ABSTRACT
During evolution, enzymes can undergo shifts in preferred substrates or in catalytic activities. An intriguing question is how enzyme function changes following horizontal gene transfer, especially for bacterial genes that have moved to animal genomes. Some insects have acquired genes that encode enzymes for the biosynthesis of bacterial cell wall components and that appear to function to support or control their obligate endosymbiotic bacteria. In aphids, the bacterial endosymbiont Buchnera aphidicola provides essential amino acids for aphid hosts but lacks most genes for remodeling of the bacterial cell wall. The aphid genome has acquired seven genes with putative functions in cell wall metabolism that are primarily expressed in the aphid cells harboring Buchnera. In analyses of aphid homogenates, we detected peptidoglycan (PGN) muropeptides indicative of the reactions of PGN hydrolases encoded by horizontally acquired aphid genes but not by Buchnera genes. We produced one such host enzyme, ApLdcA, and characterized its activity with both cell wall derived and synthetic PGN. Both ApLdcA and the homologous enzyme in Escherichia coli, which functions as an l,d-carboxypeptidase in the cytoplasmic PGN recycling pathway, exhibit turnover of PGN substrates containing stem pentapeptides and cross-linkages via l,d-endopeptidase activity, consistent with a potential role in cell wall remodeling. Our results suggest that ApLdcA derives its functions from the promiscuous activities of an ancestral LdcA enzyme, whose acquisition by the aphid genome may have enabled hosts to influence Buchnera cell wall metabolism as a means to control symbiont growth and division.
KEYWORDS: Buchnera, carboxypeptidase, cell wall, endopeptidase, enzyme promiscuity, horizontal gene transfer, multifunctional, pea aphid, peptidoglycan, symbiosis
INTRODUCTION
The near-universal ability of enzymes to perform promiscuous reactions is increasingly recognized as a starting point in the evolution of novel functions (1). Promiscuous activities become functional when, in the context of a new environment and/or mutation(s), they contribute to fitness, often via complementation of metabolic inadequacies (2–4). Host-associated bacteria that have experienced extreme genome reduction often lack essential genes or whole pathways (5), such that multifunctional enzymes derived from ancestrally promiscuous enzymes have been suggested as a likely means of compensation (6, 7). This idea is supported by examples in multiple bacterial lineages, including the mammalian pathogen Chlamydia (8, 9) and insect-associated Wolbachia (10) and Buchnera aphidicola (11) symbionts.
Alternatively, host genomes may acquire genes via horizontal-gene transfer (HGT) to supplement symbiont shortcomings. While HGT is relatively rare in eukaryotes, it occurs most often from host-associated bacteria (12) and has proven instrumental in eukaryotic evolution (13–16), most notably in the context of mitochondrial and plastid evolution (17, 18). Among insect symbioses, horizontally transferred genes (HTGs) appear to provide hosts with novel functions and, in some cases, may improve their ability to regulate or benefit from symbionts (12, 19). Recently, the compensatory nature of several mealybug HTGs has been revealed—the insect genome encodes enzymes of the peptidoglycan (PGN) synthesis pathway that symbionts lack (20), and these proteins localize within symbionts and participate in cell wall construction (21).
Aphids require Buchnera symbionts to provide essential amino acids that are missing from their exclusive diet of phloem sap. Interestingly, the pea aphid (Acyrthosiphon pisum) genome contains eight HTGs with putative functions in PGN metabolism (Fig. 1) (22, 23). Seven of these aphid HTGs appear important for symbiosis based on their increased expression in bacteriocytes (23), the specialized host cells where Buchnera reside, relative to other host tissues. In addition, RNAi knockdown of HTG expression reduces Buchnera abundance (24), and HTG expression is correlated with aphid genotypes displaying high symbiont abundances (25). Considering the close coordination of PGN metabolism with cell growth and division machinery in bacteria (26), these observations suggest that host PGN enzymes may play a role in regulating Buchnera proliferation.
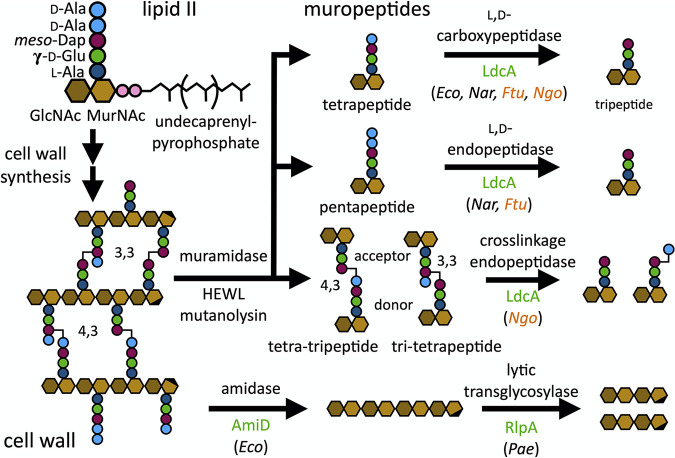
FIG 1.
Schematic representation of the enzymatic activites of previously characterized homologs of the aphid LdcA, AmiD, and RlpA enzymes toward peptidoglycan (PGN). The cell wall is assembled from lipid II, consisting of N-acetylglucosamine (GlcNAc), N-acetylmuramic acid (MurNAc), l-alanine, γ-d-glutamate, meso-diaminopimelic acid (Dap), and d-alanine. Cell wall digestion with muramidase produces muropeptides, many of which are suitable substrates for known LdcA enzyme activities, while AmiD and RlpA act on the polymeric cell wall. Enzyme reactions appear above reaction arrows, while enzyme names are shown below. Enzymes shown in green indicate those for which aphid-encoded homologs exist. Organism abbreviations, shown in parentheses, denote the species for which the reaction has been demonstrated for the enzyme homolog: Eco, E. coli (27, 67); Nar, Novosphingobium aromaticivorans (31); Ftu, Francisella tularensis (29); Ngo, Neisseria gonorrhoeae (30); and Pae, Pseudomonas aeruginosa (68). Organism abbreviations shown in orange designate enzymes capable of utilizing both muropeptides and cell wall as substrates.
Of the seven aphid HTGs implicated in symbiosis, all but one putatively function in cell wall remodeling. This gene, ldcA, encodes a homolog of the l,d-carboxypeptidase (ApLdcA) involved in PGN recycling (27), a cytoplasmic process that is absent in Buchnera but present in free-living bacteria like Escherichia coli, a close relative of Buchnera (Fig. 1) (28). While E. coli LdcA (EcLdcA) is known to utilize only solubilized PGN fragments (muropeptides), LdcA homologs from some intracellular pathogens are exported to the periplasm and display a shift in substrate tolerance, modifying the polymeric cell wall in addition to soluble muropeptides (29, 30). Furthermore, LdcA homologs exhibit multifunctionality, demonstrating endopeptidase activities in addition to their carboxypeptidase function (29–31). We hypothesized that EcLdcA might display a similar shift in activity that could enable aphids to control or support its symbionts. Specifically, endopeptidases are essential for E. coli because they are required to make space for the insertion of nascent PGN strands into the cell wall (32)—an endopeptidase may be required by Buchnera but encoded by the host. If ApLdcA is an endopeptidase, this novel function may derive from a promiscuous enzyme activity present in the ancestral enzyme.
In the present work, we investigated the hypothesis that ApLdcA displays key differences from a free-living bacterial homolog, EcLdcA. First, we provide evidence that PGN hydrolases, including an endopeptidase, are active in the aphid-Buchnera system, producing muropeptides that can be isolated from the aphid hemolymph indicative of their physiological relevance. Second, we demonstrate that ApLdcA retains its l,d-carboxypeptidase function toward soluble muropeptides and also exhibits l,d-endopeptidase activity against both stem pentapeptides and cross-linked peptidoglycan. This feature likely derives from ancestral enzyme promiscuity, since the closely related EcLdcA is also capable, albeit to a lesser extent, of l,d-endopeptidase activity. Taken together, these results reveal potential host adaptations that have capitalized on the catalytic and substrate promiscuity of LdcA in order to target symbiont PGN.
RESULTS
Complex PGN can be isolated from whole aphids.
