Abstract
Metabolic labeling of primate cells revealed the existence of phosphorylated and hypophosphorylated DNA polymerase α-primase (Pol-Prim) populations that are distinguishable by monoclonal antibodies. Cell cycle studies showed that the hypophosphorylated form was found in a complex with PP2A and cyclin E-Cdk2 in G1, whereas the phosphorylated enzyme was associated with a cyclin A kinase in S and G2. Modification of Pol-Prim by PP2A and Cdks regulated the interaction with the simian virus 40 origin-binding protein large T antigen and thus initiation of DNA replication. Confocal microscopy demonstrated nuclear colocalization of hypophosphorylated Pol-Prim with MCM2 in S phase nuclei, but its presence preceded 5-bromo-2′-deoxyuridine (BrdU) incorporation. The phosphorylated replicase exclusively colocalized with the BrdU signal, but not with MCM2. Immunoprecipitation experiments proved that only hypophosphorylated Pol-Prim associated with MCM2. The data indicate that the hypophosphorylated enzyme initiates DNA replication at origins, and the phosphorylated form synthesizes the primers for the lagging strand of the replication fork.
The initiation of chromosomal DNA replication in eukaryotes can be divided into two major independent events (reviewed in references 5 and 10). The first event takes place during the G1 phase, when a preinitiation complex is formed at the origin of replication. The complex formation requires the sequential binding of the origin recognition complex (ORC), Cdc6, and minichromosome maintenance proteins (MCM). The assembly of MCM on the chromatin plays an important role in generating a replication-competent, licensed origin. The second event occurs during the G1/S transition, when cyclin-dependent protein kinases (Cdks) as well as the Cdc7/Dbf4 kinase convert the preinitiation complex into an active replication fork by an unknown process. In addition, activation of the preinitiation complex at the G1/S transition requires sequential chromatin binding of the replication factors Cdc45, RP-A, and polymerase α-primase (Pol-Prim) (37). After the RP-A-dependent unwinding of replication origins (37), the essential initiator Pol-Prim is recruited to the unwound origin, most likely by specific protein-protein interaction with chromatin-bound Cdc45 and/or RP-A (1, 8, 19).
Pol-Prim isolated from a wide range of phylogenetic species contains a similar set of four polypeptides. The enzyme complex is composed of a 180-kDa polypeptide containing the catalytic function; a polypeptide of about 70 kDa, which is thought to be the regulatory subunit; and two polypeptides of 58 and 48 kDa associated with primase activity (reviewed in reference 38). Pol-Prim is the only enzyme capable of initiating DNA synthesis de novo by first synthesizing an RNA primer and then extending the primer by polymerization to produce a short 30-nucleotide DNA extension, which yields an RNA-DNA primer of approximately 40 nucleotides in length (reviewed in reference 38). Subsequently, in an ATP-dependent process, RF-C initiates polymerase switching that leads to recruitment of DNA polymerase δ and its auxiliary factors at the DNA primer-template junctions to synthesize the leading strands (18). During lagging strand synthesis, Pol-Prim synthesizes every RNA-DNA primer that is extended by either DNA polymerase δ or ε (35). Therefore, Pol-Prim is engaged in the initiation as well as elongation process of eukaryotic DNA replication. The unique double function of Pol-Prim makes it a likely target for cell cycle-regulating factors like Cdks that are involved in the control mechanism of DNA replication. Cell cycle-dependent phosphorylation of the 180- and 70-kDa subunits of Pol-Prim were observed in human cells at G2/M, in Schizosaccharomyces pombe in late S, and in Saccharomyces cerevisiae at G1/S (11, 25, 26). As shown, in vitro phosphorylation of Pol-Prim by purified cyclin E-Cdk2, cyclin A-Cdk2/Cdk1, and cyclin B-Cdk1 not only modified the p180 and p70 subunits, but also influenced the origin-dependent priming activity in vitro (33, 34). The authors demonstrated that cyclin A-Cdk2, but not cyclin E-Cdk2, inhibits the replication activity of human Pol-Prim in a simian virus 40 (SV40) initiation assay, whereas the activities of DNA polymerase α (Pol α) and the tightly associated primase were not impaired in simple enzyme assays (33, 34). However, the regulatory mechanism that allows primer synthesis in simple enzyme assays but inhibits origin-dependent priming activity was not elucidated.
We present evidence for the existence of two immunologically and functionally distinct populations of Pol-Prim in primate cells. Our findings suggest that two differently phosphorylated Pol-Prim populations are required for the two different priming events in eukaryotic DNA replication. We propose that only hypophosphorylated Pol-Prim is recruited to the origin of replication by specific protein-protein interaction to synthesize the first primer for the leading strand, whereas the phosphorylated form that is incompetent for origin binding synthesizes the primers for the lagging strand of the replication fork.
MATERIALS AND METHODS
Cell culture.
CV-1 cells (African green monkey kidney cell line; ATCC CL70) were cultured as exponentially growing monolayers on 145-mm-diameter plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and 2 mM glutamine at 37°C. Cultures were split 1:5 and used between passages 6 and 15. CCRF-CEM (human acute lymphoblastic T-cell leukemia line; ATCC 119-CCL) cells were maintained as suspension cells in RPMI 1640 supplemented with 10% FCS and 2 mM glutamine at 37°C. Sf9 insect cells (American Type Culture Collection) were cultured in spinner flasks, and High Five insect cells (Invitrogen) were cultured as monolayers in TC100 medium supplemented with 10% FCS and 4 mM glutamine at 27°C. Hybridomas of the HP and SJK series specific for Pol α (28; I. Dornreiter, unpublished data), PAb101 specific for SV40 T antigen (12), anti-EE specific for EE-tag, and 12CA5 specific for hemagglutinin (HA) epitope were grown as spinner cultures in RPMI 1640-DMEM (1:1) supplemented with 5% FCS and 2 mM glutamine at 37°C.
Counterflow centrifugal elutriation and FACS analysis.
Exponentially growing CEM cells were separated into eight fractions based on cell size by centrifugal elutriation with a Beckman JM25 elutriator. Approximately 2 × 109 cells in elutriation buffer (phosphate-buffered saline [PBS] supplemented with 1% FCS, 0.1% glucose, and 0.3 mM EDTA) were loaded onto a Beckman JE6b elutriator rotor (standard chamber) at a rotor speed of 2,500 rpm and buffer pump speed of 35 rpm. Fractions were collected at a rotor speed of 2,000 rpm, with small incremental increases in the pump speed. Two-hundred-milliliter fractions were collected, aliquoted (2 × 107 cells), and frozen at −80°C. Samples (106 cells) were taken to be analyzed for DNA content by fluorescence-activated cell sorting (FACS), after being fixed in 80% ethanol (−20°C) and subsequently stained with propidium iodide (50 μg/ml) containing DNase-free RNase A (10 μg/ml) in PBS for 30 min at 37°C. FACS was performed with an EPICS XL four-color flow cytometer (Coulter Electronics GmbH).
BrdU labeling and confocal microscopy.
CV-1 cells grown on glass coverslips were arrested in G0/G1 phase by incubation with isoleucine-depleted DMEM for 2 days. Transit into S phase was accomplished by exchanging the medium for complete DMEM supplemented with 10% FCS. Cell cycle synchronization was verified by FACS as described above. At appropriate times after restimulation, DNA synthesis was monitored by pulse-labeling the culture 20 min before fixation with the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) (0.1 mM; Sigma). Cells were washed once with PBS and fixed for 10 min in −20°C acetone. After rehydration in PBS, the cells were blocked with normal goat serum, incubated with the respective mouse monoclonal anti-Pol α antibodies, followed by Texas red-labeled goat anti-mouse immunoglobulin G (IgG) (Dianova). After rinsing with buffer, the probes were fixed with 1% paraformaldehyde for 10 min. DNA was denatured with 2 M HCl for 10 min at 37°C. After neutralization with buffer, the cells were incubated with normal mouse serum, and subsequently, the sites of BrdU incorporation were localized with a fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody (Boehringer Mannheim). To detect MCM2 and Pol-Prim simultaneously, S phase CV-1 cells were fixed in acetone and rehydrated in PBS, and nonspecific binding sites were blocked as described above. The prepared cells were incubated with the respective mouse monoclonal anti-p180 antibodies and rabbit polyclonal anti-MCM2 antiserum (1:400), followed by Texas red-labeled goat anti-mouse IgG (Dianova) and FITC-labeled goat anti-rabbit IgG (Dianova). Confocal images were taken with a ×63 objective on a Leica laser-scan microscope equipped with an argon-krypton laser.
Expression of recombinant proteins.
To coexpress human recombinant tetrameric Pol-Prim with dimeric PP2A, cyclin E-Cdk2, cyclin A-Cdk2, cyclin E-Cdk2/PP2A, or cyclin A-Cdk2/PP2A, 2 × 107 High Five insect cells were coinfected with each recombinant baculovirus at a multiplicity of infection of 5 and incubated for 42 to 46 h at 27°C. Collected cells were homogenized in lysis buffer A containing 50 mM HEPES (pH 7.4), 150 mM NaCl, and 0.1% NP-40 plus inhibitors (0.1 mM aprotinin, 0.1 mM leupeptin, 0.2 mM pepstatin A, 1 mM Pefabloc). Protein expression of each recombinant protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting with the appropriate antibody as described below.
Immunological reagents.
