Abstract
Since the last revision in 2015, the taxonomy of section Flavipedes evolved rapidly along with the availability of new species delimitation techniques. This study aims to re-evaluate the species boundaries of section Flavipedes members using modern delimitation methods applied to an extended set of strains (n = 90) collected from various environments. The analysis used DNA sequences of three house-keeping genes (benA, CaM, RPB2) and consisted of two steps: application of several single-locus (GMYC, bGMYC, PTP, bPTP) and multi-locus (STACEY) species delimitation methods to sort the isolates into putative species, which were subsequently validated using DELINEATE software that was applied for the first time in fungal taxonomy. As a result, four new species are introduced, i.e.A. alboluteus, A. alboviridis, A. inusitatus and A. lanuginosus, and A. capensis is synonymized with A. iizukae. Phenotypic analyses were performed for the new species and their relatives, and the results showed that the growth parameters at different temperatures and colonies characteristics were useful for differentiation of these taxa. The revised section harbors 18 species, most of them are known from soil. However, the most common species from the section are ecologically diverse, occurring in the indoor environment (six species), clinical samples (five species), food and feed (four species), droppings (four species) and other less common substrates/environments. Due to the occurrence of section Flavipedes species in the clinical material/hospital environment, we also evaluated the susceptibility of 67 strains to six antifungals (amphotericin B, itraconazole, posaconazole, voriconazole, isavuconazole, terbinafine) using the reference EUCAST method. These results showed some potentially clinically relevant differences in susceptibility between species. For example, MICs higher than those observed for A. fumigatus wild-type were found for both triazoles and amphotericin B for A. ardalensis, A. iizukae, and A. spelaeus whereas A. lanuginosus, A. luppiae, A. movilensis, A. neoflavipes, A. olivimuriae and A. suttoniae were comparable to or more susceptible as A. fumigatus. Finally, terbinafine was in vitro active against all species except A. alboviridis.
Key words: Aspergillus flavipes, Antifungal susceptibility testing, Clinical fungi, Indoor fungi, Multigene phylogeny, Soil-borne fungi, Species delimitation
Taxonomic novelties: New species: Aspergillus alboluteus F. Sklenar, Jurjević, Ezekiel, Houbraken & Hubka; Aspergillus alboviridis J.P.Z. Siqueira, Gené, F. Sklenar & Hubka; Aspergillus inusitatus F. Sklenar, C. Silva Pereira, Houbraken & Hubka; Aspergillus lanuginosus F. Sklenar & Hubka
Introduction
Aspergillus is a large genus of filamentous fungi, which currently contains 446 accepted species and this number is rapidly rising. Aspergilli have traditionally been classified into subgenera and sections, and this classification has been recently revised and updated with the addition of series rank (Houbraken et al. 2020). According to this most up-to-date overview, the accepted species are distributed over six subgenera, 27 sections, and 75 series. Thom & Church (1926) introduced the A. flavipes group and section Flavipedes was formally established by Gams et al. (1985). The section is close to sections Terrei and Jani (Kocsubé et al. 2016) and is subdivided in four series: Flavipedes, Neonivei, Olivimuriarum and Spelaei (Houbraken et al. 2020). Phylogenetic analysis performed by Peterson (2008) demonstrated the presence of undescribed species diversity and the need for a proper taxonomic revision. The section was revised by Hubka et al. (2015), who accepted 10 species, two of which, A. frequens and A. mangaliensis, are synonymous to A. micronesiensis and A. templicola, published independently during the same period (Visagie et al. 2014, Arzanlou et al. 2016). In addition, Visagie et al. (2014) introduced another species, A. capensis, a close relative of A. iizukae, isolated from house dust. Another four species were described since then, namely, A. urmiensis described from hypersaline soils in Iran (Arzanlou et al. 2016), A. suttoniae from human sputum in the USA (Siqueira et al. 2018), A. olivimuriae from olive brine in Italy (Crognale et al. 2019) and A. sakultaensis from a water sample collected in Egypt (Zohri & Al-Bedak 2020). However, the last mentioned species was not validly described [Art. 40.8, Shenzen Code], and the study does not contain sufficient data to clearly classify A. sakultaensis into the current system. The isolate was not available for this study, but the DNA sequence of the internal transcribed spacer (ITS) generated by the authors was identical with some strains of A. templicola.
The species from section Flavipedes occur globally in soil (Klich 2002) and they can also grow as endophytes (El-Elimat et al. 2014), cause food spoilage (Pitt & Hocking 2009), or contaminate the indoor environment (Visagie et al. 2014). They are also occasionally isolated from clinical samples and infrequently cause opportunistic human or animal infections (Schultz et al. 2008, Gehlot et al. 2011, Siqueira et al. 2018). Representatives of section Flavipedes are able to produce a wide range of metabolites, summarized by Frisvad & Larsen (2015). These include mycotoxins such as sterigmatocystin (Tuomi et al. 2000) and citrinin (Greenhill et al. 2008), or pharmaceutical drugs (established or potential) such as HMG CoA reductase inhibitor lovastatin (Valera et al. 2005) and antiviral xanthones (Kang et al. 2018). Section Flavipedes species are also studied for their biotechnological potential and various biological activities. For example, A. flavipes possesses the potential to act as a biocontrol agent (El-Sayed & Ali 2020), A. polyporicola and A. spelaeus may be employed in the remediation of crude oil contaminated soil (Al-Dhabaan 2021) and A. iizukae produces oxidative enzymes with an industrial application (Noman et al. 2020).
In this study, we assembled a dataset of 90 strains belonging to section Flavipedes, which were newly isolated or originated from previous studies. We re-examined species boundaries of currently known species using modern species delimitation methods and discovered several new ones. The species delimitation and phylogenetic analyses utilized the DNA sequence data of three house-keeping genes. Phenotypic variability was examined in the species related to the newly discovered ones in order to find additional support for species hypotheses. The methodology of the species delimitation analysis follows up on previous studies within the genus Aspergillus (Sklenář et al. 2017, 2020, Hubka et al. 2018a, 2018b) with notable changes in the species validation step, where we used a newly developed program DELINEATE as opposed to utilization of BPP (Bayesian Phylogenetics and Phylogeography) (Yang 2015) in previous studies.
Materials and methods
Strains
The newly isolated strains obtained from the indoor environment were isolated as described previously (Jurjević et al. 2015) and the remaining strains were mostly obtained from collaborators or culture collections. Detailed information about provenance of the strains is listed in Table 1. Dried holotype and isotype specimens of the newly described species were deposited into the herbarium of the Mycological Department, National Museum, Prague, Czech Republic (PRM) and/or into the herbarium at the Westerdijk Fungal Biodiversity Institute (CBS H; Utrecht, the Netherlands). Nomenclatural novelties and descriptions were deposited in MycoBank (Crous et al. 2004).
Table 1.
Aspergillus strains from section Flavipedes examined in this study.
| Species | Strain numbers1 | Provenance (substrate, locality, year of isolation, collector/isolator) | GenBank/EMBL accession numbers2 |
|||
|---|---|---|---|---|---|---|
| ITS rDNA | benA | CaM | RPB2 | |||
| A. alboluteus | CBS 145855T = CCF 5695T = EMSL 2420T = IFM 66815T | USA, Pennsylvania, Philadelphia, outdoor air, 2014, Ž. Jurjević | MW448663 | MW478497 | MW478511 | MW478532 |
| CBS 145859 = CCF 6201 = EMSL 3060 | USA, Florida, Seminole, A/C Vent – swab, 2015, Ž. Jurjević | MW448662 | MW478496 | MW478510 | MW478531 | |
| CBS 145854 = CCF 4916 = EMSL 2311 = IFM 66816 | USA, California, indoor air, 2005, Ž. Jurjević | MW448664 | MW478498 | MW478512 | MW478533 | |
| CCF 5849 = EMSL 2446 = IFM 66817 | USA, Tennessee, Jackson, storage room - swab, 2014, Ž. Jurjević | MW448665 | MW478499 | MW478513 | MW478534 | |
| DTO 410-I8 = CBS 147065 = CCF 6551 | Nigeria, Abia, Isiala Ngwa South, Obuba, multicrop farm, C.N. Ezekiel | MW448666 | MW478500 | MW478514 | MW478535 | |
| A. alboviridis | CBS 142665T = FMR 15175T = CCF 6049T = IFM 66819T | Spain, Balearic Islands, Mallorca, Pollença, herbivore dung, 2016, J. Gené & J.P.Z. Siqueira | LT798909 | LT798936 | LT798937 | LT798938 |
| A. ardalensis | NRRL 62824T = CCF 4031T = CCF 4426T = CMF ISB 1688T = CBS 134372T | Spain, Andalucia, Ardales, near Cueva de Doña Trinidad, soil, 2008, A. Nováková | FR733808 | HG916683 | HG916725 | HG916704 |
| IHEM 17781 | France, Giens, hospital environment, 2000, J.-Ph. Bouchara | MW448667 | LN909026 | MW478515 | MW478536 | |
| A. flavipes | NRRL 302T = CCF 3067T = IMI 171885T = ATCC 24487T = FRR 0302T | Received by Charles Thom in 1922 from Da Fonseca as Bainier’s culture of Sterigmatocystus flavipes | EF669591 | EU014085 | EF669549 | EF669633 |
| NRRL 4852 = IMI 345934 = CCF 4836 (ex-type of A. archiflavipes) | Uruguay, dead beetle, received in NRRL from CBS as Blochwitz's strain of A. archiflavipes, W. Herter | LM999909 | LM644261 | LM644241 | LM644260 | |
| A. iizukae | NRRL 3750T = CBS 541.69T = IMI 141552T = CCF 4548T | Japan, Gymna Prefecture, Fujioka, soil from stratigraphic drilling core, 1969, J. Sugiyama | EF669597 | EU014086 | EF669555 | EF669639 |
| CBS 138188T = DTO 179-E6T (ex-type of A. capensis) | South Africa, Cape Town, house dust, 2010, E. Whitfield & K. Mwange | KJ775550 | KJ775072 | KJ775279 | KP987020 | |
| CanS-34A | China, Wuhan, oilseed rape (Brassica napus), between 2008-2010 | MK072769 | MK215220 | MK215219 | MK215221 | |
| CCF 1895 | Czechia, Most (brown lignite district), soil of spoil bank, 1984, M. Černý | FR727134 | FR775336 | HG916728 | HG916707 | |
| CCF 4033 = CMF ISB 1551 = NA16 = Y14 | Czechia, Most (brown lignite district), soil of spoil bank tip, 2004, A. Nováková | FR733809 | HG916686 | HG916729 | MW478537 | |
| CCF 4032 = CMF ISB 1245 | Germany, Weissagker Berg near Cottbus, Lusatian brown lignite district, soil of spoil bank, 1999, A. Nováková | HG915894 | HG916687 | HG916730 | HG916708 | |
| CMF ISB 2544 | Romania, Dobrogea, Mangalia, soil near Movile Cave, 2011, A. Nováková | HG915895 | HG916694 | HG916731 | HG916709 | |
| CMF ISB 2417 | Romania, National Park Apuseni Mountains, Meziad Cave, earthworm casts, 2009, A. Nováková | HG915896 | HG916688 | HG916732 | HG916710 | |
| CMF ISB 2616 | Czechia, Ječmeniště, National Nature Monument, soil, 2012, A. Nováková | HG915899 | HG916689 | HG916733 | HG916711 | |
| CMF ISB 2617 | Czechia, Kolby, National Reservation Pouzdřanská step, soil, 2012, A. Nováková | HG915897 | HG916692 | HG916734 | HG916712 | |
| CMF ISB 2618 | Czechia, Kolby, National Reservation Pouzdřanská step, soil, 2012, A. Nováková | HG915898 | HG916693 | HG916735 | HG916713 | |
| CMF ISB 2619 | Czechia, Kolby, National Reservation Pouzdřanská step, earthworm casts, 2012, A. Nováková | HG915900 | HG916690 | HG916736 | HG916714 | |
| CMF ISB 2620 | Czechia, Kolby, National Reservation Pouzdřanská step, earthworm casts, 2012, A. Nováková | HG915901 | HG916691 | HG916737 | HG916715 | |
| NRRL 58963 = CCF 4843 = ZJ 1256 | USA, Illinois, indoor air of a home, 2009, Ž. Jurjević | LM644237 | LM644268 | LM644245 | MW478538 | |
| CCF 4844 = ZJ 1817 | USA, Idaho, Boise, indoor air of a home, 2012, Ž. Jurjević | LM644238 | LM644269 | LM644244 | MW478539 | |
| CCF 4845 = S746 | Romania, Movile Cave, cave sediment, 2013, A. Nováková | LM999906 | LM644270 | LM644243 | MW478540 | |
| UTHSCSA DI14-219 | USA, Illinois, human, bronchoalveolar lavage, 2012, D. Sutton | LT899477 | LT899528 | LT899579 | LT899634 | |
| FMR 15051 | Spain, Catalonia, Els Ports Natural Park, herbivore dung, 2016, J. Gené | LT899475 | LT798968 | LT899577 | LT899632 | |
| FMR 15606 | Spain, Catalonia, Els Ports Natural Park, herbivore dung, 2016, J. Gené | LT899476 | LT798969 | LT899578 | LT899633 | |
| CCF 5786 = EMSL 3408 | USA, Florida, Saint Petersburg, bedroom floor – swab, 2016, Ž. Jurjević | MW448668 | MW478501 | MW478516 | MW478541 | |
| A. inusitatus | DTO 121-G5T = CBS 147044T = CCF 6552T | Tunisia, Ras Rajel, soil in oak forest, 2009, C. Silva Pereira | MW448669 | MW478502 | MW478517 | MW478542 |
| A. lanuginosus | NRRL 4610T = IMI 350352T = CCF 4551T = IFM 66818T | Haiti, Fonds Parisien, soil | EF669604 | EU014080 | EF669562 | EF669646 |
| A. luppiae | NRRL 6326T = CBS 653.74T = CCF 4545T | France, Provence, near Aups, natural truffle soil, 1972, A.M. Luppi-Mosca | EF669617 | EU014079 | EF669575 | EF669659 |
| A. micronesiensis | CBS 138183T = DTO 267-D5T | Federated States of Micronesia, Yela of Kosrae Island, house dust, 2010, E. Whitfield & K. Mwange | KJ775548 | KJ775085 | KP987067 | KP987023 |
| NRRL 4578 = ATCC 16805 = CBS 586.65 = IMI 135423 = CCF 4555 (ex-type of A. frequens) | Haiti, soil, 1960, J. Rabel | EF669602 | EU014082 | EF669560 | EF669644 | |
| CCF 2026 | Czechia, Prague, archive material, 1986, O. Fassatiová | HG915893 | HG916684 | HG916726 | HG916705 | |
| NRRL 295 = ATCC 16814 = CBS 585.65 = IMI 135422 = CCF 4554 = FRR 0295 | USA, Minnesota, dairy products, 1933, H. Macy | EF669588 | EU014081 | EF669546 | EF669630 | |
| CCF 4005 | Czechia, Hradec Králové, hospital indoor air, 2005, V. Buchta | FR727135 | HG916685 | HG916727 | HG916706 | |
| NRRL 4263 = CCF 4556 | India, Dehradun New Forest, soil, 1955, K.B. Bakshi | EF669600 | EU014083 | EF669558 | EF669642 | |
| NRRL 286 = ATCC 1030 = FRR 0286 | Received in NNRL from Dr. J. Westerdijk (CBS) | AY373849 | LM644262 | LM644246 | LM644258 | |
| NRRL 26246 = CCF 4838 | China, soil, 1944 | LM999905 | LM644263 | LM644247 | LM644257 | |
| NRRL 58660 = CCF 4839 = ZJ 1111 | Trinidad & Tobago, indoor air of a home, 2009, Ž. Jurjević | LM644239 | LM644264 | LM644248 | MW478543 | |
| NRRL 58682 = CCF 4840 = ZJ 1132 | Puerto Rico, indoor air of a home, 2009, Ž. Jurjević | LM644240 | LM644265 | LM644251 | MW478544 | |
| NRRL 58899 = CCF 4841 = ZJ 1267 | USA, New York, indoor air of a home, 2009, Ž. Jurjević | LM999903 | LM644266 | LM644249 | MW478545 | |
| NRRL 58598 = CCF 4842 = ZJ 1038 | USA, New Jersey, indoor air of a home, 2008, Ž. Jurjević | LM999904 | LM644267 | LM644250 | MW478546 | |
| IHEM 18446 | Belgium, Brussels, floor in hospital, 2001, BCCM/IHEM collection | MW448670 | LN909029 | MW478518 | MW478547 | |
| IHEM 662 | Belgium, Brussels, indoor air in hospital, 1980, BCCM/IHEM collection | MW448671 | LN909030 | MW478519 | MW478548 | |
| IHEM 22506 = RV 21840 | Belgium, Liège, human lung, 1967, University Hospital Liège | MW448672 | LN909028 | MW478521 | MW478549 | |
| IHEM 22505 = RV 42608 | Belgium, Antwerp, human sputum (male), 1979, D. Van Vijver | MW448673 | LN909027 | MW478520 | MW478550 | |
