Abstract
Mitochondrial malfunction leads to the remodeling of myocardial energy metabolism during myocardial ischemia (MI). However, the alterations to the mitochondrial proteome profile during this period has not yet been clarified. An acute MI model was established by high position ligation of the left anterior descending artery in 8-week-old C57BL/6N mice. After 15 min of ligation, the animals were euthanized, and their hearts were collected. The myocardial ultrastructure was observed using transmission electron microscopy (TEM). The cardiac mitochondrial proteome profile was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and bioinformatics analyses. TEM showed that the outer membrane of the mitochondria was dissolved, and the inner membrane (cristae) was corrupted and broken down extensively in the MI group. The mitochondrial membrane potential was decreased. More than 1,700 mitochondrial proteins were identified by LC-MS/MS analysis, and 119 were differentially expressed. Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes functional enrichment analysis showed that endopeptidase activity regulation, the mitochondrial inner membrane, oxidative phosphorylation, the hypoxia-inducible factor-1 signaling pathway, the pentose phosphate pathway and the peroxisome proliferator-activated receptor signaling pathway were involved in the pathophysiological process in the early stage of acute MI. Extensive and substantial changes in the mitochondrial proteins as well as mitochondrial microstructural damage occur in the early stages of acute MI. In the present study, the series of proteins crucially involved in the pathways of mitochondrial dysfunction and metabolism were identified. Further studies are needed to clarify the roles of these proteins in myocardial metabolism remodeling during acute MI injury.
Keywords: acute myocardial ischemia, mitochondria, proteome, myocardial remodeling, liquid chromatography-tandem mass spectrometry
Introduction
Coronary artery-related cardiac ischemia is the most common cause of cardiomyopathy and heart failure, with high rates of morbidity and mortality worldwide (1). Coronary artery stenosis and/or occlusion lead to myocardial perfusion reduction, cardiomyocyte remodeling and fibrosis, dilated ventricles and even heart failure. Exploring the pathogenesis and regulatory mechanisms of ischemia may provide significant benefits in its diagnosis, treatment, and prognosis. Cardiac ischemia is a type of complex cellular and molecular activity that occurs at multiple levels (2-8). However, its mechanisms are yet to be fully elucidated. The heart is an organ rich in mitochondria and thus particularly vulnerable to ischemia. Due to the role of the 'energy power plant', mitochondrial homeostasis is crucial for vital cellular activities.
In the past few decades, omics technology has made a remarkable contribution to illustrating the pathogenesis and consequences of cardiac ischemia. Proteomics is a type of omics approach that focuses on systemic and dynamic molecular changes, and explores potential crosstalk between multiple molecular levels and pathways. Expression analysis via liquid chromatography-tandem mass spectrometry (LC-MS/MS) is used to investigate persistent and chronic myocardial ischemia (MI) (9-13). Transient coronary clamps cause severe cardiac ischemia and injury, accompanied by metabolic disturbances (14). Our recent unpublished experiments showed that 15 min of ligation of the left anterior descending (LAD) artery resulted in impaired mitochondrial morphology. Based on these results, it was hypothesized that mitochondrial proteome disturbance may occur in the very early stages of ischemia. In the present study, the mitochondrial proteome profiles of mice hearts that underwent short-term LAD ligation were evaluated using LC-MS/MS proteome expression analysis. More than 1,700 proteins were screened, and 119 differentially expressed proteins were identified. A number of proteins were involved in the activities and pathways that regulate oxidative phosphorylation (OXPHOS), complement activation and mitochondrial autophagy. These results indicated that mitochondrial proteomic changes may serve as potential biomarkers for a very early stage of ischemic cardiac injury and provide novel insights into the mechanism of myocardial remodeling.
Materials and methods
Animals and grouping
The animals used were 8-week-old male C57BL/6N mice and their body weights were 20-25 g. The animals were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were housed under a 12 h light/dark cycle at 22±2˚C and at a relative humidity of 40-60%. They had free access to standard mouse chow and tap water. After one week of adaptive feeding, the 24 mice were randomly divided into an acute MI group and a sham group. All experiments and procedures were approved by the Institutional Animal Care and Use Committee Review Board of the General Hospital of Ningxia Medical University (approval no. 2020-01) and complied with the guidelines of the National Institutes of Health Animal Care and Use Committee.
Acute MI model establishment
The MI mouse model was established by ligation of the LAD, following previously reported methods (15-17). The mice were anesthetized with a 2% isoflurane (RWD) by a delivery machine (Pat.4879997, VAPOMATIC™, CWE13-13000) at an airflow rate of 0.8 l/min. Deep sedation was confirmed by the absence of corneal and toe pinch reflexes. After disinfecting the surgical area, a 1-2 cm skin cut was made over the left side of the chest. After dissection and retraction of the pectoral major and minor muscles, the fourth intercostal space was exposed. A small hole was made at the fourth intercostal space with a mosquito clamp to open the pleural membrane and pericardium. Then the heart was smoothly and gently ‘popped out’ through the hole. The LAD was quickly ligated using a 6-0 silk suture ~1 mm distal to the left atrial appendage and 2 mm in width and depth. The ST-segment elevation on the ECG recording (PowerLab System, AD Instruments) was used to confirm successful occlusion of the LAD. The heart was placed back into the intrathoracic space immediately after ligation. The muscles and skin were closed. The operation in the sham group was the same, except no LAD ligation was performed. After 15 min of ligation, the animals were euthanized under anesthesia of 5% isoflurane by a delivery machine at an airflow rate of 1.0 l/min, and the left ventricles were collected immediately.
Preparation of samples for transmission electron microscopy (TEM) examination
A small piece (1 mm3) of the fresh left ventricle myocardium was rinsed in ice-cold PBS, and the buffer was then removed using clean filter paper. The tissues were fixed in 2% glutaraldehyde for 2 h, washed with 0.1 M dimethyl sodium arsenate three times, fixed in 4% osmic acid for 2 h, rinsed again in 0.1 M dimethyl sodium arsenate and successively dehydrated with 30-50-70% alcohol solutions. These processes were performed at 4˚C. The fixed tissues were infiltrated with propylene oxide and embedded in epoxy resin at 60˚C for 48 h. The tissues were then cut into ultrathin sections and observed and imaged by TEM (Hitachi HT-7800; Hitachi, Ltd.; magnifications, x1,500, x5,000 and x10,000) after staining with 2% uranyl acetate for 15 min at room temperature and lead citrate for 5 min at room temperature.
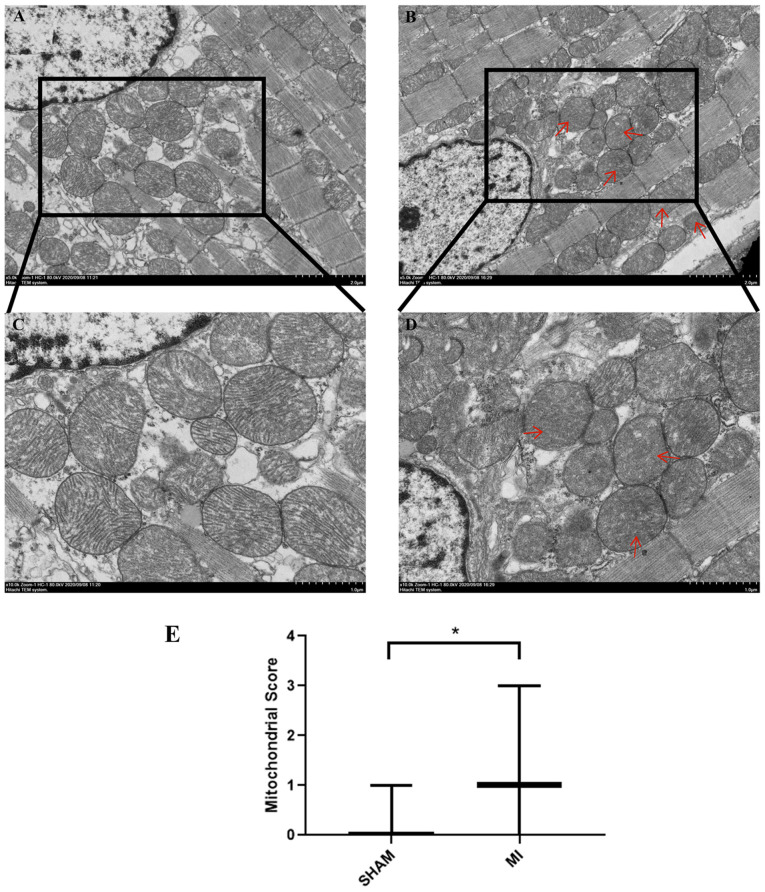
Flameng's classification method was used to quantify mitochondrial injury under TEM as previously reported (18). At the same magnification (x5,000), 5 fields were randomly selected for imaging, and ~20 mitochondria were randomly selected from each field for analysis in each mouse. According to the damage degree, the mitochondria were graded on a scale of 0 to 4 as follows: 0, normal ultrastructure of the mitochondrion; 1, normal ultrastructure of the crests and matrix but absence of granular deposits; 2, loss of matrix granules and clarification of the matrix without breaking of crests; 3, disruption of mitochondrial crests with clarification as well as condensation of the matrix; and 4, disruption of crests, and loss of integrity of the mitochondrial membranes. The final score is presented as the median and was analyzed using a Mann-Whitney U test. The experiment was repeated 3 times.
