ABSTRACT
The world was unprepared for coronavirus disease 2019 (COVID-19) and remains ill-equipped for future pandemics. While unprecedented strides have been made developing vaccines and treatments for COVID-19, there remains a need for highly effective and widely available regimens for ambulatory use for novel coronaviruses and other viral pathogens. We posit that a priority is to develop pan-family drug cocktails to enhance potency, limit toxicity, and avoid drug resistance. We urge cocktail development for all viruses with pandemic potential both in the short term (<1 to 2 years) and longer term with pairs of drugs in advanced clinical testing or repurposed agents approved for other indications. While significant efforts were launched against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in vitro and in the clinic, many studies employed solo drugs and had disappointing results. Here, we review drug combination studies against SARS-CoV-2 and other viruses and introduce a model-driven approach to assess drug pairs with the highest likelihood of clinical efficacy. Where component agents lack sufficient potency, we advocate for synergistic combinations to achieve therapeutic levels. We also discuss issues that stymied therapeutic progress against COVID-19, including testing of agents with low likelihood of efficacy late in clinical disease and lack of focus on developing virologic surrogate endpoints. There is a need to expedite efficient clinical trials testing drug combinations that could be taken at home by recently infected individuals and exposed contacts as early as possible during the next pandemic, whether caused by a coronavirus or another viral pathogen. The approach herein represents a proactive plan for global viral pandemic preparedness.
KEYWORDS: SARS-CoV-2, COVID-19, viral pandemic, pandemic preparedness, antiviral drugs, drug synergy, model-driven approach, prophylaxis, early treatment, category A-C pathogens, Ebola virus, countermeasures
INTRODUCTION
The nucleotide sequence for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that sparked the global coronavirus disease 2019 (COVID-19) pandemic, was released in January 2020. The scientific community rallied with dedication, efficiency, and skill such that novel vaccines and therapeutic antibodies demonstrated safety and efficacy in clinical trials and were authorized for administration by December 2020. Nevertheless, and despite the very recent arrival of promising antiviral drugs, the scale of death and illness as well as economic and social havoc remains unprecedented. The lack of readily available, widely implemented therapies with utility before the onset of severe complications continues to contribute to high global mortality. For the present pandemic response, and for future pandemics, whether from a coronaviruses or another virus, the scientific community must be ready with an arsenal of easily self-administered drugs (i.e., oral or inhaled) that can be tested in rapid, efficient clinical trials immediately after the causative viral agent is identified. We propose a proactive drug development strategy for high consequence viral pathogens that focuses on combinatorial approaches.
OVERVIEW: DEVELOPING ANTIVIRAL REGIMENS FOR THE NEXT GLOBAL VIRAL PANDEMICS
Since it is highly likely that pathogenic viruses will continue to spill over into humans, a major component of pandemic preparedness should be development of highly effective and widely available antiviral treatments for nonhospitalized patients. In our opinion, we must develop concurrent plans for two possible scenarios: plan A for the longer term whereby the next serious viral outbreak does not occur for ∼5 to 10 years and plan B for the dire possibility that an epidemic arises within ≤1 to 2 years, from a new CoV or from a high consequence virus currently lacking effective self-administered antiviral drugs. Both plans should focus on drug combinations, but potential input drugs would differ. Plan A would focus on the development of new chemical entities (NCEs), in particular directly acting antivirals (DAAs). Plan B could employ DAAs and host-targeting agents (HTAs), including repurposed drugs and drugs in advanced clinical testing.
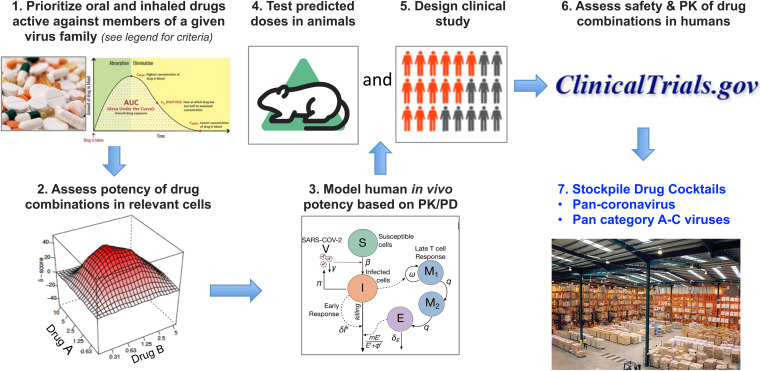
For both scenarios, a successful drug development program would be based on five key tenets (Fig. 1). The first is to prioritize oral and inhaled drugs that could be taken at home as postexposure prophylaxis or early during illness to rapidly lower viral loads and subsequent harmful immune activation. The second is to search for drug combinations to reduce emergence of drug-resistant mutants and, through multiplicative or synergistic effects, bring needed drug doses into therapeutic range and mitigate side effects associated with high doses of single drugs. The third is to prioritize drugs that are approved or in advanced clinical testing to allow a more rapid regulatory review process. Plan A with the relative luxury of time would also include drugs currently in preclinical development. The fourth is to prioritize drugs whose effective concentrations in relevant human tissue models are considerably below their toxic concentrations and in the range of attainable levels throughout the dosing interval. The fifth is to employ mathematical modeling at critical junctures of advancing to animal testing and clinical trial design. The products would be combinations of drugs that could be deployed widely and early during an outbreak as prophylaxis and early treatment. In conjunction with other nonpharmaceutical interventions, such in-home use drug cocktails could limit the burden on health care systems and thwart person-to-person virus spread by lowering viral loads and the virus’ ability to adapt to the host.
FIG 1.
A model-driven approach to develop highly potent drug combinations for global viral pandemic preparedness. The same pipeline can be used to prepare for the long term (plan A [5 to 10 years to the next outbreak]) or short term (plan B [<1 to 2 years to the next outbreak]). (Step 1) Select drugs that can be delivered orally or, for respiratory viruses, via inhalation that (i) are approved or in advanced clinical trials for plan B, with drugs in development additionally included for plan A, (ii) are active in relevant human cells, (iii) are ideally DAAs, but HTAs can also be considered, (iv) have relatively high selectivity indices (SI) (CC50/EC50), and (v) have relatively high Cmax/EC50 or maximum target tissue concentration/EC50. (Step 2) Test pairs of priority drugs (drug A and drug B) for combination effects (e.g., multiplicative or synergistic) in relevant human cells using checkerboard assays. For respiratory viruses, this should include lung cell models such as Calu3 as well as a three-dimensional (3D) culture such as lung organoids or primary human airway epithelial cells at an air-liquid interface. Drug combinations should then be prioritized for advancement based on the following: (i) drug levels needed for virus inhibition; (ii) SI; (iii) effectiveness over the entire dose-response matrix, including whether the drugs act synergistically; (iv) differing targets; (v) resistance map profiles; and (vi) other PK parameters (e.g., drug-drug interactions, side effects, half-lives, protein binding). (Step 3) Model the potential for top combinations to be potent in humans based on known PK and PD. (Step 4) Test most promising pairs of oral (and/or inhaled) drugs in small animal models. (Step 5 [concurrently with step 4]) Design clinical study. (Step 6) Conduct phase 1 trial of the drug combination. The deliverables (Step 7) will be pan-virus family oral/inhaled drug cocktails that can be stockpiled and ready for use very early following identification of the family of a pandemic-causing virus. The predesigned clinical study can be immediately implemented in the face of an on-going pandemic. The name VORTEC (Viral Outbreak Readiness Through Effective Combinations) has been suggested for the approach. The image in Step 1 (right) is from https://clinicalinfo.hiv.gov/en/glossary/pharmacokinetics. The image in Step 3 is reprinted from reference 16 with permission (© The Authors, some rights reserved; exclusive licensee AAAS. Distributed under a CC BY-NC 4.0 license [http://creativecommons.org/licenses/by-nc/4.0/]). Other images are from https://commons.wikimedia.org/wiki/Main_Page.
CURRENT STATE OF SARS-CoV-2 THERAPEUTICS
Early during the current pandemic, there were no therapeutics against SARS-CoV-2, and slight improvements in clinical outcomes were linked solely to advances in supportive care. Then, in May 2020, remdesivir (RDV), a SARS-CoV-2 polymerase inhibitor (1, 2), received an emergency use authorization (EUA) (3–8) for intravenous (IV) administration in hospitalized patients, with significant benefits noted in recent reports (3, 8). A very recent trial of IV RDV showed an 80% reduction in hospitalization (9), highlighting that treatment soon after infection is likely to be more efficacious than treatment later in the course of illness during hospitalization. Dexamethasone, an immunosuppressant without antiviral effects, also received EUA for clinical benefit in severe or critically ill hospitalized patients (10). Another set of drugs with an EUA for IV administration is RDV plus baricitinib (see below).
In November 2020, the first two monoclonal antibodies (MAbs), which target the SARS-CoV-2 spike glycoprotein, received an EUA after showing reductions in hospitalization and severe illness when dosed in clinical trials during early illness (11–15). MAbs are now available in limited settings for early use in high-risk patients, but their requirement for parenteral administration has precluded widespread implementation early during infection when therapy is most likely to be effective (16) Other limitations of MAbs include cost, manufacturing capacity, and emergence of antibody-resistant variants (17), which has already eliminated the utility of one promising MAb.
An exciting step forward is therefore the arrival of two oral SARS-CoV-2 drugs: the oral polymerase inhibitor molnupiravir (MPV, EUA approved in Europe and pending in the United States [18–21]), and the oral protease inhibitor Paxlovid (22) (PF-07321332/ritonavir; EUA pending). MPV elicited a 30% reduction in hospitalizations and death in infected high-risk people diagnosed and treated within 5 days of symptom onset (https://www.merck.com/news/merck-and-ridgeback-statement-on-positive-fda-advisory-committee-vote-for-investigational-oral-antiviral-molnupiravir-for-treatment-of-mild-to-moderate-covid-19-in-high-risk-adults/), while Paxlovid reduced hospitalizations and death by 89% when administered within 3 days of symptom onset and showed similar benefit if given within 5 days. Many other oral/inhaled anti-SARS-CoV-2 drugs are in the pipeline. Nevertheless, these pivotal strides are occurring almost 2 years into the pandemic and after 5 million people have died. During the next pandemic, be it of a novel CoV or another virus, a major priority must be to discover and widely distribute effective outpatient treatment regimens within weeks rather than years.
SCREENS FOR DRUGS TO THWART SARS-CoV-2 INFECTIONS
In the early days of the pandemic, work on small molecule antiviral drugs suffered from lack of a centralized research structure and strategic plan designed to keep pace with a rapidly expanding pandemic (23, 24). Many screens were conducted to identify drugs with activity against SARS-CoV-2 (25–32), and at least 216 small molecules approved by the Food and Drug Administration (FDA) have been reported to block SARS-CoV-2 in cell cultures. Yet most were identified in a convenient and nonrepresentative cell line (Vero E6 kidney) and later shown to have limited potency in lung epithelial cells (28, 33), the relevant cell type for SARS-CoV-2 infection and pathogenesis (34–36): SARS-CoV-2 enters Vero E6 cells by fusion in endosomes, whereas it enters lung cells by fusion at the cell surface following activation of the spike glycoprotein by the host cell surface serine protease TMPRSS2 (34–41). Many of the Vero E6 drug hits affect endosomes and were therefore not expected to block entry of SARS-CoV-2 into lung epithelial cells (38, 42). In addition, many had not completed safety and efficacy trials or are available only for IV use, making rapid widespread implementation infeasible. Many hits were also insufficiently potent against SARS-CoV-2 in lung cells in vitro at nontoxic doses; either their selective indices (SI), the ratio of half maximal efficacy to half lethal concentrations (EC50/CC50) were too low, or they would not achieve sufficient peak concentrations (Cmax) following standard dosing, reflected in a low Cmax/EC50 ratio.
These limitations contributed to the failure of many solo agents tested in clinical trials, including the highly publicized agents hydroxychloroquine (5, 30, 31) and ivermectin (43). While oral drugs to lower the risk of COVID-19-associated hospitalization and death are imminently available (18, 22) (see below), we urge the development of drug combinations to prevent the emergence of SARS-CoV-2 drug-resistant mutants (44, 45) and to potentially increase potency and breadth of coverage, and to reduce side effects.
HIGHLY EFFECTIVE DRUG COMBINATIONS AGAINST OTHER VIRUSES
We advocate for developing combinations of oral, intranasal, and inhaled drugs to prepare for emerging and reemerging viral pandemics (46–48) based on precedent with other viral infections. Treatments for chronic illnesses caused by human immunodeficiency virus (HIV) and hepatitis C virus (HCV) (49–51) are composed of two to four oral DAAs that target multiple steps in the viral life cycles, thereby inducing multiplicative or synergistic antiviral effects (50–53). The combinations reduce doses of the individual drugs needed (50), thus lowering toxic side effects. (Certain small molecules may not provide synergy but instead enhance levels of other active agents. For HIV, cobicistat and ritonavir are coformulated with integrase inhibitors and protease inhibitors, respectively, to limit metabolization of the primary agent allowing lower doses [54, 55] [also see NCT04960202].) In addition, by targeting separate steps with distinct escape mutations, successful combination regimens eliminate selection of drug-resistant viruses (56–59). While emergence of drug resistance is less certain for acute viral infections, which are usually eliminated by the acquired immune response (16), SARS-CoV-2 and influenza infections in immunocompromised hosts are notable for prolonged viral persistence at high viral loads with considerable ongoing viral mutagenesis (60–65). Thus, developing combination therapies for acute viral infections is justified, and efforts are under way for influenza virus (66), Ebola virus (47, 48, 67, 68), arenaviruses (69), and SARS-CoV-2. For Ebola virus, computational modeling (48) has suggested that combinations would provide superior in vivo activity to their single agent components.
