Summary
Ataxin-2 (Atx2) is a highly conserved RNA binding protein. Atx2 undergoes polyglutamine expansion leading to amyotrophic lateral sclerosis (ALS) or spinocerebellar ataxia type 2 (SCA2). However, the physiological functions of Atx2 in neurons remain unknown. Here, using the powerful genetics of Drosophila, we show that Atx2 is essential for normal neuronal cytoskeletal dynamics and organelle trafficking. Upon neuron-specific Atx2 loss, the microtubule and actin networks were abnormally stabilized and cargo transport was drastically inhibited. Depletion of Atx2 caused multiple morphological defects in the nervous system of third instar larvae. These include reduced brain size, impaired axon development, and decreased dendrite outgrowth. Defects in the nervous system caused loss of the ability to crawl and lethality at the pupal stage. Taken together, these data mark Atx2 as a major regulator of cytoskeletal dynamics and denote Atx2 as an essential gene in neurodevelopment, as well as a neurodegenerative factor.
Subject areas: Biological sciences, Neuroscience, Developmental neuroscience, Cellular neuroscience, Cell biology, Developmental biology
Graphical abstract
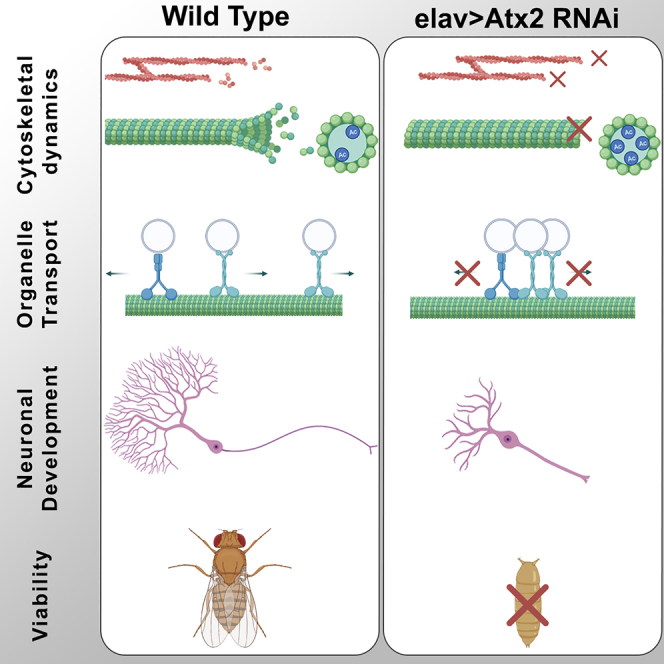
Highlights
-
•
Atx2 is a major regulator of the cytoskeleton in neurons
-
•
Atx2 is responsible for maintaining dynamic cytoskeletal networks
-
•
Atx2 depletion in the Drosophila larval CNS severely impairs organelle transport
-
•
Atx2 is necessary for correct neurite outgrowth and CNS development in Drosophila
Biological sciences; Neuroscience; Developmental neuroscience; Cellular neuroscience; Cell biology; Developmental biology
Introduction
Ataxin-2 (Atx2) is a highly conserved RNA binding protein that is known to associate with polyribosomes, coordinate RNA granule assembly, and regulate translation for synaptic plasticity (Bakthavachalu et al., 2018; McCann et al., 2011; Satterfield et al., 2002; Sudhakaran et al., 2014). Atx2 contains mRNA binding domains, intrinsically disordered domains, and a polyQ domain, shown to undergo expansion, leading to neurodegenerative diseases spinocerebellar ataxia type 2 (SCA2) and amyotrophic lateral sclerosis (ALS) (Elden et al., 2010; Riess et al., 1997). Due to its causative role in neurodegenerative diseases, there has been much interest in discerning the function of Atx2. However, while Atx2 is highly expressed in the developing central nervous system, we presently do not know how Atx2 functions in normal neurodevelopment. In one study, Atx2 knockout mice were generated (Kiehl et al., 2006). These mice displayed no obvious abnormalities, likely due to compensation by a related protein called Atx2-like. Therefore, the physiological roles of Atx2 remain undetermined. The gene encoding Atx2-like is absent in Drosophila, marking them as an ideal model organism to probe physiological Atx2 function.
Atx2 can associate with its target RNAs and stabilizes these transcripts (Satterfield and Pallanck, 2006; Singh et al., 2020). In turn, this action contributes to long-term habituation or memory formation (Bakthavachalu et al., 2018; Sudhakaran et al., 2014). Via RNA organization and control of translation, Atx2 may be a critical regulator in many cellular processes, among them cytoskeletal dynamics. For example, Atx2 has been previously indicated in actin regulation in the germline (Satterfield et al., 2002) and microtubule organization in mitosis (Gnazzo et al., 2016; Stubenvoll et al., 2016). Collectively, these observations are of note because there are strong parallels between the organization of the cytoskeleton in mitosis and in neurons (Del Castillo et al., 2020; Cheerambathur et al., 2019; Hertzler et al., 2020; Lin et al., 2012; Norkett et al., 2020; Zhao et al., 2019). In mitosis, the cytoskeleton must be reorganized to facilitate chromosome separation and actin ring contraction (Basant and Glotzer, 2018; Mishima et al., 2002). In neurons, the development of neurites is highly dependent upon the dynamics of the actin and microtubule cytoskeleton (Lu et al., 2013; Papandréou and Leterrier, 2018; Roossien et al., 2014).
We used the powerful genetics of Drosophila to probe the physiological roles of Atx2. Using this system, we show that loss of Atx2 at early developmental stages leads to “hyperstable” microtubule and actin networks, and severely reduced organelle transport. Subsequently, we show that neurodevelopment is drastically impaired and that loss of Atx2 in the nervous system is 100% lethal at the pupae stage.
Results
Ataxin-2 depletion suppresses microtubule and actin dynamics
Previous studies have shown an essential role for Atx2 in cell division via the regulation of centrosomes and the spindle midzone (Gnazzo et al., 2016; Stubenvoll et al., 2016). These structures are key microtubule organizing and containing structures respectively. Further, it is well documented that mitotic machinery is repurposed in neurons to facilitate cytoskeleton reorganization (Baas, 1999; Del Castillo et al., 2019b). While Atx2 has known roles in neurodegeneration and is highly expressed in the developing nervous system, its physiological role in neurons remains undetermined. Therefore, we hypothesized that Atx2 may be a key player in neuronal cytoskeletal dynamics at earlier developmental stages.
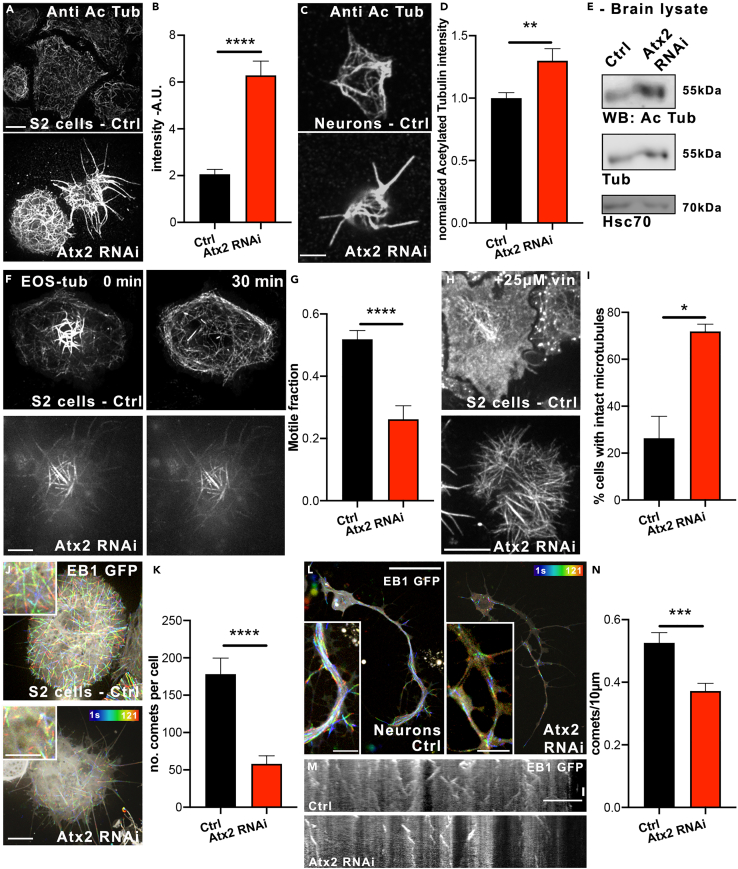
We chose to investigate the properties of microtubules in Drosophila neurons and S2 cells—an efficient system to study the cytoskeleton using dsRNA strategies (Jolly et al., 2010). We depleted Atx2 in interphase Drosophila S2 cells and carried out immunostaining for acetylated tubulin. Tubulin acetylation increases with microtubule age, so is a readout for microtubule lifetime (Janke and Magiera, 2020; Portran et al., 2017; Szyk et al., 2014). In S2 cells, we found a roughly 3-fold increase in acetylated tubulin signal upon Atx2 depletion (Figures 1A and 1B). In order to study the physiological role of Atx2 in neurons, we depleted the protein by neuron-specific expression of shRNA against Atx2 using the pan-neuronal driver elav-gal4. Crucially, this is a postmitotic driver, so any role for Atx2 in cell division is not affected. We prepared primary neuronal cultures of Drosophila third instar larvae and found acetylated tubulin signal increased around 30% in Atx2 RNAi neurons compared to control (Figures 1C and 1D). Atx2 knockdown was confirmed by Western blot of brain lysate (Figure S1). Further, we investigated acetylated tubulin levels in brain lysate from elav > Atx2 RNAi larvae brains. Consistent with our immunostaining data, western blotting demonstrated an increase in acetylated tubulin in Atx2 RNAi brain lysate compared to control (Figure 1E, expanded in Figure S1B). Taken together, these results suggest an increase in microtubule stability upon loss of Atx2 and confirm this effect is conserved between dividing and postmitotic cells.
