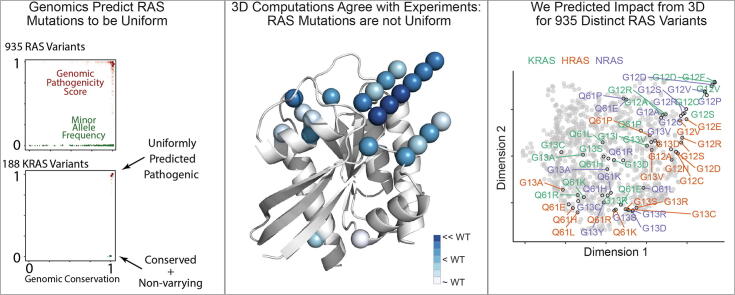
Graphical abstract
Keywords: Genomics, Protein science, Data interpretation, RAS mutation, Structural bioinformatics, Functional genomics
Highlights
-
•
RAS mutations have been non-uniformly characterized biophysically and biochemically.
-
•
We assembled the broadest-to-date resource for KRAS mutation-specific activities.
-
•
Genomics tools do not correlate with KRAS experimental data, while 3D calculations do.
-
•
Calculations indicate local unfolding propensity correlates with enzymatic activity.
Abstract
In the current study, we report computational scores for advancing genomic interpretation of disease-associated genomic variation in members of the RAS family of genes. For this purpose, we applied 31 sequence- and 3D structure-based computational scores, chosen by their breadth of biophysical properties. We parametrized our data by assembling a numerically homogenized experimentally-derived dataset, which when use in our calculations reveal that computational scores using 3D structure highly correlate with experimental measures (e.g., GAP-mediated hydrolysis RSpearman = 0.80 and RAF affinity Rspearman = 0.82), while sequence-based scores are discordant with this data. Performing all-against-all comparisons, we applied this parametrized modeling approach to the study of 935 RAS variants from 7 RAS genes, which led us to identify 4 groups of mutations according to distinct biochemical scores within each group. Each group was comprised of hotspot and non-hotspot KRAS variants, indicating that poorly characterized variants could functionally behave like pathogenic mutations. Combining computational scores using dimensionality reduction indicated that changes to local unfolding propensity associate with changes in enzyme activity by genomic variants. Hence, our systematic approach, combining methodologies from both clinical genomics and 3D structural bioinformatics, represents an expansion for interpreting genomic data, provides information of mechanistic value, and that is transferable to other proteins.
1. Introduction
RAS is one of the most studied proto-oncogenes [1], having been discovered in association with human cancers by the early 80s [2], [3], [4], [5]. Researchers soon realized that only a few positions, referred to as hotspots, contributed to neoplastic transformation when mutated. Subsequent studies showed that different mutations had distinct patterns of disease occurrence and progression, suggesting these variations reflect their underlying biochemical properties [6], [7] and the possibility for therapeutic intervention against this oncogene. However, characterization of RAS’ mutational landscape has not been uniform, with mutations assessed by different assays in different labs and using different biochemical assays or cell and animal models expressing them endogenous or exogenously. Data derived from next generation sequencing of tumors has discovered a large array of different mutations at varied frequencies. Indeed, KRAS is the most frequently mutated RAS family member, altered in 7% of TCGA samples, compared to 2.9% and 1.3% for NRAS and HRAS, respectively [8], [9]. The majority (86%) of cancer-associated hotspot mutations occur at codons G12, G13, and Q61, with a smaller number at K117 and A146. Notably, germline G12 mutations, such as G12S, also cause RASopathies, a group of distinct neurodevelopmental disorders. RASopathy mutations also occur at non-classic amino acids positions compared to the somatic hotspots [10], [11], but these non-classic alleles are also observed rarely in cancer. Thus, there is a need to better understand the effects of the KRAS mutational landscape in heritable conditions and cancer.
In the current study, we sought to develop an approach that expands, enhances, and seeks to parametrized with higher resolution, methods for interpreting genetic variants by considering gene products in their translated form - as 3D folded proteins. We report extensive characterization of RAS mutations using DNA predictive algorithms, combined with 3D structure-based and molecular mechanics calculations, as described previously [12]. This approach rendered scores that we applied to the study of 935 variants from 7 RAS genes, as a comprehensive means to assess relative similarities of mutations across different members of the RAS family. We then correlated our calculations with biochemical data, which indicated that 3D methods do capture information relevant for enzyme activity, and thus of value for drug development. Combined our results, demonstrate that the comprehensive suite of multi-level molecular scores used in this study enhances interpretation of human genetic variation not only for the evaluation of RAS and is likely applicable to other disease-causing genes families.
2. Methods
2.1. Generating a mutation-specific biochemical feature resource
We generated a resource of experimental measurements by numerically homogenizing data from across different studies and various laboratories that quantitatively have measured or described biochemical properties of mutated RAS proteins (KRAS, HRAS and NRAS), relative to the wild type (WT) (Table S1) [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]. We specifically focused on 6 biochemical assays that evaluated the rate of intrinsic hydrolysis, GAP-mediated hydrolysis, nucleotide (GDP and/ or GTP) exchange, and the binding affinities of GTP, GAP, and RAF for hotspot and non-hotspot variants. Our analysis focused on variants with the most biochemical measurements. Our final dataset consists of 23 variants that include 14 hotspot variants and 9 non-hotspot variants (at codons A18, L19, A59, T74, K117, A146 and R164Q; Table S1). For these 23 variants, if the abovementioned 6 biochemical assays were available for mutated KRAS, those measures were recorded in our resource. Otherwise, for the variants G12V/R, G13V, G13S, A59T, Q61P/L/H and A146V we obtained the measurements from mutated HRAS or Q61R for NRAS (Table S1 for detailed description).
The reported biochemical measurements of variants were often relative to the WT (Table S1). Moreover, different groups used different cell lines at different conditions for these measurements. Hence, we strategically summarized the experimental data relative to the WT in 5 categorical groups: similar to the WT (∼WT), higher impact than the WT (>WT), substantially higher impact than the WT (≫WT), lower impact than the WT (<WT), and substantially lower impact than the WT (≪WT). Individual studies’ data can show relative differences among mutations in one of these categories, but we chose to categorize so that quantitative and qualitative reports could be harmonized, expanding the number of usable mutations and measurements.
2.2. RAS computational scores
We used the experimentally derived GDP-bound structures of 7 RAS proteins from the PDB [30]: KRAS (KRAS-4B; PDB: 4obe), HRAS (4q21), NRAS (3con), MRAS (1x1r), RRAS (2fn4), RRAS2 (2ery) and RERG (2atv). For each RAS protein we identified the corresponding DNA coding regions in the human genome (GRCh38) for UniProt canonical isoforms and obtained the genetic variants somatically observed in cancer from COSMIC, genomic variants associated with congenital disease and RASopathy from ClinVar and HGMD, and variants observed in the currently healthy population from gnomAD using BioR (v5.0.0) [31] with custom scripts in R programming language [32]. As a result, we obtained total 935 variants for these 7 Ras genes. We used the pre-selected computational scores from our previous study [12] that included 6 DNA sequence-based scores and minor allele frequency (MAF), 5 protein sequence-based scores and 19 protein 3D structure-based scores. We selected these 31 scores based on the correlation among the scores for all the 935 RAS variants, and 3D scores were assessed based on their ability to capture diverse biophysical or biochemical properties. Detailed description of the 31 scores used in this study is provided in our previous study [12]. We believe these scores are unique and most efficiently cover the broadest diversity of RAS properties hence, suitable for mechanistic interpretation of the RAS hotspot and non-hotspot variants in this study.
