Abstract
Immune checkpoint blockade (ICB) therapies exhibit substantial clinical benefit in different cancers, but relatively low response rates in the majority of patients highlight the need to understand mutual relationships among immune features. Here, we reveal overall positive correlations among immune checkpoints and immune cell populations. Clinically, patients benefiting from ICB exhibited increases for both immune stimulatory and inhibitory features after initiation of therapy, suggesting that the activation of the immune microenvironment might serve as the biomarker to predict immune response. As proof-of-concept, we demonstrated that the immune activation score (ISΔ) based on dynamic alteration of interleukins in patient plasma as early as two cycles (4–6 weeks) after starting immunotherapy can accurately predict immunotherapy efficacy. Our results reveal a systematic landscape of associations among immune features and provide a noninvasive, cost-effective, and time-efficient approach based on dynamic profiling of pre- and on-treatment plasma to predict immunotherapy efficacy.
Keywords: cancer immunotherapy, immune checkpoints, immune cell population, immune activation score, noninvasive biomarker
Graphical abstract
Public summary
-
•
Reveal a systematic landscape of associations among immune features in primary, metastatic, and ICB-treated tumors
-
•
The activation of the immune microenvironment might serve as the biomarker of immunotherapy
-
•
Dynamic alteration of interleukins in patient plasma can accurately predict immunotherapy efficacy
-
•
Provide a noninvasive, cost-effective, and time-efficient approach to predict immunotherapy efficacy
Introduction
Immunotherapy provides remarkable clinical efficacy in multiple cancer types,1 including melanoma2,3 and lung cancer.4 Unfortunately, in most cancers, only a small proportion of patients benefit from immunotherapy.5 Stimulatory and inhibitory immune checkpoints are critical for maintaining in vivo immune homeostasis and regulating the type, magnitude, and duration of the immune response.6 Inhibitory immune checkpoints, including CTLA-4, PD-1/PD-L1, and LAG-3, which bind to tumor cells or tumor microenvironment ligands to attenuate T cell activity, enable tumor cells to evade immunosurveillance.7, 8, 9, 10 In contrast, stimulatory immune checkpoints, including OX40, GITR, and ICOS, enhance the activation and proliferation of effector T cells to eliminate tumors.11, 12, 13 Deeper understanding of the relationships among immune checkpoint mediators has the potential to guide identification of effective immunotherapies as well as biomarkers able to identify patients most likely to benefit from immune checkpoint blockade (ICB) therapies.
The tumor immune microenvironment (TIME) also plays critical roles in human tumorigenesis and response to immunotherapy.14, 15, 16, 17, 18 TIME involves a wide range of immune cell populations with tremendous diversity and plasticity, including numerous innate and adaptive immune cell subpopulations.19 Several immune cell populations, including activated CD8 T cells, natural killer cells, and CD4 helper 1 T cells, are associated with favorable patient prognosis.20, 21, 22 In contrast, other immune cell populations, including myeloid-derived suppressor cells (MDSCs),23 regulatory T cells (Tregs),24,25 and tumor-associated macrophages (TAMs),26 are involved in tumor immune escape25,27 and associated with worse patient prognosis.26,28,29 A comprehensive understanding of the relationship among tumor-infiltrating immune cell populations is critical to provide important insights into the mechanisms underlying immune surveillance and tumor immunotherapy.
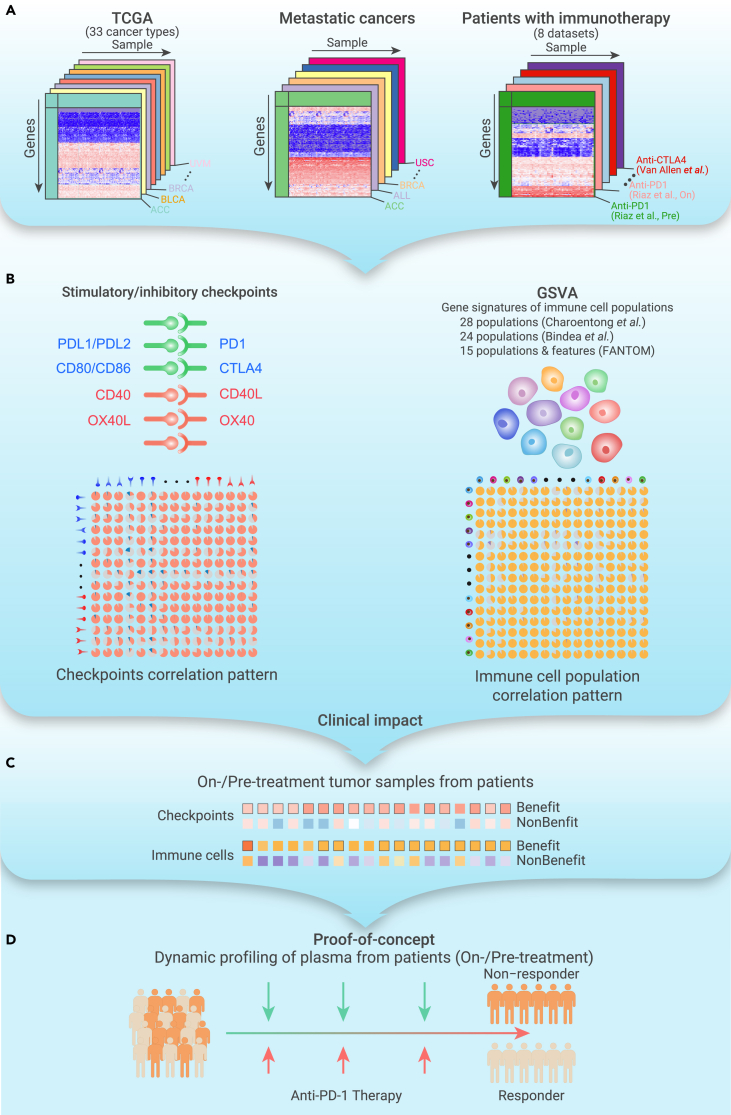
Previous studies have explored potential biomarkers to predict patient response to ICB treatment, including the expression of checkpoints (e.g., CTLA4,2 PD-1,30 PD-L1,31 PD-L22), the tumor mutation burden (TMB),32 neoantigen load, T cell-inflamed gene expression profile (GEP),33 microsatellite instability (MSI),34 and tumor immune clonality.35,36 However, recent studies demonstrated the limitations of these biomarkers.37,38 These potential biomarkers have mainly been assessed in cancer patients before ICB treatment, and it is necessary to identify powerful biomarkers for immunotherapy. A recent study demonstrated the significance of dynamic risk profiling during therapy to develop predictive biomarkers for personalized outcome prediction.39 Here, we investigated the relationships among immune features, including stimulatory and inhibitory checkpoints, as well as different tumor-infiltrating immune cell populations in primary tumors, metastatic tumors, and ICB-treated tumors (Figure 1A). We observed an overall positive correlation among these immune features (Figure 1B). Importantly, patients who benefited from ICB treatment exhibited an elevated immune microenvironment after treatment (Figure 1C). As proof-of-concept, we demonstrated that the activation of interleukins (ILs) in patient plasma after starting two cycles of immunotherapy (4–6 weeks) could predict patient response to immunotherapy (Figure 1D). This timeline is a few weeks before the traditional computed tomography (CT) (6–12 weeks), which is critical for cancer patients. Our study highlights the significance of noninvasive dynamic profiling of pre- and on-treatment biopsies in predicting immunotherapy efficacy early in the course of therapy.
Figure 1.
Overview of procedures and clinical insight of this study
(A) Sample collection from primary tumors, metastatic tumors, and patients with ICB treatment.
(B) Relationships among stimulatory and inhibitory checkpoints and relative abundance of immune cell populations.
(C) Clinical impact of dynamic profiling of patients with ICB treatment through invasive biospecimen.