| CBS 147045 = DTO 247-H3 | Mexico, Sayulita, hotel room, house dust, 2009, A. Amend & E. Whitfield & K. Mwange | KP987079 | KP987047 | KP987062 | KP987036 | |
| IMI 357699 = DTO 305-B6 = IBT 23707 (ex-type of A. sunderbaniinom. inval.) | India, West Bengal, soil | KP987084 | KP987052 | KP987069 | KP987026 | |
| UTHSCSA DI14-214 | USA, California, canine urine, 2012, D. Sutton | LT899480 | LT899529 | LT899582 | LT899637 | |
| FMR 15737 | Spain, Canary Islands, Tenerife, 2016, J. Gené | LT899479 | LT798971 | LT899581 | LT899636 | |
| A. movilensis | NRRL 62819T = CCF 4410T = CMF ISB 2614T = CBS 134395T | Romania, Mangalia, Dobrogea, soil near Movile Cave, 2011, A. Nováková | HG915904 | HG916697 | HG916740 | HG916718 |
| CBS 139559 = CCTU 749 = DTO 203-C9 = IBT 32594 | Iran, Urmia, Kabodan Island, soil, between 2011 and 2012, U. Ghosta & R. Samadi | KP987075 | KP987043 | KP987058 | KP987032 | |
| CBS 139562 = CCTU 788 = DTO 203-H3 | Iran, Urmia, Kabodan Island, soil, between 2011 and 2012, U. Ghosta & R. Samadi | KP987078 | KP987046 | KP987061 | KP987035 | |
| S1040 | Romania, soil above the Movile cave, 2014, A. Nováková | MW448674 | MW478503 | MW478522 | MW478551 | |
| A. neoflavipes | NNRL 5504T = ATCC 24484T = CBS 260.73T = IMI 171883T = IFM 40894T = CCF 4552T | Thailand, Pak Thong Chai, cellulosic material buried in forest soil, 1968, C. Klinsukont | EF669614 | EU014084 | EF669572 | EF669656 |
| A. olivimuriae | NRRL 66783T = CCF 6208T | Italy, Viterbo, olive curing brine, 2012, S. Crognale | MH298877 | MH492010 | MH492011 | MH492012 |
| A. polyporicola | NRRL 32683T = CCF 4553T | USA, Hawaii, Hilo, Alien Wet Forest Zoo, basidioma of Earliella scabrosa (Polyporales), 2003, D.T. Wicklow | EF669595 | EU014088 | EF669553 | EF669637 |
| NRRL 58570 = CCF 4828 | USA, Hawaii, Alien Wet Forest, basidioma of Rigidoporus microporus (Polyporales), 2003, D.T. Wicklow | HQ288052 | LM644274 | LM644252 | LM644254 | |
| CCF 5427 = EMSL 2612 | USA, New York, Holbrook, bedroom - settle plates, 2014, Ž. Jurjević | MW448675 | MW478504 | MW478523 | MW478552 | |
| CCF 6262 = EMSL 3169 | USA, crawled space - settle plates, 2015, Ž. Jurjević | MW448676 | MW478505 | MW478524 | MW478553 | |
| A. spelaeus | NRRL 62826T = CCF 4425T = CMF ISB 2615T = CBS 134371T | Spain, Andalusia, Nerja Cave, cave sediment, 2011, A. Nováková | HG915905 | HG916698 | HG916741 | HG916719 |
| NRRL 62827 = CCF 544 | Czechia, Bohemian Karst, Doutnáč hill near Srbsko, soil, 1961, O. Fassatiová | HG915906 | HG916699 | HG916742 | HG916720 | |
| CCF 4699 = CMF ISB 2659 | Czechia, Hostěradice, National Nature Monument U Kapličky, Allolobophora hrabei intestine, 2012, A. Nováková | HG915907 | HG916700 | HG916743 | HG916721 | |
| CCF 4679 = CMF ISB 2663 | Czechia, Ječmeniště, National Nature Monument, soil, 2012, A. Nováková | HG915908 | HG916701 | HG916744 | HG916722 | |
| CCF 4680 | Spain, Andalusia, Nerja Cave, cave sediment, 2012, A. Nováková | HG915909 | HG916702 | HG916745 | HG916723 | |
| CCF 4697 | Spain, Andalusia, Nerja Cave, cave air, 2012, A. Nováková | HG915910 | HG916703 | HG916746 | HG916724 | |
| EMSL 4874 | USA, Georgia, Sandersville, crawlspace (swab), 2018, Ž. Jurjević | MW448677 | MW478506 | MW478525 | MW478554 | |
| CCF 4886 = S716 | Spain, Andalusia, Nerja Cave, cave sediment, 2012, A. Nováková | LM999908 | LM644272 | HG916748 | LM644259 | |
| CCF 4829 = BMP 3043 | USA, Arizona, Benson, Kartchner Caverns, speleothem surface, 2008, M. Vaughan | HQ832962 | LM644273 | LM644253 | LM644255 | |
| CBS 115952 | Germany, dust, S. Ammermann | MW448678 | MW478507 | MW478526 | MW478555 | |
| UTHSCSA DI17-89 (UTHSCSA 04-3307) | USA, Missouri, human forearm, 2004, D. Sutton | LT899491 | LT899538 | LT899593 | LT899648 | |
| FMR 14606 | Spain, Balearic Islands, Mallorca, soil, 2012, J. Gené | LT899488 | LT899537 | LT899590 | LT899645 | |
| FMR 15176 | Spain, Balearic Islands, Mallorca, herbivore dung, 2016, J. Gené & J.P.Z. Siqueira | LT899489 | LT798972 | LT899591 | LT899646 | |
| FMR 15223 | Spain, Balearic Islands, Mallorca, herbivore dung, 2016, J. Gené & J.P.Z. Siqueira | LT899490 | LT798976 | LT899592 | LT899647 | |
| CCF 6263 = EMSL 4125 | USA, New Jersey, Marlton, black walnut (Juglans nigra), 2017, Ž. Jurjević | MW448679 | MW478508 | MW478527 | MW478556 | |
| CCF 6248 = EMSL 4140 | USA, New Jersey, Marlton, black walnut (Juglans nigra), 2017, Ž. Jurjević | MW448680 | MW478509 | MW478528 | MW478557 | |
| A. suttoniae | UTHSCSA DI14-215T = FMR 13523T | USA, human sputum, 2014, D. Sutton | LT899487 | LT899536 | LT899589 | LT899644 |
| A. templicola | CBS 138181T = DTO 270-C6T | Mexico, Sayulita, dust from church, 2010, E. Whitfield & K. Mwange | KJ775545 | KJ775092 | KJ775394 | KP987017 |
| CBS 138180 = DTO 267-H4 | Thailand, Bangkok, house dust, 2010, E. Whitfield & K. Mwange | KP987081 | KJ775087 | KP987064 | KP987038 | |
| NRRL 62825 = CCF 4698 = CMF ISB 2662 (ex-type of A. mangaliensis) | Romania, Mangalia, soil near Moville Cave, 2012, A. Nováková | HG915902 | HG916695 | HG916738 | HG916716 | |
| CCF 869 = NRRL 62823 | China, industrial material, 1955, V. Zánová | HG915903 | HG916696 | HG916739 | HG916717 | |
| NRRL 4893 = IMI 343701 = CCF 4846 | Japan, soil | LM999907 | LM644271 | LM644242 | LM644256 | |
| IHEM 14393 | Belgium, Charleroi, furniture in hospital 1998, BCCM/IHEM collection | MW448681 | LN909024 | MW478529 | MW478558 | |
| DK-T43978 | Denmark, Copenhagen, bronchoalveolar lavage, 2014 | MW448682 | LN909025 | MW478530 | MW478559 | |
| A. urmiensis | CBS 139558T = CCTU 742T = DTO 203-C2T = IBT 32593T | Iran, Urmia, Jade Darya (seaside), soil, 2011, U. Ghosta & R. Samadi | KP987073 | KP987041 | KP987056 | KP987030 |
| CBS 139557 = CCTU 734 = DTO 203-B3 = IBT 32597 | Iran, Jade Darya (seaside), soil, 2011, U. Ghosta & R. Samadi | KP987072 | KP987039 | KP987055 | KP987029 | |
| CBS 139766 = CCTU 743 = DTO 203-C3 = IBT 32598 | Iran, Jade Darya (seaside), soil, 2011, U. Ghosta & R. Samadi | KP987074 | KP987042 | KP987057 | KP987031 | |
Acronyms of culture collections in alphabetic order: ATCC, American Type Culture Collection, Manassas, Virginia, USA; BMP, Barry M. Pryor laboratory culture collection, Tucson, Arizona, USA; CBS, Westerdijk Fungal Biodiversity Institute (formerly Centraalbureau voor Schimmelcultures), Utrecht, the Netherlands; CCF, Culture Collection of Fungi, Department of Botany, Charles University, Prague, Czech Republic; CMF ISB, Collection of Microscopic Fungi of the Institute of Soil Biology, Academy of Sciences of the Czech Republic, České Budějovice, Czech Republic; CCTU, Culture Collection of Tabriz University, Tabriz, Iran; FMR, Faculty of Medicine, Reus, Spain; FRR, Food Fungal Culture Collection, North Ride, Australia; IFM, Collection at the Medical Mycology Research Center, Chiba University, Chiba, Japan; IHEM (BCCM/IHEM), Belgian Coordinated Collections of Micro-organisms, Fungi Collection: Human and Animal Health, Sciensano, Brussels, Belgium; IMI, CABI’s collection of fungi and bacteria, Egham, UK; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; UTHSCSA, Collection of Fungus Testing Laboratory, University of Texas, Health Science Center, San Antonio, USA.
Sequences generated in this study are designated by bold print.
Molecular studies
Total genomic DNA was isolated from 7-d-old cultures with ArchivePure DNAyeast (5 PRIME Inc., Gaithersburg, MD, USA) or NucleoSpin® Soil (Macherey–Nagel, Düren, Germany) isolation kits. The quality of the isolated DNA was verified using a NanoDrop 1 000 Spectrophotometer.
The ITS region of rDNA was amplified using forward primer ITS1 (White et al. 1990) and reverse primers NL4 (O’Donnell 1993) or ITS4 (White et al. 1990), a part of the β-tubulin gene (benA) was amplified using forward primers Bt2a (Glass & Donaldson 1995), T10 (O'Donnell & Cigelnik 1997) or Ben2f (Hubka & Kolařík 2012) and reverse primer Bt2b (Glass & Donaldson 1995), a part of the calmodulin gene (CaM) was amplified using forward primers CF1L, CF1M (Peterson 2008) or cmd5 (Hong et al. 2006) and reverse primers CF4 (Peterson 2008) or cmd6 (Hong et al. 2006) and a part of the RNA polymerase II second largest subunit gene (RPB2) was amplified using forward primer fRPB2-5F and reverse primer fRPB2-7CR (Liu et al. 1999). Various primer pairs were used for the amplification of ITS, benA and CaM loci because the sequences were generated across various research groups. Thus, it was not due to the failure of PCR with some primer combinations.
The PCR reaction volume of 25 μL contained 1.2 μL (10-20 ng) of DNA, 1 μL of both primers ( 10 μM), 0.25 μL of DreamTaq DNA Polymerase (Thermo Scientific, Waltham, MA) and 2.5 μl of DreamTaq PCR buffer and 2.5 μl of dNTP. The ITS rDNA, benA and CaM fragments were amplified using following thermal cycle profile: 93 °C/2 min; 30 cycles of 93 °C/30 s; 55 °C/30 s; 72 °C/60 s; 72 °C/10 min. Partial RPB2 gene fragments were amplified using above-mentioned cycle or touchdown thermal-cycling: 93 °C/2 min; 5 cycles of 93 °C/30 s, 65–60 °C/30 s, 72 °C/60 s; 38 cycles of 93 °C/30 s, 55 °C/30 s, 72 °C/60 s; 72 °C/10 min. PCR products were purified with ethanol and sodium acetate in a 96-well plate; 2 μL of 3 M NaOAc and 60 μL of 96 % EtOH was mixed with 20 μL of PCR product. The plate was sealed, twisted several times and incubated in the refrigerator for 20 min. After incubation, the plate was centrifuged for 30 min at 4 °C and 3 000 rpm, the supernatant was removed and 85 μL 70 % EtOH was added. The plate was centrifuged for 15 min at 4 °C and 3 000 rpm, the supernatant was subsequently removed, the pellet was dried at room temperature for 20 min and resuspended in 10 μL of H2O.
Newly obtained DNA sequences were inspected in FinchTV v. 1.4 (available online https://digitalworldbiology.com/FinchTV) and assembled in Bioedit v. 7.0.5 (Hall 1999). Multiple sequence alignments were created in MAFFT v. 7 (Katoh & Standley 2013) using the G-INS-I strategy. Sequences were deposited into GenBank with accession numbers shown in Table 1. All alignments are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.dz08kprxj.
Phylogenetic analysis and species delimitation
In order to demonstrate the phylogenetic relationships within section Flavipedes, we calculated a Maximum Likelihood (ML) tree in IQ-TREE v. 2.0 (Nguyen et al. 2015) using a concatenated alignment of all four loci as input. The inference was setup as partitioned analysis and the best-fitting model for each locus was determined using the Bayesian information criterion (BIC) in jModelTest v. 2.1.7 (Posada 2008). Selected models are listed in Table 2 together with the respective alignment statistics. To determine the branch support, the analysis ran for 1 000 bootstrap replicates.
Table 2.
Alignment characteristics and substitution models according to Bayesian Information Criterion.1
| Dataset (Series) | Locus | Alignment length | Variable sites | Parsimony informative sites | Substitution model |
|---|---|---|---|---|---|
| Flavipedes | benA | 512 | 138 | 102 | K80+G |
| CaM | 560 | 144 | 116 | TrNef+G | |
| RPB2 | 1 013 | 161 | 129 | TrNef+G | |
| ITS rDNA | 539 | 14 | 12 | F81+I | |
| Spelaei | benA | 507 | 127 | 108 | K80+G |
| CaM | 731 | 193 | 164 | TrNef+G | |
| RPB2 | 1 009 | 138 | 113 | TrNef+G | |
| ITS rDNA | 549 | 21 | 13 | HKY |
proposed by jModelTest v. 2.1.7 (Posada 2008).
For the purpose of species delimitation analyses, the dataset was split into two parts corresponding to series Flavipedes and Spelaei as designated by Houbraken et al. (2020). Series Neonivei and Olivimuriarum were excluded as these are single species series and phylogenetically distinct from series Flavipedes and Spelaei. The ITS rDNA region was excluded due to its low number of informative sites. Omission of the ITS region from the phylogenetic analysis is a common practice as discussed previously (Chen et al. 2017). The following steps were performed separately for both datasets.
The haplotype function from R v. 4.0.2 (R Core Team 2015) package pegas (Paradis 2010) was used to retain only unique sequences in alignments. The best fitting models obtained in jModelTest v. 2.1.7 (Posada 2008) are listed in Table 2.
We used one multi-locus method, STACEY (Jones 2017), and four single-locus species delimitation methods, GMYC (Fujisawa & Barraclough 2013), bGMYC (Reid & Carstens 2012), PTP and bPTP (Zhang et al. 2013), to create hypotheses about species boundaries. The multi-locus species delimitation analysis was performed in BEAST v. 2.6.3 (Bouckaert et al. 2014) add-on STACEY v. 1.2.5 (Jones 2017) with the following parameters. The chain length was set to 1 × 108 generations, the species tree prior was set to the Yule model, growth rate prior was set to lognormal distribution (M = 5, S = 2), clock rate priors for all loci were set to lognormal distribution (M = 0, S = 1), PopPriorScale prior was set to lognormal distribution (M = -7, S = 2) and relativeDeathRate prior was set to beta distribution (α = 1, β = 1 000). The output was processed with SpeciesDelimitationAnalyzer (Jones 2017). The ultrametric input trees for the GMYC method (Fujisawa & Barraclough 2013) were calculated in BEAST v. 2.6.3 (Bouckaert et al. 2014) with chain length 1 × 107 generations. The GMYC analysis was performed in R v. 4.0.2 (R Core Team 2015) with the package splits (Fujisawa & Barraclough 2013). One hundred ultrametric trees from the BEAST v. 2.6.3 inference were randomly selected using R v. 4.0.2 package ape (Paradis et al. 2004) after discarding the initial 25 % of the trees as burn-in, and then used as input for the bGMYC method. The analysis was performed in R v. 3.4.1 with package bgmyc (Reid & Carstens 2012). One thousand maximum likelihood standard bootstrap trees were calculated in IQ-TREE v. 2.0 (Nguyen et al. 2015) and used as input for PTP method. The analysis was run in Python v. 3 (van Rossum & Drake 2019) package ptp (Zhang et al. 2013). The bPTP was also performed in Python package ptp (Zhang et al. 2013). Recommended inputs for this method are trees from Bayesian inference, but without the requirement of ultrametricity. In this case, we used trees from a mcmc run in MrBayes v. 3.2.7 (Ronquist et al. 2012). Phylogenetic trees generated during STACEY analysis were used for the presentation of species delimitation results analysis. The graphical outputs were created in iTOL (Interactive Tree Of Life) (Letunic & Bork 2016).
Finally, we tested different species boundaries hypotheses in DELINEATE (Sukumaran et al. 2021). The analysis was performed according to the manual available online. Briefly, the dataset was split into hypothetical populations with “A10” analysis in BPP v. 4.3 (Yang 2015) (Supplementary Table S1). The starBEAST (Heled & Drummond 2010) implemented in BEAST v. 2.6.3 (Bouckaert et al. 2014) was used to estimate the species tree. The populations delimited by BPP were lumped into species based on the results of species delimitation methods and phenotypic characters, with several populations always left unassigned to be delimited by DELINEATE. Four models of species boundaries were set up for the Flavipedes series, 14 models for the Spelaei series and six models for both series analyzed together. The analysis was run in Python v. 3 (van Rossum & Drake 2019) package delineate (available online https://jeetsukumaran.github.io/delineate/).
