Abstract
The human β-globin genes are regulated by the locus control region (LCR), an element composed of multiple DNase I-hypersensitive sites (HS sites) located 5′ to the genes. Various functional studies indicate that the LCR confers high-level, position-independent, and copy number-dependent expression to linked globin genes in transgenic mice. However, the structural basis for LCR function is unknown. Here we show that LCR HS sites can be reconstituted in an erythroid cell-specific manner on chromatin-assembled LCR templates in vitro. Surprisingly, HS2 and HS3 are also formed with erythroid proteins in the absence of chromatin assembly, indicating that sensitivity to nucleases is not simply a consequence of nucleosome reorganization. The generation of LCR HS sites in the absence of chromatin assembly leads to the formation of S1- and KMnO4-sensitive regions in HS2 and HS3. These sites are also sensitive to S1 nuclease in erythroid cells in vivo, suggesting a distorted DNA structure in the LCR core enhancer elements. Finally, we show that RNA polymerase II initiates transcription in the HS2 and HS3 core enhancer regions in vitro. Transcription in both HS2 and HS3 proceeds in a unidirectional manner. Taken together, the data suggest that erythroid proteins interact with the core enhancer elements, distort the DNA structure, and recruit polymerase II transcription complexes. These results further our understanding of the structural basis for LCR function and provide an explanation for why the LCR core regions are so extremely sensitive to nucleases in erythroid cells.
The human β-globin locus contains five genes (ɛ, Gγ, Aγ, δ, and β), which are organized in sequential order on chromosome 11 (39). The gene order reflects the timing of expression during erythroid development, with the embryonic ɛ gene located at the 5′ end and the adult β-globin gene at the 3′ end of the locus. Developmental stage-specific expression is controlled mainly at the transcriptional level by a variety of gene-proximal or -distal cis elements. The most prominent distal regulatory element is the β-globin locus control region (LCR), located from 8 to 22 kb upstream of the ɛ-globin gene and composed of several subregions that exhibit heightened sensitivity to digestion with exogenous DNase I (HS sites) in erythroid cells (14, 18, 41). Originally, five HS sites (HS1 to HS5) were associated with LCR function, but more recently additional sites were mapped even further 5′ to the LCR (6). Many studies indicate that the human β-globin LCR is able to confer position-independent expression to linked globin genes in transgenic mice (18, 19). It is generally believed that this activity is based on the ability of the LCR to provide an open accessible chromatin structure regardless of where the transgenic locus integrates in the mouse genome. Recent results suggest that the chromatin-opening function of the LCR may not be the primary activity in the endogenous mouse or human globin locus, because the LCR can be deleted without affecting general sensitivity to DNase I (5, 36). Regardless of mechanism, virtually all studies agree that the LCR is required for conferring high-level globin gene expression throughout erythroid development.
We have previously analyzed the role of individual LCR HS core elements in the context of the whole β-globin gene locus in β-globin yeast artificial chromosome (YAC) transgenic mice (9, 10). The results of these experiments showed that in intact, single-copy transgenes, deletion of individual core HS elements dramatically reduced expression of all of the globin genes at all developmental stages. We also found that these mutations impaired the formation of DNase I hypersensitivity in the LCR and in the adult β-globin gene promoter (28).
These results support a model in which the HS sites interact to generate a higher-order structure, referred to as the LCR holocomplex (9, 10, 43). Although this model is consistent with the data from many studies addressing the function of wild-type and mutant LCRs, there is currently no direct physical evidence for the formation of an LCR holocomplex. However, recent work by Lee et al. (27) suggests that the erythroid cell-specific transcription factor EKLF (erythroid Krüppel-like factor) binds cooperatively to HS2 and HS3. In addition, Yoshida et al. (45) recently showed that the protein Bach1, which heterodimerizes with small Maf proteins and interacts with MARE (Maf-responsive element) sequences present in HS2, HS3, and HS4, is able to cross-link HS sites, thereby looping out intervening DNA sequences. These results, together with various in vitro studies, suggest that a network of protein-protein and protein-DNA interactions could mediate the formation of a larger LCR complex. Notable among these in vitro studies are observations that GATA factors are able to engage in a variety of protein-protein interactions involving EKLF, Sp1, LMO2, and Tal1 (30, 42). All of these proteins (with the exception of LMO2, which serves as a bridging molecule between GATA factors and Tal1) were shown to interact with LCR core HS sites.
The individual LCR HS core elements are 200 to 400 bp in size and harbor clusters of binding sites for erythroid cell-specific and ubiquitously expressed transcription factors (20). All of the core HS sites (HS1 to HS5) contain binding sequences for GATA factors and GC-rich elements, which can interact with Sp1 or related proteins. HS2, HS3, and HS4 also harbor MARE sequences that interact with transcription factors of the NF-E2 family (NF-E2, Nrf1, Nrf2, and Bach1). HS2 and HS3 contain potential binding sites for EKLF. E-box motifs present in HS2 were shown to be critical for HS2 function and to interact with helix-loop-helix proteins (heterodimers containing either USF or Tal1 [11]).
Several studies addressed the importance of particular transcription factors and their binding sites for the generation of DNase I hypersensitivity in individual LCR HS sites. It was shown that combinations of MARE and GATA sequences are critical components for the formation of HS2, HS3, and HS4 (35, 40). Furthermore, NF-E2 is critically involved in remodeling the nucleosome structure over the HS2 core region, where it interacts with tandemly arranged MARE sequences (1, 16). Finally, EKLF has been implicated in the formation of HS3, since this site is not formed efficiently in EKLF-deficient human β-globin locus transgenic mice (44).
To analyze the mechanism of HS site formation in the human β-globin LCR, we have developed an in vitro system to reconstitute the HS sites on chromatin-assembled templates containing the entire LCR. For these experiments, we subcloned the LCR from a human β-globin YAC by homologous recombination (gap repair) in yeast. Biotinylated LCR templates were immobilized on streptavidin-coated beads and incubated with chromatin assembly extracts in the presence of erythroid proteins. These data show that LCR HS sites are generated on chromatin in an erythroid cell-specific manner and are restricted to the core HS regions. Surprisingly, HS2 and HS3 are also formed on the LCR templates in the absence of chromatin assembly, indicating that formation of these sites does not simply reflect a change in chromatin structure. The in vitro-reconstituted HS sites are also sensitive to S1 nuclease and reveal extended regions of sensitivity to KMnO4. Finally, we show that RNA polymerase II-specific transcripts initiate within the cores of HS2 and HS3 and proceed in a directional manner.
MATERIALS AND METHODS
Subcloning of the human β-globin LCR and mutant derivatives.
The human β-globin LCR was subcloned into pRS316, a yeast episomal shuttle vector, by homologous recombination in yeast harboring the whole β-globin locus on a YAC. For generating the target construct, we isolated restriction fragments corresponding to the 5′ flanking region of HS5 (EcoRI/SacI) and to the 3′ flanking region of HS1 (SacI/XbaI) from a lambda phage library (9). These fragments were ligated into the EcoRI/XbaI site of pRS316 to generate pRSHS5/HS1. The plasmid was linearized with SacI and used to transform yeast cells harboring a 155-kb human β-globin YAC (15). After transformation, the yeast cells were plated onto selective agar plates lacking uracil (Ura− plates). Subsequently, transformants were restreaked on Ura− plates. DNA isolated from these clones was digested with PvuII, separated by electrophoresis, and transfered to nylon membranes. Hybridization was then carried out with a radiolabeled restriction fragment from pRS316 (220-bp EcoRI/PvuII) to identify clones in which the LCR was linked to the episomal vector (resulting in pRS/LCR). DNA from positive clones was isolated and used to transform electrocompetent Escherichia coli (DH10B). For subcloning a mutant LCR lacking the core enhancer of HS2, the SacI-linearized target vector pRSHS5/HS1 was used to transform yeast cells carrying a mutant β-globin YAC in which the 375-bp core enhancer of HS2 was deleted by homologous recombination (10). Selection of positive transformants and shuttling of plasmids into E. coli was carried out as described for the generation of pRS/LCR. We refer to the LCR deletion mutant as pRS/LCRΔ2. We also generated a plasmid in which the LCR was linked in cis to the adult human β-globin gene. For generating the target construct, we ligated a HincII/XbaI fragment containing the β-globin gene into the HincII/XbaI site of pRSHS5/HS1, placing the β-globin gene 3′ to the HS1 flanking region. This vector, pRSHS5/HS1-β, was used to transform yeast cells carrying wild-type human β-globin YACs. Yeast clones in which the LCR had integrated into the plasmid (referred to as pRS/LCR-β) were selected as described above, and DNA isolated from these clones was used to transform E. coli cells.
