Summary
This protocol describes the reconstitution of the filamentous Ebola virus nucleocapsid-like assembly in vitro. This is followed by solving the cryo-EM structure using helical reconstruction, and flexible fitting of the existing model into the 5.8 Å cryo-EM map. The protocol can be applied to other filamentous viral protein assemblies, particularly those with high flexibility and moderate resolution maps, which present technical challenges to model building.
For complete details on the use and execution of this profile, please refer to Su et al. (2018).
Subject areas: Cryo-EM, Microbiology, Protein Biochemistry, Protein expression and purification, Structural Biology
Graphical abstract
Highlights
-
•
Preparation of Ebola nucleocapsid-like assembly for cryo-EM
-
•
Cryo-EM helical reconstruction of flexible filamentous protein assembly
-
•
Flexible fitting of protein model into cryo-EM density at moderate resolution
This protocol describes the reconstitution of the filamentous Ebola virus nucleocapsid-like assembly in vitro. This is followed by solving the cryo-EM structure using helical reconstruction, and flexible fitting of the existing model into the 5.8 Å cryo-EM map. The protocol can be applied to other filamentous viral protein assemblies, particularly those with high flexibility and moderate resolution maps, which present technical challenges to model building.
Before you begin
The protocol below describes specific steps for recombinant Ebola nucleoprotein (eNP) expression.
Protein induction and expression
Timing: 2.5 days
-
1.
Transform 100 μL of E.coli BL21 (DE3) competent cells with 1 μL of expression plasmid (∼ 100 ng/μL) by heat shock.
-
2.
Recover cells by adding 900 μL Luria-Bertani (LB) media and incubate at 37°C for 1 h.
-
3.
Centrifuge cells at 6,000×g for 10 min. Discard supernatant.
-
4.
Resuspend cells gently in approximately 100 μL of remaining media and spread evenly on LB agar plate with 50 μg/mL ampicillin. Incubate at 37°C overnight.
-
5.
After 12–16 h, inoculate one colony into 10 mL of LB media supplemented with 50 μg/mL ampicillin, and culture at 37°C with shaking (200 rpm) for 3–5 h until it becomes turbid.
-
6.
Transfer the above culture into 2 L LB media supplemented with 50 μg/mL ampicillin and continue to culture with shaking (200 rpm) at 37°C for 4–6 h.
-
7.
When OD600 is approximately 0.6–0.7, briefly cool down media on ice for 15 min. Take 100 μL of cells for a pre-induction sample for SDS-PAGE, centrifuge for 1 min at 15,000×g, discard 75 μL of supernatant, and add 25 μL of 2× gel loading buffer. Incubate in a heat block or boiling water for 10 min and store at 4°C until ready to use. Induce protein expression with 0.5 mM IPTG and shake at 18°C for 12–14 h.
-
8.
Harvest cells by centrifugation at 6,000×g for 10 min at 10°C. Discard the supernatant.
-
9.
A cell pellet with a wet weight of 3–4 grams is resuspended in 30–40 mL of lysis buffer containing a mixture of protease inhibitors (1 μg/L leupeptin, 1 μg/L antipain, 1 μg/L benzamidine, 0.5 μg/L pepstatin).
-
10.
Take 100 μL of cells for a post-induction gel sample, centrifuge for 1 min at 15,000×g, discard 75 μL of supernatant, and add 25 μL of 2× gel loading buffer. Incubate in a heat block or boiling water for 10 min. Load 10 μL each of pre- and post-induction samples on a SDS-PAGE to determine the level of protein expression. Run at 240V for 40 min in 1× SDS-PAGE running buffer and stain with Coomassie blue .
Note: The growth status of the bacteria before adding the IPTG in this step is very important. OD600 of 0.6–0.7 at time of induction yields the highest protein levels.
Pause Point: The harvested cells can be stored at -80°C for several months.