Figure 1.
Ataxin-2 depletion suppresses microtubule dynamics
(A and B) (A) Example images showing Atx2 depletion in Drosophila S2 cells increases microtubule acetylation as quantified in (B) (Signal intensity Control = 2,061,210 ± 204,596 A.U, n = 50 cells. Atx2 RNAi = 6,286,404 ± 608,574, n = 41 cells)
(C and D) (C) Example images showing Atx2 depletion increases microtubule acetylation in Drosophila neurons in culture (scale bar, 10 μm), as quantified in (D) (Signal Intensity Control = 1.0 ± 0.04, n = 23 cells, Signal Intensity Atx2 RNAi = 1.3 ± 0.10, n = 27 cells, p = 0.01).
(E) Western blot of acetylated tubulin levels in Control and elav > Atx2 RNAi brain lysate from Drosophila third instar larvae.
(F and G) (F) Example images of EOS-tubulin in S2 cells 0 and 30mins after photoconversion (scale bar, 10 μm) as quantified in (G) (Motile fraction Control = 0.52 ± 0.03, n = 21 cells, Atx2 RNAi = 0.26 ± 0.04, n = 22 cells, p < 0.0001).
(H) Example images of tubulin staining in S2 cells showing microtubules after 25μM vinblastine treatment for 1hr. Scale bar, 10 μm.
(I) Quantification of cells with an intact microtubule network after vinblastine treatment (Control = 26.4% ± 9.4, n = 38 cells, Atx2 RNAi = 71.9% ± 3.1, n = 22 cells, p = 0.04).
(J) Temporal color code images showing motility of EB1-GFP comets in S2 cells over 120 s. Scale bar, 10 μm.
(K) Quantification of number of EB1 comets per cell (Control = 178.2 ± 21.5 comets, n = 17 cells, Atx2 RNAi = 58.0 ± 10.7, n = 15 cells, p < 0.001).
(L) Temporal color code images showing motility of EB1-GFP comets in cultured neurons over 120 s. Scale bar, 10 μm, inset, 5 μm.
(M) Kymographs showing motility of EB1 comets in neuronal processes over 120 s. Scale bar, 10 μm.
(N) Quantification of number of EB1 comets per process length (Control = 0.53 ± 0.03, n = 31, Atx2 RNAi = 0.37 ± 0.02, n = 34, p = 0.0004). Data are presented as mean ± standard error. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To investigate whether the increase in acetylated tubulin was related to an alteration in microtubule dynamics, we directly measured microtubule dynamics using Drosophila S2 cells expressing photoconvertible EOS-tubulin. UV-light was applied to a specific region to convert EOS-tubulin fluorescence from green to red and the photoconverted red signal outside the original conversion region was quantified 30 min after photoconversion. Incorporation of photoconverted tubulin subunits to microtubules located outside the original photoconverted area directly demonstrates tubulin subunit exchange. Atx2 depletion led to around a 50% decrease in photoconverted signal outside the ROI (Figures 1F and 1G, Video S1). This indicates that Atx2 is necessary for normal subunit exchange. Additionally, we investigated the stability of microtubules after treatment with a microtubule depolymerizing agent vinblastine by immunostaining for tubulin. Consistent with our previous data, we found that Atx2 depletion confers the resistance of microtubules to vinblastine—intact microtubules could be detected in the Atx2 RNAi cells, whereas microtubules in the control cells were largely depolymerized (Figures 1H and 1I).
Scale bar, 10 μm.
In order to directly assess the effect of loss of Atx2 on microtubule dynamics, we carried out live imaging of EB1 (end binding protein 1)—a marker for polymerizing microtubule plus-ends (Akhmanova and Steinmetz, 2015). Initially, we observed EB1-GFP comets in S2 cells (Figures 1J and 1K, Video S2). We found around a 65% decrease in the number of comets per cell, as indicated by the loss in comets shown in the temporal color code—a color-coded projection of each comet over time. Furthermore, we carried out the same assay in cultured larvae neurons expressing EB1-GFP (Figure 1L, Video S3). Comets in neuronal processes are demonstrated in kymographs in Figure 1M where time is projected on the Y axis, showing motile objects as diagonal lines. Upon Atx2 knockdown, we found a roughly 30% decrease in the number of comets per 10 μm (Figure 1N). Notably, the total microtubule content in these cells is unchanged, as shown by tubulin staining of extracted control and Atx2 knockdown neurons (Figures S2A and S2B). Therefore, Atx2 knockdown decreases only the dynamic microtubule population, not the total amount of microtubules. Taken together, these data show that loss of Atx2 confers increased microtubule stability and that Atx2 is essential for normal microtubule dynamics, both in cultured S2 cells, and crucially, in neurons.
Scale bar, 10 μm.
Control (elav-gal4) and Atx2 RNAi (elav > Atx2 RNAi). Scale bar, 10 μm.
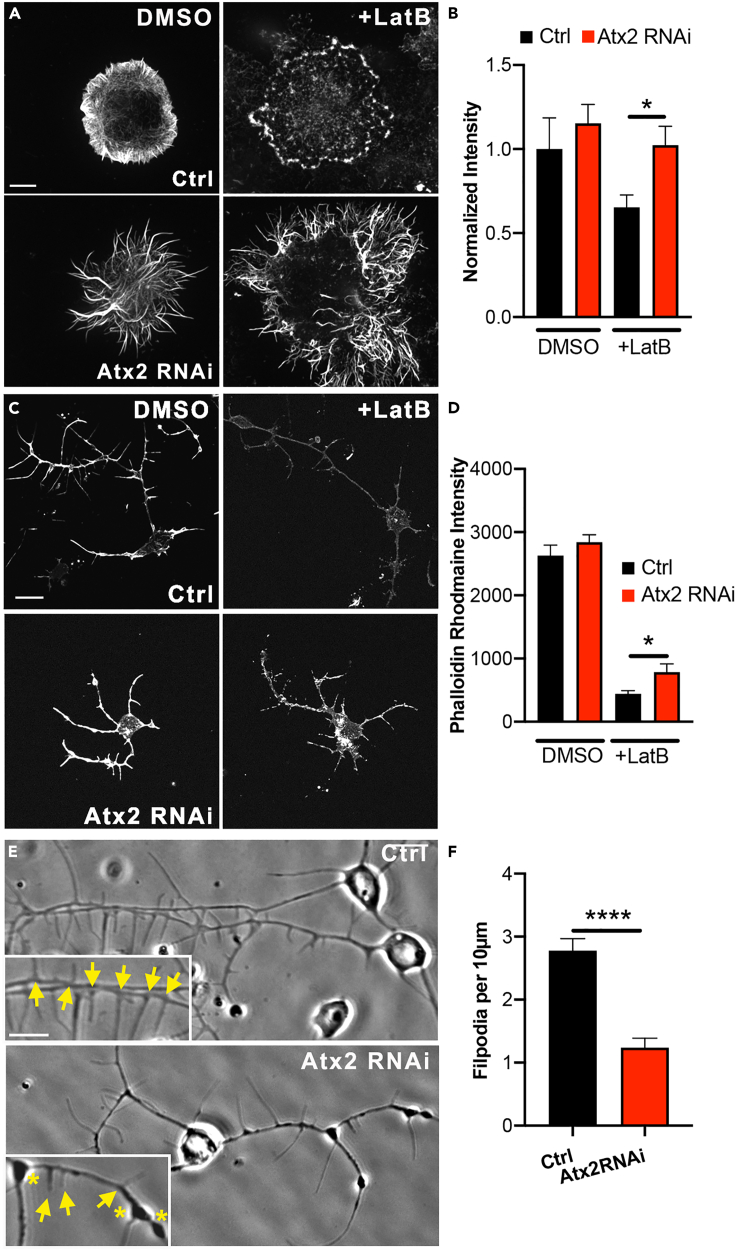
In addition to the microtubule cytoskeleton, Atx2 has been linked to the actin filament formation in Drosophila embryos (Satterfield et al., 2002). Therefore, we hypothesized that Atx2 depletion may impact actin dynamics in the nervous system. Initially, we knocked down Atx2 by RNAi in Drosophila S2 cells and stained for actin filaments using Rhodamine-phalloidin. We treated these cells with actin depolymerizing agent Latrunculin B (LatB). We found that in control cells, the F-actin signal, detected by phalloidin staining, was decreased by around 35%, indicative of actin depolymerization. However, upon Atx2 knockdown, there was no significant alteration in F-actin signal (Figures 2A and 2B). The sensitivity of F-actin to depolymerizing agent LatB decreased.
Figure 2.
Ataxin-2 depletion stabilizes the actin cytoskeleton
(A and B) (A) Example images showing phalloidin staining for F-actin in control and Atx2 RNAi treated Drosophila S2 cells treated with DMSO or 10 μM LatB for 1hr. Scale bar, 10 μm (B). Normalized Phalloidin Rhodamine signal (Control = 1.00 ± 0.19, Atx2 RNAi = 1.15 ± 0.11, Control + LatB = 0.65 ± 0.07, Atx2 RNAi + LatB = 1.02 ± 0.11, n = 15–24 cells, p = 0.03).
