Abstract
In hamsters, SARS-CoV-2 infection at the same time as or before H3N2 influenza virus infection resulted in significantly reduced influenza virus titers in the lungs and nasal turbinates. This interference may be correlated with SARS-CoV-2–induced expression of MX1.
Keywords: antiviral, Coinfection, Influenza, MX1, SARS-CoV-2
In hamsters, a previous infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interferes with a sequential infection with influenza virus (H3N2 subtype). The induction of the interferon-stimulated gene MX1induced by SARS-CoV-2 infection may play a partial role in the interference with this influenza subtype.
Simultaneous infections or coinfections with more than one respiratory virus is not uncommon in humans, particularly in hospital settings [1]. The epidemiology of disease caused by a single virus infection can be altered or influenced by a second virus infection [2]. Given the vast array of known respiratory viruses, such as influenza viruses, coronaviruses, rhinoviruses, and parainfluenza viruses, the disease outcomes for the various mixed respiratory viral infections are unclear. Some coinfections can enhance disease severity; for example, coinfection of reovirus and severe acute respiratory syndrome (SARS) coronavirus in guinea pigs leads to a more severe disease phenotype [3].
Commonly in coinfections, one virus will interfere with and suppress the replication of the other virus. This suppression can be due to competition for host factors or sites of virus replication and may be influenced by host immune responses, such as those activated by interferons and interferon-stimulated genes (ISGs) [4]. Among the best-studied families of ISGs with antiviral activity against multiple RNA viruses, including influenza viruses, are the myxovirus resistance (Mx) proteins [5]. Because SARS coronavirus 2 (SARS-CoV-2) induces strong, diffuse expression of Mx protein 1 (Mx1) in the lungs of infected hamsters [6], we decided to examine the interference of influenza virus replication in Syrian golden hamsters coinfected or previously infected with SARS-CoV-2.
METHODS
Cells and Virus
Vero E6 TMPRSS2 cells were maintained in Dulbecco minimal essential medium (MEM) with 10% fetal bovine serum in the presence of 1mg mL−1 of geneticin. Humanized Madin-Darby canine kidney (MDCK) cells were maintained in MEM containing 5% newborn calf serum in the presence of 2 µg mL−1 of puromycin and 10 µg mL−1 blasticidin. All cells were incubated at 37oC with 5% carbon dioxide and were confirmed to be mycoplasma negative by means of polymerase chain reaction testing.
An isolate of SARS-CoV-2 (B.1 lineage) from a nasopharyngeal specimen obtained from the University of Wisconsin Hospital was propagated in Vero E6 TMPRSS2 cells. Sequence analysis of the spike gene identified a glycine residue at amino acid position 614. Influenza virus A/Tokyo/UT-IMS3-1/2014 (H3N2 subtype) was propagated in humanized MDCK cells, in MEM supplemented with bovine serum albumin containing 1 µg of tosyl phenylalanyl chloromethyl ketone trypsin per milliliter. All experiments were performed in enhanced biosafety level 3 containment laboratories at the University of Wisconsin, which are approved for such use by the Centers for Disease Control and Prevention and the US Department of Agriculture.
Animal Studies and Experimental Infection of Syrian Golden Hamsters
The number of animals used in each experiment was chosen based on our previous studies with SARS-CoV-2 and influenza virus infection of hamsters, in which the sample size was sufficient to evaluate a statistically significant difference between groups. All studies were conducted under an approved protocol reviewed by the Institutional Animal Care and Use Committee at the University of Wisconsin.
In the current study, 4–5-week-old female Syrian golden hamsters (Envigo) were used. Under isoflurane anesthesia, 4 hamsters per group were inoculated with SARS-CoV-2 (103 plaque-forming units), influenza virus (106 plaque-forming units), or a mixture of both viruses. All animals were infected via intranasal inoculation, with the same concentration of virus and at the same total inoculum volume (130 µL). Animals in the control groups were inoculated with 130 µL of sterile phosphate-buffered saline (PBS).
Sample Collection
For virological and pathological analyses, animals were euthanized, and samples (lungs and nasal turbinates) were collected on the days indicated in Figure 1A–1C and Supplementary Figure 1. Frozen tissues were homogenized, and clarified supernatant was used for virus titrations. Titers performed in duplicate were determined by performing plaque assays in Vero E6 TMPRSS2 cells for SARS-CoV-2 and humanized MDCK cells for influenza virus.
Figure 1.
