing membranes for β-actin. Bio-Rad Quantity One software was use to perform the densitometric analysis (data are expressed as the fold change relative to the normalized H/V6 expression). (B) Q-PCR analysis of CXCR4 and EPO transcripts in H/2α or in H/V6 cells at different time points. Data in graphs are expressed as means ± SEM. Statistical differences were assessed by 1-way analysis of variance test with the Tukey correction for multiple comparisons or the Kruskal–Wallis test for nonparametric values. (C–E) Cell count was performed with the (C) bromodeoxyuridine (BrdU) incorporation assay, (D) Burker chamber, and (E) crystal violet techniques performed on H/2α or H/V6 at different time points. (F) Bar graph chart shows the relative quantity of the G1, G2, and S ratio in H/2α cells compared with the H/V6 control cells as means ± SD, resulting from cell-cycle analysis by flow cytometry with FCS Express 4 Flow Research Edition software. These experiments were repeated 3 separate times, and similar results were obtained.
HIF-2α Overexpression Is Associated With SerpinB3 Production in Either Human or Experimental NAFLD-Associated HCC
To confirm and detail the involvement of HIF-2α in NAFLD-associated liver carcinogenesis, we next performed an IHC analysis on human specimens obtained from a cohort of 27 well-characterized NAFLD-derived HCC patients (G2 and G3 grading) (Figure 12A). In these tumors, HIF-2α staining was detectable in 67% of the samples (18 of 27), with 11 samples (61%) showing intense positivity and 7 (39%) showing moderate staining (Figure 12A). Furthermore, 9 of 11 (82%) HCC samples with intense HIF-2α staining in the cytoplasm also were characterized by HIF-2α nuclear positivity, although the number of positive nuclei varied within patients and in different areas of the same specimens. Weak HIF-2α nuclear positivity was seen occasionally in some samples from tumors showing moderate HIF-2α cytoplasm positivity. Conversely, the nuclei of nonparenchymal cells, mainly inflammatory cells or myofibroblast-like cells in fibrotic septa, were negative for HIF-2α (Figure 12A). HIF-2α positivity was prevalent in HCCs developing in cirrhotic livers (14 of 16; 88%) compared with HCCs arising in noncirrhotic livers (3 of 10; 30%), and HIF-2α nuclear expression was associated strongly (odds ratio, 16.33; 95% CI, 2.2–121.5; P = .0085) with the presence of cirrhosis (Figure 12B). HIF-2α expression in HCCs also was associated with a trend for lower survival and earlier tumor recurrence compared with patients with undetectable HIF-2α (Figure 12C and D). However, these differences did not reach statistical significance, likely because of the limited number of patients recruited.
Figure 12.
Expression of HIF-2α in human NAFLD/NASH-related HCC patients. (A) IHC analysis of HIF-2α was performed on paraffin-embedded human liver specimens from NAFLD/NASH-related HCC patients (n = 27; grades G2–G3). Original magnification is indicated. HIF-2α expression was semiquantitatively scored blinded by a pathologist. (B) The odds ratio (OR) meta-analysis was calculated by the Fisher exact test to evaluate the strength of the association between HIF-2α expression and the HCC cirrhotic setting. (C and D) Kaplan–Meier curves of (C) survival and (D) time to recurrence according to HIF-2α expression. Statistical analysis was performed using the log-rank (Mantel–Cox) test.
IHC analysis of NAFLD-related HCCs also showed a strict association between HIF-2α and the expression of SerpinB3 (SB3), a HIF-2α–dependent cysteine–protease inhibitor that has been involved in stimulating proliferation, epithelial-to-mesenchymal transition, and invasiveness in liver cancer cells.31 In fact, 10 of 11 (90%) tumors positive for HIF-2α showed intense (n = 7) or moderate (n = 3) cytoplasmic staining for SB3 (Figure 13A), whereas the remaining HIF-2α–negative specimens were largely negative (n = 11) or weakly positive (n = 5) for SB3. In the same way, we found a strong linear correlation between HIF-2α and SB3 mRNAs (r = 0.61; P = .03; 95% CI, 0.06–0.87) in the individual NAFLD-related HCC induced in WT mice (Figure 13B), whereas SB3 levels were decreased significantly at both transcript and protein levels in the tumors from hHIF-2α–/– mice (Figure 13C and D).
Figure 13.
SB3 expression correlates with HIF-2α expression in NAFLD/NASH-related HCC patients. (A) IHC analysis of HIF-2α (left) or SB3 (right) performed on paraffin-embedded human liver specimens from NAFLD/NASH-related HCC patients (n = 27; grades G2–G3). Original magnification is indicated. SB3 expression was semiquantitatively scored blinded by a pathologist (Mann–Whitney U test). (B) Relationship between HIF-2α and SB3 mRNA in HCC tumor masses from 9 WT mice. The values represent the relative mRNA content. The correlation analysis was performed with the Pearson r test. (C) qPCR and (D) Western blot analysis for SB3 performed in HCC tumor masses from 9 WT mice or from 6 hHIF-2α–/– mice. (D) For the Western blot analysis, Bio-Rad Quantity One software was used to perform the densitometric analysis. Equal loading was evaluated by reprobing membranes for β-actin. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. The mRNA values are expressed as the fold increase over control values after normalization to the TATA box binding protein gene expression. Results are expressed as means ± SD. Boxes include the values within the 25th and 75th percentiles, whereas the horizontal bars represent the medians. The extremities of the vertical bars (10th–90th percentiles) comprise 80% of the values. (C) Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values.