(D) Proof-of-concept evidence for dynamic profiling in plasma to predict response to immunotherapy.
Results
Dynamic equilibrium of immune checkpoint mediators and cell populations in primary tumors
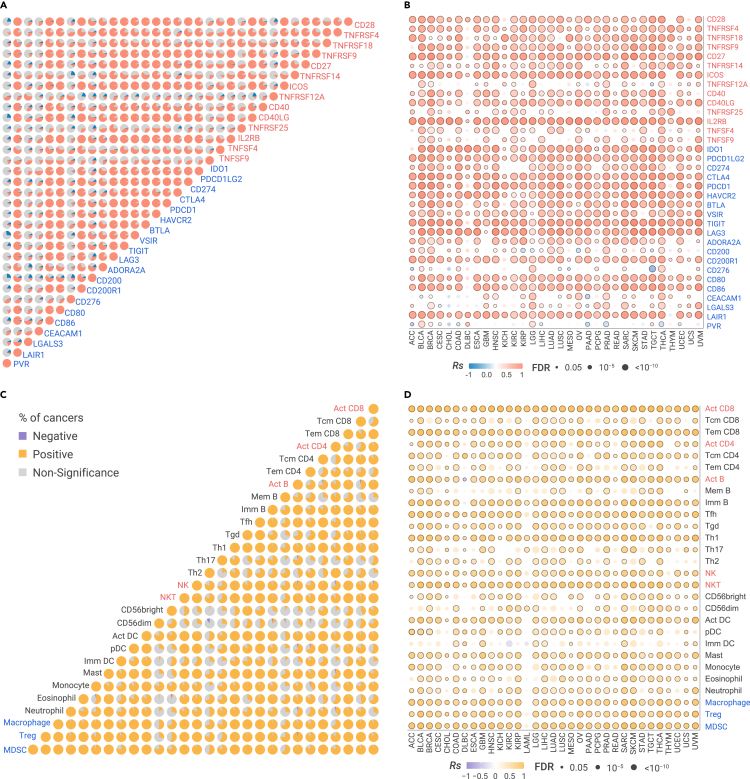
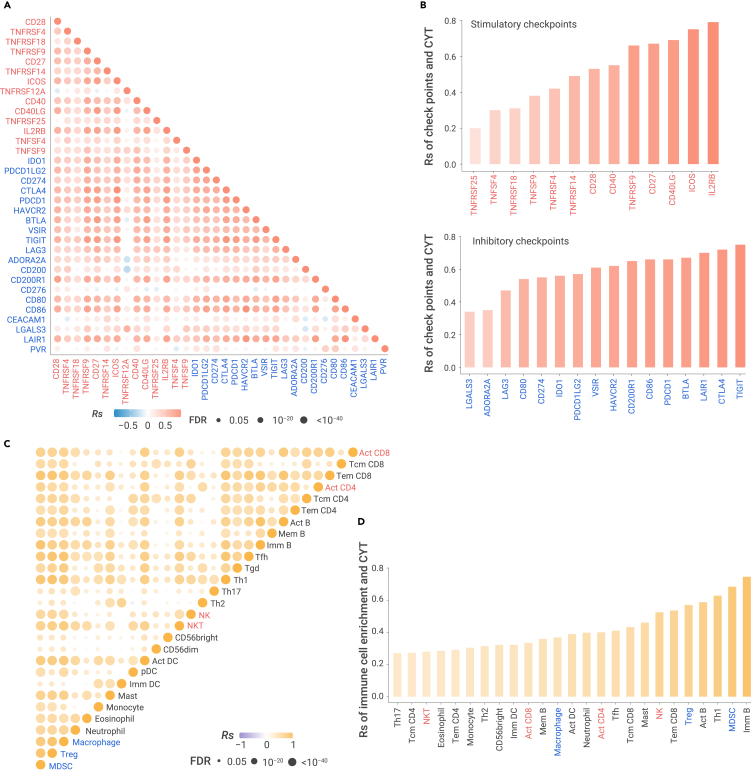
To explore the relationship between stimulatory and inhibitory immune checkpoints (Table S1) in primary tumors, we calculated pairwise correlations among the expression of 14 stimulatory and 20 inhibitory immune checkpoints40 across 33 cancer types from The Cancer Genome Atlas41 (TCGA; Figure 1; Table S2; see online Methods). We observed an overall positive correlation among checkpoints across most cancer types (|Rs| > 0.3 and false discovery rate [FDR] < 0.05; Figure 2A). The receptor/ligand pairs, either inhibitory or stimulatory checkpoints, are significantly correlated. For example, the inhibitory CTLA4/CD80 (receptor/ligand) pair is highly correlated in 32 cancer types, and the stimulatory CD28/CD80 pair is significantly correlated in 31 cancer types. Furthermore, other checkpoints beyond receptor/ligand pairs are positively correlated across multiple cancer types no matter whether they are proposed to be stimulatory or inhibitory mediators (Figure 2A). For example, inhibitory receptor CTLA4 is positively correlated with the inhibitory receptor PDCD1 (median Spearman's correlation [Rs] = 0.73), stimulatory receptor CD27 (median Rs = 0.74), inhibitory ligand PDCD1LG2 (median Rs = 0.61), and stimulatory ligand CD40LG, in 32, 32, 33, and 33 cancers, respectively. We applied partial correlation to correct tumor purity (Figure S1A) or overall immune cell infiltration (Figure S1B) as confounding factors and observed overall similar pattern. We further obtained forty independent datasets with GEPs across multiple primary tumors, including non-small cell lung cancer (NSCLC), head and neck squamous cell cancer (HNSCC), clear cell renal cell carcinoma (ccRCC), pancreatic cancer (PAAD), breast cancer (BRCA), and colorectal cancer (CRC) (see online Methods; Table S3). We observed a consistently overall positive correlation among immune checkpoints across different datasets (Figures S1C–S1F). We also examined the relationship between cytolytic activity (CYT), a proxy to reflect the ability of T cells to kill cancer cells,42 and immune checkpoints, and observed an overall positive correlation (Figure 2B). We demonstrated that CYT is not only positively correlated with most inhibitory checkpoints, but also surprisingly positively correlated with most stimulatory checkpoints.
Figure 2.
Correlation between stimulatory and inhibitory features in TCGA primary tumors
(A) The proportion of cancer types with positive (Rs > 0.2 and FDR < 0.05 in red) or negative (Rs < −0.2 and FDR < 0.05 in blue) correlation for expression of pairwise stimulatory (red) and inhibitory (blue) checkpoints in 33 cancer types.
(B) The Spearman's correlation of cytotoxic activity and the expression of checkpoints in cancers.
(C) The proportion of cancer types with positive correlation (Rs > 0.2 and FDR < 0.05 in orange) or negative correlation (Rs < −0.2 and FDR < 0.05 in purple) for relative abundance (GSVA scores) of the 28 immune cells in 33 cancer types.
(D) The Spearman's correlation of cytotoxic activity and relative abundance of immune cell populations in cancer. The sample size is listed in Table S2.