Mitochondrial isolation
Cardiac mitochondria were extracted using a mitochondrial isolation kit (Nanjing KeyGen Biotech Co., Ltd.; cat. no. KGA827) according to the manufacturer's instructions. The fresh left ventricle was immediately placed on ice, rinsed with cold saline and drained with filter paper. Then, 40-60 mg of tissue was cut into small pieces using ophthalmic scissors and transferred into a small volume (2 ml) glass homogenizer. Next, ice-cold lysis buffer (1 M Tris-HCl, 25 mM MgCl2, 0.5 M KCl) was added to the homogenizer until the accumulated volume was 6-fold that of the tissue pieces. The tissue was ground 20 times at 4˚C. After adding medium buffer (2 M sucrose), the homogenate was centrifuged at 1,200 x g for 5 min at 4˚C twice. The supernatant was collected and centrifuged at 7,000 x g for 10 min at 4˚C. The pellet contained mitochondria. Finally, the purified mitochondria were obtained by centrifugation at 9,500 x g for 5 min at 4˚C after adding suspension buffer (1 M Tris-HCl, 25 mM MgCl2, 2 M sucrose). Part of the freshly purified mitochondria was used for the respiration function study, and the rest was stored at -80˚C for proteomic analysis.
Western blotting
The mitochondrial proteins were extracted using a mitochondrial protein isolation kit (Nanjing KeyGen Biotech Co., Ltd.; cat. no. KGP850) according to the manufacturer's instructions. All steps were performed in an ice bath. Protein concentration was determined using a bicinchoninic acid (BCA) protein detection kit (Nanjing KeyGen Biotech Co., Ltd.; cat. no. KGPBCA). Proteins (40 µg/lane) were loaded and resolved on 10% SDS-PAGE and transferred to a nitrocellulose membrane. Mitochondrial transmembrane protein voltage-dependent anion-selective channel protein 1 (VDAC1) antibody (Abcam; cat. no. ab154856; 1:5,000) was used to confirm the efficiency of mitochondrial isolation and rabbit anti-GAPDH (BIOSS; cat. no. bs-2188R; 1:5,000) was used as the loading control. The nitrocellulose membrane was blocked in 10% skim milk for 1 h at 22±2˚C and then incubated with anti-VDAC1 overnight at 4˚C. The VDAC1 membrane was stripped and reblotted with GAPDH overnight at 4˚C. Horseradish peroxidase-labeled anti-rabbit secondary antibody (Abbkine, Inc.; cat. no. A21020; 1:5,000) was used to detect the primary antibodies.
Measurement of mitochondrial membrane potential (MMP)
MMP was analyzed using a fluorescent probe JC-1 assay kit (Beyotime Institute of Biotechnology; cat. no. C2006). JC-1 working solution was added to the purified mitochondria and mixed. The mixture was then added to a 96-well plate. Readings were measured using a fluorescence microplate reader (VICTOR Nivo; PerkinElmer, Inc.) at excitation and emission wavelengths of 485 and 590 nm, respectively.
Mitochondria proteome LC-MS/MS analysis
Protein digestion. The purified mitochondria were powdered in liquid nitrogen and transferred to a 5 ml centrifuge tube. Four volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail) were added to the powder and sonicated three times on ice using a high-intensity ultrasonic processor (Scientz). The remaining debris were removed by centrifugation at 12,000 x g for 10 min at 4˚C. Finally, the supernatant was collected, and the protein concentration was determined using a BCA assay. For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56˚C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in the dark. Then, the urea concentration in the protein sample was diluted to <2 M with 100 mM NH4HCO3. Finally, trypsin was added at a ratio of 1:50 trypsin: protein mass for the first digestion overnight and a ratio of 1:100 trypsin: protein mass for the second 4 h of digestion.
LC-MS/MS analysis. Tryptic peptides were dissolved in 0.1% formic acid and then loaded. The high performance liquid chromatography gradient increased from 6 to 23% (0.1% formic acid in 98% acetonitrile) over 26 min, 23 to 35% over 8 min, and increased to 80% over 3 min, then held at 80% for 3 min. On the EASY-nLC 1,000 ultra-performance LC (UPLC) system, the constant flow rate was 400 nl/min. The peptides were subjected to a nanojet ionization source followed by MS/MS in the Q Exactive™ Plus (Thermo Fisher Scientific, Inc.) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The resolution of the intact peptide detected in Orbitrap was 70,000 Da. Then, the peptides were selected with normalized collision energy setting a value of 28 for MS/MS, and the fragments were detected in the Orbitrap at a resolution of 17,500. Automatic gain control was set at 5E4. For MS scans, the m/z scan range was 350-1,800. Label-free quantification was performed as described in a previous study (19-21).
Database search and bioinformatics analysis. The MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8). The original data were obtained via database retrieval. Unsupervised principal component analysis (PCA) was used to observe the overall distribution of proteins among the samples and the stability of the analytical process. Proteins were classified by Gene Ontology (GO) annotation (22,23) into the three following categories: Cellular component, molecular function, and biological process. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to analyze pathway enrichment (24). For each category, a two-tailed Fisher's exact test was used to assess the enrichment of differentially expressed proteins vs. all identified proteins. The GO and KEGG terms with a corrected P<0.05 were considered statistically significant. The -log10(P-value) represents enrichment degree. The cluster membership was visualized using a heatmap.
Statistical analysis
All data were analyzed using SPSS v23.0 (IBM Corp) and GraphPad Prism v5.0 (GraphPad Software, Inc.). The results are presented as the mean ± standard deviation. An unpaired t-test was used to compare the means between the groups. The test significance level was α=0.05, and P<0.05 was considered to indicate a statistically significant difference. The differentially expressed proteins were screened by fold change (FC) and an unpaired t-test. The differential screening conditions were FC=1.2x and P<0.05. Both FC=0 and FC=inf belong to the difference of ‘with or without’. A two-tailed Fisher's exact test was applied to test the protein enrichment analysis, and a corrected P-value <0.05 was considered to be significant. These enrichment analyses were performed according to the database of identified proteins and a two-tailed Fisher's exact test was employed. All terms with corrected a P-value <0.05 were considered significantly enriched differentially expressed proteins. The number of experimental replications was 3.
Results
Acute MI induced mitochondrial morphological changes
First, TEM was used to examine the myocardium and mitochondrial morphology following MI. As shown in Fig. 1A and C, the sarcomeres were intact, the muscle filaments were neatly arranged, the nuclear membrane was intact (Fig. S1), and the chromatin was uniformly arranged. Mitochondria were normal and ridges were visible and not dissolved or ruptured in the sham group. The outer membrane dissolution and break down of the inner membrane (cristae) in some mitochondria was observable in the MI group (Fig. 1B and D; red arrows). The mitochondrial score in the MI group increased significantly (P<0.05) compared with the sham group (Fig. 1E). These results indicated that acute MI induced mitochondrial damage.
Figure 1.
Mitochondrial morphology observed using transmission electron microscopy. Mitochondrial morphology in the (A) Sham and (B) MI group. Scale bar, 2 µm. Mitochondrial morphology insets of the (C) Sham and (D) MI group images in (A) and (B). Scale bar, 1 µm. (E) Mitochondrial score. *P<0.05. n=3 per group. MI, myocardial ischemia.
