Summary
Pathogens including autoantigens all failed to induce systemic lupus erythematosus (SLE). We, instead, studied the integrity of host's immune response that recognized pathogen. By stimulating TCR with an antigen repeatedly to levels that surpass host's steady-state response, self-organized criticality, SLE was induced in mice normally not prone to autoimmunity, wherein T follicular helper (Tfh) cells expressing the guanine nucleotide exchange factor DOCK8 on the cell surface were newly generated. DOCK8+Tfh cells passed through TCR re-revision and induced varieties of autoantibody and lupus lesions. They existed in splenic red pulp and peripheral blood of active lupus patients, which subsequently declined after therapy. Autoantibodies and disease were healed by anti-DOCK8 antibody in the mice including SLE-model (NZBxNZW) F1 mice. Thus, DOCK8+Tfh cells generated after repeated TCR stimulation by immunogenic form of pathogen, either exogenous or endogenous, in combination with HLA to levels that surpass system's self-organized criticality, cause SLE.
Subject areas: Immunology, Immune response, Cell biology
Graphical abstract

Highlights
-
•
Autoimmunity seldom takes place under integrated steady-state immune response
-
•
Repeated invasion by pathogen, such as measles virus, is not exceptional but routine in life
-
•
DOCK8+Tfh is generated upon TCR overstimulation by pathogen beyond self-organized criticality
-
•
Newly generated DOCK8+Tfh induces autoantibodies and SLE, i.e., autoimmunity
Immunology; Immune response; Cell biology
Introduction
Autoimmunity remains a mystery since the proposal of autoimmune disease theory by Mackay and Burnet in 1949 (Dameshek et al., 1965). Classical autoimmune disease, systemic lupus erythematosus (SLE) presents with fever, organ pathologies such as erythematous skin rash or lupus nephritis, and a variety of autoantibodies including anti-double stranded DNA (anti-dsDNA) Ab, which is among the leading causes of death in young women (Yen and Singh, 2018). SLE has been suspected to be caused by some type of autoimmune mechanism, reactivation of forbidden clone, or break of tolerance (Dameshek et al., 1965). Infection with microbes such as Epstein-Barr (EB) virus that activates B cells autonomously (Pender, 2003) or with microbes having molecular mimicry with endogenous antigens (Damian, 1964) has also been suspected. Upregulation of interferonα (IFNα) is also a candidate (Hooks et al., 1979; Preble et al., 1982; Ytterberg and Schnitzer, 1982; Ronnblom et al., 1991; Shiozawa et al., 1992; Santiago-Raber et al., 2003; Mathian et al., 2005; Theofilopoulos et al., 2005; Fairhurst et al., 2008; Baccala et al., 2012; Chasset and Arnaud, 2018; Christen, 2018; Akiyama et al., 2019). Upregulated IFNα reproduced SLE in some humans and transgenic mice (Ronnblom et al., 1991; Schilling et al., 1991; Tolaymat et al., 1992; Akiyama et al., 2019) and existed in the sera of patients. However, IFNα did not induce anti-Sm Ab considered diagnostic of SLE (Schilling et al., 1991; Tolaymat et al., 1992; Akiyama et al., 2019) or often improved the autoimmunity in mice (Hron and Peng, 2004; Li et al., 2005). Further, anti-IFNα Ab therapy was scarcely effective for SLE (Khamashta et al., 2016; Kalunian et al., 2016; Furie et al., 2017). Thus, candidate pathogens including autoantigens all failed to induce, exist in, and heal upon its removal of the systemic autoimmune disease SLE.
In our study, we reproducibly induced systemic autoimmunity in mice normally not prone to autoimmune disease by repeated stimulation with an antigen. After repeated stimulation several times with antigen, T cells were rendered into anergy, whereas they recovered from anergy and began to divide and produce IL-2 upon further stimulation with antigen. This repeated stimulation led to the generation of a novel CD4 T cell type, which we designated aiCD4 T cell (autoantibody-inducing CD4 T cell), capable of directing the production of a variety of autoantibodies and the differentiation of CD8 T cells to cytotoxic T lymphocytes (CTL) via antigen cross-presentation, which subsequently induced SLE in mice (Tsumiyama et al., 2009, 2013). SLE was thus induced not by a particular antigen but by repeated TCR stimulation with a pathogen in combination with HLA that surpassed the steady-state immune response, self-organized criticality, where pathogenic aiCD4 T cells having de novo autoreactivity were newly generated. We have therefore proposed “self-organized criticality theory” as to the pathogenesis of SLE (Tsumiyama et al., 2009). The key player in this pathogenesis, the aiCD4 T cell, remained uncharacterized. In the present study, therefore, we have tried to clarify the nature and derivation of these cells by biochemical characterization, cell transfer studies in mice, and by quantification of aiCD4 T cells in the peripheral blood and tissue of patients diagnosed with SLE. We also investigated a potential therapy for SLE using an antibody directed against these cells.
Results
Identification of DOCK8+CD4 T cell as aiCD4 T cell that induces SLE
To isolate and characterize aiCD4 T cells, we first induced aiCD4 T cells and SLE in BALB/c mice by 12x repeated intraperitoneal (i.p.) immunization with ovalbumin (OVA) without adjuvants, and then, various subsets of splenic CD4 T cells, particularly those increased in 12x OVA-stimulated mice, were taken and transferred to naive recipient mice. For each subset, we assessed the generation of rheumatoid factor (RF) and anti-dsDNA Ab in the recipient mice, and in this fashion, the CD4 T cell subset containing aiCD4 T cells were focused. We finally found that these autoantibodies could be induced by transferring the CD45RBlo CD122lo PD-1+ CD4 T cell subset (Figure S1A). The membrane and cytosolic fractions were extracted from these cells and subjected to electrophoresis and silver staining. Protein bands differentially expressed were then excised and analyzed by in-gel digestion followed by mass spectrometry. We found that the guanine nucleotide exchange factor DOCK8 was uniquely expressed in the membrane fraction of this T cell subset (Figure 1A). In mice repeatedly immunized with OVA, the number of DOCK8+CD4 T cells gradually increased in the spleen up to the final 12x immunization (Figure 1B). We then isolated DOCK8+CD4 T cells from these OVA-immunized mice and transferred them to naive recipient mice. The recipient mice generated RF and anti-dsDNA Ab and developed renal disease (Figures S1B and S1C).
Figure 1.
Identification of DOCK8 as marker of aiCD4 T cell and induction of SLE by DOCK8+CD4 T cells
(A) Expression of DOCK8 on CD45RBlo CD122lo PD-1+ CD4 T cells. Membrane and cytosolic fractions of either whole T cells (Lanes 1 and 5), CD45RBhi CD122hi CD4 T cells (Lanes 2 and 6), CD45RBlo CD122lo PD-1- CD4 T cells (Lanes 3 and 7), or CD45RBlo CD122lo PD-1+ CD4 T cells (Lanes 4 and 8) from 12x OVA-immunized BALB/c mice, were subjected to 10% of polyacrylamide gel electrophoresis, transferred to Immobilon-P membrane, and stained with anti-DOCK8 Ab.
(B) Increase of DOCK8+CD4 T cells after repeated immunization with OVA without an adjuvant. Flow cytometry analyses of DOCK8+CD4 T cell from splenic CD4 T cells of BALB/c mice after repeated 12x stimulation with PBS, 6x with OVA, or 12x with OVA.
(C) DOCK8+CD4 T cells transferred into recipient mice induced autoantibodies. Rheumatoid factor (RF), anti-Sm Ab, and anti-dsDNA Ab were quantified in the sera of BALB/c mice that had been pre-immunized 8x with OVA, depleted of CD4 T cells by anti-CD4 Ab, and then inoculated with DOCK8−CD4 T cells or DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. The control group was BALB/c mice pre-immunized 8x with OVA, CD4 T cell-depleted by anti-CD4 Ab, and inoculated with CD4 T cells from 12x PBS-immunized BALB/c mice. Anti-dsDNA Ab and anti-Sm Ab titers were represented by arbitrary unit (AU). Data were represented as mean ± SEM. Statistical assessment was by Student's t-test. ∗p<0.05.
(D) Generation of CTL, i.e., IFNγ+ CD8 T cells, after transfer of DOCK8+CD4 T cells. BALB/c mice were immunized 12x with OVA, and CD4 T cells were isolated. Cells were transferred into the anti-CD4 Ab-treated recipient mice immunized 8x with OVA. Matured CTL, i.e., IFNγ+ CD8 T cells, were measured 2 weeks after booster immunization 1x with OVA (n = 5). Data were represented as mean ± SEM. Statistical assessment was by Student's t-test. ∗p<0.05.
(E) Skin lesion. a: Moderate dermatitis with dermal fibrosis, b: liquefaction degeneration (arrow) in the basal cells of epidermis, and c: panniculitis in the deep area of the dermis of mice pre-treated 8x with OVA, depleted of CD4 T cells, and inoculated with DOCK8+ T cells from 12x OVA-immunized mice. d: Discoid lupus-like advanced dermatitis observed in mice immunized 12x with OVA. e: Healed dermatitis with some fibrosis in 12x OVA-immunized mice treated with anti-DOCK8 Ab 24 h each before the 6x, 8x, 10x, and 12x OVA immunizations with OVA. H & E stain. (bar = 30μm; original magnification a, b, ×400; c, x300; d, e, x100).
(F) Lesion other than kidney or skin. a: lung interstitial pneumonitis, b: interstitial pneumonitis accompanied by angiitis, c: liver pericholangitis, d: onion-skin lesion with amyloid deposition classical to lupus in the spleen, and f: thyroiditis, accompanied by giant cells, in the BALB/c mice 8x pre-immunized with OVA, CD4 T cell-depleted, and then inoculated with DOCK8+ CD4 T cells from 12x OVA-immunized BALB/c mice. e: Classical onion-skin-like lesion with amyloid deposits in the spleen, and g: perineuritis observed in the dermis of the BALB/c mice immunized 12x with OVA. H & E stain. (bar = 30μm; original magnification ×200).
See also Figures S1–S3.
To more closely examine the role of the transferred DOCK8+CD4 T cells in the development of renal disease, we immunized BALB/c mice 8x with OVA. At this time point, nephritis-inducing CTLs had yet to mature. We then treated naive mice with anti-CD4 Ab to deplete their CD4 T cells and transferred into them, the DOCK8+CD4 T cells isolated from the spleens of BALB/c mice immunized 12x with OVA. In the recipient mice, autoantibodies were generated and IFNγ+ CD8 T cells were developed which then caused renal disease characteristic of WHO class IV and V (Weening et al., 2004) advanced disease (Figures 1C–1F and Table 1). The DOCK8+CD4 T cells capable of directing the production of a variety of autoantibodies and the differentiation of CD8 T cells into CTL to induce lupus tissue injury were also generated after 12x immunization with another antigen keyhole limpet hemocyanin (KLH) or 8x stimulation with staphylococcus enterotoxin B (SEB) which cross-linked T cell receptor (TCR) and major histocompatibilty complex (MHC) (Figure S2; Tsumiyama et al., 2009). These indicate that DOCK8+CD4 T cells generated by repeated stimulation with either antigen are capable of directing the production of a variety of autoantibodies and the differentiation of CD8 T cells into CTL leading to lupus tissue injury. Anti-dsDNA autoantibody or renal disease was not generated in DOCK8 −/− mice after 12x immunization with OVA (Figure S3).
Table 1.
Generation of renal disease in recipient mice by the transfer of DOCK8+CD4 T cells
| Donor | Transferred cell | Lupus nephritis WHO class |
|||
|---|---|---|---|---|---|
| I & II | III | IV | V | ||
| 12x PBS | CD4 T cell | 71.96 ±11.04% | 28.03 ±11.04% | 0% | 0% |
| 12x OVA | DOCK8- CD4 T cell | 68.54 ±12.39% | 29.31 ±12.75% | 2.14 ±3.36% | 0% |
| 12x OVA | DOCK8+ CD4 T cell | 18.77 ±9.40%∗∗ | 33.99 ±9.78% | 31.89 ±17.04%∗ | 15.32 ±9.17%∗ |
Glomerulonephritis in BALB/c mice that had been pre-immunized 8x with OVA, depleted of CD4 T cells by anti-CD4 Ab, and inoculated with DOCK8−CD4 T cells or DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. Control group was BALB/c mice that had been pre-immunized 8x with OVA, depleted of CD4 T cells by anti-CD4 Ab, and inoculated with CD4 T cells from 12x PBS-immunized BALB/c mice. Glomerular lesions in mice were classified according to human WHO classification (Weening et al., 2004) as follows; class I, normal glomeruli; class II, purely mesangial disease; class III, focal proliferative glomerulonephritis; class IV, diffuse proliferative glomerulonephritis; and class V, membraneous glomerulonephritis. Data were represented as mean ± SEM. Statistical assessment was by Student's t-test.
p<0.05,vis 12x OVA DOCK8- CD4 T cell and vis 12x PBS CD4 T cell.
p<0.001, vis 12x OVA DOCK8- CD4 T cell and vis 12x PBS CD4 T cell.
In addition to renal disease, transfer of DOCK8+CD4 T cells into naive mice induced dermatitis often accompanied by dermal perineuritis in 4 out of 5 recipient mice, but none in mice receiving control DOCK8−CD4 T cells (Figure 1E and Table 2; p = 0.0476 by Fisher's exact test). Panniculitis in the dermis and skin epidermal liquefaction degeneration, classical lesion of SLE, were observed in 2 of 5 DOCK8+CD4 T recipients, and none in the control mice (Figure 1E). Lung interstitial pneumonitis was seen in 4 of 5 DOCK8+CD4 T recipients, and 1 of 5 control mice (Figure 1F and Table 2). Pericholangitis with liver cell necrosis was seen in 2 of 5 DOCK8+CD4 T recipients, and none in the control (Figures 1F and Table 2). Diffuse thyroiditis was seen in 2 of 5 DOCK8+CD4 T recipients, and none in the control. Splenic perivascular fibrosis with amyloid-like deposition and classical onion skin lesion pathognomonic of SLE (Kaiser, 1942) were seen in all 5 of 5 DOCK8+CD4 T recipients, and none in the control (Figure 1F and Table 2; p = 0.0079 by Fisher's exact test). Lung interstitial pneumonitis seen in the DOCK8+CD4 T recipients was often accompanied by angiitis (Figure 1F).
Table 2.