The following primary IgGs were used: anti-human hypophosphorylated pRb monoclonal antibody (G99-549; Pharmingen); anti-cyclin E monoclonal antibody (HE12; Pharmingen); anti-cyclin A rabbit polyclonal antibody (H-432; Santa Cruz); anti-cyclin B1 monoclonal antibody (GNS-1; Pharmingen); anti-human Cdk2 rabbit polyclonal antibody specific for p33cdk2 (Upstate Biotechnology); anti-Cdk1 rabbit polyclonal antibody (Ab-1; Calbiochem); anti-PSTAIRE rabbit polyclonal antibody (Calbiochem); anti-PP2A-A rabbit polyclonal antibody (Calbiochem); methylation-sensitive anti-PP2A-C rabbit polyclonal antibody raised against C-terminal peptide corresponding to residues 298 to 309 of human PP2Ac (Ab298–309; Calbiochem); anti-PP2A-C rabbit polyclonal antibody raised against peptide corresponding to the C-terminal amino acid residues 296 to 309 of human PP2A-C (Upstate Biotechnology); and anti-MCM2 goat polyclonal antibody (Santa Cruz).
Protein manipulations.
CV-1 cells were washed in PBS, scraped from plates, and centrifuged for 5 min. CEM cells grown as a suspension culture were washed in PBS before centrifugation. Cell pellets were resuspended in lysis buffer A plus protease inhibitors as described above. Protein concentrations were determined according to the method of Bradford by using a commercial reagent with bovine serum albumin (BSA) as the standard (Bio-Rad). SDS-PAGE was carried out on 8, 10, or 12% polyacrylamide gels as indicated in the figure legends with prestained molecular weight marker proteins (Sigma). After SDS-PAGE, proteins were transferred to Immobilon membranes (Millipore), and blots were blocked in Tris-buffered saline–Tween (TBST) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% [vol/vol] Tween 20) containing 5% (wt/vol) dried milk at room temperature for 1 h. The membranes were then incubated with antibodies in TBST containing 5% dried milk as indicated in the figure legends. The secondary antibodies were either peroxidase-conjugated goat anti-rabbit, anti-mouse, or donkey anti-goat Igs (Rockland; 1:15,000) in TBST containing 5% dried milk. Detection of the protein bands was performed with an enhanced chemiluminescence reagent (Amersham) as instructed by the manufacturer. For evaluation of the methylation status of PP2A-C, immunoblots were treated for 20 min with 0.2 M NaOH at 30°C before immunodetection.
Immunoprecipitation and coimmunoprecipitation analysis.
For labeling proteins, 107 cells were incubated for 2 h with 1 mCi of [32P]orthophosphate (Amersham) at 37°C in low-phosphate medium or for 1 h with 300 μCi of [35S]methionine-cysteine in methionine-free medium. Metabolically labeled cells were lysed on ice for 30 min in lysis buffer A plus inhibitors (0.1 mM aprotinin, 0.1 mM leupeptin, 0.2 mM pepstatin, 1 mM Pefabloc, 50 mM NaF, 0.1 mM sodium orthovanadate). Pol-Prim complexes were immunoprecipitated by monoclonal anti-p180 antibodies, as indicated in the figure legends, from cell lysates that were normalized to equal amounts of protein concentrations. Immunoprecipitates were dissolved with 20 μl of 2× Laemmli sample buffer and then resolved by SDS-PAGE (10% polyacrylamide) and, for [35S]methionine labeling, fluorographed by using PPO (2,5-diphenyloxazol; Merck). For immunoprecipitation-Western blot analysis, antibodies were first covalently cross-linked to protein G-Sepharose according to a standard protocol (13). Five hundred to 1,000 μg of total-cell lysate was immunoprecipitated, subjected to SDS-PAGE, and Western blotted as described above. Sequential immunoprecipitations of Pol-Prim from crude cell lysates were performed with the respective anti-p180 monoclonal antibody bound to protein G-Sepharose. Five hundred micrograms of cell lysate was incubated with the SJK132-20 beads for 30 min at 4°C. The extract was separated from the beads by low-speed centrifugation. The procedure was repeated three times with the same anti-p180 monoclonal antibody to deplete >95% of the SJK132-20-reactive Pol-Prim from the extract, determined by measuring Pol α activity as described below, followed by two incubations with anti-p180 monoclonal antibody HP180-12. Each immunoprecipitate was subjected to SDS-PAGE (10% polyacrylamide), and the 180- and 70-kDa subunits were analyzed by Western blotting. Immunoprecipitation of recombinant Pol-Prim complexes that were coexpressed with different Cdks and/or PP2A from baculovirus-infected insect cells was performed with lysates that were normalized to equal amounts of coexpressed proteins, as determined by Western blotting.
Histone H1 kinase assay.
To test the activity of the HA-tagged recombinant kinase in the baculovirus coexpression system, 50 μg of lysates from baculovirus-infected cells was immunoprecipitated with the monoclonal anti-HA antibody 12CA5. The immunoprecipitates were assayed for kinase activity with 1 μg of H1 added as substrate in histone-kinase buffer (20 mM HEPES-KOH [pH 7.5], 1 mM dithiothreitol [DTT], 10 mM MgCl2, 4 mM EGTA, 5 mM NaF, 1 mM EDTA, 0.1 mg of BSA per ml, 0.1 mM ATP, 1 μCi of [γ-32P]ATP per assay) for 30 min at 37°C. Reactions were separated by SDS-PAGE (10% polyacrylamide), and incorporation of phosphate was visualized by autoradiography.
Mutagenesis of p180 and p70 cDNA.
To create mutations in the three potential Cdk phosphorylation sites 174, 209, and 219, which are located in the N-terminal region of human Pol α cDNA (40), the corresponding serine or threonine codons were exchanged for alanine codons by overlap extension PCR (14). In addition, 4 of the 10 putative Cdk phosphorylation sites of the regulatory 70-kDa subunit were altered to alanine (S141, S147, S152, and T156). The mutagenesis was carried out as described previously (34), and the triple p180 mutant (3xA) plus the quadruple p70 mutant (4xA) were constructed by successive repetition of this method to introduce the second, third, or fourth mutation into the cDNA.
Purification of recombinant proteins.
Recombinant human Pol-Prim complexes coexpressed with recombinant PP2Acore, cyclin E-Cdk2, cyclin A-Cdk2, cyclin E-Cdk2/PP2A, or cyclin A-Cdk2/PP2A and without additional modifying enzymes were purified from 3 × 108 baculovirus-infected insect cells on anti-p180 antibody SJK237-71–Sepharose beads as described earlier (33). DNA Pol α assays were performed on gapped duplex (“activated”) DNA as described earlier (23). One unit of Pol α activity was defined as the amount that catalyzes the incorporation of 1 nmol of dAMP into acid-insoluble material in 1 h at 37°C. The primase activity was determined by using poly(dT) single-stranded DNA (ssDNA) as the substrate and quantifying the radioactive oligoribonucleotide products with a PhosphorImager as described previously (24). One unit of primase was defined as the amount that leads to an incorporation of 1 nmol of dNMP in 1 h at 37°C by the Klenow elongation assay. Topoisomerase I and recombinant SV40 large T antigen (T Ag) were purified as described previously (21).
Initiation on φx174 ssDNA and origin-dependent SV40 initiation reactions.
Circular φx174 ssDNA served as model system with which to study the priming activity of DNA primase that is required for the initiation of Okazaki fragment synthesis. Initiation reaction mixtures contained 750 ng of φx174 ssDNA in reaction buffer (20 mM Tris-acetate [pH 7.3], 10 mM Mg-acetate, 1 mM DTT, 1 mM ATP, 0.1 mM GTP, 0.1 mM UTP, 0.01 mM CTP, 0.1 mg of BSA per ml) in the presence of 10 μCi of [α-32P]CTP (3,000 Ci/mmol; NEN). A total of 0.5 U of DNA primase of each differently phosphorylated, immunoaffinity-purified recombinant Pol-Prim complex was added to the reaction mixtures, as indicated in the figure legends, which were then incubated for 1 h at 37°C. The DNA primase activities of the various modified Pol-Prim complexes were determined as described in the section on purification of proteins. The reaction products were precipitated with 0.8 M LiCl, 10 μg of sonicated salmon sperm DNA (Sigma), and 120 μl of ethanol for 15 min on dry ice. The washed (75% ethanol–water) and dried products were dissolved in loading buffer (45% formamide, 5 mM EDTA, 0.09% xylene cyanol FF, 0.09% bromphenol blue) at 65°C for 30 min and separated by denaturing PAGE (20% polyacrylamide) for 3 to 4 h at 600 V. The reaction products were visualized by autoradiography. Origin-dependent initiation reaction mixtures (33) contained 250 ng of pUC-HS DNA, 600 ng of SV40 T Ag, 500 ng of RP-A, and 300 ng of topoisomerase I in initiation buffer (30 mM HEPES-KOH [pH 7.8], 7 mM Mg-acetate, 4 mM EGTA [pH 7.8], 5 mM NaF, 1 mM DTT, 0.2 mM UTP, 0.2 mM GTP, 0.01 mM CTP, 4 mM ATP, 40 mM creatine phosphate, 1 μg of creatine kinase, 0.2 mg of BSA per ml) in the presence of 20 μCi of [α-32P]CTP (3,000 Ci/mmol; NEN). Recombinant Pol-Prim was added as indicated in the figure legends. The reaction products were separated on 20% urea gels as described above and analyzed by autoradiography.
