Abstract
The regulatory-targeting subunit (RGL, also called GM) of the muscle-specific glycogen-associated protein phosphatase PP1G targets the enzyme to glycogen where it modulates the activity of glycogen-metabolizing enzymes. PP1G/RGL has been postulated to play a central role in epinephrine and insulin control of glycogen metabolism via phosphorylation of RGL. To investigate the function of the phosphatase, RGL knockout mice were generated. Animals lacking RGL show no obvious defects. The RGL protein is absent from the skeletal and cardiac muscle of null mutants and present at ∼50% of the wild-type level in heterozygotes. Both the level and activity of C1 protein are also decreased by ∼50% in the RGL-deficient mice. In skeletal muscle, the glycogen synthase (GS) activity ratio in the absence and presence of glucose-6-phosphate is reduced from 0.3 in the wild type to 0.1 in the null mutant RGL mice, whereas the phosphorylase activity ratio in the absence and presence of AMP is increased from 0.4 to 0.7. Glycogen accumulation is decreased by ∼90%. Despite impaired glycogen accumulation in muscle, the animals remain normoglycemic. Glucose tolerance and insulin responsiveness are identical in wild-type and knockout mice, as are basal and insulin-stimulated glucose uptakes in skeletal muscle. Most importantly, insulin activated GS in both wild-type and RGL null mutant mice and stimulated a GS-specific protein phosphatase in both groups. These results demonstrate that RGL is genetically linked to glycogen metabolism, since its loss decreases PP1 and basal GS activities and glycogen accumulation. However, PP1G/RGL is not required for insulin activation of GS in skeletal muscle, and rather another GS-specific phosphatase appears to be involved.
In recent years, the generality that the activity of the type 1 serine/threonine protein phosphatases (PP1) is dictated by the associated noncatalytic subunits has emerged. These ancillary proteins are thought to target the catalytic component (C1) to distinct subcellular locales in proximity to substrates, to confer specificity, and to regulate activity (10, 21, 33, 41). To date, more than 30 C1-binding polypeptides have been identified that direct the enzyme to a variety of subcellular structures, including glycogen (6, 24, 25, 49, 59, 60), myosin (2), ribosomes (31), nuclei (4, 13), and neuronal structures (5). A subset of C1-binding proteins includes inhibitory proteins such as inhibitors 1 and 2 (48, 67) and DARPP-32 (46).
Four C1-glycogen-targeting subunits are presently known. RGL, also called GM, was the first glycogen-binding subunit of PP1 identified (59), and the corresponding holoenzyme, PP1G/RGL, consists of the 124-kDa RGL protein (60) in association with C1. RGL is exclusively expressed in skeletal and cardiac muscle (37, 60). The NH2-terminal 240 amino acids contain binding sites for glycogen and C1 (64), whereas a hydrophobic region between residues 1063 and 1097 in the COOH terminus anchors the protein to membrane (45, 60). Of the other three glycogen-targeting subunits, GL, a 33-kDa polypeptide, is specifically expressed in liver (24), whereas PTG/R5 and R6 are ubiquitously distributed (6, 25, 49). All three share homology with the NH2-terminal region of RGL. The activity of liver PP1G/GL is controlled allosterically by glycogen phosphorylase (Ph) and by glucose-6-phosphate (G-6P) (1, 58), and expression of the GL subunit is downregulated in diabetic rats (23). Protein targeting to glycogen (PTG) interacts with glycogen-metabolizing enzymes (26) and has been implicated in insulin control of glycogen synthase (GS) (49). Overexpression of PTG in Chinese hamster ovary cells expressing the human insulin receptor increased basal and insulin-stimulated GS activity (49). Neither insulin nor forskolin induced detectable PTG phosphorylation, arguing against such a mechanism for PTG regulation. Adenovirus-mediated overexpression of GL, PTG, or RGL in primary rat hepatocytes results in basal activation of GS (12), but only in cells overexpressing GL or PTG was GS activated by insulin (27). Other studies have postulated a mechanism whereby PTG would affect PP1 activity by relieving inhibition by DARPP-32 (16). However, it has been shown that neither I-1 nor DARPP-32 is required for insulin activation of GS (53). Thus, the mechanisms for control of PTG- and R6-containing phosphatases are largely unknown.
In mammals, the major stores of glycogen are in skeletal muscle and liver. Although glycogen in these two tissues performs different functions, both pools contribute to glucose homeostasis. Approximately 80% of postprandial, insulin-stimulated glucose uptake is stored as glycogen in skeletal muscle (56). This insulin-stimulated glycogen synthesis involves activation of both glucose transport and GS (39). Glycogen metabolism is controlled in large part by the coordinated regulation of the two enzymes responsible for its synthesis and breakdown, GS and Ph. GS is inactivated by a complex, multisite phosphorylation mechanism involving several protein kinases, including GS kinase 3 (GSK-3) (38, 52). The allosteric activator G-6P can restore full activity, so that the GS activity ratio in the absence and presence of G-6P (−/+ G-6P activity ratio) is used as a kinetic index of phosphorylation state. Ph is activated by phosphorylation of a single site by phosphorylase kinase (PhK). Insulin promotes dephosphorylation and activation of GS, with no significant effect on Ph. Despite extensive investigation over the last four decades, the exact molecular mechanism(s) by which insulin activates GS in skeletal muscle is not fully understood. Epinephrine causes phosphorylation of both GS and Ph, leading to their respective inactivation and activation so that glycogen is degraded. Dephosphorylation of the key regulatory enzymes, GS, Ph, and PhK, is believed to be catalyzed primarily by glycogen-associated phosphatases (59). Also, association of RGL with C1 has been shown to enhance activity towards these three glycogen-metabolizing enzymes (33).
PP1G/RGL has been postulated to play a central role in both insulin and epinephrine control of glycogen metabolism in skeletal muscle, via phosphorylation of the RGL-targeting subunit (20, 47). Insulin control of PP1G/RGL would be mediated by the insulin-stimulated protein kinase ISPK or p90rsk which is itself regulated via the mitogen-activated protein kinase pathway (20). ISPK phosphorylates RGL at site 1 in vitro, enhancing the rate of dephosphorylation of GS and PhK, with no effect on the activity towards Ph. Treatment of rabbits with insulin was reported to increase the phosphorylation of site 1 (20). The hypothesis for control of PP1G/RGL by insulin was attractive, since this enzyme dephoshorylates GS at both the COOH- and NH2-terminal sites that are responsive in vivo to the hormone (38). However, several reports have shown that the mitogen-activated protein kinase pathway is not involved in insulin activation of GS (9, 40). Nevertheless, the results of these studies do not preclude the possibility that PP1G/RGL mediates insulin regulation of glycogen metabolism by other mechanisms.
Newer models for the control of GS activity by insulin have centered on a role for the PI-3K pathway (17, 54) and control of GSK-3, an important GS kinase whose activity is known to be inhibited by insulin. Stimulation of PI-3K by insulin would activate Akt/PKB which phosphorylates and inactivates GSK-3. Another pathway activated by insulin involves mTOR, the mammalian target for the immunosuppressant drug rapamycin (9); rapamycin has been shown to oppose insulin-mediated activation of GS in muscle and 3T3-L1 adipocytes (9, 54). Since rapamycin does not block insulin-induced inactivation of GSK-3 (18), it is possible that mTOR could control GS phosphorylation via a phosphatase.
To address the role of PP1G/RGL in the hormonal control of glycogen metabolism in a more definitive manner, we have generated RGL knockout mice. Analysis of this animal model clearly demonstrates that RGL has an important role in glycogen synthesis but is not essential for insulin activation of GS.
MATERIALS AND METHODS
Materials.
pBluescript II KS(+), pCR II, and pET15b were from Stratagene (La Jolla, Calif.), Invitrogen (Carlsbad, Calif.), and Novagen (Madison, Wis.) respectively. Aspergillus niger amyloglucosidase, yeast hexokinase, and yeast G-6P dehydrogenase were purchased from Sigma (St. Louis, Mo.). Restriction enzymes were from New England Biolabs (Beverly, Mass.). Porcine insulin was a gift from Eli Lilly Co. (Indianapolis, Ind.). The Puregene DNA isolation kit was from Gentra Systems (Minneapolis, Minn.). Taq polymerase was purchased from Promega. Radioactively labeled nucleotides, α-d-[U-14C]glucose-1-phosphate and UDP[U-14C]glucose were purchased from NEN Life Science Products (Boston, Mass.). Okadaic acid was from Roche Molecular Biochemicals (Indianapolis, Ind.). Oligonucleotides were synthesized by Life Technologies (Bethesda, Md.). Monoclonal anti-C1 antibodies that recognize all isoforms were generously provided by Jackie Vandenheede (University of Leuven, Leuven, Belgium), and anti-GS antibodies were generously provided by John C. Lawrence, Jr. (University of Virginia, Charlottesville). Antibodies to PTG and R6 were kindly provided by Alan Saltiel (Pfizer Global Research and Development, Ann Arbor, Mich.) and Patricia Cohen (University of Dundee, Dundee, United Kingdom), respectively. Anti-phosphorylated site 1 and site 2 RGL antibodies were a generous gift from Philip Cohen (University of Dundee). DNA sequencing was done at the Indiana University Biochemistry and Biotechnology Facility (Indianapolis) on an ABI PE-ABI 377XL laser sequencer using fluorescent dideoxy terminators. Antisera against mouse RGL was produced by immunizing rabbits with recombinant His-RGL(1–262) protein. Other chemicals were purchased from Sigma and Bio-Rad (Hercules, Calif.).