Acute MI decreases MMP
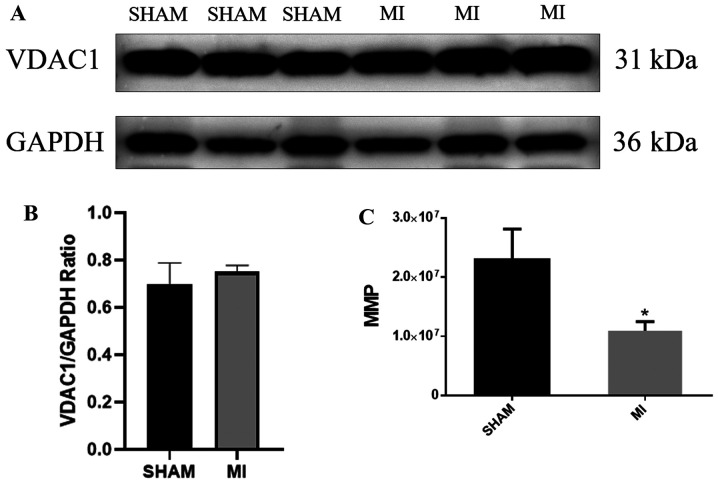
Fig. 2A shows that VDAC1 protein expression was strong (the original uncropped western blots shown in Fig. S2) and the mitochondria were efficiently isolated in both groups (Fig. 2B). The MMP was evaluated using a JC-1 assay. The results showed that the MMP of the MI group was lower than that of the sham group (Fig. 2C, P<0.05).
Figure 2.
(A) Detection of mitochondrial voltage-dependent anion-selective channel protein 1 (uncropped blots are presented in Fig. S1). (B) The total number of mitochondria did not differ significantly between the sham and MI groups. (C) MMP in the sham and MI groups. *P<0.05. n=6 per group. MI, myocardial ischemia; VDAC1, voltage-dependent anion-selective channel protein 1; MMP, mitochondrial membrane potential.
Mitochondrial proteomics
First, the expression of trusted proteins was analyzed using PCA (Fig. 3). The results showed substantial differences between the two groups. More than 1,700 trusted proteins were identified in this study. Through the analysis and synchronous comparison of myocardial mitochondria in three separated samples of the sham and MI groups, 119 differential proteins were confirmed. Overall, 27 of the 119 proteins were upregulated and 16 were downregulated compared with the sham group (Table I), and the remaining 76 proteins showed ‘yes’ or ‘no’ differences between the two groups (Tables II and III). These results identified LAD ligation/ischemia as the principal source of variance. The volcano map shows the overall distribution of the proteins (Fig. 4). The differentially expressed proteins were screened using FC=1.2 and P<0.05 as the screening conditions. The blue and red dots represent the significantly downregulated and upregulated proteins, respectively. The gray dots represent non-significant proteins. Unsupervised hierarchical clustering was used to group differentially expressed proteins according to their expression profiles. The results are shown in a heatmap (Fig. S3). Each line in the heatmap represents a protein with mean upregulation (red) or downregulation (purple) in expression within the two groups. The proteins that did not exhibit significant differences in expression are indicated in white. NADH dehydrogenase (ubiquinone) iron-sulfur protein 6 (NDUFS6), cytochrome b-c1 complex subunit 1 (UQCRC1), thrombospondin-1 (THBS1) and pyruvate dehydrogenase phosphatase regulatory subunit (PDPR) were upregulated, and dynactin subunit 2 (DCTN2) and a-kinase anchor protein 1 (AKAP1) were downregulated in the ischemic group. Cytochrome C oxidase (COX) assembly protein COX11, mitochondrion adenosine triphosphate (ATP) synthase F(0) complex subunit C2 (ATP5G2), mitochondria ubiquitin ligase activator of NF-κB1 (MUL1) were identified in the MI group and E3 ubiquitin-protein ligase UBR3 was identified in the sham group.
Figure 3.
PCA. Each square/spot represents a repetition in a grouping experiment, and different colors represent the different groups. n=3 per group. PCA, principal component analysis.
Table I.
Downregulated and upregulated proteins in the MI group.
| Accession no. | Gene | Protein description | FC | P-value | Type |
|---|---|---|---|---|---|
| Q99KJ8 | DCTN2 | Dynactin subunit 2 | 0.315 | 0.001c | Down |
| P31725 | S100A9 | Protein S100-A9 | 0.323 | 0.015a | Down |
| Q3UH59 | MYH10 | Myosin-10 | 0.370 | 0.020a | Down |
| Q62448 | EIF4G2 | Eukaryotic translation initiation factor 4γ2 | 0.423 | 0.026a | Down |
| Q8VCQ8 | CALD1 | Caldesmon 1 | 0.492 | 0.028a | Down |
| P19157 | GSTP1 | Glutathione S-transferase P1 | 0.542 | 0.010b | Down |
| O08715 | AKAP1 | A-kinase anchor protein 1, mitochondrial | 0.593 | 0.048a | Down |
| A0A0A6YX73 | PRKAR2α | cAMP-dependent protein kinase type II-α regulatory subunit | 0.619 | 0.036a | Down |
| Q9CQS4 | SLC25A46 | Solute carrier family 25 member 46 | 0.620 | 0.035a | Down |
| Q8R2P8 | KARS | Lysine-tRNA ligase | 0.635 | 0.013a | Down |
| P05132 | PRKACα | cAMP-dependent protein kinase catalytic subunit α | 0.652 | 0.047a | Down |
| P49312 | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 0.665 | 0.050a | Down |
| P55096 | ABCD3 | ATP-binding cassette sub-family D member 3 | 0.666 | <0.001c | Down |
| P14733 | LMNB1 | Lamin-B1 | 0.684 | 0.029a | Down |
| Q9ET78 | JPH2 | Junctophilin-2 | 0.745 | 0.035a | Down |
| Q6ZWN5 | RPS9 | 40S ribosomal protein S9 | 0.823 | 0.044a | Down |
| Q9D1G1 | RAB1B | Ras-related protein Rab-1B | 1.201 | 0.031a | Up |
| Q9CZ13 | UQCRC1 | Cytochrome b-c1 complex subunit 1, mitochondrial | 1.479 | 0.048a | Up |
| Q99MR8 | MCCC1 | Methylcrotonoyl-CoA carboxylase subunit α, mitochondrial | 1.496 | 0.043a | Up |
| Q8R5L1 | C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial | 1.591 | 0.027a | Up |
| Q3ULD5 | MCCC2 | Methylcrotonoyl-CoA carboxylase β chain, mitochondrial | 1.612 | 0.013a | Up |
| Q9ER35 | FN3K | Fructosamine-3-kinase | 1.654 | 0.013a | Up |
| P84096 | RHOG | Rho-related GTP-binding protein RhoG | 1.676 | 0.031a | Up |
| Q7TSQ8 | PDPR | Pyruvate dehydrogenase phosphatase regulatory subunit, mitochondrial | 1.724 | 0.046a | Up |
| Q921I1 | TF | Serotransferrin | 1.731 | 0.025a | Up |
| Q5FWK3 | ARHGAP1 | Rho GTPase-activating protein 1 | 1.800 | 0.017a | Up |
| A0A0R4J0I1 | SERPINA3K | Serine protease inhibitor A3K | 1.810 | 0.005b | Up |
| Q00897 | SERPINA1D | α-1-antitrypsin 1-4 | 1.820 | 0.026a | Up |
| Q8BJ03 | COX15 | Cytochrome c oxidase assembly protein COX15 homolog | 1.878 | 0.017a | Up |
| O08528 | HK2 | Hexokinase-2 | 1.912 | 0.015a | Up |
| P48036 | ANXA5 | Annexin A5 | 1.951 | 0.049a | Up |
| Q60994 | ADIPOQ | Adiponectin | 1.951 | 0.031a | Up |
| P07724 | ALB | Serum albumin | 1.969 | 0.031a | Up |
| P52503 | NDUFS6 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | 2.175 | 0.046a | Up |
| A0A0R4J0X5 | SERPINA1C | Alpha-1-antitrypsin 1-3 | 2.536 | 0.045a | Up |
| Q61838 | A2M | Alpha-2-macroglobulin | 2.764 | 0.019a | Up |
| O35639 | ANXA3 | Annexin A3 | 2.867 | 0.002b | Up |
| Q91VB8 | HBA-A1 | Alpha globin 1 | 3.001 | 0.037a | Up |
| P05064 | ALDOA | Fructose-bisphosphate aldolase A | 3.409 | 0.026a | Up |
| A8DUK4 | HBB-BS | β-globin | 3.730 | 0.004b | Up |
| E9QNT8 | ANK1 | Ankyrin-1 | 3.885 | 0.040a | Up |
| P01027 | C3 | Complement C3 | 4.137 | 0.044a | Up |
| Q80YQ1 | THBS1 | Thrombospondin-1 | 5.096 | 0.036a | Up |
aP≤0.05,
bP≤0.01,
cP≤0.001. Up, upregulated; Down, downregulated.