2.3. Analysis of experimental data missingness
To explore our data set of the biochemical measurements of the 23 hotspot and non-hotspot variants of KRAS and summarize the structure of the missing values therein, we used the Visualization and Imputation of Missing Values (VIM) version 6.0 [33].
2.4. Analysis of dissimilarities among experimental measures
To identify similarities and dissimilarities between the hotspot and non-hotspot variants, we used the general dissimilarity coefficient or Gower’s distance between two variants. This was the sum of all variable-specific experimental measurements from our literature-mined resource. We computed all the pairwise Gower distances between the variants in the data set using daisy for Dissimilarity Matrix Calculation as implemented in the cluster package v2.1.0 [34].
2.5. Correlation analyses
We calculated the Spearman correlation (RSpearman) among and between the experimental measurements and computational scores, and the corresponding p-values for statistical significance (via the asymptotic t approximation) using the rcorr function from Hmisc package version 4.4–1 [35].
2.6. Dimensionality reduction
For each biochemical feature from the experimental measurements, we combined the correlated computational scores for comparing among hotspot and non-hotspot variants using PHATE (Potential of Heat-diffusion for Affinity-based Trajectory Embedding). PHATE is a newly developed dimensionality reduction technique that generates a low-dimensional embedding, in a way that attempts to preserve local and global similarities to enhance interpretability of the data’s underlying structure [36]. In this study, the structure of the data indicates similarities among RAS variants, according to computational scores using sequence and 3D structure. We performed PHATE analysis using the phateR v1.0.0 package with default parameters [36].
2.7. Western Blot for pERK activity
HEK 293 KRAS mutant cells were created by Flp-In T-REx technology (Invitrogen) by co-transfecting KRAS mutant pcDNA5/FRT/TO plasmid with pOG44 plasmid and selecting for hygromycin resistant clones. Mutant cells were plated at 200,000 cells/well in 6-well plates and induced to express KRAS with 1ug/mL doxycycline the following morning. 24-hours after the addition of doxycycline, the cells were harvested with RIPA buffer. Lysates were homogenized, then cleared by centrifugation for 10 min. Protein concentration was normalized by BCA assay to 2 mg/mL. 10 mg of each sample was run on a 12% SDS PAGE gel and then transferred to a nitrocellulose membrane. Membranes were incubated with primary antibody diluted 1:000 in 5% BSA at 4 degrees overnight. Primary antibodies included: β-Actin (45 kDa) - MAB8929 R&D Systems, KRAS (21 kDa) − 05–516 MilliporeSigma, pERK (42–44 kDa) − 9101S Cell Signaling, Total ERK (42–44 kDa) − 4696S Cell Signaling. Membranes were then washed twice with TBST and incubated for 60 min in corresponding secondary antibody diluted 1:1000. After two more washes in TBST and a final wash in TBS, the membranes were imaged by chemiluminescence on a Chemidoc imager. Densitometry of the blots was performed using ImageJ software. Densitometry of pERK was normalized to total ERK levels and then fold change calculated comparing induced (+doxy) to uninduced (-doxy) samples. Densitometry of KRAS was normalized to total b-actin levels and then fold change calculated comparing induced (+doxy) to all uninduced (-doxy) samples.
3. Results
It is necessary to enhance methods for interpreting genetic variation in the RAS family because variants are known to alter enzymatic properties [14], [18], [21], [26], yet genomics-based pathogenicity predictors yield uniform calls across variants (Fig. S1). Thus, new approaches must be developed to mechanistically explain existing experimental data, and predict the effects of variants that lack experimental characterization.
Building an evidence-based resource to characterize available and missing experimental data shows that KRAS variants are not functionally equivalent
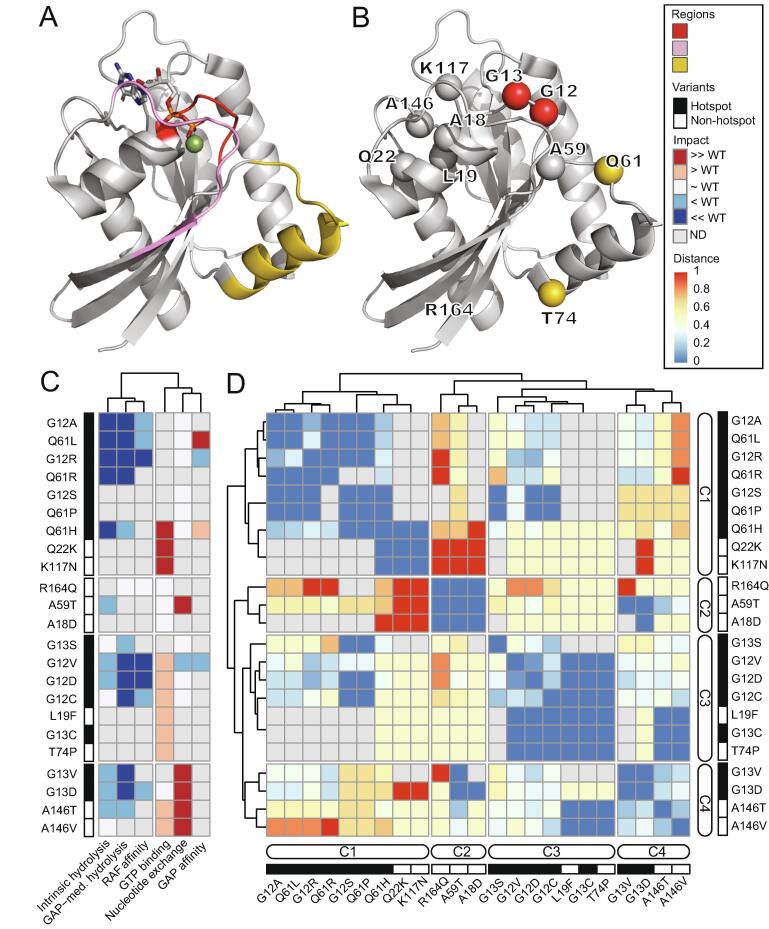
We developed a harmonized dataset for rapidly assessing mutational effects on the biochemical properties of the GTPase encoded by KRAS. We collected, processed, and categorized data derived from studying 23 KRAS hotspot and non-hotspot variants occurring at 11 amino-acid residues, distributed throughout the 3D structure (Fig. 1A and B, Table S1). These 11 residues include G12 and G13 from the P-loop (phosphate-binding), Q61 and T74 from the switch-II region (Fig. 1B), R164 in the allosteric lobe, with the remainder affecting switch-I and the nucleotide binding pocket. The properties measured include (relative to WT; see Methods) both intrinsic and GAP-mediated hydrolysis, nucleotide exchange, GTP binding, and RAF and GAP affinity (Fig. 1C). Missing data were most common for GAP affinity (82.6% of variants), RAF affinity (65.2%), GAP-mediated GTP hydrolysis (43.5%), and intrinsic GTP hydrolysis (39.1%). The most reported measurements were for both GTP binding and nucleotide exchange rates (34.8%; Fig. S2A). For six (26.1%) variants, only GTP binding measurements were available, whereas for three (13.0%), all the measurements except GAP affinity were available. Lastly, only one variant (G12V, 4.3%) had all 6 measurements (Fig. S2B). We explored the interrelationships among KRAS biochemical features by computing correlations among experimental data and observed a broad range of values (-0.76 ≤ RSpearman ≤ 1). We detect no correlation (RSpearman ∼ 0) between intrinsic GTP hydrolysis and RAF affinity, as well as between GAP-mediate hydrolysis and GTP binding. On the other hand, there was a strong and statistically significant correlation (RSpearman = 1) between RAF and GAP affinities, as well as between GTP binding and GAP affinity (Fig. S2C and Table S2). The data suggests that each feature of RAS GTPases can be altered independently of others. Therefore, the prediction of one feature is unlikely to inform about another, thus motivating the need for our broad approach.