Construction of RGL gene-disrupting targeting vector.
A 129Sv mouse genomic DNA library made in Lambda DASH II was provided by Bi Feng (Lunenfeld Research Institute, Toronto, Ontario, Canada). Screening of 8.3 × 105 plaques using rabbit RGL cDNA (3.8 kb) (60) as a probe resulted in isolation of seven positive clones, four of which contained overlapping sequences (37). Restriction mapping and sequence analysis revealed that one of the clones contained 785 nucleotides that code for exon 1, flanked by ∼5 kb upstream and 11 kb downstream. To generate the targeting vector, an 0.8-kb HindIII-EcoRV fragment, which is upstream of exon 1, was inserted into the SalI site of pBluescript II/NeoTK vector (Fig. 1) (7) which contains the phosphoglycerokinase (PGK) promoter driving the neo gene (PGK-neo) and the herpes simplex virus thymidine kinase gene (HSV-tk). A 7.0-kb BsmI-BsmI fragment downstream of exon 1 was inserted at the XhoI site of the vector. By this strategy, 785 bp coding for 262 amino acids in exon 1, 50 bp upstream of the start ATG codon, and 16 bp of intron 1 were replaced by the neomycin resistance gene. The targeting vector was linearized at the NotI site and used for electroporation into embryonic stem (ES) cells.
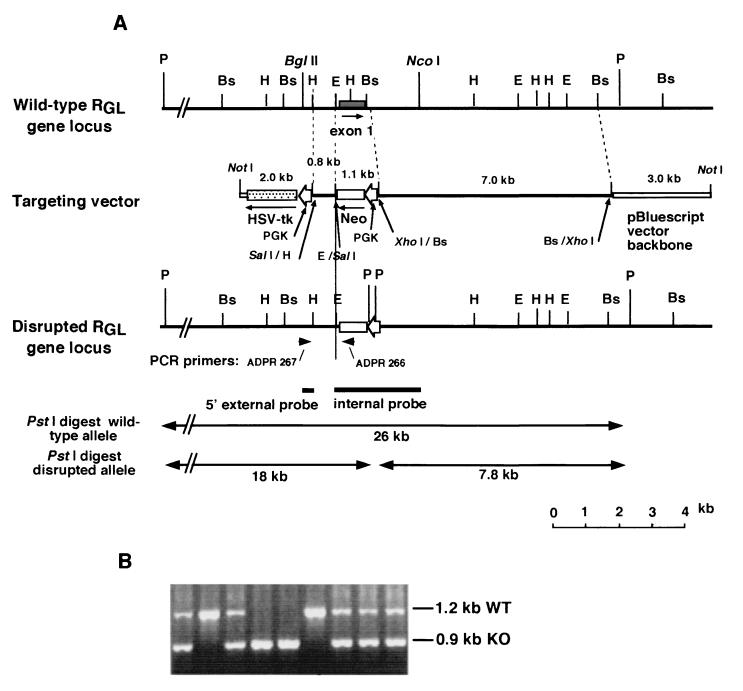
FIG. 1.
Targeted disruption of the RGL gene. (A) Maps of the mouse RGL gene locus, the targeting vector, and the disrupted RGL gene locus. Restriction sites BsmI (Bs), EcoRV (E), HindIII (H), and PstI (P) are shown. Arrowheads indicate the positions of the PCR primers (ADPR266 and ADPR267). Thick bars below the map of the disrupted locus represent the positions of the probes used for Southern blot analysis. PGK, phosphoglycerokinase gene promoter; HSV-tk, herpes simplex virus thymidine kinase gene. (B) Genotyping of wild-type, heterozygous, and homozygous RGL mutant mice. Primers from the RGL and neo genes generated 1.2 and 0.9-kb fragments for the wild type (WT) and the RGL knockout (KO) gene.
Generation of RGL knockout mice.
Culture of ES cells and isolation of homologous recombinants were performed according to standard protocols (32). Briefly, 107 CCE, E14, or R1 ES cells were transfected with 25 to 50 μg of the linearized vector. The cells were cultured under selection in G418 and ganciclovir. Resistant colonies were expanded in 98-well plates. Screening for homologous recombinants was performed by PCR methods as described previously (32). Oligonucleotides 5′-TTCTATCGCCTTCTTGACGAGTTC-3′ (ADPR266; in the neo gene) and 5′-TCCATGATGAGTACTACATCCACT-3′ (ADPR267; in the RGL gene) were used as primers (Fig. 1). Seven positive clones were identified and subjected to Southern blot analysis to confirm the structure of the targeted region. ES cell genomic DNA was digested with PstI. The 5′ external probe and the internal probe shown in Fig. 1 and the neo gene were used as probes. All seven ES cell clones were confirmed to contain the targeted disruption and used to generate RGL knockout mice, by standard procedures (32), utilizing C57BL/6J blastocysts and pseudopregnant females as foster mothers. Chimeric mice were screened by PCR and by Southern blotting for the presence of the disrupted RGL allele and mated with C57BL/6J mice to produce F1 heterozygous mice. Heterozygotes for the disrupted gene were identified by PCR using digit samples. Confirmed heterozygous F1 mice were backcrossed with C57BL/6J mice or mated with siblings to generate null mutant mice. Some of the chimeric mice were also crossed with wild-type 129SvJ, and the resulting F1 heterozygotes were backcrossed with 129SvJ to generate genetically defined mice. All mice were maintained in temperature and humidity-controlled conditions with a 12-h light–12-h dark cycle and were allowed food and water ad libitum. The RGL chimeric mice were generated by David A. Williams at Indiana School of Medicine, Indianapolis, and by Cheryl Bock at Duke Comprehensive Cancer Center, Transgenic Facility, Durham, N.C.
Southern blot analysis.
Genomic DNA from ES cells was prepared with the Puregene DNA isolation kit. Genomic DNA was isolated from mouse tail by the proteinase K digestion method. Mouse tail pieces (1 to 2 cm) were clipped and digested in a solution containing 100 mM NaCl, 10 mM Tris-HCl (pH 7.8), 1% sodium dodecyl sulfate, 200 μg of proteinase K per ml at 57°C overnight. After two chloroform-isoamyl alcohol (24:1) extractions, the DNA was precipitated with ethanol. Genomic DNA (10 to 20 μg) digested with PstI (∼5 U/μg or more) was resolved by electrophoresis in 0.5% agarose gels and transferred to nitrocellulose membranes (BA-85; Shleicher & Schuell). DNA fragment probes were radiolabeled with 32P using the Megaprime DNA labeling system (Amersham).
Western blot analysis.
For the various biochemical determinations, unless otherwise indicated, normally fed animals were sacrificed in the early afternoon. Mice were anesthetized by intraperitoneal injection of sodium pentobarbital (0.1 mg/g of body weight). Skeletal muscle of hind legs and heart were excised quickly, freeze-clamped in liquid nitrogen, and stored at −80°C till use. For Western blot analyses, frozen tissue samples were homogenized in 10 volumes (wt/vol) of a solution containing 50 mM Tris-HCl (pH 7.5) (25°C), 0.5 mM EDTA, 2 mM EGTA, 1% Triton X-100, 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone, 2 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride, and 10 μg of leupeptin per ml with a Tissue Tearer model 285–370 (Biospec Products Inc.) at maximal setting for 20 s. The homogenates were centrifuged at 600 × g for 5 min. The resultant supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins on the gels were transferred to nitrocellulose membranes which were then incubated with the appropriate antibodies. Binding of the antibody was detected either by 125I-labeled protein A or by horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence. Levels of protein expression were quantitated by densitometric scanning of the films.
Enzyme activity assays.
Ph phosphatase activity was assayed essentially as described previously (67) except that the total volume of the reaction mixture was reduced to 40 μl. The same supernatant samples used for the Western blot analysis were diluted 100-fold with a solution containing 50 mM Tris-HCl (pH 7.2), 0.2% β-mercaptoethanol, 0.1 mM EDTA, and 1 mg of bovine serum albumin per ml. Ten-microliter portions of the diluted samples were used for determination of phosphatase activity, utilizing 1 mg of [32P]Ph a per ml as a substrate in the presence or absence of 4 nM okadaic acid. One unit is defined as the amount of enzyme that releases 1 nmol of phosphate per min. For GS and Ph assays, tissue samples were homogenized in 20 volumes of buffer (50 mM Tris-HCl [pH 7.8], 10 mM EDTA, 2 mM EGTA, 100 mM NaF, 2 mM benzamidine, 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone, 50 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, and 10 μg of leupeptin per ml) using a Polytron homogenizer (Kinematica) for 20 s. After centrifugation at 3,600 × g for 5 min, 30 μl of the supernatant was used for GS and Ph assays. GS activity was determined by measuring incorporation of [14C]glucose from UDP-[14C]glucose into glycogen as described by Thomas et al. (61) in the absence or presence of 7.2 mM G-6P. Ph activity was assayed by measuring incorporation of [14C]glucose from [14C]glucose-1-phosphate (28) into glycogen in the absence or presence of 2 mM AMP. One unit of GS is the amount of enzyme that incorporates 1 μmol of [14C]glucose from UDP-[U-14C]glucose into glycogen per min, and 1 U of Ph is the amount of enzyme that incorporates 1 μmol of [14C]glucose from [U-14C]glucose-1-phosphate per min. Activity ratios represent the activity measured in the absence of the allosteric effector (G-6P for GS or AMP for Ph) divided by that in the presence of the allosteric effector.