Table II.
Proteins specific to the SHAM group.
| Accession no. | Gene | Protein descriptions | Type |
|---|---|---|---|
| Q8K0M3 | SORBS3 | Vinexin | Up |
| A2A848 | ACOX1 | Peroxisomal acyl-coenzyme A oxidase 1 | Up |
| A8Y5P4 | MAP7D1 | MAP7 domain-containing protein 1 | Up |
| A2AS45 | PKP4 | Plakophilin-4 | Up |
| A4QPC5 | CMA1 | Chymase | Up |
| D3YYT1 | GLYR1 | Putative oxidoreductase GLYR1 | Up |
| D3Z598 | LTBP4 | Latent-transforming growth factor beta-binding protein 4 | Up |
| E9Q6Q8 | TBC1D4 | TBC1 domain family member 4 | Up |
| Q5U430 | UBR3 | E3 ubiquitin-protein ligase UBR3 | Up |
| F7A1B4 | ENG | Endoglin | Up |
| O54774 | AP3D1 | AP-3 complex subunit delta-1 | Up |
| O55022 | PGRMC1 | Membrane-associated progesterone receptor component 1 | Up |
| O88587 | COMT | Catechol O-methyltransferase | Up |
| P13541 | MYH3 | Myosin-3 | Up |
| P63163 | SNRPN | Small nuclear ribonucleoprotein-associated protein N | Up |
| P37804 | TAGLN | Transgelin | Up |
| Q8BVQ9 | PSMC2 | 26S protease regulatory subunit 7 | Up |
| Q3U962 | COL5A2 | Collagen alpha-2(V) chain | Up |
| Q921L6 | CTTN | Src substrate cortactin | Up |
| Q60972 | RBBP4 | Histone-binding protein RBBP4 | Up |
| Q6P549 | INPPL1 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | Up |
| Q6ZWQ9 | MYL12A | Myosin, light chain 12A, regulatory, non-sarcomeric | Up |
| Q8BGU5 | CCNY | Cyclin-Y | Up |
| Q8VCI5 | PEX19 | Peroxisomal biogenesis factor 19 | Up |
| Q99N92 | MRPL27 | 39S ribosomal protein L27, mitochondrial | Up |
| Q9D666 | SUN1 | SUN domain-containing protein 1 | Up |
| Q9DAW9 | CNN3 | Calponin-3 | Up |
| Q9JIK5 | DDX21 | Nucleolar RNA helicase 2 | Up |
| Q9JKY7 | CYP2D22 | Cytochrome P450 CYP2D22 | Up |
| Q9JLB0 | MPP6 | MAGUK p55 subfamily member 6 | Up |
| Q9JLH8 | TMOD4 | Tropomodulin-4 | Up |
| Q9JMG7 | HDGFRP3 | Hepatoma-derived growth factor-related protein 3 | Up |
| Q9Z0P5 | TWF2 | Twinfilin-2 | Up |
| Q9Z2X1 | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | Up |
Up, upregulated; Down, downregulated.
Table III.
Proteins specific to the MI group.
| Accession no. | Gene | Protein descriptions | Type |
|---|---|---|---|
| A0A075B6A0 | IGHM | Ig mu chain C region | Up |
| A0A087WQS5 | ATP5G2 | ATP synthase F(0) complex subunit C2, mitochondrial | Up |
| E9QP56 | APOC3 | Apolipoprotein C-III | Up |
| F8VPK5 | ROCK2 | Rho-associated protein kinase; Rho-associated protein kinase 2 | Up |
| A0A2I3BPW0 | NDRG2 | N-myc downstream-regulated gene 2 protein | Up |
| A0A3B2WBH9 | TJP2 | Tight junction protein ZO-2 | Up |
| A0A494BB95 | EIF1AX | Eukaryotic translation initiation factor 1A | Up |
| A2AEX8 | FHL1 | Four and a half LIM domains protein 1 | Up |
| B7ZCL8 | MPP1 | 55 kDa erythrocyte membrane protein | Up |
| B8JJM5 | CFB | Complement factor B | Up |
| D6RGQ0 | CFH | Complement factor H | Up |
| O88986 | GCAT | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial | Up |
| F7AAP4 | ATP2B4 | Calcium-transporting ATPase | Up |
| Q91YX5 | LPGAT1 | Acyl-CoA: lysophosphatidylglycerol acyltransferase 1 | Up |
| Q8CIZ8 | VWF | von Willebrand factor; von Willebrand antigen 2 | Up |
| O55042 | SNCA | Alpha-synuclein | Up |
| O70194 | EIF3D | Eukaryotic translation initiation factor 3 subunit D | Up |
| P01837 | IGKC | Igκ chain C region | Up |
| P06684 | C5 | Complement C5 | Up |
| P07759 | SERPINA3K | Serine protease inhibitor A3K | Up |
| P19221 | F2 | Prothrombin | Up |
| P29699 | AHSG | Alpha-2-HS-glycoprotein | Up |
| P29788 | VTN | Vitronectin | Up |
| P35550 | FBL | rRNA 2-O-methyltransferase fibrillarin | Up |
| P40142 | TKT | Transketolase | Up |
| P43883 | PLIN2 | Perilipin-2 | Up |
| P49722 | PSMA2 | Proteasome subunit alpha type-2 | Up |
| Q8CE80 | CAST | Calpastatin | Up |
| P61514 | RPL37A | 60S ribosomal protein L37a | Up |
| P62835 | RAP1A | Ras-related protein Rap-1A | Up |
| Q00519 | XDH | Xanthine dehydrogenase/oxidase | Up |
| Q5SRC5 | COX11 | Cytochrome c oxidase assembly protein COX11, mitochondrial | Up |
| Q61578 | FDXR | NADPH: adrenodoxin oxidoreductase, mitochondrial | Up |
| Q6PA06 | ATL2 | Atlastin-2 | Up |
| Q6PDI5 | ECM29;AI314180 | Proteasome-associated protein ECM29 homolog | Up |
| Q8VE37 | RCC1 | Regulator of chromosome condensation | Up |
| Q7TNL9 | CHCHD10 | Coiled-coil-helix-coiled-coil-helix domain-containing 10 | Up |
| Q8BXZ1 | TMX3 | Protein disulfide-isomerase TMX3 | Up |
| Q8VCM5 | MUL1 | Mitochondrial ubiquitin ligase activator of NF-κB1 | Up |
| Q91X72 | HPX | Hemopexin | Up |
| Q99LB4 | CAPG | Macrophage-capping protein | Up |
| Q9EST5 | ANP32B | Acidic leucine-rich nuclear phosphoprotein 32 family member B | Up |
Up, upregulated; Down, downregulated.
Figure 4.
Volcano plot showing the differentially expressed proteins. The abscissa of the volcano plot is log2(FC), and the further the value from zero, the higher the difference, with upregulation on the right and downregulation on the left. The ordinate is -log10(P-value), and the further the ordinate from zero, the higher the difference. FC, fold change.
