This study establishes that efficient delivery of progranulin and prosaposin to lysosomes is regulated at the level of exit from the ER via an interaction between prosaposin and Surf4. It thus provides new insight into ER-to-Golgi trafficking of two lysosomal proteins, with neurodegenerative disease relevance.
Abstract
Progranulin is a lysosomal protein whose haploinsufficiency causes frontotemporal dementia, while homozygous loss of progranulin causes neuronal ceroid lipofuscinosis, a lysosomal storage disease. The sensitivity of cells to progranulin deficiency raises important questions about how cells coordinate intracellular trafficking of progranulin to ensure its efficient delivery to lysosomes. In this study, we discover that progranulin interactions with prosaposin, another lysosomal protein, first occur within the lumen of the endoplasmic reticulum (ER) and are required for the efficient ER exit of progranulin. Mechanistically, we identify an interaction between prosaposin and Surf4, a receptor that promotes loading of lumenal cargos into COPII-coated vesicles, and establish that Surf4 is critical for the efficient export of progranulin and prosaposin from the ER. Collectively, this work demonstrates that a network of interactions occurring early in the secretory pathway promote the ER exit and subsequent lysosomal delivery of newly translated progranulin and prosaposin.
Introduction
Heterozygous loss-of-function mutations in the granulin (GRN) gene, which encodes the progranulin protein, cause a neurodegenerative disease known as frontotemporal dementia (Baker et al., 2006; Cruts et al., 2006; Gass et al., 2006). Meanwhile, homozygous loss of progranulin results in neuronal ceroid lipofuscinosis, a lysosome storage disease with an early age of onset (Smith et al., 2012; Tanaka et al., 2014). The human progranulin protein is made up of a series of seven repeats of the cysteine-rich GRN domain and lacks identifiable similarity to known enzymes or other proteins. The linkers between these GRN domains are cleaved within lysosomes to yield individual 6–8-kDa GRN peptides (Cenik et al., 2012; Holler et al., 2017; Kao et al., 2017; Lee et al., 2017; Petkau and Leavitt, 2014; Zhou et al., 2017). Although defining the direct biochemical function of progranulin or the GRN fragments within lysosomes remains an open area of investigation, the localization of progranulin and the granulins to lysosomes, along with the lysosome storage disease arising from GRN mutations, indicates that they play a major role in supporting normal lysosome function.
Given the importance of progranulin for the maintenance of normal lysosome function, mechanisms must exist to ensure the delivery of progranulin to lysosomes. Previous studies identified sortilin as a receptor for the endocytic uptake of progranulin (Carrasquillo et al., 2010; Hu et al., 2010). Prosaposin was also identified as a progranulin-interacting protein that links progranulin to the cation-independent mannose 6-phosphate receptor (CI-M6PR) for trafficking from the TGN to endosomes, as well as to either CI-M6PR or low-density lipoprotein receptor 1 (LRP1) for endocytic uptake of progranulin (Nicholson et al., 2016; Zhou et al., 2015). Prosaposin has a modular domain organization that contains four saposin domains that are liberated by proteolytic cleavage within lysosomes. These saposins support the degradative activity of lysosomes by selectively extracting lipid-bound molecules and presenting them to specific soluble enzymes within the lysosome lumen (Kishimoto et al., 1992; Meyer et al., 2014). Prosaposin thus supports lysosome function both as a scaffold for the trafficking of progranulin to lysosomes and by serving as the precursor for lysosomal saposins.
Given that GRN-linked frontotemporal dementia arises from progranulin haploinsufficiency, increasing the expression of progranulin from a remaining WT copy of the GRN gene, gene therapy to boost GRN expression, and protein replacement therapy are all actively investigated therapeutic strategies (Zhou et al., 2021; Logan et al., 2021). However, the success of such approaches depends on a detailed understanding of the mechanisms that either stimulate the activity of progranulin/granulins within lysosomes or ensure the delivery of progranulin to its site of action within the lumen of lysosomes. To address these issues, we performed quantitative imaging and biochemical analyses of progranulin traffic through the secretory pathway, combined with genetic perturbations to candidate regulators of this process. In contrast to expectations that prosaposin promotes progranulin exit from the TGN, we discovered that progranulin exit from the ER is also highly dependent on prosaposin. This observation indicated that efflux of progranulin from the ER is not a passive process and raised questions about the mechanism for promoting the exit of progranulin and prosaposin from the ER. Investigation of the underlying mechanism led us to identify Surf4, a sorting receptor for coat protein complex II (COPII) vesicles, as a prosaposin-interacting protein that is critical for the ER-to-Golgi trafficking of progranulin and prosaposin. While mammalian Surf4 has been characterized for its ability to promote the trafficking of secreted cargoes, it was not previously implicated in the trafficking of lysosomal proteins (Emmer et al., 2018; Saegusa et al., 2018; Yin et al., 2018; Lin et al., 2020; Wang et al., 2021). Our observations support a model wherein newly translated progranulin and prosaposin interact within the lumen of the ER and bind via prosaposin to Surf4 for their packaging into COPII vesicles for delivery to the Golgi. This newly identified mechanism acts upstream of the previously defined roles for CI-M6PR, LRP1, and sortilin at the TGN and the plasma membrane that are also critical for the lysosomal delivery of progranulin and prosaposin (Carrasquillo et al., 2010; Nicholson et al., 2016; Hu et al., 2010; Zhou et al., 2015). Each of these regulated trafficking events will contribute to how efficiently progranulin-prosaposin is delivered to lysosomes and is thus of fundamental cell biological relevance; it is also of potential value for optimizing strategies to enhance progranulin trafficking for therapeutic purposes in neurodegenerative diseases.
Results
Prosaposin is required for the efficient ER exit of progranulin
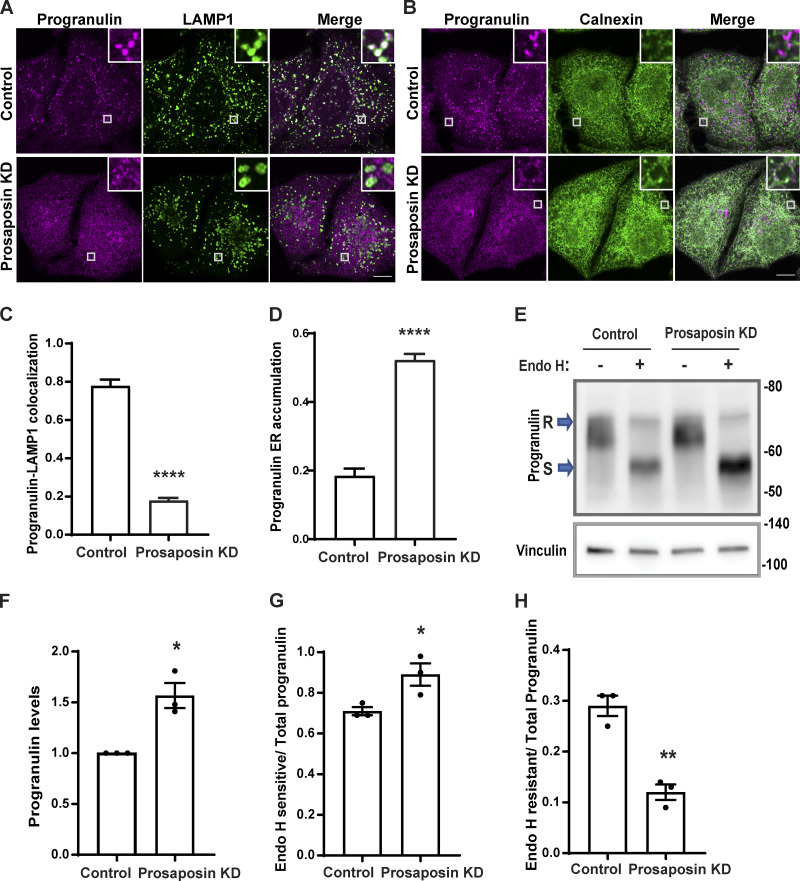
To investigate the regulatory role of prosaposin in controlling progranulin trafficking, we examined the effects of prosaposin depletion on progranulin subcellular localization. We observed that while progranulin localized to LAMP1 labeled lysosomes in control cells, progranulin was no longer enriched in lysosomes in prosaposin-depleted cells (Fig. 1, A and C; and Fig. S1, A and B), and instead, a significant portion of progranulin accumulated in the ER (Fig. 1, B and D) rather than the Golgi compartment (Fig. S1, C and D). This result did not fit with expectations arising from the proposed role for prosaposin in promoting CI-M6PR–dependent sorting of progranulin at the TGN (Zhou et al., 2015). This impaired delivery of progranulin to lysosomes was accompanied by an increase in its overall abundance (Fig. 1, E and F). In contrast to progranulin, the localization of cathepsin D, another lysosome protein, was not altered (Fig. S1, E and F). Likewise, prosaposin still localized to lysosomes in progranulin knockout (KO) cells (Fig. S1, I and J).
Figure 1.
Prosaposin is required for the efficient ER exit of progranulin. (A) Confocal images showing progranulin subcellular localization along with LAMP1-labeled late endosomes/lysosomes in control and prosaposin KD of HeLa cells. Scale bar, 10 µm; inset, 3.38 µm wide. (B) Progranulin localization along with the ER marker, calnexin, in control and prosaposin KD cells. Scale bar, 10 µm; inset, 3.38 µm wide. (C) Image quantification shows the Manders colocalization coefficient of progranulin with LAMP1 in control and prosaposin KD. Data were collected from 20 cells. Error bars show mean ± SEM; unpaired t test; ****, P < 0.0001. (D) Quantification of progranulin ER accumulation (calnexin colocalization) in control and prosaposin KD. Data were collected from 20 cells. Error bars show mean ± SEM; unpaired t test; ****, P < 0.0001. (E) Immunoblot of cell lysates from control siRNA and prosaposin siRNA transfections without (−) or with (+) Endo H enzyme treatment. S, Endo H sensitive; R, Endo H resistant. (F) Quantification of total progranulin protein levels. Total progranulin was calculated by adding Endo H–resistant fraction and Endo H–sensitive fraction. *, P < 0.05. (G and H) Ratios of the Endo H–sensitive (ER-localized) form of progranulin and the Endo H–resistant form of progranulin to total progranulin. (n = 3; mean ± SEM; unpaired t test; **, P < 0.01; *, P < 0.05).
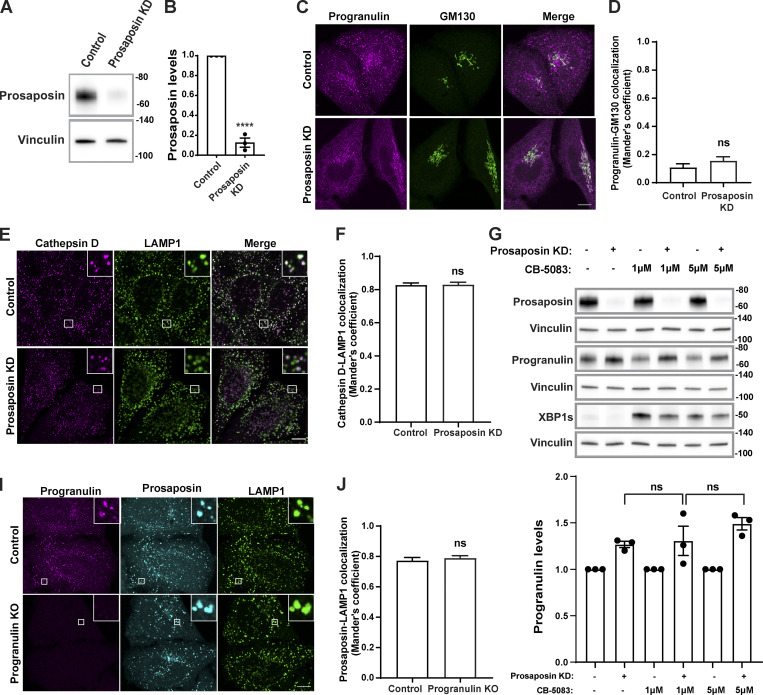
Figure S1.
