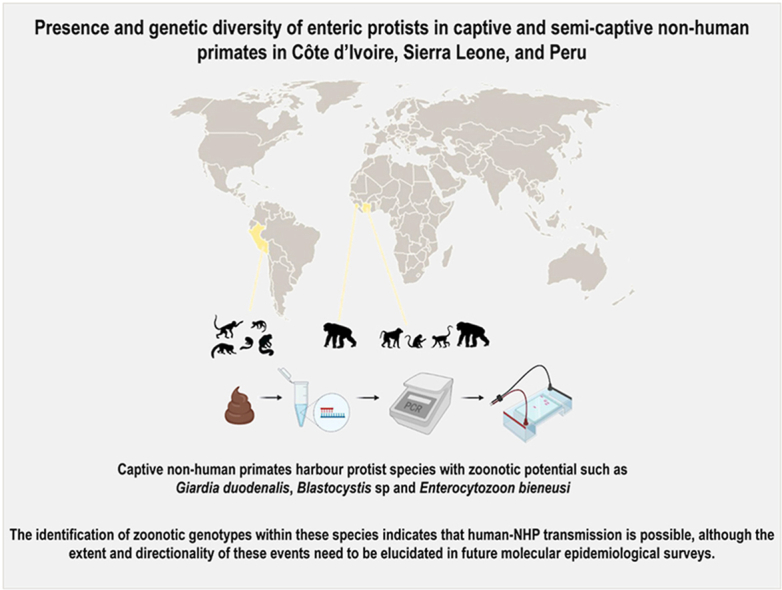
Abstract
Little information is currently available on the occurrence and genetic diversity of pathogenic and commensal protist species in captive and semi-captive non-human primates (NHP) resident in zoological gardens or sanctuaries in low- and medium-income countries. In this molecular-based study, we prospectively collected individual faecal samples from apparently healthy NHP at the Abidjan Zoological Garden (AZG) in Côte d’Ivoire, the Tacugama Sanctuary (TS) in Sierra Leone, and the Quistococha Zoological Garden (QZG) in Peru between November 2018 and February 2020. We evaluated for the presence of pathogenic (Cryptosporidium spp., Entamoeba histolytica, Giardia duodenalis, Blastocystis sp., Enterocytozoon bieneusi, Balantioides coli) and commensal (Entamoeba dispar, Troglodytella abrassarti) protist species using PCR methods and Sanger sequencing. Giardia duodenalis was the most prevalent species found (25.9%, 30/116), followed by Blastocystis sp. (22.4%, 26/116), and E. dispar (18.1%, 21/116). We detected E. bieneusi (4.2%, 1/24) and T. abrassarti (12.5%, 3/24) only on NHP from AZG. Cryptosporidium spp., E. histolytica, and B. coli were undetected at the three sampling sites investigated here. Sequence analyses revealed the presence of zoonotic sub-assemblages BIII (n = 1) in AZG and BIV (n = 1) in TS within G. duodenalis. We identified Blastocystis subtype ST3 (100%, 6/6) in AZG, ST1 (80.0%, 12/15), ST2 (6.7%, 1/15), and ST3 (13.3%, 2/15) in TS, and ST2 (80.0%, 4/5) and ST3 (20.0%, 1/5) in QZG. The only E. bieneusi isolate detected here was identified as zoonotic genotype CAF4. Our PCR-based data indicate that potentially pathogenic protist species including G. duodenalis, Blastocystis sp., E. bieneusi, and B. coli are present at variable rates in the three NHP populations investigated here. The identification of zoonotic genotypes within these species indicates that human-NHP transmission is possible, although the extent and directionality of these events need to be elucidated in future molecular surveys.
Keywords: Enteric protists, Captive non-human primates, Genotyping, Conservation, Zoonoses, Transmission
Graphical abstract
Highlights
-
•
Giardia and Blastocystis are highly prevalent in confined non-human primates.
-
•
Diarrhoea-causing Cryptosporidium and Entamoeba histolytica were undetected.
-
•
First description of Enterocytozoon bieneusi genotype CAF4 in non-human primates.
-
•
Confined non-human primates harbour protist species with zoonotic potential.
-
•
Cross-species (including human) transmission is possible in zoos and sanctuaries.
1. Introduction
Infections by enteric parasites including helminths and protists are a frequent cause of gastrointestinal disease in captive and semi-captive non-human primates (NHP) globally (Levecke, 2010). Reported clinical manifestations vary greatly from subclinical (asymptomatic) infections to acute or self-limiting diarrhoea, malabsorption, severe dehydration, granulomatous inflammation, peritonitis, and even death (Johnson-Delaney, 2009). Clinical signs and symptoms are strongly dependent on several factors including the host species considered, the parasite load, the diversity and balance of the diet, the enclosure conditions (confined environments enhance the likelihood of parasitic transmission), or stress-induced immunosuppression conditions, among others (Strait et al., 2012).
Protists commonly identified in captive and semi-captive NHP include pathogenic species of veterinary relevance such as the protozoan Cryptosporidium spp., Entamoeba histolytica, and Giardia duodenalis or the microsporidia Enterocytozoon bieneusi. Organisms of uncertain pathogenicity or commensal nature such as the stramenopile Blastocystis sp. and the ciliates Balantioides coli and Troglodytella abrassarti are also a common finding in the faeces of captive NHP (Johnson-Delaney, 2009; Medkour et al., 2020; Helenbrook and Whipps, 2021). All the above-mentioned protist species have been documented in NHP collections of zoological gardens and sanctuaries in Asian (Mul et al., 2007; Nath et al., 2012; Du et al., 2015; Adrus et al., 2019 Zhao et al., 2020), African (Munene et al., 1998; Pomajbíková et al., 2010a,b; Li et al., 2011; Mbaya and Udendeye, 2011; Ryan et al., 2012; Adetunji, 2014; Debenham et al., 2015; Kouassi et al., 2015), American (Phillips et al., 2004; Wenz et al., 2010; Milozzi et al., 2012; da Silva Barbosa et al., 2015), and European (Levecke et al., 2007; Berrilli et al., 2011; Köster et al., 2021a) countries. These studies revealed marked differences in occurrence rates depending on host species, geographical location, and detection method used. Older epidemiological surveys primarily relied on microscopy examination, a method known to underestimate the true prevalence of enteric parasites due to limited diagnostic sensitivity (Mejia et al., 2013). The advent of molecular-based methods in well-designed studies targeting multiple host species has contributed enormously to overcome this drawback. Additionally, when coupled to Sanger sequencing, PCR assays allows the accurate determination of species/genotypes, an information extremely useful to investigate transmission dynamics including the occurrence and directionality of zoonotic events. For instance, transmission of Blastocystis has been demonstrated between captive NHP and their keepers in a zoological garden in the UK (Stensvold et al., 2009).
Sequence analyses of housekeeping genes including the small subunit (ssu rRNA) or the internal transcribed spacer (ITS) region of the ribosomal RNA gene have revealed a high degree of genetic diversity within Cryptosporidium spp., G. duodenalis, Blastocystis sp., and E. bieneusi, leading to the identification of several genotypes/subtypes with marked differences in host specificity and range (Franzén et al., 2009; Stensvold et al., 2012; Li and Xiao, 2019; Garcia-R et al., 2020). Thus, the genus Cryptosporidium encompasses at least 46 valid species, with C. hominis and C. parvum causing most of the infections reported in humans and NHP globally (Ježková et al., 2021; Zahedi et al., 2021). Giardia duodenalis is constituted of eight (A to H) distinct genetic assemblages, of which zoonotic assemblages A and B are able to infect a wide diversity of mammal species including humans and NHP (Ryan and Cacciò, 2013). At least 28 subtypes (ST) have been identified within Blastocystis sp. (Higuera et al., 2021; Maloney and Santin, 2021; Maloney et al., 2021) of which ST1‒ST5, ST7‒ST11, ST13, ST15, and ST19 have been previously reported in NHP (Hublin et al., 2021). More than 500 E. bieneusi genotypes have been defined to date grouped in 11 phylogenetic groups (Li et al., 2019). Of them, at least 78 genotypes have been reported in NHP, with genotype D showing predominance (Zhong et al., 2017; Chen et al., 2019; Li et al., 2019; Karim et al., 2020; Zhao et al., 2020). Comparatively, there is much less information on the genetic variability within other protist species including E. histolytica, E. dispar, B. coli, or T. abrassarti (Ponce-Gordo and García-Rodríguez, 2021; Vallo et al., 2012; Weedall et al., 2012). This situation is likely associated to the scarcity of molecular data and the limited range of genetic markers available for typing and subtyping purposes (Gilchrist et al., 2012).