Immobilization of LCR constructs.
The LCR containing constructs (pRS/LCR, pRS/LCRΔ2, and pRS/LCR-β) were linearized with ClaI and XhoI, and the 5′ overhanging ends were filled in with biotinylated dATP, dGTP, biotinylated dCTP, biotinylated dUTP, and Klenow polymerase. The biotinylated templates were then attached to streptavidin-coated magnetic beads using a kilobase binder kit (Dynal) as recommended by the manufacturer. To monitor the efficiency of coupling, an aliquot of DNA attached to beads was digested with EcoRI and analyzed on 1% agarose gel. The DNA concentration was determined by fluorometry using a Versafluor fluorometer (Bio-Rad).
Chromatin assembly.
The preparation of Drosophila chromatin assembly extracts and the assembly of LCR constructs into chromatin was carried out essentially as described by Sandaltzopoulos et al. (38). Briefly, 600 ng of immobilized template DNA was incubated with 300 μg of Drosophila chromatin assembly extract in a reaction mixture (total volume of 70 μl) containing 10 mM HEPES-KOH, (pH 7.6), 50 mM KCl, 3 mM MgCl2 (pH 8.0), 0.5 mM EGTA (pH 8.0), 10 mM glycerolphosphate, 30 mM creatine phosphate, 30 mM ATP, 1 μg of creatine phosphokinase/ml, 10% glycerol, 1 mM dithiothreitol (DTT), and 0.2 mM phenylmethyl sulfonyl fluoride (PMSF) for 6 h at 26°C. After the chromatin assembly reaction, all unbound material was removed using a magnet (Dynal), and the beads were resuspended in buffer A (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA [pH 8.0], 10% glycerol, 10 mM glycerolphosphate, 1 mM DTT, 0.2 mM PMSF).
Preparation of protein extracts.
Extracts from two different cell types were used in the in vitro experiments: MEL (mouse erythroleukemia) cells and HeLa cells (derived from human cervical carcinoma). Protein extracts used for transcription reactions were prepared as described by Bungert et al. (8). The range of protein concentrations was between 9 and 11 mg/ml. To prepare protein extracts used for nuclease HS site and KMnO4 mapping experiments, 108 cells were pelleted by centrifugation, washed in phosphate-buffered saline (13.7 mM NaCl, 0.27 mM KCl, 0.43 mM Na2HPO4 · 7H2O, 0.14 mM KH2PO4), and resuspended in 1 ml of lysis buffer (20 mM HEPES [pH 7.6], 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF). After incubation for 3 min on ice and centrifugation at 2,000 rpm (Eppendorf Microcentrifuge) for 10 min at 4°C, the pellets were resuspended by rocking in 1 ml of nuclear extraction buffer (20 mM HEPES [pH 7.6], 20% glycerol, 400 mM NaCl, 1.5 ml MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF) for 1 h at 4°C. After centrifugation at 10,000 rpm for 10 min at 4°C, the supernatant was dialyzed for 16 h in 2 liters of dialysis buffer (20 mM HEPES [pH 7.8], 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF). The final protein concentration of different extracts was between 4 and 6 mg/ml.
Nuclease HS site mapping.
Immobilized LCR constructs (600 ng) were incubated with 100 μg of MEL or HeLa extracts for 45 min at 30°C in buffer A (total volume, 70 μl). For analyzing DNase I sensitivity on chromatin, the templates were either preincubated (prior to chromatin assembly) or postincubated with MEL or HeLa extracts for 45 min at 30°C. All unbound material was removed by incubating the reactions on a magnet for 1 min at room temperature (RT), and the beads were resuspended in 160 μl of buffer A. The samples were divided into 30-μl aliquots and digested for 1 min at RT with DNase I. For this reaction, a 30-μl mix containing increasing concentrations of DNase I (Sigma) (from 0.001 to 0.02 U for naked DNA and from 0.01 to 0.2 U for chromatin-assembled DNA) in buffer A (plus 5 mM CaCl2 and 10 mM MgCl2) was added to the samples. After addition of 20 μl of stop solution (2.5% sarcosyl, 100 mM EDTA), the samples were incubated with RNase I (1 μg/reaction) for 30 min at 37°C and subsequently overnight with 5 μl of proteinase K (50 μg/reaction) and 8 μl of 2% sodium dodecyl sulfate (SDS). After sequential phenol-chloroform and chloroform extractions, the DNA was precipitated, resuspended in 1× restriction buffer, and digested for 6 h to overnight with EcoRI. The DNA was precipitated, resuspended in Tris-EDTA (pH 7.6), loaded onto 1.3% agarose gels, and separated by electrophoresis for 6 h. After blotting to nylon membranes, the DNA was hybridized to a 32P-labeled 360-bp restriction fragment corresponding to the 3′ flanking region of HS2 (a NotI/XbaI restriction fragment from pRS306 containing the HS2 flanking regions embedding the core enhancer of HS3 [10]). The S1 sensitivity mapping experiments were carried out in the same way as described for the DNase I HS site mapping experiments except that instead of being digested with increasing concentrations of DNase I, the DNA was incubated for 2 min with S1 (Promega) (from 0.01 to 1 U in S1 nuclease reaction buffer). The samples were then processed and analyzed as described for the DNase I mapping experiment.
For micrococcal nuclease (MNase) mapping of nucleosomes, the LCR constructs were incubated with chromatin assembly extracts and, after removal of unbound material through use of a magnet, resuspended in 70 μl of buffer A. After addition of 100 μl of MNase mix (buffer A plus 5 mM CaCl and 50 U of MNase [Sigma]), 30-μl aliquots were removed (at 15, 60, and 180 s) and added to 20 μl of stop solution (2.5% sarcosyl, 100 mM EDTA [pH 8.0]). After incubation with RNase I for 30 min at 37°C, 5 μl of proteinase K (10 mg/ml) and 8 μl of SDS (2%) were added to the samples, and incubation was continued overnight at 37°C. For analyzing the efficiency of chromatin assembly, the DNA was precipitated, resuspended, and separated by electrophoresis in 1.3% agarose gels. For mapping the position of nucleosomes, the samples were prepared essentially as described for the DNase I mapping experiments except that the DNA was digested with ApaI and BglII.
The procedure for mapping DNase I and S1 sensitivity in vivo has been described previously (28). Briefly, nuclei were isolated from exponentially growing K562 (human erythroleukemia) cells, and aliquots (106 nuclei) were treated with increasing concentrations of either DNase I (0, 0.5, 2, and 10 U/ml) or S1 nuclease (0, 0.25, 1, 5, 10, and 30 U/ml). The DNA was isolated, digested with EcoRI, electrophoresed, and blotted to a nylon membrane. The radioactive probe used in Southern blotting experiments was the same as the one used in the DNase I and S1 nuclease in vitro analysis.
Mapping of single-stranded regions with KMnO4.