For determination of the effects of insulin on GS, Ph, and phosphatase activities and glycogen content, anesthetized 17 to 20-week-old RGL null mutant mice and wild-type littermates fasted for 6 h (8:00 a.m. till 2:00 p.m.) were injected in the tail vein with 2 U of insulin/kg of body weight. After 5 min, animals were sacrificed, and skeletal muscle was rapidly removed and frozen in liquid nitrogen. GS phosphatase activity was measured by utilizing partially purified phosphorylated GS and by monitoring the change in the GS −/+ G-6P activity ratio between zero time and after 15 min of incubation with muscle extract, following the procedure described by Villar-Palasi (62). Responsiveness of the animals to insulin was confirmed by determination of blood glucose levels before and after insulin administration. Under the conditions used, the blood glucose decreased by ∼40% in both groups. Protein concentration was determined by the Bradford method (14) using bovine serum albumin as standard.
Determination of glycogen content.
Samples of frozen tissues (30 to 90 mg) were hydrolyzed in 0.3 ml of 30% (wt/vol) KOH solution in a boiling water bath for 30 min. At 10 and 20 min of the incubation, tubes were shaken by hand to facilitate the digestion. After cooling to room temperature, 0.1 ml of 1 M Na2SO4 and 0.8 ml of ethanol were added, the samples were boiled again for 5 min to facilitate precipitation of glycogen and then centrifuged at 10,000 × g for 5 min. The glycogen pellet was dissolved in 0.2 ml of water, and two additional ethanol precipitations were performed. The final pellet was dried and dissolved in 0.2 ml of 0.3-mg/ml amyloglucosidase in 0.2 M sodium acetate buffer (pH 4.8) and incubated for 3 h at 40°C. The reaction mixture was then diluted two- to fivefold with water. To determine the glucose concentration, 5 μl of the diluted sample was added to 0.2 ml of the glucose assay solution which contains 0.3 M triethanolamine–KOH (pH 7.5), 1 mM ATP, 0.9 mM β-NADP, and 5 μg of G-6P dehydrogenase per ml. The absorbance at 340 nm was determined before and after addition of 1 μg of hexokinase (11). Glycogen content was expressed as micromoles of glucosyl units per gram (wet weight).
Glucose and insulin tolerance tests.
Glucose and insulin tolerance tests were performed on homozygous null mutant mice and wild-type littermates at different ages, 6 to 8, 14 to 18, and 40 to 48 weeks. For glucose tolerance tests, animals were fasted for 16 h. Glucose (2 mg/g of body weight) was administered intraperitoneally. Blood samples were collected from the tail at various times, and blood glucose was measured using a Glucometer Elite (Ames). For insulin tolerance tests, 6-h-fasted animals were injected intraperitoneally with 1 mU of porcine insulin per g. Blood glucose levels were monitored as described above for the glucose tolerance test.
Other procedures.
2-Deoxyglucose uptake in mouse diaphragm muscle was measured as previously described (57). Whole-body glucose disposal studies in euglyemic hyperinsulinemic mice were performed by Alain Baron at Indiana University, as previously described (30). Serum lactate and triglyceride levels were measured in the Immunology Core Facility of the Diabetes Research and Training Center at Indiana University.
Statistical analyses.
Statistical analyses were performed using the StatView program. Significance was assessed using the unpaired Student's t test. Results are expressed as the mean ± standard error (SE) of the number of animals indicated in the figure legends.
RESULTS
Generation of RGL knockout mice.
The strategy to generate RGL knockout mice was to disrupt the gene by replacing exon 1 which codes for the first 261 residues (785 bp) with the neomycin resistance gene (Fig. 1A). By this approach, the translational initiation codon, the C1 and glycogen-binding domains, and phosphorylation sites Ser48 and Ser67 would be removed. Even if exon 1 were skipped, the resulting mRNA starting at exon 2 would be out of frame. Furthermore, since intron 1 is far removed from the promoter (more than 27 kb), it is unlikely that a transcript could be expressed. A screen of 195 resistant cell clones gave 7 PCR positives, all of which were confirmed by Southern blotting. All seven lines were used to generate knockout mice. Twenty-four chimeras were obtained, 12 of which had high percentage of agouti coat color (75 to 100%) and were positive for the disruption by PCR analysis. Nine were males, and mating with C57BL/6 mice revealed that seven were fertile, six of which transmitted the targeted allele to their offspring. Genotyping by PCR shows a 900-bp amplified DNA fragment for the disrupted allele and a 1,235-bp fragment for the wild type (Fig. 1B). Southern blots of PstI digests indicated an 18-kb fragment for the disrupted allele and a 26-kb fragment for the wild type (data not shown). Mating of animals heterozygous for the disrupted RGL allele has generated homozygous null mutant mice.
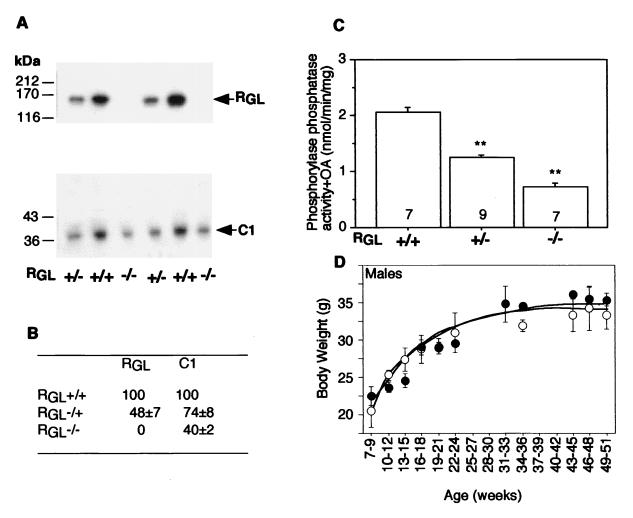
Analysis of 167 F2 mice showed that the percentages of wild-type (+/+), heterozygous (+/−), and homozygous null (−/−) RGL mice were 32, 45, and 23%, respectively. No significant difference in this ratio was observed between males and females, indicating that the RGL gene is located on an autosome and that the knockout is not lethal in utero. All animals, heterozygotes as well as homozygotes, macroscopically appeared normal with no obvious developmental or morphological defects and grew at a rate indistinguishable from that of wild-type littermates (Fig. 2D). The heart to body weight ratios are also within the normal range.
FIG. 2.
Analysis of expression of RGL and C1 in wild-type and knockout mice. (A) Skeletal muscle extracts were subjected to immunoblotting with anti-RGL (top) or anti-C1 (bottom) antibodies. (B) Quantitation by densitometric scanning of the autoradiogram shown in panel A, expressed as a percentage of the mean of the wild type ± SE (n = 6 to 8 per group). (C) Ph phosphatase activity was measured in the presence of 4 nM okadaic acid (OA) in skeletal muscle extracts as described in Materials and Methods. The number of animals in each group is indicated in each bar. Statistical significance was assessed by Student's t test (∗∗, P < 0.01). (D) Growth curves of wild-type (open circles) and RGL null mutant (closed circles) mice with 3 to 19 animals per point. The means ± SE are shown. +/+, wild type; +/−, heterozygotes, −/−, homozygote null mutants.
Analysis of PP1G/RGL expression in knockout mice and wild-type littermates.
Western immunoblotting analysis of skeletal (Fig. 2A and B) and cardiac (not shown) muscle extracts using mouse RGL antibodies indicated that the protein was absent from the homozygous animals and present at ∼50% of the wild-type level in heterozygotes. RGL-deficient mice had a 60% reduction in the level of C1 in skeletal muscle (Fig. 2A and B), and a significant 25% reduction was also observed in the heterozygous animals. These results indicate that the functional gene is disrupted, that PP1G/RGL accounts for a large proportion of PP1 in striated muscles, and that the presence of the regulatory RGL is linked to the expression level of C1. Consistent with reduced C1 protein, the type 1 phosphatase activity was also decreased, with activity towards Ph α reduced by ∼60% in the RGL null mice and by 30% in heterozygotes (Fig. 2C). The activity towards GS phosphorylated in the site 3 region was decreased by 40% in the homozygotes. Although not as pronounced, reductions in RGL, C1, and phosphatase activity were also observed in heart (data not shown), consistent with the ∼10-fold-lower level of expression of RGL in this tissue (37, 60).
Effect of ablation of PP1G/RGL on glycogen metabolism.