CURRENT PROGRESS ON DRUG COMBINATIONS AGAINST SARS-CoV-2
There have been at least 34 reports of small molecule drug combinations against SARS-CoV-2, comprising a total of 77 unique drug pairs (26, 70–100, 197–199). Many were identified in Vero E6 cells, while others were found in Calu3 lung epithelial cells. Sixty-two pairs include RDV (101), with the partner drug being arbidol, BAY-2402234, brequinar, budesonide, camostat, cepharanthine, cilostazol, clobetasol, clofazimine, cobicistat, conivaptan, dabrafenib, diltiazem, drosiprenone, emetine, ezetimibe, interferon alpha, imino sugars, IQ-1S, ivermectin, ivosidenib, lapatinib, lenvatinib, linoleic acid, mefloquine, meprednisone, MU-UNMC-2, nelfinavir, nifedipine, nimodipine, nitazoxanide, omeprazole, omipalisib, pioglitazone, quinapril, raloxifene, reserpine, rifaximin, sangivamycin, selexipag, stenoparib, sulforaphane, telmisartan, tetrandrine, tipifarnib, valdecoxib, and zafirlukast.
RDV has also been reported to synergize with six oral HCV drugs (elbasvir, grazoprevir, paritaprevir, simeprevir, vaniprevir, and velpatasvir) as well as the HCV oral combination drugs Epclusa (velpatasvir plus sofosbuvir) and Zepatier (elbasvir plus grazoprevir) (75, 79, 89). These drugs block the HCV nonstructural serine protease (HCV NS3/4A), the HCV replication-associated protein NS5A, or the HCV polymerase (NS5B). The HCV drugs likely inhibit related functions of SARS-CoV-2 proteases and replication machinery, albeit at significantly lower potency. Four HCV protease inhibitors that synergized with RDV in Vero E6 cells (simeprevir, vaniprevir, paritaprevir, and grazoprevir) block the SARS-CoV-2 papain-like protease (PLpro), a cysteine protease encoded by nsp3 of SARS-CoV-2 (75). Two HCV protease inhibitors (boceprevir and narlaprevir) that blocked the SARS-CoV-2 C-like protease (3CLpro or Mpro, a cysteine protease encoded by SARS-CoV-2 nsp5), were inhibitory on their own (EC50 for boceprevir, 20 to 40 μM; EC50 for narlaprevir, 8 to 37 μM, in Vero E6 cells) (75, 89, 102) but did not synergize with RDV (102). Experimental drugs, including brilacidin (88), a 3CLpro inhibitor (81), a RAD51 inhibitor (87), the natural product angeloylgomisin O (95), and a vitamin E derivative (97) have also been shown to synergize with RDV.
Synergistic drug pairs that do not involve a polymerase inhibitor have also been identified in Vero E6 cells, including nelfinavir plus amodiaquine (71), arbidol plus nitazoxanide (73), nitazoxanide plus emetine (73), and nelfinavir plus cepharanthine (78). Additional non-polymerase-targeted pairs found synergistic in Calu3 cells were interferon alpha plus camostat (86), interferon alpha plus nafamostat (96), and apilimod plus camostat (103). Interferon alpha plus nafamostat reduced viral loads in hamsters to a greater extent than the individual component drugs (96).
Viral polymerases are clearly good targets for DAAs, and 43 drugs have been reported to synergize with the polymerase inhibitors MPV and/or RDV. Given the goal of developing a regimen for outpatient use, we highlight in Table 1 oral and inhaled drugs that synergistically impede SARS-CoV-2 in Calu3 epithelial cells in conjunction with MPV, the oral polymerase inhibitor (18–21): three orally available drugs, the pyrimidine biosynthesis inhibitors BAY-2402234 and brequinar (77) and the HIV (aspartic) protease inhibitor nelfinavir (80), and the inhaled drug interferon alpha. We also list in Table 1 drugs reported to synergize in Calu3 cells with RDV. Although not a certainty, these drugs may also synergize with MPV, as both drugs target the SARS-CoV-2 RNA-dependent RNA polymerase (2, 104), albeit with different biochemical mechanisms (19, 105). Indeed, similar synergistic activity of RDV and MPV drug pairs was reported in two studies (77, 86). Of the 14 drugs listed, 9 are approved by the FDA, and 2 are used in Japan. Nine of the approved drugs are formulated for oral use; interferon alpha and ciclesonide are inhaled therapeutics. Two other agents appeared to synergize with RDV in Calu3 cells but were tested only at two doses: the approved oral drug mefloquine (98) and the investigational drug brilacidin (88). Of the pairs in Table 1, only the combination of MPV plus brequinar has been tested in an animal model, where it reduced viral loads and lung pathology favorably compared to either drug alone (77). Brequinar has demonstrated synergy against HCV in combination with sofosbuvir, an orally available HCV polymerase inhibitor (80).
TABLE 1.
Drugs reported to synergize with remdesivir or molnupiravir to inhibit SARS-CoV-2 infection of Calu3 lung cellsa
| Drug A | Drug B | Drug B: FDA statusb | Drug B CoV CTb | Drug B oral | Drug B targetc | Drug B: step blockedc | Reference(s)d |
|---|---|---|---|---|---|---|---|
| Remdesivir (approved for intravenous use for COVID-19) | Nelfinavir | HIV | Ph2e | Yes | M-Prof | Cleavage | 80, 94 |
| Velpatasvir | HCV | Ph2g | Yes | Pol complexf | Replication | 79 | |
| Elbasvir | HCV | No | Yes | Pol complexf | Replication | 79 | |
| Grazoprevir | HCV | No | Yes | PL-Prof | Cleavage | 79 | |
| Dabrafenib | Cancer | No | Yes | B-Raf kinase | NDh | 79 | |
| Cilostazol | Leg pain | No | Yes | PDE III | ND | 79 | |
| Nimodipine | Aneurysm | No | Yes | Ca channelsi | ND | 79 | |
| Interferon alpha | HBV, HCV | Ph3 | No | ISGs | Replicationj | 86 | |
| B02 | Preclin. | No | NA | RAD51 | ND | 87 | |
| Camostat | Preclin.k | Ph3 | Yes | TMPRSS2 | Fusion | 94 | |
| Cepharanthine | Preclin.k | No | Yes | Multiple | NDk | 94 | |
| Ciclesonide | Rhinitis | Ph3 | No | nsp3/4l, GlucR | Replicationl | 94 | |
| Brequinar | Ph2 (AML) | Ph2 | Yes | DHODHm | Replication | 77 | |
| BAY-2402234 | Ph1 (MM)n | No | Yes | DHODH | Replication | 77 | |
| Molnupiravir (EUA in Europe/pending in U.S. for oral use for COVID-19) | Nelfinavir | HIV | Ph2e | Yes | M-Prof | Cleavage | 80 |
| Interferon alpha | HBV, HCV | Ph3 | No | ISGs | Replicationj | 86 | |
| Brequinar | Ph2 (AML) | Ph2 | Yes | DHODH | Replication | 77 | |
| BAY-2402234 | Ph1 (MM)n | No | Yes | DHODH | Replication | 77 | |
Abbreviations: AML, acute myeloid leukemia; CT, clinical trial; DHODH, dihydroorotate dehydrogenase; FDA, Food and Drug Administration; GlucR, glucocorticoid receptor; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; ISGs, interferon-stimulated genes; MM, multiple myeloma; NA, not available; ND, not determined; PDE, phosphodiesterase; Ph, phase; Pol, polymerase; preclin., preclinical.
Unless specified, drugs are FDA approved for the indicated conditions. Drugs in phase 1 (Ph1) for COVID-19 (CoV), e.g., reference 97, are not listed.
Known or inferred target. Cleavage denotes polyprotein cleavage.
References are for reported synergies. Reference 79 reports 15 additional remdesivir synergies, but the cell type analyzed was not specified.
The https://biorender.com/covid-vaccine-tracker website lists a phase 2 (Ph2) study, but this was not in https://clinicaltrials.gov/.
See text and references 75, 78, and 79 for targets of nelfinavir, velpatasvir, elbasvir, and grazoprevir.
A trial (IRCT20130812014333N145) of Epclusa (sofosbuvir/velpatasvir) deemed it safe but of no apparent benefit.
Dabrafenib inhibits lymphocytic choriomeningitis virus replication (32).
L-type Ca channels.
Proposed in reference 86.
Camostat and cepharanthine are used in Japan for pancreatitis and multiple ailments, respectively (196).
Ciclesonide is an anti-inflammatory, but reference 144 supports additional action versus SARS-CoV-2 nsp3 and nsp4.
DHODH is a host enzyme required for pyrimidine biosynthesis.
The trial in patients with advanced myeloid malignancies (NCT03404726) was terminated due to lack of clinical benefit.
Two drugs are registered in clinical trials in combination with RDV: camostat (NCT04713176) and the Janus kinase inhibitor, baricitinib (NCT04401579 and NCT04693026). Although baricitinib has not been shown to synergize with RDV in vitro, it provided moderate clinical benefit when added to RDV therapy (106), supporting its EUA for hospitalized COVID-19 patients. On the basis of the results of in vitro studies, baricitinib is hypothesized to provide benefit by both antiviral and anticytokine effects (107), and its efficacy in clinical use may be independent of RDV coadministration (108). In another clinical trial (109), administration of two oral HIV protease inhibitors (lopinavir and ritonavir), an oral broad-spectrum viral replication inhibitor (ribavirin), and an injected anti-inflammatory (interferon beta-1b) reduced viral loads and improved symptoms in hospitalized patients compared to lopinavir/ritonavir alone. While these studies highlight that combinations of DAAs and immunomodulatory medicines might have a therapeutic role, their need for parenteral administration precludes easy use during early infection when DAAs have the highest potential to limit disease severity.
DESIGNING EFFECTIVE DRUG COMBINATIONS
As for oral drug combinations for patients with HIV and HCV, an ideal cocktail would contain multiple agents targeting CoV proteins, i.e., multiple DAAs. A theoretical cocktail might target the spike glycoprotein, the polymerase, and either or both viral proteases. We contend that HTAs should also be considered (47, 48, 69), perhaps to bolster pairs of DAAs. The combination of a protease inhibitor (HTA, aprotinin, given IV) targeting the host enzyme TMPRSS2, plus oral favipiravir (DAA, polymerase inhibitor) has been tested in COVID-19 patients, but the study was too small to assess clinical efficacy (110). In murine models of infection, combinations of two anti-Spike MAbs plus RDV provided benefit over single agents in some measures of COVID-19 disease (111), supporting the concept of combined targeting of virus entry and viral polymerase activity.
A current priority is to develop an organized structure to assess pairs of agents comprehensively and strategically. As shown above, there are many possible agents to consider for this purpose (112, 113). Below we propose criteria for prioritizing combinations, noting that new drugs should be incorporated into this schema as they become available.
Identify drug pairs with enhanced combinatorial potency.
Most drug combination studies involve a checkerboard assay in which various doses of drugs A and B are set in a matrix, and these defined dose mixtures (36 for a 6 × 6 matrix) are tested for inhibition of SARS-CoV-2 infection (step 2, Fig. 1). These assays assess whether drug interactions are antagonistic, neutral, additive, multiplicative, or synergistic, in increasing order of desirability. While synergy is always in theory preferable, its highest potential is in the context of single agents that have insufficient potency on their own, as may be the case for repurposed agents. Antagonistic pairs should generally be excluded from further consideration. For interstudy comparisons, it would be ideal if future studies were coordinated to employ the same virus strains, target cells, infection protocol, assay readout, reference drug pairs, and synergy scoring methodology. While many DAAs should function independently of cell type, testing in lung cell systems (e.g., Calu3 cells, human airway epithelial cells cultured at the air-liquid interface, or lung organoids [34, 114, 115]) will be especially important for drugs that target entry, which varies by cell type (28, 33–36). Promising combinations should also be tested against the most relevant emerging SARS-CoV-2 variants of concern (VOCs, e.g., omicron) and other CoVs.
SynergyFinder (https://synergyfinder.fimm.fi/) is a publicly available web tool (116) that can be used to assess whether a drug pair tested in a checkerboard assay is considered synergistic. The program can assess synergy according to Bliss independence, zero interaction potential (ZIP) or Loewe additivity mathematical models. Synergy occurs when observed potency exceeds that of the expected combination effect based on the selected synergy model. In the strictest sense, synergy denotes observed potency exceeding that predicted by a Bliss independence mathematical model, which assumes multiplicative effects of paired drugs (50). With SynergyFinder, synergy scores are calculated over the full dose matrix as well as its maximum synergistic area (MSA); scores of >10 are generally considered synergistic (69, 116, 117). MSA ZIP scores for the HCV drugs listed in Table 1 combined with either RDV or MPV, based on assays that monitored virus-induced cytopathic effect in Calu3 cells, ranged from 50 to 85, with reported MSA ZIP scores for other pairs ranging from 22 to 52 (not given for brilacidin or the pairs with pyrimidine biosynthesis inhibitors). Thus, there now exist effective methods and freely available tools to identify synergistic drug combinations.
Consider the molecular target of the drug and how the drug impinges on the SARS-CoV-2 life cycle.
A useful cocktail would contain two (or three) DAAs that target distinct CoV proteins. These may encompass drugs targeting the spike protein to block receptor binding or fusion or the enzymatic activities of its polymerase or proteases. The justification for this conclusion is based on comprehensive combinatorial testing of all licensed HIV antivirals (50). Compounds that target the same viral protein tend to have only additive effects in which the second agent adds little to overall potency. Molecules that distinctly target HIV reverse transcriptase, integrase, or protease usually have either multiplicative or synergistic effects and bypass resistance (59), though mechanisms that predict synergy rather than simply multiplicative effects are less clear from these data sets. HTAs that limit viral replication (31, 118–120) may enhance synergy of a set of DAAs and should be considered in the testing matrices.
Consider the human exposure potential of the drugs.
To leverage pairwise synergy for potential therapeutic use, the pharmacokinetic (PK) profiles of component drugs are critical. Of importance are peak and trough drug concentrations, which are determined by tissue clearance kinetics. Ideally, both drugs achieve levels that allow Bliss independence or synergy throughout the dosing interval. If there are only brief time windows of drug synergy, then synergy may have limited beneficial effects for infections such as SARS-CoV-2 that are defined by rapid replication and spread dynamics (16). For example, we demonstrated that acyclovir only partially lowers genital herpes simplex virus 2 (HSV-2) shedding rates because 6-h windows of subtherapeutic drug levels prior to redosing are sufficient for breakthrough viral replication (121).