Summary of the lesions other than kidney in the BALB/c mice 8x pre-immunized with OVA, CD4 T cell-depleted, and inoculated with DOCK8−CD4 T cells or DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice
| Transferred cell: | 12x immunized with OVA |
Treatment with Ab |
||||
|---|---|---|---|---|---|---|
| DOCK8+ CD4+ | DOCK8- CD4+ | p∗ | Control IgG | Anti- DOCK8 Ab | p∗ | |
| Skin, dermatitis | 4/5 | 0/5 | 0.0476 | 4/5 | 0/5 | 0.0476 |
| Lung, interstitial pneumonitis | 4/5 | 1/5 | 0.206 | 4/5 | 0/5 | 0.0476 |
| Liver, pericholangitis | 2/5 | 0/5 | 0.444 | 2/5 | 1/5 | 0.1 < |
| Spleen, onion skin lesion | 5/5 | 0/5 | 0.0079 | 2/5 | 1/4 | 0.1 < |
| Thyroid, thyroiditis | 2/5 | 0/5 | 0.444 | 4/5 | 0/5 | 0.0476 |
| Thymus, cortex atrophy | 0/5 | 0/5 | 0/5 | 5/5 | 0.0079 | |
Also shown were the results from anti-DOCK8 Ab treatment of 12x OVA-immunized mice: Mice received 100μg of anti-DOCK8 Ab 24 h each before the 6x, 8x, 10x, and 12x OVA immunizations with OVA. ∗Statistical analysis was by Fisher's exact test.
DOCK8+CD4 T cell is a novel T follicular helper cell, located in splenic red pulp
To clarify the nature of DOCK8+CD4 T cells, they were labeled with Au-tagged anti-DOCK8 Ab and examined by immunoelectron microscopy. These cells were observed to be relatively large lymphocytes with abundant ER and mitochondria (Figure 2A). DOCK8+CD4 T cells were CD44+, CD62L−, ICOS+ PD-1+ CXCR5mid, CD3+, and Thy1.2+, indicative of activated T follicular helper cells (Breitfeld et al., 2000; Schaerli et al., 2000) (Figures 2B, S4, and S5), in particular, the type of long-lived germinal center (GC) effector T follicular helper cells (Iyer et al., 2013; Kunzli et al., 2020) which also express FR4 and GL7 (Figure 2B). These cells also expressed Ly6C, LFA1, CCR4, CD123, and CD23 (Qi et al., 2008; Stebegg et al., 2018) (Figures 2B, S4, and S5A). DOCK8+CD4 T cells produced higher amounts of IFNγ, IL-4, IL-6, IL-17, IL-21, and IL-22 (Figure 2C), whereas IL-12 or IFNα was undetectable. Treg (regulatory T) cell or Th17 cell subsets were not increased by 12x stimulation with OVA (Figure S5B).
Figure 2.
DOCK8+ CD4 T cell as T fh cell located at splenic red pulp, and its increase in peripheral blood of patients with SLE
(A) a: Morphology of DOCK8+CD4 T cell observed by immunoelectron microscopy, stained with rabbit anti-DOCK8 Ab and Au-tagged ant-rabbit IgG Ab. (bar = 1μm; original magnification ×12,000). b,c: Nuclear membrane portions of DOCK8+ CD4 T cell, stained likewise. (bar = 1μm; original magnification ×18,000; x24,000).
(B) Markers of splenic DOCK8+CD4 T cells. Flow cytometry for ICOS, CXCR5, and PD-1 expression in splenic DOCK8+CD4 T cells taken 9 days after the final immunization of BALB/c mice immunized 12x with PBS versus OVA (Left). Flow cytometry analysis of ICOS, CXCR5, PD-1, GATA3, RORγt, T-bet, Bcl6, Ly6C, LFA1, FR4, and GL7 expression in splenic DOCK8−CD4 T cells and DOCK8+CD4 T cells isolated from 12x OVA-immunized BALB/c mice.
(C) Cytokines released in culture supernatants of 1x106/ml DOCK8+ or DOCK8- CD4 T cells following incubation with 2 μg/mL of anti-CD3 and 5 μg/mL of anti-CD28 Ab at 37°C for 24hr. Data were represented as mean ± SEM. Statistical assessment was by Student's t-test. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.005, ∗∗∗∗p<0.001.
(D) DOCK8+CD4 T cells in the autopsied spleen of untreated patient with active SLE who died before starting therapy. Tissues were fixed in 10% formalin, deparaffinized by graded xylene, and graded alcohol sequentially, and serially treated with biotinylated anti-DOCK8 monoclonal Ab and rabbit anti-IFNγ Ab (for T cell staining), alkaline phosphatase avidin D, and anti-rabbit Novolink, followed by reaction with Fast Red (Red for DOCK8+ cells) and Histogreen (Blue for T cells). S: red pulp sinus. A; central artery. P: penicillar artery. (bar = 100μm; original magnification ×100). Inset; Magnified DOCK8+CD4 T cells (Arrows). (bar = 20μm; original magnification ×400).
(E) Circulating DOCK8+CD4 T cells within CD4 T cells in the peripheral blood of patients with SLE and its relationship to the SLE disease activity index (SLEDAI). Data of control patients were depicted as; ■ rheumatoid arthritis, ▲ mixed connective tissue disease, △ polymyositis, ◆ systemic sclerosis, □ microscopic polyangiitis. Also shown was quantification of interferonα (IFNα) in sera of the same patients with SLE measured by ELISA.
See also Figures S2 and S4–S6.
In the patients with active SLE, DOCK8+CD4 T cells were located in splenic sinus, red pulp, in a disease-specific fashion (Figure 2D). This would be due to that CCR7 was not increased but CD147 and LFA1 were increased in DOCK8+CD4 T cells (Figures 2B and S4); previous study showed that CCR7 was required for T cell entry from vasculature into T cell zones and anchorage them against blood flow (Chauveau et al., 2020). Splenic sinus, red pulp, is a suitable location to encounter circulating antigens and nurse the autoreactive B cells that derive from bone marrow or shuttle between the marginal and follicular zones by expressing CD138, undergoing class-switch recombination and readily differentiating into plasma cells (William et al., 2002; Odegard et al., 2008; Arnon et al., 2013; Stebegg et al., 2018). In the spleen of 12x OVA-immunized mice, indeed, activated B cells with CD138 markers classified as plasma cells and follicular B cells were significantly increased (Figure S6). In the red pulp, DOCK8+CD4 T cells could also meet the red pulp macrophages which are highly efficient at cross-presenting antigens to T cells and maturing CTLs (Enders et al., 2020) which were essentially important for the generation of lupus tissue injuries shown previously (Tsumiyama et al., 2009, 2013).
Circulating DOCK8+CD4 T cells increase in association with SLE disease activity
Because T follicular helper cells are measurable at the periphery and their frequency reflects in vivo dynamics (La Muraglia et al., 2020), we tried to measure DOCK8+CD4 T cells in the peripheral blood of patients with SLE. Higher numbers of circulating DOCK8+CD4 T cells were found in the peripheral blood of patients, and these numbers correlated with higher SLEDAI disease activity scores (Bombardier et al., 1992) (Figure 2E). This indicates that surface expression of DOCK8 on CD4 T cells was associated with high activation levels of CD4 T cells and SLE disease activity. It was noted here that although DOCK8+CD4 T cells were significantly decreased by conventional therapy, a substantial number of patients remained with slightly increased DOCK8+CD4 T cell numbers compared with disease controls. This would be compatible with the therapeutic sequellae of SLE in which the patients require tapered but years-long continued therapy with prednisolone or immunosuppressive agents, suggesting that DOCK8+CD4 T cells, once activated, did not disappear quickly in response to conventional therapy.
TCR revision and induction of autoimmunity, characteristic of SLE
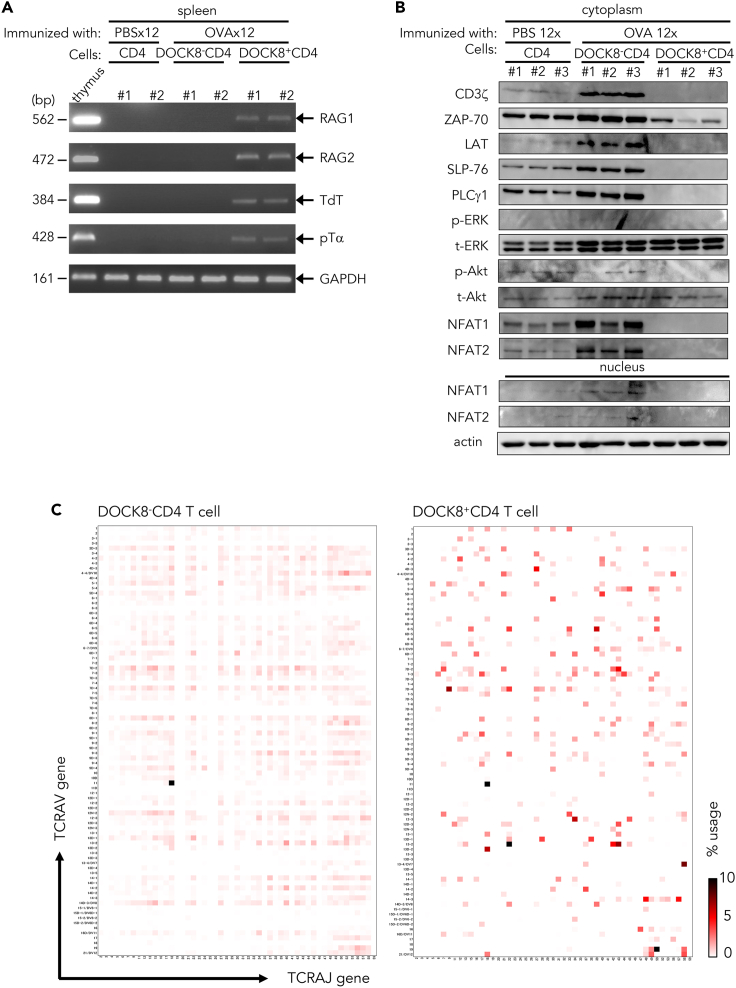
In the DOCK8+CD4 T cells, genes encoding components of V(D)J recombinase complex, recombination-activating genes 1 and 2 (RAG1/2), terminal deoxynucleotidyl transferase (TdT), and surrogate TCRα chain (pTα), were upregulated (Figure 3A). TCR basal signaling molecules, known to suppress RAG expression (Lantelme et al., 2000; Roose et al., 2003; Patra et al., 2006) such as CD3ζ, ZAP70, LAT, SLP-76, PLCγ1, ERK, Akt, and NFAT1/2 and their nuclear translocation were downregulated (Figure 3B), and the transcription factor GATA3, essential for development of T cell lineage (Ting et al., 1996) and also RAG assembly (Ho et al., 1991; Ting et al., 1996; Naik et al., 2018), was upregulated (Figure 2B). TCR repertoire analyses showed that the diversity of TCR gene usage was restricted and skewed in the direction of novel TCR repertoires, even though the stimulation was a single antigen, OVA (Figures 3C, S7, and S8, and Tables S1 and S2; supplemental information). The heatmap representation of this repertoire reveals that this novel TCR gene usage, as characterized by a combination of novel TRAV and TRAJ gene variations, was restricted to DOCK8+CD4 T cells (Figure 3C). A diversity score calculation (Shannon, 1948; Simpson, 1949), which included the inverted Simpson index, showed that the weighted arithmetic mean of the proportional abundances of TCR repertoires in the DOCK8+CD4 T cells was substantially restricted; 1.22%–5.95% of the DOCK8−CD4 T cells (Table S1; supplemental information). Unique TCR repertoires, unique sequence reads (USRs) having the combination of TRAV and TRAJ, and amino acid sequences of the CDR3 that had no identity with other sequence reads, were dominantly found in the DOCK8+CD4 T cells compared with DOCK8−CD4 T cells from the same 12x OVA-immunized mice, or compared with CD4 T cells from control mice mock-immunized with PBS (Table S2; supplemental information). A variety of autoantibodies were induced when these DOCK8+CD4 T cells from 12x OVA-immunized mice were transferred into naive mice (Figure 4). These findings together indicate that the DOCK8+CD4 T cells had undergone TCR re-revision at the periphery and induced in vivo novel antibodies, including autoantibodies.
Figure 3.
TCR revision in DOCK8+CD4 T cells
(A) Expression of V(D)J recombinase complex and related molecules in splenic CD4 T cells from 12x PBS-immunized BALB/c mice, and in DOCK8−CD4 T cells and DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. Two mice were analyzed for each group.
(B) Western blot analysis of TCR signal transduction molecules in splenic CD4 T cells from 12x PBS-immunized BALB/c mice, and in DOCK8−CD4 T cells and DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. Three mice were analyzed for each group. GAPDH, control housekeeping glyceraldehyde 3-phosphate dehydrogenase gene.
(C) TCR repertoire analysis. Heatmap visualization of TCR usage of a combination of TRV and TRJ genes in the splenic DOCK8- CD4 T cells and DOCK8+ CD4 T cells from 12x OVA-immunized mice. See also Figures S7 and S8.
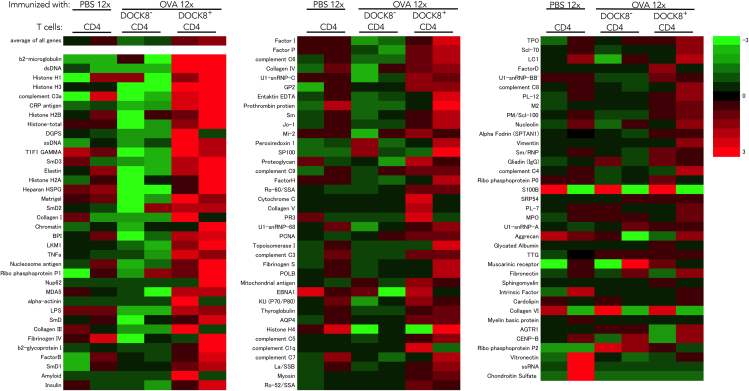
Figure 4.
Autoantibody microarray study
Autoantibodies produced in BALB/c mice pre-immunized 8x with OVA, depleted of CD4 T cells, and inoculated with DOCK8−CD4 T cells or DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. Control was autoantibodies produced in BALB/c mice pre-immunized 8x with OVA, depleted of CD4 T cells, and inoculated with CD4 T cells from 12x PBS-immunized BALB/c mice. Two mice were analyzed for each group.