Phenotypic studies
The macromorphology of colonies was studied on Czapek yeast autolysate agar (CYA; Fluka, Buchs, Switzerland), Czapek-Dox agar (CZA), malt extract agar (MEA; Oxoid, Melbourne, Australia) (Samson et al. 2014), oatmeal agar (OA; Difco, La Ponte de Claix, France) and CYA supplemented with 20 % sucrose (CY20S). The strains were inoculated in three points on 90 mm Petri dishes and incubated at 25 °C in darkness. Cardinal temperatures were determined for A. movilensis and its relatives. The strains were grown on MEA for 14 d at 10, 15, 20, 25, 30, 35, 37 and 40 °C in darkness. For the description of colony colours, we used the hexadecimal colour codes and the names were assigned according to website https://coolors.co/. Colony details were documented using an Olympus SZX2-ILLT dissecting microscope (Tokyo, Japan) equipped with an Olympus DP27 digital camera.
Micromorphological characters were observed from 14-d-old colonies grown on MEA. Every character was measured 35 times for each isolate. Lactic acid (60 %) was used as the mounting medium. Photographs were taken using an Olympus BX51 microscope with an Olympus DP72 camera. The picture processing and preparation of photographic plates was done in Adobe Photoshop CS6.
Antifungal susceptibility testing (EUCAST method)
The determination of the minimum inhibitory concentrations (MICs) of antifungal agents was carried out according to the reference European Committee on Antimicrobial Susceptibility testing (EUCAST) guidelines (E.Def 9.3.2). Pure antifungal substances were stored in aliquots at -80 °C and stock solutions prepared in DMSO (5 000 mg/L; Sigma-Aldrich, Brøndby, Denmark). Cell-culture-treated NuncTM MicroWellTM 96-Well Microplates (ThermoFisher Scientific, catalogue no. 167008) were used throughout. Microtitre plates with 2-fold dilutions were prepared using serial dilution (with two pipette tip changes) and frozen at -80 °C for at least 24 h prior to use. The following antifungal agents (final concentration ranges) were included: amphotericin B (0.004–4 mg/L; Sigma-Aldrich, Germany), itraconazole (0.004–4 mg/L and 0.016–16 mg/L; Sigma-Aldrich), posaconazole (0.004–4 mg/L; MSD, Ballerup, Denmark), voriconazole (0.004–4 mg/L and 0.016–16 mg/L; Pfizer A/S, Ballerup, Denmark and Sigma-Aldrich), isavuconazole (0.008–8 mg/L and 0.016–16 mg/L; Basilea Pharmaceutica International Ltd, Basel, Switzerland), and terbinafine (0.004–4 mg/L; Sigma-Aldrich). Two concentration ranges for some antifungals were used because the isolates were analyzed in two batches and, as the breakpoints are low, we decided to skip the highest concentrations for the second batch and move the range toward lower concentrations. Plates were incubated at 37 °C (or 25 °C for species with insufficient growth at 37 °C) for 48 h. Aspergillus fumigatus ATCC 204305 was included as quality control (Arendrup et al. 2021).
Results
The species delimitation analyses were mostly divided into two parts corresponding to the series Flavipedes and Spelaei according to Houbraken et al. (2020). The analysis of each series consisted of two steps (for detailed description see Materials and Methods).
Species delimitation and validation in the series Flavipedes
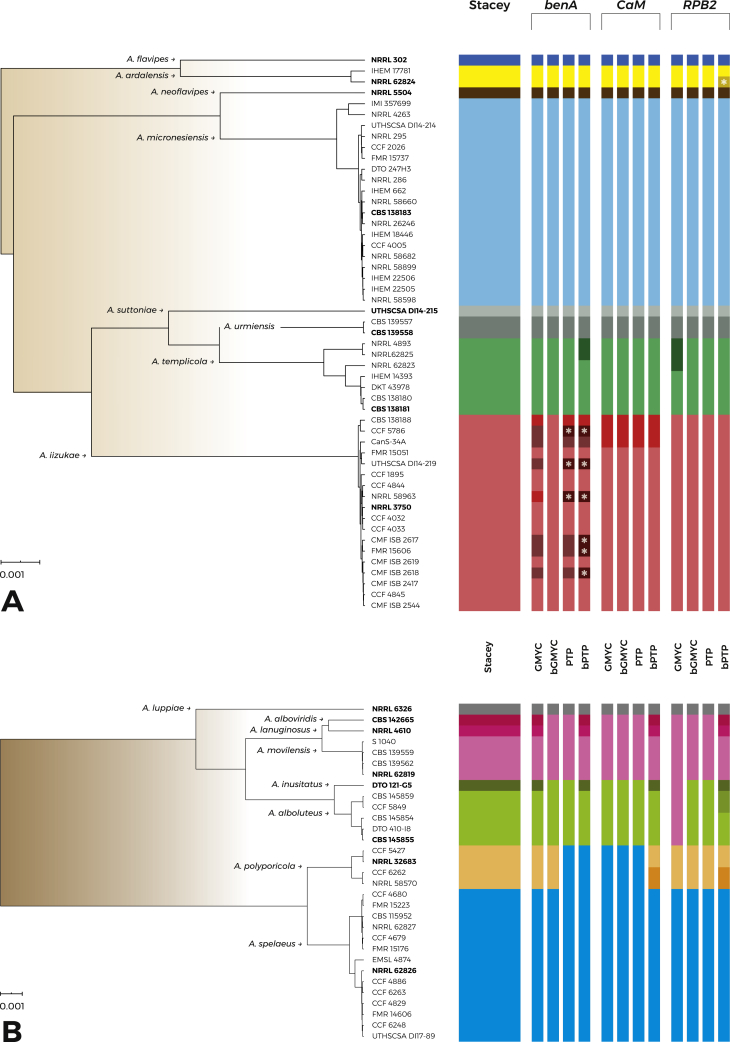
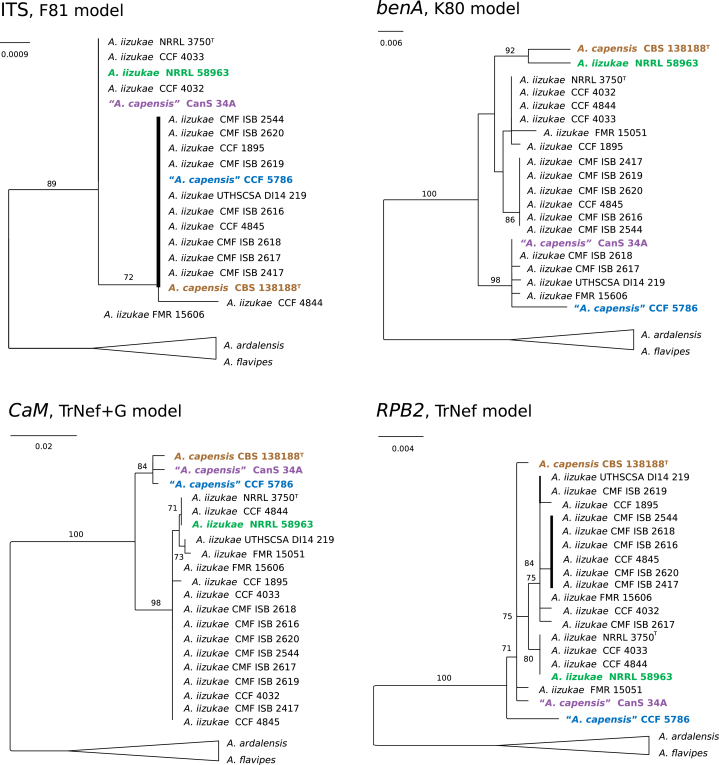
The results of various species delimitation methods in the series Flavipedes (Fig. 1A) were in broad agreement and unequivocally supported the species A. flavipes, A. neoflavipes, A. micronesiensis, A. suttoniae and A. urmiensis. Aspergillus ardalensis was split into two tentative species by bPTP using RPB2 as input, but a broader concept was supported by all other methods (Fig. 1A). Similarly, A. templicola was split into two tentative species by GMYC using RPB2 as input and by bPTP using benA as input; but all other methods resolved A. templicola as a single species. Less stable results were obtained in the node comprising A. iizukae and A. capensis. All single-locus methods delimited “A. capensis lineage” represented by CBS 138188T, CCF 5786 and CanS-34A as separate species using CaM as input. When benA was used as input, the methods showed variable results. The bGMYC method lumped A. capensis and A. iizukae together, PTP and bPTP delimited several singleton species and GMYC delimited three species (with A. capensis consisting of CBS 138188T and A. iizukae strain NRRL 58963). These results together with unstable position of particular strains in single-locus trees (Fig. 2) suggest ongoing recombination within the clade formed by A. iizukae and A. capensis.
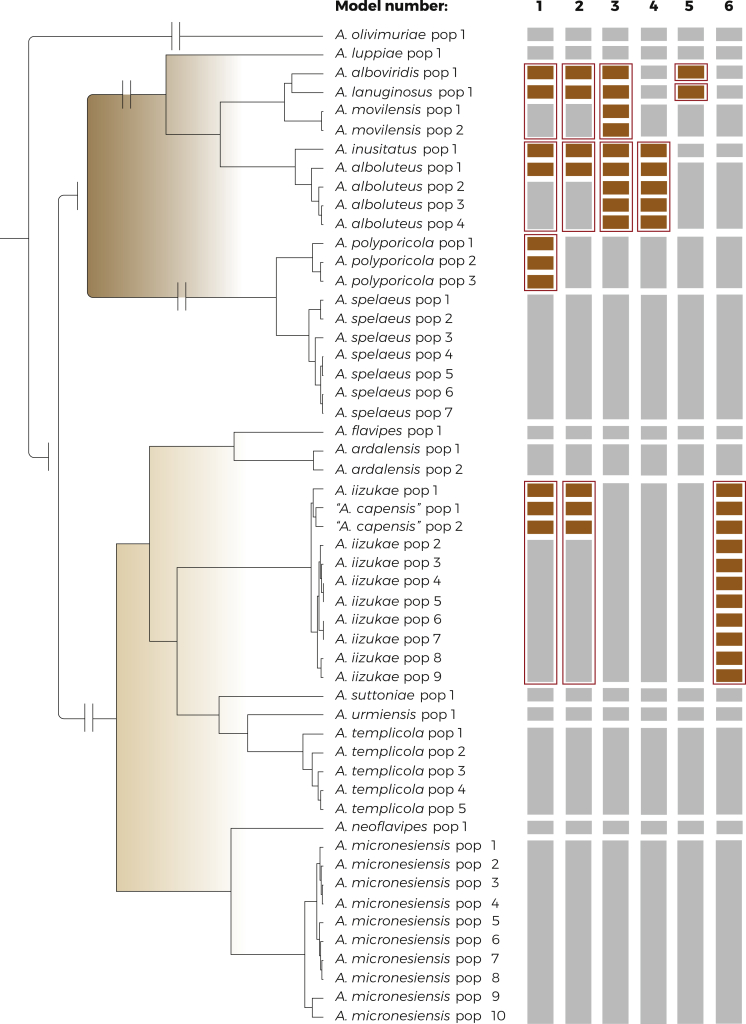
Fig. 1.
Schematic representation of species delimitation results in the series Flavipedes (A) and Spelaei (B). The analyses were based on three loci (benA, CaM, RPB2) and utilized one multi-locus method (STACEY) and four single-locus methods (GMYC, bGMYC, PTP, bPTP). Only strains with unique haplotypes were used (strains with identical sequences are represented by one tip in the tree). The results are depicted by coloured bars with different colours indicating tentative species delimited by each method. The asterisk (∗) sign designates singleton species delimited by the particular methods. Ex-type isolates are highlighted with bold font. The phylogenetic tree was calculated during STACEY analysis and is used solely for the comprehensive presentation of the results from different methods.
Fig. 2.
Single-locus Maximum Likelihood trees of clade containing isolates of Aspergillus iizuake and “A. capensis”. The trees were calculated in IQ-TREE v. 2.0 based on sequences of ITS region and benA, CaM and RPB2 loci. The ex-type isolates are designated by a superscript T. The strains with unstable position across phylogenies and thus causing incongruences are highlighted by colours.
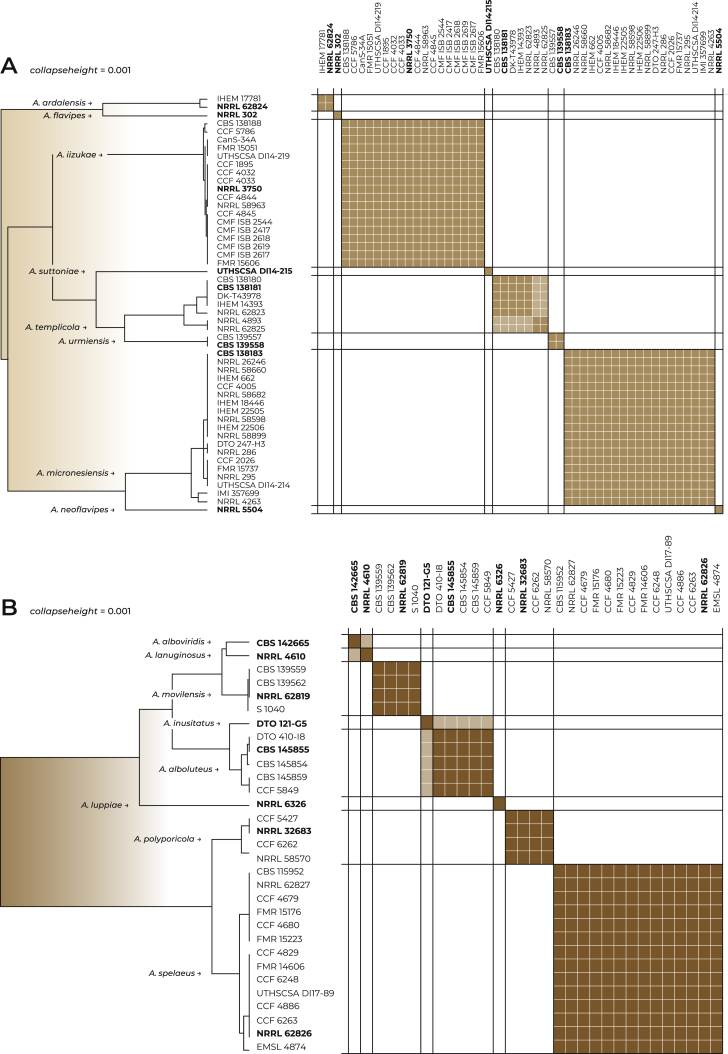
Detailed results of STACEY (Fig. 3A and Supplementary Fig. S1 with different values of collapseheight parameter) with similarity matrix displaying the probability of assignment of strains to particular species suggest that the support for splitting A. iizukae/A. capensis into two species is lower than in other species, e.g., in A. templicola or A. micronesiensis which were not split by any single-locus species delimitation methods.
Fig. 3.
The results of species delimitation by STACEY add-on of BEAST v. 2 in the series Flavipedes (A) and Spelaei (B) with the chosen collapseheight parameter = 0.001. The similarity matrices give the posterior probability of every two isolates belonging to the same multi-species coalescent cluster (tentative species). The darkest brown shade corresponds to a posterior probability of 1, while a white colour is equal to 0. The horizontal and vertical lines in the similarity matrices depict the species boundaries proposed by the analysis. Only strains with unique multilocus haplotypes were used in the analysis (strains with identical haplotype are represented by one tip in the tree). Ex-type isolates are highlighted with bold font. Presented phylogenetic trees were calculated in STACEY.
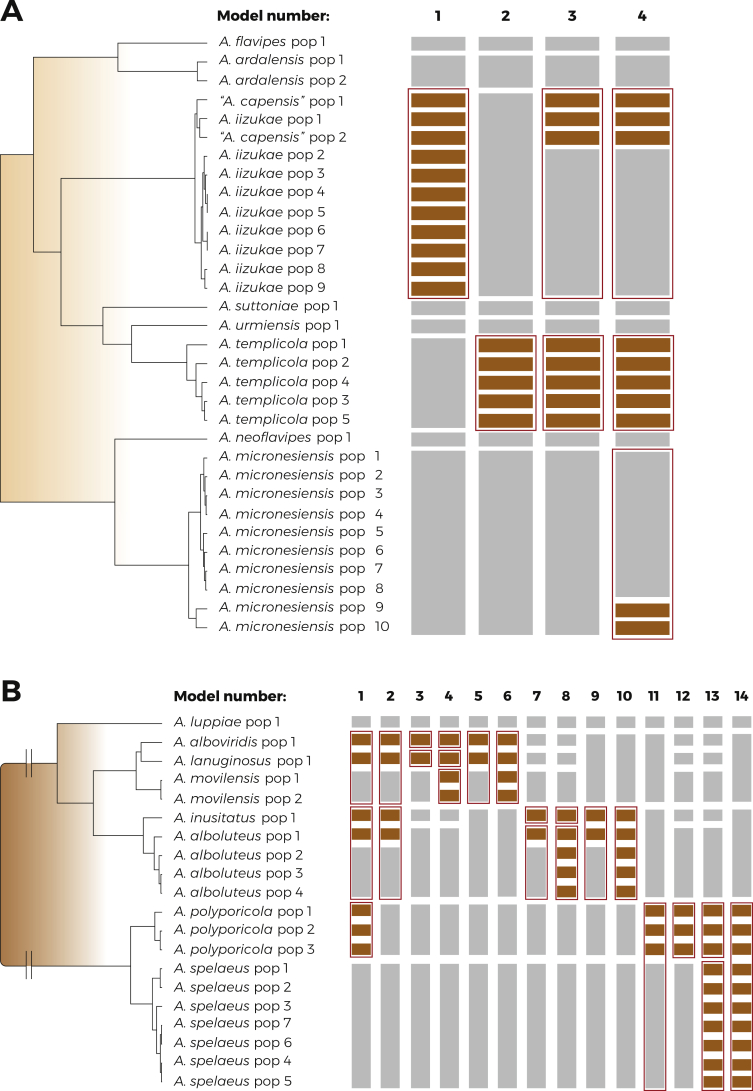
In the species validation step (Fig. 4A), we set up four different models focusing mainly on A. iizukae/A. capensis clade. The first model left all populations of A. iizukae and A. capensis delimited by BPP (Yang 2015) (Supplementary Table S1) unassigned into species, i.e., free to be delimited. These unassigned populations are depicted by brown coloured bars in the Fig. 4, while populations of other species were assigned according to the delimitation in the first step - grey bars. As a result, A. iizukae and A. capensis populations were lumped together into one species - depicted by red rectangle around bars (Fig. 4A). In the second model, populations of A. templicola were left unassigned and the results lumped them together within one species. Third and fourth model left unassigned all populations of A. templicola and several populations of “A. capensis”, A. iizukae and A. micronesiensis and the results always supported broad species definition of A. templicola, A. miconesiensis and A. iizukae (comprising A. capensis). Overall, the results supported all currently accepted species within series Flavipedes with the exception of A. capensis, which is therefore placed in synonymy with A. iizukae. This finding is also supported by morphological observations as A. iizukae and A. capensis are not distinguishable (Visagie et al. 2014).