(C) Example images showing phalloidin staining in control and elav > Atx2 RNAi neurons treated with DMSO or 10 μM LatB for 1hr. Scale bar, 10 μm.
(D) Quantification of Phalloidin Rhodamine intensity along length of neuron (Signal Intensity Control LatB = 440.4 ± 53, n = 37 cells, Signal Intensity Atx2 RNAi LatB = 786.5 ± 128, n = 27 cells, p = 0.02).
(E) Example phase-contrast images showing filopodia from neuronal processes in control and elav > Atx2 RNAi neurons. Scale bar, 10 μm.
(F) Average number of filopodia per 10μm process length (Control = 2.8 ± 0.19, n = 28 cells, Atx2 RNAi = 1.2 ± 0.15, n = 28 cells, p < 0.0001). Data are presented as mean ± standard error. ∗p < 0.05, ∗∗∗∗p < 0.0001.
To test this actin regulation in neurons, we carried out similar experiments in cultured neurons from Drosophila third instar larvae, either wild type, or after Atx2 knockdown. We demonstrated that similarly to S2 cells, actin filaments in neurons are more stable after Atx2 knockdown. As demonstrated by Rho-phalloidin staining about 40% more F-actin survived the treatment with LatB (Figures 2C and 2D). This shows that actin polymers are less sensitive to depolymerizing agents upon loss of Atx2, and so, that the actin network is less dynamic. Therefore, Atx2 is an essential actin regulator in dividing and nondividing cells.
In order to study the potential effects of increased actin stability on neuronal morphology, we chose to observe filopodia, thin rod-like protrusions rich in F-actin, essential for the generation of neurites and the growth cone (Lin et al., 1996). We prepared primary neuronal cultures from third instar larvae and carried out phase-contrast imaging to examine filopodia along neuronal processes. Upon loss of Atx2, the number of filopodia projecting from neurites decreased over 50% (Figures 2E and 2F, see arrows). This is consistent with a less dynamic actin network upon loss of Atx2, with filopodia being unable to form.
These data show that Atx2 is essential for normal actin dynamics in neurons. These effects are comparable to our data demonstrating the microtubule network is stabilized upon Atx2 knockdown, and highlight the role of Atx2 in regulating normal cytoskeletal dynamics in neurons during the development of the nervous system.
Ataxin-2 is essential for organelle transport and distribution in Drosophila neurons
Beyond neuronal morphology, both actin and microtubule networks are crucial for normal organelle transport and distribution. This is essential in neurons due to their unique architecture. Microtubules act as tracks for longer range transport of cargo by kinesin and dynein motors, whereas actin is responsible for shorter range transport and cargo anchoring by myosin motors (Kapitein et al., 2013; Lu et al., 2020; Noordstra et al., 2016; Twelvetrees, 2020; De Vos and Hafezparast, 2017). Therefore, we hypothesized that the effects of loss of Atx2 on the cytoskeleton would have a crucial impact on organelle trafficking.
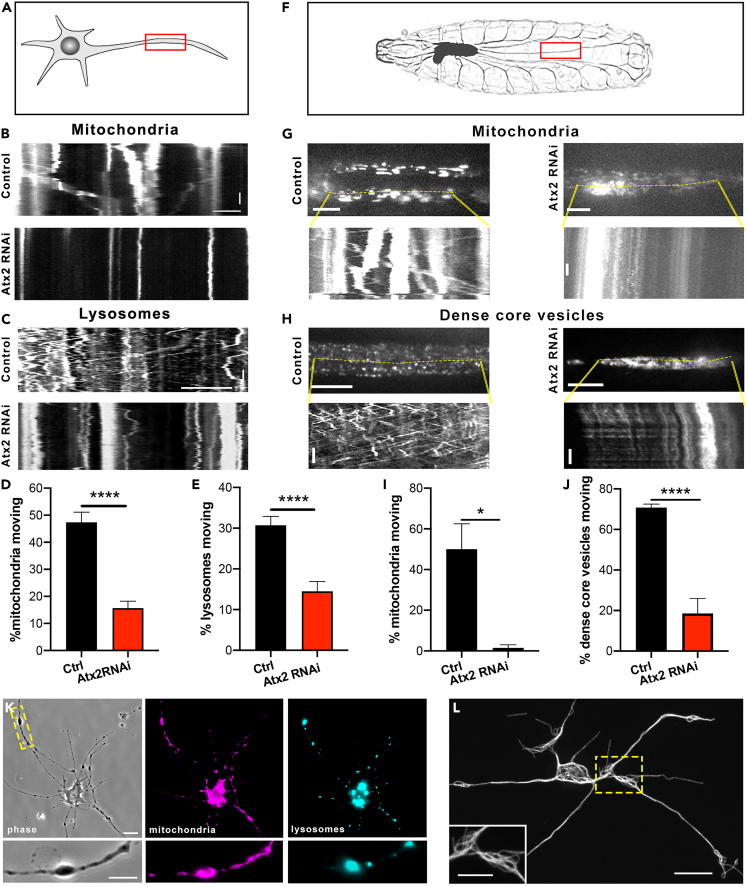
To investigate the effect of loss of Atx2 on organelle trafficking, we started by carrying out live imaging of two cargoes in Drosophila third instar larvae neurons in culture—mitochondria and lysosomes (Figure 3A). Using the neuron-specific driver elav-gal4, we labeled mitochondria using a genetically encoded mCherry fluorescent protein targeted to the outer mitochondrial membrane. Time-lapse microscopy showed a 60% reduction in motile mitochondria. This is exemplified in the kymographs in Figure 3B and in Video S4 and quantified in Figure 3D. We also studied the motility of lysosomes labeled with lysotracker. Consistent with our data for mitochondrial transport, we found a vast decrease in lysosome motility upon loss of Atx2 (Figure 3C, Video S5)—in control conditions around 30% of the organelles were moving, this dropped about two-fold in Atx2 RNAi neurons (Figure 3E).
Figure 3.
Ataxin-2 depletion inhibits organelle transport
(A) Schematic showing region of imaging in neurons in culture.
(B) Example kymographs showing mitochondria motility in processes of control and elav > Atx2 RNAi neurons. Horizontal scale bar, 10μm. Vertical scale bar, 20s.
(C) Example kymographs showing lysosome motility in processes of control and Atx2 RNAi neurons. Horizontal scale bar, 10 μm. Vertical scale bar, 20s.
(D) Quantification of motile mitochondria in control and Atx2 knockdown neurons (Control = 47% ± 3.7, n = 25 cells, Atx2 RNAi = 15.7% ± 2.5, n = 28 cells, p < 0.0001).
(E) Quantification of motile lysosomes in control and Atx2 knockdown neurons (Control = 31% ± 2.1, n = 25 cells, Atx2 RNAi = 14.5% ± 2.4, n = 29 cells, p < 0.0001).
(F–H) (F) Schematic image of a third instar larva showing segmental nerves. Example kymographs showing mitochondria motility (G) and dense-core vesicle motility (H) in segmental nerves in vivo in control and elav > Atx2 RNAi larvae. Vertical scale bars, 10s, horizontal scale bars, 10 μm for dense-core vesicles, 20 μm for mitochondria.
(I) Quantification of motile mitochondria in control and Atx2 knockdown segmental nerves in vivo (Control = 50% ± 12.5, n = 3 larvae, Atx2 RNAi = 1.5% ± 1.5, n = 3 larvae, p = 0.015).
(J) Quantification of motile DCVs in control and Atx2 knockdown segmental nerves in vivo (Control = 66% ± 1.7, n = 9 larvae, Atx2 RNAi = 18.6% ± 7.4, n = 8 larvae, p < 0.0001).
(K) Example image showing mitochondria and lysosome distribution in an elav > Atx2 RNAi cultured neuron. Scale bar, 10 μm, inset 5 μm.
(L) Example super-resolution images of microtubules in varicosities in Atx2 RNAi neurons. Scale bar 10 μm, inset 5 μm. Data are presented as mean ± standard error. ∗p < 0.05, ∗∗∗∗p < 0.0001.
Scale bar, 10 μm.
Scale bar, 10 μm.
Next, we extended these findings to investigate organelle transport in vivo in the segmental nerves of Drosophila third instar larvae. These nerves contain the axons of motor and sensory neurons which extend from the CNS into segments of the larvae body wall (Figure 3F). We studied the motility of two cargoes; in this case mitochondria and dense-core vesicles (presynaptic carriers). As in culture, we found that in vivo mitochondrial transport was drastically decreased by around 90% in Atx2 RNAi animals compared to control (Figures 3G and 3I, Video S6). Trafficking of dense-core vesicles was similarly impaired in Atx2 RNAi larvae—motile organelles decreased by around 70% (Figures 3H and 3J, Video S7). Taken together these data show Atx2 has an essential role in normal transport of multiple organelles transport in neurons in vitro and in vivo.
Scale bar, 10 μm.
Scale bar, 10 μm.
In our live imaging experiments, we noted accumulations of stationary organelles (e.g., Video S6) and we have previously shown that depleting Atx2 leads to varicosities in neuronal processes containing ER accumulations (Figure 2E asterisks) (Del Castillo et al., 2019a). Downstream of the Atx2-dependent loss of cargo transport, we investigated the distribution of organelles throughout neurons in culture. Phase-contrast imaging coupled with labeling of mitochondria and lysosomes demonstrated that these organelles could be found colocalized in the axonal varicosities (Figure 3K). Indeed, the majority of the varicosities held both of these organelles (84%), while only around 5% each of varicosities contained only one or neither of these organelles. This is consistent with a “traffic jam” leading to the accumulation of organelles at these sites due to severely impaired transport.
