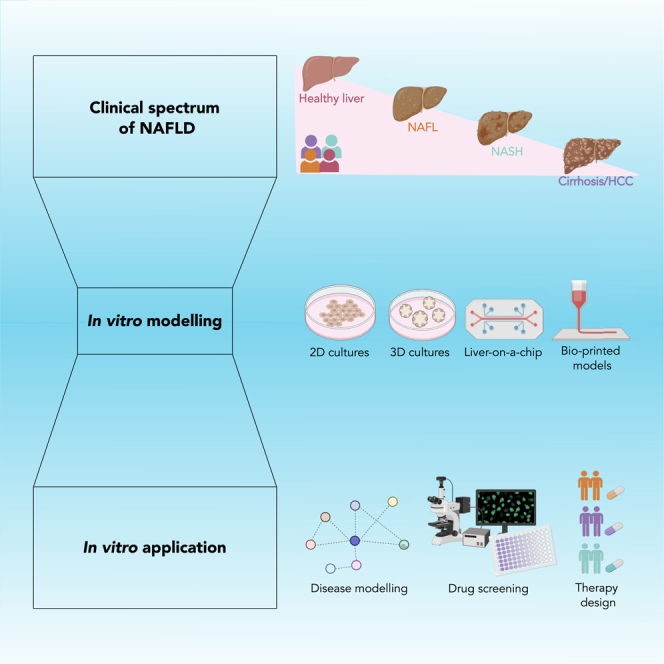
Summary
Non-alcoholic fatty liver disease (NAFLD) represents a global healthcare challenge, affecting 1 in 4 adults, and death rates are predicted to rise inexorably. The progressive form of NAFLD, non-alcoholic steatohepatitis (NASH), can lead to fibrosis, cirrhosis, and hepatocellular carcinoma. However, no medical treatments are licensed for NAFLD-NASH. Identifying efficacious therapies has been hindered by the complexity of disease pathogenesis, a paucity of predictive preclinical models and inadequate validation of pharmacological targets in humans. The development of clinically relevant in vitro models of the disease will pave the way to overcome these challenges. Currently, the combined application of emerging technologies (e.g., organ-on-a-chip/microphysiological systems) and control engineering approaches promises to unravel NAFLD biology and deliver tractable treatment candidates. In this review, we will describe advances in preclinical models for NAFLD-NASH, the recent introduction of novel technologies in this space, and their importance for drug discovery endeavors in the future.
Subject areas: Cellular physiology, Bioengineering, Cell biology
Graphical abstract
Cellular physiology; Bioengineering; Cell biology
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease, mainly affecting patients with obesity and type 2 diabetes mellitus (T2DM). NAFLD is the umbrella-term used for a range of conditions caused by a build-up of fat in the liver and encompasses isolated fatty liver (steatosis) and non-alcoholic steatohepatitis (NASH) with increasing degrees of liver inflammation, fibrosis (scarring), and potentially cirrhosis (end-stage fibrosis with vascular changes and functional impairment). Its worldwide prevalence is currently ∼25%, with 41% of the patients ultimately developing fibrosis (Younossi et al., 2016) and is projected to increase further by up to 18.3% in some developed countries by 2030 (Estes et al., 2018). Sedentary lifestyles and booming economic growth in the most developed countries sustain such trends, which see NASH becoming the leading cause of liver transplant (Zou et al., 2020).
To date, there are no licensed pharmacological treatments for NASH. While bariatric surgery is effective in a small proportion of patients (Lee et al., 2019), diet and exercise remain the cornerstone of disease management, due to evidence that a 10% body weight loss allows a reversion of steatosis, inflammation, and even fibrosis (Vilar-Gomez et al., 2015). However, the difficulty of achieving and sustaining this supports the need for disease-specific pharmacological treatment. Current medical treatment also comprises drugs for comorbidities related to NAFLD. Patients with or without T2DM may be treated with anti-diabetic medications. A recent systematic review of 18 trials (Blazina and Selph, 2019) presented evidence that supports the efficacy of some diabetes drugs in reducing liver fat and resolving NASH (e.g., pioglitazone, liraglutide), although associated weight gain with pioglitazone warrants caution. Blazina and Selph (2019) concluded that larger trials are needed to elucidate the benefits and risks of diabetes pharmacotherapy in NAFLD patients.
Given the complexity of NAFLD pathophysiology and the phenotypic heterogeneity, both genetic and environmental factors are thought to contribute to disease progression and susceptibility (Eslam et al., 2018). Several genes that participate in lipid droplet remodeling, hepatic very low density (VLDL) secretion and generation of predominant molecular species of phosphatidylinositol (PI) in cell membranes have been identified as NAFLD genetic variants. Specifically, the most well-described variants are patatin-like phospholipase domain-containing protein 3 (PNPLA3) I148M (Romeo et al., 2008), transmembrane 6 superfamily member 2 (TM6SF2) E167K (Kozlitina et al., 2014) and obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) (Mancina et al., 2016). The PNPLA3I148M variant has been linked to NASH progression and fibrosis (Valenti et al., 2010; Rotman et al., 2010; Sookoian et al., 2009) and replicated in several independent genome-wide association studies.
Increasingly, it is suggested that a ‘one size fits all’ approach to treat NAFLD might not be optimal given the heterogeneity of the disease. Moreover, early pathways that participate in NAFLD progression are not commonly investigated in clinical trials, as the predominant focus is on NASH with moderate-to-severe fibrosis or cirrhosis. A comprehensive understanding of the whole disease spectrum –and not just the later stages– could be the key to understanding this disease and its progression.
Ahead of clinical trials, in vivo models remain a crucial tool to study NAFLD. Larger animal models such as rabbits (Ogawa et al., 2010), monkeys (Hansen et al., 2017) or minipigs (Pedersen et al., 2020; Lee et al., 2009) may have greater proximity to humans but can present ethical issues, are more difficult to handle and can be time-consuming and costly. While relying mostly on rodents, in vivo modeling has shown improved human translatability in studies of NAFLD pathogenesis and evaluation of potential therapeutic targets (Hansen et al., 2020; Clapper et al., 2013). However, rodent models also have disadvantages. Indeed, the human relevance and predictivity of laboratory mice is limited by species-specific biology and genetics (Reimer et al., 2020), coupled with physiological factors that result from growth in the laboratory, as opposed to a natural environment. Critically, this can affect the microbiome of mice and their potential response to drug therapies that are subsequently funneled through to human trials (Rosshart et al., 2019). Of note, the microbiome has been strongly linked to NAFLD pathogenesis (described in more detail in the following section).
In vitro models are, in many settings, a suitable alternative for studying NAFLD, as a result of increasing sophistication and the ability to recapitulate several hallmarks of the disease. A multi-disciplinary approach to the development of in vitro models led to solutions far more complex than the traditional two-dimensional (2D) cultures (e.g., liver-on-a-chip platforms and three-dimensional (3D) models). While supporting a better understanding of the disease, these models could be used as reliable drug screening platforms. In this review, we will detail which in vitro models have been used to date for NAFLD and NASH, discussing their advantages and disadvantages. We offer our perspective on future developments and translational opportunities in this field, which will be of relevance to researchers interested in the pathophysiology of NAFLD, drug development, and bioengineering.
NAFLD pathogenesis
A detailed discussion of NAFLD pathogenesis is beyond the scope of this article; the interested reader is directed to recent comprehensive reviews (Loomba et al., 2021; Cariou et al., 2021). The current model of NAFLD-NASH pathogenesis is one of ‘substrate overload’, whereby the liver's capacity to handle the primary metabolic energy substrates (e.g., carbohydrates and fatty acids) is overwhelmed, leading to the accumulation of toxic lipid species.
Initially, the expansion of the subcutaneous adipose tissue in obese NAFLD patients leads to the accumulation of free fatty acids (FFA) in the muscles. This induces insulin resistance (IR), inhibiting glucose uptake (Brunt et al., 2015; Godoy-Matos et al., 2020). Concurrently, IR also compromises fat storage in adipocytes, causing FFA to be released into circulation and reaching the liver. Here, FFA induces hepatic IR, gluconeogenesis, very low-density lipoprotein (VLDL) release, as well as an increase of pro-inflammatory adipokines. To balance the elevated blood glucose levels, insulin, and FFA, the liver increases the storage of fatty acids and their synthesis through de novo lipogenesis, regulated by the sterol regulatory element binding protein-1 (SREBP1c) (Donnelly et al., 2005). Some sphingolipids, besides being correlated with IR, are also linked to hepatic oxidative stress and inflammation, suggesting their importance in NASH progression (Apostolopoulou et al., 2018).
The major difference between NAFLD and NASH is the presence of hepatocellular injury. In NASH, there is an increase of reactive oxygen species (ROS) and mitochondrial uncoupling, as a consequence of the metabolism of the fatty acids present in hepatocytes (Brunt et al., 2015). Hepatocyte ballooning (a special form of liver cell degeneration) has also been linked to the progression of NAFLD. These large cells with irregular cytoplasm show diminished expression of caspase-9 (conferring resistance to apoptosis) and enhanced production of Sonic hedgehog, a ligand of the hedgehog signaling pathway which promotes hepatic fibrogenesis (Hirsova and Gores, 2015). Kupffer cells (KC) and infiltrating macrophages contribute to hepatic inflammation, caused by the accumulation of fatty acids or damage-associated molecular patterns (DAMPs) released by the dying hepatocytes. They can activate toll-like receptors (TLRs) and the inflammasome, maintaining the inflammatory environment; and promote fibrosis through the activation of hepatic stellate cells (HSC). These cells play a critical role in fibrosis, and perpetuate inflammatory activity in the liver by increasing ROS and CCR5 levels, leading to extracellular matrix (ECM) deposition through stimulation by transforming growth factor β (TGFβ) (Pafili and Roden, 2020).
Gut microbiota dysbiosis (an “imbalance” in the gut microbial community) is also believed to play an important role in disease progression, as suggested by the distinctive microbiome signatures of NAFLD patients. Bacteria and their products, which reach the circulation and liver as the tight junctions of the intestinal barrier weaken, can trigger tissue and systemic inflammation. Some of these bacteria can release ethanol-like products, increasing ROS levels and inflammation, and hence promote NASH progression (Chen and Vitetta, 2020).
Although disease progression is variable in NAFLD, ∼5% will progress to cirrhosis, with an increased risk of hepatic decompensation, hepatocellular carcinoma (HCC), and liver-related mortality (Lindenmeyer and McCullough, 2018). Interestingly, HCC in NAFLD patients may also arise in non-cirrhotic liver (Huang et al., 2021). In a recent meta-analysis, non-cirrhotic NASH subjects were at greater risk of developing HCC than non-cirrhotic patients of other etiologies of liver disease (odds ratio 2.61) (Stine et al., 2018).
In vitro culture systems in NAFLD
Cell lines
Cell lines are widely used in research and drug development. When compared to in vivo models, cell lines can be easily cultured at large scale in a cost-efficient manner. Furthermore, they can be maintained longer than current NAFLD ex vivo models, which have a usual life-span of ∼5–15 days (Wu et al., 2018; Palma et al., 2019).
According to their origin, cell lines are either tumor-derived (commonly known as immortalized), primary cell lines, or pluripotent stem cells (PSC). Immortalized cell lines used for NAFLD research (e.g., HepaRG, THP-1 or LX-2), offering unlimited growth and stable phenotype, streamline standardized culture protocols and assay reproducibility (Ramboer et al., 2014). Nonetheless, mutations in immortalized cell lines can hamper translation of observations to the human in vivo condition; for instance, HepG2 is characterized by the PNPLA3 I148M mutation, which is known to affect some metabolic functions (Gunn et al., 2017). Primary human cells, isolated from liver tissue as hepatocytes or non-parenchymal cells (NPC), better resemble the in vivo phenotype (Wilkening et al., 2003). Indeed, harvesting of these cells from NAFLD patients enhanced the accuracy of drug metabolism studies (Ganji et al., 2015; Schwartz et al., 2020). However, their limited culture time and availability, especially for non-pathological (“healthy”) controls, as well as sample heterogeneity, are disadvantages (Zeilinger et al., 2016; Bell et al., 2018). When focusing on NAFLD modeling, inherent donor liver variability represents a double-edge sword, setting the basis for the investigation of genetic polymorphisms (Müller and Sturla, 2019). Lastly, PSC can be differentiated into liver-like cells from liver stem cells, induced pluripotent stem cells (iPSC) or embryonic pluripotent stem cells (ESC) (Hu and Li, 2015; Touboul et al., 2010; Takahashi et al., 2007). Cell differentiation, achieved via addition of growth factors and nutrients to the culture medium, results in phenotypic similarity to primary cell lines in addition to indefinite expansion in culture (Soret et al., 2021). Moreover, iPSC are the preferred PSC for investigation, the use of ESC being associated with ethical concerns (Zeilinger et al., 2016). Recent in vitro models with human PSC have been developed by Sinton et al. (2021); Holmgren et al. (2020); Coll et al. (2018) for NAFLD modeling. In Table 1, advantages and disadvantages of various cell lines used in NAFLD in vitro modeling are detailed.
Table 1.
In vitro cell lines that have been used to investigate NAFLD, highlighting their advantages and disadvantages
| Cell line | Description | Advantages | Disadvantages | References |
|---|---|---|---|---|
| HepG2 | Human hepatocellular carcinoma cell line |
|
|
Green et al. (2015, 2020); Gunn et al. (2017); Jennen et al. (2010); Lőrincz et al. (2021); Srivastava and Chan (2008) |
| HepG2/C3A | Clonal derivative of HepG2 |
|
|
Baquerizo et al. (2015); Flynn and Ferguson (2008); Garcia et al. (2011); Gaskell et al. (2016) |
| HepaRG | Human hepatocellular carcinoma cell line |
|
|
German and Madihally (2019); Green et al. (2020); Jennen et al. (2010); Lőrincz et al. (2021) |
| L02 | Human fetal liver cell line |
|
|
Hu et al. (2013) |
| Huh7 | Human hepatocellular carcinoma cell line |
|
|
Green et al. (2020); Gunn et al. (2017) |
| Hepa1c1c7 | Mouse hepatocellular carcinoma cell line |
|
|
El-Sayed et al. (2007) |
| AML-12 | Mouse liver cell line |
|
|
Wu et al. (1994) |
| Hepa1-6 | Mouse carcinoma cell line |
|
|
Kachlishvili et al. (2020); Lacoste et al. (2017); Urs (2018) |
| RAW264.7 | Mouse monocyte/macrophage-like cells |
|
|
Hartley et al. (2008); Taciak et al. (2018) |
| THP-1 | Human monocytic cell line |
|
|
Chanput et al. (2014) |
| Bone marrow-derived macrophages | Mouse primary macrophage cells |
|
|
Weischenfeldt and Porse (2008) |
| LX-2 | Hepatic stellate cells (HSCs) from normal liver |
|
|
Vande Bovenkamp et al. (2007); Weiskirchen et al. (2013); Xu et al. (2005) |
| T6 stellate cells | Rat primary HSCs from liver cell line |
|
|
Van de Bovenkamp et al. (2007); Vogel et al. (2000); Xu et al. (2005) |
| Primary cell lines |
|
|
|
Brandon et al. (2003); Green et al. (2020); Wilkening et al. (2003) |
| 3T3-L1 MBX | Mouse fibroblast |
|
|
ATCC; Berger and Géloën (2019) |
| 3T3-J2 | Embryonic mouse fibroblast |
|
|
Bhatia et al. (1998); Ware et al. (2021) |
| Liver sinusoidal endothelial cells (LSEC) | Primary human cell line |
|
|
German and Madihally (2019); Poisson et al. (2017) |
| Human umbilical vein endothelial cells (HUVEC) | Human non-pathological tissue |
|
|
German and Madihally (2019); Medina-Leyte et al. (2020) |
| Caco-2 | Human epithelial cells from colorectal adenocarcinoma |
|
|
Briske-Anderson et al. (1997); Poncede León-Rodríguez et al. (2019) |
| NCTC-1469 | Mouse NPC from liver cell line |
|
Asahi et al. (2010) | |
| Pluripotent stem cells (PSC) | Stem cells that can differentiate into endoderm, mesoderm or ectoderm |
|
|
Soret et al. (2021); Zeilinger et al. (2016) |
Cell culture models
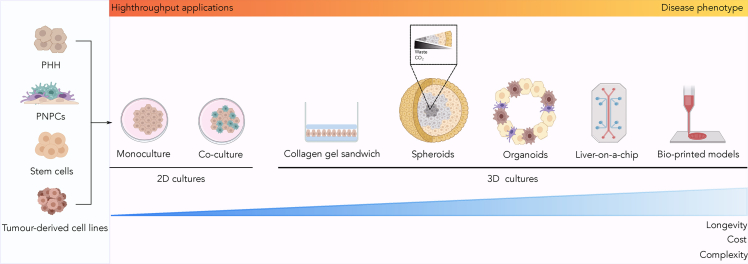
2D and 3D cultures have been extensively used in NAFLD research and in vitro modeling (Figure 1, Table S1), albeit to a different extent. We have performed a systematic review to observe the evolution and focus of in vitro models in NAFLD (Figure S1). Our systematic review indicated that researchers predominantly favored 2D monocultures (59.4%) to more complex models (2D co-cultures (14%), spheroids (9.7%), organoids (7.3%), liver-on-a-chip (7.8%), collagen gel sandwiches (1.2%), and micropatterned cultures (0.6%). When analyzing the selection of cell culture systems over time (Figure S2), we observed an increasing trend in the publication of 3D in vitro models, specifically in on-chip cultures. This suggests 3D culture systems are becoming more relevant in this field. Focusing on the large number of publications in monocultures in 2020, we note that these systems have been used in conjunction with additional in vivo experiments, or as a benchmark for more sophisticated in vitro models. As an example, the insert in Figure S2 distinguishes publications in 2020 that have used monocultures as the only in vitro experiment from those that used them in combination with other cell culture models. We speculate that technological advances, standardizing the development of more complex and affordable solutions, might underlie this observation.