YAP Influences c-Myc Activity in NAFLD-Related HCCs
Increasing evidence points out the involvement/activation of the Hippo pathway in liver carcinogenesis.32, 33, 34, 35, 36, 37 In particular, the Hippo-dependent transcriptional factor Yes-associated protein (YAP) has been shown to contribute to c-Myc activation in HCCs.33 According to Turato et al,38 SB3 can modulate c-Myc activity by inhibiting its degradation by calpain as well as by stimulating the Hippo pathway through an enhanced expression of YAP. Because we have observed that SB3 up-upregulation was strictly linked to HIF-2α stimulation in both human and rodent NAFLD-related HCCs, we next investigated whether YAP might account for high expression of c-Myc in WT tumors. We observed that YAP transcripts in individual NAFLD-derived HCCs developed in WT mice directly correlated (r = 0.666; P = .013; 95% CI, 0.18–0.89) with HIF-2α expression, whereas a significant decrease of YAP protein levels was evident in tumors from mice lacking HIF-2α (Figure 14A and B) in parallel with SB3 down-modulation. To better investigate the involvement of YAP in supporting HIF-2α–mediated carcinogenesis we went back to HepG2 cells stably overexpressing HIF-2α (H/2α cells). In this setting we observed that YAP was up-regulated in a time-dependent manner in H/2α cells (Figure 14C) and that the treatment of H/2α cells with a specific YAP small interfering RNA (siRNA) was able to reduce the expression of c-Myc at the levels observed in control cells receiving scrambled siRNA (H/V6 SC) (Figure 14D), confirming involvement of YAP in c-Myc expression by HIF-2α. From these data we propose that the activation of hepatocyte HIF-2α has a critical role in liver carcinogenesis during NAFLD evolution by promoting c-Myc activation through pathways that involve SB3 and Hyppo signaling.
Figure 14.
HIF-2α expression directly correlates with YAP. (A) Relationship between HIF-2α and YAP mRNA in HCCs from 9 WT mice. The values represent the relative mRNA content. The correlation analysis was performed with the Pearson r test. (B) Western blot analysis for YAP performed in HCCs from 9 WT mice or from 6 hHIF-2α–/– mice. For the Western blot analysis, Bio-Rad Quantity One software was used to perform the densitometric analysis. Equal loading was evaluated by reprobing membranes for β-actin. Statistical differences were assessed by the Student t test or the Mann–Whitney test for nonparametric values. (C) Western blot analysis of YAP protein levels performed on HepG2 stably transfected to overexpress HIF-2α (H/2α) or in control HepG2 cells transfected with empty vector (H/V6) at different time points. Equal loading was evaluated by reprobing membranes for vinculin. (C and D) Western blot analysis of YAP and c-Myc protein levels performed on H/2α or in control H/V6, treated or not (scramble) with a specific YAP siRNA (siYAP). Equal loading was evaluated by reprobing membranes for β-actin. Bio-Rad Quantity One software was used to perform the densitometric analysis (data are expressed as the fold change relative to the normalized H/V6 expression).
Discussion
Hypoxia and HIFs, particularly HIF-1α, have been proposed to play an important role in the progression of CLD and in the development of HCC,7, 8, 9, 10, 11,14 but the actual contribution of HIF-2α to HCC development is by far less well characterized. In particular, conflicting results have been reported on the impact of HIF-2α on liver carcinogenesis, particularly on cell survival and proliferation.15, 16, 17, 18, 19 Furthermore, although a previous study proposed HIF-2α involvement in NAFLD-related HCC,20 no definitive evidence is available to date on the actual contribution of HIF-2α in the processes leading to liver carcinogenesis.
In the present study, we took advantage of mice carrying the selective deletion of hepatocyte HIF-2α (hHIF-2α–/–) to mechanistically investigate the role of HIF-2α in NAFLD-related HCC. The use of these mice has previously allowed the demonstration of the critical contribution of hepatocyte HIF-2α in the evolution of experimental NAFLD by decreasing parenchymal injury, fatty liver, lobular inflammation, and the development of liver fibrosis.23
In this work, the induction of HCCs in hHIF-2α–/– mice by the DEN/NAFLD protocol has shown that the lack of parenchymal HIF-2α halves the number and the size of mouse HCCs compared with WT mice. Such an effect is associated with a parallel decreasing of the expression of proliferative markers PCNA and Ki67, along with an induction of p21 and p53 in cancer cells, indicating that hepatocyte HIF-2α can directly promote cancer cell proliferation and survival. These data are supported by in vitro experiments showing that HepG2 cells stably overexpressing HIF-2α (H/2α cells) display a more proliferative phenotype compared with control cells and a significant shift toward the S phase of the cell cycle. These results are in line with data linking HIF-2α with an enhanced tumor aggressiveness through the promotion of cell proliferation, stemness, and radioresistance and chemoresistance.27,28,39, 40, 41 Nonetheless, we cannot exclude that besides the boosting of cell proliferation, additional mechanisms might be involved in the procarcinogenic action of HIF-2α because we previously observed that hepatocyte HIF-2α suppression ameliorated hepatic inflammation and fibrosis in NASH livers.23 Indeed, steatohepatitis not only promotes carcinogen-induced HCCs,42 but also leads to their spontaneous development in mice fed a NASH-inducing choline-deficient diet.43,44 Moreover, the lack of hepatocyte HIF-2α can reduce both the inflammatory response, as confirmed in this study in HCCs from hHIF-2α–/– mice (Figure 6), and the recruitment of cancer-associated myofibroblasts, which contribute to the development of a permissive tumor microenvironment.45