It is important to note that synergy may provide limited added benefit in situations where a given concentration of a single agent already eliminates nearly 100% of new cell infections. This possibility highlights the need to factor in projected peak and trough concentrations of both relevant drugs relative to their EC50s to assess whether multiplicative or synergistic pharmacodynamics (PD) provides meaningful additional benefit.
For MPV, the Cmax at the dose of 800 mg given twice daily is 14 μM, while its EC50 against SARS-CoV-2 in Calu3 cells is 0.08 μM (19). This gives a Cmax/EC50 ratio of 175, meaning that a serum concentration well above its in vitro EC50 against SARS-CoV-2 is expected. Indeed, recent trial results indicate that MPV can achieve sufficient levels to lower viral replication (18), though drug synergy may further enhance its in vivo potency. As a note of caution, we have demonstrated that in vitro EC50 values often overestimate antiviral potency in vivo such that in order to suppress virus in people, 5 to 10 times higher drug levels are required than predicted by cell culture assays (122). It is also critical to consider each drug’s binding to protein in vitro and in vivo, as this can greatly influence the amount of drug available (123, 124).
Of the 11 oral drugs listed in Table 1 for which sufficient data are available, only 3 have Cmax/EC50 ratios approaching or greater than 1, which could be considered a minimal criterion for consideration due to the pharmacologic factors described above. They are camostat, nelfinavir, and brequinar, with Cmax/EC50 values of ∼1, 7, and 33, respectively. Conversely, while they are synergistic with RDV, the Cmax/EC50 ratios for the HCV drugs against SARS-2-CoV are <0.1. (These drugs are more potent against HCV.) The reported synergies of the HCV drugs with RDV do demonstrate the potential utility of combinations targeting the SARS-CoV-2 polymerase and its proteases with DAAs (e.g., MPV plus Paxlovid [18, 22]). Other new oral CoV protease inhibitors (22, 112, 113, 125–127) and polymerase inhibitors (97, 128, 129) should also be considered if they have suitable Cmax/EC50 ratios. Similarly, new TMPRSS2 inhibitors being explored (130–134) may have more suitable PK properties than camostat.
Evaluate the half-life, selective index, drug-drug interactions, and side effects of the drug.
Drug toxicity as a function of drug concentration, for example as detected with SynToxProfiler (135), should be considered, as it could limit the effectiveness of a combination found to be synergistic in vitro and otherwise suitable for human trials. In addition to known associations with end organ damage, potential positive or negative adjunct effects, such as suppression of cytokine storms and inflammation associated with serious disease, should also be considered. Expedited FDA consideration of two previously approved drugs in combination for an investigational new drug (IND) application requires preclinical evidence of lack of drug-drug interaction, favorable PK/PD, and lack of presumed overlapping toxicities.
Animal model testing.
Once a list of drug combinations is prioritized, promising cocktails should be tested in a small animal model. The model should approximate human studies in which an agent might be given as postexposure prophylaxis or as early treatment several days after viral inoculation given that efficacy may differ for these two indications for a given regimen. Readouts in animal studies should always include frequent viral load testing in addition to clinical and pathological scores such that early attempts at establishing mechanistic correlates of efficacy can be made.
Mathematical modeling for regimen optimization.
Mathematical modeling can be used throughout the development of combination antiviral regimens to maximize the likelihood of successful drug, dose, and dosing interval selection, as well as critical features of study design. Mathematical models are predicated on the concept that PK/PD equations are necessary but not sufficient for forecasting trial outcomes. Of equal importance are equations that capture viral and immune dynamics in the absence of therapy (136).
Several principles illustrate the importance of this approach. First, the timing of therapy may predict its effectiveness. In the case of SARS-CoV-2, we determined that intense innate immune responses severely limit the extent of viral replication after the first 5 days of infection (16). Therefore, agents started during the presymptomatic phase of infection, which would parallel postexposure prophylaxis in clinical practice, require higher potency to induce viral clearance than those given later when immune mechanisms assist in clearance of infected cells. Yet, because the majority of viral replication occurs early during infection and is linked to downstream aberrant inflammation, treatment given soon after development of symptoms is critical toward preventing severe outcomes. This model prediction was subsequently validated in treatment trials with MAbs, demonstrating that administration is more effective prior to hospitalization (11–13, 137).
Second, as described above, in vitro assessments of potency may overestimate a drug’s antiviral effect in people by 5- to 10-fold (122). Serial viral load measurements in animal models or in human clinical trials, coupled with mathematical modeling are vital to link plasma drug concentrations with viral kinetic outcomes. The goal is to identify the in vivo EC50, or plasma concentration of drug that eliminates 50% of cellular infection events in vivo.
Third, modeling provides necessary context for the potential benefits of combination effects that are additive, multiplicative, or synergistic. By accounting for nonlinear drug levels over time, modeling can capture the proportion of time during which levels of both drugs allow potent inhibition of cellular infection, either by virtue of single drug potency or synergy. This is vital because it can identify scenarios in which addition of a second drug is required to achieve adequate viral suppression and those in which a second agent may be unnecessary and confer unnecessary toxicity.
Finally, modeling can capture possible differential impacts of therapies in various hosts. Whereas most infected people, even those with critical illness, appear to eliminate high grade viral replication within 1 to 2 weeks (138), immunocompromised hosts can shed infectious SARS-CoV-2 at high viral loads for months (60–65), taking on a phenotype more consistent with chronic, persistent viruses like HIV. Here, the virus undergoes considerable mutation and is more likely to become resistant to small molecule DAA or MAb therapy. Immunocompromised hosts are a possible source for VOCs that have dramatically extended the duration and overall lethality of the epidemic (60). Modeling is well poised to capture the added potency required from synergistic agents in this specific clinical context.
Modeling is ideally linked to all steps in the drug development process. PD models are used to identify whether drug pairs have antagonistic, additive, multiplicative, or synergistic antiviral activity at given concentrations and to precisely recapitulate the degree of viral inhibition across all possible ranges of dual drug concentrations (48). PK models recapitulate drug levels over time in relevant animal models or humans. Models can then project the percentage of cell infection events prevented with a given regimen at a given dose. While these calculations allow for an initial estimate of in vivo benefits of synergistic or multiplicative effects, the accuracy of forecasts is limited if in vitro assays overestimate in vivo potency. We therefore fit our models to detailed virologic data from animal studies or early human clinical trials and solve for the in vivo EC50 (121, 122). We can leverage this information to synthesize PK/PD models with viral dynamics models to arrive at lowest doses and dose frequencies of drug pairs that are likely to have suppressive antiviral efficacy in human trials.
NEW DRUGS TO CONSIDER FOR COMBINATION TESTING AGAINST CoVs
While approved drugs have the advantage that FDA guidance on the development of drug combinations allows for more rapid development (https://www.fda.gov/media/80100), drugs that have passed human safety trials should also be considered as information about them becomes available. In this respect, it will be important to follow the development of AT-527 (128), other potential oral SARS-CoV-2 polymerase inhibitors (97, 129) including oral forms of RDV (139–141, 200) as well as the oral protease inhibitor GC-376 (102, 125). As new oral/inhaled drugs with the same targets but better efficacy and PK become available, they should be analyzed as substitutes in previously characterized drug synergies. Other drugs for future combination tests could be oral/inhaled variants of effective drugs currently only suitable for injection (142) and drugs of interest in other countries (https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-treatments). Inhaled drugs currently under consideration for SARS-CoV-2 include budesonide (143), ciclesonide (144), interferon alpha (86, 96), interferon beta (145), nafamostat (146), and niclosamide (147), as well as small inhalable biologics, including minibinders (148), nanobodies (149), and peptide fusion inhibitors (150). Many other potential anti-CoV drugs are also being uncovered through in silico protein docking, transcriptional profiling, and protein-protein interaction network analyses. An outpatient therapeutic regimen that combines inhaled and oral drugs is plausible. Lastly, future drug combinations could include immune boosters such as STING (stimulator of interferon genes) activators (151, 152) and may be composed of three or more drugs (49–51, 68), if enhanced synergy or PK benefit is projected. As new drug combinations are explored to combat CoVs, a unified set of mathematical equations could allow rapid updates such that new potential regimens are ranked according to their likely potency, as has been done for HIV (50).
CLINICAL CONSIDERATIONS
Executing high-quality clinical trials during both the early and mature phases of a pandemic presents challenges related to testing of drug combinations, especially for drugs with easily accessible routes of delivery. In the initial phases of an epidemic, tension between conducting research and providing clinical care when resources are constrained means that experimental therapies are often given in uncontrolled studies or as expanded access, although such use “often undermines fair access to experimental agents, [and] compromises the collection of robust data to determine the safety and efficacy of interventions” (153). During the 2014–2017 Ebola epidemic, an advisory panel to the WHO provided a seven-point list to guide conditions of investigational agent use, which urged minimal interference with the conduct of high quality clinical investigations (https://www.who.int/ebola/drc-2018/notes-for-the-record-meuri-ebola.pdf). During the COVID-19 pandemic, widespread clinical use, including through nonprescribed access, and uncontrolled studies has operationally precluded high-quality clinical trials for several repurposed agents, including hydroxycholorquine, ivermectin, and fluvoxamine (154, 155) (NCT04668950, NCT 04885530, and NCT 04510194, but see reference 156 for fluvoxamine). Numerous other small studies used various doses and combinations of proposed synergistic drugs, with each underpowered to meaningfully assess efficacy. It was difficult to meaningfully compare efficacy across disparate study regimens for pooled analysis. Lower regulatory burden improves the speed and cost at which studies on previously approved drugs can be done, but in many cases during 2020, the rapid conduct of such trials impeded progress toward interpretable data.
Presently, at the more mature stage of the pandemic, new hurdles are hampering development of oral/inhaled treatments for SARS-CoV-2. The logical positioning of combination oral drug trials is for early treatment of nonhospitalized patients with either mild symptomatic or asymptomatic infection to maximize clinical effect and prevent hospitalizations. Such potential trials now have an added hurdle because MAbs with EUA are now considered standard of care for people at moderate/high risk for hospitalization, which includes the larger share of unvaccinated adults (https://www.covid19treatmentguidelines.nih.gov/). As such, trials of oral/inhaled drug combinations can no longer ethically be placebo controlled (if conducted where MAbs are available). Because participation in clinical trials requires trust in scientific and medical institutions, persons who remain unvaccinated in places with widespread access to COVID-19 vaccines may be less likely to participate in treatment trials, although careful attention to community engagement has shown incredible success with vulnerable communities (157).
Moreover, these therapies were tested in clinical trials and authorized based on efficacy at reducing hospitalization and death. As SARS-Cov-2 vaccinations increase, an increasing proportion of infections are in vaccinated persons, who remain overwhelmingly protected from severe COVID-19 outcomes. However, ongoing studies of non-high-risk patients (vaccinated and/or without significant comorbidities) need to be evaluated for benefit in symptom reduction and/or viral shedding because hospitalizations are too uncommon an outcome to rapidly conduct studies with adequate statistical power, particularly when the comparator group includes the standard of care. For multiple other viruses, including HIV, HCV, cytomegalovirus (CMV), and HSV-2, establishment of virologic surrogate endpoints has dramatically decreased the number of people and the cost and time associated with licensure trials (158–161). As of this publication, there is no accepted definition for success in reduction of SARS-CoV-2 shedding or transmissible virus, nor are there accepted definitions for change in symptom burden or duration. Both symptoms and viral shedding are also blunted in most infections following vaccination, which additionally limits power to detect a treatment effect. In future pandemics, early trials should be designed to allow establishment of virologic surrogate endpoints. To meet this goal requires that studies include daily virologic sampling and detailed daily symptom surveys. This method will allow trialists to evaluate which viral kinetic feature (peak viral load, duration of shedding or viral area under the curve [158]) or constellation of early symptoms may be most predictive of hospitalization or death. Even if this is achieved, changes in incubation period, viral kinetics, and symptoms with different emerging VOCs may necessitate updating of these surrogate outcomes.
Finally, lack of a path toward EUA for novel drugs, or package relabeling for already approved drugs, based on potentially acceptable surrogate outcomes hampers industry-sponsored drug investigation. Drugs without efficacy as monotherapy in phase 2 studies may not progress to combination trials, despite preclinical data that would suggest potential success and may have predicted lack of success of the single agent. Significant alteration in regulatory procedures during a highly lethal pandemic is another necessary step forward.
PREPARING FOR OTHER EMERGING VIRUSES
Members of 11 virus families are considered of potential high consequence, and it is important to develop oral (and inhaled, for respiratory viruses), thermally stable, inexpensive, pan-family, drug cocktails to combat all of them (118) (Fig. 1). Common features of these category A to C (category A-C) pathogens (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens) can be exploited in therapeutic cocktail design. All families of concern contain single-stranded RNA and are inhibited by one or more of the following polymerase inhibitors: RDV, MPV, favipiravir, or galidesivir (BCX4430). The members of nine of these families, including coronaviruses, are enveloped and hence deliver their genomes into the cytoplasm to initiate replication by a stereotypical membrane fusion process mediated by a fusion glycoprotein (GP) (37, 38). Many of these (e.g., the hemagglutinins [HAs] of influenza viruses and the GPs of arenaviruses) also bind particles to the host cell surface, whereas other viruses contain a separate attachment/receptor binding protein. Both virus attachment and virus fusion are good targets for small molecule intervention (72, 162–166). As polymerase complexes are clearly excellent therapeutic targets (19, 49, 51), a “starter” pan-family cocktail could include a drug targeting a viral attachment or fusion glycoprotein and a drug targeting the viral polymerase. Flaviviruses and togaviruses also encode proteases that, like the SARS-CoV-2 proteases, process their polyproteins during virus maturation (167, 168). Proteases therefore represent additional targets for a subset of category A-C viral pathogens.
