Abstract
Summary
DeepKG is an end-to-end deep learning-based workflow that helps researchers automatically mine valuable knowledge in biomedical literature. Users can utilize it to establish customized knowledge graphs in specified domains, thus facilitating in-depth understanding on disease mechanisms and applications on drug repurposing and clinical research. To improve the performance of DeepKG, a cascaded hybrid information extraction framework is developed for training model of 3-tuple extraction, and a novel AutoML-based knowledge representation algorithm (AutoTransX) is proposed for knowledge representation and inference. The system has been deployed in dozens of hospitals and extensive experiments strongly evidence the effectiveness. In the context of 144 900 COVID-19 scholarly full-text literature, DeepKG generates a high-quality knowledge graph with 7980 entities and 43 760 3-tuples, a candidate drug list, and relevant animal experimental studies are being carried out. To accelerate more studies, we make DeepKG publicly available and provide an online tool including the data of 3-tuples, potential drug list, question answering system, visualization platform.
Availability and implementation
All the results are publicly available at the website (http://covidkg.ai/).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Clinicians and researchers need to read a large number of academic documents to understand the latest developments in their fields. However, it has become quite time-consuming and laborious to manually mine knowledge in the documents. Fortunately, blooming algorithms in natural language processing and knowledge mining provides a possible way to establish a comprehensive medical knowledge mining workflow from massive literature to the standardize knowledge graph (Lin et al., 2017). Benefiting from such powerful knowledge graph, the important information hidden in literature can be directly utilized to researchers, which may significantly improve the efficiency of the related research, such as new drug discovery (Abbas et al., 2021), drug repurposing (Berber and Doluca, 2021) and clinical decision support system (Shen et al., 2021).
For such purpose, several studies (Rotmensch et al., 2017; Sang et al., 2018; Xu et al., 2020) have been proposed around this goal based on a specific theme and knowledge graphs have been established. However, the performance of these knowledge graph construction processes and related methods is unstable in different tasks, which dramatically limits their application. In other words, the study proposed for a specific theme is difficult to achieve the same excellent performance on another topic. Technically, there are three vital parts which still have much room for improvement.
Accurate knowledge extraction will provide a solid foundation for graph construction. Lots of efforts (Crichton et al., 2017; Habibi et al., 2017) have been widely made to recognize entities and relations from biomedical literature. Without deeply optimized for the medical text, however, the performance of these methods may significantly degenerate in real-world coarse-grained text.
Potential knowledge discovery is a crucial application in the biomedical scenarios, such as drug repurposing (Zeng et al., 2020), which can be inferred by predicting the links in the 3-tuples data of ‘entity-relation-entity’. However, these traditional methods are incapable of capturing the relations in practice because of the limited representation capability (Bordes et al., 2013).
Application of workflow is a straightforward way to evidence the effectiveness of knowledge graph, including question answering (QA) system, visualization, etc. Generally, much research has been devoted to solving a single module in the stage of knowledge graph construction, not an end-to-end workflow. In practice, these methods are inherently subject to the gap of different modules, which degrades practicably.
The issues discussed above motivate to create a comprehensive workflow for knowledge graph extraction, optimization and applications. Here, we propose an end-to-end medical knowledge graph construction workflow (DeepKG) which consists of a series of optimized functional modules. In detail, DeepKG extracts knowledge accurately under a cascaded hybrid information extraction framework (CHIEF), discovers potential knowledge with a novel AutoML-based knowledge representation algorithm (AutoTransX), and provides a visual interface with search and QA system to display the constructed knowledge graph and prediction results.
2 Architecture of DeepKG framework
As shown in Figure 1, DeepKG includes three main modules: data preparation, modelling and application. In the first module, DeepKG employs a user-friendly tool for data preparation, i.e. designing the schema and 3-tuples labelling. In the latter module, DeepKG develops CHIEF to extract biomedical 3-tuples from the literature more accurately, and a biomedical database-based entity correction to refine the quality of knowledge graph. In the last module, DeepKG utilizes AutoTransX to forecast relations underlying the knowledge graph and then develops a visualization platform with a QA system to make the research results available for public straightly and readily.
Fig. 1.
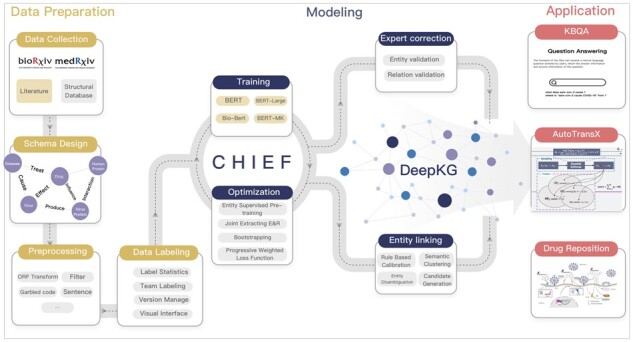
Schematic representation of DeepKG
3 The DeepKG application
Mining valuable information from biomedical literature is crucial for enlarging our knowledge of biomedical sciences, which is a challenging task given massive number of unstructured texts. Thus, researchers need effective tools to assist them establish customized knowledge graphs in specified domains, thus facilitating in-depth understanding on disease mechanisms and applications on drug repurposing and clinical research. The primary goal of DeepKG is to give an integrative platform that can facilitate the development of this field by harnessing state-of-the-art techniques.