Fig. 1.
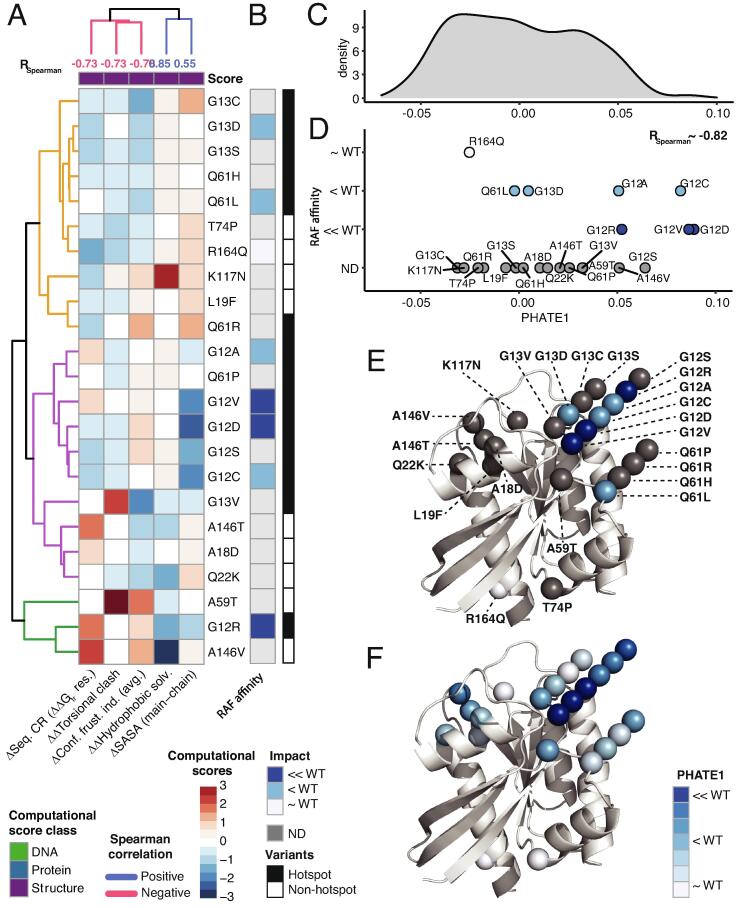
Semi-quantified biochemical measurements elucidate the similarities and differences among the hotspot and non-hotspot variants. A) 3D structure of KRAS (PDB: 4OBE) representing the sensitive regions, phosphate-binding loop (P-loop) (amino-acid 10–17), switch-I (amino acids 30–40) and switch-II (amino acids 60–76) [38]. B) 23 KRAS hot-spot and non-hotspot variants that belong to 11 amino-acid residues are projected onto the 3D structure. The amino-acid residues are colored according to the sensitive regions in (A). C) Heatmap of the semi-quantified 6 experimental measurements (representing the columns) of the 23 KRAS variants demonstrates the variable biochemical properties of mutant KRAS proteins relative to WT, and by ordering the variants (representing the rows) according to the four clusters in panel D. The complete summary data with references is in Table S1. D) The dissimilarity among the 23 KRAS variants based on the experimental assay in panel C using Gower distance. The variants with distance 0 are identical and shown in blue, whereas the maximally dissimilar variants with distance 1 are shown in red color. The dendrograms show for distinct clusters are indicated as C1, C2, C3 and C4. The hotspot and non-hotspot KRAS variants are indicated by black and white rectangles, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Because genetic variation outside of classic somatic hotspots has received relatively little attention, we next sought to quantify similarities among them by combining information across different experimental measurements (Fig. 1D). We used Gower distance which varies from 0 (identical) to 1 (maximally different). Variants fell into 4 clusters (denoted as C1 to C4; Fig. 1C and D). While, cluster assignment may change as additional measurements are made, we anticipate that major patterns among variants, that are already visually apparent, will persist. Interestingly, we first noted that hotspot mutations occurred across three clusters. Q61 alterations occur in C1, while G12A/R/S, G12V/D/C and G13C occur in C3, and G13V/D in C4. Within C1, two non-hotspot variants, Q22K and K177N, most closely associated with the hotspot Q61H for their increased GTP binding. C2 contains three non-hotspot mutations, R164Q, A59T, an dA18D, which each have a distinct profile among the 23 mutants. C3 reveal the similarities between two non-hotspot variants (L19F and T74P) and the four hotspot variants (G12V/D/C and G13C). C4 identifies similarities among A146T/V and G13V/D but differences among non-hotspot variants A146T/V and hotspot variants G12A/R/S as well as Q61L/P/H. Additionally, differences between the non-hotspot variants R164Q and the hotspot variants are not surprising, as the former behave like WT KRAS, based on available measurements. Nonetheless, our findings are especially noteworthy for hotspot variants the non-hotspot variants A59T and A146T/V, which have multiple experimental measurements. Hence, by revealing patterns of similarity between hotspot and non-hotpot variants of KRAS, this data support the fact RAS variants are not functionally equivalent [37], raising the need of a broader data science approach to scoring genomic variation in this gene.