Fig. 4.
The results of species validation using DELINEATE in the series Flavipedes (A) and Spelaei (B). The populations of each species were delimited by BPP (Supplementary Table S1) and the displayed tree was calculated in starBEAST. The bars on the right side of the tree depicts the setting and results of each model. The grey bars represent the predefined species (locked in the analysis), while the brown bars represent unassigned populations left free to be delimited. The red rectangles depict the resulting solution proposed by DELINEATE.
Species delimitation and validation in the series Spelaei
The species delimitation in the series Spelaei was less clear compared to series Flavipedes. Based on the results described below, we decided to introduce four new species, namely Aspergillus alboluteus, A. alboviridis, A. inusitatus and A. lanuginosus (see section Taxonomy).
In the first part of the analysis A. luppiae was the only species delimited by all methods without any exception (Fig. 1B). Aspergillus spelaeus and A. polyporicola were lumped together by PTP and bPTP using benA as input, and also by GMYC, bGMYC and PTP using CaM as an input. On the other hand, A. polyporicola was split into two species by bPTP using CaM and RPB2 as input. The four newly described species A. alboluteus, A. alboviridis, A. inusitatus and A. lanuginosus were all delimited by GMYC using benA and bPTP using all three genes (A. alboluteus was even divided into two species by bPTP using RPB2 as an input). The GMYC method with RPB2 as input delimited only one species in this clade, containing the four mentioned species and A. movilensis; and the remaining methods based on RPB2 delimited two species. If only these results would be considered, it might not be enough to support delimitation of five species, but there were striking morphological differences (see section Phenotype analysis) supporting the narrower species concept. This is also in agreement with the results of a multi-locus method STACEY which supported all five species in this clade, and at the same time supported the recognition of A. spelaeus and A. polyporicola as separate species (Fig. 1B). Especially the morphology of A. inusitatus does not allow its inclusion into any other species and denies the possibility of a broader concept in this clade.
Detailed results of STACEY show that when the collapseheight parameter is low enough to consider A. inusitatus a separate species (Fig. 3B and Supplementary Fig. S2B), the clade containing A. movilensis should be also split into three species (A. movilensis, A. alboviridis and A. lanuginosus). If the collapseheight parameter is too high (Supplementary Fig. S2A) to delimit A. inusitatus, also the support for A. polyporicola and A. spelaeus decreases.
In the species validation step (Fig. 4B), we set up 14 different models for testing the species hypotheses. The first model left unassigned populations of A. alboviridis, A. lanuginosus, A. inusitatus, A. polyporicola and population 1 of A. alboluteus. The results supported A. polyporicola but lumped together A. inusitatus/A. alboluteus and A. movilensis/A. alboviridis/A. lanuginosus. The results of the second model with similar setting except for predefined species status of A. polyporicola were identical. In the third and fourth model, A. inusitatus and A. alboluteus were defined as separate species. Aspergillus alboviridis and A. lanuginosus were left unassigned in the third model and in the fourth model, A. alboviridis, A. lanuginosus and A. movilensis were left unassigned. The results were similar in both cases, recognizing A. alboviridis, A. lanuginosus and A. movilensis as separate species. The fifth and sixth model were analogous to model 3 and 4, leaving A. alboviridis and A. lanuginosus (model 5); and A. alboviridis, A. lanuginosus and A. movilensis (model 6), respectively, free to be delimited. Unlike models 3 and 4, in case of models 5 and 6, A. inusitatus was predefined as a part of A. alboluteus. These models resulted in lumping of A. alboviridis, A. lanuginosus and A. movilensis into one species. The models 7–10 tested the opposite hypothesis, leaving A. inusitatus and either one population (models 7 and 9) or all populations (models 8 and 10) of A. alboluteus unassigned, and A. alboviridis and A. lanuginosus predefined either as separate species (models 7 and 8) or lumped together with A. movilensis (model 9 and 10). The final results corresponded with models 3–6. In case of A. alboviridis, A. lanuginosus and A. movilensis being defined as separate species, A. inusitatus and A. alboluteus were also delimited as separate species (models 7 and 8). If A. alboviridis, A. lanuginosus and A. movilensis were lumped together, A. inusitatus and A. alboluteus were also lumped into one species (models 9 and 10). Models 11–14 focused on A. polyporicola and A. spelaeus. Models 11–12 left all populations of A. polyporicola free to be delimited and models 13–14 left all populations of both A. polyporicola and A. spelaeus unassigned. Models 11 and 14 defined A. alboviridis and A. lanuginosus as a part of A. movilensis and A. inusitatus as a part of A. alboluteus. This resulted in lumping of A. polyporicola and A. spelaeus into one species. In models 12–13, A. alboviridis, A. lanuginosus, A. movilensis, A. inusitatus and A. alboluteus were all defined as separate species. This definition resulted in delimitation of A. polyporicola and A. spelaeus as two separate species.
Species validation in the section Flavipedes
Finally, we further validated the putative species in DELINEATE based on the combined dataset of both series using six different models (Fig. 5). The first model left unassigned A. alboviridis, A. lanuginosus, A. inusitatus, population 1 of A. alboluteus, A. polyporicola and “A. capensis” (strains CBS 138188, CanS 34A, and CCF 5786). The second model used the same assignment except for A. polyporicola. The results of both models lumped A. capensis together with A. iizukae; A. polyporicola was delimited as separate species; A. inusitatus was lumped with A. alboluteus; and A. alboviridis and A. lanuginosus were lumped with A. movilensis. The third model left unassigned A. alboviridis, A. lanuginosus, A. inusitatus and also all populations of A. movilensis and A. alboluteus. The results lumped together A. alboviridis/A. lanuginosus/A. movilensis and A. inusitatus/A. alboluteus. The fourth model left unassigned A. inusitatus and populations of A. alboluteus with A. alboviridis, A. lanuginosus and A. movilensis being defined as three separate species and it resulted in A. inusitatus/A. alboluteus lumped together. On the contrary, the fifth model left A. alboviridis and A. lanuginosus unassigned with A. inusitatus, A. alboluteus and A. movilensis predefined as separate species, and it resulted in delimitation of separate A. alboviridis and A. lanuginosus. The sixth model predefined separately all mentioned species except for populations of "A. capensis" and A. iizukae which were left free to be delimited. This model again resulted in lumping of these two species.
Fig. 5.
The results of species validation using DELINEATE for the series Flavipedes and Spelaei analyzed together. The populations of each species were delimited by BPP (Supplementary Table S1) and the displayed tree was calculated in starBEAST. The bars on the right side of the tree depicts the setting and results of each model. The grey bars represent the predefined species (locked in the analysis), while the brown bars represent unassigned populations left free to be delimited. The red rectangles depict the resulting solution proposed by DELINEATE.
Phylogeny
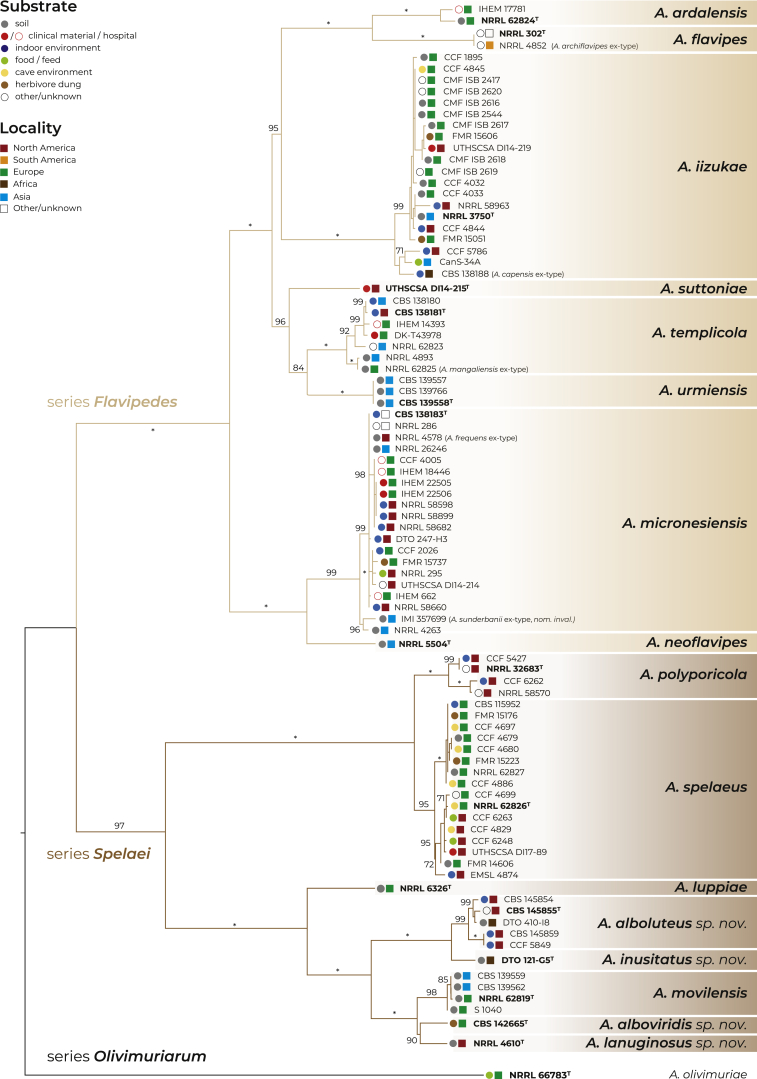
The Fig. 6 shows the best scoring Maximum likelihood (ML) tree based on the concatenated alignment of 90 strains from section Flavipedes representing known species diversity. All deep nodes gained bootstrap support of at least 95 % except for the lineage containing A. templicola and A. urmiensis. This node with support of 84 % is also the only site of incongruence between the concatenated tree and the tree generated by starBEAST during DELINEATE analysis. In the starBEAST analysis, A. iizukae formed a clade with A. suttoniae, A. templicola, and A. urmiensis (Fig. 5), while in the ML tree, A. iizukae was sister to the clade containing A. flavipes and A. ardalensis. Otherwise the topology of these two trees was identical. The only other node with lower support was the lineage comprising two singleton species, A. alboviridis and A. lanuginosus, with the bootstrap support of 90 %.
Fig. 6.
Phylogenetic relationships of the section Flavipedes members inferred by Maximum Likelihood analysis in IQ-TREE v. 2.0 using concatenated alignment of four loci ITS, (benA, CaM, RPB2). The bootstrap support values are appended to nodes (only those supported by bootstrap value of 70 % or higher are displayed) with asterisks indicating the full support. Ex-type isolates are designated by bold font and superscript T. The names of species treated as synonyms are listed in parentheses. The source and locality of isolation are indicated by coloured circles and squares, respectively.
Apart from A. capensis (ex-type strain CBS 138188), which is discussed above, several other species names are treated here as synonyms in agreement with previous studies. The ex-type strain of A. archiflavipes NRRL 4852 is a synonym of A. flavipes; A. mangaliensis with the ex-type strain NRRL 62825 is a synonym of A. templicola; A. frequens with the ex-type strain NRRL 4578 is a synonym of A. micronesiensis; and finally, an invalid name A. sunderbanii based on the strain IMI 357699 is also included in the lineage of A. micronesiensis (Fig. 6).
Before the phylogenetic analysis itself, we tested the phylogenetic position of two relatively distant species, A. olivimuriae and A. neoniveus. Aspergillus olivimuriae was conclusively placed into the section Flavipedes, and therefore we included it as an outgroup in the phylogenetic analysis, but it was excluded from the species delimitation analyses. On the other hand, the position of A. neoniveus within the section Flavipedes did not gain sufficient support and its classification within Aspergillus sections remains uncertain. These findings are in agreement with Houbraken et al. (2020), who proposed series Olivimuriarum (containing only A. olivimuriae) and series Neonivei (containing only A. neoniveus), both within sections Flavipedes. In the phylogeny based on the three-gene dataset, the series Neonivei made the section Flavipedes paraphyletic with respect to section Terrei, while in the phylogeny based on nine genes, A. neoniveus was resolved within section Flavipedes (Houbraken et al. 2020).
Phenotype analysis in relatives of A. movilensis
Selected culture and micromorphological characteristics relevant for species identification in section Flavipedes are summarized in Table 3. In the following paragraphs, we mostly focus on species related to A. movilensis because there was no clear consensus on species boundaries across molecular species delimitation methods used in this study.
Table 3.
Overview of selected phenotypic characters for section Flavipedes members.1
| Species | Growth parameters after 7 days |
Prevailing colony colours on CYA and MEA | Conidia: diam (μm)4, surface | Vesicle diam (μm) | Stipe (μm) |
Hülle cells2 | Sex. state | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYA | CZA | MEA | 37 °C | 40 °C | Length | Width | ||||||
| A. alboluteus | 15–27 | 10–20 | 13–26 | + | — | white, yellow | 2.5–3.5, smooth | (6–)9–17(–20) | (500–)800–1200(–2000) | 4–7(–10) | + | — |
| A. alboviridis | 19–21 | 9–10 | 18–23 | +3 | — | light green, white, yellow | 2.5–3.5, smooth | 12–16(–20) | 120–200(–550) | 4.5–6(–8) | + | — |
| A. ardalensis | 25–28 | 20–28 | 25–28 | + | + | pale ochreous, yellow | 2.5–3, smooth | (5–)7–19 | commonly >1000 | 3–8(–9) | + | — |
| A. flavipes | 25–35 | 25–28 | 25–35 | + | — | white, yellowish-white, pale ochreous | 2–3, smooth | 7–11 | occasionally >1000 | 3–6 | — | — |
| A. iizukae | 16–35 | 13–25 | 13–30 | + | — | yellowish-white, yellow, ochreous, brown, greyish-brown | 2–3(–3.5), smooth | (6–)14–20(–35) | 200–1500 | (3.5–)5–10(–13.5) | + | — |
| A. inusitatus | 22–23 | 8–9 | 17–19 | + | +3 | dark green, yellow | 3.5–4, echinulate | 15–18 | 250–600 | 5–6 | + | — |
| A. lanuginosus | 26–27 | 21–22 | 20–21 | + | — | white, light pinkish-brown, pale ochreous | 2.5–3, smooth | 10–12 | 600–1100 | 3.5–4.5 | — | — |
| A. luppiae | 18–20 | 17–21 | 20–22 | + | — | yellow, white | 2.5–3.5, smooth | 11–16 | 100–220(–300) | 3.5–5.5 | + | — |
| A. micronesiensis | 14–30 | 9–25 | 18–28 | + | — | yellowish-white, yellow, pale ochreous, brown, grayish-brown | (2–)2.5–3.5(–4), smooth | (4–)6–16(–31) | 250–1900 | 2–10 | + | — |
| A. movilensis | 22–25 | 19–20 | 25–30 | + | — | white, pale ochreous, light yellow-green | 2.5–3.3(–3.5), smooth | (5–)9–13(–16) | usually <400, occasionally >1000 | 3.5–6 | + | — |
| A. neoflavipes | 17–21 | 18–20 | 18–22 | + | +3 | yellow, white | (2–)2.5–3, smooth | 13–19 | 250–950 | 5–7.5 | + | + |
| A. neoniveus | 15–16 | 12–14 | 13 | — | — | yellow, white | 2–2.5, smooth | 9–11 | 150–300 | 4.5–6 | + | + |
| A. olivimuriae | 31–35 | 23–28 | 24–27 | + | — | ochreous | 2–2.5, smooth | 8–10(–15) | 100–150 | 5–6 | — | — |
| A. polyporicola | 17–27 | 14–20 | 20–32 | — | — | pale ochreous, ochreous | 2–3(–3.5), smooth | (6–)8–16(–20) | 250–1000 | 3.5–6(–9) | — | — |
| A. spelaeus | 16–30 | 6–26 | 15–32 | — | — | pale ochreous, ochreous | 2–3(–3.5), smooth | (5–)7–18(–23) | 400–1000, occasionally >1000 | 3–7(–9) | + | — |
| A. suttoniae | 24–25 | 20–22 | 24-25 | + | — | yellowish-white, ochreous | 2–3.5, smooth | (6–)12–17 | 180–420 | 4.5–6.5 | — | — |
| A. templicola | 21–32 | 21–28 | 23–30 | + | +3 | white, yellowish-white, pale ochreous | 2–3, smooth | (6–)9–23 | 120–1400 | 3.5–10 | + | — |
| A. urmiensis | 28–32 | 20–24 | 23–27 | + | — | white, ochreous | 2–3, smooth | (17–)20–23(−30) | (350–)700–850(−1330) | (5–)8–10(−12) | — | — |
"—" indicate no growth or absence of character/structure in culture.