We were interested in the microtubule structure at these “traffic jam” sites. In order to visualize microtubules at the sites, we carried out super-resolution imaging on extracted Atx2 RNAi neurons with stained for tubulin. We found that microtubules in these regions are not organized in parallel arrays, but instead were disordered and looped (Figure 3L). This disorganization of the microtubule network may prevent normal organelle transport through these sites, causing their accumulation.
The cytoskeletal and trafficking phenotypes we observe are likely dependent on the RNA transcripts regulated by Atx2. We carried out RNAseq analyses of dissected control vs Atx2 RNAi third instar larvae brains to identify differentially expressed genes that may regulate the cytoskeleton. Depletion of Atx2 induced clear alterations in the neuronal transcriptome. We identified 1636 significantly upregulated and 1684 significantly downregulated transcripts out of 16,729 tested (adjusted p value <0.05) (Figure S3A). This dataset confirms a decrease in the level of Atx2 mRNA in the Atx2 knockdown condition compared to control (Log2FC −0.97, equivalent to around a 50% decrease). Next, we grouped the differentially expressed genes according to their indexed cellular component using PANTHER gene ontology analysis (Thomas et al., 2003). The number of transcripts per group in the dataset is compared to the expected number, based on the total number of transcripts in this group in the Drosophila reference transcriptome. Therefore, any “overrepresented” cellular components can be identified according to the list of differentially expressed genes. Among enriched cellular components, cytoskeletal proteins were around 1.5 fold overrepresented (GO:0,005,856, p = 4.73 × 10−3), and components of the actin cytoskeleton are around 2 fold overrepresented (GO:0,015,629, p = 5.19 × 10−3) (Figure S3B). We further probed these two groups to identify the protein class or molecular function assigned to these transcripts and found 50% of these were “classical” cytoskeletal proteins (rather than protein modifiers), which we define as proteins annotated “PC00085” in the database (Figure S3 and Table S1). Therefore, Atx2 has the ability to regulate mRNA levels of many cytoskeleton components.
Ataxin-2 is essential for neurodevelopment, locomotion, and viability in Drosophila
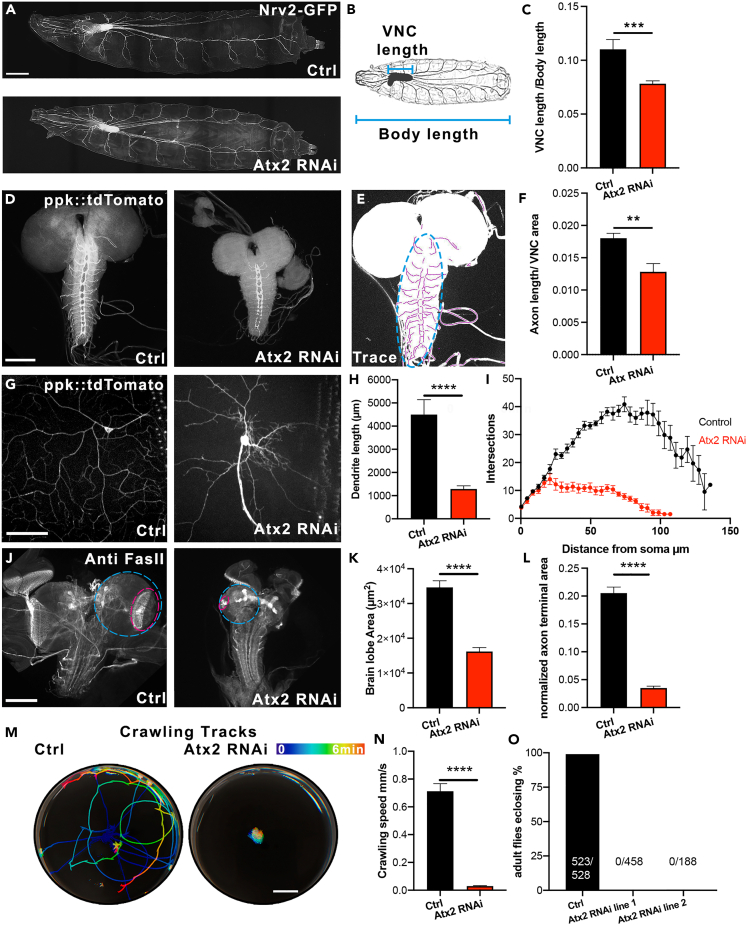
We and others have demonstrated that the cytoskeleton is one of the major drivers of neurite initiation and neuronal development (Del Castillo et al., 2015; Korobova and Svitkina, 2008; Lu et al., 2013; Norkett et al., 2020; Roossien et al., 2014; Winding et al., 2016; Yu et al., 2000). Therefore, we studied how the nervous system develops in the absence of Atx2 as a result of severely impaired cytoskeletal dynamics and trafficking. Firstly, we studied the size of the central nervous system (CNS)in intact Drosophila third instar larvae. We used the gene trap line Nrv2 GFP to endogenously label the central nervous system and segmental nerves (Figure 4A). We found that the length of the ventral nerve cord (VNC), normalized to body length, was decreased by around a third (Figures 4B and 4C). The length of the VNC was decreased while the length of the larva body was unchanged between control and Atx2 RNAi animals (Figures S4A and S4B). Therefore, Atx2 is essential for correct brain development.
Figure 4.
Ataxin-2 is essential for neurodevelopment, locomotion, and viability in Drosophila
(A–C) (A) Atx2 depletion impairs Central nervous system development. Example images of control and elav > Atx2 RNAi CNS from Drosophila third instar larvae. Scale bar 100 μm (B). Illustration showing VNC length and body length measurements (C). Length of VNC normalized to body length. (Control ratio = 0.11 ± 0.009, Atx2 RNAi ratio = 0.08 ± 0.003, n = 8 control and 10 Atx2 RNAi animals, p = 0.0006).
(D) Atx2 depletion decreases axon length. Example images of control and elav > Atx2 RNAi CNS from Drosophila third instar larvae.
(E) Illustration showing example of axon tracing (magenta) in the VNC (cyan).
(F) Axon length in VNC normalized to VNC area (Control = 0.018 ± 7x10−4, Atx2 RNAi = 0.013 ± 1.3x10−3, n = 8 control and 10 Atx2 RNAi animals, p = 0.006).
(G) Atx2 depletion decreases dendrite development. Example images of DA neurons in control and elav > Atx2 RNAi neurons. Scale bar, 50 μm.
(H) Dendrite length per cell in control and elav > Atx2 RNAi neurons (Control = 4500 μm ± 644, Atx2 RNAi = 1290 μm ± 134, n = 11 control and 11 Atx2 RNAi animals, p < 0.0001).
(I) Sholl analysis of control and elav > Atx2 RNAi neurons DA neurons showing the number of intersections with distance from soma (n = 6 control and 4 Atx2 RNAi animals).
(J) Atx2 depletion decreases brain lobe area. Example images of control and elav > Atx2 RNAi CNS from Drosophila third instar larvae. Scale bar, 100 μm.
(K) Size of brain lobes in control and elav > Atx2 RNAi brains (Control = 34,662 μm2 ± 1897, Atx2 RNAi = 16,198 μm2 ± 1128, n = 23 control and 20 Atx2 RNAi brains, p < 0.0001).
(L) Area of photoreceptor projections normalized to brain lobes in control and Atx2 RNAi brains (Control = 0.205 ± 0.01, Atx2 RNAi = 0.035 ± 0.003, n = 23 control and 20 Atx2 RNAi brains, p < 0.0001).
(M) Atx2 depletion impairs locomotion. Example crawling tracks of control and elav > Atx2 RNAi third instar larvae. Scale bar = 10 mm.
(N) Crawling velocity of control and elav > Atx2 RNAi third instar larvae (Control = 0.71 mm/s ± 0.05, Atx2 RNAi = 0.03 mm/s ± 0.003, n = 12 control and 12 Atx2 RNAi animals).
(O) Atx2 depletion is lethal at the pupae stage. Percentage of adults eclosing from pupae cases (Control = 99%, Atx2 RNAi line 1 = 0.0%, Atx2 RNAi line 2 = 0.0%). Data are presented as mean ± standard error. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We investigated the effect of the loss of Atx2 on axonal and dendritic development. To assess axon development, we dissected the CNS of these larvae and imaged the axonal projections of these neurons extending into the VNC (Figure 4D) We traced the axons in this region and found that the axon length, normalized to the area of the VNC, was significantly decreased upon Atx2 depletion (Illustrated in Figure 4E, quantified in Figure 4F). Regarding dendritic development, we labeled Class IV DA (Dendritic arborization) neurons (sensory neurons that line the Drosophila body wall) with a fluorescent reporter expressed specifically in these cells (ppk:tdTomato) (Grueber et al., 2002). Under control conditions, each neuron extended long, branched dendritic arbors which covered the whole surface of the hemi-segment, where the soma is located. However, upon loss of Atx2, we found that these dendritic arbors were drastically shrunken (Figure 4G). This effect was quantified in two ways, firstly the total dendrite length per cell was decreased by around 65% (Figure 4H), consistent with our previously published in vitro data (Del Castillo et al., 2019a). Secondly, to further describe the effect of Atx2 depletion on the shape of the dendritic arbor, we carried out Sholl analysis (Sholl, 1953). Concentric circles are drawn outwards from the soma, and the number of intersections that the dendritic arbor makes with each circle is counted as a representation of dendrite morphology. From 20 μm from the soma, the Atx2 RNAi neurons displayed many fewer intersections than control neurons and the dendritic arbors only extended to around 100 μm from the soma, whereas the dendrites of the control neurons extended around 30% more than this (Figure 4I). We also studied axon development in the central nervous system by dissecting the third instar larvae CNS and immunostaining for Fasciclin II (FasII)—an axon-specific marker. We once more found that the Atx2 RNAi CNS was drastically smaller than controls. Quantification of the brain lobes (Figure 4J, cyan) showed a roughly 2-fold decrease in area (Figure 4K). Further, we quantified the area of the retinotopic pattern—the structure representing the photoreceptor axons (Figure 4J, magenta) (Hadjieconomou et al., 2011). When normalized to the area of the brain lobe, we found this structure was decreased to around 10% of control area upon Atx2 depletion (Figure 4L). These phenotypes demonstrate that Atx2 depletion significantly impairs the outgrowth and development of both axons and dendrites. These data are consistent with our observations that depleting Atx2 has severe consequences on cytoskeletal dynamics and organelle transport.
