Abstract
SARS-CoV-2 caused the first severe pandemic of the digital era. Computational approaches have been ubiquitously used in an attempt to timely and effectively cope with the resulting global health crisis. In order to extensively assess such contribution, we collected, categorized and prioritized over 17 000 COVID-19-related research articles including both peer-reviewed and preprint publications that make a relevant use of computational approaches. Using machine learning methods, we identified six broad application areas i.e. Molecular Pharmacology and Biomarkers, Molecular Virology, Epidemiology, Healthcare, Clinical Medicine and Clinical Imaging. We then used our prioritization model as a guidance through an extensive, systematic review of the most relevant studies. We believe that the remarkable contribution provided by computational applications during the ongoing pandemic motivates additional efforts toward their further development and adoption, with the aim of enhancing preparedness and critical response for current and future emergencies.
Keywords: SARS-CoV-2, pharmacology, genomics, epidemiology, imaging, machine learning
Introduction
The ongoing COVID-19 pandemic has prompted an unprecedented research effort by the global scientific community. The urge to identify effective countermeasures against the tremendous health, economic and social impact caused by the disease led to an astounding proliferation of studies covering all the diverse aspects of the pandemic [42]. From the development of assays aimed at better understanding, the molecular mechanisms exploited by the virus to the design of epidemiological models predicting its spread, research labs around the world have produced a sheer amount of potentially fruitful knowledge, which is still growing on a daily basis at a soaring pace while we write.
COVID-19 is also the first severe pandemic of the digital era. Besides accelerating the production and spread of research literature, digital technologies produced a significant impact as investigational tools, with contributions that range from the viral sequence establishment [173] to the latest data-driven risk models that are helping governments to select the most efficient restriction measures [26]. Such results are already summarized by a number of review articles, which cover both specific application areas (like drug discovery and repositioning [108, 116, 167], preventive pharmacology [106], medical image analysis [110]) and method-oriented overviews, such as artificial intelligence (AI) applications [85, 88] and relevant software tools [73].
As valuable as these efforts are, the quantity of published studies poses a significant challenge both for researchers and for media operators striving to remain up to date with the state-of-art, and correctly inform the governments and the public. The emergency character of a pandemic crisis urges researchers to make their results timely available. In this regard, online platforms for preprint publication provide an effective shortcut [158], although the lack of a peer-review process warrants additional caution [22]. In general, the usual time span needed by the scientific community to properly digest the available literature and reach a consensus on the most promising research directions is challenged by the number of publications made available in such a short amount of time.
For these reasons, with the aim of comprehensively assessing the contribution provided by computational applications in the fight against the ongoing pandemic, we developed a software framework called computational studies about COVID-19 (CSCoV) based on automatic collection and filtering of computational studies related to COVID-19. The framework automatically gathers both articles from multiple sources and meta-data about publication and author metrics in order to prioritize the studies by predicted relevance. AI is then used to categorize the papers by topics and to predict the chances of preprint articles to pass a peer-review process. The whole framework is continuously updated with new articles and corresponding metrics. With the help of the CSCoV database and tools, we analyzed 147 346 research articles, filtered 17 269 of them involving computational approaches and assigned each of them to one of six automatically derived topics: Molecular Pharmacology and Biomarkers, Molecular Virology, Epidemiology, Healthcare, Clinical Medicine and Clinical Imaging. Finally, guided by our categorization and scoring system, we reviewed the most relevant literature within each topic, with a special focus on the computational aspects of each study.
In the following, we describe both the CSCoV framework and the most relevant literature about COVID-19 involving computational approaches at various extents. The entire database, including categorization and prioritization scores, is publicly available together with the used computational models [111].
The database of CSCoV
Article collection
We systematically collected COVID-19 related studies from four different sources (see Figure 1): PubMed for articles published in journals and arXiv, bioRxiv and medRxiv for preprint articles, gathering an initial broad collection of 147 346 papers. Studies involving computational approaches were selected based on 83 manually curated keywords appearing in the abstracts (see Supplementary Methods). We thus obtained a total of 17 269 papers, including 12 408 journal articles and 4861 preprints. The studies were published in 1655 different journals or conferences (see Supplementary Methods for additional details). Figure 2A shows a summary of the collected articles against publishing time. Some of the preprint articles originally appeared before the pandemic and have been subsequently updated for their potential application to the COVID-19 crisis. Novel preprints started to appear in January 2020, while the first journal articles date back to February. Expectedly, the ratio of journal articles versus preprint articles increased over time. On the other hand, for more than 60% of the preprints, we were not able to identify a corresponding journal publication after 70 weeks (Figure 2B) from its appearance online. Detailed statistics concerning the articles from each source are reported in Figure 2B.
Figure 1.
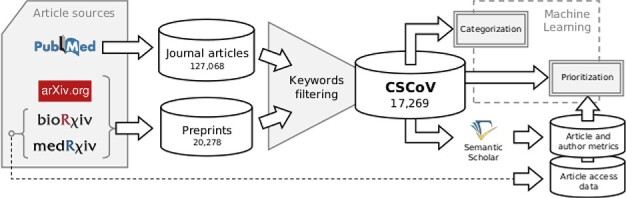
CSCoV data collection and analysis pipeline. COVID-19 related scientific papers are collected by querying 4 different sources, including both journal (PubMed) and preprint articles (arXiv, bioRxiv, medRxiv). The database of computational studies (CSCoV) is obtained by filtering the collected COVID-19 papers based on a manually curated set of 83 keywords to be matched against the article abstracts. The dataset of computational papers is then analyzed using a topic modeling method for categorization and a machine learning model together with bibliometric-based ranking for prioritization. The latter is based on additional quality metrics obtained from the Semantic Scholar website or from preprint servers.
Figure 2.
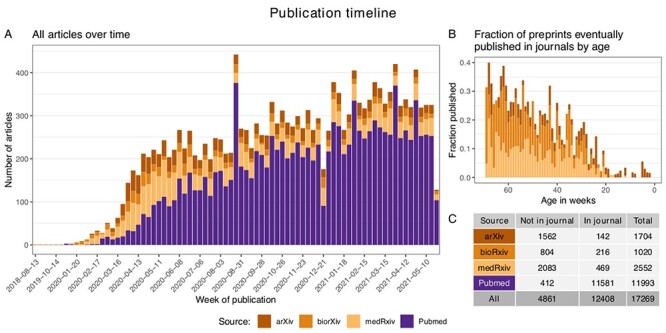
Publication timeline of all the papers in the CSCoV database. (A) Number of articles appeared weekly since January 2019 divided by source. (B) Fraction of preprint articles that eventually appeared also in journals as a function of the number of weeks from their appearance online. C) Tabular summary of the number of articles currently included in CSCoV, grouped by source and publication status.
The CSCoV database is regularly updated. Each update includes both new articles and the corresponding analytical results described in the following.
Article categorization
In order to categorize all the collected articles into topics, we trained a latent dirichlet allocation (LDA [20]) with all the paper abstracts. LDA is a generative probabilistic model that aims at modeling each document from a given collection as a mixture of latent topics, which are in turn defined by a set of words. Once the topics are obtained, each document can be assigned to one of them.
Our final solution identified six topics: Healthcare, Epidemiology, Clinical Medicine, Molecular Pharmacology and Biomarkers, Molecular Virology, and Clinical Imaging. Figure 3A summarizes the results. In the figure, the entire collection is visualized as a two-dimensional (2D) map where each point represents an article colored by the assigned topic, and similar articles based on the extracted keywords appear close to each other (see Supplementary Methods for further details). The top 10 keywords for each topic are reported in Figure 3B. Topic names were assigned based on the top keywords observed after multiple runs of the LDA algorithm.
Figure 3.
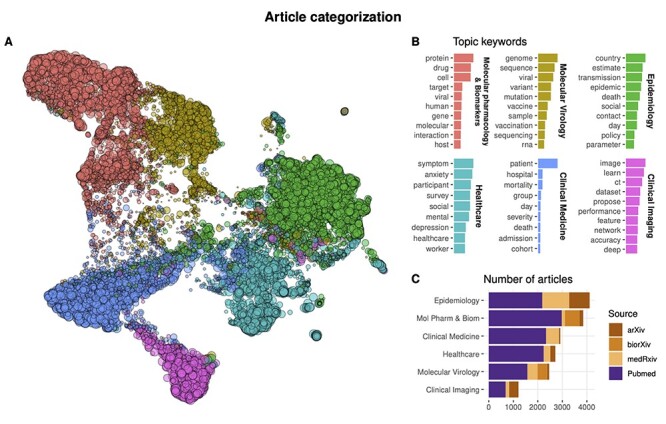
Categorization of articles in the CSCoV collection. (A) 2D visualization of the database. Each point represents an article, points proximity reflects article similarity, colors represent the extracted topic as reported in panel B. (B) Top 10 keywords in article abstracts identifying each of the six extracted topics. (C) Number of articles in CSCoV by topic.
In the map, the ‘Molecular Pharmacology and Biomarkers’ and ‘Molecular Virology’ topics lie especially close, which is expected based on drug and vaccine development efforts that make large use of the viral sequence. Interestingly, ‘Epidemiology’, ‘Healthcare’ and ‘Clinical Medicine’ are found next to each other, in a sequence that appear to arrange articles from the general population to the single patient perspective. Finally, articles in the ‘Clinical Imaging’ cluster, which are consistently driven by established Deep Learning approaches (see Subsection 3.7) constitute a well-characterized group on their own, expectedly placed next to the ‘Clinical Medicine’ cluster.