Based on data from this study, Visagie et al. (2014), Hubka et al. (2015), Arzanlou et al. (2016), Siqueira et al. (2018), Crognale et al. (2019).
Production may vary between isolates and depend on cultivation conditions.
Very restricted growth (≤ 2 mm).
Conidia of all species are globose or subglobose, and only the longer dimension is given in case of subglobose conidia.
The colony colours of species from section Flavipedes are usually yellow, white, or in shades of brown. Two species newly described in this study are different in this regard. The colour of A. alboviridis colonies on some media is light green and A. inusitatus produces dark green colonies with yellow clumps of Hülle cells on all tested media. Aspergillus inusitatus is the only species with echinulate conidia in the whole section Flavipedes. In the study of Hubka et al. (2015), the production of accessory conidia has been observed in all section Flavipedes species. We expected to find them also in the newly described species, but they were only rarely present in some A. alboluteus strains and not observed in the other species. The sexual state has been only observed in A. neoflavipes and A. neoniveus, and it is produced in a homothallic manner. All other species from the section including all newly described species are presumably heterothallic and do not produce the sexual morph in culture under conditions used in this study.
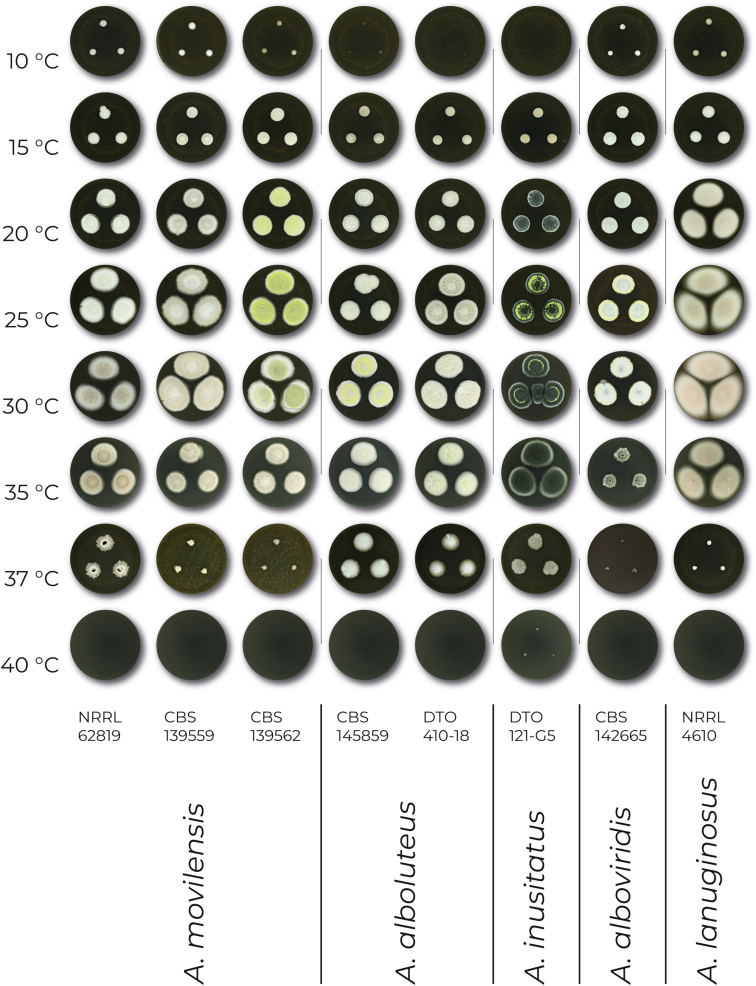
To support species hypotheses and proposal of new taxa related to A. movilensis, we determined cardinal temperatures, compared the macromorphology on five media and the micromorphology of particular strains. Cardinal temperatures were determined on MEA after 14 d of cultivation in darkness at eight different temperatures ranging from 10 °C to 40 °C. The resulting colonies are compared in Fig. 7, which demonstrates phylogenetic pattern in growth rates and abilities to grow at different temperatures. Aspergillus lanuginosus grows faster than any other species at 20, 25 and 30 °C. Aspergillus alboluteus and A. insitatus grow faster at 37 °C than the other three species and unlike the other three species they do not grow at 10 °C (or grow very restrictedly - isolate CCF 6201). Aspergillus inusitatus is the only species capable of growing at 40 °C. The colony texture of A. lanuginosus is cottony or downy on the majority of media due to the production of abundant aerial mycelium, while the colonies of the other species are rather floccose.
Fig. 7.
Temperature growth profile in the newly described species and Aspergillus movilensis after 14 d on MEA at temperatures ranging from 10 °C to 40 °C.
We observed morphological variation between the examined A. movilensis strains (Fig. 7). In general, white or grey colonies are produced, but the colonies of CBS 139562 are light green to yellow. We observed that colonies of some species change colour at suboptimal temperatures because of decreased sporulation (Fig. 7). There are differences between species in the ability to produce Hülle cells, which we observed in A. alboluteus, A. alboviridis, A. inusitatus, and A. movilensis, but they were not produced by A. lanuginosus. The length of stipes can be considered taxonomically important and may be used to distinguish A. alboviridis (mostly < 200 μm), A. movilensis (mostly < 400 μm), and A. lanuginosus (usually 600–1100 μm). Otherwise, the micromorphological characters were rather overlapping between the above-mentioned species.
Ecology
Based on the number of strains included in this study, A. micronesiensis, A. iizukae, A. spelaeus and A. templicola seem to be the most commonly encountered species. This is also in agreement with number of benA and CaM sequences deposited in GenBank for these species. In contrast to ITS region, the benA and CaM records for these species can be easily identified thanks to barcoding gap visible during BLAST analysis. The recorded numbers for benA /CaM loci in GenBank are as follows (accessed on June 10 2021) : A. micronesiensis 37 / 41, A. iizukae 24 / 23, A. spelaeus 18 / 18, and A. templicola 18 / 11.
The most common and diverse habitat for section Flavipedes members is undoubtedly the soil where 12 out of 17 species in our set of strains (excluding A. neoniveus) were found (Fig. 6). Our dataset contained five strains isolated from the hospital environment and six strains originating from clinical material. Only one of these strains belonged to the series Spelaei, specifically to A. spelaeus. The remaining strains were spread throughout the series Flavipedes, belonging to A. iizukae, A. suttoniae, A. templicola and A. micronesiensis. A significant number of species from both series originated from the indoor environment, namely A. iizukae, A. templicola, A. micronesiensis, A. polyporicola, A. spelaeus and A. alboluteus. In total three species, A. iizukae, A. spelaeus and A. alboviridis, were isolated from herbivore dung. Strains from food and feed were poorly represented in our dataset and restricted to A. iizukae, A. micronesiensis and A. spelaeus. None of the species which were represented by a high number of strains seem to be substrate specific.
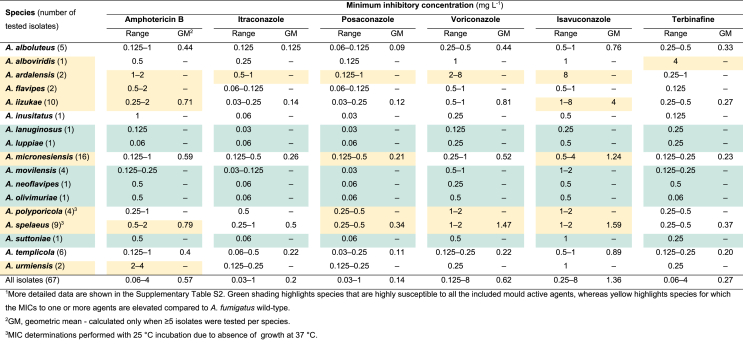
Antifungal susceptibility testing (EUCAST method)
The minimum inhibitory concentration (MIC) ranges and geometric mean (GM) values obtained by the EUCAST reference method for six antifungal agents are shown in Table 4 and more detailed results in Supplementary Table S2. Clinical breakpoints have been established for amphotericin B and the mould active azoles against A. fumigatus, which is the most common Aspergillus species in human infections. Clinical breakpoints have not been established for the Aspergillus section Flavipedes members. A general rule of thumb is that species that rarely cause disease in humans are less pathogenic, and therefore that adopting the breakpoints from the most common species in a genus for the rarer ones is clinically safe. With the caveat that some species were only represented with few strains, azole and amphotericin B MICs above the wild-type range for A. fumigatus were observed for A. ardalensis (all azoles and amphotericin B), A. micronesiensis (posaconazole and isavuconazole), A. polyporicola (posaconazole, voriconazole and isavuconazole), A. spelaeus (posaconazole, voriconazole, isavuconazole and amphotericin B), A. iizukae (isavuconazole and amphotericin B), and A. flavipes and A. urmiensis (amphotericin B) questioning the appropriateness of these drug bug combinations in clinical practice. In contrast, A. lanuginosus, A. luppiae, A. movilensis, A. neoflavipes, A. olivimuriae, and A. suttoniae were highly susceptible to amphotericin B and azoles. The MICs for the remaining species were comparable to those of A. fumigatus suggesting that these species are appropriate targets for the amphotericin B and azoles. Finally, terbinafine was active against all species except A. alboviridis.
Table 4.
Antifungal susceptibility profiles of Aspergillus section Flavipedes members determined with EUCAST E.Def.9.3 method at 37 °C.1
Taxonomy
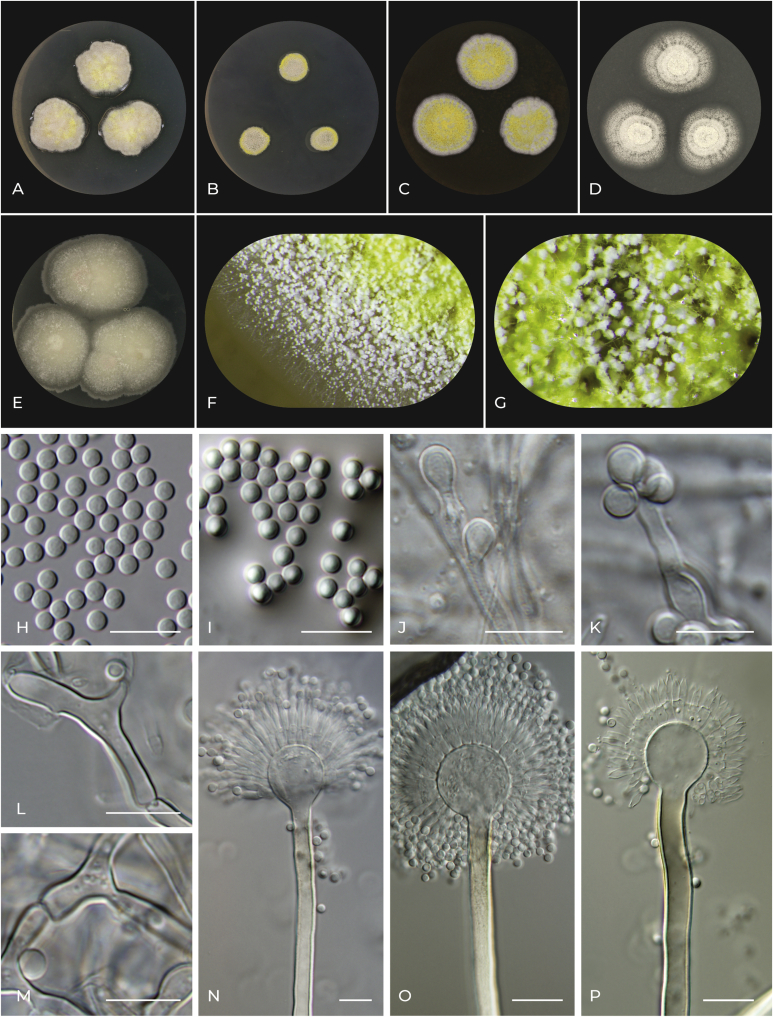
Aspergillus alboluteus F. Sklenar, Jurjević, Ezekiel, Houbraken & Hubka, sp. nov. MycoBank MB 839382. Fig. 8.
Fig. 8.
Macromorphology and micromorphology of Aspergillus alboluteus. A–E. Colonies after 14 d at 25 °C, left to right: CYA, CZA, MEA, OA and CY20S. F. Detail of colony edge on MEA. G. Detail of conidial heads and Hülle cells on MEA. H. Conidia. I. Conidia in air bubble. J, K. Accessory conidia. L, M. Hülle cells. N–P. Conidiophores. Scale bars: H–P = 10 μm.
Etymology: Named after white (sporulation) and yellow (clusters of Hülle cells) colours of the colonies on most media.
Typus: USA, Pennsylvania, Philadelphia, outdoor air, 2014, isolated by Ž. Jurjević (holotype PRM 952200, isotype PRM 952201, culture ex-type CBS 145855 = EMSL 2420 = CCF 5695 = IFM 66815).
Colony diam, 25 °C, 7 d (mm): CYA: 15–27; CZA: 10–20; MEA: 13–26; OA: 18–26; CY20S: 25–29.
Culture characteristics, 25 °C, 7 d: CYA: Colonies centrally raised; texture floccose; margin undulate to filiform; mycelial areas and sporulation white (#ffffff) to cream (#fffdd0) with icterine (#fcf75e) patches due to Hülle cell clumps; exudate absent; reverse centrally dark goldenrod (#b8860b) to light french beige (#c8ad7f), in margins dutch white (#f1ddb8). CZA: Colonies flat; texture floccose to granular; margin entire to delicately filiform; mycelial areas and sporulation white (#ffffff) with lemon yellow (#fff44f) patches due to Hülle cell clumps; exudate absent; reverse centrally flax (#eedc82), in margins naples yellow (#fada5e). MEA: Colonies slightly centrally raised; texture floccose to granular; margin entire to delicately filiform; mycelial areas and sporulation white (#ffffff) with lemon yellow (#fff44f) patches due to Hülle cell clumps; exudate absent; reverse centrally copper (#b87333) to saddle brown (#964b00), in margins bronze (#cd7f32). OA: Colonies flat to umbonate; texture granular; margin entire to delicately filiform; mycelial areas and sporulation white (#ffffff); exudate absent; reverse centrally khaki (#c3b091) to bone (#e3dac9) in margins. CY20S: Colonies flat to slightly centrally raised; texture floccose; margin slightly undulate to delicately filiform; mycelial areas linen (#faf0e6), sporulation white (#ffffff); exudate absent; reverse ecru (#c2b280).
Cardinal temperatures: Aspergillus alboluteus grows very restrictedly at 10 °C, and the optimum growth temperature is 30 °C. This species is able to grow at 37 °C but not at 40 °C (Fig. 7).
Micromorphology: Ascomata absent. Hülle cells present in strains CCF 5695 and CCF 4916 and absent in strains CCF 6201 and DTO 410-I8, elongated, branched, 20–30 μm long, forming yellow clumps. Conidial heads globose to radiate (remaining compact). Stipes smooth, brown (always hyaline under the vesicle), (500–)800–1 200(–2 000) × 4–7(–10) μm; vesicles hyaline, subglobose, (6–)9–17(–20) μm diam; metulae hyaline, cylindrical, 5–7 μm long, covering two thirds to entire surface of the vesicle; phialides hyaline, flask-shaped, 6–8 μm long. Conidia globose to subglobose, smooth, hyaline 2.5–3.5 (2.9 ± 0.1) × 2–2.5 (2.4 ± 0.1) μm. Accessory conidia absent or rare, globose to subglobose, on short, hyaline micro- to semimacronematous conidiophores.
Distinguishing characters: Aspergillus alboluteus is most closely related to A. inusitatus, but the latter is strikingly different from all related species by its green colonies and higher maximum growth temperature (40 °C). Phylogenetically, the next closest clade consists of A. alboviridis, A. lanuginosus and A. movilensis. Aspergillus alboluteus is phenotypically most similar to A. movilensis, that has similar colonies and also produces Hülle cells and accessory conidia with analogous morphology. However, these two species can be differentiated based on their conidiophore stipe lengths, vesicle diameters and colony sizes at 10 and 37 °C. The conidiophore stipes of A. movilensis rarely exceeds 400 μm, while stipes of A. alboluteus are (500–)800–1200(–2 000) μm long. The diameter of vesicles of A. movilensis rarely exceeds 13 μm, (5–)9–13(–16) μm, while vesicles of A. alboluteus are frequently larger, (6–)9–17(–20) μm. The colony diameters at 10 and 37 °C (on MEA, 14 d) slightly differ between these two species: at 10 °C, the colonies of A. movilensis attained 7 mm on average (the whole range was 6–8 mm), while colonies of A. alboluteus attained only 2 mm on average (1–2 mm); at 37 °C, A. alboluteus attained 18 mm on average (10–23 mm), while those of A. movilensis only 11 mm on average (6–19 mm).