TCR gene chromatin accessibility
aiCD4 T cells were generated following heavily repeated TCR stimulation with antigen, which released them from their anergic state, irrespective of the antigen used or genetic background, by the help of NKT cells (Figure S9; Tsumiyama et al., 2009). Thus, even repeated challenge by SEB, which cross-links TCR and MHC, and also an another antigen such as KLH, could induce the aiCD4 T cells capable of directing the production of a variety of autoantibodies and the differentiation of CD8 T cells into CTL to induce lupus tissue injury (Figures S2 and S9; Tsumiyama et al., 2009; Tsumiyama et al., 2013). Owing to these strong stimulation, DOCK8 re-localized from its normal site in the cytoplasm to the cell membrane (Cote et al., 2005; Premkumar et al., 2010; Harada et al., 2012) and the nuclear lamina (Taylor et al., 2010; Sadhukhan et al., 2014) (Figure 2A). Further, the progenitor, transforming or reprogramming factors responsible for RAG modulation and chromosomal accessibility (Bergqvist et al., 2000; Yang et al., 2003; Staal et al., 2008; Kovalovsky et al., 2009; Ippolito et al., 2014; Janssen et al., 2016; Montefiori et al., 2016; Takahashi and Yamanaka, 2016; Lee et al., 2018; Rothenberg et al., 2019; Jain et al., 2019) such as KLF4, KLF5, Zeb2, EBF1, PAX5, PAX6, Bcl11a, and its target Tcf4 were upregulated (Figure 5; Supplemental Information). Nuclear DOCK8 (Janssen et al., 2016), by binding and activating cdc42 (Taylor et al., 2010; Sadhukhan et al., 2014), can activate nuclear-WASP and upregulate H3K4 trimethyltransferase (Taylor et al., 2010; Sadhukhan et al., 2014) and thus may disrupt the association of TCR genes with nuclear lamina via actin movement, thereby enhance the chromatin accessibility to transcription factor E2-2 (Tcf4) (Figure 5). PU.1 (Spi.1), which is normally undetectable in mature T cells, was also expressed, and the genes upregulated by PU.1 such as Bcl11a, Lmo2, Mef2c, Syk, Lyn, and Cd300a, but not Bcl11b, were upregulated (Ippolito et al., 2014; Lee et al., 2018; Rothenberg et al., 2019) (Figure 5). Cell surface expression of DOCK8 should also strengthen immunological synapse formation (Cote et al., 2005; Premkumar et al., 2010; Ham et al., 2015; Janssen et al., 2016; Hashimoto-Tane and Saito, 2016) together with LFA1 or HkRP3, WIP, WASP, and PtdIns-3-kinase (Figures 2B and 5; Supplemental Information).
Figure 5.
Expression of transcriptional regulators in DOCK8+CD4 T cells
(A) Transcriptional regulators responsible for TCR revision. Western blot analysis of DOCK8 and transcriptional regulators in splenic CD4 T cells from 12x PBS-immunized BALB/c mice, and in DOCK8−CD4 T cells and DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice. Three mice were analyzed for each group.
(B) Upregulated genes in DOCK8+CD4 T cells. Results of gene expression microarray analysis, abstract from Supplemental Information (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159240). Ratio of mRNA expression of splenic DOCK8+CD4 T cells from 12x OVA-immunized BALB/c mice in comparison with splenic DOCK8−CD4 T cells from 12x OVA-immunized mice, given as a log10 scale, wherein 2, 3, 4, 5, and 6 indicated x102, x103, x104, x105, and x106, respectively.
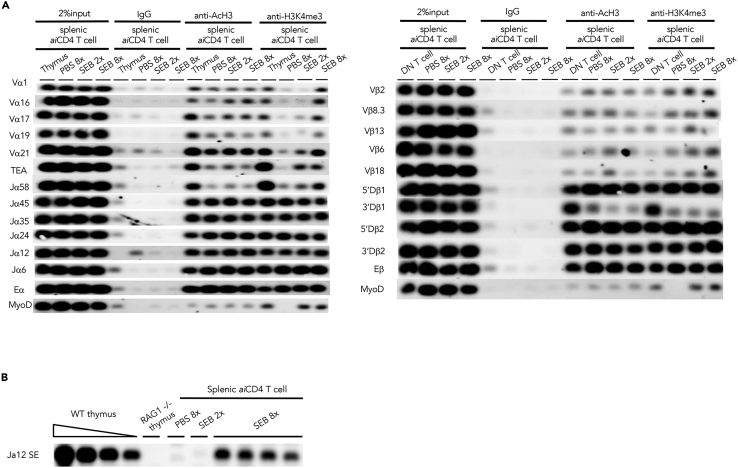
We performed chromatin-immunoprecipitation sequencing (ChIP-seq) to map histone acetylation (AcH3) and H3K4 methylation (H3K4me3) of the TCRB and TCRA genes in the DOCK8+aiCD4 T cells generated by 8x stimulation with SEB. H3K4me3 was found to be enriched at the TCRA Jα45, Jα35, Jα24, Jα12, Jα6, and Eα gene segments and TCRB 5′Dβ1, 5′Dβ2, 3′Dβ2, and Eβ gene segments of both control PBS-treated and experimental 8x SEB-stimulated Vβ8+aiCD4 T cells (Figure 6). This result is in accordance with the previous reports showing that the TCR gene recombination center is an open structure (Villey et al., 1996; Abarrategui and Krangel, 2007; Ji et al., 2010; Zacarias-Cabeza et al., 2015; Chen et al., 2018). However, in the 8x SEB-stimulated aiCD4 T cells, H3K4me3 was also found in the 5′ proximal Vα1, Vα16, Vα17, Vα21, TEA, Vβ8.3, Vβ6, and Vβ18 gene segments. These findings are in line with the result of TCR repertoire analysis in the 12x OVA-stimulated DOCK8+aiCD4 T cells that 5′ proximal Vα genes were dominantly expressed (Table S2). To confirm that V(D)J recombination took place in aiCD4 T cells, we performed ligation-mediated PCR (LM-PCR) to detect blunt-end DNA fragments harboring a rearranged coding Jα region flanked by recombination signal sequences (RSS). We identified the rearranged intermediates corresponding to TCRα joining region 12 (TCRAJ12) in the splenocytes of 8x SEB-stimulated mice (Figure 6B). Expression of the Vα and Jα genes distal from TEA in autoimmunity are compatible with the general rules showing the stepwise and coordinated proximal-to-distal progressions of Vα and Jα usage on individual TCRA alleles in CD4+ CD8+ thymocytes (Carico et al., 2017). Thus, while we were unable to show directly the contribution of transcription factor E2-2 or DOCK8 to TCR gene transactivation, the results showed that some novel TCR chromatin regions, particularly distal Vα and J genes, were accessible in the aiCD4 T cells.
Figure 6.
Enhanced chromatin accessibility in DOCK8+CD4 T cells
(A) Chromatin-immunoprecipitation sequencing (ChIP-seq) assay for V(D)J chromatin accessibility. Left panel: Histone acetylation (AcH3) and H3K4 methylation (H3K4me3) of the TCRA gene of splenic aiCD4 T cells from 8x SEB-stimulated BALB/c mice. Splenic CD4 T cells of 8x SEB-stimulated BALB/c mice which produced amply high titers of autoantibodies were assayed. TEA, TCRα promoter. Eα, TCRα enhancer. MyoD, myoblast determination 1 as negative controls. Right panel: Histone acetylation (AcH3) and H3K4 methylation (H3K4me3) of the TCRB gene of splenic aiCD4 T cells from 8x SEB-stimulated BALB/c mice as above. Eβ, TCRβ enhancer.
(B) TCRα chain revision in the splenic CD4 T cells from the mice immunized 8x with SEB producing high titers of autoantibodies was determined by LM-PCR detection of dsDNA breaks at the RSS flanking the TCRAJ12. WT, wild-type.
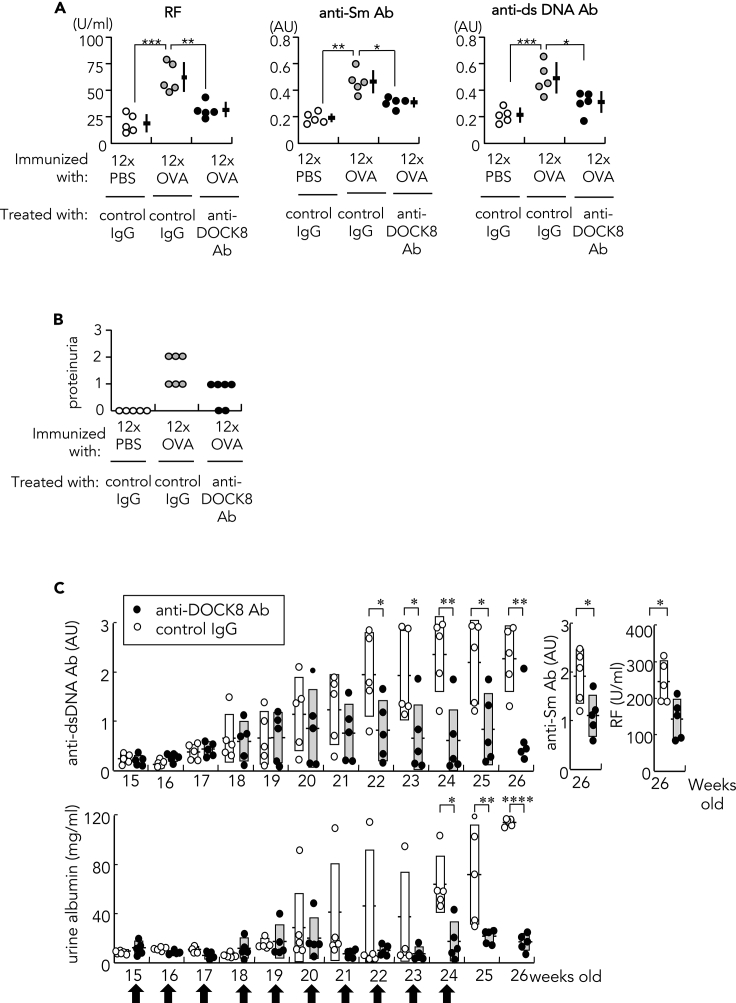
Treatment of SLE in mice with anti-DOCK8 antibody
We treated 12x OVA-immunized mice with anti-DOCK8 Ab 24 h each before the 6x, 8x, 10x, and 12x immunization with OVA, and the mice were studied 9 days after final, 12x immunization. Anti-DOCK8 Ab-treated mice developed lower titers of RF and anti-Sm and anti-dsDNA Abs compared with control isotype Ig-treated mice (Figure 7). Renal disease was significantly ameliorated and almost cured as assessed by histology (Figure 7; Table 3). Lesions of the skin (Figure 1E and Table 2), lungs, and thyroid were also significantly ameliorated by anti-DOCK8 Ab treatment (Table 2), and the thymic cortex was significantly atrophied. We also treated female (NZBxNZW) F1 mice, a classical lupus model (Andrews et al., 1978), with anti-DOCK8 Ab every 15 to 24 weeks. Autoantibodies, RF, anti-Sm Ab and anti-dsDNA Ab, and proteinuria were significantly decreased in the DOCK8 Ab-treated mice compared to controls (Figure 7C), suggesting that generation of disease in this lupus-prone mouse model was mechanistically similar to that generated in antigen-stimulated BALB/c and C57BL/6 mice.
Figure 7.
Treatment with anti-DOCK8 Ab
(A) Effect of anti-DOCK8 mAb treatment on autoantibody production in12x OVA-immunized mice. Autoantibodies, rheumatoid factor (RF), anti-Sm Ab, and anti-dsDNA Ab, were quantified in the sera of 12x PBS-immunized or 12x OVA-immunized mice: Mice received anti-DOCK8 Ab (100μg) or control rabbit IgG (100μg) 24 h each before the 6x, 8x, 10x, and 12x immunizations with OVA. Anti-Sm Ab and anti-dsDNA Ab titers were represented by arbitrary unit (AU). Data were represented as mean ± SEM. Statistical analysis was by Student's t-test; ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.005.
(B) Effects of anti-DOCK8 Ab treatment on renal disease. Proteinuria measured 9 days after the final immunization in 12x PBS-immunized or 12x OVA-immunized BALB/c mice, graded with a score of 0 (<30 mg/dL); 1 (30–99 mg/dL); 2 (100–299 mg/dL); or 3 (300–999 mg/dL).
(C) Effects of anti-DOCK8 mAb treatment on autoantibody production and renal disease in (NZBxNZW) F1 female mice. Autoantibodies, anti-dsDNA Ab, anti-Sm Ab, and rheumatoid factor (RF), and proteinuria quantified by ELISA in (NZBxNZW) F1 female mice, which were treated either with anti-DOCK8 mAb (100 μg) or control rabbit IgG (100 μg) every week from 15 to 24 weeks (arrow). Data were represented as mean ± SEM. Statistical assessment performed by Student's t-test; ∗p<0.05, ∗∗p<0.01, ∗∗∗∗p<0.001.
Table 3.
Renal lesion of BALB/c mice immunized 12x with either PBS or OVA, and treated either with anti-DOCK8 Ab (100μg) or control rabbit IgG (100μg) 24 h each before the 6x, 8x, 10x, and 12x OVA immunizations with OVA
| Immunized with | Treatment | Lupus nephritis WHO class |
|||
|---|---|---|---|---|---|
| I & II | III | IV | V | ||
| 12x PBS | Control IgG (n = 3) | 48.68 ±12.91% | 28.05 ±9.45% | 14.17 ± 3.72% | 9.08 ± 3.01% |
| 12x OVA | Control IgG (n = 6) | 7.33 ±5.12% | 18.33 ±7.34% | 24.04 ±7.35% | 50.28 ±7.62% |
| 12x OVA | Anti-DOCK8 Ab (n = 6) | 33.91 ±14.40%∗ | 36.37 ±6.98% | 19.93 ±11.64%∗∗ | 9.70 ±8.78%∗∗ |
Glomerular lesions were classified according to human WHO classification (Weening et al., 2004) as follows; class I, normal glomeruli; class II, purely mesangial disease; class III, focal proliferative glomerulonephritis; class IV, diffuse proliferative glomerulonephritis; and class V, membraneous glomerulonephritis. Data were represented as mean ± SEM. Statistical assessment was by Student's t-test.
p<0.005, vis control IgG treatment.
p<0.001, vis control IgG treatment.
Relationship to infection as a cause of SLE
Our self-organized criticality theory proposes that SLE or systemic autoimmunity is induced after repeated infection with a pathogen, in which disintegration of host immune system but not specific pathogens or antigens including autoantigens (molecular mimicry) are directly involved in its pathogenesis. Pathogens must be immunogenic, antigen-presented for autoantibody generation, and antigen cross-presented for autoimmune tissue injury, where microbial infection including molecular mimicry with autoantigens induces host's immune response upon PAMPs-TLR connection; we studied the contribution of LPS or CpG signaling to the pathogenesis of SLE. In mice repeatedly immunized with OVA or KLH with or without LPS or CpG, signaling via TLR4 or TLR9, similar but rather increased autoantibodies and renal disease were raised, which indicated that TLR signaling enhanced autoimmunity (Figure S2).
