Abstract
Objective
Diet-induced obesity is often associated with nonalcoholic fatty liver disease (NAFLD), which instigates severe metabolic disorders, including cirrhosis, hepatocellular carcinoma, and type 2 diabetes. We have shown that hepatic depletion of CREB regulated transcription co-activator (CRTC) 2 protects mice from the progression of diet-induced fatty liver phenotype, although the exact mechanism by which CRTC2 modulates this process is elusive to date. Here, we investigated the role of hepatic CRTC2 in the instigation of NAFLD in mammals.
Methods
Crtc2 liver-specific knockout (Crtc2 LKO) mice and Crtc2 flox/flox (Crtc2 f/f) mice were fed a high fat diet (HFD) for 7–8 weeks. Body weight, liver weight, hepatic lipid contents, and plasma triacylglycerol (TG) levels were determined. Western blot analysis was performed to determine Sirtuin (SIRT) 1, tuberous sclerosis complex (TSC) 2, and mammalian target of rapamycin complex (mTORC) 1 activity in the liver. Effects of Crtc2 depletion on lipogenesis was determined by measuring lipogenic gene expression (western blot analysis and qRT-PCR) in the liver as well as Oil red O staining in hepatocytes. Effects of miR-34a on mTORC1 activity and hepatic lipid accumulation was assessed by AAV-miR-34a virus in mice and Ad-miR-34a virus and Ad-anti-miR-34a virus in hepatocytes. Autophagic flux was assessed by western blot analysis after leupeptin injection in mice and bafilomycin treatment in hepatocytes. Lipophagy was assessed by transmission electron microscopy and confocal microscopy. Expression of CRTC2 and p-S6K1 in livers of human NAFLD patients was assessed by immunohistochemistry.
Results
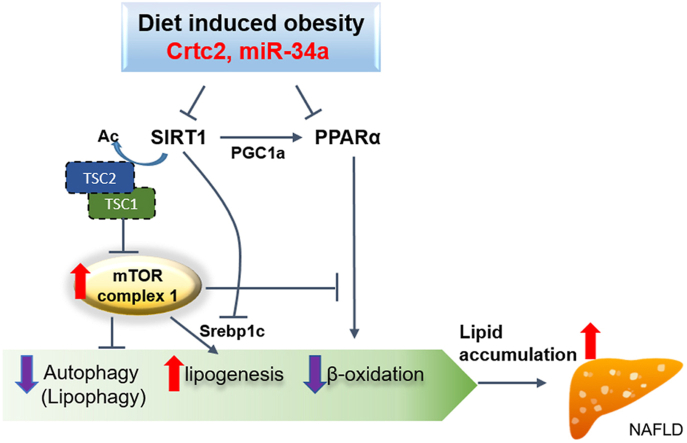
We found that expression of CRTC2 in the liver is highly induced upon HFD-feeding in mice. Hepatic depletion of Crtc2 ameliorated HFD-induced fatty liver disease phenotypes, with a pronounced inhibition of the mTORC1 pathway in the liver. Mechanistically, we found that expression of TSC2, a potent mTORC1 inhibitor, was enhanced in Crtc2 LKO mice due to the decreased expression of miR-34a and the subsequent increase in SIRT1-mediated deacetylation processes. We showed that ectopic expression of miR-34a led to the induction of mTORC1 pathway, leading to the hepatic lipid accumulation in part by limiting lipophagy and enhanced lipogenesis. Finally, we found a strong association of CRTC2, miR-34a and mTORC1 activity in the NAFLD patients in humans, demonstrating a conservation of signaling pathways among species.
Conclusions
These data collectively suggest that diet-induced activation of CRTC2 instigates the progression of NAFLD by activating miR-34a-mediated lipid accumulation in the liver via the simultaneous induction of lipogenesis and inhibition of lipid catabolism. Therapeutic approach to specifically inhibit CRTC2 activity in the liver could be beneficial in combating NAFLD in the future.
Keywords: CRTC2, mTORC1, Nonalcoholic fatty liver disease, miR-34a, Lipophagy, Lipogenesis
Abbreviations: TG, triacylglycerol; NAFLD, Non-alcoholic fatty liver disease; CRTC2, CREB regulated transcription co-activator 2; HFD, high fat diet; SIRT, Sirtuin; mTORC, mammalian target of rapamycin complex; TSC, tuberous sclerosis complex; NCD, normal chow diet; DIO, diet-induced obesity
Graphical abstract
Highlights
-
•
CRTC2 expression is elevated in livers of HFD-fed mice.
-
•
Liver-specific depletion of Crtc2 ameliorated HFD-induced fatty liver phenotype in mice.
-
•
Crtc2 knockout dampened miR-34a-mTORC1 pathway, leading to the inhibition of lipogenesis and the activation of lipophagy.
-
•
Close association of CRTC2 with mTORC1 activity was also demonstrated in the human NAFLD patients.
-
•
Diet-induced activation of CRTC2 instigates the progression of NAFLD by activating the miR-34a-mTORC1 pathway.
1. Introduction
The liver is a major organ to maintain lipid homeostasis in mammals. Genetic, epigenetic, or environmental stress perturbs the normal metabolic pathways for the lipid in the liver, resulting in the metabolic disorder that could severely affect the health of the individuals [1,2]. Non-alcoholic fatty liver disease (NAFLD) is characterized by the increased triacylglycerol (TG) in the liver, which is caused by an increased fat accumulation in hepatocytes via an imbalance between fat storage and fat mobilization [3]. This phenomenon can be stemmed from the excessive de novo lipogenesis, increased uptake of free fatty acids, decreased mobilization of fat by defects in the fatty acid β-oxidation and/or lipophagy, or the decreased ability to secrete very low density lipoprotein (VLDL), each of which can be linked to the pre-existing insulin resistance in the liver. If untreated, NAFLD could be progressed into the more severe form of diseases, such as non-alcoholic steatohepatitis (NASH), cirrhosis, and in extreme cases, hepatocellular carcinoma (HCC).
CREB regulated transcription co-activator (CRTC) is a class of transcription co-activator that promotes the transcriptional activity of basic leucine zipper type transcription factors, including CREB [4,5]. There are three family members of CRTCs, including CRTC1, CRTC2, and CRTC3. Among family members, CRTC2 is a predominant isoform in the liver. In association with CREB, CRTC2 is known to be responsible for the transcriptional regulation of hepatic gluconeogenesis [6]. CRTC2 activity is inhibited by insulin signaling via Akt-Salt inducible kinase (SIK) pathway, and the chronic activation of CRTC2 is thus thought to be causal to hyperglycemia via an increased hepatic glucose production during the insulin resistant state [7].
The role of CRTC2 in hepatic lipid metabolism has recently been delineated. Aside from its role as a transcriptional co-activator, CRTC2 was shown to interact with a coat protein complex II (COPII) subunit protein Sec31A to inhibit sterol regulatory element binding protein (SREBP) 1-mediated lipogenesis in the liver [8]. Induction of cholesterol synthesis was observed via CRTC2/FoxO1-mediated transcriptional activation of SREBP2 [9], suggesting that CRTC2 is a crucial protein in regulating both glucose and lipid metabolism in the liver. While we found that liver-specific depletion of Crtc2 did not display apparent changes in hepatic lipid metabolism under normal chow diet feeding, mice deficient in hepatic Crtc2 under high fat diet (HFD) feeding resulted in the improved metabolic homeostasis via the reduction of miR-34a expression in the liver and the subsequent induction of SIRT1/PPARα/FGF21 axis [10]. Since miR-34a or SIRT1 is involved in the regulation of lipid metabolism by controlling multiple pathways [[11], [12], [13]], we suspected that other targets of CRTC2 in the liver during HFD feeding might also be important in the observed improvement in the metabolic homeostasis.