The TIME involves a wide range of immune cell populations. To understand the relationship among immune cell populations in the microenvironment of primary tumors, we calculated the relative abundance of immune cell populations by gene set variation analysis (GSVA)43 based on their gene signatures (Table S1; see online Methods). We found that most immune cell populations are positively correlated, regardless if they are inhibitory cell populations (e.g., MDSCs, Tregs, TAMs) or stimulatory cell populations (e.g., active CD4/CD8 T cells [Act CD4/CD8], effective memory CD4/CD8 T cells [Tem CD4/CD8], natural killing cells [NKs], natural killing T cells [NKTs]) (Figure 2C). For example, Act CD8, the stimulatory cell population, is highly correlated with stimulatory cell populations, including NKTs in 32 cancers (median Rs = 0.61) and type 1 helper T cell (Th1) in 32 cancers (median Rs = 0.62). However, Act CD8 is also correlated with inhibitory cell populations, including Treg in 31 cancers (median Rs = 0.50) and MDSC in 32 cancers (median Rs = 0.70). Treg is highly correlated with inhibitory cell populations, including MDSC in 33 cancers (median Rs = 0.85) and TAM in 33 cancers (median Rs = 0.82). It is also highly correlated with stimulatory cell populations, including NK in 33 cancers (median Rs = 0.74) and Th1 in 33 cancers (median Rs = 0.84). The overall similar pattern was observed when we applied partial correlation to correct tumor purity (Figure S2A) or overall immune cell infiltration (Figure S2B) as confounding factors. These results were validated independently from other immune gene signatures, which are used to estimate the relative abundance of immune cell populations (Figures S2C and S2D), including 24 immune cell populations,44 11 immune cell populations and four immune features (MHC class I, CYT, type I IFN response, and type II IFN response).42,45 We further used MCP-counter46 and TIMER,47 two deconvolution methods, and observed a similar pattern (Figures S2E and S2F). Furthermore, CYT is highly correlated with both stimulatory and inhibitory populations (Figure 2D). Taken together, our results revealed an unexpected positive correlation among inhibitory and stimulatory immune features and immune cell populations in primary tumors, suggesting the co-regulation of stimulatory and inhibitory immune signaling pathways to maintain the dynamic equilibrium of immune microenvironment in primary tumors. Alternatively, they may reflect the coordinate recruitment of activated lymphocytes as well as populations of cells designed to maintain immune equilibrium.
Dynamic equilibrium of immune features in metastatic cancers
Currently, most ICB treatments are applied in the metastatic/advanced tumor environment, while the TCGA sample set is composed of mainly of patients at primary diagnosis. To investigate whether the correlations among immune features are also observed in metastatic cancers, we obtained transcriptional profiles from ∼1,000 metastatic tumors48 (Table S4) and combined all metastatic samples into one cohort due to the limited sample size in each cancer type. We observed that most receptor/ligand or other checkpoint pairs are highly correlated in metastatic tumors (Figure 3A). For example, CD86 is the receptor of both the inhibitory ligand CTLA4 and the stimulatory ligand CD28. CTLA4 is highly correlated with CD86 (Rs = 0.68), and CD28 is highly correlated with CD86 (Rs = 0.60). However, inhibitory receptor PDCD1 is not only correlated with other inhibitory receptors (e.g., CTLA4, Rs = 0.73; TIGIT, Rs = 0.71) and inhibitory ligands (e.g., CD80, Rs = 0.47; CD86, Rs = 0.57), but also correlated with stimulatory receptors (e.g., IL2RB, Rs = 0.72; ICOS, Rs = 0.74) and stimulatory ligands (e.g., CD40LG, Rs = 0.73; TNFSF9, Rs = 0.39). Both inhibitory/stimulatory checkpoints are highly correlated with CYT, Rs ranging from 0.3 for TNFSF4 to 0.75 for TIGIT (Figure 3B). We observed similar pattern in BRCA samples (n = 160), BRCA samples metastasized to liver (n = 68), and BRCA samples metastasized to lymph node (n = 36) (Figures S3A–S3C).
Figure 3.
Correlation among immune features in metastatic tumor
(A) Spearman's correlation of pairwise checkpoints in pan-metastatic tumors.
(B) Spearman's correlation of cytotoxic activity and the expression of stimulatory checkpoints (upper panel) and inhibitory checkpoints (bottom panel).
(C) The Spearman's correlation for the relative abundance between pairwise immune cell populations in pan metastatic tumors.
(D) The Spearman's correlation between cytotoxic activity and the relative abundance of immune cell populations. Sample information is listed in Table S4.
Furthermore, immune cell populations are highly correlated across the pan-cancer cohort of metastatic tumors (Figure 3C). For example, Act CD8 is not only positively correlated with immuno-stimulatory cell populations, such as Act CD4 (Rs = 0.59), NK (Rs = 0.33), and NKT (Rs = 0.64), but also positively correlated with immunosuppressive cell populations, such as Treg (Rs = 0.57), MDSC (Rs = 0.62), and TAM (Rs = 0.58). CYT is positively correlated with different immune cell populations, including MDSC (Rs = 0.64), Act CD4 (Rs = 0.45), and Th1 (Rs = 0.54) (Figure 3D). We observed the similar pattern in metastatic samples derived from one cancer type (Figures S3D–S3F). The positive correlations among immune cell populations in metastatic samples were validated independently from other immune gene signatures, which are used to estimate the relative abundance of immune cell populations (Figures S3G–S3H). Taken together, our results demonstrated a positive correlation among inhibitory and stimulatory immune features and immune cell populations in metastatic tumors, further suggesting a co-regulation of stimulatory and inhibitory immune signaling pathways to maintain a dynamic equilibrium of the immune microenvironment in metastatic tumors.
Dynamic equilibrium of immune checkpoints persists after ICB treatment
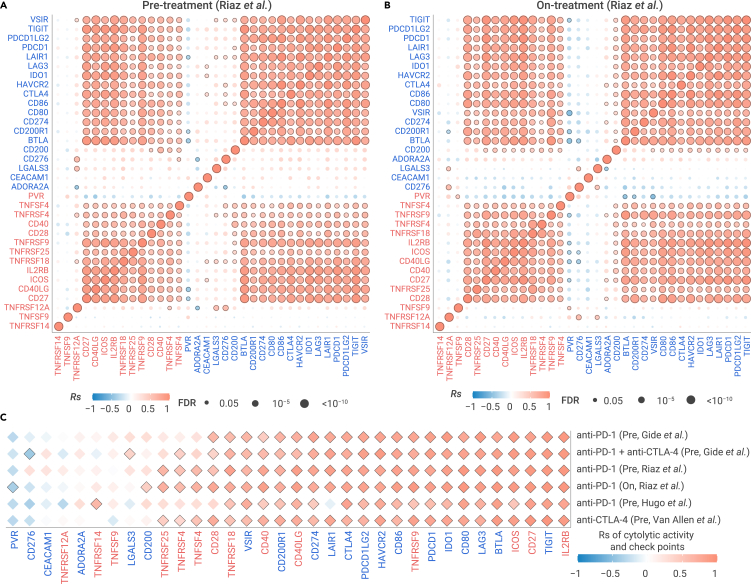
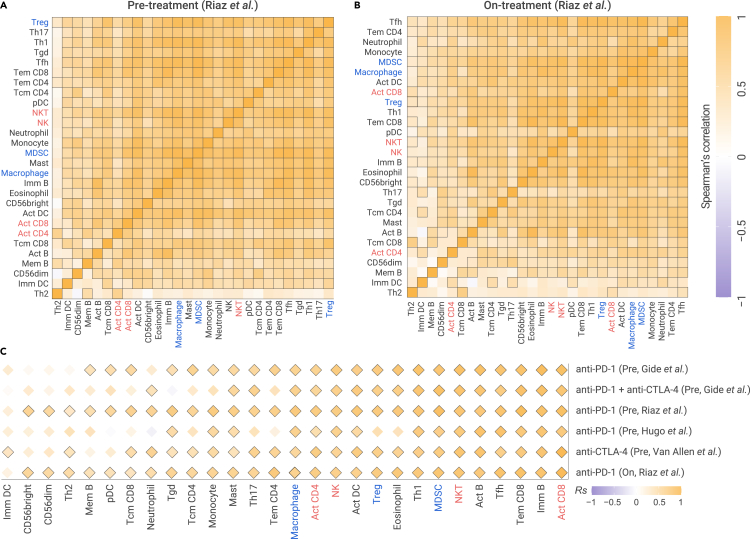
We further assessed the relationship among immune checkpoints in patients with metastatic cancer who underwent ICB treatment in five independent immunotherapy datasets (Table S5; see online Methods). We observed a strong positive correlation among stimulatory and inhibitory immune checkpoint mediators in metastatic melanoma from ICB pre-treatment patient samples.3 The inhibitory immune checkpoints are not only correlated with other inhibitory immune checkpoints, but also correlated with stimulatory immune checkpoints in anti-PD-1 pre-treatment samples (Figures 4A and S4). Stimulatory immune checkpoints are also correlated with both stimulatory and inhibitory immune checkpoints in anti-PD-1 pre-treatment samples (Figures 4A and S4A). In addition, we observed a similar positive correlation pattern for checkpoints in another four datasets (Figure S4). CYT is also highly correlated with most checkpoints in pre-treatment samples (Figure 4C). These results are consistent with our observations in primary and metastatic tumors without ICB treatment.