The relative number of articles across topics and sources also confirms an overall meaningful categorization (see Figure 3C). As expected, papers included in the arXiv collection have their larger relative shares assigned to the ‘Clinical Imaging’ and ‘Epidemiology’ groups, which largely rely on machine learning and mathematical models respectively (see Section 3). Only a negligible number of arXiv preprints is assigned to ‘Clinical Medicine’. On the other hand, bioRxiv articles are almost exclusively assigned the ‘Molecular Pharmacology and Biomarkers’ and ‘Molecular Virology’ topics, with zero articles assigned to ‘Epidemiology’ or ‘Clinical Medicine’. This is in line with the policy that was enforced by the platform maintainers since the launch of medRxiv, which requires authors to submit epidemiology- and medicine-related articles to the new server. Indeed a complimentary situation is observed for medRxiv articles, which tend to be especially associated with the ‘Epidemiology’ topic and cover the largest share of ‘Clinical Medicine’ studies. Articles collected from the PubMed collection span all the six topics, although they appear to be disproportionately fewer in the ‘Epidemiology’ topic.
Article prioritization
Collecting and categorizing 17 269 articles allowed us to obtain a meaningful general overview (see Figure 3) of the contribution provided by the research community to fight the pandemic relying on computational techniques. However, a detailed review of the published literature implies carefully reading each single article, which is only feasible through a collective, time consuming effort that is unsuitable for an emergency and rapidly evolving situation. We thus sought to establish an advantageous trade-off between extensively reviewing every articles and the urgency to timely identify the most impactful or promising research paths. To this aim, we developed additional tools to prioritize studies that are more likely to be especially relevant, as illustrated in the following subsections.
Metric-based ranking
The navigation of the sheer number of research articles constantly produced by researchers is normally facilitated by the collective work of the scientific community, which gradually digests published literature and implicitly produces signals of interest and consensus. For example, article quality has been shown to be reflected by the number of citations received [154], which in turn correlates with the number of times the article is downloaded from a hosting web server [60]. Although such indicators have many known limitations, they are commonly used by researchers as heuristics to screen scientific literature [153]. We thus took advantage of available indicators to estimate article credibility. We were able to collect the number of citations for all the papers in our collection using the Semantic Scholar online platform [50], and the number of downloads for all the preprint articles by scraping the public statistics available at bioRxiv and medRxiv. The arXiv server does not release such information.
Although the number of citations and downloads can be highly valuable for article prioritization, it is of course scarcely available for newly published studies, which are also an important target of this review. Moreover, citations and downloads are distributed differently across time. In particular, the latter appears to be especially novelty-driven, while the former extends longer over the years [60], which adds to the task difficulty. For these reasons, we sought to gather additional time-independent metrics. In particular, since a relation between the article citations and authors reputation has been shown [23], we added author data to our prioritization system. Using Semantic Scholar, we collected the number of papers published and the number of ‘Influential citations’ (see Supplementary Methods) received by each of the authors in our entire collection, which amounts to a total of 171 106 queries. The obtained scores showed similar distribution across article sources (see Figure 4A). Moreover, when comparing scores of preprints eventually published in journals against preprints of similar age that never did, we observed the former to have higher scores than the latter (see Figure 4C). By factoring in all the collected meta-data based on rank statistics (see Supplementary Methods), we were finally able to derive a score for each paper and prioritize the entire CSCoV collection accordingly. As expected, score distributions may differ across topics (see Figure 4D), which, however, we analyze independently in this paper.
Figure 4.
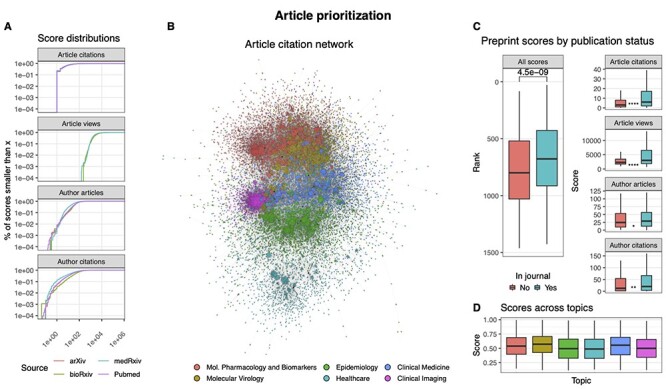
Article scoring to guide manual review. (A) Four different metrics are used, including article-related (number of citations and views) and author-related (number of published articles and citations received) scores. As shown, the scores have similar distributions across sources. (B) The article citation network is used to prioritize preprints together with their text contexts. It shows general concordance with the computed topics, as highlighted by colors. (C) Comparison of scores across two groups of preprints having similar size and age but differing in publication status. The scores for articles published in journals tend to be higher. (D) Scores are not significantly biased across topics.
Prediction of journal publication for preprint articles
Although we believe that the gathered metrics can greatly help to score the relevance of the studies in our collection, preprint articles need special caution. On one hand, it has been shown that article quality does not improve dramatically after a preprint article passes the peer-review process [16]; on the other hand, recent cases of poorly substantiated claims in COVID-19 studies that appeared on preprint servers have reminded the research community about the risks posed by the lack of proper peer review [86]. Therefore, with the aim of gaining further indication about the reliability of preprint publications, we developed a machine learning framework to predict the chances that each preprint would pass a peer-review process solely based on its contents. In particular, we fed a Deep Neural Network model with article abstracts, the full article citation network (see Figure 4B), and the topic scores previously described. Each node in the citation network represents an article in CSCoV, and there is a link between two nodes A and B if the article A cites the article B. The model learned to discriminate preprint-only articles from articles published in journals (area under the curve (AUC) = 0.76) according to a probability score (see Supplementary Methods). We used the predicted probabilities to provide an additional score to all the preprints in our database.
Article reviews by topic
Once we established the CSCoV database, we sought to exploit this resource to identify the most significant contributions provided by the research community to the fight against the pandemic. Although we used our prioritization system as a guide, we did not follow it strictly. In particular, we extracted the top 100 articles from each topic and manually reviewed them. The resulting selected articles are therefore the result of a mediation between the CSCoV recommendation system and our best judgment. Given the breadth of this review and the special focus on computational aspects, an in-depth analysis within each topic from an application field standpoint falls out of our aims. However, we cited those specialized review articles that are available in CSCoV when relevant.
Computational methodologies used in the reviewed articles are diverse, ranging from mathematical model fitting to artificial neural networks. Although we will detail them in the next Subsection, general insights can be obtained by analyzing the keywords used to build the database. Figure 5-left reports a summary including the top 10 keywords recurring within each topic, showing the importance of omics data analysis for ‘Molecular Pharmacology and Biomarkers’ and ‘Molecular Virology’, statistical models for ‘Healthcare’ and ‘Clinical Medicine’, neural network models for ‘Clinical Imaging’ and mathematical models for ‘Epidemiology’. Nonetheless, the same methodologies can of course be found across all topics.
Figure 5.
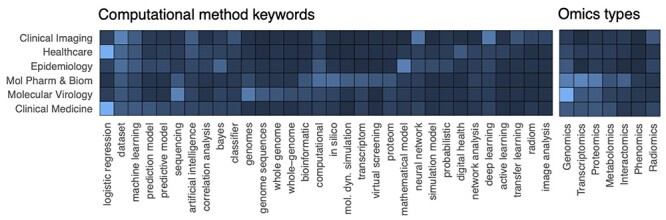
Left: association between topics and the most recurrent keywords used to select computational studies provides insights into the most used methodologies. Right: diverse ‘omics’ data types have been used across topics.
‘Omics’ data analysis, which was made possible by computational approaches, had an ubiquitous presence in COVID-19-related research. Figure 5-right reports an analysis of different omics data types across research topics. In particular, articles concerning ‘Molecular Virology’ were fundamentally driven by genomic data, while transcriptomics and proteomics approaches are also significantly present. ‘Molecular Pharmacology and Biomarkers’ is the cluster in which we observed the largest diversity of ‘omics’ data types, including transcriptomics, proteomics, genomics and interactomics. Expectedly, ‘radiomics’-related keywords emerged from the ‘Clinical Imaging’ cluster.
The next Subsection aims at establishing the beginning of the COVID-19 research endeavor according to our database. Further Subsections review the most relevant studies that we identified within each of the six topics in CSCoV. A general timeline of the main publications we reviewed is shown in Figure 6. Finally, the last Subsection proposes some of the latest preprint articles that could provide an especially relevant contribution in the near future.
Figure 6.

Timeline of the most representative reviewed studies by topic (colored dots), together with major events (black dots) related to the COVID-19 pandemic.
Early contributions
Based on the CSCoV database, we identified when the first computational studies about COVID-19 started to appear. Although a significant number of papers started to be published in February 2020, several articles appeared even earlier. These studies represent the first response by the research community to the pandemic. Here, we will mention the majority of them. As expected, most of them first appeared as preprint articles.
A few articles in the CSCoV database predate the epidemic. However they were originally unrelated with COVID-19 and later updated during the pandemic, thus we did not consider them here. By looking at single article versions, we identified the earliest published study as a preprint appeared on 19 January 2020 [32], and later published as a peer-reviewed article on 18 February [29]. The study used a mathematical model to assess the basic reproduction number of SARS-CoV-2 at 3.58. A second estimation of 2.2 obtained through stochastic simulations was published on 24 January [133]. On 31 January, a third preprint showed the use of a Bayesian framework to infer the time-calibrated phylogeny and the epidemic dynamics, resulting in an effective reproductive number of 1.1 and a most recent common ancestor dated at 7 December 2019 [186]. Contributing to the heated debate about the origins of the virus, on 27 January a preprint article showed genomic proximity with bat coronaviruses and excluded a recent recombination event based on evolutionary analysis [122]. The article appeared in a peer-reviewed journal three months later [121]. Another confirmation about the bat origin hypothesis arrived on 2 February based on RNA sequencing data analysis [173]. One of the earliest studies concerning prevention measures appeared on 28 January, confirming the risk posed by asymptomatic transmission on the basis of computational simulations [142]. Also one of the earliest studies involving pharmacological measures appeared at the end of January, attempting virtual screening for drugs inhibiting the M protease of SARS-CoV-2 [94]. The article was published in a journal one1 month later [125]. Finally, by means of epidemiological modeling, another preprint article reported on the necessity of healthcare measures, including lockdowns and universal face mask wearing, to counteract possibly disastrous consequences of the pandemic [105].