To date, the mechanisms by which HIF-2α can support HCC growth have not been fully characterized because of the interplay between HIF-1α and HIF-2α observed in HCC cell lines,15,46 and the fact that, depending on the cell context, HIF-2α overexpression could have antiproliferative and pro-apoptotic actions in HCCs.19 HIF-2α up-regulation has been reported as a common mechanism in the development of HCC resistance to the multikinase inhibitor sorafenib.18 In these settings, HIF-2α promoted cell survival by stimulating the signaling of transforming growth factor-α/epidermal growth factor receptor pathway and by inducing cyclin D1, β-catenin, and c-Myc expression.18 Here, we show that, at variance with what was observed in HCC spheroids,15 the up-regulation of HIF-2α in NAFLD-derived HCCs does not affect HIF-1α levels. Moreover, in both HepG2 cells and mouse HCCs, HIF-2α expression is associated with a stimulation in c-Myc production. Such an effect does not seem to involve signaling through the AKT, ERK1/2, JNK pathways, but appears mediated by YAP signaling. We observed, in fact, that YAP and HIF-2α transcript levels were correlated positively, and that hepatocyte HIF-2α deletion significantly affected both YAP and c-Myc content of murine tumors. Moreover, in tumors from hHIF-2α–/– mice, we detected a significant decrease of transcript levels of CCNE1 and CCNE2, 2 cyclins that have been described to have an important role for HCC progression and to synergistically impair overall survival in HCC patients.47 In line with these findings, YAP is up-regulated in H/2α cells. The capacity of YAP to sustain c-Myc activity in HCCs is consistent with the report by Xiao et al,33 who observed that c-Myc and YAP proteins are closely correlated in human liver cancers with YAP promoting c-Myc transcriptional output through c-Abl. Furthermore, Ma et al40 recently reported that HIF-2α stimulates colon cancer cell growth by up-regulating YAP activity through a mechanism independent from Src, phosphatidylinositol-3-kinase, ERK1/2, or mitogen activated protein kinase pathways. Therefore, we propose that HIF-2α activation can promote HCC growth by sustaining YAP/c-Myc interaction. Such a hypothesis does not exclude other protumorigenic actions of HIF-2α such as, for instance, the stimulation of the long noncoding RNA nuclear-enriched abundant transcript 1 enzyme, which recently was implicated in sustaining epithelial-to-mesenchymal transition and migration of HCC cells.41 Concerning the mechanisms by which HIF-2α can modulate YAP, we previously showed a role for SB3.38 Although SB3 is virtually undetectable in normal human livers, its expression is well evident in liver biopsy specimens from patients with CLD and in a fraction of HCCs.48,49 In HCC cells, SB3 is specifically regulated by HIF-2α,31 while SB3 enhances HIF-2α transcriptional activity by promoting its stabilization through the conjugation with neural precursor cell expressed developmentally down-regulated-8, induced by neural precursor cell expressed developmentally down-regulated-8–E1 activating enzyme.26
Data obtained from human NAFLD-related HCC specimens support the observations in experimental models. We have detected HIF-2α overexpression in two thirds of human HCCs developing in NAFLD patients, with a strong positive association (odds ratio, 16.33) between HIF-2α nuclear localization and HCC development in cirrhotic livers, essentially confirming data from a previous study.20 Of interest, HIF-2α activation and nuclear staining is already appreciable at the early stage of the disease (F0–F1) in approximately 70% of NAFLD patients and a similar prevalence is maintained with disease progression to fibrosis/cirrhosis (F3–F4).23 Our data also suggest that sustained high HIF-2α expression is associated with a trend for shorter survival and earlier tumor recurrence, in line with the poor patient outcome observed in other HIF-2α–expressing tumors27,28,39 and in agreement with the decreased survival previously reported in NAFLD-related HCC patients.20
Additional data obtained from the cohort of NAFLD-related HCCs analyzed in this study indicate that intranuclear HIF-2α is associated with enhanced expression of SB3. Such a prevalence of SB3 positivity in human HCC is higher than that of 22%, which previously was observed in a group of HCCs with other etiologies, mainly viral,38 indicating that SB3 induction represents a specific response to HIF-2α activation in NAFLD-associated HCCs. Consistently, SB3 is down-regulated significantly in mouse HCCs from hHIF-2α-/- mice in parallel with the decrease of both YAP and c-Myc. This suggests that SB3 indeed might be involved in modulating the HIF-2α/YAP/c-Myc axis to sustain cell growth in NAFLD-associated HCCs. Nonetheless, we cannot exclude alternative mechanisms because, for example, the orphan G-protein–coupled receptor G-protein-coupled receptor 5A has been shown to mediate HIF-2α/YAP interaction in colon cancer cells.50
In conclusion, our results indicate that HIF-2α overexpression seems to be a specific feature in NAFLD-related HCCs and might contribute significantly to sustain the tumor development in NAFLD patients. These observations, along with the notions that interference with HIF-2α counteracts HCC resistance to sorafenib29 and radiation treatment,51 suggest the possibility of using HIF-2α–blocking drugs as a therapeutic intervention for a tumor that, at present, has few curative options.
Materials and Methods
Materials
Enhanced chemiluminescence reagents and nitrocellulose membranes (Hybond-C extra) were from Amersham Pharmacia Biotech, Inc (Piscataway, NJ). The following antibodies were used: anti–HIF-1α (NB100-479) and anti–HIF-2α (NB100-122) from Novus Biologicals (Cambridge, UK); anti-CD105 (PA5-12511), anti-PCNA (PA5-27214), and anti-SB3 (PA5-30164) from ThermoFisher Scientific (Rockford, IL); anti–α-SMA (M0851) was from DAKO (Agilent, St Clara, CA); anti-SB3 (GTX32866) was from GeneTex (Irvine, CA); anti-F4/80 (14-4801-82) was from eBioscience (Affymetrix, St Clara, CA); anti-YAP (sc-15407), anti-c-MYC (sc-788), anti-SB3 (sc21767), anti-p53 (sc-6243), anti-p21 (sc-817), anti-ERK (sc-94), anti-JNK (sc-571), antivinculin (sc-73614), anti–p-Akt1/2/3 (sc-7985-R), and anti-Akt1/2/3 (sc-8312) were from Santa Cruz Biotechnology (Dallas, TX); anti-phospho-ERK (extracellular-regulated kinase, [4696]) and anti–anti-phospho-JNK (c-Jun aminoterminal kinase [9255]) were from Cell Signaling Technology (Danvers, MA); and anti–β-actin (A5441) was from Sigma Aldrich (St. Louis, MO). HiPerfect Transfection reagent was from Qiagen (Hilden, Germany), Lipofectamine 2000 was from Invitrogen-Life Technologies (Carlsbad, CA), plasmid DNA purification NucleoBond XtraMIDI was from Macherey-Nagel (Allentown, PA), and pCMV6-entry vectors were from Origene (Rockville, MD).