We studied the effect of this underdeveloped nervous system on the whole organism, initially by studying locomotion. We imaged third instar larvae crawling on agar plates and calculated the velocity. Control larvae crawled over the surface of the plate at about 0.7 mm/s. However, Atx2 knockdown larvae were largely stationary, crawling with a velocity of less than 0.1 mm/s (Video S8). This is further shown by the color-coded tracks of each larva over time (Figure 4M quantified in Figure 4N).
Crucially, we found that depleting Atx2 in neurons was lethal at the pupae stage. Animals expressing the Atx2 RNAi were not viable to adult stages (Figure 4O, control; 523/528, RNAi line 1; 0/458, RNAi line 2; 0/188 flies eclosed). There was no difference in survival between RNAi stock lines (96% viable) and controls (98% viable) when the Atx2 RNAi expression was not driven by a gal4. We found consistent results with two different Atx2 RNAi lines; therefore, this effect is specific to Atx2 knockdown rather than an off-target effect. Taken together, these data show that Atx2 is necessary for the normal development of both axons and dendrites. As a result of the loss of Atx2, the development of the nervous system is severely impaired. Subsequently, the larvae are unable to move and die during development. These data highlight the role of Atx2 as an essential gene in nervous system development and that it is absolutely required for normal cytoskeletal dynamics and cargo transport.
Discussion
Multiple works have demonstrated the involvement of mutant Atx2 in neurodegenerative conditions ALS and SCA2 (Elden et al., 2010; Riess et al., 1997). However, an understanding of the physiological roles of the RNA-binding protein in neurons is still lacking, potentially due to redundancy between Atx2 and Atx2-like in mammals. Here we use Drosophila as a model system to uncover the necessity of wild-type Atx2 for the neuronal cytoskeleton and organelle trafficking. Further to this, we show gross morphological defects in the nervous system. These neurodevelopmental defects lead to decreased locomotion of Drosophila larvae and are lethal during development, highlighting the necessity of Atx2 during development.
We show that Atx2 is essential for normal cytoskeletal dynamics, in both microtubule and actin networks. Firstly, our data show that targeted depletion of Atx2 in neurons drives an increase in microtubule acetylation, a decrease in tubulin subunit exchange, and fewer EB1 comets. Thus, we conclude microtubules shift to a more stable, less dynamic state. A role for Atx2 in regulating the microtubule network is consistent with previous reports of Atx2 having roles in centrosome organization and mitotic spindle regulation in C. elegans (Gnazzo et al., 2016; Stubenvoll et al., 2016). Secondly, we report an increase in the stability of the actin network upon Atx2 knockdown. These build upon early descriptions of Atx2 function, which showed evidence of disorganized actin bundles in retinal precursor cells, the female germline, and bristles of Atx2 null Drosophila (Satterfield et al., 2002). Our data extend previous findings by highlighting specific neuronal effects of Atx2.
Beyond the altered cytoskeletal network, we show that decreasing levels of Atx2 decreases organelle transport in culture and in vivo. Impaired organelle transport is likely a direct consequence of the cytoskeletal defects caused by Atx2. Microtubule stability and longevity would be thought to facilitate microtubule motor-based organelle transport (Godena et al., 2014; Kaul et al., 2014). However, we see a decrease in the motility of multiple cargoes. Instead, an increase in the stability of the actin network may lead to organelles being anchored or trapped by myosin motors (Kapitein et al., 2013; Lu et al., 2020). Alternatively, changes in mRNA levels of motor proteins may contribute to decreased trafficking (Figure S3 and Table S1) and would explain long-range transport defects rather than organelles only stationary in varicosities. However, the changes in transcript levels for these proteins are modest. This loss of organelle transport leads to “traffic jams,” accumulations of organelles in axonal varicosities. These axonal varicosities are also known to contain ER aggregates (Del Castillo et al., 2019a). As well as essential for development, improper organelle transport can be seen in neurodegenerative diseases, including mitochondria trapped in dystrophic neurites in an Alzheimer's disease model (Correia et al., 2016; Fiala et al., 2007), and impairments in endosome retrograde transport in ALS (Xie et al., 2015).
Finally, we demonstrate the necessity of wild-type Atx2 for normal neurodevelopment. This advances our understanding of Atx2 function beyond the information provided from the knockout mouse model by overcoming redundancy between mammalian Atx2 and Atx2-like, and by highlighting nervous system-specific effects of Atx2 loss. Neuron-specific depletion of Atx2 led to the loss of normal axon and dendrite formation in the central and peripheral nervous systems, smaller CNS (central nervous system) normalized to body size, and, ultimately locomotion deficits and death at the pupae stage. This is significant as Atx2 is typically studied as a neurodegenerative risk factor. However, our data highlight its physiological role at earlier developmental stages, yet independently of any roles in mitosis. Microtubule and actin dynamics are intricately linked with neurite outgrowth. Microtubules are crucial to transduce forces that are generated by molecular motors, which control neuronal polarization and morphology (Del Castillo et al., 2019b; Lin et al., 2012; Lu et al., 2013; Norkett et al., 2020; Roossien et al., 2014; Winding et al., 2016; Zheng et al., 2008). In neurons, in particular, actin dynamics have important consequences on development via growth cone formation and normal axon outgrowth (Lin et al., 1996; Papandréou and Leterrier, 2018). Incorrectly regulated actin dynamics may also impair the organization of the microtubule cytoskeleton. Actin is necessary to guide microtubules in the axon, at filipodia, and at the growth cone (Bradke and Dotti, 1999; Hahn et al., 2020; Korobova and Svitkina, 2008; Schaefer et al., 2008). Therefore, impaired microtubule and actin dynamics may contribute to the gross neurite extension defects we describe. Furthermore, correct cargo trafficking and delivery, mediated by microtubule tracks and the actin network, is essential for axon development and dendritic arborization (Guedes-Dias and Holzbaur, 2019; Norkett et al., 2016; Zheng et al., 2008). In turn, this impairment in organelle distribution could lead to the neuronal defects we observed with loss of Atx2 at early developmental stages.
It will be crucial to specify the mechanisms by which Atx2 can act as a major regulator of the neuronal cytoskeleton. While the mechanism is likely via translational control, protein–protein interaction remains a possibility. It is known that Atx2 does not associate directly with actin (Satterfield et al., 2002), yet Atx2 does associate with actin regulator profilin, another ALS risk factor which locates to stress granules (Figley et al., 2014; Ralser et al., 2005). However, as we show here, among the target mRNAs of Atx2 are actin and microtubule cytoskeleton components and molecular motors. These data demonstrate the ability of Atx2 to regulate multiple cytoskeletal networks. Atx2 tightly regulates RNP granules and is necessary for axon pathfinding (Bakthavachalu et al., 2018; Singh et al., 2020). It is essential for local translation for synaptic plasticity (McCann et al., 2011). Aberrant localization and translation of these transcripts could underlie the phenotypes herein reported. Further exploration of these, and our data will be invaluable in fully understanding the physiological roles of Atx2 in the neuron.
The data we present here address the physiological roles of Atx2 via a neuron-specific knockdown approach. Specifically, we study the effect of Atx2 depletion on the developing rather than mature CNS. These data complement studies of mutant, polyQ expanded Atx2 in neurodegeneration—widely believed to cause a toxic gain of function (Huynh et al., 2000). Our data are also essential to thoroughly understand the mechanism by which Atx2 knockdown ameliorates TDP-43 pathology in ALS (Becker et al., 2017). Therefore, our data are relevant to ALS disease mechanisms and therapies. Further, our use of Drosophila as a model organism circumvents the presence of Atx2-like/Ataxin2-related proteins in mammals. Curiously, recent reports of an Atx2-like knockout mouse demonstrate lethality at early developmental stages and impaired neural development as shown by thinner cortices (Key et al., 2020). These observations are consistent with our data, highlighting the conservation of roles of Atx2 and its related proteins across taxa.
