Abstract
Background
Following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection or vaccination there is significant variability between individuals in protective antibody levels against SARS-CoV-2, and within individuals against different virus variants. However, host demographic or clinical characteristics that predict variability in cross-reactive antibody levels are not well-described. These data could inform clinicians, researchers, and policymakers on the populations most likely to require vaccine booster shots.
Methods
In an institutional review board–approved prospective observational cohort study of staff at St. Jude Children’s Research Hospital, we identified participants with plasma samples collected after SARS-CoV-2 infection, after mRNA vaccination, and after vaccination following infection, and quantitated immunoglobulin G (IgG) levels by enzyme-linked immunosorbent assay to the spike receptor binding domain (RBD) from 5 important SARS-CoV-2 variants (Wuhan Hu-1, B.1.1.7, B.1.351, P.1, and B.1.617.2). We used regression models to identify factors that contributed to cross-reactive IgG against 1 or multiple viral variants.
Results
Following infection, a minority of the cohort generated cross-reactive antibodies, IgG antibodies that bound all tested variants. Those who did had increased disease severity, poor metabolic health, and were of a particular ancestry. Vaccination increased the levels of cross-reactive IgG levels in all populations, including immunocompromised, elderly, and persons with poor metabolic health. Younger people with a healthy weight mounted the highest responses.
Conclusions
Our findings provide important new information on individual antibody responses to infection/vaccination that could inform clinicians on populations that may require follow-on immunization.
Keywords: antibody response, BMI, metabolic health, SARS-CoV-2, variants of concern
There is significant variability in SARS-CoV-2 antibody levels between individuals. Yet, predictive host characteristics are not well described. Our findings provide important information on individual antibody responses to infection and vaccination that could inform clinicians on populations who may require follow-on immunization.
Since its emergence in December 2019, there have been more than 198 million cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection with over 4.2 million deaths (World Health Organization [1]). Viral evolution has caused considerable genetic change compared with the original SARS-CoV-2 Wuhan Hu-1 strain [2]. These genetic changes include substitutions within the functional domains of the viral spike (S) protein leading to altered biological properties, including increased transmissibility, enhanced binding to the human angiotensin-converting enzyme 2 (ACE2) receptor, and increasing virulence [2]. Important variants of concern (VOCs) include B.1.1.7 (Alpha), detected in the United Kingdom in September 2020; B.1.351 (Beta), detected in South Africa in May 2020; P.1 (Gamma), detected in Brazil in November 2020; and B.1.617.2 (Delta), detected in India in October 2020 [3]. As the virus continues to mutate, a better understanding of the breadth of the antibody response following infection or vaccination is critical for future vaccine design and public health interventions.
Studies have focused on assessing antibody responses to a specific VOC or set of samples—for example, convalescent patients or vaccine recipients [4–6]. These studies found reduced binding and neutralization against variants, further exemplified by a comprehensive study [7] examining antibody potency against all VOCs using convalescent samples from patients with varying disease severity, and mRNA vaccine recipients, and another showing that vaccination increases all components of the humoral response [8]. However, individual immunoglobulin G (IgG) responses differ, and recent studies showed that serum samples from convalescent patients display variable neutralization capacities [9]. It remains unclear whether specific host characteristics predict antibody breadth and magnitude against the VOCs.
To fill this knowledge gap, we used enzyme-linked immunosorbent assay (ELISA) to quantitate IgG titers against the spike receptor binding domain (RBD) from SARS-CoV-2 Wuhan Hu-1, B.1.1.7, B.1.351, B.1.617.2, and P.1 strains in plasma samples collected following infection and/or vaccination from a cohort of 399 healthcare workers enrolled in a prospective study.
METHODS
Data Availability
Authors confirm all relevant data are included in the paper and/or supplementary files.
Study Participants and Measures
The St. Jude Tracking of Viral and Host Characteristics Associated with COVID-19 Study (SJTRC; NCT04362995) is a prospective, institutional review board–approved, longitudinal cohort study of adult (≥18 years of age) employees at St. Jude Children’s Research Hospital initiated in March 2020. Participants provided written informed consent, and completed questionnaires about demographics, medical history, treatment, and symptoms, and a severity questionnaire following polymerase chain reaction (PCR)–confirmed SARS-CoV-2 infection. Participants were screened for SARS-CoV-2 by nasal swab PCR weekly when on campus, and convalescent blood was collected approximately 3–8 weeks after SARS-CoV-2 infection and/or vaccination. Blood samples were separated into cellular and plasma components within 24 hours of collection and stored at −80°C. Severity of disease was dichotomized as follows: severe or critical (dyspnea, or requiring supplemental oxygen, hospital admission, or dialysis during the COVID-19 episode) versus others. Body mass index (BMI) was self-reported, classified according to Centers for Disease Control and Prevention criteria, and dichotomized as obese or very obese (BMI ≥30 kg/m2) versus others [10]. Immunocompromise included all immunocompromising conditions as reported by participants, including autoimmune diseases, antibody deficiency, and corticosteroid use (Supplementary Table 1). Participants who had completed a full course of BNT162b2 by the data freeze were included in postvaccine cohorts.