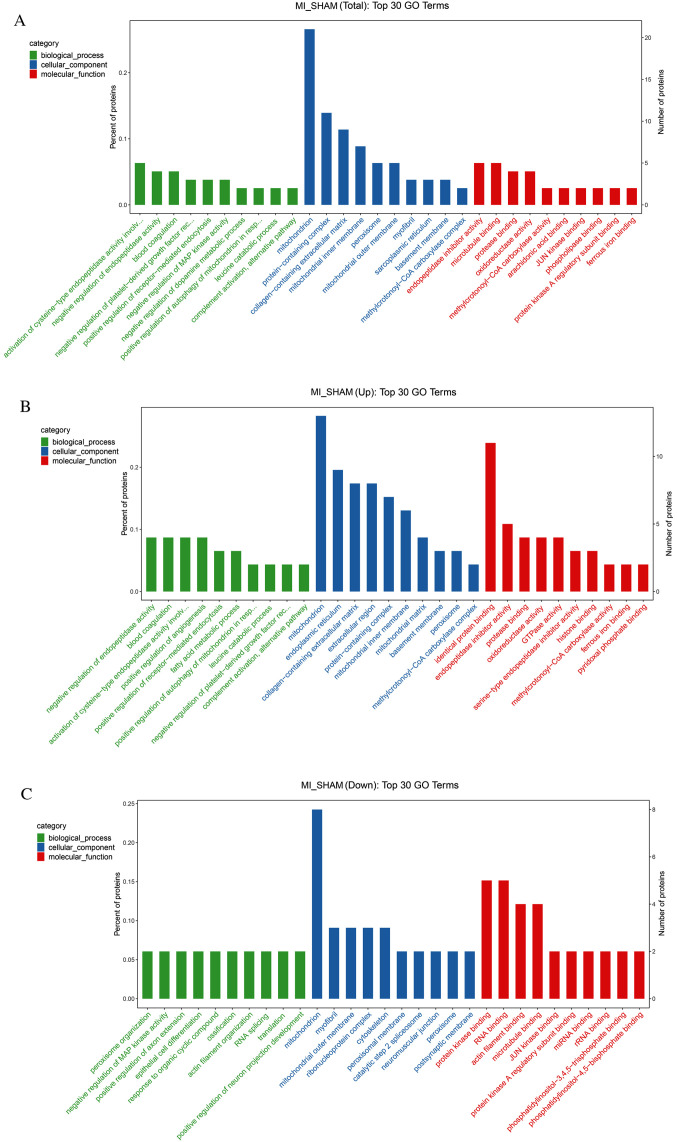
According to the GO database, the enrichment of the total, upregulated, and downregulated differentially expressed proteins were analyzed (Fig. 5A-C). For each category, a two-tailed Fisher's exact test was used to test the enrichment of differentially expressed proteins vs. all identified proteins. The GO term with a corrected P-value <0.05 was considered statistically significant. Proteins were classified by GO annotation into the following three categories: Biological processes, cellular components and molecular functions. Pathway enrichment of the top 30 GO terms of the total differentially expressed proteins is shown in Fig. 5A. The most perturbed biological processes were endopeptidase activity regulation and blood coagulation. The mitochondria, protein-containing complex, collagen-containing extracellular matrix and mitochondria inner membrane were the most relevant cellular components. Endopeptidase inhibitor activity and microtubule binding were the most relevant molecular functions.
Figure 5.
GO enrichment analysis. GO entries screened among (A) all differentially expressed proteins, (B) the upregulated proteins and (C) the downregulated proteins. GO, Gene Ontology; MI, myocardial ischemia.
For the upregulated proteins, the most relevant biological processes were the negative regulation of endopeptidase activity, blood coagulation, the activation of cysteine-type endopeptidase activity and the positive regulation of angiogenesis. The most relevant cellular components included the endoplasmic reticulum, collagen-containing extracellular matrix and extracellular region. The top four molecular functions were identical protein binding, endopeptidase inhibitor activity, protease binding and oxidoreductase activity (Fig. 5B).
For the downregulated proteins, the closely related biological processes included peroxisome organization, negative regulation of MAP kinase activity and positive regulation of axon extension. Except for mitochondria, the most highly relevant cellular components were myofibrils, the mitochondrial outer membrane, the ribonucleoprotein complex and the cytoskeleton. Closely related molecular functions included protein kinase binding, RNA binding, actin filament binding and microtubule-binding (Fig. 5C).
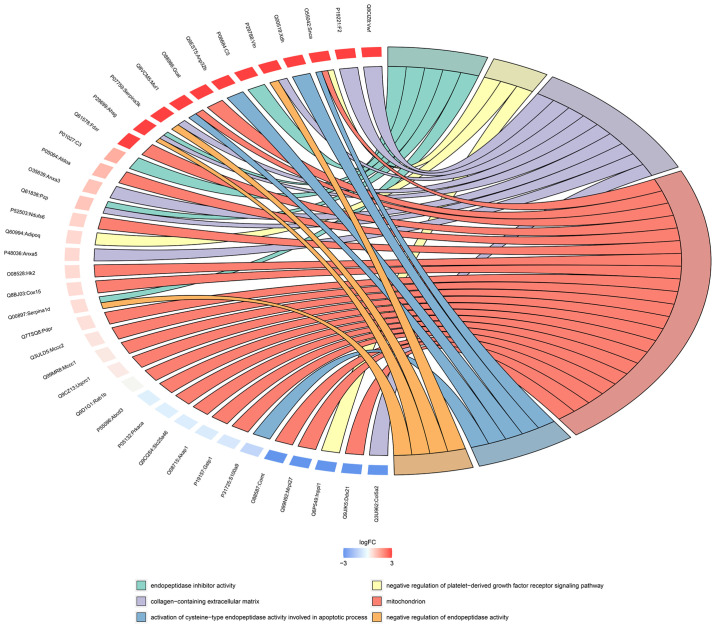
The relationship between the enriched GO terms and the differentially expressed proteins is displayed as a chord diagram in Fig. 6. A small arc on the left side of the chord diagram represents one protein. The color represents the difference: Blue indicates downregulation and red indicates upregulation. The GO terms are shown on the right-hand side of the diagram. Different colors represent different GO terms; that is, different functions. GO terms correspond to multiple differentially expressed proteins, and a protein can participate in multiple GO terms; the mitochondria and collagen-containing extracellular matrices possessed the most enriched proteins.
Figure 6.
Chord diagram of GO enrichment analysis. The protein names are on the left and the selected GO terms are on the right. Red indicates upregulated and blue indicates downregulated proteins. GO, Gene Ontology.
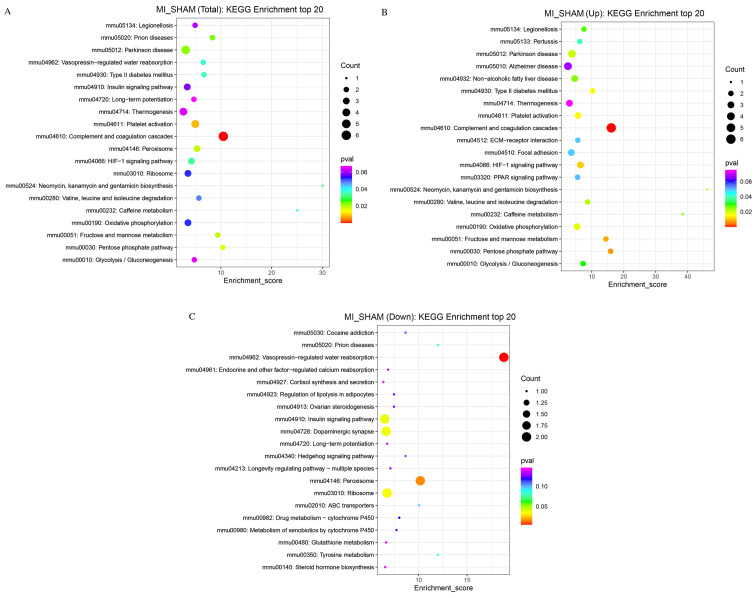
The KEGG database was used to analyze the metabolic pathways of the proteins. Biological functions related to these proteins were obtained from KEGG enrichment analysis (Fig. 7). The pathways with the most relevant differentially expressed proteins included Parkinson's disease, prion diseases, platelet activation, complement and coagulation cascades, peroxisome, hypoxia-inducible factor-1 (HIF-1) signaling, type 2 diabetes mellitus, fructose and mannose metabolism (Fig. 7A). For upregulated proteins, the most relevant biological functions included Parkinson's disease, non-alcoholic fatty liver disease, focal adhesion, complement, coagulation cascades, the peroxisome proliferator-activated receptor (PPAR) signaling pathway, OXPHOS, the pentose phosphate pathway and the HIF-1 signaling pathway (Fig. 7B). The closely related biological functions of downregulated proteins included vasopressin-regulated water reabsorption, the insulin signaling pathway, dopaminergic synapses, ribosomes, and peroxisomes (Fig. 7C).
Figure 7.
Bubble chart of the KEGG enrichment analysis. The x-axis is the enrichment score and the y-axis is the pathway information. The size of the bubbles reflects the number of differentially expressed proteins. The larger the bubble, the higher the number of differentially expressed proteins. The lower the P-value, the higher the significance of the KEGG pathway enrichment. (A) Enrichment metabolic pathways screened among all differentially expressed proteins. (B) Enriched metabolic pathways screened among upregulated proteins. (C) Enriched metabolic pathways screened among downregulated proteins. MI, myocardial ischemia; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Ischemic heart disease is the leading cause of heart failure worldwide (1). Disturbances to ATP synthesis resulting from MI leads to energy starvation in the myocardium, resulting in the remodeling of myocardial energy metabolism (25). The heart is one of the most metabolically active organs in the body (26). In the resting state, the human heart uses 6 kg of ATP per day (27). Cardiac function declines when mitochondrial OXPHOS and ATP production fail to keep up with the demand for energy consumption. Mitochondrial disorders are a sign of cardiovascular disease and heart failure (27). Early ultrastructural changes include the depletion of cytoplasmic glycogen particles, and the swelling of cardiomyocyte mitochondria can be observed after a few minutes of ischemia, although these changes are reversible if ischemia lasts no longer than 15-20 min (28). Any change in mitochondrial structure is associated with impaired mitochondrial function, such as the alteration of metabolic substrate utilization, impaired activity of the mitochondrial electron transport chain, increased formation of reactive oxygen species (ROS), and altered ion homeostasis (26). Consequently, cardiac function is affected by these adverse effects (29). Proteins are the ultimate ‘executors’ of cellular activity and function. To the best of our knowledge, the proteomic characteristics of cardiac mitochondria that suffer from short-term acute ischemia have not previously been reported.