Figure 1.
Cell sources and in vitro models used in NAFLD studies
Primary human hepatocytes (PHH), primary non-parenchymal cells (PNPC), stem cells and tumor-derived cell lines (cyan panel) can all be used to create two-dimensional (2D) and three-dimensional (3D) models for NAFLD (pink panel). 2D cultures (e.g., monocultures and co-culture) are denoted by a single or several cell types growing in a monolayer. 3D cultures (e.g., collagen gel sandwich, spheroids, organoids or liver-on-a-chip) have been recently introduced to elucidate NAFLD, and their development can rely on bio-printing technology. Self-aggregated spheroids present an accumulation of carbon dioxide (CO2) and waste in their core. Cell culture models are listed by increasing cost, longevity and complexity. While complexity correlates with ability of the model to accurately capture the disease phenotype, simpler models are better suited to high-throughput applications.
As reported above, 2D monoculture is the most established NAFLD in vitro model. In this system, steatosis induction is routinely performed by adding a mixture of unsaturated and saturated FFA to the culture medium. Typically, oleic acid (OA) and palmitic acid (PA) are selected, being the most abundant in humans (Hodson et al., 2008). According to our systematic search the most commonly used ratio for these FFA is 1:2 (PA:OA). This FFA ratio exposure emulates dietary conditions that are commonly observed in vivo, inducing hallmarks of NAFLD-NASH (e.g., cytoplasmic accumulation of TG in hepatocytes, ER stress, inflammation, and cell death) (Gomez-Lechon et al., 2007).
The low cost, ease of use, and suitability for high-throughput screening make 2D monocultures attractive for disease modeling. Here, the choice of cell sources constrains the NAFLD mechanistic studies enabled by the in vitro model. For example, Lyall et al. (2018) showed that hepatocyte-like cells derived from human ESC retained the ability to form the cytosine modification 5-hydroxy-methylcytosine (5hmC), rapidly lost when culturing HepG2, providing a means to elucidate the role of epigenetic dysregulation in NAFLD pathogenesis. Similarly, an hPSC-derived model was used to study the re-wiring of mitochondrial metabolism in early steatosis (Sinton et al., 2021). Monoculture models for NAFLD have also been valuable for investigating metabolic and fibrotic pathways (Graffmann et al., 2016; Kanda et al., 2018; Zhang et al., 2016), evaluating therapeutic compounds (Boeckmans et al., 2020; Armstrong et al., 2016; Anfuso et al., 2020), and assessing hepatic drug metabolism/toxicity (Gomez-Lechon et al., 2003).
Notwithstanding the advantages outlined above, 2D monocultures are unable to faithfully mimic in vivo NAFLD conditions due to the lack of interaction with NPC and disturbances in the extracellular environment. An improvement over this system is provided by 2D co-cultures, typically involving two cell types, such as hepatocytes and NPC, growing in a monolayer. As with monocultures, the steatotic phenotype can be induced upon treatment with FFA. Interestingly, a co-culture of Huh7 and LX-2 enabled Barbero-Becerra et al. (2015) to demonstrate the key role of hepatocyte-HSC crosstalk in the activation of HSC, a key feature of NASH. Co-cultures including macrophages (Zhong et al., 2017; Chen and Ma, 2019) have also elucidated the role of multiple inflammatory pathways in NAFLD and NASH.
NAFLD and NASH can be induced in 2D co-cultures of PHH and iPSC (Parafati et al., 2018), yet their capacity as accurate in vitro models decays with time as they lose their xenobiotic metabolism and hormone response (Gurevich et al., 2020). 3D cultures can enhance these features, allow cell-cell interactions and cell polarity (Treyer and Müsch, 2013; Godoy et al., 2013). They can provide new insights into complex physiological mechanisms and potential therapeutic targets, increasing our knowledge of NAFLD. Furthermore, they represent a suitable alternative to in vivo models at early stages of drug development (Romualdo et al., 2021; Beisner et al., 2021; Eilenberger et al., 2020). The main 3D in vitro models used for NAFLD –spheroids, organoids, and microfluidic devices– are detailed in the following section.
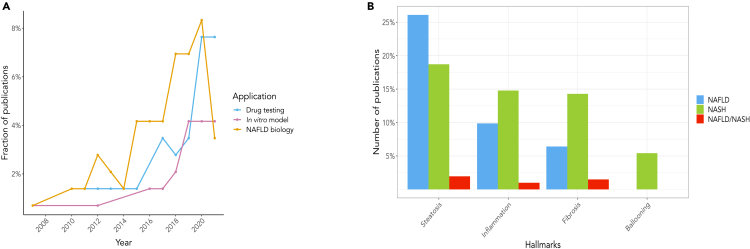
We classified the NAFLD in vitro models identified in our systematic search, according to their intended application, into NAFLD biology studies and compound testing. 47.9% of the publications relied on in vitro models to elucidate the role of different proteins, pathways or hallmarks of the disease; 18.8% of the papers detailed the development of new in vitro models to better mimic NAFLD biology and its progression; while 33.3% of the studies assessed the therapeutic potential of putative compounds and drug targets for NAFLD and NASH.
Despite a larger number of studies focused on the biology of NAFLD, the last 3 years have seen an increase in publications reporting compound testing (Figure 2A). This, together with the simultaneous decline in the study of disease mechanisms in NAFLD, suggests a possible shift in research on the condition. Specifically, recent publications leverage available knowledge on NAFLD pathogenesis to devise strategies for treatment. We explored whether this shift in the field was promoted by a greater involvement of pharma, but did not observe any direct link, as most of the investigations remain hosted in academic settings. We also noted that despite the divergence in their severity and risk of progression, comparable effort has been devoted to NAFLD (51.4% of the studies) and NASH (43.7%). Only 4.9% of the investigations proposed in vitro models to mimic both conditions. Considering the varied clinical outcomes within the disease spectrum, it would be preferable to consider both NAFLD and NASH when investigating the disease. This is of paramount importance for compound testing since the mechanism of action (and potential efficacy) of drugs will depend on the disease stage.
Figure 2.
Systematic analysis of the scientific literature on NAFLD in vitro cell culture models published between 2007 and 2021
(A) Trend over time of scientific publications centered on NAFLD in vitro models, clustered by application. Despite the majority published studies using in vitro models focused on NAFLD biology (orange), the past years have seen a marked increase in scientific articles presenting novel in vitro models (purple) or using available culture systems for compound testing (cyan). The concurrent decrease in investigations of the role of specific pathways and/or proteins in NAFLD pathogenesis suggests a shift in focus and efforts within the scientific community.
(B) Barplot showing the percentage of in vitro models that have been used to study steatosis, inflammation, fibrosis, or hepatocellular ballooning. For each disease hallmark, publications are clustered, according to their focus, in NAFLD (cyan), NASH (blue) or both NAFLD and NASH (dark blue). Steatosis is the most studied feature in NAFLD and NASH, followed by inflammation and fibrosis. Hepatocellular ballooning is the least studied feature in the scientific literature. For more information regarding the historical perspective of the evolution of in vitro models for NAFLD.
See also Figure S2.
In Figure 2B the distribution of the hallmarks of NAFLD/NASH across the studies is reported. Most publications concentrated on steatosis, followed by inflammation and fibrosis. The focus on steatosis is reflected by the predominant use of monocultures of hepatocytes. Interestingly, hepatocellular ballooning has barely been explored in these studies. Although it is difficult to objectively measure the presence of ballooned hepatocytes in patients' biopsies (Kakisaka et al., 2018), they are a pathognomonic histological feature of NASH that should be considered when studying this disease.
3D tissue culture models in NAFLD
Three-dimensional cultures are emerging as a bridge between the easy-to-use 2D cell cultures and more complex in vivo models. By maintaining hepatic cell types within a controlled microenvironment, 3D liver models better mimic the in vivo organ phenotype. From spheroids composed of a single cell type to multicellular aggregates including NPC and immune cells, 3D cultures can model the liver organotypic structure, retaining paracrine cell-cell interactions and metabolic functions for several weeks. Moreover, these models can be used to study fluid flow and perfusion when cultured on a chip. In this section, we will review the 3D culture models most recently proposed to recapitulate NAFLD. Specifically, we will detail the latest developments in spheroids, organoids, liver-on-a-chip and bio-printed platforms.
The collagen gel sandwich has been extensively used to culture liver cells (Engl et al., 2004; Sandwich, 2006; Kono et al., 1997; Choi and Choi, 2013; Wang et al., 2004). In this model, hepatocytes are seeded between two layers of collagen gel. The obtained structure better resembles the in vivo conditions, as demonstrated by reorganization of the cytoskeleton, improved morphology and polarity, and enhanced expression of liver-specific functions (Berthiaume et al., 1996). Within this model, hepatocytes maintained secretion of hepatic compounds for at least six weeks; a three-fold increase in the secretion window offered by monolayer collagen gel cultures (Dunn et al., 1991). Another collagen gel sandwich model, combining hepatocytes with liver sinusoidal endothelial cells (LSEC), mimicked the liver lobular architecture and showed stable secretion and metabolic activity for up to 4 weeks (Bale et al., 2015). While allowing long-term monitoring, the collagen gel sandwich has limitations. Indeed, when compared to 3D spheroids, a PHH-based collagen gel sandwich does not prevent hepatocyte dedifferentiation, as evidenced by perturbed glycolysis and gluconeogenesis, and shows lower sensitivity to long-term exposure to hepatotoxic compounds in cytotoxicity studies (Bell et al., 2018). Moreover, the collagen thickness may hinder cell-cell interactions (Soret et al., 2021). For these reasons, this model is seldom used to investigate NAFLD.
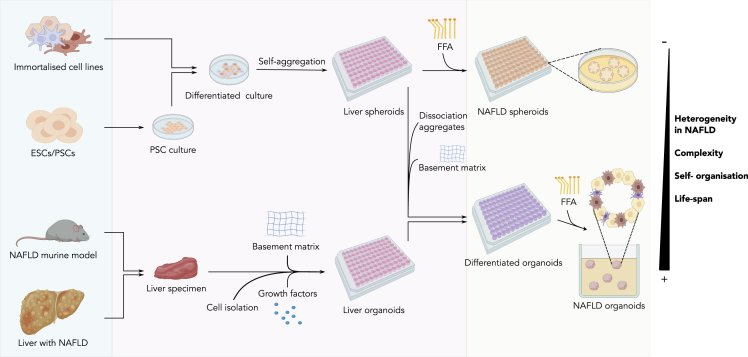
More recently, spheroids and organoids have become the 3D in vitro models of choice for studying NAFLD (Soret et al., 2021). Spheroids and organoids differ based on the origin and number of cell types in the aggregate, the culture environment or the complexity of their structure (Kang et al., 2021). In this review, we refer to spheroids as 3D structures produced by free-floating, spontaneous self-aggregation of cell lines in the presence or absence of an ECM that can recapitulate some functional aspects of the liver. In contrast, we denote organoids as 3D systems obtained by embedding tissue stem cells, progenitor cells or tissue-resident cells isolated from liver specimens in a scaffold that imposes a liver-like spatial organization, thereby enabling modeling of the organotypic structure and function (Kang et al., 2021; Gunti et al., 2021). Figure 3 details how both NAFLD spheroids and organoids can be obtained from different sources.
Figure 3.
Development protocol for spheroids and organoids
Both models can be obtained from different sources (cyan panel) -immortalized cell lines, embryonic stem cells (ESC), pluripotent stem cells (PSC), NAFLD murine models or liver from NAFLD patients. Immortalized cell lines and differentiated ESC/PSC self-aggregate to form spheroids. To induce the NAFLD phenotype, free fatty acids (FFA) supplementation is required. To form liver organoids, cells from liver specimens are extracted and isolated. Hence, they are exposed to selected growth factors in presence of a basement matrix. These liver organoids then differentiate into the specific cell lines and acquire the NAFLD phenotype upon FFA supplementation (light green panel). In addition, differentiated organoids can also be obtained from ESC/PSC or immortalized cell lines aggregated in liver spheroids. After dissociation of the aggregates and subsequent cultivation with a basement matrix, the differentiated organoids are ready to be used for NAFLD studies. The heterogeneity of the disease, its complexity, the self-organization of the cells or the life-span of the cultures is higher in organoids than in spheroids.
Spheroid models
Since the first spheroid model in the 1970s (Sutherland et al., 1971), multiple methods have been developed to establish systems of increasing sophistication. In NAFLD modeling, the most popular method for spheroid culture is liquid overlay, where cell suspensions are cultured on ultra low-adhesive surface plates and formation of spheroids can be monitored in real-time (Gunti et al., 2021). Alternative methods for spheroid assembly have been extensively reviewed in the scientific literature (Kang et al., 2021; Gilazieva et al., 2020; Ryu et al., 2019).
The simplest hepatic spheroid model consists of a single cell type, usually HepG2 or PHH. It is worth noting that spheroids support culturing of differentiated PHH for a minimum of 3–4 weeks. Upon treatment with pathological concentrations of FFA, carbohydrates, cyclosporin A or insulin, hepatic spheroids enabled induction and investigation of steatosis for up to 5 weeks (Damelin et al., 2007; Bell et al., 2016; Kozyra et al., 2018). Specifically, these models have helped to elucidate the role of hepatocytes in NAFLD (Damelin et al., 2007; Chong et al., 2020), the mechanism by which IR elicits de novo lipogenesis and lipid accumulation (Kozyra et al., 2018), the identity of putative molecular targets or the pharmacological action of novel compounds to ameliorate the disease (Chong et al., 2020; Eilenberger et al., 2020; Bell et al., 2016). Furthermore, the use of PHH in spheroids (Bell et al., 2016; Eilenberger et al., 2020) has shed light on the relevance of inter-individual variations as a determinant of susceptibility to steatosis development and progression (Dongiovanni et al., 2015). Thus, PHH-based spheroids could provide an ideal platform to demonstrate the impact of genetics on the variability in drug responses (efficacy and toxicity) among patients; the major cause of withdrawal of drugs from the market (Sim and Ingelman-Sundberg, 2011).