Limitations of the study
Our data show that neuron-specific depletion of Atx2 in Drosophila impairs proper neurodevelopment. The experimental strategy used in this work to deplete Atx2 was to express shRNA targeting Atx2 driven by elav-Gal4. One limitation of this technique is that the elav promoter starts expressing the Atx2 shRNA at embryonic stage 9, after neuronal differentiation. Therefore, there is a pool of active Atx2 protein in the neurons at initial neurodevelopmental stages. We expect that the use of an alternative strategy, which more efficiently depletes Atx2, would result even in more severe neurodevelopment defects than the ones we report in this paper. Atx2 is an mRNA binding protein, thus it is most likely that the RNAi phenotypes are caused by misregulation at the transcriptional level. We performed the RNAseq analysis of Atx2 RNAi to address this question. However, we cannot rule out the potential for the phenotypes to be dependent on Atx2 interaction partners at the protein level. Combining RNAseq with biochemical approaches such as pulldowns or proximity labeling experiments will provide new insights into the understanding of how Atx2 controls neuronal development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti FasII mouse monoclonal | DSHB | Monoclonal 1D4 |
| Anti Acetylated alpha tubulin (K40) mouse monoclonal | protein tech | Cat# 66200 |
| Anti Hsc 70 K-19 goat polyclonal | Santa Cruz | Cat# sc-1059 |
| Anti Dmel Atx2 rabbit polyclonal | Li Antibodies custom preparation | N/A |
| Anti alpha tubulin DM1a mouse monoclonal | Prepared in house | N/A |
| Anti tubulin rb polyclonal | Prepared in house | N/A |
| HRP-conjugated anti mouse | Jackson | RRID: AB_2340770 |
| HRP-conjugated anti rabbit | Jackson | RRID: AB_10015282 |
| HRP-conjugated anti goat | Jackson | RRID: AB_2340390 |
| Alexafluor 647 conjugated anti mouse IgG | Jackson | RRID: AB_2340862 |
| Chemicals, peptides, and recombinant proteins | ||
| Latrunculin B | Sigma Aldrich | Cat#L5288 CAS number 76343-94-7 |
| Vinblastine | Sigma Aldrich | Cat#V1377 CAS number 143-67-9 |
| Rhodamine conjugated Phalloidin | Thermo Fisher Scientific | Cat#R415 |
| Effectene | Qiagen | Cat#301425 |
| Critical commercial assays | ||
| TruSeq stranded mRNA kit | Illumina | Cat#20020596 |
| Deposited data | ||
| RNA seq data | This paper | GSE185473 |
| Experimental models: Cell lines | ||
| S2R+ | DGRC | RRID:CVCL_A5UM |
| Experimental models: Organisms/strains | ||
| Drosophila melanogaster Ataxin-2 RNAi TRiP line #1 (Val20, second chromosome attP40 insertion targeting CDS 561-581) TRiP.HMS02726 | Bloomington Stock Centre (BDSC) | RRID:BDSC_44102 |
| Drosophila melanogaster Ataxin-2 RNAi TRiP line #2 (Val20, third chromosome attP2 insertion targeting Atx2 CDS 474-494) TRiP.HMS01392 | Bloomington Stock Centre (BDSC) | RRID:BDSC_36114 |
| Drosophila melanogaster elavP-gal4 | Bloomington Stock Centre (BDSC) | RRID:BDSC_8760 |
| Drosophila melanogaster ppk-CD4-tdTomato | Bloomington Stock Centre (BDSC) | RRID:BDSC_35844 |
| Drosophila melanogaster UASt-mCherry.mito.OMM | Bloomington Stock Centre (BDSC) | RRID:BDSC_66532 |
| Drosophila melanogaster Ubi-EB1-GFP | S. Rogers (Shimada et al., 2006) | |
| Drosophila melanogaster D42 gal4>UAS ANF GFP | A. Lim, W. Saxton (Lim et al., 2017) | |
| Drosophila melanogaster Nrv2-GFP | Yale FlyTrap database | ZCL2903 |
| Software and algorithms | ||
| Fiji | http://fiji.sc | Version 2.0.0-rc-69/1.52p |
| Graphpad Prism | https://www.graphpad.com/scientific-software/prism/ | Version 9.1.0 (216) |
| Nikon Elements | https://www.nikon.com/products/industrial-metrology/support/download/software/imgsfw/ | Version 4.00.07 |
| PANTHER GO | http://www.pantherdb.org | Version 16.0 |
| Recombinant DNA | ||
| pMT-EOS-alpha-Tubulin | This lab (Barlan et al., 2013). | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vladimir Gelfand (v.gelfand@northwestern.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Fly stocks and plasmids
Fly stocks and crosses were cultured on standard cornmeal food at room temperature based on the Bloomington Stock Center recipe. Fly stocks used in this study are listed in the key resources table. All experiments used Atx2 shRNA line BDSC #44012 unless otherwise stated. For tubulin photoconversion experiments, S2 cells were transfected with pMT-EOS-tubulin as described in Barlan et al. (Barlan et al., 2013).
Drosophila cell culture
DrosophilaS2 cells were maintained in Insect-Xpress medium (Lonza) at 25°C. These cells came directly from DGRC and their identity was not subsequently confirmed. Transfections were carried out with Effectene (Qiagen) according to the manufacturer’s instructions. dsRNA was added to cells on days 1 and 3 and imaging was carried out on day 5. For EB1 experiments a stable S2 cell line was created expressing EB1-GFP under control of the EB1 promoter in the pMT vector. Primary neuronal cultures were prepared as previously described (Norkett et al., 2020). Briefly, brains from 3rd instar larvae were dissected in dissection saline (9.9 mM HEPES pH7.5, 137 mM NaCl, 5.4 mM KCl, 0.17 mM NaH2PO4 0.22 mM KH2PO4, 3.3 mM glucose, 43.8 mM sucrose) and tissue was dissociated using liberase-TM (Roche) for one hour with rotation and gentle mechanical dissociation. Liberase solution was removed by centrifugation (300 g 5 minutes) and washing 3x with dissection saline. Cells were plated on ConA-coated glass coverslips at a density of 2-3 brains per 100ul per coverslip and maintained in Schneiders medium supplemented with 20% FBS, 5 μg/ml insulin, 100 μg/ml Pen-Strep, 50 μg/ml Gentamycin and 10 μg/ml Tetracycline.
Methods details
Development, locomotion and survival analysis of Drosophila larvae
To assess the development of the nervous system, Atx2 RNAi was driven by elav-gal4 in flies expressing endogenously GFP tagged Nrv2, labelling the CNS. Larvae for analysis were staged based on date of egg hatching and body size to ensure the same age animals were used in control and Atx2 RNAi conditions. No differences were noted between the groups for time taken for egg hatching and body size. For locomotion assays, 10 3rd Instar larvae were washed in water for 10s. These 10 larvae were then transferred, using a fine brush, to the center of a petri dish containing 2% agarose supplemented with apple juice and charcoal powder. They were allowed to crawl for 6 minutes recorded with a Nikon D750 DSLR camera. Videos are played at 40x speed. Experiments were done at room temperature in the light. Trajectories were automatically generated using the Temporal-Color Code plugin (Physics LUT) in Fiji. For survival assays, line 44012 (which we designated Atx2 shRNA line 1), homozygous adults were crossed with elav-gal4 flies and number of adults eclosing from pupae cases was counted. For line 36114 (designated Atx2 shRNA line 2), heterozygous adults were crossed with elav-gal4 flies and number of ‘non tubby’ adults eclosing from pupae cases. Elav-gal4 flies were used as control.
Immunostaining and western blotting
Antibodies used in this study are described in the key resources table. Brains dissected from third-instar larvae in 1 × PBS, were fixed in 4% (wt/wt) formaldehyde (methanol-free) in PBT (1 × PBS, 0.1% Triton X-100) for 20 min; brains were washed five times with PBTB (PBT + 0.2% bovine serum albumin) for 10 min and blocked in 5% (vol/vol) normal goat serum for 1 h. Samples were incubated with primary anti-Fasciclin II antibody (1:40) at 4°C overnight, washed and incubated with FITC conjugated anti-mouse secondary (1:200) for 2 h at room temperature (RT). Finally, samples were washed with PBT for 10 min five times before mounting in Mowiol.
For immunostaining of S2 cells and cultured neurons, cells were fixed 5 mins in methanol (S2 cells) or 1% glutaraldehyde and quenched in 2 mg/ml sodium borohydride (neurons). Cells were permeabilised and blocked in wash buffer (1% BSA, 0.1% Triton X-100 in TBS) for 30mins and incubated with primary antibody in wash buffer (Anti-Acetylated tubulin 1:100-1:250, DM1a 1:1000). Coverslips were washed 3x in wash buffer and incubated with secondary antibodies (1:200). For extraction, cells were incubated in extraction buffer (1% Triton X-100, 1uM taxol, 30% glycerol in BRB-80) for 2 minutes, then in extraction buffer with 1% glutaraldehyde for 2 minutes, then 1% glutaraldehyde in PBS for 3 minutes followed by reduction as above. For western blotting, dissected third instar larvae brains or S2 cell lysate were prepared in 1% SDS, boiled in laemmli buffer and analyzed by SDS-PAGE on 8% acrylamide gels. After electrophoresis, transfer onto nitrocellulose membrane was carried out and blocking was performed in 4% milk in PBS-T. Western blotting was performed using advansta western bright quantum substrate and Licor Imagequant system. To analyze acetylated tubulin levels, membranes were first incubated with anti-acetylated tubulin antibody and developed with HRP- conjugated mouse secondary. The membrane was stripped using stripping buffer (2% SDS, 0.625M Tris pH6.8, 0.8% beta-mercaptoethanol) for 30 minutes at 37°C, washed 2x 5 minutes in PBS-T and reblocked. Then membranes were incubated with anti-tubulin antibody and developed with HRP- conjugated rabbit secondary.