CRITICAL: The lysis buffer composition and purification method is important for obtaining high-quality nucleoprotein assemblies that will lead to high-resolution cryo-EM structures. The salt concentration, pH, and other components of the lysis buffer should be experimentally determined in order to ensure that the desired nucleoprotein assembly is obtained.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E.coli BL21 (DE3) | Agilent | Cat#200131 |
| Chemicals, peptides, and recombinant proteins | ||
| eNP-2 (25-457) recombinant proteins | Leung et al., 2015; Su et al., 2018 | N/A |
| uranyl acetate | Electron Microscopy Sciences | Cat#22400 |
| Tobacco Etch Virus protease (TEV Protease) | New England Biolabs | Cat#P8112S |
| Deposited data | ||
| Cryo-EM map for eNP-2 | This Paper | https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-7343 |
| Flexible fitted models of eNP-2 to cryoEM map | Su et al., 2018 | N/A |
| Recombinant DNA | ||
| Plasmid pMBP-His6TEV eNP-2 | Leung et al., 2015; Su et al., 2018 | N/A |
| Software and algorithms | ||
| Origin | OriginLab | https://www.originlab.com (Version 7) |
| PRISM | GraphPad | https://www.graphpad.com/ (Version 7) |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/ |
| EMAN2 | Tang et al., 2007 | http://blake.bcm.edu/emanwiki/EMAN2 |
| Relion2 |
Scheres, 2012; He and Scheres., 2017 |
www2.mrc-lmb.cam.ac.uk/relion |
| Motioncorr v2.1 | Li et al., 2013 | https://github.com/jianglab/motioncorr |
| CTFFIND4 | Rohou and Grigorieff., 2015 | http://grigoriefflab.janelia.org/ctf |
| EMAN | Ludtke et al., 1999 | http://blake.bcm.edu/emanwiki/EMAN1 |
| IHRSR | Egelman, 2007 | https://cryoem.ucsd.edu/wikis/software/start.php?id=ihrsr |
| Segger and ProMod plugin in UCSF Chimera |
Pintilie et al., 2010 Pintilie et al., 2016 |
https://github.com/gregdp/segger |
| MassMatrix | Xu and Freitas, 2009 | http://www.massmatrix.bio/ |
| HDExaminer | Sierra Analytics, Modesto, CA | http://massspec.com/hdexaminer/ |
| MDFF | Trabuco et al., 2009 | http://www.ks.uiuc.edu/Research/mdff/ |
| ProMod | Peitsch, 1996 | https://pypi.org/project/promod/ |
| PSIPRED | McGuffin et al., 2000 | http://bioinf.cs.ucl.ac.uk/psipred/ |
| Other | ||
| Cu grids with continuous carbon film | Ted Pella | Cat#01754-F |
| Quantifoil R 1.2/1.3 Cu grids (with continuous carbon film | Quantifoil | Cat#Q325CR1.3 |
Materials and equipment
Lysis Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 2 mL |
| NaCl (5 M) | 1 M | 20 mL |
| Imidazole (5 M) | 20 mM | 0.4 mL |
| 2-mercaptoethanol (BME) (14.3 M) | 5 mM | 0.35 mL |
| ddH2O | n/a | 77.25 mL |
| Total | n/a | 100 mL |
Filter and sterilize with a 0.22 μm filter. The lysis buffer should be made fresh and pre-cooled at 4°C before use. The lysis buffer is recommended to store no more than a week to avoid contamination.
PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| Na2HPO4 (0.2 M) | 10 mM | 50 mL |
| KH2PO4 (0.2 M) | 2 mM | 10 mL |
| NaCl (5 M) | 137 mM | 27.4 mL |
| KCl (0.2 M) | 2.7 mM | 13.5 mL |
| ddH2O | n/a | 899.1 mL |
| Total | n/a | 1,000 mL |
Add concentrated hydrochloric acid to adjust the pH to 7.4, and then add deionized water to make the solution volume to 1,000 mL. After autoclaving, The buffer can be stored at room temperature for no more than a week to avoid contamination.
Note: There are no divalent cations in the above PBS Buffer. If necessary, 1 mM CaCl2 and 0.5 mM MgCl2 can be added to the formula.
SDS-PAGE loading buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 6.8 (1 M) | 250 mM | 1.25 mL |
| Sodium dodecyl sulfate (SDS) | 10% (W/V) | 0.50 g |
| Bromophenol blue (BPB) | 0.5% (W/V) | 0.025 g |
| Glycerol | 50% (V/V) | 2.50 mL |
| 2-mercaptoethanol (BME) (14.3 M) | 5% (V/V) | 0.25 mL |
| ddH2O | n/a | to 5 mL |
| Total | n/a | 5 mL |
Store at room temperature for about a month.
5×SDS-PAGE running buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 0.125 M | 15.1 g |
| Glycine | 1.25 M | 94 g |
| SDS | 0.5% (W/V) | 5 g |
| ddH2O | n/a | to 1000 mL |
| Total | n/a | 1000 mL |
Before use, dilute with ddH2O to 1×SDS-PAGE running buffer. Store at room temperature for about a month.
Ni6FF column elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 2 mL |
| NaCl (5 M) | 1 M | 20 mL |
| Imidazole (5 M) | 200 mM | 4 mL |
| 2-mercaptoethanol (BME) (14.3 M) | 5 mM | 0.35 mL |
| ddH2O | n/a | 73.65 mL |
| Total | n/a | 100 mL |
Filter and sterilize with a 0.22 μm filter. Store at room temperature for about a week.
Wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 2 mL |
| NaCl (5 M) | 50 mM | 1 mL |
| 2-mercaptoethanol (BME) (14.3 M) | 5 mM | 0.35 mL |
| ddH2O | n/a | 96.65 mL |
| Total | n/a | 100 mL |
Filter and sterilize with a 0.22 μm filter. Store at room temperature for about a week.
Amylose column elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 2 mL |
| NaCl (5 M) | 50 mM | 1 mL |
| Maltose | 1% | 1 g |
| 2-mercaptoethanol (BME) (14.3 M) | 5 mM | 0.35 mL |
| ddH2O | n/a | 95.65 mL |
| Total | n/a | 100 mL |
Filter and sterilize with a 0.22 μm filter. Store at room temperature for about a week.
Storage buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 2 mL |
| NaCl (5 M) | 500 mM | 10 mL |
| Tris(2-carboxyethyl)phosphine (TCEP) (1 M) | 2 mM | 0.2 mL |
| ddH2O | n/a | 87.8 mL |
| Total | n/a | 100 mL |
Filter and sterilize with a 0.22 μm filter. Store at room temperature for about a week.
Step-by-step method details
Purification of recombinant Ebola nucleoprotein
This protocol is optimized for preparation of Ebola virus nucleoprotein (eNP) constructs, among which eNP-2 (residue 25–457) formed nucleocapsid-like assembly for cryo-EM study (Su et al., 2018).