We first sought to understand the role that host HTGs may play in Buchnera PGN metabolism by characterizing Buchnera’s cell wall. Cell wall PGN is comprised of repeating units of β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) disaccharide with a short stem peptide attached to the MurNAc lactyl moiety via amide bond (33). In Gram-negative bacteria, the stem peptide typically consists of five amino acids: l-Ala, γ-d-glutamate, meso-diaminopimelic acid (mDap), d-Ala, and d-Ala (pentapeptide). Synthesis of PGN, the major constituent of the cell wall (also called murein), begins in the cytoplasm with the multistep construction of lipid II and transitions to the periplasm where the cell wall, or murein sacculus, is assembled from lipid II by a series of additional enzymes (33). Cell wall remodeling is necessary for bacterial growth and division, for antibiotic resistance, for repair of the cell wall, and for the insertion of outer membrane proteins (34, 35). Bacteria additionally recycle PGN by importing muropeptides, the products of cell wall remodeling events, into the cytoplasm to be shunted back into lipid II biosynthesis (35). A wide range of chemical modifications are possible during these processes, leading to distinct cell wall compositions among even closely related bacterial species (34–36). The Buchnera genome has a greatly reduced repertoire of cell wall synthesis and remodeling genes relative to E. coli, including a lack of any typical carboxypeptidases or cross-linkage endopeptidases. We thus anticipated that evidence of these enzyme activities, such as trimmed stem peptides, might be absent or limited in the Buchnera cell wall. Buchnera is not culturable outside its aphid host, but we were able to purify PGN directly from aphids. We subjected A. pisum homogenate to high-performance liquid chromatography (HPLC) and screened fractions for muropeptides using nano ultrahigh performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) and a novel proteomics-based approach for the automated identification of muropeptides (37).
To compare Buchnera PGN with that of its closest free-living relative, we also analyzed E. coli PGN derived from digestion of the isolated sacculus with the muramidase mutanolysin. The E. coli cell wall exhibits a range of chemical modifications, including GlcNAc-anhydro-MurNAc (G-aM) disaccharides at the termini of PGN strands, stem peptide cross-linkages that give the cell wall its mesh-like architecture, and a low level of substitution of stem peptide d-Ala residues for noncanonical l- or d-amino acids (NCLAAs and NCDAAs, respectively). We detected a similarly complex assortment of muropeptides from aphids with both GlcNAc-MurNAc (GM) and G-aM glycans, variable stem peptide lengths and sequences, and diverse cross-linked compounds, including those derived from three strands of peptidoglycan (Fig. 2; see also Fig. S1A and B in the supplemental material). We analyzed fractions from two distinct homogenate treatments—both were sonicated to lyse Buchnera cells, while one was additionally treated with hen egg-white lysozyme (HEWL) to digest the cell wall. Untreated and lysozyme-treated samples had largely similar PGN profiles and sonication alone is insufficient to shear glycan chains into small units (38), suggesting that soluble muropeptides are produced in aphids in the absence of exogenously added lysozyme.
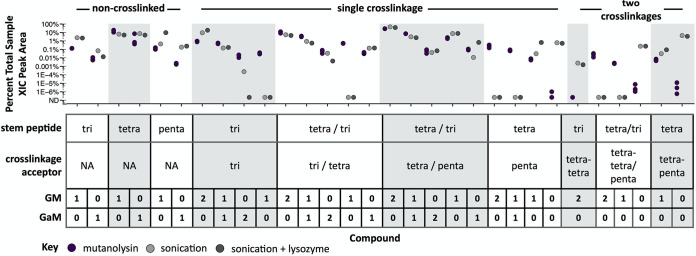
FIG 2.
Composition of Buchnera-derived cell wall fragments purified from A. pisum. Aphid homogenate was successively filtered to components of ≤10 kDa and subjected to LC-MS/MS analysis. Muropeptide compounds were identified from MS/MS spectra and extracted-ion chromatogram (XIC) peak areas determined using Byonic and Byologic softwares, respectively (Protein Metrics). Areas were baseline subtracted and normalized by sample, such that the data shown is the percentage of total PGN represented by each compound. Untreated mutanolysin-derived E. coli muropeptides are shown for comparison (purple). Two treatments were used without replicates: aphid homogenate was sonicated to lyse Buchnera cells (light gray) or additionally treated with lysozyme to digest Buchnera cell walls (dark gray). The table describes the compound structure: PGN compounds vary by stem peptide sequence and glycan (GM = GlcNAc-MurNAc, GaM = GlcNAc-anhydro-MurNAc). For distinct compounds that are equivalent in mass (differing either in stem peptide sequence or cross-linkage type), we were unable to quantify each compound abundance independently, because the two compounds could not be chromatographically resolved—such structural isomers were integrated together, and their sequences are reported with variable residues shown separated by backslashes within parentheses, such that the same relative position within parentheses refers to the sequence of one isomer.
Composition of Buchnera-derived and LdcA-treated E. coli muropeptides as determined by LC-MS/MS. (A and B) Noncanonical non-cross-linked (A) and singly cross-linked (B) muropeptides identified from A. pisum homogenate (Fig. 2). Untreated E. coli mutanolysin-derived muropeptides are shown for comparison (purple). Two treatments were used without replicates: aphid homogenate was either sonicated to lyse Buchnera cells (light gray) or sonicated and additionally treated with lysozyme to digest Buchnera cell walls (dark gray). (C and D) Protein fractions generated during IMAC purification of recombinantly expressed A. pisum LdcA (C) and E. coli LdcA (D). For each SDS-PAGE gel, fractions analyzed included the insoluble pellet (P), soluble cell lysate (L), Ni-NTA flowthrough (FT), lysis buffer wash (W1), HEPES buffer wash (W2), and HEPES-buffered elutions of increasing imidazole concentrations: 50 mM (E1), 100 mM (E2), 200 mM (E3), and 500 mM (E4). (E to G) Noncanonical non-cross-linked (E), noncanonical singly cross-linked (F), and canonical doubly cross-linked (G) muropeptides after treatment of mutanolysin-derived E. coli muropeptides (purple) with EcLdcA (blue) and ApLdcA (green) (Fig. 3). All stem peptide sequences consist of l-Ala, γ-d-Glu, and meso-l-Dap, followed by the single-letter amino acid code indicated in the compound table. The absence of an amino acid indicates that the tripeptide is the complete stem-peptide sequence. Glycan substituents include GlcNAc-MurNAc (GM) and GlcNAc-anhydro-MurNAc (GaM). Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The composition of muropeptides in aphid homogenate implicates several host and/or symbiont PGN enzymes in their origin. Muropeptides containing terminal G-aM (including G-aM itself) are products of lytic transglycosylases such as ApRlpA (Fig. 1) or Buchnera’s MltA and MltE enzymes, nonhydrolytic enzymes that fragment PGN chains (35, 39). On the other hand, GM-substituted muropeptides could be produced by hydrolytic muramidases, like the two endogenous invertebrate (i-type) lysozymes (NCBI gene IDs 100160909 and 100168424) that are more highly expressed in bacteriocytes relative to other host tissues (40). We detected muropeptides containing tripeptide and tetrapeptide stems, which are likely derived from carboxypeptidase and/or endopeptidase activities (Fig. 1). Since the Buchnera genome encodes no recognizable l,d-carboxypeptidases or endopeptidases and LdcA homologs from intracellular pathogens collectively exhibit both of these functions (29, 30), it is possible that ApLdcA could be responsible for producing these stem peptides in Buchnera.
Aphid PGN also includes cross-linked PGN compounds containing 4,3- and 3,3-cross-linkages (Fig. 1). The 4,3-cross-linkages, which predominate in E. coli, are likely formed by the d,d-transpeptidases PBP1B and PBP3 encoded by Buchnera during cell wall synthesis, but 3,3-cross-linkages typically require the l,d-transpeptidases YnhG and YcbB (41), which Buchnera spp. lack. Because some cross-linked muropeptides are equivalent in mass and yet display distinct cross-linkage types (i.e., 4,3-cross-linked tetra-tripeptide and 3,3-cross-linked tri-tetrapeptide), these could not be definitively distinguished by their MS/MS fragmentation patterns, as described by Bern et al. (37). However, the presence of tri-tripeptide cross-linked stems, which contain only 3,3-cross-linkages, indicates that both 4,3- and 3,3-cross-linkage types are represented among PGNs derived from aphid homogenate (Fig. 2). In addition, while E. coli PGN is devoid of cross-linked stem peptides lacking any glycan substituents, we observed glycanless cross-linked stem peptides among muropeptides from aphid homogenate that are likely products of ApAmiD or Buchnera’s AmiB amidase (Fig. 2).