Here we found that hepatic expression of CRTC2 is induced upon HFD-feeding in mice, with concomitant activation of mTORC1. Depletion of hepatic Crtc2 ameliorated HFD-induced NAFLD phenotypes, concomitant with the reduced mTORC1 activity. We found that SIRT1-mediated deacetylation and activation of TSC2 is pronounced upon depletion of hepatic Crtc2, resulting in the inhibition of mTORC1 pathway in this setting. We confirmed that the increased activity of CRTC2, miR-34a, and mTORC1 is significantly associated with the NAFLD patients in humans. These data collectively suggest that the diet-induced activation of CRTC2 instigates the progression of NAFLD by activating miR-34a-mediated lipid accumulation in the liver.
2. Materials and methods
2.1. Animal experiments
All animal experiments were performed in a specific pathogen-free facility at the Central Laboratory Animal Research Center, Korea University (12:12-h light–dark cycle, maintained at 22 °C), based on the protocols approved by the Korea University Institutional Animal Care and Use Committee (IACUC #: KUIACUC-2018-0031).
Information regarding liver-specific Crtc2 knockout mice has been described [10]. For the non-alcoholic fatty liver disease mouse model, male mice were fed a 60% high fat diet (D12492, Research Diets) from 7 weeks of age. Duration of HFD feeding is described in figure legends. To measure autophagic flux in vivo, mice were deprived of food and injected intraperitoneally with leupeptin (40 mg/kg) for 4 h before being sacrificed.
Pair-feeding experiments were performed as described previously [14]. Briefly, male mice (16 weeks old) were individually housed in cages under the standard conditions. Mice were fed 2.4 g of HFD per day for 7 weeks. To reduce stress, each mouse was housed with enrichment. Body weight was measured every week to ensure the effect of the pair-feeding regimen.
2.2. Viruses
Adenovirus expressing constitutive active form of CRTC2 (Ad-Crtc2Δ) and Adeno-associated virus expressing miR-34a (Ad-miR-34a) have been described previously [10,15]. 2 × 108 pfu (per mouse) of purified virus was injected into the tail vein. After 7 days, mice were sacrificed, and the isolated liver was analyzed.
To generate the lentiviral vector encoding an shRNA that targeted a miR-34a, oligo pairs harboring the sense and antisense sequences were synthesized, annealed, and cloned in the AgeI-and EcoRI-digested pLKO.1 vector (Addgene plasmid #10879) by ligation. The shRNA sense and antisense sequences are: 5′-CCGGTGTGGCAGTATCTTAGCTGATTATCTTCCTGTCAGACAACCAGCTAAGACACTGCCATTTTTG-3′ and 5′-AATTCAAAAAGTGGCAGTATCTTAGCTGATTATCTTCCTGTCAGACAACCAGCTAAGACACTGCCAA-3’. Lentivirus was produced and packaged in HEK293T cells as described previously [16].
2.3. In vitro cell cultures
Hepatocytes were isolated from male C57BL/6N mice at 8–12 weeks of age [10]. Collagenase perfusion was briefly performed through the portal vein. After the isolation, the perfused liver was minced and was subject to percoll gradient to purify hepatocytes. Cells were then maintained in medium 199 (M199; Sigma–Aldrich) with 10% fetal bovine serum (FBS, HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 nM of dexamethasone. After the attachment, the cells were then treated with adenoviruses (Ad-GFP and Ad-miR-34a) as shown in the figure legends. The AML12 (alpha mouse liver 12) cells were purchased from ATCC, USA (ATCC® CRL-2254™) and maintained as described in the standard protocol (https://www.atcc.org/products/all/CRL-2254.aspx#culturemethod).
2.4. Western blot analysis
Western blot analysis was performed using 20–90 μg of whole liver lysates. The liver lysates were prepared by homogenization in RIPA buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1% Triton x-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 5% glycerol, 50 mM NaF, 5 mM beta-glycerophosphate, and 1 mM PMSF) with a cocktail of protease inhibitors (GenDEPOT). The samples were boiled for 5 min in the SDS sample buffer (50 mM Tris -HCl pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 5% 2-mercaptoethanol) and resolved in the SDS PAGE gel. After transferring the proteins to NC membranes (GE) using the Trans-Blot Turbo transfer system (Bio-Rad), the blots were blocked with 5% non-fat milk in TBS-T (1X Tris buffered Saline with 0.5% Tween-20). The blots were incubated with the primary antibodies overnight at 4 °C. After washes with TBS-T and incubation with HRP-conjugated secondary antibodies (Abcam), the blots were developed using ECL (Bio-Rad). Band density was quantified by using the Image J software (NIH). Specific conditions for antibodies used in the western blot analysis are summarized in Table S1.
2.5. RNA extraction and quantitative RT-PCR
Total RNA was isolated using QIAzol (Qiagen) and miRNeasy kit (Qiagen) according to the manufacturer's protocols. Complimentary DNA was synthesized by using Goscript Reverse transcription system (Promega) as described in the manufacturer's protocol. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using a CFX connect real-time system (Bio-Rad). mRNA levels were normalized to ribosomal L32 or Gapdh. For microRNA quantification, total RNA was reverse transcribed by using the miScript II RT kit (Qiagen). Primers specific for mouse-miR-34a were purchased from Qiagen. The values were normalized to RNU6B. Sequences for gene-specific primers are shown in Table S2.
2.6. Histological analysis
Liver was isolated from mice and was fixed with 10% neutral formalin (Sigma). Histological changes were examined by hematoxylin and eosin (H&E) staining. Neutral lipid accumulation in the liver was analyzed by LipidTox (Thermo Scientific, H34476) staining according to the manufacture's protocol. DAPI was used for counterstain. Immunohistochemistry (IHC) was performed to analyze CRTC2, p-S6K1, and F4/80 protein expression in the liver using specific antibodies (see Supporting Table S1 for specific information). Mayer's hematoxylin was used as a counterstain. Slides were observed with a light microscope (Leica DMi 8). Liver fibrosis was assessed by Picro-Sirius Red Stain Kit (Abcam, ab150681) according to the manufacturer's protocol.
2.7. Confocal microscopy
Mouse primary hepatocytes were plated on confocal dishes and infected with various adenoviruses such as control, miR-34a, GFP-LC3, and tf-LC3 for 3 days, before being induced for autophagy by starvation (with EBSS media) with or without SRT1720. Cells were fixed with 3% paraformaldehyde (PFA) for 10 m at room temperature (RT). The cells were washed with PBS and then blocked with 10% donkey serum and 0.3% Triton X-100 for 1 h at RT, followed by incubation for overnight at 4 °C with anti-Lamp1 antibody, and reacted with anti-rat IgG (Alexa Fluor 488) and LipidTox dye for 1 h at RT. The nucleus was stained with DAPI for 5 m at RT. Cells were then mounted after washing 3x with 0.1% BSA in PBS. Fluorescence images were obtained using an LSM800 confocal laser scanning microscope (Carl Zeiss).