Human Subjects
For this study, we analyzed liver specimens from 27 NAFLD patients with HCC (Edmonson–Steiner grades G2 and G3), referring to the Division of Gastro-Hepatology of the University of Turin. All samples were collected at the time of resection or transplantation. All subjects gave informed consent to the analysis and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and was approved by the Ethics Committee of the Azienda Ospedaliera Universitaria Città della Salute (Torino, Italy). The clinical and biochemical features of the patients are reported in Table 1.
Table 1.
Clinical and Biochemical Characterization of NAFLD Carrying HCC Patients Investigated
| Demographic data | |
| Patients, male/female, n | 27 (25/2) |
| Age, y | 71 (49–86) |
| BMI | 28.2 (22.3–34.6) |
| Clinical data | |
| Hypertension | 88.9% |
| Dyslipidemia, TG >150/HDL (<40 males/<50 females) | 70.4% |
| Diabetes mellitus | 85.2% |
| CHILD | A (59.3%) |
| Resection | 9/14 (64.3%) |
| OLT | 6 (22.2%) |
| MELD | 9 (6–14) |
| Biochemical data | |
| Triglycerides, mg/dL (nv, 50–150) | 111 (77–155) |
| AST, U/L (nv, 5–40) | 37 (17–83) |
| ALT, U/L (nv, 5–40) | 37 (13–86) |
| γ-GT, U/L (nv, 5–45) | 111 (14–307) |
| Bilirubin, U/L | 0.9 (0.3–2.5) |
| AFP, ng/mL (nv, <10) | 131.6 (1–2919) |
| Albumin, g/L | 4.1 (3.3–4.8) |
| Histologic data | |
| Steatosis score | 0 (18.5%) 1 (70.4%) 2 (11.1%) 3 (3.7%) |
| Ballooning score | 1 (45.5%) |
| Fibrosis score | 1 (40.9%) |
| Cirrhosis | 59.3% |
| NAS score | 1–3 (27.3%) |
| Oncologic data | |
| Nodules, n | 1 (1–3) |
| Dimensions, mm | 73 (7–180) |
| Edmondson–Steiner grading, 1–4 | 9 (6–14) |
NOTE. The values are expressed as medians and interquartile range. For histologic scores the range of variability is included.
AFP, alfa-fetoprotein; ALT, aspartate aminotransferase; AST, alanine aminotransferase; BMI, body mass index; γ-GT, γ-glutamyl transpeptidase; HDL, high density lipoprotein; MELD, Mayo End stage Liver Disease; nv, normal value; OLT, orthotopic liver transplantation; NAS, NASH Activity score; TG, triglycerides.
Animal Experimentation
Mice carrying a hepatocyte-specific deletion of HIF-2α (hHIF-2α-/- mice) were obtained by breeding HIF-2αfl/fl C57BL/6 mice with mice on the same genetic background expressing the Cre-recombinase under the control of the albumin promoter (Alb/Cre+/+ mice) (Jackson Laboratories, Bar Harbor, ME).23 NAFLD-associated liver carcinogenesis was induced in male hHIF-2α-/- mice (n = 6) and related control sibling littermates not carrying the HIF-2α deletion (WT, n = 9), with an established experimental protocol involving a single administration of DEN (25 mg/kg body weight, intraperitoneally) at the age of 2 weeks, followed by feeding with a CDAA diet (Laboratorio Dottori Piccioni, Gessate, Italy) for 25 weeks starting from the age of 6 weeks.24 At the time of death, the livers of the animals were collected, measured, photographed, and the number of visible HCC tumor masses on the surface of the livers was counted and measured with a caliper. For each animal, the 2 biggest tumor masses were isolated and collected for specific analysis. In preliminary experiments, 8-week-old male hHIF-2α-/- mice (n = 8) and related control sibling littermates not carrying the HIF-2α deletion (WT, n = 8) were fed with the corresponding choline-sufficient control diet for 12 or 24 weeks. The experiments complied with national ethical guidelines for animal experimentation and the experimental protocols were approved by the Italian Ministry of Health.
Cell Lines and Culture Conditions
HepG2 cells (American Type Culture Collection, Manassas, VA) were used and maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 25 μg/mL amphotericin B, as previously reported.31 The pCMV6-based mammalian expression vectors, empty (used as a control) and encoding HIF-2α (OriGene), were used to generate and select HepG2 cells stably overexpressing HIF-2α.31 HepG2 cells were seeded and then transfected 24 hours later with 10 μg of each vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). HIF-2α expression of the generated stable transfectants was carefully characterized, after which the cell lines carrying the empty vector (H/V6) and overexpressing HIF-2α (H/2α) then were used for the experiment described.
H/2α and control cells containing the empty pCMV6 vector (H/V6) were grown in Dulbecco’s modified Eagle medium under normoxic conditions to obtain the desired subconfluence level (65%–70%).
Western Blot Analysis
Total cell/tissue lysates, obtained as previously described,23,26,31 were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 12%, 10%, or 7.5% acrylamide gels, incubated with the desired primary antibodies, then with peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulins in Tris-buffered saline–Tween containing 2% (wt/vol) nonfat dry milk, and finally developed with the enhanced chemiluminescence reagents according to the manufacturer’s instructions. Sample loading was evaluated by reblotting the same membrane with antibodies raised against β-actin or vinculin.
Quantitative Real-Time PCR
RNA extraction, complementary DNA synthesis, and quantitative real-time polymerase chain reaction (Q-PCR) reactions were performed on cell samples, murine liver specimens, and on 2 HCC tumor masses isolated from each murine liver as previously described.23,26 mRNA levels were measured by Q-PCR, using the SYBR (BIO-RAD, Hercules, CA) green method as described.23 More details and oligonucleotide sequences of primers used for Q-PCR are available in Table 2.
Table 2.