To illustrate the DeepKG clearly, the essential part is explained in depth. First, given some biomedical literature as input, the data section outputs a set of biomedical 3-tuples labelled by experts. Following the entities and relations of schema defined by users, the semantic materials after preprocessing are labelled by experts using the user-friendly labelling tool, including team labelling, hard examples labelling, label statistic and version manager. Second, the modelling module in DeepKG generates enormous biomedical 3-tuples inferred by CHIEF using the labelled semantic materials from literature and entity names from structure database. CHIEF mainly consists of entity supervised pre-training, subject recognition and subject guide triplet extraction; all of them are also optimized for biomedical to achieve higher accuracy. CHIEF use the structure of CASEL (Wei et al., 2020) and optimize it to achieve the better subject guided triplet extraction(shown in Appendix 1, Supplementary Fig. S1b). Third, a cleaner knowledge graph is generated from the filtered biomedical 3-tuples after entity correction, including entity linking and expert correction. Fourth, to infer more valuable information from a knowledge graph, AutoTransX is applied to optimize the combination of candidate operations and explore the optimal model to represent the relations in the biomedical knowledge graph accurately. And the comparison experiments shown its superiority (shown in Appendix 1 and 2, Supplementary Fig. S2a and b). Finally, in the purpose of helping scientists and clinicians to use knowledge graph, a QA and visualization platform is built to make the knowledge graph publicly available.
In order to evaluate the effectiveness of DeepKG, the case about COVID-19 is taken as an example. First, after filtering semantic material from literature in CORD-19 (Wang et al., 2020), 1904 sentences are labelled by senior doctors using triplet labelling tool in DeepKG. Second, the hand-crafted 2546 entities and 3830 relations are gotten (Supplementary Table S1) as inputs of the CHIEF in DeepKG. By inferring 3-tuples from 144 900 scholarly literature in CORD-19 dataset, a knowledge graph dataset, including 361 524 entities and 945 048 relations (Supplementary Table S1), is generated. After correcting the knowledge graph raw dataset by expert and entity linking in DeepKG, a cleaner biomedical dataset including 7980 entities and 43 760 relations is obtained to construct a COVID-19 knowledge graph. Third, a potential drug list (e.g. allopurinol, berberine, etc.) which may inhibit the SARS-CoV-2 is inferred from the COVID-19 KG by AutoTransX. Specifically, the potential mechanism of allopurinol and berberine is demonstrated in biological pathways (shown in Supplementary Fig. S2c). Finally, a visualization platform with a QA system is developed to ease the use of COVID-19 KG, and all the results are publicly available at the website (http://covidkg.ai/).
4 Conclusions
DeepKG, as a comprehensive modular workflow, can integrate and mine knowledge from massive literature corpus, which promises a broader application prospect for transplantation to any new biomedical domain. Meanwhile, we redesign and standardize the knowledge graph construction process to promote more convenient application. To facilitate use, different parts with clear input and output information are integrated modularly, which can be utilized assembly and freely. We also have developed a complete platform covering the whole process, including the visual interface for corpus labelling, a search result display and a QA system. The application is freely available to everyone, and can be accessed from http://covidkg.ai/.
Supplementary Material
Acknowledgement
None of this material has been published or is under consideration for publication elsewhere.
Funding
This work was supported by the technology project ‘Public service platform for artificial intelligence aided diagnosis in medical and health industry’ of Ministry of Industry and Information Technology of the Peoples Republic of China [2020-0103-3-1].
Conflict of Interest: We declare that all authors have no conflict of interest
References
- Abbas K. et al. (2021) Application of network link prediction in drug discovery. BMC Bioinform., 22, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berber B., Doluca O. (2021) A comprehensive drug repurposing study for COVID19 treatment: novel putative dihydroorotate dehydrogenase inhibitors show association to serotonin–dopamine receptors. Brief. Bioinform., 22, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes A. et al. (2013) Translating embeddings for modeling multi-relational data. In: Proceedings of the 26th International Conference on Neural Information Processing Systems, Sydney, Australia, Vol. 2, pp. 2787–2795. [Google Scholar]
- Crichton G. et al. (2017) A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinform., 18, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi M. et al. (2017) Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics, 33, i37–i48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. et al. (2017) Learning entity and relation embeddings for knowledge resolution. Proc. Comput. Sci., 108, 345–354. [Google Scholar]
- Rotmensch M. et al. (2017) Learning a health knowledge graph from electronic medical records. Sci. Rep., 7, 5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang S. et al. (2018) SemaTyP: a knowledge graph based literature mining method for drug discovery. BMC Bioinform., 19, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. et al. (2021) Adam Landman, clinical decision support system, using expert consensus-derived logic and natural language processing, decreased sedation-type order errors for patients undergoing endoscopy. J. Am. Med. Inf. Assoc., 28, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. (2020) CORD-19: the Covid-19 open research dataset. arXiv:2004.10706v2.
- Wei Z. et al. (2020) A novel cascade binary tagging framework for relational triple extraction. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pp. 1476–1488.
- Xu J. et al. (2020) Building a PubMed knowledge graph. Sci. Data, 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. et al. (2020) Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res., 19, 4624–4636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