Timing: 2 days
CRITICAL: Protein purification is conducted at 4°C to reduce protein degradation. Addition of protease inhibitors (1 μg/L leupeptin, 1 μg/L antipain, 1 μg/L benzamidine, 0.5 μg/L pepstatin) to buffers will also help to limit protein degradation.
-
1.
Lyse cells using an EmulsiFlex-C5 homogenizer (Avestin).
-
2.
Centrifuge lysates at 30,000×g at 4°C for 40 min. Gently transfer the supernatant to a clean container.
Note: Before proceeding to the next step, filter the supernatant with a 1.2 μm filter to avoid clogging the column.
-
3.Purify eNP-2 using a series of affinity and ion exchange chromatographic columns.
-
a.Load the supernatant on a 15 mL Ni6FF column (Cytiva) equilibrated in lysis buffer. Wash the column with 3 column volumes of lysis buffer and then elute the bound protein with 3 column volumes of elution buffer.
-
b.Take the eluted sample and load onto a 15 mL amylose column (NEB). Wash the column with 3 column volumes of wash buffer and then elute the bound protein with 3 column volumes of amylose column elution buffer .
-
c.Apply eluted protein to a 8 mL Source15Q ion exchange column (Cytiva). Wash column with 3 column volumes of Source15Q ion exchange column wash buffer. Elute protein with a linear gradient of NaCl up to 1 M in 5 column volumes.
-
a.
-
4.
The maltose-binding protein and six consecutive histidine residues (MBP-His6) tag is cleaved with TEV protease to obtain the target protein with three tag-derived residues (GHM) at the N-terminus. Add TEV enzyme and target protein at a ratio of 1:100 (protein concentration is approximately determined by A280 absorbance), and digest for 12–16 h at 25°C.
-
5.
Dilute the digested protein sample with wash buffer to conductivity of approximately 8 mS/cm, load the sample onto a Source15Q ion exchange column to separate undigested protein, MBP-His6 tag, and Tev from the cleaved protein. Elute protein with a linear gradient of NaCl up to 1 M in 5 column volumes.
Note: Additional steps onto an amylose affinity column and/or Ni column can be used here to further eliminate any remnants of fusion protein or MBP-His6 tag.
-
6.
Concentrate eNP-2 protein and load onto a 120 mL 10/300 SD200 gel filtration column (Cytiva) for final purification with 1 column volume of storage buffer.
-
7.
eNP-2 protein elutes in two peaks. Peak 1 that elutes earlier (∼52 mL) usually contains eNP-2 protein oligomers, assembly, or aggregates whereas peak 2 contains monomeric eNP-2. Peak 1 is collected and concentrated using a 30 kDa ultrafiltration centrifuge tube (Millpore Amicon® Ultra) to a final concentration of 10 mg/mL as determined by NanoDrop One (Thermo Fisher Scientific).
-
8.
Finally, the purity of eNP-2 assembly is determined to be about >95% by a Coomassie blue stained SDS-PAGE gel, and the purified eNP-2 assembly is promptly used for negative stain and cryo-EM studies. The sample can be stored at 4°C for no more than a week. It is not recommended to freeze the sample for storage as freeze and thaw may disrupt protein assembly.
Negative stain
Timing: 0.5–1 h
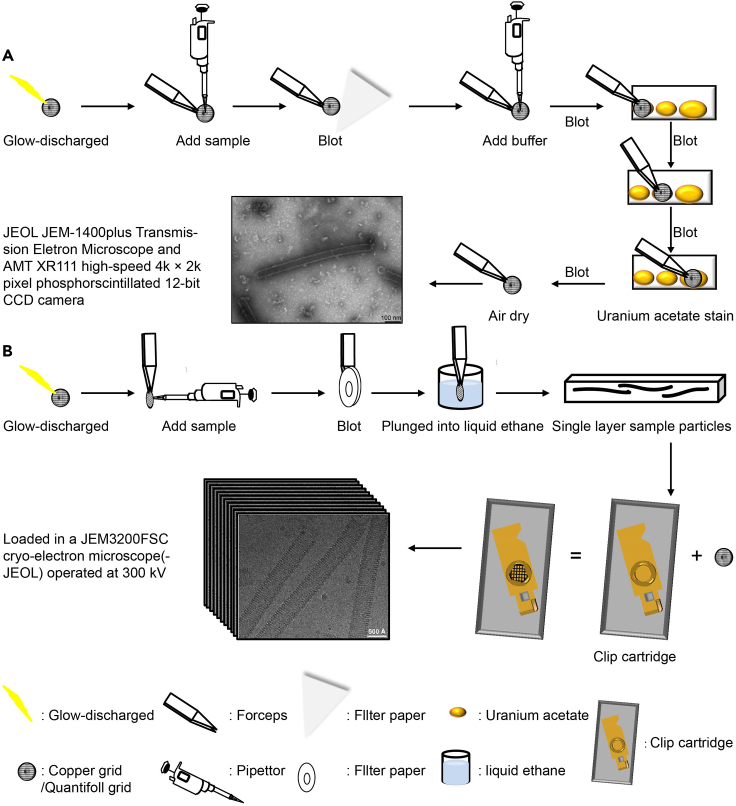
This step describes observation of eNP-2 assembly under negative stain at room temperature (Figure 1A).
-
9.