This molecular-based study aims at providing novel data on the occurrence and genetic diversity of enteric protist species of pathogenic and commensal nature in captive and semi-captive NHP in three different epidemiological scenarios in two African (Côte d’Ivoire and Sierra Leone) and one South American (Peru) countries. Information provided here is expected to improve our current knowledge on the epidemiology of these microorganisms and their zoonotic potential in NHP.
2. Materials and methods
Ethical statement
This study was carried out in accordance with the International Guiding Principles for Biomedical Research Involving Animals issued by the Council for International Organization of Medical Sciences and the International Council for Laboratory Animal Science (RD 53/2013). This study has been approved by Ethics Committee of the Health Institute Carlos III on December 17, 2018 under the reference number CEI PI 90_2018-v2. Written informed consent was obtained from zookeepers that volunteered to participate in the survey.
2.1. Study area in côte d’Ivoire
We collected faecal samples (n = 24) from captive NHP at the Abidjan Zoological Garden (AZG, Abidjan, Côte d’Ivoire) between April–August 2019. The AZG include 230 individuals belonging to 48 mammalian, reptilian, and avian species. Collected faecal samples belonged to six different genera of NHP including Cercocebus (n = 3), Cercopithecus (n = 1), Chlorocebus (n = 2), Erythrocebus (n = 3), Pan (n = 11), and Papio (n = 4). NHP of the same species were kept in specific enclosures without contact with other NHP.
2.2. Study area in Sierra Leone
We collected faecal samples (n = 67) from captive chimpanzees at Tacugama Sanctuary (TS) in Sierra Leona between November and December 2018. The Sanctuary is placed within the forest of Western Peninsula National Park, but Sanctuary chimpanzees have no contact with the wild ones in the park because they are isolated by fences. The chimpanzees sampled are living in enclosures, with access to forested fenced areas of surfaces smaller than 1 ha. They have daily proximity to humans during feeding (food is brought from the market) and regular veterinarian treatments.
2.3. Study area in Peru
We collected faecal samples (n = 25) from captive NHP at the Quistococha Zoological Garden (QZG, Iquitos, Peru) in February 2020. The QZG extends over 369 ha of secondary rainforest and includes individuals belonging to near 70 mammalian, reptilian, and avian species. Collected faecal samples belonged to seven different genera of NHP including Ateles (n = 5), Callicebus (n = 1), Cebus (n = 4), Lagothrix (n = 3), Pithecia (n = 1), Saimiri (n = 3), and Sapajus (n = 8). NHP of the same species were kept in specific enclosures without contact with other NHP. Additionally, we collected individual stool samples from three zookeepers in close contact with NHP that volunteered to participate in the study.
2.4. Faecal samples conservation
We collected 5–10 g from the inner core of faeces with sterile plastic spatulas and stored them in 70% ethanol for conservation and transport to the Parasitology Reference and Research Laboratory, Spanish National Centre for Microbiology, Majadahonda (Spain) for further processing and downstream molecular testing. We collected information on NHP species, enclosure conditions, sampling date, and faecal consistency (diarrhoeal versus non-diarrhoeal). No information was available regarding sex and gender, as faecal specimens could not be traced back to individual animals.
2.5. DNA extraction and purification
We isolated genomic DNA from about 200 mg of each faecal specimen by using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, except that samples mixed with InhibitEX buffer were incubated for 10 min at 95 °C. Extracted and purified DNA samples were eluted in 200 μL of PCR-grade water and kept at 4 °C until further molecular analysis.
2.6. Molecular detection of Cryptosporidium spp
We assessed the presence of Cryptosporidium spp. using a nested-PCR protocol to amplify a 587-bp fragment of the ssu rRNA gene of the parasite (Tiangtip and Jongwutiwes, 2002). Amplification reactions (50 μL) included 3 μL of DNA sample and 0.3 μM of the primer pairs CR-P1/CR-P2 in the primary reaction and CR-P3/CPB-DIAGR in the secondary reaction (Table S1). Both PCR reactions were carried out as follows: one step of 94 °C for 3 min, followed by 35 cycles of 94 °C for 40 s, 50 °C for 40 s and 72 °C for 1 min, concluding with a final extension of 72 °C for 10 min.
2.7. Molecular differential detection of Entamoeba histolytica and Entamoeba dispar
We carried out detection and differential diagnosis between pathogenic E. histolytica and non-pathogenic E. dispar by a qPCR method targeting a 172-bp fragment of the gene codifying the ssu rRNA gene of the E. histolytica/E. dispar complex (Verweij et al., 2003a; Gutiérrez-Cisneros et al., 2010). Amplification reactions (25 μL) consisted of 3 μL template DNA, 0.5 μM of the primer set Ehd-239F/Ehd-88R, 0.2 μM of each TaqMan® probe (Table S1), and TaqMan® Gene Expression Master Mix (Applied Biosystems, CA, USA). Detection of parasitic DNA was performed on a Corbett Rotor GeneTM 6000 real-time PCR system (Qiagen) using an amplification protocol consisting of an initial hold step of 2 min at 55 °C and 15 min at 95 °C followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C.
2.8. Molecular detection and characterization of Giardia duodenalis
We conducted G. duodenalis DNA detection using a real-time PCR (qPCR) method targeting a 62-bp region of the gene codifying the ssu rRNA gene of the parasite (Verweij et al., 2003b). Amplification reactions (25 μL) consisted of 3 μL of template DNA, 0.5 μM of each primer Gd-80F and Gd-127R, 0.4 μM of probe (Table S1), and 12.5 μL TaqMan® Gene Expression Master Mix (Applied Biosystems). Cycling conditions and data analysis were as described above for the detection of E. histolytica/E. dispar.
We subsequently assessed G. duodenalis isolates that tested positive by qPCR by sequence-based multi-locus genotyping of the genes encoding for the glutamate dehydrogenase (gdh) (Read et al., 2004), β-giardin (bg) (Lalle et al., 2005), and triose phosphate (tpi) (Sulaiman et al., 2003) proteins of the parasite. We conducted amplifications by semi-nested and nested PCR protocols using specific primer pairs (Table S1).
2.9. Molecular detection and characterization of Blastocystis sp
We identified Blastocystis sp. by a direct PCR protocol targeting the ssu rRNA gene of the parasite (Scicluna et al., 2006). The assay uses the pan-Blastocystis, barcode primer pair RD5/BhRDr to amplify a PCR product of ∼600 bp. Amplification reactions (25 μL) included 5 μL of template DNA and 0.5 μM of each primer (Table S1). Amplification conditions consisted of one-step of 95 °C for 3 min, followed by 30 cycles of 1 min each at 94, 59 and 72 °C, with an additional 2 min final extension at 72 °C.
2.10. Molecular detection and characterization of Enterocytozoon bieneusi
We conducted E. bieneusi detection by a nested PCR protocol to amplify the ITS region as well as portions of the flanking large and small subunit of the ribosomal RNA gene as previously described (Buckholt et al., 2002). We used the outer EBITS3/EBTIS4 and inner EBITS1/EBITS2.4 primer sets (Table S1) to generate a final PCR product of 390 bp, respectively. PCR reactions (50 μL) consisted of 1 μL of template DNA and 0.2 μM of each primer. Cycling conditions for the primary PCR consisted of one step of 94 °C for 3 min, followed by 35 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s, and elongation at 72 °C for 40 s), with a final extension at 72 °C for 10 min. Conditions for the secondary PCR were identical to the primary PCR except only 30 cycles were carried out with an annealing temperature of 55 °C.
2.11. Molecular detection of Balantioides coli
We carried out B. coli detection by a direct PCR assay to amplify the complete ITS1–5.8s-rRNA–ITS2 region and the last 117 bp (3′ end) of the ssu-rRNA sequence of this ciliate using the primer set B5D/B5RC (Ponce-Gordo et al., 2011). PCR reactions (25 μL) consisted of 2 μL of template DNA and 0.4 μM of each primer (Table S1). PCR conditions were as follows: 94 °C for 10 min; 30 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min, and a final extension for 5 min at 72 °C.
2.12. Molecular detection of Troglodytella spp
Hitherto, we aimed to detect the ciliate mutualist Troglodytella spp. by a direct PCR method targeting a 401-bp fragment of the ITS region of the rDNA (ITS1-5.8S rDNA-ITS2) (Vallo et al., 2012). PCR reactions (25 μL) contained 2 μL of template DNA and 0.8 μM of each primer ssu-end/LSU-start (Table S1). Conditions of PCR for ITS amplification were initial denaturation for 2 min at 94 °C, 35 cycles of 45 s at 94 °C, 45 s at 50 °C, and 90 s at 72 °C, and terminal elongation for 5 min at 72 °C.