Receptor Binding Domain (RBD) ELISA
Purified SARS-CoV-2 Wuhan Hu-1, B.1.1.7, B.1.351, P.1, and B.1.617.2 RBD proteins, expressed in 293T cells as described [11, 12], were diluted to a concentration of 1.5 µg/mL in phosphate-buffered saline (PBS) and added at 50 µL per well to a 96-well ELISA plate (Immulon 4 HBX; ThermoFisher Scientific). The ELISA plates were sealed and allowed to incubate at 4ºC overnight, as described [12]. The next day the coating solution was removed, and the plates were blocked at room temperature using 3% nonfat milk prepared in PBS (200 µL/well) for 1–4 hours. While the plates were being blocked, the samples were prepared by diluting the heat-inactivated plasma 1:50 in 1% fat dry milk. Following blocking, the milk was removed, and the plates were washed 3 times with PBS containing 0.1% Tween-20 (PBST) at 200 µL per well. The diluted plasma was added to the blocked plate at 50 µL per well and incubated for 1.5 hours at room temperature and then removed and washed 3 times with 200 µL 0.1% PBST. Goat anti-human IgG horseradish peroxidase (HRP) conjugated secondary antibody (ThermoFisher Scientific) was diluted 1:2500 in 1% fat dry milk and 50 µL was added to each well of the washed plate and incubated at room temperature for 30 minutes. Following the incubation period, the secondary antibody was removed, and the plate was washed 3 times with 0.1% PBST. SIGMAFAST OPD (o-phenylenediamine dihydrochloride; Sigma-Aldrich) substrate was prepared directly before use and added at 50 µL per well for exactly 8 minutes. The OPD substrate was stopped by adding 50 µL of 3M hydrochloric acid. The optical density at 490 nm (OD490) was measured using a Synergy 4 (BioTek) plate reader. SARS-CoV-2 reactive human monoclonal antibody CR3022 [11] at 1:5000, 1:25 000, 1:125 000, and 1:625 000 dilutions and plasma from naturally infected donors at a 1:50 dilution and known negative, prepandemic plasma samples diluted at 1:50 were used as controls. The background value was set at an OD490 of 0.15, as previously described [11]. Samples with an OD490 2 times greater than background, or 0.3 or greater, were considered positive as this value has been suggested as a surrogate for the presence of neutralizing antibodies in the same assay [12].
Adiponectin and Leptin ELISA
Adiponectin and leptin concentrations were measured in heat-inactivated plasma diluted at 1:50 or 1:5000 using the R&D Systems Leptin and Adiponectin DuoSet ELISA, respectively, per the manufacturer’s protocol. An adiponectin to leptin ratio greater than 1.0 was defined as at low risk for adipose tissue dysfunction or metabolic syndrome, more than 0.5 to less than 1.0 as moderate risk, and less than 0.5 as high risk [11, 13–15]. For the statistical analyses, the ratio was dichotomized as high risk versus others.
Statistical Analysis
This study comprised a convenience subcohort of 399 participants from the larger study, with 581 relevant samples selected to represent important antigen-exposure paradigms. Although not prospectively powered for this objective, this analysis had greater than 80% power to detect an OD fold-change of 1.3 or greater or 0.77 or less, using a t test with a coefficient of variation of 1 and a ratio of 2:1 vaccination versus infection only [16]. Initial descriptive analysis estimated medians and interquartile ranges (IQRs) of OD values in each group, proportions with error bars for categorical variables, and means with standard deviations for continuous variables. Chi-square test or a univariate logistical model was implemented to compare categorical variables if the count was not sparse; otherwise, Fisher’s exact test was used. Wilcoxon signed-rank test was performed to compare continuous measures. After descriptive analysis, OD values were natural log-transformed. Ward’s hierarchical agglomerative clustering method [17] using Spearman’s correlation as the distance measure for transformed OD values and using Euclidean distance for covariates was applied to examine the pattern of OD values. Infection or vaccination was a split variable in clustering. Multivariate linear effects models on natural log-transformed OD values were used to investigate associations of host characteristics with antibody levels. Host characteristics, including gender, age, race, ethnicity, immunocompromise, BMI classification, adiponectin to leptin (Adp:Lep) ratio, patient contact, time since infection diagnosis or vaccination completion, infection severity, and time of infection (before or after 1 October 2020), were considered as covariate candidates. The date cutoff of 1 October 2020 was selected arbitrarily as being between the first and second epidemic waves in Memphis, Tennessee. The elastic net model selection method [18], which extends the LASSO method [19] and ridge regression [20], was carried out with the Bayesian information criterion (BIC) [21] applied. For analysis of cross-reactivity only, samples with an OD value of 0.3 or greater were considered positive as this value has been suggested as a surrogate for the presence of neutralizing antibodies in the same assay [22]. Cross-reactivity was defined as the presence of antibodies (OD ≥0.3) against Wuhan Hu-1 or B.1.1.7, plus either B.1.351 or P.1 variants. Multivariate logistic models were implemented to analyze cross-reactivity, with a similar model selection strategy. Multivariate linear or logistic mixed models were also attempted, but were not included because estimates were very close, and the BIC consistently favored the former. A 2-sided P value less than .05 was considered significant. Statistical analyses were performed in SAS software, version 9.4 (SAS Institute, Inc.), and R software, version 4.0.2 (R Foundation for Statistical Computing).
RESULTS
Cohort Selection
Participants were enrolled into SJTRC, a prospective longitudinal cohort study of adult (≥18 years of age) employees at St. Jude Children’s Research Hospital between March 2020 and March 2021. Participants were screened for SARS-CoV-2 by nasal swab PCR approximately weekly or if symptomatic and, of 1315 participants, 191 had documented SARS-CoV-2 infection prior to June 2021. (Supplementary Table 1). The included subcohort of 399 participants included in this analysis comprised 120 with infection alone, 237 with vaccination alone, and 42 who had vaccination after infection. Subcohort characteristics are shown in Table 1 and Supplementary Tables 1 and 2 and were similar to the overall cohort.
Table 1.