Apply 2 μL of eNP-2 sample at 1 mg/mL in PBS to a glow-discharged (40 s) copper grid coated with continuous carbon film (Ted Pella) and wait for 1 min.
-
10.
Blot the grid from the side with a piece of filter paper for 5–10 s.
-
11.
Apply 4 μL PBS to the grid and repeat step 10.
-
12.
Stain with 2% uranyl acetate (UA). Prepare three droplets of roughly 20, 20, and 60 μL UA on a piece of parafilm. Touch the grid to first two 20 μL UA droplets sequentially for 3 s and repeat step 10 in between. Then wash the grid in the 60 μL UA droplet for 30 s, repeat step 10 and airdry the grid before loading into the microscope.
Note: You may speed up the air-dry procedure by gently swinging the grid in air.
Alternatives: Other staining reagent such as uranyl formate (UF) can be used as alternatives in negative stain.
CRITICAL: UA (and UF) is radioactive and toxic, safety precautions should be taken to reduce inhalation and contamination by using the fume hood (for aqueous solution preparation), wearing the appropriate PPE, and ensuring proper disposal procedure for radioactive waste.
-
13.
Load the negative stained grid from step 12 into the room temperature side-entry holder of JEOL JEM-1400plus Transmission Electron Microscope operating at 80 kV and recorded with an AMT XR111 high-speed 4k × 2k pixel phosphor-scintillated 12-bit CCD camera at 30,000× magnification (corresponding to a pixel size of 4 Å).
Figure 1.
Schematic diagram of the sample preparation process
(A) Negative stain sample preparation, scale bar 100 nm.
(B) Cryo-EM sample preparation, scale bars 500 Å.
Cryo-EM sample preparation
Timing: 1–1.5 h
This step describes standard cryo-EM sample preparation procedure using Mark IV Vitrobot (Figure 1B). Before starting, liquid ethane is prepared using a designed foam container with liquid nitrogen surrounded as coolant, a spider-like metal piece that conducts heat, and a blue button ready for grid storage (Figure 2A). Quantifoil R 1.2/1.3 Cu grids (200 mesh) are glow discharged for 40 s (Figure 2B). Mark IV Vitrobot is set to 4°C and 100% humidity in the chamber, one blot for 3 s, no wait time, no drain time, zero offset. These parameters can be adjusted based on the resulting ice thickness and distribution (e.g., blot time and offset can be increased when ice is too thick; glow discharged time can be increased when ice distribution is not even).
CRITICAL: Ethane gas is flammable and explosive at room temperature, safety precautions should be taken to ensure complete closure of the ethane pressure gas tank everytime after usage. Ethane’s melting point is higher than nitrogen, which will lead to solidification of ethane under liquid nitrogen temperature. Formation of white solid in liquid ethane container is a sign for reaching ethane’s melting point and ready for vitrification. One should avoid accumulating too much ethane solid that will bend the grid during plunge freezing.
Note: When too much ethane solid is observed, one can use a room temperature spider-like metal piece up-side down to warm up and remove excess ethane solid.
-
14.
Glow discharged grid is loaded on the tweezer into the chamber, liquid ethane container is raised to the bottom of the chamber (Figure 2C).
-
15.
Apply 2 μL of eNP-2 assembly sample to the grid from the side of the chamber (Figure 2D).
-
16.
The grid is blotted for 3 s and plunged into liquid ethane (Figure 2E).
-
17.
The liquid ethane container is lowered. Now transfer the grid to the button for storage until future use.
Note: Detailed cryo-EM sample preparation procedure with videos was previously described (Meyerson et al., 2011). Other resources can also be found online.
Pause Point: Frozen grids can be stored in liquid nitrogen for years before loading onto the microscope.
Figure 2.
Cryo-EM sample preparation in practice
(A) Liquid nitrogen cools down ethane in the middle metal container using a spider-like piece for heat conduction in a designed foam container. The blue button is for grid storage.
(B) The grids are glow discharged.
(C) The grid is loaded on the tweezer and lifted into the MarkIV vitrobot chamber, the liquid ethane container is lifted to the bottom of the chamber.
(D) Load the sample from one side of the chamber onto the grid, blot the grid with filter papers.
(E) Plunge the grid into liquid ethane and transfer to the blue grid button for storage.
Cryo-EM data acquisition
Timing: 8 days
This step describes manual cryo-EM single particle data acquisition for eNP-2 assembly, which is necessary due to scarcity of helical tubes found under the microscope.
Note: For samples that appear to be more concentrated, automated data acquisition may be used with latest data acquisition software (e.g. EPU2, Tomo4, SerialEM, etc.), which may significantly reduce the timing estimated for data collection, whereas percentage of high quality images may decrease so further selection is required in the later data processing steps.
Pause Point: Depending on the microscope schedule, data acquisition can be paused at any time and the grid can be recovered for future use.
-
18.
Load frozen grids into JEM3200FSC (JEOL) operated at 300 kV with 70 μm condenser lens aperture, 60 μm objective lens aperture, spot size 1, and with 30 eV in-column energy filter.
-
19.