By co-culturing hepatocytes with NPC, beyond enhancing hepatocyte function in vitro, researchers can generate liver-like structures that recover the relevant mechanistic steps in NAFLD-NASH evolution (e.g. inflammation, HSC activation, and fibrogenesis). Hepatoma cell lines have been co-cultured with primary HSC to define pathways linked to liver steatosis and fibrosis (Salloum et al., 2021; Hurrell et al., 2020). Specifically, these co-culture systems showed enhanced collagen production, activation of HSC, upregulation of genes associated with fibrosis progression – such as collagen type I alpha I (COL1A1), TIMP metallopeptidase inhibitor 1 (TIMP1) or actin alpha 2 (ACTA2), and induction of pro-inflammatory cytokines. Bell et al. (2016) also co-cultured PHH-based spheroids with KC, HSC, and biliary cells, and highlighted that the co-culture systems enable modeling of an inflammatory response, as measured by an increase in IL-6 secretion. Another example of a multicellular spheroid model has been proposed by Kachlishvili et al. (2020). The researchers combined a hepatocyte cell line with macrophages to study the effect of low-dose trans-resveratrol on lipid metabolism and pro-inflammatory processes. They showed a decrease in lipid accumulation due to enhanced poly(ADP-ribose) polymerase (PARP) activity, following treatment with the compound. As previous studies in monocultures hinted at an environment and cell-type dependent effect of resveratrol on metabolic pathways (Lan et al., 2017), the study by Kachlishvili et al. (2020) demonstrated the relevance of 3D spheroids for investigating the mechanism of action of drug candidates.
Spheroid models have further enhanced our understanding of genetic variants linked to NAFLD. In Tanaka et al. (2021), HepG2-HSC spheroids were used to evaluate the effect of the MBOAT7 variant on inflammation in NAFLD. Silencing MB0AT7 expression with siRNA, the authors demonstrated a positive correlation between the presence of the variant and collagen expression, activation of HSC and expression of inflammatory and profibrotic cytokines. A similar system proved relevant to investigate the most common genetic variant in NAFLD patients, PNPLA3 I148M (Pingitore et al., 2019), and study the mechanisms involved in the early stages of fibrogenesis induced by lipid accumulation. The genetically predisposed spheroids were characterized by high fat accumulation and collagen production. This 3D model also captured antifibrotic effects following exposure to the compounds liraglutide and elafibranor. The same model was later selected as a screening platform for small molecule inhibitors, leading to the identification of momelotinib as a putative drug against the PNPLA3 I148M variant (Schwartz et al., 2020). PHH spheroids were additionally used to study the effect of the TM6SF2 E167K variant on steatosis establishment and progression in vitro (Prill et al., 2019). While the mutation led to higher intracellular lipids and upregulation of cholesterol biosynthesis and de novo lipogenesis, downregulation of genes implicated in NAFLD (e.g., ACADS, PNPLA2) revealed discrepancies with observed in vivo pathogenic mechanisms. However, the conclusions drawn from this study are limited by the small sample size exhibiting the TM6SF2 E167K variant (2 out of 5 PHH donors); a common challenge when working with PHH.
Despite their potential for disease modeling and drug efficacy testing, stem cell-based models are still mainly used in 2D cultures. Holmgren et al. (2020) assembled PSC-derived hepatocytes and HSC in spheroids, thereby avoiding the nuisance factor of spontaneous HSC activation induced by the stiffness of the ECM in 2D co-cultures (Wells, 2005). Gene expression analysis revealed similarities with PHH in glucose, lipid and cholesterol metabolism, reinforcing its suitability for NAFLD research. However, limited urea secretion and downregulation of enzymes involved in phase I and II drug metabolism hindered its use for drug screening studies.
Notwithstanding their simplicity and low cost, spheroid models offer limited control over their assembly and size when cultured in standard non-adhesive plates, presenting a scalability and reproducibility issue for liver spheres. Indeed, clusters of spheroids tend to merge into larger aggregates, challenging cell viability in culture by hampering both the intake of nutrients and oxygen diffusion (Underhill and Khetani, 2018a). To overcome this drawback, protocols detailing automated assembly of liver spheres have been published (Meseguer-Ripolles et al., 2021). In addition, novel approaches have incorporated the inclusion of hydrogel scaffolds, which direct the assembly of spatially isolated, controlled-size spheroids, in the development protocol (Ryu et al., 2019).
Organoid models
Liver organoids are 3D culture models obtained by embedding tissue stem cells, progenitor cells or tissue-resident cells isolated from liver specimens within a hydrogel ECM. Their development protocol allows tighter control over the spatial organization of the multicellular aggregate, better approximating the heterogeneity and organotypic structure observed in vivo (Gunti et al., 2021); once the exposure to growth factors and support matrices enabling differentiation of multiple cell types has been defined. Organoids supersede the ability of 2D cultures to model (patho)physiological processes, spanning cell differentiation, migration and cell-cell interactions. Furthermore, they retain genomic stability and, owing to wide expansion in culture, are suitable for long-term storage and high-throughput screening of drugs (Soret et al., 2021).
Murine-derived organoids have been widely used to model the NAFLD phenotype. After feeding wild-type or genetically modified mice with high-fat containing or specialized diets (such as the methionine and choline-deficient diet), researchers can harvest liver tissue for cell isolation and culture the cells within a hydrogel ECM stimulated with selected stem cell media or growth factors (Broutier et al., 2016). Recently, Elbadawy et al. (2020) modeled NAFLD progression via murine organoids assembled with hepatocytes and activated HSC isolated from animals at three distinct disease stages (mild, moderate and severe NASH) (Elbadawy et al., 2020). In doing so, Elbadawy et al. established a model to investigate the effect of genetic stability and lipid metabolism on NAFLD progression. The authors noticed larger size and enhanced release of pro-inflammatory cytokines in early stage organoid models, compared to later stage ones. Other murine organoids have been used to decipher liver metabolism and its response to therapeutic compounds. For example, Pittol et al. (2020)explored the dependency of metabolic functions on Farnesoid X receptor (FXR) isoforms; Sano et al. (2021) elucidated the associations among free amino acids, FFA and the Keap1-Nrf2 system – involved in the principal protective response to oxidative and electrophilic stresses (Turpaev, 2013) – in lipotoxic hepatocytes, while Liu et al. (2021) investigated the use of uridine to ameliorate NAFLD. Despite notable successes, murine-based organoids can only approximate the pathophysiology of human NAFLD (Kim et al., 2020), suggesting a human organoid model might provide a more suitable research platform.
Several approaches for the development of human organoids to model NAFLD (and its extra-hepatic organ involvement) have recently been published. Ouchi et al. (2019) created a model from human PSC by co-differentiating epithelial and stromal lineages to form spheroids, later embedded in Matrigel and cultured with retinoic acid and maturation media. This protocol successfully yielded NPC including HSC-like cells, Kupffer-like cells and biliary stem cells; as well as hepatocyte-like and cholangiocyte-like cells, as confirmed by single-cell transcriptomics and FACS analysis. Upon FFA exposure, the organoids acquired a NAFLD phenotype denoted by steatosis, inflammation, hepatocyte ballooning and collagen production; hallmarks of NASH. Gurevich et al. (2020) opted instead for iPSC-derived cryopreservable hepatocytes sourced from NASH donors for organoid development. When co-cultured with physiologically-relevant ratios of isogenic HSC-like and Kupffer-like cells on collagen-coated ultra-low binding plates, the hepatocytes assembled in organoids amenable to observation for over 10 days. This system, characterized by the formation of bile canaliculi, proved useful for drug metabolism studies. In alignment with scientific evidence (Teo et al., 2014), efficient hepatocyte differentiation was promoted via glycogen synthase kinase 3 (GSK3) inhibition, which is known to induce endoderm differentiation. Similarly to Ouchi et al. (2019), treatment with FFA induced a dose-dependent accumulation of intracellular lipids and establishment of a NASH phenotype. McCarron et al. (2021) instead developed bipotent ductal organoids derived from end-stage NASH patient liver explants, with normal donor livers as controls. Strikingly, NASH patient-derived organoids were highly diverse in terms of metabolic and pro-inflammatory pathways and response to drugs. Moreover, the model was characterized by increased sensitivity to apoptosis, increased FFA induced lipid accumulation, and reduced albumin secretion– all typical features of advanced NASH. Another hepatic organoid model was developed using posterior foregut and hepatic endoderm. FFA induction elicited increasing lipid peroxidation and ROS. Interestingly, this 3D model highlighted disruption of the bile canalicular network with disease progression (Ramli et al., 2020).
Several organoid models recapitulating NAFLD phenotypes have used bulk RNA sequencing (RNA-seq) to identify gene signatures and pathways associated with the disease (McCarron et al., 2021; Elbadawy et al., 2020; Beisner et al., 2021; Ouchi et al., 2019). Yet, only a few have included flow cytometry (Ouchi et al., 2019; Gurevich et al., 2020; Ramli et al., 2020; Abbey et al., 2020) or single-cell RNA-seq (Ouchi et al., 2019) in their research to determine the proportion of the different cell types present in these organoids. Quantifying such ratio might inform protocols offering tighter control over the development of these 3D systems, and help address two open challenges: heterogeneity and low reproducibility (Garreta et al., 2020).
The organoid technology, especially when based on human stem cells, holds great promise for investigating NAFLD pathophysiology due to the ability to overcome confounding factors introduced by animal models. Thanks to enhanced stability and complexity, organoid models offer a viable platform for future drug development pipelines. The establishment of new cryopreservation (Gurevich et al., 2020) and reproducibility (Nuciforo and Heim, 2020) standards will certainly aid in unlocking the potential of organoids to offer a preclinical alternative to animal models in the near future. Indeed, validated organoids could provide an easy to image and more accurate model of human NAFLD. Recently, Takebe et al. (2017) targeted organoids reproducibility by establishing a technological platform to manufacture standardized liver buds starting from 3 committed progenitors on a large scale (>108).
We note that the bioengineering field is boosting efforts toward controlling organoids' cellular complexity, providing vascular networks, and enhancing organoid maturation (Garreta et al., 2020). For example, Guye et al. (2016) showcased the use of heterologous GATA-binding factor 6 (GATA6) expression to trigger co-differentiation events within an iPSC population and obtain, within 2 weeks, a complex tissue with liver-like phenotype. In Velazquez et al. (2021), engineering of gene regulatory networks in PSC-derived human liver organoids enabled advancement of maturity and vascularization of the obtained in vitro system. In addition, gene editing using CRISPR/Cas9 has also been used in bioengineering to alter and study the function of specific genes. For example, Abbey et al. (2020) deleted Tribbles-1 (TRIB1) – a gene associated with NAFLD, in human iPSC which were then used to create hepatic spheroids.
Liver-on-a-chip
Despite having proved informative for the study of NAFLD and complex diseases, a major drawback of the static 3D cultures is the lack of long-term control over fluid shear stress, nutrient/gas exchange or waste removal. To enable the automated control of such factors, microfluidic technology and liver-on-a-chip approaches have been adopted to model the liver microenvironment (Underhill and Khetani, 2018b). While the cost and complexity of microfluidic setups constitutes a barrier to their diffusion in laboratories, the standardized fabrication of chips favors reproducibility, a hurdle in organoid research.
Several liver-on-a-chip systems have recently been proposed to study NAFLD. The first appearing in the scientific literature approximated the liver architecture and microvasculature by culturing HepG2 cells in parallel microchannels mimicking the endothelial barrier (Gori et al., 2016). This microfluidic device demonstrated the ability of FFA perfusion to recapitulate steatosis. Since then, the combination of multiple cell types made on-chip cultures amenable to the study of cell-cell interactions, better reflecting the complexity of the hepatic microenvironment. Suurmond et al. (2019) co-cultured HUVEC with GelMa-encapsulated HepG2 to investigate NAFLD pathogenesis. Similarly to Gori et al., their system, distinguished by high cell viability and homogeneous cell distribution, enabled monitoring of steatosis establishment. Inclusion of KC in this in vitro model led to enhanced ROS expression and proinflammatory cytokine release. While HepG2 are a widely used cell line to investigate regulation of drug-metabolizing enzymes, PHH are the preferred choice in biotransformation studies, thanks to their in vivo-like functionality (Wilkening et al., 2003).
In light of their similarities to PHH, researchers increasingly based in vitro models on HepaRG cells. Suurmond et al. (2019) highlighted that by substituting HepG2 with HepaRG cells, their liver-on-a-chip displayed elevated levels of proinflammatory cytokines and improved reversibility of steatosis. HepaRG, in conjunction with KC, HUVEC and HSC encapsulated in GelMA, were used in the microfluidic model proposed by Cho et al. (2021). Through a 21-day life-span, the system recapitulated NASH features and showed neovascularization. Freag et al. (2021) also developed an on-chip model by co-culturing primary human hepatic cell lines (hepatocytes, KC, HSC and LSEC). In this system, lipotoxic conditions successfully generated the NASH phenotype, including hepatocellular ballooning. Furthermore, a PHH-based liver-on-a-chip modeled the progressive intracellular lipid accumulation and reduction in cytochrome P activity observed in NAFLD patient samples (Kostrzewski et al., 2020). Interestingly, the presence of the PNPLA3 I148M variant in three of the donor samples allowed the authors to highlight that this variant can worsen the NASH phenotype through effects on HSC.
Despite the numerous advantages of liver-on-a-chip platforms to study NAFLD, the liver is not the only organ involved in the disease, with adipose tissue (Stojsavljević et al., 2014) and the gut (Scarpellini et al., 2014) known to play a key role in its progression. For this reason, it is important to study the interactions between multiple organs, and their contribution to disease establishment and progression. Multi-organ systems, also known as body-on-a-chip, are emerging as technological platforms to study the efficacy and safety of drugs (Picollet-D'hahan et al., 2021; Chen et al., 2020; Skardal et al., 2020). As most of the lipids accumulated in the liver are attributed to dietary intake, Lee and Sung (2018) focused on the gut-liver axis and developed an on-chip device generating hepatic steatosis. Their gut-liver-on-a-chip device consists of two chambers, incorporating Caco-2 (gut) and HepG2 (liver) cells, separated by a porous membrane and a scaffold on the intestinal part. Using this device, Lee and Sung showed that butyrate, a compound known to enhance the gut barrier function, attenuated lipid accumulation; a benefit not observed when the drug was tested on hepatocytes alone. Recently, Slaughter et al. (2021) developed a body-on-a-chip with adipose and liver tissue, to evaluate the mechanisms involved in NAFLD development and serve as a drug screening platform. The authors were able to validate the indirect influence of adipocytes on hepatocytes in NAFLD progression. At the cost of greater complexity, the examples above hint that body-on-a-chip devices may offer a better approximation of the in vivo NAFLD environment.
3D bio-printed liver-like tissues
Three-dimensional bio-printing is another promising technique for the development of in vitro models. 3D structures, including liver-like tissues, are generated via layer-by-layer, spatially controlled deposition of biological materials, biochemicals and cells (van Grunsven, 2017). A comparative analysis of the three most established methods to generate 3D bio-printed liver structures (e.g., inkjet-based, extrusion-based, and photocuring-based bioprinting), is extensively reviewed elsewhere (Murphy and Atala, 2014; Ma et al., 2020). Despite notable advantages, a challenge to the development and widespread use of 3D bio-printed structures is the selection of an appropriate bio-ink. Indeed, the material must show compatibility with both cells and the printing process, while retaining physicochemical –and therefore functional– properties analogous to the organ under consideration. Alginate, gelatin, and collagen are among the most routinely used materials for this purpose.