2.8. Transmission electron microscopy
Liver tissues were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 1 h at 4 °C. After washing in phosphate buffer, the samples were postfixed with 1% osmium tetroxide plus 1.5% potassium ferrocyanide in 0.1 M phosphate buffer for 1 h on ice in the dark and then washed again with phosphate buffer. The samples were then embedded in Epon 812 after dehydration in an ethanol series. Polymerization was conducted using pure resin at 70 °C for 2 days. Ultrathin sections (70 nm) were obtained with an ultramicrotome (UltraCut-UCT, Leica, Vienna, Austria), which were then collected in Formvar-coated copper grids.
After staining with uranyl acetate and lead citrate, the sections were examined by a Bio-HVEM system (JEM-1400Plus at 120 kV and JEM-1000BEF at 1000 kV).
2.9. Triacylglycerol levels
Total liver lipids were extracted according to the Folch method with a slight modification [17]. Mouse liver was briefly homogenized with chloroform/methanol solution (2:1, v/v) and was centrifuged at room temperature. The supernatant was then washed with 1/5 volumes of 0.9% NaCl and was again centrifuged. After discarding the upper phase, the remaining solution was evaporated under the vacuum. Hepatic and plasma TG contents were measured using an enzymatic colorimetric assay kit (Wako Chemicals).
2.10. Isotope tracing for lipid synthesis
Isolated mouse primary hepatocyte cultured in 6-well plates. Isotopic labeling was performed by feeding cells with M199 media supplemented with 20 mM [U–13C]-glucose (Cambridge Isotope Laboratories, Inc, CLM-1396-PK), 2 mM [U–13C]-glutamine (Cambridge Isotope Laboratories, Inc, CLM-1822-PK), 10% fetal bovine serum, dialyzed, US origin (Thermofisher Scientific, cat#: 26400044), and 1% Penicillin-Streptomycin (Gibco, cat#:15140148) for 24 h. Then, the cells were washed with ice-cold PBS for twice and quenched in liquid nitrogen. Frozen cells are extracted with 240 μl of 50:30:20 (v/v/v) methanol:acetonitrile:water solution per each well in 6-well plates separately at −20 °C. The metabolite extractions were stored at −80 °C for at least 1 h, followed by centrifugation, twice at 17500 rpm for 20 m to obtain protein-free metabolite extraction. To quantify the metabolite labeling distribution, liquid chromatography-mass spectrometry was performed on an Agilent 1290 Infinity LC and an Agilent 6490 Triple Quadrupole MS system equipped with Agilent Jet Stream ESI source (Agilent Technologies, USA). LC separations were carried out on Waters BEH C18 column (100 × 2.1 mm, particle size 1.7 μm, Waters, USA) for palmitate analysis. The binary gradient system comprised 10 mM ammonium acetate in acetonitrile/water 40/60 (v/v) (solvent A) and 10 mM ammonium acetate in acetonitrile:isopropanol 10/90 (v/v) (solvent B). The linear gradient used for elution and equilibrating the initial gradient for subsequent runs was as follows: 40–65% B from 0 to 5 m, 65–70% B from 5 to 12 m, 70–99% B from 12 to 15 m, 99% B from 15 to 17 m, and 40% B from 17.1 to 20 m. For Acetyl-CoA analysis, LC separations were carried out on XBridge® BEH Amide column (150 × 2.1 mm, particle size 2.5 μm, Waters, USA). The binary gradient system comprised water, 5% acetonitrile, and 5% 1 M ammonium acetate (v/v/v) (solvent A) and acetonitrile, 5% water, and 5% 1 M ammonium acetate (v/v/v) (solvent B). Mobile phase was set to pH 9.0. The linear gradient used for elution and equilibrating the initial gradient for subsequent runs was as follows: 80% B from 0 to 1 m, 80-60% B from 1 to 9 m, 60% B from 9 to 15 m, 80% B from 15.5 to 30 m. To avoid non-labeled palmitate contamination from plastic ware, we modeled the labeling of palmitate derived from malonyl-CoA. Mole Percent Enrichment (MPE) was calculated as described previously [18].
2.11. Fatty acid uptake assay
Endpoint fluorescence of fatty acid uptake in mouse primary hepatocytes in the presence of insulin (100 nM) was measured using QBT fatty acid uptake assay kit (Molecular devices, R8132). Data represent mean RFU±SEM of quadruplicate wells.
2.12. Oil-Red O staining
Frozen sections of livers and hepatocytes were fixed in 10% formalin for 30 m and washed three times with PBS buffer. Samples were washed with 60% isopropanol and incubated with Oil-Red O staining solution for 20 m and washed with PBS twice. For nuclear staining, hematoxylin was added and washed with PBS. A light microscope (Leica DMi 8) was used to acquire images. For the quantification of lipid accumulation in cells, 1 ml of isopropanol was added to each well, gently mixed for 30 m and the absorbance at 490 nm was measured with a 96-well plate reader.
2.13. Human study
A total of 20 patients were enrolled in this study at Soon Chun Hyang University Hospital Bucheon. The study was performed in accordance with the ethical guidelines of the 1975 Helsinki declaration. The study protocol was approved by the Soon Chun Hyang University Hospital Institutional Review Board (IRB#: SCHBC 2017-05-020-002) and informed consent was obtained from all patients.
All 20 subjects were stratified to control and NAFLD according to the steatosis severity using Fibroscan steatosis scores and histological scores. Men who consumed >30 g of alcohol per day and women who consumed >20 g of alcohol per day were excluded. Comprehensive clinical and biochemical assessments were performed, and liver biopsy samples were flash-frozen in liquid nitrogen for further analysis. The subject characteristics are listed in Table S3.
2.14. Quantification of data
IHC image quantification was performed with a color deconvolution method by separating the brown color of DAB from the hematoxylin blue color using Image J (Fiji). The LC3 dot images were quantified using the default ‘Analyze particles’ plugin in image J (Fiji). For co-localization analysis, pixel-intensity-based Pearson's correlation coefficient (R) between two channels was calculated using Image J plugin called ‘coloc2’. The western blots were quantified using the Image J.
2.15. Statistics
Data were analyzed using Prism (GraphPad 7.0) and presented as mean ± SEM or SD. Statistical significance was determined using the unpaired two-tailed Student's t test with Welch's correction for single variables for comparing two values or one-way ANOVA, followed by Tukey's multiple comparisons for comparing more than two values. P values of <0.05 were considered to be statistically significant and were presented as ∗; p < 0.05, ∗∗; p < 0.01, or ∗∗∗; p < 0.001.
3. Results
3.1. Depletion of Crtc2 in the liver interferes with HFD-mediated induction of mTORC1 pathway
HFD-feeding promotes hepatic insulin resistance in part by an enhanced lipid accumulation in the liver. As a master regulator of cell growth and anabolic process, the mammalian target of rapamycin complex 1 (mTORC1) has been shown to regulate hepatic lipid metabolism [19]. In line with this idea, we observed that HFD feeding dramatically induced mTORC1 signaling pathway, as shown by increased p-S6K1 levels in western blot analysis (Supplementary Fig. S1A). We also observed an increased CRTC2 expression under HFD feeding compared with the normal chow diet (NCD) feeding, suggesting that the increased CRTC2 level in the liver could be correlated with the enhanced mTORC1 signaling pathway under HFD feeding.