The members of 9 of the 11 category A-C virus families (arena-, bunya-, calci-, filo-, flavi-, orthomyxo-, picorna-, rhabdo-, and togaviruses) enter cells through endosomes (38), and hence, the endosomal pathway is a target for their therapeutic intervention (169, 170). Indeed, many drug screens have uncovered endosome-targeting drugs against these pathogens. Endosomal features that can be targeted are virus particle internalization (e.g., via clathrin or macropinocytosis) as well as endosome trafficking, maturation, and composition, including low pH, cathepsin proteases, Ca2+ and other ions, and specific lipids (171–173). Even Nipah virus and Hendra virus, which are category C paramyxoviruses that fuse at the plasma membrane, require a low-pH-activated endosomal cathepsin to process their fusion glycoproteins and form infectious particles (174). Furthermore, endosome-targeted drugs could inhibit infections by CoVs in tissues outside the lung (34–36).
While DAAs are considered ideal based on their generally higher potency and selective indices, we posit that targeting host proteins critically involved in the viral life cycle, including drugs that target endosomes for most category A-C viruses, should be considered in combination therapies, especially if we are preparing for the short-term (plan B) for endosome-entering viruses. From a screen of 78 unique pairs, we identified several demonstrating clear synergistic activity against Ebola virus in cell cultures (47). Based on PK and other considerations, two pairs (bepridil plus sertraline and sertraline plus toremifene) were further evaluated for eventual testing in an oral formulation in mice against a lethal Ebola virus challenge (48). The components of these pairs are approved, endosome-affecting drugs; each had previously been shown to protect mice (50 to 100%) in a lethal challenge model when given intraperitoneally (46, 175). In the same study (48), we showed, through mathematical modeling of PK, PD and Ebola viral dynamics, that compared to their individual constituents, both synergistic drug pairs have superior potential to reduce viral loads in humans (48). It will be interesting to see how these drug pairs function compared to their individual components when given orally to mice infected with Ebola virus. Moreover, all three compounds in these drug pairs have also been shown to bind to a pocket in the Ebola virus GP, thereby affecting its stability (176). Hence, some endosome-targeting drugs may also act directly on viral glycoproteins (69, 176–179), i.e., be both HTAs and DAAs. Other host factors to consider for targeting include host proteases that prime viral glycoproteins for fusion (37, 38, 119, 120), other host proteins involved in virus entry (180), host cell kinases (181–184), and host proteins involved in viral RNA production (185, 186), nuclear export (187, 188), and virus assembly and egress (189, 190). Given common viral infection strategies, the possibility exists for cross-family drug cocktails.
While investigators are tailoring novel drugs to specific viral proteins, we urge that concurrent work proceed to develop cocktails against all category A-C viruses, which might comprise approved (repurposed) drugs or combinations of advanced clinical stage and repurposed drugs. If a new pandemic emerges in the next few years or if resistant VOCs emerge during the current pandemic, individual suboptimal agents with good projected efficacy based on mathematical modeling assessment of synergy and projected drug levels could be tested rapidly, in combinations, in clinical trials. Even a cocktail with incomplete suppression of viral replication could have a clinical benefit: for Ebola virus (191, 192) and SARS-CoV-2 (193), a 1-log-unit-lower viral load has been associated with survival. Reductions in viral loads may also have public health benefits by reducing transmission rates and the opportunity for new variants.
CONCLUSIONS
SARS-CoV-2 is projected to remain in circulation in the human population, and novel CoVs may spill over from animals to humans (194, 195). An urgent goal is to develop inexpensive oral and/or inhaled regimens to test at the inception of the next outbreak. Drug combinations should be considered in this pursuit to limit drug resistance and enhance potency (Fig. 1). Moreover, the entire antiviral drug arsenal requires significant boosting for global viral pandemic preparedness. It is our opinion that this effort could be significantly augmented by carefully designed drug combination studies, at both the preclinical and clinical stages.
ACKNOWLEDGMENTS
Awards from the NIH (AI114776 [J.M.W.], AI121129 [J.T.S.], and K23AI129659 [R.B.I.]), the Estonian research council (MOBTT39 to D.K.), the Sigrid Jusélius Foundation (T.A.), and the Washington Research Foundation (S.J.P.) funded work in our laboratories.
We thank Samuel Green for critical comments on the text and help with the figure.
Contributor Information
Judith M. White, Email: jw7g@virginia.edu.
Joshua T. Schiffer, Email: jschiffe@fredhutch.org.
Stephen J. Polyak, Email: polyak@uw.edu.
Vinayaka R. Prasad, Albert Einstein College of Medicine
REFERENCES
- 1.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, et al. 2016. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. 2020. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M-D, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group Members . 2020. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med 383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garibaldi BT, Wang K, Robinson ML, Zeger SL, Bandeen-Roche K, Wang M-C, Alexander GC, Gupta A, Bollinger R, Xu Y. 2021. Comparison of time to clinical improvement with vs without remdesivir treatment in hospitalized patients with COVID-19. JAMA Netw Open 4:e213071. doi: 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Solidarity Trial Consortium, Pan H, Peto R, Henao-Restrepo A-M, Preziosi M-P, Sathiyamoorthy V, Abdool Karim Q, Alejandria MM, Hernández García C, Kieny M-P, Malekzadeh R, Murthy S, Reddy KS, Roses Periago M, Abi Hanna P, Ader F, Al-Bader AM, Alhasawi A, Allum E, Alotaibi A, Alvarez-Moreno CA, Appadoo S, Asiri A, Aukrust P, Barratt-Due A, Bellani S, Branca M, Cappel-Porter HBC, Cerrato N, Chow TS, Como N, Eustace J, García PJ, Godbole S, Gotuzzo E, Griskevicius L, Hamra R, Hassan M, Hassany M, Hutton D, Irmansyah I, Jancoriene L, Kirwan J, Kumar S, Lennon P, Lopardo G, Lydon P, Magrini N, Maguire T, Manevska S, Manuel O, McGinty S, et al. 2021. Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N Engl J Med 384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn M-Y, Nahass RG, Chen Y-S, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wei X, Gaggar A, Brainard DM, Towner WJ, Muñoz J, Mullane KM, Marty FM, Tashima KT, Diaz G, Subramanian A, GS-US-540-5773 Investigators . 2020. Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med 383:1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mozaffari E, Chandak A, Zhang Z, Liang S, Thrun M, Gottlieb RL, Kuritzkes DR, Sax PE, Wohl DA, Casciano R, Hodgkins P, Haubrich R. 2021. Remdesivir treatment in hospitalized patients with COVID-19: a comparative analysis of in-hospital all-cause mortality in a large multi-center observational cohort. Clin Infect Dis doi: 10.1093/cid/ciab875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckland MS, Galloway JB, Fhogartaigh CN, Meredith L, Provine NM, Bloor S, Ogbe A, Zelek WM, Smielewska A, Yakovleva A, Mann T, Bergamaschi L, Turner L, Mescia F, Toonen EJM, Hackstein C-P, Akther HD, Vieira VA, Ceron-Gutierrez L, Periselneris J, Kiani-Alikhan S, Grigoriadou S, Vaghela D, Lear SE, Török ME, Hamilton WL, Stockton J, Quick J, Nelson P, Hunter M, Coulter TI, Devlin L, CITIID-NIHR COVID-19 BioResource Collaboration, MRC-Toxicology Unit COVID-19 Consortium, Bradley JR, Smith KGC, Ouwehand WH, Estcourt L, Harvala H, Roberts DJ, Wilkinson IB, Screaton N, Loman N, Doffinger R, Lyons PA, Morgan BP, Goodfellow IG, Klenerman P, Lehner PJ, Matheson NJ, Thaventhiran JED. 2020. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun 11:6385. doi: 10.1038/s41467-020-19761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill JA. 2021. Remdesivir for the treatment of high-risk non-hospitalized individuals with COVID-19: a randomized, double-blind, placebo-controlled trial. IDWeek 2021, abstr LB1.
- 10.RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. 2021. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med 384:693–704. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators . 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, Shawa I, Adams AC, Van Naarden J, Custer KL, Shen L, Durante M, Oakley G, Schade AE, Sabo J, Patel DR, Klekotka P, Skovronsky DM, BLAZE-1 Investigators . 2021. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med 384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dougan M, Nirula A, Azizad M, Mocherla B, Gottlieb RL, Chen P, Hebert C, Perry R, Boscia J, Heller B, Morris J, Crystal C, Igbinadolor A, Huhn G, Cardona J, Shawa I, Kumar P, Adams AC, Van Naarden J, Custer KL, Durante M, Oakley G, Schade AE, Holzer TR, Ebert PJ, Higgs RE, Kallewaard NL, Sabo J, Patel DR, Dabora MC, Klekotka P, Shen L, Skovronsky DM, BLAZE-1 Investigators . 2021. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N Engl J Med 385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien MP, Forleo-Neto E, Musser BJ, Isa F, Chan K-C, Sarkar N, Bar KJ, Barnabas RV, Barouch DH, Cohen MS, Hurt CB, Burwen DR, Marovich MA, Hou P, Heirman I, Davis JD, Turner KC, Ramesh D, Mahmood A, Hooper AT, Hamilton JD, Kim Y, Purcell LA, Baum A, Kyratsous CA, Krainson J, Perez-Perez R, Mohseni R, Kowal B, DiCioccio AT, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Weinreich DM, Covid-19 Phase 3 Prevention Trial Team . 2021. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N Engl J Med 385:1184–1195. doi: 10.1056/NEJMoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta A, Gonzalez-Rojas Y, Juarez E, Crespo Casal M, Moya J, Falci DR, Sarkis E, Solis J, Zheng H, Scott N, Cathcart AL, Hebner CM, Sager J, Mogalian E, Tipple C, Peppercorn A, Alexander E, Pang PS, Free A, Brinson C, Aldinger M, Shapiro AE. 2021. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody Sotrovimab. N Engl J Med 385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 16.Goyal A, Cardozo-Ojeda EF, Schiffer JT. 2020. Potency and timing of antiviral therapy as determinants of duration of SARS-CoV-2 shedding and intensity of inflammatory response. Sci Adv 6:eabc7112. doi: 10.1126/sciadv.abc7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 18.Fischer W, Eron JJ, Holman W, Cohen MS, Fang L, Szewczyk LJ, Sheahan TP, Baric R, Mollan KR, Wolfe CR, Duke ER, Azizad MM, Borroto-Esoda K, Wohl DA, Loftis AJ, Alabanza P, Lipansky F, Painter WP. 2021. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv 10.1101/2021.06.17.21258639. [DOI]
- 19.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. 2020. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox RM, Wolf JD, Plemper RK. 2021. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol 6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahl A, Gralinski LE, Johnson CE, Yao W, Kovarova M, Dinnon KH, Liu H, Madden VJ, Krzystek HM, De C, White KK, Gully K, Schäfer A, Zaman T, Leist SR, Grant PO, Bluemling GR, Kolykhalov AA, Natchus MG, Askin FB, Painter G, Browne EP, Jones CD, Pickles RJ, Baric RS, Garcia JV. 2021. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Dantonio A, Di L, Eng H, Ferre R, Gajiwala KS, Gibson SA, Greasley SE, Hurst BL, Kadar EP, Kalgutkar AS, Lee JC, Lee J, Liu W, Mason SW, Noell S, Novak JJ, Obach RS, Ogilvie K, Patel NC, Pettersson M, Rai DK, Reese MR, Sammons MF, Sathish JG, Singh RSP, Steppan CM, Stewart AE, Tuttle JB, Updyke L, Verhoest PR, Wei L, Yang Q, Zhu Y. 2021. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 23.Dolgin E. 2021. The race for antiviral drugs to beat COVID - and the next pandemic. Nature 592:340–343. doi: 10.1038/d41586-021-00958-4. [DOI] [PubMed] [Google Scholar]
- 24.Nature Research. 2021. Funders, now is the time to invest big in COVID drugs. Nature 592:326. doi: 10.1038/d41586-021-00960-w. [DOI] [PubMed] [Google Scholar]
- 25.Jeon S, Ko M, Lee J, Choi I, Byun SY, Park S, Shum D, Kim S. 2020. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother 64:e00819-20. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, Burgstaller-Muehlbacher S, De Jesus PD, Teriete P, Hull MV, Chang MW, Chan JF-W, Cao J, Poon VK-M, Herbert KM, Cheng K, Nguyen T-TH, Rubanov A, Pu Y, Nguyen C, Choi A, Rathnasinghe R, Schotsaert M, Miorin L, Dejosez M, Zwaka TP, Sit K-Y, Martinez-Sobrido L, Liu W-C, White KM, Chapman ME, Lendy EK, Glynne RJ, Albrecht R, Ruppin E, Mesecar AD, Johnson JR, Benner C, Sun R, Schultz PG, Su AI, García-Sastre A, Chatterjee AK, Yuen K-Y, Chanda SK. 2020. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weston S, Coleman CM, Haupt R, Logue J, Matthews K, Li Y, Reyes HM, Weiss SR, Frieman MB. 2020. Broad anti-coronavirus activity of Food and Drug Administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J Virol 94:e01218-20. doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dittmar M, Lee JS, Whig K, Segrist E, Li M, Kamalia B, Castellana L, Ayyanathan K, Cardenas-Diaz FL, Morrisey EE, Truitt R, Yang W, Jurado K, Samby K, Ramage H, Schultz DC, Cherry S. 2021. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep 35:108959. doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakowski MA, Beutler N, Wolff KC, Kirkpatrick MG, Chen E, Nguyen T-TH, Riva L, Shaabani N, Parren M, Ricketts J, Gupta AK, Pan K, Kuo P, Fuller M, Garcia E, Teijaro JR, Yang L, Sahoo D, Chi V, Huang E, Vargas N, Roberts AJ, Das S, Ghosh P, Woods AK, Joseph SB, Hull MV, Schultz PG, Burton DR, Chatterjee AK, McNamara CW, Rogers TF. 2021. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat Commun 12:3309. doi: 10.1038/s41467-021-23328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sourimant J, Aggarwal M, Plemper RK. 2021. Progress and pitfalls of a year of drug repurposing screens against COVID-19. Curr Opin Virol 49:183–193. doi: 10.1016/j.coviro.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao K, Tzou PL, Nouhin J, Bonilla H, Jagannathan P, Shafer RW. 2021. SARS-CoV-2 antiviral therapy. Clin Microbiol Rev 34:e00109-21. doi: 10.1128/CMR.00109-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan W, Zhu S, Li S, Shang W, Zhang R, Li H, Liu W, Xiao G, Peng K, Zhang L. 2021. High-throughput screening of an FDA-approved drug library identifies inhibitors against arenaviruses and SARS-CoV-2. ACS Infect Dis 7:1409–1422. doi: 10.1021/acsinfecdis.0c00486. [DOI] [PubMed] [Google Scholar]