Molecular pharmacology and biomarkers
From the inception of the pandemic, the development of effective treatments and vaccines has been one of the main hopes to effectively fight it. In recent years, computational tools supported researchers both in the former [152] and in the latter [117]. During this pandemic, it has been proposed that further efforts should be directed toward the definition of reliable computational pipelines in order to be more prepared for the next one [51], particularly given the potential of AI-based approaches [192]. Many studies assigned to this topic do not focus specifically on the identification of treatments, but also on the elucidation of molecular biomarkers that underlie potential treatments or disease mechanisms. They also span multiple omics data types, such as transcriptomics, proteomics, genomics, interactomics and metabolomics (see Figure 5-right).
Drug discovery and repositioning
One of the most promising approaches to cope with health emergencies is the repositioning of already approved drugs to quickly respond to new outbreaks. This specific topic has been covered by a number of reviews also included in our database [3, 192]. Computational approaches have been used to prioritize potentially effective small molecules in a variety of ways, such as using molecular docking simulations and network-based drug repurposing, even with ad hoc SARS-CoV-2-related software tools provided to the community [84]. Docking simulations have been used to propose the efficacy of statins [43], recently confirmed by clinical data analysis [61]. In another study, remdesivir was identified through molecular docking and proposed as a potentially effective treatment for its ability to target the RNA-dependent RNA polymerase (RdRp) [41]. The drug has later been approved by the US Food and Drug Administration, although recommended against by the World Health Organization (WHO) [100]. Among network-based computational approaches, on 16 March, one integrative drug repurposing methodology was published to discover potential drugs in interactomic approach [191]. A total of 16 repurposable drugs (e.g. melatonin, mercaptopurine and sirolimus) were prioritized and further validated in human cell lines and three potential drug combinations (e.g. sirolimus plus dactinomycin, mercaptopurine plus melatonin and toremifene plus emodin) were identified. Another study used a knowledge graph based on 24 million PubMed papers to identify 41 drugs, among which dexamethasone, a glucocorticoid whose efficacy has been confirmed in hospitalized patients [57] and niclosamide, an anthelmintic recently proposed for repurposing on the basis of its ability to suppress the calcium-activated ion channel TMEM16F activity [15].
A number of in vitro studies used metabolomics and transcriptomics data with the aim of identifying novel therapeutics [52, 63]. For example, using metabolomics profiling, spermidine, mk-2206 and niclosamide were shown to exert antiviral effects in VeroFM cells [52], while transcriptomics analysis was used to identify imatinib and mycophenolic acid as inhibitors of SARS-CoV-2 in hpsc-derived lung organoids [63].
Besides direct drug discovery, many computational applications have provided insights about related molecular mechanisms. For example, one study reported the use of machine learning with chemoinformatics data to classify drugs and predict target specificity [138]. Among others, the study classified chloroquine and its highly debated [107] derivative hydroxychloroquine as non-specific drugs. Structural and molecular modeling was used to further investigate the two molecules helping to understand their mode of actions [44]. Molecular dynamics simulations have also been used to study the binding mechanism of remdesivir to RdRp, suggesting that the small molecule could act as a SARS-CoV-2 RNA-chain terminator, thus stopping RNA replication [184].
Vaccines and antibody therapies
One of the biggest breakthroughs in fighting the pandemic has been the development of vaccines. The efficiency of computational approaches has been advocated as a means to speed up vaccine and therapeutic antibodies for the COVID-19 emergency [140]. Among the papers we found of particular significance, one used a combinatorial machine learning approach to evaluate and optimize peptide vaccine formulations [94]. Many other studies in this context focused on the identification of epitopes and on the essential understanding of COVID-19 immunological mechanisms (see Subsection 3.2.3). Another work used machine learning-based reverse vaccinology tools, namely Vaxign and Vaxign-ML, to predict vaccine targets, supporting that a cocktail containing structural and non-structural proteins can be effective through the stimulation of complementary immune responses.
A number of proteomic studies have been dedicated to the identification of epitopes. Based on a tool named VirScan, a high-throughput method to analyze epitopes of antiviral antibodies in human sera, 800 SARS-CoV-2 epitopes were identified, 10 of which were considered likely recognized by neutralizing antibodies. Furthermore, XGBoost was used to predict SARS-CoV-2 exposure from the output of VirScan [146].
Besides high-throughput approaches, other studies focused on specific interactions. For example, using structural modeling, a specific conformation of CR3022, a neutralizing antibody isolated from a SARS patient, was demonstrated to be required in order for it to bind a cross-reactive SARS-CoV-2 epitope [179]. In fact, prior knowledge about the SARS-CoV virus has been largely exploited. In one study, detection of sequence homology has been used to identify conserved regions between SARS-CoV and SARS-CoV-2. Epitope prediction was then performed using BepiPred 2, a random forest, sequence-based algorithm and Discotope, which relies on structural modeling [56]. Finally, an interesting application of computational methods in animal modeling is also worth of mention. Using sequence analysis and structural modeling, it was possible to identify a panel of adaptive mutations in a mouse-adapted SARS-CoV-2 strain potentially associated with increased virulence [58].
Molecular biomarkers
Understanding the pathogenicity mechanisms of COVID-19 is important for the development of effective drugs, vaccines and antibody therapies, or to characterize the disease. Many transcriptomics studies were conducted to elucidate infection risk and mechanisms by evaluating the expression of angiotensin-converting enzyme 2 (ACE2) in different organs, such as lungs, heart, kidneys, intestines, brain and testicles [7, 67, 120, 168, 181, 182]. Other studies use transcriptomics to analyze the immune response among COVID-19 patients to discover biomarkers. For example, release of excessive cytokine, such as CCL2/MCP-1, CXCL10/IP-10, CCL3/MIP-1A and CCL4/MIP1B were suggested as biomarkers for COVID-19 pathogenesis [118, 175, 193]. Transcriptomics analysis was also used to identify type I interferon deficiency as a biomarker of COVID-19 severity [62].
Proteomic data analysis was also widely employed for biomarker discovery. For example, one study analyzed the cellular infection profile of SARS-CoV-2 on human cell-cultures using mass spectroscopy (MS). Central cellular pathways such as translation, splicing, carbon metabolism, protein homeostasis and nucleic acid metabolism were reported to be reshaped after SARS-CoV-2 infection. Moreover, two inhibitors, 2-deoxy-d-Glucose, which blocks glycolysis, and NMS-873, which affects protein homeostasis, were found effective against viral replication in vitro [12]. Another study also used MS to analyze phosphorylation and perturbations in protein abundance, suggesting inhibition of the p38, CK2, CDK, AXL and PIKFYVE kinases as antiviral mechanisms [14]. Biomarkers were also used for automatic classification. One study profiled plasma proteomics of COVID-19 cases and reported 11 plasma proteins as biomarkers for severity. Using a machine-learning-based pipeline, the authors found that plasma levels of ORM1, ORM2, S100A9, CRP, AZGP1, CFI, SERPINA3/ACT and LCP1/LPL were elevated in severe COVID-19 conditions, while levels of FETUB, CETP and PI16 were reduced [147]. Using a similar approach with multi-omics data, a preprint article highlighted the specificities of proteomic and metabolomic responses to COVID-19 in younger patients, identifying potential children-specific markers [162]. Besides MS, sequence and structural analysis approaches were also used for proteomic studies. For example, different cytokine profiling and inflammatory signaling after SARS-CoV-2 infection were described in this way [112].
Finally, metabolomic data were also employed in a number of studies. For example, circulating lipids, such as phosphatidylcholine 14:0_22:6 and 16:1_22:6, and phosphatidylethanolamine 18:1_20:4 were identified as potential COVID-19 biomarkers [8].
Molecular virology
Unsurprisingly, articles identified as related to Molecular Virology appear next to the Molecular Pharmacology and Biomarkers cluster (see Figure 3), which make large use of sequence and structural analysis especially based on genomics data (see Figure 5-right). Molecular virology studies significantly marked the progressive understanding of SARS-CoV-2, with the most important contribution arguably being the already mentioned (see Subsection 3.1) sequencing of its complete genome in January 2020 (published in February 2020 [173]). Using RNA sequencing and phylogenetic analysis, the study also showed that the new virus is closely related to a group of SARS-like coronaviruses found in bats. Based on the growing collections of viral sequences in online repositories, it became soon possible to identify reliable RT-PCR targets by identifying conserved sequences across multiple strains [36] and develop the effective molecular diagnostic tools. In April 2020, proteomics study of human leukocyte antigen susceptibility map for SARS-CoV-2 [113] and high-resolution transcriptome and epitranscriptome map of the SARS-CoV-2 were also reported [81]. Strongly linked to pharmacological studies, molecular virology research played a pivotal role in helping to shed light on the virus origin, detect novel variants and track the local and global evolution of the pandemic, as summarized in the next Subsections.
