Abstract
An abridged prion protein (PrP) molecule of 106 amino acids, designated PrP106, is capable of forming infectious miniprions in transgenic mice (S. Supattapone, P. Bosque, T. Muramoto, H. Wille, C. Aagaard, D. Peretz, H.-O. B. Nguyen, C. Heinrich, M. Torchia, J. Safar, F. E. Cohen, S. J. DeArmond, S. B. Prusiner, and M. Scott, Cell 96:869–878, 1999). We removed additional sequences from PrP106 and identified a 61-residue peptide, designated PrP61, that spontaneously adopted a protease-resistant conformation in neuroblastoma cells. Synthetic PrP61 bearing a carboxy-terminal lipid moiety polymerized into protease-resistant, β-sheet-enriched amyloid fibrils at a physiological salt concentration. Transgenic mice expressing low levels of PrP61 died spontaneously with ataxia. Neuropathological examination revealed accumulation of protease-resistant PrP61 within neuronal dendrites and cell bodies, apparently causing apoptosis. PrP61 may be a useful model for deciphering the mechanism by which PrP molecules acquire protease resistance and become neurotoxic.
A wealth of evidence contends that the infectious pathogen causing the prion diseases, also referred to as spongiform encephalopathies, is solely comprised of PrPSc, the pathogenic isoform of the prion protein (21–23). Both PrPSc and its normal cellular counterpart, PrPC, are encoded by a cellular gene (2, 19). Physical and molecular characterization of PrPSc and PrPC has failed to reveal any chemical differences between the two isoforms (32). However, PrPSc acquires distinctive conformational characteristics upon its conversion from PrPC. Whereas PrPC is soluble in most detergents and can be easily digested by proteases, PrPSc is insoluble in detergents and maintains a protease-resistant core, designated PrP27-30, which polymerizes into amyloid (25). Spectroscopic studies show that PrPC contains 3% β-sheet and 42% α-helix, whereas PrPSc displays 43% β-sheet and 30% α-helix (20, 28). Recent structural nuclear magnetic resonance studies of recombinant PrP molecules provide evidence for three α-helices (residues 144 to 154, 172 to 193, and 200 to 227), two short β-strand regions (residues 129 to 131 and 161 to 163), and a relatively unstructured N terminus (residues 23 to 128) (4, 10, 15, 26, 27).
Detailed structural investigations of full-length PrPSc have been hampered by the insolubility, heterogeneity, and complexity of PrPSc molecules in preparations of infectious prions. These difficulties prompted us to identify the smallest fragment of PrP that can adopt conformations resembling PrPSc, propagate infectivity, and kill neurons. In its mature form, mouse PrP consists of 208 amino acids and possesses two carbohydrate side chains plus a glycophosphatidyl inositol (GPI) anchor. At this point, the smallest identified PrPSc molecule is a 106-amino-acid prion protein with both an N-terminal truncation (Δ23–88) and an internal deletion (Δ141–176), designated PrP106, which can form infectious miniprions containing protease-resistant PrPSc106 (33). Infectious PrP106 miniprions lack almost half of the amino acid residues present in full-length PrPSc but retain both glycosylation sites and the GPI anchor.
In order to identify more precisely the structural components of the PrP molecule responsible for the pathogenic properties of prions and to facilitate structural studies of PrPSc, we sought to design a miniprion even smaller than PrPSc106. Using PrP106 as a starting point, we extended the Δ141–176 internal deletion towards both the N and C termini. We tested these abridged PrP molecules for formation of protease-resistant conformers by expression in murine neuroblastoma cells. In this way, we identified a lipid-anchored 61-amino-acid PrP peptide, PrP61, which spontaneously adopted an insoluble, protease-resistant conformation. We then characterized the biophysical properties of synthetic PrP61 peptide and investigated the neuropathologic effects of PrP61 expression in transgenic mice.
MATERIALS AND METHODS
Explanation of nomenclature.
MHM2 is a full-length chimeric construct that differs from wild-type mouse PrP (MoPrP) at positions 108 and 111 (31). Substitution at these positions with the corresponding residues (109 and 112, respectively) from the Syrian hamster PrP (SHaPrP) sequence creates an epitope for the anti-PrP 3F4 monoclonal antibody (MAb) (13), which does not recognize wild-type MoPrP and hence facilitates specific detection of the transgene by Western blotting.
Mature MHM2 and MoPrP comprise residues 23 to 230 after processing, because residues 1 to 22 are removed by a signal peptidase and residues 231 to 254 are removed during addition of a GPI anchor. PrP106 refers to a truncated MHM2 molecule in which residues 23 to 88 and 141 to 176 have been removed. PrP106 can also be designated MHM2(Δ23–88,Δ141–176). PrP61 is MHM2 (Δ23–88,Δ141–221).
Construction of DNA plasmids and transgenic (Tg) mice.
Most of the new constructs described in this report were created by standard cassette mutagenesis of PrP cDNA constructs using oligonucleotides purchased from Gibco-BRL. Silent site mutagenesis was used to produce an AvrII cloning site to create deletions starting at residue 127. A similar strategy using BstEII, MluI, and StuI was used in conjunction with XbaI to create deletions ending at residues 186, 205, and 221, respectively. DNA sequencing (Perkin-Elmer, Foster City, Calif.) with T7 and SP6 primers was used to verify the sequence of every new insert. The mutagenized PrP inserts were removed from psp72 by digestion with BglII/XhoI and subcloned into BamHI/XhoI-digested pSPOX.neo vector (31) to create pSPOX N2a cell expression plasmids. Qiagen maxiprep columns were used to purify pSPOX expression plasmids for transfection experiments.
CosTet(PrP61) cosmid was generated in a two-step process from pSPOX[MHM2(Δ23–88,Δ141–221)]. First, a 400-bp KpnI/XhoI insert from pSPOX[MHM2 (Δ23–88,Δ141–221)] was ligated into KpnI/XhoI-digested psp72(SalI)MHM2 vector (31). Second, the SalI/XhoI PrP insert from the modified psp72(SalI)PrP construct was cloned into SalI-digested cosSHa.Tet. Microinjection, breeding, and screening of Tg animals was performed as previously described (30).
Expression in neuroblastoma cells and preparation of brain homogenates.