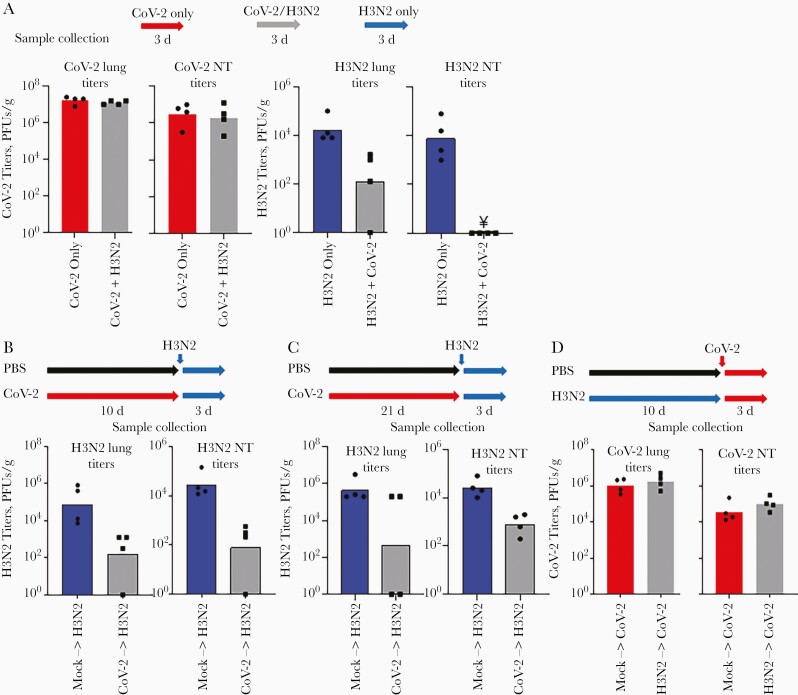
Simultaneous and serial infection studies. A, Groups of hamsters were infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) only, influenza virus (H3N2), or simultaneously with influenza virus and SARS-CoV-2 (CoV-2). Viral titers in the lungs and nasal turbinates (NTs) were determined 3 days after infection. Note: ¥ represents titers below the limit of detection (10 plaque-forming units [PFUs]/g. B, C, For serial infection studies, groups of hamsters were first inoculated with phosphate-buffered saline (PBS; mock) or infected with SARS-CoV-2 and then infected with influenza virus after 10 (B) or 21 (C) days. D, In addition, groups of hamsters were first inoculated with PBS (mock) or infected with influenza virus and then infected with SARS-CoV-210 days later. Viral titers in the lungs and NTs were determined 3 days after influenza infection.
The remaining lung tissues were fixed in 10% neutral buffered formalin and then embedded in paraffin. Tissue sections (3 µm) were cut and mounted onto glass slides for either standard hematoxylin-eosin staining or immunohistochemical staining for viral antigens. A rabbit polyclonal antibody for the SARS nucleocapsid (ANT-180; Prospec), a rabbit polyclonal antibody for type A influenza virus nucleoprotein (NP) (prepared in our laboratory), or a mouse monoclonal antibody against Mx1 (clone M143; Merck Millipore) were used for antigen staining; reactions were visualized using 3,3’-diaminobenzidine tetrahydrochloride and the Dako Envision system.
Visual observations of lung lobe inflammation were scored from 0 to 4, depending on severity. Each lung lobe was analyzed for degree of inflammation and scored as 0 (none; 0%), 1 (minimal; <10%), 2 (mild; 10%–50%), 3 (moderate; >50%), or 4 (severe; score 3 plus edema or/and hemorrhage) by a scorer not blinded to the group allocation.
Data and Statistical Analyses
Viral titers from animals are expressed as scatterplots with bars and individual data points obtained using GraphPad Prism 9 software. Statistical analyses between 2 groups were performed using unpaired Student t tests. The data that support these findings of this study are available from the corresponding authors on reasonable request.
RESULTS
Syrian hamsters are a useful animal model for studying both influenza A virus [7] and SARS-CoV-2 [8] infections because they allow robust replication of both viruses in their respiratory tissues. Using this animal model, we performed simultaneous or serial infection studies with the influenza virus strain A/Tokyo/UT-IMS3-1/2014 (H3N2 subtype) and a SARS-CoV-2 isolate from the B.1 lineage with glycine at amino acid position 614 in its spike protein.
In the first study, groups of animals (4 hamsters per group) were infected by intranasal inoculation (130 µL per animal) with SARS-CoV-2 (103 plaque-forming units [PFUs]), influenza virus (106 PFUs), or both viruses simultaneously at the same concentrations and volume (Figure 1A). Three days after infection, comparing coinfected animals with those infected with SARS-CoV only, there was no significant differences in the ability of SARS-CoV-2 to replicate in the lungs (P = .37) or nasal turbinates (P = .70) (Figure 1A). However, influenza virus replication was impaired in the group of coinfected animals; influenza virus titers were reduced in the lungs (P = .03) and undetectable in the nasal turbinates (P < .001) relative to those in the group of hamsters infected with only influenza virus (Figure 1A).