Origin of SARS-CoV-2
On the same day in which results about the similarity of SARS-CoV-2 to other bat coronaviruses were published [173], another article appeared that analyzed the similarity of SARS-CoV-2 to SARS-CoV, with 79.6% sequence identity and the same cell entry receptor (ACE2) identified [190]. The highest similarity was found against another bat coronavirus, RaTG13, with 96% identity. Andersen et al. [4] summarized the notable genome features of SARS-CoV-2 as mutations in the receptor-binding domain (RBD)and having polybasic furin cleavage site and O-linked glycans. They discussed three theories on the origin of this virus, (i) natural selection in an animal host before transfer to human, (ii) natural selection in humans after zoonotic transfer and (iii) laboratory selection and inadvertent release outside. Based on the genomic features observed, they concluded the latter to be implausible. Based on a population genetics-phylogenetics approach, another study used the full sequences of 52 SARS-CoV-2 strains to analyze selective events that accompanied the divergence of SARS-CoV-2 from RaTG13, concluding that the two viruses are likely to share a common ancestor [19]. Moreover, Zhou et al. [188] found a new bat virus (RmYN02) with 93.3% sequence identity to SARS-CoV-2. Its spike protein contains an insertion of multiple amino acids at the S1/S2 cleavage site, which is also observed in SARS-CoV-2 but not other betacoronaviruses, pointing again to natural evolution. Using phylogenetic dating methods, Boni et al. [13] assessed the divergence between SARS-CoV-2 and RaTG13 to possibly have happened as early as 1969. They also concluded that, given the large number of existing bat coronaviruses and their mutation rate, global surveillance systems employing genomic tools to identify and characterize pathogens in human disease are highly needed. Despite the many studies based on the viral genomic sequence, the origins of SARS-CoV-2 are still a major source of public debate while we write [102].
Identification of variants
Another fundamental use of genomic analysis is to track ‘variants of concern’, which pose a serious threat both for their potentially higher lethality or infectivity, and for the unknown efficacy of existent vaccines in contrasting them [37]. On 9 March 2020, a study employed metatranscriptome sequencing of samples from patients and controls and found that the number of intrahost variants was as much as 51, with a median of 1–4 in SARS-CoV-2-infected patients [143]. This indicates the in vivo evolution of SARS-CoV-2 after infection. During this pandemic, several transmissive variants have been reported. For example, the variant B.1.1.7 was first detected in southeast England in September 2020 and quickly became the dominant lineage in the country. Through genome analysis, 17 non-synonymous mutations and deletions in B.1.1.7 were identified, 8 of which in the spike protein, including N501Y, occurring at a key contact residue of the RBD [128]. In December 2020, the variant B.1.351 was reported from Eastern Cape Province, South Africa and characterized as carrying nine non-synonymous mutations, three of which at key sites in the RBD (K417N, E484K and N501Y) [156]. The P.1 variant was first detected in north Brazil in December 2020, and three RBD mutations, K417T, E484K and N501Y, were also identified in this lineage [45]. The 501Y mutation, present in all of the three lineages, has been reported to potentially cause an increased transmission rate, up to 70% [40]. Variants of concern are still constantly monitored across countries in an attempt to anticipate novel threats [172].
Tracking international spread of SARS-CoV-2
A constant effort is also being devoted to track the virus spread at the regional, national and global level since the first epidemic outbreak. In February 2020, Park et al. [123] analyzed the first COVID-19 case in Korea using phylogenetic analysis and found that it clustered together with other SARS-CoV-2 sequences reported from Wuhan. Subsequently, by fitting a molecular clock model, Zehender et al. [180] analyzed the viral sequences isolated from three patients in the first outbreak of COVID-19 in Italy and concluded that the virus was present in Italy weeks before the first case was reported in 21 February 2020. De Jesus et al. [77] analyzed six cases of early reports in Brazil by combining phylogenetic analysis with self-reported travel history. Their results suggested multiple independent importations from Italy at the beginning of the Brazilian COVID-19 outbreak, further contributing to understand the dynamics of the pandemic. In the meantime, nine viral genomes from early patients in the United States were sequenced and analyzed [46]. Through a combination of genome epidemiology and travel pattern analysis, it was found that coast-to-coast spread had occurred, thus highlighting an urgent need for national surveillance. On the other hand, Lu et al. [98], who analyzed 53 genomes from infected individuals in Guangdong, China, showed that the large majority of viral infections in the province were caused by multiple importations, thus concluding that the surveillance and intervention measures taken had been effective. They also recommended careful interpretation of phylogenetic trees built in the early phase of the pandemic and suggested that epidemiological information should be combined with genomic data for more reliable results. In this regard, a related study has been conducted by Lemey et al. [91], who integrated travel history data in Bayesian phylogeographic inference. The study analyzed 282 SARS-CoV-2 genomes, 64 of which included travel history data, concluding that more realistic spreading hypotheses and higher predictive accuracy could be obtained as compared to using sample locations only.
Epidemiology
Although the ‘Molecular Pharmacology and Biomarkers’ and ‘Molecular Virology’ clusters discussed above mostly assume a molecular perspective, the remaining four clusters shift the focus toward the actual impact of the pandemic, from epidemiological considerations to individual patient care. Here, we review the main studies in the ‘Epidemiology’ cluster.
Understanding epidemiological features and transmission dynamics of the pandemic is crucial to inform intervention policies [183], such as coordinating screening and containment strategies, anticipating the viral spread, and ensuring optimal use of resources to reduce morbidity and mortality [78]. During the COVID-19 pandemic, governments across the world have relied on epidemiological models to help guide their decisions [1]. For example, results from a study by the Imperial College in early March 2020 significantly influenced the country’s response strategies [1, 47]. The following Subsections describe major studies in the areas of epidemiological parameters estimation and assessment of non-pharmaceutical interventions (NPI).
Epidemiological parameters estimation
Critical epidemiological parameters influencing the spread of a virus include the ability of sustained human-to-human transmission, the basic reproduction number 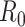 , and the incubation period. In order to estimate such parameters, a large variety of methods have been used, including susceptible, infectious and recovered models; phylogenetic analysis; statistical simulations; agent-based models; network models; etc. Nonetheless, their objectives and contexts are generally homogeneous, therefore we summarized all of them by systematically collecting methodologies, dataset details, specific applications and resource availability. All this information is summarized in Supplementary Table 4. The most relevant studies are discussed below.
, and the incubation period. In order to estimate such parameters, a large variety of methods have been used, including susceptible, infectious and recovered models; phylogenetic analysis; statistical simulations; agent-based models; network models; etc. Nonetheless, their objectives and contexts are generally homogeneous, therefore we summarized all of them by systematically collecting methodologies, dataset details, specific applications and resource availability. All this information is summarized in Supplementary Table 4. The most relevant studies are discussed below.
During the initial phase of the COVID-19 outbreak, human-to-human transmission by a novel coronavirus was confirmed, also based on Sanger sequencing and phylogenetic analysis, in a family of six COVID-19 patients, only five of which had been in Wuhan between 29 December 2019 and 4 January 2020 [25]. Another research analyzed the first 425 confirmed cases detected in Wuhan based on parametric model fitting of epidemiological information, reporting evidence of human-to-human transmission among close contacts dating back to December 2019 [92].
The basic reproduction number 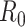 is the average number of secondary cases generated by an infected person. Many simulations were conducted to estimate its value across different countries. Most early studies reported a mean
is the average number of secondary cases generated by an infected person. Many simulations were conducted to estimate its value across different countries. Most early studies reported a mean 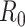 to be within the range of 2 to 3 using epidemic data from China [48, 92, 134]. However, another estimate based on a bats-hosts-reservoir-people transmission network model resulted in a value of 3.58 [30]. As the virus spread across Europe, further early
to be within the range of 2 to 3 using epidemic data from China [48, 92, 134]. However, another estimate based on a bats-hosts-reservoir-people transmission network model resulted in a value of 3.58 [30]. As the virus spread across Europe, further early 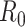 estimates were published from Italy (2.43–3.10) [39] and England (2.8–3.10) [96]. The value of
estimates were published from Italy (2.43–3.10) [39] and England (2.8–3.10) [96]. The value of 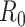 is studied continuously, as environmental and viral features evolve, and more data become available.
is studied continuously, as environmental and viral features evolve, and more data become available.
Another crucial epidemiological parameter is the viral incubation period i.e. the time interval between infection and occurrence of the first symptoms. Incubation period estimations impact important public health activities, such as active monitoring, surveillance and control [90]. Studies showed that the median incubation period for COVID-19 is approximately 5 days, which is similar to other coronaviruses, such as SARS and MERS [90, 92, 157, 183]. Moreover, the mean serial interval, that is the time interval between the first symptoms in the primary patient and the beginning of symptoms in the next infected patients, has also been studied. By analyzing 8579 cases from 30 provinces excluding Hubei in China, one early study found such interval to be slightly shorter than the mean incubation period, thus indicating a risk of asymptomatic transmission [183]. The ability of the virus to transmit without causing visible symptoms has been a prominent cause of its dramatic spread.
NPI assessment
Epidemiological models have been widely used to estimate the efficacy of NPI, such as case isolation, contact tracing, social distancing and lockdowns [78]. As for the previous Subsection, we reported a detailed summary of the used approaches in Supplementary Table 4 and will discuss a selection below.
On 28 February 2020, based on a stochastic transmission model, it was reported that highly effective contact tracing and case isolation could be enough to control a new COVID-19 outbreak within 3 months, as long as less than 1% asymptomatic cases occur [66]. However, asymptomatic transmission was later estimated at 6% by another study concluding that only widely used digital contact-tracing apps could possibly control the epidemic [48]. Although digital contact-tracing apps have raised concerns about privacy issues, they are widely accepted by some of the countries that achieved the best results at flattening the COVID-19 cases curve [72]. On this subject, Keeling et al. [79] conducted a detailed survey including information on social encounters from 5800 UK respondents, coupled with predictive models of contact tracing and control. The study concluded that the UK definition of contact as a permanence of at least 15 min within 2 meters is appropriate. However, according to the study, it also places a significant burden on health services, thus timely case detection and quarantine remain necessary to ensure the success of contact tracing.