In the present study, cardiac mitochondrial injury, such as outer membrane dissolution, inner membrane (cristae) loss and rupture were observed in the MI group 15 min after LAD ligation. However, the cardiomyocyte membrane, sarcomeres and nuclei remained intact. Decreased MMP measurements also showed damaged mitochondria in the MI group. These results indicated that mitochondria are extremely sensitive to ischemia and may be a potential marker of ischemia onset. Moreover, the results suggested that proteome disturbances are inevitable during this period. To elucidate the proteome changes, the proteome expression profile of the left ventricle mitochondria was evaluated using LC-MS/MS analysis. More than 1,700 proteins were screened, and 119 differentially expressed proteins were confirmed, with 27 upregulated and 16 downregulated proteins in the MI group. In addition, 34 proteins were present in the sham group and 42 in the MI group. Once the differential proteins were identified, GO annotation and the KEGG pathway enrichment analyses were performed to elucidate the proteins' function. According to the GO annotation, the pathway categories include cellular component, molecular function and biological process. Several differentially expressed proteins were involved in the activities and pathways of OXPHOS, complement activation, positive regulation of mitochondrial autophagy, and oxidative dismutase activity. These proteins included numerous upregulated proteins with FC ≥1.47, such as UQCRC1, pyruvate dehydrogenase phosphatase regulatory subunit, rho GTPase-activating protein 1, NDUFS6, Annexin A3 and A5, complement 1Q subcomponent-binding and protein complement C3, fructose-bisphosphate aldolase A and ankyrin-1, and the downregulated dynactin subunit 2, myosin 10, A-kinase anchor protein 1, cAMP-dependent protein kinase type II-α regulatory subunit and cAMP-dependent protein kinase catalytic subunit α. Interestingly, a number of proteins were present only in the sham or MI group. For example, UBR3, cAMP-dependent protein kinase type II-α regulatory subunit (PRKAR2α), A-kinase anchor protein 1 (AKAP1, mitochondrial), and Junctophilin-2 (JPH2) were expressed only in the sham group. These genes were primarily involved in inflammatory response, microtubule binding and protease binding. Dozens of the proteins were related to cellular components. ATP5G2, calcium-transporting ATPase, COX11, adrenodoxin oxidoreductase and MUL1 were expressed in the MI group only. Some of the proteins that were present in the MI group are subunits of the mitochondrial respiratory chain complex. The ‘presence’ difference between the sham and MI groups indicated that mitochondrial proteome disturbance was extensive and distinct as early as minutes after MI occurrence.
To the best of our knowledge, there are no studies that have elucidated the relationship between vasopressin-regulated water reabsorption, dopaminergic synapses, peroxisomes and MI. Studies regarding the relevance of Parkinson's disease, non-alcoholic fatty liver disease and prion diseases to MI are lacking. Platelet activation, coagulation dysfunction and abnormal glucose metabolism may be involved in MI. THBS1, involved in coagulation dysfunction, also underwent the most substantial changes. The link between these pathways and MI remains to be fully elucidated and requires further investigation. Interestingly, OXPHOS, the HIF-1 signaling pathway, pentose phosphate pathway and PPAR signaling pathway are closely related to electron transport and energy metabolism, suggesting that the disorder of energy metabolism in cardiomyocytes is the initial pathophysiological feature of acute MI (30-32).
Mitochondrial adaptation plays a vital role in protecting key cellular components from ischemic damage. In addition, this is the main entry point for electrons to enter the process of OXPHOS, and is helpful in establishing the proton gradient needed for a large number of cells to synthesize ATP (33-34). Complex I (ubiquinone oxidoreductase) is the largest complex of the mitochondrial respiratory chain, which is composed of >40 subunits and is located in the mitochondrial inner membrane. Mutations that lead to complex I defects are the most common cause of neurodegeneration, myopathies and heart failure (35-37). Complex I is the most vulnerable enzyme, and the first to be damaged (38). The most common diagnosis in patients with energy production disorders is complex I activity deficiency (39). NDUFS6 is a highly conserved subunit of complex I (40,41). NDUFS4 and NDUFS6 are crucial to the final step of complex I assembly and may provide a connection point for the integration of other subunits (42). Knockout of the NDUFS6 gene can lead to complex I instability and functional defects (43). The deletion of NDUFS6 increases ROS production and damages mitochondria through complex I deficiency (44). A previous study established NDUFS6gt/gt mice with complex I deficiency, which revealed decreased NDUFS6 protein expression levels (43). The mice developed cardiomyopathy and heart failure. Isolated heart studies have shown that resting contractile function is severely impaired even in NDUFS6gt/gt mice without heart failure. UQCRC1 is a subunit of complex III in the respiratory chain, is encoded by nuclear DNA and consists of 480 amino acids (45-48). Disrupting one UQCRC1 allele resulted in decreased UQCRC1 mRNA and protein expression in mice. In addition, complex III formation, activity, and ATP content were decreased in the brain at baseline. Under ischemic conditions, the brain tissue showed decreased MMP and ATP content as well as increased free radical levels compared with wild-type mice. These results indicated that UQCRC1 plays a key role in complex III (49-51). Overexpression of UQCRC1 can enhance the activity of complex III in mice (52). Specific loss of UQCRC1 in cells of epithelial origin can lead to mitochondrial dysfunction. The overexpression of UQCRC1 helps to maintain the mitochondrial function of H9C2 cells and regulates apoptosis-related proteins by mediating the PI3K/Akt/GSK-3b pathway to protect H9C2 cells from simulated ischemia/reperfusion injury (53). These studies suggest that UQCRC1 plays an important role in mitochondrial function, and may have a protective effect on cardiomyocytes. In the present study, short-term cardiac ischemia caused significant upregulation of NDUFS6 and UQCRC1 compared with the sham group. This might be compensatory regulation for protecting the heart from metabolic dysfunction due to mitochondrial injury. COX consists of 13 subunits; the subunits COX I, COX II and COX III are encoded by mitochondrial DNA (54-56). COX11 is an endogenous mitochondrial membrane protein that is essential for the assembly of active COX (57,58). The ATP synthase F (0) complex consists of subunits C1 (ATP5G1), C2 (ATP5G2) and C3 (ATP5G3) (59). A previous study identified a substantial increase in ATP5G1 in a rat model of ischemic heart failure (60). Only ATP5G2 was identified in the hearts of the ischemic mice. However, its effect on mitochondrial hemostasis needs to be further elucidated.
In conclusion, comprehensive and significant mitochondrial proteomic changes occurred after a short period of acute MI. However, the screened proteins should be confirmed by parallel reaction monitoring. Due to the limited output of mitochondrial protein in one mouse heart, the differential proteins will be validated by ELISA and/or PCR in blood samples in the future. There are still other limitations in the present study; for example, detection of serum CKMB level is necessary for confirming establishment of the MI model and will enrich the data. Among the differentially expressed proteins, the mitochondrial respiratory chain protein is the major focus of our lab. These differential proteins are involved in cellular component, oxidative phosphorylation and ubiquitination, which may regulate the post-translational modification of mitochondrial proteins. Illustrating their downstream effects and possible molecular mechanisms by overexpression/knockdown, western blotting, co-immunoprecipitation and confocal microscopy analysis in a hypoxic cardiomyocyte model will be performed in our future studies.
In conclusion, short-term acute ischemia can result in apparent cardiac mitochondrial damage and extensive disturbances to the proteome. A series of altered proteins are closely correlated with the pathways involved in OXPHOS, complement activation and mitochondrial autophagy. The results of the present study showed that mitochondrial changes may be regarded as biomarkers of the very early stages of ischemic cardiac injury. In addition, this study provides novel insights into the mechanism of myocardial remodeling targeting the mitochondrial proteome.
Supplementary Material
Acknowledgements
We would like to thank Miss Yipin Han (Bloomberg School of Public Health, Department of Epidemiology-Cardiovascular and Clinical Epidemiology, Johns Hopkins University, Baltimore USA) and Mr. Mewand Khan (School of Clinical Medicine, Ningxia Medical University, Yinchuan, Ningxia, China) for their suggestions and discussions.