The template DNA (600 ng of immobilized LCR constructs) was incubated with HeLa or MEL protein extracts (100 μg) for 45 min at 30°C in a total volume of 70 μl (in buffer A). After adding KMnO4 (final concentration, 4 mM), the samples were incubated 5 min at RT. The reaction was stopped by the addition of 200 μl of stop buffer (consisting of 6% β-mercaptoethanol, 2% SDS, and 100 mM EDTA). The samples were then incubated with 1 μg of RNase for 30 min at 37°C. After the addition of 20 μl of proteinase K (10-mg/ml stock solution) and 3 μl of SDS (20%) the samples were incubated overnight at 37°C. After sequential phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol extractions the DNA was precipitated and resuspended in 4 μl of H2O. After addition of 1 μl of 32P-labeled primers (HS2US, 5′ GCATCCTCATCTCTGATTAAATAAGC 3′; HS2DS, 5′ GTCACATTCTGTCTCAGGCATCCAT 3′; HS3 US, 5′ TGGTGTGCCAGATGTGTCTA 3′; HS3DS, 5′ GCTGCTATGCTGTGCCTCCC 3′) and 45 μl of PCR mix (5 μl of 10× Taq polymerase buffer, 1 μl of 100 mM deoxynucleoside triphosphate, 1 μl of 1M MgCl2, 0.25 μl of Taq polymerase, and 37.75 μl of H2O), DNA was amplified by primer extension-PCR (eight cycles of 1 min at 92°C, 1 min at 60°C, and 1 min at 72°C) and then analyzed by electrophoresis in 8% sequencing gels and autoradiography.
In vitro transcription assayed by primer extension.
A typical transcription reaction mixture contained 300 ng of immobilized template (pRS/LCR or pRS/LCR-β), 4 μl of transcription buffer (Promega), 3 mM MgCl2, 40 μM ribonucleoside triphosphates, 0.5 μl of RNasin (40 U/μl), and 70 μg of protein extract (55 μg of MEL protein extract/15 μg of HeLa protein extract) in a final volume of 25 μl and was incubated for 60 min at 30°C. After the addition of 175 μl of stop buffer (Promega) and phenol-chloroform-isoamyl alcohol extraction, nucleic acids were precipitated with 500 μl of ethanol for 15 min on dry ice and pelleted by centrifugation. The pellets were resuspended in 5 μl of primer extension buffer (Promega) and 5 μl of nuclease-free water. After addition of 1 μl of 32P-labeled primer, the samples were denatured (10 min at 70°C and 5 min on ice) and incubated for 20 min at 55°C and 10 min at RT. After addition of 9 μl of buffer containing 5 μl of primer extension buffer, 2.8 mM sodium pyrophosphate, 12 ng of actinomycin D, 0.5 μl of RNasin, and 1 μl of avian myeloblastosis virus reverse transcriptase (1 U/μl; Promega), the reaction mixtures were incubated for 60 min at 42°C. The primers used in the primer extension assay were the same as those described for the KMnO4 mapping experiments with the exception of the HS3 downstream primer (HS3 DS∗), which has the sequence 5′ CATGTCTGCCCTCTACTCATGG 3′. The primer for analyzing transcription of the β-globin gene has the sequence 5′ CGGCAGACTTCTCCTCAGGAGTCAGGTG 3′. After precipitation, the samples were resuspended in 5 μl of loading dye (Promega), loaded on 8% sequencing gels, separated by electrophoresis, and subjected to autoradiography.
RESULTS
Subcloning and immobilization of the human β-globin LCR.
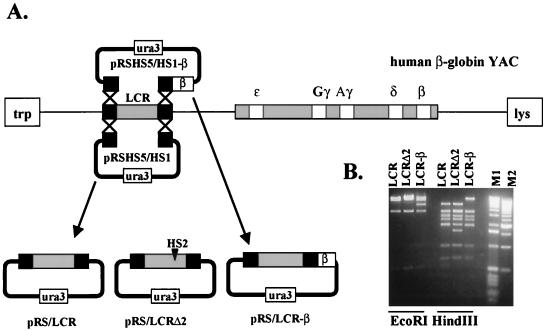
The LCR was subcloned from yeast cells carrying human β-globin YACs by homologous recombination (Fig. 1; Materials and Methods). A yeast episomal plasmid was generated by ligating the 5′ flanking region of HS5 and the 3′ flanking region of HS1 into pRS316. The plasmid was linearized with SacI and used to transform yeast cells carrying the human β-globin YAC (A201 F4 [15]). Clones able to grow on Ura− plates were restreaked, and DNA was isolated from single colonies. The DNA was digested with PvuII, subjected to electrophoresis, blotted to nylon membrane, and hybridized to a 220-bp EcoRI/PvuII restriction fragment from pRS316. Plasmids that incorporated sequences from the human β-globin LCR were transformed into E. coli. DNA from transformed cells was analyzed by restriction digestion with EcoRI and HindIII. Using this strategy, we were able to generate pRS constructs containing the wild-type LCR, a mutant LCR lacking the core enhancer of HS2, and the wild-type LCR linked to the adult human β-globin gene (pRS/LCR, pRS/LCRΔ2, and pRS/LCR-β, respectively [Fig. 1B]).
FIG. 1.
Subcloning of wild-type and mutant human β-globin LCRs by homologous recombination and gap repair in yeast carrying β-globin YACs. (A) Strategy for subcloning the human β-globin LCR into the yeast episomal vector pRS316. HS5 5′ and HS1 3′ fragments were ligated into pRS316 to generate pRSHS5/HS1. This plasmid was linearized with SacI and used to transform yeast cells carrying a wild-type human β-globin YAC. Plasmids that have adopted the whole LCR, as determined by Southern blotting experiments, were used to transform E. coli cells. For subcloning the LCR and a linked β-globin gene, the β-globin gene was ligated 3′ to the HS1 3′ flanking region, creating pRSHS5/HS1-β. This plasmid was then used to transform yeast cells carrying wild-type human β-globin YACs. The LCR mutant lacking the 375-bp HS2 core enhancer (pRS/LCRΔ2) was subcloned by transforming yeast carrying a β-globin YAC with a deletion of HS2 (10) with pRSHS5/HS1. (B) Restriction enzyme analysis of pRS subclones containing the wild-type or HS2-deficient LCR or the LCR with a linked β-globin gene. DNA was isolated from E. coli carrying the various pRS subclones (indicated on top) and digested with EcoRI or HindIII as indicated. The expected fragment sizes for pRS/LCR digested with EcoRI are 10.4, 3.3, and 0.5 kb, along with a 9-kb fragment containing part of the LCR and the vector. pRS/LCR-β harbors an additional 5-kb fragment containing the β-globin gene (lane 3). The restriction fragments generated by digesting pRS/LCR with HindIII are 3.3 kb (two fragments), 2.7 kb, 2.3 kb, 1.9 kb (two fragments), and 1 kb, along with a 5.1-kb fragment, containing also the rest of the vector. pRS/LCRΔ2 lacks one of the two 1.9-kb HindIII fragments and contains instead a novel 1.5-kb fragment lacking the HS2 core. M1, 1-kbp ladder, M2, 500-bp ladder.
For immobilization, plasmids pRS/LCR, pRS/LCRΔ2, and pRS/LCR-β were linearized, biotinylated, and bound to streptavidin-coated magnetic beads overnight at RT (Materials and Methods).
Erythroid cell-specific reconstitution of DNase I HS sites in vitro.
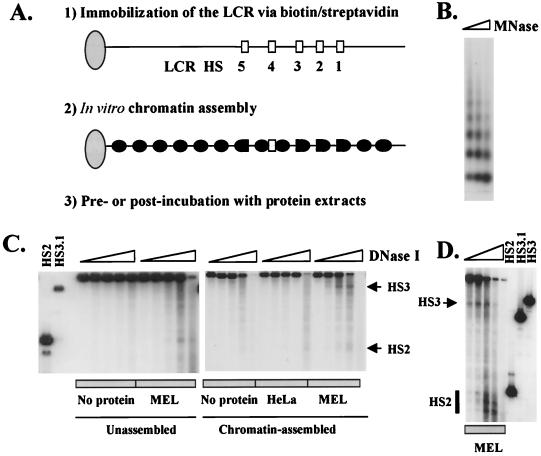
To test whether formation of HS sites could be reconstituted on chromatin-assembled LCR templates in vitro, pRS/LCR was assembled into chromatin using an extract from Drosophila embryos enriched in histones and nucleosome assembly factors (Fig. 2) (38). Chromatin assembly was monitored by MNase, which digests nucleosomal DNA preferentially in the linker region, releasing mono- or oligonucleosomal fragments. As shown in Fig. 2B, the 22-kb LCR template is efficiently assembled into chromatin, revealing a typical nucleosome ladder with a repeat length of about 200 bp. For reconstitution of DNase I HS sites, the LCR was first assembled into chromatin and then postincubated with MEL or HeLa cell extracts, after which all unbound proteins were removed by incubating the beads on a magnet (Fig. 2C). Chromatin assembly was performed as described above. After DNase I followed by EcoRI digestion, which releases a 10.4-kb fragment containing HS2, HS3, and HS4, the DNA was blotted and hybridized to a 32P-labeled probe corresponding to the 3′ flanking region of HS2.
