Metastasis is the leading cause of death in cancer patients. We discovered that proteolytic processing of endothelial TNFR1 by the metalloprotease ADAM17 is essential for metastases formation. Based on our findings, we present a novel therapeutic approach to target metastasis.
Abstract
Metastasis is the major cause of death in cancer patients. Circulating tumor cells need to migrate through the endothelial layer of blood vessels to escape the hostile circulation and establish metastases at distant organ sites. Here, we identified the membrane-bound metalloprotease ADAM17 on endothelial cells as a key driver of metastasis. We show that TNFR1-dependent tumor cell–induced endothelial cell death, tumor cell extravasation, and subsequent metastatic seeding is dependent on the activity of endothelial ADAM17. Moreover, we reveal that ADAM17-mediated TNFR1 ectodomain shedding and subsequent processing by the γ-secretase complex is required for the induction of TNF-induced necroptosis. Consequently, genetic ablation of ADAM17 in endothelial cells as well as short-term pharmacological inhibition of ADAM17 prevents long-term metastases formation in the lung. Thus, our data identified ADAM17 as a novel essential regulator of necroptosis and as a new promising target for antimetastatic and advanced-stage cancer therapies.
Introduction
Tumor metastasis, a process where circulating tumor cells (CTCs) disseminate to distant organs, is a major cause of cancer-related death. CTCs establish metastases after extravasation from blood (or lymph) vessels and subsequently invade the target tissue, thereby escaping immune surveillance mechanisms within the circulation. CTC extravasation involves adhesion to microvascular endothelium, elevation of endothelial permeability, and transendothelial migration into the underlying tissue. All steps during tumor cell (TC) extravasation can be induced either by paracrine factors secreted from the TCs or by interacting hematopoietic cells, such as macrophages, granulocytes, or platelets (Strilic and Offermanns, 2017). Several of these factors, such as C-C motif chemokine 2 (CCL2; Wolf et al., 2012) or ATP (Schumacher et al., 2013) signal via G protein–coupled receptors on either endothelial cells (ECs) or myeloid cells.
Release of TNF from macrophages promotes both hematogenic and lymphogenic tumor metastasis (Kim et al., 2009; Ji et al., 2014). TNF-induced engagement of TNF receptor 1 (TNFR1) results in intracellular formation of the membrane-bound complex I of TNFR1 signaling. Complex I is a large multiprotein complex that includes recruitment of the adaptor proteins TNFR1-associated with death domain (TRADD), TNFR-associated factor 2, receptor interacting kinase 1 (RIPK1), and the E3 ubiquitin ligases cellular inhibitor of apoptosis proteins 1 and 2 (Brenner et al., 2015; Peltzer and Walczak, 2019). Ubiquitination of RIPK1 and other signal proteins of complex I is essential for the recruitment of kinase complexes, which finally results in phosphorylation of inhibitor of NF-κB (IκB) and subsequent gene induction through nuclear translocation of NF-κB. Notably, complex I disassembly and RIPK1 deubiquitination induces a switch from TNFR1 prosurvival signaling to induction of cell death. This can either be mediated via formation of a cytosolic complex II resulting in pro–caspase-8 recruitment, activation, and execution of apoptosis or via formation of a heteroamyloid complex called necrosome. The necrosome consists of the adaptor protein Fas-associated with death domain (FADD) and RIPK1 and 3. The latter phosphorylates the pseudokinase mixed-lineage kinase domain-like (MLKL), leading to the induction of cell death via MLKL-mediated pore formation and subsequent necrosis (Brenner et al., 2015; Peltzer and Walczak, 2019). By contrast, TNFR2 is lacking a death domain in its cytoplasmic domain and, therefore, is unable to directly induce cell death. TNFR2 engagement, rather, results in the activation of NF-κB signaling (Brenner et al., 2015). The affinity of TNFR2 for membrane-bound TNF was demonstrated to be higher than for soluble TNF (Grell et al., 1995). Interestingly, while TNFR1 is ubiquitously expressed, TNFR2 expression is restricted to hematopoietic and ECs (Brenner et al., 2015).
Induction of EC necroptosis and concomitant release of death-associated molecular patterns is discussed as a mechanism of how CTCs breach the endothelial barrier (Strilic et al., 2016; Hänggi et al., 2017; Hou et al., 2019; Yang et al., 2019; Patel et al., 2020). However, the detailed mechanism underlying endothelial barrier opening during TC extravasation is still incompletely understood. Identification of inducers and mediators of this process could potentially establish therapeutic targets against metastasis.
Proteolytic release of ectodomains, a process termed ectodomain shedding, is an irreversible posttranslational mechanism that regulates protein function, intracellular signaling, and provision of signals to neighboring cells. Members of the disintegrin and metalloprotease (ADAM) family are major mediators of ectodomain shedding (Reiss and Saftig, 2009). The family member ADAM17 is a membrane-bound metalloprotease involved in the shedding of a plethora of transmembrane proteins, including growth factors, cytokines, and receptor molecules (Reiss and Saftig, 2009).
Activation of ADAM17 has been linked to the engagement of G protein–coupled receptors, as well as to p38 and ERK MAPKs (Xu et al., 2012). Mice with genetic deficiency in Adam17 are embryonically lethal (Peschon et al., 1998). In addition, ADAM17-inactivating mutations are associated with severe pathologies in humans (Blaydon et al., 2011), mainly caused by impaired release of epidermal growth factor receptor (EGFR) ligands. We previously generated viable ADAM17 hypomorphic (ADAM17ex/ex) mice exhibiting only ∼5% residual Adam17 expression in all tissues, allowing us to study the impact of ADAM17 deficiency in vivo (Chalaris et al., 2010). ADAM17 is involved in the generation of a chronic inflammatory state, thereby predisposing to cancer development, since it proteolytically cleaves and releases TNF and IL-6R (Scheller et al., 2011). Overexpression of ADAM17 in TCs of primary tumors (Murphy, 2008) is often observed and predominantly linked to autocrine EGFR activation on TCs. So far, the impact of ADAM17 on CTC extravasation and subsequent formation of metastases has not been addressed. Given that proteolytic release of TNF is predominantly mediated by ADAM17, we hypothesized that ADAM17 might be essentially involved in CTC extravasation and metastases formation through regulation of the TNF/TNFR signaling cascade.
In the present study, we discovered that proteolytic processing of TNFR1 by ADAM17 is essential for the induction of cellular necroptosis. We demonstrate that as such, ADAM17 is crucial for TC-induced EC death and acts as a facilitator of TC extravasation and metastases formation. Consequently, acute treatment of mice with the antagonistically acting recombinant prodomain (PD) of ADAM17 and thereby blocking ADAM17 catalytic activity was sufficient to prevent TC extravasation and formation of metastatic colonies in the long term. Therefore, ADAM17 is a novel and promising target for advanced-stage cancer therapy.
Results
Microenvironmental ADAM17 promotes metastasis
We employed a murine model of hematogenic metastasis in which syngeneic TCs were i.v. injected into immunocompetent mice, resulting in the formation of experimental lung metastases. To investigate the impact of microenvironmental ADAM17 on TC extravasation and metastatic colony formation, we used hypomorphic ADAM17ex/ex mice in which residual ADAM17 expression is lowered to 5% compared with WT control mice (Fig. 1 A and Fig. S1 A). We injected either Lewis lung carcinoma (LLC) or B16F1 melanoma TCs into both ADAM17ex/ex mice and WT animals (Fig. 1 B). While ADAM17 was markedly reduced in the metastatic microenvironment of ADAM17ex/ex mice, the inoculated TCs were still ADAM17 proficient (Fig. 1 A and Fig. S1 B). Using TCs with stable β-galactosidase (β-Gal; LacZ) expression, we detected significantly reduced numbers of LLC TCs 6 h after injection in the lungs of ADAM17ex/ex mice compared with controls (Fig. 1 C), indicating reduced TC extravasation in these mice. The number of extravasated B16F1 TCs in ADAM17ex/ex mice was reduced to a similar extent (Fig. 1 D). Importantly, we also observed a striking effect on metastases formation, irrespective of the tumor entity, 3 wk after TC inoculation, where ADAM17ex/ex mice displayed a drastically decreased tumor burden (Fig. 1, E and F). Consequently, ADAM17-deficient mice were significantly protected from metastasis-induced death (Fig. 1 G).
Figure 1.
Host ADAM17 promotes metastasis to the lung. (A) Expression of ADAM17 (red lines) in the metastatic microenvironment of WT and hypomorphic ADAM17ex/ex mice in the experimental model used in this study. Note that ADAM17 expression is not altered on injected TCs. (B) Experimental outline for experiments in C–F. Syngeneic TCs are injected i.v. into the tail vein of the indicated mouse lines. TCs expressing LacZ are used in C and D, and mice were sacrificed 6 h after TC injection, while parental TCs are used in E and F, and mice were sacrificed 3 wk after TC injection. (C and D) Extravasated TCs were quantified by β-Gal staining 6 h after TC injection. Representative microscopic images are shown. Scale bars represent 50 μm. n = 3–4 mice/group. *, P < 0.05 (C); P = 0.12 (D) by two-sided Wilcoxon rank sum test with continuity correction. (E and F) 21 d after TC injection, lung metastases were counted. Representative lung images 21 d after TC injection are shown. Scale bar indicates 5 mm. n = 8–9 mice/group (E) and 6 mice/group (F). *, P < 0.05; **, P < 0.01 by unpaired two-tailed bootstrap t test based on 5,000 Monte Carlo simulations. (G) Kaplan–Meier survival curves for WT and ADAM17ex/ex mice demonstrate that ADAM17 is essential for metastasis-induced death. n = 15 mice/group. ****, P < 0.0001. Data represent mean ± SEM. p.i., postinjection.
Figure S1.
Host ADAM17 promotes metastasis. (A) Immunoblot analysis of whole-lung tissue lysate from WT and ADAM17ex/ex mice. (B) Immunoblot analysis of whole-cell lysate of the indicated cell lines. NIH3T3 served as control for ADAM17 expression and was not used in the in vivo studies. (C) Confocal images of human lung metastases sections stained for the indicated markers. Scale bar indicates 25 μm. (D) Bone marrow–derived macrophages (BMDMs) and bone marrow–derived neutrophils (BMDNs) from WT and ADAM17ex/ex mice were incubated with supernatant from cultured LLC cells, and secretion of TNF was determined by ELISA. n = 3 mice/group. *, P < 0.05 by unpaired two-tailed Mann–Whitney U test. (E) Expression of ADAM17 (red outline) in different cell types of LysM-Cre::ADAM17flox/flox (ADAM17ΔMC) mice with the indicated genotype and treatment. (F) BMDMs from ADAM17ΔMC mice were stimulated with 1 μg/ml LPS, and TNF secretion was assessed by ELISA. n = 3 mice/group. **, P < 0.01 by unpaired two-tailed Mann–Whitney U test. (G) Experimental outline as performed in H and I. (H and I) WT and ADAM17ΔMC mice were i.v. injected with 5 × 105 LLC or B16F1 TCs. After 21 d, lung metastases were counted. Representative lung images are shown. Scale bars indicate 5 mm. Shown are the results of one representative out of three independent experiments. n = 3 mice/group (H), n = 3 (control), and n = 4 (ADAM17ΔMC; mice/group (I); unpaired two-tailed Mann–Whitney U test. Data represent mean ± SEM. ca, carcinoma; nd, not detectable; p.i., postinjection.