To investigate a potential role of CRTC2 in the control of mTORC1 pathway under HFD-mediated diet-induced obesity (DIO) conditions, we utilized liver-specific Crtc2 knockout mice. As shown before, liver-specific Crtc2 knockout (Crtc2LKO) mice showed no apparent differences in hepatic lipid contents under the normal chow diet (NCD) compared with the control (Supplementary Fig. S1B and [10]). In addition, no changes were shown in liver weight between the two genotypes under the NCD, although the overall bodyweight tended to be lower upon hepatic depletion of Crtc2 compared with the control (Supplementary Figs. S1C and S1D). These data suggest that depletion of hepatic Crtc2 per se might not perturb the lipid metabolism at least under the NCD. Unlike the case for NCD, Crtc2LKO mice displayed a reduction in body weight and liver weight, together with the decreased accumulation of lipid in the liver compared with Crtc2f/f mice upon HFD feeding (Figure 1A and Supplementary Figs. S1E and S1F). In line with these data, plasma TG levels were reduced in Crtc2LKO mice compared with the control (Supplementary Fig. S1F). To eliminate a potential effect of the overall adiposity on hepatic lipid accumulation by HFD, we placed Crtc2f/f mice and Crtc2LKO mice under pair feeding (HFD) for 7 weeks. In the pair feeding conditions, we were still able to observe a reduced lipid accumulation in livers of Crtc2LKO mice compared with the control (Supplementary Fig. S2). We observed a significant accumulation of macrophages in the liver of HFD-fed control mice, but not that of 7 week HFD-fed Crtc2LKO mice, as evidenced by the F4/80 staining (Figure 1B). Depletion of hepatic Crtc2 greatly reduced expression of pro-inflammatory cytokine genes such as TNFα and IL-1β in the livers of HFD-fed mice, showing that pro-inflammatory macrophage accumulation was inhibited upon liver-specific depletion of Crtc2 in mice (Figure 1C). These data suggest that the depletion of hepatic Crtc2 could inhibit the progression of NAFLD in mice.
Figure 1.
Hepatic depletion of Crtc2 protects against diet-induced hepatic lipid accumulation. A. Liver sections of Crtc2f/f or Crtc2LKO mice (8 week-HFD) were stained with hematoxylin and eosin (H&E, top). Scale bar: 20 μm. Frozen sections were also stained with LipidTox (red) to detect the neutral lipid (middle). Relative intensity of the lipid content was calculated by Image J software (bottom). Data represent mean ± SEM (n = 11 per group) ∗∗∗p < 0.001. B. Histological staining of F4/80 (brown) by using livers of Crtc2f/f or Crtc2LKO mice (7-week HFD) (left). Scale bar: 100 μm. Quantitative analysis was also shown (right). Data represent mean ± SEM (n = 3–5 per group). ∗p < 0.05. C. The mRNA levels of macrophage-specific genes were shown by qRT-PCR in the liver tissues of Crtc2f/f or Crtc2LKO mice (7 week-HFD). Data represent mean ± SEM (n = 6–7 per group). ∗p < 0.05. D. Western blot analysis showing hepatic protein levels of Crtc2f/f or Crtc2LKO mice (7 week-HFD) under fasted or refed conditions (left). Quantitation of key bands in the refed condition was shown (right). Data represent mean ± SEM (n = 4 per group) ∗p < 0.05, ∗∗p < 0.01.
We observed that the mTORC1 signaling pathway was decreased, as shown by the reduced p-S6K1 and p-4E-BP1 in the livers of HFD-fed Crtc2LKO mice compared with the wild type control (Figure 1D). A recent study suggested that CREB-CRTC2 may rather interfere with the mTORC1 signaling pathway by mediating transcriptional activation of PER2, a potential inhibitor of mTORC1 [20]. While we observed that PER2 expression was indeed reduced upon depletion of Crtc2 in the liver, mTORC1 activity was also reduced in the liver of Crtc2LKO mice compared with the control under HFD feeding. These data indicated that CREB/CRTC2-PER2 axis may not be critical in the regulation of mTORC1 activity at least under the HFD feeding. We suspected that HFD-mediated induction of CRTC2 could be causal to the enhanced mTORC1 pathway in DIO conditions.
3.2. CRTC2-miR-34a axis controls mTORC1 pathway in the liver
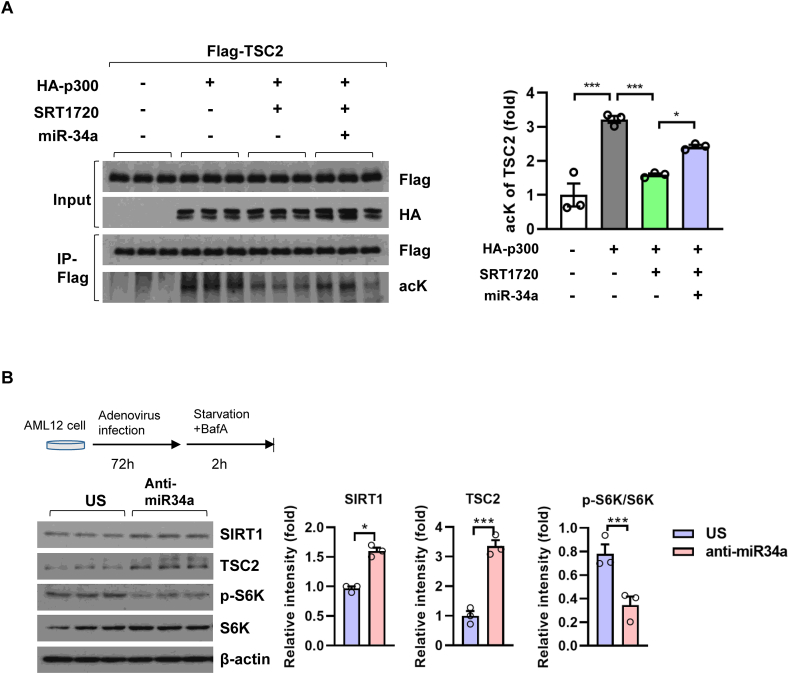
We have previously shown that CREB-CRTC2 transcriptional machinery is responsible for the HFD-feeding induced activation of miR-34a in the liver [10]. As a result, miR-34a expression was significantly reduced in livers of Crtc2LKO mice compared with the control under HFD feeding (Supplementary Fig. S3A), leading to the activation of fatty acid β-oxidation and FGF21 expression via the SIRT1-PPARα axis [10]. However, alterations in fatty acid β-oxidation or FGF21 expression per se cannot directly promote the activation of mTORC1 by CRTC2 under HFD-feeding in the liver, prompting us to investigate a potential direct link between miR-34a and mTORC1 pathway. SIRT1 was shown to be involved in the down-regulation of mTORC1 signaling, in part via deacetylation and the resultant activation of TSC2, a potent inhibitor of mTORC1 [21,22]. We attempted to investigate whether miR-34a-dependent regulation of mTORC1 signaling is mediated through the SIRT1/TSC2 axis. We were able to confirm the previous finding by showing that acetylation of TSC2 by p300 was reduced in response to the treatment of SRT1720, an activator of SIRT1 (Supplementary Fig. S3B). Furthermore, while the total TSC2 levels were increased, acetylation of TSC2 was reduced in Crtc2-depleted liver compared with the control, suggesting that increased expression of SIRT1 due to the reduced miR-34a levels led to the inhibition of mTORC1 pathway via the stimulation of TSC2 deacetylation and its stabilization in this setting (Figure 2A). We also observed a small but significant increase in TSC1 levels upon Crtc2 depletion, perhaps due to the enhanced stability of TSC1/2 complex.