Oligonucleotide sequences of primers used for Q-PCR
| Primer | Forward | Reverse |
|---|---|---|
| Murine CNNE1 | 5’-CCCTGGGATGATAATTCAGC-3’ | 5’-TCTGGGTGGTCTGATTTTCC-3’ |
| Murine CNNE2 | 5’-TCTGTGCATTCTAGCCATCG-3’ | 5’-GTCATCCCATTCCAAACCTG-3’ |
| Murine C-MYC | 5’-CTGTGGAGAAGAGGCAAACC-3’ | 5’-TTGTGCTGGTGAGTGGAGAC-3’ |
| Human CXCR4 | 5’-TCCATTCCTTTGCCTCTTTTGC-3’ | 5’-ACGGAAACAGGGTTCCTTCAT-3’ |
| Murine CXCR4 | 5’-TGGAACCGATCAGTGTGAGT-3’ | 5’-TTGCCGACTATGCCAGTCAA-3’ |
| Murine Cyp2e1 | 5’-TGGGGAAACAGGGTAATGAG-3’ | 5’-GTGCACAGCCAATCAGAAAG-3’ |
| Human EPO | 5’-GAGCCCAGAAGGAAGCCATC-3’ | 5’-GCGGAAAGTGTCAGCAGTGA-3’ |
| Murine EPO | 5’-CAGCCACCAGAGACCCTTC-3’ | 5’-ACATCAATTCCTTCTGAGCTCCC-3’ |
| Murine FLK1 | 5’-GGCGGTGGTGACAGTATCTT-3’ | 5’-GTCACTGACAGAGGCGATGA-3’ |
| Murine F4/80 | 5’-GTACAGATGGGGGATGACCAC-3’ | 5’-GACTGAGTTAGGACCACAAGGTGAG-3’ |
| Human GAPDH | 5’-TGGTATCGTGGAAGGACTCATGAC-3’ | 5’-ATGCCAGTGAGCTTCCCGTTCAGC-3’ |
| Murine HIF-1α | 5’-TCAAGTTCAGCAACGTGGAAG-3’ | 5’-TATCGAGGCTGTGTCGACTG-3’ |
| Murine HIF-2α | 5’-AGAGCTGAGGAAGGAGAAATC-3’ | 5’-ATGTGTCCGAAGGAAGCTG-3’ |
| Murine IRF-4 | 5’-GCAGCTCACTTTGGATGACA-3’ | 5’-CCAAACGTCACAGGACATTG-3’ |
| Murine Ki67 | 5’-CATGCAAACCCTCACACTTG-3’ | 5’-GCTGGTTCCAATTTCTGAGC-3’ |
| Murine matrix metalloprotease 9 | 5’-CGTCGTGATCCCCACTTACT-3’ | 5’-AACACACAGGGTTTGCCTTC-3’ |
| Murine PCNA | 5’-CTGTGCAAAGAATGGGGTGAA-3’ | 5’-AGCAAACGTTAGGTGAACAGG-3’ |
| Murine PD-L1 | 5’-AATGCTGCCCTTCAGATCAC-3’ | 5’-TCAGCGTGATTCGCTTGTAG-3’ |
| Murine SB3 | 5’-TTTTACACAAGTCCTTTGTGGAGG-3’ | 5’-CTGGACACATGGAAGAGACACCAC-3’ |
| Murine TBP | 5’-CACATCACAGCTCCCCACCA-3’ | 5’-AGCGGAGAAGATGCTGGAAAC-3’ |
| Murine VE-cadherin | 5’-ATTGAGACAGACCCCAAACG-3’ | 5’-TTCTGGTTTTCTGGCAGCTT-3’ |
| Murine VEGF-A | 5’-CAGGCTGCTGTAACGATGAA-3’ | 5’-TTTCTTGCGCTTTCGTTTTT-3’ |
| Murine YAP | 5′-TCAGACAACAACATGGCAGGA-3′ | 5′-TTCATGGCTGAAGCCGAGTT-3′ |
| Murine αSMA | 5’-CTGACAGAGGCACCACTGAA-3’ | 5’-CATCTCCAGAGTCCAGCACA-3’ |
αSMA, alfa smooth muscle actin; EPO, erythropoietin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRF-4, interferon regulatory factor - 4; PD-L1, programmed death-ligand 1; TBP, TATA box binding protein; VE, vascular endothelial; VEGF, vascular endothelial growth factor.
Immunohistochemistry, Sirius Red Staining, and Histomorphometric Analysis
Paraffin-embedded human liver specimens and/or murine liver specimens used in this study were immunostained as previously reported.23,26,31 Briefly, paraffin sections (4-μm thick), mounted on poly-L-lysine–coated slides, were incubated with the monoclonal antibody against HIF-1α (dilution, 1:100 vol/vol), HIF-2α (dilution, 1:200 vol/vol), α-SMA (dilution, 1:400, vol/vol), PCNA (dilution, 1:400 vol/vol), F4/80 (dilution, 1:500 vol/vol), and SB3 (dilution, 1:50 vol/vol). After blocking endogenous peroxidase activity with 3% hydrogen peroxide and performing microwave antigen retrieval, primary antibodies were labeled using the EnVision, Horseradish-Peroxidase–Labeled System (DAKO, Santa Clara, CA) and visualized by 3’-diaminobenzidine substrate. Collagen deposition was evidenced by Picro-Sirius Red staining as previously described,23 and quantification of fibrosis in the murine liver was performed by histomorphometric analysis using a digital camera and a bright-field microscope to collect images that then were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD).
Cell Proliferation Assays
Proliferation of H/V6 or H/2α cells was evaluated by crystal violet assay by seeding cells in a 96-well plate at a density of 104 cells per well for up to 72 hours. At the desired time, the medium was removed, and the cells were washed twice with phosphate-buffered saline, once with distilled water, and then stained with 0.5% (wt/vol) crystal violet solution for 20 minutes. After washing with water, the crystal violet was solubilized with 50 μL of 10% acetic acid solution, and absorbance was measured at 595–650 nm using a microplate reader (SpectraMAX M3; Molecular Devices, Sunnyvale, CA). The proliferative capacity of H/V6 or H/2α cells was confirmed further by bromodeoxyuridine incorporation assay using a colorimetric kit supplied by Roche Diagnostic (Indianapolis, IN, 11647229001).