Microscopy and image analysis
To image DA neurons or S2 cells we used a Nikon Eclipse U2000 inverted microscope equipped with a Yokogawa CSU10 spinning disk head with a perfect focus system and a 40x 1.30 or 100x 1.45 oil immersion lens. Images were acquired using Evolve EMCCD (Photometrics) controlled by Nikon Elements 4.00.07 software. For EB1 imaging in S2 cells images were acquired at 1 fps. For organelle imaging in larvae, larvae were immobilised and frame acquisition was at 3 fps (dense core vesicles) or 0.5 fps (mitochondria). Fixed and immunostained CNS samples and neurons were imaged on a Nikon Ti2 inverted microscope equipped with Yokogawa CSU-W1 spinning disk driven by Nikon Elements 5.20 using a 20X or 100x 1.45 oil immersion lens and CMOS prime 95B sensor (Photometrics). Whole larvae were immobilized by heat treatment at 70°C for 1 minute and mounted in 100% glycerol. Samples were imaged with a 10x 0.30 dry lens and six fields of view were stitched together in Nikon Elements. For microtubule organization images, this set-up was used with a Live-SR (Gataca systems). Live imaging of lysosomes and EB1 comets in neurons was carried out at 1fps. For photoconversion experiments we applied 405 nm light from a light-emitting diode light source (89 North Heliophor) for 5 s, using an adjustable diaphragm to constrain light and illuminate only a specific region of interest. After photoconversion, images were collected every ten minutes for 30 minutes. Imaging of filopodia, organelle distribution in culture and mitochondrial trafficking was carried out on a Nikon Ti inverted microscope with a Hamamatsu Orca Flash sensor.
Image analysis was carried out in Fiji unless otherwise stated. Total neurite length in DA neurons was measured using the ‘curve tracing’ v.0.3.5 plugin for ImageJ (https://github.com/ekatrukha/CurveTrace) and Sholl analysis plugins (https://github.com/tferr/ASA/blob/Sholl_Analysis-4.0.1). Total axon length of DA neurons in the VNC was measured using the ‘curve tracing’. VNC area, length and body length were measured manually. Tubulin motile fraction was calculated as ‘photoconverted signal outside original photoconversion zone / original photoconverted signal’ at 30 mins after photoconversion. Numbers and motility of EB1 comets, mitochondria and lysosomes in culture or in vivo was carried out by generating kymographs and analyzed using the ‘kymobutler’ software (Jakobs et al., 2019). Motile organelles were defined as those that moved more than 2 μm over the course of the video. Comets that moved more than 1 μm over the course of the video were counted. EB1 comets in S2 cells were counted using Nikon elements analysis software (v5.20). Numbers of filopodia and organelles in varicosities were scored manually.
RNAseq
For RNAseq analysis, libraries from control or Atx2 RNAi 3rd instar larvae brains were prepared using TruSeq stranded mRNA kit (Illumina). 3 control and 3 Atx2 RNAi libraries were prepared, generating 6 samples. The stranded mRNA-seq was conducted in the Northwestern University NUSeq Core Facility. Briefly, total RNA samples were checked for quality using RINs generated from Agilent Bioanalyzer 2100. RNA quantity was determined with Qubit fluorometer. The Illumina TruSeq Stranded mRNA Library Preparation Kit was used to prepare sequencing libraries of high-quality RNA samples (RIN>7). The Kit procedure was performed without modifications. This procedure includes mRNA enrichment and fragmentation, cDNA synthesis, 3’ end adenylation, Illumina adapter ligation, library PCR amplification and validation. Illumina HiSeq 4000 NGS Sequencer was used to sequence the libraries with the production of single-end 50 bp reads. The quality of reads, in FASTQ format, was evaluated using FastQC. Reads were trimmed to remove Illumina adapters from the 3’ ends using cutadapt (Martin, 2011). Trimmed reads were aligned to the Drosophila melanogaster genome (BDGP6.93) using STAR (Dobin et al., 2013). Read counts for each gene were calculated using htseq-count (Anders et al., 2015) in conjunction with a gene annotation file for BDGP6.93 obtained from Ensembl (http://useast.ensembl.org/index.html). Normalization and differential expression were calculated using DESeq2 that employs the Wald test (Love et al., 2014). The cutoff for determining significantly differentially expressed genes was an FDR-adjusted p-value less than 0.05 using the Benjamini-Hochberg method (Data S1).
Following identification of significantly differentially expressed genes, the PANTHER Gene ontology database was used to identify cellular components over- and under-represented in our data set based on the Drosophila transcriptome. From the cytoskeleton and actin cytoskeleton nodes, genes were further sorted according to their protein class and those ascribed ‘cytoskeleton’ (PC00085) were identified for representation of differential expression (Table S1).
Quantification and statistical analysis
Statistical significance between two groups was determined using the unpaired, nonparametric Mann–Whitney U test. Data analyses were performed with Prism v6 (GraphPad Software). Statistical significance is presented as ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001. Data in Figure legends are presented as mean ± standard error.
Acknowledgments
We thank members of the Gelfand lab and Dr. M. Gelfand for helpful discussion. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Drs. A. Lim, W. Saxton and S. Rogers for Drosophila lines. This work was supported by the Northwestern University NUSeq Core Facility. Research reported in this study was supported by National Institute of General Medical Sciences Grants R01GM052111 and R35GM131752 (to V.I.G.).
Author contributions
Conceptualization, UdC, RN and VG; Methodology, UdC, RN; Formal Analysis, UdC, RN; Investigation, UdC, RN, AS, WL; Writing original draft, RN; Writing – review and editing, UdC, RN, VIG; Visualization, UdC, RN; Supervision, VIG; Funding Acquisition, VIG.
Declaration of interests
The authors have nothing to disclose.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103536.
Contributor Information
Urko del Castillo, Email: urd4002@med.cornell.edu.
Vladimir I. Gelfand, Email: vgelfand@northwestern.edu.
Supplemental information
Data and code availability
-
•
RNA-seq data is available at GEO, accession number GSE185473. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All data associated with this work are included in this article and the appendix.
References
- Akhmanova A., Steinmetz M.O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P.W. Microtubules and neuronal polarity: lessons from mitosis. Neuron. 1999;22:23–31. doi: 10.1016/s0896-6273(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Bakthavachalu B., Huelsmeier J., Sudhakaran I.P., Hillebrand J., Singh A., Petrauskas A., Thiagarajan D., Sankaranarayanan M., Mizoue L., Anderson E.N., et al. RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron. 2018;98:754–766.e4. doi: 10.1016/j.neuron.2018.04.032. [DOI] [PubMed] [Google Scholar]
- Barlan K., Lu W., Gelfand V.I. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 2013;23:317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basant A., Glotzer M. Spatiotemporal regulation of RhoA during cytokinesis. Curr. Biol. 2018;28:R570–R580. doi: 10.1016/j.cub.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L.A., Huang B., Bieri G., Ma R., Knowles D.A., Jafar-Nejad P., Messing J., Kim H.J., Soriano A., Auburger G., et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradke F., Dotti C.C. The role of local actin instability in axon formation. Science. 1999;283:1931–1934. doi: 10.1126/science.283.5409.1931. [DOI] [PubMed] [Google Scholar]
- Cheerambathur D.K., Prevo B., Chow T.L., Hattersley N., Wang S., Zhao Z., Kim T., Gerson-Gurwitz A., Oegema K., Green R., et al. The kinetochore-microtubule coupling machinery is repurposed in sensory nervous system morphogenesis. Dev. Cell. 2019;48:864–872.e7. doi: 10.1016/j.devcel.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia S.C., Perry G., Moreira P.I. Mitochondrial traffic jams in Alzheimer’s disease - pinpointing the roadblocks. Biochim. Biophys. Acta - Mol. Basis Dis. 2016;1862:1909–1917. doi: 10.1016/j.bbadis.2016.07.010. [DOI] [PubMed] [Google Scholar]
- De Vos K.J., Hafezparast M. Neurobiology of axonal transport defects in motor neuron diseases: opportunities for translational research? Neurobiol. Dis. 2017;105:283–299. doi: 10.1016/j.nbd.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo U., Lu W., Winding M., Lakonishok M., Gelfand V.I. Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr. Biol. 2015;25:200–205. doi: 10.1016/j.cub.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo U., Gnazzo M.M., Sorensen Turpin C.G., Nguyen K.C.Q., Semaya E., Lam Y., de Cruz M.A., Bembenek J.N., Hall D.H., Riggs B., et al. Conserved role for Ataxin-2 in mediating endoplasmic reticulum dynamics. Traffic. 2019;20:436–447. doi: 10.1111/tra.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo U., Norkett R., Gelfand V.I. Unconventional roles of cytoskeletal mitotic machinery in neurodevelopment. Trends Cell Biol. 2019;29:901–911. doi: 10.1016/j.tcb.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo U., Müller H.A.J., Gelfand V.I. Kinetochore protein Spindly controls microtubule polarity in Drosophila axons. Proc. Natl. Acad. Sci. U S A. 2020;117:12155–12163. doi: 10.1073/pnas.2005394117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C., Feinberg M., Peters A., Barbas H. Mitochondrial degeneration in dystrophic neurites of senile plaques may lead to extracellular deposition of fine filaments. Brain Struct. Funct. 2007;212:195–207. doi: 10.1007/s00429-007-0153-1. [DOI] [PubMed] [Google Scholar]