Figure 4.
Correlation of immune checkpoints in metastatic tumors with ICB treatment
(A and B) The Spearman's correlation of pairwise immune checkpoints in metastatic melanomas with anti-PD-1 in pre-treatment samples (A) and on-treatment samples (B).
(C) The Spearman's correlation of CYT and expression of checkpoints in five ICB pre-treatment datasets. Sample information is listed in Table S5.
We further examined the relationship among these immune checkpoints in metastatic melanoma from patients after ICB treatment.3 Surprisingly, these checkpoints remained positively correlated in on-treatment samples (Figure 4B). For example, PDCD1, the inhibitory checkpoint, is correlated with 15 inhibitory checkpoints, including TIGIT (Rs = 0.95) and LAG3 (Rs = 0.94), and is correlated with 11 stimulatory checkpoints, including IL2RB (Rs = 0.96) and CD27 (Rs = 0.94). The stimulatory checkpoint mediator ICOS is correlated with 11 other stimulatory checkpoints, including IL2RB (Rs = 0.87) and CD27 (Rs = 0.85), and is correlated with 15 inhibitory checkpoints, including TIGIT (Rs = 0.91) and PDCD1 (Rs = 0.88). We further observed a positive correlation between both inhibitory and stimulatory checkpoints and CYT in ICB on-treatment patient samples (Figure 4C). For example, CYT is correlated with inhibitory checkpoint PDCD1 (Rs = 0.91) and also correlated with stimulatory checkpoint CD27 in anti-PD-1 on-treatment samples (Rs = 0.89). Taken together, our results demonstrated that positive correlation among inhibitory and stimulatory immune checkpoints features persists during ICB treatment, suggesting a strong co-regulation of immune signaling pathways to maintain a dynamic equilibrium of the immune microenvironment.
Dynamic equilibrium of immune cell populations remains after ICB treatment
We further assessed relationship among immune cell populations in patients with metastatic cancer who underwent ICB treatment. Both immunosuppressive cell populations and immunostimulatory cell populations are highly correlated in pre-treatment samples (Figure 5A), which is consistent with our analysis in TCGA primary tumors and metastasis tumors. These positive correlations were observed in ICB pre-treatment samples of metastatic melanoma in four other independent datasets (Figure S5). We observed similar positive correlations using Bindea et al. gene signatures (Figure S6) and Fantom5 gene signatures (Figure S7) for immune cell populations. Further, we found that most stimulatory and inhibitory immune cell populations are also highly correlated with CYT in samples with anti-CTLA-4 plus anti-PD-1 treatment, anti-CTLA-4 treatment, and anti-PD-1 pre-treatment (Figure 5C).
Figure 5.
The relative abundance of immune cell populations in metastatic tumors with ICB treatment
(A and B) The Spearman's correlation of pairwise immune cell populations in metastatic melanomas with anti-PD-1 in pre-treatment samples (A) and on-treatment samples (B).
(C) The Spearman's correlation of CYT and the relative abundance of immune cell populations in five ICB pre-treatment datasets. Sample information is listed in Table S5.
Importantly these immune cell populations remained positively correlated in ICB on-treatment samples.3 Immunosuppressive cell populations, including MDSCs, Tregs, and TAMs, are highly correlated with each other (Figure 5B). Immune-stimulatory cell populations, including Act CD4/CD8, Tem CD4/CD8, NK, and NKT, are highly correlated with each other (Figure 5B). For example, MDSC is correlated with Treg in anti-PD-1 on-treatment samples (Rs = 0.92), and Act CD8 is correlated with NKT in anti-PD-1 on-treatment samples (Rs = 0.74). In addition, the immunosuppressive cell populations and immune-stimulatory cell populations are also highly correlated, instead of anti-correlated. For example, MDSC is correlated with Act CD8 in anti-PD-1 on-treatment samples (Rs = 0.79). We further observed positive correlation between both stimulatory and immunosuppressive cell populations and CYT in ICB on-treatment samples (Figure 5C). For example, CYT correlated with the stimulatory immune cell NKT (Rs = 0.76) and also correlated with suppressed immune cell MDSC in anti-PD-1 on-treatment samples (Rs = 0.76). Taken together, our results demonstrated positive correlations among the relative abundance of stimulatory and immunosuppressive cell populations in ICB on-treatment tumor samples, suggesting the dynamic equilibrium of immune cell populations in the immune microenvironment.
Patients with activation of the immune microenvironment benefited from ICB treatment
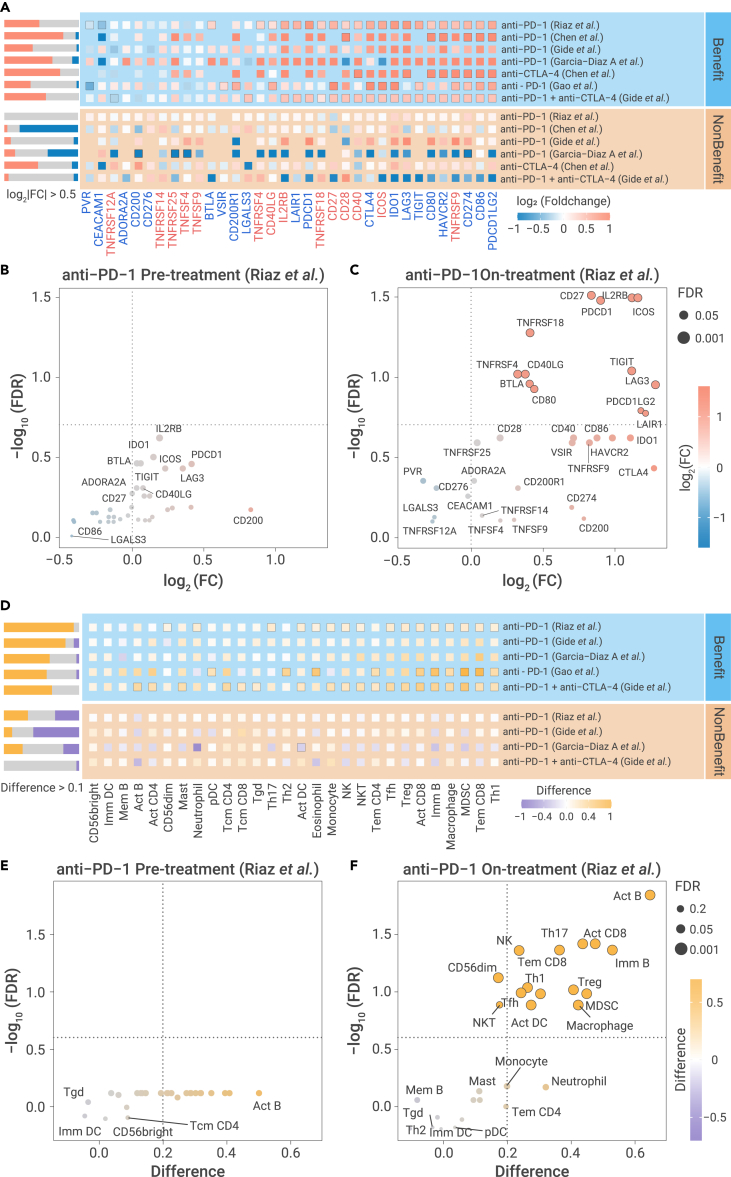
To examine whether dynamic molecule profiles provide useful clinical insight, we next sought to compare alterations of immune microenvironment between pre-treatment and on-treatment biopsies in patients treated with anti-PD-1 treatment.3 Thirteen inhibitory checkpoints, including PDCD1, LAG3, and IDO1, and eight stimulatory checkpoints, including TNFRSF4, ICOS, and TNFRSF9, showed significant upregulation in the benefit group defined as partial/complete responders (PR/CR) or stable disease (SD). In contrast, no checkpoint showed upregulation in the non-benefit group defined as progressive disease (PD) (Figure 6A). We further evaluated the inhibitory and stimulatory checkpoints between on-treatment and pre-treatment tumor samples for different ICB treatments (Table S5). We consistently observed activation of both stimulatory and inhibitory checkpoints among the group of patients who benefited from ICB therapy, while there is no significant activation among the non-benefit group (Figure 6A). We next sought to determine whether this difference exist in pre-treatment samples. Strikingly, we did not observe any significant difference between the benefit and non-benefit groups before ICB treatment (Figure 6B), suggesting a similar baseline expression level of checkpoints in both groups of patients. In contrast, most checkpoints, including PDCD1, ICOS, IL2RB, and CD27, are significantly higher in the benefit group than in the non-benefit group among ICB on-treatment samples taken 23–29 days following the commencement of immunotherapy (Figure 6C). Another dataset taken from tumors treated 7–14 days following the commencement of ICB also demonstrated that the majority of immune checkpoints are significantly higher in the benefit group than non-benefit group (Figure S8A). To be noticed, in the pre-treatment of this dataset and other datasets, we found most immune checkpoints showed nonsignificant difference of mRNA expression between the benefit and non-benefit groups (Figure S8B), suggesting the mRNA level of these checkpoints may not be a good biomarker in the pre-treatment samples. We also found previous biomarkers, including TMB, neoantigen load, and GEP, only show significant difference between the benefit and non-benefit groups in limited datasets (Figure S9), suggesting the necessity to identify powerful biomarkers for immunotherapy. Therefore, the upregulation of immune checkpoints at early time points may serve as a potential biomarker to identify patients who will benefit from ICB treatment.