Figure 2.
Depletion of Crtc2 repressed mTORC1 activity via an activation of TSC2. A. Effects of hepatic Crtc2 knockout on SIRT1 expression as well as TSC2 and its acetylation levels in livers of Crtc2f/f or Crtc2LKO mice (7-week HFD) (left). Quantitation of SIRT1, TSC1, TSC2 and acetyl TSC2 from western blot analysis was also shown (right). Data represent mean ± SEM (n = 5 per group). ∗p < 0.05, ∗∗p < 0.01. B. Effects of miR-34a on mTORC1 activity in the liver. AAV-GFP or AAV-miR-34a were infected in HFD-fed Crtc2f/f mice or Crtc2LKO mice for 4 weeks (under 10 week-HFD). Representative Western blot analysis were shown to monitor protein levels of CRTC2, SIRT1, TSC2, S6K1 or p-S6K1. β-Actin level was shown as a loading control (left). Quantitation of p-S6K1/S6K1 and TSC2/β -actin from western blot analysis was also shown (right). Data represent mean ± SEM (n = 4 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. C. Liver sections of Crtc2f/f;+ AAV-GFP, Crtc2LKO;+ AAV-GFP, Crtc2LKO;+ AAV-miR-34a mice (as in Figure 2B) were stained with hematoxylin and eosin (H&E, top). Scale bar: 100 μm. Frozen sections were also stained with Oil red O (red) to detect the neutral lipid (bottom). Representative image was shown.
We wanted to further verify the notion that miR-34a controlled mTORC1 activity in the liver. Indeed, overexpression of miR-34a countered the effect of hepatic Crtc2 depletion on mTORC1 activity, as evidenced by reduced TSC2 expression and subsequent restoration of p-S6K1 levels in the liver (Figure 2B). As a result, reduced hepatic lipid accumulation in Crtc2LKO mice was restored to the level of Crtc2f/f mice upon ectopic expression of miR-34a (Figure 2C and Supplementary Fig. S3C). The effect of miR-34a on mTORC1 signaling was also confirmed in AML12 hepatic cells (Supplementary Fig. S3D). Furthermore, miR-34a could specifically reverse SRT1720-mediated deacetylation of TSC2, showing that miR-34a-mediated activation of mTORC1 is mediated in part via the regulation of SIRT1-TSC2 axis (Figure 3A). We observed that inhibition of miR-34a by ectopic expression of anti-miR-34a enhanced SIRT1 expression with a concomitant reduction in p-S6K1 levels, confirming our hypothesis that miR-34a controls mTORC1 activity via the regulation of SIRT1 in the liver (Figure 3B).
Figure 3.
miR-34a promoted mTORC1 activity via a repression of SIRT1-TSC2 axis. A. Western blot analysis showing effects of miR-34a and/or SRT1720 (5 μM, 4h) on p300-mediated acetylation status of TSC2. Representative data of three independent experiments were shown (left). Quantitation of acetyl TSC2 from western blot analysis was also shown (right). Data represent mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. B. Effects of anti-miR-34a on mTORC1 activity in AML12 hepatocytes. Cells were infected with negative control (Ad-US) or Ad-anti-miR-34a for 3 days (top, left, scheme). Western blot analysis showing protein levels of SIRT1, TSC2, p-S6K1 and S6K1 (bottom, left). Quantitation of SIRT1, p-S6K1/S6K1, and TSC2 was shown (right). Data represent mean ± SEM (n = 3 per group). ∗p < 0.05.
3.3. Activation of SIRT1 counter-regulates the mTORC1 pathway, resulting in the reduced lipid accumulation in Crtc2-depleted hepatocytes
Reduced mTORC1 signaling pathway may dampen the lipogenic pathway by decreased action of PPARγ and/or SREBP-1c [23]. Although Han et al. reported an inhibitory role of CRTC2 in SREBP-1c activity, their work mostly relied on systemic Crtc2 knockout mouse model mainly in the normal chow diet setting [8]. Because of the tissue-specific role of CRTC2 in controlling metabolic pathways in various tissues [15,24,25], we thought it was necessary to delineate the role of CRTC2 in hepatic lipogenesis by using liver-specific Crtc2 knockout mouse model. We observed a decreased expression of both PPARγ and SREBP-1c, transcriptional regulators for lipogenesis, as well as that of their target genes in lipid metabolism in livers of HFD-fed Crtc2LKO mice compared with the control (Figure 4A, B). The effect of Crtc2 depletion on lipogenic gene expression was negated by the overexpression of miR-34a (Supplementary Fig. S4A). Expression of ChREBP, another transcriptional regulator for lipogenesis, is also reduced in Crtc2-depleted liver under HFD (Figure 4B), perhaps reflecting the role of SIRT1 in the regulation of these transcription factors [12]. In line with this result, we observed a tendency of reduced lipogenesis and a pronounced decrease in lipid accumulation in Crtc2 depleted hepatocytes compared with the control (Figure 4C and Supplementary Fig. S4B). Although we observed a decreased expression of CD36 in livers of HFD-fed Crtc2LKO mice compared with the control, we did not observe a significant changes in fatty acid uptake in hepatocytes from both genotypes (Supplementary Fig. S4C). Collectively, these data suggest that depletion of Crtc2 inhibits hepatic lipogenesis.
Figure 4.
Hepatic depletion of Crtc2 inhibits lipogenic pathway in the liver. A. Western blot analysis showing hepatic protein levels of Crtc2f/f or Crtc2LKO mice (7 week-HFD) under fasted or refed conditions (left). Quantitation of key bands in the refed condition was shown (right). Data represent mean ± SEM (n = 4 per group) ∗∗p < 0.01. B. Effect of chronic depletion of hepatic Crtc2 in mice on genes involved in the lipid uptake, the formation of lipid droplets, and lipogenesis. Data represent mean ± SEM (n = 4–7 per group). ∗p < 0.05, ∗∗p < 0.01. C. Primary hepatocytes from Crtc2f/f or Crtc2LKO mice were incubated for 24 h in low glucose media and then either maintained further in the same media for 48h or switched to high glucose plus insulin media for 48h. Oil red o staining was performed to measure hepatic lipid accumulation. Quantitation was also shown. Data represent mean ± SEM (n = 3 per group). ∗∗∗p < 0.001.
Activation of the mTORC1 pathway was also shown to interfere with autophagic flux in various cell types including hepatocytes [26]. Since we observed a decreased mTORC1 pathway in HFD-fed Crtc2 depleted liver compared with the control (Figure 1D), we suspected that depletion of Crtc2 in the liver may promotes autophagy. We observed an increased autophagic flux in livers of Crtc2LKO mice compared with the control as measured by the changes in LC3-II levels upon leupeptin injection in HFD-fed mice (Figure 5A). While p62 levels appeared to be higher in livers of Crtc2LKO mice compared with the control, leupeptin treatment further elevated p62 levels in Crtc2-depleted liver, showing that autophagic flux was enhanced in this setting. Conversely, overexpression of Crtc2 in the liver reduced autophagic flux in mice, corroborating our hypothesis that CRTC2-mTORC1 axis could inhibit autophagy in the liver (Figure 5B). Ectopic expression of miR-34a in hepatic cells reduced autophagic flux in part via the regulation of the SIRT1/mTORC1-mediated pathway as shown by the measurement of the differential accumulation of LC3-II levels upon treatment of lysosome inhibitor Bafilomycin A1 (BafA) (Figure 5C and Supplementary Fig. S5A). Conversely, we observed that inhibition of miR-34a enhanced autophagic flux in cultured hepatocytes (Figure 5D). Confocal microscopic analysis was also confirmed the inhibitory effect of miR-34a on the formation of autophagosomes and autolysosomes (Supplementary Fig. S5B).