RESULTS
In vivo identification of two differently phosphorylated Pol-Prim populations by monoclonal anti-p180 antibodies.
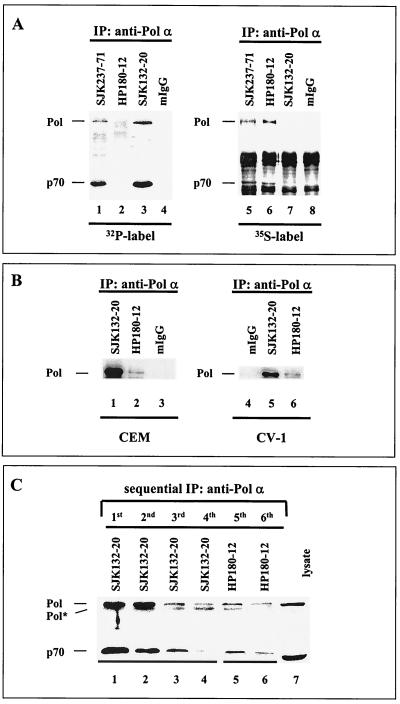
Each of the p180 and p70 subunits of human Pol-Prim harbor 10 S/T-P motifs in their protein sequence that are potential Cdk1/Cdk2 phosphorylation sites (4, 40; Dornreiter, unpublished). Consequently, Cdk-dependent modification generates various phosphorylated populations of the tetrameric human Pol-Prim in vivo and in vitro (25, 33, 34). To identify such Pol-Prim populations in vivo, monoclonal anti-Pol α antibodies were tested with immunoprecipitation assays. Monoclonal anti-p180 antibodies SJK132-20, -237-71, and -287-38 (28) and HP180-7, -180-12, and -180-35 (Dornreiter, unpublished) were used in the immunoprecipitation assays. Asynchronous CEM cells were pulse-labeled with [32P]orthophosphate to probe for the phosphorylated Pol-Prim or with [35S]methionine-cysteine to identify the newly synthesized and non- or hypophosphorylated form. Autoradiography demonstrated that the 32P-labeled 180- and 70-kDa Pol α subunits were immunoprecipitated by SJK132-20 (Fig. 1A, lane 3), whereas the 35S incorporation into SJK132-20-immunoprecipitated Pol-Prim was not detectable (Fig. 1A, lane 7). In contrast, the antibody HP180-12 immunoprecipitated newly synthesized Pol-Prim, which contained large amounts of 35S label (Fig. 1A, lane 6), but 32P was not incorporated into the p180 and p70 subunits (Fig. 1A, lane 2). All other tested monoclonal antibodies of the SJK and HP series immunoprecipitated 32P- and 35S-labeled Pol-Prim (Fig. 1A, lanes 1 and 5) (data not shown). Sequential immunoprecipitation experiments demonstrated that the monoclonal antibody HP180-12, which precipitates very small amounts of Pol-Prim from different primate cell lysates (Fig. 1B, lanes 2 and 6), recognizes the remaining SJK132-20–Pol-Prim complex (Fig. 1C). The results demonstrate the identification of two anti-p180 monoclonal antibodies: SJK132-20 preferentially detects the phosphorylated and abundant Pol-Prim, whereas HP180-12 exclusively binds to a non- or hypophosphorylated population of Pol-Prim that is newly synthesized.
FIG. 1.
Two distinct populations of human Pol-Prim can be distinguished by monoclonal anti-p180 antibodies in vivo. (A) Asynchronously growing CEM cells were metabolically labeled with [32P]orthophosphate for 2 h (lanes 1 to 4) or with [35S]methionine-cysteine for 1 h (lanes 5 to 8). Equal amounts of protein were used to immunoprecipitate (IP) the Pol-Prim complexes. Immunoprecipitates were separated by SDS-PAGE (8% polyacrylamide) and visualized by autoradiography. (B) Cell lysates (500 μg) from asynchronously growing CEM and CV-1 cells were used to immunoprecipitate Pol-Prim with monoclonal anti-p180 antibodies SJK132-20 (lanes 1 and 5) and HP180-12 (lanes 2 and 6) or normal mouse IgG (mIgG; lanes 3 and 4). Immunoprecipitates were separated by SDS-PAGE (10% polyacrylamide) and analyzed by Western blotting with anti-Pol α monoclonal antibody HP180-7 (1:5). (C) Pol-Prim was sequential immunoprecipitated from CV-1 total-cell lysate (500 μg) four times on SJK132-20–Sepharose beads (lanes 1 to 4) and subsequently two times on HP180-12–Sepharose beads (lanes 5 and 6). Immunoprecipitates were separated by SDS-PAGE (10% polyacrylamide), and the p180 and p70 subunits were detected with monoclonal anti-Pol α antibody HP180-7 (1:5) and anti-p70 antiserum (1:5,000). One hundred micrograms of lysate from CV-1 cells was used as a positive control for the expression of the p180 and p70 subunits of Pol-Prim (lane 7). Pol* indicates a 165-kDa degradation product of the 180-kDa subunit of Pol-Prim.
Cyclin A-Cdk-dependent phosphorylation of Pol-Prim abolishes the immunoreactivity of anti-p180 monoclonal antibody HP180-12.
To further analyze whether phosphorylation and dephosphorylation of Pol-Prim affect the immunoreactivity of the monoclonal anti-p180 antibodies SJK132-20 and HP180-12, recombinant human Pol-Prim was baculovirus coexpressed with different cyclin-Cdks and the protein serine/threonine phosphatase 2A (PP2A). PP2A is a trimeric holoenzyme consisting of a catalytic subunit (C) and a structural subunit (A) plus a variable regulatory subunit (B) (reviewed in reference 39), with the latter determining the substrate specificity (15). For the baculovirus coexpression experiments, only the PP2A core was used, which consists of the A and C subunits. Coexpression of PP2A with cyclin A-Cdk, cyclin B-Cdk1, or cyclin E-Cdk2 and vice versa did not inhibit the activity of the Cdks (Fig. 2A) or PP2A (data not shown).
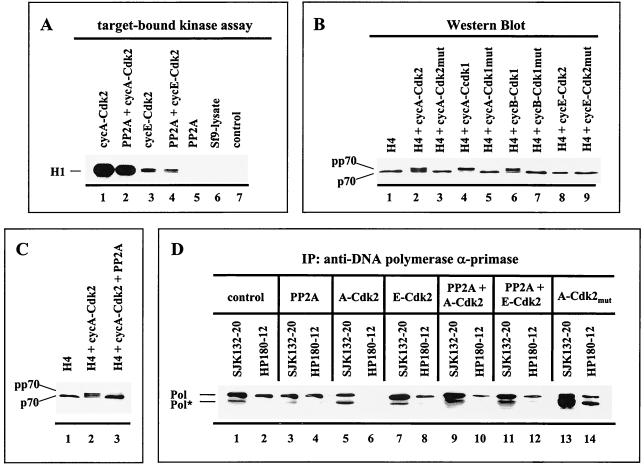
FIG. 2.
Cyclin A- and cyclin B-dependent kinases induce a p70 mobility shift that is reversible with PP2A and correlates with the abrogation of the immunoreactivity of the monoclonal anti-Pol α antibody HP180-12. (A) Fifty micrograms of lysates from baculovirus-infected cells was immunoprecipitated (IP) with the monoclonal antibody 12CA5 specific for the HA-tagged kinase and tested for kinase activity with 1 μg of H1 added as a substrate (lanes 1 to 4). 12CA5-immunoprecipitates, which expressed recombinant PP2A only (lane 5), and mock-infected insect cells (lane 6) were analyzed in an analogous target-bound kinase assay. The control reaction mixture consisted of kinase buffer and H1 (lane 7). Reactions were separated by SDS-PAGE (10% polyacrylamide) and visualized by autoradiography. (B) Five micrograms of lysates that express the four subunits of Pol-Prim (H4) in the absence (lane 1) or presence of the indicated recombinant Cdks (lanes 2 to 9) was subjected to SDS-PAGE (8% polyacrylamide). The p70 subunit was detected by Western blotting with an anti-p70 antiserum (1:5,000). (C) Recombinant Pol-Prim was coexpressed with the modifying enzymes as indicated. Five micrograms of lysates was separated by SDS-PAGE (8% polyacrylamide), and p70 was detected as described above. (D) Recombinant Pol-Prim (lanes 1 and 2) coexpressed with modifying enzymes, as indicated in lanes 3 to 14, was immunoprecipitated (IP) with anti-p180 monoclonal antibodies SJK132-20 and HP180-12. Immunoprecipitates obtained from equal amounts of protein from total-cell lysates were immunoblotted and analyzed with monoclonal anti-p180 antibody HP180-7 (1:5). Pol* indicates a 165-kDa degradation product of Pol α.