Stock cultures of N2a and ScN2a cells were maintained in minimal essential medium with Earle's salts plus 10% fetal bovine serum, 10% Glutamax (Gibco BRL), 100 U of penicillin, and 100 μg of streptomycin per ml. Cells from a single confluent 100-mm dish were trypsinized and split into 10 separate 60-mm dishes containing Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum, 10% Glutamax, 100 U of penicillin, and 100 μg of streptomycin per ml (supplemented DMEM) 1 day prior to transfection. For each construct, 15 μg of DNA was resuspended in 150 μl of sterile HEPES-buffered saline on the day of transfection. The DNA solution was then mixed with an equal volume of 333 μg of DOTAP (Boehringer Mannheim, Indianapolis, Ind.) per ml in HEPES-buffered saline in Falcon 2059 tubes and was incubated at room temperature for 10 min to allow formation of DNA-lipid complexes. Supplemented DMEM (2.5 ml) was added to the mixture, and this was then pipetted onto drained cell monolayers. The following day, the medium containing DNA-lipid complexes was removed and replaced with fresh supplemented DMEM.
Three days after transfection, cells were harvested by lysis in 1.2 ml of 20 mM Tris buffer (pH 8.0) containing 100 mM NaCl, 0.5% NP-40, and 0.5% deoxycholate. Nuclei were removed from the lysate by centrifugation at 2,000 rpm for 5 min. This lysate typically had a total protein concentration of 0.5 mg/ml measured by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). For samples not treated with proteinase K, 40 μl of whole lysate (representing 20 μg of total protein) was mixed with 40 μl of 2× sodium dodecyl sulfate (SDS) sample buffer. For proteinase K digestion, 1 ml of lysate was incubated with either (i) 20 μg of proteinase K (total protein/enzyme ratio, 25:1) per ml for 1 h at 37°C or (ii) 7 μg of proteinase K (total protein/enzyme ratio, 71:1) per ml for 30 min at 37°C, depending on the experimental paradigm. Proteolytic digestion was terminated by the addition of 8 μl of 0.5 M phenymethylsulfonyl fluoride in absolute ethanol. Samples were then centrifuged for 75 min in a Beckman TLA-45 rotor (Fullerton, Calif.) at 100,000 × g at 4°C. The pellet was resuspended by repeated pipetting in 80 μl of 1× SDS sample buffer. The entire sample (representing 0.5 mg of total protein before digestion) was boiled for 5 min and cleared by centrifugation for 1 min at 14,000 rpm in a Beckman ultrafuge. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in 1.5-mm 15% polyacrylamide gels (14) or 16% Tricine gels (Novex) as indicated.
Brain homogenates (10% [wt/vol]) in sterile phosphate-buffered saline (PBS) were prepared by repeated extrusion through syringe needles of successively smaller size, as previously described (29). Homogenates were adjusted to 1 mg of protein per ml in 100 mM NaCl–1 mM EDTA–0.5% sodium deoxycholate–0.5% Triton X-100–50 mM Tris-HCl (pH 7.5). Proteinase K (Boehringer Mannheim) was added to 0.5 ml of adjusted homogenate to achieve a ratio of total protein to enzyme of 50:1. After incubation at 37°C for 1 h, proteolytic digestion was terminated by the addition of 8 μl of 0.5 M phenylmethylsulfonyl fluoride in absolute ethanol. Samples were then centrifuged for 75 min in a Beckman TLA-45 rotor at 100,000 × g at 4°C. The pellet was resuspended by repeated pipetting in 80 μl of 1× SDS sample buffer. Undigested samples were prepared by mixing equal volumes of adjusted homogenate and 2× sample buffer.
Western blotting.
Following electrophoresis, Western blotting was performed as previously described (29). Membranes were blocked with 5% nonfat milk protein in PBST (calcium- and magnesium-free PBS plus 0.1% Tween 20) for 1 h at room temperature. Blocked membranes were incubated with primary 3F4 MAb at a 1:5,000 dilution in PBST overnight at 4°C. Following incubation with primary antibody, membranes were washed 3× for 10 min in PBST, incubated with horseradish peroxidase-labeled anti-mouse immunoglobulin G secondary antibody (Amersham Life Sciences, Arlington Heights, Ill.) diluted 1:5,000 in PBST for 25 min at room temperature, and washed again 3× for 10 min in PBST. After chemiluminescent development with ECL reagent (Amersham) for 1 to 15 min, blots were sealed in plastic covers and exposed to ECL Hypermax film (Amersham). Films were processed automatically in a Konica film processor.
Peptide synthesis.
The peptides were synthesized on Rink Amide MBHA resin (Novabiochem, La Jolla, Calif.) using an automated Applied Biosystems (Perkin-Elmer) 433A synthesizer. All amino acids and resins were purchased from Novabiochem, and all other reagents and solvents were obtained from Perkin-Elmer. Highly optimized-fluorenylmethoxy carbonyl (Fmoc) chemical protocols were used based on previously described procedures (1a) with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and hydroxybenzotriazole activation, and a capping procedure was performed with N-(2-chlorobenzyloxycarbonyloxy) succinimide (1b). All syntheses were performed at 0.25-mmol scales, and single coupling cycles were used throughout. A lysine residue, orthogonally protected with a methyl trityl group, was added at the C terminus to allow the incorporation of myristic acid. At the end of the synthesis and before removal of the N-terminal Fmoc-protecting group, the peptidyl resin was treated with 1% trifluoroacetic acid (TFA) in dichloromethane. The resulting TFA salt was neutralized with 5% diisopropylethylamine in dichloromethane. Myristic acid was preactivated as the mixed anhydride with dicyclohexylcarbodiimide and hydroxybenzotriazole before being coupled to the deprotected peptidyl resin. After overnight coupling, the resin was washed to remove excess reagents before removal of the Fmoc group with 20% piperidine in N-methyl pyrrolidone.
Deprotection of the peptide and its cleavage from the resin was achieved using 95% TFA containing the scavengers ethandithiol, thioanisole, and thiophenol (1:2:2). The cleavage reaction was performed at room temperature for 4 h. The cleaved peptide was precipitated in dry diethyl ether, centrifuged down, and allowed to air dry.
Purification of the crude peptides was performed on either C4 or C18 Vydac (Hesperia, Calif.) semipreparative reversed-phase high-pressure liquid chromatography columns (250 by 10 mm), which were equilibrated at 50°C using a Rainin (Varian, San Jose, Calif.) 200 system equipped with a single wavelength detector. Fractions were analyzed on Vydac C4 or C18 analytical columns (150 by 4.6 mm). Separations were achieved using linear gradients of 0 to 100% solvent B for 180 or 30 min at a flow rate of either 3 or 1 ml/min, respectively. Solvent A was water–0.045% TFA, and solvent B was acetonitrile–0.036% TFA. Detection was at 220 nm. The identity of the peptides was confirmed by electrospray mass spectrometry using a Sciex (Perkin-Elmer) model 300 mass spectrometer.
Bioassay for prion infectivity.