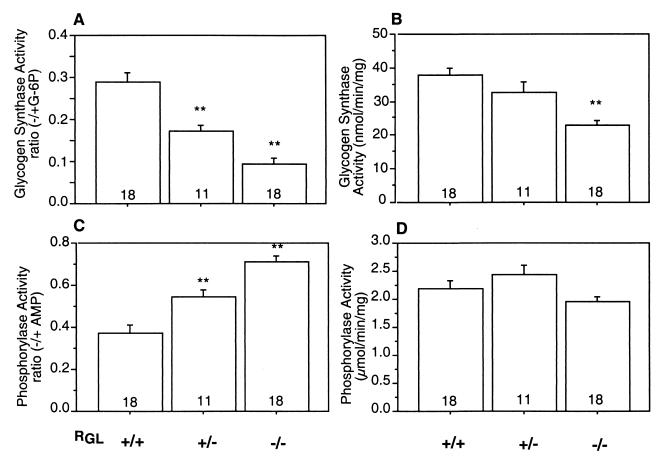
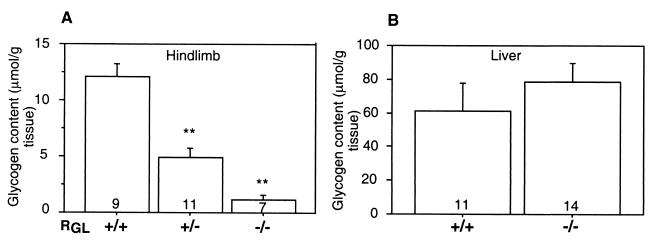
To determine the effect of PP1G/RGL deficiency on the glycogen-metabolizing enzymes, GS and Ph activities were measured in skeletal muscle extracts. The GS −/+ G-6P activity ratio was reduced from 0.3 in the wild-type littermates to 0.1 in the RGL null mutants and to 0.17 in the heterozygotes (Fig. 3A). Conversely, the activity state of Ph, measured as the activity ratio in the absence and presence of AMP, increased from 0.37 in the wild type to 0.7 in the homozygous mutant mice and to 0.54 in the heterozygotes (Fig. 3C). The total GS activity, as determined by activity measured in the presence of G-6P, was also reduced by ∼30% (Fig. 3B), as was the protein level monitored by Western immunoblotting. The total Ph activity, measured in the presence of AMP, was unaffected in both heterozygous and homozygous mutants (Fig. 3D). Similar effects were observed in five independent knockout lines of mixed BL57 and 129Sv background and in both male and female animals, indicating that there are no gender differences. Consistent with the altered GS and Ph activities, the glycogen content was dramatically decreased in the null mutant mice of both genetic backgrounds to ∼10% of the wild-type level and to ∼40% in the heterozygous littermates (Fig. 4A). Similar changes in GS and Ph activities and glycogen levels were observed in null mice generated from F3 heterozygotes obtained after backcrossing the chimeras and subsequent generations with 129Sv mice, suggesting that the effects of the RGL ablation are independent of the genetic background. PP1/RGL is therefore a significant GS and Ph phosphatase, determining the phosphorylation state and activity of the glycogen-metabolizing enzymes. The extremely low glycogen level in the RGL null mutant may indirectly be responsible for the reduced amount of GS protein. All the GS in skeletal muscle is bound to glycogen whose low content in the knockout mice may limit GS association, leading to lower stability and enhanced degradation. It is interesting though, that even in the RGL null mutants, all of the GS present was bound to glycogen. In contrast to GS, only 50% of the Ph in the muscle is bound to glycogen, so that its stability may be less dependent on association with the polysaccharide.
FIG. 3.
GS and Ph activity in skeletal muscle of wild-type and RGL knockout mice (A) GS activity was assayed in the absence (−) and presence (+) of G-6P in supernatant (after centrifugation at 3,600 × g for 5 min) of skeletal muscle from wild-type (+/+), RGL heterozygous (+/−), and homozygous (−/−) mice. (B) Total GS activity measured in the presence of 7.2 mM G-6P, expressed as nanomoles per minute per milligram of protein. (C) Ph activity was assayed in the absence (−) and presence (+) of 2 mM AMP. (D) Total Ph activity measured in the presence of AMP, expressed as micromoles per minute per milligram of protein. The means ± SE are shown. Statistical significance was assessed by Student's t test (∗∗, P < 0.01). The number of animals analyzed is shown inside the bars.
FIG. 4.
Glycogen content in skeletal muscle and liver. Glycogen content in skeletal muscle (A) and liver (B) of wild-type (+/+), RGL heterozygous (+/−), and homozygous (−/−) mice was determined by an alkaline lysis method as described in Materials and Methods. Glycogen concentration is expressed as micromoles of glycosyl units per gram (wet weight). The means ± SE are shown. Statistical significance was assessed by Student's t test (∗∗, P < 0.01). The number of animals analyzed is shown inside the bars.
Since glycogen accumulation was impaired in skeletal muscle, its level in the liver was measured to determine whether this organ might have been compensating. As shown in Fig. 4B, the glycogen levels in the livers of the null mutants were not significantly different from those of the wild-type mice.
Glucose and insulin responsiveness in RGL knockout mice.
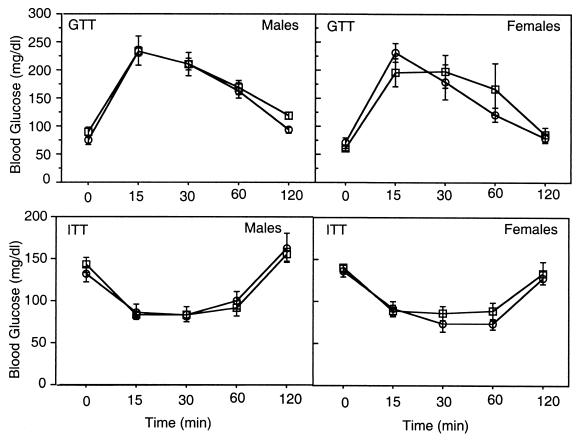
Since ablation of RGL impairs glycogen accumulation in skeletal muscle, reportedly a major organ for glucose disposal, a critical question was whether the RGL null mutant animals were hyperglycemic and/or insulin resistant. Three approaches were taken to address this issue. First, blood glucose levels were measured in both fasted and fed male and female mice of different ages. As shown in Fig. 5 and Table 1, the basal blood glucose levels for 14- to 18-week-old mice were, as expected, lower after overnight fasting, but no significant differences were detected between wild-type and knockout mice, in either males or females. Similar results were obtained with animals 6 to 8 or 40 to 48 weeks old (not shown). Second, we evaluated the glucose and insulin tolerance after challenging with a bolus of either glucose (2 mg/g) or insulin (1 U/kg). The time course of blood glucose clearance after glucose administration was indistinguishable between the wild-type and knockout mice (Fig. 5). Similarly, the blood glucose response to insulin treatment was the same in wild-type littermates and knockout animals. Again these results were the same for males and females and for animals of different ages and with a 129Sv background (not shown). Third, the basal and insulin-stimulated whole-body glucose disposal in euglycemic hyperinsulinemic clamped conscious mice (30) was very similar in knockout and wild-type animals. In addition, no significant differences were observed in serum insulin, lactate, and triglyceride levels (Table 1).
FIG. 5.
Glucose and insulin tolerance tests. Glucose tolerance tests (GTT) were performed on 14- to 18-week-old 16-h-fasted, male and female wild-type (open circles) and RGL null mutant (open squares) mice. Insulin tolerance tests (ITT) were performed a week later on the same mice except that they were fasted only for 6 h from 8:00 a.m. till 2:00 p.m. Six wild-type and eight null mutant males and four wild-type and 4 null mutant females were used.
TABLE 1.
Comparison of blood component levels in wild-type and RGL knockout micea
| Blood component level | Wild-type littermates
|
RGL null mutants
|
||
|---|---|---|---|---|
| Male | Females | Male | Female | |
| Blood glucose, fasted (mg/dl) | 74.8 ± 7.3 | 70.0 ± 9.7 | 88.9 ± 9.4 | 60.5 ± 4.7 |
| Blood glucose, fed (mg/dl) | 131.3 ± 9.4 | 135.8 ± 7.0 | 142.8 ± 9.0 | 140.0 ± 4.9 |
| Serum insulin (μg/ml) | 0.72 ± 0.14 | 0.46 ± 0.07 | 0.62 ± 0.11 | 0.46 ± 0.10 |
| Serum lactate (mM) | 5.93 ± 0.86 | 4.77 ± 0.63 | 5.90 ± 1.49 | 5.16 ± 0.45 |
| Serum triglycerides (mg/dl) | 94.4 ± 5.3 | 76.5 ± 12.6 | 98.5 ± 15.4 | 88.0 ± 9.3 |
Mice were 12 to 24 weeks old. The number of animals per group was 4 to 8. No significant differences were observed between the values for comparable groups of wild-type and knockout animals using Student's t test.
Although the older mice had not demonstrated any obvious metabolic defects when fed a normal diet, we investigated the possibility that a high-fat diet could provoke abnormalities in glucose disposal. Both male and female, wild-type and knockout mice were subjected for a period of 20 weeks to a high-fat diet (Biosev) in which 57% of the caloric intake is derived from fat. Body weight, blood glucose levels, and food consumption were monitored weekly. Weight gain (∼50%), increases in blood glucose levels (∼30%), and food intake (no significant increase) were not different between the knockout and wild-type littermate animals. Glucose and insulin tolerance curves at the end of the feeding period were similar in both groups, although the response was blunted compared to that before the high-fat diet, indicating that all animals had become somewhat insulin resistant.
Insulin control of glycogen metabolism in RGL knockout mice and wild-type littermates.