- Figley M.D., Bieri G., Kolaitis R.-M., Taylor J.P., Gitler A.D. Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J. Neurosci. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnazzo M.M., Uhlemann E.M.E., Villarreal A.R., Shirayama M., Dominguez E.G., Skop A.R. The RNA-binding protein ATX-2 regulates cytokinesis through PAR-5 and ZEN-4. Mol. Biol. Cell. 2016;27:3052–3064. doi: 10.1091/mbc.E16-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godena V.K., Brookes-Hocking N., Moller A., Shaw G., Oswald M., Sancho R.M., Miller C.C.J., Whitworth A.J., De Vos K.J. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber W.B., Jan L.Y., Jan Y.N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Guedes-Dias P., Holzbaur E.L.F. Axonal transport: driving synaptic function. Science. 2019;366:eaaw9997. doi: 10.1126/science.aaw9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D., Timofeev K., Salecker I. A step-by-step guide to visual circuit assembly in Drosophila. Curr. Opin. Neurobiol. 2011;21:76–84. doi: 10.1016/j.conb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Hahn I., Voelzmann A., Parkin J., Fuelle J., Slater P.G., Lowery L.A., Sanchez-Soriano N., Prokop A. Tau, XMAP215/Msps and Eb1 jointly regulate microtubule polymerisation and bundle formation in axons. BioRxiv. 2020;17:e1009647. doi: 10.1371/journal.pgen.1009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzler J.I., Simonovitch S.I., Albertson R.M., Weiner A.T., Nye D.M.R., Rolls M.M. Kinetochore proteins suppress neuronal microtubule dynamics and promote dendrite regeneration. Mol. Biol. Cell. 2020;31:2125–2138. doi: 10.1091/mbc.E20-04-0237-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh D.P., Figueroa K., Hoang N., Pulst S.M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- Jakobs M.A., Dimitracopoulos A., Franze K. Kymobutler, a deep learning software for automated kymograph analysis. Elife. 2019;8:e42288. doi: 10.7554/eLife.42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020;21:307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- Jolly A.L., Kim H., Srinivasan D., Lakonishok M., Larson A.G., Gelfand V.I. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc. Natl. Acad. Sci. 2010;107:12151–12156. doi: 10.1073/pnas.1004736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L.C., Van Bergeijk P., Lipka J., Keijzer N., Wulf P.S., Katrukha E.A., Akhmanova A., Hoogenraad C.C. Myosin-V opposes microtubule-based cargo transport and drives directional motility on cortical actin. Curr. Biol. 2013;23:828–834. doi: 10.1016/j.cub.2013.03.068. [DOI] [PubMed] [Google Scholar]
- Kaul N., Soppina V., Verhey K.J. Effects of α-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophys. J. 2014;106:2636–2643. doi: 10.1016/j.bpj.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J., Harter P.N., Sen N.E., Gradhand E., Auburger G., Gispert S. Mid-gestation lethality of atxn2l-ablated mice. Int. J. Mol. Sci. 2020;21:1–24. doi: 10.3390/ijms21145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl T.R., Nechiporuk A., Figueroa K.P., Keating M.T., Huynh D.P., Pulst S.M. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- Korobova F., Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A., Rechtsteiner A., Saxton W.M. Two kinesins drive anterograde neuropeptide transport. Mol. Biol. Cell. 2017;28:3542–3553. doi: 10.1091/mbc.E16-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., Espreafico E.M., Mooseker M.S., Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- Lin S., Liu M., Mozgova O.I., Yu W., Baas P.W. Mitotic motors coregulate microtubule patterns in axons and dendrites. J. Neurosci. 2012;32:14033–14049. doi: 10.1523/JNEUROSCI.3070-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Fox P., Lakonishok M., Davidson M.W., Gelfand V.I. Initial neurite outgrowth in Drosophila neurons is driven by kinesin-powered microtubule sliding. Curr. Biol. 2013;23:1018–1023. doi: 10.1016/j.cub.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Lakonishok M., Liu R., Billington N., Rich A., Glotzer M., Sellers J.R., Gelfand V.I. Competition between kinesin-1 and myosin-v defines drosophila posterior determination. Elife. 2020;9:e54216. doi: 10.7554/eLife.54216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011;17:10. [Google Scholar]
- McCann C., Holohan E.E., Das S., Dervan A., Larkin A., Lee J.A., Rodrigues V., Parker R., Ramaswami M. The ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. U S A. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Noordstra I., Liu Q., Nijenhuis W., Hua S., Jiang K., Baars M., Remmelzwaal S., Martin M., Kapitein L.C., Akhmanova A. Control of apico–basal epithelial polarity by the microtubule minus-end-binding protein CAMSAP3 and spectraplakin ACF7. J. Cell Sci. 2016;129:4278–4288. doi: 10.1242/jcs.194878. [DOI] [PubMed] [Google Scholar]
- Norkett R., Modi S., Birsa N., Atkin T.A., Ivankovic D., Pathania M., Trossbach S.V., Korth C., Hirst W.D., Kittler J.T. DISC1-dependent regulation of mitochondrial dynamics controls the morphogenesis of complex neuronal dendrites. J. Biol. Chem. 2016;291:613–629. doi: 10.1074/jbc.M115.699447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkett R., Del Castillo U., Lu W., Gelfand V.I. Ser/Thr kinase Trc controls neurite outgrowth in Drosophila by modulating microtubule-microtubule sliding. Elife. 2020;9:e52009. doi: 10.7554/eLife.52009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandréou M.J., Leterrier C. The functional architecture of axonal actin. Mol. Cell. Neurosci. 2018;91:151–159. doi: 10.1016/j.mcn.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Portran D., Schaedel L., Xu Z., Théry M., Nachury M.V. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell Biol. 2017;19:391–398. doi: 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M., Nonhoff U., Albrecht M., Lengauer T., Wanker E.E., Lehrach H., Krobitsch S. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum. Mol. Genet. 2005;14:2893–2909. doi: 10.1093/hmg/ddi321. [DOI] [PubMed] [Google Scholar]
- Riess O., Laccone F.A., Gispert S., Schöls L., Zühlke C., Menezes Vieira-Saecker A.M., Herlt S., Wessel K., Epplen J.T., Weber B.H.F., et al. SCA2 trinucleotide expansion in German SCA patients. Neurogenetics. 1997;1:59–64. doi: 10.1007/s100480050009. [DOI] [PubMed] [Google Scholar]
- Roossien D.H., Lamoureux P., Miller K.E. Cytoplasmic dynein pushes the cytoskeletal meshwork forward during axonal elongation. J. Cell Sci. 2014;127:3593–3602. doi: 10.1242/jcs.152611. [DOI] [PubMed] [Google Scholar]
- Satterfield T.F., Pallanck L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- Satterfield T.F., Jackson S.M., Pallanck L.J. A drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162:1687–1702. doi: 10.1093/genetics/162.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.W., Schoonderwoert V.T.G., Ji L., Mederios N., Danuser G., Forscher P. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell. 2008;15:146–162. doi: 10.1016/j.devcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y., Yonemura S., Ohkura H., Strutt D., Uemura T. Polarized transport of frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell. 2006;10:209–222. doi: 10.1016/j.devcel.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Sholl D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Singh A., Huelsmeier J., Kandi A.R., Pothapragada S.S., Hillebrand J., Petrauskas A., Agrawal K., Krishnan R.T., Thiagarajan D., VijayRaghavan K., et al. Antagonistic roles for Ataxin-2 structured and disordered domains in RNP condensation. BioRxiv. 2020;10:e60326. doi: 10.7554/eLife.60326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenvoll M.D., Medley J.C., Irwin M., Song M.H. ATX-2, the C. elegans ortholog of human ataxin-2, regulates centrosome size and microtubule dynamics. PLoS Genet. 2016;12:e1006370. doi: 10.1371/journal.pgen.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakaran I.P., Hillebrand J., Dervan A., Das S., Holohan E.E., Hülsmeier J., Sarov M., Parker R., VijayRaghavan K., Ramaswami M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E99–E108. doi: 10.1073/pnas.1309543111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A., Deaconescu A.M., Spector J., Goodman B., Valenstein M.L., Ziolkowska N.E., Kormendi V., Grigorieff N., Roll-Mecak A. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 2014;157:1405–1415. doi: 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees A.E. The lifecycle of the neuronal microtubule transport machinery. Semin. Cell Dev. Biol. 2020;107:74–81. doi: 10.1016/j.semcdb.2020.02.008. [DOI] [PubMed] [Google Scholar]
- Winding M., Kelliher M.T., Lu W., Wildonger J., Gelfand V.I. Role of kinesin-1–based microtubule sliding in Drosophila nervous system development. Proc. Natl. Acad. Sci. 2016;113:E4985–E4994. doi: 10.1073/pnas.1522416113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhou B., Lin M.Y., Wang S., Foust K.D., Sheng Z.H. Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron. 2015;87:355–370. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Cook C., Sauter C., Kuriyama R., Kaplan P.L., Baas P.W. Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J. Neurosci. 2000;20:5782–5791. doi: 10.1523/JNEUROSCI.20-15-05782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Oztan A., Ye Y., Schwarz T.L. Kinetochore proteins have a post-mitotic function in neurodevelopment. Dev. Cell. 2019;48:873–882.e4. doi: 10.1016/j.devcel.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wildonger J., Ye B., Zhang Y., Kita A., Younger S.H., Zimmerman S., Jan L.Y., Jan Y.N. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat. Cell Biol. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bar, 10 μm.
Scale bar, 10 μm.
Control (elav-gal4) and Atx2 RNAi (elav > Atx2 RNAi). Scale bar, 10 μm.
Scale bar, 10 μm.
Scale bar, 10 μm.
Scale bar, 10 μm.
Scale bar, 10 μm.
Data Availability Statement
-
•
RNA-seq data is available at GEO, accession number GSE185473. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All data associated with this work are included in this article and the appendix.