Evaluation of prosaposin- and progranulin-depleted cells. (A) Immunoblot evaluation of prosaposin protein levels in cells transfected with control and prosaposin siRNAs. Vinculin was used as a loading control. (B) Quantification of prosaposin levels normalized to vinculin (n = 3 independent experiments; mean ± SEM; unpaired t test; ****, P < 0.0001). (C) Confocal immunofluorescence images showing progranulin and GM130 (cis-Golgi) localization in control and prosaposin siRNA–treated cells. Scale bar, 10 µm. (D) Manders colocalization coefficients for progranulin and GM130 in control and prosaposin KD cells. Data were collected from n = 20 cells. Error bars show mean ± SEM. Mann–Whitney U test. (E) Confocal immunofluorescence images show cathepsin D and LAMP1 localization in control and prosaposin siRNA–treated cells. Scale bar, 10 µm; inset, 5 µm wide. (F) Manders colocalization coefficients for cathepsin D with LAMP1 in control and prosaposin KD cells (20 cells per condition). Error bars show mean ± SEM. Unpaired t test. (G) Immunoblot evaluation of progranulin and spliced XBP1 protein levels in control and prosaposin KD cells treated for 7 h with the indicated concentrations of CB-5083, an ERAD inhibitor. Immunoblot quantification shows that progranulin levels are not significantly increased in prosaposin KD cells treated with CB-5083 compared with DMSO control. Progranulin levels were normalized to vinculin. n = 3 independent experiments; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. (I) Confocal immunofluorescence images showing localization of prosaposin and LAMP1 (lysosomes) in control and progranulin KO cells. Scale bar, 10 µm; inset, 3.38 µm wide. (J) Manders colocalization coefficients for prosaposin with LAMP1 in control and progranulin KO cells. n = 20 cells. Error bars show mean ± SEM. Unpaired t test.
To further test for a defect in the trafficking of newly made progranulin out of the ER, we tested for sensitivity to endoglycosidase H (Endo H), a bacterial enzyme that selectively recognizes and deglycosylates proteins with the high mannose and hybrid N-linked glycans found on newly translated proteins within the ER, while the glycans on proteins that have passed through the medial Golgi become resistant to Endo H (Dunphy and Rothman, 1983). We found that most of the full-length progranulin is in the Endo H–sensitive form in control cells (Fig. S2, A and B). Consistent with an accumulation of progranulin in the ER, prosaposin depletion was accompanied by an increase in total progranulin levels that was paralleled by an increase in the Endo H–sensitive fraction and a decrease in the Endo H–resistant fraction (Fig. 1, E–H).
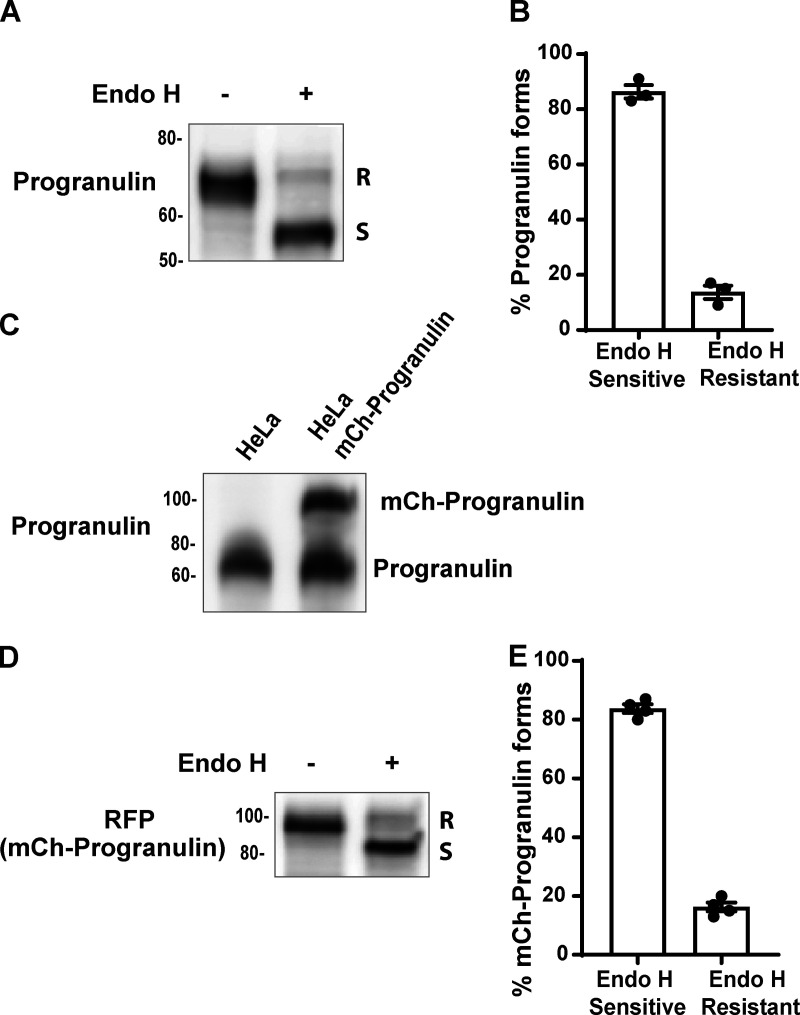
Figure S2.
Full-length progranulin is predominantly sensitive to Endo H. (A) Immunoblot analysis of Endo H sensitivity of the endogenous progranulin in cell lysates collected from control HeLa cells. (B) Blot quantification shows the percentage of progranulin forms present in control cells. n = 3 experiments; mean ± SEM. (C) Immunoblot showing the levels of stably expressed mCherry-progranulin in comparison to endogenous progranulin levels in HeLa cells. (D) Immunoblot analysis of mCherry-progranulin forms upon Endo H digestion of cell lysates collected from HeLa cells stably expressing mCherry-progranulin. (E) Immunoblot quantification showing the percentage of mCherry-progranulin forms present. n = 4 experiments; mean ± SEM.
Cells respond to abnormal protein buildup in the ER via activation of the unfolded protein response (UPR) and ER-associated decay (ERAD; Qi et al., 2017). To assess the UPR, we measured levels of the spliced form of XBP1 and found that the ER accumulation of progranulin was not accompanied by UPR activation in prosaposin-depleted cells (Fig. S1 G). As a positive control for ER stress, we observed that XBP1 splicing was altered following treatment with CB-5083, which inhibits p97 AAA ATPase, a key regulator of ERAD pathway (Anderson et al., 2015; Fig. S1 G). Even under these conditions of ER stress, progranulin levels were not significantly increased (Fig. S1 G). These results support a selective impact of prosaposin depletion on progranulin accumulation in the ER and argue that this arises largely from trafficking defects rather than as an indirect response to ER stress.
Prosaposin interactions with progranulin begin in the ER
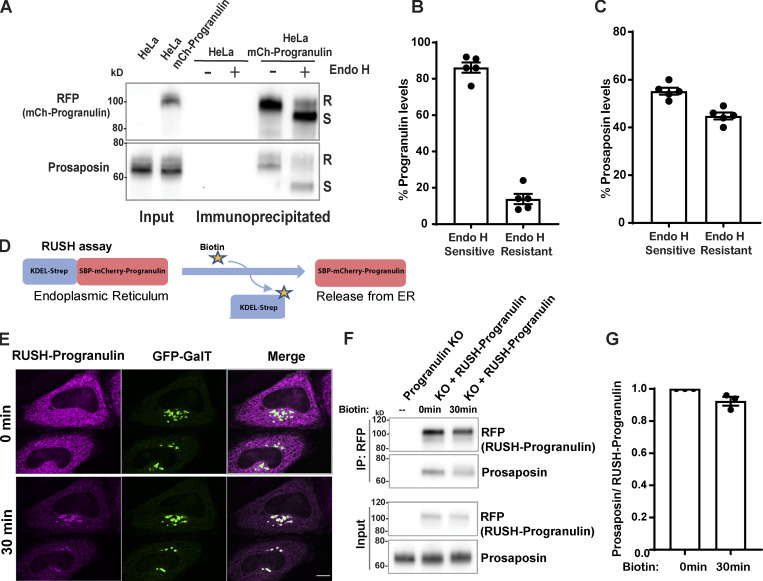
As prosaposin depletion causes the buildup of progranulin in ER, we hypothesized that progranulin interactions with prosaposin must be initiated early in the secretory pathway. To identify the subcellular location of such interactions, we used cells that stably express mCherry-tagged progranulin at near-endogenous levels (Fig. S2 C; Nguyen et al., 2018). We confirmed that this reporter behaved similarly to the endogenous protein in that the majority of the full-length protein was Endo H sensitive (Fig. S2, D and E). Endo H sensitivity assays on immunoprecipitates from cells stably expressing mCherry-tagged progranulin at near-endogenous levels (Fig. S2 C) revealed the robust presence of Endo H-sensitive (ER form) prosaposin in complex with progranulin (Fig. 2, A–C).
Figure 2.
Progranulin interactions with prosaposin begin in the ER. (A) Endo H–treated or untreated IP fractions of mCherry-progranulin. (B and C) Quantification of Endo H–sensitive and –resistant forms of progranulin and prosaposin in IP fractions (n = 5 independent experiments; mean ± SEM). (D) RUSH strategy: mCherry-progranulin fused to SBP is retained in the ER via its interaction with KDEL-streptavidin. Addition of biotin disrupts this interaction and causes the synchronous release of SBP-mCherry-progranulin from the ER. (E) Localization of RUSH-progranulin at 0 and 30 min after biotin addition. HeLa cells were cotransfected with RUSH-progranulin and GFP-GalT (Golgi marker) 1 d before imaging. Scale bar, 10 µm. (F) Immunoblot of prosaposin in IP fractions of RUSH-progranulin at 0 and at 30 min after biotin treatment. (G) Quantification of immunoblot shows the ratio of prosaposin to RUSH-progranulin at 0 and 30 min after biotin treatment (n = 3 independent experiments; mean ± SEM).
To further test for progranulin–prosaposin interaction within the ER, we employed a combination of the retention using selective hooks (RUSH) strategy (Boncompain et al., 2012) with immunoprecipitation (IP). To this end, we generated an mCherry-tagged progranulin reporter protein fused to Streptavidin-binding peptide (SBP-mCherry-progranulin, referred to as RUSH-progranulin) and coexpressed it with KDEL-Streptavidin to achieve biotin-regulated retention of RUSH-progranulin in the ER (Fig. 2 D). Before the addition of biotin, RUSH-progranulin was retained in the ER compartment (Fig. 2 E). Following the addition of biotin, RUSH-progranulin was released from the ER and was concentrated in the Golgi compartment at 30 min (Fig. 2 E) before eventually reaching the lysosomes (Fig. S3 A). We next evaluated the RUSH-progranulin interactions with endogenous prosaposin without biotin and 30 min after adding biotin and found prosaposin interacting similarly with both the ER and Golgi localized pools of progranulin (Fig. 2, F and G). Collectively, these results establish that the prosaposin–progranulin interaction begins in the ER compartment and that the formation of this protein complex facilitates the ER exit of progranulin.
Figure S3.
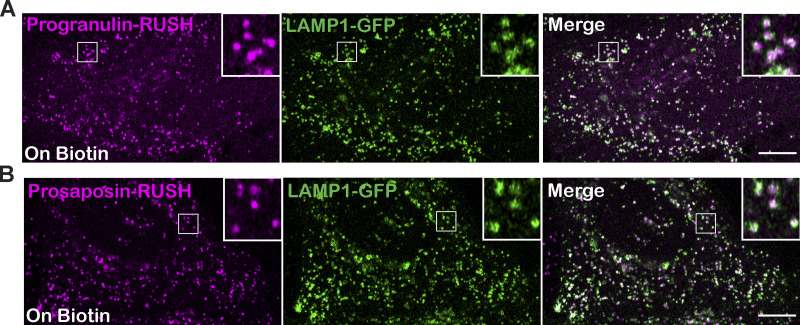
Lysosomal delivery of prosaposin-RUSH and progranulin-RUSH reporter proteins. (A and B) Confocal live-cell images show the localization of progranulin-RUSH (A) and prosaposin-RUSH (B) proteins to LAMP1-GFP–labeled late endosomes and lysosomes in HeLa cells treated with biotin overnight. Scale bar, 10 µm; inset, 4.2 µm wide.