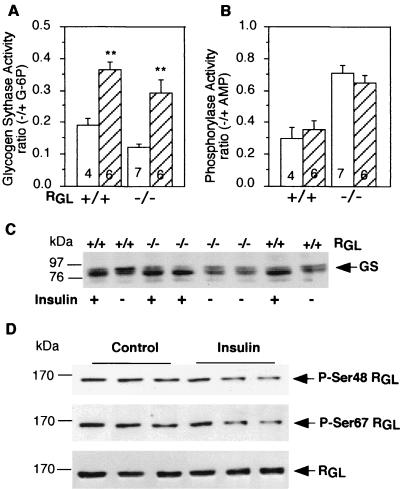
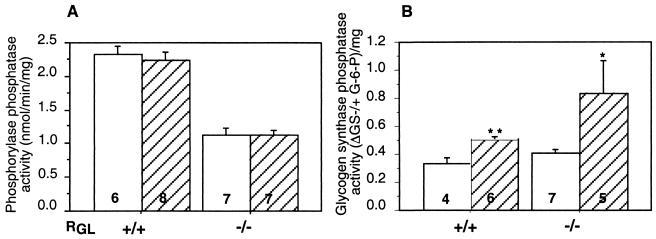
Muscle PP1G has been postulated to play a key role in the insulin-induced activation of glycogen synthase. In order to address this crucial issue, wild-type and RGL knockout mice were injected intravenously with 2 U of insulin/kg for 5 min. Insulin treatment resulted in a twofold activation of skeletal muscle GS in both control and RGL-deficient mice (Fig. 6A), with no changes in glycogen Ph (Fig. 6B). Glycogen content was also increased more than twofold in the knockout mice (Fig. 7A). In absolute amount, the increases in glycogen in wild-type and mutant mice were very similar. In the wild-type animals, the final level (including the relatively small increment) was not statistically significantly different from the high initial level. It should be noted that such a short time of insulin stimulation would not cause large changes in glycogen accumulation. Western immunoblotting also showed that insulin treatment increased the electrophoretic mobility of GS in muscle extracts from wild-type and RGL-deficient mice, consistent with dephosphorylation of the protein (Fig. 6C). This finding confirms that the increase in GS activity was not due to carryover of allosteric effector G-6P from the extract into the assay (which involves more than 100-fold dilution). From these results, it is clear that both GS and glycogen syntheses are still stimulated by insulin in the absence of RGL. Even though RGL was not required for insulin activation of GS, we investigated whether it was phosphorylated in response to the hormone. Western immunoblotting with phospho-specific antibodies against phosphorylated site 1 or site 2 (63) revealed that the phosphorylation of neither was altered by insulin treatment, at times when GS was activated (Fig. 6D). Quantitation of blots from five animals in each group gave values of 89.7 ± 9.2 (site 1) or 88.1 ± 9.5 (site 2), expressed as a percentage of the non-insulin-treated samples normalized for total RGL present.
FIG. 6.
GS and Ph activity in insulin-treated mice. (A and B) GS and Ph activity ratios were measured in skeletal muscle extracts as described in the legend to Fig. 3. The mice were treated with insulin (hatched bars) or not treated with insulin (open bars). Statistical significance was assessed by Student's t test (∗∗, P < 0.01). The number of animals analyzed is shown inside the bars. (C) Muscle extracts from control and insulin-treated mice were immunoblotted with anti-GS antibodies. (D) Muscle extracts from wild-type mice, control and insulin treated, were immunoblotted with antibodies that specifically recognize RGL phosphorylated at Ser48 (P-Ser48), RGL phosphorylated at Ser67 (P-Ser67), or nondiscriminating antibodies (RGL). Three representative independent samples per treatment are shown. +/+, wild type; −/−, RGL null mutant mice.
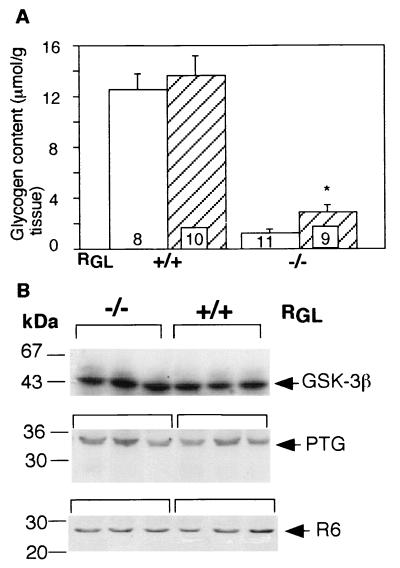
FIG. 7.
Insulin-stimulated glycogen accumulation and GSK-3, PTG, and R6 levels in wild-type and RGL knockout mice. (A) Glycogen content was measured as described in the legend to Fig. 4 in wild-type (+/+) and RGL homozygous knockout (−/−) mice with (hatched bars) and without (open bars) insulin treatment. The number of experiments is indicated inside the bars. Statistical significance was assessed by Student's t test (∗, P < 0.05). (B) Three representative samples from wild-type and RGL knockout mice were analyzed by immunoblotting with antibodies against GSK-3β, PTG, and R6. The numbers to the left show molecular size markers (in kilodattons).
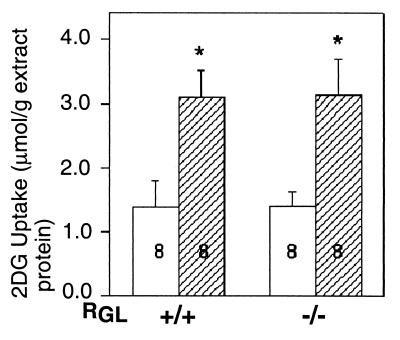
Since the RGL knockout mice responded normally to insulin stimulation, one possibility is altered expression of some of the other components of the insulin signaling pathway, such as GSK-3, or the other two known glycogen-associated PP1-binding proteins, PTG and R6. As shown in Fig. 7B, Western blot analyses indicated that the levels of PTG, R6, and GSK-3β were unchanged in the knockout animals. It has also been reported that overexpression of RGL in L6 cultured cells resulted in a twofold increase in insulin-stimulated glucose transport (50). We therefore investigated whether the RGL deficiency affected glucose uptake by in vitro analysis of isolated hemidiaphragm muscles. The results revealed no differences in basal and insulin-stimulated 2-deoxyglucose uptake between wild-type and knockout mice (Fig. 8).
FIG. 8.
Insulin stimulation of 2-deoxy[2-3H]glucose (2DG) uptake in hemidiaphragms from control and RGL knockout mice. Hemidiaphragms were incubated at 37°C for 15 min with or without 250 mU of insulin per ml. The hemidiaphragms were then incubated with insulin (hatched bars) or without insulin (open bars) in medium containing 5 mM 2DG for 10 min. The results presented are the mean values ± SE of eight experiments. Statistical significance was assessed by Student's t test (∗, P < 0.02).
Insulin-stimulated GS phosphatase activity in wild-type and RGL knockout mice.
If RGL is not required for insulin activation of GS, some other mechanism must be operating. Activation of GS could result from inhibition of protein kinase activity or activation of a GS phosphatase distinct from PP1G/RGL. Decreased GSK-3 activity, by itself, cannot account for activation of GS (39), so we determined whether an insulin-stimulated phosphatase was present in RGL knockout mice. With Ph as a substrate, no increase in activity was detected in either mutant mice or wild-type littermates (Fig. 9A). However, utilizing partially purified GS in an assay that couples dephosphorylation with changes in the activity ratio (62), we detected a phosphatase activity that was stimulated ∼2-fold by insulin in both wild-type and RGL knockout mice (Fig. 9B). Not only is PP1G/RGL dispensable for insulin activation of GS, but a distinct insulin-activated phosphatase is present in mouse skeletal muscle. This enzyme is not PP2A, since it was detectable in the presence of 4 nM okadaic acid, conditions that completely inhibit 2A activity. The phosphatase is unlikely to be a divalent cation-requiring enzyme such as PP2C or PP2B, because the extracts and the assays contained 2 mM EGTA. Thus, we believe that it is a type 1 form. It is also unlikely to be the PTG-associated phosphatase, since this enzyme acts on Ph as substrate (15), whereas the insulin-stimulated form appears to be specific for GS.
FIG. 9.
Insulin-stimulated GS phosphatase. (A) Ph phosphatase in skeletal muscle extracts of non-insulin-treated (open bars) and insulin-treated (hatched bars) mice was measured as described in the legend to Fig. 4. (B) GS phosphatase was measured by the dephosphorylation-activation coupled assay as described in Materials and Methods. The results are expressed as the difference (Δ) between the GS −/+ G-6P activity ratios measured after 15 min of incubation of GS with tissue extract and that at zero time, normalized by protein concentration. Statistical significance was assessed by Student's t test (∗, P < 0.05; ∗∗, P < 0.01). The number of animals analyzed is indicated inside the bars.
DISCUSSION
The most important outcome of this study is to demonstrate that RGL is genetically linked to striated muscle glycogen accumulation but is not essential for insulin control of glycogen metabolism. Analyses of the knockout mice demonstrate that RGL directs phosphatase activity towards glycogen-metabolizing enzymes since GS activity is reduced and Ph activity is enhanced in the null mutant mice. Correspondingly, muscle glycogen stores are significantly reduced to only 10% of those in wild-type animals. However, despite the impaired glycogen accumulation, the animals remain normoglycemic and are insulin responsive.