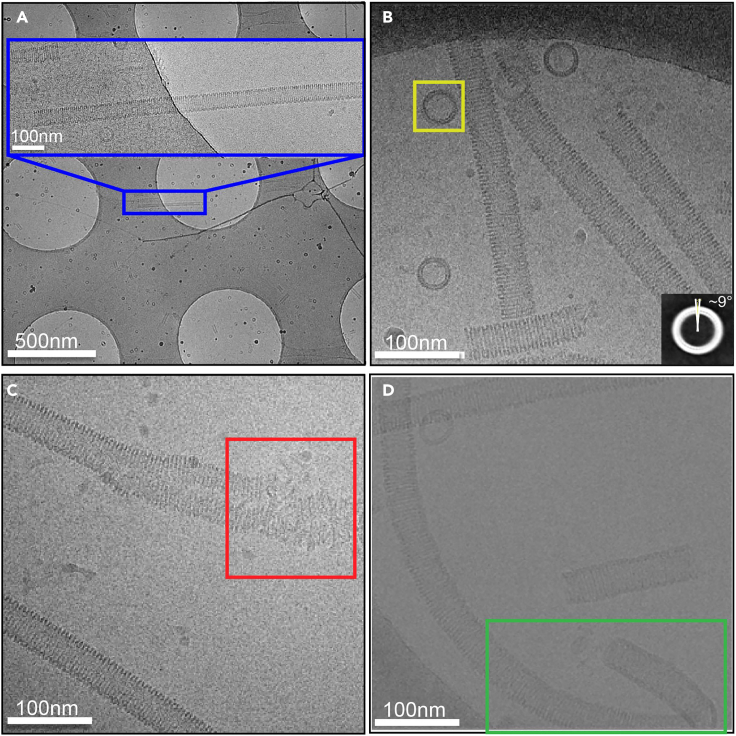
Set search mode magnification 3,000× for eNP-2 helical tube visualization (Figure 3A), and imaging mode magnification to 30,000× (corresponding to a calibrated sampling of 1.2 Å per pixel) for data acquisition (Figure 3B).
Note: Imaging parameters change with different microscopes, the parameters in this protocol are practical determined for JEM3200FSC, whereas different parameters may be used for other microscopes. Only regions with proper ice thickness are selected for data acquisition. Thin ice results in disruption of eNP-2 assembly (Figure 3C), whereas thick ice allows additional flexibility of the eNP-2 assembly (Figure 3D). See troubleshooting for suggestions on how to optimize ice thickness in sample freezing and screening.
-
20.
Micrographs are recorded with a K2 Summit direct electron device (Gatan) operating in super resolution mode at a recording rate of 5 raw frames per second and exposure rate of 3 e-/Å2 per second. The total exposure time is 8 s, yielding 40 frames per movie stack and a total dose of 24 e-/Å2.
-
21.
Defocus values are set to -1.0 to -2.0 μm and a total of 1,266 images are collected manually in two sessions.
Figure 3.
Cryo-EM sample preparation condition screening
(A) eNP-2 helical tube visualization at low magnifications (blue box), scale bars 500 nm and 100 nm.
(B) Representative cryo-EM micrograph that shows side views of long helical tubes and top views of short helical tubes or rings (yellow box), scale bar 100 nm.
(C) Thin ice disrupts a helical tube (red box), scale bar 100 nm.
(D) Thick ice allows additional flexibility of a helical tube (green box), scale bar 100 nm.
Helical reconstruction
Timing: 4 days
This step describes data processing and helical reconstruction of the cryo-EM micrographs for eNP-2 assembly. In general, cryo-EM data processing requires CPU and GPU computing resources (e.g., workstation, cluster). The timing given in this step is estimated based on a workstation with 40 CPU cores and 8 GPUs. Using more computing resource decreases the amount of time in this step.
Note: The timing specified here is typical for helical reconstruction with known helical parameters. Additional days to weeks may be needed if the helical parameters need to be determined from scratch.
Pause Point: This process can be paused at any time.
-
22.
Import the raw movie frames into Relion 2.1 (He and Scheres, 2017; Scheres, 2012).
Note: This protocol is carried out in Relion 2.1, in which the helical reconstruction suite is first introduced. Minor changes have since been made in later versions such as Relion 3 and Relion 4.
-
23.
Micrographs are motion corrected using Motioncorr v2.1 (Li et al., 2013) with default parameters except that “Number of patches X, Y” is set to “5, 5” and “Iter” is set to 10 (15 min).
-
24.
Contrast transfer function (CTF) is estimated using CTFFIND4 (Rohou and Grigorieff, 2015) with the corresponding microscope parameters and other default parameters except that “Amount of astigmatism” is set to 1000 (8 min), 1,113 micrographs with visible thon rings at better than 6 Å resolution are selected (2 min).
-
25.
Helical tubes in the selected micrographs are manually selected using e2helixboxer.py in EMAN2.12 (Tang et al., 2007) with “helix-width” set to 400. The output txt files that contain start and end point coordinates of each helical tube are imported in Relion 2.1 (2–3 h).
-
26.
A total of 200,691 segments is extracted in Relion 2.1 with “Particle box size” set to 528, “Re-scaled size” set to 132, “Tube diameter” set to 500, “Number of asymmetrical units (ASUs)” set to 5, “Helical rise” set to 2.65, and other parameters remain default. This corresponds to 1,003,455 ASUs (10 min).
-
27.
Two-dimensional (2D) class averaging is performed with “Number of classes” set to 200, “mask diameter” set to 600, “Limit resolution E-step” set to 10, “Tube diameter” set to 500, and other parameters remain default (12 h). Good 2D class averages and corresponding particles are selected (3 min).
Note: Here we also included some top-viewed ring-like particles in 2D class averages to provide additional information to guide helical parameter determination as described in step 29 (Figure 3B bottom right).