Relationship to interferonα(IFNα) as a cause of SLE
We observed that IFN response genes were also upregulated in DOCK8+CD4 T cells (Figure S10; supplemental information). Recent studies have shown that activation of enhancer-like promoter are preferentially associated with the expression of housekeeping or stress response genes by default, including IFN response genes (Dao et al., 2017; Dao and Spicuglia, 2018). Upon chronic infection, T cells shift toward a T follicular helper phenotype and upregulate IFN signature genes (Crawford et al., 2014; Vella et al., 2017). Tcf4, which was upregulated in DOCK8+CD4 T cells, is a transcription factor specific for plasmacytoid dendritic cells (pDC) that promotes expression of the pDC-enriched genes such as SpiB, Irf8, and Zeb2 (Cisse et al., 2008; Keles et al., 2014; Sisirak et al., 2014) (Figures 5 and S10, supplemental information). Microarray analyses show that DOCK8+CD4 T cells share some characteristics of pDC (Figures 5 and S10); however, pDC numbers per se were not increased in 12x OVA-immunized mice (Figure S10). Serum IFNα was also not increased in the sera of 12x OVA- or 8x SEB-stimulated mice (data not shown), or in most of the SLE patients studied (Figure 2E). Serum IFNα was slightly higher in 3 treated patients with SLE (indicated by ○ in Figure 2E), whereas their serum levels were still below the levels detectable in patients with active SLE (Shiozawa et al., 1992), as well as in the mice in which we could induce SLE by the several-fold upregulation of IFNα (Akiyama et al., 2019). Two out of 3 patients with SLEDAI score of 0 and 3.5 (Figure 2E) had been treated with conventional therapy and potentially could have had higher IFNα levels at an earlier time, which might have induced SLE. Overall, however, serum IFNα was not significantly increased in the patients studied here, even though the disease mechanism described here should enhance IFNα generation. Thus, while highly increased IFNα can induce SLE (Akiyama et al., 2019) in at least some patients, IFNα appears to be increased mostly in association with the disease mechanism shown in the present study, i.e., chronic stimulation by pathogen, and in most cases, exert a disease-modifying, but not causative, role in manifestations such as erythematous skin rash, neuropsychiatric disorders, or renal disease (Shiozawa et al., 1992; Akiyama et al., 2019).
Discussion
These results show that systemic autoimmune disease, SLE, is caused not by the autoimmune mechanism but by repeated infection by a pathogen. Causative pathogen may differ individually depending on host's MHC (major histocompatibility complex) (Tsumiyama et al., 2009); however, for autoimmunity to develop, TCR must be stimulated strongly beyond host's steady-state immune response, i.e., self-organized criticality, wherein we found that a previously uncharacterized immune responses took place. CD4 T cells were resuscitated from pathogen-induced anergy and matured into the Tfh cells that expressed guanine nucleotide exchange factor DOCK8 on the cell surface and acquired autoreactivity via TCR re-revision at the periphery. These DOCK8+ Tfh cells subsequently induced a variety of autoantibodies and SLE.
Host's immune system is equipped with safeguard mechanisms, and thus, it is rational that autoimmunity does not occur within the steady-state immune response. Indeed, all pathogens previously proposed including autoantigens failed to induce systemic autoimmunity except for some mutations (Linterman et al., 2009). Living organisms are, however, constantly exposed to a variety of pathogens including non-fatal viruses that can invade the host if they circumvent host's CTL defense (Zinkernagel, 1996; Rose, 2017), as exemplified by the recent re-emergence of measles infection in a subpopulation of Japanese young adults who circumvented vaccination against the virus. These repeated rounds of infection, however, may be unapparent at the time of re-infection if CTL memory has lapsed. Such antigens, if presented in combination with the appropriate host's HLA, may maximally stimulate the host's TCR to levels that surpass system's self-organized criticality.
Previous studies have shown that, upon encounter with peripherally expressed antigens such as mouse mammary tumor virus (Mtv-8) or influenza hemagglutinin (HA), the CD4 T cells reactive against them are deleted or anergized (St. Rose et al., 2009; Higdon et al., 2014). However, further repeated stimulation causes a few cells to be resuscitated from anergy, proliferate, and pass through TCR re-revision at the periphery, acquiring a T follicular helper-like phenotype (St. Rose et al., 2009; Crawford et al., 2014; Higdon et al., 2014; Vella et al., 2017). Because these cells no longer react against the same antigens and reside in a restricted space, splenic GC, immune response will cease (Higdon et al., 2014). However, if further stimulated by an “immunogenic” form of antigen such virus as influenza HA, the once-anergized T cells will begin to proliferate (St. Rose et al., 2009). The SLE-inducing DOCK8+ Tfh cells are generated in this fashion, and the newly generated DOCK8-expressing Tfh cells reside in the splenic red pulp, a suitable location encountering circulating antigens and nursing autoreactive B cells (William et al., 2002; Odegard et al., 2008; Arnon et al., 2013) and also efficiently cross-presenting antigens to T cells and maturing CTL (Enders et al., 2020), which was essentially important for generating lupus tissue injuries (Tsumiyama et al., 2013). Therefore, repeated stimulation with “immunogenic” antigen, either exogenous or endogenous, generates DOCK8-expressing Tfh cells which then induce autoantibodies and SLE. Induction of SLE by repeated stimulation with endogenous viral, “immunogenic form” of antigen is compatible with the molecular mimicry theory of autoimmunity (Damian, 1964; Fujinami et al., 1983; Oldstone, 2014; Rose, 2017). However, for this theory to be valid, host's steady-state immune response must be disintegrated beforehand, as shown in the present study.
Because SLE is unique in that a large variety of autoantibodies, sometimes exceeding 100 types, are generated in an individual (Yaniv et al., 2015; Olsen et al., 2017), generation of autoantibodies and SLE as a result of TCR re-revision at the periphery seems rational as compared with imaging reactivation of forbidden clones.
Limitations of the study
The cause of autoimmunity or SLE remained unknown since the proposal of “autoimmne disease theory” in 1949 by Mackay and Burnet (Dameshek et al., 1965). Extensive studies since then all failed to show autoreactivity including autoantigens as the cause of SLE. Self-attacking is dangerous and such characteristic must not be maintained genetically. It then follows that interaction with surroundings such as infection should be the cause of autoimmunity. However, despite extensive investigation no pathogens have been identified yet. We instead thought that, from the viewpoint of T cell stimulation, because pathogen is antigen-presented on HLA, causative pathogens might differ from person to person depending on HLA. We thus tested the robustness of host's immune response that responds against pathogen by repeatedly immunizing with an antigen, and found that if TCR stimulation with antigen plus HLA surpassed immune system's self-organized criticality, autoimmunity results in. Repeated infection with pathogen is indeed not exceptional or rather routine in life, wherein signs and symptoms of infection are often unapparent because cTL response is ignored or down in memory at re-infection (Nathan, 2010; Zinkernagel, 1996). The limitation of the present study is that, while repeated infection is clinically likely, we did not clarified all the pathway or manner of infection other than “repeated infection” that leads to overstimulation of immune system and generation of DOCK8+Tfh cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-B220-PE Ab | Biolegend | Cat#103208 RRID; AB_312993 |
| Anti-BST2-FITC Ab | Biolegend | Cat#127008 RRID; AB_2028462 |
| Anti-Bcl6-PE Ab | eBioscience Thermo Fisher Scientific | Cat#12-5760-80 RRID; AB_10852866 |
| Anti-CD3 Ab | Biolegend | Cat#100302 RRID; AB_312667 |
| Anti-CD3-PE/Cy7 Ab | Biolegend | Cat#100320 RRID; AB_312685 |
| Anti-CD4 Ab | Biolegend | Cat#100402 RRID; AB_312687 |
| Anti-human CD4-PE Cy7 Ab | Biolegend | Cat#300511 RRID; AB_314079 |
| Anti-mouse CD4-FITC Ab | Biolegend | Cat#100510 RRID; AB_312913 |
| Anti-CD4-PerCP/Cy5.5 Ab | Biolegend | Cat#100540 RRID; AB_893326 |
| Anti-CD5-FITC Ab | Biolegend | Cat#100605 RRID; AB_312734 |
| Anti-CD11c-APC Ab | BD Biosciences | Cat#550261 RRID; AB_398460 |
| Anti-CD16/32 Ab | Biolegend | Cat#101302 RRID; AB_312801 |
| Anti-CD21-FITC Ab | Biolegend | Cat#123407 RRID; AB_940403 |
| Anti-CD23-PE Ab | Biolegend | Cat#101607 RRID; AB_312832 |
| Anti-CD23-Alexa Fluor 647 Ab | Biolegend | Cat#101611 RRID; AB_493479 |
| Anti-CD25-PE Ab | Biolegend | Cat#10200 RRID; AB_312857 |
| Anti-CD27-FITC Ab | Biolegend | Cat#124207 RRID; AB_1236463 |
| Anti-CD28 mAb | Biolegend | Cat#102102 RRID; AB_312867 |
| Anti-CD43 activation-associated glycoform-APC Ab | Biolegend | Cat#121213 RRID; AB_528806 |
| Anti-CD44-FITC Ab | Biolegend | Cat#103005 RRID; AB_312956 |
| Anti-human CD45RB-Alexa Fluor 488 Ab | eBioscience Thermo Fisher Scientific | Cat#53-9458-82 RRID; AB_2574448 |
| Anti-CD62L-APC Ab | Biolegend | Cat#104411313098 RRID; AB_ |
| Anti-CD80-FITC Ab | Biolegend | Cat#104705 RRID; AB_313126 |
| Anti-CD83-FITC Ab | Biolegend | Cat#121505 RRID; AB_572008 |
| Anti-CD86-PE Ab | Biolegend | Cat#105008 RRID; AB_313157 |
| Anti-human CD122-APC Ab | Biolegend | Cat#339007 RRID; AB_2248891 |
| Anti-CD123-PE Ab | Biolegend | Cat#106005 RRID; AB_2124403 |
| Anti-CD138-PE Ab | Biolegend | Cat#142503 RRID; AB_10915989 |
| Anti-CD147-FITC Ab | Biolegend | Cat#123705 RRID; AB_1227513 |
| Anti-CCR4-PE/Cy7 Ab | Biolegend | Cat#131213 RRID; AB_2074506 |
| Anti-CXCR5-FITC Ab | Biolegend | Cat#145520 RRID; AB_25622866 |
| Anti-CXCR5-APC Ab | Biolegend | Cat#145506 RRID; AB_2561970 |
| Anti-CD19-PE/Cy7 Ab | Biolegend | Cat#115519 RRID; AB_313654 |
| Anti-DOCK8 rabbit Ab | Proteintech | Cat#11622-1-AP RRID; AB_10216360 |
| Au-goat anti-rabbit IgG H&L Ab | Abcam | Cat#ab27234 RRID; AB_954427 |
| Anti-fluorescein-ALP Ab | Amersham Pharmacia | Cat#RPN3690 RRID; AB_ |
| Anti-Foxp3-APC Ab | eBioscience | Cat#14-5773-80 RRID; AB_467575 |
| Anti-FR4-FITC Ab | Biolegend | Cat#125005 RRID; AB_1134204 |
| Anti-GATA3-Alexa Fluor 488 Ab | eBioscience | Cat#53-9966-41 RRID; AB_2574492 |
| Anti- GL7-FITC Ab | Biolegend | Cat#144603 RRID; AB_2561696 |
| Anti-GL7-PE Ab | Biolegend | Cat#144607 RRID; AB_2562295 |
| Anti-ICOS-FITC Ab | Biolegend | Cat#313505 RRID; AB_416329 |
| Anti-acetyl Histone H3 Ab | Millipore Sigma | Cat#06-599 RRID; AB_2115283 |
| Anti-trimethyl Histone H3 (Lys4) Ab | Millipore Sigma | Cat#07-473RRID; AB_1977252 |
| Anti-Histone H3 (trimethyl K4) Ab. ChIP grade | Abcam | Cat#ab8580 RRID; AB_306649 |
| Anti-rabbit IgG-PE Ab | Biolegend | Cat#406421 RRID; AB_2563484 |
| Anti-rabbit IgG-FITC Ab | Biolegend | Cat#406403 RRID; AB_893531 |
| Anti-rabbit IgG-HRP Ab | Cappel Laboratories | Cat#55641 |
| Anti-mouse IgG-Cy3 Ab | Biolegend | Cat#405309 RRID; AB_893530 |
| Anti-human IgG Fc-PE Ab | Biolegend | Cat#409304 RRID;AB_10895907 |
| Anti-IgM-FITC Ab | Biolegend | Cat#406505 RRID; AB_315055 |
| Anti-IFNγ rabbit Ab | Abcam | Cat#ab96575 RRID; AB_306649 |
| Anti-LFA1-APC Ab | Biolegend | Cat#141009 RRID; AB_2564305 |
| Anti-Ly6C-FITC Ab | BD Pharmingen | Cat#561085 RRID; AB_10584332 |
| Anti-PD-1-APC Ab | Biolegend | Cat#135210 RRID; AB_2159183 |
| Anti-RORγt-APC Ab | eBioscience Thermo Fisher Scientific | Cat#17-6981-82 RRID; AB_2573254 |
| Anti-Siglec H-PE Ab | Biolegend | Cat#129605 RRID; AB_1227763 |
| Anti-T-bet-Alexa Fluor 647 Ab | Biolegend | Cat#644803 RRID; AB_15955573 |
| Anti-Thy1.2-APC Ab | Biolegend | Cat#105311 RRID; AB_313182 |
| Anti-TCRβ-APC Ab | Biolegend | Cat#109211 RRID; AB_313434 |
| Anti-TLR7 Ab | BD Biosciences | Cat#565557 RRID; AB_2739295 |
| Anti-Vβ8 TCR-PE Ab | BD Biosciences | Cat#553862 RRID; AB_395098 |
| Chemicals, peptides, and recombinant proteins | ||
| Alpha-galactosylceramide (αGalCer) | Kirin Brewery Co. | Gift |
| Alkaline phosphatase avidin D | Vector Laboratories | Cat#A-2100 |
| Animal-free blocker | Vector Laboratories | Cat#SP5030 |
| AccuCheck counting beads | Life Technologies | Cat#PCB100 |
| Brefeldin A | Sigma-Aldrich | Cat#B7651 |
| Calf thyms DNA | Worthington Biochem | Cat#DNA |
| CFSE | Thermo Fisher Scientific | Cat#C34570 |
| CpG | Invitrogen | Cat#tlrl-2395 |
| Dynabeads Protein G | VERITAS | Cat#DB10003 |
| E.coli DNA polymerase | Invitrogen | Cat#18010017 |
| E.coli DNA ligase | Invitrogen | Cat#18052019 |