Figure 6.
Clinical impact of dynamic profiling of immune features in pre-treatment and on-treatment patients
(A) Alterations of checkpoint expression between on-treatment and pre-treatment samples in benefit group (upper panel) and non-benefit group (bottom panel) in six ICB treatment datasets. Text label (red: stimulatory checkpoint; blue: inhibitory checkpoints). Black box of dots indicates the significant difference (FDR < 0.2).
(B and C) The differential expression of inhibitory and stimulatory immune checkpoints between benefit group and non-benefit group in pre-treatment (B) and on-treatment (C). Black circle of dots indicates significant difference (FDR < 0.2). Benefit group denotes patients with PR/CR and SD under ICB treatment, and non-benefit group denotes patients with PD under ICB treatment.
(D) The alteration of relative abundance of 28 immune cell populations between on-treatment and pre-treatment samples in benefit group (upper panel) and non-benefit group (bottom panel) in five ICB treatment datasets. Black box of dots indicates the significant difference (FDR < 0.2).
(E and F) Difference of relative abundance of immune cell populations between benefit group and non-benefit group in pre-treatment (E) and on-treatment (F). Black circle of dots indicates the significant difference (FDR < 0.2). Statistical analysis is performed by two-sided Student's t test.
Furthermore, the relative abundance of immune cell populations increased significantly in on-treatment samples compared with paired pre-treatment samples in the benefit group for active immune cell populations, including Act CD8, NK, and Tem CD8, and suppressive immune cell populations, including TAM, MDSC, and Treg. Very little significant change was observed in the non-benefit group (Figure 6D). This dynamic change in immune cell populations was validated by four other independent datasets, including one dataset with anti-CTLA-4 plus anti-PD-1 treatment,49 two other datasets with anti-PD-1 treatment,49,50 and one dataset with anti-PD-1 treatment51 (Figure 6D). The relative abundance of immune cell populations quantified by two other independent gene signatures demonstrated a similar pattern (Figures S10A and S10B). Interestingly, in pre-treatment samples, there is not a significant difference in the relative abundance of either the stimulatory or inhibitory immune cell populations between the patients who did and did not benefit from anti-PD-1 treatment (Figure 6E). In on-treatment samples, taken 23–29 days following the commencement of immunotherapy, stimulatory immune cell populations, including Act B, Act CD8, NK, and Th17, and suppressive immune cell populations, including Treg and MDSC, are significantly higher in the benefit group than the non-benefit group (Figure 6F). In addition, the majority of immune cell populations showed significantly upregulated relative abundance in the benefit group compared with the non-benefit group in ICB early on-treatment samples49 (Figure S10C). Most of the stimulatory or inhibitory immune cell populations showed nonsignificant difference in the relative abundance between the benefit patients and non-benefit patients, suggesting the individual cell populations may not be a good biomarker in pre-treatment samples (Figure S10D). Our results demonstrated that the dynamic infiltration of multiple types of immune cell populations during immunotherapy may be a key biomarker of immunotherapy benefit.
A proof-of-concept study of noninvasive profiling to predict immunotherapy benefit in NSCLC patients
Through above comprehensive analysis, we demonstrated that these immune checkpoints and cell populations are highly correlated, suggesting that the activation of the immune microenvironment, instead of individual immune checkpoints and/or immune cell populations, might serve as the biomarker to predict immune response. Our observation aligns well with previous studies to demonstrate the fundamental immunologic mechanisms that activating immune pathways track with immune infiltration,42,52, 53, 54 but from a novel perspective of comprehensive relationships among immune checkpoints and immune cell populations. Based on this knowledge, we aim to develop a simple, rapid, and cost-effective approach to evaluate the activation of immune microenvironment. To further prove our findings that dynamic alteration of immune features during immunotherapy can accurately predict immunotherapy benefit, we collected a cohort of 22 NSCLC patients with anti-PD-1 treatment (cohort 1, Table S6). Despite the recent emergence of PD-L1 protein expression as a predictive biomarker in cancer immunotherapy,55,56 it could not predict the response in our cohort (Figure S12).