Purified peptides were lyophilized to remove TFA and acetonitrile and were dissolved at a concentration of 1 mg/ml in sterile PBS without calcium or magnesium plus 5 mg of bovine serum albumin per ml. Ten-percent brain homogenates in PBS were prepared by repeated extrusion through syringe needles of successively smaller size, from 18 to 22 gauge. New, sterile, individually packaged needles, syringes, and tubes were used. All work was carried out in laminar flow hoods to avoid cross-contamination. Mice of either sex aged 7 to 10 weeks were inoculated intracerebrally with 30 μl of either 1 mg of peptide per ml or 1% brain homogenate in calcium- and magnesium-free PBS plus 5 mg of bovine serum albumin per ml. Inoculation was carried out with a 27-gauge disposable hypodermic needle inserted into the right parietal lobe. After inoculation, mice were examined daily for neurological dysfunction. Standard diagnostic criteria were used to identify animals affected by scrapie (1c, 24). In each group, some animals whose deaths were imminent were sacrificed, and their brains were removed for histologic and biochemical analysis.
Circular dichroism spectroscopy.
Spectra were collected at room temperature with a spectropolarimeter using a 0.1-cm path length. Spectra of 5 scans for each protein were accumulated, and buffer spectra obtained under identical conditions were subtracted.
Fourier transform infrared spectroscopy.
SHa synPrP61-Kma was dissolved at 2 mg/ml in 20 mM Na acetate (pH 5.5)–150 mM NaCl in D2O. Fourier transform infrared resonance (FTIR) analysis was performed as previously described (35). Samples without addition of NaCl polymerized upon dissolution in D2O-containing buffer. This isotope effect precluded the analysis of nonpolymerized peptide by FTIR.
Negative-stain electron microscopy.
Negative staining was done on carbon-coated 600-mesh copper grids which were glow-discharged prior to staining. Five-microliter samples were adsorbed for up to 1 min and then were stained with freshly filtered 2% ammonium molybdate. After being dried, the samples were viewed with a Jeol JEM 100CX II electron microscope at 80 kV at a standard magnification of ×40,000. The magnification was calibrated using negatively stained catalase crystals.
Congo red dye binding assay.
SHa synPrP61-Kma polymers were spun down from 20 mM Na acetate (pH 5.5)–150mM NaCl at 100,000 × g for 1 h at 20°C (Beckman Optima ultracentrifuge and Rotor TLA55). The pellet was washed with phosphate buffer (100 mM Na phosphate [pH 7.4]–150 mM NaCl) and spun down again for 15 min. The pellet was then stained with 100 μM Congo red dye in phosphate buffer for 1.5 h on an end-over-end rotator. After being spun down again for 15 min, the pellet was washed once each with phosphate buffer and H2O. After the final spin, the pellet was resuspended in a small volume of H2O, placed on a glass slide, and air dried. Samples were examined with a Leica microscope equipped with a set of polarization filters.
RESULTS
A 61-amino acid PrP peptide spontaneously adopts a protease-resistant conformation in neuroblastoma cells.
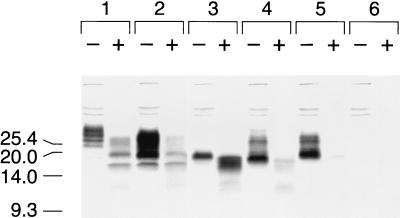
We previously reported the unexpected finding that PrP106 exhibits a moderate degree of protease resistance when expressed in uninfected N2a neuroblastoma cells (33). Furthermore, the relative protease resistance of three PrP deletion mutant polypeptides expressed in N2a cells, MHM2(Δ23–88,Δ141–155), MHM2(Δ23–88,Δ141–164), and MHM2(Δ23–88,Δ141–176) (PrP106), increased as the internal deletion increased in size (33). In contrast, neither MHM2(Δ23–88,Δ127–164) nor MHM2(Δ23–88,Δ127–176) was resistant to relatively mild proteinase K digestion for 30 min at 37°C using a total protein/enzyme ratio of 71:1 in either N2a or scrapie-infected ScN2a cells (Fig. 1).
FIG. 1.
Expression of PrP deletion constructs in scrapie-infected neuroblastoma cells. Scrapie-infected (ScN2a) cells were transfected transiently with the pSPOX expression vector carrying modified PrP genes as noted below. Lane 1, MHM2; lane 2, MHM2(Δ23–88); lane 3, MHM2(Δ23–88,Δ141–176); lane 4, MHM2(Δ23–88,Δ127–176); lane 5, MHM2(Δ23–88,Δ127–164); lane 6, mock transfection. Minus symbols denote undigested control sample, and plus symbols designate the pellet fraction of sample subjected to limited proteolysis with 7 μg of proteinase K per ml for 30 min at 37°C, corresponding to a total protein/proteinase K ratio of 71:1. Units are apparent molecular sizes based on migration of protein standards in kilodaltons.
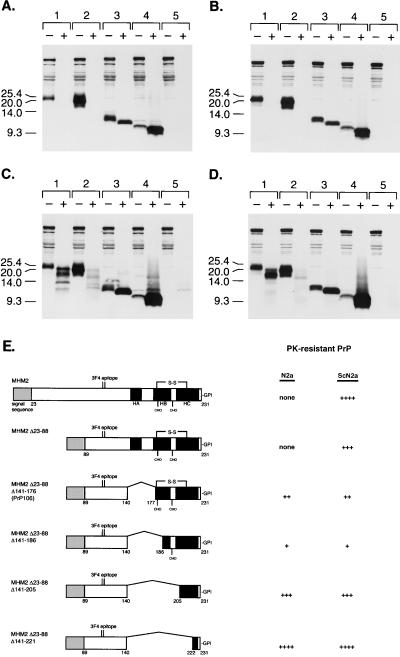
From these data, we hypothesized that residues 89 to 140 might represent a core which manifests inherent protease resistance when PrP structure is destabilized by removal of C-terminal sequences. We therefore designed a new series of PrP deletion mutants in which the region 89 to 140 was attached directly to GPI-anchored C termini of varying lengths. When these mutant PrPs were expressed in N2a cells and subjected to stringent proteinase K digestion for 1 h at a total protein/enzyme ratio of 25:1, neither PrP106 nor MHM2(Δ23–88,Δ141–186,C213A) displayed significant protease resistance (Fig. 2A, lane pairs 1 and 2). However, the shorter molecules MHM2(Δ23–88,Δ141–205,C213A) and MHM2(Δ23–88,Δ141–221) were both resistant to this stringent level of proteinase K digestion (Fig. 2A, lane pairs 3 and 4). Under milder digestion conditions, it became apparent that MHM2(Δ23–88,Δ141–186,C213A) is far more protease sensitive than PrP106 (Fig. 2C, lane pairs 1 and 2), whereas MHM2(Δ23–88,Δ141–221) appears to be slightly more protease resistant than MHM2(Δ23–88,Δ141–205,C213A) (Fig. 2C, lane pairs 3 and 4). Since formation of these protease-resistant conformers occurred spontaneously and did not require preexisting prions, we were not surprised to find that similar results were obtained when the same constructs were expressed in scrapie-infected ScN2a cells (Fig. 2B and D). The results are summarized schematically in Fig. 2E.