- 33.Ko M, Jeon S, Ryu W, Kim S. 2021. Comparative analysis of antiviral efficacy of FDA‐approved drugs against SARS‐CoV‐2 in human lung cells. J Med Virol 93:1403–1408. doi: 10.1002/jmv.26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittaker GR, Daniel S, Millet JK. 2021. Coronavirus entry: how we arrived at SARS-CoV-2. Curr Opin Virol 47:113–120. doi: 10.1016/j.coviro.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach P-Y. 2021. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J 40:e107821. doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White JM, Whittaker GR. 2016. Fusion of enveloped viruses in endosomes. Traffic 17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci USA 102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. 2020. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tummino TA, Rezelj VV, Fischer B, Fischer A, O’Meara MJ, Monel B, Vallet T, White KM, Zhang Z, Alon A, Schadt H, O’Donnell HR, Lyu J, Rosales R, McGovern BL, Rathnasinghe R, Jangra S, Schotsaert M, Galarneau JR, Krogan NJ, Urban L, Shokat KM, Kruse AC, García-Sastre A, Schwartz O, Moretti F, Vignuzzi M, Pognan F, Shoichet BK. 2021. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camprubí D, Almuedo-Riera A, Martí-Soler H, Soriano A, Hurtado JC, Subirà C, Grau-Pujol B, Krolewiecki A, Muñoz J. 2020. Lack of efficacy of standard doses of ivermectin in severe COVID-19 patients. PLoS One 15:e0242184. doi: 10.1371/journal.pone.0242184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szemiel AM, Merits A, Orton RJ, MacLean OA, Pinto RM, Wickenhagen A, Lieber G, Turnbull ML, Wang S, Furnon W, Suarez NM, Mair D, da Silva Filipe A, Willett BJ, Wilson SJ, Patel AH, Thomson EC, Palmarini M, Kohl A, Stewart ME. 2021. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog 17:e1009929. doi: 10.1371/journal.ppat.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou S, Hill CS, Sarkar S, Tse LV, Woodburn BMD, Schinazi RF, Sheahan TP, Baric RS, Heise MT, Swanstrom R. 2021. β-d-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J Infect Dis 224:415–419. doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johansen LM, DeWald LE, Shoemaker CJ, Hoffstrom BG, Lear-Rooney CM, Stossel A, Nelson E, Delos SE, Simmons JA, Grenier JM, Pierce LT, Pajouhesh H, Lehár J, Hensley LE, Glass PJ, White JM, Olinger GG. 2015. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med 7:290ra89. doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- 47.Dyall J, Nelson EA, DeWald LE, Guha R, Hart BJ, Zhou H, Postnikova E, Logue J, Vargas WM, Gross R, Michelotti J, Deiuliis N, Bennett RS, Crozier I, Holbrook MR, Morris PJ, Klumpp-Thomas C, McKnight C, Mierzwa T, Shinn P, Glass PJ, Johansen LM, Jahrling PB, Hensley LE, Olinger GG, Thomas C, White JM. 2018. Identification of combinations of approved drugs with synergistic activity against ebola virus in cell cultures. J Infect Dis 218:S672–S678. doi: 10.1093/infdis/jiy304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finch CL, Dyall J, Xu S, Nelson EA, Postnikova E, Liang JY, Zhou H, DeWald LE, Thomas CJ, Wang A, Xu X, Hughes E, Morris PJ, Mirsalis JC, Nguyen LH, Arolfo MP, Koci B, Holbrook MR, Hensley LE, Jahrling PB, Schmaljohn C, Johansen LM, Olinger GG, Schiffer JT, White JM. 2021. Formulation, stability, pharmacokinetic, and modeling studies for tests of synergistic combinations of orally available approved drugs against Ebola virus in vivo. Microorganisms 9:566. doi: 10.3390/microorganisms9030566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, Emini EA, Chodakewitz JA, Deutsch P, Holder D, Schleif WA, Condra JH. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med 337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 50.Jilek BL, Zarr M, Sampah ME, Rabi SA, Bullen CK, Lai J, Shen L, Siliciano RF. 2012. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med 18:446–451. doi: 10.1038/nm.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shafran SD, Shaw D, Charafeddine M, Agarwal K, Foster GR, Abunimeh M, Pilot-Matias T, Pothacamury RK, Fu B, Cohen E, Cohen DE, Gane E. 2018. Efficacy and safety results of patients with HCV genotype 2 or 3 infection treated with ombitasvir/paritaprevir/ritonavir and sofosbuvir with or without ribavirin (QUARTZ II-III). J Viral Hepat 25:118–125. doi: 10.1111/jvh.12782. [DOI] [PubMed] [Google Scholar]
- 52.Shen L, Rabi SA, Sedaghat AR, Shan L, Lai J, Xing S, Siliciano RF. 2011. A critical subset model provides a conceptual basis for the high antiviral activity of major HIV drugs. Sci Transl Med 3:91ra63. doi: 10.1126/scitranslmed.3002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, Siliciano RF. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med 14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandwani A, Shuter J. 2008. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Ther Clin Risk Manag 4:1023–1033. doi: 10.2147/tcrm.s3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman EM, Worley MV, Unger NR, Gauthier TP, Schafer JJ. 2015. Cobicistat: review of a pharmacokinetic enhancer for HIV infection. Clin Ther 37:1876–1893. doi: 10.1016/j.clinthera.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 56.Lehár J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF, Staunton JE, Jin X, Lee MS, Zimmermann GR, Borisy AA. 2009. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol 27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Y-S, Williamson PR, Zheng W. 2019. Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr Opin Pharmacol 48:92–98. doi: 10.1016/j.coph.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenbloom DIS, Hill AL, Rabi SA, Siliciano RF, Nowak MA. 2012. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med 18:1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sampah MES, Shen L, Jilek BL, Siliciano RF. 2011. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc Natl Acad Sci USA 108:7613–7618. doi: 10.1073/pnas.1018360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. 2021. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med 385:562–566. doi: 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. 2020. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY-T, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Truong TT, Ryutov A, Pandey U, Yee R, Goldberg L, Bhojwani D, Aguayo-Hiraldo P, Pinsky BA, Pekosz A, Shen L, Boyd SD, Wirz OF, Röltgen K, Bootwalla M, Maglinte DT, Ostrow D, Ruble D, Han JH, Biegel JA, Li M, Huang C, Sahoo MK, Pannaraj PS, O’Gorman M, Judkins AR, Gai X, Bard JD. 2021. Persistent SARS-CoV-2 infection and increasing viral variants in children and young adults with impaired humoral immunity. medRxiv 10.1101/2021.02.27.21252099. [DOI] [PMC free article] [PubMed]
- 64.Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, CITIID-NIHR BioResource COVID-19 Collaboration, COVID-19 Genomics UK (COG-UK) Consortium, Sharrocks K, Blane E, Modis Y, Leigh KE, Briggs JAG, van Gils MJ, Smith KGC, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth CJR, McCoy LE, Gupta RK. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JM, Eguia R, Zost SJ, Choudhary S, Wilson PC, Bedford T, Stevens-Ayers T, Boeckh M, Hurt AC, Lakdawala SS, Hensley SE, Bloom JD. 2019. Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. Elife 8:e49324. doi: 10.7554/eLife.49324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belardo G, Cenciarelli O, La Frazia S, Rossignol JF, Santoro MG. 2015. Synergistic effect of nitazoxanide with neuraminidase inhibitors against influenza A viruses in vitro. Antimicrob Agents Chemother 59:1061–1069. doi: 10.1128/AAC.03947-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bekerman E, Neveu G, Shulla A, Brannan J, Pu S-Y, Wang S, Xiao F, Barouch-Bentov R, Bakken RR, Mateo R, Govero J, Nagamine CM, Diamond MS, De Jonghe S, Herdewijn P, Dye JM, Randall G, Einav S. 2017. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J Clin Invest 127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun W, He S, Martínez-Romero C, Kouznetsova J, Tawa G, Xu M, Shinn P, Fisher E, Long Y, Motabar O, Yang S, Sanderson PE, Williamson PR, García-Sastre A, Qiu X, Zheng W. 2017. Synergistic drug combination effectively blocks Ebola virus infection. Antiviral Res 137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herring S, Oda JM, Wagoner J, Kirchmeier D, O’Connor A, Nelson EA, Huang Q, Liang Y, DeWald LE, Johansen LM, Glass PJ, Olinger GG, Ianevski A, Aittokallio T, Paine MF, Fink SL, White JM, Polyak SJ. 2021. Inhibition of arenaviruses by combinations of orally available approved drugs. Antimicrob Agents Chemother 65:e01146-20. doi: 10.1128/AAC.01146-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choy K-T, Wong AY-L, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PP-H, Huang X, Peiris M, Yen H-L. 2020. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res 178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ianevski A, Yao R, Fenstad MH, Biza S, Zusinaite E, Reisberg T, Lysvand H, Løseth K, Landsem VM, Malmring JF, Oksenych V, Erlandsen SE, Aas PA, Hagen L, Pettersen CH, Tenson T, Afset JE, Nordbø SA, Bjørås M, Kainov DE. 2020. Potential antiviral options against SARS-CoV-2 infection. Viruses 12:642. doi: 10.3390/v12060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD, Kavanagh Williamson M, Shoemark DK, Garzoni F, Staufer O, Milligan R, Capin J, Mulholland AJ, Spatz J, Fitzgerald D, Berger I, Schaffitzel C. 2020. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bobrowski T, Chen L, Eastman RT, Itkin Z, Shinn P, Chen CZ, Guo H, Zheng W, Michael S, Simeonov A, Hall MD, Zakharov AV, Muratov EN. 2021. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol Ther 29:873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ordonez AA, Bullen CK, Villabona-Rueda AF, Thompson EA, Turner ML, Davis SL, Komm O, Powell JD, D’Alessio FR, Yolken RH, Jain SK, Jones-Brando L. 2021. Sulforaphane exhibits in vitro and in vivo antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses. bioRxiv 10.1101/2021.03.25.437060. [DOI] [PMC free article] [PubMed]
- 75.Bafna K, White K, Harish B, Rosales R, Ramelot TA, Acton TB, Moreno E, Kehrer T, Miorin L, Royer CA, García-Sastre A, Krug RM, Montelione GT. 2021. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep 35:109133. doi: 10.1016/j.celrep.2021.109133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raymonda MH, Ciesla JH, Monaghan M, Leach J, Asantewaa G, Smorodintsev-Schiller LA, Lutz MM, Schafer XL, Takimoto T, Dewhurst S, Munger J, Harris IS. 2020. Pharmacologic profiling reveals lapatinib as a novel antiviral against SARS-CoV-2 in vitro. bioRxiv [DOI] [PMC free article] [PubMed]
- 77.Schultz DC, Johnson RM, Ayyanathan K, Miller J, Whig K, Kamalia B, Dittmar M, Weston S, Hammond HL, Dillen C, Castellana L, Lee JS, Li M, Lee E, Constant S, Ferrer M, Thaiss CA, Frieman MB, Cherry S. 2021. Pyrimidine biosynthesis inhibitors synergize with nucleoside analogs to block SARS-CoV-2 infection. bioRxiv 10.1101/2021.06.24.449811. [DOI] [PMC free article] [PubMed]
- 78.Ohashi H, Watashi K, Saso W, Shionoya K, Iwanami S, Hirokawa T, Shirai T, Kanaya S, Ito Y, Kim KS, Nomura T, Suzuki T, Nishioka K, Ando S, Ejima K, Koizumi Y, Tanaka T, Aoki S, Kuramochi K, Suzuki T, Hashiguchi T, Maenaka K, Matano T, Muramatsu M, Saijo M, Aihara K, Iwami S, Takeda M, McKeating JA, Wakita T. 2021. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 24:102367. doi: 10.1016/j.isci.2021.102367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyenla X, Wehri E, Van Dis E, Biering SB, Yamashiro LH, Stroumza J, Dugast-Darzacq C, Graham T, Stanley S, Schaletzky J. 2020. Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells. bioRxiv 10.1101/2020.09.18.302398. [DOI] [PMC free article] [PubMed]
- 80.Ianevski A, Yao R, Biza S, Zusinaite E, Mannik A, Kivi G, Planken A, Kurg K, Tombak E-M, Ustav M, Shtaida N, Kulesskiy E, Jo E, Yang J, Lysvand H, Løseth K, Oksenych V, Aas PA, Tenson T, Vitkauskienė A, Windisch MP, Fenstad MH, Nordbø SA, Ustav M, Bjørås M, Kainov DE. 2020. Identification and tracking of antiviral drug combinations. Viruses 12:1178. doi: 10.3390/v12101178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boras B, Jones RM, Anson BJ, Arenson D, Aschenbrenner L, Bakowski MA, Beutler N, Binder J, Chen E, Eng H, Hammond H, Hammond J, Haupt RE, Hoffman R, Kadar EP, Kania R, Kimoto E, Kirkpatrick MG, Lanyon L, Lendy EK, Lillis JR, Logue J, Luthra SA, Ma C, Mason SW, McGrath ME, Noell S, Obach RS, O’Brien MN, O’Connor R, Ogilvie K, Owen D, Pettersson M, Reese MR, Rogers TF, Rossulek MI, Sathish JG, Shirai N, Steppan C, Ticehurst M, Updyke LW, Weston S, Zhu Y, Wang J, Chatterjee AK, Mesecar AD, Frieman MB, Anderson AS, Allerton C. 2020. Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. bioRxiv 10.1101/2020.09.12.293498. [DOI]
- 82.Son J, Huang S, Zeng Q, Bricker TL, Case JB, Zhou J, Zang R, Liu Z, Chang X, Harastani HH, Chen L, Castro MFG, Zhao Y, Kohio HP, Hou G, Fan B, Niu B, Guo R, Rothlauf PW, Bailey AL, Wang X, Shi P-Y, Martinez ED, Whelan SPJ, Diamond MS, Boon ACM, Li B, Ding S. 2020. Nitazoxanide and JIB-04 have broad-spectrum antiviral activity and inhibit SARS-CoV-2 replication in cell culture and coronavirus pathogenesis in a pig model. bioRxiv 10.1101/2020.09.24.312165. [DOI] [PMC free article] [PubMed]
- 83.Yuan S, Yin X, Meng X, Chan J, Ye Z-W, Riva L, Pache L, Chan CC-Y, Lai P-M, Chan C, Poon V, Matsunaga N, Pu Y, Yuen C-K, Cao J, Liang R, Tang K, Sheng L, Du Y, Xu W, Sze K-H, Zhang J, Chu H, Kok K-H, To K, Jin D-Y, Sun R, Chanda S, Yuen K-Y. 2020. Clofazimine is a broad-spectrum coronavirus inhibitor that antagonizes SARS-CoV-2 replication in primary human cell culture and hamsters. Res Sq 10.21203/rs.3.rs-86169/v1.