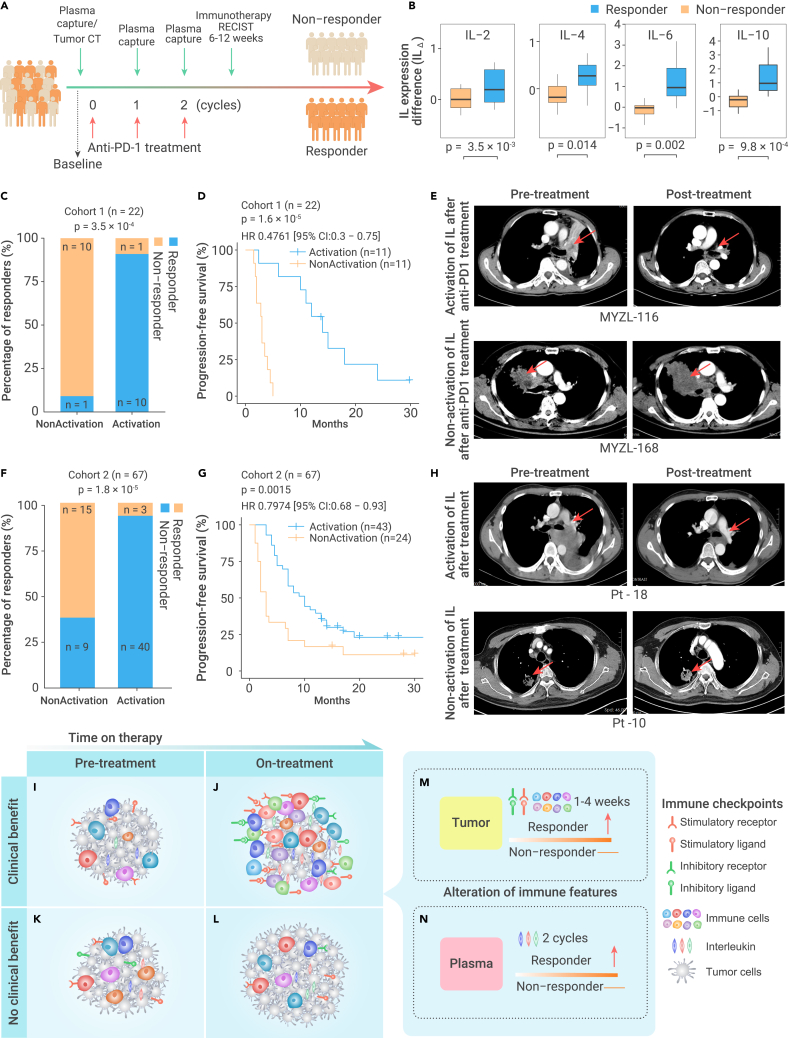
To develop a noninvasive approach, we aim to identify potential biomarkers to reflect the activation of immune microenvironment. We collected plasma from these patients 1 day before each cycle of anti-PD-1 therapy (Figure 7A). We focused on the protein expression levels of four ILs, IL-2, IL-4, IL-6, and IL-10, for the reason that immune cells produce a wide spectrum of ILs during the stimulation of immune system.57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68 Gene set enrichment analysis (GSEA)69 revealed that the IL gene set was significantly enriched in on-treatment samples compared with paired pre-treatment samples in the benefit group in four independent ICB treatment cohorts (Figure S11A). In addition, ILs are highly positively correlated with checkpoints and CYT (Figures S11B and S11C). More importantly, the four ILs are included in the Human Th1/2/17 Cytokine kit, which is a very standard cytokine kit that is widely used for patients.70,71 During the anti-PD-1 therapy, we observed the overall activation of IL proteins at cycles 1 and 2 of anti-PD-1 treatment in anti-PD-1 responders, while there was almost no change for ILs in non-responders (Figures S11A and S11B). Indeed, all four IL proteins showed significant upregulation at cycle 2 of anti-PD-1 therapy in responders (Figure 7B). Furthermore, upregulation of each IL protein during anti-PD-1 therapy showed significantly negative correlation with tumor size (Figure S12E). To assess the activation of immune system, we designed an immune score (IS) based on these four ILs, where ISΔ > 0 was designated as immune activation and ISΔ ≤ 0 was designated as immune nonactivation (see materials and methods; Table S7). Among 22 patients recruited, 11 patients with immune activation (ISΔ > 0) at cycle 2 of anti-PD-1 treatment, and 10 of these 11 patients (90.9%) were responders to anti-PD-1 treatment. This is significantly higher than the percentage of patients without immune activation (ISΔ ≤ 0; 10/11 versus 1/11; Figure 7C). These patients also had significantly better progression-free survival (PFS) (log rank test, p = 1.6 × 10−5, Figure 7D). Radiographic results demonstrated that patient MYZL-116, with immune activation in the plasma after 6 weeks of anti-PD-1 treatment, had 95% reduction in tumor diameter, while patient MYZL-168, without immune activation, had a 200% increase in tumor diameter (Figure 7E). To assess whether the activation score is an independent predictor, we considered age, sex, the expression of PD-L1, and ISΔ as variables to perform the multivariate Cox regression analysis, we found that the ISΔ is an independent predictor significantly associated with better prognosis (Figure S12F). We further performed multivariate Cox regression analysis by considering different drugs and IS as variables to avoid the effect of different drugs, and also found that ISΔ is an independent predictor (Figure S12G). It is critical to investigate at earlier timepoints for the potential biomarker. We further examined the levels of ILs at cycle 1 (2–3 weeks) and observe the upregulated ILs, but unfortunately did not reach the statistical significance (Figure S12H).
Figure 7.
Dynamic profiling of immune features to predict response to immunotherapy
(A) Schematic of sample collection for lung cancer patients with anti-PD-1 therapy alone or chemotherapy and anti-PD-1 combination therapy.
(B) IL expression difference (ILΔ) at cycle 2 of anti-PD-1 therapy is significantly higher in responders (n = 11) than non-responders (n = 11). p value was determined by paired Mann-Whitney-Wilcoxon test. The boxes indicate the median ±1 quartile, with the whiskers extending from the hinge to the smallest or largest value within 1.5× IQR from the box boundaries.
(C–H) Immune activation (ISΔ) can accurately predict the patient response to anti-PD-1 therapy in patient cohort 1 (C–E) and cohort 2 (F–H). (C and F) Patients with immune activation (ISΔ > 0) after ICB are more likely to respond to anti-PD-1 treatment. p value was determined by Fisher's test. (D and G) Patients with immune activation (ISΔ > 0) demonstrate significantly better PFS. p value was determined by log rank test. (E and H) Tumor size decreases significantly in patients with immune activation (ISΔ > 0) based on CT image.
(I–N) Schematic of immune features in patients (I and J) with benefit or (K and L) without benefit from ICB treatment at (I and K) pre-treatment and (J and L) on-treatment. (M) The activation of immune checkpoints and immune cell abundance in tumor microenvironment can predict immunotherapy benefit after 1–4 weeks of ICB therapy. (N) The activation of ILs in patient plasma at cycle 2 can predict immune benefit of ICB therapy (4–6 weeks).
Considering the small sample size (n = 22) of our patient cohort, we further collected an independent cohort of 67 patients with lung cancer with anti-PD-1 treatment alone or combination treatment of chemotherapy and anti-PD-1 (cohort 2, Table S8). In cohort 2, the PD-L1 protein expression of PD-L1 could not predict the response of cancer immunotherapy (Figures S13A–S13D). Consistently, all four IL proteins are significantly upregulated at cycle 2 of anti-PD-1 therapy in responders (Figure S13E; Table S9), and the alteration of these four IL proteins also negatively correlated with the shrink of tumor (Figure S13F). Among these 67 patients 40 of 43 patients with immune activation at cycle 2 (93.0%) were responders to anti-PD-1 treatment, which is significantly higher than the percentage of patients without immune activation (ISΔ ≤ 0; 9/24 versus 40/43, Figure 7F). In addition, patients with immune activation have significantly better PFS (Figure 7G) and significantly reduced tumor size in radiographic analysis (Figure 7H). We performed the survival analysis for NSCLC and SCLC patients in activation and nonactivation group, respectively, and found that immune activation is associated with better PFS in both cancer types (Figure S14G and S14H). We further performed multivariate Cox regression analysis and demonstrated that ISΔ is an independent predictor by considering the confounding factors, including age, sex, the expression of PD-L1, and different drug treatments (Figures S14I and S14J). Taken together, our results provide the first proof-of-concept evidence that the activation of ILs in an early treatment stage may accurately predict the response to immunotherapy.
Discussion
With the recent remarkable successes with cancer immunotherapies, there is a great need to reveal mutual relationships among immune features in the tumor microenvironment to improve the understanding of ICB and to identify effective combinations and biomarkers. Here, we revealed a systematic landscape of associations among immune features through multiple public datasets for the first time, thereby providing a comprehensive perspective for investigating TIME in immunotherapy. Our results demonstrated an overall positive relationship among immune features in the TIME before and after immunotherapy, suggesting the co-regulation of infiltration of various immune checkpoints and cell populations in the tumor-immune system. These results could alternatively indicate that processes leading to immune activation result in the subsequent induction of regulatory processes designed to bring the immune system into equilibrium. In the static environment of a tumor biopsy, this presents as correlations between both activation and inactivation events.