Surf4 is required for efficient trafficking of progranulin and prosaposin out of the ER
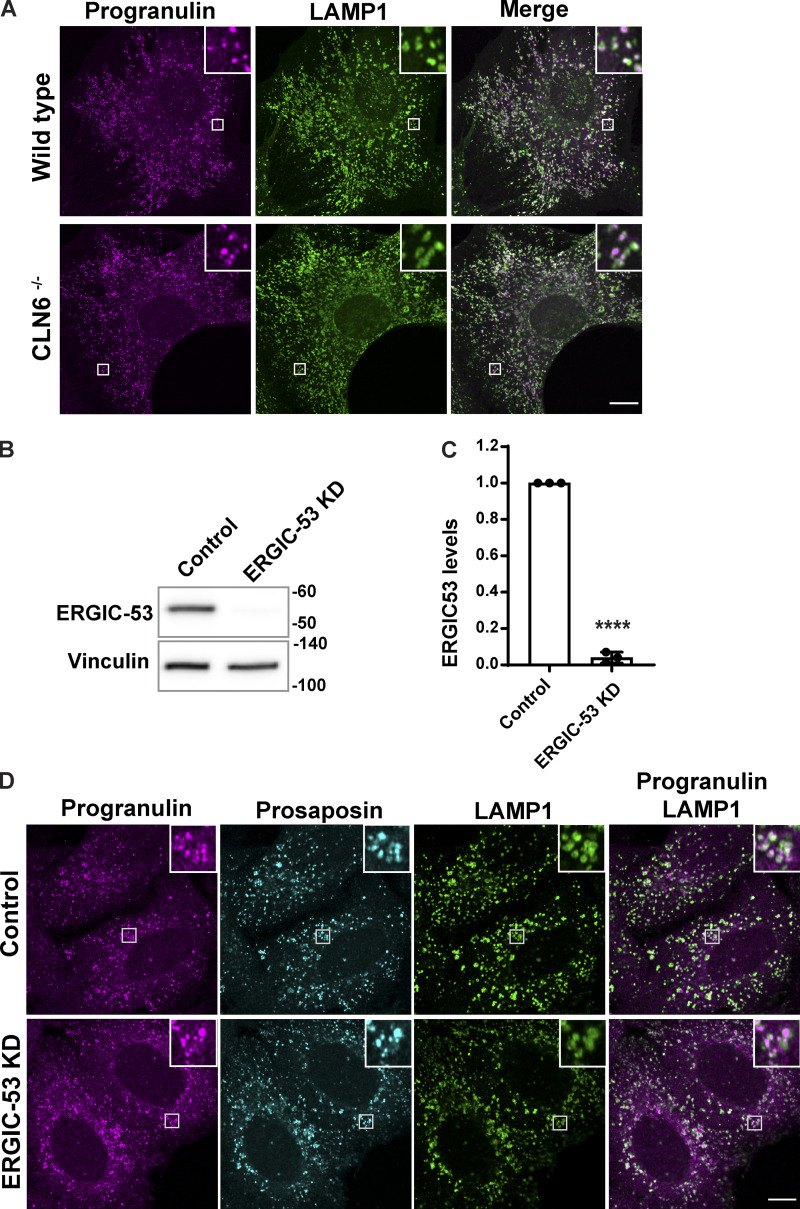
A significant fraction of newly made proteins are thought to exit the ER via the process of “bulk flow,” wherein they are nonselectively captured within the fluid phase of COPII vesicles that bud from the ER and deliver their contents to the cis-Golgi (Barlowe and Helenius, 2016). However, the dependence of progranulin on prosaposin for its ER exit suggested that the passive process of bulk flow is not sufficient to explain their trafficking and, instead, suggested the need for binding to a receptor to promote their trafficking out of the ER. In contrast to bulk flow, a subset of ER lumenal proteins are selectively recruited to budding COPII vesicles via transmembrane cargo receptors (Gomez-Navarro and Miller, 2016). So far, only a few ER cargo receptors have been identified. Of these, ERGIC-53 and Surf4 are best characterized for their ability to promote the ER export of lumenal proteins. More recently, CLN6 was shown to recruit some lysosomal enzymes and present them to CLN8 for ER export (Bajaj et al., 2020; di Ronza et al., 2018). However, we ruled out a role for CLN6 in the trafficking of progranulin based on our observation of normal lysosome localization of progranulin in fibroblasts from the nclf mutant mouse that lacks CLN6 (Fig. S4 A; Wheeler et al., 2002). Although progranulin and prosaposin are glycoproteins and thus candidate cargoes for ERGIC-53, which recognizes clients based on their glycosylation (Appenzeller et al., 1999), we found that progranulin and prosaposin were still lysosome localized in ERGIC-53–depleted cells (Fig. S4, B–D).
Figure S4.
CLN6 and ERGIC-53 are not required for the localization of progranulin to lysosomes. (A) Confocal immunofluorescence images showing progranulin localization to LAMP1-positive late endosomes and lysosomes in embryonic fibroblasts from WT and homozygous CLN6 mutant MEFs. Scale bar, 10 µm; inset, 3.38 µm wide. (B) Immunoblot shows the ERGIC-53 protein levels in control and ERGIC-53 siRNA–transfected cells. (C) Quantification of immunoblots where the graph shows ERGIC-53 levels normalized to loading control (n = 3 independent experiments; mean ± SEM; unpaired t test; ****, P < 0.0001). (D) Immunofluorescence images showing progranulin and prosaposin localization similar to LAMP1-labeled endosomes/lysosomes in control and ERGIC-53 KD cells. Scale bar, 10 µm; inset, 5 µm wide.
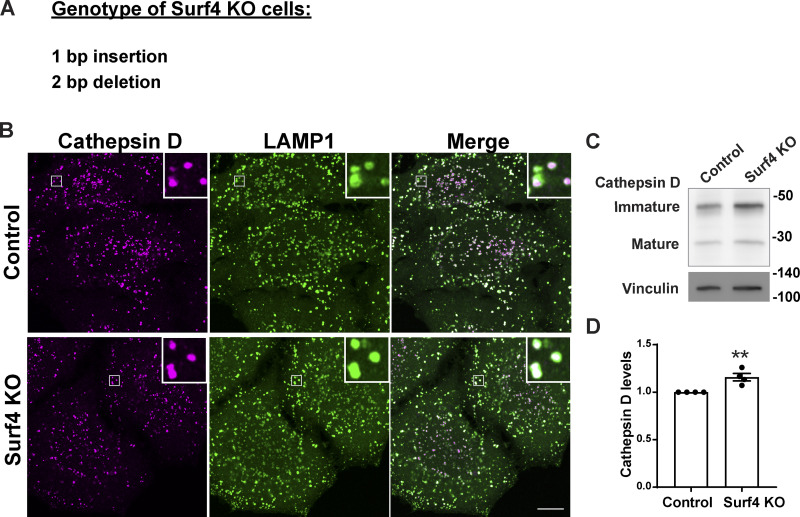
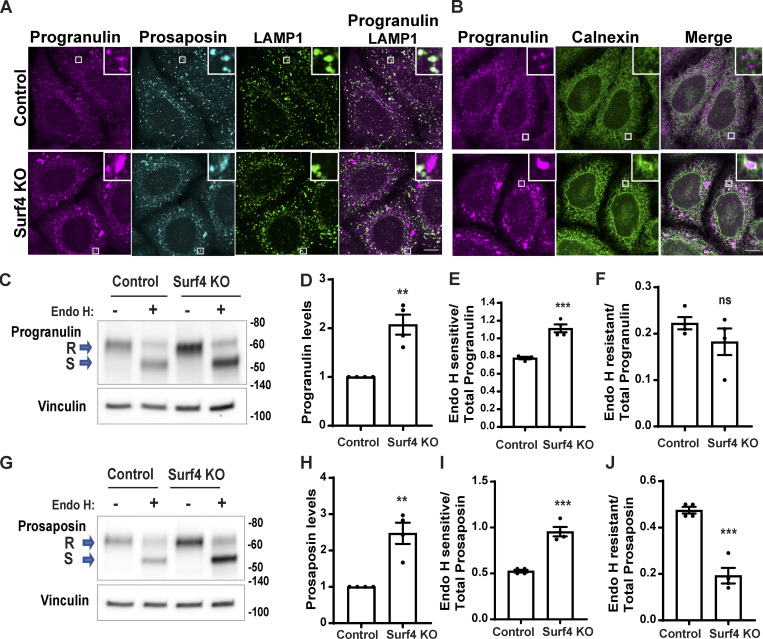
To test the role of Surf4 in progranulin/prosaposin trafficking out of the ER, we generated Surf4 KO cells by CRISPR/Cas9 genome editing (Fig. S5 A). In these cells, progranulin and prosaposin were not enriched in lysosomes and instead built up in the ER (Fig. 3, A and B). Endo H sensitivity assays revealed an increased abundance of the Endo H–sensitive (ER resident) form and a decrease in the Endo H–resistant form of both progranulin and prosaposin in the Surf4 KO cells (Fig. 3, C–J). In contrast, although cathepsin D protein levels were also slightly elevated in the Surf4 KO cells (possibly reflecting changes in lysosome homeostasis arising from progranulin and prosaposin delivery defects), cathepsin D localization to lysosomes was unaffected (Fig. S5, B–D). Altogether, these experiments show that Surf4 is selectively required for the efficient delivery of progranulin and prosaposin to lysosomes and point to a key role for Surf4 in their ER export. Although Surf4 was previously shown to function as a receptor for certain secreted soluble cargos such as PCSK9, apolipoproteins, erythropoietin, and dentin sialophosphoprotein, its role in the trafficking of lysosome proteins was previously unknown (Emmer et al., 2018; Saegusa et al., 2018; Yin et al., 2018; Lin et al., 2020; Wang et al., 2021).
Figure S5.
Cathepsin D trafficking is normal in the absence of Surf4. (A) Genotyping of Surf4 KO cells identified frameshift causing mutations (1-bp insertion and 2-bp deletion). (B) Confocal immunofluorescence images of the localization of cathepsin D and LAMP1 in control and Surf4 KO cells. Scale bar, 10 µm; inset, 3.38 µm wide. (C) Immunoblot of cathepsin D protein levels in control and Surf4 KO cells. Vinculin was used as a loading control. (D) Quantification of cathepsin D levels from four independent experiments (mean ± SEM; unpaired t test; **, P < 0.01).
Figure 3.
Surf4 is required for the trafficking of both progranulin and prosaposin. (A) Confocal images of progranulin and prosaposin in control and Surf4 KO cells along with LAMP1-labeled lysosomes. Scale bar, 10 µm; inset, 3.38 µm wide. (B) Confocal images of progranulin and calnexin (ER protein) in control and Surf4 KO cells. Scale bar, 10 µm; inset, 3.38 µm wide. (C) Immunoblot analysis for progranulin in control versus Surf4 KO cell lysates without (−) or with (+) Endo H treatment. (D–F) Quantification of immunoblots in C. (G) Immunoblot analysis for prosaposin in control versus Surf4 KO lysates. S, Endo H sensitive; R, Endo H resistant. (H–J) Quantification of immunoblots in G. In C–J, n = 4 independent experiments; mean ± SEM; unpaired t test; **, P < 0.01; ***, P < 0.001.