Characteristics of Included Participants
| SARS-CoV-2 Infection Only (n = 120) | SARS-CoV-2 Vaccination Only (n = 237) | Vaccination After SARS-CoV-2 Infection (n = 42) | ||||
|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | |
| Gender | ||||||
| Female | 94 | (78.3%) | 186 | (78.5%) | 34 | (81%) |
| Male | 26 | (21.7%) | 51 | (21.5%) | 8 | (19%) |
| Age, mean (SD), years | 41.2 (11.2) | 47 (12.3) | 45.8 (12.2) | |||
| Race (self-reported) | ||||||
| White/Caucasian | 89 | (74.2%) | 214 | (90.3%) | 36 | (85.7%) |
| Black/African American | 18 | (15%) | 13 | (5.5%) | 4 | (9.5%) |
| Asian | 8 | (6.7%) | 6 | (2.5%) | 1 | (2.4%) |
| Other | 4 | (3.3%) | 3 | (1.3%) | 0 | (0%) |
| Not reported | 1 | (0.8%) | 1 | (0.4%) | 1 | (2.4%) |
| Hispanic ethnicity | 5 | (4.2%) | 6 | (2.5%) | 1 | (2.4%) |
| Immunocompromise | 6 | (5%) | 9 | (3.8%) | 1 | (2.4%) |
| Patient contact | ||||||
| None | 66 | (55%) | 62 | (26.2%) | 13 | (31%) |
| Indirect | 12 | (10%) | 42 | (17.7%) | 7 | (16.7%) |
| Direct | 42 | (35%) | 133 | (56.1%) | 22 | (52.4%) |
| BMI classification | ||||||
| Underweight (<18.5 kg/m2) | 1 | (0.8%) | 5 | (2.1%) | 1 | (2.4%) |
| Normal weight (18.5–<25.0 kg/m2) | 34 | (28.3%) | 97 | (40.9%) | 13 | (31%) |
| Overweight (25.0 to <30 kg/m2) | 30 | (25%) | 72 | (30.4%) | 12 | (28.6%) |
| Obese (30 to <40 kg/m2) | 35 | (29.2%) | 50 | (21.1%) | 14 | (33.3%) |
| Very obese (≥40 kg/m2) | 11 | (9.2%) | 8 | (3.4%) | 2 | (4.8%) |
| Not reported | 9 | (7.5%) | 5 | (2.1%) | (0%) | |
| Adiponectin to leptin ratio | ||||||
| Normal (≥1) | 32 | (26.7%) | 88 | (37.1%) | 11 | (26.2%) |
| Abnormal (0.5 to <1) | 15 | (12.5%) | 35 | (14.8%) | 10 | (23.8%) |
| Very abnormal (<0.5) | 73 | (60.8%) | 113 | (47.7%) | 21 | (50%) |
| Not available | 0 | (0%) | 1 | (0.4%) | 0 | (0%) |
| Infection severity | ||||||
| Asymptomatic | 10 | (8.3%) | N/A | 2 | (4.8%) | |
| Mild–moderate | 67 | (55.8%) | N/A | 33 | (78.6%) | |
| Severe | 36 | (30%) | N/A | 7 | (16.7%) | |
| Critical | 2 | (1.7%) | N/A | 0 | (0%) | |
| Not reported | 5 | (4.2%) | N/A | 0 | (0%) | |
Abbreviations: BMI, body mass index; N/A, not applicable; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation.
Most participants with SARS-CoV-2 infection had mild or moderate illness with viruses derived from Wuhan-like viruses or B.1 derivative lineages that did not contain known markers of substantial antigenic effect (Supplementary Table 9). Severe or critical coronavirus disease 2019 (COVID-19) was associated with immunocompromise (odds ratio [OR] = 6.9; P = .016) and obesity (BMI ≥30 kg/m2, OR = 2.64; P = .0085). Lower Adp:Lep ratio also correlated with disease severity (Table 2); 70% of participants with critical or severe disease had Adp:Lep less than 0.5, signifying adipose tissue dysfunction. There was no association with gender, race, age, ethnicity, or patient contact (Table 2 and Supplementary Table 3).
Table 2.
Risk Factors for Severe or Critical COVID-19
| Nonsevere (n = 112) | Severe or Critical (n = 45) | Not Reported (n = 5) | |||||
|---|---|---|---|---|---|---|---|
| Characteristics | n | (%) | n | (%) | n | (%) | P a |
| Gender | .29 | ||||||
| Female | 86 | (76.8%) | 38 | (84.4%) | 4 | (80%) | |
| Male | 26 | (23.2%) | 7 | (15.6%) | 1 | (20%) | |
| Age, mean (SD), years | 43.2 (12.0) | 40.7 (11.0) | 39.0 (4.1) | .32 | |||
| Race (self-reported) | .36b | ||||||
| White/Caucasian | 88 | (78.6%) | 33 | (73.3%) | 4 | (80%) | |
| Black/African American | 13 | (11.6%) | 9 | (20%) | 0 | (0%) | |
| Asian | 6 | (5.4%) | 2 | (4.4%) | 1 | (20%) | |
| Others | 3 | (2.7%) | 1 | (2.2%) | 0 | (0%) | |
| Not reported | 2 | (1.8%) | 0 | (0%) | 0 | (0%) | |
| Hispanic ethnicity | 5 | (4.5%) | 1 | (2.2%) | 0 | (0%) | .67 |
| BMI classification | .0087c | ||||||
| Underweight (<18.5 kg/m2) | 2 | (1.8%) | 0 | (0%) | 0 | (0%) | |
| Normal (18.5 to <25.0 kg/m2) | 37 | (33%) | 8 | (17.8%) | 2 | (40%) | |
| Overweight (25.0 to <30 kg/m2) | 32 | (28.6%) | 9 | (20%) | 1 | (20%) | |
| Obese (30 to <40 kg/m2) | 32 | (28.6%) | 17 | (37.8%) | 0 | (0%) | |
| Very obese (≥40 kg/m2) | 6 | (5.4%) | 7 | (15.6%) | 0 | (0%) | |
| Not reported | 3 | (2.7%) | 4 | (8.9%) | 2 | (40%) | |
| Adiponectin to leptin ratio | .0436 | ||||||
| Normal (≥1) | 34 | (30.4%) | 8 | (17.8%) | 1 | (20%) | |
| Abnormal (0.5 to <1) | 18 | (16.1%) | 5 | (11.1%) | 2 | (40%) | |
| Very abnormal (<0.5) | 60 | (53.6%) | 32 | (71.1%) | 2 | (40%) | |
| Immunocompromise | 2 | (1.8%) | 5 | (11.1%) | 0 | (0%) | .016 |
| Patient contact | .88 | ||||||
| None | 52 | (46.4%) | 22 | (48.9%) | 5 | (100%) | |
| Indirect | 13 | (11.6%) | 6 | (13.3%) | 0 | (0%) | |
| Direct | 47 | (42%) | 17 | (37.8%) | 0 | (0%) | |
Participants who did not report were excluded from comparisons. Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; SD, standard deviation.