To detect the Cdk-catalyzed phosphorylation and PP2A-catalyzed dephosphorylation of Pol-Prim in the coexpression system, an electrophoretic mobility shift assay was used as described previously (34). Coexpression of Pol-Prim with cyclin A-Cdk2, cyclin A-Cdk1, or cyclin B-Cdk1 yielded a phosphorylation-induced slower-migrating form of the p70 subunit, designated pp70 (Fig. 2B, lanes 2, 4, and 6). In contrast, the slower-migrating pp70 was not observed when Pol-Prim was coexpressed with cyclin E-Cdk2 (Fig. 2B, lane 8) or the respective inactive Cdk mutants (Fig. 2B, lanes 3, 5, and 7). The phosphorylation-induced shift of p70 was completely removed when PP2A was present in the cyclin A-Cdk2 or -Cdk1 and cyclin B-Cdk1 coexpression experiments (Fig. 2C, lane 3) (data not shown), indicating that PP2A removes specifically Cdk-incorporated phosphate groups from serine/threonine residues of Pol-Prim. The same results were obtained when commercially available purified PP2A was added to the phosphorylated Pol-Prim complexes (data not shown).
The results demonstrate that modification of Pol-Prim with cyclin A- or cyclin B-Cdk produces a higher phosphorylated population, whereas cyclin E-Cdk2 or PP2A generates a hypophosphorylated form. Therefore, the system was used to verify the observed in vivo correlation between the phosphorylation status of Pol-Prim and the reactivity of the monoclonal antibodies HP180-12 and SJK132-20. Immunoprecipitation experiments demonstrated that addition of PP2A reduced the binding of SJK132-20 slightly, but had no effect on HP180-12 (Fig. 2D, lanes 1 to 4). Phosphorylation of Pol-Prim by cyclin A-Cdk2 or cyclin E-Cdk2 had no notable influence on the reactivity of SJK132-20 (Fig. 2D, lanes 1, 5, and 7). However, coexpression with cyclin A-Cdk2 completely abolished the immunoreactivity of HP180-12, whereas cyclin E-Cdk2-catalyzed phosphorylation of Pol-Prim caused a modest decrease in the reactivity of this antibody (Fig. 2D, lanes 2, 6, and 8). Addition of recombinant PP2A to cyclin A-Cdk2-coexpressed Pol-Prim partially restored the reactivity of HP180-12 (Fig. 2D, lanes 6 and 10). To confirm the impact of the cyclin A-Cdk-catalyzed phosphorylation of Pol-Prim on the antibody's immunoreactivity, the replicase was coexpressed with mutant cyclin A-Cdk2 or -Cdk1. As expected, the presence of inactive cyclin A-Cdk did not alter the immunoreactivity of the monoclonal antibody HP180-12 (Fig. 2D, lane 14) (data not shown). The results are consistent with the in vivo immunoprecipitation results depicted in Fig. 1A and suggest that in comparison to SJK132-20, antibody HP180-12 does not recognize cyclin A- or cyclin B-Cdk-phosphorylated Pol-Prim.
In G1, only hypophosphorylated Pol-Prim associates with PP2A.
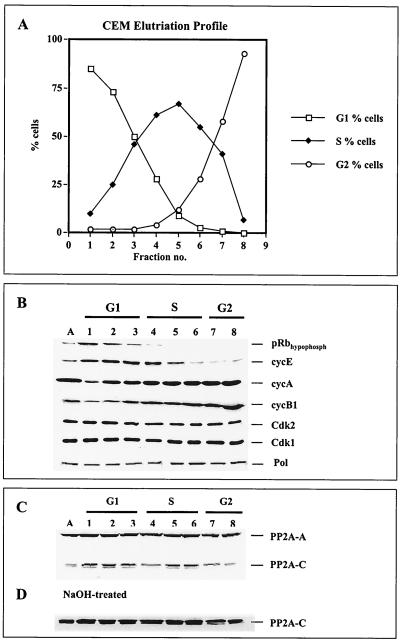
Monoclonal anti-p180 antibodies SJK132-20 and HP180-12 were used in coimmunoprecipitation assays to determine the in vivo-interacting enzymes in question that modulate the replicase during different stages of the cell cycle. CEM cells were fractionated into substages of the cell cycle by counterflow centrifugal elutriation. The efficiency of the elutriation was monitored by FACS analysis (Fig. 3A) and confirmed by Western blot analysis of such cell cycle-regulating factors as retinoblastoma tumor suppressor protein Rb (pRb); cyclins E, A, and B1; Cdk2; Cdk1; and PP2A. As expected, Fig. 3B shows hypophosphorylated pRb exclusively in G1 and G1/S, a steady increase in cyclins A and B1, a decrease in cyclin E in S phase, and a constant expression level of the kinases Cdk2 and Cdk1. Consistent with earlier reports (11, 20, 36), the level of Pol α did not change significantly during the cell cycle (Fig. 3B).
FIG. 3.
Analysis of cell cycle-regulating factors in elutriated human CEM cells. (A) Exponentially growing CEM cells were separated by centrifugal elutriation. From each fraction, 106 cells were stained with propidium iodide and analyzed by FACS for DNA content. (B) Total-cell lysates (100 μg) from each fraction (lanes 1 to 8) were separated by SDS-PAGE (10% polyacrylamide) and analyzed by immunoblotting with anti-hypophosphorylated pRb (1:2,000), anti-cyclin E (1:500), anti-cyclin A (1:2,500), anti-cyclin B1 (1:1,000), anti-Cdk2 (1:1,000), and anti-Cdk1 (1:1,000) antibodies plus anti-p180 antibody HP180-7 (1:5). (C) The same samples (lanes 1 to 8) were used to analyze the PP2A core by immunoblotting. The structural A and catalytic C subunits were detected with anti-PP2A-A antiserum (1:5,000) and methylation-sensitive anti-PP2A-C antiserum (C-terminus Ab298–309, 1:5,000). (D) Immunoblot of the same samples pretreated with 0.2 M NaOH as described in Materials and Methods. PP2A-C was detected with the methylation-sensitive anti-PP2A-C antiserum as described above.
An interesting observation was made when the PP2A levels during different stages of the cell cycle were examined. The structural A subunit of PP2A was constitutively present throughout the cell cycle as described before (27). In contrast, the amount of the catalytic C subunit decreased in early S phase, returned to its initial level in mid-S phase, and declined again in G2 phase (Fig. 3C). Treatment of the blotting membrane with alkali that leads to demethylation of proteins increased the C-terminal immunoreactivity of PP2A-C for the methylation-sensitive anti-PP2A-C antibody, and the protein level of PP2A-C was constant throughout the cell cycle (Fig. 3D).
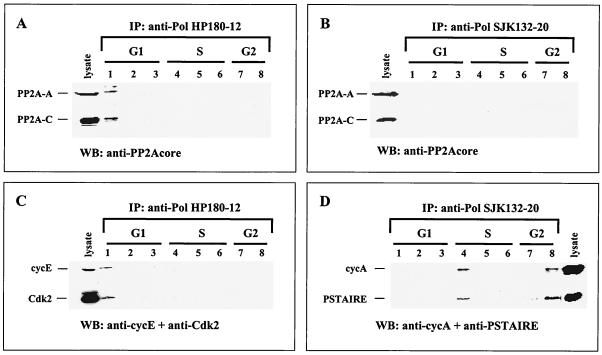
To test for possible cell cycle-dependent association of PP2A with Pol-Prim, HP180-12 and SJK132-20 immune complexes were examined with antibodies specific for the A and C subunits of the trimeric PP2A. The data revealed the presence of the two PP2A core subunits complexed to the HP180-12-reactive Pol-Prim in G1 (Fig. 4A, lane 1). Western blot analysis of the Pol-Prim-associated PP2A also gave a strong signal of the catalytic PP2A subunit in G1 when the methylation-sensitive anti-PP2A-C antibody was used (data not shown), which indicates that Pol-Prim interacts with demethylated PP2A. The phosphatase was not detected in the SJK132-20-derived immune complexes in any substages of the cell cycle (Fig. 4B).
FIG. 4.
Pol-Prim populations interact with PP2A and Cdks in a cell cycle-dependent manner. Cell lysates from elutriated CEM cells (Fig. 3A) were immunoprecipitated (IP) with anti-Pol α antibody HP180-12, specific for the hypophosphorylated form, and anti-Pol α antibody SJK132-20, specific for the phosphorylated enzyme. The immunoprecipitates were separated by SDS-PAGE (12% polyacrylamide), and Pol-Prim-associated proteins were analyzed by Western blotting with antibodies as indicated. Lanes 1 to 8 correspond to the elutriated fractions that were used for the coimmunoprecipitation experiments. Cell lysates (100 μg) from asynchronously growing CEM cells were used as positive controls for protein expression. (A) HP180-12-derived precipitates were analyzed with anti-PP2A-A (1:5,000) and nonmethylation-sensitive anti-PP2A-C (1:1,000) antibodies for associated PP2A (lane 1). (B) SJK132-20-reactive Pol-Prim complexes were probed for associated PP2A as mentioned above. (C) The HP180-12-coimmunoprecipitated PSTAIRE kinase was identified as Cdk2 (lane 1) by using an anti-Cdk2 polyclonal antibody (1:1,000). The same blotting membrane was incubated with a monoclonal anti-cyclin E antibody (1:500). (D) SJK132-20-obtained immunoprecipitates were analyzed with anti-PSTAIRE polyclonal antibody (1:250). The SJK132-20-coimmunoprecipitated PSTAIRE kinase (lanes 4 and 8) could not be identified in more detail. The same immunoblot was incubated with an anti-cyclin A antiserum (1:2,500).
In G1, hypophosphorylated Pol-Prim is complexed to cyclin E-Cdk2, whereas the phosphorylated replicase associates with cyclin A-dependent-kinase in S and G2.