Note: The initial 2D class averages did not yield distinguished protein features (Figure 4A). We used e2proc2d.py in EMAN2 to clip 192 pixels along the tube axis in the middle of the particles. The orientation alignment of the clipped particles focus on the central region that result in 2D class averages with improved protein features (Figure 4B).
Figure 4.
2D class averages of eNP-2 helical tube segments
(A) Initial 2D class averages showed no high-resolution features.
(B) 2D class averages of masked particles showed improved high-resolution features.
-
28.
Prepare initial model in e2initialmodel.py in EMAN2 with default parameters using selected 2D class average images as input (10 min).
Note: Alternatively, an existing cryo-EM helical reconstruction map with similar diameter low-pass filtered to 60 Å, or simply a cylindrical density with the same diameter generated in EMAN2 and other software can be used as initial models.
-
29.Determine helical parameters (3 days)
-
a.Fourier transform of a few segments (normally < 100) can be coherently summed to generate a layer line image (Figure 5A) (5 min).
-
b.The layer line indexing result can be generated by selection rule and used as initial guess for helical parameter refinement. In this eNP-2 assembly case, we estimated there were approximately 40 ASUs per helical turn based on 2D averaging result of the assembly top views (Figure 3B, bottom right), which was used as input for the selection rule (Stewart, 1988) and resulted in -8.96° in twist, corresponding to 40.2 ASUs per helical turn (Figure 5B). (10 min).
-
c.A subset of 20,000 segments is used for helical parameter refinement in EMAN (Ludtke et al., 1999) integrated with iterative helical real space reconstruction (IHRSR) (Egelman, 2007). We performed three-dimensional (3D) auto-refinement using six different helical parameters as initial input, which correspond to 39.2, 40.2, 41.2, 42.2, 43.2 and 44.2 ASUs per turn. Helical reconstruction of 42.2 ASUs per helical turn reveals best high-resolution features in 2D projection along Z-axis, whereas 2D projections of 3D reconstructions using other helical parameters show spike-like defects on inner side of the tube (39.2 and 40.2 ASUs per turn), on outer side of the tube (43.2 and 44.2 ASUs per turn), or less high-resolution features (41.2 ASUs per turn) (Figure 5C). The final refined helical parameters are 2.65 Å in rise and -8.53° in twist (see also troubleshooting for alternates) (2–3 days).
-
a.
-
30.
Selected particles from 2D class averages are subjected to 3D auto-refinement in Relion 2.1 with “Mask diameter” set to 600, “Initial angular sampling” set to 1.8, “Local searches from auto-sampling” set to 0.9, “Tube diameter-inner, outer” set to 250, 500, “Number of asymmetrical units” set to 5, “Initial twist, rise” set to -8.53, 2.65, “Twist search-min, max, step” set to -8.74, -8.33, 0.004, “Rise search-min, max, step” set to 2.59, 2.71, 0.001 and other parameters remain default (3–4 days).
-
31.
The auto-refinement result is subjected to postprocess in Relion 2.1 using the default parameters except that the “inimask_threshold” is set to 0.008 (5 min).
Figure 5.
Determination of helical parameters
(A) Layer line indexing of eNP-2 helical tube, whereas l is the layer line number, n is order of the layer line, blue dashed lines form the reciprocal lattice.
(B) Using the selection rule l=tn+um with input of l and n will yield the results of the number of ASUs (u) in the number of helical turns (t).
(C) Refinement of helical parameters using six different ASU per turn numbers. The red arrow points to the spike-like defects on inner side of the tube. The yellow arrow points to the spike-like defects on outer side of the tube.
Segmentation and modeling
Timing: 1 days
-
32.
The reconstructed density of the helical tube was segmented using the Segger plugin in UCSF Chimera (Pettersen et al., 2004; Pintilie et al., 2010, 2016).
-
33.
Smoothing and grouping (5 steps, step size 1) was applied to get an approximate segmentation of each eNP-2 molecule (Figures 6A and 6B).
-
34.
The crystal structures of several eNP-2 (PDB: 4YPI) were then rigidly fitted by alignment to segments corresponding to individual eNP-2 (Figures 6C–6E) (Kirchdoerfer et al., 2015).
-
35.
Secondary structure elements in the fitted model matched those visible in the density, indicating correct fits (Figure 6F).
Figure 6.
Segmentation and modeling steps
(A) Cryo-EM reconstruction at 5.8 Å.
(B) Segger segmentation result.
(C) Extracted segmentation for one protein monomer in the asymmetric unit of two monomers.
(D) Existing monomeric protein model.
(E) Fitting of the model to the segments of one protein monomer.
(F) Fitting of the model to the cryo-EM density of one protein monomer.
(G) Extraction of additional segments based on the model.
(H) Fitting of the model to the newly extracted cryo-EM density of one protein monomer.
(I) Extraction of additional segments based on the model of the other protein monomer in the asymmetric unit.
(J) Fitting of the model to the cryo-EM density of the other protein monomer in the asymmetric unit.
Flexible fitting and modeling
Timing: 1 days
-
36.
First, the rigidly fitted crystal structure was flexibly fitted to the corresponding extracted density of each eNP-2 in one ASUs using MDFF (Trabuco et al., 2009); this moves helices a21 and a22 and the b-hairpin motif into the density. MDFF was run 10 times starting with the crystal model, which produces 10 slightly different results (Figures 6G and 6H).