Importantly, patients who benefitted from immunotherapy tend to have overall upregulation of both immune stimulatory and inhibitory features in on-treatment biopsies compared with pre-treatment biopsies (Figures 7I–7K), while patients who did not benefit from immunotherapy showed no significant immune alterations after treatment (Figures 7K and 7L). Furthermore, there is no significant difference for most immune checkpoints and immune cell populations in the pre-treatment samples between patients who benefited and patients who did not benefit from immunotherapy, suggesting that it is difficult to identify patients who will benefit from treatment based on the pre-treatment samples alone. Although a single pre-treatment biomarker would be useful, a recent novel concept demonstrated the clinical utility of dynamic biomarkers, especially for the complex system that may be characterized by multiple phases.72 Our study highlights the possibility of predicting immunotherapy response by comparing pre-treatment and on-treatment biospecimens for multiple different immune criteria. We showed that an on-treatment invasive biospecimen as early as 7–14 days after initiation of therapy could provide important information regarding which patients are likely to benefit (Figure 7M). Furthermore, we demonstrated a noninvasive approach for predicting immunotherapy benefit by examining the activation of ILs in patient plasma as early as two cycles (4–6 weeks) after treatment starts in two independent cohorts, which is cost effective and time efficient and provides a novel paradigm for identifying biomarkers to predict immunotherapy efficacy (Figure 7N). Those patients do not have the activation of IS; they may still remain not responsive based on our follow up data for up to 32 months. Considering that follow-up responses to immunotherapy may need to be evaluated through imaging at least 6–12 weeks according to guidelines for immunotherapy from The Response Evaluation Criteria in Solid Tumors (iRECIST),73 our approach can evaluate the response of patients several weeks before the traditional CT scan, which is critical for cancer patients. Mechanistically, the benefit from immunotherapy may not correlate with the induction of either positive or negative mediators, but rather with the immune activation or recruitment of activated immune cells. While it is straightforward to understand why evidence for positive immune regulators or effectors would correlate with benefit, it is more difficult to explain associations with negative regulators. It may be that any signal of an ability to activate the immune system is associated with an improved outcome and that positive events subsequently activate negative regulators to re-establish homeostasis. Nevertheless, the underlying mechanisms need to be investigated in further works.
Our study is limited by the expression level of checkpoints, and the abundance of the immune cell populations are estimated based on gene signatures. Considering challenges to capture the identity of immune checkpoints and immune cell populations in transcriptomes, further technologies, and approaches are necessary to validate our observations. Currently, the single-cell transcriptome profiles of tumors are limited in few patients, which prevent us from accessing the associations among different immune features across patients. Furthermore, the single-cell RNA sequencing (scRNA-seq) and fluorescence-activated cell sorting (FACS) normally assumed all-cell populations summed up as 100%. In the real tumor microenvironment, the number of immune cells may be increased or reduced with different percentages. This may further require dynamic single-cell analysis of immune contexture, as it changes under the influence of ICB. In this situation, the assumption that all-cell populations summed up as 100% through scRNA-seq and/or FACS may not be appropriate to reflect this phenomenon. Nevertheless, we developed a simple, rapid, and cost-effective approach to evaluate the activation of immune microenvironment in this pilot study. Further investigations in large-scale patient samples and patients with other cancer types (e.g., melanoma) for immunotherapy trials, especially at an earlier time point, are necessary.
Materials and methods
We obtained gene expression data from TCGA, Gene Expression Omnibus (GEO), and European Bioinformatics Institute (EBI) for primary, metastasis, and tumor with ICB treatment. We also collected immune checkpoint genes and immune cell populations from previous publications.40 We recruited two independent patient cohorts from Hunan Cancer Hospital and analyzed radiographic images and molecular features in patients with immunotherapy. All tissue samples were collected in compliance with the informed consent policy. The study protocol was approved by the Institutional Review Board of Hunan Cancer Hospital (SBQLL-2019-035). More information is available in the supplemental information file.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (no. 2019YFA0111600 and no. 2019YFE0120800 to H.L.), the National Natural Science Foundation of China (no. 81971487 to Y.Y., no. 31800979 to H.L., no. 81902149 to Q.G., and no. 82102891 to X.K.). the Natural Science Foundation of China for outstanding Young Scholars (no. 82022060 to H.L.), the Shanghai Pujiang Program (no. 20PJ1412800 to Y.Y.), the Natural Science Foundation of Shanghai (no. 20ZR1472900 to Y.Y.), the Natural Science Foundation of Hunan Province for outstanding Young Scholars (no. 2019JJ30040 to H.L.), the Natural Science Foundation of Hunan Province of China (no. 2018SK2082 to H.L.), and the Scientific Research Project of Hunan Health and Family Planning Commission (no. B20180855 to H.L.). We thank LeeAnn Chastain for editorial assistance.
Author contributions
L.H. conceived and supervised the project. Y.Y. and L.H. designed the research. Y.Y., D.X., B.R., and Z.Z. performed data analysis. N.Y., Y.Z., Q.G., H.L., and X.C. obtained patient data with immunotherapy. Q.G. and K.X. performed experimental operation, Y.Y., Q.H., C.S., B.Z., L.W., C.L., J.G., Y.L., S.H.L., L.D., H.L., G.B.M., and L.H. interpreted the results. Y.Y., S.H.L., L.D., G.B.M., and L.H. wrote the manuscript.
Declaration of interests
G.B.M. notes personal fees for consulting from Abbvie, Amphista, AstraZeneca, Chrysallis Biotechnology, GSK, ELlipses Pharma, ImmunoMET, Ionis, Lilly, Medacorp, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, Zentalis Pharmaceuticals; stock options from Catena Pharmaceuticals, ImmunoMet, SignalChem, Tarveda, Turbine; Licensed Technology of HRD assay to Myriad Genetics, and DSP patents with Nanostring. All other authors declare no competing interests.
Published Online: December 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2021.100194.
Contributor Information
Hong Liu, Email: hongliu1014@csu.edu.cn.
Xiang Chen, Email: chenxiangck@126.com.
Gordon B. Mills, Email: millsg@ohsu.edu.
Leng Han, Email: leng.han@tamu.edu.
Lead contact website
Supplemental information
Sample information for metastatic tumors from the Database of Genotypes and Phenotypes under accession number phs000673.v2.p1, including SRA ID, biospecimen site, histological type, and patient gender.