P value compares severe or critical vs asymptomatic or mild–moderate.
White/Caucasian vs others.
Compares obese and very obese vs not obese.
Characteristics Associated With Generation of Cross-protective IgG During Infection
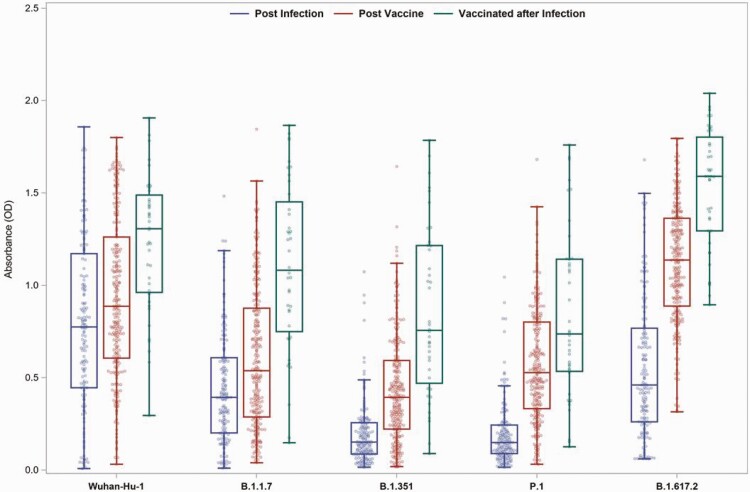
To define factors underlying the magnitude and breadth of the IgG response against Wuhan Hu-1, B.1.1.7, B.1.351, B.1.617.2, and P.1 RBDs produced in response to infection, vaccination, or vaccination postinfection, 581 evaluable samples were tested for IgG against the RBD of SARS-CoV-2 variants by ELISA. ODs less than 0.3 were considered negative for the cross-reactivity analysis. Median IgG titers against SARS-CoV-2 variants were highest in samples collected after completion of SARS-CoV-2 vaccination, with titers against Wuhan Hu-1 and B.1.617.2 higher than the other 3 VOCs (Figure 1).
Figure 1.
Receptor binding domain antibodies against SARS-CoV-2 variants following infection, vaccination, or vaccination after infection. The samples with a valid absorbance (OD) value for groups of participants who had SARS-CoV-2 infection, vaccination, or vaccination following infection are shown; 428 samples from 384 individuals collected >14 days and ≤90 days after the diagnosis or the final dose of the vaccine were included. The box plots with median, upper, and lower quartiles and minimum and maximum values for postinfection samples with an OD value from individuals who were infected, postvaccination samples from individuals who were only vaccinated, and postvaccination samples from individuals vaccinated after infection are shown in blue, red, and green, respectively. Abbreviations: OD, optical density; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Host Characteristics Associated With Broadly Reactive IgG Responses After Infection Include Poor Metabolic Health
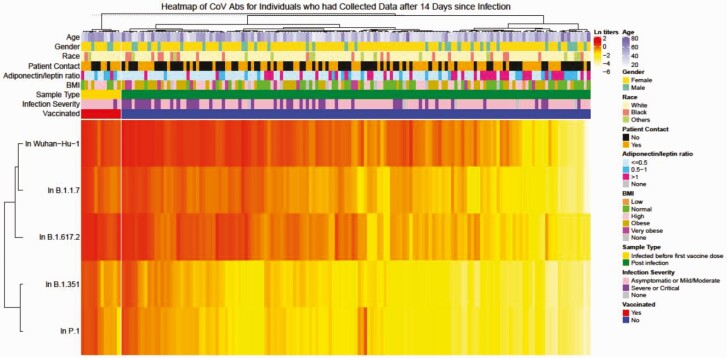
Hierarchical clustering was used to identify factors underlying the cross-reactive antibodies. We classified individual VOC RBD IgG titers to infection (Figure 2) and vaccination (Figure 3) and examined host characteristics associated with high-magnitude, cross-reactive IgG responses. Following SARS-CoV-2 infection, time since infection (>14 days vs ≤14 days), more severe illness, and receipt of any SARS-CoV-2 vaccine were positively associated with higher IgG against all SARS-CoV-2 RBDs (Figure 2, Supplementary Table 4, and Supplementary Figure 1) or, in the case of disease severity, 3 of 4 VOCs: B.1.351, B.1.617.2, and P.1 (Figure 4 and Supplementary Table 5).
Figure 2.
Heatmap of receptor binding domain antibody levels against SARS-CoV-2 variants after infection. Natural log-transformed OD values in all samples collected >14 days after a SARS-CoV-2 infection were clustered with Ward’s hierarchical agglomerative clustering method using Spearman’s correlation as the distance measure for transformed OD values and Euclidean distance for all covariates was applied. SARS-CoV-2 vaccination was used as a split variable in clustering. Abbreviations: Abs, antibodies; BMI, body mass index; CoV, coronavirus; ln, natural log; OD, optical density; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
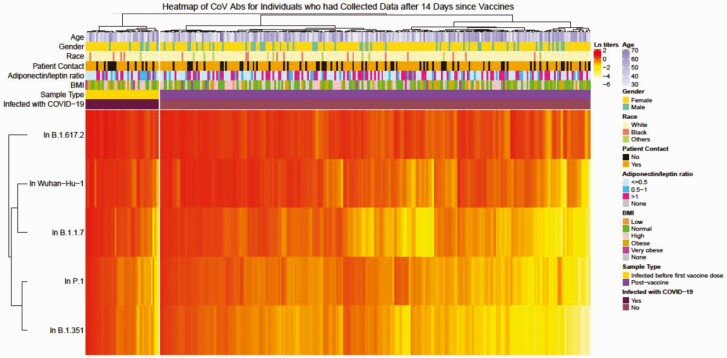
Figure 3.