| Extraction buffer I & II | EMB Bioscience | Cat#539790 |
| Epon 812 | TAAB | Cat#T023 |
| FCS (fetal calf serum) | BioWest | Cat#S1820-500 |
| Fast red | Nichirei Bioscience | Cat#415261 |
| 16-pad FAST slides | EMD Milipore | Cat#Z721115-20EA |
| Histogreen | LINARIS | Cat#E109 |
| Hybond N+ membrane | GE Healthcare | Cat#RPN1210B |
| Ionomycin | Sigma-Aldrich | Cat#10634 |
| Immobilon-P membrane | Milipore | Cat#IPVH00010 |
| KAPA HiFi DNA polymerase | Kapa Biosystems | Cat#KK2101 |
| KLH (keyhole limpet hemocyanin) | Sigma-Aldrich | Cat#H7017 |
| Lead staining solution | Sigma-Aldrich | Cat#18-0875-2-25ML-J |
| LPS (lipopolysaccharide) | Sigma-Aldrich | Cat#L4516 |
| Anti-rabbit IgG MicroBeads | Miltenyi Biotech | Cat#130-048-602 |
| Mouse CD4 MicroBeads | Miltenyi Biotech | Cat#130-049-201 |
| Nonidet P-40 (NP-40) | MP Biochemicals | Cat#198595 |
| Nylon membrane | Roche Diagnostic | Cat#1 417 240 001 |
| Normal rabbit IgG | Invitrogen | Cat#10500C |
| Nonimmunized rabbit IgG | Cappel Laboratories | Cat#55944 |
| Ovalbumin (OVA) | Sigma-Aldrich | Cat#A5503 |
| OsO4 | Nisshin EM | Cat#300-1 |
| PMA (phorbol myristate acetate) | Sigma-Aldrich | Cat#P1585 |
| Protein G magnetic beads | Invitrogen | Cat#100-03D |
| Peanut agglutinin (PNA)-fluorescein | Vector Laboratories | Cat#FL-1071 |
| Polyacrylamide gel | Sigma-Aldrich | Cat#GF90890466 |
| Protease inhibitor cocktail | EMD Milipore | Cat#539790 |
| Proteinase K | TaKaRa Bio Inc. | Cat#9033 |
| RNase A | Qiagen | Cat#19101 |
| RNase H | Invitrogen | Cat#18021071 |
| Sph I restriction enzyme | TaKaRa Bio Inc. | Cat#1246 |
| Superscript III reverse transcriptase | Invitrogen | Cat#18081044 |
| Saponin | Sigma-Aldrich | Cat#S4521 |
| Sm antigen | ImmunoVision | Cat#SMA-3000 |
| SEB (staphylococcus enterotoxin B) | Toxin Technologies | Cat#BT202 |
| SEA (staphylococcus enterotoxin A) | Toxin Technologies | Cat#TA101 |
| T4 DNA polymerase | Invitrogen | Cat#18005025 |
| T4 DNA ligase | TaKaRa Bio Inc. | Cat#2011A |
| T4 DNA ligase | Roche Diagnostic | Cat#481220 |
| Tween 20 | FUJIFILM Wako | Cat#167-11515 |
| Uranyl acetate | EM Sciences | Cat#22400 |
| Critical commercial assays | ||
| Mouse IFNγ ELISA kit | Biolegend | Cat#430804 |
| Mouse Il-2 ELISA kit | Biolegend | Cat#431004 |
| Mouse IL-4 ELISA kit | Biolegend | Cat#431104 |
| Mouse IL-6 ELISA kit | Biolegend | Cat#431304 |
| Mouse IL-10 ELISA kit | Biolegend | Cat#431414 |
| Mouse IL-17 ELISA kit | Biolegend | Cat#432504 |
| Mouse IL21 ELISA kit | eBioscience Thermo Fisher Scientific | Cat#88-8210-22 RRID 2575209 |
| Mouse IL-22 ELISA kit | eBioscience Thermo Fisher Scientific | Cat#88-7422-22; RRID 2572119 |
| Mouse IL-23 ELISA kit | Biolegend | Cat#433707 |
| Mouse TGFβ ELISA kit | eBioscience Thermo Fisher Scientific | Cat#88-8350-22; RRID 2575209 |
| Mouse TNFα ELISA kit | eBioscience Thermo Fisher Scientific | Cat#88-7324-22; RRID 2575076 |
| Biotin labelling kit-HN2 | Dojindo Molecular Technologies | Cat#LK03 |
| Cell Trace CFSE cell proliferation kit | Thermo Fisher Scientific | Cat#C34554 |
| CDP-Star chemiluminiscence | Amersham Pharmacia | Cat#RPN3690 |
| Dynabeads M280-Tosylactivated | Thermo Fisher Scientific | Cat#14203 |
| Expi 293 system | Thermo Fisher Scientific | Cat#A14635 |
| Foxp3/transcription factor staining buffer set | eBioscience | Cat#00-5523-00 |
| MACS Beads mouse CD4 isolation kit | Miltenyi Biotech | Cat#130-104-454 |
| MinElute Reaction Cleanup kit | Qiagen | Cat#28206 |
| MaxiSorp immunotubes | Thermo Fisher Scientific | Cat#444474 |
| Nextera XT Index kit v2 | Illumine | Cat#FC-131-1002 |
| ProteoExtract Subcellular Proteome Extraction kit | EMD Milipore | Cat#539790 |
| Peroxidase detection system | Leica Biosystems | Cat#RE7110-k |
| RF ELISA kit | FUJIFILM Wako | Cat#AKRRG-101 |
| RNeasy Mini kit | Qiagen | Cat#74106 |
| SurePrint G3 mouse GE Microassay | Agilent Tchnology | Cat#8x60K ver2.0 |
| QIA quick PCR purification kit | Qiagen | Cat#28104 |
| Deposited data | ||
| Supplemental information; ShunichiShiozawa (2021), “TCRA and TCRB repertoire analysis Results”, Mendeley Data, V1, doi:https://doi.org/10.17632/7r5pjbz793.1 | This paper, Mendeley Data | https://data.mendeley.com//datasets/7r5pjbz793/2 |
| Gene expression microarray analysis Results of DOCK8+CD4 T cells, DOCK8-CD4 T cells of 12x OVA-immunized mice and CD4 T cells of 12x PBS-immunized mice | This paper, NCBI’s Gene Expression Omnibus | GEO Series accession number GSE159240. |
| Oligonucleotides | ||
| Primer and probe sequences used in CHIP assay | This paper | Table S3 |
Resource availability
Lead contact
Further requests on data and code should be directed to and will be fulfilled by the Lead Contact, Shunichi Shiozawa (shiozawa@port.kobe-u.ac.jp).
Materials availability
There are restriction to the availability of monoclonal anti-DOCK8 Ab due to our planning to furher refine them for clinical use. If strongly wished to use them, conditions such as MTA agreement are required.
Experimental model and subject details
Mice
BALB/c, B10.D2, C57BL/6 and NZB/W F1 female mice were obtained from Japan SLC, Inc., Hamamatsu, Japan. CD1d knockout (KO) female mice of BALB/c background (C.129S2-Cd1tm1Gru/J) were purchased from Jackson Laboratory (Bar Harbor, ME). DOCK8 -/- female mice of C57BL/6 background were provided by Dr. Fukui (Harada et al., 2012). All animal experiments were conducted in accordance with the guidelines of the Kyushu University Institution Review Board (Permission number A29-174-1) and the Review Board for Experimental Studies of the Institute for Rheumatic Diseases (Permission number A2017-001). All immunization study started at 7∼8 weeks old age.
Patients
Human blood samples were studied in accordance with the guidelines of the Kyushu University Institution Review Board for Human Genome/gene research (Protocol 711-00, approved on March 21, 2017). Patients with systemic lupus erythematosus (female/male:40/3, mean age±SD 43.4±16.7) met the revised American College of Rheumatology (ACR) classification criteria (Tan et al., 1982).
Method details
Animal studies
BALB/c female mice (8 weeks old) or C57BL/6 mice (8 weeks old) were immunized with 500μg OVA, 100μg KLH or PBS by means of i.p. injection every 5days for a total of 12 injections. Mice were analyzed 9 days after the final immunization. Mice were also immunized with 25μg SEB, 25μg SEA or PBS by means of i.p. injection every 5days for a total of 8 injections, and analyzed thereafter.
To isolate and characterize aiCD4 T cells, we first induced aiCD4 T cells and SLE in BALB/c mice by 12x repeated intraperitoneal (i.p.) immunization with OVA without adjuvants, and then, various subsets of splenic CD4 T cells, particularly those increased in 12x OVA-stimulated mice (Miyazaki et al., 2013), were taken and transferred to naive recipient mice. For each subset we assessed the generation of rheumatoid factor (RF) and anti-dsDNA Ab in the recipient mice, and in these fashion, the CD4 T cell subset containing aiCD4 T cells was focused (Miyazaki and Shiozawa, 2013).
For adoptive cell transfer into previously-immunized mice, BALB/c mice were first immunized 8x with OVA, and 24hr after the final immunization, mice were administered 200μg anti-CD4 Ab (GK1.5) to deplete CD4 T cells. Four days later, DOCK8+ CD4 T cells from the mice immunized 12x with OVA or the mice immunized 12x with KLH were transferred into the CD4 T-depleted mice. The recipient mice received single i.p. injection of 500μg OVA or 100μg KLH 24h after cell transfer and were analyzed 2 weeks later. For adoptive cell transfer study which tested the ability of SEB-stimulated aiCD4 T cells to induce lupus renal disease, BALB/c mice were first immunized 8x with OVA, and 24hr after the final immunization, mice were administered 200μg anti-CD4 Ab (GK1.5) to deplete CD4 T cells. Four days later, CD4 T cells from the mice immunized 8x with 25μg SEB were transferred into the CD4 T-depleted mice. The recipient mice received a single i.p. injection of 25μg SEB 24h after cell transfer and were analyzed 2 weeks later.
For the SEB immunization study, BALB/c or CD1d KO female mice (8-15 weeks old) were immunized with 25μg SEB, 25μg SEA, or 5μg αGC by means of i.p. injection every 5days for a total of 2 or 8 injections. Mice were bled 3 hr after 2nd and 8th immunization, and serum IL-2 and autoantibodies were measured by ELISA. To evaluate T cell division, cells (1x107/ml) were incubated with 10μM carboxyfluorescein diacetate succinimidyl ester (CFSE) in PBS for 10min and an equal volume of fetal calf serum was added to quench the reaction. After washing, CFSE-labeled cells (1x105/200μl/well) were stimulated with 2μg/ml of plate-bound anti-CD3 mAb and 5μg/ml of plate-bound anti-CD28 Ab for 72hr, and the number of mitotic events was measured by flow cytometry (FACSCalibur, Becton Dickinson, San Jose, CA).
For therapeutic Ab treatment, BALB/c mice immunized with OVA were treated with rabbit anti-DOCK8 Ab (100μg) or control rabbit IgG (100μg) 24 hr before each of the 6x, 8x, 10x and 12x OVA immunizations, and analyzed 2 weeks later. The (NZBxNZW) F1 female mice (8 weeks old) were treated either with anti-DOCK8 Ab (100μg) or control rabbit IgG (100μg) every 15 to 24 weeks, and blood and urine samples were analyzed weekly.
Mass spectrometry analysis
Cytosolic and membrane proteins were extracted from CD45RBhi CD122hi CD4 T cells, CD45RBlo CD122lo PD-1- CD4 T cells and CD45RBlo CD122lo PD-1+ CD4 T cells that had been harvested from spleens of mice immunized 12x with OVA, as well as control CD4 T cells derived from the spleen of mice treated 12x with PBS. One hundred million cells were reacted with Extraction Buffer I containing 1% protease inhibitor cocktail (ProteoExtract Subcellular Proteome Extraction kit) for 10 min at 4°C, and the supernatants containing cytosolic proteins were taken. The pellets were reacted with Extraction Buffer II containing 1% protease inhibitor cocktail (ProteoExtract Subcellular Proteome Extraction kit) for 30 min at 4°C, and subjected to centrifugation at 5,500 x g for 10 min, and the supernatants were taken as the membrane protein fraction. Ten μg total protein per sample was fractionated by electrophoresis in a 10% polyacrylamide gel, and the gel was stained with silver nitrate. Representative portions in the gel were cut out and digested with trypsin. Digested peptides were eluted with 0.1% formic acid and subjected to LC-MS/MS analysis, performed on a Q-Tof 2 quadrupole/time-of-flight hybrid mass spectrometer (Waters Co, Milford, MA) interfaced with a CapLC capillary reverse-phase LC system. The eluted peptides were sprayed directly into the mass spectrometer. MS/MS data were acquired by MassLynx software (Micromass) and converted into a single text file (containing the observed precursor peptide m/z, the fragment ion m/z and intensity value) by ProteinLynx software (Micromass). The file was analyzed using the Matrix Science Mascot MS/MS Ion Search software (http://www.matrixscience.com) to search and assign the obtained peptides to the NCBI non-redundant database. Proteins (10μg each) were again subjected to electrophoresis in 10% polyacrylamide gel, transferred to an Immobilon-P membrane and stained sequentially with specific Ab and labelled secondary Ab.
In vitro autoantibody and cytokine assay
Sera were assayed 9days after final priming by ELISA for RF, anti-dsDNA Ab (Worthington Biochemical Co., Lakewood, NJ), or anti-Sm Ab using Sm antigen (ImmunoVision, Springdale, AR). Isolated DOCK8+CD4 T cell CD4 T cells (1x106/ml) were stimulated with 2 μg/ml anti-CD3 Ab and 5 μg/ml anti-CD28 Ab at 37°C for 24hr, and the culture supernatants were assayed for cytokines by ELISA.
Flow cytometry and cell sorting
Lymphocyte surface markers of T and B cells of mice and human peripheral blood were assayed using anti-CD4-FITC Ab, anti-CD4-PerCP/Cy5.5 Ab or respective Abs after blocking non-specific FcR binding with anti-mouse CD16/32 Ab. For DOCK8 staining, lymphocytes were treated with rabbit anti-DOCK8 Ab followed by staining with anti-rabbit IgG-FITC Ab or anti-rabbit IgG-PE Ab. For intracellular staining, DOCK8-stained cells were fixed and permeabilized in fixation/permeabilization solution and permeabilization buffer from the Foxp3/Transcription Factor Staining Buffer Set. They were next stained with respective Abs against Foxp3, T-bet, GATA3 and RORγt. Cells were washed once, resuspended in PBS and analyzed on a FACSCallibur flow cytometer (BD Biosciences, San Jose, CA). To detect intracellular IFNγ, cells (1x106/ml) were stimulated with 50 ng/ml phorbol myristate acetate and 500 ng/ml ionomycin in the presence of 10 μg/ml brefeldin A. After 4 h, cells were stained with anti-CD8 Ab, followed by fixation with 2% formaldehyde, permeabilization with 0.5% saponin and stained for IFNγ.