Extensive research has been carried out on bio-printed liver structures (Norona et al., 2016; Smith et al., 2004; van Grunsven, 2017). To the best of our knowledge, however, only one 3D bio-printed system has been used to generate the NAFLD phenotype (Kizawa et al., 2017). In this study, a pathological liver tissue was bio-printed using cells harvested from genetically obese Zucker rats with NAFLD. Specifically, the Kenzan method (Moldovan et al., 2017) was used to create a scaffold-free bio-printed structure from liver spheroids composed of rat hepatocytes and mouse fibroblasts. The authors observed high lipid content over 23 days, suggesting that the pathological conditions could be retained and monitored long-term. Despite being designed to investigate drug-induced liver injury, the sophistication of the bio-printed structure proposed by Nguyen et al. (2016) makes it an ideal candidate for the study of NAFLD and NASH. This two-compartment system includes NPC – HUVEC and HSC – seeded at the boundary of each compartment, filled with PHH.
In parallel, due to the shortage of donor organs and inherent risks, researchers are exploring alternative solutions to liver transplant in patients with advanced liver diseases. Emerging approaches in development include liver-like structures bio-printed from HepG2 (Jeon et al., 2017; Ide et al., 2020) or PHH (Kim et al., 2017, 2018) that could pave the way to artificial livers in the future.
3D bio-printing technology may also enhance the assembly of liver-on-a-chip systems, which present challenges such as biological culture preparation (Bhatia and Ingber, 2014). The 3D bio-printed organ-on-a-chip of Lee and Cho (2016) was devised to overcome limitations such as spatial heterogeneity and the difficulty of providing multiple types of ECM environments when studying cell–ECM interactions (Yeon and Park, 2007). HepG2 cells encapsulated in collagen type I, and HUVEC encapsulated in gelatin were used to construct 2D and 3D hydrogel-based on-chip devices. Similarly, Bhise et al. (2016) developed a liver-on-a-chip with bio-printed HepG2/C3A spheroids. The viability of both models, their extensive life-span and similarities with intact liver tissue, makes them promising systems for both NAFLD research and drug screening pipelines.
Taken together, although use in NAFLD research is limited, these studies highlight the potential use of 3D bio-printing. However, to enable more widespread use, the costs and specialized knowledge and expertise required to establish and maintain this technology will need to be addressed (van Grunsven, 2017).
Use of in vitro models in NAFLD drug development pipelines
The majority of clinical trials have focused on patients with NASH and bridging fibrosis, who have an increased risk of developing cirrhosis and adverse outcomes. Depending on the drug mechanism of action, the primary endpoint of interventional trials comprises either NASH resolution without worsening of fibrosis or a minimum of one-stage fibrosis improvement without worsening of NASH. Yet, most clinical trials have been unsuccessful and 4 out of 5 NASH treatments reaching phase III were recently terminated. This suggests monotherapies –individual drugs– might be insufficient to tackle the complex interaction between metabolic, inflammatory and fibrotic pathways that characterize this disease. As a result, evaluation of drug combinations is becoming a major focus of Pharma NASH pipelines. However, despite more than twenty years of intense research activity, no breakthrough has occurred. Indeed, the phase II ATLAS study (NCT03449446) failed to meet the primary endpoint, except for the firsocostat-cilofexor combination, currently subject of further investigation (Gilead Sciences, 2019). While a detailed discussion of the pharmaceutical compounds that have been part of clinical trials does not fall under the scope of this review, we point to extensive recent reviews that address this topic (Fallowfield et al., 2021; Campbell et al., 2021; Guirguis et al., 2021). In order to facilitate the reader's understanding of this section, we have provided two tables that summarize the most relevant information of the ongoing (Table 2) and the terminated/completed (Table 3) phase III clinical trials for pharmaceutical compounds in NAFLD.
Table 2.
Ongoing phase III clinical trials in NAFLD and NASH
| Compound/company | Study design | Mechanism of action | Intervention/control | Key inclusion criteria | Key exclusion criteria | Status |
|---|---|---|---|---|---|---|
| Obeticholic acid (OCA)/Intercept pharmaceuticals | REGENERATE. NASH patients (NCT02548351). Primary outcomes: ≥1 stage fibrosis improvement and no worsening of NASH or NASH resolution without worsening of fibrosis | Farnesoid X (FXR) receptor agonist | – OCA 10 mg/day (N= 330) – OCA 25 mg/day (N= 324) – Placebo (N= 312) |
– NASH diagnosed by liver biopsy with 3 key histological features of NASH CRN – F2/F3 or F1 with BMI≥30/type 2 diabetes/ALT>1.5 x ULN – Stable body weight |
– Model of end stage liver disease (MELD) score >12 – ALT≥10x ULN – HbA1c>9.5% – Other liver diseases – Liver transplant – Cirrhosis – BMI>45 kg/m2 |
Ongoing. Current results: – NASH resolution not met (p= 0.18/p= 0.13) (OCA 10 mg/OCA 25 mg) – Fibrosis improvement met (p= 0.045/p= 0.0002) |
| Aramchol/Galmedresearch and development, Ltd. | ARMOR. NASH (NCT04104321). Primary outcomes: ≥1 stage improvement fibrosis without worsening of NASH or NASH resolution without worsening fibrosis | Staroyl coenzyme A desaturase 1 inhibitor | –Aramchol 300 mg/day – Placebo (N total= 2000) |
– NASH diagnosed by liver biopsy – NAS≥4 with at least 1 on each NAS component – F2/F3 – BMI 25–40 kg/m2 – AST>20 IU/L |
– Cirrhosis – MELD score>12 – Other liver diseases – Weight loss>5% in last 3 months – Bariatric surgery – Treatment with anti-diabetic medications |
Ongoing |
| Resmetirom (MGL-3196)/Madrigal pharmaceuticals, Inc. | MAESTRO-NAFLD1 NAFLD patients (NCT04197479). Primary outcomes: assess the effect of daily oral admin. | Thyroid hormone receptor-β agonist | –Resmetirom 80 mg/day –Resmetirom open label or double-blinded 100 mg/day – Placebo (N total= 700) |
Suspected or confirmed NAFLD or NASH | – Other liver diseases – Cirrhosis – Bariatric surgery – HbA1c≥9% – GLP-1/vit. E/pioglitazone therapy – MELD>12 – ALT>250 U/L – Weight change>5% |
Ongoing |
| Resmetirom (MGL-3196)/Madrigal pharmaceuticals, Inc. | MAESTRO-NASH. NASH patients (NCT03900429). Primary outcomes: NASH resolution with <2 points in NAS and no worsening of fibrosis | Thyroid hormone receptor-β agonist | –Resmetirom 80 mg/day –Resmetirom 100 mg/day – Placebo (N total= 2000) |
Biopsy proven NASH with NAS≥4 and at least 1 on each component and F1/F2/F3 | – Other liver diseases – Cirrhosis – Bariatric surgery – HbA1c≥9– GLP-1/vit. E/pioglitazone therapy – MELD>12 – ALT>250 U/L |
Ongoing |
| Resmetirom (MGL-3196)/Madrigal pharmaceuticals, Inc. | MAESTRO-NAFLD-OLE. NAFLD patients (NCT04951219). Primary outcomes: assess the effect of daily oral admin. | Thyroid hormone receptor-β agonist | –Resmetirom 80 mg/day first 12 w followed by open-label –Resmetirom 100 mg/day –Resmetirom 100 mg/day first 12 w followed by open-label –Resmetirom 100 mg/day – Open-label Resmetirom 100 mg (N total= 1000) |
– Participated in MAESTRO-NAFLD-1 – Liver biopsy with NAS = 3 and F2/F3 or NAS≥4 in all NAS components and F1, PRO-C3≤14 |
– Other liver diseases – HCC |
Ongoing |
| Lanifibranor (IVA337)/Inventiva Pharma | NATiV3: NASH patients with F2/F3 (NCT04849728). Primary outcomes: resolution of NASH and improvement of fibrosis of ≥1 according to NASH CRN (Part 1). Assess the effect on delaying NASH disease progression measured by a composite endpoint that includes progression to cirrhosis, liver-related clinical outcome events, or all-cause death | Pan-PPAR agonist | –Lanifibranor 800 mg/day –Lanifibranor1200 mg/day – Placebo (N total= 2000) |
– Liver biopsy with steatosis score ≥1, act. score A3/A4 and F2/F3 according to Steatosis-Activity-Fibrosis (SAF) – Stable dose of GLP-1, vit. E or statins – No weight change ≥5% |
– Other chronic liver disease – HCC or cirrhosis – HbA1c>9% – Bariatric surgery |
Ongoing |
| Semaglutide/Novo Nordisk A/S | NASH patients with F2/F3 (NCT04822181). Primary outcomes: resolution of NASH and no worsening of fibrosis or improvement of fibrosis and no worsening of NASH (Part 1). Assess the effect on delaying NASH disease progression measured by a composite endpoint that includes progression to cirrhosis, liver-related clinical outcome events, or all-cause death | Glucagon-like peptide-1 receptor agonist (GLP-1 RA) | – Semaglutide 1 subcutaneous admin./week – Placebo (N total= 1200) |
– Liver biopsy with NASH – F2/F3 according to NASH CRN System – NAS score≥4 with 1 score in steatosis, inflammation and ballooning |
– Other liver diseases – Treatment with pioglitazone, vit. E or other glucose-lowering agents |
Ongoing |
Table 3.
Terminated and completed phase III clinical trials in NAFLD and NASH
| Compound/Company or academic lead | Study design | Mechanism of action | Intervention/control | Key inclusion criteria | Key exclusion criteria | Status |
|---|---|---|---|---|---|---|
| Metadoxine/Hospital General de Mexico | Non-diabetic patients with NAS>3 (NCT02541045). Primary outcome: Improvement of NAS | –↓Oxidative stress – Restoration NADH, ATP and GSH – Prevention increase collagen –↓TNF secretion |
–Metaxodine 500 mg/day – Placebo (N total= 108) |
– Non-diabetic patients – BMI≥25 – Proof of liver steatosis with ultrasonography – NAS≥3 with at least 1 point on each – Without fibrosis or fibrosis stage ≤ F2 according to NASH CRN classification |
– Cirrhosis – Other liver diseases – Uncontrolled chronic disease, hypothyroidism or hyperthyroidism |
Suspended due to lack of financial resources |
| Rimonabant (SR141716)/Sanofi | NASH patients with T2DM (NCT00577148) and without (NCT00576667) | Cannabinoid-1 receptor blocker | – Rimonabant 20 mg/day – Placebo (N total= 89 for NCT00577148and N= 165 for NCT00576667) |
NASH patients | – T1DM for NCT00577148 or type I/II diabetes for NCT00576667 – Other chronic liver disease – Previous or current HCC – Previous bariatric surgery |
Terminated by company decision taken in light of demands by certain national health authorities |
| Cenicriviroc (CVC)/AbbVie | AURORA: NASH patients with liver fibrosis (NCT03028740) Primary outcomes: Improvement of ≥1 stage in fibrosis according to NASH CRN System without worsening of NASH (Part 1); and improvement of histopathological progression, liver-related clinical outcomes and all-cause mortality (Part 2) | Chemokine 2 and 5 receptor antagonist | – CVC 150 mg/day – Placebo (N total= 1779) |
– Proof NASH based on liver biopsy – Subjects in part 1: F2/F3 per the NASH CRN System based on liver biopsy. Subjects in Part 2: F3 per the NASH CRN System, based on liver biopsy. |
– Other liver diseases or serious infections – Liver transplantation – HbA1c>10% at Screening – Weight reduction ≥7% through bariatric surgery in the past 5 years or bariatric surgery planned during study – Malignancy within past 5 years or ongoing, other than basal cell carcinoma or resected noninvasive cutaneous squamous cell carcinoma – GLP-1 agonist, DPP-4 inhibitor, SGLT2 and/or SGLT1 inhibitor, TZD for ¡6 mo. before screening |
Terminated due to lack of efficacy in Part I |
| Elafibranor (GTF505)/Genfit | RESOLVE-IT: NASH patients with fibrosis (NCT02704403). Primary outcomes: NASH resolution without worsening of fibrosis; composite long-term outcome composed of all-cause mortality, cirrhosis, liver-related clinical outcome | Peroxisome proliferator-activated receptor-α and peroxisome proliferator-activated receptor-δ agonist | – Elafibranor 120 mg/day – Placebo (N total= 2157) |
– BMI≤45 kg/m2 – NASH confirmation by liver biopsy with at least 1 in each component NAS score – NAS score≥4 – Fibrosis stage≥1 or <4 according to the NASH CRN system |
– Known heart failure – Bariatric surgery – Uncontrolled hypertension – T1DM patients – HbA1c>9% – Weight loss >5% within last 6 months – Compensated and decompensated cirrhosis – Other chronic liver diseases |
Terminated due to failure to meet primary efficacy endpoint |
| Selonsertib (SEL/GS-4997)/Gilead Sciences | NASH patients and F3 (NCT03053050) Primary outcomes: ≥1 stage improvement in fibrosis according to NASH CRN System and event-free survival | ASK-1 inhibitor | – SEL 6 mg/day – SEL 18 mg/day – Placebo (N total= 808) |
– Liver biopsy NASH and F3 according to NASH CRN System – HbA1c≥9.5– ALT ≤ x8 ULN |
– MELD score >12 – Other liver diseases – Liver transplantation – HCC |
Terminated due to lack of efficacy |
| Diamel/Catalysis SL | NASH with insulin resistance (NCT00820651). Primary outcome: histological improvement | – Antioxidant – Biocatalyst –↓ Gastrointestinal absorption glucose |
–Diamel 660 mg/x2 every 8h – Placebo: hypocaloric diet of 1620 kcal daily (N total= 158) |
Histological diagnosis of NASH | – Other liver disease – Decompensated cirrhosis – Fasting glucose levels >250 mg/dL |
Completed (no results available) |
| Pioglitazone or vit. E/National Institute of diabetes and Digestive and Kidney diseases (NIDDK) | PIVENS: NASH (NCT00063622). Primary outcomes: improvement of ≥1 hepatocyte ballooning, NAS≤3 or decrease of 2 points and no worsening of fibrosis | – Vit. E: antioxidant – Pioglitazone: thiazolidinedione that targets insulin resistance and adipose tissue dysfunction |
– Pioglitazone 30 mg/day (N= 80) – Vit. E 800 IU/day (N= 84) – Placebo (N= 83) |
NASH based on liver biopsy | – Adults with diabetes – Other liver or cardiovascular diseases – Cirrhosis |
Completed. Significant difference for vit. E (p= 0.001), but not with pioglitazone (p= 0.04). Improvement in serum ALT and AST (p <0.001), steatosis (p= 0.005/p= 0.001) (vit. E/pioglitazone) and inflammation (p= 0.02/p= 0.004); but not fibrosis scores (p= 0.24/p= 0.12). (Pre-specified level of significance was p= 0.025) |
| Losartan/Newcastle-upon-Tyme Hospitals NHS Trust | FELINE: NASH patients (NCT01051219). Primary outcome: change in histological fibrosis stage (NASH CRN system) | Angiotensin II receptor type 1 (AT1) antagonist | – Losartan 50 mg/day (N= 15) – Placebo (N= 17) |
NASH and fibrosis stage 1–3 NASH CRN System | – Use of ACEI or ARBs in past year – Change in diabetes regimen in last 3 months – Weight loss >50% in last 6 months |
Study was terminated early due to slow recruitment, but patients were allowed to complete the study if they wanted to. No significant results |
| Oltipraz/PharmaKing | NAFLD patients (NCT02068339). Primary outcomes: change in liver fat assessed by MRS | AMP-activated protein kinase (AMPK) activator | –Oltipraz 90 mg/day –Oltipraz 120 mg/day – Placebo (N total= 283) |
– NAFLD patients – Abnormal ALT/AST |
– Cirrhosis – AST/ALT >2 ratio – T1DM/T2DM – Other liver diseases/cancer – Vit. E consumption – Bariatric surgery within 6 months |
Completed. No results available |
| Pentoxifylline (PO TID)/NortwesternUniversity | NASH patients (NCT00267670). Primary outcomes: Improvement of ALT≥30% change | Non-specific phosphodiesterase inhibitor | – PO TID 400 mg/day (N= 19) – Placebo (N= 7) |
– Steatosis score ≥1 – ALT ≥ x1.5 ULN – HgbA1c<7% |
– Decompensated cirrhosis – Current anti-diabetic, anti-TNF-α or vit. E medication |
Completed. No significant change in ALT (p= 0.08) |
In vitro cell culture assays have been used to support pre-clinical development of candidate therapies and to validate the utility of new cellular models. For instance, obeticholic acid (OCA) has been tested in human iPSC-derived hepatocytes (Parafati et al., 2018), a co-culture of human hepatoma cells (Huh7) and HSC (LX-2) (Anfuso et al., 2020) and an organotypic human hepatocyte system (Dash et al., 2017). Boeckmans et al. (2020) investigated the properties of several PPAR agonists in PHH, HepaRG, HepG2, LX-2 and human skin stem cell-derived hepatic cells. Other studies, include the use of human HSC (HSC-T6 and LX-2) to determine the efficacy of selonsertib in liver fibrosis (Yoon et al., 2020) or primary rat HSC to evaluate the effects of pioglitazone in vitro (Kawaguchi et al., 2004).