Heatmap of receptor binding domain antibody levels against SARS-CoV-2 variants after vaccination. Natural log-transformed OD values in all samples collected >14 days after SARS-CoV-2 vaccination were clustered with Ward’s hierarchical agglomerative clustering method using Spearman’s correlation as the distance measure for transformed OD values and Euclidean distance for all covariates was applied. SARS-CoV-2 infection was used as a split variable in clustering. Abbreviations: Abs, antibodies; BMI, body mass index; CoV, coronavirus; COVID-19, coronavirus disease 2019; ln, natural log; OD, optical density; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
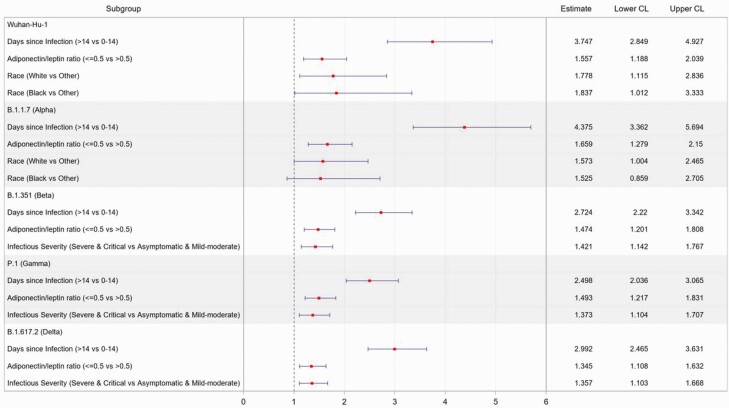
Figure 4.
Multivariate analysis of predictors of receptor binding domain antibody levels against SARS-CoV-2 variants after infection. Multivariate linear models on natural log-transformed OD values were carried out among samples collected from 1 to 281 days after diagnosis with a SARS-CoV-2 infection without subsequent vaccination. Host characteristics, including gender, age, race, ethnicity, immunocompromised or not, BMI classification, adiponectin to leptin ratio, patient contact, time since infection diagnosis, infection severity, and time of infection (before or after 1 October 2020), were considered as covariate candidates. The elastic net model selection method was carried out with the BIC. The estimate was expressed in fold-change in the antibody level. Point estimates and 95% confidence intervals were transformed back to the original OD scale. Abbreviations: BIC, Bayesian information criterion; BMI, body mass index; CL, confidence limit; OD, optical density; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
A low Adp:Lep, indicative of poor metabolic health, was also associated with higher cross-reactive IgG titers (Figure 2, Supplementary Table 4, and Supplementary Figure 1). We hypothesized that this was related to the increased disease severity observed in this group (Table 2). However, the effect of adipose tissue dysfunction on IgG levels was not mediated by severity of infection, as the effect size was not changed by adjustment for infection severity in the multivariable model (Figure 4) or by subgroup analyses stratified by infection severity. Attempts to link this to viral titers or viral shedding time were also unsuccessful. Participants who self-identified as White or Black race had higher IgG responses to Wuhan Hu-1 and B1.1.7, but not to B1.351, P.1, or B.1.617.2, compared with self-reported “other race.”
Finally, we examined the host characteristics associated with a positive response (OD value ≥0.3) against either Wuhan Hu-1 or B.1.1.7, plus either B.1.351 or P.1, to assess what factors were important in facilitating a robust cross-reactive response. Days since infection (>14 days vs ≤14 days), vaccination after infection, and higher disease severity were highly significant, whereas lower Adp:Lep was marginally associated (Supplementary Table 6). A similar association was noted when considering a positive response (OD value ≥0.3) against either Wuhan Hu-1 or B.1.617.2.
Overall, people with high levels of cross-reactive IgG following infection were more likely to have been vaccinated after infection, have had severe disease, have adipose tissue dysfunction, or self-identify as White or Black race. Other factors, including gender and immunocompromised status, were not associated with IgG levels against any of the variant RBDs tested.
Vaccination Increases Cross-reactive IgG Levels
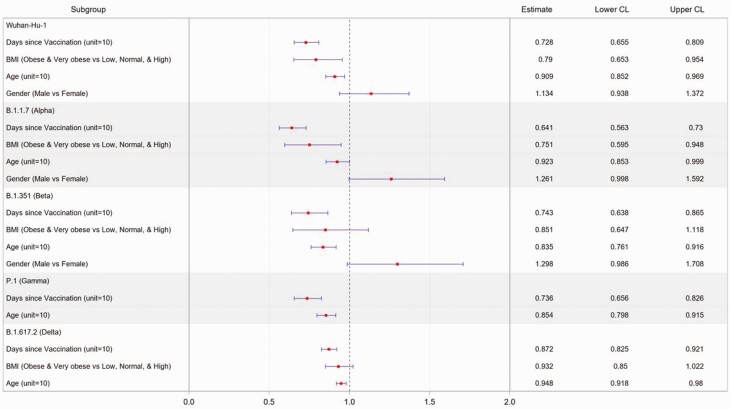
Vaccination increased cross-reactive IgG levels in all populations. Younger people with healthy weight and more recent vaccination had the highest levels of cross-reactive IgG against all SARS-CoV-2 VOCs (Figure 5 and Supplementary Tables 7 and 8). Specifically, after vaccination, obesity was associated with significantly lower levels of antibodies against RBD from Wuhan Hu-1 and B.1.1.7, but not B.1.351, P.1, or B.1.617.2, and time since vaccination and older age were both significantly associated with lower levels of antibody for all of the VOCs and cross-reactivity defined previously (Figure 5). Of interest, people with self-reported mild to moderate immunocompromise did not have reduced antibody responses to vaccines for any of the VOCs.