FIG. 2.
Expression of PrP deletion constructs in control and scrapie-infected neuroblastoma cells. Control (N2a) or scrapie-infected (ScN2a) cells were transfected transiently with the pSPOX expression vector carrying modified PrP genes as noted below. (A) Uninfected N2a cells digested with 20 μg of proteinase K per ml for 1 h at 37°C; (B) infected ScN2a cells digested with 20 μg of proteinase K per ml for 1 h at 37°C; (C) uninfected N2a cells digested with 7 μg of proteinase K per ml for 30 min at 37°C; (D) infected ScN2a cells digested with 7 μg of proteinase K per ml for 30 min at 37°C. Minus symbols denote undigested control sample, and plus symbols designate the pellet fraction of sample subjected to limited proteolysis as specified above. Samples that were digested with either 7 or 20 μg of proteinase K per ml correspond to a total protein/proteinase K ratio of 71:1 or 25:1, respectively. SDS-PAGE was performed on 16% Tricine gels (Novex). Western blotting was performed with 3F4 MAb as described in Materials and Methods. Paired sample lanes are numbered as follows: lane 1, MHM2(Δ23–88,Δ141–176), referred to as PrP106; lane 2, MHM2(Δ23–88,Δ141–186,C213A); lane 3, MHM2(Δ23–88,Δ141–205,C213A); lane 4, MHM2(Δ23–88,Δ141–221); lane 5, mock transfection. Units are apparent molecular sizes based on migration of protein standards in kilodaltons. (E) Schematic comparison of MHM2(Δ23–88) deletion mutants. Darkened areas correspond to α-helices determined in the nuclear magnetic resonance structure of PrP90 to -231 (10). None, no fragments detected with MAb 3F4 after digestion with 7 μg of proteinase K per ml for 30 min at 37°C; ++, fragments detected after digestion with 7 μg of proteinase K per ml for 30 min at 37°C but not after digestion with 20 μg of proteinase K per ml for 1 h at 37°C; ++++, intense 3F4 immunoreactive fragments seen even after digestion with 20 μg of proteinase K per ml for 1 h at 37°C. HA, α-helix A; HB, α-helix B; HC, α-helix C; CHO, carbohydrate; S-S, disulfide bond.
Synthetic myristylated PrP61 peptide forms amyloid in vitro.
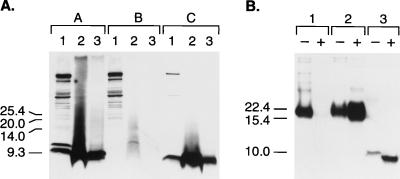
Because deletion of the two C-terminal helices is accompanied by loss of the N-linked glycosylation sites, we reasoned that we might be able to chemically synthesize an analog of MHM2(Δ23–88,Δ141–221), which we designated PrP61. To do this, we synthesized a PrP61 peptide with an additional lysine residue at the C terminus. Inclusion of this lysine allowed us to chemically attach a myristic acid moiety to the C terminus in an effort to mimic the naturally occurring GPI anchor. The resulting synthetic molecule, synPrP61-Kma, was then mixed with a crude lysate of untransfected N2a cells and subjected to proteinase K digestion. Since the myristyl group is not identical in mass to the naturally occurring GPI anchor, the mobility of undigested synPrP61-Kma was slightly faster than the mobility of undigested MHM2(Δ23–88,Δ141–221) expressed in N2a cells (Fig. 3A, compare lanes A1 and C1). Nonetheless, it was apparent that under both mild and stringent conditions of protease digestion, synPrP61-Kma was insoluble and displayed a resistance to protease digestion similar to that of gene-encoded MHM2(Δ23–88,Δ141–221) expressed in N2a cells (Fig. 3A, compare lanes C2 and C3 with lanes A2 and A3).
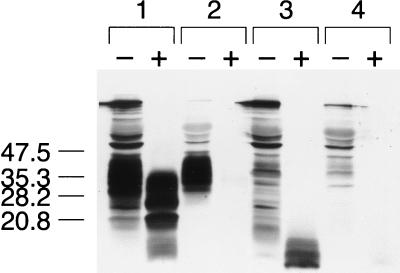
FIG. 3.
Protease digestion of MHM2(Δ23–88,Δ141–221). (A) Lanes A1 through A3, N2a cells transfected with pSPOX[MHM2(Δ23–88,Δ141–221)]; lanes B1 through B3, mock-transfected N2a cells; lanes C1 through C3, 0.4 μg of myristylated, synthetic MHM2(Δ23–88,Δ141–221) peptide per ml mixed with untransfected N2a cell lysate (0.5 mg of total protein per ml). Lanes A1, B1, and C1, untreated whole-cell lysates; lanes A2, B2, and C2, pellet fractions of lysates digested with 7 μg of proteinase K per ml for 30 min at 37°C; lanes A3, B3, and C3, pellet fractions of lysates digested with 20 μg of proteinase K per ml for 1 h at 37°C. SDS-PAGE was performed on 16% Tricine gels (Novex). Western blotting was performed with 3F4 MAb at 1:5,000 dilution. After processing, lanes 1 and 2 were exposed for 1 min and lane 3 was exposed for 15 min to Hypermax film (Amersham Life Sciences). Units are apparent molecular sizes based on migration of protein standards in kilodaltons. (B) Proteinase K digestion of brain homogenates from Tg mice. Lane 1, 60-day-old, uninoculated Tg(PrP106)Prnp0/0 mouse; lane 2, 65-day-old, scrapie-infected Tg(PrP106)Prnp0/0 mouse; lane 3, 48-day-old, spontaneously ataxic Tg(PrP61)Prnp0/0 mouse. Minus symbols denote undigested control sample, and plus symbols designate the pellet fraction of sample subjected to digestion with 20 μg of proteinase K per ml for 1 h at 37°C. Units are apparent molecular sizes based on migration of protein standards in kilodaltons.