Animals homozygous for disruption of the RGL gene have a substantial (60%) decrease in overall muscle C1 protein and PP1 activity, indicating that a phosphatase containing RGL represents a significant proportion of the enzyme in skeletal muscle. Conversely, targeted overexpression of RGL in the skeletal muscle of mice resulted in an increase in the levels of C1 protein (22; Y. Suzuki and A. A. DePaoli-Roach, unpublished results). These observations argue that the basal level of C1 protein depends on its association with regulatory subunits. Although the level of the protein can be controlled at different stages, we favor a mechanism involving stabilization of the protein. This hypothesis is supported by the observation that overexpression of C1 in COS1 cells produces mostly insoluble protein associated with lysosomal structures, whereas coexpression with either RGL or inhibitor 2 (data not shown) generates higher levels of soluble and active enzyme. In fact, the proposed chaperone function of the phosphatase inhibitor 2 (3) most likely reflects a similar phenomenon whereby formation of a cytosolic form of PP1, the ATP-Mg-dependent phosphatase, also stabilizes C1.
Our results raise some critical questions about the role of muscle glycogen in blood glucose homeostasis (19, 56). Glycogen deposition in muscle is widely perceived to be the primary fate of ingested glucose. With this premise, one might have predicted that animals impaired in their ability to accumulate glycogen in muscle would become hyperglycemic. This is not the case, suggesting that other mechanisms for glucose homeostasis must be operating. One important consideration, however, is the degree to which studies of mice are an accurate reflection of human metabolism and physiology, and we cannot exclude the possibility that skeletal muscle glycogen accumulation in humans has a different impact on blood glucose levels than in rodents. This caveat, of course, is valid for all of the numerous genetically altered mouse models that have recently been constructed to probe insulin action and whole-body glucose metabolism (for recent reviews see references 35 and 36). Shulman and colleagues (56) in fact have concluded that muscle glycogen is the predominant fate of ingested glucose in humans, at least under the conditions of hyperglycemic hyperinsulinemic clamps, and that impaired glycogen synthesis correlates with type 2 diabetes. They are also of the opinion that glucose transport is the rate-limiting step for glycogen accumulation, which is defective in the diabetic condition. The RGL knockout mice, however, have normal basal and insulin-stimulated glucose transport but reduced glycogen content supporting the view that the glycogen-metabolizing enzymes play critical roles in glycogen accumulation (8, 39). It will be interesting to examine mouse models in which muscle glycogen accumulation is completely defective or in which insuling signaling to GS is specifically ablated. For example, genetic elimination of the novel insulin-stimulated GS phosphatase described in this work might achieve the latter goal.
Glucose is not being accumulated as excess fat, since lean and fat masses as determined by dexascan are similar in wild-type and RGL knockout mice, as is the respiratory quotient (VCO2/VO2) over a 24-h period (data not shown). Liver glycogen is not elevated, so the liver is not compensating. Since food consumption is no different between wild-type and RGL null animals, a simplistic question relates to the fate of the ingested calories. However, the apparent paradox is minimized if one considers that the caloric equivalent of the muscle glycogen in the mouse is less than 2% of a daily ∼15-kcal intake, based on a glycogen content of 2.5 mg/g for a 25-g mouse. Therefore, muscle glycogen in mice does not actually represent a significant caloric reserve compared to food consumption. Glycogen is continuously used for local contractile activity and is replaced by incoming glucose, directly after meals, or indirectly from the liver via glycogenolysis or gluconeogenesis during fasting. The RGL null mutant mice appear to have a lower glycogen set point, consistent with GS being inactivated and Ph activated. As demonstrated by the normal basal and insulin-stimulated 2-deoxyglucose uptake, glucose enters the cells normally and presumably is stored as glycogen. However, hyperactive Ph may cause rapid degradation, resulting in a lower steady-state glycogen concentration. Thus, insulin stimulation promotes glucose uptake normally in the RGL knockout mice, and the overall lower glycogen content is most likely due to the altered activities of GS and/or Ph.
The studies with the RGL-deficient mice clearly show that the muscle-specific PP1G, although important for basal glycogen accumulation, is dispensable for insulin-induced activation of GS. Together with our finding of an insulin-stimulated phosphatase both in the knockout and wild-type animals, there is compelling evidence that an enzyme distinct from PP1G/RGL is responsible for activation of GS. Thus, the mechanism of an insulin-induced phosphorylation of RGL and activation of the associated phosphatase can be discounted, consistent with a recent report (63) that in rat skeletal muscle phosphorylation of Ser48 is not increased in response to insulin. Furthermore, since our initial communication (22) that GS was activated by insulin in RGL-deficient mice, other reports argue against a role for RGL in insulin control of glycogen metabolism (43, 66). Our studies are not in agreement with the report that overexpression or depletion of RGL in L6 cells affected activation of GS and glucose uptake by insulin (50). The observation that neither insulin activation of GS nor basal or insulin-stimulated glucose uptake is altered in the RGL null mice indicates that PP1G is not involved in regulation of glucose transport. Rasmussen et al. (51) were also unable to reproduce the effect of overexpressed RGL protein on glucose transport.
Recently it has been proposed that RGL acts as a scaffold that binds both GS and C1 and that this association is required for GS activation and for β-adrenergic control of GS (42). Our animal studies show that activation of GS can occur in the absence of RGL, indicating that association with RGL is not essential for control of GS activity in vivo. Furthermore, even in the RGL null mutant muscle, all the GS is still bound to glycogen implying that RGL is not needed for this association.
The work presented suggests that a phosphatase distinct from PP1G/RGL mediates insulin activation of GS. Although the identity of this insulin-stimulated phosphatase is not known, it is likely to be a novel enzyme. We can exclude the possibility that it is a type 2A, 2B, or 2C enzyme, since it was detected in the presence of 4 nM okadaic acid and 2 mM EGTA, conditions that would inhibit the activity of those forms. Preliminary data indicate that the phosphatase activity was not detectable in the presence of 200 nM okadaic acid, suggesting that it is a type 1 enzyme. Obvious candidates would be enzymes containing the other two glycogen-binding subunits, PTG and R6. No regulatory mechanisms for these proteins have been identified so far. Overexpression of PTG increased both basal and insulin-stimulated GS activity (49). However, we consider it unlikely that the insulin-stimulated phosphatase activity we detected is associated with PTG or R6, because their expression is not restricted to insulin-responsive tissues. In addition and most importantly, insulin was shown to stimulate the PTG phosphatase activity towards Ph (15), whereas the insulin-stimulated phosphatase we identified appears to be specific for GS. Insulin does not affect phosphorylase activity, consistent with the inability to detect stimulation of phosphatase activity towards Ph (Fig. 9). Neither PTG nor R6 functionally compensates for the loss of RGL, since loss of RGL results in low basal muscle glycogen storage and PTG and R6 protein levels are not upregulated in the knockout mice. The implication is that the various glycogen-associated phosphatases are not redundant, even though PTG and R6 are present in skeletal muscle at ∼10% of the level of RGL (37, 60). Therefore, the insulin-stimulated protein phosphatase may be a form of PP1, involving association of C1 with some other regulatory subunit, either novel or already known in some other context.
Several studies over the last few years have attempted to link mutations in the human RGL (PPP1R3) gene with type 2 diabetes. However, the frequency of the initially reported polymorphism at codon 905 (29) was shown to be identical in normal and diabetic populations. More recently, adenovirus-mediated expression of Asp905 and Tyr905 human RGL in L6 cells indicated no differences in glycogen synthesis (51). Another substitution at codon 883 and a variant in an ATTTA motif in the 3′ untranslated region of the human RGL gene have also been found (66). Polymorphisms in the ATTTA element appear to correlate with lower expression of the protein, insulin resistance, and type 2 diabetes (65). At least on the basis of our studies showing that mice lacking RGL are neither diabetic nor insulin resistant, one might question whether subtle modifications of the human protein would cause an aberrant phenotype. Most notably, the small reduction in the level of RGL observed in subjects carrying the ATTTA spacing polymorphism is unlikely to be responsible for the defective insulin responses associated with diabetes. Similarly, it is not likely that putative alterations of RGL function in the other coding variants are critically involved in the etiology of type 2 diabetes.
If RGL is not required for insulin activation of GS or glucose transport, a key question is whether it is involved in other regulatory processes. Is it necessary or involved in the β-adrenergic control of glycogen metabolism? A main function of glycogen in skeletal muscle is to provide fuel for contraction. Since the glycogen content is extremely low in the RGL knockout mice, one can ask whether these animals will be able to sustain exercise and whether RGL is involved in the neuronal activation of GS, such as occurs during contraction and exercise. RGL has also been proposed to control β-adrenergic-induced phosphorylation of phospholamban in heart (45, 63). Since this phosphorylation reduces the inhibitory effect of phospholamban on the sarcoplasmic reticulum-associated Ca2+-ATPase (34), it may be that the RGL knockout mice have an increased cardiac performance, a phenotype observed in the phospholamban null mice (44). Work is in progress to address these important and interesting questions.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health Grant DK36569 and by a research grant from the American Diabetes Association.