-
37.
The resulting 10 structures were then put into ProMod to create an average model and to calculate uncertainty in residue positions (Peitsch, 1996).
-
38.
For one of the eNP-2, the loop and helix a23 were then added based on the segments and density nearby (Figure 6I). The entire sequence of the protein was obtained by BLAST with the sequence of the modeled residues. This entire sequence contained unmodeled residues at the C terminal end. The sequence was input into secondary structure prediction method PSIPRED (McGuffin et al., 2000). A helix was predicted in the unmodeled region, consisting of 19 residues, separated from the modeled helix a22 by a loop of 8 residues. The residues predicted to form a helix were modeled into the tubular density using Build Structure in Chimera.
-
39.
The loop connecting a22 and the new a23 was then also modeled within Chimera.
-
40.
The resulting model was flexibly fitted 10 times to the density using MDFF; Promod was then applied to the 10 resulting models to estimate the average model and uncertainties at each residue position (Figure 6J).
Expected outcomes
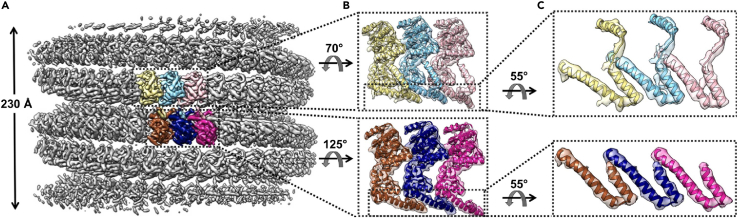
Following this protocol will allow sample preparation, cryo-EM helical reconstruction at moderate to high resolution, and model building for Ebola nucleocapsid-like helical assembly, and likely for other viral protein helical assemblies. We used this protocol to determine the cryo-EM structure of the Ebola nucleocapsid-like assembly at 5.8 Å resolution (Figure 7A), and subsequently build model into the cryo-EM density (Figure 7B) to reveal the assembly interaction interfaces (Figure 7C).
Figure 7.
Cryo-EM reconstruction of the eNP-2 assembly reveals the interaction interface
(A) Cryo-EM structure of the eNP-2 assembly at 5.8 Å resolution.
(B) Zoom in views of three adjacent ASUs from each strand.
(C) Zoom in views of the assembly interaction interfaces.
Quantification and statistical analysis
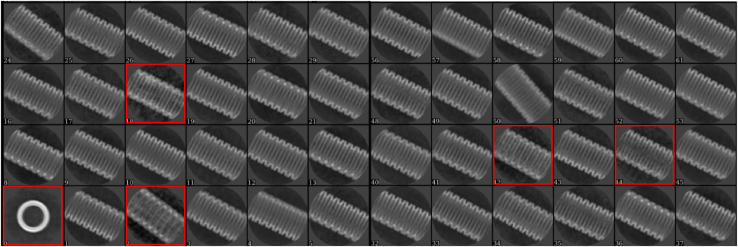
In this protocol during the cryo-EM data processing helical reconstruction (step 27), 169,526 segments belonging to the good 2D classes with clear protein features on the outside of the tube and relative clean background were selected, whereas 31,165 segments belonging to the bad 2D classes with blurred protein features and more noisy background were rejected (Figure 8).
Figure 8.
The result of 2D class average allows rejection of bad particles (red boxes)
Limitations
The limitations of this protocol are as follow:
Manual acquisition of the cryo-EM data requires significant amount of time and effort. This is due to the scarcity of high-quality (relatively straight and long) eNP-2 helical assembly on the grid. In case of samples with abundant high-quality filaments, automated data collection may be used to either increase data size that may lead to improved resolution, or achieve the same resolution with decreased amount of time.
The reported 5.8 Å resolution of eNP-2 assembly makes de novo modeling extremely challenging. Model building and flexible fitting in this protocol requires that the ASU model is already known. In other cases when ASU model is not known, cryo-EM structures at higher resolution that allow de novo modeling will be needed. Alternatively, the rapidly emerging predictive methods of protein structures assisted by deep learning algorithms may aid the generation of preliminary models for further refinement (Jumper et al., 2021; Baek et al., 2021).
Troubleshooting
Problem 1
The protein cannot be folded correctly and forms an inclusion body (related to protein induction and expression; steps 7).
Potential solution
The main cause of inclusion body formation is the fast expression of protein. In the process of protein purification, some methods are generally adopted to avoid inclusion body formation. For example, decrease the induction temperature to 16°C–25°C; decrease the IPTG concentration to 0.01–0.1 mM and extend the induction time; switch to other promoter sequences to slow down the rate of expression. In addition, fusion protein such as GST and MBP can be introduced for coexpression to increase solubility.
Problem 2
Lack of filamentous assembly in negative staining (related to negative stain; steps 9–13).
Potential solution
Formation of filamentous viral protein assembly are often affected by purification conditions. High salt conditions (2–3 M NaCl) can remove nucleic acids that nonspecifically bind to proteins and enhance salt bridge contacts that may help to rigidify the protein assembly (Su et al., 2018). Whereas low salt conditions (100–200 mM NaCl) help to maintain protein-nucleic acid complexes that may yield different assembly conformations (Sugita et al., 2018) Purification conditions can be optimized to obtain more rigid filaments.
Problem 3
Scarcity of high-quality filaments under cryo-EM condition (related to cryo-EM data acquisition; steps 18–21).