Surf4 is required for the efficient ER exit of prosaposin and progranulin
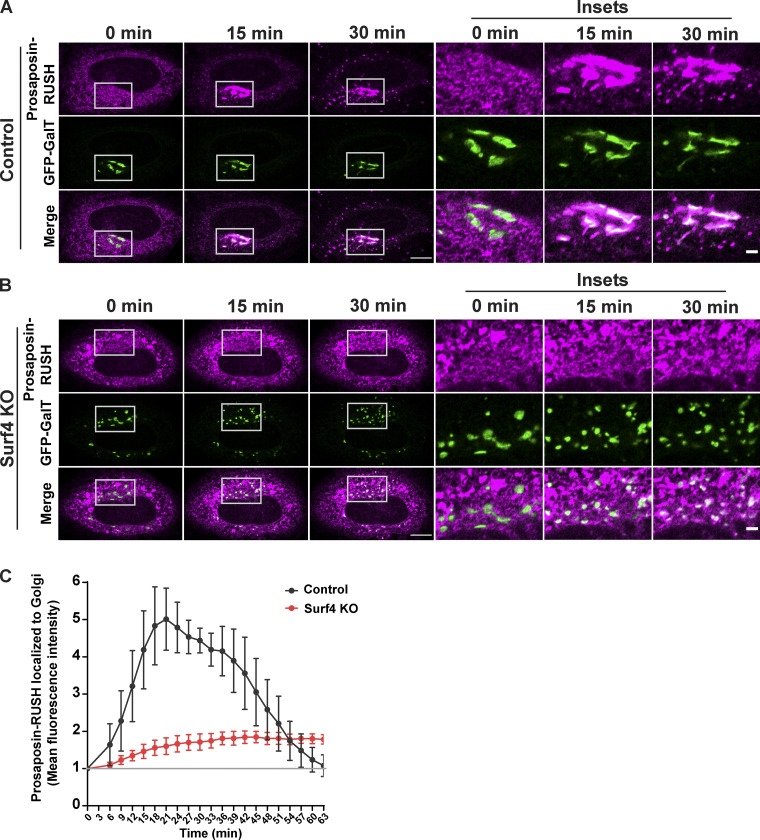
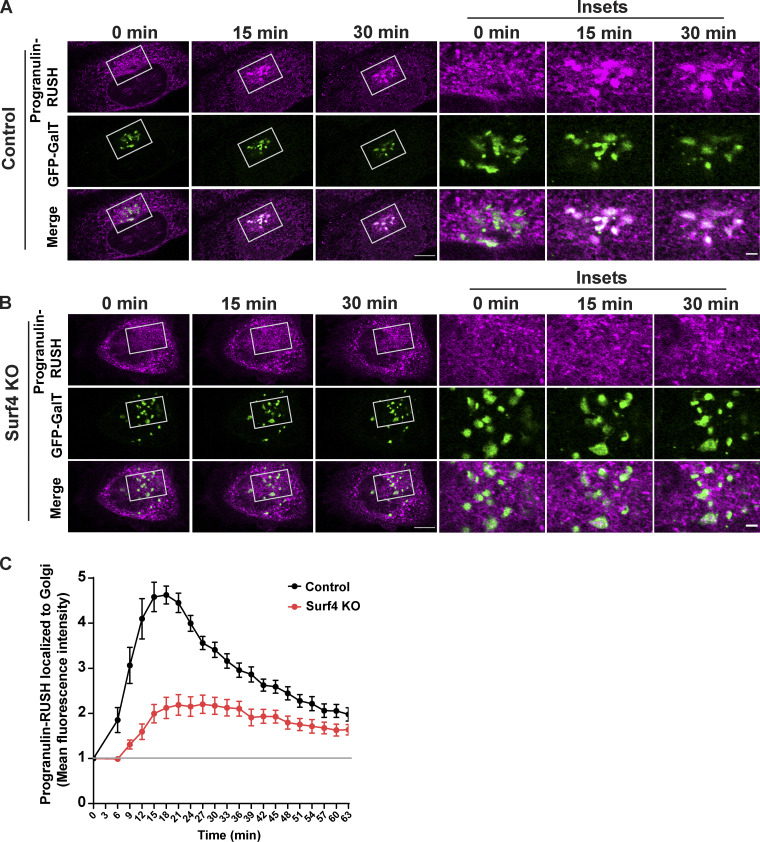
Steady-state localization of prosaposin and progranulin shows that these proteins build up in the ER in the absence of Surf4. However, calculating the extent of cargo trafficking defects at steady state is obscured by continuous fractional transfer of cargo. To more directly test the effects of Surf4 loss on the ER-to-Golgi trafficking kinetics of prosaposin and progranulin, we employed the RUSH strategy combined with live cell imaging (Boncompain et al., 2012). As expected, prosaposin-RUSH was retained in the ER before the addition of biotin (Fig. 4 A) and was delivered to LAMP1-GFP–labeled lysosomes in the presence of biotin (Fig. S3 B). In control cells, prosaposin-RUSH was concentrated in the Golgi compartment ∼20 min after ER release, while in Surf4 KO cells, prosaposin-RUSH remained in the ER and failed to enrich at the Golgi following biotin addition (Fig. 4, A–C; and Videos 1 and 2). Progranulin-RUSH also trafficked from the ER to lysosomes (Fig. S3 A) and was highly dependent on Surf4 for ER-to-Golgi trafficking (Fig. 5, A–C; and Videos 3 and 4). Collectively, these quantitative analyses of the dynamic trafficking of prosaposin and progranulin establish a critical role for Surf4 in their delivery from the ER to the Golgi. Notably, the strong reduction in the rate of ER-to-Golgi traffic of prosaposin-RUSH in Surf4 KO cells demonstrates the extent to which Surf4 prioritizes the ER export of prosaposin and, by extension, progranulin. We also observed fragmentation of the Golgi in Surf4 KO cells (GFP–β1,4-galactosyltransferase [GFP-GalT] images in Figs. 4 B and 5 B), consistent with previous reports indicating the role of Surf4 in the maintenance of Golgi morphology (Mitrovic et al., 2008). Given that cathepsin D was still delivered to lysosomes in the Surf4 KO cells and previous reports that Golgi fragmentation does not affect the passage of small cargos, it is unlikely that the trafficking defects of progranulin and prosaposin in Surf4 KO cells is secondary to this change in Golgi morphology (Lavieu et al., 2014).
Figure 4.
Surf4 is required for the efficient exit of prosaposin from the ER. (A and B) Live-cell imaging of prosaposin-RUSH traffic from ER in control and Surf4 KO cells at 0-, 15-, and 30-min time points after biotin addition; scale bar, 10 µm. Magnified insets show the localization of prosaposin to GFP-GalT–labeled Golgi; inset scale bar, 2 µm. (C) Quantification of the delivery of prosaposin-RUSH to the Golgi in control and Surf4 KO cells. The mean fluorescence intensities of prosaposin-RUSH localized to the Golgi (defined by GalT-GFP signal) after biotin addition were normalized to the mean fluorescence intensities before biotin (baseline marked by gray line) and plotted at the indicated times of biotin treatment. Data were collected from four independent experiments with n = 9–14 cells per experiment; error bars show mean ± SEM.
Video 1.
Prosaposin-RUSH trafficking from the ER in control cells. Confocal microscopy shows trafficking of prosaposin-RUSH (magenta) from the ER in control cells beginning 6 min after biotin addition. eGFP-GalT labels the Golgi (green). Images were acquired at 3-min intervals. Video plays at 2 frames/s with time display in h:min format. Scale bar, 10 µm.
Video 2.
Prosaposin-RUSH trafficking from the ER in Surf4 KO cells. Confocal microscopy shows trafficking of prosaposin-RUSH (magenta) from the ER in Surf4 KO cells beginning 6 min after biotin addition. eGFP-GalT labels the Golgi (green). Images were acquired at 3-min time intervals. Video plays at 2 frames/s with time display in h:min format. Scale bar, 10 µm.
Figure 5.
Surf4 is required for the efficient exit of progranulin from the ER. (A and B) Live-cell imaging of progranulin-RUSH traffic from ER for control and Surf4 KO cells at 0-, 15-, and 30-min time points after biotin addition; scale bar, 10 µm. Magnified insets show the localization of progranulin to GFP-GalT–labeled Golgi; inset scale bar, 2 µm. (C) Quantification of the delivery of progranulin-RUSH traffic to the Golgi in control and Surf4 KO cells. The mean fluorescence intensities of progranulin-RUSH within the Golgi (defined by GalT-GFP signal) at the indicated time points after biotin addition were normalized to the mean fluorescence intensities before biotin (baseline marked by gray line) and plotted at the indicated times of biotin treatment. Data were collected from 10 cells; error bars show mean ± SEM.
Video 3.
Progranulin-RUSH trafficking from the ER in control cells. Confocal microscopy shows trafficking of progranulin-RUSH (magenta) from the ER in control cells beginning 6 min after biotin addition. eGFP-GalT labels the Golgi compartment (green). Images were acquired at 3-min intervals. Video plays at 2 frames/s with time display in h:min format. Scale bar, 10 µm.
Video 4.
Progranulin-RUSH trafficking from the ER in Surf4 KO cells. Confocal microscopy shows trafficking of progranulin-RUSH (magenta) from the ER in Surf4 KO cells beginning 6 min after biotin addition (images were acquired at 3-min intervals). eGFP-GalT labels the Golgi compartment (green). Video plays at 2 frames/s with time display in h:min format. Scale bar, 10 µm.
Prosaposin binds Surf4 in the ER and facilitates the ER exit of progranulin–prosaposin complex
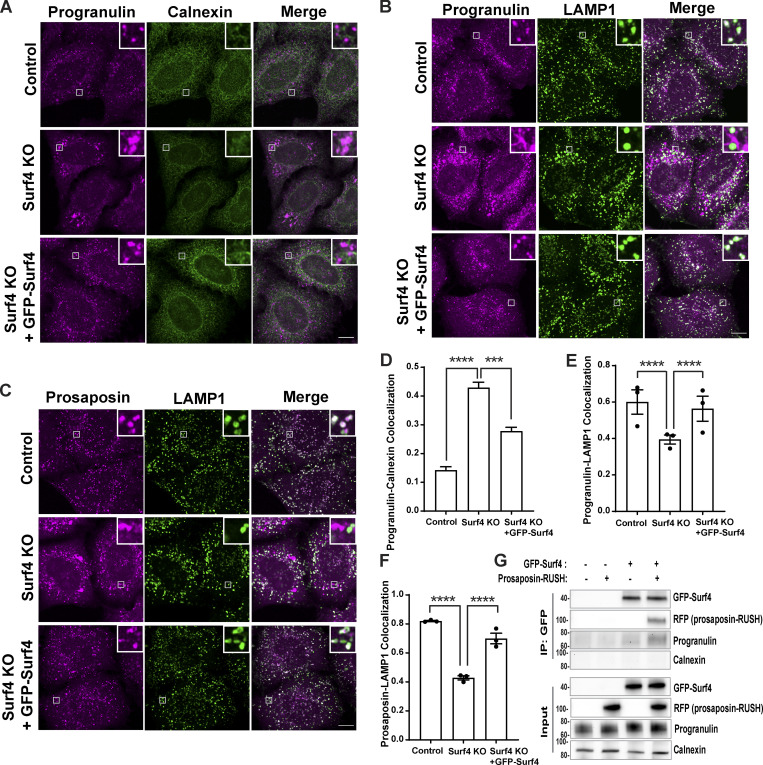
Consistent with its role in controlling traffic between the ER and cis-Golgi (Emmer et al., 2018; Mitrovic et al., 2008), GFP-Surf4 localized predominantly to the ER, and its expression reversed the ER accumulation phenotype of progranulin and prosaposin in Surf4 KO cells and restored their lysosome localization (Fig. 6, A–F). To further test whether Surf4 functions as a receptor for the progranulin–prosaposin complex, we next performed IP experiments. Following reversible cross-linking reaction with dithiobis succinimidyl propionate (Hermanson, 2008), we found that ER-retained prosaposin-RUSH copurified with Surf4 and that expression of prosaposin-RUSH increased the abundance of progranulin in the Surf4 IP (Fig. 6 G). The absence of calnexin in the GFP-Surf4 IPs served as a negative control for the specificity of the interactions of progranulin and prosaposin with Surf4 (Fig. 6 G). These results support a model wherein prosaposin promotes the ER export of progranulin by acting as a linker between progranulin and Surf4.