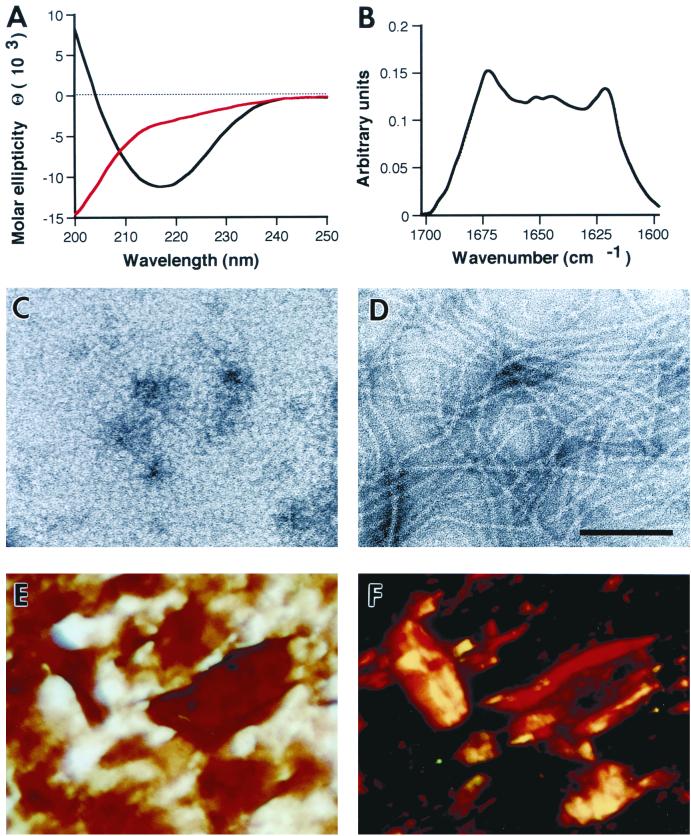
We then explored the biophysical characteristics of pure synPrP61-Kma. Using circular dichroism and FTIR spectroscopy, we observed that synPrP61-Kma spontaneously adopted a predominantly β-sheet structure at a physiological salt concentration of 150 mM NaCl (Fig. 4A, black line, and B). In contrast, acetylated synPrP61-Kac lacking the myristyl group folded into a random coil structure (data not shown), suggesting that the lipid attachment was required to achieve this β-sheet structure. When examined by negative-stain electron microscopy, synPrP61-Kma suspended in 150 mM NaCl polymerized into 5- to 10-nm fibrils (Fig. 4D). These fibrils bound the Congo red dye (Fig. 4E and F) and exhibited yellow-green birefringence when examined by polarized light, indicating that they had formed amyloid rods. In contrast to its amyloidogenic behavior at physiological salt concentrations, synPrP61-Kma failed to polymerize (Fig. 4C) and adopted a random coil structure (Fig. 4A, red line) at low ionic strength.
FIG. 4.
Characterization of SHa synPrP61-Kma polymers. (A) Far-UV circular dichroism spectra of SHa synPrP61-Kma in 20 mM Na acetate, pH 5.5, with (black line) or without (red line) addition of 150 mM NaCl. (B) FTIR spectrum of SHa synPrP61-Kma in 20 mM Na acetate (pD 5.5)–150 mM NaCl in D2O. (Transfer into D2O-containing buffers resulted in peptide polymerization without the addition of salt.) Deconvolution of the spectrum yielded estimates of 25% α-helix and 41% β-sheet. (C and D) Negative-stain electron micrographs of SHa synPrP61-Kma in 20 mM Na acetate, pH 5.5, with (D) or without (C) addition of 150 mM NaCl; each was negatively stained with 2% ammonium molybdate. Bar = 100 nm. (E and F) Congo red dye binding assay on SHa synPrP61-Kma in 20 mM Na acetate (pH 5.5)–150 mM NaCl. Bright-field illumination (E) and crossed polarizers (F) are shown.
Because the biophysical properties of synPrP61-Kma resemble PrPSc, we investigated whether PrP61 peptides could initiate an infectious prion disease in rodents. We intracerebrally inoculated Syrian hamsters, CD-1 mice, Tg(SHaPrP)Prnp0/0 mice, Tg(MoPrP) mice, Tg(MoPrP P101L)Prnp0/0 mice, and Tg(PrP106)Prnp0/0 mice with a single 30-μg dose of synPrP61-Kma containing either Syrian hamster, mouse, or mouse P101L sequences. However, no animals have developed prion disease >300 days after inoculation (Table 1).
TABLE 1.
Bioassays in wild-type rodents and Tg mice expressing wild-type and mutant PrP molecules
| Host | Inoculum | Age of onset or incubation period (days ± SEM) | n/n0a |
|---|---|---|---|
| Syrian hamsters | None | >300 | 0/10 |
| Sc237 hamster prions | 74 ± 1 | 52/56 | |
| SHa synPrP61-Kma peptide | >300 | 0/6 | |
| Tg(SHaPrP)3922HOZ/Prnp0/0 mice | None | >500 | 0/10 |
| Sc237 hamster prions | 45 ± 2 | 8/8 | |
| SHa synPrP61-Kma peptide | >500 | 0/10 | |
| CD-1 mice | None | >500 | 0/10 |
| RML murine prions | 132 ± 1 | 29/29 | |
| Mo synPrP61-Kma peptide | >410 | 0/10 | |
| Mo synPrP61(P101L)-Kma peptide | >410 | 0/9 | |
| Tg(MoPrP-A)4053/Prnp0/0 mice | None | >700 | 0/20 |
| RML murine prions | 50 ± 2 | 16/16 | |
| Mo synPrP61-Kma peptide | >500 | 0/10 | |
| Mo synPrP61(P101L)-Kma peptide | >500 | 0/5 | |
| Tg(MoPrP P102L)196/Prnp0/0 mice | None | 613 ± 34 | 9/32 |
| RML murine prions | 229 ± 6 | 6/6 | |
| Mo synPrP61-Kma peptide | >50∗ | 0/10 | |
| Mo synPrP61(P101L)-Kma peptide | >410 | 0/8 | |
| Tg(PrP106)4290/Prnp0/0 mice | None | >500 | 0/7 |
| RML murine prions | 300 ± 22 | 8/8 | |
| PrP106 murine prions | 66 ± 3 | 40/40 | |
| Mo synPrP61-Kma peptide | >450 | 0/9 | |
| Mo synPrP61(P101L)-Kma peptide | >450 | 0/9 |
n, number of mice that developed disease; n0, number of inoculated mice. An asterisk represents an ongoing experiment.
Tg mice expressing PrP61 develop spontaneous neurodegeneration.