EC-derived ADAM17 promotes tumor extravasation and metastatic seeding
We hypothesized a reduction in vascular leakage as the reason for the impaired TC extravasation in the absence of ADAM17. 6 h after TC injection, we observed a transient increase of vascular permeability in mice, as determined by FITC-dextran egress, which was significantly lower in ADAM17ex/ex mice than in WT control (Fig. 2, A and B) and declined to baseline levels after 24 h in both mouse variants (data not shown). We concluded that EC-derived ADAM17 might control transendothelial migration of CTCs. In agreement with this assumption, we observed that migration of fluorescently labeled TCs through a monolayer of HUVECs and human lung microvascular ECs (HMVEC-L) was significantly reduced upon siRNA-mediated ADAM17 suppression in ECs (Fig. 2 C and Fig. S2 A). To test the hypothesis that EC-derived ADAM17 controls TC extravasation in vivo, we generated mice with an EC-specific ADAM17 deficiency (ADAM17iΔEC; Fig. 2 D; and Fig. S2, C–E) using tamoxifen-inducible Cdh5-CreERT2 mice (Sörensen et al., 2009). Indeed, TC extravasation was significantly lowered in ADAM17iΔEC mice (Fig. 2, E–G), and tumor burden was reduced to a similar extent as in ADAM17ex/ex mice 3 wk after TC inoculation (Fig. 2, H and I). We detected ADAM17 expression in ECs of human lung metastases irrespective of the primary tumor entity (Fig. S1 C), which is compatible with a critical role of endothelial ADAM17 in human metastasis. Taken together, while previous reports have observed a role of TC-derived ADAM17 for primary tumor growth, we demonstrate here that EC-derived ADAM17 in the metastatic microenvironment is essential for TC extravasation and the establishment of metastatic colonies.
Figure 2.
TC-induced endothelial permeability is promoted by ADAM17. (A) Experimental outline as performed in B. Parental LLC or B16F1 TCs were i.v. injected into the indicated mouse strains. 6 or 24 h later, FITC-labeled dextran was i.v. injected 10 min before sacrificing. (B) Confocal images of lung sections stained for the indicated markers. FITC+ area was quantified as a measure of vascular permeability. Scale bar indicates 25 μm. n = 3 mice/group (top), n = 4 mice/group (bottom), with six images per section quantified. *, P < 0.05 by one-way ANOVA on ranks with Dunn’s post hoc test. (C) Transendothelial migration of CFSE-labeled MDA-MB231 TCs through monolayers of HUVEC or HMVEC-L with siRNA-mediated knockdown of ADAM17. Shown is the quantification of six (HUVEC) or three (HMVEC-L) independent experiments. *, P < 0.05; ****, P < 0.0001 by unpaired two-tailed t test with Welch’s correction. (D) Tamoxifen (TAM) injection into Cdh5-CreERT2::ADAM17flox/flox mice results in ADAM17iΔEC. Note that all other cells in the metastatic microenvironment express ADAM17. (E) Experimental outline as performed in F–I. To induce ADAM17iΔEC, mice received four consecutive tamoxifen injections. Control animals were injected with vehicle (EtOH/oil). 7 d later, TCs were injected i.v. into the tail vein of the indicated mouse lines. TCs expressing LacZ were used in F and G, and mice were sacrificed 6 h after TC injection, while parental TCs were used in H and I and mice were sacrificed 3 wk after TC injection. (F and G) Extravasated TCs were quantified by β-Gal staining. Representative microscopic images are shown. Scale bar indicates 50 μm. n = 4–6 mice/group. *, P < 0.05; **, P < 0.01 by two-factorial ANOVA with group as fixed factor (F) or signed rank test based on 5,000 Monte Carlo simulations (G). (H and I) 3 wk after TC injection, lung metastases were counted. Representative lung images 21 d after TC injection are shown. Scale bar indicates 5 mm. n = 4–5 mice/group. *, P < 0.05; ***, P < 0.001 by unpaired two-tailed Student’s t test with Welch’s correction. Data represent mean ± SEM. Ctrl, control; p.i., postinjection; rel. relative.
Figure S2.
Genetic deficiency of ADAM17 and TNFR1 in human and murine ECs. (A) Immunoblot analysis of HUVECs with siRNA-mediated suppression of ADAM17 or TNFR1, respectively. (B) Immunoblot analysis of MLECs with siRNA-mediated suppression of ADAM17. (C) Strategy for MACS-based isolation of murine lung ECs. (D) Representative FACS plots after different purification steps during murine lung EC isolation. (E) ADAM17 expression of the indicated cell populations was assessed by immunoblot analysis and indicates specific ADAM17 deficiency in lung ECs in tamoxifen (TAM)-treated Cdh5-CreERT2::ADAM17flox/flox mice. Ctrl, control; SSC-A, side scatter area; FSC-A, forward scatter area; FSC-H, forward scatter height.
ADAM17 confers TC-induced EC death
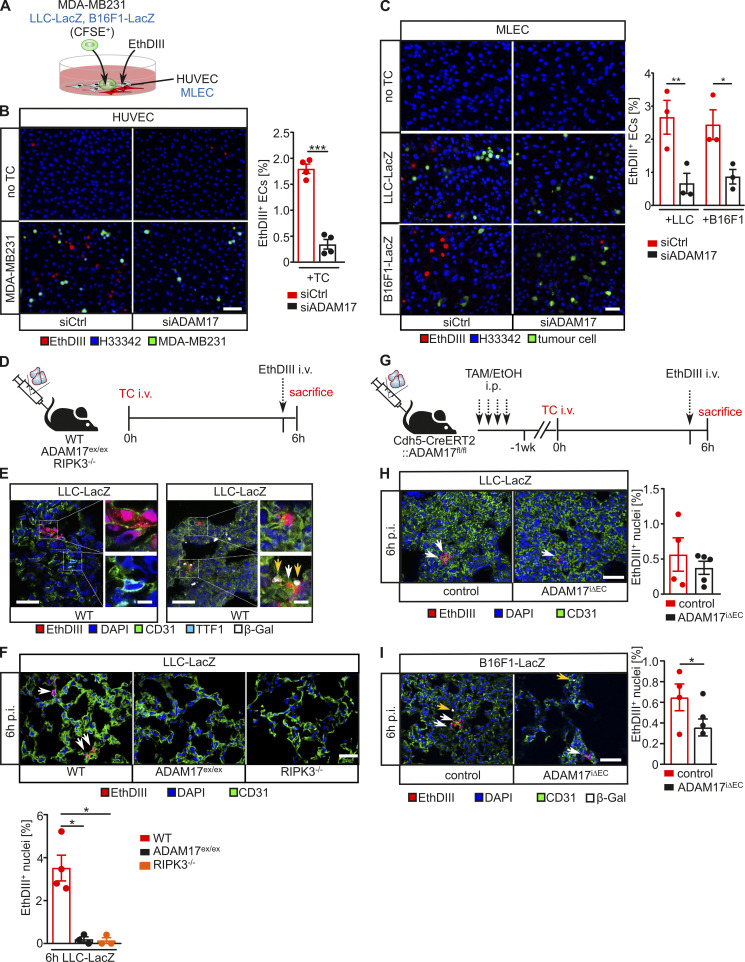
TC-induced EC death is one mechanism that is discussed to mediate vascular permeability preceding TC extravasation (Hänggi et al., 2017; Hou et al., 2019; Yang et al., 2019; Patel et al., 2020). We therefore assessed whether TC-dependent EC death induction depends on ADAM17 on the basis of ethidium homodimer III (EthDIII) uptake into cells with impaired membrane integrity, as previously described (Strilic et al., 2016). Cocultivation of HUVECs with fluorescently labeled MDA-MB231 breast cancer cells (Fig. 3 A) resulted in a significantly lower level of EthDIII incorporation into HUVECs with siRNA-mediated suppression of ADAM17 compared with ADAM17-competent control cells (Fig. 3 B). Similar results were obtained when LacZ-expressing B16F1 or LLC murine TCs were cultivated on murine microvascular lung ECs (MLECs; Fig. 3 C and Fig. S2 B). Accordingly, EthDIII incorporation into CD31+ lung ECs but not into TTF1+ pneumocytes 6 h after TC injection was significantly reduced in ADAM17ex/ex mice, almost to the same level as in necroptosis-deficient RIPK3−/− mice (Fig. 3, D–F). Consistent with the assumption of a cell-autonomous effect of ADAM17, EC-specific ADAM17 deficiency in ADAM17iΔEC mice was sufficient to suppress TC-induced EC death during TC extravasation (Fig. 3, G–I).
Figure 3.
ADAM17 is essential for TC-induced EC death. (A) HUVECs (human) or MLECs (murine) with indicated siRNA-mediated knockdown were cocultured with CFSE-labeled MDA-MB231 (human) or B16F1-LacZ or LLC-LacZ (murine) TCs, respectively, and cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. (B) Shown are representative confocal images of cocultures of MDA-MB231 on HUVECs with the indicated genetic deficiency and quantification of four independent experiments with three technical replicates and three images per replicate analyzed. Scale bar indicates 100 μm. ***, P < 0.001 by unpaired two-tailed Mann–Whitney U test. (C) Shown are representative confocal images of LLC-LacZ or B16F1-LacZ cocultured on MLECs with the indicated genetic deficiency and the quantification of three independent experiments with three technical replicates and three images per replicate analyzed. Scale bar indicates 100 μm. *, P < 0.05; **, P < 0.01 by one-way ANOVA with Tukey’s post hoc test. (D) Experimental outline as performed in E and F. LacZ-expressing LLC TCs were i.v. injected into the indicated mouse strains. 6 h later, EthDIII was i.v. injected 5 min before sacrificing. (E) Representative confocal image of lung sections for the indicated markers indicates cell death in CD31+ ECs but not in TTF1+ type II pneumocytes. Scale bar indicates 25 μm in the overview and 5 μm in the enlarged view. White arrows indicate EthDIII+CD31+ cells, and yellow arrows indicate β-Gal+ TCs. (F) Pulmonary EthDIII+ ECs were quantified from confocal images of lung sections stained for the indicated markers. Scale bar indicates 50 μm. White arrows indicate EthDIII+CD31+ cells. n = 3 mice/group, with three to six sections per mouse and at least three images per section analyzed. *, P < 0.05 by one-way ANOVA on ranks with Dunn’s post hoc test. (G) Experimental outline as performed in H and I. To induce EC-specific ADAM17 deficiency, mice received four consecutive tamoxifen (TAM) injections. Control animals were injected with vehicle (EtOH/oil). 7 d later, LacZ-expressing LLC TCs were i.v. injected into the indicated mouse strains. 6 h after TC injection, EthDIII was i.v. injected 5 min before sacrificing. (H and I) Pulmonary EthDIII+ ECs were quantified from confocal images of lung sections stained for the indicated markers. Scale bar indicates 50 μm. White arrows indicate EthDIII+CD31+ cells, and yellow arrows indicate β-Gal+ TCs. n = 3–5 mice/group with three to six sections per mouse and at least three images per section analyzed. *, P < 0.05, unpaired two-tailed bootstrap t test based on 5,000 Monte Carlo simulations (H) or generalized linear model based on log-normal distributions (I). Data represent mean ± SEM. Ctrl, control; p.i., postinjection.
Both EC- and TC-derived TNF promote TC extravasation and metastasis
TNF secreted from myeloid cells was previously demonstrated to promote lung metastasis (Kim et al., 2009). We therefore hypothesized that ADAM17-mediated TNF release from myeloid cells regulates metastasis. However, while we detected a significant reduction of TNF release from ADAM17-deficient bone marrow–derived macrophages and neutrophils (Fig. S1, D–F), metastases burden was not reduced in mice exhibiting a myeloid-specific ADAM17 deficiency (Fig. S1, G–I). We therefore concluded that myeloid-derived TNF is dispensable for metastasis.
In contrast, siRNA-mediated suppression of TNF in both human (Fig. 4 A) and murine ECs (Fig. 4 B) reduced TC-induced cell death in vitro, indicating that EC-derived TNF promotes TC-induced EC death (Fig. 4 C). In line with the assumption that TNFR1 mediates TNF-induced cell death signaling, we observed significantly reduced TC-induced EC death in vitro in HUVECs with siRNA-mediated suppression of TNFR1 (Fig. 4 D). Consequently, the number of extravasated TCs in vivo was significantly lower in TNFR1−/− mice compared with WT animals (Fig. 4, E and F).