Limitations to social activities to reduce contacts have also been widely studied and adopted. Measures like school and workplace closures or limiting gatherings have reduced the risk of overwhelming the health systems and bought more time for treatment and vaccine development, although at the cost of economic downturn [5]. For this reason, assessing the benefit-cost ratio of each intervention has been an objective of paramount importance, and many research efforts have been devoted to it. For example, in March 2020 a preprint article reported a comparison between one-time and intermittent social distancing scenarios in the United States based on a mathematical model [82]. The study concluded that adoption of the former could have delayed the epidemic peak eventually exacerbating the load on critical care services. The study contributed to the debate on the actual implementation of social distancing measures, which can take diverse forms. Another study, for example, used an agent-based model to analyze the effects of self-isolation, and in particular its impact on intensive care unit (ICU) occupancy in Canada [145]. According to the study, even with a self-isolation ratio of 40%, the need for ICU beds would still exceed the total supply in the country, suggesting once again the need for multiple combined interventions. On the other hand, cautious but timely lifting of social restrictions is also necessary in order to reduce their social burden. Therefore, the consequences of lifting NPIs have also been thoroughly studied. Hoertel et al. [68], for example, used a stochastic agent-based model to simulate the COVID-19 epidemic in France and analyze the potential impact of lifting a national lockdown. Although the study found a rebound to be almost certain, it also concluded that other measures, such as social distancing, mask-wearing and shielding of vulnerable people would still prevent the overwhelming of the critical care services.
Healthcare
In our embedding, the next cluster of studies concerns public health and related services. One of the greatest dangers posed by the ongoing pandemic has been the overwhelming of the national healthcare systems, constituting the main rationale behind policies aimed at ‘flattening the epidemic curve’. Among the top papers in our collection, a significant number in the ‘Healthcare’ group were devoted to studying the psychological burden on healthcare workers (HCWs) and lay people, the digital technologies to speed up the response by the healthcare system, and literature mining tools aiming at keeping researchers and practitioners up to date with latest knowledge.
Psychological burden on HCWs and the public
During the battle against COVID-19, HCWs played the role of front-line fighters. Many factors, such as the ever-increasing number of patients, the overwhelming workloads, the shortage of specific drugs or equipment, have contributed to place a significant psychological burden on them, prompting researchers to study such secondary effects of the pandemic. In our database, most articles on the subject collected data digitally through online surveys and analyzed them using logistic regression-based models. Given the uniformity of the approaches, we summarized the most representative ones in Supplementary Table 5, reporting the geographical area of each study, the number of individuals surveyed, the specific aim and the used method. We describe a few representative examples in the following.
In two studies, multivariate regression models were used to assess psychological impact on Wuhan HCWs based on 5062 [195] and 1577 [114] surveyed subjects respectively. Factors like concomitant chronic diseases, history of mental disorders and family members or relatives confirmed or suspected positive were found associated with stress, anxiety and depression. Conversely, social and professional support was confirmed to exert protective effects. The need for psychological support was also underlined by another study on 1257 HCWs in China, especially among women and nurses [87]. Similar results were also reported from other countries. For example, an online platform was used to gather data from 1379 HCWs in Italy [137]. The study concluded that younger age and female sex were associated with posttraumatic stress symptoms, depression, anxiety and high perceived stress. Frontline HCWs were associated with posttraumatic stress symptoms, while nurses and health care assistants were more likely to endorse severe insomnia.
Besides the direct psychological burden on front-line HCWs, the general public has also been psychologically affected by severe intervention measures such as isolation and social distancing, which impose changes in routines and may favor anxiety and depression [21]. Many studies have been conducted to assess their impact. For example, Mazza et al. [103] tried to identify risk and protective factors for psychological distress in Italy. Using an online survey platform, they collected data from 2766 individuals. Multivariate ordinal logistic regression highlighted an association between female gender, negative affect and detachment with higher levels of depression, anxiety and stress. In general, psychological effects of the pandemic may involve a complex system of factors, such as the fear of getting infected, the worry about socioeconomic costs, xenophobic attitudes, and compulsive checking and reassurance seeking [155]. Some of these conditions may be ameliorated not only by psychological interventions, but also through behavioral attitudes. One study found that physical activity following the WHO guidelines may be beneficial in this sense [97]. Finally, sentiment analysis has also been used to assess the emotional state of the public in response to the pandemic. For example, one study analyzed 105+ million tweets in six languages (English, Spanish, Arabic, French, Italian and Chinese) using deep learning language models to identify positive, negative and complex (like joking) expressions. They found that early tweets were dominated by a mixture of joking with anxious/pessimistic/annoyed feelings, which shifted toward positive states (optimistic, thankful and empathetic) as the pandemic came under control [185].
Digital technologies for rapid response
Digital technologies in the public health response to the pandemic are being harnessed worldwide with diverse applications [18]. Also using online data collection and analysis platforms, initiatives such as the epidemic intelligence from open sources by the WHO aim at early detection, verification, assessment and communication of public health threats based on publicly available data [171]. Rapid response strategies have been proposed using mobile applications in diverse contexts, such as self-reporting through online surveys [2, 130], digital contact tracing through Bluetooth-based proximity detection [48], or even remote healthcare services. For example, one study demonstrated vital signs measurement based on a convolutional neural network (CNN) model including an attention module to analyze image data acquired from the device’s camera [95].
COVID-19 research and public health
The surge in COVID-19 research publishing has posed both the opportunity and the challenge of exploiting continuously updated scientific knowledge with the aim of timely implementing state-of-art interventions [42]. In this context, AI approaches have been widely applied to help identify relevant literature, including the present work. On the other hand, crowd-based manually curated approaches to create annotated literature datasets for machine learning algorithms have been proposed as well [71]. In this regard, one notable effort has been made to automatically produce and update the COVID-19 research dataset (CORD-19) database, which include 500 000 scientific articles directly or indirectly related to SARS-CoV-2 [164]. This dataset has been specifically created for researchers to apply natural language processing algorithms and develop information retrieval and hypothesis generation approaches. Toward this aim, the TREC-COVID initiative was launched to build a test set and assess the ability of algorithms to rank CORD-19 papers based on their relevance to COVID-19-related topics [135, 161]. Based on the CORD-19 dataset, other works constructed knowledge graphs [170], also with applications to drug discovery [165].
Clinical medicine
Most studies belonging to the ‘Clinical Medicine’ cluster analyze data directly obtained from patients. They usually apply statistical analysis approaches, which are well assessed in medical literature and better suited in a context of low dimensional data. However, a number of studies also make use of omics data analysis (see Figure 5-right) mostly for the identification of biomarkers with clinical applications. For example, transcriptomics data were used to characterize critically ill patients leukocytes [139]; proteomics data were used to characterize SARS-CoV-2 neurotropism [160]; metabolomics data were used to identify prognostic biomarkers [38]. One study integrated all of these three data types to identify molecular markers in peripheral blood and plasma samples of COVID-19 patients [32]. Biomarkers identification is a common theme within the ‘Clinical Medicine’ cluster, as shown in the remainder of this Subsection.
Prognostics
The vast majority of the top-scoring papers in the ‘Clinical Medicine’ topic concerns the assessment of risk and the identification of risk factors for COVID-19 patients. Many of them use classical logistic regression-based methods on internally collected data from hospitals and clinics [94, 187], in one case even producing an online risk calculator for the public [76]. One notable study proposed an online platform for the collection of large pseudonymised health records from English subjects at the national level, accompanied by open source statistical data analysis software [169]. The tool identified being male, greater age, diabetes and severe asthma among the most prominent COVID-19 risk factors based on the health records of ˜17 000 000 English adults, including ˜11 000 COVID-19 patients. Concerning model complexity, it is worth mentioning one study that used an XGBoost model as a best-in-class approach to assess the performance of a generalized additive model combined with LASSO regression, which is more interpretable at the cost of slightly lower predictive power [83].
As opposed to identifying risk factors, other studies have focused on investigating some of the known ones in particular. For example, one paper supports that low levels of vitamin D concentration, which have been a concern also linked to lockdown measures [24], has little effect on the risk of infection based on the analysis of clinical data for ˜348 000 patients, 449 of which had confirmed COVID-19 infection [64]. Other such studies have investigated risk associations to diverse factors, such as smoking status based on Bayesian meta-analyses [149], hyperglycaemia using logistic regression [125] and even generic variants through whole-exome sequencing analysis [159].
Finally, among the highest rated papers in the ‘Clinical Medicine’ category, it is worth mentioning a systematic review, which underlines both the importance of prognostic models and the urgency to improve their reliability [174].
Clinical sign identification and clinical trials
Most other articles in the cluster are devoted to the identification of COVID-19 patients clinical signs and to the assessment of treatement efficacy. Of note, some articles dealing with clinical signs use imaging techniques like computed tomography (CT) [11, 173, 178]; however, they focus on the statistical analysis of manually extracted features, as opposed to direct computational segmentation or classification as described in Subsection 3.7.
Finally, our database picked up a few clinical trial studies that specifically refer to modeling tools. For example, a retrospective study on 13 981 patients with COVID-19 in the Hubei Province, China, based on a mixed-effect Cox model, found a reduction of all-cause mortality from 5.2 to 9.4% in the subgroup of 1219 patients treated with statins [186]. A living systematic review uses a Bayesian network meta-analysis of clinical data from 85 trials (at time of publication) to monitor treatment efficacy, confirming a large uncertainty in the outcomes of highly discussed drugs, such as remdesivir, azithromycin, hydroxychloroquine and tocilizumab [148].
Clinical imaging
The area of Clinical Imaging conceptually falls under the broader field of Clinical Medicine. However, due to the large number of specifically dedicated papers in our database, most of them sharing Deep Learning techniques, they formed a well-characterized cluster on their own (see Figure 3). A number of studies in this area use the term ‘radiomics’ to refer to the computational extraction of features from biomedical images [74, 89] (see Figure 5-right). Thoracic computer tomography (CT) and chest X-ray imaging have played an important role during this pandemic as easily accessible tools to diagnose COVID-19, monitor therapeutic efficacy and assess patients for discharge. In China, for example, portable chest X-ray devices are used in point-of-care testing, especially to monitor immobile, critically ill patients on a daily basis [93].