In vivo human Pol-Prim is phosphorylated in a cell cycle-dependent manner (25). In vitro the replicase is phosphorylated by recombinant cyclin E-Cdk2 and cyclin A-Cdk2/Cdk1, as well as cyclin B1-Cdk1 (33, 34), and by coexpression of Cdks in the baculovirus system (Fig. 2B). The cyclin-Cdks that phosphorylate Pol-Prim in vivo have not been identified yet. Therefore, we wanted to determine the associated cyclin-Cdks that modulate Pol-Prim during different stages of the cell cycle. Pol-Prim complexes were immunoprecipitated from elutriated CEM cell lysates (Fig. 3A) with the phosphorylation-specific antibodies HP180-12 and SJK132-20. Western blot analysis with an anti-PSTAIRE antibody showed that the HP180-12 and SJK132-20-reactive Pol-Prim populations coimmunoprecipitated with a PSTAIRE kinase in a cell cycle-dependent manner (data not shown). Since Cdk1, Cdk2, and Cdk3 all contain the PSTAIRE motif, we wanted to identify the associated PSTAIRE kinase by probing the immune complexes with the appropriate anti-Cdk antibodies. The HP180-12-reactive Pol-Prim-associated PSTAIRE kinase was identified as Cdk2 (Fig. 4C, lane 1), whereas none of the available PSTAIRE kinase-specific antibodies and additional non-PSTAIRE kinase-specific antibodies reacted with the SJK132-20-associated PSTAIRE kinase (Fig. 4D, lanes 4 and 8).
In parallel, all HP180-12- and SJK132-20-derived immune complexes were probed for the kinase-associated cyclins A, E, and B1. The HP180-12-associated Cdk2 was complexed to cyclin E only in G1 (Fig. 4C, lane 1), whereas the SJK132-20-associated PSTAIRE kinase was exclusively found in a complex with cyclin A in the S (Fig. 4D, lane 4) and G2 (lane 8) phases. Despite the ability of cyclin B-Cdk1 to phosphorylate Pol-Prim in vitro (33, 34) (Fig. 2B, lane 6), no in vivo interaction was observed (data not shown). In summary, the HP180-12-reactive Pol-Prim associates with PP2A and cyclin E-Cdk2 in G1, whereas the SJK132-20-reactive Pol-Prim interacts with a cyclin A-PSTAIRE-kinase in S and G2.
PP2A-catalyzed dephosphorylation of cyclin A-Cdk-inactivated Pol-Prim restores the origin-dependent initiation activity of the human replicase in vitro.
Cyclin A-Cdk-phosphorylated Pol-Prim is initiation inactive in an origin-dependent replication assay (34). Therefore, we wanted to test whether PP2A-catalyzed dephosphorylation of cyclin A-Cdk-phosphorylated Pol-Prim restores the in vitro origin-dependent initiation activity. To test this hypothesis, recombinant human Pol-Prim was coexpressed with PP2A, cyclin E-Cdk2, cyclin E-Cdk2/PP2A, cyclin A-Cdk2, or cyclin A-Cdk2/PP2A and immunoaffinity purified with anti-p180 monoclonal antibody SJK237-71, which does not discriminate between phosphorylated and hypophosphorylated Pol-Prim (Fig. 1A, lanes 1 and 5).
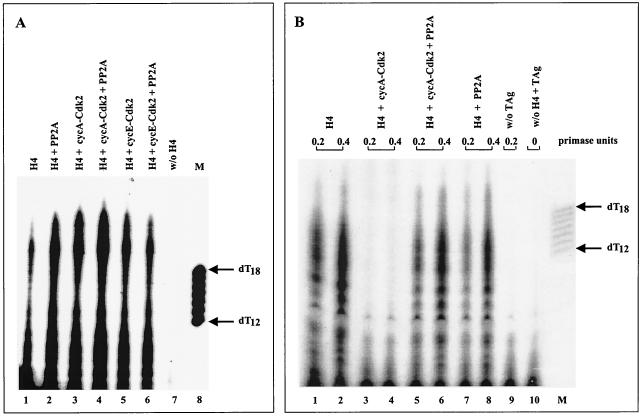
The differently phosphorylated human Pol-Prim complexes displayed no inhibited priming activity on single-stranded circular φx174 template, which was used as a model system to study the enzyme's priming ability during the elongation process of the lagging strand synthesis (Fig. 5A). In contrast, different data were obtained with a plasmid DNA containing the SV40 origin as a substrate for an in vitro initiation assay. In agreement with previous results (33, 34), cyclin A-Cdk2-phosphorylated Pol-Prim did not initiate DNA replication (Fig. 5B, lanes 3 and 4). However, coexpression of Pol-Prim with cyclin A-Cdk2 and PP2A fully restored the origin-dependent initiation activity of the replicase (Fig. 5B, compare lanes 1 and 2 with 5 and 6). No significant difference in the initiation efficiency was observed with PP2A (Fig. 5B, compare lanes 1 and 2 with 7 and 8)-, cyclin E-Cdk2-, or cyclin E-Cdk2/PP2A-modified Pol-Prim (data not shown). The results demonstrate that cyclin A-Cdk2-mediated phosphorylation does not affect the basic priming activity of the replicase that is needed for the lagging strand synthesis, but inactivates the origin-dependent initiation activity of Pol-Prim; dephosphorylation by PP2A fully restores this specific activity.
FIG. 5.
Cyclin A-Cdk2 phosphorylation of Pol-Prim inhibits the origin-dependent initiation activity that is reactivated by PP2A. (A) Equal amounts (0.5 U of DNA primase) of differently phosphorylated purified Pol-Prim complexes (H4) were assayed for the ability to synthesize primers at φx174 ssDNA that serves as a model system for lagging strand synthesis. Products were separated on 20% urea gels and analyzed by autoradiography (lanes 1 to 6). The radioactive material that is detectable in the absence of Pol-Prim is shown in lane 7. The arrows on the right indicate the length of 5′-end-labeled oligo(dT12–18) marker (lane M). (B) Equal amounts (0.2 and 0.5 U of DNA primase) of the same Pol-Prim complexes were tested in an origin-dependent initiation SV40 DNA replication assay and analyzed as described above. The initiation products of untreated Pol-Prim (H4) are shown in lanes 1 and 2. Cyclin A-Cdk2-phosphorylated (lanes 3 and 4), cyclin A-Cdk2/PP2A-modified (lanes 5 and 6), and PP2A-dephosphorylated Pol-Prim (lanes 7 and 8) complexes were incubated and analyzed under identical conditions. Control reactions were carried out in the absence of SV40 T Ag (lane 9) or Pol-Prim (lane 10). The arrows on the right indicate the length of 5′-end-labeled oligo(dT12–18) marker (lane M).
Cyclin A-Cdk2-dependent phosphorylation of Pol-Prim abrogates the interaction with the origin-binding protein SV40 T Ag and thus initiation of DNA replication.
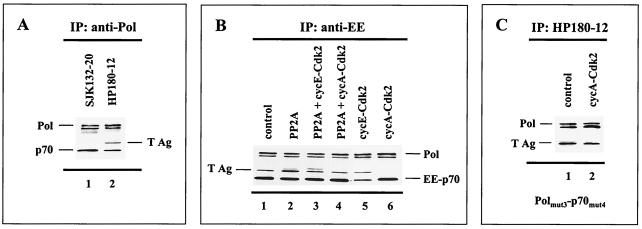
Pol-Prim has no affinity for double-stranded DNA; its binding to the origin of replication depends upon the interaction with an origin-binding protein or complex. In the SV40 system, loading of Pol-Prim into the origin requires the interaction with the viral origin-binding protein T Ag in the preinitiation complex (9). To elucidate the mechanism that leads to inhibition of the origin-dependent initiation activity of Pol-Prim upon cyclin A-Cdk-dependent phosphorylation, we tested whether the phosphorylation status of the replicase determines complex formation with T Ag and thus initiation of DNA replication. To generate the hypophosphorylated and phosphorylated Pol-Prim populations, recombinant human Pol-Prim and T Ag were baculovirus coexpressed with modifying enzymes as indicated in Fig. 6B. Immunoprecipitation of Pol-Prim from the baculovirus control expression, where no additional modifying enzyme was included, demonstrated that T Ag-complexed Pol-Prim was only detectable with the monoclonal antibody HP180-12 (Fig. 6A, lane 2), but not with SJK132-20 (Fig. 6A, lane 1). The data indicate that T Ag interacts exclusively with the hypophosphorylated Pol-Prim population.
FIG. 6.