Cell-Cycle Analysis
Cell-cycle analysis was performed essentially as recently reported.26 Briefly, H/V6 and H/2α cells were seeded in culture plates (105 cells/well, 35 mm diameter), for up to 72 hours. At the indicated time point, cells were trypsinized, centrifuged at 1000 rpm for 10 minutes, and fixed with ethanol (ETOH, 70%), then treated with RNase for 30 minutes (final concentration, 0.4 mg/mL), and stained with propidium iodide (final concentration, 0.184 mg/mL). The cell cycle was analyzed by flow cytometry (Accuri C6 flow cytometer; Becton Dickinson, Milan, Italy) and quantified with FCS Express 4 Flow Research Edition software (De Novo Software, Pasadena, CA).
YAP Silencing by Small RNA Interference
RNA interference experiments to knock down YAP expression in H/V6 or H/2α were performed using siRNA duplex and HiPerfect Transfection reagent (Qiagen Italia, Milano, Italy) according to the manufacturer’s instructions for up to 72 hours, as previously described.34 The following target sequence was used for YAP: 5’-CAGGTGATACTATCAACCAAA-3’.
Data Analysis and Statistical Calculations
Statistical analyses were performed with GraphPad Prism 6.01 statistical software (GraphPad Software, San Diego, CA) using 1-way analysis of variance testing with the Tukey correction for multiple comparisons, or the Student t test or the Mann–Whitney test for nonparametric values. Significance was taken at the 5% level. Normality distribution was assessed by the Kolmogorov–Smirnov algorithm. Associations were estimated using the Pearson correlation and the Fisher exact test for the contingency analysis. Kaplan–Meier curves of survival and time to recurrence were estimated using the log-rank (Mantel–Cox) test. The data from cell culture experiments represent the means ± SEM of at least 3 independent experiments.
All authors had access to the study data and reviewed and approved the final manuscript.
CRediT Authorship Contributions
Beatrice Foglia (Conceptualization: Supporting; Formal analysis: Equal; Investigation: Equal; Methodology: Lead; Supervision: Equal; Writing – review & editing: Supporting)
Salvatore Sutti (Conceptualization: Supporting; Formal analysis: Equal; Investigation: Equal; Methodology: Lead; Supervision: Equal; Writing – review & editing: Supporting)
Stefania Cannito (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Chiara Rosso (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Marina Maggiora (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Riccardo Autelli (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Erica Novo (Formal analysis: Supporting; Investigation: Supporting)
Claudia Bocca (Formal analysis: Supporting; Investigation: Supporting)
Gianmarco Villano (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Naresh Naik Ramavath (Formal analysis: Supporting; Investigation: Supporting)
Ramy Younes (Formal analysis: Supporting; Investigation: Supporting)
Ignazia Tusa (Formal analysis: Supporting; Investigation: Supporting)
Elisabetta Rovida (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Patrizia Pontisso (Conceptualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Elisabetta Bugianesi (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Emanuele Albano (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Maurizio Parola (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding The research leading to these results was funded by the Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 2014 ID 15274 project (M.P.); European Union’s Horizon 2020 research and innovation program grant 634413 for the Elucidating Pathways of Steatohepatitis project (E.B.); The (Cassa di Risparmio delle Province Lombarde) CariPLO Foundation grant 2011-0470 (E.A. and M.P.); The University of Torino (E.N. and M.P.); and the University of Padova project CPDA110795 (P.P.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., Wong V.W.-S., Yilmaz Y., George J., Fan J., Vos M. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 2.McPherson S., Hardy T., Henderson E., Burt A.D., Day C.P., Anstee Q.M. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J Hepatol. 2015;62:1148–1155. doi: 10.1016/j.jhep.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Golabi P., de Avila L., Minhui Paik J., Srishord M., Fukui N., Qiu Y., Burns L., Afendy A., Nader F. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J Hepatol. 2019;71:793–801. doi: 10.1016/j.jhep.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Torres D.M., Harrison S.A. Nonalcoholic steatohepatitis and noncirrhotic hepatocellular carcinoma: fertile soil. Semin Liver Dis. 2012;32:30–38. doi: 10.1055/s-0032-1306424. [DOI] [PubMed] [Google Scholar]
- 5.Younes R., Bugianesi E. Should we undertake surveillance for HCC in patients with NAFLD? J Hepatol. 2018;68:326–334. doi: 10.1016/j.jhep.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Younossi Z., Stepanova M., Ong J.P., Jacobson I.M., Bugianesi E., Duseja A., Eguchi Y., Wong V.W., Negro F., Yilmaz Y., Romero-Gomez M., George J., Ahmed A., Wong R., Younossi I., Ziayee M., Afendy A. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Global Nonalcoholic Steatohepatitis Council. Clin Gastroenterol Hepatol. 2019;17:748–755.e3. doi: 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 7.Nath B., Szabo G. Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology. 2012;55:622–633. doi: 10.1002/hep.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson G.K., Tennant D.A., McKeating J.A. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: current understanding and future directions. J Hepatol. 2014;61:1397–1406. doi: 10.1016/j.jhep.