Stem peptides may include noncanonical residues in place of one or both terminal d-Ala residues. NCDAAs are introduced to stem peptides either during lipid II synthesis via racemase enzymes or during cell wall synthesis via l,d-transpeptidases (42) and play a role in regulating PGN composition (42–44). NCLAAs found within stem peptides derive from covalent attachment of outer membrane proteins, such as murein lipoprotein (Lpp) in E. coli (45) and can be detected following proteolytic digest (46). Though our approach is incapable of discerning amino-acid stereochemistry, we observed that 33.1% of all E. coli detected muropeptides contain atypical amino acids, which are present in both non-cross-linked (see Fig. S1A) or cross-linked stem peptides (see Fig. S1B and Table S1). Some noncanonical stem peptides reach proportions similar to that of canonical tri- or pentapeptides. Among these, AE-mDap-K and AE-mDap-KR stem peptides are derived from Lpp. In contrast, only 10.7% of aphid muropeptides contain noncanonical amino acids, and a much lower diversity of stem sequences is represented. Despite this, we detected AE-mDap-K and AE-mDap-KR stems in aphids (see Fig. S1A and B). This observation is unexpected for two reasons: (i) the Buchnera genome lacks the gene encoding Lpp, as well as any homologs of the three l,d-transpeptidases, in E. coli that cross-links Lpp to PGN (ldtA to ldtC) (41), and (ii) unlike the E. coli sacculus, aphid PGN samples were not treated with proteases. Most Gram-negative bacteria lack Lpp homologs—in these species, the cell wall is covalently linked to different outer membrane proteins (46), suggesting the same may be true for Buchnera. While the source of AE-mDap-KR muropeptides in aphid homogenate is unclear, these molecules are abundant and are likely part of a specific and significant process in Buchnera.
Differences in average percent abundance of total PGN per compound between mutanolysin-digest of E. coli sacculus (E. coli), muropeptides isolated from A. pisum homogenate (Buchnera), and E. coli PGN treated with EcLdcA (EcLdcA) and ApLdcA (ApLdcA). Download Table S1, DOCX file, 0.07 MB (74.8KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Soluble muropeptides purified from aphid homogenate are derived from the Buchnera cell wall and reflect the collective enzymatic activities of both (i) cell wall remodeling and (ii) any downstream processing of soluble remodeling products. Our results show that endopeptidase and l,d-carboxypeptidase activities are involved in one or both of these processes. Furthermore, we found that Buchnera PGN is essentially as complex as that of E. coli, notwithstanding the limited set of PGN enzymes. This level of complexity also suggests that Buchnera muropeptides are not exhaustively degraded by symbiont or host PGN enzymes, indicating that these enzymes may play a more specific role in sculpting the Buchnera cell wall architecture.
Characterization of LdcA activities using a multisubstrate, automated assay.
Next, we investigated the reactions of ApLdcA and EcLdcA in vitro. We expressed the ldcA genes in E. coli and purified recombinant ApLdcA and EcLdcA proteins by immobilized metal-affinity chromatography (IMAC) (see Fig. S1C and D). We then treated muropeptides derived from mutanolysin digestion of E. coli cell walls with the recombinant proteins (Fig. 3A). These compounds are similar to the substrates normally encountered by EcLdcA in the cytoplasm, consisting of at most a single glycan substituent per stem peptide. In addition, the complexity of E. coli PGN in terms of the abundance and diversity of cell wall modifications allows for simultaneous evaluation of a wide range of potential enzyme substrates. Treated muropeptides were reduced with sodium borohydride and desalted by HPLC before applying the same proteomics-based approach used above to identify individual muropeptide compounds and evaluate LdcA activity toward each potential substrate (Fig. 3B). The same quantity of an identical preparation of E. coli sacculus was used for each reaction, such that, for a given compound, relative differences in abundance between treatments are essentially quantitative (see Table S1).
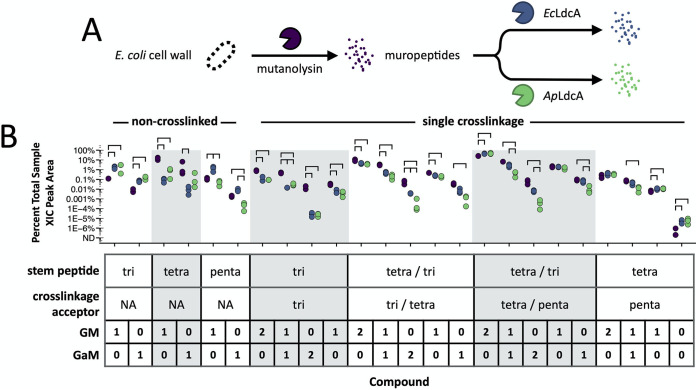
FIG 3.
Comparison of the muropeptide composition and abundance of soluble PGN substrates following treatment with EcLdcA and ApLdcA. (A) Mutanolysin-derived E. coli muropeptides were treated with EcLdcA and ApLdcA and subjected to the same LC-MS/MS analysis used for the data in Fig. 2. (B) Areas were baseline subtracted and normalized by sample. For each compound, the data shown are the average percentages of total PGN from three replicates (see Table S1). Comparisons between treatments were made using Tukey’s HSD test with adjustment for false discovery. All bars shown represent P values of <0.05.
Relative to untreated E. coli muropeptides, we observed reduced tetrapeptide abundance for both EcLdcA and ApLdcA with concurrent increases in the amount of tripeptides, demonstrating that the l,d-carboxypeptidase activity of these enzymes can be readily detected by our approach as a decrease in substrate and an accumulation of product (Fig. 3B; see also Table S1). Interestingly, we observed a decrease in pentapeptide abundance for ApLdcA and an increase for EcLdcA. The former suggests that ApLdcA acts as an l,d-endopeptidase that converts pentapeptides directly to tripeptides, an activity previously reported for LdcA homologs from Novosphingobium aromaticivorans and Francisella tularensis (29, 31). An explanation for the latter observation is described in the following section. We also detected a lower abundance of several noncanonical tetrapeptide monomers for both enzymes (see Fig. S1E). When these are taken into account, ApLdcA generally shows less turnover of non-cross-linked muropeptides than does EcLdcA, likely indicating a reduced preference for these substrates relative to EcLdcA.
Unlike the cell wall fragments that EcLdcA might encounter during PGN recycling, mutanolysin-derived muropeptides contain cross-linked stem peptides that would normally be hydrolyzed by endopeptidases prior to being imported into the cytoplasm. Though EcLdcA is not known to hydrolyze cross-linkages, we observed a decrease in the abundance of nearly all cross-linked compounds for both EcLdcA and ApLdcA-treated muropeptides, including both 4,3- and 3,3-cross-linked stem peptides (Fig. 3B). Among cross-linked compounds with two glycan substituents (one on each stem peptide), we observed greater turnover of substrates containing G-aM over GM glycans (2 G-aM > 1 G-aM and 1 GM > 2 GM) (Fig. 3B). Cross-linked compounds containing only one glycan (on either stem peptide), likely resulting from partial amidase activity, are generally less affected by either LdcA enzyme than di-substituted compounds, though G-aM-containing muropeptides are still preferred over those with GM (Fig. 3B). We found the same trends among some noncanonical cross-linked stem peptides (see Fig. S1G). In general, ApLdcA treatment decreased the abundance of cross-linked muropeptides to a greater extent than EcLdcA (Fig. 3B; see also Fig. S1F and G and Table S1).
Collectively, these results demonstrate that ApLdcA can act as an l,d-endopeptidase on pentapeptide substrates and that both EcLdcA and ApLdcA are capable of hydrolyzing PGN cross-linkages, an activity previously not reported for EcLdcA (47).
LdcA is an l,d-endopeptidase toward both stem pentapeptides and cross-linkages.
We next sought to validate the results of our proteomic analysis of LdcA activity and to characterize the transformations for each LdcA enzyme with authentic synthetic muropeptide samples. The activities of the LdcA enzymes were assayed with each of 11 authentic PGN substrates produced by multistep chemical syntheses developed previously in our laboratory (Table 1; see also Fig. S2) (48, 49). Three different types of peptide were used to assess each activity observed above for LdcA—tetrapeptide for the l,d-carboxypeptidase activity, pentapeptide for the l,d-endopeptidase activity, and 4,3-cross-linked tetra-tripeptide for cross-linkage endopeptidase activity (Fig. 1). Substrate glycans also varied in length and composition (Table 1). Reactions were monitored by UPLC-MS, with products identified by an analysis of retention times, high-resolution mass measurements, and MS/MS spectra (Fig. 4 and 5; see also Fig. S3 to S6). Comparisons of reaction products to authentic synthetic standards were made whenever possible (Fig. 4 and 5; see also Fig. S3 to S6), and negative controls were included for most synthetic endopeptidase substrates (see Fig. S7 at https://figshare.com/articles/figure/Figure_S7_Negative_controls/16823347). In this section, we refer to specific substrates and products with a lowercase “s” and “p,” respectively, preceding a number referring to the structures shown in Fig. S2 to S6.
TABLE 1.