Since PrP61 shares many of the biochemical and physical properties of PrPSc, we expressed MHM2(Δ23–88,Δ141–221) in Tg mice lacking endogenous PrP (Prnp0/0). We were unable to identify any founder animals expressing high levels of PrP61, presumably because these animals died in utero. However, three separate founder mice were identified that expressed PrP61 at levels lower than 1× normal Syrian hamster PrP. All three of these mice spontaneously developed ataxia at 21, 48, and 120 days of age, respectively. These Tg(PrP61)Prnp0/0 mice developed rapidly progressive neurological disease, and all three animals died within one to two days of the onset of ataxia. Neuropathological examination revealed a loss of pyramidal cells in the proximal two-thirds of the CA1 region of the hippocampus, degenerating neurons in the distal one-third of CA1 (Fig. 5D) and scattered necrotic neurons throughout the cerebral cortex and thalamus. The dead cells were easily recognized by their extremely shrunken and dark nuclei. Neuronal cell loss was accompanied by marked astrocytic gliosis (Fig. 5B), punctate accumulation of PrP-immunoreactive deposits within dendrites and cell bodies (Fig. 5C), and positive in situ staining for apoptosis (Fig. 5D). The PrP deposits did not bind Congo red (results not shown), suggesting that PrP61 molecules do not form mature amyloid when expressed in Tg mice. Nonetheless, when analyzed biochemically, PrP61 in the brains of Tg(PrP61)Prnp0/0 mice was insoluble and was resistant to digestion by proteinase K (Fig. 3B). Homogenates prepared from spontaneously sick Tg(PrP61)Prnp0/0 mice containing protease-resistant PrP61 have not transmitted the disease to Tg(PrP106)Prnp0/0 mice (which do not develop spontaneous disease) >200 days after inoculation.
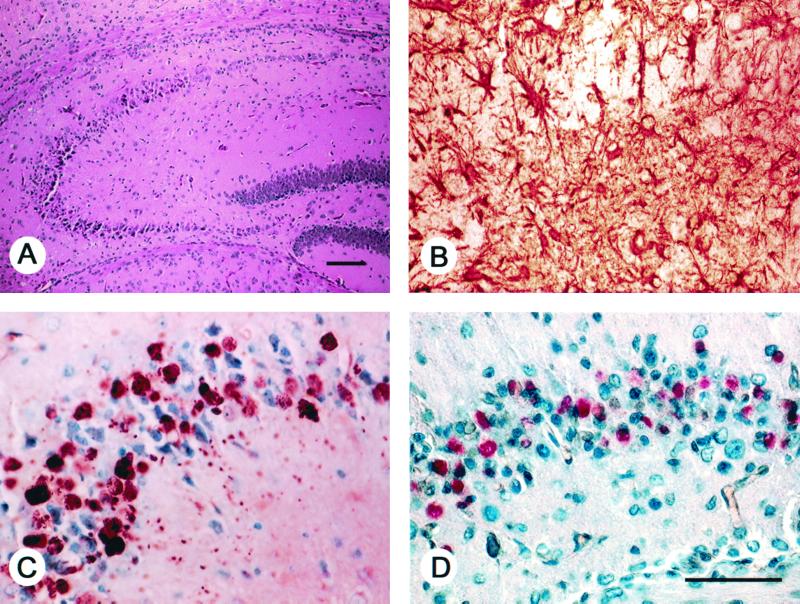
FIG. 5.
Neuropathological changes in ataxic Tg(PrP61) mice. Nerve cell loss and degeneration in the hippocampus of a spontaneously sick, 21-day-old Tg(PrP61)Prnp0/0 mouse. Bars, 100 μm (A) and 50 μm (D). Panels B through D are identical in magnification. (A) Hematoxylin and eosin stain shows loss of pyramidal cells in the proximal CA1 region and degenerating neurons in the distal CA1 region. (B) Anti-glialacidic fibrillary protein immunostain of the distal CA1 region shows intense reactive astrocytic gliosis. (C) Intraneuronal cytoplasmic PrP61 inclusions in neurons of the distal CA1 region revealed by the hydrolytic autoclaving method for protease-resistant PrP using MAb 3F4 as previously described (33). (D) In situ end labeling utilizing the Apoptag kit (Intergen) as previously described (1). Positive labeling of cells is suggestive of apoptosis. No apoptosis was seen in sections from age-matched, non-Tg Prnp0/0 mice.
To assess whether the neurotoxic effects of PrP61 could be suppressed by coexpression of full-length PrP, we generated Tg(PrP61)Prnp+/+ mice by injecting CosTet(PrP61) DNA into wild-type FVB mouse oocytes. Three separate Tg(PrP61)Prnp+/+ founders developed ataxia and died between 20 and 60 days of age with neuropathology similar to that seen in Tg(PrP61)Prnp0/0 mice. These results indicate that the PrP61-induced neurotoxicity cannot be prevented by coexpression of full-length MoPrP.
PrP61 is purged from ScN2a cells by exposure to PPI.
ScN2a cells can be selectively purged of PrPSc by treatment with branched polyamines, which render PrPSc protease sensitive (34). To determine whether protease-resistant PrP61 would be similarly eliminated from cells by exposure to a branched polyamine, we incubated ScN2a cells expressing PrP61 with polypropyleneimine (PPI) generation 4.0 for 4 h. Exposure to PPI purged the ScN2a cells of protease-resistant PrP61 in a manner similar to that for bona fide PrPSc (Fig. 6).
FIG. 6.
Treatment of ScN2a cells with PPI. ScN2a cells were transfected transiently with the pSPOX expression vector encoding either the full-length MHM2 or the MHM2(Δ23–88,Δ141–221) sequence. Three days after transfection cells were incubated with either control medium or medium plus 150 μg of PPI generation 4.0 per ml for 4 h. Cells were then harvested and lysed as described in Materials and Methods. Minus symbols denote undigested control lysate, and plus symbols designate the pellet fraction of lysate subjected to limited proteolysis with 20 μg of proteinase K per ml for 1 h at 37°C, corresponding to a total protein/proteinase K ratio of 25:1. Units are apparent molecular sizes based on migration of protein standards in kilodaltons. Lane 1, MHM2 control; lane 2, MHM2 plus PPI; lane 3, PrP61 control; lane 4, PrP61 plus PPI.
DISCUSSION
A peptide model for structural studies of PrPSc.
We have engineered a molecule less than one-third the size of full-length PrP which spontaneously adopts a conformation similar to PrPSc. PrP61 forms amyloid, is insoluble in nondenaturing detergents, displays the same level of resistance as PrPSc to digestion by proteinase K, and displays susceptibility to PPI. When expressed at low levels in Tg mice, PrP61 caused a spontaneous, fatal neurological disease characterized by PrP accumulation and neuronal apoptosis. Unfortunately, a limiting feature of PrP61 as a model PrPSc molecule is that we have been unable to demonstrate that it is infectious. However, PrP61 may prove to be a useful model for structural studies for several reasons. First, it is a neurotoxic PrP molecule that possesses many of the biophysical characteristics associated with PrPSc. Therefore, PrP61 can be used to investigate the structural basis of these characteristics. Second, because PrP61 is short and lacks carbohydrate side chains, it can be synthesized in large amounts, obviating the need to purify PrPSc from infected animals. Third, PrP61 does not display molecular heterogeneity caused by differential glycosylation.