Additionally, fibrogenic spheroids have been used to test the efficacy of putative drugs (e.g., sorafenib (Romualdo et al., 2021), lanifibranor, elafibranor, or cenicriviroc (Hurrell et al., 2020)) in mediating resolution of NASH, thereby validating the role of spheroids for drug screening.
Beyond being suitable models to elucidate NAFLD pathogenesis, liver-on-a-chip systems also play a key role in drug development. To date, Gori et al. (2016), Freag et al. (2021), Cho et al. (2021), Du et al. (2021) and Gori et al. (2021) have used liver-on-a-chip devices to test the ability of polyphenols, OCA, elafibranor, and pirfenidone to ameliorate NAFLD. Interestingly, while the disease-modifying effects of elafibranor were shown in a liver-on-a-chip assay, this did not translate to clinical efficacy in a pivotal phase III NASH trial. As a consequence, the drug program was recently terminated (GENFIT, 2020). These contrasting in vitro versus in vivo observations indicate that preclinical NAFLD models still have a long way to go before reliably simulating human NAFLD and predicting patient responses to therapies.
Translation of in vitro platforms
The challenge of identifying an efficacious treatment for the disease is evidenced by the litany of recent clinical trial failures that have encompassed a diverse range of targets. Notwithstanding, many synthetic drugs are currently in development as monotherapy or combination therapy regimens. The availability of reliable human-based in vitro models would help to address this challenge, by both enhancing our understanding of NAFLD progression and predicting the therapeutic response to drug candidates. So far, in vitro models have only managed to recapitulate specific aspects of the in vivo disease phenotype. Indeed, the establishment of multi-dimensional systems that reflect the combined effects of genetics, sex, comorbidities and inter-organ cross-talk remains a pipe dream. Mounting evidence suggests, however, that the complexity of in vitro models will be key to screen drugs and to predict clinically relevant therapeutic responses. This is illustrated in Lee and Sung (2018), where the beneficial effect of butyrate on lipid accumulation only became apparent in a gut-liver-on-chip, but was not detected when the compound was tested using hepatocytes alone.
An ideal NAFLD in vitro model used for drug screening should satisfy biological fidelity and usability requirements. Biological fidelity depends on the selection of appropriate liver cells (parenchymal cells and NPC) and their aggregation in a structure enabling cell-cell and cell-ECM interactions. Future trends will likely see a transition from immortalized cell lines to PSC, due to their almost unlimited supply, life-span and similarity to in vivo human phenotype (Hay et al., 2007; Gao and Liu, 2017). For the observed response to be shaped by inter-organ cross-talk, these systems need to be linked with intestinal, pancreatic and adipose tissue models. 3D liver structures cultured in perfusion devices are quickly developing to meet these standards. Usability of such models relies instead on their lifetime (e.g., a minimum of 2–3 weeks of observation are needed for cytotoxicity testing) and the availability of specialized know-how for their operation.
To ensure clinical relevance, NAFLD in vitro models require validation against known features of in vivo human pathogenic processes or responses to therapeutic agents. Such validation could be performed via an ‘omics’ approach, which can evaluate the degree of pathological equivalence between an in vitro model and human liver samples at the molecular level, as measured by differentially expressed genes and enriched pathways. Such a strategy has been adopted by Ramli et al. (2020) and Feaver et al. (2016).
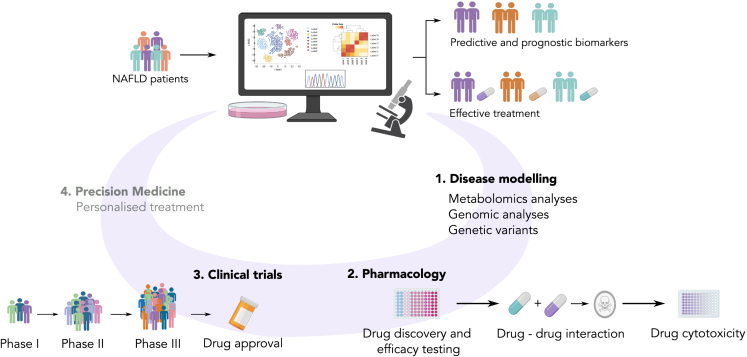
Alternatively, models could be assessed by their ability to reproduce results obtained in ongoing clinical trials. For example, Hurrell et al. (2020) demonstrated the potential of their system as a drug screening platform by testing pharmaceuticals that were in phase II or phase III clinical development. However, robust human validation and critical comparative analyses are rarely performed, with most researchers focusing on specific genes and pathways of interest (e.g., the expression level of TGF-α, α-SMA or TNF-β eliciting fibrosis and inflammation) when designing in vitro systems for NAFLD. Ideally, extensively validated platforms would allow more reliable pharmacodynamic-pharmacokinetic and toxicity assays of candidate compounds for NAFLD treatment, de-risking subsequent clinical trials. Several drugs that are currently in phase III have undergone preliminary in vitro testing. For instance, Aramchol was tested in bioassays on PHH, HSC, and pHSC (Fernández-Ramos et al., 2020; Bhattacharya et al., 2021). While not using 3D in vitro models, the authors selected multiple cell lines to investigate genes and pathways modulated by Aramchol (e.g., downregulation of SCD1, COL1A1, and ACTA2; upregulation of PPARG). This cost-effective strategy benchmarks a putative drug according to multiple aspects of NAFLD, before proceeding to test its efficacy in suitable cohorts of patients. Similarly, in vitro studies with OCA in co-cultures of Huh7 and HSC (Anfuso et al., 2020) or PHH collagen gel sandwich (Dash et al., 2017), have delineated mechanisms of action including modulation of collagen and MMP2 and 9 activity, as well as its possible toxicological effects. Figure 4 shows the different potential applications of in vitro models in translational research for NAFLD.
Figure 4.
Potential applications of in vitro models in translational research for NAFLD
The combination of ‘omics’ and cell cultures has the potential to enable the study NAFLD patients ‘in-the-dish’, resulting in the design of effective treatment or identification of predictive and prognostic biomarkers. In vitro NAFLD models, combined with metabolomic and genomic analyses/genetic variant studies, permit the identification of pharmacological targets and susceptible patient subpopulations. Once a putative drug – or combination of drugs – has been indicated as a candidate NAFLD therapy, several pharmacological investigations test the efficacy, cytotoxicity, and possible interactions between compounds. Drugs that are successfully triaged are funneled to clinical evaluation. The ultimate goal –which has not yet been achieved– would employ a precision medicine approach (gray) to the design of patient-tailored NAFLD treatments.
Despite the advantages they offer, 3D in vitro models remain expensive and inaccessible to new users. Indeed, liver-on-a-chip and bio-printed liver-like tissues are quite complex and steep learning curves are associated with their operation. Ideally, technological advancements and the establishment of rigorous standards will enable these models to become a concrete alternative to animal models in the near future.
Future perspectives
Emerging in vitro human-based models have enhanced our understanding of NAFLD evolution and disease mechanisms. However, open challenges remain. Among these, the most prominent are the ability of the model to encompass the whole spectrum of the disease, to reproduce the inter-individual variability observed in NAFLD patients and capture the interaction between multiple organs involved in the pathogenesis of the disease. We expect that in the near future complex in vitro models, such as body-on-a-chip based on iPSC or PHH, will become the platform of choice to mimic NAFLD establishment and progression. Inclusion of physiologically-based toxicokinetic computer modeling to study ADME (absorption, distribution, metabolism, and excretion) in these systems will likely represent a further step toward the replication of the human liver microenvironment.
The availability of validated preclinical models will streamline the adoption of a quantitative approach to (a) the identification of NAFLD etiology and potential therapeutic targets, and (b) the rational design of pharmacological treatments. These aims are increasingly pursued in a biological systems control perspective, wherein a disease arises due to failure of multi-scale control mechanisms ensuring robustness of biological processes to internal/environmental perturbations. Using this approach, mathematical models and gene regulatory networks reconstructed from ‘omics’ data provide new insights on the establishment of diseases; and the design of personalized effective treatments can be framed as an engineering control problem. For example, Liao et al. (2020) used a kinetic model of hepatic fructose metabolism to simulate the effect of excessive dietary uptake of this sugar on dyslipidemia observed in early NAFLD patients. The authors identified fructokinase as a molecular target of the fructose pathway and predicted its suppression would revert lipid accumulation. Van Riel et al. (2020) used instead a machine learning algorithm, informed by metabolomic and transcriptomic time-series, to predict the metabolic response induced by treatment with a Liver X Receptor (LXR) agonist. Currently, 3D models are mainly used for biological studies due to their cost and low-throughput. Tackling such limitations will prompt adoption of these in vitro models by the pharmaceutical industry. Indeed, within drug development pipelines, these models would allow only the most promising compounds to funnel toward clinical trials, with obvious economic benefit. If used in combination with disease-relevant ex vivo models (e.g., human precision-cut liver slices) (Palma et al., 2019), in vitro models might help to reduce the (over)reliance on in vivo models for drug efficacy and toxicological studies.
We expect complex in vitro platforms to be increasingly integrated with ‘big data’ for compound testing, identification of therapeutic targets, and personalized medicine applications. Precision medicine approaches would enable, first, the clustering of NAFLD patients by disease stage and/or genetic variants (e.g., PNPLA3) and the development of in vitro models from patient-derived iPSC on which to assess safety and efficacy of candidate therapies. Second, these studies could identify novel companion biomarkers of NAFLD progression or regression, thereby supporting the design of non-invasive diagnostic tests.
In conclusion, we have provided a comprehensive overview of various in vitro models proposed to study the pathophysiology of NAFLD. The different models available for this disease have specific advantages and disadvantages. Accordingly, the choice of model may depend on the particular context of use.
Acknowledgments
M.J.R. is an iCASE PhD student funded by the Medical Research Council and Galecto Biotech. F.M. is supported by an EPSRC Innovation Fellowship (EP/S001921/1) and an EPSRC New Investigator Award (EP/R035350/1). L.B. is supported by EPSRC funding EP/P017134/1-CONDSYC.
Author contributions
M.J.R. searched and screened the articles for the systematic analysis according to title and abstract. M.J.R. and L.B. screened the full-text and extracted the data from the retrieved publications. M.J.R performed the statistical analyses. M.J.R, L.B., J.A.F., and F.M. interpreted the results. M.J.R. and L.B. drafted the manuscript and designed the figures. All authors edited, reviewed, and approved the final manuscript.
Declaration of interests
JA Fallowfield has served as a consultant or advisory board member for Redx Pharma, Novartis, Ferring Pharmaceuticals, Macrophage Pharma, Galecto Biotech, Caldan Therapeutics, Cypralis Ltd, Rallybio, Tectonic Therapeutic, River 2 Renal Corp.,Gilde Healthcare, Guidepoint, Techspert.io and has received research grant funding from Novartis and Intercept Pharmaceuticals. M Jimenez-Ramos MRC iCASE PhD studentship is partially funded by Galecto Biotech.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103549.
Supplemental information
This table includes the title of the publication, the year it was published, and the journal where it was published.