FIG. 2.
Reconstitution of DNase I HS sites in chromatin-assembled LCR templates. (A) Outline of the general strategy for immobilizing the LCR on magnetic beads (step 1), the chromatin assembly reaction (step 2), and the incubation with erythroid and non-erythroid cell extracts (step 3). (B) MNase digestion pattern of chromatin-assembled pRS/LCR. pRS/LCR was attached to the beads and incubated with Drosophila chromatin assembly extract (Materials and Methods). The beads were washed on a magnet and digested with MNase for 15, 30, or 60 s, after which the DNA was isolated and separated in a 1.3% agarose gel. The DNA was then blotted to a nylon membrane and hybridized to a 32P-labeled fragment corresponding to the 3′ region of HS2. (C) DNase I HS site mapping of unassembled and chromatin-assembled pRS/LCR. For analyzing DNase I sensitivity in the unassembled construct, pRS/LCR was incubated in buffer A (No protein) or in buffer A with 100 μg of MEL protein extract (MEL) for 45 min at 30°C and then digested with increasing concentrations of DNase I. For analyzing DNase I sensitivity in chromatin-assembled templates, pRS/LCR was first assembled into chromatin as described for panel B and then incubated in buffer A containing no protein, 100 μg of HeLa protein extract (HeLa), or 100 μg of MEL protein extract (MEL) for 45 min at 30°C. The isolated DNA was then digested with EcoRI, processed, and analyzed as described for panel B. (D) DNase I hypersensitivity mapping on chromatin-assembled pRS/LCR preincubated with MEL protein extracts. pRS/LCR was incubated with 100 μg of protein extract from MEL cells (MEL) for 45 min at 30°C prior to chromatin assembly and then digested with increasing concentrations of DNase I. The samples were processed and analyzed as described for panel C. In lanes HS2, HS3, and HS3.1, restriction fragments marking the positions of the HS2 and HS3 core enhancers were included (HS2, EcoRI/HindIII, marking the 5′ end of HS2; HS3, EcoRI/SpeI, marking the 5′ end of HS3; HS3.1, EcoRI/ScaI, corresponding to the 3′ end of HS3).
Neither the naked DNA nor the chromatin-assembled LCR template revealed hypersensitivity to DNase I in the absence of a protein extract (Fig. 2C). After incubation of the chromatin-assembled template with HeLa extract, a weak HS site is detectable near HS2. In contrast, after incubation with MEL extracts, two regions of DNase I hypersensitivity, one localizing to HS3 and the other localizing to HS2, are detectable. It is noteworthy that the formation of HS sites is not very efficient when the LCR is incubated with MEL extracts after chromatin assembly. As shown in Fig. 2D, when the LCR is first incubated with MEL extracts and then assembled into chromatin, hypersensitivity to DNase I in HS2 and HS3 is significantly stronger, and there is also much less sensitivity between the core enhancer regions. The fact that there is little DNase I sensitivity between the core HS sites is consistent with in vivo observations demonstrating that hypersensitivity in the LCR is restricted to the core regions in erythroid cells (28). These results demonstrate that two important aspects of LCR HS site formation can be recapitulated on chromatin-assembled templates in vitro: (i) HS site formation is erythroid cell specific, and (ii) hypersensitivity is almost exclusively localized to the core enhancer regions. It is important to note that the quality of the HeLa extract used in these studies is comparable to the quality of the MEL cell extract. Both extracts exhibit comparable binding activities to an Sp1 binding site in electrophoretic mobility shift assays (data not shown).
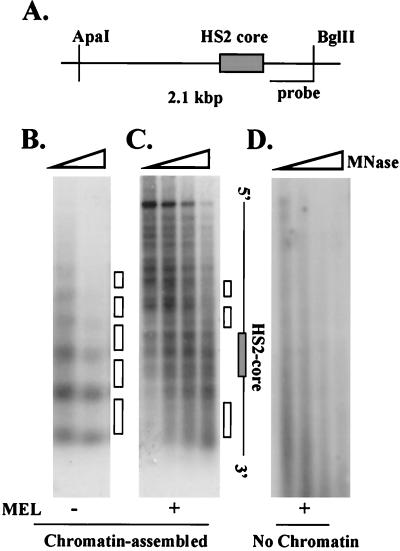
The positions of nucleosomes on chromatin-assembled LCR templates in the HS2 core region were next mapped by indirect end labeling. The immobilized LCR was first incubated with chromatin assembly extracts, and all unbound material was removed with a magnet. After MNase digestion, the DNA was digested with ApaI and BglII and analyzed by Southern blotting using a 32P-labeled probe derived from the 3′ end of the restriction fragment (Fig. 3A). As shown in Fig. 3B, chromatin assembly leads to the formation of a regular array of positioned nucleosomes over the HS2 core and flanking sequences. Preincubation of the LCR with MEL protein extracts disturbs this regular nucleosomal array (Fig. 3C). Incubation of the LCR with MEL extracts alone does not lead to the appearance of any nucleosomal structure on the template, indicating that under these conditions the LCR is not assembled into chromatin (Fig. 3D). These data indicate that erythroid proteins bind to the core enhancer fragment of HS2 and prevent the association of nucleosomes with this region. HS2 flanking regions continue to display a nucleosomal pattern if the LCR is preincubated with erythroid proteins and subsequently assembled into chromatin. Taken together, the data suggest that only the HS2 core enhancer is rendered nucleosome-free by prior incubation with erythroid proteins.
FIG. 3.
Erythroid proteins change the pattern of MNase digestion in the core region of HS2 on chromatin-assembled LCR constructs. (A) Representation of the HS2 region indicating the positions of restriction sites and the 32P-labeled probe used to map the positions of nucleosomes by indirect end labeling. (B) The LCR was attached to magnetic beads and assembled into chromatin. Aliquots were digested with MNase for 15, 45, or 90 s. The DNA was isolated, digested with ApaI and BglII, subjected to electrophoresis, blotted to a nylon membrane, and hybridized to a 32P-labeled probe from the 3′ flanking region of HS2 (the white boxes in panels B and C indicate the positions of nucleosomes). (C) The immobilized LCR template was incubated with 100 μg of MEL protein extract for 45 min at 30°C. The subsequent chromatin assembly and analysis of MNase digestion pattern were performed as described for panel B. (D) The immobilized LCR construct was incubated for 45 min at 30°C with 100 μg of MEL cell extract. The MNase digestion pattern was analyzed as described for panel B.
Generation of erythroid cell-specific LCR HS sites in the absence of chromatin assembly.
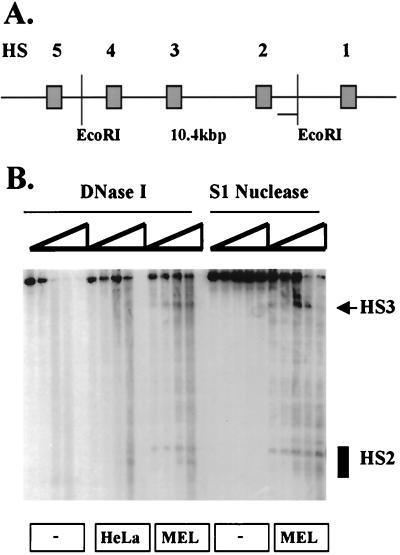
As a control for the experiment shown in Fig. 2, the immobilized LCR template was incubated with proteins from HeLa or MEL cells without prior chromatin assembly and then analyzed with DNase I. Surprisingly, we found that the same sites generated on the chromatin-assembled template are also generated on naked DNA (Fig. 4B). It should be noted that the MEL and HeLa cell extracts used in these studies are likely to contain proteins associated with chromatin or with the regulation of chromatin structure. However, these proteins are not capable of forming a nucleosomal structure over the LCR (Fig. 3D and data not shown) and thus do not generate nucleosomal templates.