Antibody dilutions used were 1:5,000 for surface staining with PNA-fluorescein, 1:1,000 for surface staining with anti-GL7-PE Ab, 1:200 for surface staining with anti-CD4-FITC Ab or anti-CD4-PerCP/Cy5.5 Ab, and 1:100 for the staining with the other Abs.
For cell isolation, CD4 T cells, DOCK8+ T cells were isolated from spleens of mice to >90% purity using Ab-coated MACS beads. Cell numbers were determined using AccuCheck Counting Beads according to manufacturer's instructions.
Pathological studies
Spleens from patients with SLE and controls obtained at autopsy were fixed in 10% formalin and embedded in paraffin. Paraffin sections of 4μm thickness were deparaffinized by graded xylene and graded alcohol sequentially, treated with 0.01M citric acid in an autoclave at 121°C for 10min, and blocked with Animal-free Blocker for 10min. Specimens were then treated with biotinylated anti-DOCK8 monoclonal Ab diluted at 1:200 and rabbit anti-interferonγ Ab (for T cells) diluted at 1:1,000 at 4°C for 24hr. After blocking endogenous peroxidase with hydrogen peroxide RT for 15min, the specimens were reacted serially with ALP-avidin D and anti-rabbit Novolink RT for 30min, followed by Fast red and Histogreen staining RT for 40min and 2min, respectively. After washing, the specimens were air dried, embedded, and observed under microscope.
For immunoelectron microscopic studies, CD4 T cells were isolated from the spleens of 12x OVA-immunized mice to >90% purity using Ab-coated MACS beads, pelleted by centrifugation, washed, and stained with rabbit anti-DOCK8 Ab followed by Au-goat anti-rabbit IgG Ab RT for 4hr. They were then washed with PBS, fixed in 1% OsO4 for 2 hr at 4°C, and then dehydrated in graded alcohol and embedded in Epon 812. Sections 90 nm thick were cut using an LKB ultramicrotome, counter-stained with uranyl acetate and lead, and observed under a Hitachi H-7600 electron microscope for Au-stained cells. Renal Pathology was classified according to SLE human renal disease classification (Weening et al., 2004).
RT-PCR for RAG, TdT and pTα
Total RNA was reversely transcribed to cDNA and amplified by PCR. The products were fractionated by electrophoresis and transferred to nylon membranes. The membranes were hybridized to fluorescein end-labelled probes and visualized by alkaline phosphatase ALP-anti-fluorescein Ab and Gene Images CDP-Star chemiluminescence reaction. The primers and probes were: 5′-CCAAGCTGCAGACATTCTAGCACTC-3′ (forward), 5′-CAACATCTGCCTTCACGTCGATCC-3′ (reverse) and 5′-AACATGGCTGCCTCCTTGCCGTCTACCCT-3′ (probe) for RAG1 (Huang et al., 2002); 5′-CACATCCACAAGCAGGAAGTACAC-3′ (forward), 5′-GGTTCAGGGACATCTCCTACTAAG-3′ (reverse) and 5′-GCAATCTTCTCTAAAGATTCCTGCTACCT-3′ (probe) for RAG2 (Huang et al., 2002); 5′-GAACAACTCGAAGAGCCTTCC-3′ (forward), 5′-CAAGGGCATCCGTGAATAGTTG-3′ (reverse) and 5′-ATTCGGTCACCCACATTGTGGCAGAGAAC-3′ (probe) for TdT; 5′-CAACTGGGTCATGCTTCTCC-3′ (forward), 5′-TGGCTGTCGAAGATTCCC-3′ (reverse) and 5′-CCGTCTCTGGCTCCACCCATCACACTGCT-3′ (probe) for pTα.
Next-generation sequencing of TCR genes
One microgram total RNA from DOCK8+CD4 T cells, DOCK8-CD4 T cells, and CD4 T cells were each converted into complementary DNA (cDNA) with Superscript III reverse transcriptase. BSL-18E primer containing polyT18 (5′-AAAGCGGCCGCATGCTTTTTTTTTTTTTTTTTTVN-3′) and anSphI site was used for cDNA synthesis. Double stranded (ds)-cDNA was synthesized using E.coli DNA polymerase I, E.coli DNA ligase and RNase H. The ds-cDNA was then blunted with T4 DNA polymerase (Invitrogen). P10EA/P20EA adapter (5′-GGGAATTCGG-3′/5′-TAATACGACTCCGAATTCCC-3′) was ligated to the 5′end of the ds-cDNA followed by digestion with SphI restriction enzyme (Kitaura et al., 2016). After removing adaptor and primer by MinElute Reaction Cleanup kit, PCR was performed using either specific for TCRα–chain constant region (mCA1; 5′-TCATGTCCAGCACAGTTTTG-3′) or TCRβ–chain constant region (mCB1; 5′-AGGATTGTGCCAGAAGGTAG-3′) and P20EA adaptor using KAPA HiFi DNA Polymerase. Next, a second PCR was performed using the P20EA and 2nd PCR primers for TCRα (mCA2; 5′-GTTTTCGGCACATTGATTTG-3′) or TCRβ (mCB2; 5′-TTGTAGGCCTGAGGGTCC-3′). Amplicons were subsequently prepared by amplifying the second PCR products using P22EA-ST1-R Tag primers (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTAATACGACTCCGAATTCCC-3′) and mCA-ST1-R Tag primer for TCRα (5′-TCGTCGGCAGCGTCAGAT GTGTATAAGAGACAGGTGGTACACAGCAGGTTCT-3′) or mCB-ST1-R Tag primer for TCRβ (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTTGGGTGGAGTCACATTT-3′). After PCR amplification, index (barcode) sequences were added by amplification with the Nextera XT Index kit v2 setA or setD. The indexed amplicon products were mixed in equal molar concentrations and quantified by a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA). Sequencing was performed using the Illumina Miseq paired-end platform (2x300bp). All the paired-end reads were classified by index sequences. All sequence reads were classified by MID Tag sequences. Artificially added sequences and sequences with low quality scores were removed from both terminals of sequence reads using software installed on the 454 sequencing system (Kitaura et al., 2016). The remaining sequences were used for assignment of TRAV and TRAJ for TCRα sequences, and TRBV and TRBJ for TCRβ sequences. Sequences were assigned by determining highest identity in a data set of reference sequences for all TRAV, TRAJ, TRBV and TRBJ genes including pseudogenes and open reading frame (ORF) reference sequences available from the international ImMunoGeneTics information system® (IMGT) database (http://www.imgt.org). Data processing, assignment, and data aggregation were automatically performed using repertoire analysis software RG (Repertoire Genesis Inc., Osaka, Japan), which implemented a program for sequence homology searches using BLATN, an automatic aggregation program, a graphics program for gene usage, and CDR3 length distribution (Bolstad et al., 2003). Sequence identities at the nucleotide level between query and entry sequences were automatically calculated by carefully optimizing for respective repertoires for parameters that increased sensitivity and accuracy; i.e. E-value threshold, minimum kernel, and high-scoring segment pair (HSP) score. CDR3 sequences were defined as the sequences ranging from a conserved Cysteine at position 104 (Cys104) in the IMGT nomenclature to a conserved Phenylalanine (Phe118) or Tryptophan (Trp118) at position 118, and these, together with the next Glycine (Gly119), were translated into the deduced amino acid sequences (Kitaura et al., 2017). A unique sequence read (USR) was defined as a sequence read having no identity with other sequence reads in TRV, TRJ and deduced CDR3 amino acid sequences. The copy number of identical USR was automatically counted by the RG software in each samples and ranked by copy number. Percentage frequencies of sequence reads within TRAV, TRAJ, TRBV and TRBJ genes to total sequence reads were calculated. To estimate TCR diversity in deep sequencing data, several diversity indices were used, including Shannon-Weaver index H’ and inverted Simpson index 1/λ. U, unique read and copy numbers were used for species and individuals in these equations (Shannon, 1948; Simpson, 1949).
Gene expression microarray assay
Total RNA was extracted with the RNeasy Mini Kit, and total RNA was quantified by absorbance at 260 nm using a Nano Drop 2000 spectrophotometer (Thermo Fisher Scientific, MA. USA). RNA samples were quantified by an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and the quality was confirmed with the Experion System (Bio-Rad Laboratories, Hercules, CA). The mRNA expression profile was determined using a SurePrint G3 mouse GE Microarray, and assay was performed by Cell Innovator Inc. (Fukuoka, Japan). Relative hybridization intensities and background hybridization values were calculated using Agilent Feature Extraction Software (9.5.1.1). Raw signal intensities and Flags of two samples were calculated from hybridization intensities using gProcessedSignal and spot information using gIsSaturated module in Bioconductor software (Gentleman et al., 2004). The data was log2-transformed and normalized by quantile algorithm with ‘preprocessCore’ library package (Bolstad et al., 2003). Probes named ‘P’ flag were set at least in one sample, and Z-scores (Quackenbush, 2002) and ratios (non-log scaled fold-change) from normalized signal intensities of each probe for comparison between control and experiment sample were calculated. The genes for which the Z score and the ratio were above 2.0 and 1.5, respectively, were considered up-regulated. In contrast, the genes for which the Z scores and ratios were below −2.0 and 0.66, respectively, were considered down-regulated. Data Analysis consisted of Flag criteria on GeneSpring Software which classified the results as A: absent, indicating that the feature was not-positive and significant and that the feature was not above background; M: marginal, indicating that the feature was not uniform, saturated and a population outlier; P: present, indicating values other than A or M.
Autoantibody microarray assay
Antigens diluted to optimal concentration were spotted in duplicate and random fashion using Microgrid II micro-arrayer onto Nitrocellulose-coated 16-pad FASTTM slides. A 16-section frame was applied to each slide to separate individual assays. After washing with blocking PBS buffer containing 1% bovine serum albumin and Tween 20, sera diluted 1:100 with blocking buffer were added, and incubated with Cy3-anti-mouse IgG Ab. The arrays were washed with buffer, and the frames were removed. The slides were immersed in PBS and then spun dry. A Genepix 4000B scanner with 532nm laser wavelength was used to generate Tif images for analysis.
Chromatin immunoprecipitation (CHIP) assay
Forty million cells were suspended in 1ml RPMI, to which 0.1ml of 11% formaldehyde was added and the tube rocked RT for 5 min. Next, 0.1ml of 1.5M glycine was added to terminate crosslinking. Cells were pelleted (3K x g, 5 min, 4°C), and dissolved in 1ml FACS solution (PBS containing 2%FBS and 0.05% NaN3) and washed. After the second wash, cells were suspended in 0.2ml SDS lysis buffer (50mM Tris-HCl, pH 8.0, 10mM EDTA, and 1%SDS) containing 1% protease inhibitor cocktail on ice for 10 min. Chromosomal DNA was sheared using a Bioruptor UCD-200 on ice for 10 min. After centrifugation at 14K x g, 4°C for 10min, the supernatant was transferred to a 2ml siliconized tube. The chromatin sample was stored by addition of 0.1vol 50% glycerol at −80°C. Fifty μl Dynabeads Protein G were washed twice with Blocking solution (PBS containing 0.5% BSA), dissolved in 0.5ml of the same, and loaded with 5μg of anti-acetyl histone H3 (acetylated H3K9, K14) Ab, anti-trimethyl-Histone H3 Ab (Lys4), or normal rabbit IgG. After loading, the pellets were washed twice with Blocking solution. For ChIP analyses, Dynabeads Protein G were diluted with 1.8ml ChIP dilution buffer at 4°C. Chromatin derived from 2 million cells was used for each ChIP experiment with each antibody. The chromatin samples were then added to the rabbit Ig-loaded beads and rocked at 4°C for 6hr. Beads were spun down at 15K x g, 4°C for 2min, and 2ml supernatants were divided into 2 tubes, from which 980μl was transferred to new tubes containing a second batch of rabbit Ig-loaded beads, and rocked 1 hr at 4°C. These were next washed 5x with RIPA buffer (50mM HEPES-KOH, pH7.6, 500mM LiCl, 1mM EDTA, pH8.0, 1% NP-40, and 0.7% sodium deoxycholate) and once with TE buffer (10mM Tris-HCl, pH8.0, 1mM EDTA, and 50mM NaCl). Twenty μl was used as 1% Input. Beads were reacted with 210μl of ChIP direct elution buffer (10mM Tris-HCl, 300mM NaCl, 5mM EDTA, and 0.5% SDS) at 65°C for 30min with rotation. After centrifugation at 15K x g, RT for 1min, 200μl of supernatant was incubated at 65°C. The 2%input chromatin was treated identically. Two hundred μl of the supernatant was mixed with RNase A (final 0.2mglml), rotated, spun down and incubated at 37°C for 30min. Subsequently, proteinase K (final 0.2mg/ml) was added, the sample rotated, spun down and incubated at 55°C for 1hr. DNA was then extracted using the QIA quick PCR purification kit, and reacted with TCR locus primers (Table S3) by PCR at 94°C for 10min, followed by PCR amplification at 94°C 30s, 58°C 1min, and 72°C 1min for 35 cycles, and incubation at 72°C for 10min. PCR products (10μl) were applied to a 2% agarose gel. Input chromatin samples were diluted so that IP and input samples gave approximately equal qPCR signals if 2% of the region of interest were present in the IP sample.