We thank Suzanne Baugh and Paula Ladd for their technical assistance in some of the work presented. We are indebted to David A. Williams and Cheryl Bock for generating the RGL chimeric mice. We are grateful to Alain Baron (Amylin San Diego, Calif.) and Mark Heiman (Eli Lilly Co.) for performing glucose clamping and determining body composition and metabolic rates, respectively. We thank Alan Saltiel (Pfizer Global Research and Development), Patricia Cohen and Philip Cohen (University of Dundee) for their generous gifts of PTG, R6, and phospho-RGL antibodies, respectively. We are particularly grateful to Peter J. Roach and Robert A. Harris for discussion of the work and for criticism of the manuscript.
REFERENCES
- 1.Alemany S, Cohen P. Phosphorylase a is an allosteric inhibitor of the glycogen and microsomal forms of rat hepatic protein phosphatase-1. FEBS Lett. 1986;198:194–202. doi: 10.1016/0014-5793(86)80404-5. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D, MacDougall L K, Sola M M, Ikebe M, Cohen P. The control of protein phosphatase-1 by targetting subunits. The major myosin phosphatase in avian smooth muscle is a novel form of protein phosphatase-1. Eur J Biochem. 1992;210:1023–1035. doi: 10.1111/j.1432-1033.1992.tb17508.x. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D R, Street A J, Cohen P, Cohen P T. Inhibitor-2 functions like a chaperone to fold three expressed isoforms of mammalian protein phosphatase-1 into a conformation with the specificity and regulatory properties of the native enzyme. Eur J Biochem. 1993;213:1055–1066. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 4.Allen P B, Kwon Y G, Nairn A C, Greengard P. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J Biol Chem. 1998;273:4089–4095. doi: 10.1074/jbc.273.7.4089. [DOI] [PubMed] [Google Scholar]
- 5.Allen P B, Ouimet C C, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong C G, Browne G J, Cohen P, Cohen P T. PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1. FEBS Lett. 1997;418:210–214. doi: 10.1016/s0014-5793(97)01385-9. [DOI] [PubMed] [Google Scholar]
- 7.Askew G R, Doetschman T, Lingrel J B. Site-directed point mutations in embryonic stem cells: a gene-targeting tag-and-exchange strategy. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azpiazu I, Manchester J, Skurat A V, Roach P J, Lawrence J C., Jr Control of glycogen synthesis is shared between glucose transport and glycogen synthase in skeletal muscle fibers. Am J Physiol Endocrinol Metab. 2000;278:E234–E243. doi: 10.1152/ajpendo.2000.278.2.E234. [DOI] [PubMed] [Google Scholar]
- 9.Azpiazu I, Saltiel A R, DePaoli-Roach A A, Lawrence J C. Regulation of both glycogen synthase and PHAS-I by insulin in rat skeletal muscle involves mitogen-activated protein kinase-independent and rapamycin-sensitive pathways. J Biol Chem. 1996;271:5033–5039. doi: 10.1074/jbc.271.9.5033. [DOI] [PubMed] [Google Scholar]
- 10.Barford D, Das A K, Egloff M P. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 11.Bergmeyer H U, Berndt E, Schmidt F, Stork H. Determination with hexakinase and glucose-6-phosphate dehydrogenase. In: Bergmeyer H U, editor. enzymatic analysis. 2nd ed. Vol. 3. New York, N.Y: Academic Press; 1974. pp. 1196–1201. [Google Scholar]
- 12.Berman H K, O'Doherty R M, Anderson P, Newgard C B. Overexpression of protein targeting to glycogen (PTG) in rat hepatocytes causes profound activation of glycogen synthesis independent of normal hormone- and substrate-mediated regulatory mechanisms. J Biol Chem. 1998;273:26421–26425. doi: 10.1074/jbc.273.41.26421. [DOI] [PubMed] [Google Scholar]
- 13.Beullens M, Van Eynde A, Bollen M, Stalmans W. Inactivation of nuclear inhibitory polypeptides of protein phosphatase-1 (NIPP-1) by protein kinase A. J Biol Chem. 1993;268:13172–13177. [PubMed] [Google Scholar]
- 14.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Brady M J, Bourbonais F J, Saltiel A R. The activation of glycogen synthase by insulin switches from kinase inhibition to phosphatase activation during adipogenesis in 3T3–L1 cells. J Biol Chem. 1998;273:14063–14066. doi: 10.1074/jbc.273.23.14063. [DOI] [PubMed] [Google Scholar]
- 16.Brady M J, Nairn A C, Saltiel A R. The regulation of glycogen synthase by protein phosphatase 1 in 3T3–L1 adipocytes. Evidence for a potential role for DARPP-32 in insulin action. J Biol Chem. 1997;272:29698–29703. doi: 10.1074/jbc.272.47.29698. [DOI] [PubMed] [Google Scholar]
- 17.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 18.Cross D A, Watt P W, Shaw M, van der Kaay J, Downes C P, Holder J C, Cohen P. Insulin activates protein kinase B, inhibits glycogen synthase kinase-3 and activates glycogen synthase by rapamycin-insensitive pathways in skeletal muscle and adipose tissue. FEBS Lett. 1997;406:211–215. doi: 10.1016/s0014-5793(97)00240-8. [DOI] [PubMed] [Google Scholar]
- 19.DeFronzo R A, Jacot E, Jequier E, Maeder E, Wahren J, Felber J P. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 20.Dent P, Lavoinne A, Nakielny S, Caudwell F B, Watt P, Cohen P. The molecular mechanism by which insulin stimulates glycogen synthesis in mammalian skeletal muscle. Nature. 1990;348:302–308. doi: 10.1038/348302a0. [DOI] [PubMed] [Google Scholar]
- 21.DePaoli-Roach A A, Park I K, Cerovsky V, Csortos C, Durbin S D, Kuntz M J, Sitikov A, Tang P M, Verin A, Zolnierowicz S. Serine/threonine protein phosphatases in the control of cell function. Adv Enzyme Regul. 1994;34:199–224. doi: 10.1016/0065-2571(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 22.DePaoli-Roach A A, Suzuki Y, Lanner C, Zhang H, Scrimgeour A, Lawrence J C. Glycogen metabolism in transgenic mice overexpressing or deficient in the glycogen/sarcoplasmic reticulum-associated phosphatase, PP1G. Diabetes. 1999;48:A25. [Google Scholar]
- 23.Doherty M J, Cadefau J, Stalmans W, Bollen M, Cohen P T. Loss of the hepatic glycogen-binding subunit (GL) of protein phosphatase 1 underlies deficient glycogen synthesis in insulin-dependent diabetic rats and in adrenalectomized starved rats. Biochem J. 1998;333:253–257. doi: 10.1042/bj3330253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doherty M J, Moorhead G, Morrice N, Cohen P, Cohen P T. Amino acid sequence and expression of the hepatic glycogen-binding (GL)-subunit of protein phosphatase-1. FEBS Lett. 1995;375:294–298. doi: 10.1016/0014-5793(95)01184-g. [DOI] [PubMed] [Google Scholar]
- 25.Doherty M J, Young P R, Cohen P T. Amino acid sequence of a novel protein phosphatase 1 binding protein (R5) which is related to the liver- and muscle-specific glycogen binding subunits of protein phosphatase 1. FEBS Lett. 1996;399:339–343. doi: 10.1016/s0014-5793(96)01357-9. [DOI] [PubMed] [Google Scholar]
- 26.Fong N M, Jensen T C, Shah A S, Parekh N N, Saltiel A R, Brady M J. Identification of binding sites on protein targeting to glycogen for enzymes of glycogen metabolism. J Biol Chem. 2000;275:35034–35039. doi: 10.1074/jbc.M005541200. [DOI] [PubMed] [Google Scholar]
- 27.Gasa R, Jensen P B, Berman H K, Brady M J, DePaoli-Roach A A, Newgard C B. Distinctive regulatory and metabolic properties of glycogen-targeting subunits of protein phosphatase-1 (PTG, GL, GM/RGl) expressed in hepatocytes. J Biol Chem. 2000;275:26396–26403. doi: 10.1074/jbc.M002427200. [DOI] [PubMed] [Google Scholar]
- 28.Gilboe D P, Larson K L, Nuttall F Q. Radioactive method for the assay of glycogen phosphorylases. Anal Biochem. 1972;47:20–27. doi: 10.1016/0003-2697(72)90274-6. [DOI] [PubMed] [Google Scholar]
- 29.Hansen L, Hansen T, Vestergaard H, Bjorbaek C, Echwald S M, Clausen J O, Chen Y H, Chen M X, Cohen P T, Pedersen O. A widespread amino acid polymorphism at codon 905 of the glycogen-associated regulatory subunit of protein phosphatase-1 is associated with insulin resistance and hypersecretion of insulin. Hum Mol Genet. 1995;4:1313–1320. doi: 10.1093/hmg/4.8.1313. [DOI] [PubMed] [Google Scholar]
- 30.Hebert L F, Jr, Daniels M C, Zhou J, Crook E D, Turner R L, Simmons S T, Neidigh J L, Zhu J S, Baron A D, McClain D A. Overexpression of glutamine:fructose-6-phosphate amidotransferase in transgenic mice leads to insulin resistance. J Clin Investig. 1996;98:930–936. doi: 10.1172/JCI118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano K, Ito M, Hartshorne D J. Interaction of the ribosomal protein, L5, with protein phosphatase type 1. J Biol Chem. 1995;270:19786–19790. doi: 10.1074/jbc.270.34.19786. [DOI] [PubMed] [Google Scholar]