Structural requirements for protease-resistant PrP fragments.
Previously, we reported that expression of PrP106 in uninfected neuroblastoma cells led to accumulation of PrP*106, which was identified by virtue of its partial resistance to limited proteolysis (33). Formation of PrP*106 appeared to require removal of α-helix A, as well as an intervening sequence between α-helices A and B, since a series of internal deletions from Δ141–155 to Δ141–176 resulted in a corresponding progressive increase in protease resistance (33). In this study, we extended the internal deletion even further towards the C terminus. When the internal deletion invaded α-helix C, the resulting PrP fragments displayed protease resistance similar to that of wild-type PrPSc (Fig. 2). In contrast, extending the internal deletion towards the N terminus resulted in complete loss of protease resistance.
We conclude from these data that a subset of residues 89 to 140 must be required to form protease-resistant PrP. Interestingly, synthetic peptides with sequences encompassed within this region form filaments in vitro (18) and can induce native, full-length PrP to become protease resistant in vitro (12). These observations and those reported here suggest that the region comprised of residues 89 to 140 inherently prefers to adopt a protease-resistant conformation but may be prevented from doing so in full-length PrPC by the stabilizing influence of structured regions, such as α-helices A and C. Perhaps, an important step in the conformation change from PrPC to PrPSc involves destabilization of these α-helices, allowing residues 89 to 140 to fold into a protease-resistant conformation.
Our data also demonstrate that the C-terminal lipid moiety contributes to the formation of protease-resistant PrP fragments. Without lipid, synthetic PrP61 adopted a protease-sensitive, random coil conformation. When myristic acid was covalently attached to the C terminus, synPrP61-Kma spontaneously adopted a protease-resistant, amyloidogenic conformation rich in β-sheet.
Neurodegeneration mediated by PrP deletion mutants.
We observed that expression of PrP61 at low levels in Prnp0/0 mice led to a rapidly fatal neurological disease characterized by PrP accumulation, neuronal degeneration, and reactive gliosis (Fig. 5). Previously, Shmerling et al. found that Prnp0/0 mice expressing MoPrP(Δ32–121) and MoPrP(Δ32–134) displayed spontaneous ataxia accompanied by localized neurodegeneration and accumulation of PrP in cerebellar granule cells. The ataxic phenotype of these mice could be rescued by coexpression of full-length PrP (D. Shmerling, M. Fischer, T. T. Blättler, I. Hegy, S. Brandner, A. Aguzzi, and C. Weissmann. Abstr. 29th Annu. Meet. Union Swiss Soc. Exp. Biol., abstr. A46, 1997). In a different study, Muramoto et al. demonstrated that expression of MHM2(Δ23–88,Δ177–200) or MHM2(Δ23–88,Δ201–217) in Prnp0/0 mice generated spontaneous diseases with massive accumulation of mutant PrP in the endoplasmic reticulum of neurons (17). It is unlikely that the neurodegenerative diseases caused by expression of different PrP deletion mutants all share a common mechanism, since there appear to be distinct patterns of pathology. It is noteworthy that the pattern of neurodegeneration in spontaneously ill Tg(PrP61)Prnp0/0 mice closely resembles that seen in scrapie-infected Tg(PrP106)Prnp0/0 mice (33). In both cases, a marked degeneration of hippocampal pyramidal cells was observed (Fig. 5). So whereas N-terminal and C-terminal PrP deletion mutants may provide good models for granule cell neurodegeneration and neuronal storage disorders, respectively, PrP61 may be the best PrP deletion mutant to use as a model for PrPSc-associated neurotoxicity. Correspondingly, among PrP deletion mutants that cause neurodegeneration, PrP61 is the only one that is protease resistant.
One of the features of the Tg(PrP61)Prnp0/0 mouse neurodegenerative model that is suggested by our data is neuronal apoptosis. Apoptosis is one of the features of prion diseases, seen in scrapie-infected sheep brain (6), scrapie-infected mice (8, 16), fatal familial insomnia (5), and Creutzfeldt-Jakob disease (9). Neuronal apoptosis was also observed in Tg mice expressing a mutant PrP with 14 octapeptide repeats whose human homologue is associated with an inherited prion dementia (3). Furthermore, a synthetic amyloidogenic peptide containing the PrP residues 106 to 126 has been shown to induce apoptosis in primary mouse cell cultures (7). More work is required to determine the mechanism by which neuronal apoptosis is triggered by PrPSc and PrP61 peptides.
New approaches arising from this work.
PrP61, a neurotoxic molecule that shares many of the biophysical characteristics of PrPSc and can be synthesized chemically, may find application in areas of research other than structural biology. For instance, a simple in vitro assay has previously been described that can be used to identify chemical compounds which render PrPSc susceptible to lysosomal hydrolases (34). Our data suggest that PrP61 peptide could be substituted for bona fide PrPSc as the substrate in these assays (Fig. 6), thereby decreasing the cost of screening for new therapeutic compounds.
Another potential application of PrP61 is the chemical synthesis of infectious prions. Recently, it has been demonstrated that a refolded, β-sheet peptide, MoPrP(89–143,P101L), initiates a prion disease in Tg196 mice expressing low levels of MoPrP(P102L) (11). synPrP61-Kma resembles MoPrP(89–143,P101L) in sequence but also possesses a C-terminal lipid moiety. This lipid moiety, which mimics the natural GPI anchor of PrPSc, is significant because it promotes acquisition of protease resistance and formation of amyloid. Although synPrP61-Kma was not infectious under the conditions tested, it may be possible to generate spontaneous prion infectivity from synthetic peptides resembling PrP61 by making substitutions in the amino acid sequence or by changing the lipid group. Alternatively, it may be possible to infect animals with synPrP61-Kma either by changing the refolding protocol or by delivering the peptide continuously into the brains of experimental animals using infusion pumps.
In conclusion, using a strategy of deletion mutagenesis, we have engineered a 61-amino acid PrP peptide that adopts a protease-resistant conformation. When expressed in Tg mice, this peptide accumulated in neuronal dendrites and cell bodies, causing an apoptotic neurodegenerative disease with an extremely early age of onset. A synthetic analog of this peptide polymerized into amyloid fibrils in vitro, adopted a conformation rich in β-sheet, and exhibited a resistance to protease digestion similar to that of wild-type PrPSc in infectious prion preparations. PrP61 may be a useful model for future research.
ACKNOWLEDGMENTS
We thank Chris Petromilli, Conny Heinrich, Darlene Groth, and Patrick Culhane for their expert contributions.
This work was supported by grants from the National Institutes of Health (NS14069, AG02132, and AG10770) and the American Health Assistance Foundation as well as a gift from the Leila and Harold Mathers Foundation. Surachai Supattapone was supported by the Burroughs Wellcome Fund Career Development Award and an NIH Clinical Investigator Development Award (K08 NS02048-02). Mass spectrometry was carried out in the UCSF Mass Spectrometry Facility, supported by NIH NCRR RR01614.