FIG. 4.
Erythroid cell-specific generation of LCR DNase I and S1 nuclease HS sites in the absence of chromatin assembly. (A) Representation of the human β-globin LCR indicating the positions of EcoRI sites that were used to map hypersensitivity in HS2 and HS3. (B) The LCR (600 ng) was attached to magnetic beads and incubated for 45 min at 30°C with no protein, with 100 μg of HeLa extract, or with 100 μg of MEL protein extract, as indicated. Subsequently, aliquots were digested with increasing concentrations of either DNase I or S1 nuclease, as indicated. The DNA was digested with EcoRI, subjected to electrophoresis, blotted to a nylon membrane, and then hybridized to a 32P-labeled probe corresponding to the 3′ region of HS2.
Formation of HS3 in the absence of chromatin assembly is erythroid cell specific, as this site is not formed when the template is incubated with protein extracts from HeLa cells. In contrast to HS3, there are two prominent DNase I HS sites corresponding to the core of HS2. Both of these sites are formed in the presence of proteins from erythroid cells, whereas only the weaker of the HS2-specific subsites is formed in the presence of HeLa extracts. As shown in Fig. 3D, incubation of the LCR template with MEL extracts does not lead to the formation of a nucleosomal ladder in the absence of chromatin assembly; thus, hypersensitivity in the LCR induced by erythroid proteins does not reflect the rearrangement of nucleosomes.
We next explored the possibility that erythroid proteins bind to the LCR core enhancer regions and distort the DNA, thereby rendering these sites highly sensitive to DNase I. To examine whether DNase I hypersensitivity in HS2 and HS3 in the absence of chromatin is due to distortion of the DNA or caused by the generation of single-stranded regions, S1 nuclease sensitivity was analyzed in the LCR after incubation with MEL extracts. S1 nuclease is a single-strand-specific enzyme but also digests DNA that adopts non-B DNA conformations (34). Figure 4B shows that the same regions exhibiting DNase I sensitivity in HS2 and HS3 are also sensitive to S1 nuclease. Similar to what we observed for sensitivity to DNase I, S1 sensitivity is restricted to the core enhancer regions; furthermore, the generation of S1 sensitivity in HS2 and HS3 is also erythroid cell specific, as it is not generated in the presence of protein extracts from HeLa cells (data not shown). These results indicate that erythroid cell-specific protein complexes interact with the core enhancer regions and distort or unwind the DNA, thereby rendering these sites highly sensitive to nucleases.
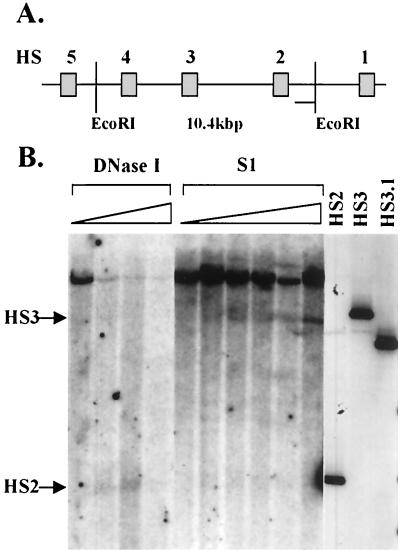
To eliminate the possibility that the formation of S1 nuclease-sensitive sites in the LCR is due to an in vitro artifact, we mapped S1 sensitivity in the LCR in K562 cells, a human erythroleukemia cell line expressing the embryonic β-like globin genes (Fig. 5). The results of these experiments show that HS2 and HS3 are also sensitive to S1 nuclease in erythroid cells in vivo. Importantly, S1 nuclease sensitivity colocalizes with DNase I sensitivity in HS2 and HS3. Two observations may be of interest. First, it appears that sensitivity to S1 nuclease in vivo is weaker in HS2 than it is in HS3. Second, the parental 10.4-kbp band disappears completely with higher concentrations of DNase I but not with increasing concentrations of S1 nuclease, possibly indicating that S1 nuclease sensitivity in the LCR is restricted to only a fraction of cells. Together, these data show that one more (and novel) aspect of HS site formation in the human β-globin LCR is recapitulated in our in vitro system.
FIG. 5.
DNase I and S1 nuclease sensitivity in HS2 and HS3 in erythroid cells in vivo. Nuclei were isolated from K562 cells and incubated with increasing concentrations of either DNase I or S1 nuclease. The DNA was isolated and digested with EcoRI. After gel electrophoresis, the DNA was hybridized to a radioactively labeled probe corresponding to the 3′ flanking region of HS2. (A) Diagrammatic representation of the LCR indicating the locations of EcoRI sites used to map HS2 and HS3. (B) DNase I and S1 nuclease sensitivity in the human β-globin LCR. Lanes HS2, HS3, and HS3.1 represent restriction fragments marking the position of the HS2 5′ end (HS2) and the HS3 5′ (HS3) and 3′ (HS3.1) ends (generated as described in the legend to Fig. 2D).
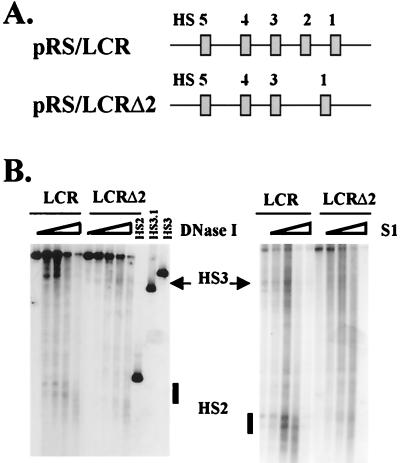
In vivo studies indicate that individual HS sites cooperate to generate a functional LCR (9, 10). To test whether cooperativity between LCR HS sites is required for the generation of DNase I or S1 nuclease sensitivity in the core regions, HS site formation in an LCR mutant lacking the core enhancer of HS2 was analyzed. As shown in Fig. 6, after deletion of HS2, DNase I and S1 nuclease HS sites are no longer formed around HS2. Although HS3 is still formed in the absence of HS2, the efficiency of HS site formation in this region is clearly diminished, suggesting that HS site formation between HS2 and HS3 may be cooperative.
FIG. 6.
Deletion of the HS2 core enhancer eliminates HS site formation in HS2 and impairs HS site formation in HS3. (A) Representation of the LCR and a mutant LCR lacking the 375-bp core enhancer of HS2 (Fig. 1). (B) DNase I and S1 nuclease HS site formation in wild-type (pRS/LCR) and mutant (pRS/LCRΔ2) LCRs. Immobilized templates (600 ng) were incubated with 100 μg of MEL cell extract and processed exactly as described in the legend to Fig. 4. Lanes HS2, HS3.1, and HS3 represent marker fragments corresponding to the HS2 5′, HS3 3′, and HS3 5′ regions, respectively.
Transcription of the HS2 and HS3 core enhancers.
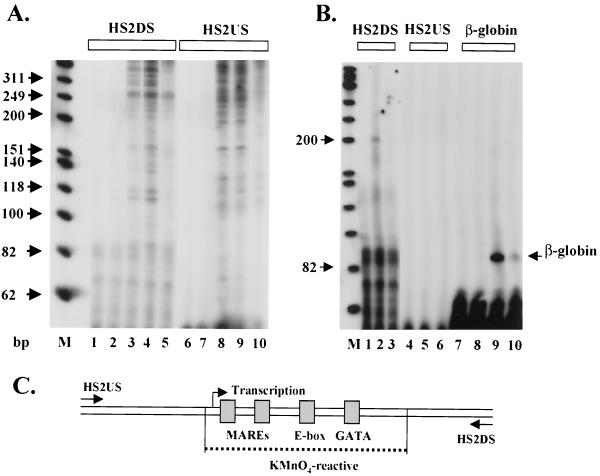
To determine whether the S1-sensitive sites in HS2 and HS3 represent extended regions of single-stranded DNA, we analyzed sensitivity to KMnO4 in HS2 and HS3 after incubating the LCR with HeLa or MEL cell extracts. Permanganate reacts with and oxidizes thymine residues in regions of single-stranded DNA, leading to strand breaks. The strand breaks induced by KMnO4 can be monitored by primer extension-PCR (Materials and Methods). KMnO4 has previously been used to map single-stranded DNA in promoter regions and to localize stalled polymerases (26, 31). As shown in Fig. 7A, incubation of the LCR with MEL extracts leads to the generation of an extended region of KMnO4 reactivity in the core of HS2. This region is about 130 bp long and encompasses the MARE sequences, a GATA site, and an E box, previously shown to interact with helix-loop-helix proteins and to be required for HS2 function (11). We did not detect KMnO4-reactive sites in HS2 after incubation of the LCR with HeLa extracts, despite the DNase I mapping experiments showing that a subsite in HS2 is formed by HeLa extracts (Fig. 4B). These data show that the HeLa-induced sensitivity in HS2 does not represent a region of DNA that is distorted or unwound because it does not react with KMnO4. The control experiments analyzing KMnO4 reactivity in the absence of protein extracts, or primer extension products in the absence of KMnO4, indicate that a subregion in HS2 is specifically rendered single stranded by erythroid proteins.