Ligation-mediated PCR(LM-PCR)
Genomic DNA (1 μg) was ligated to 20 pmols BW linker, which was made by annealing the oligonucleotides BW-1 and BW-2, in 50μL of reaction buffer containing 66mM Tris-HCl (pH7.5), 5mM MgCl2, 5mM DTT, 1mM ATP and 2.5 U of T4 DNA ligase for 16 hours at 16°C. Ligated DNA was diluted with an equal volume of a buffer containing 10 mM Tris (pH8.3), 50 mM KCl, 0.5% Nonidet P-40 and 0.5% Tween20 and heated for 15 min at 95°C. Ligated DNA (100 ng) was amplified for 12 cycles with the locus-specific primer for first PCR and linker-specific primer BW-1H. For nested LM-PCR reactions, 1μL of the products was then amplified for another 27 cycles with the locus-specific primer for nested PCR and BW-1H. The LM-PCR products were fractionated by electrophoresis and transferred to Hybond N+ membranes, Buckinghamshire, England). The membranes were hybridized to alkaline phosphatase (ALP)-labeled probes and visualized by Gene Images CDP-Star chemiluminescence reaction. The sequences of the linkers and linker-specific primers were; 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ for BW-1 linker, 5′-GAATTCAGATC -3′ for BW-2 linker and 5′-CCGGGAGATCTGAATTCCAC -3′ for BW-1H primer. The sequences of the locus-specific primers and probes were as follows: 5′-TTACAAGAACTCCCTCTGCCTCTC-3′ for first PCR, 5′-AAGACACACTGGCTAGAGTTCTGGT-3′ for nested PCR and 5′AGATGCCTAGGCTTCTGTAAAGGTGTCACCTGCAGTGGAATTCAG-3′ (probe) for signal end (SE) of Jα2 (Jα2 SE); 5′-AGGAGATGCAAGCAAAGGAGTT-3′ for first PCR, 5′-AGGCAGAATTCGACTTGAGTAAG G-3′ for nested PCR and 5′-ACCTGTGGGTGTTTTTGACTGACTAAGAAACACTGTGGAATTCAG-3′ (probe) for Jα12 SE; 5′-GAACACAGAAGAAGGCCCAATATC-3′ for first PCR, 5′-AAAAGGAGATCAGAGAGGGGTGTT-3′ for nested PCR and 5′-CTGGGGACGCCATTTTGTAGACGTGTTTGTCACAGTGGAATTCAG-3′ (probe) for Jα24 SE; 5′-GCCTCTGTGGTCTAGTGTTTCTCA-3′ for first PCR, 5′-TGAGAACTTTACCCAGGAGGAAGA-3′ for nested PCR and 5′-CTCTGAGGAAGTTTTTGTTGTAGAGTCAGCCACTGTGGAATTCAG-3′ (probe) for Jα35; 5′-GTCAAAGCACCCAGAATAAAGAGG-3′ for first PCR, 5′-CCTGCACTATCACTGACTGTTTCC-3′ for nested PCR and 5′-CCTTACCTTGGTTTATGTAGAGACACAGAACACTGTGGAATTCAG-3′ (probe) for Jα40 SE; 5′-CAATGGAGGGAAACAACACTAGC-3′ for first PCR, 5′-AGAAGCTAGGGAATCAGGGAGAAT-3′ for nested PCR and 5′-GATGCCACGAGTTTTTGCAAAGCCCTTCAGTGCAGTGGAATTCAG-3′ (probe) for Jα58 SE.
Anti-DOCK8 monoclonal Ab
Recombinant GST/His-tagged mouse DOCK8 encompassing the DHR1 domain (N-terminal 561-730 amino acids; Uniprot accession number Q8C147), was expressed in E. coli. After capturing on glutathione sepharose column, His-tagged mouse DOCK8 was eluted by protease-digestion, and fractionated with size-exclusion chromatography. Using this purified antigen, anti-DOCK8 antibodies were generated from non-immune phage antibody libraries developed in KAN Research Institute. Purified His-tagged mouse DOCK8 was immobilized on Dynabeads M280-Tosylactivated, and monoclonal single chain Fvs (scFvs) were selected by 3x repeated reaction with the beads, followed by interaction with His-tagged mouse DOCK8 coated onto MaxiSorp immunotubes. Monoclonal scFvs picked from the 3rd and 4th rounds of selection were expressed as bacterial supernatants. Positive scFvs were identified by ELISA reactive against His-tagged mouse DOCK8. ELISA-positive scFvs obtained were sequenced and genetically converted to human IgG format. Anti-DOCK8 mAbs were expressed in the Expi293 system and purified by Protein A resin. The anti-DOCK8 IgG mAbs thus obtained were validated by size-exclusion chromatography, ELISA reactive against His-tagged mouse DOCK8561-730, and intracellular staining of mouse CD4+ T cells by flow-cytometry. Finally, mAb generated were used for staining DOCK8+CD4 T cells from SLE patients and control individuals. DOCK8+CD4 T cells in the peripheral blood were stained with anti-DOCK8 mAb and anti-human IgGFc Ab, and DOCK8+CD4 T cell fraction within the each patient's CD4 T cells were compared with individual SLE disease activity indices (SLEDAI) (Bombardier et al., 1992).
Quantification and statistical analysis
Fisher's exact test was used for pathology results, and Student's t-test was used for experimental results.
Acknowledgments
We thank Prof. Yoshinori Fukui, Medical Institute of Bioregulation, Kyushu University for kindly providing DOCK8 −/− mice, Prof. Nancy J. Olsen, Penn State M.S. Hershey Medical Center, for kindly arranging autoantibody microarray, Prof. Takashi Sawai, Sendai Open Hospital, and Dr. Hiroshi Ishii, Oita Red Cross Hospital, for patients' samples under written consent, Dr. Marc Lamphier for English edition, and Mitsufumi Uemura, Institute for Rheumatic Disease and Aperat, for graphic art-works.
This study was supported by the grant of the Institute for Rheumatic Diseases, and the grant-in-aid 25515003, 17659301, 13204059, 11557026, 12204074, 13204059, and the Global Center of Excellence (COE) Program grant to S.S. and 18K06933 to K.T. of the Ministry of Education, Culture, Sports, Science and Technology of Japan; the New Industry Research Organization grant 0003 to S.S.; the grant of the Contract Development Program of Japan Science and Technology Organization to S.S.
Author contributions
Conceptualization, S.S.; Methodology, S.S.; Investigation, S.S., K.T., Y.M., K.U., K.S., T.N., H.M., A.D., M.T., M.I., M.K., Y.F., C.S., Q-Z.L., K.M., Y.M., T.E., S.T., Y.H., and M.M.; Resources, S.S., T.H., T.M., M.O., T.Y., H.K., K.M., and K.S.; Writing, S.S.; Project Administration, S.S.; Funding Acquisition, S.S. and K.T.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103537.
Supplemental information
Data and code availability
Supplemental information; Next-generation sequencing (NGS) data of TCRA and TCRB repertoire analysis in splenic DOCK8+ CD4 T cells of x12 OVA-immunized BALB/c mice, the splenic DOCK8- CD4 T cells of x12 OVA-immunized mice, and the CD4 T cells of x12 PBS-immunized mice were deposited in online at https://data.mendeley.com//datasets/7r5pjbz793/21 and are publicly available as of the date of publication.
The data on gene expression microarray data which compared among splenic DOCK8+ CD4 T cells of x12 OVA-immunized BALB/c mice, the splenic DOCK8- CD4 T cells of x12 OVA-immunized mice, and the CD4 T cells of x12 PBS-immunized mice, were deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE159240 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159240).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abarrategui I., Krangel M.S. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J. 2007;26:4380–4390. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama C., Tsumiyama K., Uchimura C., Honda E., Miyazaki Y., Sakurai K., Miura Y., Hashiramoto A., Shiozawa S. Conditional upregulation of IFN-α alone is sufficient to induce systemic lupus erythematosus. J. Immunol. 2019;203:835–843. doi: 10.4049/jimmunol.1801617. [DOI] [PubMed] [Google Scholar]
- Andrews B.S., Eisenberg R.A., Theofilopoulos A.N., Izui S., Wilson C.B., McConahey P.J., Murphy E.D., Roths J.B., Dixon F.J. Spontaneous murine lupus-like syndromes.Clinical and immunopathological manifestations in several strains. J. Exp. Med. 1978;148:1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon T.I., Horton R.M., Grigorova L.L., Cyster J.G. Visualization of splenic marginal zone B-cell suttling and follicular B-cell egress. Nature. 2013;493:684–690. doi: 10.1038/nature11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R., Gonzalez-Quintial R., Schreiber R.D., Lawson B.R., Kono D.H., Theofilopoulos A.N. Anti-IFN-α/β receptor antibody treatment ameliorates disease in lupus-predisposed mice. J. Immunol. 2012;189:5976–5984. doi: 10.4049/jimmunol.1201477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist I., Eriksson M., Saarikettu J., Eriksson B., Corneliussen B., Grundstrom T., Holmberg D. The basic helix-loop-helix transcription factor E2-2 is involved in T lymphocyte development. Eur. J. Immunol. 2000;30:2857–2863. doi: 10.1002/1521-4141(200010)30:10<2857::AID-IMMU2857>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irrizary R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bombardier C., Gladman D.D., Urowitz M.B., Caron D., Chang C.H. Derivation of the SLEDAI. A disease activity index for lupus patients.The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- Breitfeld D., Ohl L., Kremmer E., Ellwart J., Sallusto F., Lipp M., Folster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carico Z.M., Choudhury K.R., Zhang B., Zhuang Y., Krangel M.S. Tcrd rearrangement redirects a processive TCRA recombination program to expand the Tcrα repertoire. Cell Rep. 2017;19:2157–2173. doi: 10.1016/j.celrep.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasset F., Arnaud L. Targeting interferons and their pathways in systemic lupus erythematosus. Autoimmun. Rev. 2018;17:44–52. doi: 10.1016/j.autrev.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Chauveau S., Pirgova G., Cheng H.-W., De Martin A., Zhou F.Y., Wideman S., Rittscher J., Ludewig B., Amon T.I. Visualization of T cell migration in the spleen reveals a network of perivascular pathways that guide entry into T zones. Immunity. 2020;52:794–807. doi: 10.1016/j.immuni.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Luperchio T.R., Wong X., Doan E.B., Byrd A.T., Choudhury K.R., Reddy K.L., Krangel M.S. A lamina-associated domain border governs nuclear lamina interactions, transcription, and recombination of the Tcrb locus. Cell Res. 2018;25:1729–1740. doi: 10.1016/j.celrep.2018.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen U. Pathogen infection and autoimmune disease. Clin.Exp.Immunol. 2018;195:10–14. doi: 10.1111/cei.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweler C., den Hollander N.S., Kant S.G., et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J.-F., Motoyama A.B., Bush J.A., Vuori K. A novel and evolutionarily conserved PtdIns (3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A., Angelosanto J.M., Kao C., DOering T.A., Odorizzi P.M., Barnett B.E., Wherry E.J. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dameshek W., Witebsky E., Milgrom F. Autoimmunity: experimental and clinical aspects. Ann. NY Acad. Sci. 1965;124:1–980. [Google Scholar]
- Damian R.T. Molecular mimicry: antigen sharing parasite and host and its consequences. Am. Nat. 1964;98:129–149. [Google Scholar]
- Dao L.T.M., Gallindo-Albarran A.O., Castro-Mondragon J.A., Andrieu-Soler C., Medina-Rivera A., Souaid C., Charbonnier G., Griffon A., Vanhille L., Stephen T., et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 2017;49:1073–1081. doi: 10.1038/ng.3884. [DOI] [PubMed] [Google Scholar]
- Dao L.T.M., Spicuglia S. Transcriptional regulation by promoters with enhancer function. Transcription. 2018;5:307–314. doi: 10.1080/21541264.2018.1486150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders M., Franken L., Philipp M.-S., Kessler N., Baumgart A.-K., Eichler M., Wiertz E.J.H., Garbi N., Kurts C. Splenic red pulp macrophages cross-prime early effector CTL that provide rapid defense against viral infections. J. Immunol. 2020;204:87–100. doi: 10.4049/jimmunol.1900021. [DOI] [PubMed] [Google Scholar]
- Fairhurst A.M., Mathian A., Connolly J.E., Wang A., Gray H.F., George T.A., Boudreaux C.D., Zhou X.J., Li Q.Z., et al. Systemic IFN-alpha drives kidney nephritis in B6.Sle123 mice. Eur. J. Immunol. 2008;38:1948–1960. doi: 10.1002/eji.200837925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinami R.S., Oldstone M.B., Wroblewska Z., Frankel M.E., Koprowski H. Molecular mimicry in viral infection: cross-reaction of measles virus phosphoprotein or of herpes simplex virus protein with human intermediate filaments. Proc. Natl. Acad. Sci. U S A. 1983;80:2346–2350. doi: 10.1073/pnas.80.8.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furie R., Khamashta M., Merrill J.T., Werth V.P., Kalunian K., Brohawn P., Illei G.G., Drappa J., Wang L., Yoo S., CD1013 Study Investigators Anifrolumab, an anti-interferon-α receptor monoclonal antibody, in moderate-to-severe systemic lupus erythematosus. Arthritis Rheumatol. 2017;69:376–386. doi: 10.1002/art.39962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.C., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H., Huynh W., Schoon R.A., Vale R.D., Billadeau D.D. HkRP3 is a microbubule-binding protein regulating lytic granule clustering and NK cell killing. J. Immunol. 2015;194:3984–3996. doi: 10.4049/jimmunol.1402897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y., Tanaka Y., Terasawa M., Pieczyk M., Habiro K., Katakai T., Hanawa-Suetsugu K., Kukimoto-Niino M., Nishizaki T., Shirouzu M., et al. DOCK8 is a Cdc42 activator critical for intestinal dendritic cell migration during immune responses. Blood. 2012;119:4451–4461. doi: 10.1182/blood-2012-01-407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Tane A., Saito T. Dynamic regulation of TCR-microclusters and the microsynapse for T cell activation. Front. Immunol. 2016;7:255. doi: 10.3389/fimmu.2016.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon L.E., Deets K.A., Friesen T.J., Sze K.-Y., Fink P.J. Receptor revision in CD4 T cells is influenced by follicular helper T cell formation and germinal center interactions. Proc. Natl. Acad. Sci. U S A. 2014;111:5652–5657. doi: 10.1073/pnas.1321803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho I.C., Vorhees P.N., Martin N., Oakley B.K., Tsai S.-F., Orkin S.H., Leiden M. Human GATA3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor α gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks J.J., Moutsopoulos H.M., Gais S.A., Stahl N.I., Decker J.L., Notkins A.L. Immune interferon in the circulation of patients with autoimmune disease. N. Engl. J. Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- Hron J.D., Peng S.L. Type I IFN protects against murine lupus. J. Immunol. 2004;173:2134–2142. doi: 10.4049/jimmunol.173.3.2134. [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Golub R., Wu G.E., Kanagawa O. Superantigen-induced TCRα locus secondary rearrangement: role in tolerance induction. J. Immunol. 2002;168:3259–3265. doi: 10.4049/jimmunol.168.7.3259. [DOI] [PubMed] [Google Scholar]