Figure 5.
CRTC2-miR-34a-mTORC1 axis is critical in limiting autophagic flux. A. Effects of chronic Crtc2 depletion on autophagy in the liver. In vivo autophagic flux was estimated by using lysosomal inhibitor, leupeptin, in Crtc2f/f or Crtc2LKO mice (8 week-HFD). General scheme for the measurement of in vivo autophagic flux (top, left). Western blot analysis demonstrating changes in LC3-II as well as p62 levels was shown (bottom, left). Quantification of net autophagic flux calculated by differences in band intensity of LC3-II between groups injected with leupeptin and PBS (right). Data represent mean ± SD (n = 3 per group). ∗p < 0.05. B. Effects of constitutively active CRTC2 (CRTC2Δ) overexpression on LC3-II and p62 levels in mouse liver. Mice under the NCD diet were infected with either Ad-GFP or Ad-Crtc2 for 7 days before being sacrificed for the analysis. In vivo autophagic flux was estimated using the lysosomal inhibitor leupeptin. General scheme for the measurement of in vivo autophagic flux (top, left). Western blot analysis demonstrating changes in LC3-II as well as p62 levels was shown (bottom, left). Quantification of net autophagic flux calculated by differences in band intensity of LC3-II between groups injected with leupeptin and PBS (right). Data represent mean ± SD (n = 3 per group). ∗p < 0.05. C. Effects of miR-34a on autophagy in AML12 hepatocytes. General scheme for the measurement of in vitro autophagic flux (top, left). Cells were infected with either Ad-GFP or Ad-miR-34a and then cultured in the complete media or EBSS, with or without BafA (100 nM) for 2 h. Western blot analysis demonstrating changes in LC3-II levels was shown (bottom, left). Quantification of net autophagic flux calculated by differences in band intensity of LC3-II between control and BafA-treated cells (right). Data represent mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01. D. Effects of anti-miR-34a on autophagy in AML12 hepatocytes. Cells were infected with either Ad-US or Ad-anti-miR-34a and then cultured in the complete media or EBSS, with or without BafA (100 nM) for 2 h. Western blot analysis demonstrating changes in LC3-II levels was shown (bottom, left). Quantification of net autophagic flux calculated by differences in band intensity of LC3-II between control and BafA-treated cells (right). Data represent mean ± SEM (n = 3 per group). ∗p < 0.05.
Lipophagy, a selective autophagy of lipid droplets, is one of the main mechanisms for controlling hepatic lipid contents. We observed an increased formation of autophagic vacuoles (AV) mainly containing lipids in livers of HFD-fed Crtc2LKO mice compared with the control (Figure 6A), suggesting that the depletion of Crtc2 promoted an increased lipophagy in the liver. This notion was indeed confirmed by the microscopic analysis which demonstrated the increased co-localization of lipid droplet with lysosome as measured by the staining the livers of HFD-fed mice with LipidTox and antibody against LAMP1 (Supplementary Fig. S6A). The effect of miR-34a on lipophagy was further tested in cultured hepatocytes. We found that LC3 puncta formation as well as co-localization of LC3 and lipid droplet was reduced in primary hepatocytes expressing miR-34a compared with the control (Supplementary Fig. S6B). We observed a dramatic accumulation of lipid droplet in primary hepatocytes upon palmitate loading by ectopic expression of miR-34a, which was partially reversed by the treatment of SIRT1 activator SRT1720, presumably due to the reduced lipophagy (Figure 6B). This idea was indeed confirmed by the reduced co-localization of LAMP1 and lipid droplets in miR-34a expressing hepatocytes, suggesting that miR-34a-dependent regulation of hepatic lipid accumulation is also mediated in part through lipophagy pathway (Figure 6C). While ectopic expression of constitutively active CRTC2 enhanced lipid accumulation and reduced the colocalization of LAMP1 and lipid droplets, anti-miR-34a reversed this phenomenon, corroborating our hypothesis that CRTC2-miR-34a axis is critical in the regulation of lipophagy in hepatocytes (Supplementary Figs. S7A and S7B). These data unequivocally suggest that miR-34a promoted the activation of mTORC1 pathway to enhance hepatic lipid accumulation by the activation of lipogenesis and the inhibition of autophagic flux.
Figure 6.
CRTC2-miR-34a-mTORC1 axis inhibits lipophagic flux. A. Liver tissues from 4-h-fasted Crtc2f/f or Crtc2LKO mice (8-week HFD) were fixed, and processed for transmission electron microscopy analysis (left, red arrow; lipid droplet containing lysosome structure). Percent of autophagic vacuoles containing lipid droplet among total lipid droplets were shown (right). Data represent mean ± SEM (n = 12 per group). ∗∗∗p < 0.001. B. Effect of miR-34a on hepatic lipid accumulation in mouse primary hepatocytes. General scheme for the experiment (left). Cells were infected with either Ad-GFP or Ad miR-34a and then treated with either SRT1720 (5 μM) or DMSO control in the presence of either BSA or BSA conjugated palmitate (150 μM) for 4 h. Confocal microscopic analysis of lipid droplet (Red, LipidTox) and the nucleus (Blue, DAPI) were shown (middle). Scale bar: 20 μm. Quantitation for the content of lipid droplets were shown (right). Data represent mean ± SEM (n = 5 per group). ∗∗∗p < 0.001. C. Effect of miR-34a on hepatic lipophagy in mouse primary hepatocytes. General scheme for the experiment (left). Cells were infected with either Ad-GFP or Ad miR-34a and then treated with either SRT1720 (5 μM) or DMSO control in the presence of either BSA or BSA conjugated palmitate (150 μM) for 4 h. Confocal microscopic analysis showing a co-localization of lipid droplet (Red, LipidTox) and lysosome (Green, LAMP1) were shown (middle). Blue, DAPI, Scale bar: 10 μm. Quantitation for the content of lipid droplets (top, right) or lipid droplet containing lysosome (bottom, right) is shown. Data represent mean ± SEM (n = 4–8 per group). ∗p < 0.05, ∗∗∗p < 0.001.
3.4. Hepatic mTORC1 activity is induced in NAFLD
After observing that increased mTORC1 activity could be causal to the miR-34a-mediated lipid accumulation in hepatocytes, we wanted to determine whether CRTC2 and mTORC1 pathway is indeed induced in the human patients of NAFLD. We observed an elevated expression of TNFα in the livers of human NAFLD patients, showing a sign of increased inflammation in that setting (Figure 7A). We found that miR-34a expression was also highly induced in the livers of human NAFLD patients compared with the control (Figure 7B), as was the case for mouse models (Figure 7C). The strong correlation between CRTC2 and mTORC1 pathway on NAFLD was demonstrated by immunohistochemistry which shows that human NAFLD patients with severe fibrosis (as observed by Sirius red staining) exhibited increased CRTC2 and p-S6K1 expression (Figure 7D). Lipogenic gene expression was also elevated in livers of NAFLD patients compared with the control (Figure 7E). Overall, these data suggest that the diet-induced activation of CRTC2 instigates the progression of NAFLD by activating miR-34a-mediated lipid accumulation in the liver.