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Lefere S., Van Steenkiste C., Verhelst X., Van Vlierberghe H., Devisscher L., Geerts A. Hypoxia-regulated mechanisms in the pathogenesis of obesity and non-alcoholic fatty liver disease. Cell Mol Life Sci. 2016;73:3419–3431. doi: 10.1007/s00018-016-2222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Lou T. Hypoxia inducible factors in hepatocellular carcinoma. Oncotarget. 2017;8:46691–46703. doi: 10.18632/oncotarget.17358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKeown S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radiol. 2014;87:20130676. doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majmundar A.J., Wong W.J., Simon M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schito L., Semenza G.L. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Luo D., Wang Z., Wu J., Jiang C., Wu J. The role of hypoxia inducible factor-1 in hepatocellular carcinoma. Biomed Res Int. 2014;2014:409272. doi: 10.1155/2014/409272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menrad H., Werno C., Schmid T., Copanaki E., Deller T., Dehne N., Brune B. Roles of hypoxia-inducible factor-1alpha (HIF-1alpha) versus HIF-2alpha in the survival of hepatocellular tumor spheroids. Hepatology. 2010;51:2183–2192. doi: 10.1002/hep.23597. [DOI] [PubMed] [Google Scholar]
- 16.He C., Sun X.P., Qiao H., Jiang X., Wang D., Jin X., Dong X., Wang J., Jiang H., Sun X. Downregulating hypoxia-inducible factor-2alpha improves the efficacy of doxorubicin in the treatment of hepatocellular carcinoma. Cancer Sci. 2012;103:528–534. doi: 10.1111/j.1349-7006.2011.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H.X., Xu Y., Yang X.R., Wang W.M., Bai H., Shi R.Y., Naya S.K., Devbhandari R.P., He Y., Zhu Q.-F., Sun Y.-F., Hu B., Khan M., Anders R.A., Fan J. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/E2F transcription factor 1-dependent apoptotic pathway. Hepatology. 2013;57:1088–1097. doi: 10.1002/hep.26188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao D., Zhai B., He C., Tan G., Jiang X., Pan S., Dong X., Wei Z., Ma L., Qiao H., Jiang H., Sun X. Upregulation of HIF-2alpha induced by sorafenib contributes to the resistance by activating the TGF-alpha/EGFR pathway in hepatocellular carcinoma cells. Cell Signal. 2014;26:1030–1039. doi: 10.1016/j.cellsig.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 19.Yang S.L., Liu L.P., Niu L., Sun Y.F., Yang X.R., Fan J., Ren J.-W., Chen G.G., Lai P.B.S. Downregulation and pro-apoptotic effect of hypoxia-inducible factor 2 alpha in hepatocellular carcinoma. Oncotarget. 2016;7:34571–34581. doi: 10.18632/oncotarget.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Huang J., Li Z., Gong Y., Zou B., Liu X., Ding L., Li P., Zhu Z., Zhang B., Guo H., Cai C., Li J. HIF-2α upregulation mediated by hypoxia promotes NAFLD-HCC progression by activating lipid synthesis via the PI3K-AKT-mTOR pathway. Aging. 2019;11:10839–10860. doi: 10.18632/aging.102488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rankin E.B., Rha J., Selak M.A., Unger T.L., Keith B., Liu Q., Haase V.H. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29:4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu A., Taylor M., Xue X., Matsubara T., Metzger D., Chambon P., Gonzalez F.J., Shah Y.M. Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 2011;54:472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morello E., Sutti S., Foglia B., Novo E., Cannito S., Bocca C., Rajsky M., Bruzzì S., Abate M.L., Rosso C., Bozzola C., David E., Bugianesi E., Albano E., Parola M. Hypoxia-inducible factor 2α drives nonalcoholic fatty liver progression by triggering hepatocyte release of histidine-rich glycoprotein. Hepatology. 2018;67:2196–2214. doi: 10.1002/hep.29754. [DOI] [PubMed] [Google Scholar]
- 24.Ma C., Kesarwala A.H., Eggert T., Medina-Echeverz J., Kleiner D.E., Jin P., Stroncek D.F., Terabe M., Kapoor V., ElGindi M., Han M., Thornton A.M., Zhang H., Egger M., Luo J., Felsher D.W., McVicar D.W., Weber A., Heikenwalder M., Greten T.F. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salomao M., Remotti H., Vaughan R., Siegel A.B., Lefkowitch J.H., Moreira R.K. The steatohepatitic variant of hepatocellular carcinoma and its association with underlying steatohepatitis. Hum Pathol. 2012;43:737–746. doi: 10.1016/j.humpath.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Cannito S., Foglia B., Villano G., Turato C., Delgado T.C., Morello E., Pin F., Novo E., Napione L., Quarta S., Ruvoletto M., Fasolato S., Zanus G., Colombatto S., Lopitz-Otsoa F., Fernández-Ramos D., Bussolino F., Sutti S., Albano E., Martínez-Chantar M.L., Pontisso P., Parola M. SerpinB3 differently up-regulates hypoxia inducible factors-1α and -2α in hepatocellular carcinoma: mechanisms revealing novel potential therapeutic targets. Cancers (Basel) 2019;11:E1933. doi: 10.3390/cancers11121933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh M.Y., Lemos R., Jr., Liu X., Powis G. The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 2011;71:4015–4027. doi: 10.1158/0008-5472.CAN-10-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keith B., Johnson R.S., Simon M.C. HIF-1α and HIF-2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F., Dong X., Lv H., Xiu P., Li T., Wang F., Xu Z., Li J. Targeting hypoxia-inducible factor-2α enhances sorafenib antitumor activity via β-catenin/C-Myc-dependent pathways in hepatocellular carcinoma. Oncol Lett. 2015;10:778–784. doi: 10.3892/ol.2015.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Méndez-Blanco C., Fondevila F., García-Palomo A., González-Gallego J., Mauriz J.L. Sorafenib resistance in hepatocarcinoma: role of hypoxia-inducible factors. Exp Mol Med. 2018;50:134. doi: 10.1038/s12276-018-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannito S., Turato C., Paternostro C., Biasiolo A., Colombatto S., Cambieri I., Quarta S., Novo E., Morello E., Villano G., Fasolato S., Musso T., David E., Tusa I., Rovida E., Autelli R., Smedile A., Cillo U., Pontisso P., Parola M. Hypoxia up-regulates SERPINB3 through HIF-2α in human liver cancer cells. Oncotarget. 2015;6:2206–2221. doi: 10.18632/oncotarget.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S., Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019;61:64–71. doi: 10.1016/j.ceb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Xiao W., Wang J., Ou C., Zhang Y., Ma L., Weng W., Pan Q., Sun F. Mutual interaction between YAP and c-Myc is critical for carcinogenesis in liver cancer. Biochem Biophys Res Commun. 2013;439:167–172. doi: 10.1016/j.bbrc.2013.08.071. [DOI] [PubMed] [Google Scholar]