2.13. PCR and gel electrophoresis standard procedures
We carried out all the direct, semi-nested, and nested PCR protocols described above on a 2720 Thermal Cycler (Applied Biosystems). Reaction mixes included 2.5 units of MyTAQ™ DNA polymerase (Bioline GmbH, Luckenwalde, Germany), and 5 × MyTAQ™ Reaction Buffer containing 5 mM dNTPs and 15 mM MgCl2. The specific DNA primer and probe sequences used in the present study were detailed in Table S1. We routinely used laboratory-confirmed positive and negative DNA samples of human and animal origin for each parasitic species investigated as controls and included them in each round of PCR. We visualized PCR amplicons on 1.5–2% D5 agarose gels (Conda, Madrid, Spain) stained with Pronasafe (Conda) or Gel Red (Biotium, Fremont, California, USA) nucleic acid staining solutions. We used a 100 bp DNA ladder (Boehringer Mannheim GmbH, Baden-Wurttemberg, Germany) for the sizing of obtained amplicons.
2.14. Sequence analyses
We directly sequenced positive-PCR products in both directions using appropriate internal primer sets (Table S1). We conducted DNA sequencing by capillary electrophoresis using the BigDye® Terminator chemistry (Applied Biosystems) on an ABI PRISM 3130 automated DNA sequencer. We visually inspected the obtained chromatograms for quality control and for detecting the presence of ambiguous (double peak) positions. Sequences obtained in this study were deposited in GenBank under accession numbers MZ501256–MZ501257 (Giardia duodenalis), MZ496540–MZ496545 (Blastocystis sp.), MZ502643 (Enterocytozoon bieneusi), and MZ502644 (Troglodytella abrassarti).
3. Results
3.1. Occurrence of enteric protist species
We report the PCR-based occurrence rates of enteric protist species identified in the captive and semi-captive NHP population investigated in the present study in Table 1. We present the full dataset of the study including diagnostic and molecular genotyping results in Table S2. All participating NHPs had no apparent gastrointestinal clinical manifestations at sampling, and the consistency of their excrements were formed.
Table 1.
Prevalence of enteric parasite and commensal protists identified in the captive and semi-captive non-human primate populations investigated at Abidjan Zoological Garden (AZG) in Côte d’Ivoire, Tacugama Sanctuary (TS) in Sierra Leone, and Quistococha Zoological Garden (QZG) in Peru.
| AZG, Côte d’Ivoire (n = 24) |
TS, Sierra Leone (n = 67) |
QZG, Peru (n = 25) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Pos. (n) | % | 95% CI | Pos. (n) | % | 95% CI | Pos. (n) | % | 95% CI |
| Cryptosporidium spp. | 0 | 0.0 | – | 0 | 0.0 | – | 0 | 0.0 | – |
| E. histolytica | 0 | 0.0 | – | 0 | 0.0 | – | 0 | 0.0 | – |
| E. dispar | 13 | 54.2 | 32.8–74.5 | 1 | 1.5 | 0.04–8.4 | 7 | 28.0 | 12.1–49.4 |
| G. duodenalis | 7 | 29.2 | 12.6–51.1 | 22 | 32.8 | 21.9–45.4 | 1 | 4.0 | 0.1–20.4 |
| Blastocystis sp. | 6 | 25.0 | 9.8–46.7 | 15 | 22.4 | 13.1–34.2 | 5 | 20.0 | 0.1–20.4 |
| E. bieneusi | 1 | 4.2 | 0.1–21.1 | 0 | 0.0 | – | 0 | 0.0 | – |
| B. coli | 0 | 0.0 | – | 0 | 0.0 | – | 0 | 0.0 | – |
| T. abrassarti | 3 | 12.5 | 26.6–46.7 | 0 | 0.0 | – | 0 | 0.0 | – |
At AZG (Côte d’Ivoire), the most common pathogenic protozoa identified was G. duodenalis (29.2%), followed by Blastocystis sp. (25.0%), and E. bieneusi (4.2%). Non-pathogenic E. dispar and T. abrassarti were found in 54.2% and 12.5% of samples, respectively (Table 1). Giardia duodenalis was detected in members of all six NHP genera present at AZG, and Blastocystis sp. in three (Cercocebus, Pan, and Papio), whereas we detected E. bieneusi only in a sooty mangabey (Cercocebus atys). Regarding commensal protists, E. dispar colonized members of all six NHP genera present at AZG, but T. abrassarti was detected in chimpanzees (Pan troglodytes) only (Table S2).
At TS (Sierra Leone), we identified two pathogenic protist species including G. duodenalis (32.8%) and Blastocystis sp. (22.4%) and a single commensal species (E. dispar, 1.5%) infecting or colonizing the chimpanzee population at this sanctuary (Table 1).
We observed a similar diversity of enteric protist species, although at different frequency rates for some of them, in NHP at IZG (Peru). Thus, G. duodenalis (4.0%) and Blastocystis sp. (20.0%) were the only species of pathogenic significance found, and E. dispar (28.0%) the only commensal species identified (Table 1). We detected G. duodenalis in a monk saki (Pithecia monachus), and Blastocystis sp. in four out of the five genera of NHP (Ateles, Callicebus, Lagothrix, and Pithecia) present at IZG. Entamoeba dispar colonized members of three NHP genera including Ateles, Callicebus, and Lagothrix (Table S2). Regarding human stool samples, we identified the presence of Blastocystis sp. in one of the three zookeepers from IZG Peru that participated in the study. The three individuals tested negative for the remaining enteric protist species investigated here.
We did not detect pathogenic Cryptosporidium spp., E. histolytica, and B. coli in any of the three NHP populations surveyed, whereas T. abrassarti was not circulating among NHP at TS (Sierra Leone) and QZG (Peru) (Table 1). Of note, DNA samples from three chimpanzees at AZG (Côte d’Ivoire) cross-reacted with T. abrassarti when tested by PCR for the presence of B. coli (Table S2). Similarly, DNA samples from 10 chimpanzees (nine from TS Sierra Leone and one from AZG Côte d’Ivoire) cross reacted with parasitic protozoan flagellates of the genus Tetratrichomonas when tested by PCR for the presence of T. abrassarti (Table S2).
3.2. Molecular characterization of enteric protist species
The 30 DNA isolates that yielded a positive result for G. duodenalis by qPCR in the present study (seven from AZG Côte d’Ivoire, 22 from TS Sierra Leone, and one from QZG Peru) generated cycle threshold (Ct) values ranging from 25.4 to 39.5 (median: 34.4). We successfully amplified only two of them corresponding to chimpanzee samples from AZG Côte d’Ivoire (Ct value: 26.6) and TS Sierra Leone (Ct value: 25.4), respectively, at the gdh locus, but not at the bg or tpi loci. Sequence analyses revealed the presence of sub-assemblage BIII (AZG Côte d’Ivoire) and sub-assemblage BIV (TS Sierra Leone) (Table 2).
Table 2.
Diversity, frequency, and molecular features of Giardia duodenalis, Blastocystis sp., Enterocytozoon bieneusi, and Troglodytella abrassarti isolates identified in the NHP populations investigated in the present study.
| Species | Genotype | Sub-genotype | No. isolates | Locus | Reference sequence | Stretch | Single nucleotide polymorphisms | GenBank ID |
|---|---|---|---|---|---|---|---|---|
| Giardia duodenalis | B | BIII | 1a | gdh | AF069059 | 41–428 | C87T, G215A, A414G | MZ501256 |
| B | BIV | 1b | gdh | L40508 | 85–496 | T183C, G359R, T387C, C396T, C432T | MZ501257 | |
| Blastocystis sp. | ST1 | Allele 8 | 9b | ssu rRNA | MZ182327 | 32–606 | None | MZ496540 |
| ST1 | Alleles 7 + 8 | 3b | ssu rRNA | MZ182327 | 5–606 | G176R | MZ496541 | |
| ST2 | Allele 11 | 1c | ssu rRNA | MF669067 | 1–604 | None | MZ496542 | |
| ST2 | Alleles 11 + 12 | 2c | ssu rRNA | MF669067 | 53–580 | G178R, C476G | MZ496543 | |
| ST2 | Unknown | 1c | ssu rRNA | – | – | – | – | |
| ST2 | Unknown | 1b | ssu rRNA | – | – | – | – | |
| ST3 | Allele 30 | 4a | ssu rRNA | MN338079 | 4–593 | A260T | MZ496544 | |
| ST3 | Allele 34 | 1c | ssu rRNA | KY929101 | 13–553 | None | MZ496545 | |
| ST3 | Unknown | 2a | ssu rRNA | – | – | – | – | |
| ST3 | Unknown | 2b | ssu rRNA | – | – | – | – | |
| Enterocytozoon bieneusi | CAF4 | – | 1 | ITS | DQ683749 | 1–242 | None | MZ502643 |
| Troglodytella abrassarti | ‒ | ‒ | 3 | ITS | EU680311 | 1–418 | T82C | MZ502644 |
Gdh: Glutamate dehydrogenase; ITS: Internal transcribed spacer; ssu rRNA: Small subunit ribosomal RNA.