Cyclin A-Cdk-dependent phosphorylation of Pol-Prim abrogates complex formation with SV40 T Ag. Human Pol-Prim and T Ag were baculovirus coexpressed with modifying recombinant enzymes as indicated. One hundred to 200 μg of protein extract was used for the coimmunoprecipitation (IP) assays. The precipitates were subjected to SDS-PAGE (10% polyacrylamide) followed by Western blot analysis. (A) Pol-Prim complexes that were coexpressed with T Ag were precipitated by anti-p180 monoclonal antibodies SJK132-20 (lane 1) and HP180-12 (lane 2). The p180 (Pol) and p70 (p70) subunits of precipitated Pol-Prim plus coimmunoprecipitating T Ag were detected with monoclonal antibodies anti-p180 HP180-7 (1:5), anti-p70 (1:5, 000), and anti-T Ag Pab101 (1:10). (B) Complex formation of recombinant Pol-Prim with T Ag in the absence (lane 1) or presence of coexpressed PP2A (lane 2), PP2A/cyclin E-Cdk2 (lane 3), PP2A/cyclin A-Cdk2 (lane 4), cyclin E-Cdk2 (lane 5), and cyclin A-Cdk2 (lane 6) was investigated by immunoprecipitation with the monoclonal antibody anti-EE for the EE-tagged p70 subunit. Immunoprecipitates were analyzed with monoclonal antibodies anti-p180 HP180-7 (1:5) and anti-EE (1:10). The presence of coimmunoprecipitating T Ag was detected as described above. (C) Complex formation of the Pol α(3xA) and p70(4xA) phosphorylation mutants with T Ag in the absence (lane 1) or presence (lane 2) of cyclin A-Cdk2 was investigated with the monoclonal p180 antibody HP180-12, which is specific for its hypophosphorylated antigen. The presence of mutant Pol α and coimmunoprecipitating T Ag was detected with monoclonal antibodies anti-p180 HP180-7 (1:5) and anti-T Ag Pab101 (1:10).
Since phosphorylation of Pol-Prim influences the immunoreactivity of HP180-12 and SJK132-20 in vivo (Fig. 1A) and in vitro (Fig. 2D), an EE-tagged p70 subunit and the anti-EE monoclonal antibody were used to investigate whether the phosphorylation status of Pol-Prim affects the complex formation with T Ag. Figure 6B demonstrates that the coexpression of Pol-Prim and T Ag with PP2A, Cdks, and PP2A/Cdks had no impact on the assembly of the Pol-Prim complex. The observed cyclin A-Cdk2-generated slower-migrating form of pp70 (Fig. 2A) was not resolved in this SDS gel system. In the next step, the Pol-Prim immune complexes were probed with an anti-T Ag monoclonal antibody. Coexpression with PP2A or cyclin E-Cdk2 did not abrogate the binding of T Ag to Pol-Prim (Fig. 6B, lanes 2 and 5). However, the complex formation of Pol-Prim with T Ag was completely eradicated when cyclin A-Cdk2 was present in the baculovirus coexpression system (Fig. 6B, lane 6). Due to the fact that PP2A catalyzed the dephosphorylation of cyclin A-Cdk2-phosphorylated Pol-Prim (Fig. 2C, lane 3) and as a result restored the origin-dependent initiation activity of the replicase (Fig. 5B, lanes 5 and 6), we tested the effect of PP2A on the abolished Pol-Prim–T Ag interaction. As expected, the complex formation of Pol-Prim with T Ag was established when PP2A was present in the cyclin A-Cdk2 coexpression experiment (Fig. 6B, lane 4).
To investigate whether the cyclin A-Cdk2-induced abrogation of the Pol-Prim–T Ag interaction is indeed caused by phosphorylation of the Pol-Prim complex, mutations in the Cdk phosphorylation sites of the 180- and 70-kDa phosphoproteins were introduced. As shown before, the T Ag-binding site of Pol α lies within amino acid (aa) region 195 to 313 of the N terminus (9). Hence, the two putative Cdk phosphorylation sites (S209 and T219) in this particular region and an adjacent site (T174) were exchanged with alanine. A target-bound kinase assay with the Pol α–glutathione S-transferase (GST) fusion protein C (aa 195 to 313) (11) used as a substrate demonstrated that these Cdk sites are phosphorylated by cyclin A-Cdk2 in vitro (data not shown). In addition, 4 of the 10 putative Cdk phosphorylation sites of the regulatory p70 subunit, which are phosphorylated by cyclin A-Cdk2 in vitro (34), were altered to alanine (S141, S147, S152, and T156) as described previously (34). The coexpression of the mutated replicase with T Ag in the presence of cyclin A-Cdk2 not only eliminated the phosphorylation-induced shift of p70 (data not shown), but also yielded Pol-Prim-associated T Ag (Fig. 6C, lane 2). Moreover, Pol-Prim could be immunoprecipitated in the presence of cyclin A-Cdk2 with the monoclonal antibody HP180-12, which is specific for the hypophosphorylated form of the replicase (Fig. 6C, lane 2). The data strongly suggest that cyclin A-Cdk-dependent phosphorylation of Pol-Prim inhibited the origin-dependent initiation activity of the replicase due to prevention of the complex formation with the viral origin-binding protein T Ag. In conclusion, only hypophosphorylated Pol-Prim is competent for T Ag binding and therefore origin firing.
Only hypophosphorylated Pol-Prim does not colocalize with sites of active DNA synthesis but colocalizes and binds to MCM2.
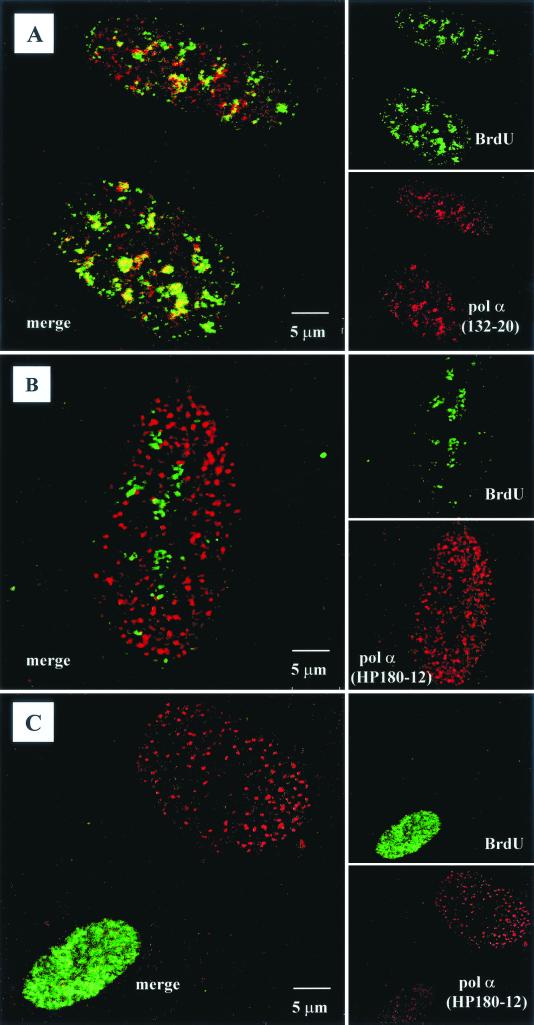
To investigate the different roles of the two distinct phosphorylated Pol-Prim populations in mammalian cells, we determined the spatial relationship between the sites of DNA synthesis and the presence of the two immunologically distinct Pol-Prim complexes by confocal microscopy with synchronized CV-1 cells. For synchronization, CV-1 cells were arrested by isoleucine withdrawal in G0/G1. In S phase cells (10 to 16 h postrelease), sites of active DNA synthesis were visualized by incorporation of the nucleotide analog BrdU, followed by labeling with an FITC-conjugated anti-BrdU antibody. The BrdU signal varied from densely packed dots to larger speckles that clearly colocalized with accumulations of the phosphorylated form of SJK132-20-reactive Pol-Prim (Fig. 7A). In contrast, a dot-like distribution of the HP180-12-reactive Pol-Prim complex within the nucleus was observed, and this pattern was most prominent in cells that exhibited a weak BrdU signal (Fig. 7B). In those cells, the dots representing hypophosphorylated Pol-Prim were restricted to areas of the nucleus devoid of a BrdU signal (Fig. 7B). In nuclei that presented an increased BrdU signal, labeling with the Pol α antibody HP180-12 diminished, but was present in specific dots that did not colocalize with the BrdU signal (Fig. 7C). Therefore, we assumed that the hypophosphorylated form of Pol-Prim might be located at chromosomal origins where DNA synthesis was not yet initiated and is implicated in the initiation of bidirectional DNA replication. In contrast, the phosphorylated enzyme might be involved in the initiation process of the discontinuous lagging strand DNA synthesis.
FIG. 7.
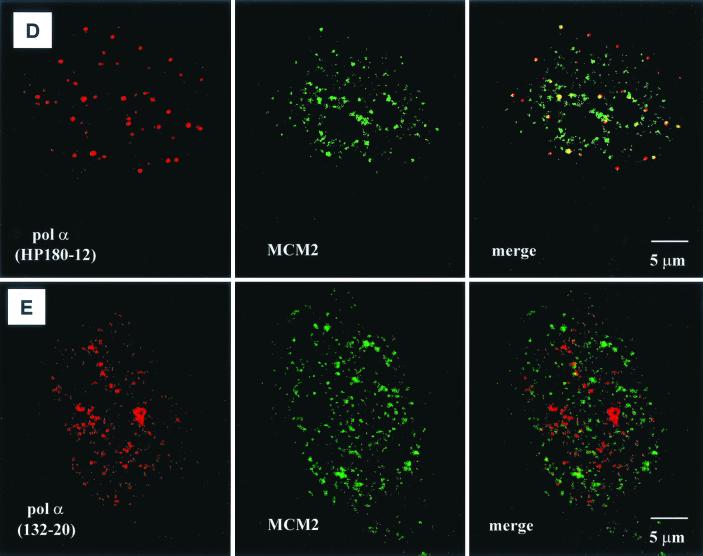
Localization of SJK132-20- and HP180-12-reactive Pol-Prim populations in replicating CV-1 cells by confocal microscopy. (A) Phosphorylated Pol-Prim colocalized with sites of active DNA synthesis in S phase cells identified by BrdU incorporation (right site, upper panel). Accumulations of the SJK132-20-reactive and therefore phosphorylated Pol-Prim (right site, lower panel) colocalized with the BrdU signal (left site). (B) Hypophosphorylated Pol-Prim detected with anti-p180 monoclonal antibody HP180-12 (right site, lower panel) does not colocalize with sites of active DNA synthesis in S phase cells (right site, upper panel). The merged image showed that hypophosphorylated Pol-Prim is not present at sites of DNA replication (left site). (C) Late-S-phase cells, identified by an increased number of large BrdU foci (right site, upper panel), showed a sharp reduction of the HP180-12 signal (right site, lower panel, and left site, merged image). (D) Hypophosphorylated Pol-Prim colocalized with MCM2 in early-S-phase cells. The merged image showed 50% colocalization of HP180-12-reactive Pol-Prim with the MCM2 speckles (right panel). (E) The merged image illustrates that the phosphorylated Pol-Prim (SJK132-20 reactive) does not colocalize with MCM2 (right panel).