Figure 4.
EC-derived TNF is necessary for TC-induced EC death. (A) HUVECs with siRNA-mediated TNF knockdown were cocultured with CFSE-labeled MDA-MB231 TCs, and cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images and the quantification of three independent experiments. Three technical replicates per experiment with three to four images per replicate were analyzed. Scale bar indicates 50 μm. ****, P < 0.0001 by semiparametric generalized linear model based on log-normal distributions. (B) Murine MLECs with siRNA-mediated TNF knockdown were cocultured with the indicated CFSE-labeled murine TCs. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images and the quantification of four independent experiments. Three technical replicates per experiment with three to four images per replicate were analyzed. Scale bar indicates 50 μm. *, P < 0.05 by unpaired two-tailed Mann–Whitney U test. (C) Contact with TCs might induce an autocrine soluble TNF loop in ECs, leading to EC death. (D) HUVECs with indicated siRNA-mediated knockdown were cocultured with CFSE-labeled MDA-MB231 TCs, and cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images of three independent experiments. Three technical replicates per experiment with three to four images per replicate were analyzed. Scale bar indicates 50 μm. ***, P < 0.001 by one-way ANOVA on ranks with Dunn’s post hoc test. (E) Experimental outline as performed in F. LacZ-expressing LLC TCs were i.v. injected into the indicated mouse strains, and TC extravasation was assessed after 6 h. (F) Extravasated TCs 6 h after TC injection were quantified by β-Gal staining. Representative microscopic images are shown. Scale bar indicates 50 μm. n = 3 mice/group. ***, P < 0.001 by unpaired Student’s t test. Data represent mean ± SEM. Ctrl, control; p.i., postinjection.
We furthermore assessed whether TC-derived TNF contributes to metastasis in our model (Fig. 5 A). We detected low-level TNF on the surface (Fig. 5 B) and in the supernatant (Fig. S4, A and B) of all three TC lines used in this study. We therefore generated LacZ-expressing LLC and B16F1 TCs with a genetic deficiency in TNF using CRISPR/Cas9. We targeted Tnf exon 1 with a multiguide approach (Fig. S3 A), which resulted in a 110-bp deletion (Fig. S3, B–D), leading to a premature stop codon (Fig. S3 D). TC-specific TNF deficiency did not alter TC-induced MLEC death in an in vitro coculture assay (Fig. S3 E). Similarly, TC-induced EC death in vivo was not altered in the absence of TC-derived TNF (Fig. 5, C and D). In contrast, TC extravasation was significantly reduced in the absence of TC-derived TNF (Fig. 5, C and E). Consequently, metastasis formation was largely reduced in the absence of TC-derived TNF 3 wk after injection (Fig. 5 F), suggesting that TC survival in vivo depends on a low-level TNF autocrine loop. We indeed observed a slightly increased susceptibility toward TNF-induced cell death in the absence of TC-derived TNF for LLC-LacZ cells (Fig. 5 G).
Figure 5.
TC-derived TNF is dispensable for TC-induced EC death but required for metastasis. (A) TCs can produce membrane-bound or soluble TNF-α that can directly bind to TNFR1 on ECs to induce EC death and promote endothelial permeability. (B) TCs used in this study express very-low-level TNF as assessed by flow cytometry using anti-TNF antibodies. (C) Experimental outline as performed in D–G. TNF-deficient (TNFko) LacZ–expressing LLC or B16F1 TCs were generated by CRISPR/Cas9 (Fig. S3). Parental or TNF-deficient (TNFko) LacZ-expressing LLC or B16F1 TCs were i.v. injected into the indicated mouse strains. 6 h later, EthDIII was i.v. injected 5 min before sacrificing, where indicated (E and F). 21 d after injection, lung metastases were determined (G). (D) Pulmonary EthDIII+ ECs were quantified from confocal images of lung sections stained for the indicated markers. Scale bar indicates 50 μm. White arrows indicate EthDIII+ ECs. n = 4–5 mice/group; unpaired two-tailed Mann–Whitney U test. (E) Extravasated TCs 6 h after TC injection were quantified by β-Gal staining. Representative microscopic images are shown. Scale bar indicates 100 μm. Black arrows indicate extravasated TCs. n = 4–5 mice/group. ****, P < 0.0001 by unpaired two-tailed generalized linear model based on γ-distributions. (F) 3 wk after TC injection, lung metastases were counted. Representative lung images 21 d after TC injection are shown. n = 4–5 mice/group. **, P < 0.01 by two-tailed Mann–Whitney U test. (G) LLC-LacZ TCs with the indicated genotype were incubated where indicated with 100 μM zVAD 15 min before stimulation with 50 ng/ml TNF. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images and the quantification of four independent experiments with three technical replicates and three to four images per replicate analyzed. Scale bar indicates 50 μm. *, P < 0.05 by Welch’s ANOVA with Tukey’s post hoc test. Data represent mean ± SEM. p.i., postinjection.
Figure S4.
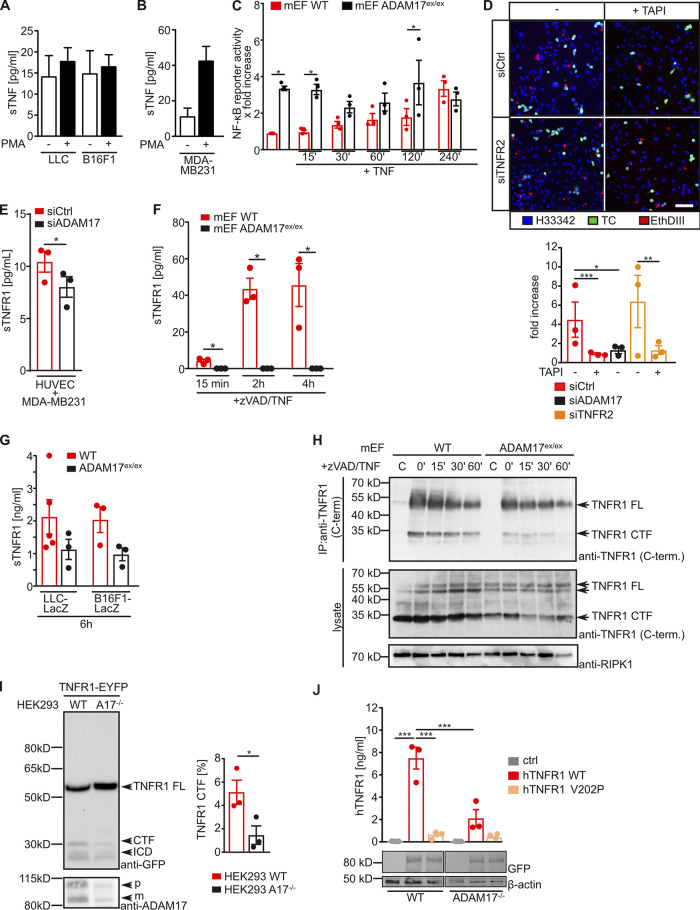
TNFR1 ectodomain is released via ADAM17-mediated proteolysis. (A and B) The indicated TCs were mock treated or stimulated with 500 nM PMA, and soluble TNF (sTNF)-α in the supernatant was determined by ELISA. Shown is the quantification of three independent experiments. Unpaired one-tailed Mann–Whitney U test. (C) WT and ADAM17ex/ex mEFs were transfected with an NF-κB luciferase reporter plasmid, stimulated with 200 ng/ml TNF-α for the indicated time points, and luciferase activity was determined according to standard procedures. *, P < 0.05 by one-way ANOVA on ranks with Bonferroni’s post hoc test. (D) HUVECs with indicated siRNA-mediated knockdown were cocultured with CFSE-labeled MDA-MB231 TCs in the absence or presence of 250 nM TAPI-1. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images of three independent experiments with three technical replicates per experiment and three to four images per replicate analyzed. Scale bar indicates 50 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA on ranks with Dunn’s post hoc test. (E) HUVECs with indicated siRNA-mediated knockdown were cocultured with MDA-MB231 cells, and sTNFR1 in the supernatant was determined by ELISA. The quantification of three independent experiments is shown. *, P < 0.05 by unpaired two-tailed Student’s t test. (F) WT or ADAM17ex/ex mEFs were stimulated for the indicated time points with 100 μM zVAD and 50 ng/ml TNF-α. Release of sTNFR1 was assessed by ELISA. The quantification of three independent experiments is shown. *, P < 0.05 by one-way ANOVA on ranks with Bonferroni’s post hoc test. (G) LacZ-expressing LLC TCs were i.v. injected into the indicated mouse strains. 6 h later, mice were sacrificed, and plasma levels of sTNFR1 were determined by ELISA. n = 3–5 mice/group; unpaired two-tailed Mann–Whitney U test. (H) WT and ADAM17ex/ex mEFs were stimulated with zVAD/TNF-α for the indicated time points and subjected to cell lysis. TNFR1 was isolated by immunoprecipitation (IP) and subsequently analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. (I) TNFR1-EYFP was overexpressed in parental (WT) or ADAM17-deficient HEK293T cells (A17−/−), and TNFR1 proteolysis was assessed by SDS-PAGE and immunoblotting using the indicated antibodies. Shown is one representative blot and the quantification of the C-terminal fragment (CTF) formation of three independent experiments. *, P < 0.05 by unpaired two-tailed Mann–Whitney U test. (J) WT HEK293T cells or HEK293T cells with CRISPR/Cas9-mediated ADAM17−/− were transfected with the indicated TNFR1 variants, and the generation of sTNFR1 was determined by ELISA. ***, P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data represent mean ± SEM. C/Ctrl, control; C-term., C-terminus; FL, full length; ICD, intracellular domain.
Figure S3.
CRISPR/Cas9-mediated generation of TNF-deficient murine TCs. (A) Targeting and genotyping strategy. Exon 1 of murine Tnf is shown. Arrows below indicate targeting area of sgRNAs. Combination of cleavage events can result in the deletion of a 110-bp fragment. Arrows above indicate PCR primers resulting in a 500-bp PCR product of unmodified Tnf. (B) PCR-based genotyping of pool of targeted murine TCs indicates 88% (LLC-LacZ) and 87% (B16F1-LacZ) insertion/deletion (indel) formation. (C) PCR-based genotyping of single-cell clones used in this study demonstrates deletion of a 110-bp fragment within Tnf. (D) Sanger sequencing of Tnf exon 1 confirms 110-bp deletion, resulting in amino acid deletion, a frameshift, and the appearance of a premature stop codon after a total of 24 amino acids in targeted TCs. (E) TNF-deficient LacZ-expressing B16F1 TCs were labeled with CSFE and cocultured on murine MLECs. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images of three to four independent experiments with three technical replicates and three to four images per replicate. Scale bar indicates 50 μm; unpaired one-tailed Mann–Whitney U test. Data represent mean ± SEM. fw, forward.
We next assessed whether pharmacological blockade of TNF signaling prevents TC extravasation. TC-induced EC death in vitro in the HUVEC/MDA-MB231 coculture model was significantly reduced when TNF was neutralized by binding to etanercept, a recombinant form of the soluble human TNFR2 ectodomain fused to a human Fc-tag (Fig. 6, A and B; Moreland et al., 1997). TC-induced EC death in vivo was significantly lowered when etanercept was administered to mice 5 min before TC injection (Fig. 6, C and D). Consequently, TC extravasation was significantly reduced but not completely absent when TNF was neutralized by etanercept (Fig. 6 E).
Figure 6.