References
- Abbey D., Elwyn S., Hand N.J., Musunuru K., Rader D.J. Self-organizing human induced pluripotent stem cell hepatocyte 3D organoids inform the biology of the pleiotropic trib1 gene. Hepatol.Commun. 2020;4:1316–1331. doi: 10.1002/hep4.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfuso B., Tiribelli C., Adorini L., Rosso N. Obeticholic acid and INT-767 modulate collagen deposition in a NASH in vitro model. Sci.Rep. 2020;10:1–12. doi: 10.1038/s41598-020-58562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulou M., Gordillo R., Koliaki C., Gancheva S., Jelenik T., De Filippo E., Herder C., Markgraf D., Jankowiak F., Esposito I., et al. Specific hepatic sphingolipids relate to insulin resistance, oxidative stress, and inflammation in nonalcoholic steatohepatitis. Diabetes Care. 2018;41:1235–1243. doi: 10.2337/dc17-1318. [DOI] [PubMed] [Google Scholar]
- Armstrong M.J., Hull D., Guo K., Barton D., Hazlehurst J.M., Gathercole L.L., Nasiri M., Yu J., Gough S.C., Newsome P.N., et al. Glucagon-like peptide 1 decreases lipotoxicity in non-alcoholic steatohepatitis. J.Hepatol. 2016;64:399–408. doi: 10.1016/j.jhep.2015.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahi J., Kamo H., Baba R., Doi Y., Yamashita A., Murakami D., Hanada A., Hirano T. Bisphenol a induces endoplasmic reticulum stress-associated apoptosis in mouse non-parenchymal hepatocytes. Life Sci. 2010;87:431–438. doi: 10.1016/j.lfs.2010.08.007. [DOI] [PubMed] [Google Scholar]
- ATCC https://www.atcc.org/products/crl-324
- Bale S.S., Golberg I., Jindal R., McCarty W.J., Luitje M., Hegde M., Bhushan A., Usta O.B., Yarmush M.L. Long-term coculture strategies for primary hepatocytes and liver sinusoidal endothelial cells. Tissue Eng.Part C: Methods. 2015;21:413–422. doi: 10.1089/ten.TEC.2014.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquerizo A., Bañares R., Saliba F. Transplantation of the Liver; Elsevier BV; 2015. Current Clinical Status of the Extracorporeal Liver Support Devices; pp. 1463–1487. [DOI] [Google Scholar]
- Barbero-Becerra V.J., Giraudi P.J., Chavez-Tapia N.C., Uribe M., Tiribelli C., Rosso N. The interplay between hepatic stellate cells and hepatocytes in an in vitro model of NASH. Toxicol. Vitro. 2015;29:1753–1758. doi: 10.1016/J.TIV.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Beisner J., Filipe Rosa L., Kaden-Volynets V., Stolzer I., Günther C., Bischoff S.C. Prebiotic inulin and sodium butyrate attenuate obesity-induced intestinal barrier dysfunction by induction of antimicrobial peptides. Front.Immunol. 2021;12:1975. doi: 10.3389/fimmu.2021.678360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.C., Hendriks D.F., Moro S.M., Ellis E., Walsh J., Renblom A., Puigvert L.F., Dankers A.C., Jacobs F., Snoeys J., et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci.Rep. 2016;6:1–13. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.C., Dankers A.C., Lauschke V.M., Sison-Young R., Jenkins R., Rowe C., Goldring C.E., Park K., Regan S.L., Walker T., et al. Comparison of hepatic 2D sandwich cultures and 3D spheroids for long-term toxicity applications: a multicenter study. Toxicol. Sci. 2018;162:655–666. doi: 10.1093/toxsci/kfx289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E., Géloën A. Adipocytes as lipid sensors of oleic acid transport through a functional Caco-2/HT29-MTX intestinal barrier. Adipocyte. 2019;8:83–97. doi: 10.1080/21623945.2019.1580842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiaume F., Moghe P.V., Toner M., Yarmush M.L. Effect of extracellular matrix topology on cell structure, function, and physiological responsiveness: hepatocytes cultured in a sandwich configuration. Faseb J. 1996;10:1471–1484. doi: 10.1096/fasebj.10.13.8940293. [DOI] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat.Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Bhatia S., Balis U., Yarmush M., Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: role of homotypic cell interactions. Biotechnol.Prog. 1998;14:378–387. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Basta B., Mato J.M., Craig A., Fernández-Ramos D., Lopitz-Otsoa F., Tsvirkun D., Hayardeny L., Chandar V., Schwartz R.E., et al. Aramchol downregulates stearoyl CoA-desaturase 1 in hepatic stellate cells to attenuate cellular fibrogenesis. JHEP Rep. 2021;3:100237. doi: 10.1016/j.jhepr.2021.100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhise N.S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M., Lang Q., Zhang Y.S., Shin S.R., Calzone G., et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- Blazina I., Selph S. Diabetes drugs for nonalcoholic fatty liver disease: a systematic review. Syst.Rev. 2019;8:1–13. doi: 10.1186/s13643-019-1200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmans J., Natale A., Rombaut M., Buyl K., Rogiers V., De Kock J., Vanhaecke T., Rodrigues R.M. Anti-nash drug development hitches a lift on ppar agonism. Cells. 2020;9:37. doi: 10.3390/cells9010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon E.F., Raap C.D., Meijerman I., Beijnen J.H., Schellens J.H. An update on in vitro test methods in human hepatic drug biotransformation research: pros and cons. Toxicol.Appl.Pharmacol. 2003;189:233–246. doi: 10.1016/S0041-008X(03)00128-5. [DOI] [PubMed] [Google Scholar]
- Briske-Anderson M.J., Finley J.W., Newman S.M. The influence of culture time and passage number on the morphological and physiological development of Caco-2 cells. Proc.Soc.Exp. Biol.Med. 1997;214:248–257. doi: 10.3181/00379727-214-44093. [DOI] [PubMed] [Google Scholar]
- Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.-K., Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat.Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- Brunt E.M., Wong V.W.-S., Nobili V., Day C.P., Sookoian S., Maher J.J., Bugianesi E., Sirlin C.B., Neuschwander-Tetri B.A., Rinella M.E. Nonalcoholic fatty liver disease. Nat.Rev. Dis.Prim. 2015;1:1–22. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- Campbell P., Symonds A., Barritt IV A.S. Therapy for nonalcoholic fatty liver disease: current options and future directions. Clin.Therapeut. 2021;43:500–517. doi: 10.1016/j.clinthera.2021.01.021. [DOI] [PubMed] [Google Scholar]
- Cariou B., Byrne C.D., Loomba R., Sanyal A.J. Nonalcoholic fatty liver disease as a metabolic disease in humans: a literature review. Diabetes Obes.Metabol. 2021;23:1069–1083. doi: 10.1111/dom.14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanput W., Mes J.J., Wichers H.J. Thp-1 cell line: an in vitro cell model for immune modulation approach. Int.Immunopharm. 2014;23:37–45. doi: 10.1016/j.intimp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ma K. Nlrc4 inflammasome activation regulated by TNF-α promotes inflammatory responses in nonalcoholic fatty liver disease. Biochem.Biophys. Res. Commun. 2019;511:524–530. doi: 10.1016/j.bbrc.2019.02.099. [DOI] [PubMed] [Google Scholar]
- Chen J., Vitetta L. Gut microbiota metabolites in NAFLD pathogenesis and therapeutic implications. Int. J. Mol. Sci. 2020;21:5214. doi: 10.3390/ijms21155214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yang Y., Ueno H., Esch M.B. Body-in-a-cube: a microphysiological system for multi-tissue co-culture with near-physiological amounts of blood surrogate. Microphysiol.Syst. 2020;4 doi: 10.21037/mps-19-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-J., Kim H.-J., Lee K., Lasli S., Ung A., Hoffman T., Nasiri R., Bandaru P., Ahadian S., Dokmeci M.R., et al. Bioengineered multicellular liver microtissues for modeling advanced hepatic fibrosis driven through non-alcoholic fatty liver disease. Small. 2021;17:2007425. doi: 10.1002/smll.202007425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.J., Choi D. Successful mouse hepatocyte culture with sandwich collagen gel formation. J. Kor. Surg. Soc. 2013;84:202–208. doi: 10.4174/jkss.2013.84.4.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L.-W., Tsai C.-L., Yang K.-C., Liao C.-C., Hsu Y.-C. Targeting protein palmitoylation decreases palmitate-induced sphere formation of human liver cancer cells. Mol. Med. Rep. 2020;22:939–947. doi: 10.3892/mmr.2020.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapper J.R., Hendricks M.D., Gu G., Wittmer C., Dolman C.S., Herich J., Athanacio J., Villescaz C., Ghosh S.S., Heilig J.S., et al. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am. J. Physiol. Gastrointest.Liver Physiol. 2013;305:G483–G495. doi: 10.1152/ajpgi.00079.2013. [DOI] [PubMed] [Google Scholar]
- Coll M., Perea L., Boon R., Leite S.B., Vallverdú J., Mannaerts I., Smout A., El Taghdouini A., Blaya D., Rodrigo-Torres D., et al. Generation of hepatic stellate cells from human pluripotent stem cells enables in vitro modeling of liver fibrosis. Cell Stem Cell. 2018;23:101–113. doi: 10.1016/j.stem.2018.05.027. [DOI] [PubMed] [Google Scholar]
- Damelin L.H., Coward S., Kirwan M., Collins P., Selden C., Hodgson H.J. Fat-loaded HepG2 spheroids exhibit enhanced protection from pro-oxidant and cytokine induced damage. J. Cell. Biochem. 2007;101:723–734. doi: 10.1002/jcb.21229. [DOI] [PubMed] [Google Scholar]
- Dash A., Figler R., Blackman B., Marukian S., Collado M., Lawson M., Hoang S., Mackey A., Manka D., Cole B., et al. Pharmacotoxicology of clinically-relevant concentrations of obeticholic acid in an organotypic human hepatocyte system. Toxicol. Vitro. 2017;39:93–103. doi: 10.1016/j.tiv.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongiovanni P., Romeo S., Valenti L. Genetic factors in the pathogenesis of nonalcoholic fatty liver and steatohepatitis. Biomed. Res. Int. 2015;2015:460190. doi: 10.1155/2015/460190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin.Invest. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K., Li S., Li C., Li P., Miao C., Luo T., Qiu B., Ding W. Modeling nonalcoholic fatty liver disease on a liver lobule chip with dual blood supply. Acta Biomater. 2021;134:228–239. doi: 10.1016/j.actbio.2021.07.013. [DOI] [PubMed] [Google Scholar]
- Dunn J.C., Tompkins R.G., Yarmush M.L. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol.Prog. 1991;7:237–245. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- Eilenberger C., Selinger F., Rothbauer M., Lin Y., Limbeck A., Schädl B., Grillari J., Kavok N.S., Klochkov V.K., Malyukin Y.V., et al. Cytotoxicity, retention, and anti-inflammatory effects of a CeO2 nanoparticle-based supramolecular complex in a 3D liver cell culture model. ACS Pharmacol. Transl. Sci. 2020;4:101–106. doi: 10.1021/acsptsci.0c00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed W.M., Aboul-Fadl T., Roberts J.C., Lamb J.G., Franklin M.R. Murine hepatoma (Hepa1c1c7) cells: a responsive in vitro system for chemoprotective enzyme induction by organoselenium compounds. Toxicol.Vitro. 2007;21:157–164. doi: 10.1016/j.tiv.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbadawy M., Yamanaka M., Goto Y., Hayashi K., Tsunedomi R., Hazama S., Nagano H., Yoshida T., Shibutani M., Ichikawa R., et al. Efficacy of primary liver organoid culture from different stages of non-alcoholic steatohepatitis (NASH) mouse model. Biomaterials. 2020;237:119823. doi: 10.1016/j.biomaterials.2020.119823. [DOI] [PubMed] [Google Scholar]
- Engl T., Boost K.A., Leckel K., Beecken W.-D., Jonas D., Oppermann E., Auth M.K., Schaudt A., Bechstein W.-O., Blaheta R.A. Phosphorylation of hepatocyte growth factor receptor and epidermal growth factor receptor of human hepatocytes can be maintained in a (3D) collagen sandwich culture system. Toxicol.Vitro. 2004;18:527–532. doi: 10.1016/j.tiv.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Eslam M., Valenti L., Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J. Hepatol. 2018;68:268–279. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Estes C., Anstee Q.M., Arias-Loste M.T., Bantel H., Bellentani S., Caballeria J., Colombo M., Craxi A., Crespo J., Day C.P., et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J. Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- Fallowfield J.A., Jimenez-Ramos M., Robertson A. Emerging synthetic drugs for the treatment of liver cirrhosis. Expert Opin.Emerg. Drugs. 2021:1–15. doi: 10.1080/14728214.2021.1918099. [DOI] [PubMed] [Google Scholar]
- Feaver R.E., Cole B.K., Lawson M.J., Hoang S.A., Marukian S., Blackman B.R., Figler R.A., Sanyal A.J., Wamhoff B.R., Dash A. Development of an in vitro human liver system for interrogating nonalcoholic steatohepatitis. JCI insight. 2016;1:e90954. doi: 10.1172/jci.insight.90954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Ramos D., Lopitz-Otsoa F., Delacruz-Villar L., Bilbao J., Pagano M., Mosca L., Bizkarguenaga M., Serrano-Macia M., Azkargorta M., Iruarrizaga-Lejarreta M., et al. Arachidyl amido cholanoic acid improves liver glucose and lipid homeostasis in nonalcoholic steatohepatitis via ampk and mtor regulation. World J. Gastroenterol. 2020;26:5101. doi: 10.3748/wjg.v26.i34.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn T.J., Ferguson M.S. Multiendpoint mechanistic profiling of hepatotoxicants in HepG2/C3A human hepatoma cells and novel statistical approaches for development of a prediction model for acute hepatotoxicity. Toxicol. Vitro. 2008;22:1618–1631. doi: 10.1016/j.tiv.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Freag M.S., Namgung B., Reyna Fernandez M.E., Gherardi E., Sengupta S., Jang H.L. Human nonalcoholic steatohepatitis on a chip. Hepatol.Commun. 2021;5:217–233. doi: 10.1002/hep4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji S.H., Kashyap M.L., Kamanna V.S. Niacin inhibits fat accumulation, oxidative stress, and inflammatory cytokine IL-8 in cultured hepatocytes: impact on non-alcoholic fatty liver disease. Metabolism. 2015;64:982–990. doi: 10.1016/j.metabol.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Gao X., Liu Y. A transcriptomic study suggesting human iPSC-derived hepatocytes potentially offer a better in vitro model of hepatotoxicity than most hepatoma cell lines. Cell Biol. Toxicol. 2017;33:407–421. doi: 10.1007/s10565-017-9383-z. [DOI] [PubMed] [Google Scholar]
- Garcia M.C., Amankwa-Sakyi M., Flynn T.J. Cellular glutathione in fatty liver in vitro models. Toxicol. Vitro. 2011;25:1501–1506. doi: 10.1016/j.tiv.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Garreta E., Kamm R.D., de Sousa Lopes S.M.C., Lancaster M.A., Weiss R., Trepat X., Hyun I., Montserrat N. Rethinking organoid technology through bioengineering. Nat. Mater. 2020:1–11. doi: 10.1038/s41563-020-00804-4. [DOI] [PubMed] [Google Scholar]
- Gaskell H., Sharma P., Colley H.E., Murdoch C., Williams D.P., Webb S.D. Characterization of a functional C3A liver spheroid model. Toxicol. Res. 2016;5:1053–1065. doi: 10.1039/c6tx00101g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GENFIT GENFIT: announces results from interim analysis of RESOLVE-IT phase 3 trial of elafibranor in adults with NASH and fibrosis. 2020. https://ir.genfit.com/news-releases/news-release-details/genfit-announces-results-interim-analysis-resolve-it-phase-3/
- German C.L., Madihally S.V. Type of endothelial cells affects HepaRG cell acetaminophen metabolism in both 2D and 3D porous scaffold cultures. J. Appl. Toxicol. 2019;39:461–472. doi: 10.1002/jat.3737. [DOI] [PubMed] [Google Scholar]
- Gilazieva Z., Ponomarev A., Rutland C., Rizvanov A., Solovyeva V. Promising applications of tumor spheroids and organoids for personalized medicine. Cancers. 2020;12:2727. doi: 10.3390/cancers12102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilead Sciences I. Gilead announces topline results from phase 2 atlas study in patients with bridging fibrosis (F3) and compensated cirrhosis (f4) due to nonalcoholic steatohepatitis (NASH) 2019. https://www.gilead.com/news-and-press/press-room/press-releases/2019/12/gilead-announces-topline-results-from-phase-2-atlas-study-in-patients-with-bridging-fibrosis-f3-and-compensated-cirrhosis-f4-due-to-nonalcoholic-s
- Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Boettger J., et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and adme. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Matos A.F., Júnior W.S.S., Valerio C.M. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetol. Metab. Syndr. 2020;12:1–20. doi: 10.1186/s13098-020-00570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lechon M., Donato M., Castell J., Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr. Drug Metab. 2003;4:292–312. doi: 10.2174/1389200033489424. [DOI] [PubMed] [Google Scholar]