Potential solution
This is likely caused by low amounts of filaments and improper ice thickness, which may be solved by changing the grid surface properties in addition to blotting parameters. Firstly, other types of grids such as UltrAufoil with Au film instead of Quantifoil with carbon film could be used. Secondly, additional supporting films such as continuous carbon film or graphene (oxide) could be used to both increase filament absorption onto the grid and better control of ice thickness. Lastly, recently versions of data collection software such as EPU2 have integrated ice quality classification at low magnification, which may assist finding proper ice thickness with more high-quality filaments.
Problem 4
Correct helical parameters could not be determined after layer line indexing and refinement by 3D reconstructions (related to helical reconstruction; step 29).
Potential solution
While the searching range could always be expanded in order to find the correct helical parameters, this is always very time consuming. An alternative way to obtain helical parameters is direct single particle reconstructions without applying helical parameters using packages such as Relion, CryoSPARC, EMAN2, etc., and find out the helical parameters if the final reconstruction was at high enough resolution (normally at 10 Å or better). Though, it must be mentioned that this method requires high-quality particles of various views other than side views, such as top and tilted views.
Problem 5
Flexible fitting and modeling. When rigid-fitting the model using SegFit, the correct fit may not be found initially. This can be idenfitied if secondary structures in the fitted model do not match those in the map. When flexible fitting with MDFF, it is also possible that the helices which are not matching the density do not move into the corresponding density (related to flexible fitting and modeling; step 36).
Potential solution
To find the correct fit when doing rigid fitting with SegFit, the rotational search can be used (instead of the principal axes approach which is faster) to improve the chances that the correct fit is found; furthermore the number of rotations tested can be increased in the SegFit dialog, e.g., from 100 to 200. If the helices do not move into density when applying MDFF, a larger gradient force can be applied, e.g., increasing it from the default 0.3–0.6.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhaoming Su (zsu@scu.edu.cn).
Materials availability
The unique reagents generated in this study are available from the lead contacts with a completed Materials Transfer Agreement upon request.
Acknowledgments
This work was supported by Sichuan University start-up funding 20822041D4057 and Natural Science Foundation of China (NSFC) 82041016 and 32070049 to Z.S. and by National Institutes of Health (P41GM103832, R01GM079429, P01AI120943, and S10OD021600 to W.C.; P01AI120943 to G.K.A. and D.W.L., R01AI123926 to G.K.A., and R01AI107056 to D.W.L.).
Author contributions
Z.S., J.B., G.D.P., D.W.L., W.C., and G.K.A. developed the protocol, all authors contributed to writing the protocol.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Gaya K. Amarasinghe, Email: gamarasinghe@wustl.edu.
Daisy W. Leung, Email: dwleung@wustl.edu.
Zhaoming Su, Email: zsu@scu.edu.cn.
Data and code availability
The cryo-EM map and associated atomic coordinate model have been deposited in the wwPDB OneDep System under EMD accession code EMD-7343 and PDB ID code 6C54.
References
- Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., Wang J., Cong Q., Kinch L.N., Schaeffer R.D., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman E.H. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- He S., Scheres S.H.W. Helical reconstruction in RELION. J. Struct. Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer Robert N., Abelson Dafna M., Li S., Wood Malcolm R., Saphire Erica O. Assembly of the ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep. 2015;12:140–149. doi: 10.1016/j.celrep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Borek D., Luthra P., Binning J.M., Anantpadma M., Liu G., Harvey I.B., Su Z., Endlich-Frazier A., Pan J., et al. An intrinsically disordered peptide from ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein-RNA interactions. Cell Rep. 2015;11:376–389. doi: 10.1016/j.celrep.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Meyerson J.R., White T.A., Bliss D., Moran A., Bartesaghi A., Borgnia M.J., de la Cruz M.J., Schauder D., Hartnell L.M., Nandwani R., et al. Determination of molecular structures of HIV envelope glycoproteins using cryo-electron tomography and automated sub-tomogram averaging. JoVE (Journal of Visualized Experiments) 2011;58:e2770. doi: 10.3791/2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch M.C. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie G., Chen D.H., Haase-Pettingell C.A., King J.A., Chiu W. Resolution and probabilistic models of components in CryoEM maps of mature P22 bacteriophage. Biophys. J. 2016;110:827–839. doi: 10.1016/j.bpj.2015.11.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie G.D., Zhang J., Goddard T.D., Chiu W., Gossard D.C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Computer image processing of electron micrographs of biological structures with helical symmetry. J. Electron Microsc. Tech. 1988;9:325–358. doi: 10.1002/jemt.1060090404. [DOI] [PubMed] [Google Scholar]
- Su Z., Wu C., Shi L., Luthra P., Pintilie G.D., Johnson B., Porter J.R., Ge P., Chen M., Liu G., et al. Electron cryo-microscopy structure of ebola virus nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell. 2018;172:966–978.e912. doi: 10.1016/j.cell.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y., Matsunami H., Kawaoka Y., Noda T., Wolf M. Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution. Nature. 2018;563:137–140. doi: 10.1038/s41586-018-0630-0. [DOI] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Trabuco L.G., Villa E., Schreiner E., Harrison C.B., Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Freitas M.A. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The cryo-EM map and associated atomic coordinate model have been deposited in the wwPDB OneDep System under EMD accession code EMD-7343 and PDB ID code 6C54.