3.1. 3D structure-based scores explain experimental differences among 23 KRAS variants
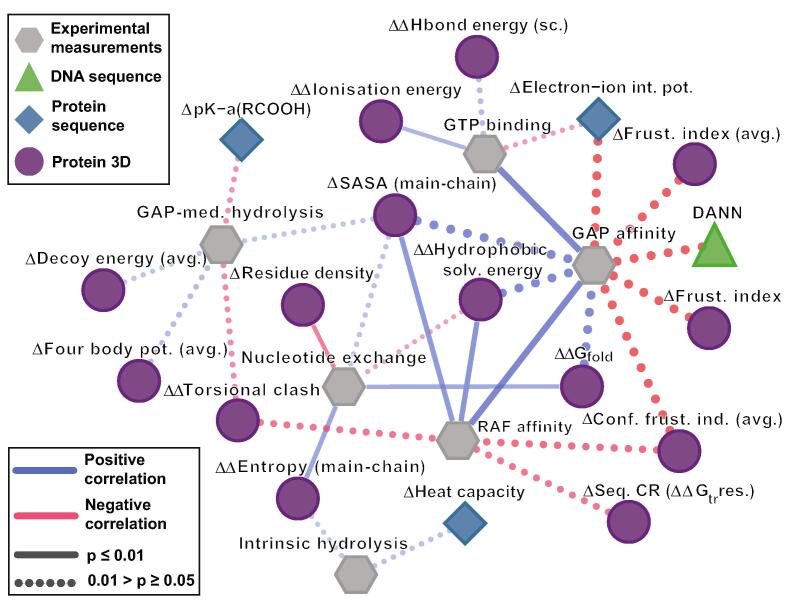
We evaluated the congruency between experimental measurements and computational scores derived from calculations at multiple molecular levels – genomic, protein sequence, and protein 3D structure. Specifically, we computed the pairwise Spearman correlation (RSpearman) between each experimental measurement and 31 computational scores (Fig. S3A) across the 23 KRAS variants. We excluded MAF and conservation as these scores each had identical values across the 23 KRAS variants (Figs. S3A and B). We filtered using p-value < 0.05 and obtained 18 computational scores that are highly correlated (RSpearman > 0.51) with experimental measurements, consisting of one DNA sequence, three protein sequence, and fourteen 3D structure-based scores (Fig. 2 and Table S3). This finding is important to consider since genomic scores are generally based on DNA sequence features, and therefore mostly incapable of capturing nuanced impact of mutations on specific biochemical properties compared to the 3D structure-based scores.
Fig. 2.
Structure-based scores dominate the experiment guided mechanistic interpretation of the hotspot and non-hotspot variants. The correlation between the experimental measurements (in grey color) and the computational scores indicated by different shapes and colors green, blue and purple from multiple molecular levels DNA sequence, protein sequence and protein 3D structure, respectively for the 23 KRAS variants are shown as a network diagram. In the network diagram the thickness of the edges indicates absolute Spearman correlation value (|RSpearman|), where higher correlation is indicated by thicker edge. Note that the correlations among the computational scores are not shown for clarity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The fourteen 3D scores that correlate with experimental biochemical measures are physically interrelated. They coordinate around five molecular properties (folding entropy, folding stability, local packing and energetic stability, and residue level folding cooperativity) that represent different contributions to the probability of local unfolding or conformational rearrangement. For example, the change from WT in main-chain SASA correlated with GAP-mediated hydrolysis rate (RSpearman = 0.55; p-value = 0.05), nucleotide exchange rate (0.56; 0.03), GAP affinity (0.95; 0.05) and RAF affinity (0.82; 0.01; Fig. 2). This observation supports the inference that changes in SASA caused by mutation impacts KRAS activity through modulation of GAP-mediated hydrolysis and nucleotide exchange rates, GAP and RAF affinities. Similarly, protein folding stability (ΔΔGfold) positively correlate with nucleotide exchange rate (RSpearman = 0.71; p-value = 0.00) and GAP affinity (0.95; 0.05), further suggesting that stability changes for mutated proteins impacts both properties. These findings directly show the strong association between the 3D scores and the measurements for key biochemical properties of mutated KRAS, beyond what is currently available from genomic scores, enabling us to better interpret mechanisms of dysfunction underlying distinct KRAS variants.
We modeled all six experimental parameters from our biochemical feature resource using the same procedure, with GAP-mediated hydrolysis and RAF affinity having the strongest associations with the derived scores. Thus, we first detail the marginal and composite scores for these two features in the next sections, and then discuss the remaining four, followed by overall patterns across the dataset.
3.2. 3D structure-based scores reveal mechanisms of disruption of GAP-mediated hydrolysis of the 23 KRAS variants
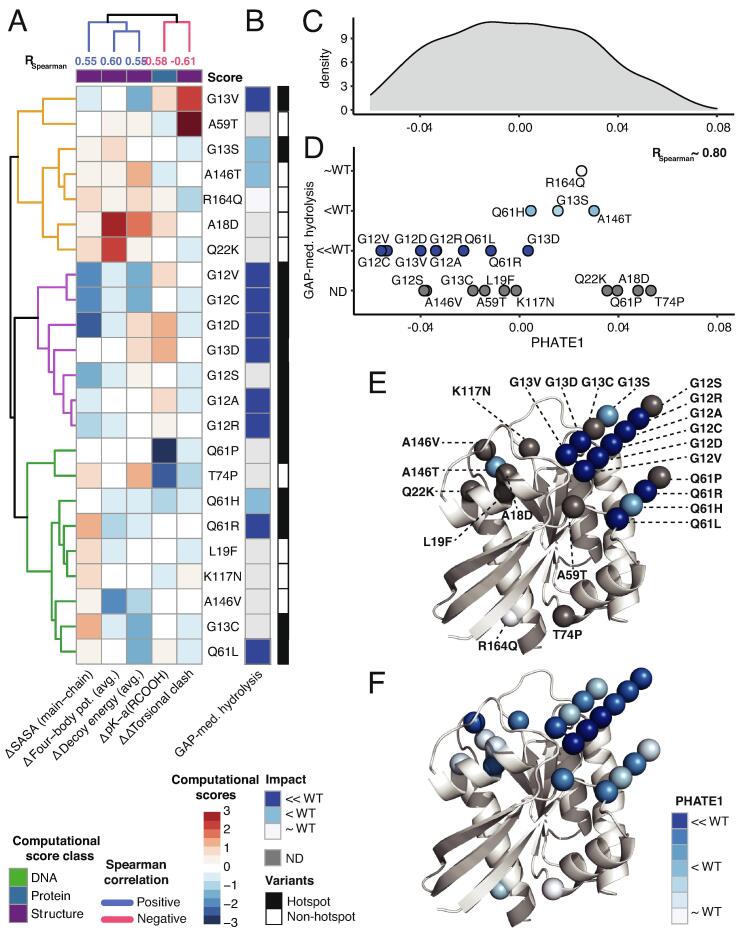
We used dimensionality reduction to derive a composite score from the multiple individual scores that correlated with GAP-mediated hydrolysis. Our goal was to prioritize similarities or differences in potential underlying mechanisms of dysfunction among variants, congruent with the available experimental evidence. We used two approaches leveraging computational scores for the 23 KRAS variants that correlate with experimental measurements of GAP-mediated hydrolysis: 1) pattern analysis using k-clustering and 2) unsupervised dimensionality reduction for all RAS variants.