Funding Statement
Funding: This study was supported by the National Natural Science Foundation (grant no. 81660045) and Ningxia Natural Science Foundation (grant no. 2020AAC02037).
Availability of data and materials
Additional details regarding the LC-MS/MS analysis are available in the public database proteomecentral.proteomexchange.org/cgi/GetDataset? (accession no. PXD026207; lingfeng_zeng@163.com, Password: sUdIhJj4). The datasets used and/or analyzed during the current study are also available from the corresponding author on reasonable request.
Authors' contributions
JW and JH confirm the authenticity of all the raw data. JW and YF collected the tissues, prepared the specimens, and performed the western blotting and statistical analysis. QL and FX extracted the mitochondria and performed the western blotting. JW, RH and RY established the mouse model. JH and JW designed the experiments. JW and JH wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All experiments and procedures were approved by the Institutional Animal Care and Use Committee Review Board of the General Hospital of Ningxia Medical University (approval no. 2020-01) and complied with the guidelines of the National Institutes of Health Animal Care and Use Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Farthing DE, Farthing CA, Xi L. Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: From bench to point-of-care. Exp Biol Med (Maywood) 2015;240:821–831. doi: 10.1177/1535370215584931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shvedova M, Anfinogenova Y, Atochina-Vasserman EN, Schepetkin IA, Atochin DN. c-Jun N-terminal kinases (JNKs) in myocardial and cerebral ischemia/reperfusion injury. Front Pharmacol. 2018;9(715) doi: 10.3389/fphar.2018.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oyama Y, Bartman CM, Gile J, Sehrt D, Eckle T. The circadian PER2 enhancer nobiletin reverses the deleterious effects of midazolam in myocardial ischemia and reperfusion injury. Curr Pharm Des. 2018;24:3376–3383. doi: 10.2174/1381612824666180924102530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Škrlec I, Milić J, Steiner R. The impact of the circadian genes CLOCK and ARNTL on myocardial infarction. J Clin Med. 2020;9(484) doi: 10.3390/jcm9020484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavo N, Lukovic D, Zlabinger K, Zimba A, Lorant D, Goliasch G, Winkler J, Pils D, Auer K, Jan Ankersmit H, et al. Sequential activation of different pathway networks in ischemia-affected and non-affected myocardium, inducing intrinsic remote conditioning to prevent left ventricular remodeling. Sci Rep. 2017;7(43958) doi: 10.1038/srep43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Meana M, Abellán A, Miró-Casas E, Garcia-Dorado D. Opening of mitochondrial permeability transition pore induces hypercontracture in Ca2+ overloaded cardiac myocytes. Basic Res Cardiol. 2007;102:542–552. doi: 10.1007/s00395-007-0675-y. [DOI] [PubMed] [Google Scholar]
- 7.Santos-Gallego CG, Vahl TP, Goliasch G, Picatoste B, Arias T, Ishikawa K, Njerve IU, Sanz J, Narula J, Sengupta PP, et al. Sphingosine-1-phosphate receptor agonist fingolimod increases myocardial salvage and decreases adverse postinfarction left ventricular remodeling in a porcine model of ischemia/reperfusion. Circulation. 2016;133:954–966. doi: 10.1161/CIRCULATIONAHA.115.012427. [DOI] [PubMed] [Google Scholar]
- 8.Saito T, Nah J, Oka SI, Mukai R, Monden Y, Maejima Y, Ikeda Y, Sciarretta S, Liu T, Li H, et al. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest. 2019;129:802–819. doi: 10.1172/JCI122035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquet S, Yin X, Sicard P, Clark J, Kanaganayagam GS, Mayr M, Marber MS. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial infarction by proteomics analysis. Mol Cell Proteomics. 2009;8:2687–2699. doi: 10.1074/mcp.M900176-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim N, Lee Y, Kim H, Joo H, Youm JB, Park WS, Warda M, Cuong DV, Han J. Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics. 2006;6:1237–1249. doi: 10.1002/pmic.200500291. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Zhang Y, Wu Q, Xie D, Dai W, Zhang X, Yang Z, Wang D. Integrative analyses of myocardial lipidome and proteome implicate mitochondrial dysfunction in lethal ventricular tachyarrhythmia (LVTA) induced by acute myocardial ischemia (AMI) J Proteomics. 2019;197:14–22. doi: 10.1016/j.jprot.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Cao TH, Jones DJL, Voors AA, Quinn PA, Sandhu JK, Chan DC, Parry HM, Mohan M, Mordi IR, Sama IE, et al. Plasma proteomic approach in patients with heart failure: Insights into pathogenesis of disease progression and potential novel treatment targets. Eur J Heart Fail. 2020;22:70–80. doi: 10.1002/ejhf.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Li C, Chuo W, Liu Z, Ouyang Y, Li D, Han J, Wu Y, Guo S, Wang W. Integrated proteomic and metabolomic analysis reveals the NADH-mediated TCA cycle and energy metabolism disorders based on a new model of chronic progressive heart failure. Mol Biosyst. 2013;9:3135–3145. doi: 10.1039/c3mb70263d. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Zhou B, Zheng Y, Feng K, Rao L, Zhang J, Xin J, Zhang B, Zhang L. Time course proteomic profile of rat acute myocardial infarction by SELDI-TOF MS analysis. Int J Cardiol. 2009;131:225–233. doi: 10.1016/j.ijcard.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Gao E, Koch WJ. doi: 10.1007/978-1-62703-505-7_17. A novel and efficient model of coronary artery ligation in the mouse. Wound Regeneration and Repair. Springer, Clifton, NJ, pp299-311, 2013. [DOI] [PubMed] [Google Scholar]
- 16.Lu L, Ma J, Sun M, Wang X, Gao E, Lu L, Ren J, Yang L, Yang J. Melatonin ameliorates MI-induced cardiac remodeling and apoptosis through a JNK/p53-dependent mechanism in diabetes mellitus. Oxid Med Cell Longev. 2020;2020(1535201) doi: 10.1155/2020/1535201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Ceholski DK, Liang L, Fish K, Hajjar RJ. Variability in coronary artery anatomy affects consistency of cardiac damage after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2017;313:H275–H282. doi: 10.1152/ajpheart.00127.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flameng W, Borgers M, Daenen W, Stalpaert G. Ultrastructural and cytochemical correlates of myocardial protection by cardiac hypothermia in man. J Thorac Cardiovasc Surg. 1980;79:413–424. [PubMed] [Google Scholar]
- 19.Wang W, Zhou X, Cui F, Shi C, Wang Y, Men Y, Zhao W, Zhao J. Proteomic analysis on exosomes derived from Patients' sera infected with Echinococcus granulosus. Korean J Parasitol. 2019;57:489–497. doi: 10.3347/kjp.2019.57.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Huang Y, Wang X, Chen Y. Label-free LC-MS/MS proteomics analyses reveal proteomic changes accompanying MSTN KO in C2C12 cells. BioMed Res Int. 2019;2019(7052456) doi: 10.1155/2019/7052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium: Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. The Gene Ontology Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M. Post-Genome Informatics. Oxford University Press, Oxford, NY, 2000. [Google Scholar]
- 25.van Bilsen M, Smeets PJ, Gilde AJ, van der Vusse GJ. Metabolic remodelling of the failing heart: The cardiac burn-out syndrome? Cardiovasc Res. 2004;61:218–226. doi: 10.1016/j.cardiores.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Brown DA, Perry JB, Allen ME, Sabbah HN, Stauffer BL, Shaikh SR, Cleland JG, Colucci WS, Butler J, Voors AA, et al. Expert consensus document: Mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol. 2017;14:238–250. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson MT, Grimsrud PA, Lai L, Draper JA, Fisher-Wellman KH, Narowski TM, Abraham DM, Koves TR, Kelly DP, Muoio DM. Extreme acetylation of the cardiac mitochondrial proteome does not promote heart failure. Circ Res. 2020;127:1094–1108. doi: 10.1161/CIRCRESAHA.120.317293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frangogiannis NG. Pathophysiology of myocardial infarction. Compr Physiol. 2015;5:1841–1875. doi: 10.1002/cphy.c150006. [DOI] [PubMed] [Google Scholar]