- 84.Shionoya K, Yamasaki M, Iwanami S, Ito Y, Fukushi S, Ohashi H, Saso W, Tanaka T, Aoki S, Kuramochi K, Iwami S, Takahashi Y, Suzuki T, Muramatsu M, Takeda M, Wakita T, Watashi K. 2021. Mefloquine, a potent anti-severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) drug as an entry inhibitor in vitro. Front Microbiol 12:651403. doi: 10.3389/fmicb.2021.651403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeffreys L, Pennington SH, Duggan J, Breen A, Jinks J, Ardrey A, Donnellan S, Patterson EI, Hughes GI, Hong WD, O’Neill PM, Aljayyoussi G, Owen A, Ward SA, Biagini G. 2020. Remdesivir-ivermectin combination displays synergistic interaction with improved in vitro antiviral activity against SARS-CoV-2. bioRxiv 10.1101/2020.12.23.424232. [DOI] [PMC free article] [PubMed]
- 86.Ianevski A, Yao R, Zusinaite E, Lello LS, Wang S, Jo E, Yang J, Ravlo E, Wang W, Lysvand H, Løseth K, Oksenych V, Tenson T, Windisch MP, Poranen M, Nieminen AI, Nordbø SA, Fenstad MH, Grødeland G, Aukrust P, Trøseid M, Kantele A, Vitkauskiene A, Legrand N, Merits A, Bjørås M, Kainov DE. 2021. Interferon alpha-based combinations suppress SARS-CoV-2 infection in vitro and in vivo. bioRxiv 10.1101/2021.01.05.425331. [DOI] [PMC free article] [PubMed]
- 87.Biering SB, Van Dis E, Wehri E, Yamashiro LH, Nguyenla X, Dugast-Darzacq C, Graham TGW, Stroumza JR, Golovkine GR, Roberts AW, Fines DM, Spradlin JN, Ward CC, Bajaj T, Dovala D, Schulze-Gamen U, Bajaj R, Fox DM, Ott M, Murthy N, Nomura DK, Schaletzky J, Stanley SA. 2021. Screening a library of FDA-approved and bioactive compounds for antiviral activity against SARS-CoV-2. ACS Infect Dis 7:2337−2351. 10.1021/acsinfecdis.1c00017. doi: 10.1021/acsinfecdis.1c00017. [DOI] [PubMed] [Google Scholar]
- 88.Bakovic A, Risner K, Bhalla N, Alem F, Chang TL, Weston WK, Harness JA, Narayanan A. 2021. Brilacidin demonstrates inhibition of SARS-CoV-2 in cell culture. Viruses 13:271. doi: 10.3390/v13020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gammeltoft KA, Zhou Y, Duarte Hernandez CR, Galli A, Offersgaard A, Costa R, Pham LV, Fahnøe U, Feng S, Scheel TKH, Ramirez S, Bukh J, Gottwein JM. 2021. Hepatitis C virus protease inhibitors show differential efficacy and interactions with remdesivir for treatment of SARS-CoV-2 in vitro. Antimicrob Agents Chemother 65:e0268-20. doi: 10.1128/AAC.02680-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shytaj IL, Fares M, Lucic B, Gallucci L, Tolba MM, Zimmermann L, Ayoub AT, Cortese M, Neufeldt CJ, Laketa V, Chlanda P, Fackler OT, Boulant S, Bartenschlager R, Stanifer M, Savarino A, Lusic M. 2021. The FDA-approved drug cobicistat synergizes with remdesivir to inhibit SARS-CoV-2 replication. bioRxiv 10.1101/2021.03.09.434219. [DOI] [PMC free article] [PubMed]
- 91.Pohl MO, Busnadiego I, Marrafino F, Wiedmer L, Hunziker A, Fernbach S, Glas I, Moroz-Omori EV, Hale BG, Caflisch A, Stertz S. 2021. Combined computational and cellular screening identifies synergistic inhibition of SARS-CoV-2 by lenvatinib and remdesivir. J Gen Virol 102:001625. doi: 10.1099/jgv.0.001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pizzorno A, Padey B, Dubois J, Julien T, Traversier A, Dulière V, Brun P, Lina B, Rosa-Calatrava M, Terrier O. 2020. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res 181:104878. doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Imamura K, Sakurai Y, Enami T, Shibukawa R, Nishi Y, Ohta A, Shu T, Kawaguchi J, Okada S, Hoenen T, Yasuda J, Inoue H. 2021. iPSC screening for drug repurposing identifies anti-RNA virus agents modulating host cell susceptibility. FEBS Open Bio 11:1452–1464. doi: 10.1002/2211-5463.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ko M, Chang SY, Byun SY, Ianevski A, Choi I, Pham Hung d’Alexandry d’Orengiani A-L, Ravlo E, Wang W, Bjørås M, Kainov DE, Shum D, Min J-Y, Windisch MP. 2021. Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. Viruses 13:651. doi: 10.3390/v13040651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cao J, Liu Y, Zhou M, Dong S, Jia X, Lan X, Zhang Y, Guo J, Xiao G, Wang W. 2021. Screening of botanical drugs against SARS-CoV-2 entry. bioRxiv 10.1101/2021.06.03.447021. [DOI] [PMC free article] [PubMed]
- 96.Ianevski A, Yao R, Lysvand H, Grødeland G, Legrand N, Tenson T, Bjørås M, Kainov DE. 2021. Nafamostat-interferon-alpha combination suppresses SARS-CoV-2 infection in vitro and in vivo. bioRxiv 10.1101/2021.06.16.448653. [DOI] [PMC free article] [PubMed]
- 97.Pacl HT, Tipper JL, Sevalkar RR, Crouse A, Crowder C, UAB Precision Medicine Institute, Ahmad S, Ahmad A, Holder GD, Kuhlman CJ, Chinta KC, Nadeem S, Green TJ, Petit CM, Steyn AJC, Might M, Harrod KS. 2021. Water-soluble tocopherol derivatives inhibit SARS-CoV-2 RNA-dependent RNA polymerase. bioRxiv 10.1101/2021.07.13.449251. [DOI]
- 98.Sacramento CQ, Fintelman-Rodrigues N, Dias SSG, Temerozo JR, Da Silva ADPD, da Silva CS, Ferreira AC, Mattos M, Soares VC, Pereira-Dutra F, Miranda MD, Barreto-Vieira DF, da Silva MAN, Santos SS, Torres M, Rajoli RKR, Paccanaro A, Owen A, Bou-Habib DC, Bozza PT, Souza TML. 2021. Unlike Chloroquine, mefloquine inhibits SARS-CoV-2 infection in physiologically relevant cells and does not induce viral variants. bioRxiv 10.1101/2021.07.21.451321. [DOI] [PMC free article] [PubMed]
- 99.Jang WD, Jeon S, Kim S, Lee SY. 2021. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc Natl Acad Sci USA 118:e2024302118. doi: 10.1073/pnas.2024302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin W, Stokes JM, Eastman RT, Itkin Z, Zakharov AV, Collins JJ, Jaakkola TS, Barzilay R. 2021. Deep learning identifies synergistic drug combinations for treating COVID-19. Proc Natl Acad Sci USA 118:e2105070118. doi: 10.1073/pnas.2105070118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fu L, Ye F, Feng Y, Yu F, Wang Q, Wu Y, Zhao C, Sun H, Huang B, Niu P, Song H, Shi Y, Li X, Tan W, Qi J, Gao GF. 2020. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun 11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kreutzberger AJB, Sanyal A, Ojha R, Pyle JD, Vapalahti O, Balistreri G, Kirchhausen T. 2021. Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. J Virol 95:e00975-21. doi: 10.1128/JVI.00975-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gordon CJ, Tchesnokov EP, Schinazi RF, Götte M. 2021. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem 297:100770. doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Robson F, Khan KS, Le TK, Paris C, Demirbag S, Barfuss P, Rocchi P, Ng W-L. 2020. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell 79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalil AC, Patterson TF, Mehta AK, Tomashek KM, Wolfe CR, Ghazaryan V, Marconi VC, Ruiz-Palacios GM, Hsieh L, Kline S, Tapson V, Iovine NM, Jain MK, Sweeney DA, El Sahly HM, Branche AR, Regalado Pineda J, Lye DC, Sandkovsky U, Luetkemeyer AF, Cohen SH, Finberg RW, Jackson PEH, Taiwo B, Paules CI, Arguinchona H, Erdmann N, Ahuja N, Frank M, Oh M-D, Kim E-S, Tan SY, Mularski RA, Nielsen H, Ponce PO, Taylor BS, Larson L, Rouphael NG, Saklawi Y, Cantos VD, Ko ER, Engemann JJ, Amin AN, Watanabe M, Billings J, Elie M-C, Davey RT, Burgess TH, Ferreira J, Green M, Makowski M, et al. 2021. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N Engl J Med 384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stebbing J, Sánchez Nievas G, Falcone M, Youhanna S, Richardson P, Ottaviani S, Shen JX, Sommerauer C, Tiseo G, Ghiadoni L, Virdis A, Monzani F, Rizos LR, Forfori F, Avendaño Céspedes A, De Marco S, Carrozzi L, Lena F, Sánchez-Jurado PM, Lacerenza LG, Cesira N, Caldevilla Bernardo D, Perrella A, Niccoli L, Méndez LS, Matarrese D, Goletti D, Tan Y-J, Monteil V, Dranitsaris G, Cantini F, Farcomeni A, Dutta S, Burley SK, Zhang H, Pistello M, Li W, Romero MM, Andrés Pretel F, Simón-Talero RS, García-Molina R, Kutter C, Felce JH, Nizami ZF, Miklosi AG, Penninger JM, Menichetti F, Mirazimi A, Abizanda P, Lauschke VM. 2021. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci Adv 7:eabe4724. doi: 10.1126/sciadv.abe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marconi VC, Ramanan AV, de Bono S, Kartman CE, Krishnan V, Liao R, Piruzeli MLB, Goldman JD, Alatorre-Alexander J, de Cassia Pellegrini R, Estrada V, Som M, Cardoso A, Chakladar S, Crowe B, Reis P, Zhang X, Adams DH, Ely EW, COV-BARRIER Study Group . 2021. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir Med doi: 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hung IF-N, Lung K-C, Tso EY-K, Liu R, Chung TW-H, Chu M-Y, Ng Y-Y, Lo J, Chan J, Tam AR, Shum H-P, Chan V, Wu AK-L, Sin K-M, Leung W-S, Law W-L, Lung DC, Sin S, Yeung P, Yip CC-Y, Zhang RR, Fung AY-F, Yan EY-W, Leung K-H, Ip JD, Chu AW-H, Chan W-M, Ng AC-K, Lee R, Fung K, Yeung A, Wu T-C, Chan JW-M, Yan W-W, Chan W-M, Chan JF-W, Lie AK-W, Tsang OT-Y, Cheng VC-C, Que T-L, Lau C-S, Chan K-H, To KK-W, Yuen K-Y. 2020. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ivashchenko AA, Azarova VN, Egorova AN, Karapetian RN, Kravchenko DV, Krivonos NV, Loginov VG, Poyarkov SV, Merkulova EA, Rosinkova OS, Savchuk NP, Topr MA, Simakina EN, Yakubova EV, Ivachtchenko AV. 2021. Effect of Aprotinin and Avifavir® combination therapy for moderate COVID-19 patients. Viruses 13:1253. doi: 10.3390/v13071253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martinez DR, Schäfer A, Leist SR, Li D, Gully K, Yount B, Feng JY, Bunyan E, Porter DP, Cihlar T, Montgomery SA, Haynes BF, Baric RS, Nussenzweig MC, Sheahan TP. 2021. Prevention and therapy of SARS-CoV-2 and the B.1.351 variant in mice. Cell Rep 36:109450. doi: 10.1016/j.celrep.2021.109450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tian D, Liu Y, Liang C, Xin L, Xie X, Zhang D, Wan M, Li H, Fu X, Liu H, Cao W. 2021. An update review of emerging small-molecule therapeutic options for COVID-19. Biomed Pharmacother 137:111313. doi: 10.1016/j.biopha.2021.111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MÁ, Urquiza J, Ramírez D, Alonso C, Campillo NE, Martinez A. 2020. COVID-19: drug targets and potential treatments. J Med Chem 63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 114.Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, Aoued H, Tharp GM, Pellegrini KL, Manfredi C, Sorscher E, Mainou B, Lobby JL, Kohlmeier JE, Lowen AC, Shi P-Y, Menachery VD, Anderson LJ, Grakoui A, Bosinger SE, Suthar MS. 2020. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J Virol 94:e00985-20. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beumer J, Geurts MH, Lamers MM, Puschhof J, Zhang J, van der Vaart J, Mykytyn AZ, Breugem TI, Riesebosch S, Schipper D, van den Doel PB, de Lau W, Pleguezuelos-Manzano C, Busslinger G, Haagmans BL, Clevers H. 2021. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat Commun 12:5498. doi: 10.1038/s41467-021-25729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ianevski A, Giri AK, Aittokallio T. 2020. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yadav B, Wennerberg K, Aittokallio T, Tang J. 2015. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J 13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meganck RM, Baric RS. 2021. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat Med 27:401–410. doi: 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- 119.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, Braga L, Williamson MK, Hassard JA, Staller E, Hanley B, Osborn M, Giacca M, Davidson AD, Matthews DA, Barclay WS. 2021. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 120.Hoffmann M, Kleine-Weber H, Pöhlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schiffer JT, Swan DA, Corey L, Wald A. 2013. Rapid viral expansion and short drug half-life explain the incomplete effectiveness of current herpes simplex virus 2-directed antiviral agents. Antimicrob Agents Chemother 57:5820–5829. doi: 10.1128/AAC.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schiffer JT, Swan DA, Magaret A, Corey L, Wald A, Ossig J, Ruebsamen-Schaeff H, Stoelben S, Timmler B, Zimmermann H, Melhem MR, Van Wart SA, Rubino CM, Birkmann A. 2016. Mathematical modeling of herpes simplex virus-2 suppression with pritelivir predicts trial outcomes. Sci Transl Med 8:324ra15. doi: 10.1126/scitranslmed.aad6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boffito M, Back DJ, Flexner C, Sjö P, Blaschke TF, Horby PW, Cattaneo D, Acosta EP, Anderson P, Owen A. 2021. Toward consensus on correct interpretation of protein binding in plasma and other biological matrices for COVID-19 therapeutic development. Clin Pharmacol Ther 110:64–68. doi: 10.1002/cpt.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smith DA, Di L, Kerns EH. 2010. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat Rev Drug Discov 9:929–939. doi: 10.1038/nrd3287. [DOI] [PubMed] [Google Scholar]