- 34.Tao J., Calvisi D.F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., Monga S.P.S. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perra A., Kowalik M.A., Ghiso E., Ledda-Columbano G.M., Di Tommaso L., Angioni M.M., Raschioni C., Testore E., Roncalli M., Giordano S., Columbano A. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61:1088–1096. doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 36.Moon H., Cho K., Shin S., Kim D.Y., Han K.H., Ro S.W. High risk of hepatocellular carcinoma development in fibrotic liver: role of the Hippo-YAP/TAZ signaling pathway. Int J Mol Sci. 2019;20:581. doi: 10.3390/ijms20030581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu C., Tabas I., Schwabe R.F., Pajvani U.B. Maladaptive regeneration - the reawakening of developmental pathways in NASH and fibrosis. Nat Rev Gastroenterol Hepatol. 2021;18:131–142. doi: 10.1038/s41575-020-00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turato C., Cannito S., Simonato D., Villano G., Morello E., Terrin L., Quarta S., Biasiolo A., Ruvoletto M., Martini A., Fasolato S., Zanus G., Cillo U., Gatta A., Parola M., Pontisso P. SerpinB3 and Yap interplay increases Myc oncogenic activity. Sci Rep. 2015;5:17701. doi: 10.1038/srep17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borovski T., De Sousa E., Melo F., Vermeulen L., Medema J.P. Cancer stem cell niche: the place to be. Cancer Res. 2011;71:634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 40.Ma X., Zhang H., Xue X., Shah Y.M. Hypoxia-inducible factor 2α (HIF-2α) promotes colon cancer growth by potentiating Yes-associated protein 1 (YAP1) activity. J Biol Chem. 2017;292:17046–17056. doi: 10.1074/jbc.M117.805655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X., Zhang Y., Liu Y., Fang L., Li L., Sun J., Pan Z., Xin W., Huang P. HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem. 2018;119:3247–3256. doi: 10.1002/jcb.26481. [DOI] [PubMed] [Google Scholar]
- 42.Park E.J., Lee J.H., Yu G.Y., He G., Ali S.R., Holzer R.G., Osterreicher C.H., Takahashi H., Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Minicis S., Agostinelli L., Rychlicki C., Sorice G.P., Saccomanno S., Candelaresi C., Giaccari A., Trozzi L., Pierantonelli I., Mingarelli E., Marzioni M., Muscogiuri G., Gaggini M., Benedetti A., Gastaldelli A., Guido M., Svegliati-Baroni G. HCC development is associated to peripheral insulin resistance in a mouse model of NASH. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf M.J., Adili A., Piotrowitz K., Abdullah Z., Boege Y., Stemmer K., Ringelhan M., Simonavicius N., Egger M., Wohlleber D., Lorentzen A., Einer C., Schulz S., Clavel T., Protzer U., Thiele C., Zischka H., Moch H., Tschöp M., Tumanov A.V., Haller D., Unger K., Karin M., Kopf M., Knolle P., Weber A., Heikenwalder M. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Baglieri J., Brenner D.A., Kisseleva T. The role of fibrosis and liver-associated fibroblasts in the pathogenesis of hepatocellular carcinoma. Int J Mol Sci. 2019;20:1723. doi: 10.3390/ijms20071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong X.X., Qiu X.Y., Hu D.X., Chen X.Q. Advances in hypoxia-mediated mechanisms in hepatocellular carcinoma. Mol Pharmacol. 2017;92:246–255. doi: 10.1124/mol.116.107706. [DOI] [PubMed] [Google Scholar]
- 47.Sonntag R., Giebeler N., Nevzorova Y.A., Bangen J.M., Fahrenkamp D., Lambertz D., Haas U., Hu W., Gassler N., Cubero F.J., Müller-Newen G., Abdallah A.T., Weiskirchen R., Ticconi F., Costa I.G., Barbacid M., Trautwein C., Liedtke C. Cyclin E1 and cyclin-dependent kinase 2 are critical for initiation, but not for progression of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2018;115:9282–9287. doi: 10.1073/pnas.1807155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guido M., Roskams T., Pontisso P., Fassan M., Thung S.N., Giacomelli L., Sergio A., Farinati F., Cillo U., Rugge M. Squamous cell carcinoma antigen in human liver carcinogenesis. J Clin Pathol. 2008;61:445–447. doi: 10.1136/jcp.2007.051383. [DOI] [PubMed] [Google Scholar]
- 49.Turato C., Vitale A., Fasolato S., Ruvoletto M., Terrin L., Quarta S., Ramirez Morales R., Biasiolo A., Zanus G., Zali N., Tan P.S., Hoshida Y., Gatta A., Cillo U., Pontisso P. SERPINB3 is associated with TGF-β1 and cytoplasmic β-catenin expression in hepatocellular carcinomas with poor prognosis. Br J Cancer. 2014;110:2708–2715. doi: 10.1038/bjc.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenhough A., Bagley C., Heesom K.J., Gurevich D.B., Gay D., Bond M., Collard T.J., Paraskeva C., Martin P., Sansom O.J., Malik K., Williams A.C. Cancer cell adaptation to hypoxia involves a HIF-GPRC5A-YAP axis. EMBO Mol Med. 2018;10:e8699. doi: 10.15252/emmm.201708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertout J.A., Majmundar A.J., Gordan J.D., Lam J.C., Ditsworth D., Keith B., Brown E.J., Nathanson K.L., Simon M.C. HIF-2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci U S A. 2009;106:14391–14396. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]