REFERENCES
- 1.Adle-Biassette H, Levy Y, Colombel M, Poron F, Natchev S, Keohane C, Gray F. Neuronal apoptosis in HIV infection in adults. Neuropathol Appl Neurobiol. 1995;21:218–227. doi: 10.1111/j.1365-2990.1995.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 1a.Ball H L, Mascagni P. Chemical protein synthesis and purification: a methodology. Int J Peptide Protein Res. 1996;48:31–47. doi: 10.1111/j.1399-3011.1996.tb01104.x. [DOI] [PubMed] [Google Scholar]
- 1b.Ball H L, Mascagni P. Application of reversible biotinylated label for directed immobilisation of synthetic peptides and proteins: isolation of ligates from crude cell lysates. J Peptide Sci. 2000;3:252–260. doi: 10.1002/(SICI)1099-1387(199707)3:4%3C252::AID-PSC100%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 1c.Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Linkage of prion protein and scrapie incubation time genes. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 2.Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith J M, Garon C, Haase A. Identification of scraple prion protein-specific mRNA in scrapie-infected and uninfected brain. Nature. 1985;315:331–333. doi: 10.1038/315331a0. [DOI] [PubMed] [Google Scholar]
- 3.Chiesa R, Drisaldi B, Quaglio E, Migheli A, Piccardo P, Ghetti B, Harris D A. Accumulation of protease-resistant prion protein (PrP) and apoptosis of cerebellar granule cells in transgenic mice expressing a PrP insertional mutation. Proc Natl Acad Sci USA. 2000;97:5574–5579. doi: 10.1073/pnas.97.10.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorandeu A, Wingertsmann L, Chretien F, Delisle M B, Vital C, Parchi P, Montagna P, Lugaresi E, Ironside J W, Budka H, Gambetti P, Gray F. Neuronal apoptosis in fatal familial insomnia. Brain Pathol. 1998;8:531–537. doi: 10.1111/j.1750-3639.1998.tb00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairbairn D W, Carnahan K G, Thwaits R N, Grigsby R V, Holyoak G R, O'Neill K L. Detection of apoptosis induced DNA cleavage in scrapie-infected sheep brain. FEMS Microbiol Lett. 1994;115:341–346. doi: 10.1111/j.1574-6968.1994.tb06661.x. [DOI] [PubMed] [Google Scholar]
- 7.Forloni G, Angeretti N, Chiesa R, Monzani E, Salmona M, Bugiani O, Tagliavini F. Neurotoxicity of a prion protein fragment. Nature. 1993;362:543–546. doi: 10.1038/362543a0. [DOI] [PubMed] [Google Scholar]
- 8.Giese A, Groschup M H, Hess B, Kretzschmar H A. Neuronal cell death in scrapie-infected mice is due to apoptosis. Brain Pathol. 1995;5:213–221. doi: 10.1111/j.1750-3639.1995.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 9.Gray F, Chretien F, Adle-Biassette H, Dorandeu A, Ereau T, Delisle M B, Kopp N, Ironside J W, Vital C. Neuronal apoptosis in Creutzfeldt-Jakob disease. J Neuropathol Exp Neurol. 1999;58:321–328. doi: 10.1097/00005072-199904000-00002. [DOI] [PubMed] [Google Scholar]
- 10.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko K, Ball H L, Wille H, Zhang H, Groth D, Torchia M, Tremblay P, Safar J, Prusiner S B, DeArmond S J, Baldwin M A, Cohen F E. A synthetic peptide initiates Gerstmann-Sträussler-Scheinker (GSS) disease in transgenic mice. J Mol Biol. 2000;295:997–1007. doi: 10.1006/jmbi.1999.3386. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko K, Peretz D, Pan K-M, Blochberger T, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Prion protein (PrP) synthetic peptides induce cellular PrP to acquire properties of the scrapie isoform. Proc Natl Acad Sci USA. 1995;32:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Solution structure of Syrian hamster prion protein rPrP(90–231) Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 16.Lucassen P J, Williams A, Chung W C, Fraser H. Detection of apoptosis in murine scrapie. Neurosci Lett. 1995;198:185–188. doi: 10.1016/0304-3940(95)11995-9. [DOI] [PubMed] [Google Scholar]
- 17.Muramoto T, DeArmond S J, Scott M, Telling G C, Cohen F E, Prusiner S B. Heritable disorder resembling neuronal storage disease in mice expressing prion protein with deletion of an α-helix. Nat Med. 1997;3:750–755. doi: 10.1038/nm0797-750. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen J T, Inouye H, Baldwin M A, Fletterick R J, Cohen F E, Prusiner S B, Kirschner D A. X-ray diffraction of scrapie prion rods and PrP peptides. J Mol Biol. 1995;252:412–422. doi: 10.1006/jmbi.1995.0507. [DOI] [PubMed] [Google Scholar]
- 19.Oesch B, Westaway D, Wälchli M, McKinley M P, Kent S B H, Aebersold R, Barry R A, Tempst P, Teplow D B, Hood L E, Prusiner S B, Weissmann C. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 20.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 22.Prusiner S B. Prion diseases. In: Nathanson N, Ahmed R, Gonzalez-Scarano F, Griffin D, Holmes K, Murphy F A, Robinson H L, editors. Viral pathogenesis. New York, N.Y: Raven Press; 1996. pp. 855–911. [Google Scholar]
- 23.Prusiner S B. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 24.Prusiner S B, Cochran S P, Groth D F, Downey D E, Bowman K A, Martinez H M. Measurement of the scrapie agent using an incubation time interval assay. Ann Neurol. 1982;11:353–358. doi: 10.1002/ana.410110406. [DOI] [PubMed] [Google Scholar]
- 25.Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 26.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 27.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 28.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 29.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 30.Scott M, Groth D, Foster D, Torchia M, Yang S-L, DeArmond S J, Prusiner S B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 31.Scott M R, Köhler R, Foster D, Prusiner S B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Structural analysis of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 33.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen H-O B, Heinrich C, Torchia M, Safar J, Cohen F E, DeArmond S J, Prusiner S B, Scott M. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 34.Supattapone S, Nguyen H-O B, Cohen F E, Prusiner S B, Scott M R. Elimination of prions by branched polyamines and implications for therapeutics. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wille H, Zhang G-F, Baldwin M A, Cohen F E, Prusiner S B. Separation of scrapie prion infectivity from PrP amyloid polymers. J Mol Biol. 1996;259:608–621. doi: 10.1006/jmbi.1996.0343. [DOI] [PubMed] [Google Scholar]