In our first approach, we examined patterns among five computational scores (one protein sequence and four 3D structure-based scores) that correlate with the GAP-mediated hydrolysis rate (Fig. 3A and B). All four structure-based scores relate to local stability, and therefore we expect their alteration by mutations to modulate local unfolding. We identified three groups of variants, according to these scores. One cluster contained p-loop mutations, another contained switch-II mutations plus G13C and selected non-hotspot variants (e.g. A146V and K117N), and the third contained only non-hotspot variants plus G13V. The composition of mutations differs among these clusters, and patterns according to changes in multiple structure-based stability scores. Thus, computational scores reveal patterns among hotspot and non-hotspot variants’ GAP-mediate hydrolysis that may be driven by differences in local unfolding probabilities.
Fig. 3.
3D structure and protein sequence-based scores unravel the mechanism of disruption of GAP-mediated hydrolysis. A) Five computational scores (4 3D structure and 1 protein sequence -based) for the 23 KRAS variants that strongly correlate with the GAP-mediated hydrolysis measurements. The computational scores from protein sequence (blue) and 3D structure (purple) are represented as z-scores from high (red) and low (blue), with upper dendrogram colored by correlation with GAP-mediated hydrolysis measurements. B) Semi-quantified measurements of the GAP-mediated hydrolysis of the KRAS variants based on the similarity to the WT. Next, we performed 1D PHATE analysis of the 935 variants from 7 RAS genes using the 5 correlated computational scores from (A) for mechanistic interpretation of GAP-mediated hydrolysis. C) 1D probability density plot of PHATE1 for all the 935 RAS variants. D) Scattered plot of the 23 KRAS variants (out of 935) along the PHATE1 coordinate that show RSpearman ∼ 0.80 with the GAP-mediated hydrolysis measurements. The variants are colored and separated vertically based on their GAP-mediated hydrolysis value. E) The 23 KRAS variants are projected onto the 3D structure and colored according to experimentally measured GAP-mediated hydrolysis values, or F) PHATE1 values from panel D. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In our second approach, we used PHATE dimensionality reduction to score overall similarities among the 935 variants from 7 RAS genes. We performed 1D PHATE (PHATE1) analysis of the 5 computational scores that correlated with GAP-mediated hydrolysis (see Fig. 3C-F). The composite summary score from PHATE1 across (Fig. 3C) provides a probability density distribution from, which we projected the 23 KRAS variants; they were largely ordered by their GAP-mediated hydrolysis rate (Fig. 3D). Moreover, PHATE1 and experimental measurements display a better correlation (RSpearman ∼ 0.80) than any individual computational score (|RSpearman|≤0.61; Fig. 3E and F) and were able to predict relative levels for mutations that have not been experimentally measured. These results are additionally important since there are conflicting reports on the effects of G12D [21], [26], [27] and weather G13D behaves more like WT compared to G12D [14]. In this study, we rectified conflicts by favoring the data more supported by the allele’s incidence rate in cancer. Yet, our calculations may help to clarify the intrinsic effect of each allele by interpolating among the data generated independently between different laboratories, and to prioritize experiments for future validation studies. Among variants with missing GAP-mediated hydrolysis measurements, the PHATE1 values suggest, for example, that G12S has comparable effects to G12A/R and relatively lower effect for G13C compared to G13V. A146V associates to higher relative GAP-mediated hydrolysis than A146T, which is like G12A/R/S and G13V. Mutant-to-mutant correlations show that L19F, A59T, and K117N compare to G13D and Q61H. The remaining variants (A18D, Q22K and T74P) appear like WT (Fig. 3F). These findings indicate that mutated KRAS proteins may modulate GAP binding by changing the probability of locally unfolded conformations, which will in turn change GAP-mediated hydrolysis rates.
3.3. 3D structure-based scores reveal that changes in RAF-affinity underlies the dysfunction of 23 KRAS variants
We investigated how the 23 KRAS variants change RAF affinity by first identifying the computational scores exhibiting strongest associations with this property (Fig. 4), all of which were from 3D structure and emphasize hydrophobic and entropic properties. By observing the patterns of these computational scores (Fig. 4A and B), we identified non-hotspot variants with similar profiles and RAF affinity as hot-spot variants. We find T74P and R164Q resembled hotspot variants G13C/D/S and Q61H/L. On the other hand, the non-hotspot variants L19F and K117N more closely resembled Q61R, A18D, Q22K, and A146T resembled G13V, and A59T and A146V clustered with G12R. In contrast, we observed the remaining hotspot variants, G12C/D/V/A/S and Q61P, in a separate cluster. PHATE1 analysis of all 935 variants from 7 RAS genes show high correlation (RSpearman ∼ -0.82) with experimental RAF affinity measurements (Fig. 4C and D). We used PHATE1 values to compare the degree of predicted RAF affinity with experiment (Fig. 4E and F), the largest difference being between G12C and G12R. Overall, these predictions agreed with experimental data and showed much stronger associations with direct empirical measurement than using genomic scores alone.
Fig. 4.
Structure-based scores reveal differential mechanism of dysfunction of RAF affinity among the hotspot and non-hotspot variants. A) Pattern of the five 3D structure-based computational scores (as z-scores) for the 23 KRAS variants that B) strongly correlate with semi-quantified RAF affinity measurements (visualizations as in Fig. 3). C) Next, we performed 1D PHATE analysis of the 935 variants from 7 RAS genes using the 5 computational scores for RAF affinity, and summarize using probability density across all the 935 variants. D) Scatter plot of the 23 variants (out of 935 variants) along the PHATE1 coordinate that show RSpearman ∼ 0.85 with the RAF affinity measurements. The 23 variants are colored and separated vertically based on RAF affinity, and classic hotspot mutations are on the upper tail of the distribution. The 23 KRAS variants are projected onto the 3D structure and colored according to E) RAF affinity and F) PHATE1 values.
Additionally, we predicted relative RAF affinity changes for amino acid substitutions lacking experimental measurements (15 out of 23; Fig. 4B). These include G12S, which we predict has a decreased RAF affinity like other G12 variants, and G13V, Q61P, A59T, and A146V which we predict to have intermediate decreases in RAF affinity. On the other hand, the variants Q61H, A18D and Q22K show relatively smaller changes, closer to WT levels than the other mutations assessed. The remaining variants (G13C/S, Q61R, L19F, T74P, K117N, and R164Q) are predicted to have RAF affinity comparable to WT. Our analysis identified biophysical properties (changes in folding propensity and solvation energies) that distinguish RAF affinities among RAS mutants better than genomic scores.