Pharmacological inhibition of TNF impairs TC-induced EC death and tumor extravasation. (A) Etanercept is a recombinant protein fusion of the ECD of murine TNFR2 and a human IgG Fc portion that binds to TNF and thereby blocks TNF signaling. (B) HUVECs were cocultured with CFSE-labeled MDA-MB231 TCs in the presence or absence of etanercept. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake and confocal microscopy. Shown are representative images of three independent experiments with three technical replicates and three to four images per replicate analyzed. Scale bar indicates 50 μm. *, P < 0.05 by one-way ANOVA on ranks with Dunn’s post hoc test. (C) Experimental outline as performed in D and E. Where indicated, mice were injected i.v. with PBS or etanercept 5 min before TC injection. LacZ-expressing LLC TCs were i.v. injected into WT mice. 6 h later, EthDIII was i.v. injected 5 min before sacrificing where indicated. (D) Pulmonary EthDIII+ ECs were quantified from confocal images of lung sections stained for the indicated markers. Scale bar indicates 50 μm. n = 5–6 mice/group. *, P < 0.05 by unpaired two-tailed bootstrap t test based on 5,000 Monte Carlo simulations. (E) Extravasated TCs 6 h after TC injection were quantified by β-Gal staining. Representative microscopic images are shown. Scale bar indicates 50 μm. n = 5–6 mice/group. *, P < 0.05 by unpaired two-tailed generalized linear model based on log-Tweedie distributions. Data represent mean ± SEM. p.i., postinjection.
Taken together, while TC-derived TNF seems to be essential for CTC survival, EC-derived TNF is crucial for TC-induced EC death. Therefore, TNF is a druggable target for the prevention of tumor metastasis.
ADAM17 is essential for TNFR1-mediated necroptosis
On the basis of the cell-autonomous effect of ADAM17 in ECs for TC-induced cell death, we next hypothesized that ADAM17 activity is a general prerequisite for TNF-induced cell death. We therefore induced necroptosis in WT or ADAM17ex/ex murine embryonic fibroblasts (mEFs) by administration of TNF and the pancaspase inhibitor carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (zVAD), as previously described (Lafont et al., 2018). TNF/zVAD-induced cell death was drastically impaired in ADAM17ex/ex mEFs (Fig. 7 A), indicating that ADAM17 is required for TNFR1-induced necroptosis. Also in HUVECs, TNF/zVAD-induced cell death was significantly reduced when ADAM17 expression was suppressed (Fig. 7 B). This led to the hypothesis that canonical NF-κB signaling might prevent cell death induction in the absence of ADAM17. Phosphorylation of IκB by IκB kinase induces ubiquitin-dependent proteasomal degradation of IκB and subsequent translocation of a NF-κB heterodimer, such as p50/p65, to the nucleus to enable transcription of NF-κB target genes (Napetschnig and Wu, 2013). In line with a protective role of NF-κB signaling in ADAM17-deficient cells, we observed comparable levels of cell death induction in control cells and HUVECs with siRNA-mediated suppression of ADAM17 upon inhibition of IκB kinase with the small molecule TPCA-1 (Fig. 7 B). This correlated with ligand-independent p65 phosphorylation in ADAM17ex/ex mEFs, while in WT mEFs, p65 phosphorylation was only detectable upon TNF stimulation (Fig. 7 C). Interestingly, ligand-dependent IκB phosphorylation and concomitant IκB degradation were only observed in WT mEFs, while IκB levels remained unchanged in ADAM17ex/ex mEFs (Fig. 7 C). Nuclear localization of the NF-κB subunit p65 and activation of an NF-κB reporter construct was significantly elevated in ADAM17ex/ex mEFs compared with WT mEFs (Fig. 7 D and Fig. S4 C). In the same line, NF-κB target genes, such as Tnf, Cflar, and Birc3, were constitutively elevated in ADAM17ex/ex mEFs compared with WT mEFs, which showed increased NF-κB target gene levels only after TNF stimulation (Fig. 7 E). Taken together, our data suggest a constitutive noncanonical NF-κB activation in the absence of ADAM17.
Figure 7.
ADAM17 is essential for TNF-mediated necroptotic cell death. (A) mEFs with the indicated genotype were incubated with 100 μM zVAD 15 min before stimulation with 50 ng/ml TNF. Cells with impaired membrane integrity were determined after 6 h by EthDIII uptake. Shown are representative images of three independent experiments with three technical replicates per experiment and three to four images per replicate analyzed. Scale bar indicates 200 μm. ***, P < 0.001 by one-way ANOVA on ranks with Dunn’s post hoc test. (B) HUVECs with indicated siRNA-mediated knockdown were incubated with 100 μM zVAD and TPCA-1, if indicated, 15 min before stimulation with 50 ng/ml TNF. Cells with impaired membrane integrity were determined after 4 h by EthDIII uptake. Shown are representative images of three to five independent experiments with three technical replicates per experiment and three to four images per replicate analyzed. Scale bar indicates 200 μm. *, P < 0.05 by one-way ANOVA on ranks with Dunn’s post hoc test. (C) mEFs with the indicated genotype were stimulated with 200 ng/ml modified tandem affinity purification–tagged TNF for the indicated time points and subsequently lysed. Cell lysates were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Immunoblots of one representative experiment and the quantification of three independent experiments are shown. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 by generalized estimating equation models with least significant difference post hoc tests with Bonferroni corrections. (D) mEFs with the indicated genotype were grown on coverslips, and phosphorylation and nuclear translocation of the NF-κB component p65 was determined by immunofluorescence and confocal microscope. Shown are representative microscopic images and the quantification of nuclear signals from eight (p65) or three (pp65) independent experiments. Scale bars indicate 25 μm. *, P < 0.05; ***, P < 0.001 by unpaired two-tailed bootstrap t test. (E) mEFs with the indicated genotype were stimulated with 200 ng/ml TNF for the indicated time points, and expression of the indicated NF-κB target genes were assessed by quantitative RT-PCR from six independent experiments. *, P < 0.05; **, P < 0.01 by unpaired two-tailed bootstrap t test based on 5,000 Monte Carlo simulations. Data represent mean ± SEM. Ctrl, control.
Proteolytic processing of TNFR1 is essential for cell death induction
We next tested whether TNFR2 mediates protective effects in the absence of ADAM17. We therefore suppressed TNFR2 expression in HUVECs by siRNA and simultaneously inhibited ADAM17 activity using the hydroxamate-type metalloproteinase inhibitor TAPI-1. TC-induced EC death was only slightly increased when TNFR2 expression was downregulated by siRNA (Fig. S4 D). In contrast, inhibition of ADAM17 activity by TAPI-1 resulted in impaired TC-induced EC death to the same extent as siRNA-mediated suppression of ADAM17 in both TNFR2-proficient HUVECs and those with siRNA-mediated suppression of TNFR2 (Fig. S4 D). We therefore hypothesized that instead, proteolytic processing of TNFR1 by ADAM17 might be essential for TC-induced EC death. Accordingly, levels of soluble TNFR1 (sTNFR1) in the supernatant of HUVECs with ADAM17 suppression (Fig. S4 E), cocultivated with TCs, as well as in the supernatant of TNF-stimulated mEFs (Fig. S4 F), were significantly reduced. Furthermore, sTNFR1 plasma levels 6 h after TC injection were reduced in ADAM17ex/ex mice (Fig. S4 G). In line with these data, appearance of a TNFR1 C-terminal fragment was reduced in ADAM17ex/ex mEFs (Fig. S4 H), as well as in ADAM17-deficient HEK293 cells (Fig. S4 I). To test our hypothesis, we generated ADAM17 cleavage-resistant TNFR1 mutants (Fig. 8 A) by substituting the P1′ valine at the ADAM17 cleavage site by proline, as previously described (Brakebusch et al., 1994). When transfected into HEK293 cells, proteolytic processing of TNFR1 V202P was almost completely impaired and to the same extent as processing of TNFR1 WT in ADAM17-deficient HEK293 cells (Fig. S4 K). While expression of both TNFR1 variants was comparable in reconstituted TNFR1−/− mEFs (Fig. 8 B), cell surface levels of TNFR1 V202P were slightly elevated compared with TNFR1 WT (Fig. 8 C). In line with our hypothesis, TNF/zVAD-induced necroptosis was significantly reduced in TNFR1 V202P–reconstituted mEFs and lowered to a similar extent as in ADAM17ex/ex mEFs (Fig. 8 D). Accordingly, ligand-induced RIPK3 phosphorylation was impaired in TNFR1 V202P-reconstituted mEFs (Fig. 8 E). These data indicate that ADAM17-mediated ectodomain shedding of TNFR1 is important for TNF-induced cell death. In support of this notion, we observed a strongly reduced association of RIPK1 and cleaved caspase 8 to the adaptor protein FADD (Fig. 8 F), indicating impaired TNFR1 complex II and necrosome formation.
Figure 8.
Proteolytic processing of TNFR1 by ADAM17 and γ-secretase enables cell death induction. (A) Amino acid sequence around the ADAM17 cleavage site in human and murine TNFR1. Substitution of the P1’ valine to proline in the cleavage-resistant TNFR1 V202P variant is indicated in red. (B) TNFR1−/− mEFs were retrovirally reconstituted with the indicated FLAG-hTNFR1 variants, and expression was verified by SDS-PAGE and immunoblotting using the indicated antibodies. (C) Cell surface levels of the indicated FLAG-hTNFR1 variants stably expressed in TNFR1−/− mEFs were determined by flow cytometry using anti-TNFR1 antibodies; ctrl (control) indicates unstained cells. (D) TNFR1−/− mEFs reconstituted with the indicated TNFR1 variants or ADAM17ex/ex mEFs were incubated with 100 μM zVAD 15 min before stimulation with 50 ng/ml TNF. Cells with impaired membrane integrity were detected by EthDIII uptake after 4 h. Shown are representative images and the quantification of three independent experiments with three replicates and three to four images per replicate analyzed. Scale bar indicates 50 μm. *, P < 0.05; ***, P < 0.001 by generalized estimating equation model. (E) TNFR1−/− mEFs reconstituted with the indicated TNFR1 variants were incubated with 100 μM zVAD or mock-treated 15 min before stimulation with 50 ng/ml TNF for 2 h. Cells were subsequently lysed, and lysates were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. One representative immunoblot and the quantification of five independent experiments are shown. *, P < 0.05 by generalized estimating equation model. (F) WT or ADAM17ex/ex mEFs were incubated with 100 μM zVAD 15 min before stimulation with 1 μg/ml TNF for the indicated time points and subsequently lysed. FADD was isolated by immunoprecipitation, and coassociated proteins were identified by SDS-PAGE and immunoblotting using the indicated antibodies. One representative immunoblot and the quantification of three independent experiments are shown. Asterisk indicates Ig light chains. **, P < 0.01 by generalized estimation equation model. (G) TNFR1 ectodomain shedding by ADAM17 might be followed by intramembrane proteolysis mediated by the PSEN-containing γ-secretase complex, which would result in the liberation of a C-terminal TNFR1 fragment. (H) mEFs with the indicated genotype were incubated with 100 μM zVAD 15 min before stimulation with 50 ng/ml TNF. Cells with impaired membrane integrity were detected by EthDIII uptake after 4 h. Shown are representative images and the quantification of three independent experiments with three technical replicates per experiment and three to four images per replicate analyzed. **, P < 0.01; unpaired two-tailed Student’s t test. Data represent mean ± SEM. assoc. associated; cl. casp.8, cleaved caspase 8; IP, immunoprecipitation; rel., relative; stim., stimulated; TM, transmembrane; VP, V202P.
Ectodomain shedding of transmembrane proteins is often followed by regulated intramembrane proteolysis (RIP). The γ-secretase complex with its catalytic subunits presenilin (PSEN) 1 and 2 is a major mediator of RIP (Oikawa and Walter, 2019) and is involved in TNFR1 processing (Fig. 8 G; Chhibber-Goel et al., 2016). In support of this notion, we observed that TNF/zVAD-induced necroptosis was significantly reduced in PSEN1+/−2−/− mEFs (Fig. 8 H). Taken together, our data suggest that ADAM17-mediated ectodomain shedding and subsequent γ-secretase-mediated RIP of TNFR1 is a prerequisite for TNF-induced cell death.