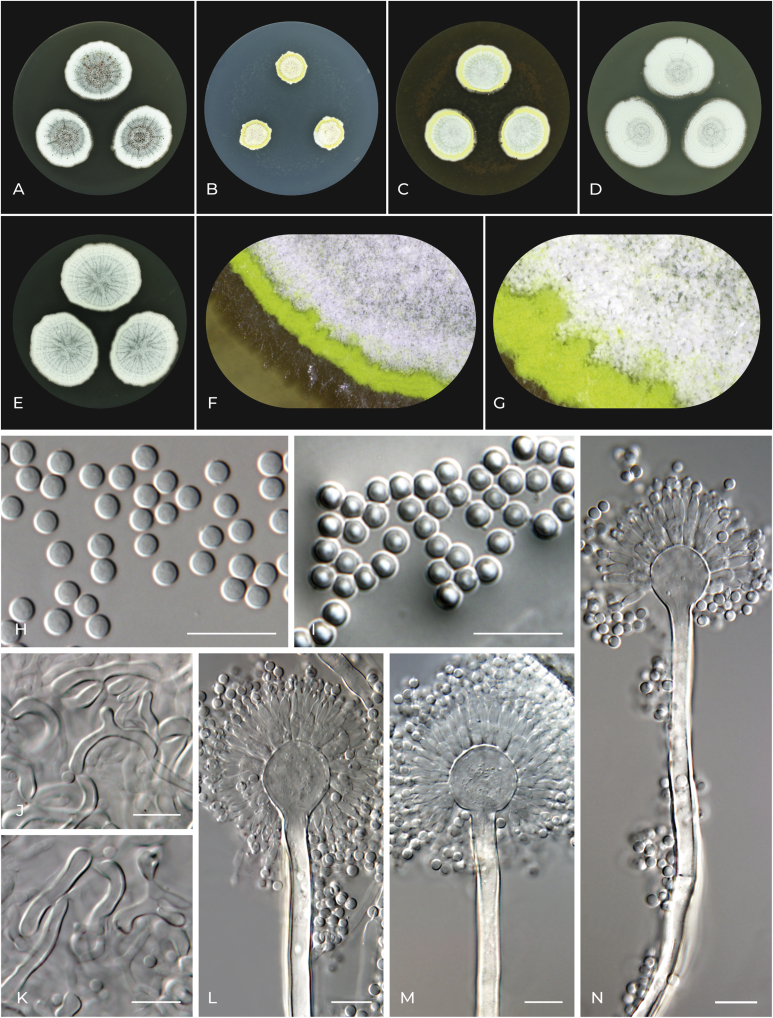
Aspergillus alboviridis J.P.Z. Siqueira, Gené, F. Sklenar & Hubka, sp. nov. MycoBank MB 821808. Fig. 9.
Fig. 9.
Macromorphology and micromorphology of Aspergillus alboviridis. A–E. Colonies after 14 d at 25 °C, left to right: CYA, CZA, MEA, OA and CY20S. F, G. Detail of colony edge, conidial heads and Hülle cells on MEA. H. Conidia. I. Conidia in air bubble. J, K. Hülle cells. L–N. Conidiophores. Scale bars: H–N = 10 μm.
Etymology: Refers to the white and green colony colour.
Typus: Spain, Balearic Islands, Mallorca, Pollença, herbivore dung, 2016, isolated by J. Gené and J.P.Z. Siqueira (holotype CBS H-23128, isotype PRM 954607, culture ex-type CBS 142665 = FMR 15175 = CCF 6049 = IFM 66819).
Colony diam, 25 °C, 7 d (mm): CYA: 19–21; CZA: 9–10; MEA: 18–23; OA: 18–19; CY20S: 23–24.
Culture characteristics, 25 °C, 7 d: CYA: Colonies centrally raised; texture floccose; margin entire; sporulation centrally green sheen (#6eaea1) to white (#ffffff) in margins; large clear droplets of exudate on the colony surface; reverse centrally gold metallic (#d4af37) to flax (#eedc82) in margins. CZA: Colonies convex; texture floccose; margin slightly undulate; sporulation centrally pale spring bud (#ecebbd) to white (#ffffff) in margins with canary (#ffff9a) circle close to margin formed by Hülle cells; clear droplets of exudate on the colony surface; reverse centrally satin sheen gold (#ce9d41) to gold crayola (#e6be8a), in margins dutch white (#f1ddb8) to bone (#e3dac9). MEA: Colonies flat to umbonate; texture floccose; margin entire to filiform; sporulation centrally cambridge blue (#a3c1ad) to white (#ffffff) in margins with canary (#ffff9a) circle close to margin formed by Hülle cells; exudate absent; reverse centrally saddle brown (#964b00) to ochre (#cc7722), in margins earth yellow (#e1a95f). OA: Colonies slightly umbonate; texture floccose to granular, margin entire to delicately filiform; sporulation cenrally middle blue green (#8dd9cc) to white (#ffffff); clear droplets of exudate on the surface in the colony center; reverse centrally liver chestnut (#987456) to satin sheen gold (#ce9d41), in margins flax (#eedc82). CY20S: Colonies centrally raised, wrinkled; texture centrally velutinous to floccose in margins due to conidial heads; margin slightly undulate to filiform; sporulation centrally polished pine (#5da493) to white (#ffffff) in margins; exudate absent; reverse centrally antique bronze (#665d1e) to vegas gold (#c5b358), in margins dutch white (#f1ddb8).
Cardinal temperatures: Aspergillus alboviridis grows restrictedly at 10 °C, and the optimum growth temperature is around 25–30 °C. This species is able to grow very restrictedly at 37 °C (Fig. 7).
Micromorphology: Ascomata absent. Hülle cells elongated, frequently curved and branched, 20–30 μm long, forming yellow clumps. Conidial heads globose to compactly columnar. Stipes smooth, hyaline or brown (always hyaline under the vesicle), 120–200(–550) × 4.5–6(–8) μm; vesicles hyaline, globose to subglobose, 12–16(–20) μm diam; metulae hyaline, cylindrical, 6–7(–9) μm long, covering three quarters to entire surface of the vesicle; phialides hyaline, flask-shaped, 6–7.5(–8.5) μm long. Conidia globose to subglobose, smooth, hyaline 2.5–3.5 (2.9 ± 0.1) × 2–3 (2.5 ± 0.1). Accessory conidia not observed.
Distinguishing characters: Aspergillus alboviridis is most closely related to A. lanuginosus and A. movilensis. The colony colour of A. alboviridis is on some media in shades of green, most prominently on CYA, but also on MEA, OA and CY20S. Green coloured sporulation is not observed in A. lanuginosus and A. movilensis. Aspergillus alboviridis produces Hülle cells unlike A. lanuginosus. In contrast to A. movilensis, production of accessory conidia was not observed in A. alboviridis.
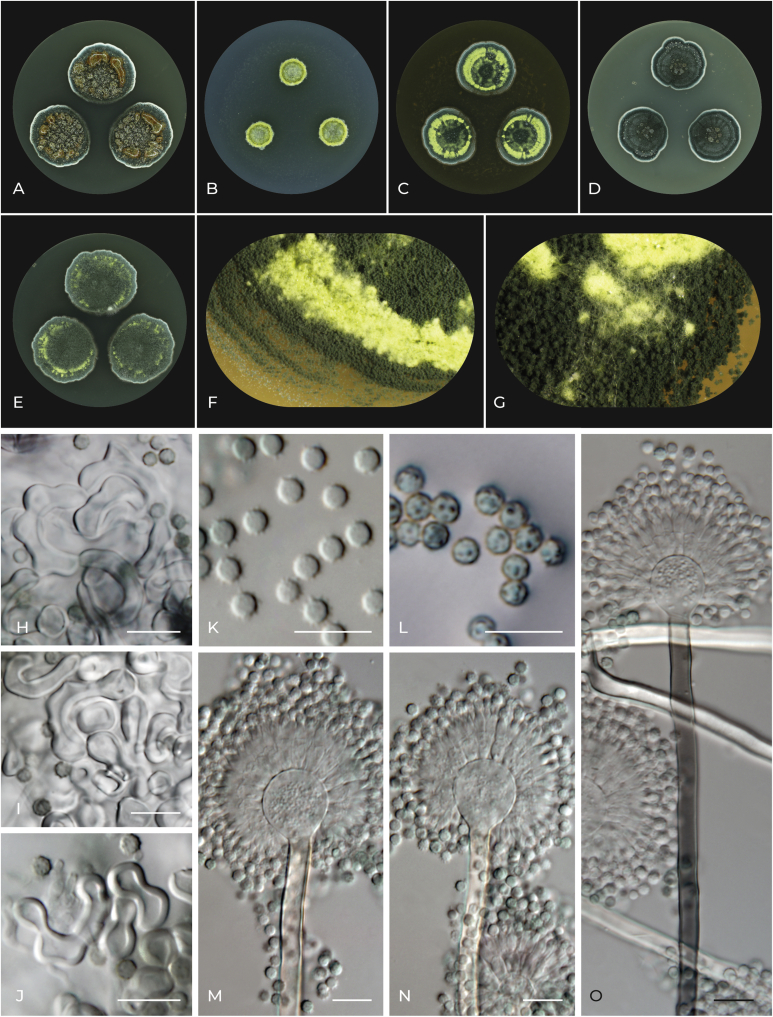
Aspergillus inusitatus F. Sklenar, C. Silva Pereira, Houbraken & Hubka, sp. nov. MycoBank MB 839383. Fig. 10.
Fig. 10.
Macromorphology and micromorphology of Aspergillus inusitatus. A–E. Colonies after 14 d at 25 °C, left to right: CYA, CZA, MEA, OA and CY20S. F. Detail of colony edge on MEA. G. Detail of conidial heads and Hülle cells on MEA. H–J. Hülle cells. K. Conidia. L. Conidia in air bubble. M–O. Conidiophores. Scale bars: H–O = 10 μm.
Etymology: Name refers to the strikingly different colony morphology in comparison with other members of section Flavipedes.
Typus: Tunisia, Ras Rajel, soil in oak forest, 2009, isolated by C. Silva Pereira (holotype PRM 954606, culture ex-type DTO 121-G5 = CBS 147044 = CCF 6552).
Colony diam, 25 °C, 7 d (mm): CYA: 22–23; CZA: 8–9; MEA: 17–19; OA: 17–18; CY20S: 25–26.
Culture characteristics, 25 °C, 7 d: CYA: Colonies flat, densely covered with exudate droplets; texture floccose; margin delicately undulate or delicately filiform; sporulation bottle green (#006a4e), in margins maximum blue green (#30bfbf) to white (#ffffff); large clear or golden brown (#996515) droplets of exudate on the entire surface of the colony, small clear droplets in margins; reverse centrally satin sheen gold (#cba135), in margins flax (#eedc82). CZA: Colonies umbonate with raised edge; texture floccose, cottony in central area; margin delicately undulate to filiform; sporulation middle blue green (#8dd9cc) to cadmium green (#006b3c) or white (#ffffff), canary (#ffff9a) ring at the colony edge formed by clumps of Hülle cells; exudate absent; reverse centrally olive green (#b5b35c), in margins vegas gold (#c5b358) to flax (#eedc82). MEA: Colonies centrally raised, richly permeated with clumps of Hülle cells; texture floccose; margin slightly undulate to delicately filiform; sporulation british racing green (#004225) to bottle green (#006a4e) to maximum blue green (#30bfbf) with clear boundaries between sectors, in margins middle blue green (#8dd9cc) to white (#ffffff), canary (#ffff9a) circle in the middle of the colony formed by Hülle cells; exudate absent; reverse centrally metallic sunburst (#9c7c38) to saddle brown (#964b00), in margins sage (#bcb88a). OA: Colonies centrally raised, densely covered with exudate droplets; texture floccose; margin entire to delicately filiform; sporulation cadmium green (#006b3c) to middle blue green (#8dd9cc), in margins deep jungle green (#004b49) to maximum blue green (#30bfbf) to white; clear droplets of exudate mainly in the colony center, scarcely on the edge; reverse centrally straw (#e4d96f) to artichoke (#8f9779), in margins sage (#bcb88a). CY20S: Colonies centrally raised; texture floccose with cottony patches; margin delicately undulate to delicately filiform; sporulation british racing green (#004225) to bottle green (#006a4e), in margins middle blue green (#8dd9cc) to white (#ffffff), canary (#ffff9a) circle close to the colony edge formed by Hülle cells; exudate absent; reverse centrally straw (#e4d96f) to vegas gold (#c5b358), in margins flax (#eedc82).
Cardinal temperatures: Aspergillus inusitatus grows at 15 °C but does not grow at 10 °C. The optimum growth temperature is 30–35 °C. This species is able to grow restrictedly at 40 °C but not at 42 °C (Fig. 7).
Micromorphology: Ascomata absent. Hülle cells elongated, frequently curved or branched, 20–30 μm long or subglobose to ovate, 9–12 × 8–10 μm, forming yellow clumps. Conidial heads compactly radiate. Stipes smooth, hyaline or dark brown (always hyaline under the vesicle), 250–600 × 5–6 μm; vesicles hyaline, subglobose to pyriform, 15–18 μm diam; metulae hyaline, cylindrical, 6.5–7.5 μm long, usually covering the entire surface of the vesicle; phialides hyaline, flask-shaped, 7.5–8.5 μm long. Conidia subglobose, echinulate, fern green (#4f7942), 3.5–4 (3.6 ± 0.2) × 3–3.5 (3.1 ± 0.2) μm. Accessory conidia not observed.
Distinguishing characters: Aspergillus inusitatus is most closely related to A. alboluteus. Phylogenetically, the next closest clade consists of A. alboviridis, A. lanuginosus and A. movilensis. Aspergillus inusitatus differs strikingly from all these species by its dark green colony colour. The optimum growth temperature of A. alboluteus, A. alboviridis, A. lanuginosus and A. movilensis is around 25–30 °C, while the optimum growth temperature of A. inusitatus is 30–35 °C. Unlike the four above-mentioned species, A. inusitatus is able to grow at 40 °C. Furthermore, unlike all other species in the section Flavipedes, A. inusitatus produces echinulate conidia.
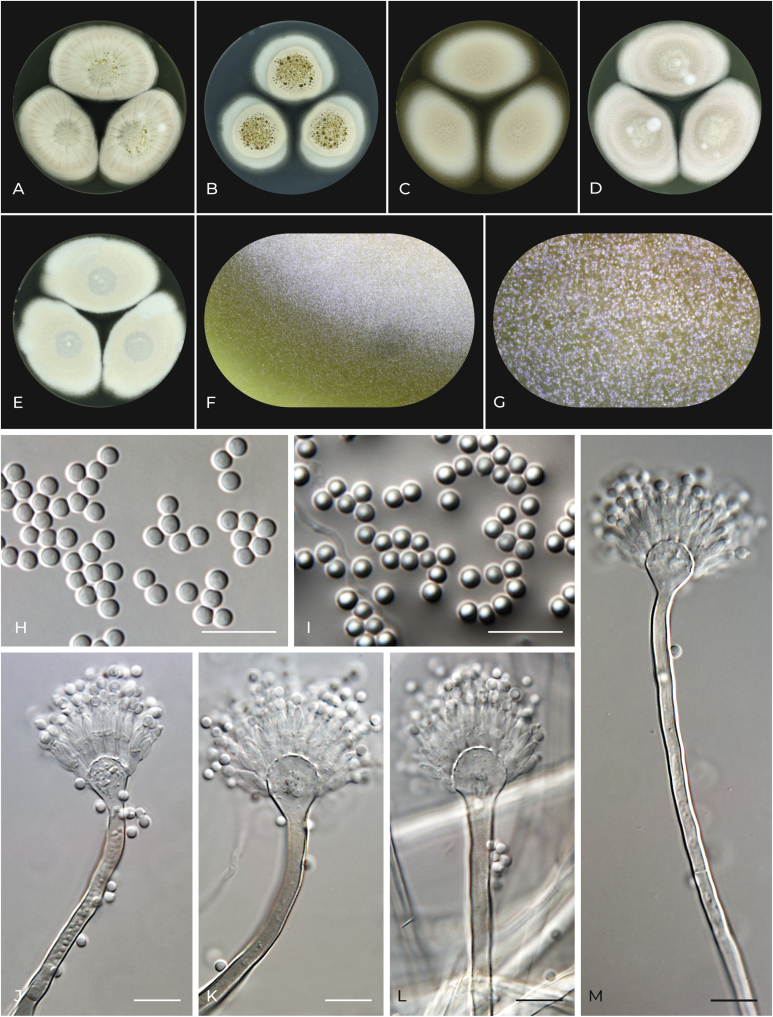
Aspergillus lanuginosus F. Sklenar & Hubka, sp. nov. MycoBank MB 839384. Fig. 11.
Fig. 11.
Macromorphology and micromorphology of Aspergillus lanuginosus. A–E. Colonies after 14 d at 25 °C, left to right: CYA, CZA, MEA, OA and CY20S. F. Detail of colony edge on MEA. G. Detail of conidial heads on MEA. H. Conidia. I. Conidia in air bubble. J–M. Conidiophores. Scale bars: H–M = 10 μm.
Etymology: Refers to the relatively rich production of aerial mycelium making the colonies downy on some media.
Typus: Haiti, Fonds Parisien, soil, unknown year of collection, unknown isolator (holotype PRM 954608, isotype PRM 954609, culture ex-type NRRL 4610 = IMI 350352 = CCF 4551 = IFM 66818).
Colony diam, 25 °C, 7 d (mm): CYA: 26–27; CZA: 21–22; MEA: 20–21; OA: 25–26; CY20S: 28–29.