AZG Côte d’Ivoire.
TS Sierra Leone.
QZG Peru.
We confirmed a total of 26 isolates as Blastocystis-positive by Sanger sequencing. Of them, six were from NHP (three chimpanzees, two baboons and a mangabey) at AZG Côte d’Ivoire, 15 from chimpanzees at TS Sierra Leone, and the remaining five from NHP (two brown woolly monkeys, a monk saki, a white-bellied spider monkey, and a yellow-handed titi monkey) at QZG Peru. We identified all six samples from AZG Côte d’Ivoire as ST3 (Table 2). Allele 30 was the most common (66.7%, 4/6) genetic variant found within ST3, whereas no allelic information was available for the remaining two ST3 isolates due to insufficient sequence quality.
We identified three Blastocystis STs circulating among chimpanzees at TS Sierra Leone including ST1 (80.0%, 12/15), ST2 (6.7%, 1/15), and ST3 (13.3%, 2/15). We confirmed allele 8 as the most prevalent genetic variant within ST1 (75%, 9/12), followed by mixed infections (determined by the presence of a clear double peak at chromatogram inspection) involving alleles 7 + 8 (25%, 3/12). ST2 and ST3 isolates could not be analysed at the allelic level due to suboptimal sequence quality associated with one or more miscalled nucleotide positions (Table 2).
We found ST2 as the most prevalent Blastocystis subtype present in NHP at QZG Peru (80.0%, 4/5) followed by ST3 (20.0%, 1/5). Alleles 11 (25%, 1/4), alleles 11 + 12 (50.0%, 2/4) were identified within ST2. The allelic form of the fourth ST2 isolate could not be determined due to insufficient sequence quality. The only isolate belonging to ST3 was characterised as allele 34 (Table 2). The zookeeper that tested positive to Blastocystis was colonized by ST1 allele 4, not present in any of the NHP investigated in this zoological garden.
Of note, additional 37 isolates (nine from AZG Côte d’Ivoire, and 28 from TS Sierra Leone) yielded amplicons of the expected size but in the form of faint bands on gel that produced poor quality, unreadable sequences. We conservatively considered these isolates as Blastocystis-negative.
We detected E.bieneusi only in a sooty mangabey from AZG Côte d’Ivoire. Sequence analysis of the ITS region confirmed this isolate as genotype CAF4 (Table 2).
Finally, we identified the presence of T. abrassarti in three captive chimpanzees at AZG Côte d’Ivoire. All three ITS sequences were identical among them but differed by a single SNP (T82C) from reference sequence EU680311 (Table 2).
4. Discussion
Present molecular-based epidemiological study provides novel data on the occurrence of eukaryotic intestinal (including parasite and commensal) species in captive and semi-captive NHP populations in two low-income (Côte d’Ivoire and Sierra Leone) and one middle-income (Peru) countries.
Cryptosporidium spp. are well known diarrhoea-causing pathogens in all major classes of vertebrates. In this study, Cryptosporidium infections were apparently absent in the surveyed NHP collections. In African NHP populations, Cryptosporidium spp. have been reported at 0–9% by conventional microscopy in the Republic of Congo and Tanzania (Gillespie et al., 2009; Gonzalez-Moreno et al., 2013), and at 0–21% by PCR in Senegal and Tanzania (Parsons et al., 2015; Renelies-Hamilton et al., 2019; Köster et al., 2021b). No studies have been conducted on the occurrence of Cryptosporidium spp. in Peruvian NHP populations to date. Remarkably, molecular epidemiological surveys carried out in the last decade have demonstrated that Cryptosporidium infections in NHP are primarily caused by C. hominis, the species most reported in human cryptosporidiosis cases globally (Widmer et al., 2020).
As in the case of Cryptosporidium spp., E. histolytica infections were undetected in the three NHP populations investigated. It is still unclear to which extent NHP are suitable hosts for E. histolytica and, if yes, what the actual pathogenic significance of the infection is. Our PCR-based data agreed with the negative results obtained in similar studies conducted in zoo-kept NHP in Spain (Köster et al., 2021a) and wild chimpanzee populations in Senegal and Tanzania (Jirků-Pomajbíková et al., 2016; Köster et al., 2021b). These surveys showed that wild chimpanzees harboured Entamoeba species (e.g. E. dispar) similar to those occurring in humans, but no pathogenic species (e.g. E. histolytica) were detected. Opposite results have also been reported: a PCR-based prevalence of E. histolytica of 11–34% has been observed in apparently healthy chimpanzees and baboons in Tanzania (Deere et al., 2019), whereas a recent study using a next generation sequencing approach revealed the presence of the parasite at low proportions in free-living chimpanzees and gorillas in Cameroon (Vlčková et al., 2018). Although a clear link between E. histolytica and the occurrence of clinical manifestations could not be established in those surveys, other studies have confirmed outcomes similar to those of human invasive amebiasis in laboratory chimpanzees (Miller and Bray, 1966) and diarrhoeal episodes in infected wild lemurs from Madagascar (Ragazzo et al., 2018). Additionally, two captive spider monkeys presenting with amoebic dysentery and a mandrill with diarrhoea have been also reported in a French zoo (Verweij et al., 2003c). In this study, non-pathogenic E. dispar was detected at variable (28–54% in zoo NHP, 1.5% in semi-captive chimpanzees) rates. These data are well in agreement with the figures reported in other captive (Regan et al., 2014; Köster et al., 2021a) or free-living (Lau et al., 2013; Jirků-Pomajbíková et al., 2016; Köster et al., 2021b) NHP populations globally.
Information on the occurrence and genetic diversity of G. duodenalis in African and South American NHP populations is scarce (Squire and Ryan, 2017; Rivero et al., 2020; Fantinatti et al., 2021). The parasite has been reported at prevalences of 2–67% in free-living gorillas from Central African Republic, Rwanda, and Uganda (Graczyk et al., 2002; Sak et al., 2013; Hogan et al., 2014), colobus monkeys from Ghana and Uganda (Teichroeb et al., 2009; Johnston et al., 2010), and wild chimpanzees from the Republic of Congo, Guinea Bissau, and Senegal (Gillespie et al., 2009; Sá et al., 2013; Renelies-Hamilton et al., 2019). In South American countries, G. duodenalis has been identified in 40–76% of semi-captive and wild howler monkeys in Argentina and Brazil (Volotão et al., 2008; Kowalewski et al., 2011). No information on the presence of the parasite is currently available in NHP from Peru. As in the case of our study, most of the above-mentioned surveys did not report the occurrence of clinical manifestations (including diarrhoea) in the investigated animals, suggesting that G. duodenalis infections were primarily asymptomatic. In the present study, G. duodenalis was more prevalently found in African (29–33%) than in Peruvian (4%) sites, probably reflecting differences in host species, infection sources and/or management procedures of captive/semi-captive animals. Regarding molecular data, we found zoonotic sub-assemblages BIII and BIV in two resident chimpanzees at AZG Côte d’Ivoire and TS Sierra Leone. Both G. duodenalis sub-assemblages have also been reported in mountain gorillas in Rwanda (Hogan et al., 2014) and wild howler monkeys and captive NHP in Brazil (Volotão et al., 2008; Soares et al., 2011). The low genotyping success rate (6.7%, 2/30) obtained here can be explained by the limited sensibility of the single-copy genes (gdh, bg, and tpi) used for that purpose, which is in sharp contrast with the high sensitivity associated with the multi-copy ssu rRNA gene used in qPCR detection. This fact is consistent with the low parasite burden (indicated by high qPCR Ct values) and apparent absence of clinical manifestations (suggested by the formed consistency of the faecal material) typically found in the surveyed NHP populations.