Reactions of synthetic PGNs with ApLdcA or EcLdcA, given as the percentage of product formed following treatment of different peptide (column headers) plus glycan (rows) substrates with enzyme (columns)
| Glycana | Tetrapeptide (Ala-Glu-mDap=Ala) |
Pentapeptide (Ala-Glu-mDap=Ala-Ala) |
Tetra-tripeptide (4,3) (Ala-Glu-mDap-Ala=mDap-Glu-Ala) |
|||
|---|---|---|---|---|---|---|
| ApLdcA | EcLdcA | ApLdcA | EcLdcA | ApLdcA | EcLdcA | |
| mM | 100 | 100 | 75 | 38 | 8/35 ∗4/6 |
7/22 ∗3/6 |
| aM | 100 | 100 | 84 | 41 | 18/66 | 13/29 |
| G-mM | NA | NA | 41/80 ∗0/12 |
18/56 ∗0/3 |
NA | NA |
| G-aM | 100 | 100 | 43/89 ∗0/5 |
8/25 ∗0/0 |
NA | NA |
| GMG-mM | NA | NA | 37/58 ∗0/12 |
14/51 ∗0/3 |
NA | NA |
| GMG-aM | 100 | 100 | NA | NA | NA | NA |
Product formation was determined after 2 h (single value) or at 2 and 24 h (values separated by “/”, respectively). Values preceded by an asterisk (∗) represent the percentage of N-acetyl muramyl l-Ala amidase product observed. For glycans, mM indicates that the MurNAc C-1 hydroxyl is replaced by β-OCH3, and aM indicates 1,6-anhydro-MurNAc. “NA” indicates where substrates were not available.
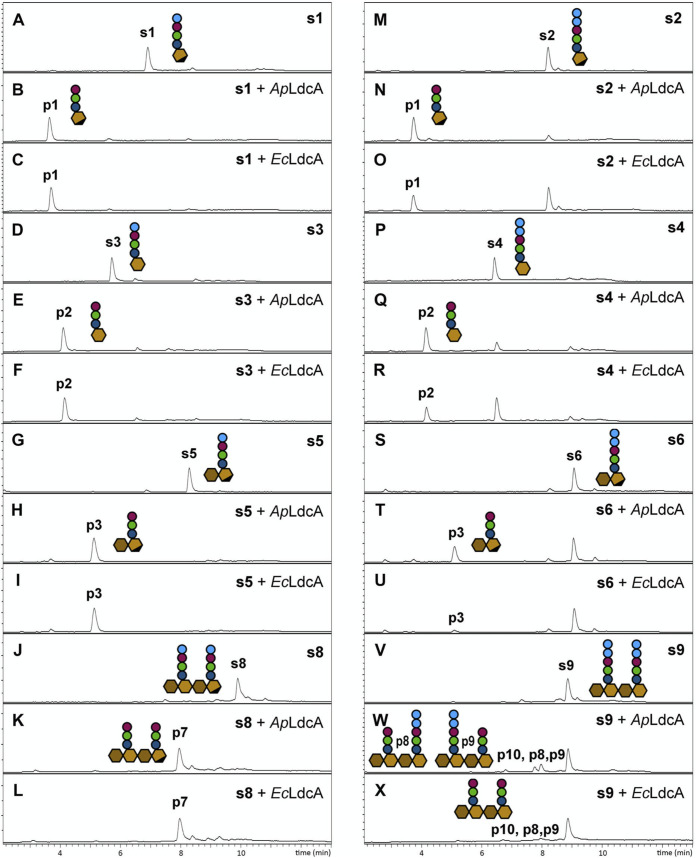
FIG 4.
Single-substrate assays demonstrating the l,d-carboxypeptidase and l,d-endopeptidase activities of LdcA enzymes on stem tetrapeptides (A to L) and pentapeptides (M to X), respectively. LC-MS traces are shown for each reaction. Chemical structures of substrates (preceded by an “s”) are shown in Fig. S2. MS data are shown in Fig. S3 to S5.
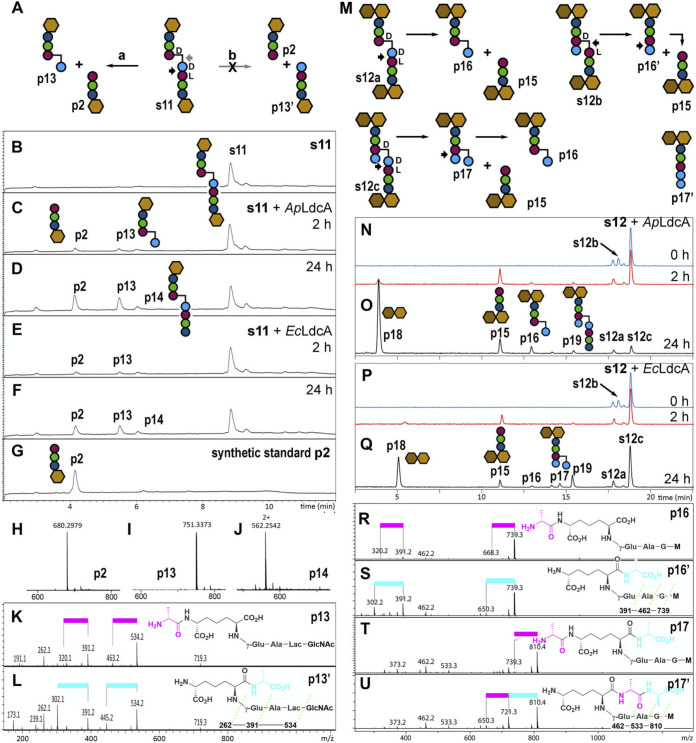
FIG 5.
Single-substrate assays demonstrating the l,d-endopeptidase activity of LdcA enzymes on cross-linked muropeptides. (A) Two hydrolyzable bonds in s11 indicated with black and gray arrows correspond to routes “a” and “b,” respectively. (B to G) LC-MS traces of LdcA reactions with s11. (H to J) Mass spectra of s11 reaction products p2, p13, and p14. (K and L) Collision-induced dissociation (CID) MS/MS spectra of p13 and p13′ confirm hydrolysis of s11 by route “a.” The pink bar indicates the loss of Ala from the N terminus (71 Da), while the blue bar indicates the loss of Ala from the C terminus (89 Da). (M to Q) Reactions of LdcA with s12 (M) and the resulting substrate and product LC-MS traces (N to Q). (R to U) CID MS/MS spectra of detected route “a” products (p16 and p17) and potential route “b” products (p16′ and p17′). Further LC-MS/MS data are shown in Fig. S6.
(A) Synthetic peptidoglycans (s1 to s11) and purified muropeptides (s12, a mixture of s12a, s12b, and s12c) used in this study. (B) Stem peptide chemical structures of l,d-carboxypeptidase and l,d-endopeptidase substrates and their corresponding cartoon representations. Each substrate shares the moiety within the dashed box. The white hexagon represents the variable glycan substitutions of substrates s1 to s12 (MurNAc, anhydro-MurNAc, GlcNAc-MurNAc, and GlcNAc-anhydro-MurNAc). Download FIG S2, TIF file, 0.7 MB (734.9KB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A, E, K, and O) Reactions of LdcA with monosaccharide compounds s1 to s4. LC-MS traces for substrates and products of reactions with s1 (B to D), s2 (F to I), s3 (L to N), and s4 (P to S). (J and T) Mass spectra of substrates, corresponding reaction products, and available synthetic standards. The m/z values shown for all MS peaks represent [M + H]+ ions. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A, M) Reactions of LdcA with cross-linked compounds s10 and s12, the latter representing a mixture of s12a, s12b, and s12c. The bond hydrolyzed by LdcA is shown in red. Two hydrolysable bonds in s10 indicated with green arrows correspond to reactions “a” and “b” of Fig. 5A, respectively. LC-MS traces for substrates and products of reactions with s10 (B to G) and s12 (N to T) are shown. (H to J, U) Mass spectra of substrate and reaction products. (K and L) CID MS/MS spectra of products p12 and s1. The pink bar indicates the loss of Ala at the N terminus (71 Da), while the blue bar indicates that of Ala at the C terminus (89 Da). The route “a” products were detected after treatment of s10 with LdcA (K). For each MS and MS/MS spectrum, the m/z values shown represent [M + H]+ ions unless labeled with “2+”, indicating an [M + 2H]2+ ion, and except for p18, which is an [M+Na]+ ion. Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All four tetrapeptide substrates, including three non-cross-linked muropeptides (s1, s3, and s5) and both stem peptides of a glycan-linked dimer (s8), were hydrolyzed completely to the corresponding tripeptide products by both EcLdcA and ApLdcA (Table 1, Fig. 4A to L; see also Fig. S3 to S5). Pentapeptide substrates (s2, s4, s6, s7, and s9), though not as rapidly consumed as tetrapeptide substrates, were also converted to tripeptide products by both EcLdcA and ApLdcA, with ApLdcA demonstrating higher turnover ability than EcLdcA with each substrate (Table 1 and Fig. 4M to X; see also Fig. S3 to S5). This disparity appears even more pronounced with disaccharide-pentapeptides (s6 and s7; Table 1; see also Fig. 4S to U) and glycan-linked pentapeptide dimers (s9; Table 1 and Fig. 4V to X; see also Fig. S5) than with monosaccharide pentapeptides (s2 and s4; Table 1 and Fig. 4M to R; see also Fig. S3). No products were detected in 24-h control reactions of pentapeptide substrates with either heat-inactivated enzymes or bovine serum albumin (BSA; see Fig. S7 at https://figshare.com/articles/figure/Figure_S7_Negative_controls/16823347), indicating that noncatalytic degradation does not occur under the conditions employed. It is possible, however, that trace amounts of unidentified peptidoglycan-degrading enzymes were copurified with the recombinant LdcA enzymes, such that products accumulated only after long incubation times. Tandem MS fragmentation patterns of tetra- and pentapeptide substrates and products are shown in Fig. S5.