Figure 6.
Surf4 binds prosaposin–progranulin complex in the ER and facilitates ER exit. (A) Immunofluorescence images of endogenous progranulin and calnexin (ER marker) in control, Surf4 KO, and Surf4 KO cells expressing GFP-Surf4. Scale bar, 10 µm; inset, 3.38 µm wide. (B and C) Immunofluorescence images of endogenous progranulin and prosaposin along with LAMP1 (late endosomes and lysosomes) in control, Surf4 KO, and Surf4 KO cells expressing GFP-Surf4. Scale bar, 10 µm; inset, 3.38 µm wide. (D) Quantification of progranulin accumulation in ER (calnexin colocalization) in Surf4 KO cells. Data were collected from n = 25–32 cells. Error bars show mean ± SEM. Kruskal–Wallis test with Dunn’s multiple comparisons test; ***, P < 0.001; ****, P < 0.0001. (E and F) Quantification of Manders colocalization coefficients for progranulin and prosaposin with LAMP1 in control and Surf4 KO cells. Data were collected from three independent experiments with n = 20 cells quantified per experiment. Error bars show mean ± SEM. One-way ANOVA with Sidak’s multiple comparisons test; ****, P < 0.0001. (G) Anti-GFP-Surf4 IPs support an interaction with prosaposin and show that prosaposin enhances the abundance of endogenous progranulin in the complex with Surf4. Similar results were observed in three independent experiments.
Prosaposin N-terminus region interacts with Surf4 for efficient ER exit
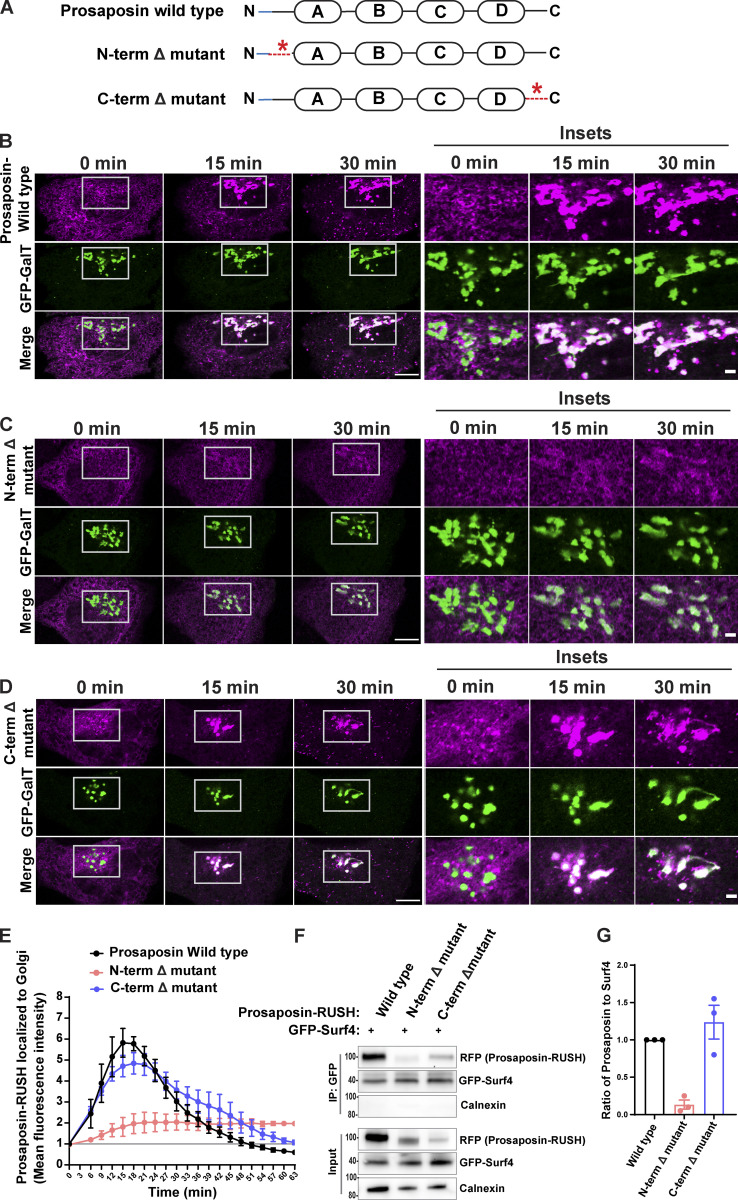
Having established that prosaposin interacts with Surf4 in the ER, we next investigated the basis for this interaction. Prosaposin has four saposin domains that function as enzyme cofactors in lysosomes (Fig. 7 A). In addition, prosaposin contains conserved N- and C-terminal regions that were previously proposed to be involved in promoting protein traffic (Patthy, 1991). We tested this by individually deleting the N- or C-terminal regions. Using RUSH assays, we found that the prosaposin N-terminal deletion mutant remained in the ER and failed to enrich in the Golgi following biotin addition, while the C-terminal deletion mutant (although not well expressed) trafficked like WT prosaposin (Fig. 7, B–E). Deletion of the prosaposin N-terminus but not the C-terminus furthermore resulted in reduced interaction with Surf4 (Fig. 7, F and G). These results indicate that the N-terminus of prosaposin is critical for both interaction with Surf4 and efficient ER exit of prosaposin.
Figure 7.
Prosaposin N-terminus region is required for interaction with Surf4 and ER exit. (A) Organization of prosaposin showing saposin domains A–D in rectangles. The blue area indicates the signal sequence of prosaposin, and dashed red lines indicate sites of N- and C-terminal deletions. (B–D) Live-cell imaging of prosaposin-RUSH traffic from ER to Golgi of WT, N-terminal Δ mutant, and C-terminal Δ mutant; scale bar, 10 µm. Magnified insets show the localization of prosaposin to GFP-GalT–labeled Golgi; inset scale bar, 2 µm. (E) Time-dependent delivery of prosaposin-RUSH traffic to the Golgi (defined by GFP-GalT signal) for prosaposin-WT, N-terminal Δ mutant, and C-terminal Δ mutant. The mean fluorescence intensities of prosaposin-RUSH localized to the Golgi at each time point after biotin addition were normalized to the mean fluorescence intensities before biotin (baseline marked by gray line). Data were collected from three independent experiments with n = 9–12 cells per experiment; error bars show mean ± SEM. (F) Immunoblots from anti-GFP-Surf4 IPs show a reduction of the N-terminally deleted prosaposin (N-terminal Δ mutant) in the IP fraction of GFP-Surf4, in comparison with C-terminal Δ mutant of prosaposin. Note that while the overall levels of the prosaposin C-terminal Δ mutant are lower than the WT, the proportion that interacts with Surf4 is similar. (G) Quantification of the ratio of prosaposin-RUSH to GFP-Surf4 in IP fractions. Prosaposin-RUSH levels in IP fractions were normalized to input samples. n = 3 independent experiments; error bars show mean ± SEM.
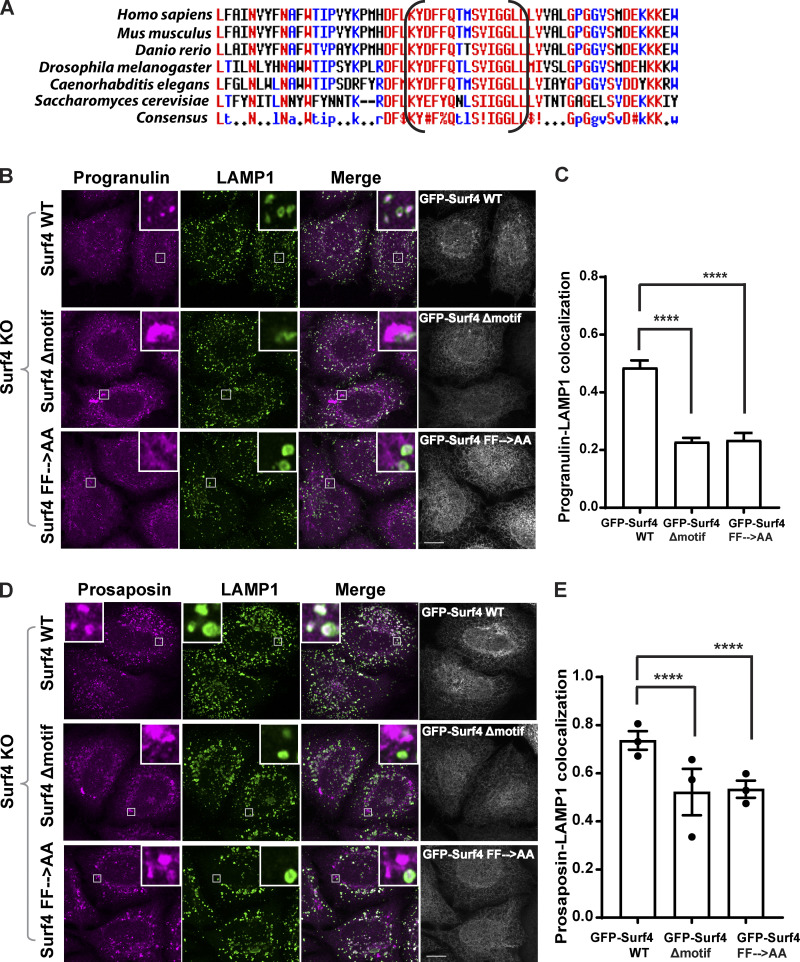
A C-terminal motif is critical for Surf4-dependent ER exit of progranulin and prosaposin
Surf4 is thought to promote efflux of cargoes from the ER by recruiting them into COPII-coated vesicles (Barlowe, 2003; Gomez-Navarro and Miller, 2016). Erv29p, the budding yeast homologue of Surf4, was experimentally determined to possess four transmembrane domains with both N- and C-termini localized to the cytoplasm (Foley et al., 2007). Although Surf4 lacks a canonical COPII-interacting motif such as a pair of phenylalanines at the very C-terminus found in other cargo receptors such as ERGIC-53/LMAN1 and p24 proteins (Nie et al., 2018; Nufer et al., 2002; Barlowe, 2003), a striking feature arising from our analysis of the Surf4 C-terminus was the presence of a 20–amino acid segment whose sequence is highly conserved across multiple distantly related species (Fig. 8 A). To test the requirement for this conserved motif in Surf4-dependent traffic of progranulin/prosaposin, we generated GFP-Surf4 with a 15–amino acid deletion within this region (Surf4Δmotif) and found that in contrast to the WT GFP-Surf4, the GFP-tagged Surf4Δmotif failed to restore lysosome localization of progranulin and prosaposin in Surf4 KO cells (Fig. 8, B–E). A GFP-Surf4 mutant wherein a pair of adjacent phenylalanines within the conserved motif was changed to alanines (GFP-Surf4 FF→AA) also failed to rescue the lysosome localization of progranulin and prosaposin (Fig. 8, B–E). Together, these results indicate that the ability of Surf4 to support ER export is dependent on a conserved region within its cytoplasmic C-terminus. We speculate that this region might play a role in promoting COPII interactions.
Figure 8.