- 125.Dampalla CS, Zheng J, Perera KD, Wong L-YR, Meyerholz DK, Nguyen HN, Kashipathy MM, Battaile KP, Lovell S, Kim Y, Perlman S, Groutas WC, Chang K-O. 2021. Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc Natl Acad Sci USA 118:e2101555118. doi: 10.1073/pnas.2101555118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vandyck K, Deval J. 2021. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr Opin Virol 49:36–40. doi: 10.1016/j.coviro.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morita T, Miyakawa K, Jeremiah SS, Yamaoka Y, Sada M, Kuniyoshi T, Yang J, Kimura H, Ryo A. 2021. All-trans retinoic acid exhibits antiviral effect against SARS-CoV-2 by inhibiting 3CLpro activity. Viruses 13:1669. doi: 10.3390/v13081669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Good SS, Westover J, Jung KH, Zhou X-J, Moussa A, La Colla P, Collu G, Canard B, Sommadossi J-P. 2021. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob Agents Chemother 65:e02479-20. doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sourimant J, Lieber CM, Aggarwal M, Cox RM, Wolf JD, Yoon J-J, Toots M, Ye C, Sticher Z, Kolykhalov AA, Martinez-Sobrido L, Bluemling GR, Natchus MG, Painter GR, Plemper RK. 2 December 2021. 4′-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science. 10.1126/science.abj5508. [DOI] [PMC free article] [PubMed]
- 130.Carpinteiro A, Gripp B, Hoffmann M, Pöhlmann S, Hoertel N, Edwards MJ, Kamler M, Kornhuber J, Becker KA, Gulbins E. 2021. Inhibition of acid sphingomyelinase by ambroxol prevents SARS-CoV-2 entry into epithelial cells. J Biol Chem 296:100701. doi: 10.1016/j.jbc.2021.100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bradfute SB, Ye C, Clarke EC, Kumar S, Timmins GS, Deretic V. 2020. Ambroxol and ciprofloxacin show activity against SARS-CoV2 in Vero E6 cells at clinically-relevant concentrations. bioRxiv 10.1101/2020.08.11.245100. [DOI]
- 132.Sun YJ, Velez G, Parsons D, Li K, Ortiz M, Sharma S, McCray PB, Bassuk AG, Mahajan VB. 2021. TMPRSS2 structure-phylogeny repositions Avoralstat for SARS-CoV-2 prophylaxis in mice. bioRxiv 10.1101/2021.01.04.425289. [DOI] [PMC free article] [PubMed]
- 133.Shapira T, Monreal IA, Dion SP, Jager M, Désilets A, Olmstead AD, Vandal T, Buchholz DW, Imbiakha B, Gao G, Chin A, Rees WD, Steiner T, Nabi IR, Marsault E, Sahler J, August A, Van de Walle G, Whittaker GR, Boudreault P-L, Aguilar HC, Leduc R, Jean F. 2021. A novel highly potent inhibitor of TMPRSS2-like proteases blocks SARS-CoV-2 variants of concern and is broadly protective against infection and mortality in mice. bioRxiv 10.1101/2021.05.03.442520. [DOI]
- 134.Mahoney M, Damalanka VC, Tartell MA, Chung DH, Lourenço AL, Pwee D, Mayer Bridwell AE, Hoffmann M, Voss J, Karmakar P, Azouz NP, Klingler AM, Rothlauf PW, Thompson CE, Lee M, Klampfer L, Stallings CL, Rothenberg ME, Pöhlmann S, Whelan SPJ, O’Donoghue AJ, Craik CS, Janetka JW. 2021. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc Natl Acad Sci USA 118:e2108728118. doi: 10.1073/pnas.2108728118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ianevski A, Timonen S, Kononov A, Aittokallio T, Giri AK. 2020. SynToxProfiler: an interactive analysis of drug combination synergy, toxicity and efficacy. PLoS Comput Biol 16:e1007604. doi: 10.1371/journal.pcbi.1007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 137.ACTIV-3/TICO LY-CoV555 Study Group, Lundgren JD, Grund B, Barkauskas CE, Holland TL, Gottlieb RL, Sandkovsky U, Brown SM, Knowlton KU, Self WH, Files DC, Jain MK, Benfield T, Bowdish ME, Leshnower BG, Baker JV, Jensen J-U, Gardner EM, Ginde AA, Harris ES, Johansen IS, Markowitz N, Matthay MA, Østergaard L, Chang CC, Davey VJ, Goodman A, Higgs ES, Murray DD, Murray TA, Paredes R, Parmar MKB, Phillips AN, Reilly C, Sharma S, Dewar RL, Teitelbaum M, Wentworth D, Cao H, Klekotka P, Babiker AG, Gelijns AC, Kan VL, Polizzotto MN, Thompson BT, Lane HC, Neaton JD. 2021. A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N Engl J Med 384:905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 139.Schooley RT, Carlin AF, Beadle JR, Valiaeva N, Zhang X-Q, Clark AE, McMillan RE, Leibel SL, McVicar RN, Xie J, Garretson AF, Smith VI, Murphy J, Hostetler KY. 2021. Rethinking remdesivir: synthesis, antiviral activity and pharmacokinetics of oral lipid prodrugs. bioRxiv 10.1101/2020.08.26.269159. [DOI] [PMC free article] [PubMed]
- 140.Schäfer A, Martinez DR, Won JJ, Moreira FR, Brown AJ, Gully KL, Kalla R, Chun K, Du Pont V, Babusis D, Tang J, Murakami E, Subramanian R, Barrett KT, Bleier BJ, Bannister R, Feng JY, Bilello JP, Cihlar T, Mackman RL, Montgomery SA, Baric RS, Sheahan TP. 2021. Therapeutic efficacy of an oral nucleoside analog of remdesivir against SARS-CoV-2 pathogenesis in mice. bioRxiv 10.1101/2021.09.13.460111. [DOI]
- 141.Cao L, Li Y, Yang S, Li G, Zhou Q, Sun J, Xu T, Yang Y, Zhu T, Huang S, Ji Y, Cong F, Luo Y, Zhu Y, Luan H, Zhang H, Chen J, Liu X, Wang P, Yu Y, Xing F, Ke B, Zheng H, Deng X, Zhang W, Li C, Zhang Y, Zhao J, Zhang X, Guo D. 2021. The adenosine analogue prodrug ATV006 is orally bioavailable and has potent preclinical efficacy against SARS-CoV-2 and its variants. bioRxiv 10.1101/2021.10.13.463130. [DOI] [PMC free article] [PubMed]
- 142.White KM, Rosales R, Yildiz S, Kehrer T, Miorin L, Moreno E, Jangra S, Uccellini MB, Rathnasinghe R, Coughlan L, Martinez-Romero C, Batra J, Rojc A, Bouhaddou M, Fabius JM, Obernier K, Dejosez M, Guillén MJ, Losada A, Avilés P, Schotsaert M, Zwaka T, Vignuzzi M, Shokat KM, Krogan NJ, García-Sastre A. 2021. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heinen N, Meister TL, Klöhn M, Steinmann E, Todt D, Pfaender S. 2021. Antiviral effect of budesonide against SARS-CoV-2. Viruses 13:1411. doi: 10.3390/v13071411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsuyama S, Kawase M, Nao N, Shirato K, Ujike M, Kamitani W, Shimojima M, Fukushi S. 2020. The inhaled steroid ciclesonide blocks SARS-CoV-2 RNA replication by targeting the viral replication-transcription complex in cultured cells. J Virol 95:e01648-20. doi: 10.1128/JVI.01648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, Gabbay FJ, Davies DE, Holgate ST, Ho L-P, Clark T, Djukanovic R, Wilkinson TMA, Inhaled Interferon Beta COVID-19 Study Group . 2021. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med 9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li K, Meyerholz DK, Bartlett JA, McCray PB. 2021. The TMPRSS2 inhibitor nafamostat reduces SARS-CoV-2 pulmonary infection in mouse models of COVID-19. mBio 12:e00970-21. doi: 10.1128/mBio.00970-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Backer V, Sjöbring U, Sonne J, Weiss A, Hostrup M, Johansen HK, Becker V, Sonne DP, Balchen T, Jellingsø M, Sommer MOA. 2021. A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: a broad spectrum antiviral candidate for treatment of COVID-19. Lancet Reg Health Eur 4:100084. doi: 10.1016/j.lanepe.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Case JB, Chen RE, Cao L, Ying B, Winkler ES, Johnson M, Goreshnik I, Pham MN, Shrihari S, Kafai NM, Bailey AL, Xie X, Shi P-Y, Ravichandran R, Carter L, Stewart L, Baker D, Diamond MS. 2021. Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe 29:1151−1161.e5. doi: 10.1016/j.chom.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, et al. 2020. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.de Vries RD, Schmitz KS, Bovier FT, Predella C, Khao J, Noack D, Haagmans BL, Herfst S, Stearns KN, Drew-Bear J, Biswas S, Rockx B, McGill G, Dorrello NV, Gellman SH, Alabi CA, de Swart RL, Moscona A, Porotto M. 2021. Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science 371:1379–1382. doi: 10.1126/science.abf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu W, Reyes HM, Yang JF, Li Y, Stewart KM, Basil MC, Lin SM, Katzen J, Morrisey EE, Weiss SR, You J. 2021. Activation of STING signaling pathway effectively blocks human coronavirus infection. J Virol 95:e00490-21. doi: 10.1128/JVI.00490-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li M, Ferretti M, Ying B, Descamps H, Lee E, Dittmar M, Lee JS, Whig K, Kamalia B, Dohnalová L, Uhr G, Zarkoob H, Chen Y-C, Ramage H, Ferrer M, Lynch K, Schultz DC, Thaiss CA, Diamond MS, Cherry S. 2021. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci Immunol 6:eabi9007. doi: 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.National Academies of Sciences, Engineering, and Medicine, Health and Medicine Division, Board on Health Sciences Policy, Board on Global Health, Committee on Clinical Trials During the 2014–2015 Ebola Outbreak. 2017. Integrating clinical research into epidemic response: the Ebola experience. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 154.Barnabas RV, Brown ER, Bershteyn A, Stankiewicz Karita HC, Johnston C, Thorpe LE, Kottkamp A, Neuzil KM, Laufer MK, Deming M, Paasche-Orlow MK, Kissinger PJ, Luk A, Paolino K, Landovitz RJ, Hoffman R, Schaafsma TT, Krows ML, Thomas KK, Morrison S, Haugen HS, Kidoguchi L, Wener M, Greninger AL, Huang M-L, Jerome KR, Wald A, Celum C, Chu HY, Baeten JM, Baeten JM, Hydroxychloroquine COVID-19 PEP Study Team . 2021. Hydroxychloroquine as postexposure prophylaxis to prevent severe acute respiratory syndrome coronavirus 2 infection: a randomized trial. Ann Intern Med 174:344–352. doi: 10.7326/M20-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lenze EJ, Mattar C, Zorumski CF, Stevens A, Schweiger J, Nicol GE, Miller JP, Yang L, Yingling M, Avidan MS, Reiersen AM. 2020. Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID-19: a randomized clinical trial. JAMA 324:2292–2300. doi: 10.1001/jama.2020.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Reis G, Dos Santos Moreira-Silva EA, Silva DCM, Thabane L, Milagres AC, Ferreira TS, Dos Santos CVQ, de Souza Campos VH, Nogueira AMR, de Almeida APFG, Callegari ED, de Figueiredo Neto AD, Savassi LCM, Simplicio MIC, Ribeiro LB, Oliveira R, Harari O, Forrest JI, Ruton H, Sprague S, McKay P, Glushchenko AV, Rayner CR, Lenze EJ, Reiersen AM, Guyatt GH, Mills EJ, TOGETHER investigators . 2021. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the TOGETHER randomised, platform clinical trial. Lancet Glob Health doi: 10.1016/S2214-109X(21)00448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Andrasik MP, Broder GB, Wallace SE, Chaturvedi R, Michael NL, Bock S, Beyrer C, Oseso L, Aina J, Lucas J, Wilson DR, Kublin JG, Mensah GA. 2021. Increasing Black, Indigenous and People of Color participation in clinical trials through community engagement and recruitment goal establishment. PLoS One 16:e0258858. doi: 10.1371/journal.pone.0258858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duke ER, Williamson BD, Borate B, Golob JL, Wychera C, Stevens-Ayers T, Huang M-L, Cossrow N, Wan H, Mast TC, Marks MA, Flowers ME, Jerome KR, Corey L, Gilbert PB, Schiffer JT, Boeckh M. 2021. CMV viral load kinetics as surrogate endpoints after allogeneic transplantation. J Clin Invest 131:e133960. doi: 10.1172/JCI133960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murray JS, Elashoff MR, Iacono-Connors LC, Cvetkovich TA, Struble KA. 1999. The use of plasma HIV RNA as a study endpoint in efficacy trials of antiretroviral drugs. AIDS 13:797–804. doi: 10.1097/00002030-199905070-00008. [DOI] [PubMed] [Google Scholar]