(A, E, and R) Reactions of LdcA with disaccharide compounds s5 to s7. LC-MS traces for substrates and products of reactions with s5 (B to D), s6 (F to K), and s7 (S to X). (L to Q, Y) Mass spectra of substrates, reaction products, and synthetic standards. The m/z values shown for all MS peaks represent [M + H]+ ions. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and E) Reactions of LdcA with dimeric compounds s8 and s9. LC-MS traces for substrates and products of reactions with s8 (B to D) and s9 (F to L). (M to S) Mass spectra of substrates and reaction products. The m/z values shown in panels M to S represent [M + 2H]2+ ions. The glycan GmM represents GlcNAc-MurNAc with the C-1 hydroxyl of MurNAc replaced by β-OCH3. (T) Structure elucidation of reaction substrates and products by CID MS/MS. The m/z values shown in T represent [M + H]+ ions. The red bar indicates the loss of CH4O (32 Da). Download FIG S5, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We confirmed that LdcA exhibits cross-linkage endopeptidase activity and found that LdcA carries out a reaction that is not performed by any known endopeptidase. For 4,3-cross-linked tetra-tripeptide substrates (s10 and s11), there are two potential hydrolysable bonds (Fig. 5; see also Fig. S6), both of which yield tripeptide and tetrapeptide products. While the tripeptide product from either reaction is identical, the tetrapeptide products, though equivalent in mass, differ by the position of d-Ala on either the mDap side chain (Fig. 5A; see also Fig. S6, route “a”) or the main chain (Fig. 5A; see also Fig. S6, route “b”). The two products are readily differentiated from their MS/MS fragmentation patterns, revealing that both EcLdcA and ApLdcA proceed through route “a” (Fig. 5K and L; see also Fig. S6), reinforcing the specificity of LdcA for cleavage of l,d-amide bonds. This specificity distinguishes LdcA endopeptidase activity from that of the d,d-endopeptidase activity of Pseudomonas aeruginosa penicillin-binding protein 4, which we previously showed turns over the same cross-linked substrate by route “b” (49). ApLdcA was more active than EcLdcA toward both cross-linked substrates (s10 and s11; Table 1 and Fig. 5B to G; see also Fig. S6), suggesting some differences in glycan preference. No substrate degradation or product formation was observed when 4,3-cross-linked substrates were treated with heat-inactivated enzymes or BSA (see Fig. S7 at https://figshare.com/articles/figure/Figure_S7_Negative_controls/16823347).
In our automated analysis, we observed a decreased abundance of 3,3-cross-linked tri-tripeptide compounds following LdcA treatment (Fig. 3B), but did not have synthetic 3,3-cross-linked substrates readily available to confirm this reaction. To address this remaining question, we purified a mixture of cross-linked substrates from mutanolysin-derived E. coli muropeptides (s12), containing 4,3-cross-linked tetra-tripeptide (s12a), 3,3-cross-linked tri-tetrapeptide (s12b), and 4,3-cross-linked tetra-tetrapeptide (s12c) in a ratio of ∼1:1:5, respectively. Figure 5M illustrates the reaction of LdcA with these three substrates. Reaction mixtures showed complete turnover of the 3,3-cross-linked s12b and partial turnover of 4,3-cross-linked compounds s12a and s12c, demonstrating that both ApLdcA and EcLdcA are more active against 3,3-cross-linkages than 4,3-cross-linkages (Fig. 5N to Q; see also Fig. S6). MepK is the only other enzyme known to display l,d-endopeptidase activity toward 3,3-cross-linkages (50). MepK is also capable of cleaving 4,3-cross-linkages, but via d,d-endopeptidase activity (Fig. 5M, route “b”) (50). Reaction of the 4,3-cross-linked substrates with LdcA was evident by the accumulation of route “a”-type tetrapeptide (p16) and pentapeptide (p17) products (Fig. 5N to Q). We confirmed that p16 and p17 are not the mass-equivalent route “b” products (p16′ and p17′) by comparison of their MS/MS fragmentation patterns (Fig. 5R to U). While p16 represents a reaction end product, p17 can be further converted to p16 by the l,d-carboxypeptidase activity of LdcA. Reactions with ApLdcA showed accumulation of p16 only, while EcLdcA treatment produced more p17 than p16 (Fig. 5N to Q; see also Fig. S6), suggesting that the conversion of p17 to p16 proceeds more slowly for EcLdcA than ApLdcA. This result may explain why the proportion of stem pentapeptides in mutanolysin-derived E. coli PGN decreased after treatment with ApLdcA but increased for EcLdcA (Fig. 3B).
Taken together, these results confirm the majority of our conclusions from the proteomic analysis of LdcA treatment on E. coli PGN (Fig. 3), demonstrating that: (i) EcLdcA and ApLdcA act as l,d-carboxypeptidases with un-cross-linked stem tetrapeptides and as l,d-endopeptidases with both 4,3- and 3,3-stem peptide cross-linkages, and (ii) ApLdcA exhibits increased turnover of endopeptidase substrates relative to EcLdcA (Table 1). The LdcA homolog from N. gonorrhoeae is also capable of hydrolyzing 3,3-cross-linkages in vitro; although the exact bond cleavage was not determined in that case, this finding suggests that the endopeptidase activity of this enzyme exhibits the same specificity for l,d-amide bonds that we observed for EcLdcA and ApLdcA (30). In addition, whereas our automated analysis successfully identified ApLdcA as an l,d-endopeptidase with regard to un-cross-linked stem pentapeptides, single substrate analysis revealed that EcLdcA, too, exhibits this activity (Table 1).
DISCUSSION
Despite its role in PGN recycling, a cytoplasmic process involving soluble muropeptides, there is growing evidence that LdcA is an efficient starting point for the evolution of novel enzyme activities that modify the cell wall (29, 30). In aphids, the use of LdcA in their symbiotic association with Buchnera implies a derived ability of ApLdcA to target symbiont cell walls. We found by biochemical characterization that both EcLdcA and ApLdcA exhibit l,d-carboxypeptidase and l,d-endopeptidase activities. The endopeptidase activities of EcLdcA might be considered promiscuous, since the cytoplasmic EcLdcA does not encounter cross-linked stem peptides in nature. In contrast, ApLdcA is likely to encounter cross-linked Buchnera PGN because the enzyme is produced by the host, outside of symbiont cells. Whether ApLdcA is transported into the Buchnera periplasm, where it directly remodels the cell wall, or is present in the bacteriocyte cytoplasm and acts on soluble muropeptides released during Buchnera cell wall remodeling is unknown. There is evidence of host protein transport to Buchnera for at least one host HTG, RlpA4 (51), although this enzyme contains a eukaryotic signal peptide that is absent from ApLdcA (23). Because the endopeptidase activities of EcLdcA are shared by ApLdcA but are likely physiologically relevant in aphids, our results suggest an evolutionary link between the inherent enzyme promiscuity of EcLdcA and the higher turnover of endopeptidase substrates by some extant LdcA enzymes, including ApLdcA.
Catalytically promiscuous enzyme reactions vary greatly in magnitude among related proteins, sometimes approaching a level of efficiency similar to that of their primary functions (52). In addition, many models of protein evolution emphasize enzymatic tradeoffs between promiscuous and primary activities (1). Nonetheless, ApLdcA and some other multifunctional LdcA enzymes retain a high level of l,d-carboxypeptidase activity (Fig. 3) (29–31). Thus, enzyme function is not easily distinguished from promiscuity without a demonstrable role in biology. We provide evidence that each catalytic activity of ApLdcA plays a role in the aphid-Buchnera symbiosis, implying selection for multifunctionality. Some of the muropeptides identified from aphid homogenate are likely the products of Buchnera cell wall digestion by ApLdcA, implying its functionality in vivo (Fig. 2). Specifically, tripeptide monomers can only be formed from tetrapeptides or by the cleavage of stem peptide cross-linkages via l,d-carboxypeptidase or endopeptidase activities, respectively, both of which can be catalyzed by ApLdcA and are not encoded elsewhere in the Buchnera or host genomes. Though additional enzyme functions cannot be ruled out for the other PGN-modifying enzymes that are retained in the Buchnera genome, our data indicate that these roles can be fulfilled by the catalytic functions of ApLdcA.