- Gomez-Lechon M.J., Donato M.T., Martínez-Romero A., Jiménez N., Castell J.V., O’Connor J.-E. A human hepatocellular in vitro model to investigate steatosis. Chem. Biol.Interact. 2007;165:106–116. doi: 10.1016/j.cbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Gori M., Simonelli M.C., Giannitelli S.M., Businaro L., Trombetta M., Rainer A. Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS One. 2016;11:e0159729. doi: 10.1371/journal.pone.0159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori M., Giannitelli S.M., Zancla A., Mozetic P., Trombetta M., Merendino N., Rainer A. Quercetin and hydroxytyrosol as modulators of hepatic steatosis: a NAFLD-on-a-chip study. Biotechnol.Bioeng. 2021;118:142–152. doi: 10.1002/bit.27557. [DOI] [PubMed] [Google Scholar]
- Graffmann N., Ring S., Kawala M.-A., Wruck W., Ncube A., Trompeter H.-I., Adjaye J. Modeling nonalcoholic fatty liver disease with human pluripotent stem cell-derived immature hepatocyte-like cells reveals activation of plin2 and confirms regulatory functions of peroxisome proliferator-activated receptor alpha. Stem Cell. Dev. 2016;25:1119–1133. doi: 10.1089/scd.2015.0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.J., Johnson D., Amin H.D., Sivathondan P., Silva M.A., Wang L.M., Stevanato L., McNeil C.A., Miljan E.A., Sinden J.D., et al. Characterization of lipid metabolism in a novel immortalized human hepatocyte cell line. Am. J. Physiol. Endocrinol.Metab. 2015;309:E511–E522. doi: 10.1152/ajpendo.00594.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.J., Parry S.A., Gunn P.J., Ceresa C.D., Rosqvist F., Piché M.-E., Hodson L. Studying non-alcoholic fatty liver disease: the ins and outs of in vivo, ex vivo and in vitro human models. Horm. Mol. Biol. Clin. Invest. 2020;41 doi: 10.1515/hmbci-2018-0038. [DOI] [PubMed] [Google Scholar]
- Guirguis E., Grace Y., Bolson A., DellaVecchia M.J., Ruble M. Emerging therapies for the treatment of nonalcoholic steatohepatitis: a systematic review. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2021;41:315–328. doi: 10.1002/phar.2489. [DOI] [PubMed] [Google Scholar]
- Gunn P.J., Green C.J., Pramfalk C., Hodson L. In vitro cellular models of human hepatic fatty acid metabolism: differences between Huh7 and HepG2 cell lines in human and fetal bovine culturing serum. Physiol. Rep. 2017;5:e13532. doi: 10.14814/phy2.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunti S., Hoke A.T., Vu K.P., London N.R. Organoid and spheroid tumor models: techniques and applications. Cancers. 2021;13:874. doi: 10.3390/cancers13040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I., Burton S.A., Munn C., Ohshima M., Goedland M.E., Czysz K., Rajesh D. iPSC-derived hepatocytes generated from NASH donors provide a valuable platform for disease modeling and drug discovery. Biol. Open. 2020;9:bio055087. doi: 10.1242/bio.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using gata6. Nat. Commun. 2016;7:1–12. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B.C., Liang Z., Sun F., Yang Z., Tang C., Chen Z., Yubo S., Yao Z., Wu M., Chen Y., et al. Nonalcoholic fatty liver disease (nafld) in obese rhesus monkeys provides the first animal model that accurately reflects the human condition. FASEB J. 2017;31:895–896. doi: 10.1096/fasebj.31.1_supplement.895.6. [DOI] [Google Scholar]
- Hansen H.H., Ægidius H.M., Oró D., Evers S.S., Heebøll S., Eriksen P.L., Thomsen K.L., Bengtsson A., Veidal S.S., Feigh M., et al. Human translatability of the gan diet-induced obese mouse model of non-alcoholic steatohepatitis. BMC Gastroenterol. 2020;20:1–12. doi: 10.1186/s12876-020-01356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J.W., Evans L.H., Green K.Y., Naghashfar Z., Macias A.R., Zerfas P.M., Ward J.M. Expression of infectious murine leukemia viruses by RAW264.7 cells, a potential complication for studies with a widely used mouse macrophage cell line. Retrovirology. 2008;5:1–6. doi: 10.1186/1742-4690-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D.C., Zhao D., Ross A., Mandalam R., Lebkowski J., Cui W. Direct differentiation of human embryonic stem cells to hepatocyte-like cells exhibiting functional activities. Clon.Stem Cell. 2007;9:51–62. doi: 10.1089/clo.2006.0045. [DOI] [PubMed] [Google Scholar]
- Hirsova P., Gores G.J. Ballooned hepatocytes, undead cells, sonic hedgehog, and vitamin e: therapeutic implications for nonalcoholic steatohepatitis. Hepatology. 2015;61:15–17. doi: 10.1002/hep.27279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson L., Skeaff C.M., Fielding B.A. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog.Lipid Res. 2008;47:348–380. doi: 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Holmgren G., Ulfenborg B., Asplund A., Toet K., Andersson C.X., Hammarstedt A., Hanemaaijer R., Küppers-Munther B., Synnergren J. Characterization of human induced pluripotent stem cell-derived hepatocytes with mature features and potential for modeling metabolic diseases. Int. J. Mol. Sci. 2020;21:469. doi: 10.3390/ijms21020469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Li L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein & cell. 2015;6:562–574. doi: 10.1007/s13238-015-0180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Yang T., Li C., Zhang L., Li M., Huang W., Zhou P. Vol. 45. Elsevier; 2013. Human fetal hepatocyte line, L-02, exhibits good liver function in vitro and in an acute liver failure model; pp. 695–700. (Transplantation Proceedings). [DOI] [PubMed] [Google Scholar]
- Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of nafld-related hcc: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021;18:223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurrell T., Kastrinou-Lampou V., Fardellas A., Hendriks D.F., Nordling Å., Johansson I., Baze A., Parmentier C., Richert L., Ingelman-Sundberg M. Human liver spheroids as a model to study aetiology and treatment of hepatic fibrosis. Cells. 2020;9:964. doi: 10.3390/cells9040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide I., Nagao E., Kajiyama S., Mizoguchi N. A novel evaluation method for determining drug-induced hepatotoxicity using 3D bio-printed human liver tissue. Toxicol. Mech. Methods. 2020;30:189–196. doi: 10.1080/15376516.2019.1686795. [DOI] [PubMed] [Google Scholar]
- Jennen D.G., Magkoufopoulou C., Ketelslegers H.B., van Herwijnen M.H., Kleinjans J.C., van Delft J.H. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- Jeon H., Kang K., Park S.A., Kim W.D., Paik S.S., Lee S.-H., Jeong J., Choi D. Generation of multilayered 3D structures of HepG2 cells using a bio-printing technique. Gut Liver. 2017;11:121. doi: 10.5009/gnl16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlishvili T., Ksovreli M., Gabruashvili D., Museridze M., Bezhuashvili M., Zaalishvili G., Kulikova N. Low-dose trans-resveratrol induce poly (ADP)-ribosylation-dependent increase of the PPAR-γ protein expression level in the in vitro model of non-alcoholic fatty liver disease. Mol. Biol. Rep. 2020;47:8331–8337. doi: 10.1007/s11033-020-05863-z. [DOI] [PubMed] [Google Scholar]
- Kakisaka K., Suzuki Y., Fujiwara Y., Abe T., Yonezawa M., Kuroda H., Ishida K., Sugai T., Takikawa Y. Evaluation of ballooned hepatocytes as a risk factor for future progression of fibrosis in patients with non-alcoholic fatty liver disease. J. Gastroenterol. 2018;53:1285–1291. doi: 10.1007/s00535-018-1468-9. [DOI] [PubMed] [Google Scholar]
- Kanda T., Matsuoka S., Yamazaki M., Shibata T., Nirei K., Takahashi H., Kaneko T., Fujisawa M., Higuchi T., Nakamura H., et al. Apoptosis and non-alcoholic fatty liver diseases. World J. Gastroenterol. 2018;24:2661. doi: 10.3748/wjg.v24.i25.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-M., Kim D., Lee J.-H., Takayama S., Park J.Y. Engineered microsystems for spheroid and organoid studies. Adv. Healthc. Mater. 2021;10:2001284. doi: 10.1002/adhm.202001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K., Sakaida I., Tsuchiya M., Omori K., Takami T., Okita K. Pioglitazone prevents hepatic steatosis, fibrosis, and enzyme-altered lesions in rat liver cirrhosis induced by a choline-deficient l-amino acid-defined diet. Biochem.Biophys. Res. Commun. 2004;315:187–195. doi: 10.1016/j.bbrc.2004.01.038. [DOI] [PubMed] [Google Scholar]
- Kim Y., Kang K., Jeong J., Paik S.S., Kim J.S., Park S.A., Kim W.D., Park J., Choi D. Three-dimensional (3D) printing of mouse primary hepatocytes to generate 3D hepatic structure. Ann. Surg. Treat. Res. 2017;92:67–72. doi: 10.4174/astr.2017.92.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kang K., Yoon S., Kim J.S., Park S.A., Kim W.D., Lee S.B., Ryu K.-Y., Jeong J., Choi D. Prolongation of liver-specific function for primary hepatocytes maintenance in 3D printed architectures. Organogenesis. 2018;14:1–12. doi: 10.1080/15476278.2018.1423931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Koo B.-K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizawa H., Nagao E., Shimamura M., Zhang G., Torii H. Scaffold-free 3D bio-printed human liver tissue stably maintains metabolic functions useful for drug discovery. Biochem.Biophys. Rep. 2017;10:186–191. doi: 10.1016/j.bbrep.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y., Yang S., Roberts E.A. Extended primary culture of human hepatocytes in a collagen gel sandwich system. In Vitro Cell Dev. Biol. Anim. 1997;33:467–472. doi: 10.1007/s11626-997-0065-7. [DOI] [PubMed] [Google Scholar]
- Kostrzewski T., Maraver P., Ouro-Gnao L., Levi A., Snow S., Miedzik A., Rombouts K., Hughes D. A microphysiological system for studying nonalcoholic steatohepatitis. Hepatol.Commun. 2020;4:77–91. doi: 10.1002/hep4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjærg-Hansen A., Vogt T.F., Hobbs H.H., Cohen J.C. Exome-wide association study identifies a tm6sf2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyra M., Johansson I., Nordling Å., Ullah S., Lauschke V.M., Ingelman-Sundberg M. Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-32722-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste B., Raymond V.-A., Cassim S., Lapierre P., Bilodeau M. Highly tumorigenic hepatocellular carcinoma cell line with cancer stem cell-like properties. PLoS One. 2017;12:e0171215. doi: 10.1371/journal.pone.0171215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Weikel K.A., Cacicedo J.M., Ido Y. Resveratrol-induced amp-activated protein kinase activation is cell-type dependent: lessons from basic research for clinical application. Nutrients. 2017;9:751. doi: 10.3390/nu9070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Cho D.-W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16:2618–2625. doi: 10.1039/C6LC00450D. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Sung J.H. Gut–liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol.Bioeng. 2018;115:2817–2827. doi: 10.1002/bit.26793. [DOI] [PubMed] [Google Scholar]
- Lee L., Alloosh M., Saxena R., Van Alstine W., Watkins B.A., Klaunig J.E., Sturek M., Chalasani N. Nutritional model of steatohepatitis and metabolic syndrome in the ossabaw miniature swine. Hepatology. 2009;50:56–67. doi: 10.1002/hep.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Doumouras A.G., Yu J., Brar K., Banfield L., Gmora S., Anvari M., Hong D. Complete resolution of nonalcoholic fatty liver disease after bariatric surgery: a systematic review and meta-analysis. Clin.Gastroenterol.Hepatol. 2019;17:1040–1060. doi: 10.1016/j.cgh.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Liao Y., Davies N.A., Bogle I.D.L. Computational modeling of fructose metabolism and development in NAFLD. Front. Bioeng.Biotechnol. 2020;8:762. doi: 10.3389/fbioe.2020.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmeyer C.C., McCullough A.J. The natural history of nonalcoholic fatty liver disease–an evolving view. Clin. Liver Dis. 2018;22:11–21. doi: 10.1016/j.cld.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xie C., Zhai Z., Deng Z.-y., De Jonge H.R., Wu X., Ruan Z. Uridine attenuates obesity, ameliorates hepatic lipid accumulation and modifies the gut microbiota composition in mice fed with a high-fat diet. Food Funct. 2021;12:1829–1840. doi: 10.1039/D0FO02533J. [DOI] [PubMed] [Google Scholar]
- Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz T., Deák V., Makk-Merczel K., Varga D., Hajdinák P., Szarka A. The performance of HepG2 and HepaRG systems through the glass of acetaminophen-induced toxicity. Life. 2021;11:856. doi: 10.3390/life11080856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall M.J., Cartier J., Thomson J.P., Cameron K., Meseguer-Ripolles J., O’Duibhir E., Szkolnicka D., Villarin B.L., Wang Y., Blanco G.R., et al. Modelling non-alcoholic fatty liver disease in human hepatocyte-like cells. Phil. Trans. R. Soc. B: Biol. Sci. 2018;373:20170362. doi: 10.1098/rstb.2017.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Wu Y., Li Y., Aazmi A., Zhou H., Zhang B., Yang H. Current advances on 3D-bioprinted liver tissue models. Adv. Healthc. Mater. 2020;9:2001517. doi: 10.1002/adhm.202001517. [DOI] [PubMed] [Google Scholar]
- Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R., Borén J., Montalcini T., Pujia A., Wiklund O., et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of european descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarron S., Bathon B., Conlon D.M., Abbey D., Rader D.J., Gawronski K., Brown C.D., Olthoff K.M., Shaked A., Raabe T.D. Functional characterization of organoids derived from irreversibly damaged NASH patient liver. Hepatology. 2021;74:1825–1844. doi: 10.1002/hep.31857. [DOI] [PubMed] [Google Scholar]
- Medina-Leyte D.J., Domínguez-Pérez M., Mercado I., Villarreal-Molina M.T., Jacobo-Albavera L. Use of human umbilical vein endothelial cells HUVEC) as a model to study cardiovascular disease: a review. Appl. Sci. 2020;10:938. doi: 10.3390/app10030938. [DOI] [Google Scholar]
- Meseguer-Ripolles J., Kasarinaite A., Lucendo-Villarin B., Hay D.C. Protocol for automated production of human stem cell derived liver spheres. STAR Protoc. 2021;2:100502. doi: 10.1016/j.xpro.2021.100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan N.I., Hibino N., Nakayama K. Principles of the kenzan method for robotic cell spheroid-based three-dimensional bioprinting. Tissue Eng. B: Rev. 2017;23:237–244. doi: 10.1089/ten.teb.2016.0322. [DOI] [PubMed] [Google Scholar]
- Müller F.A., Sturla S.J. Human in vitro models of nonalcoholic fatty liver disease. Curr.Opin.Toxicol. 2019;16:9–16. doi: 10.1016/j.cotox.2019.03.001. [DOI] [Google Scholar]
- Murphy S.V., Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Nguyen D.G., Funk J., Robbins J.B., Crogan-Grundy C., Presnell S.C., Singer T., Roth A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PloS one. 2016;11:e0158674. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norona L.M., Nguyen D.G., Gerber D.A., Presnell S.C., LeCluyse E.L. Editor’s highlight: modeling compound-induced fibrogenesis in vitro using three-dimensional bioprinted human liver tissues. Toxicol. Sci. 2016;154:354–367. doi: 10.1093/toxsci/kfw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuciforo S., Heim M.H. JHEP Reports; 2020. Organoids to Model Liver Disease; p. 100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Fujii H., Yoshizato K., Kawada N. A human-type nonalcoholic steatohepatitis model with advanced fibrosis in rabbits. Am. J. Pathol. 2010;177:153–165. doi: 10.2353/ajpath.2010.090895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi R., Togo S., Kimura M., Shinozawa T., Koido M., Koike H., Thompson W., Karns R.A., Mayhew C.N., McGrath P.S., et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. CellMetab. 2019;30:374–384. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pafili K., Roden M. Nonalcoholic fatty liver disease (NAFLD) from pathogenesis to treatment concepts in humans. Mol. Metab. 2020;50:101122. doi: 10.1016/j.molmet.2020.101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma E., Doornebal E.J., Chokshi S. Precision-cut liver slices: a versatile tool to advance liver research. Hepatol. International. 2019;13:51–57. doi: 10.1007/s12072-018-9913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parafati M., Kirby R.J., Khorasanizadeh S., Rastinejad F., Malany S. A nonalcoholic fatty liver disease model in human induced pluripotent stem cell-derived hepatocytes, created by endoplasmic reticulum stress-induced steatosis. Dis. Models Mech. 2018;11:dmm033530. doi: 10.1242/dmm.033530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen H.D., Galsgaard E.D., Christoffersen B.Ø., Cirera S., Holst D., Fredholm M., Latta M. Nash-inducing diets in göttingen minipigs. J. Clin. Exp. Hepatol. 2020;10:211–221. doi: 10.1016/j.jceh.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picollet-D’hahan N., Zuchowska A., Lemeunier I., Le Gac S. Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021;39:788–810. doi: 10.1016/j.tibtech.2020.11.014. [DOI] [PubMed] [Google Scholar]