We found Blastocystis sp. at a remarkably similar prevalence rates (20–25%) in the three NHP populations investigated here. This finding would suggest that Blastocystis is a common component of the NHP gut microbiota regardless of the host and geographical area. The infection/colonization rates found in the present study are in the lower range of those typically reported in African NHP populations including chimpanzees, gorillas, and colobus and vervet monkeys. Blastocystis sp. has been documented at a prevalence of 22% by conventional microscopy in Cameroon (Drakulovski et al., 2014), and of 6–100% by PCR in Senegal, the Republic of Congo, and Tanzania (Petrášová et al., 2011; Renelies-Hamilton et al., 2019; Köster et al., 2021b; Menu et al., 2021). Regarding genetic diversity, ST1 has been demonstrated as the most prevalent Blastocystis subtype circulating in free-living NHP in Africa, accounting for 80–100% of the isolates characterised in wild chimpanzees in Senegal (Renelies-Hamilton et al., 2019; Köster et al., 2021b) and Tanzania (Petrášová et al., 2011), although in the former country few animals carried also ST2 and ST3. Interestingly, a recent molecular study has evidenced that approximately nine out of 10 ST1 isolates identified in Senegalese wild chimpanzees carried the allele 8 of the protist, likely representing a Blastocystis genetic variant particularly adapted to infect/colonize African Western chimpanzees (Köster et al., 2021b). This seems to be the case also of the semi-captive chimpanzee population resident at TS Sierra Leona in the present study, where allele 8 was identified (alone or in combination with other alleles) in 80% (12/15) of the animals carrying Blastocystis ST1. In contrast, ST1 was absent in captive NHP resident at AZG Côte d’Ivoire, where all Blastocystis-positive animals were infected/colonized by ST2 or ST3. These discrepancies in the frequencies of genetic variants may be due to differences in diet quality, management, or exposure to sources of infection including those of anthroponotic nature. Very few studies have described Blastocystis STs in Central and South American NHP populations. Thus, howler monkeys have been found infected/colonized by ST4 in Colombia (Ramírez et al., 2014), and by ST8 in Ecuador (Helenbrook et al., 2015a). However, all captive NHP resident at QZG Peru with a Blastocystis-positive result harboured ST2 and ST3, a pattern more closely related to the one found at AZG Côte d’Ivoire. Of interest, the only zookeeper that tested positive to Blastocystis in this institution was infected/colonized by ST1 allele 4, a genetic variant not detected in any of the NHP investigated in this zoological garden.
Enterocytozoon bieneusi, the dominant member of the pathogenic microsporidian species, may cause diarrhoea and other gastrointestinal manifestations in humans and domestic and wild animals (Santín and Fayer, 2011). Reported prevalences for such organism in captive NHP populations are in the range of 4%–68%, with most studies coming from China (reviewed in Leśniańska and Perec-Matysiak, 2017). Very little information is currently available on the epidemiology of E. bieneusi in African or South American NHP. The parasite has been reported in 12.3% (29/235) of captive, apparently healthy olive baboons (Papio anubis) in Kenya, with juvenile individuals harbouring the bulk of the infections (Li et al., 2011). In the present study, E. bieneusi was detected in a sooty mangabey resident at AZG Côte d’Ivoire (overall prevalence: 4.2%). Recent phylogenetic analysis of valid E. bieneusi genotypes at the ITS locus have allowed the recognition of 11 major genetic groups (Groups 1 to 11) of which Groups 1 and 2 include genotypes with potential for zoonotic or cross-species transmission (Li et al., 2019). Near 78 E. bieneusi ITS genotypes have been reported from various NHP species to date (Leśniańska and Perec-Matysiak, 2017; Chen et al., 2019; Li et al., 2019; Karim et al., 2020; Zhao et al., 2020). Most of E. bieneusi genotypes identified in NHP belong to Group 1 (D, EbpA, EbpC, Type IV, O, Peru11, etc.) and Group 2 (BEB4, BEB6, I, J, etc.). In addition, other genotypes included in Group 5 (KB-6 and PtEb XII), Group 6 (Gorilla 3 and KB-5) and Group 7 (CM18 and XH) have been found only in NHP, suggesting that these genetic variants seem to be adapted to this host (Li et al., 2019). Our sequence analysis revealed that the E. bieneusi infection detected in a zoo mangabey was caused by the extremely rare genotype CAF4. Indeed, CAF4 has only been identified to date in four HIV-positive patients in Gabon and five HIV-negative individuals in Cameroon (Breton et al., 2007). Genotype CAF4 belongs to Group 5 of E. bieneusi, which contains genotypes adapted to humans and NHP (Li et al., 2019). Genotype CAF4 was thought to be human-specific, but this study confirms its presence in a new host. These data strongly suggest that CAF4 may represent a zoonotic genotype circulating among human and NHP populations in Western Africa. More research is needed to elucidate the epidemiology of E. bieneusi in low-income countries, including the identification of suitable hosts and transmission pathways.
Balantioides coli is largely considered a non-pathogenic gut commensal, although fatal ulcerative colitis by this ciliate has been described in captive gorillas (Lankester et al., 2008). This parasite is fairly common in captive and semi-captive great apes, with prevalence rates ranging 14–68% in chimpanzees and gorillas living in sanctuaries in Cameroon and Kenya (Pomajbíková et al., 2010b), or 9% in captive chimpanzees in Nigeria (Mbaya and Udendeye, 2011). In captive and wild-trapped olive baboons, vervets and sykes in Kenya, prevalence ranged from 23 to 53% (Munene et al., 1998). Surprisingly, B. coli was not detected in the chimpanzee populations investigated in our study; at present we do not have a clear explanation for this result. In relation to the non-detection of the parasite in New World NHP, it is an expected result, since B. coli is considered rare or absent in New World monkeys (Levecke et al., 2007). Low prevalences of B. coli cysts have been reported in captive Saimiri sciureus, Leontopithecus sp., Cebus albifrons and Aotus nigriceps (Guerrero et al., 2012; da Silva Barbosa et al., 2015), and in wild Alouatta palliata (Helenbrook et al., 2015b).
The colonic mutualist T. abrassarti is believed to specifically colonize African great apes as most records are from chimpanzees, bonobos, and gorillas (Goussard et al., 1983; Vallo et al., 2012). However, the finding of this ciliate protist in lesser apes of the genus Hylobates suggests that the range of competent host species may be wider than initially anticipated (O'Donoghue et al., 1993). Microscopy-based epidemiological studies have previously reported the presence of T. abrassarti at rates ranging from 0 to 100% and 60–100% in captive and wild chimpanzees, respectively (Muehlenbein, 2005; Pomajbíková et al., 2010a). The fact that T. abrassarti does not form environmentally resistant cysts and rapidly degrading trophozoites are the transmissible stage present in faeces has important diagnostic consequences. PCR-based methods could not be the best choice for the detection of T. abrassarti if genomic DNA from fresh faecal material is not readily available. This limitation may explain the low occurrence rates, or even apparent absence, of T. abrassarti typically reported in molecular studies investigating captive or wild NHP (Köster et al., 2021a,b). This is also the case of the present survey, where T. abrassarti was found at relative low rates (12.5%) in captive chimpanzees at AZG Côte d’Ivoire but was absent in semi-captive chimpanzees at TS Sierra Leone. The later result is in contrast with previous microscopy-based results showing that T. abrassarti is naturally circulating among the resident chimpanzee population in that very same location (Irbis et al., 2008).
5. Conclusions
This is, to date, the first molecular-based epidemiological survey investigating the presence and genetic diversity of protist parasite and commensal species in captive and semi-captive NHP populations conducted in Côte d’Ivoire, Sierra Leone, and Peru. Among well-recognized pathogens, G. duodenalis was found at high occurrence rates in the African sites, but not in the Peruvian one. In contrast, Blastocystis sp. was present at similar rates in the three NHP populations investigated here. Cryptosporidium spp., E. histolytica, and B. coli were undetected in the three sites. Enterocytozoon bieneusi genotype CAF4 was only detected in a captive sooty mangabey in Côte d’Ivoire, being the first report of this genetic variant in NHP. The identification of zoonotic genetic variants (sub-assemblages BIII and BIV within G. duodenalis, ST1-ST3 within Blastocystis sp., and CAF4 within E. bieneusi) suggest that human-NHP transmission is possible, although the frequency, extent, and directionality of these events remain to be elucidated in subsequent molecular epidemiological surveys.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Dr Mónica Santín (Agricultural Research Service, United States Department of Agriculture, Beltsville, USA) for her assistance with the identification of E. bieneusi genotypes. This study was funded by the Health Institute Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness under project PI16CIII/00024. David González-Barrio is the recipient of a ‘Sara Borrell’ postdoctoral fellowship (CD19CIII/00011) funded by the Spanish Ministry of Science, Innovation and Universities. Alejandro Dashti is the recipient of a PFIS contract (FI20CIII/00002) funded by the Spanish Ministry of Science and Innovation and Universities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2021.12.004.
Contributor Information
Francisco Ponce-Gordo, Email: pponce@farm.ucm.es.