We performed similar interpretations for GTP binding activity (Fig. S4A), intrinsic hydrolysis (Fig. S4B), nucleotide exchange (Fig. S5A), and GAP affinity (Fig. S5B). We identified pattern of highly correlated computational scores associated with the measurements for these KRAS variants. However, for GTP binding activity and nucleotide exchange rate, the correlations between PHATE1 (using the associated computational scores as input) and experimental measurements were low (RSpearman ∼ 0.28 and 0.05, respectively). For intrinsic hydrolysis, we identified two highly correlated computational scores. On the other hand, GAP affinity showed a strong correlation with 8 computational scores (RSpearman ∼ 0.95), which most likely is due to the large number of missing measurements (19 out of 23 variants). These results underscore that more data is needed to complete our understanding of the structure–function relationship for mutations that encode dysfunctional RAS enzymes. Thus, combined, the use of 3D structure-based scores enhances the information available from genomics over existing methods that are based only on sequences, or those which incorporate a more limited repertoire of structural evaluations.
3.4. Deriving a more uniform and mechanistic interpretation of all RAS variants
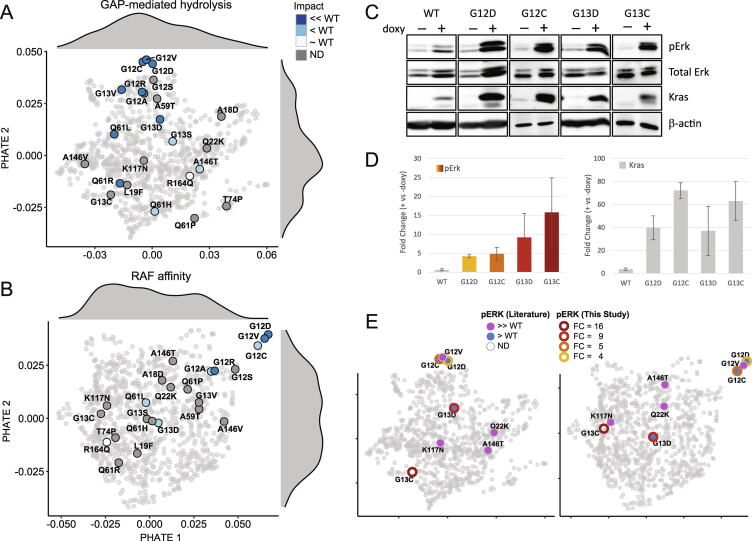
To visualize the relative changes in GAP-mediated hydrolysis and RAF affinity across all 935 RAS family variants, we extended PHATE to 2D and followed a similar procedure as above. 2D PHATE for GAP-mediated hydrolysis (Fig. 5A) and RAF affinity (Fig. 5B), identify mutations with the largest experimental changes occupied tails of the value distribution, supporting that computational scores can distinguish which mutations have the largest effect on the enzyme. G12 KRAS hotspot variants occupy a distinct region while G13 and Q61 KRAS hotspot variants are more broadly distributed. This observation indicates that G12 alterations are specific vulnerabilities to the enzyme and impart relatively unique changes, while other mutations have more varied changes. To validate these predictions, we added information about the downstream effect of activated RAS using pERK levels. We first identified four representative alleles that spanned the range of values from our composite scores of both biochemical features (G12C/D and G12C/D). We generated inducible human cell lines carrying each of these alleles and completed Western blot analyses (Fig. 5C). This work revealed that each mutation conveyed a different level of downstream activity, yet with G12D and G12C having similar levels of induction and G13 mutations having higher induction with higher variance (Fig. 5D). These data were concordant with the ranking of these alleles from our composite scores. We also gathered pERK data from the literature (Table S4) and mapped both types of pERK data onto the PHATE spaces (Fig. 5E), again demonstrating concordant ranking of the mutation. Together, these data indicate the potential for structure-based scores to enhance existing genomics scores with data of mechanistic value.
Fig. 5.
Mechanistic interpretation of all RAS variants from experimental measurements. We performed 2D PHATE analysis to identify spatial patterns for interpreting the variable degree of changes across 935 RAS variants. A) 935 RAS variants are projected on the 2D PHATE space using the 5 computational scores that strongly correlate with the GAP-mediated hydrolysis rate of the 23 KRAS variants that are colored with the semi-quantified measurements in the plot (see Fig. 3A-B). Marginal distributions along PHATE1 and PHATE2 are also shown. B) A similar 2D PHATE plot of 935 RAS variants using the 5 computational scores highly correlated with the semi-quantified RAF affinity measurements of the 23 KRAS variants that are indicated in the plot (see Fig. 4A-B). C) From the patterns among RAS mutations, we selected four KRAS alleles that spanned the range of PHATE values for functional validation. Representative Western blots are shown for pERK, a critical downstream marker of RAS activity. D) We quantified pERK levels downstream of each mutant after 24hr induction of each allele (see Methods). E) Using the PHATE spaces defined in panels (A) and (B), we show the pERK data derived from the literature (Table S4) and from this study. Downstream activity changes for these KRAS alleles are highly concordant with their ranking according to computed scores.
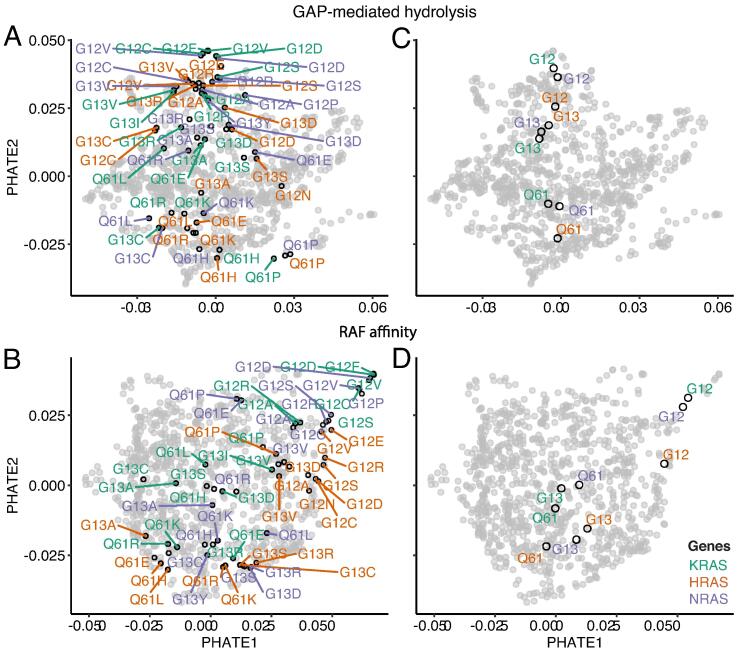
We investigated whether the above large-scale patterns characterize all KRAS, HRAS and NRAS hotspot variants, by projecting the amino acid substitutions at G12, G13 and Q61 and observed an overall similar pattern, but with differences between the RAS proteins (Fig. 6A and B). To quantify and better visualize the differences by RAS and hotspot location, we calculated the centroid of the G12, G13 and Q61 variants for each RAS (Fig. 6C and D). Interestingly, G12 variants especially from KRAS and NRAS, cluster independently of other mutations. Hotspot location was a distinct feature for GAP-mediated hydrolysis independent of RAS protein. G13 and Q61 alterations were more intermixed for RAF affinity, but with a clear trend for HRAS mutations to have relative differences from NRAS and KRAS. From these patterns among hotspot variants, we inferred certain amino acid substitutions have the same mechanisms across RAS proteins, while other substitutions are modulated by intrinsic biochemical properties of individual RAS genes [38]. These computational observations are important since differences between RAS proteins are evident, both by disease incidence and mutational biases, but the GTP-binding domains, which catalyzes GTP hydrolysis and mediates downstream signaling, are 95% identical between them. In fact, the sequences between KRAS, NRAS, and HRAS are identical in the effector lobes (residues 1–86) and exhibit small differences in allosteric lobes (residues 87–166). Therefore, the information from linear sequence is insufficient to characterize the subtle similarities or differences among the hotspot variants, but 3D structure-based scores appear to identify signals from the allosteric lobe that links into mutation-specific effect differences.