Several LdcA enzymes from bacteria have now been biochemically characterized and, in conjunction with our own results, some patterns emerge that hint at the evolutionary origin of enhanced endopeptidase functions within the LdcA family. The A. pisum ldcA gene is thought to have been acquired by an ancient aphid ancestor from Wolbachia-like bacteria (22), which are frequently associated with arthropod hosts as intracellular pathogens or facultative symbionts (53). Given that all other LdcA enzymes known to target cell wall and display endopeptidase activity are derived from intracellular bacteria, we hypothesize that the ldcA gene originally acquired by ancestral aphids already exhibited these abilities and provided an immediate advantage to the insect host, possibly enabling aphids to establish the high degree of control over Buchnera that exists today. An analysis of crosswise promiscuity within the LdcA family that includes both extant and ancestrally reconstructed enzymes of free-living, intracellular, and aphid origins could be used to test this idea.
Besides ApLdcA, aphids harbor six other HTGs with putative roles in PGN remodeling and symbiosis: amiD, encoding an amidase, and rlpA1 to rlpA5, all encoding lytic transglycosylases (Fig. 1). Together with ApLdcA, they possess each of the necessary enzyme functions required for PGN remodeling in free-living bacteria (54). Based on their high levels of bacteriocyte-specific expression (23), their importance for both aphid and symbiont growth (24), and their correlation with high symbiont abundance among distinct aphid genotypes (25), these genes appear to contribute to Buchnera proliferation. HTGs could be involved in releasing cell wall fragments that mediate host-microbe interactions (55) or in degrading those fragments as a means for hosts to curb their own immune response to indigenous microbiota (56, 57). However, because aphids lack PGRPs (58), it seems unlikely that Buchnera cell wall fragments affect aphid hosts at all. Though it is possible that some other aphid signaling pathway has been coopted for recognition of Gram-negative PGN, we propose an alternative hypothesis in which host control of PGN metabolism enables aphids to regulate symbiont PGN metabolism and regulate their growth and/or cell division.
Multifunctionality is apparently common among individual PGN hydrolase domains (29, 30, 59–61), suggesting that multifunctionality in the other aphid HTGs or in Buchnera’s own PGN remodeling enzymes may exist. For example, Buchnera contains AmiB and the typically nonenzymatic NlpD. In E. coli, NlpD is the designated activator of AmiC, while EnvC, missing in Buchnera, activates AmiA and AmiB (62). In Waddlia chondrophila and Chlamydia pneumoniae, NlpD acts as a bifunctional d,d-carboxypeptidase and d,d-endopeptidase, independent of any amidase (59, 60). In addition, E. coli PBP1B exhibits d,d-carboxypeptidase activity under acidic conditions and in the presence of its activator, LpoB (61)—both of these proteins are encoded by Buchnera. Thus, in addition to ApLdcA, either Buchnera NlpD or PBP1B could contribute to the production of trimmed stem peptides among muropeptides from aphid homogenate (Fig. 2). In another example of multifunctionality, Buchnera lacks Alr and DadX, each capable of producing d-Ala for lipid II biosynthesis via alanine-racemase activity, but contains GlyA, to which the alanine-racemase activity of C. pneumoniae has been attributed (63). Our results support the idea that aphid PGN hydrolases are involved in Buchnera PGN metabolism, but further interrogation of these pathways is required to understand how host enzymes contribute. To add another layer of complexity, Buchnera symbionts of different aphid species vary greatly in PGN gene repertoire (64, 65), which could translate to substantial differences in cell wall and/or enzyme chemistry depending on their metabolic needs.
In conclusion, aphids encode each of the three enzyme functions required for PGN remodeling. While both EcLdcA and ApLdcA behave as endopeptidases, ApLdcA exhibits enhanced endopeptidase activity toward pentapeptides and cross-linked dimers, revealing adaptations in this enzyme specific to the aphid-Buchnera system. Host acquisition and retention of these HTGs throughout aphid evolution reflect their importance in symbiosis. Potentially, their acquisition by aphid ancestors was instrumental in establishing control over symbionts that has since evolved further in extant aphids.
MATERIALS AND METHODS
Purification of PGN.
Buchnera muropeptides were purified from homogenates of 7-day-old fourth-instar A. pisum nymphs by successive filtration. The filtrate was subjected to HPLC, and the collected fractions were subjected to proteomic analysis. Whole E. coli DH5α murein sacculus was purified following the methods of Desmarais et al. (66). More details are provided in the supplemental Materials and Methods (see Text S1).
Supplemental results: additional single-substrate enzyme assay results reporting the minor accumulation of amidase products in LdcA enzyme reactions. Supplemental methods: detailed descriptions of the methods used in this work. Download Text S1, DOCX file, 0.08 MB (84.3KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein production and purification.
The A. pisum and E. coli ldcA genes were amplified by PCR using primers in Table S2, then cloned into the pET-28b plasmid. E. coli Rosetta (DE3) was transformed with plasmid and induced with isopropyl β-d-1-thioglactopyranoside (IPTG). Proteins were purified from cell pellets by (i) lysis with HEWL and high-pressure homogenization, (ii) binding to Ni-NTA IMAC resin, and (iii) elution with imidazole.
Primers used to construct plasmids for recombinant protein expression. Tm values were calculated for the specific DNA polymerase used for each primer set. Sequences in boldface denote restriction-enzyme recognition sites, while underlined sequences highlight the target sequence used to calculate the Tm. Download Table S2, DOCX file, 0.06 MB (61.2KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteomics-based muropeptide analysis.
E. coli sacculus was digested with mutanolysin (Sigma-Aldrich) and the resulting muropeptides treated with sodium borohydride and desalted using HPLC. Muropeptides were incubated with LdcA enzyme and then desalted again by HPLC. Treated samples were subjected to nano LC-MS/MS analysis following the methods of Bern et al. (37). Individual PGN compounds were identified and quantified from MS and MS/MS spectra using Byonic and Byologic software, respectively (Protein Metrics). All MS data and associated Byonic and Byologic files are available for download via the mass spectrometry database MassIVE (MSV000087634). Data transformation, statistical comparison, and plotting were accomplished using custom R scripts, available along with raw and transformed data at GitHub (https://github.com/smit4227/ApLdcA_proteomics).
Single-substrate enzyme assays.
Synthetic PGN substrates (s1 to s11) used in this study were synthesized using previously reported methods (48, 49). The muropeptide mixture s12 was purified from mutanolysin-derived E. coli muropeptides by HPLC. Reactions of LdcA enzymes with synthetic PGNs were stopped at different time points (2, 8, and 24 h) and analyzed by UPLC-MS. Reactions of LdcA enzymes with s12 were carried out under the same conditions, except that reaction mixtures were reduced with sodium borohydride after treatment.
ACKNOWLEDGMENTS
This study was supported by NIH awards F32GM126706 (to T.E.S.), R35GM131738 (to N.A.M.), and R35GM131685 (to S.M.) and NSF award 1551092 (to N.A.M.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank the DNA Sequencing and Proteomics Facilities at the University of Texas at Austin for Sanger sequencing and mass spectrometry services, the lab of Andreas Matouschek for use of lab equipment, the lab of David Taylor for assistance with protein purification by FPLC, and Marshall Bern and James Moore for providing academic licenses for Protein Metrics softwares Byonic and Byologic.
T.E.S., N.A.M., and S.M. conceptualized the research idea. T.E.S., M.L., and S.M. developed the methodology. T.E.S. and M.L. performed all experimental investigations. M.L., D.H., and M.D.P. provided the resources used in this study in the form of synthesis of PGN substrates (M.L. and D.H.) and LC-MS instrumentation and operation (M.D.P.). T.E.S. performed all other formal analyses. N.A.M. and S.M. supervised all research activities. T.E.S. and M.L. prepared figures and tables for data visualization. T.E.S. wrote the original draft of the manuscript with contributions from M.L. and M.D.P., while review and editing was completed by T.E.S., M.L., M.D.P., S.M., and N.A.M.
Contributor Information
Thomas E. Smith, Email: smit4227@gmail.com.
Joerg Graf, University of Connecticut.