Pharmacological inhibition of ADAM17 is sufficient to block metastasis
We further sought to explore whether pharmacological inhibition of ADAM17 is sufficient to block TC-induced EC death in vivo and, in consequence, CTC extravasation and metastasis. To this end, we used the ADAM17 inhibitor TAPI-1 and the recently developed, highly specific inhibitory recombinant ADAM17 PD (Fig. 9 A; Wong et al., 2016). Proteolytic removal of the inhibitory PD from the ADAM17 proenzyme in the Golgi during maturation renders ADAM17 catalytically competent (Scheller et al., 2011; Wong et al., 2015). ADAM17 PD shares almost no homology with other PDs of the ADAM or matrix metalloproteinase family, making recombinant ADAM17 PD a suitable candidate for highly specific ADAM17 inhibition (Wong et al., 2016). TAPI-1 or ADAM17 PD impaired ADAM17 activity in vitro at an half maximal inhibitory concentration (IC50) of 128.0 ± 0.9 nM and 110.8 ± 1.6 nM, respectively (Fig. S5, A–D). We administered a single dose of TAPI-1 or ADAM17 PD i.v. 5 min before TC injection (Fig. 8 B) and observed significantly reduced lung EC necroptosis 6 h after TC injection (Fig. 9 C). Accordingly, TC extravasation was significantly reduced in inhibitor-treated animals (Fig. 9 D). Consequently, injection of a single dose of ADAM17 PD before TC injection had long-term effects in mice. Mice with ADAM17 PD injection showed a markedly reduced tumor burden in the lung, and partially in liver and kidney (Fig. S5 E), 3 wk after TC injection (Fig. 9 E). Taken together, short-term pharmacological inhibition of ADAM17 activity is sufficient to prevent long-term metastatic seeding to the lung.
Figure 9.
Pharmacological inhibition of ADAM17 prevents metastasis to the lung. (A) Schematic of ADAM17 PD–mediated inhibition of ADAM17. (B) Experimental outline as performed in C–E. WT animals were i.v. injected with 12 mg/kg BW TAPI-1 or 1 mg/kg BW ADAM17 PD 10 min before i.v. injection with 5 × 105 parental LLC or B16F1 TCs (E) or LacZ-expressing LLC TCs (C and D). 6 h later, EthDIII was i.v. injected 10 min before sacrificing (C); extravasated TCs were quantified by β-Gal staining (D); and 21 d after injection, lung metastases were determined (E). (C) Confocal images of lung sections 6 h after TC injection, stained for the indicated markers. Scale bar indicates 100 μm. n = 6–7 mice/group (PBS), n = 3 mice/group (TAPI), and n = 8 mice/group (ADAM17 PD). *, P < 0.05; ***, P < 0.001; one-way ANOVA on ranks with Dunn’s post hoc test. (D) Representative microscopic images 6 h after TC injection. Scale bar indicates 50 μm. n = 6–7 mice/group (PBS), n = 3 mice/group (TAPI), and n = 8 mice/group (ADAM17 PD). **, P < 0.01; ***, P < 0.001; one-way ANOVA on ranks with Dunn’s post hoc test. (E) Lung images 21 d after TC injection are shown. Scale bar indicates 5 mm. n = 3–5 mice/group. ***, P < 0.001; unpaired two-tailed Student’s t test. Data are mean ± SEM. p.i., postinjection.
Figure S5.
IC50 of TAPI-1 and ADAM17 PD were analyzed using recombinant catalytic domain of ADAM17 and a quenched fluorogenic TGF-α–based substrate peptide. (A–D) The relative velocity determined by linear regression and IC50 values calculated using four-parameter logistic curve fitting. One representative experiment (A and B) and the quantification of three independent experiments are shown. (E) WT animals were i.v. injected with 1 mg/kg BW ADAM17 PD 10 min before i.v. injection with 5 × 105 B16F1 TCs, and 21 d after injection, liver and kidney metastases were determined. Liver and kidney images 21 d after TC injection are shown. Scale bars indicate 5 mm. n = 4–5 mice/group. *, P < 0.05 by unpaired two-tailed Student’s t test. Data represent mean ± SEM. p.i., postinjection; rel., relative.
Discussion
Here, we describe a previously unknown essential role of ADAM17 in the metastatic microenvironment, leading to a novel concept of antimetastatic therapy. We demonstrate that efficient formation of metastases depends on extravasation of TCs, which requires ADAM17 activity in ECs. Mechanistically, TC- and TNF-induced cellular necroptosis largely depend on ADAM17-mediated proteolytic processing of death receptor TNFR1. Genetic loss of ADAM17 in ECs or inhibition of ADAM17 catalytic activity is therefore sufficient to (i) prevent TC-induced EC necroptosis, (ii) lower TC extravasation, and (iii) protect from metastases formation in two syngeneic hematogenic metastasis models.
Albeit tumor metastasis is the major cause of death in cancer patients, there are currently no clinically approved drugs available that specifically target steps of the metastatic cascade (Strilic and Offermanns, 2017). This is also due to the fact that beneficial effects of therapeutic inhibition of a particular pathway during metastasis are difficult to predict. For example, anti-CCL2 monotherapy reduces metastasis of mammary carcinoma but promotes tumor progression after cessation of therapy (Bonapace et al., 2014). Therefore, development of novel therapeutic concepts targeting tumor metastasis is highly warranted. Multiple pathways are implicated in the induction of endothelial permeability and TC extravasation (Reymond et al., 2013; Strilic and Offermanns, 2017), including soluble factors released from CTCs or hematopoietic cells, such as CCL2 (Wolf et al., 2012) or ATP (Schumacher et al., 2013), that act through engagement of G protein–coupled receptors. Certain subsets of G protein–coupled receptors have been linked to activation of ADAM17 (Inoue et al., 2012).
Previous reports suggested a pivotal contribution of the TNFR superfamily (TNFRSF) members TNFR1 (Ji et al., 2014) and death receptor 6 (Strilic et al., 2016) to lymphogenic and hematogenic metastasis, respectively. TC-mediated induction of EC necroptosis via TNFRSF signaling is discussed to promote TC metastasis (Strilic et al., 2016; Hänggi et al., 2017; Hou et al., 2019; Yang et al., 2019; Patel et al., 2020), and metastasis was shown to be reduced in mice with EC-specific RIPK3 deficiency (Strilic et al., 2016; Yang et al., 2019) and in mice with kinase-inactive RIPK1 (Hänggi et al., 2017; Hou et al., 2019). However, these findings are also controversially discussed (Patel et al., 2020). Furthermore, it should be noted that i.v. injection of TCs, which is a widely used model of TC extravasation and hematogenic metastasis (Abdul Pari et al., 2021), has its limitations because it might be influenced by passive cancer cell trapping in the microvasculature or additional unknown effects.
Here, we show that hematogenic metastasis depends on TNF signaling via TNFR1 and concomitant induction of cell death in ECs. We show that during TC extravasation, TNF seems to be provided primarily by ECs. By contrast, TCs seem to exploit autocrine TNF signaling to escape from TNF-induced cell death in the circulation. It is therefore not surprising that TC-induced EC death and concomitant metastasis is impaired upon TNF neutralization.
We furthermore provide evidence that hematogenic metastasis relies on ADAM17 on ECs and that TC-induced and TNF-mediated EC death is promoted by cell-autonomous ADAM17 activity. ADAM proteases, notably ADAM9 and ADAM10, were shown to accelerate necroptosis through ectodomain shedding of cell adhesion proteins (Cai et al., 2016). Furthermore, we have previously suggested that ADAM17 is involved in the induction of necroptosis (Fuchslocher Chico et al., 2018). Here, we demonstrate that TNFR1-induced cell death relies on proteolytic TNFR1 ectodomain release by ADAM17. Recently, a hexagonal arrangement of TNFR1 trimers on the plasma membrane mediated at least partially via its extracellular domains (ECDs) was proposed (Vanamee and Faustman, 2018; Vanamee and Faustman, 2020). In this model, the intracellular domains of individual TNFR1 trimers are separated largely enough to adapt a higher-order signaling complex mediated by TNFR-associated factor dimerization and trimerization interfaces (Napetschnig and Wu, 2013; Vanamee and Faustman, 2020).
We propose that proteolytic processing of TNFR1 by ADAM17, and subsequently by the γ-secretase complex, relieves the membrane-mediated hexagonal structural constraints, thereby destabilizing the membrane-proximal receptor complex (Fig. 10 A). Subsequently, RIPK1 is liberated and upon deubiquitination, forms either the cytosolic complex IIa/b or in the presence of inactive pro-caspase-8, the RIPK1/RIPK3-containing necrosome to induce cell death via apoptosis or necroptosis, respectively. Interestingly, insertion of a tobacco etch virus protease cleavage site into the juxtamembrane region of death receptor 5 allowed for cell death induction upon tobacco etch virus protease–mediated removal of the ECD (Pan et al., 2019), suggesting that proteolytic removal of the ECD might be a general prerequisite for cell death induction by TNFRSF members.
Figure 10.
Model of ADAM17-mediated TNFR1-dependent EC death during TC extravasation. (A) Interaction of TCs with ECs induces endothelial TNF secretion. Soluble TNF (sTNF) might also be provided by inflammatory cells, such as T cells. Engagement of TNFR1 is followed by ADAM17-mediated ectodomain cleavage. The TNFR1 membrane stub is further subjected to intramembrane proteolysis PSEN1/2. Proteolytic processing of TNFR1 enables dissociation of TNFR1 complex I, assembly of complex II or the necrosome, and induction of EC death. (B) In the absence of endothelial ADAM17 or if ADAM17 enzymatic activity is inhibited, TNFR1 signaling is restricted to complex I formation and prosurvival signaling via canonical NF-κB activation. Furthermore, accumulation of transmembrane TNF (tmTNF) enhances TNFR2 signaling and activation of noncanonical NF-κB signaling. Consequently, ECs are protected from TNF-induced cell death.
We demonstrate that in the absence of ADAM17, levels of membrane-bound TNF increase, leading to enhanced TNFR2 engagement and subsequent noncanonical NF-κB activation (Fig. 10 B). Therefore, ADAM17 fine-tunes TNF responses via TNFR1 and TNFR2. During metastasis, ADAM17 promotes EC death by switching TNFR1/2-mediated prosurvival signaling toward TNFR1-mediated cell death signaling. A similar role for ADAM17 in regulating a TNFR1-to-TNFR2 balance might be involved in atherosclerosis (Nicolaou et al., 2017) and liver fibrosis (Sundaram et al., 2019).
There is compelling evidence that expression of ADAM17 on TCs provides protumorigenic signals, e.g., by the release of EGFR ligands (Murphy, 2008). We recently demonstrated that ADAM17 activity on tumor-associated macrophages is needed to promote intestinal tumorigenesis via IL-6 transsignaling (Schmidt et al., 2018). Furthermore, diagnostic or therapeutic manipulation of primary tumors by fine-needle biopsy, tumor irradiation, or tumor surgery were associated with an increase in CTCs and with an increased risk of metastasis (Martin et al., 2017). This might be, at least in part, a result of the induction of a local inflammatory response with increased release of inflammatory cytokines, likely involving the proteolytic activity of ADAM17. Inhibition of ADAM17 as adjuvant therapy in patients with advanced-stage cancer might therefore lower the risk of metastasis. We indeed observed that ADAM17 is strongly expressed on ECs in lungs of patients suffering from pulmonary metastatic spread of different solid tumors, suggesting that activation of endothelial ADAM17 is a general phenomenon during metastasis (Fig. S1 C).