The general workflow of imaging-based diagnostics can be divided into three phases: pre-scan preparation, image acquisition, and disease diagnosis [196]. After raw data are acquired, images are stored in a picture archiving and communication systems. In the diagnosis phase, specifically trained radiologists inspect the images to assess the presence of COVID-19-related features. The integration of AI in the loop can help speeding up this step, potentially producing a significant impact in emergent situations.
CT image segmentation and classification
Application of AI in CT imaging can be generally divided into two main tasks: segmentation and classification. In chest CT image segmentation, deep learning models are employed to extract a target region of interest (ROI), such as lesion and lung lobes, and quantify the corresponding morphological features. We summarized studies belonging to this category in Supplementary Table 6, which includes references, methodology, number of patients, target ROI, data availability and performance scores. We mention some of the most representative studies below.
One intriguing approach proposed a weakly supervised model, which embeds a generative adversarial network (GAN) model [54] within the segmentation framework. This model is trained to replace the lesion with normal features until the image is classified as generated from a healthy patient, thus implicitly learning abnormal morphologies. This allowed to perform effective training even with a single voxel-level annotated COVID-19 patient CT scan [176]. Another innovative study used both a novel preprocessing method and a novel deep learning model for segmentation and quantification. Specifically, in order to deal with training data shortage, the proposed framework included a CT scan simulator for generating new images by interpolating existing ones over different time points. Moreover, the 3D segmentation problem was decomposed into three 2D problems, significantly reducing complexity and improving accuracy, as demonstrated over multi-country, multi-hospital, and multi-machine datasets [189].
Other approaches use human-in-the-loop strategies, in which radiologists correct the results of automatic segmentation, thus speeding up the learning process. One such example called VB-Net is based on a modified 3D CNN combining a V-Net [104] model with a bottle-neck structure [141]. Another one, called COPLE-Net, uses an adaptive self-learning framework with a noise-robust Dice loss that is suitable for noisy labels [163]. Finally, Chen et al. [31] designed a modified U-Net [136] model, achieving the highest performance among those we reviewed.
Besides segmenting ROIs, many studies have also been proposed to detect COVID-19 using chest CT images, reporting impressively high accuracy. As before, we summarized the main features of many top-scoring articles in this category in tabular form (see Supplementary Table 7) and mention some of them in the following.
Many studies employed segmentation models, such as U-Net [136] or its variations, to extract features before classification. This was the case for Gozes et al. [55] (U-Net) and Chen et al. [27] (UNet++ [194]), both of which also used a pre-trained ResNet-50 [65] model for classification, achieving 0.996 and 0.989 accuracy, respectively. Other works applied deep transfer learning models without using segmentation. For example, Jaiswal et al. [75] used a DenseNet201 architecture [70] and Pathak et al. [124] used a ResNet [65]. Among alternative approaches, Wang et al. [166] combined graph convolution models with convolutional neural networks and proposed a new model named FGCNet, which achieved 0.971 accuracy.
X-ray image classification
Due to the nature of X-ray images, segmentation is less used in this context. In fact, all the top papers in our database concerning X-ray imaging are devoted to classification tasks, aiming at COVID-19 diagnosis. Also, in this case, we produced a detailed tabular summary, which includes references, dataset details, learning model, application, data availability and accuracy scores (see Supplementary Table 8).
Among the top-performing models, Ozturk et al. [119] built on a previous You Only Look Once architecture named DarkNet [132], achieving 0.981 accuracy. Another work by Brunese et al. [17] used a model based on VGG-16 [150], which yielded an accuracy of 0.980. A number of studies focus on overcoming data limitation as a fundamental prerequisite to improve classification performance. For example, a work by Khalifa et al. [80] employed GAN for data augmentation and ResNet18 [65] for classification, achieving an F1-score of 0.990. On the other hand, Rajaraman et al. [127] trained a custom CNN and a selection of ImageNet pre-trained models on publicly available X-ray images and then applied transfer learning on COVID-19 images achieving 0.990 accuracy. Finally, Nour et al. [115] used a 5-layer CNN model for feature extraction and other machine learning models such as k-nearest neighbor, support vector machine (SVM), and decision trees for classification. In their results, the combination of CNN model and SVM achieved the highest accuracy (0.990).
In addition to diagnosis, automatic severity assessment has also been proposed through X-ray image analysis. Cohen et al. [35] created a geographic extent score (ranging 0–8) and lung opacity score (ranging 0–6) based on the evaluation from three experts, and used them to train a DenseNet model [70]. Their results show that the model can regress the two scores with 1.14 and 0.78 mean absolute error, respectively.
Latest contributions
In order to systematically review the latest research trends, we extracted the most recent (March–May 2021), top-scoring preprint articles that were also predicted to pass a peer review process with the highest probability. By reviewing such articles, we observed a shift of focus toward the monitoring of emergent SARS-CoV-2 variants, the evaluation of approved vaccines efficacy in real-world settings, and the follow-up of patients to investigate post-COVID-19 syndrome. We report a selection of representative articles in the following. Regardless of our effort to select the most potentially impactful literature, the mentioned results need to be considered unconfirmed.
Emergent variants of SARS-CoV-2
Based on sequencing techniques and bioinformatic analyses, several emergent variants of SARS-CoV-2 are being identified around the world (see Subsection 3.3.2). For example, variant P.3, carrying multiple mutations in the Spike protein, has been reported from the Philippines. These mutations could possibly impact the interactions of the Spike protein with the ACE2 receptor and neutralizing antibodies [9]. B.1.617 lineage is found to be the predominant clade in Maharashtra, India, with accumulation of convergent mutations [33]. Variant B.1.616 was identified in Western France. It is reported to have higher lethality and to be poorly detectable by RT-PCR on nasopharyngeal samples [49]. Besides, another study identified multiple N-terminal domain (NTD) and RBD mutations of SARS-CoV-2 associated with reduced antibody neutralization from an immunosuppressed patient with tacrolimus, steroids and convalescent plasma therapy [28]. It provides an evidence that immunocompromised patients with convalescent plasma therapy are potential breeding grounds for immune-escape mutants.
Effectiveness of vaccines in a real-world setting
Despite the benefits of vaccine clinical trials, their accuracy is limited by subject recruitment restrictions and sample size [177]. Evaluation in real-world settings is therefore necessary to obtain more detailed safety and efficacy estimations [144]. One of the most urgent needs is the evaluation of vaccine efficacy against SARS-CoV-2 variants [10, 126, 129, 144], which has been the subject of several recent studies. This is obtained through data analysis of large clinical datasets against sequence variants. For example, researchers in Oxfordshire, UK evaluated the effectiveness of Pfizer-BioNTech BNT162b2, Oxford-AstraZeneca ChAdOx1 and immunity after natural infection, against the B.1.1.7 variant in 13109 HCWs [99]. They found that natural infection with detectable anti-spike antibodies and two vaccine doses both provide robust protection. Besides, better understanding vaccine adverse effects is also important, both to minimize their impact on patients and to cope with vaccination hesitancy [131]. Researchers in the UK surveyed 974 HCWs with prior COVID-19 infections and compared those with and without a COVID-19 history using two-way analysis of covariance and a logistic regression model to evaluate the adverse effects following BNT162b2 vaccination [131]. They found that previous infection in absence of long COVID symptoms (see next Subsection) was associated with an increased risk of self-reported adverse events among the respondents. Besides, researchers in India assessed the outcomes of 515 HCWs completed two doses of Covishiel ChAdOx1-nCOV and Covaxin BBV-152 vaccines using logistic regression [151]. Both showed good immune response after two doses, which is good news in the war against COVID-19.
Elucidating the post-COVID syndrome
A portion of COVID-19 patients continue to experience persistent symptoms after being discharged from hospitals. This condition is known as post-COVID syndrome or ‘long COVID’ [34, 53, 69, 101]. To better characterize the syndrome, a follow-up study was conducted with 958 non-hospitalized patients in Germany, mostly with mild COVID-19 symptoms [6]. The study assessed the predictors of long-term symptoms, finding that 12.8% of the patients were affected by shortness of breath, anosmia, ageusia and fatigue at four or seven months after infection. Another nationwide cohort study in Germany followed 8679 hospitalized patients and analyzed risk factors [59]. It found a considerable long-term mortality of 29.6% in all subjects and readmission rates of 26.8% among 6235 discharged patients. Coagulopathy, body mass index 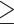 40 and age were reported to be risk factors for 180-day mortality. Finally, a study compared the CpGs number, telomere length, and ACE2 and DPP4 expression between 117 COVID-19 survivors and 144 non-infected volunteers using pyrosequencing [109]. The results showed a significant telomere shortening (3.03–10.67 Kb) and ACE2 expression decreasing in the COVID-19 survivors.
40 and age were reported to be risk factors for 180-day mortality. Finally, a study compared the CpGs number, telomere length, and ACE2 and DPP4 expression between 117 COVID-19 survivors and 144 non-infected volunteers using pyrosequencing [109]. The results showed a significant telomere shortening (3.03–10.67 Kb) and ACE2 expression decreasing in the COVID-19 survivors.
Discussion and conclusions
The ongoing COVID-19 pandemic has impacted all aspects of society and ignited an unprecedented global research effort. With the last severe pandemic being the Spanish flu, which dates back to 1918, COVID-19 is the first one to occur in a digitized world. With computers being ubiquitously used in modern societies, they are also expected to constitute a novel tool to fight global health emergencies, providing faster data and knowledge sharing, advanced analytical tools, and more accurate forecasting capabilities.