Fig. 6.
Each RAS gene and hotspot site has a fingerprint. All the G12, G13 and Q61 variants from KRAS (green), HRAS (red) and NRAS (purple) out of the 935 variants are indicated on the 2D PHATE space generated for predicting changes in A) GAP-mediated hydrolysis and B) RAF affinity. The median position of each hotpsot, across mutations observed at each position, and for C) GAP-mediated hydrolysis and D) RAF affinity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study we present the broadest-to-date resource for KRAS mutational effects on enzymatic properties from the literature, spanning 23 distinct alterations and six biochemical measurements, which demonstrates the functional heterogeneity of RAS mutations. We identified that 3D structure-based calculations correlate with enzymatic activity much better than well-established DNA scores. DNA scores are primarily based on sequence conservation and functional information of the genome; while informative and extensively used in predicting pathogenicity in clinical workflows, they provide little data of mechanistic value for interpreting variants at the molecular level. Structure-based and additional molecular calculations can enhance DNA scores because they provide information about features that are not considered when developing genomics annotations [12], [39], [40]. Thus, we believe the high specificity of 3D scores will make them a vital component to research into oncogenic variation, understanding of differential drugability of each mutation, and translation of genomic data into clinical knowledge.
While the effects of classic hotspot mutations are generally accepted, as we gathered data from literature, we identified some discrepancies among these measurements. For example, KRAS G12D and in HEK 293 T/17 cells had 4.5-fold higher GTP-bound level compared to WT [26], whereas similar measurements in MCF10A cells showed no change [21]. From another study by Hunter et al. [14], the same KRAS G12D variant showed 4.8-fold lower RAF affinity relative to WT, while Poulin et al. [27] reported that KRAS G12D variant exhibited similar RAF affinity as WT. These differences may be due to feedback mechanisms where cells compensate for changes in KRAS activity by modulating other RAS enzymes, downstream effectors, or upstream receptors. Thus, future study is needed to better understand the context specificity of even classic hotspot variation, which may be enhanced by mechanistic and structure-based data, as in the current study.
Limitations on the availability of the biochemical measurements for KRAS variants need to be considered, especially for variants with only single experimental measurements. Filling in the missing data in the future, may modify the associations among experimental and computational scores that we have identified (Fig. 2), but our study demonstrates that 3D protein structure-based scores are key in providing insight into mechanisms underlying altered enzymatic activities of the mutated protein (Fig. 3, Fig. 4). Finally, we note that intrinsic differences for each RAS-family protein are observed, despite the highly similar sequences among them. Thus, mechanistic information is encoded in 3D structures which can also inform about baseline differences among WT enzyme isoforms. Computations between RAS isoforms could play a role in future studies to explain differential disease incidence across the repertoire of KRAS, HRAS and NRAS mutations in different types of human cancers.
We analyzed RAS at the molecular level to investigate how mutations affect distinct biochemical properties. One limitation of our approach is resolution of the experimental data, which we chose as semi-quantitative to harmonize across different reports. Further, RAS proteins are known to activate different signaling pathways (e.g. RAF/MAPK and PI3K/AKT/mTOR) depending on particular cell types, tissues and their subcellular localization [20], [41]. Also, hotspot mutations surround the nucleotide-binding site and earlier studies suggested that the nucleotide exchange may alter the affinity of mutant RAS proteins for downstream effectors proteins [14], [42]. For scoring, we focused on biochemical experimental measurements on the RAS protein, rather than downstream effects which are mediated by additional factors. However, we also considered pERK, which is a critical downstream effect of RAS activation and one of the only measurements, the other being pMEK, recommended by a recent RASopathy expert panel [43]. We found that our composite scores, primarily leveraging 3D structural features, ranked KRAS mutations concordant with experimental measures of pERK induction. Finally, due to the limited availability of systematic and quantitative experimental measurements of the RAS variants, we were unable to perform an ideal supervised machine learning approach to classify the variants based on biochemical properties. We are actively pursuing studies of the KRAS enzyme to gather the requisite data to train and test more robust models. Thus, our current study establishes that 3D structural data brings added value to genomic pathogenicity predictors for precision oncology, and future work combining 3D modeling and quantitative systems analyses of signaling networks will better determine the phenotypic outcome differences among RASopathy and cancer-associated RAS mutations [44].
5. Conclusions
Genomic scores are commonly used in clinical workflows, but they cannot discriminate among RAS mutations that have different biochemical properties. Thus, we used a broad repertoire of protein sequence and 3D structure-based scores to enhance genomic scores with more nuance of the translated gene product. We demonstrated that these additional scores explain differences among biochemical measurements of KRAS, that classic hotspot and non-hotspot mutations alike can have similarly altered profiles, and that these profiles associate with enzymatic properties. The current study lays the groundwork for our multi-level structural bioinformatic approach, which we believe will be informative to precision oncology efforts to combat RAS alteration, and be generalizable to other proteins for increasing researchers’ ability to interpret the effects of human genetic variation.
Funding
This research was completed in part with computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin. This project is funded in part by the Advancing a Healthier Wisconsin Endowment at the Medical College of Wisconsin. This publication was supported in part by The Linda T. and John A. Mellowes Endowed Innovation and Discovery Fund, the Genomic Sciences and Precision Medicine Center of Medical College of Wisconsin, and NIH R01-DK52913.
Role of the funder
The funder had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclaimer
The views expressed are those of the authors and do not necessarily reflect the position or policy of the Funders.
Author contributions
MZ and RU conceptualized the study. MZ and ST contributed to methodology and software. MZ, ST, ND, and AM applied formal analyses, data visualization, and data curation. AM and EL conducted experiments. MZ and ST wrote the original draft. MZ, ST, ND, and RU contributed to revision and editing. MZ oversaw project administration. RU acquired funding.