- 32.Hogen B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 33.Hubbard M J, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 34.Kadambi V J, Kranias E G. Phospholamban: a protein coming of age. Biochem Biophys Res Commun. 1997;239:1–5. doi: 10.1006/bbrc.1997.7340. [DOI] [PubMed] [Google Scholar]
- 35.Kadowaki T. Insights into insulin resistance and type 2 diabetes from knockout mouse models. J Clin Investig. 2000;106:459–465. doi: 10.1172/JCI10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahn B B, Flier J S. Obesity and insulin resistance. J Clin Investig. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanner, C., Y. Suzuki, C. Bi, H. Zhong, L. D. Cooper, and A. A. DePaoli-Roach. Gene structure and expression of the regulatory subunit, RGL, of the glycogen-associated type 1 protein phosphatase, PP1G. Arch. Biochem. Biophys., in press. [DOI] [PubMed]
- 38.Lawrence J C, Jr, Hiken J F, DePaoli-Roach A A, Roach P J. Hormonal control of glycogen synthase in rat hemidiaphragms. Effects of insulin and epinephrine on the distribution of phosphate between two cyanogen bromide fragments. J Biol Chem. 1983;258:10710–10719. [PubMed] [Google Scholar]
- 39.Lawrence J C, Jr, Roach P J. New insights into the role and mechanism of glycogen synthase activation by insulin. Diabetes. 1997;46:541–547. doi: 10.2337/diab.46.4.541. [DOI] [PubMed] [Google Scholar]
- 40.Lazar D F, Wiese R J, Brady M J, Mastick C C, Waters S B, Yamauchi K, Pessin J E, Cuatrecasas P, Saltiel A R. Mitogen-activated protein kinase kinase inhibition does not block the stimulation of glucose utilization by insulin. J Biol Chem. 1995;270:20801–20807. doi: 10.1074/jbc.270.35.20801. [DOI] [PubMed] [Google Scholar]
- 41.Lee E Y, Zhang L, Zhao S, Wei Q, Zhang J, Qi Z Q, Belmonte E R. Phosphorylase phosphatase: new horizons for an old enzyme. Front Biosci. 1999;4:D270–D285. doi: 10.2741/lee. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Brautigan D L. Glycogen synthase association with the striated muscle glycogen-targeting subunit of protein phosphatase-1. Synthase activation involves scaffolding regulated by beta-adrenergic signaling. J Biol Chem. 2000;275:26074–26081. doi: 10.1074/jbc.M003843200. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Brautigan D L. Insulin-stimulated phosphorylation of the protein phosphatase-1 striated muscle glycogen-targeting subunit and activation of glycogen synthase. J Biol Chem. 2000;275:15940–15947. doi: 10.1074/jbc.M909303199. [DOI] [PubMed] [Google Scholar]
- 44.Luo W, Grupp I L, Harrer J, Ponniah S, Grupp G, Duffy J J, Doetschman T, Kranias E G. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 45.MacDougall L K, Jones L R, Cohen P. Identification of the major protein phosphatases in mammalian cardiac muscle which dephosphorylate phospholamban. Eur J Biochem. 1991;196:725–734. doi: 10.1111/j.1432-1033.1991.tb15871.x. [DOI] [PubMed] [Google Scholar]
- 46.Nairn A C, Hemmings H C, Jr, Walaas S I, Greengard P. DARPP-32 and phosphatase inhibitor-1, two structurally related inhibitors of protein phosphatase-1, are both present in striatonigral neurons. Neurochem. 1988;50:257–262. doi: 10.1111/j.1471-4159.1988.tb13258.x. [DOI] [PubMed] [Google Scholar]
- 47.Nakielny S, Campbell D G, Cohen P. The molecular mechanism by which adrenalin inhibits glycogen synthesis. Eur J Biochem. 1991;199:713–722. doi: 10.1111/j.1432-1033.1991.tb16175.x. [DOI] [PubMed] [Google Scholar]
- 48.Oliver C J, Shenolikar S. Physiologic importance of protein phosphatase inhibitors. Front Biosci. 1998;3:D961–D972. doi: 10.2741/a336. [DOI] [PubMed] [Google Scholar]
- 49.Printen J A, Brady M J, Saltiel A R. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- 50.Ragolia L, Begum N. The effect of modulating the glycogen-associated regulatory subunit of protein phosphatase-1 on insulin action in rat skeletal muscle cells. Endocrinology. 1997;138:2398–2404. doi: 10.1210/endo.138.6.5194. [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen S K, Hansen L, Frevert E U, Cohen P T, Kahn B B, Pedersen O. Adenovirus-mediated expression of a naturally occurring Asp905Tyr variant of the glycogen-associated regulatory subunit of protein phosphatase-1 in L6 myotubes. Diabetologia. 2000;43:718–722. doi: 10.1007/s001250051369. [DOI] [PubMed] [Google Scholar]
- 52.Roach P J. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 53.Scrimgeour A G, Allen P B, Fienberg A A, Greengard P, Lawrence J C., Jr Inhibitor-1 is not required for the activation of glycogen synthase by insulin in skeletal muscle. J Biol Chem. 1999;274:20949–20952. doi: 10.1074/jbc.274.30.20949. [DOI] [PubMed] [Google Scholar]
- 54.Shepherd P R, Nave B T, Siddle K. Insulin stimulation of glycogen synthesis and glycogen synthase activity is blocked by wortmannin and rapamycin in 3T3–L1 adipocytes: evidence for the involvement of phosphoinositide 3-kinase and p70 ribosomal protein-S6 kinase. Biochem J. 1995;305:25–28. doi: 10.1042/bj3050025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shulman G I. Cellular mechanisms of insulin resistance. J Clin Investig. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shulman G I, Rothman D L, Jue T, Stein P, DeFronzo R A, Shulman R G. Quantitation of muscle glycogen synthesis in normal subjects and subjects with non-insulin-dependent diabetes by 13C nuclear magnetic resonance spectroscopy. N Engl J Med. 1990;322:223–228. doi: 10.1056/NEJM199001253220403. [DOI] [PubMed] [Google Scholar]
- 57.Smith R L, Lawrence J C., Jr Insulin action in denervated rat hemidiaphragms. Decreased hormonal stimulation of glycogen synthesis involves both glycogen synthase and glucose transport. J Biol Chem. 1984;259:2201–2207. [PubMed] [Google Scholar]
- 58.Stalmans W, Cadefau J, Wera S, Bollen M. New insight into the regulation of liver glycogen metabolism by glucose. Biochem Soc Trans. 1997;25:19–25. doi: 10.1042/bst0250019. [DOI] [PubMed] [Google Scholar]
- 59.Stralfors P, Hiraga A, Cohen P. The protein phosphatases involved in cellular regulation. Purification and characterisation of the glycogen-bound form of protein phosphatase-1 from rabbit skeletal muscle. Eur J Biochem. 1985;149:295–303. doi: 10.1111/j.1432-1033.1985.tb08926.x. [DOI] [PubMed] [Google Scholar]
- 60.Tang P M, Bondor J A, Swiderek K M, DePaoli-Roach A A. Molecular cloning and expression of the regulatory (RGL) subunit of the glycogen-associated protein phosphatase. J Biol Chem. 1991;266:15782–15789. [PubMed] [Google Scholar]
- 61.Thomas J A, Schlender K K, Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 62.Villar-Palasi C. Effect of glucose phosphorylation on the activation by insulin of skeletal muscle glycogen synthase. Biochim Biophys Acta. 1995;1244:203–208. doi: 10.1016/0304-4165(95)00006-w. [DOI] [PubMed] [Google Scholar]
- 63.Walker K S, Watt P W, Cohen P. Phosphorylation of the skeletal muscle glycogen-targetting subunit of protein phosphatase 1 in response to adrenaline in vivo. FEBS Lett. 2000;466:121–124. doi: 10.1016/s0014-5793(99)01771-8. [DOI] [PubMed] [Google Scholar]
- 64.Wu J, Liu J, Thompson I, Oliver C J, Shenolikar S, Brautigan D L. A conserved domain for glycogen binding in protein phosphatase-1 targeting subunits. FEBS Lett. 1998;439:185–191. doi: 10.1016/s0014-5793(98)01371-4. [DOI] [PubMed] [Google Scholar]
- 65.Xia J, Scherer S W, Cohen P T, Majer M, Xi T, Norman R A, Knowler W C, Bogardus C, Prochazka M. A common variant in PPP1R3 associated with insulin resistance and type 2 diabetes. Diabetes. 1998;47:1519–1524. doi: 10.2337/diabetes.47.9.1519. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto-Honda R, Honda Z, Kaburagi Y, Ueki K, Kimura S, Akanuma Y, Kadowaki T. Overexpression of the glycogen targeting (G(M)) subunit of protein phosphatase-1. Biochem Biophys Res Commun. 2000;275:859–864. doi: 10.1006/bbrc.2000.3391. [DOI] [PubMed] [Google Scholar]
- 67.Yang J, Hurley T D, DePaoli-Roach A A. Interaction of inhibitor-2 with the catalytic subunit of type 1 protein phosphatase. Identification of a sequence analogous to the consensus type 1 protein phosphatase-binding motif. J Biol Chem. 2000;275:22635–22644. doi: 10.1074/jbc.M003082200. [DOI] [PubMed] [Google Scholar]