FIG. 7.
Transcription and formation of an extended single-stranded region in the HS2 core enhancer. (A) Mapping of KMnO4 sensitivity in the HS2 core. Immobilized pRS/LCR (600 ng) was incubated with either no protein (lanes 1 and 6) or 100 μg of protein extract (HeLa, lanes 2 and 7; MEL, lanes 3, 4, 5, 8, 9, and 10). The samples were incubated with KMnO4 (4 mM, except lanes 5 and 10, which contained no KMnO4) for 5 min at RT. The DNA was extracted and analyzed by primer extension-PCR using primers specific for the HS2 upstream and downstream regions (see Materials and Methods for details). Lane M represents a 32P-labeled marker (Promega). The PCR products were analyzed on 8% sequencing gels. (B) Primer extension analysis of transcripts initiating in the HS2 core enhancer. The immobilized LCR template (pRS/LCR or pRS/LCR-β) was incubated with either no protein (lanes 1, 4, 7, and 8) or 70 μg of protein extract (55 μg of MEL/15 μg of HeLa; lanes 2, 3, 5, 6, 9, and 10) for 60 min at 30°C. Lanes 3, 6, and 10 contained 2 μg of α-amanitin per ml. The RNA was isolated and analyzed by primer extension using primers specific for the upstream and downstream regions of HS2 as well as a primer specific for the β-globin gene (lane M depicts the same marker as in panel A). (C) Summary of the results of the KMnO4 and transcription mapping experiments in the HS2 core enhancer. Shown are transcription factor binding sites localized in the KMnO4-sensitive region (dotted line) as well as the position of the transcription start site in HS2.
Single-stranded DNA regions are often found within promoters of transcribed genes. To test whether the core enhancer elements are transcribed in vitro, we incubated the LCR with protein extracts and then monitored the synthesis of RNA by primer extension (Fig. 7B). As a control in these experiments, transcription of the β-globin gene in a construct in which the β-globin gene is linked to the LCR (pRS/LCR-β) was analyzed simultaneously. As shown in Fig. 7B, the β-globin gene linked to the LCR is efficiently transcribed in vitro. Using a primer that specifically hybridizes to the downstream region of HS2, we also detected transcripts initiating within the HS2 core enhancer. A major transcription start site mapped to a region 5′ to the two MARE sequences; this transcript proceeds in a 5′-to-3′ direction (Fig. 7B, lane 2). We detected no transcripts in HS2 that proceed in the opposite direction (Fig. 7B, lanes 4 to 6). Taken together, the results indicate that transcription initiates within HS2 and proceeds in a unidirectional (5′-to-3′) manner. To determine which polymerase is responsible for this transcription, RNA synthesis was analyzed in the presence of α-amanitin. Transcription in HS2 is inhibited by low concentrations of α-amanitin, demonstrating that generation of the HS2 unidirectional transcript is mediated by RNA polymerase II (Fig. 7B, lane 3), as is the β-globin transcript (Fig. 7B, lane 10).
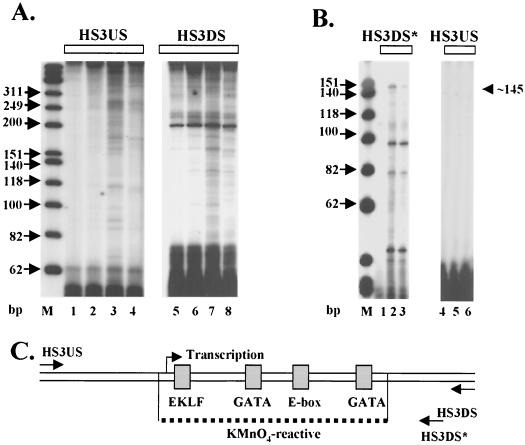
KMnO4-sensitive regions were also analyzed in HS3 (Fig. 8A). Similar to the data obtained from the analysis of HS2, an extended region of about 90 to 100 bp in HS3 was sensitive to KMnO4 after incubation with MEL cell extracts. This region in the core of HS3 encompasses a putative binding site for EKLF (or Sp1), an E box, and two GATA sites. It should be noted that there is a higher background in the control experiments using the HS3-specific primers than what was observed in the experiments using the HS2-specific primers. We have focused our attention to only those sites that are highly sensitive to KMnO4 and erythroid cell specific.
FIG. 8.
Transcription and formation of an extended KMnO4-sensitive region in the HS3 core enhancer. (A) Mapping of KMnO4 sensitivity in HS3. pRS/LCR was incubated with either no protein (lanes 1 and 5) or 100 μg of protein extract (HeLa, lanes 2 and 6; MEL, lanes 3, 4, 7, and 8) and treated with KMnO4 (except for lanes 4 and 8) as described in the legend to Fig. 7. (B) Primer extension analysis of transcription initiating in the HS3 core enhancer. Immobilized LCR templates were incubated with either no protein (lanes 1 and 4) or 70 μg of protein extract (55 μg of MEL/15 μg HeLa; lanes 2, 3, 5, and 6) for 60 min at 30°C. Lanes 3 and 6 also contained 2 μg of α-amanitin per ml. The RNA was isolated and analyzed by primer extension using primers hybridizing to the HS3 5′ region (HS3US) or HS3 3′ region (HS3DS∗; note that this primer is different from the one used to map KMnO4 sensitivity in HS3, HS3DS). (C) Diagram summarizing the results of the KMnO4 and transcription mapping experiments in the HS3 core enhancer. Shown are transcription factor binding sites localized in the KMnO4-sensitive region (dotted line) as well as the position of the transcription initiation site.
Given that we detected transcripts initiating within HS2, we were interested in examining whether HS3 is also transcribed. For this series of experiments, the LCR was incubated with protein extracts and analyzed for RNA synthesis by primer extension. As shown in Fig. 8B, a major transcript that was sensitive to 2 μg of α-amanitin per ml and proceeded in a 5′-to-3′ direction was detected. The smaller primer extension products seen in Fig. 8A were still detectable in reaction mixtures incubated with α-amanitin. These smaller products do not represent transcripts generated by RNA polymerase III, because they were not inhibited by millimolar concentrations of α-amanitin (data not shown). The polymerase II-specific transcript in HS3 maps to a position 5′ to a putative EKLF binding site. No transcripts were detected initiating in HS3 and proceeding in a 3′-to-5′ direction (HS3US [Fig. 8, lanes 4 to 6]).
DISCUSSION
The human β-globin LCR has long been the subject of intense study. Recent advances in analyzing the function of the LCR in the endogenous mouse β-globin locus or in transgenic mice carrying the whole human globin locus on YAC or cosmid constructs clearly established an important functional role of the LCR in mediating high-level expression of all the β-like globin genes throughout erythroid development (7, 12). Although the LCR in the endogenous mouse locus does not appear to be required for the generation of an open, DNase I-accessible chromatin structure, it is quite clear that the human β-globin LCR has the ability to provide an open accessible chromatin structure in transgenic mice in a position-independent manner (21). Despite these advances, however, the mechanism and structural basis for LCR function are poorly understood. We have established conditions that allow us to reconstitute and analyze the formation of HS sites in the human β-globin LCR in vitro. We wished to analyze HS site formation in the context of the entire LCR because previous results indicated that the HS sites synergize to establish a fully functional LCR (9, 10). Therefore, these studies were initiated by subcloning the LCR and mutant derivatives from yeast carrying β-globin YACs by homologous recombination and gap repair. We then established conditions to first attach the LCR constructs to magnetic beads via biotin-streptavidin and to then assemble the DNA into chromatin using Drosophila chromatin assembly extracts (4, 38).