David Carmena, Email: dacarmena@isciii.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adetunji V.E. Prevalence of gastrointestinal parasites in primates and their keepers from two zoological gardens in Ibadan, Nigeria. Sokoto J. Vet. Sci. 2014;12:25–30. doi: 10.4314/sokjvs.v12i2.5. [DOI] [Google Scholar]
- Adrus M., Zainudin R., Ahamad M., Jayasilan M.A., Abdullah M.T. Gastrointestinal parasites of zoonotic importance observed in the wild, urban, and captive populations of non-human primates in Malaysia. J. Med. Primatol. 2019;48:22–31. doi: 10.1111/jmp.12389. [DOI] [PubMed] [Google Scholar]
- Berrilli F., Prisco C., Friedrich K.G., Di Cerbo P., Di Cave D., et al. Giardia duodenalis assemblages and Entamoeba species infecting non-human primates in an Italian zoological garden: zoonotic potential and management traits. Parasites Vectors. 2011;4:199. doi: 10.1186/1756-3305-4-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton J., Bart-Delabesse E., Biligui S., Carbone A., Seiller X., et al. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 2007;45:2580–2589. doi: 10.1128/JCM.02554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholt M.A., Lee J.H., Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002;68:2595–2599. doi: 10.1128/aem.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhao J., Li N., Guo Y., Feng Y., et al. Genotypes and public health potential of Enterocytozoon bieneusi and Giardia duodenalis in crab-eating macaques. Parasites Vectors. 2019;12:254. doi: 10.1186/s13071-019-3511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Barbosa A., Pissinatti A., Dib L.V., de Siqueira M.P., Cardozo M.L., et al. Balantidium coli and other gastrointestinal parasites in captives non-human primates of the Rio de Janeiro, Brazil. J. Med. Primatol. 2015;44:18–26. doi: 10.1111/jmp.12140. [DOI] [PubMed] [Google Scholar]
- Debenham J.J., Atencia R., Midtgaard F., Robertson L.J. Occurrence of Giardia and Cryptosporidium in captive chimpanzees (Pan troglodytes), mandrills (Mandrillus sphinx) and wild Zanzibar red colobus monkeys (Procolobus kirkii) J. Med. Primatol. 2015;44:60–65. doi: 10.1111/jmp.12158. [DOI] [PubMed] [Google Scholar]
- Deere J.R., Parsons M.B., Lonsdorf E.V., Lipende I., Kamenya S., et al. Entamoeba histolytica infection in humans, chimpanzees and baboons in the Greater Gombe Ecosystem, Tanzania. Parasitology. 2019;146:1116–1122. doi: 10.1017/S0031182018001397. [DOI] [PubMed] [Google Scholar]
- Drakulovski P., Bertout S., Locatelli S., Butel C., Pion S., et al. Assessment of gastrointestinal parasites in wild chimpanzees (Pan troglodytes troglodytes) in southeast Cameroon. Parasitol. Res. 2014;113:2541–2550. doi: 10.1007/s00436-014-3904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S.Z., Zhao G.H., Shao J.F., Fang Y.Q., Tian G.R., et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling Mountains. Kor. J. Parasitol. 2015;53:395–402. doi: 10.3347/kjp.2015.53.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantinatti M., Gonçalves-Pinto M., Lopes-Oliveira L.A.P., Da-Cruz A.M. Epidemiology of Giardia duodenalis assemblages in Brazil: there is still a long way to go. Mem. Inst. Oswaldo Cruz. 2021;115 doi: 10.1590/0074-02760200431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O., Jerlström-Hultqvist J., Castro E., Sherwood E., Ankarklev J., et al. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-R J.C., Cox M.P., Hayman D.T.S. Comparative genetic diversity of Cryptosporidium species causing human infections. Parasitology. 2020;147:1532‒1537. doi: 10.1017/S0031182020001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist C.A., Ali I.K., Kabir M., Alam F., Scherbakova S., et al. Multilocus Sequence Typing System (MLST) reveals a high level of diversity and a genetic component to Entamoeba histolytica virulence. BMC Microbiol. 2012;12:151. doi: 10.1186/1471-2180-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie T.R., Morgan D., Deutsch J.C., Kuhlenschmidt M.S., Salzer J.S., et al. A legacy of low-impact logging does not elevate prevalence of potentially pathogenic protozoa in free-ranging gorillas and chimpanzees in the Republic of Congo: logging and parasitism in African apes. EcoHealth. 2009;6:557–564. doi: 10.1007/s10393-010-0283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Moreno O., Hernandez-Aguilar R.A., Piel A.K., Stewart F.A., Gracenea M., et al. Prevalence and climatic associated factors of Cryptosporidium sp. infections in savanna chimpanzees from Ugalla, Western Tanzania. Parasitol. Res. 2013;112:393–399. doi: 10.1007/s00436-012-3147-8. [DOI] [PubMed] [Google Scholar]
- Goussard B., Collet J.Y., Garin Y., Tutin C.E., Fernandez M. The intestinal entodiniomorph ciliates of wild lowland gorillas (Gorilla gorilla gorilla) in Gabon, West Africa. J. Med. Primatol. 1983;12:239–249. [PubMed] [Google Scholar]
- Graczyk T.K., Bosco-Nizeyi J., Ssebide B., Thompson R.C., Read C., et al. Anthropozoonotic Giardia duodenalis genotype (assemblage) A infections in habitats of free-ranging human-habituated gorillas, Uganda. J. Parasitol. 2002;88:905–909. doi: 10.1645/0022-3395(2002)088[0905:agdgaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Guerrero F.M., Serrano-Martínez E., Tantaleán M., Quispe M., Casas G. Identificación de parásitos gastrointestinales en primates no humanos del Zoológico Parque Natural de Pucallpa. Perú. Rev. Investig. Vet. Perú. 2012;23:469–476. [Google Scholar]
- Gutiérrez-Cisneros M.J., Cogollos R., López-Vélez R., Martín-Rabadán P., Martínez-Ruiz R., et al. Application of real-time PCR for the differentiation of Entamoeba histolytica and E. dispar in cyst-positive faecal samples from 130 immigrants living in Spain. Ann. Trop. Med. Parasitol. 2010;104:145–149. doi: 10.1179/136485910x12607012373759. [DOI] [PubMed] [Google Scholar]
- Helenbrook W.D., Shields W.M., Whipps C.M. Characterization of Blastocystis species infection in humans and mantled howler monkeys, Alouatta palliata aequatorialis, living in close proximity to one another. Parasitol. Res. 2015;114:2517–2525. doi: 10.1007/s00436-015-4451-x. [DOI] [PubMed] [Google Scholar]
- Helenbrook W.D., Wade S.E., Shields W.M., Stehman S.V., Whipps C.M. Gastrointestinal parasites of Ecuadorian mantled howler monkeys (Alouatta palliata aequatorialis) based on fecal analysis. J. Parasitol. 2015;101:341–350. doi: 10.1645/13-356.1. [DOI] [PubMed] [Google Scholar]
- Helenbrook W.D., Whipps C.M. Molecular characterization of Blastocystis in captive and free-ranging new world primates. Platyrrhini. Acta Parasitol. 2021 doi: 10.1007/s11686-021-00397-1. https://doir.org/10.1007/s11686-021-00397-1 (in press) [DOI] [PubMed] [Google Scholar]
- Higuera A., Herrera G., Jimenez P., García-Corredor D., Pulido-Medellín M., et al. Identification of multiple Blastocystis subtypes in domestic animals from Colombia using amplicon-based next generation sequencing. Front. Vet. Sci. 2021;8:732129. doi: 10.3389/fvets.2021.732129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan J.N., Miller W.A., Cranfield M.R., Ramer J., Hassell J., et al. Giardia in mountain gorillas (Gorilla beringei beringei), forest buffalo (Syncerus caffer), and domestic cattle in Volcanoes National Park, Rwanda. J. Wildl. Dis. 2014;50:21–30. doi: 10.7589/2012-09-229. [DOI] [PubMed] [Google Scholar]
- Hublin J.S.Y., Maloney J.G., Santin M. Blastocystis in domesticated and wild mammals and birds. Res. Vet. Sci. 2021;135:260–282. doi: 10.1016/j.rvsc.2020.09.031. [DOI] [PubMed] [Google Scholar]
- Irbis C., Garriga R., Kabasawa A., Ushida K. Phylogenetic analysis of Troglodytella abrassarti isolated from chimpanzees (Pan troglodytes verus) in the wild and in captivity. J. Gen. Appl. Microbiol. 2008;54:409–413. doi: 10.2323/jgam.54.409. [DOI] [PubMed] [Google Scholar]
- Ježková J., Limpouchová Z., Prediger J., Holubová N., Sak B., et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: cryptosporidiidae), the species adapted to the nutria (Myocastor coypus) Microorganisms. 2021;9:813. doi: 10.3390/microorganisms9040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirků-Pomajbíková K., Čepička I., Kalousová B., Jirků M., Stewart F., et al. Molecular identification of Entamoeba species in savanna woodland chimpanzees (Pan troglodytes schweinfurthii) Parasitology. 2016;143:741–748. doi: 10.1017/s0031182016000263. [DOI] [PubMed] [Google Scholar]