In order to analyze the impact of computational applications to the ongoing pandemic from a scientific point of view, we built a large database of research articles covering diverse aspects of the emergency, but all sharing the use of computational tools. Due to the complexity of the constructed database, we in turn used computational approaches to guide our review. The main topics we identified i.e. Molecular Pharmacology and Biomarkers, Molecular Virology, Epidemiology, Healthcare, Clinical Medicine and Clinical Imaging highlight the fundamental role of computational applications in supporting critical activities such as scientific discovery, clinical practices and institutional decision making in diverse areas of the ongoing crisis. We believe that our approach guided by automatic collection, categorization and prioritization of research articles can help to deal with publication bursts that are expected during emergencies such as the COVID-19 crisis.
During the pandemic, computational approaches have been used at various extents, from facilitating statistical data analysis of large datasets and gain a better understanding of the rapidly changing situation, to the construction of sophisticated machine learning models for automating, accelerating and/or guiding biomedical tasks. For some applications, such as those within the fields of genomics or structural chemistry, computers are now established tools routinely used through well assessed and standardized analytical pipelines. In other areas, such as drug repurposing or vaccine development, they are widely used as tools for candidate prioritization or hypothesis generation. In clinical applications such as automatic diagnostics and prognostics, computational models proved to be potentially effective, although the special caution required by patient care warrants further assessment and development toward fruitful integrating within healthcare systems [174]. Simulations based on mathematical models have been widely used to forecast viral spread or the effect of NPI measures. Governments around the world have relied on the results of such models to make informed decisions with vast socioeconomic effects. The immense impact that mathematical modeling can have in this area prompts the scientific community to strive for more reliability and wise use of available data, which can be particularly fragmented and inconsistent at the beginning of a global health emergency [78]. In other areas, such as the assessment of treatments efficacy and risk factors, computational approaches have supported classical statistical analyses of large datasets collected and managed through digital platforms.
While we write, the COVID-19 pandemic still poses a global threat. Nonetheless, extraordinary successes have been achieved by the scientific community in understanding the SARS-CoV-2 mechanisms and countermeasures. The contribution of computational sciences in this endeavor has been remarkable. In this regard, we believe that the experience gathered during the COVID-19 pandemic should lay the foundation for objectives that reach beyond the end of the current crisis.
Key Points
A software framework was developed to automatically collect 17 269 computational studies related to COVID-19 from multiple sources, including PubMed and preprint servers.
Using an AI model, articles were automatically categorized into clusters corresponding to six topics: Molecular Virology, Molecular Pharmacology and Biomarkers, Epidemiology, Healthcare, Clinical Medicine and Clinical Imaging.
All the studies were ranked using bibliometric information and a Deep Neural Network model was developed to predict the chance for preprint articles to pass peer review.
The developed framework was used as a guide throughout an extensive and detailed manual review, which demonstrates the huge impact of computational approaches during the COVID-19 global crisis.
Supplementary Material
Francesco Napolitano is a research scientist at Structural and Functional Bioinformatics Group, Computational Bioscience Research Center, KAUST.
Xiaopeng Xu is a PhD student at Structural and Functional Bioinformatics Group, Computational Bioscience Research Center, KAUST.
Xin Gao is a professor at Computational Bioscience Research Center, KAUST and the principal investigator of Structural and Functional Bioinformatics Group.
Contributor Information
Francesco Napolitano, Computational Bioscience Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Makkah, Saudi Arabia.
Xiaopeng Xu, Computational Bioscience Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Makkah, Saudi Arabia.
Xin Gao, Computational Bioscience Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Makkah, Saudi Arabia.
Data availability statement
The CSCoV database is publicly available from Zenodo repository: https://zenodo.org/record/5495823 [111]. The code used in the study is publicly available from the GitHub repository: https://github.com/SFB-KAUST/covid-review.
Author contributions statement
F.N. collected and categorized the articles. F.N. and X.X. reviewed the articles by topic. X.X. retrieved and reviewed the latest contributions. F.N., X.X. and X.G. wrote and reviewed the manuscript.
Funding
This work was supported by grants from KAUST under the award number BAS/1/1624-01, FCC/1/1976-18-01, FCC/1/1976-23-01, FCC/1/1976-25-01, FCC/1/1976-26-01, REI/1/4473-01-01, URF/1/4352-01-01, and REI/1/4742-01-01.
References
- 1. Adam D. Special report: The simulations driving the world’s response to covid-19. Nature 2020; 580(7802):316–9. [DOI] [PubMed] [Google Scholar]
- 2. Allen WE, Altae-Tran H, Briggs J, et al. Population-scale longitudinal mapping of covid-19 symptoms, behaviour and testing. Nat Hum Behav 2020; 4(9):972–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altay O., Mohammadi E., Lam S., Turkez H., Boren J., Nielsen J., Uhlen M., and Mardinoglu A.. Current Status of COVID-19 Therapies and Drug Repositioning Applications, jul 2020. [DOI] [PMC free article] [PubMed]
- 4. Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of sars-cov-2. Nat Med 2020; 26(4):450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson RM, Heesterbeek H, Klinkenberg D, et al. How will country-based mitigation measures influence the course of the covid-19 epidemic? The lancet 2020; 395(10228): 931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Augustin M, Schommers P, Stecher M, et al. Recovered not restored: Long-term health consequences after mild covid-19 in non-hospitalized patients medRxiv. 2021.
- 7. Baig AM, Khaleeq A, Ali U, et al. Evidence of the covid-19 virus targeting the cns: tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem Nerosci 2020; 11(7):995–8. [DOI] [PubMed] [Google Scholar]
- 8. Barberis E, Timo S, Amede E, et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sars-cov-2. Int J Mol Sci 2020; 21(22):8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bascos NAD, Mirano-Bascos D, Saloma CP. Structural analysis of spike protein mutations in the sars-cov-2 p. 3 variant bioRxiv. 2021.
- 10. Bernal JL, Andrews N, Gower C, et al. Effectiveness of covid-19 vaccines against the b. 1.617. 2 variant medRxiv. 2021. [DOI] [PubMed]
- 11. Bhayana R, Som A, Li MD, et al. Abdominal imaging findings in COVID-19: Preliminary observations. Radiology oct 2020; 297(1):E207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bojkova D, Klann K, Koch B, et al. Proteomics of sars-cov-2-infected host cells reveals therapy targets. Nature 2020; 583(7816):469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boni MF, Lemey P, Jiang X, et al. Evolutionary origins of the sars-cov-2 sarbecovirus lineage responsible for the covid-19 pandemic. Nat Microbiol 2020; 5(11):1408–17. [DOI] [PubMed] [Google Scholar]
- 14. Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of sars-cov-2 infection. Cell 2020; 182(3):685–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Braga L, Ali H, Secco I, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike-induced syncytia. Nature apr 2021;1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brainard J. Do preprints improve with peer review? A little, one study suggests. Science mar 2020. [Google Scholar]
- 17. Brunese L, Mercaldo F, Reginelli A, et al. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from x-rays. Comput Methods Programs Biomed 2020; 196:105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Budd J, Miller BS, Manning EM, et al. Digital technologies in the public-health response to covid-19. Nat Med 2020; 26(8):1183–92. [DOI] [PubMed] [Google Scholar]
- 19. Cagliani R, Forni D, Clerici M, et al. Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. J Virol 2020; 94(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campbell JC, Hindle A, Stroulia E. Latent Dirichlet Allocation: Extracting Topics from Software Engineering Data. The Art and Science of Analyzing Software Data 2015; 3:139–59. [Google Scholar]
- 21. Campos JADB, Martins BG, Campos LA, et al. Early psychological impact of the covid-19 pandemic in brazil: A national survey. J Clin Med 2020; 9(9):2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carneiro CFD, Queiroz VGS, Moulin TC, et al. Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature. Research Integrity and Peer Review 2020; 5(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castillo C, Donato D, Gionis A. Estimating number of citations using author reputation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Vol. 4726LNCS. Berlin, Heidelberg: Springer, 2007, 107–17. [Google Scholar]
- 24. Chakhtoura M, Napoli N, El Hajj Fuleihan G. Commentary: Myths and facts on vitamin D amidst the COVID-19 pandemic. Metabolism: Clinical and Experimental 2020; 109:154276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The lancet 2020; 395(10223):514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang S, Pierson E, Koh PW, et al. Mobility network models of COVID-19 explain inequities and inform reopening. Nature 2020. [DOI] [PubMed] [Google Scholar]
- 27. Chen J, Wu L, Zhang J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci Rep 2020; 10(1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen L, Zody MC, Mediavilla JR, et al. Emergence of multiple sars-cov-2 antibody escape variants in an immunocompromised host undergoing convalescent plasma treatment medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 29. Chen TM, Rui J, Wang QP, et al. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty 2020; 9(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen T-M, Rui J, Wang Q-P, et al. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty 2020; 9(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X, Yao L, Zhang Y. Residual attention u-net for automated multi-class segmentation of covid-19 chest ct images arXiv preprint arXiv:2004.05645. 2020.