Data availability
Input data are publicly available. The scores produced by this study are available in our Supplemental Information.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.12.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Harvey J.J. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204(4963):1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M., Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 3.Cooper G.M. Cellular transforming genes. Science. 1982;217(4562):801–806. doi: 10.1126/science.6285471. [DOI] [PubMed] [Google Scholar]
- 4.Marshall C.J., Hall A., Weiss R.A. A transforming gene present in human sarcoma cell lines. Nature. 1982;299(5879):171–173. doi: 10.1038/299171a0. [DOI] [PubMed] [Google Scholar]
- 5.Bos J.L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 6.Haigis K.M. KRAS alleles: the devil is in the detail. Trends Cancer. 2017;3(10):686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubbert S., Bollag G., Lyubynska N., Nguyen H., Kratz C.P., Zenker M., et al. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27(22):7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1. [DOI] [PMC free article] [PubMed]
- 9.Sanchez-Vega F., Mina M., Armenia J., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337 e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubbert S., Zenker M., Rowe S.L., Böll S., Klein C., Bollag G., et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38(3):331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 11.Bertola D.R., Pereira A.C., Brasil A.S., Albano L.M.J., Kim C.A., Krieger J.E. Further evidence of genetic heterogeneity in Costello syndrome: involvement of the KRAS gene. J Hum Genet. 2007;52(6):521–526. doi: 10.1007/s10038-007-0146-1. [DOI] [PubMed] [Google Scholar]
- 12.Tripathi S, Dsouza NR, Urrutia R, et al. Structural Bioinformatics Enhances Mechanistic Interpretation of Genomic Variation, Demonstrated Through the Analyses of 935 Distinct RAS Family Mutations. Bioinformatics 2020; 10.1093/bioinformatics/btaa972. [DOI] [PMC free article] [PubMed]
- 13.Smith M.J., Neel B.G., Ikura M. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci U S A. 2013;110(12):4574–4579. doi: 10.1073/pnas.1218173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter J.C., Manandhar A., Carrasco M.A., Gurbani D., Gondi S., Westover K.D. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13(9):1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 15.Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 16.Killoran R.C., Smith M.J. Conformational resolution of nucleotide cycling and effector interactions for multiple small GTPases determined in parallel. J Biol Chem. 2019;294(25):9937–9948. doi: 10.1074/jbc.RA119.008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gideon P., John J., Frech M., et al. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol. 1992;12(5):2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith G., Bounds R., Wolf H., Steele R.J.C., Carey F.A., Wolf C.R. Activating K-Ras mutations outwith 'hotspot' codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102(4):693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solman M, Ligabue A, Blazevits O, et al. Specific cancer-associated mutations in the switch III region of Ras increase tumorigenicity by nanocluster augmentation. Elife 2015;4:e08905. [DOI] [PMC free article] [PubMed]
- 20.Munoz-Maldonado C., Zimmer Y., Medova M. A comparative analysis of individual RAS mutations in cancer biology. Front Oncol. 2019;9:1088. doi: 10.3389/fonc.2019.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolze B., Reinhart S., Bulllinger L., Fröhling S., Scholl C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep. 2015;5(1) doi: 10.1038/srep08535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Der C.J., Finkel T., Cooper G.M. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44(1):167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 23.Haigis K.M., Kendall K.R., Wang Y., Cheung A., Haigis M.C., Glickman J.N., et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feig L.A., Cooper G.M. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8(6):2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akagi K., Uchibori R., Yamaguchi K., Kurosawa K., Tanaka Y., Kozu T. Characterization of a novel oncogenic K-ras mutation in colon cancer. Biochem Biophys Res Commun. 2007;352(3):728–732. doi: 10.1016/j.bbrc.2006.11.091. [DOI] [PubMed] [Google Scholar]
- 26.Tyner JW, Erickson H, Deininger MW, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood 2009;113(8):1749-55. [DOI] [PMC free article] [PubMed]
- 27.Poulin E.J., Bera A.K., Lu J., Lin Y.-J., Strasser S.D., Paulo J.A., et al. Tissue-specific oncogenic activity of KRAS(A146T) Cancer Discov. 2019;9(6):738–755. doi: 10.1158/2159-8290.CD-18-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janakiraman M., Vakiani E., Zeng Z., Pratilas C.A., Taylor B.S., Chitale D., et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70(14):5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacal J.C., Aaronson S.A. Activation of ras p21 transforming properties associated with an increase in the release rate of bound guanine nucleotide. Mol Cell Biol. 1986;6(12):4214–4220. doi: 10.1128/mcb.6.12.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman H.M., Bhat T.N., Bourne P.E., et al. The Protein Data Bank and the challenge of structural genomics. Nat Struct Biol. 2000;7(Suppl):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 31.Kocher JP, Quest DJ, Duffy P, et al. The Biological Reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics 2014;30(13):1920-2. [DOI] [PMC free article] [PubMed]
- 32.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing (http://www.R-project.org). 2014. In.
- 33.Kowarik A., Templ M. Imputation with the R Package VIM. J Stat Softw. 2016;74(7):1–16. [Google Scholar]
- 34.Maechler M, Rousseeuw P, Struyf A, et al. cluster: Cluster Analysis Basics and Extensions. https://CRAN.R-project.org/package=cluster. In. v2.1.0 ed; 2021.
- 35.Harrell F. Hmisc: Harrell Miscellaneous. https://cran.r-project.org/package=Hmisc. In; 2020.
- 36.Moon K.R., van Dijk D., Wang Z., Gigante S., Burkhardt D.B., Chen W.S., et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol. 2019;37(12):1482–1492. doi: 10.1038/s41587-019-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobbs G.A., Der C.J. RAS mutations are not created equal. Cancer Discov. 2019;9(6):696–698. doi: 10.1158/2159-8290.CD-19-0406. [DOI] [PubMed] [Google Scholar]
- 38.Johnson C.W., Reid D., Parker J.A., Salter S., Knihtila R., Kuzmic P., et al. The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J Biol Chem. 2017;292(31):12981–12993. doi: 10.1074/jbc.M117.778886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu B., Scott A.D., Sengupta S., Bailey M.H., Batra P., Ning J., et al. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat Genet. 2016;48(8):827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimoto A., Okada Y., Boroevich K.A., Tsunoda T., Taniguchi H., Nakagawa H. Systematic analysis of mutation distribution in three dimensional protein structures identifies cancer driver genes. Sci Rep. 2016;6(1) doi: 10.1038/srep26483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocks O., Peyker A., Bastiaens P.IH. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr Opin Cell Biol. 2006;18(4):351–357. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Voice J.K., Klemke R.L., Le A., Jackson J.H. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem. 1999;274(24):17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 43.Gelb B.D., Cavé H., Dillon M.W., Gripp K.W., Lee J.A., Mason-Suares H., et al. ClinGen's RASopathy Expert Panel consensus methods for variant interpretation. Genet Med. 2018;20(11):1334–1345. doi: 10.1038/gim.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiel C., Serrano L. Structure-energy-based predictions and network modelling of RASopathy and cancer missense mutations. Mol Syst Biol. 2014;10:727. doi: 10.1002/msb.20145092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Input data are publicly available. The scores produced by this study are available in our Supplemental Information.