- Johnson-Delaney C.A. Parasites of captive nonhuman primates. Vet. Clin. North Am. Exot. Anim. Pract. 2009;12:563–581. doi: 10.1016/j.cvex.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Johnston A.R., Gillespie T.R., Rwego I.B., McLachlan T.L., Kent A.D., et al. Molecular epidemiology of cross-species Giardia duodenalis transmission in western Uganda. PLoS Neglected Trop. Dis. 2010;4:e683. doi: 10.1371/journal.pntd.0000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M.R., Rume F.I., Rahman A., Zhang Z., Li J., et al. Evidence for zoonotic potential of Enterocytozoon bieneusi in its first molecular characterization in captive mammals at Bangladesh National Zoo. J. Eukaryot. Microbiol. 2020;67:427–435. doi: 10.1111/jeu.12792. [DOI] [PubMed] [Google Scholar]
- Köster P.C., Dashti A., Bailo B., Muadica A.S., Maloney J.G., et al. Occurrence and genetic diversity of protist parasites in captive non-human primates, zookeepers, and free-living sympatric rats in the Córdoba Zoo Conservation Centre, Southern Spain. Animals (Basel) 2021;11:700. doi: 10.3390/ani11030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster P.C., Renelies-Hamilton J., Dotras L., Llana M., Vinagre-Izquierdo C., et al. Molecular detection and characterization of enteric and hematic parasites in wild chimpanzees (Pan troglodytes verus) in Senegal. Animals (Basel) 2021;11:3291. doi: 10.3390/ani11113291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouassi R.Y., McGraw S.W., Yao P.K., Abou-Bacar A., Brunet J., et al. Diversity and prevalence of gastrointestinal parasites in seven non-human primates of the Taï National Park, Côte d'Ivoire. Parasite. 2015;22:1. doi: 10.1051/parasite/2015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalewski M.M., Salzer J.S., Deutsch J.C., Raño M., Kuhlenschmidt M.S., et al. Black and gold howler monkeys (Alouatta caraya) as sentinels of ecosystem health: patterns of zoonotic protozoa infection relative to degree of human-primate contact. Am. J. Primatol. 2011;73:75–83. doi: 10.1002/ajp.20803. [DOI] [PubMed] [Google Scholar]
- Lalle M., Pozio E., Capelli G., Bruschi F., Crotti D., et al. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lankester F., Mätz-Rensing K., Kiyang J., Jensen S.A., Weiss S., et al. Fatal ulcerative colitis in a western lowland gorilla (Gorilla gorilla gorilla) J. Med. Primatol. 2008;37:297–302. doi: 10.1111/j.1600-0684.2008.00287.x. [DOI] [PubMed] [Google Scholar]
- Lau Y.L., Anthony C., Fakhrurrazi S.A., Ibrahim J., Ithoi I., et al. Real-time PCR assay in differentiating Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii infections in Orang Asli settlements in Malaysia. Parasites Vectors. 2013;6:250. doi: 10.1186/1756-3305-6-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leśniańska K., Perec-Matysiak A. Wildlife as an environmental reservoir of Enterocytozoon bieneusi (Microsporidia) – analyses of data based on molecular methods. Ann. Parasitol. 2017;63:265–281. doi: 10.17420/ap6304.113. [DOI] [PubMed] [Google Scholar]
- Levecke B. Ghent University; Ghent (Belgium): 2010. The Importance of Gastrointestinal Protozoa in Captive Non-human Primates. [dissertation Thesis] [Google Scholar]
- Levecke B., Dorny P., Geurden T., Vercammen F., Vercruysse J. Gastrointestinal protozoa in non-human primates of four zoological gardens in Belgium. Vet. Parasitol. 2007;148:236–246. doi: 10.1016/j.vetpar.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Li W., Feng Y., Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019;35:436–451. doi: 10.1016/j.pt.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Li W., Kiulia N.M., Mwenda J.M., Nyachieo A., Taylor M.B., et al. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J. Clin. Microbiol. 2011;49:4326–4329. doi: 10.1128/jcm.05051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xiao L. Multilocus sequence typing and population genetic analysis of Enterocytozoon bieneusi: host specificity and its impacts on public health. Front. Genet. 2019;10:307. doi: 10.3389/fgene.2019.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney J.G., Jang Y., Molokin A., George N.S., Santin M. Wide genetic diversity of Blastocystis in white-tailed deer (Odocoileus virginianus) from Maryland, USA. Microorganisms. 2021;9:1343. doi: 10.3390/microorganisms9061343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney J.G., Santin M. Mind the gap: new full-length sequences of Blastocystis subtypes generated via Oxford Nanopore Minion sequencing allow for comparisons between full-length and partial sequences of the small subunit of the ribosomal RNA gene. Microorganisms. 2021;9:997. doi: 10.3390/microorganisms9050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbaya A.W., Udendeye U.J. Gastrointestinal parasites of captive and free-roaming primates at the Afi Mountain Primate Conservation Area in Calabar, Nigeria and their zoonotic implications. Pakistan J. Biol. Sci. 2011;14:709–714. doi: 10.3923/pjbs.2011.709.714. [DOI] [PubMed] [Google Scholar]
- Medkour H., Amona I., Laidoudi Y., Davoust B., Bitam I., et al. Parasitic infections in African humans and non-human primates. Pathogens. 2020;9:561. doi: 10.3390/pathogens9070561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia R., Vicuña Y., Broncano N., Sandoval C., Vaca M., et al. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am. J. Trop. Med. Hyg. 2013;88:1041–1047. doi: 10.4269/ajtmh.12-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menu E., Davoust B., Mediannikov O., Akiana J., Mulot B., et al. Occurrence of ten protozoan enteric pathogens in three non-human primate populations. Pathogens. 2021;10:280. doi: 10.3390/pathogens10030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Bray R.S. Entamoeba histolytica infections in the chimpanzee (Pan satyrus) J. Parasitol. 1966;52:386–388. [Google Scholar]
- Milozzi C., Bruno G., Cundom E., Mudry M.D., Navone G.N. Intestinal parasites of Alouatta caraya (Primates, Ceboidea): preliminary study in semi-captivity and in the wild in Argentina. Mastozool. Neotrop. 2012;19:163–178. [Google Scholar]
- Muehlenbein M.P. Parasitological analyses of the male chimpanzees (Pan troglodytes schweinfurthii) at ngogo, kibale national park, Uganda. Am. J. Primatol. 2005;65:167–179. doi: 10.1002/ajp.20106. [DOI] [PubMed] [Google Scholar]
- Mul I.F., Paembonan W., Singleton I., Wich S.A., van Bolhuis H.G. Intestinal parasites of free-ranging, semicaptive, and captive Pongo abelii in Sumatra, Indonesia. Int. J. Primatol. 2007;28:407–420. doi: 10.1007/s10764-007-9119-7. [DOI] [Google Scholar]
- Munene E., Otsyula M., Mbaabu D.A., Mutahi W.T., Muriuki S.M., et al. Helminth and protozoan gastrointestinal tract parasites in captive and wild-trapped African non-human primates. Vet. Parasitol. 1998;78:195–201. doi: 10.1016/s0304-4017(98)00143-5. [DOI] [PubMed] [Google Scholar]
- Nath B.G., Islam S., Chakraborty A. Prevalence of parasitic infection in captive non human primates of Assam State Zoo, India. Vet. World. 2012;5:614–616. doi: 10.5455/VETWORLD.2012.614-616. [DOI] [Google Scholar]
- O'Donoghue P.J., Gasser R.B., Tribe A. New host record for the entodiniomorphid ciliate, Troglodytella abrassarti, from siamangs (Hylobates syndactylus) Int. J. Parasitol. 1993;23:415–418. doi: 10.1016/0020-7519(93)90020-y. 1993. [DOI] [PubMed] [Google Scholar]
- Parsons M.B., Travis D., Lonsdorf E.V., Lipende I., Roellig D.M., et al. Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášová J., Uzlíková M., Kostka M., Petrželková K.J., Huffman M.A., et al. Diversity and host specificity of Blastocystis in syntopic primates on Rubondo Island, Tanzania. Int. J. Parasitol. 2011;41:1113–1120. doi: 10.1016/j.ijpara.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Phillips K.A., Haas M.E., Grafton B.W., Yrivarren M. Survey of the gastrointestinal para-sites of the primate community at Tambopata National Reserve, Peru. J. Zool. 2004;264:149–151. doi: 10.1017/S0952836904005680. [DOI] [Google Scholar]
- Pomajbíková K., Petrzelková K.J., Profousová I., Petrásová J., Kisidayová S., et al. A survey of entodiniomorphid ciliates in chimpanzees and bonobos. Am. J. Phys. Anthropol. 2010;142:42–48. doi: 10.1002/ajpa.21191. [DOI] [PubMed] [Google Scholar]