- 29.Kolb AL, Corridon PR, Zhang S, Xu W, Witzmann FA, Collett JA, Rhodes GJ, Winfree S, Bready D, Pfeffenberger ZJ, et al. Exogenous gene transmission of isocitrate dehydrogenase 2 mimics ischemic preconditioning protection. J Am Soc Nephrol. 2018;29:1154–1164. doi: 10.1681/ASN.2017060675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentillon C, Li D, Duan M, Yu WM, Preininger MK, Jha R, Rampoldi A, Saraf A, Gibson GC, Qu CK, et al. Targeting HIF-1α in combination with PPARα activation and postnatal factors promotes the metabolic maturation of human induced pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 2019;132:120–135. doi: 10.1016/j.yjmcc.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Chen P, Li Y, Ardell C, Der T, Shohet R, Chen M, Wright GL. HIF-1α in heart: Protective mechanisms. Am J Physiol Heart Circ Physiol. 2013;305:H821–H828. doi: 10.1152/ajpheart.00140.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibb AA, Lorkiewicz PK, Zheng YT, Zhang X, Bhatnagar A, Jones SP, Hill BG. Integration of flux measurements to resolve changes in anabolic and catabolic metabolism in cardiac myocytes. Biochem J. 2017;474:2785–2801. doi: 10.1042/BCJ20170474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Tucker EJ, Compton AG, Calvo SE, Thorburn DR. The molecular basis of human complex I deficiency. IUBMB Life. 2011;63:669–677. doi: 10.1002/iub.495. [DOI] [PubMed] [Google Scholar]
- 35.Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsui H, Ide T, Kinugawa S. Mitochondrial oxidative stress, DNA damage, and heart failure. Antioxid Redox Signal. 2006;8:1737–1744. doi: 10.1089/ars.2006.8.1737. [DOI] [PubMed] [Google Scholar]
- 37.Elurbe DM, Huynen MA. The origin of the supernumerary subunits and assembly factors of complex I: A treasure trove of pathway evolution. Biochim Biophys Acta. 2016;1857:971–979. doi: 10.1016/j.bbabio.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Triepels RH, Van Den Heuvel LP, Trijbels JM, Smeitink JA. Respiratory chain complex I deficiency. Am J Med Genet. 2001;106:37–45. doi: 10.1002/ajmg.1397. [DOI] [PubMed] [Google Scholar]
- 39.Forbes JM, Ke BX, Nguyen TV, Henstridge DC, Penfold SA, Laskowski A, Sourris KC, Groschner LN, Cooper ME, Thorburn DR, et al. Deficiency in mitochondrial complex I activity due to Ndufs6 gene trap insertion induces renal disease. Antioxid Redox Signal. 2013;19:331–343. doi: 10.1089/ars.2012.4719. [DOI] [PubMed] [Google Scholar]
- 40.Kmita K, Wirth C, Warnau J, Guerrero-Castillo S, Hunte C, Hummer G, Kaila VR, Zwicker K, Brandt U, Zickermann V. Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc Natl Acad Sci USA. 2015;112:5685–5690. doi: 10.1073/pnas.1424353112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Guo L, Han S, Chen L, Li C, Zhang Z, Hong Y, Zhang X, Zhou X, Jiang D, et al. Adult mesenchymal stem cell ageing interplays with depressed mitochondrial Ndufs6. Cell Death Dis. 2020;11(1075) doi: 10.1038/s41419-020-03289-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ke BX, Pepe S, Grubb DR, Komen JC, Laskowski A, Rodda FA, Hardman BM, Pitt JJ, Ryan MT, Lazarou M, et al. Tissue-specific splicing of an Ndufs6 gene-trap insertion generates a mitochondrial complex I deficiency-specific cardiomyopathy. Proc Natl Acad Sci USA. 2012;109:6165–6170. doi: 10.1073/pnas.1113987109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulte U, Arretz M, Schneider H, Tropschug M, Wachter E, Neupert W, Weiss H. A family of mitochondrial proteins involved in bioenergetICS and biogenesis. Nature. 1989;339:147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- 45.Shan W, Li J, Xu W, Li H, Zuo Z. Critical role of UQCRC1 in embryo survival, brain ischemic tolerance and normal cognition in mice. Cell Mol Life Sci. 2019;76:1381–1396. doi: 10.1007/s00018-019-03007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unni S, Thiyagarajan S, Srinivas Bharath MM, Padmanabhan B. Tryptophan oxidation in the UQCRC1 subunit of mitochondrial complex III (Ubiquinol-Cytochrome C Reductase) in a mouse model of myodegeneration causes large structural changes in the complex: A molecular dynamics simulation study. Sci Rep. 2019;9(10694) doi: 10.1038/s41598-019-47018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis M, Whitely T, Turnbull DM, Mendelow AD. doi: 10.1007/978-3-7091-6837-0_17. Selective impairments of mitochondrial respiratory chain activity during aging and ischemic brain damage. Brain Edema X. Springer, Berlin, pp56-58, 1997. [DOI] [PubMed] [Google Scholar]
- 48.Moro MA, Almeida A, Bolaños JP, Lizasoain I. Mitochondrial respiratory chain and free radical generation in stroke. Free Radic Biol Med. 2005;39:1291–1304. doi: 10.1016/j.freeradbiomed.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Fan H, He Z, Huang H, Zhuang H, Liu H, Liu X, Yang S, He P, Yang H, Feng D. Mitochondrial quality control in cardiomyocytes: A critical role in the progression of cardiovascular diseases. Front Physiol. 2020;11(252) doi: 10.3389/fphys.2020.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol Cell Biol. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibanuma M, Inoue A, Ushida K, Uchida T, Ishikawa F, Mori K, Nose K. Importance of mitochondrial dysfunction in oxidative stress response: A comparative study of gene expression profiles. Free Radic Res. 2011;45:672–680. doi: 10.3109/10715762.2011.564169. [DOI] [PubMed] [Google Scholar]
- 52.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 53.Yi T, Wu X, Long Z, Duan G, Wu Z, Li H, Chen H, Zhou X. Overexpression of ubiquinol-cytochrome c reductase core protein 1 may protect h9c2 cardiac cells by binding with Zinc. BioMed Res Int. 2017;2017(1314297) doi: 10.1155/2017/1314297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J Biol Chem. 2005;280:22664–22669. doi: 10.1074/jbc.M414077200. [DOI] [PubMed] [Google Scholar]
- 55.Sun M, Zuo X, Li R, Wang T, Kang YJ. Vascular endothelial growth factor recovers suppressed cytochrome c oxidase activity by restoring copper availability in hypertrophic cardiomyocytes. Exp Biol Med (Maywood) 2014;239:1671–1677. doi: 10.1177/1535370214541910. [DOI] [PubMed] [Google Scholar]
- 56.Palmer DN, Fearnley IM, Medd SM, Walker JE, Martinus RD, Bayliss SL, Hall NA, Lake BD, Wolfe LS, Jolly RD. Lysosomal storage of the DCCD reactive proteolipid subunit of mitochondrial ATP synthase in human and ovine ceroid lipofuscinoses. Adv Exp Med Biol. 1989;266:211–223. doi: 10.1007/978-1-4899-5339-1_15. [DOI] [PubMed] [Google Scholar]
- 57.Li HS, Zhang JY, Thompson BS, Deng XY, Ford ME, Wood PG, Stolz DB, Eagon PK, Whitcomb DC. Rat mitochondrial ATP synthase ATP5G3: Cloning and upregulation in pancreas after chronic ethanol feeding. Physiol Genomics. 2001;6:91–98. doi: 10.1152/physiolgenomics.2001.6.2.91. [DOI] [PubMed] [Google Scholar]
- 58.Liu T, Chen L, Kim E, Tran D, Phinney BS, Knowlton AA. Mitochondrial proteome remodeling in ischemic heart failure. Life Sci. 2014;101:27–36. doi: 10.1016/j.lfs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadosky KM, Willis MS. The story so far: Post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation. Am J Physiol Heart Circ Physiol. 2012;302:H515–H526. doi: 10.1152/ajpheart.00703.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Wang SP, Shao Q, Li PF, Sun Y, Luo LZ, Yan XQ, Fan ZY, Hu J, Zhao J, et al. Brain-derived neurotrophic factor mimetic, 7,8-dihydroxyflavone, protects against myocardial ischemia by rebalancing optic atrophy 1 processing. Free Radic Biol Med. 2019;145:187–197. doi: 10.1016/j.freeradbiomed.2019.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional details regarding the LC-MS/MS analysis are available in the public database proteomecentral.proteomexchange.org/cgi/GetDataset? (accession no. PXD026207; lingfeng_zeng@163.com, Password: sUdIhJj4). The datasets used and/or analyzed during the current study are also available from the corresponding author on reasonable request.