References
- 1.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Allen E.M., Miao D., Schilling B., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riaz N., Havel J.J., Makarov V., et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellmann M.D., Ciuleanu T.-E., Pluzanski A., et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marin-Acevedo J.A., Dholaria B., Soyano A.E., et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J. Hematol. Oncol. 2018;11:39. doi: 10.1186/s13045-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruhashi T., Okazaki I.mi., Sugiura D., et al. LAG-3 inhibits the activation of CD4 + T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat. Immunol. 2018;19:1415–1426. doi: 10.1038/s41590-018-0217-9. [DOI] [PubMed] [Google Scholar]
- 8.Bilir C., Sarisozen C. Indoleamine 2,3-dioxygenase (IDO): only an enzyme or a checkpoint controller? J. Oncol. Sci. 2017;3:52–56. [Google Scholar]
- 9.Yu X., Harden K., Gonzalez L.C., et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 10.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanmamed M.F., Pastor F., Rodriguez A., et al. Agonists of co-stimulation in cancer immunorapy directed against CD137, OX40, GITR, CD27, CD28, and ICOS. Semin. Oncol. 2015;42:640–655. doi: 10.1053/j.seminoncol.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Yard W.N. OX40: structure and function - what questions remain? Am. J. Respir. Cell Mol. Biol. 2018;59:437–447. [Google Scholar]
- 13.Aspeslagh S., Postel-Vinay S., Rusakiewicz S., et al. Rationale for anti-OX40 cancer immunotherapy. Eur. J. Cancer. 2016;52:50–66. doi: 10.1016/j.ejca.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Y., Kuang X., Xie Z., et al. Small-molecule MMP2/MMP9 inhibitor SB-3CT modulates tumor immune surveillance by regulating PD-L1. Genome Med. 2020;12:1–19. doi: 10.1186/s13073-020-00780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y., Ying J., Li L., et al. Sex-associated molecular differences for cancer immunotherapy. Nat. Commun. 2020;11:1779. doi: 10.1038/s41467-020-15679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing Y., Liu J., Ye Y., et al. Multi-omics prediction of immune-related adverse events during checkpoint immunotherapy. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-18742-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing Y., Zhang Y., Wang J., et al. Association between sex and immune-related adverse events during immune checkpoint inhibitor therapy. J. Natl. Cancer Inst. 2021;113:1396–1404. doi: 10.1093/jnci/djab035. [DOI] [PubMed] [Google Scholar]
- 19.Thorsson V., Gibbs D.L., Brown S.D., et al. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridman W.H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 21.Pasero C., Gravis G., Granjeaud S., et al. Highly effective NK cells are associated with good prognosis in patients with metastatic prostate cancer. Oncotarget. 2015;6:14360–14373. doi: 10.18632/oncotarget.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peranzoni E., Lemoine J., Vimeux L., et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti – PD-1 treatment. Proc. Natl. Acad. Sci. U S A. 2018;115:4041–4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umansky V., Blattner C., Gebhardt C., Utikal J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines. 2016;4:36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka A., Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaudhary B., Elkord E. Regulatory T Cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. Vaccines. 2016;4:28. doi: 10.3390/vaccines4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L., Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J. Hematol. Oncol. 2017;10:58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toor S.M., Elkord E. Therapeutic prospects of targeting myeloid-derived suppressor cells and immune checkpoints in cancer. Immunol. Cell Biol. 2018;96:888–897. doi: 10.1111/imcb.12054. [DOI] [PubMed] [Google Scholar]
- 28.Tesi R.J. MDSC; the most important cell you have never heard of. Trends Pharmacol. Sci. 2019;40:4–7. doi: 10.1016/j.tips.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Shang B., Liu Y., Jiang S.J., Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paré L., Pascual T., Seguí E., et al. Association between PD1 mRNA and response to anti-PD1 monotherapy across multiple cancer types. Ann. Oncol. 2018;29:2121–2128. doi: 10.1093/annonc/mdy335. [DOI] [PubMed] [Google Scholar]
- 31.Hellmann M.D., Nathanson T., Rizvi H., et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33:843–852. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samstein R.M., Lee C., Shoushtari A.N., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cristescu R., Mogg R., Ayers M., et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quiroga D., Lyerly H.K., Morse M.A. Deficient mismatch repair and the role of immunotherapy in metastatic colorectal cancer. Curr. Treat. Options Oncol. 2016;17:41. doi: 10.1007/s11864-016-0414-4. [DOI] [PubMed] [Google Scholar]
- 35.Arakawa A., Vollmer S., Tietze J., et al. Clonality of CD4+ blood T cells predicts longer survival with CTLA4 or PD-1 checkpoint inhibition in advanced melanoma. Front. Immunol. 2019;10:1–12. doi: 10.3389/fimmu.2019.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuben A., Zhang J., Chiou S.H., et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat. Commun. 2020;11:603. doi: 10.1038/s41467-019-14273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKean W.B., Moser J.C., Rimm D., Hu-Lieskovan S. Biomarkers in precision cancer immunotherapy: promise and challenges. Am. Soc. Clin. Oncol. Educ. B. 2020;40:e275–e291. doi: 10.1200/EDBK_280571. [DOI] [PubMed] [Google Scholar]
- 38.Lu Z., Peng Z., Liu C., et al. Current status and future perspective of immunotherapy in gastrointestinal cancers. The Innovation. 2020;1:100041. doi: 10.1016/j.xinn.2020.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz D.M., Esfahani M.S., Scherer F., et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell. 2019;178:699–713. doi: 10.1016/j.cell.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auslander N., Zhang G., Lee J.S., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018;24:1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein J.N., Collisson E.A., Mills G.B., et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooney M.S., Shukla S.A., Wu C.J., et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bindea G., Mlecnik B., Tosolini M., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 45.The FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becht E., Giraldo N.A., Lacroix L., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Severson E., Pignon J.-C., et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:1–16. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson D.R., Wu Y., Lonigro R.J., et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gide T.N., Quek C., Menzies A.M., et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell. 2019;35:238–255. doi: 10.1016/j.ccell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Diaz A., Shin D.S., Moreno B.H., et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J., Ward J.F., Pettaway C.A., et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017;23:551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirosh I., Izar B., Prakadan S.M., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jerby-Arnon L., Shah P., Cuoco M.S., et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175:984–997. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu D., Schilling B., Liu D., et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019;25:1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibney G.T., Weiner L.M., Atkins M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17 doi: 10.1016/S1470-2045(16)30406-5. PE542–E551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 57.Boyman O., Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H., Wang Y., Hwang E.S., He Y.W. Interleukin-10: an immune-activating cytokine in cancer immunotherapy. J. Clin. Oncol. 2016;34:3576–3578. doi: 10.1200/JCO.2016.69.6435. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka T., Narazaki M., Kishimoto T. Il-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zamorano J., Rivas M.D., Pérez-G M. Interleukin-4: a multifunctional cytokine. Inmunologia. 2003;22:215–224. [Google Scholar]
- 61.Sun Z., Ren Z., Yang K., et al. A next-generation tumor-targeting IL-2 preferentially promotes tumor-infiltrating CD8+ T-cell response and effective tumor control. Nat. Commun. 2019;10:3874. doi: 10.1038/s41467-019-11782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalia V., Sarkar S. Regulation of effector and memory CD8 T Cell differentiation by IL-2-A balancing. Front. Immunol. 2018;9:2987. doi: 10.3389/fimmu.2018.02987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao J., Hu Y., Li Y., et al. Involvement of JNK signaling in IL4-induced M2 macrophage polarization. Exp. Cell Res. 2017;357:155–162. doi: 10.1016/j.yexcr.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Park S.J., Lee K.P., Kang S., et al. Sphingosine 1-phosphate induced anti-atherogenic and atheroprotective M2 macrophage polarization through IL-4. Cell. Signal. 2014;26:2249–2258. doi: 10.1016/j.cellsig.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:1–16. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher D.T., Appenheimer M.M., Evans S.S. The two faces of IL-6 in the tumor microenvironment. Semin. Immunol. 2014;26:38–47. doi: 10.1016/j.smim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dennis K.L., Blatner N.R., Gounari F., Khazaie K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr. Opin. Oncol. 2013;25:637–645. doi: 10.1097/CCO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu T., Hu E., Xu S., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwase S., Murakami T., Saito Y., Nakagawa K. Steep elevation of blood interleukin-6 (IL-6) associated only with late stages of cachexia in cancer patients. Eur. Cytokine Netw. 2004;15:312–316. [PubMed] [Google Scholar]
- 71.Jaglal M.V., Laber D.A., Arnold F.W., et al. Obtaining routine blood cultures during interleukin-2-containing therapy is unnecessary. Am. J. Clin. Oncol. Cancer Clin. Trials. 2009;32:429–431. doi: 10.1097/COC.0b013e31819142c7. [DOI] [PubMed] [Google Scholar]
- 72.Joost Lesterhuis W., Bosco A., Millward M.J., et al. Dynamic versus static biomarkers in cancer immune checkpoint blockade: unravelling complexity. Nat. Rev. Drug Discov. 2017;16:264–272. doi: 10.1038/nrd.2016.233. [DOI] [PubMed] [Google Scholar]
- 73.Seymour L., Bogaerts J., Perrone A., et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–e152. doi: 10.1016/S1470-2045(17)30074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information for metastatic tumors from the Database of Genotypes and Phenotypes under accession number phs000673.v2.p1, including SRA ID, biospecimen site, histological type, and patient gender.