Culture characteristics, 25 °C, 7 d: CYA: Colonies centrally raised; texture floccose; margin delicately filiform; sporulation centrally pale spring bud (#ecebbd) to beige (#f5f5dc) to champagne pink (#f1ddcf) in margins; clear droplets of exudate on the surface in the colony center; reverse centrally orange peel (ff9f00) to dutch white (f1ddb8) to gold crayola (e6be8a), in margins linen (#faf0e6). CZA: Colonies centrally raised; texture floccose; margin delicately filiform; sporulation centrally champagne pink (#f1ddcf), white (#ffffff) in margins; large amount of clear droplets of exudate on the surface in the colony center; reverse centrally camel (#c19a6b) to wheat (#f5deb3), in margins cream (#fffdd0). MEA: Colonies umbonate; texture cottony to floccose; margin delicately filiform; sporulation centrally champagne pink (#f1ddcf) to white (#ffffff) in margins; exudate absent; reverse centrally golden brown (#996515) to dark goldenrod (#b8860b), in margins maize crayola (#f2c649). OA: Colonies umbonate; texture floccose, cottony in the center; margin delicately filiform; sporulation centrally champagne pink (#f1ddcf) to white (#ffffff) in margins; exudate absent; reverse centrally antique bronze (#665d1e) to bistre brown (#967117), in margins flax (#eedc82). CY20S: Colonies flat to umbonate; texture floccose with small cottony patches in central areas; margin delicately filiform; sporulation centrally opal (#aac4c4) to linen (#faf0e6), in margins white (#ffffff); exudate absent; reverse centrally vegas gold (#c5b358) to bistre brown (#967117), in margins medium champagne (#f3e5ab).
Cardinal temperatures: Aspergillus lanuginosus grows at 10 °C, and the optimum growth temperature is 30 °C. This species is able to grow restrictedly at 37 °C but not at 40 °C (Fig. 7).
Micromorphology: Ascomata absent. Hülle cells absent. Conidial heads globose to radiate (remaining compact). Stipes smooth, hyaline or light brown (always hyaline under the vesicle), 600–1 100 × 3.5–4.5 μm; vesicles hyaline, subglobose to pyriform, 10–12 μm diam; metulae hyaline, cylindrical, 5–5.5 μm long, covering two thirds of the vesicle; phialides hyaline, flask-shaped, 5.5–6.5 μm long. Conidia globose to subglobose, smooth, hyaline 2.5–3 (2.7 ± 0.1) × 2–2.5 (2.3 ± 0.1) μm. Accessory conidia not observed.
Distinguishing characters: Aspergillus lanuginosus is most closely related to A. alboviridis and A. movilensis. In contrast to these two species, A. lanuginosus does not produce Hülle cells, its stipes are longer (A. lanuginosus 600–1 100 μm, A. alboviridis usually <200 μm, A. movilensis usually <400 μm) and the colony texture of A. lanuginosus on some media (MEA, OA, CY20S) is cottony, at least in the colony center, unlike the other species with mostly floccose colony texture.
Notes: The ex-type strain of A. lanuginosus NRRL 4610 was treated as A. carneus (section Terrei) by Raper & Fennell (1965). The phenotypic differences between ex-type strains of A. movilensis and A. lanuginosus were also observed by Hubka et al. (2015) who provisionally treated both strains as A. movilensis. These two strains (NRRL 4610 and NRRL 62819) also showed unique PCR fingerprinting pattern using the phage M13-core oligonucleotide primer and primer 834t (Hubka et al. 2015).
Aspergillus iizukae Sugiy., J. Fac. Sci. Univ. Tokyo, Sect. 3, Bot. 9: 390. 1967. MycoBank MB 326636.
Synonym: Aspergillus capensis Visagie et al., Stud. Mycol. 78: 105. 2014. MycoBank MB 809193.
Discussion
Species delimitation
In this study, we followed up on previous studies dealing with species delimitation in the genus Aspergillus (Sklenář et al. 2017, 2020, Hubka et al. 2018a, 2018b). The species delimitation step employing four single-locus and one multi-locus method was performed in the same way as in Sklenář et al. (2020). In contrast to our previous studies, here we presented the results of STACEY in the form of similarity matrices showing the assignments of individuals into species using several different values of the collapseheight parameter. This parameter is critical for the delimitation and its value is chosen arbitrarily. Our goal was to find the lowest value of the parameter which correctly delimits the species with indisputable species boundaries. These “reference species” differ from their close relatives clearly by phenotypic characters and/or were conclusively delimited by vast majority of single-locus delimitation methods. In the series Flavipedes, we considered A. ardalensis, A. flavipes, A. suttoniae, A. templicola, and A. urmiensis as these “reference species”. The results of species delimitation methods in series Spelaei were less stable and only A. luppiae and A. inusitatus were regarded as “reference species”. Then we checked how the results changed depending on the increase or decrease of the collapseheight parameter. For series Flavipedes, we concluded that a sensible value is 1 × 10-3. Using this value, all currently known species except A. capensis were supported, with A. templicola having some probability of splitting into two species (Fig. 3A). A higher value of the collapseheight parameter (5 × 10-3) resulted in excessive lumping, supporting only four species in the series (Supplementary Fig. S1A). When we used a lower value (1 × 10-4), A. templicola and A. micronesiensis were split into several species, but A. capensis still formed one species together with strains of A. iizukae (Supplementary Fig. S1B).
The same value of the collapseheight parameter (1 × 10-3) was applied on the series Spelaei. Using this value, STACEY supported all species related to A. movilensis, i.e., A. alboviridis, A. lanuginosus, A. movilensis, A. inusitatus and A. alboluteus (Fig. 3B). All species were also delimited when a slightly lower value (7.5 × 10-4) was used, but at the same time this value resulted in increased support for additional splitting of A. polyporicola and A. spelaeus (Supplementary Fig. S2B). On the other hand, slightly higher value (2 × 10-3) resulted in lumping of A. alboluteus with A. inusitatus, and lower support for delimitation of A. movilensis/A. alboviridis/A. lanuginosus, and also A. polyporicola/A. spelaeus as separate species (Supplementary Fig. S2A).
The four single-locus methods gave mostly similar results within the series Flavipedes. The incongruences in the results were present among strains of A. iizukae and A. capensis, which are morphologically indistinguishable and the latter was described solely based on phylogeny (Visagie et al. 2014). Especially the results of single-locus delimitation methods based on the benA gene together with STACEY results indicate that recombination is present in this clade. This fact is also evident in the single gene phylogenies (Fig. 2) proving that recognition of A. capensis is not supported by genealogical concordance phylogenetic species recognition concept (GCPSR) (Taylor et al. 2000).
The results of single-locus methods within series Spelaei were more variable. The clade consisting of A. spelaeus and A. polyporicola was delimited as one species by 5/12 methods, as two separate species by 5/12 methods, and as three species by 2/12 methods (Fig. 1B). Aspergillus movilensis clade consisting of A. alboviridis, A. lanuginosus, A. movilensis, A. inusitatus and A. alboluteus was delimited as one species by 1/12 methods, two species by 7/12 methods, five species by 3/12 methods, and six species 1/12 methods. In this ambiguous situation, our conclusions were significantly influenced by the presence of species-specific characters. Most importantly, A. inusitatus was conspicuously different compared to all other species within the section (Fig. 7, Table 3) and was also considered one of the “reference species” for STACEY method. In all cases where A. inusitatus was delimited as separate species, A. alboviridis and A. lanuginosus gained support.
The GMYC method is sometimes considered prone to over-splitting (Lohse 2009, Talavera et al. 2013), but it did not over-split the datasets in our previous study on the section Nidulantes (Sklenář et al. 2020), neither the dataset in this study. The method delimiting the highest number of species was bPTP and the most conservative method (resulting in the lowest number of delimited species) was bGMYC.
Species validation
For the species validation step, we employed the program DELINEATE, which is used for the first time to delimit species within the fungal kingdom. Until now, the most commonly used software for species validation was BPP (Bayesian Phylogenetics and Phylogeography) (Yang 2015). In the past studies on Aspergillus, this method was not very useful for distinguishing between different hypotheses on species boundaries, because its results are usually very benevolent, supporting even more splits than proposed by species delimitation methods. This excessive acceptance of splitting scenarios was also noted in other organisms and described by Sukumaran & Knowles (2017), who proposed the usage of the protracted speciation model by software for species validation. DELINEATE is the first program to challenge BPP as a choice for species validation.
In this study, BPP was only used as a tool for dividing the individuals from a given dataset into populations (Supplementary Table S1). The starBEAST was then used to calculate the “species tree” based on the populations. Finally, DELINEATE separated the populations from this tree into species. In contrast to BPP, at least some populations in DELINEATE need to be assigned to species (ideally those whose classification into species is clear) and others are left open to be delimited which offers opportunities to test a number of hypotheses. The limiting factor of the method are high computational demands that increase significantly with the number of unassigned populations. At the same time, a high total number of populations (unassigned and assigned into species) does not hinder the DELINEATE computation, but it can cause problems in starBEAST during species tree calculation.
Similarly to STACEY, where we applied several values of the collapseheight parameter, we generated several different models to be tested with DELINEATE. The results were again more complex in the series Spelaei, where we tested 14 models compared to only four models in series Flavipedes. The analysis of the model with all species in A. movilensis clade unassigned was not computationally feasible. Therefore, we had to generate separate models with different parts of the clade being unassigned (Fig. 4B, Fig. 5). When we left A. alboviridis, A. lanuginosus and A. inusitatus unassigned, as a result, they were lumped with their sister species. When A. inusitatus was defined as a separate species, A. alboviridis and A. lanuginosus were also separated from A. movilensis. Analogously, when A. alboviridis and A. lanuginosus were designated as separate species, A. inusitatus was also separated from A. alboluteus. Both parts of the analysis showed mixed support for the delimitation of A. inusitatus as separate species. But if A. inusitatus was designated as separate species, A. alboviridis and A. lanuginosus were consistently delimited as separate species too.
In the series Flavipedes, we mainly focused on the validation of species limits in the clade containing A. iizukae and A. capensis (Fig. 4A, Fig. 5). The results of all models were consistent. Aspergillus capensis was always lumped together with A. iizukae. Therefore, we consider A. capensis as a synonym of A. iizukae. Aspergillus templicola and A. micronesiensis were always delimited in the same way as suggested by majority of the species delimitation methods.
Furthermore, seven models of the whole section were generated to be tested in DELINEATE (Fig. 5). The results were congruent with the analyses performed separately in both series with one exception. The fourth model lumped A. inusitatus with A. alboluteus even though A. alboviridis and A. lanuginosus were specified as separate species. Based on our study, DELINEATE proved to give more relevant results which were more in agreement with putative species delimited in the first step of analysis compared to very benevolent BPP.
Similarly to our previous studies, we could not fully comply with the assumption of the species delimitation methods, i.e. not including species without variability (singleton species). It is a common issue in this kind of analyses and it has been discussed before (Ahrens et al. 2016). Singleton species are frequently described within genus Aspergillus with the general idea of the endeavor to describe all discovered diversity. The possibility of the singleton species (A. alboviridis, A. lanuginosus and A. inusitatus) being part of other closely related species was tested, but the results supported the splitting scenarios.
In this study, we enriched the analyses by addition of new strains and thus variability, resulting in more robust and accurate knowledge of the species boundaries in comparison with the previous studies on the section Flavipedes. In general, the data supported currently known species and in one case demonstrated recombination leading to synonymization of A. capensis with A. iizukae. A similar case occurred for instance in A. parafelis and A. pseudofelis (section Fumigati) which were described based on several strains with very limited genetic variability (Sugui et al. 2014) and later put into synonymy with A. felis when higher number of strains was available for analyses (Hubka et al. 2018a). The support for delimitation of A. polyporicola and A. spelaeus as separate species was also ambiguous (Fig. 1B). These two species were proposed by Hubka et al. (2015) based on the differences in their ecology and massive production of accessory conidia by A. polyporicola in contrast to A. spelaeus. In this study, we obtained new strains demonstrating that the ecological factor is not relevant as a taxonomic character for A. polyporicola, which was considered by Hubka et al. (2015) as a fungicolous species. We also examined the presence and abundance of accessory conidia in a broader set of A. spelaeus and A. polyporicola strains and we did not find any relevant differences between these two species – accessory conidia were rather rare or absent in all newly examined strains. Aspergillus polyporicola and A. spelaeus can be therefore considered cryptic species and there is a significant chance for their synonymization in the future when a larger dataset will be available in terms of the number of strains or loci. But in the present study, the splitting scenario gained overall higher support. There is also no conflict in the topology of single-gene phylogenies and both species are supported by GCPSR (data not shown).
Incorporating multispecies coalescent model-based methods into taxonomy of Aspergillus
The polyphasic approach is currently a standard for species delimitation in Aspergillus. It always involves the assessment of phylogenetic data, with the GCPSR most commonly claimed to be used as a method of choice for delineation of species boundaries. It is however seldomly used methodologically properly as originally described by Dettman et al. (2003). Using the methods employed in this study, the final decision regarding the species boundaries remains partly subjective as other components of the polyphasic approach are still included in the decision-making process. The species delimitation part based on molecular data is however largely free from subjectivity and the methods force the taxonomists to take a fresh and unbiased look at the species limits. Furthermore, these coalescent-based methods are at least to some degree able to accommodate phenomena such as incomplete lineage sorting, recombination, or non-reciprocal monophyly (Edwards 2009). We do not expect or encourage these methods to replace the polyphasic approach, but they should enhance and strengthen it.
Several studies including this study highlighted the fact that the intraspecific genetic variability in Aspergillus is probably higher than expected and that species range sizes are unequal across aspergilli (Geiser et al. 2007, Hubka et al. 2018a). Consequently, it can be expected that the species numbers in some extensively studied groups with a narrow species concept will be reduced in the future. The resulting taxonomy should become clearer, which is demanded by user community given the number of studies (from clinical, biotechnological, or industrial background) struggling with the identification of cryptic Aspergillus species in the various species complexes (Negri et al. 2014, Pantelides et al. 2017, D'hooge et al. 2019, Imbert et al. 2019, Mincuzzi et al. 2020).
The ultimate future goal is to bring the majority of species in Aspergillus to the same level from the population genetics standpoint, which will certainly result in a delimitation of new species and synonymization of some others, with the synonymization being more likely in species groups which have been extensively studied and have a narrower species concept. An important prerequisite for these steps is a shift of taxonomy from a discovery phase, where species are described based on a small number of strains, to a phase where species limits are assessed based on large numbers of strains from different localities. It is already possible to obtain comprehensive datasets from databases for some species or species groups such as A. fumigatus, series Nigri, Versicolores, Flavi or others which can be re-analyzed using similar approaches.
Another important factor relevant for species concept in Aspergillus is the inevitable advent of species delimitation based on whole-genome sequences (Matute & Sepúlveda 2019). In this aspect, it is crucial not to exploit this vast source of genetic variability to flood the genus with cryptic species most likely corresponding to natural populations of a broader species, but to utilize it to learn more about the mechanisms of species boundaries formation and preservation, eventually leading to the establishment of meaningful rules for species delimitation (biologically meaningful species concept).
Ecology and clinical relevance
The ecology and clinical relevance of section Flavipedes members has been extensively reviewed by Hubka et al. (2015). The members of the section are distributed worldwide in soil, especially in tropical and subtropical regions (Klich 2002, Domsch et al. 2007, Choochuay et al. 2017). Some representatives are osmotolerant and xerophilic and they are frequently isolated from arid or saline environments, e.g. sea sand (Lee et al. 2016), saline soil (Kang et al. 2018) or salterns (Chung et al. 2019). There are also several reports on species adopting life strategy of plant endophytes (Luyen et al. 2019, Qin et al. 2019, El-Hawary et al. 2020). Other studies report another type of symbiosis of these fungi, namely with marine animals, sea cucumbers (Nerva et al. 2019) and marine sponges (Wiese et al. 2011). The same authors, Nerva et al. (2019), also reported that A. spelaeus harbors mycovirus Aspergillus spelaeus polymycovirus 1 (AsPMV1). Another habitat where section Flavipedes members are present is the cave environment. Numerous A. spelaeus strains used in this study originated from cave air or sediment and there are also reports of A. iizukae and A. movilensis isolated from caves (Hubka et al. 2015, Nováková et al. 2018). Similarly to many other Aspergilli, species from section Flavipedes spoil various food and feed (Pitt & Hocking 2009) and are frequently isolated from the indoor environment (Visagie et al. 2014, Sánchez Espinosa et al. 2021, this study). Even though hospitals can be also considered indoor environments, we highlighted these strains separately in Fig. 6, because some species have the potential to be pathogenic and hospital environment monitoring has usually a different purpose than monitoring indoor fungi in other buildings.
Some section Flavipedes members are uncommon human and animal pathogens. The majority of well documented cases of aspergillosis were attributed to A. flavipes and comprise diverse clinical manifestation including cutaneous aspergillosis (Barson & Ruymann 1986), onychomycosis (Gehlot et al. 2011), otomycosis (Stuart & Blank 1955), osteomyelitis (Roselle & Baird 1979), diskospondylitis (Schultz et al. 2008), pulmonary aspergillosis (Katou et al. 1999) and cerebral aspergillosis (Masih et al. 2016). However, the species identification of these strains according to the current taxonomy is mostly unclear except for the isolate associated with onychomycosis (GenBank EU515154) which represents A. micronesiensis. Even though there are sequences available for the strain VPCI 631/P/15 (GenBank KX455808, KX455766), a cause of cerebral aspergillosis (Masih et al. 2016), their information is contradictory. The benA sequences represent A. flavipes s. str., while the CaM sequence represents A. micronesiensis. These two species are phylogenetically distant (Fig. 6) and one of these sequences was therefore most likely incorrectly deposited. Apart from mentioned cases, A. micronesiensis was also reported by Siqueira et al. (2018) from canine urine sample. The same authors also reported A. iizukae and A. suttoniae from human sputum and bronchoalveolar lavage, respectively, and A. spelaeus from human forearm. However, detailed information about clinical relevance of these isolates was not included. In this study, we examined two strains (IHEM 22505 and IHEM 22506) originally identified as A. flavipes from human sputum and lung, respectively, which were reidentified as A. micronesiensis. Both strains were isolated from patients in Belgium and preserved in BCCM/IHEM collection. Another clinical strain examined here was obtained from bronchoalveolar lavage of a Danish patient and was identified as A. templicola. The available anamnestic data for these cases are limited and do not allow confirmation of the etiological relevance of strains.