To further analyze whether the hypophosphorylated form of Pol-Prim might be located at origins where DNA synthesis was not yet initiated, confocal studies with an anti-MCM2 antibody were performed. MCM2 is a component of a mammalian prereplication complex (reviewed in reference 16) that is loaded onto early- and late-replicating chromatin throughout G1 and rapidly excluded from active replication forks during S phase (6, 31). Synchronized CV-1 cells were used in double-labeling experiments to monitor the presence of HP180-12-reactive Pol-Prim and MCM2 in early-S-phase nuclei (10 h postrelease). As observed before, the hypophosphorylated Pol-Prim appeared as a discrete dot-like pattern (Fig. 7D, left panel), whereas MCM2 showed a more diffuse distribution and existed as well in larger speckles within the nucleus (Fig. 7D, middle panel). After extraction of the nucleus, only the larger speckles of MCM2 remained, which indicates that the speckles reflect MCM2 that is tightly associated with chromatin (data not shown). Unfortunately, the extraction method could not be used to obtain a more distinct MCM2 signal, because the loosely chromatin-bound Pol-Prim was also removed from the nucleus (data not shown). In nonextracted early-S-phase nuclei, the merged image shows that approximately 50% of the HP180-12-reactive Pol-Prim population colocalizes only with the larger MCM2 speckles (Fig. 7D, right panel). This finding suggests that the yellow dots might reflect the location of MCM2 and hypophosphorylated Pol-Prim at the time of origin firing. In contrast, the phosphorylated and SJK132-20-reactive Pol-Prim that was found at active sites of DNA replication never colocalized with the MCM2 speckles (Fig. 7E, right panel).
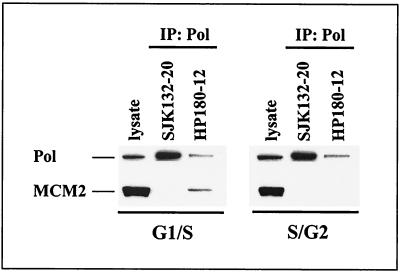
This finding was strengthened by immunoprecipitation experiments that were carried out with elutriated CEM cells. Fractions 4 and 6, which correspond to early- and late-S-phase cells (Fig. 3A), respectively, were used to immunoprecipitate Pol-Prim with phosphorylation-specific anti-p180 antibodies SJK132-20 and HP180-12. Figure 8 demonstrates that only HP180-12-reactive and therefore hypophosphorylated Pol-Prim interacts with MCM2 exclusively in early-S-phase cells, but not in S/G2 phase cells. In contrast, SJK132-20-reactive and therefore phosphorylated Pol-Prim was not found complexed to MCM2. The data strongly suggest diverse roles for the two differently phosphorylated and immunologically distinct Pol-Prim populations in mammalian DNA replication.
FIG. 8.
MCM2 interacts exclusively with hypophosphorylated Pol-Prim in G1/S. Fractions 4 (G1/S) and 6 (S/G2) from elutriated CEM cells (Fig. 3A) were immunoprecipitated (IP) with anti-p180 antibodies as indicated. Five hundred micrograms of protein extract was used to precipitate phosphorylated Pol-Prim with SJK132-20. One thousand micrograms of protein extract from G1/S and 2,000 μg from S/G2 cells were used to immunoprecipitate hypophosphorylated Pol-Prim with HP180-12. Cell lysates (50 μg) from each fraction were used as positive controls for protein expression (lysate). The precipitates were subjected to SDS-PAGE (8% polyacrylamide) and analyzed by Western blotting. The presence of p180 (Pol) and coimmunoprecipitating MCM2 was detected with monoclonal antibody anti-p180 HP180-7 (1:5) and polyclonal antibody anti-MCM2 (1:200).
DISCUSSION
Metabolic labeling of primate cells and in vitro phosphorylation studies revealed the existence of phosphorylated and hypophosphorylated Pol-Prim populations that can be distinguished by monoclonal anti-p180 antibodies (Fig. 1A and 2D). The significance of the presence of two distinguishable Pol-Prim populations in vivo was underlined by the finding that they interact physically and functionally with PP2A and different Cdks in a cell cycle-dependent manner. In S and G2, a cyclin A-PSTAIRE kinase bound exclusively to the phosphorylated form of Pol-Prim (Fig. 4D). The associated kinase could not be identified with available anti-Cdk antibodies. One possible explanation is that the respective Cdk epitope is modified posttranslationally and therefore is not recognized by these antibodies. Alternatively, the SJK132-20-reactive Pol-Prim population interacts with a novel, yet unknown cyclin A-dependent PSTAIRE kinase.
In G1, only HP180-12-reactive and therefore hypophosphorylated Pol-Prim was found in a complex with cyclin E-Cdk2 and PP2A (Fig. 4A and C). We have not yet identified the variable regulatory B subunit of the Pol-Prim-complexed PP2A, which mediates the substrate specificity of the phosphatase (3, 15) and is involved in the intracellular localization of the enzyme (39). Recently, Lin et al. (17) presented evidence that PP2A is essential for the initiation, but not for the elongation, of DNA replication. Despite binding of the prereplication components ORC, Cdc6, and MCM to chromatin, DNA replication was not initiated in PP2A-depleted Xenopus extracts (17). The data indicate that PP2A might be necessary for the assembly of an initiation-competent prerecognition complex or for the binding of additional initiation factors. As shown, replication of SV40 DNA requires PP2A-catalyzed dephosphorylation of specific serine residues within the origin-binding protein T Ag (32). In addition, among several heterotrimeric forms of PP2A, only one form activated the origin-binding and -unwinding properties of T Ag (3). Therefore, dephosphorylation of Pol-Prim by an unknown heterotrimeric form of PP2A might be essential to activate the origin competence of the replicase in G1. The observed methylation of PP2A-C in S (Fig. 3C, lane 4) indicates an interconversion of the regulatory and substrate specificity-mediating B subunit during the G1/S transition (41), which might lead to a change in substrate specificity towards Pol-Prim. The interconversion of PP2A at the G1/S boundary could be part of a mechanism that prevents dephosphorylation and consequently reactivation of origin-competent Pol-Prim after the cyclin A-dependent kinase-inactivating phosphorylation step. This conclusion is supported by the fact that cyclin A-Cdk-inactivated Pol-Prim is reactivated by PP2A-catalyzed dephosphorylation to initiate SV40 DNA replication in vitro (Fig. 5B, lanes 5 and 6).
Eukaryotic viral initiator proteins like T Ag interact directly with and recruit Pol-Prim to the origin of replication (4, 7–9). Accordingly, a correlation between the phosphorylation status of Pol-Prim and the ability to interact with SV40 T Ag was investigated. We could demonstrate that only HP180-12, which is specific for the hypophosphorylated replicase immunoprecipitated the T Ag-complexed recombinant human Pol-Prim readily from baculovirus lysates (Fig. 6A, lane 2) and SV40-infected CV-1 cells (data not shown). However, the phosphorylated and SJK132-20-reactive Pol-Prim was never found in a complex with T Ag in vitro (Fig. 6A, lane 1) or in vivo (data not shown). We could show that cyclin A-Cdk-dependent phosphorylation of Pol-Prim abrogated the interaction of Pol-Prim with T Ag (Fig. 6B, lane 6). Importantly, PP2A-catalyzed dephosphorylation of cyclin A-Cdk-phosphorylated Pol-Prim reestablished the complex formation with T Ag (Fig. 6B, lane 4). To prove that cyclin A-Cdk phosphorylation of Pol-Prim and not phosphorylation of T Ag is responsible for the abolished complex formation, a mutant Pol-Prim was used. In the presence of active cyclin A-Cdk, the mutant Pol α(3xA)-p70(4xA) primase was recognized by the phosphorylation-sensitive monoclonal anti-p180 antibody HP180-12 and indeed complexed to T Ag (Fig. 6C, lane 2). Previously, two populations of murine Pol-Prim were identified that differ in their affinity for polyomavirus (Py) T Ag and their ability to initiate Py origin-dependent DNA synthesis (22). Only a small fraction of the total Pol α activity that was retained by the Py T Ag column initiated Py origin-dependent DNA synthesis. In contrast, the Pol-Prim activity in the nonbound murine fraction did not initiate DNA replication. The authors speculated that the two Pol-Prim populations might differ in posttranslational modification, which could explain the different properties of the murine replicase. We have presented evidence that the phosphorylation status of human Pol-Prim determines the specific protein-protein interaction with the origin-binding protein T Ag and thus initiation of DNA replication.