Given the inhibition of influenza virus replication in the simultaneous infection study, we next performed serial infection studies, focusing on the potential inhibition of influenza virus by SARS-CoV-2. Groups of hamsters (4 hamsters per group) were first infected with SARS-CoV-2 or inoculated with PBS (Figure 1B). Ten days later, a time period after infection associated with no to limited SARS-CoV-2 replication in the lungs of different animals but at which strong innate immune and inflammatory responses are still present [6, 8, 9], the animals were infected with influenza virus. Influenza virus replication in the lungs and nasal turbinates was significantly impaired (P = .02 and P = .01, respectively) in hamsters previously infected with SARS-CoV-2 compared with control hamsters inoculated with PBS and then infected with influenza virus (Figure 1B).
Interestingly, 21 days after the first infection with SARS-CoV-2 (a time period after infection at which we anticipated that the host responses to infection would have cleared), hamsters infected with influenza virus also showed impairment of virus replication in the nasal turbinates (P = .002) but no significant reduction of virus replication in the lungs (P = .10), although 2 of the 4 animals in this group had no detectable virus replication in the lungs (Figure 1C). In contrast, when animals were first infected with influenza virus and then infected with SARS-CoV-2 after 10 days ( consistent with the interval between infections in the above-described experiment), the previous infection with influenza virus had no effect on the replication of SARS-CoV-2 in the respiratory tissues tested (P = .46 for lung and P = .24 for nasal turbinates); Figure 1D).
To gain further insight into the mechanism of SARS-CoV-2 interference of influenza virus infection, we examined lung pathology. Hematoxylin-eosin staining revealed notable inflammation in the lungs of hamsters that were infected with SARS-CoV-2 first, even 10 days after infection relative to control animals inoculated with PBS (Supplementary Figure 1 and Supplementary Table 1), which is consistent with previously published findings [9], while immunohistochemical staining of the lung tissues revealed no SARS-CoV-2 nucleocapsid antigen in any animals (Supplementary Figure 1; SARS-CoV-2 only).
Three days after influenza virus infection, influenza virus NP antigen was detected in the lungs of all control animals inoculated with PBS 10 days before infection. We noted light inflammation in the lungs around the bronchi, corresponding to the NP antigen staining in the bronchial epithelia (Supplementary Table 1 and Supplementary Figure 1; PBS + H3N2). The inflammation in the lungs after influenza infection was not exacerbated by the previous SARS-CoV-2 infection (Supplementary Table 1), but influenza NP antigen was less abundant (Supplementary Figure 1; SARS-CoV-2 + H3N2), consistent with a reduction in influenza virus titers. With animals previously infected with SARS-CoV-2 10 days earlier, influenza virus NP antigen was less abundant (Supplementary Figure 1; SARS-CoV-2 + H3N2) and found in only 50% of the animals (Supplementary Table 1); the animals that were viral antigen positive by immunohistochemistry were also live virus positive (Figure 1B).
It has been reported that SARS-CoV-2–infected hamsters express detectable levels of the antiviral gene MX1 in their lungs [6]. Because replication of influenza viruses is restricted by this cellular ISG [10], we examined the expression of MX1 10 days after the SARS-CoV-2 infection and after the sequential infection with influenza virus. While there was no detectable staining of Mx1 in the lungs of control hamsters inoculated with PBS (Figure 2A), Mx1 was still detectable even 10 days after SARS-CoV-2 infection (Figure 2B), which was similar to previously reported findings [6]. Infection of PBS control hamsters with influenza virus induced the strongest expression of MX1 at day 3 after infection (Figure 2C). However, this increase in MX1 expression 3 days after influenza virus infection was not apparent in hamsters previously infected (10 days earlier) with SARS-CoV-2 (compare Figure 2C and 2D). This result most likely reflects the suppression of influenza virus replication by several host factors, including MX1, that were induced by the previous SARS-CoV-2 infection. Further research is needed to fully understand the contribution of MX1 to this phenomenon.
Figure 2.
Immunohistochemical staining of myxovirus resistance protein 1 (Mx1) in lung tissues. A, B, Representative images of Mx1 staining from fixed lung tissues collected 10 days after inoculation with phosphate-buffered saline (PBS) only (A) or infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) only (B). C, D, In a serial infection study, animals were first inoculated with PBS (C) or SARS-CoV-2 (D) and then infected with influenza virus 10 days later. Mx1 staining was performed on fixed lung tissues collected 3 days after influenza virus infection. (Scale bars represent 100 µm.).