Figure 5.
Multivariate analysis of predictors of receptor binding domain antibody levels against SARS-CoV-2 variants after vaccination. Multivariate linear models on natural log-transformed OD values were carried out among SARS-CoV-2–naïve participants with complete vaccination. Samples were collected 25 to 54 days after completion of vaccine course. Host characteristics, including gender, age, race, ethnicity, immunocompromised or not, BMI classification, adiponectin to leptin ratio, patient contact, and time since the final dose of the vaccine, were considered as covariate candidates. The elastic net model selection method was carried out with the BIC. The estimate was expressed in fold-change in the antibody level. Point estimates and 95% confidence intervals were transformed back to the original OD scale. Abbreviations: BIC, Bayesian information criterion; BMI, body mass index; CL, confidence limit; OD, optical density; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The host characteristics that resulted in lower VOC IgG responses postvaccination included obesity, time since vaccination, and older age (Figure 5). Further work is needed to define the mechanism(s) for the reduced IgG levels.
DISCUSSION
This study addresses a critical, unresolved question about IgG responses to SARS-CoV-2 infection and/or vaccination at an individual level—defining the characteristics associated with a lower cross-reactive IgG response against SARS-CoV-2 VOCs. We determined that strong cross-reactive antibody responses after infection were positively associated with the following: 14 or more days since diagnosis, greater disease severity, and counterintuitively, poor metabolic health. Vaccination increased the magnitude and breadth of the antibody response, especially with past infection. However, in contrast to infection alone, the only other significant positive predictors of cross-reactive antibody response following vaccination were younger age and healthy BMI. This provides a snapshot of host characteristics associated with differential SARS-CoV-2 antibody responses. The results show the importance of vaccination and identify populations who have a high likelihood of requiring vaccine boosting to maintain protective antibody levels.
Strengths of the study include the cohort size; focus on hospital staff; detection of mild cases through regular screening with SARS-CoV-2 PCR; direct comparison between infection, vaccination, and vaccination postinfection; and statistical analysis to define the population least likely to mount broadly protective IgG responses [7, 8].
The characteristics associated with the magnitude and breadth of IgG responses to SARS-CoV-2 VOCs differed between infection and vaccination. Following infection alone, we showed a clear association of IgG levels with disease severity and time since infection, which is in keeping with the literature [23, 24]. Published studies have shown that antibodies from late convalescence or following more severe disease exhibited increased neutralization potency to VOCs, suggesting persistence of cross-neutralizing plasma IgG levels. Age, gender, and immunocompromise had no significant effect on IgG levels after infection. If confirmed in other studies, the finding that moderate immunocompromise was not associated with decreased antibody responses to any of the variants would be reassuring for immunocompromised individuals. The lack of association between disease severity and gender was surprising since a male bias in COVID-19 mortality has been reported [25]. This may be due to the limited number of participants with severe disease. There was also no sex bias associated with overall difference in IgG responses, consistent with 1 other study [26]. Although we found an association between race and IgG levels, the construct is confounded by many social factors, so it is difficult to determine the relevance or mechanism of this finding.
We also demonstrated that high BMI or a low Adp:Lep, indicating adipose tissue dysfunction and metabolic syndrome, was associated with more severe disease, which parallels other studies [27, 28]. However, following infection only, poor metabolic health (Adp:Lep <0.5) was associated with higher IgG responses against VOCs. This was an independent factor unrelated to disease severity, and is consistent with other postconvalescent wild-type SARS-CoV-2 neutralizing antibody levels in individuals with metabolic syndrome comorbidities [29]. The reason for the association is unclear. Increased systemic inflammation caused by the metabolic syndrome might lead to higher antibody responses or, alternatively, these individuals might shed virus longer, leading to increased immune stimulation [30].
Vaccination resulted in plasma antibodies against VOCs that are mostly comparable to the anti-RBD ELISA titers against the original Wuhan Hu-1 strain. While the levels of cross-reactive IgG against RBD from multiple SARS-CoV-2 variants following vaccination were significantly associated with younger age, healthy weight, being a male, and more recent vaccination, vaccination improved IgG levels in all groups. Although the finding that being overweight was associated with lower antibody responses to most variants following vaccination is in stark contrast to antibody levels following infection in participants with the metabolic syndrome, it is clearly consistent with other data suggesting that patients with obesity have poorer responses to other vaccines, such as influenza [31].
Overall, this study identifies factors influencing the magnitude and breadth of the IgG response to the spike RBD of 4 important SARS-CoV-2 VOCs. Limitations include the relatively low number of patients with critical illness or immunocompromised status, completion prior to widespread emergence of the Delta variant, and use of ELISA rather than neutralization titers. However, we have previously demonstrated that samples with an ELISA OD less than 0.3 have no neutralizing activity [12]. Further, this OD cutoff was defined in the original protocol as being the difference between samples that would be positive and negative against the spike protein [11]. These data could help prioritize vaccines and boosters for individuals expected to have the lowest antibody levels.
Conclusions
This study showed that vaccination improved IgG levels in all groups, highlighting the importance of vaccinating vulnerable populations, such as persons with poor metabolic health or high BMI, as well as those who had PCR-confirmed infection, to provide more broadly protective antibody responses to SARS-CoV-2 VOCs. However, older people and those with higher BMI are most likely to have lower antibody levels and could be prioritized for booster shots to retain broadly protective antibody levels. Overall, these studies highlight the importance of vaccinating vulnerable populations against COVID-19, including those with poor metabolic health or high BMI, immunocompromising conditions, and those who have previously had PCR-confirmed infection, to provide more broadly cross-reactive antibody responses to SARS-CoV-2 VOCs.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
St. Jude Investigative Team. Department of Biostatistics, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: Tomi Mori; Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: Diego R. Hijano, Hana Hakim, Ronald H. Dallas, Valerie Cortez, Ana Vazquez-Pagan, Richard J. Webby, Thomas Fabrizio, Jamie Russell-Bell; Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: David C. Brice, Ashley Castellaw, Resha Bajracharya, Brandi L. Clark, Lee-Ann Van de Velde, Walid Awad, Taylor L. Wilson, Allison M. Kirk; Department of Hematology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: Jason Hodges; Department of Global Pediatric Medicine, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: James Sparks; Office of Quality and Patient Care, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: David E. WIttman; Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: Randall T. Hayden; Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA: James Hoffman.