Figure 7.
Increased hepatic CRTC2 and mTORC1 are associated with human cases of NAFLD. A. Hepatic levels of TNFα were determined in the control or NAFLD patients. Data represent mean ± SEM (n = 5 per group). ∗p < 0.05. B. Hepatic levels of miR-34a were determined in the control or NAFLD patients. Data represent mean ± SEM (n = 5 per group). ∗∗p < 0.01. C. Expression of miR-34a was determined in the livers of mice fed either NCD or HFD for 8 weeks. Data represent mean ± SD (n = 3–4 per group). ∗∗p < 0.01. D. Representative immunohistochemical analysis showing the expression of CRTC2 and p-S6K1 in the livers of control and NAFLD patients. Sirius red staining was also shown (left). Quantitation was also shown (right). Data represent mean ± SEM (n = 10–11 per group). ∗p < 0.05, ∗∗∗p < 0.001. E. Hepatic expression of lipogenic genes were determined in the control or NAFLD patients. Data represent mean ± SEM (n = 5 per group). ∗p < 0.05. F. A proposed model for the CRTC2-instigated progression of NAFLD. Under DIO conditions, elevated expression of CRTC2 promotes increased miR-34a in the liver, leading to the activation of the mTORC1 pathway via the increased acetylation and the degradation of TSC2 by inhibiting SIRT1. Reduced SIRT1 activity and the resultant mTORC1 activity is critical in the initial progression of NAFLD by promoting hepatic steatosis via activation of lipogenesis as well as inhibition of lipophagy and fatty acid β-oxidation.
4. Discussion
Activation of hepatic CRTC2 under diet-induced or genetic mouse models of obesity has been linked to the dysregulation of glucose homeostasis in part via an uncontrolled enhancement of gluconeogenesis and the stimulation of hepatic insulin resistance [7,27]. Previously, we have demonstrated that the depletion of hepatic Crtc2 is beneficial in alleviating the symptoms of metabolic diseases by inhibiting the progression of fatty liver and promoting energy metabolism via the reduction of miR-34a under HFD-mediated obesity and insulin resistant state in mice [10]. We demonstrated that miR-34a is a direct transcriptional target of CREB-CRTC2 in the liver, explaining the increased expression of miR-34a in DIO conditions that is common to both rodents and humans. Depletion of Crtc2 in the liver subsequently reduced the expression of miR-34a, leading to the increased protein expression of SIRT1 and PPARα, well characterized targets for miR-34a. We found that increased FGF21, a transcriptional target of PPARα, is mainly responsible for the improved energy homeostasis in Crtc2 LKO mice under DIO conditions. However, since FGF21 levels were usually higher in DIO conditions both in rodents and humans, perhaps reflecting a relative FGF21-resistant state [28], we decided to look for the additional pathways that are affected by miR-34a in the liver. We found that the mTORC1 pathway was significantly reduced in the livers of Crtc2LKO mice compared with the control under HFD-feeding, prompting us to look for the potential link between miR-34a and mTORC1 pathway in the liver. A recent report showed that SIRT1 could specifically reduce the activity of mTORC1 by deacetylation and activation of TSC2 [21,22], an inhibitor of mTORC1 pathway, suggesting that increased miR-34a could specifically induce mTORC1 signaling pathway under the DIO setting by reducing SIRT1 expression and the subsequent inhibition of TSC2 expression.
mTORC1 coordinates cellular energy metabolism not only by promotion of anabolic pathways, but also by inhibition of catabolic pathways in mammals [26]. Most notably, mTORC1 suppresses catabolic pathways of macromolecules by inhibiting autophagy [29]. Activation of mTORC1 under nutrient-abundant state is shown to inhibit autophagy by phosphorylation and inactivation of key components of autophagy including Unc-51 like autophagy activating kinase (ULK) 1 and 2 as well as ATG13 and autophagy/beclin 1 regulator 1 (AMBRA1). In this regard, mTORC1 directly opposes the action of AMPK in that mTORC1-mediated phosphorylation of ULK1 (at Ser757) prevents a distinct phosphorylation of ULK1 at Ser317 and Ser777 by AMPK [30]. Autophagy is not only critical in the regulation of proteostasis but also important in the proper regulation of intracellular organelles including lipid droplet. Hyperactivation of mTORC1 under the nutrient-rich state in the liver could result in the accumulation of hepatic TG by simultaneous activation of lipogenesis (via the activation of SREBP-1c and PPARγ among other factors) and inhibition of lipid clearance by limiting lipophagic flux.
Despite the increased autophagic flux, we did not observe changes in expression of key genes in the autophagy pathway such as Atg5, Atg7, Atg16l1, and Tfeb upon hepatic depletion of Crtc2 in HFD-fed mice, [10]. This result appears to be in complete disagreement with the study by Kemper and colleague, who showed that CREB-CRTC2 is critical in the transcriptional regulation of key genes in the autophagy [31]. We would suspect that the presence of a potential compensatory mechanism may be involved in the transcriptional regulation of genes in the autophagy pathway in the chronic absence of Crtc2 in the liver, especially under the HFD. We found that hepatic depletion of Crtc2 led to the increased expression of PPARα [10], which was also shown to enhance transcriptional activation of key genes in the autophagy such as Atg5 and Atg7 in another recent study [32]. Based on these observations, we would conclude that chronic depletion of Crtc2 in the liver promotes increased autophagic flux without specific changes in expression of individual genes involved in the autophagy.
While our work was under peer review, a recent study showed a paradoxical role of mTORC1 in promoting lipophagy in the liver under the physiological conditions [33]. In their work, the authors utilized HFD feeding for only 4 weeks deliberately to study the importance of mTORC1 pathway in the Plin3-dependent lipophagy before the onset of NAFLD in mice. While the work delineated the role of Plin3 in hepatic lipophagy by siRNA and shRNA against Plin3 in vitro and in vivo, they relied on rapamycin to inhibit mTORC1 activity without utilizing genetic manipulation of mTORC1 machinery. Although rapamycin is widely considered as a specific inhibitor for mTORC1 (via rapamycin-FKBP12 binding to mTOR kinase domain within mTORC1), prolonged treatment of rapamycin has been shown to inhibit mTORC2 as well, by sequestering uncomplexed mTOR from the potential mTORC2 complex [34,35]. Since we did not directly test the involvement of mTORC1 in the control of lipid metabolism in Crtc2-deficient mouse liver either, more delicate genetic study is necessary to unequivocally test the role of mTORC1 in the control of lipophagy in vivo setting, especially in the pathological state, which should be considered as a follow-up study.
Based on the current data and our previous work, we would propose the following model (Figure 7F). The nutrient stress induced the increased expression of CRTC2, which in turn promotes elevation of miR-34a in the liver. miR-34a-mediated repression of SIRT1 led to the decreased PGC-1α/PPAR-α pathway, resulting in the reduced fatty acid β-oxidation. At the same time, diminished SIRT1-TSC2 axis led to the activation of mTORC1, promoting the increased lipogenesis via the induction of SREBP-1c and the reduced lipophagic flux in DIO conditions. Thus, inhibition of the CRTC2-miR-34a pathway could alleviate the progression of NAFLD and the related diseases by effectively impeding lipid accumulation in the liver.