The antifungal susceptibility patterns in section Flavipedes are largely unknown and restricted to few clinical isolates without clear species identification (A. flavipes s. l.) and examined by various methods (Del Carmen Serrano et al. 2003, Martin-Mazuelos et al. 2003, Masih et al. 2016). In this study, we evaluated antifungal susceptibilities to six antifungal agents in a large set of reliably identified strains across section Flavipedes. Our results showed notable differences in susceptibility pattern between the species as the MIC distributions for itraconazole, posaconazole, and isavuconazole spanned six two-fold dilutions and those for amphotericin B, voriconazole and terbinafine spanned seven two-fold dilutions across the 67 strains. In general, species-specific MIC distributions for wild type strains span three two-fold dilutions when tested in a single laboratory with a single batch of plates. Therefore, the difference in susceptibility among the species included herein cannot be explained by inherent variation of the test but must reflect intrinsic differences in susceptibility among the members of the Aspergillus section Flavipedes. About half of the species were susceptible to all agents whereas the remaining species were resistant to one or several agents. Therefore, species identification and susceptibility testing is important in clinical infections.
Conclusions
The revision of section Flavipedes led to an increase of its known variability. We performed phylogenetic and phenotypic analyses utilizing strains from previous studies and newly isolated strains. As a result, four species were newly described and one species, A. capensis, was put in synonymy. So far, the species from the series have been known to produce white, yellow, and brown colonies. The newly described species extend this heterogeneity, namely, the A. inusitatus deviates significantly by its dark green colonies and also colonies of A. alboviridis are light green on some media. The species delimitation analysis performed here builds upon recent studies on the genus Aspergillus and expands the spectrum of methods in the species validation step. Antifungal susceptibility across species diversity of the section Flavipedes showed elevated MICs to amphotericin B or azole derivates in significant part of the tested species, especially in those that are potentially clinically relevant. This finding emphasizes the need for species identification and susceptibility testing in clinical strains.
Acknowledgements
The project was supported by Czech Ministry of Health (grant NU21-05-00681) and the Charles University Research Centre program no. 204069. František Sklenář was supported by the project of Charles University Grant Agency (GAUK 140520). We are grateful to Jan Karhan for the help with graphical adjustments of analysis outputs. We thank Milada Chudíčková and Lenka Zídková for their invaluable assistance in the laboratory. Vit Hubka is grateful for the support from the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research in Japan (Standard). This study was partially supported by the Grant-in-aid for JSPS research fellow (No. 20F20772).
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2021.100120.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahrens D., Fujisawa T., Krammer H.J., et al. Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology. 2016;65:478–494. doi: 10.1093/sysbio/syw002. [DOI] [PubMed] [Google Scholar]
- Al-Dhabaan F.A. Mycoremediation of crude oil contaminated soil by specific fungi isolated from Dhahran in Saudi Arabia. Saudi Journal of Biological Sciences. 2021;28:73–77. doi: 10.1016/j.sjbs.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Jørgensen K.M., Hanemaaijer N., et al. ISO standard 20776-1 or serial 2-fold dilution for antifungal susceptibility plate preparation: that is the question! Journal of Antimicrobial Chemotherapy. 2021;76:1793–1799. doi: 10.1093/jac/dkab088. [DOI] [PubMed] [Google Scholar]
- Arzanlou M., Samadi R., Frisvad J.C., et al. Two novel Aspergillus species from hypersaline soils of The National Park of Lake Urmia, Iran. Mycological Progress. 2016;15:1081–1092. [Google Scholar]
- Barson W.J., Ruymann F.B. Palmar aspergillosis in immunocompromised children. Pediatric Infectious Disease. 1986;5:264–268. doi: 10.1097/00006454-198603000-00021. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Heled J., Kuhnert D., et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10 doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen Serrano M Del, Valverde-Conde A., Chávez M., et al. In vitro activity of voriconazole, itraconazole, caspofungin, anidulafungin (VER002, LY303366) and amphotericin B against Aspergillus spp. Diagnostic Microbiology and Infectious Disease. 2003;45:131–135. doi: 10.1016/s0732-8893(02)00507-2. [DOI] [PubMed] [Google Scholar]
- Chen A.J., Hubka V., Frisvad J.C., et al. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology. 2017;88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choochuay J., Xu X., Rukachaisirikul V., et al. Curvularin derivatives from the soil-derived fungus Aspergillus polyporicola PSU-RSPG187. Phytochemistry Letters. 2017;22:122–127. [Google Scholar]
- Chung D., Kim H., Choi H.S. Fungi in salterns. Journal of Microbiology. 2019;57:717–724. doi: 10.1007/s12275-019-9195-3. [DOI] [PubMed] [Google Scholar]
- Crognale S., Pesciaroli L., Felli M., et al. Aspergillus olivimuriaesp. nov., a halotolerant species isolated from olive brine. International Journal of Systematic and Evolutionary Microbiology. 2019;69:2899–2906. doi: 10.1099/ijsem.0.003575. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A., et al. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Dettman J.R., Jacobson D.J., Taylor J.W. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003;57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- D’hooge E., Becker P., Stubbe D., et al. Black aspergilli: a remaining challenge in fungal taxonomy? Medical Mycology. 2019;57:773–780. doi: 10.1093/mmy/myy124. [DOI] [PubMed] [Google Scholar]
- Domsch K.H., Gams W., Anderson T.-H. IHW-Verlag. Eching; Germany: 2007. Compendium of soil fungi. [Google Scholar]
- Edwards S.V. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- El-Elimat T., Raja H.A., Graf T.N., et al. Flavonolignans from Aspergillus iizukae, a fungal endophyte of milk thistle (Silybum marianum) Journal of Natural Products. 2014;77:193–199. doi: 10.1021/np400955q. [DOI] [PubMed] [Google Scholar]
- El-Hawary S.S., Moawad A.S., Bahr H.S., et al. Natural product diversity from the endophytic fungi of the genus Aspergillus. RSC Advances. 2020;10:22058–22079. doi: 10.1039/d0ra04290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A.S.A., Ali G.S. Aspergillus flavipes is a novel efficient biocontrol agent of Phytophthora parasitica. Biological Control. 2020;140:104072. [Google Scholar]
- Frisvad J.C., Larsen T.O. Chemodiversity in the genus Aspergillus. Applied Microbiology and Biotechnology. 2015;99:7859–7877. doi: 10.1007/s00253-015-6839-z. [DOI] [PubMed] [Google Scholar]
- Fujisawa T., Barraclough T.G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Systematic Biology. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W., Christensen M., Onions A.H.S., et al. In: Advances in Penicillium and Aspergillus Systematics. Samson R.A., Pitt J.I., editors. Plenum Press; New York: 1985. Infrageneric taxa of Aspergillus; pp. 55–62. [Google Scholar]
- Gehlot P., Purohit D.K., Singh S.K. Molecular diagnostics of human pathogenic Aspergillus species. Indian Journal of Biotechnology. 2011;10:207–211. [Google Scholar]
- Geiser D.M., Klich M.A., Frisvad J.C., et al. The current status of species recognition and identification in Aspergillus. Studies in Mycology. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhill A.R., Blaney B.J., Shipton W.A., et al. Mycotoxins and toxigenic fungi in sago starch from Papua New Guinea. Letters in Applied Microbiology. 2008;47:342–347. doi: 10.1111/j.1472-765X.2008.02439.x. [DOI] [PubMed] [Google Scholar]
- Hall T.A. Nucleic Acids Symposium Series. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- Heled J., Drummond A.J. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-B., Cho H.-S., Shin H.-D., et al. Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Kocsubé S., Visagie C.M., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Barrs V., Dudová Z., et al. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia. 2018;41:142–174. doi: 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kolařík M. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specifity testing and taxonomical consequences. Persoonia. 2012;29:1–10. doi: 10.3767/003158512X658123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Jurjević Ž., et al. Polyphasic data support the splitting of Aspergillus candidus into two species; proposal of A. dobrogensis sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2018;68:995–1011. doi: 10.1099/ijsem.0.002583. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Kolařík M., et al. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Janisect. nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- Imbert S., Normand A.C., Gabriel F., et al. Multi-centric evaluation of the online MSI platform for the identification of cryptic and rare species of Aspergillus by MALDI-TOF. Medical Mycology. 2019;57:962–968. doi: 10.1093/mmy/myz004. [DOI] [PubMed] [Google Scholar]
- Jones G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology. 2017;74:447–467. doi: 10.1007/s00285-016-1034-0. [DOI] [PubMed] [Google Scholar]
- Jurjević Ž., Kubátová A., Kolařík M., et al. Taxonomy of Aspergillus section Petersoniisect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution. 2015;301:2441–2462. [Google Scholar]
- Kang H.-H., Zhang H.-B., Zhong M.-J., et al. Potential antiviral xanthones from a coastal saline soil fungus Aspergillus iizukae. Marine Drugs. 2018;16:449. doi: 10.3390/md16110449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou K., Nanjou K., Kawakami M. A case of chronic necrotizing pulmonary aspergillosis due to Aspergillus flavipes associated with pulmonary fibrosis. Journal of the Japanese Respiratory Society. 1999;37:938–942. [PubMed] [Google Scholar]
- Klich M.A. Biogeography of Aspergillus species in soil and litter. Mycologia. 2002;94:21–27. [PubMed] [Google Scholar]
- Kocsubé S., Perrone G., Magistà D., et al. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Park M.S., Lim Y.W. Diversity of marine-derived Aspergillus from tidal mudflats and sea sand in Korea. Mycobiology. 2016;44:237–247. doi: 10.5941/MYCO.2016.44.4.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lohse K. Can mtDNA barcodes be used to delimit species? A response to Pons et al. (2006) Systematic Biology. 2009;58:439–442. doi: 10.1093/sysbio/syp039. [DOI] [PubMed] [Google Scholar]
- Luyen N.D., Huong L.M., Thi Hong Ha T., et al. Aspermicrones A-C, novel dibenzospiroketals from the seaweed-derived endophytic fungus Aspergillus micronesiensis. Journal of Antibiotics. 2019;72:843–847. doi: 10.1038/s41429-019-0214-8. [DOI] [PubMed] [Google Scholar]
- Martin-Mazuelos E., Pemán J., Valverde A., et al. Comparison of the Sensititre YeastOne colorimetric antifungal panel and Etest with the NCCLS M38-A method to determine the activity of amphotericin B and itraconazole against clinical isolates of Aspergillus spp. Journal of Antimicrobial Chemotherapy. 2003;52:365–370. doi: 10.1093/jac/dkg384. [DOI] [PubMed] [Google Scholar]
- Masih A., Singh P.K., Kathuria S., et al. Clinically significant rare Aspergillus species in a referral chest hospital, Delhi, India: molecular and MALDI TOF identification and their antifungal susceptibility profiles. Journal of Clinical Microbiology. 2016;54:2354–2364. doi: 10.1128/JCM.00962-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D.R., Sepúlveda V.E. Fungal species boundaries in the genomics era. Fungal Genetics and Biology. 2019;131:103249. doi: 10.1016/j.fgb.2019.103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincuzzi A., Ippolito A., Montemurro C., et al. Characterization of Penicilliums.s. and Aspergillus sect. Nigri causing postharvest rots of pomegranate fruit in Southern Italy. International Journal of Food Microbiology. 2020;314:108389. doi: 10.1016/j.ijfoodmicro.2019.108389. [DOI] [PubMed] [Google Scholar]
- Negri C., Gonçalves S., Xafranski H., et al. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. Journal of Clinical Microbiology. 2014;52:3633–3640. doi: 10.1128/JCM.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerva L., Forgia M., Ciuffo M., et al. The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Research. 2019;273:197737. doi: 10.1016/j.virusres.2019.197737. [DOI] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H.A., Haeseler A von, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman E., Al-Gheethi A.A., Talip B.A., et al. Oxidative enzymes from newly local strain Aspergillus iizukae EAN605 using pumpkin peels as a production substrate: optimized production, characterization, application and techno-economic analysis. Journal of Hazardous Materials. 2020;386:121954. doi: 10.1016/j.jhazmat.2019.121954. [DOI] [PubMed] [Google Scholar]
- Nováková A., Hubka V., Valinová Š., et al. Cultivable microscopic fungi from an underground chemosynthesis-based ecosystem: a preliminary study. Folia Microbiologica. 2018;63:43–55. doi: 10.1007/s12223-017-0527-6. [DOI] [PubMed] [Google Scholar]
- O'Donnell K. In: The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. Reynolds D.R., Taylor J.W., editors. CAB International; Wallingford: 1993. Fusarium and its near relatives; pp. 225–236. [Google Scholar]
- O'Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Pantelides I.S., Aristeidou E., Lazari M., et al. Biodiversity and ochratoxin A profile of Aspergillus section Nigri populations isolated from wine grapes in Cyprus vineyards. Food Microbiology. 2017;67:106–115. doi: 10.1016/j.fm.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Hocking A.D. Springer; London, UK: 2009. Fungi and food spoilage. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Qin J., Lyu A., Zhang Q., et al. Strain identification and metabolites isolation of Aspergillus capensis CanS-34A from Brassica napus. Molecular Biology Reports. 2019;46:3451–3460. doi: 10.1007/s11033-019-04808-5. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: a language and environment for statistical computing. [Google Scholar]
- Raper K.B., Fennell D.I. Williams & Wilkins; Baltimore, MD, USA: 1965. The genus Aspergillus. [Google Scholar]
- Reid N.M., Carstens B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evolutionary Biology. 2012;12:196. doi: 10.1186/1471-2148-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Mark P van der, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselle G.A., Baird I.M. Aspergillus flavipes group osteomyelitis. Archives of Internal Medicine. 1979;139:590–592. [PubMed] [Google Scholar]
- Rossum G van, Drake F.L. Python Software Foundation; 2019. Python language reference, version 3. [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J., et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Espinosa K.C., Almaguer Chávez M., Duarte-Escalante E., et al. Phylogenetic identification, diversity, and richness of Aspergillus from homes in Havana, Cuba. Microorganisms. 2021;9:115. doi: 10.3390/microorganisms9010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R.M., Johnson E.G., Wisner E.R., et al. Clinicopathologic and diagnostic imaging characteristics of systemic aspergillosis in 30 dogs. Journal of Veterinary Internal Medicine. 2008;22:851–859. doi: 10.1111/j.1939-1676.2008.0125.x. [DOI] [PubMed] [Google Scholar]
- Siqueira J.P.Z., Wiederhold N., Gené J., et al. Cryptic Aspergillus from clinical samples in the USA and description of a new species in section Flavipedes. Mycoses. 2018;61:814–825. doi: 10.1111/myc.12818. [DOI] [PubMed] [Google Scholar]
- Sklenář F., Ž Jurjević, Peterson S.W., et al. Increasing the species diversity in the Aspergillus section Nidulantes: six novel species mainly from the indoor environment. Mycologia. 2020;112:342–370. doi: 10.1080/00275514.2019.1698923. [DOI] [PubMed] [Google Scholar]
- Sklenář F., Ž Jurjević, Zalar P., et al. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology. 2017;88:161–236. doi: 10.1016/j.simyco.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart E.A., Blank F. Aspergillosis of the ear; a report of twenty-nine cases. Canadian Medical Association Journal. 1955;72:334–337. [PMC free article] [PubMed] [Google Scholar]
- Sugui J.A., Peterson S.W., Figat A., et al. Genetic relatedness versus biological compatibility between Aspergillus fumigatus and related species. Journal of Clinical Microbiology. 2014;52:3707–3721. doi: 10.1128/JCM.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J., Knowles L.L. Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:1607–1611. doi: 10.1073/pnas.1607921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sukumaran J., Holder M.T., Knowles L.L. Incorporating the speciation process into species delimitation. PLoS Computational Biology. 2021;17:e1008924. doi: 10.1371/journal.pcbi.1008924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Dincă V., Vila R. Factors affecting species delimitations with the GMYC model: insights from a butterfly survey. Methods in Ecology and Evolution. 2013;4:1101–1110. [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S., et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Thom C., Church M.B. Williams & Wilkins; Baltimore, MD, USA: 1926. The Aspergilli. [Google Scholar]
- Tuomi T., Reijula K., Johnsson T., et al. Mycotoxins in crude building materials from water-damaged buildings. Applied and Environmental Microbiology. 2000;66:1899–1904. doi: 10.1128/aem.66.5.1899-1904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera H.R., Gomes J., Lakshmi S., et al. Lovastatin production by solid state fermentation using Aspergillus flavipes. Enzyme and Microbial Technology. 2005;37:521–526. [Google Scholar]
- Visagie C.M., Hirooka Y., Tanney J.B., et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T.D., Lee S.B., et al. In: PCR–Protocols and Applications–a Laboratory Manual. Innis M., Gelfand G., Sninsky J., White T., editors. Academic Press; San Diego: 1990. Amplification and direct sequencing of ribosomal RNA genes and the internal transcribed spacer in fungi; pp. 315–322. [Google Scholar]
- Wiese J., Ohlendorf B., Blümel M., et al. Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Marine Drugs. 2011;9:561–585. doi: 10.3390/md9040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. The BPP program for species tree estimation and species delimitation. Current Zoology. 2015;61:854–865. [Google Scholar]
- Zhang J., Kapli P., Pavlidis P., et al. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohri A.-N.A., Al-Bedak O. Aspergillus sakultaensis, a new species in section Flavipedes isolated from Sohag Governorate, Egypt. Journal of Environmental Studies. 2020;20:21–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.