Further insight into the mechanism that regulates loading of Pol-Prim into origins and consequently initiation of DNA replication was derived from confocal studies. Confocal microscopy showed that only the phosphorylated Pol-Prim population is present at sites of DNA synthesis (Fig. 7A), whereas the hypophosphorylated form is detectable in the nucleus before the onset of DNA replication or in replicating cells at sites at which DNA replication was not yet initiated (Fig. 7B). Essential initiation factors like MCM4, MCM7, Cdc45, and RP-A dissociate from the origin after initiation and move with the eukaryotic DNA replication fork (2, 30). Therefore, we chose MCM2 as an origin marker, because in mammalian cells, the factor is rapidly excluded from active replication forks during S phase and consequently is not associated with engaged replication forks (6, 31). In replicating cells, the merged image shows approximately 50% colocalization of HP180-12-reactive Pol-Prim with the MCM2 speckles (Fig. 7D, right panel). We conclude that the yellow dots might reflect origin-bound hypophosphorylated Pol-Prim and MCM2 at the time of origin firing, whereas the noncolocalizing hypophosphorylated Pol-Prim could be located at origins at the time after firing and MCM2 displacement to synthesize the first RNA-DNA primer for the leading strand. Since it was not possible to extract the nucleus to obtain a more distinct MCM2 pattern, noncolocalizing MCM2 might reflect the chromatin-unbound fraction. This assumption is based on the fact that at the G1/S border, about 50% of MCM2 is chromatin bound, whereas the other half is present in the soluble nucleosolic fraction (2, 29). The colocalization data were supported by the finding that only hypophosphorylated Pol-Prim binds to the origin marker MCM2, as shown by coimmunoprecipitation experiments (Fig. 8). Complex formation of MCM2 with HP180-12-reactive Pol-Prim was detected only in early-S-phase cells, but not in late-S-phase cells. Data indicate that after origin firing formation of new MCM2-Pol-Prim complexes might be prevented by cyclin A-Cdk2 phosphorylation of Pol-Prim as it was observed for the Pol-Prim interaction with viral origin-binding protein T Ag (Fig. 6B, lane 6).
In conclusion, we suggest that the small fraction of hypophosphorylated Pol-Prim that colocalized and interacted with the origin-binding protein MCM2 is required exclusively for the initiation of origin-dependent DNA replication, but not for the elongation of previously engaged replication forks. However, the BrdU-colocalizing and abundant phosphorylated form of Pol-Prim that did not colocalize or associate with MCM2 synthesizes the primers for the Okazaki fragments on the lagging strand of the replication fork.
ACKNOWLEDGMENTS
We are grateful to T. S.-F. Wang for the recombinant baculovirus encoding human Pol α, B. Hemmings for recombinant baculovirus encoding PP2A, S. E. Kearsey for MCM2 antisera, G. Walter for providing the anti-EE hybridoma, and K. Weisshart for purified human RP-A.
The financial support of the Deutsche Forschungsgemeinschaft (De212/8–2, Na190/6–3, Na190/8–1, and Na190/12–1), Deutsche Krebshilfe (10–1417-De), and NATO (CRG920123) is gratefully acknowledged. The IMB is financially supported by Freistaat Thüringen and Bundesministerium für Bildung und Forschung. The Heinrich-Pette-Institute is financially supported by Freie und Hansestadt Hamburg and Bundesministerium für Gesundheit.
REFERENCES
- 1.Aparicio O M, Stout A M, Bell S P. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Cegielska A, Shaffer S, Derua R, Goris J, Virshup D M. Different oligomeric forms of protein phosphatase 2A activate and inhibit simian virus 40 DNA replication. Mol Cell Biol. 1994;14:4616–4623. doi: 10.1128/mcb.14.7.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins K L, Russo A A R, Tseng B Y, Kelly T J. The role of the 70 kDa subunit of human DNA polymerase α in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diffley J F X. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrova D S, Todorov I T, Melendy T, Gilbert D M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dornreiter I, Höβ A, Arthur A K, Fanning E. SV40 T antigen binds directly to the catalytic subunit of DNA polymerase α. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dornreiter I, Erdile F L, Gilbert I, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase α-primase with replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornreiter I, Copeland W C, Wang T S-F. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta A, Bell S P. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 11.Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurney E G, Harrison R O, Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigen and for similarities among nonviral T antigens. J Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, New York, N.Y.: Cold Spring Harbor Press; 1988. [Google Scholar]
- 14.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis—a practical approach. New York, N.Y: Oxford University Press; 1991. pp. 217–249. [Google Scholar]
- 15.Kamibayashi C, Estes R, Licksteig R L, Yang S I, Craft C, Mumby M C. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J Biol Chem. 1994;269:20139–20148. [PubMed] [Google Scholar]
- 16.Kearsey S E, Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 17.Lin X-H, Walter J, Scheidtmann K, Ohst K, Newport J, Walter G. Protein phosphatase 2A is required for the initiation of chromosomal DNA replication. Proc Natl Acad Sci USA. 1998;95:14693–14698. doi: 10.1073/pnas.95.25.14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maga G, Stucki M, Spadari S, Hübscher U. DNA polymerase switching. I. Replication factor C displaces DNA polymerase α prior to PCNA loading. J Mol Biol. 2000;295:791–801. doi: 10.1006/jmbi.1999.3394. [DOI] [PubMed] [Google Scholar]
- 19.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazawa H, Izumi M, Tada S, Tadada R, Masutani M, Ui M, Hanaoka F. Molecular cloning of the cDNAs for the four subunits of mouse DNA polymerase α-primase complex and their gene expression during cell proliferation and the cell cycle. J Biol Chem. 1993;268:8111–8122. [PubMed] [Google Scholar]
- 21.Moarefi I F, Small D, Gilbert I, Höpfner M, Randall S K, Schneider C, Russo A A R, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moses K, Prives C. A unique subpopulation of murine DNA polymerase α/primase specifically interacts with polyomavirus T antigen and stimulates DNA replication. Mol Cell Biol. 1994;14:2767–2776. doi: 10.1128/mcb.14.4.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasheuer H-P, Grosse F. Immunoaffinity-purified DNA polymerase α displays novel properties. Biochemistry. 1987;26:8458–8466. doi: 10.1021/bi00399a064. [DOI] [PubMed] [Google Scholar]
- 24.Nasheuer H-P, Grosse F. DNA polymerase α-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J Biol Chem. 1988;263:8981–8988. [PubMed] [Google Scholar]
- 25.Nasheuer H-P, Moore A, Wahl A F, Wang T S-F. Cell cycle-dependent phosphorylation of human DNA polymerase α. J Biol Chem. 1991;266:7893–7903. [PubMed] [Google Scholar]
- 26.Park H, Davis R, Wang T S-F. Studies of Schizosaccharomyces pombe DNA polymerase α at different stages of the cell cycle. Nucleic Acids Res. 1995;23:4337–4344. doi: 10.1093/nar/23.21.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruediger R, van Wart Hood J E, Mumby M, Walter G. Constant expression and activity of protein phosphatase 2A in synchronized cells. Mol Cell Biol. 1991;11:4282–4285. doi: 10.1128/mcb.11.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka S, Hu S-Z, Wang T S-F, Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J Biol Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 29.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 30.Tercero J A, Labib K, Diffley J F. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todorov I T, Attaran A, Kearsey S E. BM28, a human member of the MCM2–3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virshup D M, Cegielska A, Russo A, Kelly T J, Shaffer S. The initiation of SV40 DNA replication is controlled by a phosphorylation-dephosphorylation cycle. Adv Protein Phosphatases. 1993;7:271–293. [Google Scholar]
- 33.Voitenleitner C, Fanning E, Nasheuer H-P. Phosphorylation of DNA polymerase α-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene. 1997;14:1611–1615. doi: 10.1038/sj.onc.1200975. [DOI] [PubMed] [Google Scholar]
- 34.Voitenleitner C, Rehfuess C, Hilmes M, O'Rear L, Liao P-C, Gage D A, Ott R, Nasheuer H-P, Fanning E. Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol Cell Biol. 1999;19:646–656. doi: 10.1128/mcb.19.1.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 36.Wahl A F, Geis A M, Spain B H, Wong S W, Korn D, Wang T S-F. Gene expression of human DNA polymerase α during cell proliferation and the cell cycle. Mol Cell Biol. 1988;8:5016–5025. doi: 10.1128/mcb.8.11.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of cdc45, RPA, and DNA polymerase α. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang T S-F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 39.Wera S, Hemmings B A. Serine/threonine protein phosphatases. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller M W, Wang T S-F. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu T, Matsuzawa S-I, Mizuno Y, Kamibayashi C, Mumby M C, Andjelkovic N, Hemmings B A, Onoe K, Kikuchi K. The interconversion of protein phosphatase 2A between PP2A1 and PP2A0 during retinoic acid-induced granulocytic differentiation and a modification on the catalytic subunit in S phase of HL-60 cells. Arch Biochem Biophys. 1997;339:210–217. doi: 10.1006/abbi.1996.9835. [DOI] [PubMed] [Google Scholar]