Identification of a cytoplasmic motif of Surf4 that is critical for the trafficking of progranulin and prosaposin. (A) A stretch of 15 amino acids within the cytoplasmic C-terminus of Surf4 proteins is conserved across species. The region shown in brackets was deleted to generate the GFP-Surf4Δmotif mutant. Amino acids with high consensus (MultAlin) shown in red, the low consensus in blue, and neutral in black. (B and D) Immunofluorescence images of progranulin (B) and prosaposin and LAMP1 (D) in Surf4 KO cells expressing GFP-Surf4 WT, GFP-Surf4Δmotif, and GFP-Surf4 FF→AA (mutation of the diphenylalanine within the conserved motif). Scale bar, 10 µm; inset, 3.38 µm wide. (C) Manders colocalization coefficients of progranulin with LAMP1. Data were collected from 20 cells. Error bars show mean ± SEM. Kruskal–Wallis test with Dunn’s multiple comparisons test; ****, P < 0.0001. (E) Manders colocalization coefficients of prosaposin with LAMP1. Data were collected from three independent experiments with 20 cells. Error bars show mean ± SEM. Kruskal–Wallis test with Dunn’s multiple comparisons test; ****, P < 0.0001.
Discussion
Protein sorting within successive compartments in the secretory pathway is essential for ensuring the degradative activity of lysosomes by controlling the delivery of hydrolases and their regulatory factors (Braulke and Bonifacino, 2009). It has long been known that an important regulated step supporting the delivery of lysosome lumenal proteins takes place at the TGN via their interactions with proteins such as M6PRs that promote sorting into the endolysosomal pathway (Ghosh et al., 2003). However, in this study, we establish that lysosomal delivery of progranulin is also strongly influenced by sequential interactions that occur within the ER. First, export of progranulin from the ER is strongly promoted by the interaction between prosaposin and progranulin. Second, we identified Surf4, a receptor known to recruit lumenal cargos into COPII vesicles, as a major facilitator of ER export of the progranulin–prosaposin complex. These discoveries demonstrate how a series of protein–protein interactions within the ER prioritizes the trafficking of progranulin and prosaposin and has a major impact on their delivery to lysosomes.
Our discovery of a role for Surf4 in promoting the ER exit of progranulin and prosaposin parallels recent studies that have identified roles for CLN6 and CLN8 proteins in the sorting of other lysosomal proteins at the ER (Bajaj et al., 2020; di Ronza et al., 2018). Like mutations in the GRN gene, CLN6 and CLN8 mutations also cause the lysosome storage disease known as neuronal ceroid lipofuscinosis (Gao et al., 2002; Ranta et al., 1999). As progranulin does not depend on the CLN6/8-dependent export mechanism (Fig. S3), the Surf4-dependent trafficking of progranulin and prosaposin from the ER represents a distinct mechanism that enhances ER export of progranulin and prosaposin beyond what can be achieved via bulk flow. Although Surf4 was not previously demonstrated to sort mammalian lysosomal cargos, the budding yeast Surf4 homologue known as Erv29 supports trafficking of carboxypeptidase Y to the vacuole (equivalent to the mammalian lysosome; Belden and Barlowe, 2001). We observed a selective role for Surf4 in the trafficking of progranulin/prosaposin versus cathepsin D, although it remains to be determined to what extent Surf4 contributes to the trafficking of other lysosome proteins and whether additional ER-based sorting pathways exist for lysosomal proteins.
Although bulk flow has been proposed as a major mechanism to explain how lumenal proteins exit the ER, one reason for receptors to promote the efflux of specific cargos from the ER is to prioritize the export of cargos with the propensity to form aggregates (Barlowe and Helenius, 2016). Such a mechanism has been characterized for a subset of extracellular matrix proteins that depend on Surf4 (Yin et al., 2018). Consistent with a need for prioritizing the ER exit of progranulin and prosaposin, we observed that they both exhibited focal accumulations in the ER of Surf4 KO cells (Fig. 3, A and B). Although the basis for these local accumulations remains unknown, one possible factor is that the very cysteine-rich nature of progranulin and prosaposin could create a vulnerability for the formation of nonspecific disulfide bonds if their abundances exceed the capacity for protein disulfide isomerases to handle them.
The four saposin domains that are liberated from prosaposin by lysosomal proteases promote the breakdown of several species of lipids by extracting them from membranes and presenting them to specific hydrolases (Kishimoto et al., 1992; Meyer et al., 2014). For example, saposin C presents glucosylsphingosine and glucosylceramide to glucocerebrosidase (encoded by the GBA gene; Schnabel et al., 1991). Mutations in GBA cause Gaucher’s disease (homozygous) and confer significant Parkinson’s disease risk (heterozygous; Avenali et al., 2020). Therefore, beyond our focus on the relationship between progranulin and prosaposin, the Surf4 dependence of prosaposin trafficking may have relevance for understanding these diseases and for therapies based on promoting the activity of glucocerebrosidase.
In summary, we have established that efficient ER exit and lysosomal availability of progranulin depends on an interaction with prosaposin that begins soon after their cotranslational insertion into the ER. The complex of progranulin and prosaposin subsequently depends on an interaction between prosaposin and Surf4 for efficient efflux from the ER (and eventual delivery to lysosomes). Thus, although it has previously been proposed that receptor-independent bulk flow is sufficient to support the egress of many lumenal proteins from the ER, progranulin and prosaposin are particularly dependent on Surf4 to prioritize their export. This dependence of progranulin and prosaposin on Surf4 parallels (but is distinct from) other recent reports of lysosomal hydrolases relying on CLN6 and CLN8 proteins for their ER export (Bajaj et al., 2020; di Ronza et al., 2018). Beyond defining an important mechanism for the intracellular traffic of progranulin and prosaposin, our findings have implications for therapeutic strategies to increase progranulin and prosaposin levels in diseases arising from their deficiency.
Materials and methods
Plasmids and DNA cloning
EGFP-GalT was a gift from Jennifer Lippincott-Schwartz (Howard Hughes Medical Institute Janelia Research Campus, Ashburn, VA; plasmid 11929; Addgene; RRID:Addgene_11929; Cole et al., 1996). Str-KDEL_SBP-mCherry-GPI was a gift from Franck Perez (Institut Curie, Paris, France; plasmid 65295; Addgene; RRID:Addgene_65295; Boncompain et al., 2012). New RUSH plasmids for progranulin and prosaposin were prepared using Gibson assembly (E2611S; NEB). For RUSH-progranulin (Str-KDEL_SBP-mCherry-progranulin), human progranulin was PCR amplified from AAVS1_Puro_PGK_mCherry-progranulin (Nguyen et al., 2018), and the plasmid backbone was amplified from Str-KDEL_SBP-mCherry-GPI (plasmid 65295; Addgene; RRID:Addgene_65295) which supports coexpression (via an internal ribosome entry site linker) of the Str-KDEL and the SBP-fusion protein of interest. The IL-2 signal sequence that was present in the original mCherry-progranulin plasmid was later replaced with the progranulin signal sequence by site-directed mutagenesis (Q5 site directed mutagenesis kit; NEB). For cloning of prosaposin-RUSH (Str-KDEL_Prosaposin-SBP-mCherry), prosaposin was amplified from human prosaposin cDNA (pCMV6-XL5-PSAP, SC118405; Origene); SBP-mCherry insert and the plasmid backbone were amplified from Str-KDEL_SBP-mCherry-GPI (65295; Addgene) in parallel. Prosaposin-RUSH N-terminal Δ mutant construct was generated by the deletion of an 18–59-aa region of prosaposin N-terminus from prosaposin-RUSH plasmid using site-directed mutagenesis. Prosaposin-RUSH C-terminal Δ mutant plasmid was generated by the deletion of a 488–524-aa region of prosaposin C-terminus from prosaposin-RUSH plasmid using site-directed mutagenesis. For cloning of progranulin-RUSH (Str-KDEL_Progranulin-SBP-mCherry), progranulin was amplified from RUSH-progranulin; SBP-mCherry insert and vector were amplified from Str-KDEL_SBP-mCherry-GPI (65295; Addgene). The eGFP-Surf4 plasmid (pLV[Exp]-Puro-EF1A>eGFP-3xGS-hSurf4) was designed to fuse eGFP to the N-terminus of human Surf4 (GenBank accession no. NM_033161.4) in a mammalian lentiviral gene expression vector (VectorBuilder). eGFP-Surf4 Δmotif and eGFP-Surf4 FF→AA mutant plasmids were generated from the eGFP-Surf4 plasmid by site-directed mutagenesis. The CRISPR px459 plasmid (62988; Addgene) was kindly provided by Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA). For expression of mCherry-progranulin from the AAVS1 safe harbor locus, the hCas9 and gRNA_AAVS1-T2 plasmids were gifts from George Church (Harvard Medical School, Boston, MA; plasmid 41815; Addgene; RRID:Addgene_41815; and plasmid 41818; Addgene; RRID:Addgene_41818; Mali et al., 2013). pcDNA3 was from Invitrogen. Oligonucleotide primer sequences are summarized in Table S1.
Cell culture
HeLa cells and mouse embryonic fibroblasts (MEFs) were maintained in DMEM supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin mix in a humidified 37°C incubator with 5% CO2. HeLa cells were seeded at 100,000 cells in 2 ml medium per well in a six-well dish; transfected with 333 ng each of hCas9 (plasmid 41815; Addgene; Mali et al., 2013), gRNA_AAVS1-T2 (plasmid 41818; Addgene; Mali et al., 2013), and AAVS1_Puro_PGK_mCherry-progranulin (Nguyen et al., 2018) plasmids added to the 100 µl of Opti-MEM and 3 µl Fugene 6 transfection mix; and preincubated for 20 min. 48 h after transfection, the transfected cells were selected with puromycin (2 µg/ml) for 2 wk to isolate the surviving cells that stably expressed mCherry-progranulin. An immortalized line of CLN6 mutant MEFs (as well as a WT littermate controls) was generated by serial passaging of primary MEFs from the nclf line of spontaneous mutant mice obtained from The Jackson Laboratory (Todaro and Green, 1963; Gao et al., 2002).
Generation of KO cells by CRISPR/Cas9 gene editing
For the generation of Surf4 KO HeLa cells, sgRNA targeting exon 3 of Surf4 gene (5′-GGTGTTGAGCAGGAACTTCG-3′) was cloned into mammalian expression vector pX459 and confirmed by DNA sequencing (Ran et al., 2013). 0.4 µg of plasmid was added to 21 µl Opti-MEM reduced serum medium (Gibco) and 1.2 µl Fugene 6 (Promega) transfection mix and incubated for 15 min at room temperature, before adding to HeLa cells plated in 1 well of a 24-well plate. The next day, the transfected cells were selected with puromycin (2 µg/ml) for 48 h. The cells were plated at low density to isolate clones derived from single cells. To identify indels near the sgRNA target region, genomic DNA was isolated (Quick Extract DNA extraction solution; Lucigen); amplified by PCR, followed by blunt-end ligation of the amplified DNA into TOPO cloning vector (Zero Blunt TOPO PCR cloning kit; Invitrogen); and transformed into TOP10 Escherichia coli. 18 colonies were screened by isolating plasmids, and indel mutations were confirmed by sequencing using the M13 forward sequencing primer. Generation of progranulin KO HeLa cells by CRISPR ribonucleoprotein electroporation was described previously (Nguyen et al., 2018), and indel mutations (1-bp insertion) were confirmed by DNA sequencing. DNA sequencing was performed at the Keck DNA sequencing facility, Yale School of Medicine.
siRNA transfection
For siRNA transfection, 7.5 µl of 20 µM siRNA was added to 500 µl Opti-MEM reduced serum medium (Gibco), followed by the addition of 5 µl Lipofectamine RNAi max reagent (Invitrogen), and gently mixed. After incubating the mix for 15 min at room temperature, HeLa cells were plated at 100,000 cells in 2 ml medium per well in a six-well dish and transfected for 72 h before further analysis by immunostaining and immunoblotting. Prosaposin siRNA (5′-GGCCGACAUAUGCAAGAACUAUATC-3′), ERGIC-53 siRNA 1 (5′-GUGAAUACUUCUAAUUGUUUCCUTT-3′), ERGIC-53 siRNA 3 (5′-GGCUUUACACCAGAUAAAAAUGATT-3′), and negative control siRNA (5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′) were purchased from IDT. For ERGIC-53 knockdown (KD), equal volumes of ERGIC-53 siRNA 1 and 3 (each 3.75 µl of 20 µM siRNA) were added to the siRNA transfection mix protocol as described above, and 72 h after siRNA transfection, cells were analyzed by immunoblotting and immunostaining.