- 160.Foster GR, Afdhal N, Roberts SK, Bräu N, Gane EJ, Pianko S, Lawitz E, Thompson A, Shiffman ML, Cooper C, Towner WJ, Conway B, Ruane P, Bourlière M, Asselah T, Berg T, Zeuzem S, Rosenberg W, Agarwal K, Stedman CAM, Mo H, Dvory-Sobol H, Han L, Wang J, McNally J, Osinusi A, Brainard DM, McHutchison JG, Mazzotta F, Tran TT, Gordon SC, Patel K, Reau N, Mangia A, Sulkowski M, ASTRAL-2 Investigators, ASTRAL-3 Investigators . 2015. Sofosbuvir and velpatasvir for HCV genotype 2 and 3 infection. N Engl J Med 373:2608–2617. doi: 10.1056/NEJMoa1512612. [DOI] [PubMed] [Google Scholar]
- 161.Agyemang E, Magaret AS, Selke S, Johnston C, Corey L, Wald A. 2018. Herpes simplex virus shedding rate: surrogate outcome for genital herpes recurrence frequency and lesion rates, and phase 2 clinical trials end point for evaluating efficacy of antivirals. J Infect Dis 218:1691–1699. doi: 10.1093/infdis/jiy372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hoffman LR, Kuntz ID, White JM. 1997. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol 71:8808–8820. doi: 10.1128/JVI.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bodian DL, Yamasaki RB, Buswell RL, Stearns JF, White JM, Kuntz ID. 1993. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry 32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 164.Narkhede YB, Gonzalez KJ, Strauch E-M. 2021. Targeting viral surface proteins through structure-based design. Viruses 13:1320. doi: 10.3390/v13071320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xia S, Lan Q, Zhu Y, Wang C, Xu W, Li Y, Wang L, Jiao F, Zhou J, Hua C, Wang Q, Cai X, Wu Y, Gao J, Liu H, Sun G, Münch J, Kirchhoff F, Yuan Z, Xie Y, Sun F, Jiang S, Lu L. 2021. Structural and functional basis for pan-CoV fusion inhibitors against SARS-CoV-2 and its variants with preclinical evaluation. Signal Transduct Target Ther 6:288. doi: 10.1038/s41392-021-00712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet L-M, Zeng Q, Tahan S, Droit L, Ilagan MXG, Tartell MA, Amarasinghe G, Henderson JP, Miersch S, Ustav M, Sidhu S, Virgin HW, Wang D, Ding S, Corti D, Theel ES, Fremont DH, Diamond MS, Whelan SPJ. 2020. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Luo D, Vasudevan SG, Lescar J. 2015. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res 118:148–158. doi: 10.1016/j.antiviral.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 168.Das PK, Puusepp L, Varghese FS, Utt A, Ahola T, Kananovich DG, Lopp M, Merits A, Karelson M. 2016. Design and validation of novel chikungunya virus protease inhibitors. Antimicrob Agents Chemother 60:7382–7395. doi: 10.1128/AAC.01421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mazzon M, Ortega-Prieto AM, Imrie D, Luft C, Hess L, Czieso S, Grove J, Skelton JK, Farleigh L, Bugert JJ, Wright E, Temperton N, Angell R, Oxenford S, Jacobs M, Ketteler R, Dorner M, Marsh M. 2019. Identification of broad-spectrum antiviral compounds by targeting viral entry. Viruses 11:176. doi: 10.3390/v11020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamauchi Y, Helenius A. 2013. Virus entry at a glance. J Cell Sci 126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 171.Drews K, Calgi MP, Harrison WC, Drews CM, Costa-Pinheiro P, Shaw JJP, Jobe KA, Nelson EA, Han JD, Fox T, White JM, Kester M. 2019. Glucosylceramidase maintains influenza virus infection by regulating endocytosis. J Virol 93:e00017-19. doi: 10.1128/JVI.00017-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. 2010. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog 6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nelson EA, Dyall J, Hoenen T, Barnes AB, Zhou H, Liang JY, Michelotti J, Dewey WH, DeWald LE, Bennett RS, Morris PJ, Guha R, Klumpp-Thomas C, McKnight C, Chen Y-C, Xu X, Wang A, Hughes E, Martin S, Thomas C, Jahrling PB, Hensley LE, Olinger GG, White JM. 2017. The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Negl Trop Dis 11:e0005540. doi: 10.1371/journal.pntd.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pager CT, Craft WW, Patch J, Dutch RE. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Johansen LM, Brannan JM, Delos SE, Shoemaker CJ, Stossel A, Lear C, Hoffstrom BG, Dewald LE, Schornberg KL, Scully C, Lehár J, Hensley LE, White JM, Olinger GG. 2013. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med 5:190ra79. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ren J, Zhao Y, Fry EE, Stuart DI. 2018. Target identification and mode of action of four chemically divergent drugs against Ebolavirus infection. J Med Chem 61:724–733. doi: 10.1021/acs.jmedchem.7b01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hulseberg CE, Fénéant L, Szymańska-de Wijs KM, Kessler NP, Nelson EA, Shoemaker CJ, Schmaljohn CS, Polyak SJ, White JM. 2019. Arbidol and other low-molecular-weight drugs that inhibit Lassa and Ebola viruses. J Virol 93:e02185-18. doi: 10.1128/JVI.02185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kadam RU, Wilson IA. 2017. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci USA 114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schafer A, Xiong R, Cooper L, Nowar R, Lee H, Li Y, Ramirez BE, Peet NP, Caffrey M, Thatcher GRJ, Saphire EO, Cheng H, Rong L. 2021. Evidence for distinct mechanisms of small molecule inhibitors of filovirus entry. PLoS Pathog 17:e1009312. doi: 10.1371/journal.ppat.1009312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sarute N, Cheng H, Yan Z, Salas-Briceno K, Richner J, Rong L, Ross SR. 2021. Signal-regulatory protein alpha is an anti-viral entry factor targeting viruses using endocytic pathways. PLoS Pathog 17:e1009662. doi: 10.1371/journal.ppat.1009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM, Batra J, Richards AL, Stevenson E, Gordon DE, Rojc A, Obernier K, Fabius JM, Soucheray M, Miorin L, Moreno E, Koh C, Tran QD, Hardy A, Robinot R, Vallet T, Nilsson-Payant BE, Hernandez-Armenta C, Dunham A, Weigang S, Knerr J, Modak M, Quintero D, Zhou Y, Dugourd A, Valdeolivas A, Patil T, Li Q, Hüttenhain R, Cakir M, Muralidharan M, Kim M, Jang G, Tutuncuoglu B, Hiatt J, Guo JZ, Xu J, Bouhaddou S, Mathy CJP, Gaulton A, Manners EJ, Félix E, et al. 2020. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Drayman N, DeMarco JK, Jones KA, Azizi S-A, Froggatt HM, Tan K, Maltseva NI, Chen S, Nicolaescu V, Dvorkin S, Furlong K, Kathayat RS, Firpo MR, Mastrodomenico V, Bruce EA, Schmidt MM, Jedrzejczak R, Muñoz-Alía MÁ, Schuster B, Nair V, Han K-Y, O’Brien A, Tomatsidou A, Meyer B, Vignuzzi M, Missiakas D, Botten JW, Brooke CB, Lee H, Baker SC, Mounce BC, Heaton NS, Severson WE, Palmer KE, Dickinson BC, Joachimiak A, Randall G, Tay S. 2021. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science 373:931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Saul S, Karim M, Huang P-T, Ghita L, Chiu W, Kumar S, Bhalla N, Leyssen P, Cohen CA, Huie K, Tindle C, Sibai M, Pinsky BA, Das S, Ghosh P, Dye JM, Solow-Cordero DE, Jin J, Jochmans D, Neyts J, Narayanan A, De Jonghe S, Einav S. 2021. Discovery of pan-ErbB inhibitors protecting from SARS-CoV-2 replication, inflammation, and lung injury by a drug repurposing screen. bioRxiv 10.1101/2021.05.15.444128.
- 184.Meyer B, Chiaravalli J, Gellenoncourt S, Brownridge P, Bryne DP, Daly LA, Grauslys A, Walter M, Agou F, Chakrabarti LA, Craik CS, Eyers CE, Eyers PA, Gambin Y, Jones AR, Sierecki E, Verdin E, Vignuzzi M, Emmott E. 2021. Characterising proteolysis during SARS-CoV-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat Commun 12:5553. doi: 10.1038/s41467-021-25796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Weston S, Baracco L, Keller C, Matthews K, McGrath ME, Logue J, Liang J, Dyall J, Holbrook MR, Hensley LE, Jahrling PB, Yu W, MacKerell AD, Frieman MB. 2020. The SKI complex is a broad-spectrum, host-directed antiviral drug target for coronaviruses, influenza, and filoviruses. Proc Natl Acad Sci USA 117:30687–30698. doi: 10.1073/pnas.2012939117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Williams CG, Jureka AS, Silvas JA, Nicolini AM, Chvatal SA, Carlson-Stevermer J, Oki J, Holden K, Basler CF. 2021. Inhibitors of VPS34 and fatty-acid metabolism suppress SARS-CoV-2 replication. Cell Rep 36:109479. doi: 10.1016/j.celrep.2021.109479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kashyap T, Murray J, Walker CJ, Chang H, Tamir S, Hou B, Shacham S, Kauffman MG, Tripp RA, Landesman Y. 2021. Selinexor, a novel selective inhibitor of nuclear export, reduces SARS-CoV-2 infection and protects the respiratory system in vivo. Antiviral Res 192:105115. doi: 10.1016/j.antiviral.2021.105115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jorquera PA, Mathew C, Pickens J, Williams C, Luczo JM, Tamir S, Ghildyal R, Tripp RA. 2019. Verdinexor (KPT-335), a selective inhibitor of nuclear export, reduces respiratory syncytial virus replication in vitro. J Virol 93:e01684-18. doi: 10.1128/JVI.01684-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. 2014. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol 88:7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wan JJ, Brown RS, Kielian M. 2020. Berberine chloride is an alphavirus inhibitor that targets nucleocapsid assembly. mBio 11:e01382-20. doi: 10.1128/mBio.01382-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lanini S, Portella G, Vairo F, Kobinger GP, Pesenti A, Langer M, Kabia S, Brogiato G, Amone J, Castilletti C, Miccio R, Zumla A, Capobianchi MR, Di Caro A, Strada G, Ippolito G, INMI-EMERGENCY EBOV Sierra Leone Study Group . 2015. Blood kinetics of Ebola virus in survivors and nonsurvivors. J Clin Invest 125:4692–4698. doi: 10.1172/JCI83111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vernet M-A, Reynard S, Fizet A, Schaeffer J, Pannetier D, Guedj J, Rives M, Georges N, Garcia-Bonnet N, Sylla AI, Grovogui P, Kerherve J-Y, Savio C, Savio-Coste S, de Séverac M-L, Zloczewski P, Linares S, Harouna S, Abdoul BM, Petitjean F, Samake N, Shepherd S, Kinda M, Koundouno FR, Joxe L, Mateo M, Lecine P, Page A, Tchamdja TM, Schoenhals M, Barbe S, Simon B, Tran-Minh T, Longuet C, L’Hériteau F, Baize S. 2017. Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea. JCI Insight 2:e88864. doi: 10.1172/jci.insight.88864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, Nadkarni G, Glicksberg BS, Houldsworth J, Cordon-Cardo C. 2020. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med 8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Telenti A, Arvin A, Corey L, Corti D, Diamond MS, García-Sastre A, Garry RF, Holmes EC, Pang P, Virgin HW. 2021. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- 195.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. 2020. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bailly C. 2019. Cepharanthine: an update of its mode of action, pharmacological properties and medical applications. Phytomedicine 62:152956. doi: 10.1016/j.phymed.2019.152956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stone NE, Jaramillo SA, Jones AN, Vazquez AJ, Martz M, Versluis LM, Raniere MO, Nunnally HE, Zarn KE, Nottingham R, Ng KR, Sahl JW, Wagner DM, Knudsen S, Settles EW, Keim P, French CT. 2021. Stenoparib, an inhibitor of cellular Poly(ADP-Ribose) polymerase, blocks replication of the SARS-CoV-2 and HCoV-NL63 human coronaviruses in vitro. MBio 12:e03495-20. doi: 10.1128/mBio.03495-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Acharya A, Pandey K, Thurman M, Klug E, Trivedi J, Sharma K, Lorson CL, Singh K, Byrareddy SN. 2021. Discovery and evaluation of entry inhibitors for SARS-CoV-2 and its emerging variants. J Virol 95:e01437-21. doi: 10.1128/JVI.01437-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bennett RP, Postnikova EN, Eaton BP, Cai Y, Yu S, Smith CO, Liang J, Zhou H, Kocher GA, Murphy MJ, Smith HC, Kuhn JH. 2021. Sangivamycin is highly effective against SARS-CoV-2 in vitro and has favorable drug properties. JCI Insight. doi: 10.1172/jci.insight.153165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cox RM, Wolf JD, Lieber CM, Sourimant J, Lin MJ, Babusis D, DuPont V, Chan J, Barrett KT, Lye D, Kalla R, Chun K, Mackman RL, Ye C, Cihlar T, Martinez-Sobrido L, Greninger AL, Bilello JP, Plemper RK. 2021. Oral prodrug of remdesivir parent GS-441524 is efficacious against SARS-CoV-2 in ferrets. Nat Commun 12:6415. doi: 10.1128/mBio.03495-20. [DOI] [PMC free article] [PubMed] [Google Scholar]