- Pingitore P., Sasidharan K., Ekstrand M., Prill S., Lindén D., Romeo S. Human multilineage 3D spheroids as a model of liver steatosis and fibrosis. Int. J. Mol. Sci. 2019;20:1629. doi: 10.3390/ijms20071629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittol J.M.R., Milona A., Morris I., Willemsen E.C., van der Veen S.W., Kalkhoven E., van Mil S.W. FXR isoforms control different metabolic functions in liver cells via binding to specific dna motifs. Gastroenterology. 2020;159:1853–1865. doi: 10.1053/j.gastro.2020.07.036. [DOI] [PubMed] [Google Scholar]
- Poisson J., Lemoinne S., Boulanger C., Durand F., Moreau R., Valla D., Rautou P.-E. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J. Hepatol. 2017;66:212–227. doi: 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Ponce de León-Rodríguez M.d.C., Guyot J.-P., Laurent-Babot C. Intestinal in vitro cell culture models and their potential to study the effect of food components on intestinal inflammation. Crit. Rev. Food Sci. Nutr. 2019;59:3648–3666. doi: 10.1080/10408398.2018.1506734. [DOI] [PubMed] [Google Scholar]
- Prill S., Caddeo A., Baselli G., Jamialahmadi O., Dongiovanni P., Rametta R., Kanebratt K.P., Pujia A., Pingitore P., Mancina R.M., et al. The TM6SF2 E167K genetic variant induces lipid biosynthesis and reduces apolipoprotein b secretion in human hepatic 3D spheroids. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-47737-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboer E., De Craene B., De Kock J., Vanhaecke T., Berx G., Rogiers V., Vinken M. Strategies for immortalization of primary hepatocytes. J. Hepatol. 2014;61:925–943. doi: 10.1016/j.jhep.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramli M.N.B., Lim Y.S., Koe C.T., Demircioglu D., Tng W., Gonzales K.A.U., Tan C.P., Szczerbinska I., Liang H., Soe E.L., et al. Human pluripotent stem cell-derived organoids as models of liver disease. Gastroenterology. 2020;159:1471–1486. doi: 10.1053/j.gastro.2020.06.010. [DOI] [PubMed] [Google Scholar]
- Reimer K.C., Wree A., Roderburg C., Tacke F. New drugs for NAFLD: lessons from basic models to the clinic. Hepatol. Int. 2020;14:8–23. doi: 10.1007/s12072-019-10001-4. [DOI] [PubMed] [Google Scholar]
- Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A., Boerwinkle E., Cohen J.C., Hobbs H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdo G.R., Da Silva T.C., de Albuquerque Landi M.F., Morais J.Á., Barbisan L.F., Vinken M., Oliveira C.P., Cogliati B. Sorafenib reduces steatosis-induced fibrogenesis in a human 3D co-culture model of non-alcoholic fatty liver disease. Environ. Toxicol. 2021;36:168–176. doi: 10.1002/tox.23021. [DOI] [PubMed] [Google Scholar]
- Rosshart S.P., Herz J., Vassallo B.G., Hunter A., Wall M.K., Badger J.H., McCulloch J.A., Anastasakis D.G., Sarshad A.A., Leonardi I., et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365:eaaw4361. doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman Y., Koh C., Zmuda J.M., Kleiner D.E., Liang T.J., CRN N. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (pnpla3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu N.-E., Lee S.-H., Park H. Spheroid culture system methods and applications for mesenchymal stem cells. Cells. 2019;8:1620. doi: 10.3390/cells8121620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum S., Jeyarajan A.J., Kruger A.J., Holmes J.A., Shao T., Sojoodi M., Kim M.-H., Zhuo Z., Shroff S.G., Kassa A., et al. Fatty acids activate the transcriptional coactivator YAP1 to promote liver fibrosis via p38 mitogen-activated protein kinase. Cell. Mol. Gastroenterol. Hepatol. 2021;12:1297–1310. doi: 10.1016/j.jcmgh.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandwich C.G. Rat hepatocyte cultures. Cytochrome P450 Protoc. 2006;320:247. doi: 10.1385/1-59259-998-2:247. [DOI] [Google Scholar]
- Sano A., Kakazu E., Hamada S., Inoue J., Ninomiya M., Iwata T., Tsuruoka M., Sato K., Masamune A. Steatotic hepatocytes release mature vldl via methionine and tyrosine metabolism in a Keap1-Nrf2 dependent manner. Hepatology. 2021;74:1271–1286. doi: 10.1002/hep.31808. [DOI] [PubMed] [Google Scholar]
- Scarpellini E., Lupo M., Iegri C., Gasbarrini A., De Santis A., Tack J. Intestinal permeability in non-alcoholic fatty liver disease: the gut-liver axis. Rev. Recent Clin.Trials. 2014;9:141–147. doi: 10.2174/1574887109666141216104334. [DOI] [PubMed] [Google Scholar]
- Schwartz B.E., Rajagopal V., Smith C., Cohick E., Whissell G., Gamboa M., Pai R., Sigova A., Grossman I., Bumcrot D., et al. Discovery and targeting of the signaling controls of PNPLA3 to effectively reduce transcription, expression, and function in pre-clinical NAFLD/NASH settings. Cells. 2020;9:2247. doi: 10.3390/cells9102247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S.C., Ingelman-Sundberg M. Pharmacogenomic biomarkers: new tools in current and future drug therapy. Trends Pharmacol. Sci. 2011;32:72–81. doi: 10.1016/j.tips.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Sinton M.C., Meseguer-Ripolles J., Lucendo-Villarin B., Wernig-Zorc S., Thomson J.P., Carter R.N., Lyall M.J., Walker P.D., Thakker A., Meehan R.R., et al. A human pluripotent stem cell model for the analysis of metabolic dysfunction in hepatic steatosis. IScience. 2021;24:101931. doi: 10.1016/j.isci.2020.101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A., Aleman J., Forsythe S., Rajan S., Murphy S., Devarasetty M., Zarandi N.P., Nzou G., Wicks R., Sadri-Ardekani H., et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. 2020;12:025017. doi: 10.1088/1758-5090/ab6d36. [DOI] [PubMed] [Google Scholar]
- Slaughter V.L., Rumsey J.W., Boone R., Malik D., Cai Y., Sriram N.N., Long C.J., McAleer C.W., Lambert S., Shuler M.L., et al. Validation of an adipose-liver human-on-a-chip model of NAFLD for preclinical therapeutic efficacy evaluation. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-92264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M., Stone A.L., Parkhill R.L., Stewart R.L., Simpkins M.W., Kachurin A.M., Warren W.L., Williams S.K. Three-dimensional bioassembly tool for generating viable tissue-engineered constructs. Tissue Eng. 2004;10:1566–1576. doi: 10.1089/ten.2004.10.1566. [DOI] [PubMed] [Google Scholar]
- Sookoian S., Castaño G.O., Burgueño A.L., Gianotti T.F., Rosselli M.S., Pirola C.J. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret P.-A., Magusto J., Housset C., Gautheron J. In vitro and in vivo models of non-alcoholic fatty liver disease: a critical appraisal. J. Clin. Med. 2021;10:36. doi: 10.3390/jcm10010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Chan C. Application of metabolic flux analysis to identify the mechanisms of free fatty acid toxicity to human hepatoma cell line. Biotechnol.Bioeng. 2008;99:399–410. doi: 10.1002/bit.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine J.G., Wentworth B.J., Zimmet A., Rinella M.E., Loomba R., Caldwell S.H., Argo C.K. Systematic review with meta-analysis: risk of hepatocellular carcinoma in non-alcoholic steatohepatitis without cirrhosis compared to other liver diseases. Aliment.Pharmacol.Ther. 2018;48:696–703. doi: 10.1111/apt.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojsavljević S., Palčić M.G., Jukić L.V., Duvnjak L.S., Duvnjak M. Adipokines and proinflammatory cytokines, the key mediators in the pathogenesis of nonalcoholic fatty liver disease. World J. Gastroenterol.WJG. 2014;20:18070. doi: 10.3748/wjg.v20.i48.18070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland R.M., McCredie J.A., Inch W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971;46:113–120. [PubMed] [Google Scholar]
- Suurmond C.-A.E., Lasli S., van den Dolder F.W., Ung A., Kim H.-J., Bandaru P., Lee K., Cho H.-J., Ahadian S., Ashammakhi N., et al. In vitro human liver model of nonalcoholic steatohepatitis by coculturing hepatocytes, endothelial cells, and kupffer cells. Adv. Healthc. Mater. 2019;8:1901379. doi: 10.1002/adhm.201901379. [DOI] [PubMed] [Google Scholar]
- Taciak B., Białasek M., Braniewska A., Sas Z., Sawicka P., Kiraga Ł., Rygiel T., Król M. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PloS one. 2018;13:e0198943. doi: 10.1371/journal.pone.0198943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Kimura M., Yoshizawa E., Ayano S., Koido M., Funayama S., Nakanishi N., Hisai T., Kobayashi T., et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Shimanaka Y., Caddeo A., Kubo T., Mao Y., Kubota T., Kubota N., Yamauchi T., Mancina R.M., Baselli G., et al. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut. 2021;70:180–193. doi: 10.1136/gutjnl-2020-320646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Valdez I.A., Dirice E., Kulkarni R.N. Comparable generation of activin-induced definitive endoderm via additive wnt or bmp signaling in absence of serum. Stem Cell Rep. 2014;3:5–14. doi: 10.1016/j.stemcr.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touboul T., Hannan N.R., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Di Santo J., et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Treyer A., Müsch A. Hepatocyte polarity. Compr.Physiol. 2013;3:243. doi: 10.1002/cphy.c120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpaev K. Keap1-Nrf2 signaling pathway: mechanisms of regulation and role in protection of cells against toxicity caused by xenobiotics and electrophiles. Biochemistry (Moscow) 2013;78:111–126. doi: 10.1134/S0006297913020016. [DOI] [PubMed] [Google Scholar]
- Underhill G.H., Khetani S.R. Advances in engineered human liver platforms for drug metabolism studies. Drug Metab.Dispos. 2018;46:1626–1637. doi: 10.1124/dmd.118.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill G.H., Khetani S.R. Bioengineered liver models for drug testing and cell differentiation studies. Cell. Mol. Gastroenterol.Hepatol. 2018;5:426–439. doi: 10.1016/j.jcmgh.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs S. 2018. Hepa 1-6–a Murine Model of Hepatocellular Carcinoma.https://drugdevelopment.labcorp.com/industry-solutions/oncology/preclinical/tumor-spotlights/hepa-1-6-a-murine-model-of-hepatocellular-carcinoma.html [Google Scholar]
- Valenti L., Al-Serri A., Daly A.K., Galmozzi E., Rametta R., Dongiovanni P., Nobili V., Mozzi E., Roviaro G., Vanni E., et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin i148m polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- Van de Bovenkamp M., Groothuis G., Meijer D., Olinga P. Liver fibrosis in vitro: cell culture models and precision-cut liver slices. Toxicol.Vitro. 2007;21:545–557. doi: 10.1016/j.tiv.2006.12.009. [DOI] [PubMed] [Google Scholar]
- van Grunsven L.A. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 2017;121:133–146. doi: 10.1016/j.addr.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Van Riel N.A., Tiemann C.A., Hilbers P.A., Groen A.K. Metabolic modelling combined with machine learning integrates longitudinal data and identifies the origin of LXR-induced hepatic steatosis. Front. Bioeng.Biotechnol. 2020;8:1545. doi: 10.3389/fbioe.2020.536957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J.J., LeGraw R., Moghadam F., Tan Y., Kilbourne J., Maggiore J.C., Hislop J., Liu S., Cats D., de Sousa Lopes S.M.C., et al. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 2021;12:41–55. doi: 10.1016/j.cels.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar-Gomez E., Martinez-Perez Y., Calzadilla-Bertot L., Torres-Gonzalez A., Gra-Oramas B., Gonzalez-Fabian L., Friedman S.L., Diago M., Romero-Gomez M. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology. 2015;149:367–378. doi: 10.1053/j.gastro.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Vogel S., Piantedosi R., Frank J., Lalazar A., Rockey D.C., Friedman S.L., Blaner W.S. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J. Lipid Res. 2000;41:882–893. doi: 10.1016/S0022-2275(20)32030-7. [DOI] [PubMed] [Google Scholar]
- Wang Y.-J., Liu H.-L., Guo H.-T., Wen H.-W., Liu J. Primary hepatocyte culture in collagen gel mixture and collagen sandwich. World J. Gastroenterol. 2004;10:699. doi: 10.3748/wjg.v10.i5.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware B.R., Liu J.S., Monckton C.P., Ballinger K.R., Khetani S.R. Micropatterned coculture with 3T3-J2 fibroblasts enhances hepatic functions and drug screening utility of HepaRG cells. Toxicol. Sci. 2021;181:90–104. doi: 10.1093/toxsci/kfab018. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J., Porse B. Bone marrow-derived macrophages (BMM): isolation and applications. Cold Spring Harbor Protoc. 2008;2008 doi: 10.1101/pdb.prot5080. pdb–prot5080. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R., Weimer J., Meurer S.K., Kron A., Seipel B., Vater I., Arnold N., Siebert R., Xu L., Friedman S.L., et al. Genetic characteristics of the human hepatic stellate cell line LX-2. PloS one. 2013;8:e75692. doi: 10.1371/journal.pone.0075692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R.G. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J. Clin. Gastroenterol. 2005;39:S158–S161. doi: 10.1097/01.mcg.0000155516.02468.0f. [DOI] [PubMed] [Google Scholar]
- Wilkening S., Stahl F., Bader A. Comparison of primary human hepatocytes and hepatoma cell line HepG2 with regard to their biotransformation properties. Drug Metab.Dispos. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- Wu J.C., Merlino G., Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Roberto J.B., Knupp A., Kenerson H.L., Truong C.D., Yuen S.Y., Brempelis K.J., Tuefferd M., Chen A., Horton H., et al. Precision-cut human liver slice cultures as an immunological platform. J.Immunol.Methods. 2018;455:71–79. doi: 10.1016/j.jim.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Hui A., Albanis E., Arthur M., O’byrne S., Blaner W., Mukherjee P., Friedman S., Eng F. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeon J.H., Park J.-K. Microfluidic cell culture systems for cellular analysis. Biochip J. 2007;1:17–27. [Google Scholar]
- Yoon Y.-C., Fang Z., Lee J.E., Park J.H., Ryu J.-K., Jung K.H., Hong S.-S. Selonsertib inhibits liver fibrosis via downregulation of ask1/mapk pathway of hepatic stellate cells. Biomol.Ther. 2020;28:527. doi: 10.4062/biomolther.2020.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- Zeilinger K., Freyer N., Damm G., Seehofer D., Knöspel F. Cell sources for in vitro human liver cell culture models. Exp. Biol. Med. 2016;241:1684–1698. doi: 10.1177/1535370216657448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Y., Yuan W.-G., He P., Lei J.-H., Wang C.-X. Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J.Gastroenterol. 2016;22:10512. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao L., Wang Y., Zhang C., Liu J., Wang P., Zhou W., Yang P., Varghese Z., Moorhead J.F., et al. Cluster of differentiation 36 deficiency aggravates macrophage infiltration and hepatic inflammation by upregulating monocyte chemotactic protein-1 expression of hepatocytes through histone deacetylase 2-dependent pathway. Antioxid.Redox Signal. 2017;27:201–214. doi: 10.1089/ars.2016.6808. [DOI] [PubMed] [Google Scholar]
- Zou B., Yeo Y.H., Nguyen V.H., Cheung R., Ingelsson E., Nguyen M.H. Prevalence, characteristics and mortality outcomes of obese, nonobese and lean NAFLD in the United States, 1999-2016. J. Intern. Med. 2020;288:139–151. doi: 10.1111/joim.13069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table includes the title of the publication, the year it was published, and the journal where it was published.