- Pomajbíková K., Petrželková K.J., Profousová I., Petrášová J., Modrý D. Discrepancies in the occurrence of Balantidium coli between wild and captive African great apes. J. Parasitol. 2010;96:1139–1144. doi: 10.1645/GE-2433.1. [DOI] [PubMed] [Google Scholar]
- Ponce-Gordo F., Fonseca-Salamanca F., Martínez-Díaz R.A. Genetic heterogeneity in internal transcribed spacer genes of Balantidium coli (Litostomatea, Ciliophora) Protist. 2011;162:774–794. doi: 10.1016/j.protis.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Ponce-Gordo F., García-Rodríguez J.J. Balantioides coli. Res. Vet. Sci. 2021;135:424–431. doi: 10.1016/j.rvsc.2020.10.028. [DOI] [PubMed] [Google Scholar]
- Ragazzo L.J., Zohdy S., Velonabison M., Herrera J., Wright P.C., et al. Entamoeba histolytica infection in wild lemurs associated with proximity to humans. Vet. Parasitol. 2018;249:98–101. doi: 10.1016/j.vetpar.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez L.V., Bautista D.C., Corredor A.F., Flórez A.C., et al. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014;22:223–228. doi: 10.1016/j.meegid.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Read C.M., Monis P.T., Thompson R.C. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect. Genet. Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Regan C.S., Yon L., Hossain M., Elsheikha H.M. Prevalence of Entamoeba species in captive primates in zoological gardens in the UK. PeerJ. 2014;2:e492. doi: 10.7717/peerj.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelies-Hamilton J., Noguera-Julian M., Parera M., Paredes R., Pacheco L., et al. Exploring interactions between Blastocystis sp., Strongyloides spp. and the gut microbiomes of wild chimpanzees in Senegal. Infect. Genet. Evol. 2019;74:104010. doi: 10.1016/j.meegid.2019.104010. [DOI] [PubMed] [Google Scholar]
- Rivero M.R., Feliziani C., De Angelo C., Tiranti K., Salomon O.D., et al. Giardia spp., the most ubiquitous protozoan parasite in Argentina: human, animal and environmental surveys reported in the last 40 years. Parasitol. Res. 2020;119:3181–3201. doi: 10.1007/s00436-020-06853-7. [DOI] [PubMed] [Google Scholar]
- Ryan U., Cacciò S.M. Zoonotic potential of Giardia. Int. J. Parasitol. 2013;43:943–956. doi: 10.1016/j.ijpara.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Ryan S.J., Brashares J.S., Walsh C., Milbers K., Kilroy C., et al. A survey of gastro-intestinal parasites of Olive Baboons (Papio anubis) in human settlement areas of Mole National Park, Ghana. J. Parasitol. 2012;98:885–888. doi: 10.1645/ge-2976.1. [DOI] [PubMed] [Google Scholar]
- Sá R.M., Petrášová J., Pomajbíková K., Profousová I., Petrželková K.J., et al. Gastrointestinal symbionts of chimpanzees in Cantanhez National Park, Guinea-Bissau with respect to habitat fragmentation. Am. J. Primatol. 2013;75:1032–1041. doi: 10.1002/ajp.22170. [DOI] [PubMed] [Google Scholar]
- Sak B., Petrzelkova K.J., Kvetonova D., Mynarova A., Shutt K.A., et al. Long-term monitoring of microsporidia, Cryptosporidium and Giardia infections in western lowland gorillas (Gorilla gorilla gorilla) at different stages of habituation in dzanga sangha protected areas, Central African Republic. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santín M., Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011;90:363–371. doi: 10.1016/j.rvsc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Scicluna S.M., Tawari B., Clark C.G. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Soares R.M., de Souza S.L., Silveira L.H., Funada M.R., Richtzenhain L.J., et al. Genotyping of potentially zoonotic Giardia duodenalis from exotic and wild animals kept in captivity in Brazil. Vet. Parasitol. 2011;180:344–348. doi: 10.1016/j.vetpar.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Squire S.A., Ryan U. Cryptosporidium and Giardia in Africa: current and future challenges. Parasites Vectors. 2017;10:195. doi: 10.1186/s13071-017-2111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold C.R., Alfellani M., Clark C.G. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect. Genet. Evol. 2012;12:263‒273. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Alfellani M.A., Nørskov-Lauritsen S., Prip K., Victory E.L., et al. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Strait K., Else J.G., Eberhard M.L. Nonhuman Primates in Biomedical Research. second ed. Elsevier Inc.; Cambridge, MA, USA: 2012. Parasitic diseases of non-human primates. [Google Scholar]
- Sulaiman I.M., Fayer R., Bern C., Gilman R.H., Trout J.M., et al. Triose phosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichroeb J.A., Kutz S.J., Parkar U., Thompson R.C., Sicotte P. Ecology of the gastrointestinal parasites of Colobus vellerosus at Boabeng-Fiema, Ghana: possible anthropozoonotic transmission. Am. J. Phys. Anthropol. 2009;140:498–507. doi: 10.1002/ajpa.21098. [DOI] [PubMed] [Google Scholar]
- Tiangtip R., Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health. 2002;7:357–364. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- Vallo P., Petrželková K.J., Profousová I., Petrášová J., Pomajbíková K., et al. Molecular diversity of entodiniomorphid ciliate Troglodytella abrassarti and its co-evolution with chimpanzees. Am. J. Phys. Anthropol. 2012;148:525–533. doi: 10.1002/ajpa.22067. [DOI] [PubMed] [Google Scholar]
- Verweij J.J., Oostvogel F., Brienen E.A., Nang-Beifubah A., Ziem J., et al. Prevalence of Entamoeba histolytica and Entamoeba dispar in northern Ghana. Trop. Med. Int. Health. 2003;8:1153–1156. doi: 10.1046/j.1360-2276.2003.01145.x. [DOI] [PubMed] [Google Scholar]
- Verweij J.J., Schinkel J., Laeijendecker D., van Rooyen M.A., van Lieshout L., et al. Real-time PCR for the detection of Giardia lamblia. Mol. Cell. Probes. 2003;17:223–225. doi: 10.1016/s0890-8508(03)00057-4. [DOI] [PubMed] [Google Scholar]
- Verweij J.J., Vermeer J., Brienen E.A., Blotkamp C., Laeijendecker D., et al. Entamoeba histolytica infections in captive primates. Parasitol. Res. 2003;90:100–103. doi: 10.1007/s00436-002-0808-z. [DOI] [PubMed] [Google Scholar]
- Vlčková K., Kreisinger J., Pafčo B., Čížková D., Tagg N., et al. Diversity of Entamoeba spp. in African great apes and humans: an insight from Illumina MiSeq high-throughput sequencing. Int. J. Parasitol. 2018;48:519–530. doi: 10.1016/j.ijpara.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Volotão A.C., Júnior J.C., Grassini C., Peralta J.M., Fernandes O. Genotyping of Giardia duodenalis from southern Brown howler monkeys (Alouatta clamitans) from Brazil. Vet. Parasitol. 2008;158:133–137. doi: 10.1016/j.vetpar.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Weedall G.D., Clark C.G., Koldkjaer P., Kay S., Bruchhaus I., et al. Genomic diversity of the human intestinal parasite Entamoeba histolytica. Genome Biol. 2012;13:R38. doi: 10.1186/gb-2012-13-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz A., Heymann E.W., Petney T.N., Taraschewski H.F. The influence of human settlements on the parasite community in two species of Peruvian tamarin. Parasitology. 2010;137:675–684. doi: 10.1017/s0031182009991570. [DOI] [PubMed] [Google Scholar]
- Widmer G., Köster P.C., Carmena D. Cryptosporidium hominis infections in non-human animal species: revisiting the concept of host specificity. Int. J. Parasitol. 2020;50:253–262. doi: 10.1016/j.ijpara.2020.01.005. [DOI] [PubMed] [Google Scholar]
- Zahedi A., Bolland S.J., Oskam C.L., Ryan U. Cryptosporidium abrahamseni n. sp. (Apicomplexa: cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae) Exp. Parasitol. 2021;223:108089. doi: 10.1016/j.exppara.2021.108089. [DOI] [PubMed] [Google Scholar]
- Zhao W., Zhou H., Jin H., Sun L., Li P., et al. Genotyping of Enterocytozoon bieneusi among captive long-tailed macaques (Macaca fascicularis) in Hainan Province: high genetic diversity and zoonotic potential. Acta Trop. 2020;201:105211. doi: 10.1016/j.actatropica.2019.105211. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Li W., Deng L., Song Y., Wu K., et al. Multilocus genotyping of Enterocytozoon bieneusi derived from nonhuman primates in southwest China. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