Using recombinant inhibitory ADAM17 PD, we present here a novel therapeutic concept to prevent metastatic seeding. ADAM17 PD shares almost no homology with other PDs of the ADAM or matrix metalloproteinase family, making it a suitable candidate for specific ADAM17 inhibition (Wong et al., 2016). While many antitumor agents suppress growth of the primary tumor, they enhance metastatic seeding. Inhibition of ADAM17 with the highly specific ADAM17 PD might therefore be superior to previous treatment regimen, as it inhibits both growth of the primary tumor (Schmidt et al., 2018) and metastatic seeding. Future development of recombinant ADAM17 PD for the treatment of advanced-stage cancer is therefore highly warranted. Taken together, we identified the transmembrane metalloprotease ADAM17 as a novel key player of cellular necroptosis and as a promising target for the prevention of tumor metastasis.
Materials and methods
Compounds, antibodies, and reagents
PBS (#D8537), Hoechst 33342, and FITC-labeled 40-kD dextran (#FD40S) were purchased from Sigma-Aldrich. TAPI-1 (#BML-PI134) was from Enzo Life Sciences. EthDIII (#B-40050) was from Biotium/Hölzel Diagnostika. TNFR1 (#DY-425) ELISAs were from R&D Systems. CFSE was from eBiosciences. zVAD-fmk (#N-1510) was from Bachem. TPCA-1 (#Cay-15115) was from Biomol/Cayman Chemical. Human and mouse TNF was purchased from ImmunoTools.
Cell lines and reagents
LLC and B16F1 cells were from Cell Lines Service. MDA-MB231 were provided by Dr. Nina Hedemann (University Hospital Kiel, Kiel, Germany). HUVECs were from PromoCell. HMVEC-L were from Lonza. LLC-LacZ, B16F1-LacZ, ADAM17-deficient HEK293T cells, ADAM17ex/ex mEFs, and PSEN1+/−/2−/− mEFs were described previously (Tournoy et al., 2004; Chalaris et al., 2010; Riethmueller et al., 2016). TNF−/− mEFs (Peltzer et al., 2014) and TNFR1−/− mEFs (Pfeffer et al., 1993) were described previously. HUVECs and HMVEC-L were cultivated in EGM-2 (#C-22011; PromoCell) and EGM-MV2 (#C-2212; PromoCell), respectively. All other cell lines were cultivated in DMEM + 10% FCS. All cells were incubated at 37°C, 5% CO2 and 95% humidity. Cell lines were tested routinely at least every 3 mo for Mycoplasma contamination by PCR. For more detailed information on cell lines or reagents, see supplemental tables.
Generation of TNF-deficient murine TCs
Murine TC lines B16F1-LacZ and LLC-LacZ were transfected with single guide RNAs (sgRNAs; Table S4) and recombinant Cas9 using the Neon electroporation device (Thermo Fisher Scientific) according to manufacturer’s instructions. For detailed settings, see Table S5. Cells were subsequently plated in fresh culture medium. After 3 d, single-cell suspensions were seeded. Single-cell clones and the polyclonal pool were analyzed by PCR-based genotyping. In brief, genomic DNA was isolated using GeneJET Genomic DNA isolation kit (Thermo Fisher Scientific), and exon 5 was PCR amplified. PCR amplicons were excised, gel extracted using GeneJET Gel Extraction Kit (Thermo Fisher Scientific), and verified by Sanger sequencing.
Isolation of primary murine lung ECs
Tamoxifen-injected Cdh5-CreERT2::ADAM17flox/flox or control mice were sacrificed, and lungs were first manually cut into small pieces and further subjected to digestion by collagenase type II (STEMCELL Technologies) for 30 min at 37°C in a shaking water bath. After 15 min and at the end of the digestion, tissue was passed through a 21G syringe. The digestion was stopped with DMEM plus 10% FCS, and cells were further filtered through cell strainers (100 μm, 70 μm, 40 μm). The resulting cell suspension was centrifuged (500 g, 5 min, 4°C) twice. The pellets were combined and washed in PBS. Cells were treated with Fc-block in MACS buffer (0.5% BSA, 2 mM EDTA in PBS without Ca2+) and further stained with anti-CD31-FITC, anti-CD45-PE, and anti-Terr119-PE for 30 min, shaking at room temperature (for detailed antibody usage see Table S2). Depletion of PE+ cells was performed with anti-PE magnetic beads and LD columns according to the manufacturer’s instructions. Subsequently, the PE-depleted suspension was subjected to positive selection of FITC+ cells with anti-FITC magnetic beads and LS columns (see Table S1 for further details). The PE+, PE-FITC double-negative, and FITC+ fractions were collected, and purity was determined by flow cytometry on a FACSCanto II system. Samples were lysed in radioimmunoprecipitation assay buffer and boiled for 5 min with 5× Laemmli buffer. SDS-PAGE and immunoblot analysis followed as described above.
Animal models
RIPK3−/− mice were originally obtained from Genentech (Newton et al., 2004). Hypomorphic ADAM17 (ADAM17ex/ex; Chalaris et al., 2010; Schmidt et al., 2018) and TNFR1−/− mice (Pfeffer et al., 1993) have been described previously. LysM-Cre::ADAM17flox/flox (ADAM17ΔMC) and Cdh5-CreERT2::ADAM17flox/flox (ADAM17iΔEC) mice were homozygous for the floxed ADAM17 allele (Horiuchi et al., 2007) and heterozygous for the Cre recombinase under the control of the lysozyme M (Clausen et al., 1999) and the tamoxifen-inducible cadherin 5 promoter (Sörensen et al., 2009), respectively. Cdh5-CreERT2::ADAM17flox/flox mice were injected i.p. with 25 mg/kg body weight (BW) 4-OH-tamoxifen (Sigma-Aldrich) in EtOH/oil, for 4 d consecutively to induce recombination of the floxed ADAM17 exon 2. Control mice received the same amount of injections with EtOH/oil only. All mice were bred on a C57BL/6 background and housed under controlled conditions (specific pathogen free, 22°C, 12-h day–night cycle) and fed standard laboratory chow ad libitum. Animal experiments were conducted according to national and European animal regulations and have been approved by Department of Consumer Protection and Veterinary Affairs at the Ministry of Energy, Agriculture, the Environment, Nature and Digitalization of the Schleswig Holstein State Government (V242-74639/2016 [75-6/15], V243-65407/2015 [75-6/11], V242-70603/2017 [53-5/16]).
Human living donor samples
Tissue specimen of human pulmonary metastases and adjacent peritumoral tissue derived from either primary colorectal or renal cell carcinoma were supplied by the Kiel Biomaterial Bank of the Comprehensive Cancer Center, University Medical Center Schleswig-Holstein. Informed written consent was obtained from the patients, and the sample collection and analyses in this study were approved by the ethics committee of the Medical Faculty of Kiel University (Ref. No. 110/99).
Metastasis model
5 × 105 LLC or B16F1 cells in 250 μl sterile PBS (#D8537; Sigma-Aldrich) were injected i.v. into the lateral tail vein. For inhibitory experiments, TAPI-1 or ADAM17 PD were injected i.v. 5 min before TC injection. ADAM17 PD and TAPI-1 were injected at a concentration of 1 mg/kg BW and 12 mg/kg BW, respectively. To assess endothelial permeability, mice were injected with 200 μl 2.5 mg/ml FITC-labeled 40-kD dextran (#FD40S; Sigma-Aldrich) 10 min before sacrificing.
For the identification of necroptotic ECs, EthDIII (#B-40050; Biotium/Hölzel Diagnostika) was injected i.v. at a concentration of 300 μM (in 50 μl PBS) 5 min before sacrificing the mice.
Immunohistochemistry and immunofluorescence
Tissue specimens of human pulmonary metastases and adjacent peritumoral tissue derived from either primary colorectal or renal cell carcinoma were obtained from patients’ fresh, unfixed surgical resectates; split by pathologists into tumor tissue and peritumoral nonmalignant tissue; and snap frozen in liquid N2 and stored at the biobank at −80°C.
Murine tissue specimens were snap frozen in optimal cutting temperature (OCT) Tissue-Tek compound (Plano GmbH) and cut into 8-μm-thick sections. Frozen human tissue specimens were embedded in OCT Tissue-Tek compound and cut into 8-μm-thick sections. Sections were fixed in acetone:methanol (1:1 vol/vol) for 2 min and subjected to immunofluorescent staining according to standard procedures. Antibody specifications are listed in Table S2. LacZ-expressing cells were identified in frozen tissue sections by β-Gal enzymatic assay.
Flow cytometry
Murine LLC-LacZ and B16F1-LacZ cells were stimulated with 100 nM PMA, 1.3 μM ionomycin, and 10 μM brefeldin A for 4 h at 37°C. Subsequently, cells were trypsinized and fixed in 3.7% paraformaldehyde for 10 min at 37°C. Washed cells were permeabilized in 90% methanol on ice for 30 min and after washing, stained with a-TNF-α-APC (#17-7321-81, clone MP6-XT22, 1:80; eBioscience) or isotype control for 1 h at room temperature in the dark.
Human MDA-MB231 cells were detached with citrate buffer (135 mM KCl, 15 mM sodium citrate), washed in PBS, and blocked with 10% FCS in 1% BSA in PBS for 10 min on ice. Subsequently, membrane-bound TNF-α was stained with primary antibody a-TNF-α (1:16; R&D Systems) for 1 h on ice and with secondary anti-goat-AF488 (A11055, 1:200; Life Technologies) for 30 min on ice in the dark. All samples were acquired in 1% BSA in PBS on a FACSCanto II system and analyzed with FlowJo version 10.06 software.
mRNA isolation and quantitative RT-PCR
Activation of NF-κB–dependent genes upon TNF stimulation in ADAM17ex/ex mEFs and control mEFs was analyzed in 5 × 105 cells per condition. For mRNA isolation and quantitative PCR, 16 h after seeding, cells were stimulated with 200 ng/ml TNF in DMEM without serum for the indicated time points. Cells were subsequently washed in PBS, and RNA was isolated with the NucleoSpin RNA Kit according to the manufacturer’s instructions. 1 μg of RNA was used for cDNA synthesis using oligo-(dT)18 primers and RevertAid Reverse Transcription Kit (#EP0441; Thermo Fisher Scientific) according to the manufacturer’s instructions. Data of the quantitative PCR were acquired on a Roche LightCycler 480 using Roche Universal Probes.
β-Gal staining and analysis
Lung tissue was snap frozen in OCT Tissue-Tek compound and cut into 8-μm-thick sections. Sections were fixed in 0.2% glutaraldehyde for 10 min at 4°C. Slides were then washed three times in LacZ washing buffer (PBS, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40) at room temperature. Subsequently, sections were circled with a PAP pen (DAKO) and incubated at 37°C overnight with staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.5 mg/ml X-Gal in LacZ washing buffer) in a humidified chamber. The next day, slides were washed for 5 min in PBS and double-distilled H2O, respectively. Sections were counterstained with Nuclear Fast Red (Sigma-Aldrich) according to manufacturer’s instructions and mounted with Histokitt II (Carl Roth). Tissue sections were imaged with a Hamamatsu NanoZoomer digital slide scanner, and β-Gal+ cells on the complete section were quantified using ImageJ.