Treatment with ERAD inhibitor, CB-5083
48 h after prosaposin siRNA transfection, HeLa cells were treated with either DMSO control or CB-5083 (19311; Cayman Chemical Co.) for 7 h before isolating protein lysates for immunoblot analysis.
RUSH live-cell imaging and quantification
HeLa cells were plated at 100,000 cells in 2 ml medium onto 35-mm MatTek glass-bottom dishes and transfected with 0.9 µg RUSH plasmid and 0.1 µg eGFP-GalT or LAMP1-GFP using 100 µl Opti-MEM and 3 µl Fugene 6 transfection reagent (Promega) 1 d before imaging. Live-cell imaging was performed in an environment-controlled chamber set at 37°C and 5% CO2 on a Zeiss LSM880 confocal laser scanning microscope with Airy scan using a Plan Apochromat 63× objective (NA 1.4), a 32-channel gallium arsenide phosphide photomultiplier tube detector, and 488-, 561-, and 633-nm laser lines. Image acquisition was controlled with Zen imaging software from Zeiss. Time-lapse images were acquired at 3-min intervals for 1 h after the addition of biotin (40 µM) to the medium. Images were quantified using ImageJ (Schneider et al., 2012). To quantify the Golgi localization of prosaposin-RUSH and progranulin-RUSH proteins, the eGFP-GalT–labeled Golgi region was masked by setting up an intensity threshold. In this masked region, the fluorescence intensities of mCherry-tagged RUSH proteins were quantified at 3-min time intervals after the addition of biotin and were normalized to images taken before the addition of biotin.
Transient transfection of plasmids
For transient transfection of eGFP-Surf4 plasmids, cells were plated on 12-mm no. 1 glass coverslips at 20,000 cells in 0.5 ml medium per well in a 24-well dish and transfected with 0.25 µg plasmid using 25 µl Opti-MEM and 0.75 µl Fugene 6 transfection mix that was preincubated for 20 min. 48 h after transfection, cells were used for immunostaining.
Immunostaining, immunofluorescence, and image quantification
For immunostaining, cells seeded on 12-mm no. 1 glass coverslips (Carolina Biological Supply) were fixed for 30 min at room temperature with 8% PFA in 0.1 M sodium phosphate buffer (pH 7.2) that was added 1:1 to the cell culture medium. Cells were then rinsed once with PBS and permeabilized for 15 min with 0.1% saponin prepared in PBS, followed by incubation with blocking buffer (3% BSA and 0.1% saponin in PBS) for 20 min. Cells were incubated with primary antibodies overnight at 4°C and secondary antibodies for 30 min at room temperature. Primary and secondary antibodies were diluted in blocking buffer. The coverslips were washed three times with PBS containing 0.1% saponin after incubation with antibodies and mounted on glass slides using Prolong Gold mounting medium (Invitrogen).
The primary antibodies used for the immunostaining were mouse anti-LAMP1 (H4A3; DSHB), rabbit anti-saposin C (clone H-81, SC-32587; Santa Cruz), goat anti-progranulin (AF2420; R&D Systems), rabbit anti-calnexin (C5C9; Cell Signaling Technology), mouse anti-GM130 (610822; BD Transduction Laboratories), goat anti-cathepsin D (AF1014; R&D Systems), sheep anti-mouse progranulin (AF2557; R&D Systems), and mouse anti-LAMP1 (1D4B; DSHB). Alexa Fluor–conjugated secondary antibodies used for the immunostaining were purchased from Invitrogen Life Technologies.
Images were acquired with the Zeiss LSM880 confocal laser scanning microscope equipped as described above. Images were quantified for the colocalization of progranulin-LAMP1, prosaposin-LAMP1, cathepsin D–LAMP1, and progranulin-GM130. Near-spherical structures such as progranulin, prosaposin, LAMP1, cathepsin D, and GM130 were quantified to find overlap in the localization by calculating the Manders coefficient using the measure correlation module of CellProfiler (Carpenter et al., 2006) and by setting the Otsu automatic intensity thresholding for puncta of size >0.294 µm.
Because ER structure forms a network, the Manders coefficient was calculated using the automatic threshold strategy of the measure correlation module in CellProfiler. Using this method, ER accumulations of progranulin were quantified for the colocalization of calnexin-progranulin.
IP and immunoblotting
To prepare protein lysates, cells were washed twice with cold PBS and scraped off the dish using cold lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) along with protease and phosphatase inhibitors (Complete, Mini-EDTA free; PhosSTOP) and centrifuged at 14,000 rpm for 6 min at 4°C to collect the supernatant, leaving behind the insoluble pellet. For IP, protein lysates were added to RFP-Trap beads (Chromotek) and incubated for 1 h at 4°C on a rotating shaker. Beads were then washed four times with lysis buffer and eluted in 2× Laemmli buffer containing 1% 2-mercaptoethanol (Sigma-Aldrich) by heating at 95°C for 3 min.
For IP of eGFP-Surf4 protein complexes, Surf4 KO HeLa cells plated on 10-cm dishes were transiently transfected with pcDNA3, prosaposin-RUSH, eGFP-Surf4, and prosaposin-RUSH+eGFP-Surf4 plasmids. Equal amounts of prosaposin-RUSH and eGFP-Surf4 plasmids were used in each dish. In dishes where prosaposin-RUSH or eGFP-Surf4 were transfected alone, empty pcDNA3 was added to make up the total amount of plasmid DNA to 7 µg per dish. 7 µg plasmid was added to 700 µl Opti-MEM and 21 µl Fugene 6 transfection mix and incubated for 20 min before adding to the cells. 1 d after transfection, cells were rinsed twice with PBS, and protein cross-linking reaction was performed by incubating with freshly prepared 0.5 mM dithiobis succinimidyl propionate (Thermo Fisher Scientific) in PBS for 30 min at room temperature followed by quenching the reaction with 20 mM Tris, pH 7.4, for 10 min. Cells were rinsed twice with cold PBS, lysed in cold lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 1% NP-40) along with protease and phosphatase inhibitors (Complete Mini-EDTA free, PhosSTOP), and centrifuged at 14,000 rpm for 6 min at 4°C to collect the supernatant, leaving behind the insoluble pellet. IP of GFP-Surf4 protein complexes was performed using GFP-Trap (Chromotek) similarly to the RFP-Trap IP experiment described above. To detect GFP-Surf4 on immunoblots, protein samples were denatured in Laemmli buffer for 10 min at room temperature before running on SDS-PAGE gels.
For IP of eGFP-Surf4 protein complexes with prosaposin-RUSH deletion mutants, Surf4 KO HeLa cells plated on 10-cm dishes were transiently transfected with eGFP-Surf4 along with prosaposin-RUSH WT, prosaposin-RUSH N-terminal Δ mutant, and prosaposin-RUSH C-terminal Δ mutant plasmids. Equal amounts of prosaposin-RUSH and eGFP-Surf4 plasmids were used in each dish. 1 d after transfection, IP experiments were performed as described above.
Proteins were separated on SDS-PAGE gels (4–15% gradient Mini-PROTEAN TGX precast polyacrylamide gels; Bio-Rad) and transferred to nitrocellulose membranes using wet blot transfer system (Bio-Rad). Membranes were blocked with 5% nonfat dry milk prepared in TBST (Tris-buffered saline containing 0.1% Tween-20) for 1 h at room temperature followed by incubation with primary antibodies diluted in 5% BSA prepared in TBST at 4°C overnight. The next day, membranes were incubated with the corresponding HRP-conjugated secondary antibodies diluted in 5% nonfat dry milk in TBST for 1 h at room temperature. Membranes were washed three times with TBST after antibody incubations. Membranes were developed using ECL chemiluminescence substrate for HRP to detect protein signal (Thermo Fisher Scientific) using a Versadoc imaging system (Bio-Rad). Immunoblots were quantified using ImageJ.
The primary antibodies used for the immunoblotting were rabbit anti-saposin C (clone H-81, SC-32587; Santa Cruz), goat anti-progranulin (AF2420; R&D Systems), rabbit anti-calnexin (C5C9; Cell Signaling Technology), mouse anti-vinculin (clone vin-11-5, v4505; Sigma-Aldrich), rabbit anti-RFP (600-401-379; Rockland Immunochemicals), anti-GFP-HRP (600-103-215; Rockland Immunochemicals), anti-ERGIC-53 (ab125006; Abcam), and anti-XBP1s (clone 143F, 647501; BioLegend). The secondary antibodies used were anti-biotin-HRP (7075P5; Cell Signaling Technology), anti-rabbit HRP (7074s; Cell Signaling Technology), anti-mouse HRP (7076s; Cell Signaling Technology), and anti-goat HRP (31402; Invitrogen).
Endo H assays
Protein lysates were prepared in a lysis buffer containing 1% Triton X-100 as described above. 20 µg of protein lysates were incubated with glycoprotein denaturation buffer at 95°C for 10 min and treated with 0.3 µl Endo H (P0702; NEB) enzyme in glyco-buffer 3 for 1 h at 37°C. After Endo H digestion, samples were denatured in Laemmli buffer by heating at 95°C for 5 min and resolved on SDS-PAGE gel for immunoblotting as above. To perform Endo H assays on immunoprecipitated samples, beads were eluted in glycoprotein denaturation buffer at 95°C for 10 min followed by Endo H treatment as described above. Total Endo H–treated protein was quantified from immunoblots by adding Endo H–resistant fraction to Endo H–sensitive fraction.
Protein sequence analysis
Human Surf4 protein sequence was analyzed for amino acid sequence conservation across species using MultAlin algorithm (Corpet, 1988). To identify protein motifs, human Surf4 protein sequence was scanned against the PROSITE collection of motifs using ScanProsite (https://prosite.expasy.org/scanprosite/; de Castro et al., 2006; Sigrist et al., 2013).
Statistical analysis
All statistical tests were performed using Prism (GraphPad). All data are represented as mean ± SEM. Normality tests were performed to check the Gaussian distribution of data points (Shapiro–Wilk normality test for sample size n ≤ 5 and D’Agostino and Pearson omnibus K2 test for sample size ≥ 20). Parametric tests (two-tailed unpaired t tests or one-way ANOVA) were used for normally distributed data, and nonparametric tests (Mann–Whitney U or Kruskal–Wallis test) were used for non–normally distributed data. Statistical tests performed are indicated along with P values in figure legends.
Online supplemental material
Fig. S1 shows evaluation of prosaposin- and progranulin-depleted cells. Fig. S2 shows that full-length progranulin is predominantly sensitive to Endo H. Fig. S3 shows the lysosomal delivery of prosaposin-RUSH and progranulin-RUSH reporter proteins. Fig. S4 shows that CLN6 and ERGIC-53 are not required for the localization of progranulin to lysosomes. Fig. S5 shows that cathepsin D trafficking is normal in the absence of Surf4. Table S1 lists sequences of the oligonucleotide primers used for PCR amplification of genomic DNA. Video 1 shows prosaposin-RUSH trafficking from the ER in control cells. Video 2 shows prosaposin-RUSH trafficking from the ER in Surf4 KO cells. Video 3 shows progranulin-RUSH trafficking from the ER in control cells. Video 4 shows progranulin-RUSH trafficking from the ER in Surf4 KO cells.
Supplementary Material
lists oligonucleotide primer sequences used in this study.
Acknowledgments
We are grateful for microscopy resources provided by the Yale Program in Cellular Neuroscience, Neurodegeneration and Repair imaging facility. We appreciate efforts by Jin Meng to characterize CLN6 mutant fibroblasts in our laboratory. Taylor Skibitcky provided key administrative and organizational support.
This research was supported by grants from the Bluefield Project to Cure FTD (S. Devireddy and S.M. Ferguson), the Parkinson’s Disease Foundation (S.M. Ferguson), and the National Institutes of Health (AG062210 to S.M. Ferguson).