Notes
Acknowledgments. The authors thank the members of the Thomas, McGargill, and Schultz-Cherry labs for technical assistance and feedback on the work. The authors also thank Gang Wu and the Center for Applied Bioinformatics at St. Jude along with Michael Meagher, Timothy Lockey, and the St. Jude Good Manufacturing Practice (GMP) facility, and Matthew Lear and Charles Mullighan at the St. Jude Biorepository. The authors also thank Tamanna Shamrin and Rishi Kodela for creation and management of the clinical database.
Financial support. This work was supported by The American Lebanese Syrian Associated Charities, the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH) under Department of Health and Human Services (HHS) contract HHSN27220140006C for the St. Jude Center of Excellence for Influenza Research and Surveillance, by the NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00052 to S. S.-C. and P. G. T. J. H. E. is supported by the American Society of Hematology Scholar Award. Work in the Krammer laboratory is partially funded by the NIAID CIVIC contract 75N93019C00051, NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS; contract number HHSN272201400008C), by the generous support of the Cohen Foundation, the JPB Foundation, and the Open Philanthropy Project (research grant 2020-215611 [5384]), and by anonymous donors. F. K. reports receiving the following support: NIH (Centers of Excellence for Influenza Research and Response (CEIRR; 75N93021C00014); National Cancer Institute, NIH, under contract number 75N91019D00024, task order no. 75N91020F00003; and research funding from Pfizer for the development of animal models for SARS-CoV-2. M. A. M. reports the following support: NIH/NIAID (3U01AI144616-02S1). L. T. reports the following funding: National Cancer Institute (5P30CA021765-42; Biostatistics Shared Resources). P. G. T. reports the following funding: NIAID (5U01AI144616 and U01AI150747).
Potential conflicts of interest. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays (the “Serology Assays”) and Newcastle disease virus-based SARS-CoV-2 vaccines which name F. K. as a co-inventor; F. K. also notes the following, which could be perceived as a conflict of interest: He has previously published work on influenza virus vaccines with S. Gilbert (University of Oxford); has consulted for Curevac, Merck, and Pfizer (before 2020); is currently consulting for Pfizer, Seqirus, and Avimex; his laboratory is collaborating with Pfizer on animal models of SARS-CoV-2; his laboratory is collaborating with N. Pardi at the University of Pennsylvania on mRNA vaccines against SARS-CoV-2; his laboratory was working in the past with GlaxoSmithKline on the development of influenza virus vaccines and 2 of his mentees have recently joined Moderna; F. K. reports royalties/licenses for Avimex; payment/honoraria for several academic lectures over the past 2 years. E. K. A. reports patents on methods for treating or reducing the severity of a viral infection. J. H. E. reports grants/support to their institution from the following: Pfizer, Eli Lily and Company, Forma Therapeutics, Global Blood Therapeutics, National Institutes of Health, and American Society of Hematology; reports personal consulting fees from Global Blood Therapeutics, Emmaus Life Sciences, Daiichi Sankyo, and Agios; personal payments/honoraria for educational events from Global Blood Therapeutics; personal payments from the National Institutes of Health for serving on a Data Safety and Monitoring Board/Advisory Board; receipt of medical writing support from Global Blood Therapeutics. P. G. T. reports the following funding: NIAID 5R01AI128805-05, 1R01AI154470, 75N93021C00016, 75N93021C00018, 5R01AI121832, 5R01AI136514, and 5R01AI35025; reports personal consulting fees from Johnson and Johnson, Cytoagents, and Immunoscape; reports personal payments for educational events from Illumina–Future Genomic Advances, Yale University, CZ Biohub, PACT Pharma, UCSD, Tufts, University of Arizona, Mt. Sinai, UMass, OSU, Korean Association of Immunology, SISMID, University of Washington, MSKCC, Washington University, University of Missouri, Fred Hutch, UNM, Illumina Single Cell, and Cincinnati Childrens; reports travel support for the following: NIH Study Section, Midsummer Immunology, NIAID CEIRR, CEIRS, and CIVIC meetings; GRC, 10X Users Symposium, Illumina Symposia; reports the following licenses/patents: Methods for Treating or Reducing the Severity of a Viral Infection, US20170304293A1 Coordinated metabolic reprogramming in response to productive viral infections, and Cloning and expression system for T-cell receptors (license payments made to them and their institution). S. S.-C. reports the following support: NIAID 5R01AI125392-05, 5U01AI144616-03S1, 5R01AI140766-03, 5R01AI123144-05, 75N93021C00016, 75N93021C00018, 3R21AI135254-02S1, and 5R21AI135254-02; consulting fees from CIDARA (personal fees), Versatope, and GenoTwin; payments/honoraria for educational activities for American Philosophical Society (paid to University of Wisconsin), NIAID workshops (several; paid to Wayne State), NEDL/Boston University (paid to the University of Nebraska); travel support for the following events: GRC, NIH Study Section, ASM PSAC, and NIAID CEIRR, CEIRS, and CIVIC meetings; participation in DMB 16-0092; and serving as Chair, PSAC, for the American Society for Microbiology. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Li Tang, Department of Biostatistics, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Sean Cherry, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Elaine I Tuomanen, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Ericka Kirkpatrick Roubidoux, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Chun Yang Lin, Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Kim J Allison, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Ashleigh Gowen, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Pamela Freiden, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
E Kaitlynn Allen, Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Yin Su, Department of Biostatistics, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Aditya H Gaur, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Jeremie H Estepp, Department of Hematology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA; Department of Global Pediatric Medicine, St. Jude Children’s Research Hospital, Memphis, Tennessee, USAand.