TNFR1 complex II isolation
For TNFR1 complex II isolation, 107 control or ADAM17ex/ex mEFs were starved overnight and stimulated the next day with 20 μM zVAD and 1 μg/ml TNF for the indicated time periods. zVAD was administered 15 min before TNF. Stimulation was stopped by removal of medium. Cells were lysed in coimmunoprecipitation buffer (50 mM Hepes, 150 mM HCl, 1 mM EDTA, 2 mM EGTA, 10% glycerol, 0.5% NP-40, 2 mM KCl) supplemented with protease and phosphatase inhibitors (#11836170001, #4906837001; Roche). Removed medium with stimulus was centrifuged (1,400 rpm for 3 min), and pellets of dying cells were combined with cell lysates. Lysates were cleared by centrifugation and concentrations adjusted. For immunoprecipitation, protein G agarose (#16-266; Millipore) was washed in HNGT buffer (20 mM Hepes, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100) and incubated with a-FADD antibody (1.4 μg/time point, G-4, #sc-271748; Santa Cruz Biotechnology) for 2 h rolling at room temperature. Prepared beads were subsequently incubated with 1–2 mg of protein lysate, rolling overnight at 4°C. The next day, beads were washed with HNGT buffer supplemented with protease and phosphatase inhibitors and heated to 60°C for 30 min with 2× reducing Laemmli buffer. Immunoprecipitates and untreated lysates were analyzed by SDS-PAGE and immunoblotting as described above.
Immunoblotting
Where indicated, cells were starved overnight in DMEM without serum before stimulation with 200 ng/ml TNF for the indicated time periods. Samples were lysed in radioimmunoprecipitation buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 mM EGTA, 0.5% NP-40) supplied with 50 mM NaF and protease and phosphatase inhibitors. Proteins were separated by electrophoresis on 10% SDS gels and transferred to polyvinylidenfluoride membranes. Membranes were incubated with primary antibodies overnight at 4°C and with horseradish peroxidase–conjugated secondary antibodies at room temperature for 30 min. An enhanced chemiluminescence substrate kit (Thermo Fisher Scientific) was used for detection.
ELISA
Mouse TNFR1 (#DY425), human TNFR1 (#DY225), mouse TNF (#DY410), and human TNF (#DY210) (R&D Systems) ELISAs were performed according to the manufacturer’s instructions.
Transmigration assays
HUVECs or HMVEC-L were cultured for 2 d and subsequently seeded at 5 × 104 into collagen I–coated 24-well inserts with a pore diameter of 8 μm (Sarstedt). On the next day, ECs were transfected twice with 2 pmol siRNA (Table S3) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) on 2 consecutive days. For the transendothelial migration experiments, 5 × 103 MDA-MB231 cells were stained with 8 μM CFSE and seeded on top of the ECs. Transmigrated cells that passed the membrane were imaged 6 h later on a Leica DMi8 inverted fluorescent microscope and quantified using ImageJ.
In vitro cell death assays
To assess cell death in vitro, 104 HUVECs were seeded in collagen I–coated lumox 96-well plates (Sarstedt). Subsequently, cells were transfected twice with 2 pmol siRNA/well (Table S2) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) on 2 consecutive days. For coculture experiments, 1.5 × 103 MDA-MB231 cells were stained with 8 μM CFSE (Thermo Fisher Scientific) and seeded on top of the ECs in endothelial growth medium. To trigger cell death independent of TCs, ECs or mEFs were treated with 100 µM zVAD-fmk (#N-1510; Bachem) either alone or together with 5 μM TPCA-1 (#Cay-15115; Biomol/Cayman Chemical) 10 min before TNF (50 ng/ml; ImmunoTools) stimulation. After incubation for 6 h, the cells were stained with 2 μM Hoechst 33342 (Sigma-Aldrich) and 1.6 μM EthDIII (Biotium/Hölzel Diagnostika) and subsequently imaged on an Olympus Fluoview 1000 confocal laser scanning microscope equipped with an incubation chamber at 37°C, 5% CO2 and 55% humidity. EthDIII+ ECs were quantified with ImageJ and expressed as a percentage of all ECs.
Protease activity assay
Inhibitory effect of ADAM17 PD on ADAM17 activity was determined by a peptide-based activity assay. 50 nM recombinant human ADAM17 catalytic domain was mixed with increasing concentrations of either TAPI-1 (#BML-PI134; Enzo Life Sciences) or recombinant ADAM17 PD. After incubation for 10 min at 37°C, 5 μM PEPDAB014 (BioZyme) ADAM17 substrate peptide was added. Alternatively, cells were washed once with PBS and incubated for 30 min with the indicated inhibitors at 37°C and 5% CO2 before the addition of the indicated stimuli and 5 μM PEPDAB014 substrate peptide. Subsequently, the change in fluorescence intensity was recorded every 30 s for 18 min. The initial linear slope was determined by linear regression and plotted against the inhibitor concentrations. IC50 values were calculated by four-parameter nonlinear curve fitting using GraphPad Prism software.
Data analysis and statistics
Data were checked for consistency and normality by using normal probability plots and Kolmogorov–Smirnov tests. Independent Student’s t test with and without the assumption of variance homogeneity were used, and Levene’s test was used to test for variance homogeneity. Bootstrap t tests with and without assumption of variance homogeneity based on 5,000 Monte Carlo simulations were also used. In case of log-normal or γ-distributed data, semiparametric generalized linear models based on log-normal and γ-distributions were used. Mann–Whitney U test was used to compare two independent groups nonparametrically. ANOVA and Welch’s ANOVA were used to test for fixed factors, and a random-effects model was also used. Generalized estimating equation models with various distributions (normal, log-normal, γ, and Tweedie) were used for continuously distributed variables, and corresponding least significant difference tests were used for pairwise comparisons. The robust estimator for the covariance matrix was used.
When using linear regression analysis, the initial linear slope was determined by and plotted against the inhibitor concentrations. IC50 values were calculated by four-parameter nonlinear curve fitting. All reported tests were two sided, and two-sided P < 0.05 was considered statistically significant. All statistical analyses in this article were performed by use of NCSS 10 software (NCSS, LLC), Statistica 13 (Hill and Lewicki, 2006), SPSS version 27 for Windows (IBM Corporation), RStudio (RStudio Team, 2020), Mathematica version 12 (Wolfram Research), and GraphPad Prism 8.0.0 for Windows (GraphPad Software). In case of experimental reproduction and replication, details of statistical analyses for the computation of an a priori power analysis are given as means, sample sizes, and SEs.
Online supplemental material
Fig. S1 shows that host ADAM17 promotes metastasis. Fig. S2 shows genetic deficiency of ADAM17 and TNFR1 in human and murine ECs. Fig. S3 shows CRISPR/Cas9-mediated generation of TNF-deficient murine TCs. Fig. S4 shows that the TNFR1 ectodomain is released via ADAM17-mediated proteolysis. Fig. S5 shows an analysis of IC50 of TAPI-1 and ADAM17 PD using recombinant catalytic domain of ADAM17 and a quenched fluorogenic TGF-α–based substrate peptide. Table S1 lists the reagents and resources used in this study. Table S2 lists the antibodies for immunoblotting, immunofluorescence, FACS, and immunoprecipitation used in this study. Table S3 lists siRNAs used in this study. Table S4 lists sgRNAs used in this study. Table S5 lists pulse settings during Neon electroporation.
Supplementary Material
lists the reagents and resources used in this study.
lists the antibodies for immunoblotting, immunofluorescence, FACS, and immunoprecipitation used in this study.
lists siRNAs used in this study.
lists the sgRNAs used in this study.
lists pulse settings during Neon electroporation.
Acknowledgments
We thank Konrad Aden and Bimba Hoyer (University Medical Center Schleswig-Holstein) for the provision of etanercept. We thank Silke Horn and Fabian Neumann for excellent technical assistance. We are grateful to the members of the Victor-Hensen animal facility at the Christian-Albrechts-University Kiel. We thank Frank D. Böhmer for fruitful discussions and critical reading of the manuscript. We are grateful to Arne Bathke (Department of Mathematics, Paris-Lodron-University Salzburg) for discussions on statistical issues.
This work was supported by the Deutsche Forschungsgemeinschaft grants SFB841 (project C1 to D. Schmidt-Arras and S. Rose-John, C3 to E. Galun) and SFB877 (project number 125440785, project A1 to S. Rose-John, projects A3 and Z3 to P. Saftig, projects A9 and A15 to C. Becker-Pauly, and project B2 to D. Adam), the Australian Technology Network/Deutscher Akademischer Austauschdienst (project-based personnel exchange program to D. Schmidt-Arras, 57315783), the Medical Faculty of the Christian-Albrechts-University Kiel (to J. Bolik and D. Schmidt-Arras, F358901), and the Cluster of Excellence “Inflammation at Interfaces” to S. Rose-John. The Biomaterial Bank of the Comprehensive Cancer Center is a member of the biobanking network Pop-Gen 2.0 Network, Medical Faculty, Christian-Albrechts-University Kiel, and was supported by Federal Ministry of Education and Research grant 01EY1103.
Author contributions: D. Schmidt-Arras conceived the project, supervised experimentation, performed the data analysis, and wrote the manuscript. J. Bolik, F. Krause, M. Stevanovic, M. Gandraß, and J. Fritsch performed most experiments, performed the data analysis, and contributed to the manuscript writing. I. Thomsen, S. Schacht, and E. Rieser performed the experiments. J. Bergmann, C. Röder, C. Schafmayer, and J.-H. Egberts isolated, archived, and provided human lung tissue specimens. R. Wichert and C. Becker-Pauly produced and provided recombinant ADAM17 catalytic domain. W. Schneider-Brachert provided TNFR1 expression plasmids. R. Barikbin provided TNFR1−/− mice. D. Adams provided material, scientific insight, and RIPK3−/− mice. P. Saftig provided PSEN-deficient mEFs. M. Voss contributed to the generation of TNF-deficient TCs. R. Lucius contributed to the tissue analysis. M. Müller and N. Schumacher performed the animal experimentation and data analysis. B. Strilic provided HMVEC-L and scientific insight and contributed to the manuscript writing. W. Hitzl performed the data analysis and statistics. A. Krüger provided LacZ-expressing TCs and scientific insight. E. Galun and S. Schacht provided scientific insight. I. Sagi generated ADAM17 PD and contributed to the data analysis. H. Walczak provided material, analyzed data, and contributed to writing the manuscript. S. Rose-John provided scientific insight, analyzed data, and contributed to manuscript writing. All authors edited and approved the manuscript.