The authors declare no competing financial interests.
Author contributions: S. Devireddy and S.M. Ferguson designed experiments. S. Devireddy performed all experiments. S. Devireddy and S.M. Ferguson prepared the manuscript.
References
- Anderson, D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S., et al. 2015. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell. 28:653–665. 10.1016/j.ccell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller, C., Andersson H., Kappeler F., and Hauri H.P.. 1999. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1:330–334. 10.1038/14020 [DOI] [PubMed] [Google Scholar]
- Avenali, M., Blandini F., and Cerri S.. 2020. Glucocerebrosidase Defects as a Major Risk Factor for Parkinson’s Disease. Front. Aging Neurosci. 12:97. 10.3389/fnagi.2020.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj, L., Sharma J., di Ronza A., Zhang P., Eblimit A., Pal R., Roman D., Collette J.R., Booth C., Chang K.T., et al. 2020. A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer. J. Clin. Invest. 130:4118–4132. 10.1172/JCI130955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 442:916–919. 10.1038/nature05016 [DOI] [PubMed] [Google Scholar]
- Barlowe, C. 2003. Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol. 13:295–300. 10.1016/S0962-8924(03)00082-5 [DOI] [PubMed] [Google Scholar]
- Barlowe, C., and Helenius A.. 2016. Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu. Rev. Cell Dev. Biol. 32:197–222. 10.1146/annurev-cellbio-111315-125016 [DOI] [PubMed] [Google Scholar]
- Belden, W.J., and Barlowe C.. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 294:1528–1531. 10.1126/science.1065224 [DOI] [PubMed] [Google Scholar]
- Boncompain, G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Braulke, T., and Bonifacino J.S.. 2009. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 1793:605–614. 10.1016/j.bbamcr.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Carpenter, A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo, M.M., Nicholson A.M., Finch N., Gibbs J.R., Baker M., Rutherford N.J., Hunter T.A., DeJesus-Hernandez M., Bisceglio G.D., Mackenzie I.R., et al. 2010. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am. J. Hum. Genet. 87:890–897. 10.1016/j.ajhg.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik, B., Sephton C.F., Kutluk Cenik B., Herz J., and Yu G.. 2012. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 287:32298–32306. 10.1074/jbc.R112.399170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, N.B., Smith C.L., Sciaky N., Terasaki M., Edidin M., and Lippincott-Schwartz J.. 1996. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 273:797–801. 10.1126/science.273.5276.797 [DOI] [PubMed] [Google Scholar]
- Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts, M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J., et al. 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 442:920–924. 10.1038/nature05017 [DOI] [PubMed] [Google Scholar]
- de Castro, E., Sigrist C.J.A., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., and Hulo N.. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34:W362–W365. 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Ronza, A., Bajaj L., Sharma J., Sanagasetti D., Lotfi P., Adamski C.J., Collette J., Palmieri M., Amawi A., Popp L., et al. 2018. CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis. Nat. Cell Biol. 20:1370–1377. 10.1038/s41556-018-0228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy, W.G., and Rothman J.E.. 1983. Compartmentation of asparagine-linked oligosaccharide processing in the Golgi apparatus. J. Cell Biol. 97:270–275. 10.1083/jcb.97.1.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer, B.T., Hesketh G.G., Kotnik E., Tang V.T., Lascuna P.J., Xiang J., Gingras A.C., Chen X.W., and Ginsburg D.. 2018. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. eLife. 7:e38839. 10.7554/eLife.38839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, D.A., Sharpe H.J., and Otte S.. 2007. Membrane topology of the endoplasmic reticulum to Golgi transport factor Erv29p. Mol. Membr. Biol. 24:259–268. 10.1080/09687860601178518 [DOI] [PubMed] [Google Scholar]
- Gao, H., Boustany R.M.N., Espinola J.A., Cotman S.L., Srinidhi L., Antonellis K.A., Gillis T., Qin X., Liu S., Donahue L.R., et al. 2002. Mutations in a novel CLN6-encoded transmembrane protein cause variant neuronal ceroid lipofuscinosis in man and mouse. Am. J. Hum. Genet. 70:324–335. 10.1086/338190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass, J., Cannon A., Mackenzie I.R., Boeve B., Baker M., Adamson J., Crook R., Melquist S., Kuntz K., Petersen R., et al. 2006. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet. 15:2988–3001. 10.1093/hmg/ddl241 [DOI] [PubMed] [Google Scholar]
- Ghosh, P., Dahms N.M., and Kornfeld S.. 2003. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4:202–212. 10.1038/nrm1050 [DOI] [PubMed] [Google Scholar]
- Gomez-Navarro, N., and Miller E.. 2016. Protein sorting at the ER-Golgi interface. J. Cell Biol. 215:769–778. 10.1083/jcb.201610031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson, G.T. 2008. Homobifunctional Crosslinkers. Second edition. Academic Press, Cambridge, MA. 1232 pp., 10.1016/B978-0-12-370501-3.00004-7 [DOI] [Google Scholar]
- Holler, C.J., Taylor G., Deng Q., and Kukar T.. 2017. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro. 4:ENEURO.0100-17.2017. 10.1523/ENEURO.0100-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., Padukkavidana T., Vægter C.B., Brady O.A., Zheng Y., Mackenzie I.R., Feldman H.H., Nykjaer A., and Strittmatter S.M.. 2010. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 68:654–667. 10.1016/j.neuron.2010.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, A.W., McKay A., Singh P.P., Brunet A., and Huang E.J.. 2017. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18:325–333. 10.1038/nrn.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto, Y., Hiraiwa M., and O’Brien J.S.. 1992. Saposins: structure, function, distribution, and molecular genetics. J. Lipid Res. 33:1255–1267. 10.1016/S0022-2275(20)40540-1 [DOI] [PubMed] [Google Scholar]
- Lavieu, G., Dunlop M.H., Lerich A., Zheng H., Bottanelli F., and Rothman J.E.. 2014. The Golgi ribbon structure facilitates anterograde transport of large cargoes. Mol. Biol. Cell. 25:3028–3036. 10.1091/mbc.e14-04-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.W., Stankowski J.N., Chew J., Cook C.N., Lam Y.-W., Almeida S., Carlomagno Y., Lau K.-F., Prudencio M., Gao F.-B., et al. 2017. The lysosomal protein cathepsin L is a progranulin protease. Mol. Neurodegener. 12:55. 10.1186/s13024-017-0196-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z., King R., Tang V., Myers G., Balbin-Cuesta G., Friedman A., McGee B., Desch K., Ozel A.B., Siemieniak D., et al. 2020. The Endoplasmic Reticulum Cargo Receptor SURF4 Facilitates Efficient Erythropoietin Secretion. Mol. Cell. Biol. 40:e00180-20. 10.1128/MCB.00180-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, T., Simon M.J., Rana A., Cherf G.M., Srivastava A., Davis S.S., Low R.L.Y., Chiu C.-L., Fang M., Huang F., et al. 2021. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell. 184:4651–4668.e25. 10.1016/j.cell.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R.C., Giddens M.M., Coleman B.M., and Hall R.A.. 2014. The protective role of prosaposin and its receptors in the nervous system. Brain Res. 1585:1–12. 10.1016/j.brainres.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic, S., Ben-Tekaya H., Koegler E., Gruenberg J., and Hauri H.P.. 2008. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell. 19:1976–1990. 10.1091/mbc.e07-10-0989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A.D., Nguyen T.A., Zhang J., Devireddy S., Zhou P., Karydas A.M., Xu X., Miller B.L., Rigo F., Ferguson S.M., et al. 2018. Murine knockin model for progranulin-deficient frontotemporal dementia with nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA. 115:E2849–E2858. 10.1073/pnas.1722344115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, A.M., Finch N.A., Almeida M., Perkerson R.B., van Blitterswijk M., Wojtas A., Cenik B., Rotondo S., Inskeep V., Almasy L., et al. 2016. Prosaposin is a regulator of progranulin levels and oligomerization. Nat. Commun. 7:11992. 10.1038/ncomms11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, C., Wang H., Wang R., Ginsburg D., and Chen X.W.. 2018. Dimeric sorting code for concentrative cargo selection by the COPII coat. Proc. Natl. Acad. Sci. USA. 115:E3155–E3162. 10.1073/pnas.1704639115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nufer, O., Guldbrandsen S., Degen M., Kappeler F., Paccaud J.P., Tani K., and Hauri H.P.. 2002. Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J. Cell Sci. 115:619–628. 10.1242/jcs.115.3.619 [DOI] [PubMed] [Google Scholar]
- Patthy, L. 1991. Homology of the Precursor of Pulmonary Surfactant-associated Protein SP-B with Prosaposin and Sulfated Glycoprotein 1. J. Biol. Chem. 266:6035–6037. 10.1016/S0021-9258(18)38079-7 [DOI] [PubMed] [Google Scholar]
- Petkau, T.L., and Leavitt B.R.. 2014. Progranulin in neurodegenerative disease. Trends Neurosci. 37:388–398. 10.1016/j.tins.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Qi, L., Tsai B., and Arvan P.. 2017. New insights into the physiological role of endoplasmic reticulum-associated degradation. Trends Cell Biol. 27:430. 10.1016/j.tcb.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta, S., Zhang Y., Ross B., Lonka L., Takkunen E., Messer A., Sharp J., Wheeler R., Kusumi K., Mole S., et al. 1999. The neuronal ceroid lipofuscinoses in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat. Genet. 23:233–236. 10.1038/13868 [DOI] [PubMed] [Google Scholar]
- Saegusa, K., Sato M., Morooka N., Hara T., and Sato K.. 2018. SFT-4/Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J. Cell Biol. 217:2073–2085. 10.1083/jcb.201708115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel, D., Schröder M., and Sandhoff K.. 1991. Mutation in the sphingolipid activator protein 2 in a patient with a variant of Gaucher disease. FEBS Lett. 284:57–59. 10.1016/0014-5793(91)80760-Z [DOI] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., and Xenarios I.. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41(D1):D344–D347. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F., et al. 2012. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 90:1102–1107. 10.1016/j.ajhg.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Chambers J.K., Matsuwaki T., Yamanouchi K., and Nishihara M.. 2014. Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol. Commun. 2:78. 10.1186/s40478-014-0078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro, G.J., and Green H.. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299–313. 10.1083/jcb.17.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Wang H., Xu B., Huang D., Nie C., Pu L., Zajac G.J.M., Yan H., Zhao J., Shi F., et al. 2021. Receptor-Mediated ER Export of Lipoproteins Controls Lipid Homeostasis in Mice and Humans. Cell Metab. 33:350–366.e7. 10.1016/j.cmet.2020.10.020 [DOI] [PubMed] [Google Scholar]
- Wheeler, R.B., Sharp J.D., Schultz R.A., Joslin J.M., Williams R.E., and Mole S.E.. 2002. The gene mutated in variant late-infantile neuronal ceroid lipofuscinosis (CLN6) and in nclf mutant mice encodes a novel predicted transmembrane protein. Am. J. Hum. Genet. 70:537–542. 10.1086/338708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Garcia M.R., Novak A.J., Saunders A.M., Ank R.S., Nam A.S., and Fisher L.W.. 2018. Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol. 16:e2005140. 10.1371/journal.pbio.2005140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Sun L., Bastos de Oliveira F., Qi X., Brown W.J., Smolka M.B., Sun Y., and Hu F.. 2015. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol. 210:991–1002. 10.1083/jcb.201502029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Paushter D.H., Feng T., Sun L., Reinheckel T., and Hu F.. 2017. Lysosomal processing of progranulin. Mol. Neurodegener. 12:62. 10.1186/s13024-017-0205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Kukar T., and Rademakers R.. 2021. Lysosomal Dysfunction and Other Pathomechanisms in FTLD: Evidence from Progranulin Genetics and Biology. Adv. Exp. Med. Biol. 1281:219–242. 10.1007/978-3-030-51140-1_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists oligonucleotide primer sequences used in this study.