- Ippolito G.C., Dekker J.D., Wang Y.-H., Lee B.-K., Shaffer A.L., III, Lin J., Wall J.K., Lee B.-S., Staudt L.M., Liu Y.-J., et al. Dendritic cell fate is determined by BCL11A. Proc. Natl. Acad. Sci. U S A. 2014;111:E998–E1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S.S., Latner D.R., Zilliox M.J., McCausland M., Akondy R.S., Penaloza-Master P., Hale J.S., Ye L., Mahammed A.-U.-R., Yamaguchi T.,S., et al. Identification of novel markers for mouse CD4+ T follicular helper cells. Eur. J. Immunol. 2013;43:3219–3232. doi: 10.1002/eji.201343469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Hartert K., Tadros S., Fiskus W., Havranek O., Ma M.C.J., Bouska A., Heavican T., Kumar D., Deng Q., et al. Targetable genetic alterations of TCF4 (E2-2) drive immunoglobulin expression in diffuse large B-cell lymphoma. Sci. Transl. Med. 2019;11:eaav5599. doi: 10.1126/scitranslmed.aav5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen E., Tohme M., Hedayat M., Leick M., Kumari S., Ramesh N., Massaad M.J., Ullas J.S., Azcutla V., Goodnow C.C., et al. A DOCK8-WIP-WASp complex links T cell receptors to the actin cytoskeleton. J. Clin. Invest. 2016;126:3837–3851. doi: 10.1172/JCI85774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Resch W., Corbett E., Yamane A., Casellas R., Schatz D.G. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser I.H. The specificity of periarterial fibrosis of the spleen in disseminated lupus erythematosus. Bull. Johns Hopkins Hosp. 1942;71:31–43. [Google Scholar]
- Kalunian K.C., Merrill J.T., Maciuca R., McBride J.M., Townsend M.J., Wei X., Davis J.C., Jr., Kennedy W.P. A phase II study of the efficacy and safety of rotalizumab (rhuMAb interferon-α) in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2016;75:196–202. doi: 10.1136/annrheumdis-2014-206090. [DOI] [PubMed] [Google Scholar]
- Keles S., Jabara H.H., Reisli I., McDonald D.R., Barlan I., Hanna-Wakim R., Dbaibo G., Lefranc G. Plasmacytoid dendritic cell depletion in DOCK8 deficiency: rescue of severe herpetic infections with IFN-α 2b therapy. J. Allergy Clin. Immunol. 2014;133:1753–1755. doi: 10.1016/j.jaci.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamashta M., Merrill T.T., Werth V.P., Furie R., Kalunian K., Illei G.G., Drappa J., Wang L., Greth W., CD1067 Study Investigators Sifaimumab, an anti-interferon-α monoclonal antibody, in moderate to severe systemic lupus erythematosus: a randomised, double-blind, placebo-controlled study. Ann. Rheum. Dis. 2016;75:1909–1916. doi: 10.1136/annrheumdis-2015-208562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaura K., Shini T., Matsutani T., Suzuki R. A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) and repertoires and identifying potential new invariant TCR chains. BMC Immunol. 2016;17:38. doi: 10.1186/s12865-016-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaura K., Yamashita H., Ayabe H., Shini T., Matsutani T., Suzuki R. Different somatic hypermutation levels among antibody subclasses sequencing-based antibody repertoire analysis. Front. Immunol. 2017;8:389. doi: 10.3389/fimmu.2017.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D., Yu Y., Dose M., Emmanouilidou A., Konstantinou T., Germer K., Aghajani K., Guo Z., Mandal M., Gounari F. β−catenin/Tcf determines the outcome of thymic selection in response to αβTCR signaling. J. Immunol. 2009;183:3873–3884. doi: 10.4049/jimmunol.0901369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzli M., Schreiner T.C., Pereboom N., Swarnalekha L.C., Litzler J., Lotscher Y.I., Ertuna J., Roux F., Geier R.P., Jakob T., et al. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci.Immunol. 2020;5:eaay5552. doi: 10.1126/sciimmunol.aay5552. [DOI] [PubMed] [Google Scholar]
- La Muraglia G.M., II, Wagener M.E., Ford M.L., Badell I.R. Circulating T follicular helper cells are a biomarker of humoral alloreactivity and predict donor-specific antibody formation after transplantation. Am. J. Transplant. 2020;20:75–87. doi: 10.1111/ajt.15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantelme E., Palermo B., Granziero L., Mantovani S., Campanelli R., Monafo V., Lazavecchia A., Giachino C. Recombinase-activating gene expression and V(D)J recombination in CD4+CD3low mature T lymphocytes. J. Immunol. 2000;164:3455–3459. doi: 10.4049/jimmunol.164.7.3455. [DOI] [PubMed] [Google Scholar]
- Lee B.-S., Lee B.-K., Lyer V.R., Sleckman B.P., Shaffer A.L., III, Ippolito G.C., Tucker H.O., Dekker J.D. Corrected and republished from: BCL11A is a critical component of a transcriptional network that activates RAG expression and V(D)J recombination. Mol. Cell Biol. 2018;38 doi: 10.1128/MCB.00362-17. e00362–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.Z., Zhou J., Wandstrat A.E., Carr-Johnson F., Branch V., Karp D.R., Mohan C., Li J., Liu Y., Xie C., et al. Deficiency of type I interferon contributes to Sle2-associated component lupus phenotypes. Arthritis Rheum. 2005;52:3063–3072. doi: 10.1002/art.21307. [DOI] [PubMed] [Google Scholar]
- Linterman M.A., Rigby R.J., Wong R.K., Yu D., Brink R., Cannons J.L., Schwartzberg P.L., Cook M.C., Walters G.D., Vinuesa C.G. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathian A., Weinberg A., Gallegos M., Banchereau J., Koutouzov S. IFN-α induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1, but not in Balb/c mice. J. Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Shiozawa S. In vivo cell transfer assay to detect autoreactive T cell subsets. Methods Mol. Biol. 2013;1142:49–53. doi: 10.1007/978-1-4939-0404-4_6. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Tsumiyama K., Yamane T., Ito M., Shiozawa S. Expansion of PD-1-positive effector CD4 T cells in an experimental model of SLE: contribution to the self-organized criticality theory. Kobe J. Med. Sci. 2013;59:E64–E71. [PubMed] [Google Scholar]
- Montefiori L., Wuerffel R., Roqueiro D., Lajoie B., Guo C., Gerasimova T., De S., Wood W., Becker K.G., Dekker J., et al. Extremely long-range chromatin loops link topolotical domains to facilitate a diverse antibody repertoire. Cell Rep. 2016;14:896–906. doi: 10.1016/j.celrep.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik A.K., Byrd A.T., Lucander A.C.K., Krangel M.S. Hierarchical assembly and disassembly of a transcriptionally active RAG locus in CD4+CD8+ thymocytes. J. Exp. Med. 2018;216:231–243. doi: 10.1084/jem.20181402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Non-resolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Odegard J.M., Marks B.R., DiPlacido L.D., Poholek A.C., Kono D.H., Dong C., Flavell R.A., Craft J. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone M.B. Molecular mimicry: its evolution from concept to mechanism as a cause of autoimmune disease. Monoclon.Antib.Immunodiagn.Immunother. 2014;33:158–165. doi: 10.1089/mab.2013.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen N.J., Choi M.Y., Fritzler M.J. Emerging technologies in autoantibody testing for rheumatic diseases. Arthritis Res. Ther. 2017;19:172. doi: 10.1186/s13075-017-1380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A.K., Drewes T., Engelmann S., Chuvpito S., Kishi H., Hunig T., Serfling E., Bommhardt U.H. PKB rescues calcineurin/NFAT-induced arrest of Rag expression and pre-T cell differentiation. J. Immunol. 2006;177:4567–4576. doi: 10.4049/jimmunol.177.7.4567. [DOI] [PubMed] [Google Scholar]
- Pender M.P. Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune diseases. Trends Immunol. 2003;24:584–588. doi: 10.1016/j.it.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Preble O.T., Black R.J., Friedman R.J., Klippel J.H., Vilcek J. Systemic lupus erythematosus: presence in human serum of an unusual acid-labile leukocyte interferon. Science. 1982;216:429–431. doi: 10.1126/science.6176024. [DOI] [PubMed] [Google Scholar]
- Premkumar L., Bobkov A.A., Patel M., Jaroszewski L., Bankston L.A., Stec B., Vuori K., Cote J.-F., Liddington R.C. Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs) J. Biol. Chem. 2010;285:13211–13222. doi: 10.1074/jbc.M110.102517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Cannons J.L., Klauschen F., Schwartzberg P.L., Germain R.N. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat. Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Ronnblom L.E., Alm G.V., Oberg K.E. Autoimmunity after-interferon therapy for malignant carcinoid tumors. Ann. Intern. Med. 1991;115:178–183. doi: 10.7326/0003-4819-115-3-178. [DOI] [PubMed] [Google Scholar]
- Rose N.R. Negative selection, epitope mimicry and autoimmunity. Curr.Opin.Immunol. 2017;49:51–55. doi: 10.1016/j.coi.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Roose J.P., Diehn M., Tomlinson M.G., Lin J., Alizadeh A.A., Botstein D., Brown P.O., Weiss A. T cell receptor-independent basal signaling via Erk and Abl kinases suppresses RAG gene expression. PLoS Biol. 2003;1:E53. doi: 10.1371/journal.pbio.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V., Hosokawa H., Ungerback J. Mechanisms of action of hematopoietic transcription factor PU.1 in initiation of T-cell development. Front. Immunol. 2019;10:228. doi: 10.3389/fimmu.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan S., Sarkar K., Taylor M., Candotti F., Vyas Y.M. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J. Immunol. 2014;193:150–160. doi: 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber M.-L., Baccala R., Haraldsson K.M., Choubey D., Stewart T.A., Kono D.H., Theofilopoulos A.N. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J. Exp. Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P., Willimann K., Lang A.B., Lipp M., Loetscher P., Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling P.J., Kurzrock R., Kantarjian H., Gutterman J.U., Talpaz M. Development of systemic lupus erythematosus after interferon therapy for chronic myelogenous leukemia. Cancer. 1991;68:1536–1537. doi: 10.1002/1097-0142(19911001)68:7<1536::aid-cncr2820680713>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Shannon C.A. Mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- Shiozawa S., Kuroki Y., Kim Y., Hirohata S., Ogino T. Interferon-alpha in lupus psychosis. Arthritis Rheum. 1992;35:417–422. doi: 10.1002/art.1780350410. [DOI] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:588. [Google Scholar]
- Sisirak V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D’Agati V., Elkon K.B., Reizis B. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 2014;211:1969–1976. doi: 10.1084/jem.20132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal F.J.T., Luis T.C., Tiemessen M.M. WNT signaling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- St. Rose M.-C., Qui H.Z., Bandyopadhyay S., Mihalyo M.A., Hagymasi A.T., Clark R.B., Adler A.J. The E3 ubiquitin ligase Cbl-b regulates expansion but not functional activity of self-reactive CD4 T cells. J. Immunol. 2009;183:4975–4983. doi: 10.4049/jimmunol.0901243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebegg M., Kumar S.D., Silva-Cayetano A., Fonseca V.R., Linterman M.A., Graca L. Regulation of the germinal center response. Front. Immunol. 2018;9:2469. doi: 10.3389/fimmu.2018.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tan E.M., Cohen A.S., Fries J.F., Masi A.T., McShane D.J., Rothfield N.F., Schaller J.G., Talal N., Winchester R.J. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- Taylor M.D., Sadhukhan S., Kottangada P., Ramgopal A., Sarkar K., D’Silva S., Selvakumar A., Candotti F., Vyas Y.M. Nuclear role of WASp in the pathogenesis of dysregulated Th1 immunity in human Wiscott-Aldrich syndrome. Sci. Transl. Med. 2010;2:37ra44. doi: 10.1126/scitranslmed.3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Ting C.-N., Olson M.C., Barton K.P., Leiden J.M. Transcription factor GATA3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Tolaymat A., Leventhal B., Sakarcan S., Kashima H., Monteiro C. Systemic lupus erythematosus in a child receiving long-term interferon therapy. J. Pediatr. 1992;120:429–432. doi: 10.1016/s0022-3476(05)80913-8. [DOI] [PubMed] [Google Scholar]
- Tsumiyama K., Miyazaki Y., Shiozawa S. Self-organized criticality theory of autoimmunity. PLoS One. 2009;4:8382. doi: 10.1371/journal.pone.0008382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumiyama K., Hashiramoto A., Takimoto M., Tsuji-Kawahara S., Miyazawa M., Shiozawa S. IFN-γ-producing effector CD8 T lymphocytes cause immune glomerular injury by recognizing antigen presented as immune comples on target tissue. J. Immunol. 2013;191:91–96. doi: 10.4049/jimmunol.1203217. [DOI] [PubMed] [Google Scholar]
- Vella L.A., Herati R.S., Wherry E.J. CD4+ T cell differentiation in chronic viral infections: the Tfh perspective. Trends Mol. Med. 2017;23:1072–1087. doi: 10.1016/j.molmed.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villey I., Caillol D., Selz F., Ferrier P., de Villartay J.-P. Defect in rearrangement of the most 5’ TCR-Jα following targeted deletion of T early α (TEA): implications for TCRα locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- Weening J.J., D'Agati V.D., Schwartz M.M., Seshan S.V., Alpers C.E., Appel G.B., Balow J.E., Bruijn J.A., Cook T., Ferrario F., et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J. Am. Soc. Nephrol. 2004;15:241–250. doi: 10.1097/01.asn.0000108969.21691.5d. [DOI] [PubMed] [Google Scholar]
- William J., Euler C., Christensen S., Shlomchik M.J. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- Yang X.O., Doty R.T., Hicks J.S., Willerford D.M. Regulation of T-cell receptor Dβ1 promoter by KLF5 through reiterated GC-rich motifs. Blood. 2003;101:4492–4499. doi: 10.1182/blood-2002-08-2579. [DOI] [PubMed] [Google Scholar]
- Yaniv G., Twig G., Shor D.B.-A., Furer A., Sherer Y., Mozes O., Komisar O., Slonimsky E., Klang E., Lotan E., et al. A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients. Autoimmun. Rev. 2015;14:75–79. doi: 10.1016/j.autrev.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Yen E.Y., Singh R.R. Lupus-an unrecognized leading cause of death in young females: a population-based study using nation-wide death certificates, 2000-2015. Arthritis Rheumatol. 2018;70:1251–1255. doi: 10.1002/art.40512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg S.R., Schnitzer T.J. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25:401–406. doi: 10.1002/art.1780250407. [DOI] [PubMed] [Google Scholar]
- Zacarias-Cabeza J., Belhocine M., Vanhille L., Cauchy P., Koch F., Pekowska A., Fenouil R., Bergon A., Gut M., Gut I., et al. Transcription-dependent generation of a specialized chromatin structure at the TCRβ locus. J. Immunol. 2015;194:3432–3443. doi: 10.4049/jimmunol.1400789. [DOI] [PubMed] [Google Scholar]
- Zinkernagel R.M. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental information; Next-generation sequencing (NGS) data of TCRA and TCRB repertoire analysis in splenic DOCK8+ CD4 T cells of x12 OVA-immunized BALB/c mice, the splenic DOCK8- CD4 T cells of x12 OVA-immunized mice, and the CD4 T cells of x12 PBS-immunized mice were deposited in online at https://data.mendeley.com//datasets/7r5pjbz793/21 and are publicly available as of the date of publication.
The data on gene expression microarray data which compared among splenic DOCK8+ CD4 T cells of x12 OVA-immunized BALB/c mice, the splenic DOCK8- CD4 T cells of x12 OVA-immunized mice, and the CD4 T cells of x12 PBS-immunized mice, were deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE159240 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159240).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.