References
- Abdul Pari, A.A., Singhal M., and Augustin H.G.. 2021. Emerging paradigms in metastasis research. J. Exp. Med. 218:e20190218. 10.1084/jem.20190218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydon, D.C., Biancheri P., Di W.-L., Plagnol V., Cabral R.M., Brooke M.A., van Heel D.A., Ruschendorf F., Toynbee M., Walne A., et al. 2011. Inflammatory skin and bowel disease linked to ADAM17 deletion. N. Engl. J. Med. 365:1502–1508. 10.1056/NEJMoa1100721 [DOI] [PubMed] [Google Scholar]
- Bonapace, L., Coissieux M.-M., Wyckoff J., Mertz K.D., Varga Z., Junt T., and Bentires-Alj M.. 2014. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 515:130–133. 10.1038/nature13862 [DOI] [PubMed] [Google Scholar]
- Brakebusch, C., Varfolomeev E.E., Batkin M., and Wallach D.. 1994. Structural requirements for inducible shedding of the p55 tumor necrosis factor receptor. J. Biol. Chem. 269:32488–32496. 10.1016/S0021-9258(18)31661-2 [DOI] [PubMed] [Google Scholar]
- Brenner, D., Blaser H., and Mak T.W.. 2015. Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 15:362–374. 10.1038/nri3834 [DOI] [PubMed] [Google Scholar]
- Cai, Z., Zhang A., Choksi S., Li W., Li T., Zhang X.-M., and Liu Z.-G.. 2016. Activation of cell-surface proteases promotes necroptosis, inflammation and cell migration. Cell Res. 26:886–900. 10.1038/cr.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalaris, A., Adam N., Sina C., Rosenstiel P., Lehmann-Koch J., Schirmacher P., Hartmann D., Cichy J., Gavrilova O., Schreiber S., et al. 2010. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J. Exp. Med. 207:1617–1624. 10.1084/jem.20092366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber-Goel, J., Coleman-Vaughan C., Agrawal V., Sawhney N., Hickey E., Powell J.C., and McCarthy J.V.. 2016. γ-Secretase activity is required for regulated intramembrane proteolysis of tumor necrosis factor (TNF) receptor 1 and TNF-mediated pro-apoptotic signaling. J. Biol. Chem. 291:5971–5985. 10.1074/jbc.M115.679076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277. 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Fuchslocher Chico, J., Falk-Paulsen M., Luzius A., Saggau C., Ruder B., Bolik J., Schmidt-Arras D., Linkermann A., Becker C., Rosenstiel P., et al. 2018. The enhanced susceptibility of ADAM-17 hypomorphic mice to DSS-induced colitis is not ameliorated by loss of RIPK3, revealing an unexpected function of ADAM-17 in necroptosis. Oncotarget. 9:12941–12958. 10.18632/oncotarget.24410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell, M., Douni E., Wajant H., Löhden M., Clauss M., Maxeiner B., Georgopoulos S., Lesslauer W., Kollias G., Pfizenmaier K., and Scheurich P.. 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 83:793–802. 10.1016/0092-8674(95)90192-2 [DOI] [PubMed] [Google Scholar]
- Hänggi, K., Vasilikos L., Valls A.F., Yerbes R., Knop J., Spilgies L.M., Rieck K., Misra T., Bertin J., Gough P.J., et al. 2017. RIPK1/RIPK3 promotes vascular permeability to allow TC extravasation independent of its necroptotic function. Cell Death Dis. 8:e2588. 10.1038/cddis.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T., and Lewicki P.. 2006. Statistics: Methods and Applications. StatSoft, Tulsa, OK. [Google Scholar]
- Horiuchi, K., Kimura T., Miyamoto T., Takaishi H., Okada Y., Toyama Y., and Blobel C.P.. 2007. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 179:2686–2689. 10.4049/jimmunol.179.5.2686 [DOI] [PubMed] [Google Scholar]
- Hou, J., Ju J., Zhang Z., Zhao C., Li Z., Zheng J., Sheng T., Zhang H., Hu L., Yu X., et al. 2019. Discovery of potent necroptosis inhibitors targeting RIPK1 kinase activity for the treatment of inflammatory disorder and cancer metastasis. Cell Death Dis. 10:493. 10.1038/s41419-019-1735-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, A., Ishiguro J., Kitamura H., Arima N., Okutani M., Shuto A., Higashiyama S., Ohwada T., Arai H., Makide K., and Aoki J.. 2012. TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nat. Methods. 9:1021–1029. 10.1038/nmeth.2172 [DOI] [PubMed] [Google Scholar]
- Ji, H., Cao R., Yang Y., Zhang Y., Iwamoto H., Lim S., Nakamura M., Andersson P., Wang J., Sun Y., et al. 2014. TNFR1 mediates TNF-α-induced tumour lymphangiogenesis and metastasis by modulating VEGF-C-VEGFR3 signalling. Nat. Commun. 5:4944. 10.1038/ncomms5944 [DOI] [PubMed] [Google Scholar]
- Kim, S., Takahashi H., Lin W.-W., Descargues P., Grivennikov S., Kim Y., Luo J.-L., and Karin M.. 2009. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 457:102–106. 10.1038/nature07623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont, E., Draber P., Rieser E., Reichert M., Kupka S., de Miguel D., Draberova H., von Mässenhausen A., Bhamra A., Henderson S., et al. 2018. TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol. 20:1389–1399. 10.1038/s41556-018-0229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, O.A., Anderson R.L., Narayan K., and MacManus M.P.. 2017. Does the mobilization of circulating tumour cells during cancer therapy cause metastasis? Nat. Rev. Clin. Oncol. 14:32–44. 10.1038/nrclinonc.2016.128 [DOI] [PubMed] [Google Scholar]
- Moreland, L.W., Baumgartner S.W., Schiff M.H., Tindall E.A., Fleischmann R.M., Weaver A.L., Ettlinger R.E., Cohen S., Koopman W.J., Mohler K., et al. 1997. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N. Engl. J. Med. 337:141–147. 10.1056/NEJM199707173370301 [DOI] [PubMed] [Google Scholar]
- Murphy, G. 2008. The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 8:929–941. 10.1038/nrc2459 [DOI] [PubMed] [Google Scholar]
- Napetschnig, J., and Wu H.. 2013. Molecular basis of NF-κB signaling. Annu. Rev. Biophys. 42:443–468. 10.1146/annurev-biophys-083012-130338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K., Sun X., and Dixit V.M.. 2004. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell Biol. 24(4):1464–1469. 10.1128/MCB.24.4.1464-1469.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou, A., Zhao Z., Northoff B.H., Sass K., Herbst A., Kohlmaier A., Chalaris A., Wolfrum C., Weber C., Steffens S., et al. 2017. Adam17 deficiency promotes atherosclerosis by enhanced TNFR2 signaling in mice. Arterioscler. Thromb. Vasc. Biol. 37:247–257. 10.1161/ATVBAHA.116.308682 [DOI] [PubMed] [Google Scholar]
- Oikawa, N., and Walter J.. 2019. Presenilins and γ-secretase in membrane proteostasis. Cells. 8:E209. 10.3390/cells8030209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, L., Fu T.-M., Zhao W., Zhao L., Chen W., Qiu C., Liu W., Liu Z., Piai A., Fu Q., et al. 2019. Higher-order clustering of the transmembrane anchor of DR5 drives signaling. Cell. 176:1477–1489.e14. 10.1016/j.cell.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S., Webster J.D., Varfolomeev E., Kwon Y.C., Cheng J.H., Zhang J., Dugger D.L., Wickliffe K.E., Maltzman A., Sujatha-Bhaskar S., et al. 2020. RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases. Cell Death Differ. 27:161–175. 10.1038/s41418-019-0347-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer, N., and Walczak H.. 2019. Cell death and inflammation - a vital but dangerous liaison. Trends Immunol. 40:387–402. 10.1016/j.it.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Peltzer, N., Rieser E., Taraborrelli L., Draber P., Darding M., Pernaute B., Shimizu Y., Sarr A., Draberova H., Montinaro A., et al. 2014. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9(1):153–165. 10.1016/j.celrep.2014.08.066 [DOI] [PubMed] [Google Scholar]
- Peschon, J.J., Slack J.L., Reddy P., Stocking K.L., Sunnarborg S.W., Lee D.C., Russell W.E., Castner B.J., Johnson R.S., Fitzner J.N., et al. 1998. An essential role for ectodomain shedding in mammalian development. Science. 282:1281–1284. 10.1126/science.282.5392.1281 [DOI] [PubMed] [Google Scholar]
- Pfeffer, K., Matsuyama T., Kündig T.M., Wakeham A., Kishihara K., Shahinian A., Wiegmann K., Ohashi P.S., Krönke M., and Mak T.W.. 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 73:457–467. 10.1016/0092-8674(93)90134-C [DOI] [PubMed] [Google Scholar]
- Reiss, K., and Saftig P.. 2009. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 20:126–137. 10.1016/j.semcdb.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Reymond, N., d’Água B.B., and Ridley A.J.. 2013. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer. 13:858–870. 10.1038/nrc3628 [DOI] [PubMed] [Google Scholar]
- Riethmueller, S., Ehlers J.C., Lokau J., Düsterhöft S., Knittler K., Dombrowsky G., Grötzinger J., Rabe B., Rose-John S., and Garbers C.. 2016. Cleavage Site Localization Differentially Controls Interleukin-6 Receptor Proteolysis by ADAM10 and ADAM17. Sci. Rep. 6:25550. 10.1038/srep25550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . 2020. RStudio: Integrated Development for R. http://www.rstudio.com (accessed January 9, 2020).
- Scheller, J., Chalaris A., Garbers C., and Rose-John S.. 2011. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 32:380–387. 10.1016/j.it.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Schmidt, S., Schumacher N., Schwarz J., Tangermann S., Kenner L., Schlederer M., Sibilia M., Linder M., Altendorf-Hofmann A., Knösel T., et al. 2018. ADAM17 is required for EGF-R-induced intestinal tumors via IL-6 trans-signaling. J. Exp. Med. 215:1205–1225. 10.1084/jem.20171696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, D., Strilic B., Sivaraj K.K., Wettschureck N., and Offermanns S.. 2013. Platelet-derived nucleotides promote tumor-cell transendothelial migration and metastasis via P2Y2 receptor. Cancer Cell. 24:130–137. 10.1016/j.ccr.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Sörensen, I., Adams R.H., and Gossler A.. 2009. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 113:5680–5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Strilic, B., and Offermanns S.. 2017. Intravascular survival and extravasation of TCs. Cancer Cell. 32:282–293. 10.1016/j.ccell.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Strilic, B., Yang L., Albarrán-Juárez J., Wachsmuth L., Han K., Müller U.C., Pasparakis M., and Offermanns S.. 2016. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature. 536:215–218. 10.1038/nature19076 [DOI] [PubMed] [Google Scholar]
- Sundaram, B., Behnke K., Belancic A., Al-Salihi M.A., Thabet Y., Polz R., Pellegrino R., Zhuang Y., Shinde P.V., Xu H.C., et al. 2019. iRhom2 inhibits bile duct obstruction-induced liver fibrosis. Sci. Signal. 12:eaax1194. 10.1126/scisignal.aax1194 [DOI] [PubMed] [Google Scholar]
- Tournoy, J., Bossuyt X., Snellinx A., Regent M., Garmyn M., Serneels L., Saftig P., Craessaerts K., De Strooper B., and Hartmann D.. 2004. Partial loss of presenilins causes seborrheic keratosis and autoimmune disease in mice. Hum. Mol. Genet. 13(13):1321–1331. 10.1093/hmg/ddh151 [DOI] [PubMed] [Google Scholar]
- Vanamee, É.S., and Faustman D.L.. 2018. Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal. 11:eaao4910. 10.1126/scisignal.aao4910 [DOI] [PubMed] [Google Scholar]
- Vanamee, É.S., and Faustman D.L.. 2020. On the TRAIL of better therapies: understanding TNFRSF structure-function. Cells. 9:E764. 10.3390/cells9030764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, M.J., Hoos A., Bauer J., Boettcher S., Knust M., Weber A., Simonavicius N., Schneider C., Lang M., Stürzl M., et al. 2012. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer Cell. 22:91–105. 10.1016/j.ccr.2012.05.023 [DOI] [PubMed] [Google Scholar]
- Wong, E., Maretzky T., Peleg Y., Blobel C.P., and Sagi I.. 2015. The functional maturation of a disintegrin and metalloproteinase (ADAM) 9, 10, and 17 requires processing at a newly identified proprotein convertase (PC) cleavage site. J. Biol. Chem. 290:12135–12146. 10.1074/jbc.M114.624072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E., Cohen T., Romi E., Levin M., Peleg Y., Arad U., Yaron A., Milla M.E., and Sagi I.. 2016. Harnessing the natural inhibitory domain to control TNFα converting enzyme (TACE) activity in vivo. Sci. Rep. 6:35598. 10.1038/srep35598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Liu J., Sakaki-Yumoto M., and Derynck R.. 2012. TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci. Signal. 5:ra34. 10.1126/scisignal.2002689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Joseph S., Sun T., Hoffmann J., Thevissen S., Offermanns S., and Strilic B.. 2019. TAK1 regulates endothelial cell necroptosis and tumor metastasis. Cell Death Differ. 26:1987–1997. 10.1038/s41418-018-0271-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the reagents and resources used in this study.
lists the antibodies for immunoblotting, immunofluorescence, FACS, and immunoprecipitation used in this study.
lists siRNAs used in this study.
lists the sgRNAs used in this study.
lists pulse settings during Neon electroporation.