REFERENCES
- 1.Glasner ME, Truong DP, Morse BC. 2020. How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation. FEBS J 287:1323–1342. doi: 10.1111/febs.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD. 2010. Three serendipitous pathways in Escherichia coli can bypass a block in pyridoxal‐5′‐phosphate synthesis. Mol Syst Biol 6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juárez-Vázquez AL, Edirisinghe JN, Verduzco-Castro EA, Michalska K, Wu C, Noda-García L, Babnigg G, Endres M, Medina-Ruíz S, Santoyo-Flores J, Carrillo-Tripp M, Ton-That H, Joachimiak A, Henry CS, Barona-Gómez F. 2017. Evolution of substrate specificity in a retained enzyme driven by gene loss. Elife 6:e22679. doi: 10.7554/eLife.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontrelli S, Fricke RCB, Teoh ST, Laviña WA, Putri SP, Fitz-Gibbon S, Chung M, Pellegrini M, Fukusaki E, Liao JC. 2018. Metabolic repair through emergence of new pathways in Escherichia coli. Nat Chem Biol 14:1005–1009. doi: 10.1038/s41589-018-0149-6. [DOI] [PubMed] [Google Scholar]
- 5.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 6.Kelkar YD, Ochman H. 2013. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193:303–307. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton MS, Arcus VL, Gerth ML, Patrick WM. 2018. Enzyme evolution: innovation is easy, optimization is complicated. Curr Opin Struct Biol 48:110–116. doi: 10.1016/j.sbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Adams NE, Thiaville JJ, Proestos J, Juárez-Vázquez AL, McCoy AJ, Barona-Gómez F, Iwata-Reuyl D, de Crécy-Lagard V, Maurelli AT. 2014. Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. mBio 5:e01378-14–e01314. doi: 10.1128/mBio.01378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liechti G, Singh R, Rossi PL, Gray MD, Adams NE, Maurelli AT. 2018. Chlamydia trachomatis dapF encodes a bifunctional enzyme capable of both d-glutamate racemase and diaminopimelate epimerase activities. mBio 9:e00204-18. doi: 10.1128/mBio.00204-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferla MP, Brewster JL, Hall KR, Evans GB, Patrick WM. 2017. Primordial‐like enzymes from bacteria with reduced genomes. Mol Microbiol 105:508–524. doi: 10.1111/mmi.13737. [DOI] [PubMed] [Google Scholar]
- 11.Price DR, Wilson AC. 2014. A substrate ambiguous enzyme facilitates genome reduction in an intracellular symbiont. BMC Biol 12:110. doi: 10.1186/s12915-014-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husnik F, McCutcheon JP. 2018. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol 16:67–79. doi: 10.1038/nrmicro.2017.137. [DOI] [PubMed] [Google Scholar]
- 13.Jackson DJ, Macis L, Reitner J, Wörheide G. 2011. A horizontal gene transfer supported the evolution of an early metazoan biomineralization strategy. BMC Evol Biol 11:238. doi: 10.1186/1471-2148-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, Novelli J, Jiang D, Dailey HA, Landmann F, Ford L, Taylor MJ, Carlow CKS, Kumar S, Foster JM, Slatko BE. 2013. Interdomain lateral gene transfer of an essential ferrochelatase gene in human parasitic nematodes. Proc Natl Acad Sci USA 110:7748–7753. doi: 10.1073/pnas.1304049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wybouw N, Dermauw W, Tirry L, Stevens C, Grbić M, Feyereisen R, Van Leeuwen T. 2014. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife 3:e02365. doi: 10.7554/eLife.02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelomi M, Danchin EGJ, Heckel D, Wipfler B, Bradler S, Zhou X, Pauchet Y. 2016. Horizontal gene transfer of pectinases from bacteria preceded the diversification of stick and leaf insects. Sci Rep 6:26388. doi: 10.1038/srep26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams KL, Palmer JD. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet E 29:380–395. doi: 10.1016/S1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 19.Nakabachi A. 2015. Horizontal gene transfers in insects. Curr Opin Insect Sci 7:24–29. doi: 10.1016/j.cois.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson ACC, von Dohlen CD, Fukatsu T, McCutcheon JP. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Bublitz DC, Chadwick GL, Magyar JS, Sandoz KM, Brooks DM, Mesnage S, Ladinsky MS, Garber AI, Bjorkman PJ, Orphan VJ, McCutcheon JP. 2019. Peptidoglycan production by an insect-bacterial mosaic. Cell 179:703–712.e7. doi: 10.1016/j.cell.2019.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikoh N, Nakabachi A. 2009. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol 7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikoh N, McCutcheon JP, Kudo T, Miyagishima S-y, Moran NA, Nakabachi A. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet 6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung SH, Jing X, Luo Y, Douglas AE. 2018. Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem Mol Biol 95:55–63. doi: 10.1016/j.ibmb.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Smith TE, Moran NA. 2020. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc Natl Acad Sci USA 117:2113–2121. doi: 10.1073/pnas.1916748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Templin MF, Ursinus A, Höltje J-V. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J 18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otten C, Brilli M, Vollmer W, Viollier PH, Salje J. 2018. Peptidoglycan in obligate intracellular bacteria. Mol Microbiol 107:142–163. doi: 10.1111/mmi.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zellner B, Mengin-Lecreulx D, Tully B, Gunning WT, Booth R, Huntley JF. 2021. A Francisella tularensis l,d-carboxypeptidase plays important roles in cell morphology, envelope integrity, and virulence. Mol Microbiol 115:1357–1378. doi: 10.1111/mmi.14685. [DOI] [PubMed] [Google Scholar]
- 30.Lenz JD, Hackett KT, Dillard JP. 2017. A single dual-function enzyme controls the production of inflammatory NOD agonist peptidoglycan fragments by Neisseria gonorrhoeae. mBio 8:e01464-17. doi: 10.1128/mBio.01464-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das D, Hervé M, Elsliger M-A, Kadam RU, Grant JC, Chiu H-J, Knuth MW, Klock HE, Miller MD, Godzik A, Lesley SA, Deacon AM, Mengin-Lecreulx D, Wilson IA. 2013. Structure and function of a novel ld-carboxypeptidase A involved in peptidoglycan recycling. J Bacteriol 195:5555–5566. doi: 10.1128/JB.00900-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 33.Benedetti SD, Fisher JF, Mobashery S. 2021. Bacterial cell wall: morphology and biochemistry, p 167–204. In Goldman E, Green LH (ed), Practical handbook of microbiology, 4th ed. Taylor & Francis, New York, NY. [Google Scholar]
- 34.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dik DA, Fisher JF, Mobashery S. 2018. Cell-wall recycling of the Gram-negative bacteria and the nexus to antibiotic resistance. Chem Rev 118:5952–5984. doi: 10.1021/acs.chemrev.8b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner RD, Vollmer W, Foster SJ. 2014. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bern M, Beniston R, Mesnage S. 2017. Towards an automated analysis of bacterial peptidoglycan structure. Anal Bioanal Chem 409:551–560. doi: 10.1007/s00216-016-9857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verwer RW, Beachey EH, Keck W, Stoub AM, Poldermans JE. 1980. Oriented fragmentation of Escherichia coli sacculi by sonication. J Bacteriol 141:327–332. doi: 10.1128/jb.141.1.327-332.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dik DA, Marous DR, Fisher JF, Mobashery S. 2017. Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit Rev Biochem Mol Biol 52:503–542. doi: 10.1080/10409238.2017.1337705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen AK, Moran NA. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA 108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. 2008. Identification of the l,d-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol 190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. 2011. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J 30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. 2009. d-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández SB, Dörr T, Waldor MK, Cava F. 2020. Modulation of peptidoglycan synthesis by recycled cell wall tetrapeptides. Cell Rep 31:107578. doi: 10.1016/j.celrep.2020.107578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun V, Sieglin U. 1970. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. Eur J Biochem 13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 46.Sandoz KM, Moore RA, Beare PA, Patel AV, Smith RE, Bern M, Hwang H, Cooper CJ, Priola SA, Parks JM, Gumbart JC, Mesnage S, Heinzen RA. 2021. β-Barrel proteins tether the outer membrane in many Gram-negative bacteria. Nat Microbiol 6:19–26. doi: 10.1038/s41564-020-00798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leguina JI, Quintela JC, de Pedro MA. 1994. Substrate specificity of Escherichia coli ld‐carboxypeptidase on biosynthetically modified muropeptides. FEBS Lett 339:249–252. doi: 10.1016/0014-5793(94)80425-7. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. 2013. Reactions of the three AmpD enzymes of Pseudomonas aeruginosa. J Am Chem Soc 135:4950–4953. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee M, Hesek D, Blázquez B, Lastochkin E, Boggess B, Fisher JF, Mobashery S. 2015. Catalytic spectrum of the penicillin-binding protein 4 of Pseudomonas aeruginosa, a nexus for the induction of β-lactam antibiotic resistance. J Am Chem Soc 137:190–200. doi: 10.1021/ja5111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chodisetti PK, Reddy M. 2019. Peptidoglycan hydrolase of an unusual cross-link cleavage specificity contributes to bacterial cell wall synthesis. Proc Natl Acad Sci USA 116:7825–7830. doi: 10.1073/pnas.1816893116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima S. 2014. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol 24:R640–R641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 52.Khersonsky O, Tawfik DS. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 53.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 54.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 55.Irazoki O, Hernandez SB, Cava F. 2019. Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol 10:500. doi: 10.3389/fmicb.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maire J, Vincent-Monégat C, Balmand S, Vallier A, Hervé M, Masson F, Parisot N, Vigneron A, Anselme C, Perrin J, Orlans J, Rahioui I, Da Silva P, Fauvarque M-O, Mengin-Lecreulx D, Zaidman-Rémy A, Heddi A. 2019. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc Natl Acad Sci USA 116:5623–5632. doi: 10.1073/pnas.1821806116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Troha K, Nagy P, Pivovar A, Lazzaro BP, Hartley PS, Buchon N. 2019. Nephrocytes remove microbiota-derived peptidoglycan from systemic circulation to maintain immune homeostasis. Immunity 51:625–637. doi: 10.1016/j.immuni.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, de Vos M, Duncan EJ, Evans JD, Gabaldón T, Ghanim M, Heddi A, Kaloshian I, Latorre A, Moya A, Nakabachi A, Parker BJ, Pérez-Brocal V, Pignatelli M, Rahbé Y, Ramsey JS, Spragg CJ, Tamames J, Tamarit D, Tamborindeguy C, Vincent-Monegat C, Vilcinskas A. 2010. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol 11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frandi A, Jacquier N, Théraulaz L, Greub G, Viollier PH. 2014. FtsZ-independent septal recruitment and function of cell wall remodelling enzymes in chlamydial pathogens. Nat Commun 5:4200. doi: 10.1038/ncomms5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klöckner A, Otten C, Derouaux A, Vollmer W, Bühl H, De Benedetti S, Münch D, Josten M, Mölleken K, Sahl H-G, Henrichfreise B. 2014. AmiA is a penicillin target enzyme with dual activity in the intracellular pathogen Chlamydia pneumoniae. Nat Commun 5:4201. doi: 10.1038/ncomms5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egan AJF, Biboy J, van’t Veer I, Breukink E, Vollmer W. 2015. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc B 370:20150031. doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uehara T, Parzych KR, Dinh T, Bernhardt TG. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J 29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Benedetti S, Bühl H, Gaballah A, Klöckner A, Otten C, Schneider T, Sahl H-G, Henrichfreise B. 2014. Characterization of serine hydroxymethyltransferase GlyA as a potential source of d-alanine in Chlamydia pneumoniae. Front Cell Infect Microbiol 4:19. doi: 10.3389/fcimb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong RA, Park H, Moran NA. 2019. Genome evolution of the obligate endosymbiont Buchnera aphidicola. Mol Biol Evol 36:1481–1489. doi: 10.1093/molbev/msz082. [DOI] [PubMed] [Google Scholar]
- 65.Atwal S, Chuenklin S, Bonder EM, Flores J, Gillespie JJ, Driscoll TP, Salje J. 2021. Discovery of a diverse set of bacteria that build their cell walls without the canonical peptidoglycan polymerase aPBP. mBio 12:e01342-21. doi: 10.1128/mBio.01342-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desmarais SM, Cava F, de Pedro MA, Huang KC. 2014. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp 2014:e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uehara T, Park JT. 2007. An anhydro-N-acetylmuramyl-l-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J Bacteriol 189:5634–5641. doi: 10.1128/JB.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jorgenson MA, Chen Y, Yahashiri A, Popham DL, Weiss DS. 2014. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol Microbiol 93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of Buchnera-derived and LdcA-treated E. coli muropeptides as determined by LC-MS/MS. (A and B) Noncanonical non-cross-linked (A) and singly cross-linked (B) muropeptides identified from A. pisum homogenate (Fig. 2). Untreated E. coli mutanolysin-derived muropeptides are shown for comparison (purple). Two treatments were used without replicates: aphid homogenate was either sonicated to lyse Buchnera cells (light gray) or sonicated and additionally treated with lysozyme to digest Buchnera cell walls (dark gray). (C and D) Protein fractions generated during IMAC purification of recombinantly expressed A. pisum LdcA (C) and E. coli LdcA (D). For each SDS-PAGE gel, fractions analyzed included the insoluble pellet (P), soluble cell lysate (L), Ni-NTA flowthrough (FT), lysis buffer wash (W1), HEPES buffer wash (W2), and HEPES-buffered elutions of increasing imidazole concentrations: 50 mM (E1), 100 mM (E2), 200 mM (E3), and 500 mM (E4). (E to G) Noncanonical non-cross-linked (E), noncanonical singly cross-linked (F), and canonical doubly cross-linked (G) muropeptides after treatment of mutanolysin-derived E. coli muropeptides (purple) with EcLdcA (blue) and ApLdcA (green) (Fig. 3). All stem peptide sequences consist of l-Ala, γ-d-Glu, and meso-l-Dap, followed by the single-letter amino acid code indicated in the compound table. The absence of an amino acid indicates that the tripeptide is the complete stem-peptide sequence. Glycan substituents include GlcNAc-MurNAc (GM) and GlcNAc-anhydro-MurNAc (GaM). Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differences in average percent abundance of total PGN per compound between mutanolysin-digest of E. coli sacculus (E. coli), muropeptides isolated from A. pisum homogenate (Buchnera), and E. coli PGN treated with EcLdcA (EcLdcA) and ApLdcA (ApLdcA). Download Table S1, DOCX file, 0.07 MB (74.8KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Synthetic peptidoglycans (s1 to s11) and purified muropeptides (s12, a mixture of s12a, s12b, and s12c) used in this study. (B) Stem peptide chemical structures of l,d-carboxypeptidase and l,d-endopeptidase substrates and their corresponding cartoon representations. Each substrate shares the moiety within the dashed box. The white hexagon represents the variable glycan substitutions of substrates s1 to s12 (MurNAc, anhydro-MurNAc, GlcNAc-MurNAc, and GlcNAc-anhydro-MurNAc). Download FIG S2, TIF file, 0.7 MB (734.9KB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A, E, K, and O) Reactions of LdcA with monosaccharide compounds s1 to s4. LC-MS traces for substrates and products of reactions with s1 (B to D), s2 (F to I), s3 (L to N), and s4 (P to S). (J and T) Mass spectra of substrates, corresponding reaction products, and available synthetic standards. The m/z values shown for all MS peaks represent [M + H]+ ions. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A, M) Reactions of LdcA with cross-linked compounds s10 and s12, the latter representing a mixture of s12a, s12b, and s12c. The bond hydrolyzed by LdcA is shown in red. Two hydrolysable bonds in s10 indicated with green arrows correspond to reactions “a” and “b” of Fig. 5A, respectively. LC-MS traces for substrates and products of reactions with s10 (B to G) and s12 (N to T) are shown. (H to J, U) Mass spectra of substrate and reaction products. (K and L) CID MS/MS spectra of products p12 and s1. The pink bar indicates the loss of Ala at the N terminus (71 Da), while the blue bar indicates that of Ala at the C terminus (89 Da). The route “a” products were detected after treatment of s10 with LdcA (K). For each MS and MS/MS spectrum, the m/z values shown represent [M + H]+ ions unless labeled with “2+”, indicating an [M + 2H]2+ ion, and except for p18, which is an [M+Na]+ ion. Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A, E, and R) Reactions of LdcA with disaccharide compounds s5 to s7. LC-MS traces for substrates and products of reactions with s5 (B to D), s6 (F to K), and s7 (S to X). (L to Q, Y) Mass spectra of substrates, reaction products, and synthetic standards. The m/z values shown for all MS peaks represent [M + H]+ ions. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and E) Reactions of LdcA with dimeric compounds s8 and s9. LC-MS traces for substrates and products of reactions with s8 (B to D) and s9 (F to L). (M to S) Mass spectra of substrates and reaction products. The m/z values shown in panels M to S represent [M + 2H]2+ ions. The glycan GmM represents GlcNAc-MurNAc with the C-1 hydroxyl of MurNAc replaced by β-OCH3. (T) Structure elucidation of reaction substrates and products by CID MS/MS. The m/z values shown in T represent [M + H]+ ions. The red bar indicates the loss of CH4O (32 Da). Download FIG S5, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental results: additional single-substrate enzyme assay results reporting the minor accumulation of amidase products in LdcA enzyme reactions. Supplemental methods: detailed descriptions of the methods used in this work. Download Text S1, DOCX file, 0.08 MB (84.3KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to construct plasmids for recombinant protein expression. Tm values were calculated for the specific DNA polymerase used for each primer set. Sequences in boldface denote restriction-enzyme recognition sites, while underlined sequences highlight the target sequence used to calculate the Tm. Download Table S2, DOCX file, 0.06 MB (61.2KB, docx) .
Copyright © 2021 Smith et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.