Maureen A McGargill, Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Paul G Thomas, Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Stacey Schultz-Cherry, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Joshua Wolf, Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
St. Jude Investigative Team:
Tomi Mori, Diego R Hijano, Hana Hakim, Ronald H Dallas, Valerie Cortez, Ana Vazquez-Pagan, Richard J Webby, Thomas Fabrizio, Jamie Russell-Bell, David C Brice, Ashley Castellaw, Resha Bajracharya, Brandi L Clark, Lee Ann Van de Velde, Walid Awad, Taylor L Wilson, Allison M Kirk, Jason Hodges, James Sparks, David E WIttman, Randall T Hayden, and James Hoffman
References
- 1. World Health Organization. Coronavirus disease (COVID-19) pandemic. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 2 August 2021.
- 2. Krause PR, Fleming TR, Longini IM, et al. SARS-CoV-2 variants and vaccines. N Engl J Med 2021; 385:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization. Tracking SARS-CoV-2 variants. Available at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 2 August 2021.
- 4. Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438–43. [DOI] [PubMed] [Google Scholar]
- 5. Stamatatos L, Czartoski J, Wan YH, et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science 2021; 372:1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dejnirattisai W, Zhou D, Supasa P, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021; 184:2939–54, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caniels TG, Bontjer I, van der Straten K, et al. Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination. medRxiv, doi: 10.1101/2021.05.26.21257441, 2021, preprint, not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z, Muecksch F, Schaefer-Babajew D, et al. Naturally enhanced neutralizing breadth to SARS-CoV-2 after one year. bioRxiv, doi: 10.1101/2021.05.07.443175, 2021, preprint, not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noval MG, Kaczmarek ME, Koide A, et al. Antibody isotype diversity against SARS-CoV-2 is associated with differential serum neutralization capacities. Sci Rep 2021; 11:5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention. Defining adult overweight and obesity. Available at: https://www.cdc.gov/obesity/adult/defining.html. Accessed 2 August 2021.
- 11. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wohlgemuth N, Whitt K, Cherry S, et al. ; St. Jude Investigative Team. . Choosing the right tool for the job: a comprehensive assessment of serological assays for SARS-CoV-2 as surrogates for authentic virus neutralization. medRxiv, doi: 10.1101/2021.04.14.21255399, 2021, preprint, not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fruhbeck G, Catalan V, Rodriguez A, Gomez-Ambrosi J.. Adiponectin-leptin ratio: a promising index to estimate adipose tissue dysfunction. Relation with obesity-associated cardiometabolic risk. Adipocyte 2018; 7:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vega GL, Grundy SM.. Metabolic risk susceptibility in men is partially related to adiponectin/leptin ratio. J Obes 2013; 2013:409679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skvarca A, Tomazic M, Blagus R, Krhin B, Janez A.. Adiponectin/leptin ratio and insulin resistance in pregnancy. J Int Med Res 2013; 41:123–8. [DOI] [PubMed] [Google Scholar]
- 16. East 6. Statistical software for the design, simulation and monitoring clinical trials. Cambridge, MA: Cytel, Inc, 2020. [Google Scholar]
- 17. Ward JH Jr. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 1963; 58:236–44. [Google Scholar]
- 18. Zou H, Hastie T.. Regularization and variable selection via the elastic net. J Roy Stat Soc Ser B (Stat Method) 2005; 67:301–20. [Google Scholar]
- 19. Tibshirani R. Regression shrinkage and selection via the lasso. JR Statist Soc B 1996; 58: 267–88. [Google Scholar]
- 20. Hoerl A, Kennard R.. Ridge regression. In: Kotz S, Lloyd Johnson N, Read CB, eds. Encyclopedia of statistical sciences. Vol 8. New York: Wiley, 1988:129–36. [Google Scholar]
- 21. Schwarz G. Estimating the dimension of a model. Ann Statis 1978; 6:461–4. [Google Scholar]
- 22. Wohlgemuth N, Whitt K, Cherry S, et al. ; St. Jude Investigative Team, et al. An assessment of serological assays for SARS-CoV-2 as surrogates for authentic virus neutralization. Microbiol Spectr 2021; 9:e0105921. doi: 10.1128/Spectrum.01059-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pirofski LA. Disease severity and durability of the SARS-CoV-2 antibody response: a view through the lens of the second year of the pandemic. Clin Infect Dis 2021; 73:e1345–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moriyama S, Adachi Y, Sato T, et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity 2021; 54:1841– 52.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL.. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol 2020; 20:442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang B, Cai Y, Li N, et al. Sex-based clinical and immunological differences in COVID-19. BMC Infect Dis 2021; 21:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flerlage T, Boyd DF, Meliopoulos V, Thomas PG, Schultz-Cherry S.. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat Rev Microbiol 2021; 19:425–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith M, Honce R, Schultz-Cherry S.. Metabolic syndrome and viral pathogenesis: lessons from influenza and Coronaviruses. J Virol 2020; 94:e00665–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Racine-Brzostek SE, Yang HS, Jack GA, et al. Postconvalescent SARS-CoV-2 IgG and neutralizing antibodies are elevated in individuals with poor metabolic health. J Clin Endocrinol Metab 2021; 106:e2025–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Islam SMRU, Akther T, Sultana S, Munshi SU.. Persistence of SARS-CoV-2 RNA in a male with metabolic syndrome for 72 days: a case report. SAGE Open Med Case Rep 2021; 9:2050313X-1989492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neidich SD, Green WD, Rebeles J, et al. Increased risk of influenza among vaccinated adults who are obese. Int J Obes (Lond) 2017; 41:1324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Authors confirm all relevant data are included in the paper and/or supplementary files.