Reconstitution of nuclease HS sites on chromatin-assembled LCR templates in vitro.
The initial experiments showed that HS sites can be reconstituted on chromatin-assembled, immobilized LCR templates in vitro. Two significant observations are of note: (i) formation of hypersensitivity is erythroid cell specific, and (ii) hypersensitivity in the LCR is principally restricted to the 200- to 400-bp core enhancer regions. These results show that two aspects of HS site formation observed in vivo can be recapitulated in this in vitro system. HS site formation in HS2 and HS3 was more efficient when the LCR was preincubated with MEL protein extracts prior to chromatin assembly than in the postincubation experiments (compare Fig. 2C and D). In addition, the formation of an apparently nucleosome-free HS2 core is also more efficient when the LCR is preincubated with erythroid proteins prior to chromatin assembly (Fig. 3). An explanation for these observations could be that once the chromatin structure is formed over the LCR, erythroid proteins cannot efficiently displace nucleosomes. It could be argued that MEL cells may not contain activities required for the remodeling of chromatin structure over the LCR. However, nucleosome remodeling complexes have been purified from MEL cells and shown to change the chromatin structure at the β-globin promoter (2). A plausible alternative explanation is that an active LCR is established by the interaction of proteins or protein complexes prior to the formation of chromatin structure. The binding of erythroid proteins to the core enhancers could prevent the association of a repressive chromatin structure over the LCR. Data published by Milot et al. (32) provide evidence that mutations in the LCR may affect the activity of the globin locus in transgenic mice in a way such that the globin genes are expressed for only a short time after cell division and become silenced because a mutant LCR may no longer be able to resist heterochromatization. In vivo studies of transcription factor binding and chromatin structure in the Drosophila hsp70 gene promoter showed that although the transcription factors were displaced from the promoter in mitotic chromatin, a characteristic DNase I HS site was still detectable (29). These results were interpreted to mean that a noncanonical chromatin conformation was maintained at the hsp70 promoter during mitosis, which then allows the reassembly of a functional promoter during interphase. All these data are in agreement with a model proposed by Felsenfeld (13), according to which regulatory elements, like the human β-globin LCR, are structurally prepared by the binding of proteins after replication, and these proteins may then confer resistance to the formation of repressive chromatin.
The MNase mapping experiments shown in Fig. 3 suggest that the region around HS2 is assembled into an array of positioned nucleosomes. Furthermore, it appears that preincubation of the LCR with protein extracts from MEL cells prior to chromatin assembly changes the MNase pattern in the core HS region but does not seem to prevent nucleosomes from associating with the flanking regions. Although the conclusion that the HS2 core remains free of nucleosomes should be treated with caution, it is clear that the nucleosomal pattern in the 5′ region of HS2 is different in the presence or absence of MEL cell extracts, suggesting that erythroid proteins change the position of nucleosomes in this region (compare Fig. 3B and C).
Nuclease HS site formation in the absence of chromatin assembly.
It is generally believed that DNase I HS sites represent regions in chromatin in which nucleosomes are displaced, excluded, or modified. It was therefore surprising to us that DNase I hypersensitivity in HS2 and HS3 is also formed in an erythroid cell-specific manner in the absence of chromatin assembly. This result indicates that it is not simply the rearrangement of nucleosomes that renders these sites highly sensitive to nucleases. Similar to what was observed on chromatin-assembled DNA, hypersensitivity in the unassembled LCR construct is erythroid cell specific and tightly limited to the core enhancer regions. It could be argued that hypersensitivity in the core regions simply reflects the binding of transcription factors, as in vitro footprinting studies often show that sites immediately flanking protected regions reveal higher sensitivity to DNase I. However, these data cannot be compared to the long-range HS site mapping experiments in our study. There are many transcription factor binding sites in the LCR, not just in the core enhancer elements (23), yet our results show that hypersensitivity to DNase I is mainly restricted to the HS2 and HS3 core regions.
We hypothesized that a specific combination of protein binding sites in the HS2 and HS3 core enhancers could recruit protein complexes that change the topology of the DNA and render these sites highly sensitive to nucleases. In support of this hypothesis, we found that the same regions in HS2 and HS3 that reveal sensitivity to DNase I are also sensitive to S1 nuclease, indicating that the DNA in the core regions is distorted or rendered single stranded by erythroid proteins. Importantly, we found that HS2 and HS3 are also sensitive to S1 nuclease in K562 cells in vivo (Fig. 5). In contrast to the in vitro analysis, in which both HS2 and HS3 are equally sensitive to S1, it appears that HS3 is more sensitive to S1 nuclease than HS2 in K562 cells. The difference could be due to higher-order chromatin structure or to developmental stage-specific differences, as the in vitro experiments were performed with an extract from erythroid cells revealing an adult specific pattern of globin gene expression (MEL cells).
To further explore the extent of single stranded regions in vitro, we mapped KMnO4 reactivity in the HS2 and HS3 core enhancers. In HS2, an extended region of about 130 bp is rendered sensitive to KMnO4 after incubation with MEL cell extract. This region encompasses the tandem MARE sequences, two GATA sites, and an E-box motif. The KMnO4-sensitive region in HS3 is about 90 to 100 bp long and extends over a putative EKLF binding site as well as two GATA sites and an E-box motif. Several mechanisms could lead to the formation of S1 nuclease- and KMnO4-sensitive regions in the core enhancer. First, erythroid protein complexes could assemble at the core enhancer regions and distort or unwind the DNA. Second, nuclear protein complexes like 13S condensin have DNA-dependent ATPase activities and change the topology of the DNA, which in turn could create regions sensitive to S1 nuclease or KMnO4 (24). Finally, it is also possible that the assembly of protein complexes could recruit transcription complexes, which would lead to the generation of single-stranded regions in the core enhancer.
Transcription of LCR core HS sites.
To test whether transcripts could be detected in the HS2 and HS3 core enhancer regions, we assayed for RNA synthesis by primer extension. In HS2, a specific transcript that initiates just upstream of the two MARE sequences was detected. Transcription is unidirectional, proceeding in a 5′-to-3′ direction (i.e., we were unable to detect transcripts initiating in HS2 and proceeding in a 3′-to-5′ orientation). This result is in agreement with previously published experiments showing that unidirectional transcripts initiate in HS2 in erythroid cells (25). Transcription in HS3 is initiated 5′ to a putative EKLF binding site and also proceeds in a 5′-to-3′ direction. Furthermore, we show that transcription of both HS2 and HS3 core enhancers is mediated by RNA polymerase II. How the RNA polymerase II transcription complex is recruited to the core enhancers is not known, but our in vitro system is ideally suited to analyze the regulatory sequences that recruit these transcription complexes. There is no obvious TATA sequence in the vicinity of the putative transcription start site in HS2 or HS3. However, it is worth noting that an E box is located about 50 bp downstream of the initiation site in HS2 and about 30 bp downstream of the transcription start site in HS3. E-box binding proteins have previously been implicated in transcription complex assembly on a variety of polymerase II-transcribed genes (37), including genes specifically expressed in erythroid cells (8). In this respect, it is interesting that an E box is also located in the downstream promoter-initiator region of the adult β-globin gene (K. M. Leach et al., unpublished data). The E-box motif in HS3 is not phylogenetically conserved (20). Comparison of the effects of HS3 deletions in the endogenous mouse locus and in the transgenic human β-globin locus points to functional differences between the human and mouse HS3 enhancers. Deletion of HS3 in the murine locus leads to a mild reduction of adult β-globin gene (mouse βmaj) expression (22), whereas deletion of the human HS3 enhancer dramatically reduced expression of both the embryonic ɛ-globin and adult β-globin genes (9, 33). Thus, the E box could confer a unique function to the human HS3 enhancer.