DISCUSSION
In humans, coinfections of influenza virus and SARS-CoV-2 have been documented [11, 12]. In the current study, we demonstrated that Syrian golden hamsters are an ideal animal model to examine the consequences of simultaneous and serial infection of SARS-CoV-2 and influenza virus, as well as the host factors (eg, antiviral genes like MX1) that are induced by one virus to suppress the other virus. Our group and others [6] have shown that MX1 is induced by SARS-CoV-2 infection, although other ISGs may also play a role in the inhibition of influenza virus infection during coinfection and serial infection with SARS-CoV-2. These other host factors should be explored in more detail both in cell culture systems, such as human air organoids, and animal models including knockout mice. The long-term persistence of SARS-CoV-2 RNA in tissues after infection has been noted in human lung tissue [13] and may trigger activation of the interferon pathway, leading to persistent induction of ISGs, including MX1. Although our data suggest that the induction of MX1 by SARS-CoV-2 restricts influenza virus infection, the induction of MX1 by influenza virus had no effect on SARS-CoV-2 replication, an observation of potential antagonism of MX1 function by SARS-CoV-2 that should be further studied.
The outcome of coinfections and serial infections with SARS-CoV-2 and influenza virus may depend on the influenza virus subtype. While we found interference of H3N2 influenza virus replication by SARS-CoV-2, Zhang et al [14] reported more severe disease outcomes and higher influenza virus titers in hamsters infected with a pandemic H1N1 influenza virus and SARS-CoV-2. This difference in results might be due to better replication of H1N1 influenza viruses in hamsters relative to H3N2 influenza viruses or stronger resistance of pandemic 2009 H1N1 influenza viruses to the antiviral gene MX1 [15]. The outcome of coinfection could also be dependent on the SARS-CoV-2 variant used to infect hamsters, a variable not explored in the current study but worth investigating in future studies, given the diverse panel of SARS-CoV-2 variants that has emerged.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Susan Watson for scientific editing.
Financial support. This work was supported by the Center for Research on Influenza Pathogenesis, which is funded by the National Institutes of Allergy and Infectious Diseases (grant HHSN272201400008C to Y. K.) and by the Japan Agency for Medical Research and Development (grants JP20fk0108104 and JP20wm0125008 to T. S.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Goka EA, Vallely PJ, Mutton KJ, Klapper PE.. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect 2015; 143:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waner JL. Mixed viral infections: detection and management. Clin Microbiol Rev 1994; 7:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang L, He C, Lei M, et al. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol 2005; 24:485–90. [DOI] [PubMed] [Google Scholar]
- 4. Kumar N, Sharma S, Barua S, Tripathi BN, Rouse BT.. Virological and immunological outcomes of coinfections. Clin Microbiol Rev 2018; 31:e00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhelst J, Hulpiau P, Saelens X.. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 2013; 77:551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tostanoski LH, Wegmann F, Martinot AJ, et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med 2020; 26:1694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iwatsuki-Horimoto K, Nakajima N, Ichiko Y, et al. Syrian hamster as an animal model for the study of human influenza virus infection. J Virol 2018; 92:e01693-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai M, Iwatsuki-Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci USA 2020; 117:16587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia LF. Immune response, inflammation, and the clinical spectrum of COVID-19. Front Immunol 2020; 11:1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horisberger MA. Interferons, Mx genes, and resistance to influenza virus. Am J Resp Crit Care 1995; 152:S67–71. [DOI] [PubMed] [Google Scholar]
- 11. Azekawa S, Namkoong H, Mitamura K, Kawaoka Y, Saito F.. Co-infection with SARS-CoV-2 and influenza A virus. IDCases 2020; 20:e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lew S, Manes P, Smith B.. Coinfection with SARS-CoV-2 and influenza A virus in a 32-year-old man. Am J Case Rep 2020; 21:e926092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ceulemans LJ, Khan M, Yoo SJ, et al. Persistence of SARS-CoV-2 RNA in lung tissue after mild COVID-19. Lancet Respir Med 2021; 9:e78–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang ANJX, Lee ACY, Chan JFW, et al. Coinfection by severe acute respiratory syndrome coronavirus 2 and influenza A(H1N1)pdm09 virus enhances the severity of pneumonia in golden Syrian hamsters. Clin Infect Dis 2021; 72:E978–E992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X.. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 2012; 86:13445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.