Author contributions
H.S.H. and S.H.K. conceived the idea and developed the study design. H.S.H., S.H.J, Y. K., D. C., T. H. and E. A. performed experiments, and S.G.K., Y.S.K., S.H.J. and D.H.L. provided and analyzed the human NAFLD samples. E.M. performed TEM analysis. H.S.H., S.G.K, S.H.J, H.S.K., J.K.S., G.S.H., K.W.C., and S.H.K. interpreted data, and H.S.H., K.W.C. and S.H.K wrote the manuscript.
Acknowledgement
We would thank Dr. Hyuna Noh for the critical advice regarding the pair-feeding study. This research was supported by the National Research Foundation of Korea Grant funded by the Korean Government (MSIT) (NRF-2015R1A5A1009024, NRF-2019M3A9D5A01102794, and NRF-2021R1A2C3003435 (to S.H.K.) and NRF-2019R1A2C1084684 (to K.W.C.)). H.S.H. was supported by NRF-2018R1A6A3A11043165. S.H.K. was also supported by a grant from Korea University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101402.
Contributor Information
Kae Won Cho, Email: kwcho@sch.ac.kr.
Seung-Hoi Koo, Email: koohoi@korea.ac.kr.
Conflict of interest
The authors have declared no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Arab J.P., Arrese M., Trauner M. Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annual Review of Pathology: Mechanisms of Disease. 2018;13:321–350. doi: 10.1146/annurev-pathol-020117-043617. [DOI] [PubMed] [Google Scholar]
- 2.Eslam M., George J. Genetic contributions to NAFLD: leveraging shared genetics to uncover systems biology. Nature Reviews Gastroenterology & Hepatology. 2020;17(1):40–52. doi: 10.1038/s41575-019-0212-0. [DOI] [PubMed] [Google Scholar]
- 3.Koo S.H. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clinical and Molecular Hepatology. 2013;19(3):210–215. doi: 10.3350/cmh.2013.19.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature Reviews Molecular Cell Biology. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conkright M.D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J.B., et al. TORCs: transducers of regulated CREB activity. Molecular Cell. 2003;12(2):413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Koo S.H., Flechner L., Qi L., Zhang X., Screaton R.A., Jeffries S., et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 7.Dentin R., Liu Y., Koo S.H., Hedrick S., Vargas T., Heredia J., et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449(7160):366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 8.Han J., Li E., Chen L., Zhang Y., Wei F., Liu J., et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature. 2015;524(7564):243–246. doi: 10.1038/nature14557. [DOI] [PubMed] [Google Scholar]
- 9.Li Y., Song Y., Zhao M., Guo Y., Yu C., Chen W., et al. A novel role for CRTC2 in hepatic cholesterol synthesis through SREBP-2. Hepatology. 2017;66(2):481–497. doi: 10.1002/hep.29206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han H.S., Choi B.H., Kim J.S., Kang G., Koo S.H. Hepatic Crtc2 controls whole body energy metabolism via a miR-34a-Fgf21 axis. Nature Communications. 2017;8(1):1878. doi: 10.1038/s41467-017-01878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi S.E., Fu T., Seok S., Kim D.H., Yu E., Lee K.W., et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12(6):1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemper J.K., Choi S.E., Kim D.H. Sirtuin 1 deacetylase: a key regulator of hepatic lipid metabolism. Obesity. 2013;91:385–404. doi: 10.1016/B978-0-12-407766-9.00016-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J., Padhye A., Sharma A., Song G., Miao J., Mo Y.Y., et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. Journal of Biological Chemistry. 2010;285(17):12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellacott K.L., Morton G.J., Woods S.C., Tso P., Schwartz M.W. Assessment of feeding behavior in laboratory mice. Cell Metabolism. 2010;12(1):10–17. doi: 10.1016/j.cmet.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.H., Wen X., Cho H., Koo S.H. CREB/CRTC2 controls GLP-1-dependent regulation of glucose homeostasis. The FASEB Journal. 2018;32(3):1566–1578. doi: 10.1096/fj.201700845R. [DOI] [PubMed] [Google Scholar]
- 16.Park J., Yoon Y.S., Han H.S., Kim Y.H., Ogawa Y., Park K.G., et al. SIK2 is critical in the regulation of lipid homeostasis and adipogenesis in vivo. Diabetes. 2014;63(11):3659–3673. doi: 10.2337/db13-1423. [DOI] [PubMed] [Google Scholar]
- 17.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 18.Green C.R., Wallace M., Divakaruni A.S., Phillips S.A., Murphy A.N., Ciaraldi T.P., et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nature Chemical Biology. 2016;12(1):15–21. doi: 10.1038/nchembio.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caron A., Richard D., Laplante M. The roles of mTOR complexes in lipid metabolism. Annual Review of Nutrition. 2015;35:321–348. doi: 10.1146/annurev-nutr-071714-034355. [DOI] [PubMed] [Google Scholar]
- 20.Wu R., Dang F., Li P., Wang P., Xu Q., Liu Z., et al. The circadian protein Period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metabolism. 2019;29(3):653–667. doi: 10.1016/j.cmet.2018.11.006. e656. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh H.S., McBurney M., Robbins P.D. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5(2) doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Aguilar A., Guillen C., Nellist M., Bartolome A., Benito M. TSC2 N-terminal lysine acetylation status affects to its stability modulating mTORC1 signaling and autophagy. Biochimica et Biophysica Acta. 2016;1863(11):2658–2667. doi: 10.1016/j.bbamcr.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Li S., Brown M.S., Goldstein J.L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proceedings of the National Academy of Sciences of the U S A. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchet E., Van de Velde S., Matsumura S., Hao E., LeLay J., Kaestner K., et al. Feedback inhibition of CREB signaling promotes beta cell dysfunction in insulin resistance. Cell Reports. 2015;10(7):1149–1157. doi: 10.1016/j.celrep.2015.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon Y.S., Liu W., Van de Velde S., Matsumura S., Wiater E., Huang L., et al. Activation of the adipocyte CREB/CRTC pathway in obesity. Communications Biology. 2021;4(1) doi: 10.1038/s42003-021-02735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu D., Oh K.J., Jo H.Y., Hedrick S., Kim Y.N., Hwang Y.J., et al. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase. LIPIN1. Cell Metabolism. 2009;9(3):240–251. doi: 10.1016/j.cmet.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Fisher F.M., Chui P.C., Antonellis P.J., Bina H.A., Kharitonenkov A., Flier J.S., et al. Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes. 2010;59(11):2781–2789. doi: 10.2337/db10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y.C., Guan K.L. mTOR: a pharmacologic target for autophagy regulation. Journal of Clinical Investigation. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J.M., Wagner M., Xiao R., Kim K.H., Feng D., Lazar M.A., et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Macia M., Santos-Ledo A., Leslie J., Paish H.L., Collins A.L., Scott R.S., et al. A mammalian target of rapamycin-perilipin 3 (mTORC1-Plin3) pathway is essential to activate lipophagy and protects against hepatosteatosis. Hepatology. 2021 doi: 10.1002/hep.32048. [DOI] [PubMed] [Google Scholar]
- 34.Ricoult S.J.H., Manning B.D. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Reports. 2013;14(3):242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamming D.W., Ye L., Katajisto P., Goncalves M.D., Saitoh M., Stevens D.M., et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science (New York, N Y ) 2012;335(6076):1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.