HS site formation in HS2 and HS3 is not inhibited by the presence of α-amanitin (data not shown), indicating that transcription per se is not required for HS site formation. At this point we cannot rule out the possibility that the formation of a transcription complex, a step not inhibited by α-amanitin, contributes to the generation of single-stranded regions and HS site formation. However, it appears more likely that the generation of DNase I- and S1-sensitive regions in HS2 and HS3 precedes transcription. In other words, protein complexes that assemble on the individual HS sites may change the topology of the DNA and may represent attachment sites for RNA polymerase II transcription complexes. The in vivo analysis of LCR transcription in the human β-globin locus (3, 17) showed that LCR transcripts are detectable in only a small fraction of erythroid cells, which supports the notion that these transcripts may play only a transient role in setting up an active locus. Data presented by Gribnau et al. (17) suggest that transcription of LCR and intergenic regions is important for chromatin opening because the appearance of intergenic transcripts correlates with the localization of nuclease-sensitive domains. Another possibility is that the LCR contains multiple attachment sites for RNA polymerase II transcription complexes and these are somehow delivered to the individual globin gene promoters. Although the globin genes appear to have stronger promoters (at least in vitro) and are expected to recruit transcription complexes more efficiently than the core enhancer elements of the LCR, it is possible that in vivo the promoter regions may not be as accessible as the LCR. An indication for this may be that the LCR HS sites are much more sensitive to nucleases than are the individual globin gene promoters (14, 28, 41). In this respect, a distorted DNA conformation in the LCR core enhancer elements could contribute to higher accessibility and more efficient recruitment of polymerase II transcription complexes in vivo. For example, it is possible that changes in DNA topology over the core enhancer elements in vivo could prevent the association of repressive chromatin, thereby allowing transcription complexes to be recruited preferentially to the LCR core regions. Current models view the LCR as a unit in which individual HS sites interact via protein-protein interactions. Such a large protein-DNA complex may be very efficient in recruiting transcription complexes and chromatin remodeling factors, and these activities could be delivered to individual globin gene promoters by a tracking or looping mechanism (7, 12).
Studies addressing the DNA structure in genes reactivated after mitosis came to the conclusion that transcription initiation sites in genes scheduled for reactivation display a distorted DNA conformation, whereas start sites of those genes that remain repressed are undistorted (31). It was argued that protein-dependent conformational changes in the DNA structure could mark genes for reexpression. Similarly, conformational changes in the β-globin LCR could also mark regions that remain active after mitosis, before chromatin structure is completely reassembled. This hypothesis is consistent with our observation that HS site formation in chromatin is more efficient when the LCR is preincubated with erythroid proteins. We are currently in the process of analyzing the changes in DNA structure and bound proteins in the LCR during the cell cycle.
In summary, our data indicate that the mechanisms leading to HS site formation in the human β-globin LCR involve reorganization of chromatin structure as well as the generation of S1- and KMnO4-sensitive regions in the core enhancers. These results further our understanding of the structural basis for LCR function and also have implications for the structure and function of other regulatory elements in the globin locus as well as other loci.
ACKNOWLEDGMENTS
We are grateful to Gail Green for expert technical assistance. We thank Mike Kilberg and Thomas Yang (University of Florida) for critically reading the manuscript.
This work was supported by grants from the NIH (HL24415 to J.D.E.; DK 52356 to J.B.) and from the Howard Hughes Medical Institute (Research Resources Program, University of Florida, to J.B.).
REFERENCES
- 1.Armstrong J A, Emerson B M. NF-E2 disrupts chromatin structure at β-globin locus control region hypersensitive site 2 in vitro. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 3.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker P B, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender M, Bulger M, Close J, Groudine M. Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice does not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 6.Bulger M, von Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 8.Bungert J, Kober I, Düring F, Seifart K H. Transcription factor eUSF is an essential component of isolated transcription complexes on the duck histone H5 gene and it mediates the interaction of TFIID with a TATA-deficient promoter. J Mol Biol. 1992;223:885–898. doi: 10.1016/0022-2836(92)90250-n. [DOI] [PubMed] [Google Scholar]
- 9.Bungert J, Davé U, Lim K-C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 10.Bungert J, Tanimoto K, Patel S, Liu Q, Fear M, Engel J D. Hypersensitive site 2 specifies a unique function within the human β-globin locus control region to stimulate globin gene transcription. Mol Cell Biol. 1999;19:3062–3072. doi: 10.1128/mcb.19.4.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elnitski L, Miller W, Hardison R. Conserved E-boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region: role of basic helix-loop-helix proteins. J Biol Chem. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- 12.Engel J D, Tanimoto K. Looping, linking and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 13.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 14.Forrester W C, Takegawa S, Papayannopoulos T, Stamatoyannopoulos G, Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaensler K M L, Burmeister M, Brownstein B H, Taillon-Miller P, Myers R M. Physical mapping of yeast artificial chromosomes containing sequences from the human β-globin gene region. Genomics. 1991;10:976–984. doi: 10.1016/0888-7543(91)90188-k. [DOI] [PubMed] [Google Scholar]
- 16.Gong Q H, McDowell J C, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the epsilon globin gene by 5′ hypersensitive site 2 of the β-globin locus control region. Mol Cell Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 18.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 19.Grosveld F. Activation by locus control regions? Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 20.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;20:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 21.Higgs D. Do LCRs open chromatin domains? Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 22.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Analysis of mice containing a targeted deletion of β-globin locus control region hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson J D, Miller W, Hardison R C. Sequences within and flanking hypersensitive sites 3 and 2 of the β-globin locus control region required for synergistic versus additive interaction with the ɛ-globin gene promoter. Nucleic Acids Res. 1996;24:4327–4335. doi: 10.1093/nar/24.21.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensins: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 25.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 27.Lee J-S, Lee C-H, Chung J H. The β-globin promoter is important for recruitment of erythroid Krüppel-like factor to the locus control region in erythroid cells. Proc Natl Acad Sci USA. 1999;96:10051–10055. doi: 10.1073/pnas.96.18.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Lim K-C, Engel J D, Bungert J. Individual LCR hypersensitive sites cooperate to generaté an open chromatin domain spanning the human β-globin locus. Genes Cells. 1998;3:415–430. doi: 10.1046/j.1365-2443.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Balbàs M A, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 30.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelotti E F, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 32.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 33.Navas P A, Peterson K R, Li Q, Skarpidi E, Rohde A, Shaw S E, Clegg C H, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol. 1998;17:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill D, Bornschlegel K, Flamm M, Castle M, Bank A. A DNA-binding factor in adult hematopoietic cells interacts with a pyrimidine-rich domain upstream from the human delta-globin gene. Proc Natl Acad Sci USA. 1991;88:8953–8957. doi: 10.1073/pnas.88.20.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomerantz O, Goodwin A J, Joyce T, Lowrey C H. Conserved elements containing NF-E2 and tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human β-globin locus control region. Nucleic Acids Res. 1998;26:5684–5691. doi: 10.1093/nar/26.24.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not for the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 38.Sandaltzopoulos R, Blank T, Becker P B. Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm drosophila chromatin. EMBO J. 1994;15:373–379. doi: 10.1002/j.1460-2075.1994.tb06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatoyannopoulos G, Nienhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhuis A W, Majerus P, Varmus H, editors. The molecular basis of blood diseases. W. B. Philadelphia, Pa: Saunders; 1994. pp. 107–155. [Google Scholar]
- 40.Stamatoyannopoulos J A, Goodwin A, Joyce T, Lowrey C H. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuan D, Solomon W, Li Q, London I M. The “β-like globin gene domain” in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the Tal1, E47, GATA-1 and Ldb1/NL1 proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 44.Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, Fraser P. The role of EKLF in human β-globin gene competition. Genes Dev. 1996;10:2894–2920. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida C, Tokumasu F, Hohmura K I, Bungert J, Hayashi N, Nagasawa T, Engel J D, Yamamoto M, Takeyasu K, Igarashi K. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells. 1999;4:643–655. doi: 10.1046/j.1365-2443.1999.00291.x. [DOI] [PubMed] [Google Scholar]