- 32. Chen Y-MY, Zheng Y, Yu Y, et al. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J 2020; 39(24):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cherian S, Potdar V, Jadhav S, et al. Convergent evolution of sars-cov-2 spike mutations, l452r, e484q and p681r, in the second wave of covid-19 in maharashtra, india bioRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 34. Chopra V, Flanders SA, O’Malley M, et al. Sixty-day outcomes among patients hospitalized with covid-19. Ann Intern Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cohen JP, Dao L, Roth K, et al. Predicting covid-19 pneumonia severity on chest x-ray with deep learning. Cureus 2020; 12(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-ncov) by real-time rt-pcr. Eurosurveillance 2020; 25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cyranoski D. Alarming COVID variants show vital role of genomic surveillance, jan, 2021. [DOI] [PubMed]
- 38. Danlos F-X, Grajeda-Iglesias C, Durand S, et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death & Disease 2021 12:3 2021; 12(3):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D’Arienzo M, Coniglio A. Assessment of the sars-cov-2 basic reproduction number, r0, based on the early phase of covid-19 outbreak in italy. Biosafety and Health 2020; 2(2):57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of sars-cov-2 lineage b. 1.1. 7 in england. Science 2021; 372(6538). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci 2020; 253:117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Else H. How a torrent of COVID science changed research publishing - in seven charts, 2020. [DOI] [PubMed]
- 43. Encinar JA, Menendez JA. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2’-O-Methylation of Viral RNA. Viruses 2020; 12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fantini J, Di Scala C, Chahinian H, et al. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents 2020; 55(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Faria NR, Claro IM, Candido D, et al. Genomic characterisation of an emergent sars-cov-2 lineage in manaus: preliminary findings. Virological 2021. [Google Scholar]
- 46. Fauver JR, Petrone ME, Hodcroft EB, et al. Coast-to-coast spread of sars-cov-2 during the early epidemic in the united states. Cell 2020; 181(5):990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ferguson N, Laydon D, Nedjati-Gilani G, et al. Report 9: Impact of non-pharmaceutical interventions (npis) to reduce covid19 mortality and healthcare demand. Imperial College London 2020; 10(77482):491–7. [Google Scholar]
- 48. Ferretti L, Wymant C, Kendall M, et al. Quantifying sars-cov-2 transmission suggests epidemic control with digital contact tracing. Science 2020; 368(6491). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fillatre P, Dufour MJ, Behillil S, et al. A new sars-cov-2 variant poorly detected by rt-pcr on nasopharyngeal samples, with high lethality, 2021. [DOI] [PMC free article] [PubMed]
- 50. Fricke S. Semantic scholar. J Med Libr Assoc 2018; 106(1): 145–7. [Google Scholar]
- 51. Galindez G, Matschinske J, Rose TD, et al. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nature Computational Science 2021; 1(1):33–41. [DOI] [PubMed] [Google Scholar]
- 52. Gassen NC, Papies J, Bajaj T, et al. Analysis of sars-cov-2-controlled autophagy reveals spermidine, mk-2206, and niclosamide as putative antiviral therapeutics BioRxiv, 2020.
- 53. Goërtz YM, Van Herck M, Delbressine JM, et al. Persistent symptoms 3 months after a sars-cov-2 infection: the post-covid-19 syndrome? ERJ open research 2020; 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks arXiv preprint arXiv:1406.2661. 2014.
- 55. Gozes O, Frid-Adar M, Greenspan H, et al. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis arXiv preprint arXiv:2003.05037. 2020.
- 56. Grifoni A, Sidney J, Zhang Y, et al. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host and Microbe 2020; 27(4):671–680.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. T. R. C. Group . Dexamethasone in Hospitalized Patients with Covid-19. New England Journal of Medicine 2021; 384(8):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gu H, Chen Q, Yang G, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020; 369(6511). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guenster C, Busse R, Spoden M, et al. 6-month follow up of 8679 hospitalized covid-19 patients in germany: A nationwide cohort study medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 60. Guerrero-Bote VP, Moya-Anegón F. Relationship between downloads and citations at journal and paper levels, and the influence of language. Scientometrics 2014; 101(2):1043–65. [Google Scholar]
- 61. Gupta A, Madhavan MV, Poterucha TJ, et al. Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat Commun 2021; 12(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type i interferon activity and inflammatory responses in severe covid-19 patients. Science 2020; 369(6504):718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Han Y, Yang L, Duan X, et al. Identification of candidate covid-19 therapeutics using hpsc-derived lung organoids BioRxiv, 2020.
- 64. Hastie CE, Mackay DF, Ho F, et al. Vitamin D concentrations and COVID-19 infection in UK Biobank. Diabetes Metab Syndr 2020; 14(4):561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, 770–8.
- 66. Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling covid-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 2020; 8(4):e488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hikmet F, Méar L, Edvinsson Å, et al. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 2020; 16(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoertel N, Blachier M, Blanco C, et al. A stochastic agent-based model of the sars-cov-2 epidemic in france. Nat Med 2020; 26(9):1417–21. [DOI] [PubMed] [Google Scholar]
- 69. Huang C, Huang L, Wang Y, et al. 6-month consequences of covid-19 in patients discharged from hospital: a cohort study. The Lancet 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, 4700–8.
- 71. Huang T-H, Huang C-Y, Ding C-KC, et al. Coda-19: Using a non-expert crowd to annotate research aspects on 10,000+ abstracts in the covid-19 open research dataset. In: Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, 2020.
- 72. Huang Y, Sun M, Sui Y. How Digital Contact Tracing Slowed Covid-19 in East Asia. Harvard Business Review Digital Article, pages 2020; 1–8. [Google Scholar]
- 73. Hufsky F, Lamkiewicz K, Almeida A, et al. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief Bioinform 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ito R, Iwano S, Naganawa S. A review on the use of artificial intelligence for medical imaging of the lungs of patients with coronavirus disease 2019. Diagn Interv Radiol 2020; 26(5):443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jaiswal A, Gianchandani N, Singh D, et al. Classification of the covid-19 infected patients using densenet201 based deep transfer learning. Journal of Biomolecular Structure and Dynamics 2020;1–8. [DOI] [PubMed] [Google Scholar]
- 76. Jehi L, Ji X, Milinovich A, et al. Individualizing Risk Prediction for Positive Coronavirus Disease 2019 Testing: Results From 11,672 Patients. Chest 2020; 158(4):1364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jesus JGD, Sacchi C, Candido DDS, et al. Importation and early local transmission of covid-19 in brazil, 2020. Revista do Instituto de Medicina Tropical de Sao Paulo 2020; 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jewell NP, Lewnard JA, Jewell BL. Predictive mathematical models of the covid-19 pandemic: underlying principles and value of projections. JAMA 2020; 323(19):1893–4. [DOI] [PubMed] [Google Scholar]
- 79. Keeling MJ, Hollingsworth TD, Read JM. Efficacy of contact tracing for the containment of the 2019 novel coronavirus (covid-19). J Epidemiol Community Health 2020; 74(10):861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khalifa NEM, Taha MHN, Hassanien AE, et al. Detection of coronavirus (covid-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset arXiv preprint arXiv:2004.01184. 2020.
- 81. Kim D, Lee J-Y, Yang J-S, et al. The architecture of sars-cov-2 transcriptome. Cell 2020; 181(4):914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kissler SM, Tedijanto C, Lipsitch M, et al. Social distancing strategies for curbing the covid-19 epidemic medRxiv. 2020.
- 83. Knight SR, Ho A, Pius R, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. The BMJ 2020; 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kong R, Yang G, Xue R, et al. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020; 36(20):5109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kumar A, Gupta PK, Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. Diabetes Metab Syndr 2020; 14(4):569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kwon D. How swamped preprint servers are blocking bad coronavirus research. Nature 2020; 581(7807):130–1. [DOI] [PubMed] [Google Scholar]
- 87. Lai J, Ma S, Wang Y, et al. Factors associated with mental health outcomes among health care workers exposed to coronavirus disease 2019. JAMA Netw Open 2020; 3(3):e203976–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lalmuanawma S., Hussain J., and Chhakchhuak L.. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: A review, oct 2020. [DOI] [PMC free article] [PubMed]
- 89. Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14(12):749–62. [DOI] [PubMed] [Google Scholar]
- 90. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (covid-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020; 172(9):577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lemey P, Hong SL, Hill V, et al. Accommodating individual travel history and unsampled diversity in bayesian phylogeographic inference of sars-cov-2. Nat Commun 2020; 11(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li Q, Guan X, Wu P, et al. Early transmission dynamics in wuhan, china, of novel coronavirus–infected pneumonia. New England journal of medicine 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liang T. Handbook of covid-19 prevention and treatment. The First Affiliated Hospital, Zhejiang University School of Medicine Compiled According to Clinical Experience 2020; 68. [Google Scholar]
- 94. Liu W, Tao ZW, Wang L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl) 2020; 133(9): 1032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu X, Fromm J, Patel S, et al. Multi-task temporal shift attention networks for on-device contactless vitals measurement arXiv preprint arXiv:2006.03790. 2020.
- 96. Liu Y, Tang JW, Lam TT. Transmission dynamics of the covid-19 epidemic in england. Int J Infect Dis 2021; 104:132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. López-Bueno R, Calatayud J, Ezzatvar Y, et al. Association between current physical activity and current perceived anxiety and mood in the initial phase of covid-19 confinement. Front Psych 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lu J, du Plessis L, Liu Z, et al. Genomic epidemiology of sars-cov-2 in guangdong province, china. Cell 2020; 181(5): 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lumley SF, Rodger G, Constantinides B, et al. An observational cohort study on the incidence of sars-cov-2 infection and b. 1.1. 7 variant infection in healthcare workers by antibody and vaccination status medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 100. Mahase E. Covid-19: US approves remdesivir despite WHO trial showing lack of efficacy. BMJ 2020; 371:m4120. [Google Scholar]
- 101. Mandal S, Barnett J, Brill SE, et al. ‘long-covid’: a cross-sectional study of persisting symptoms, biomarker and imaging abnormalities following hospitalisation for covid-19. Thorax 2021; 76(4):396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maxmen A. Divisive COVID ‘lab leak’ debate prompts dire warnings from researchers. Nature 2021; 594(7861):15–6. [DOI] [PubMed] [Google Scholar]
- 103. Mazza C, Ricci E, Biondi S, et al. A nationwide survey of psychological distress among italian people during the covid-19 pandemic: immediate psychological responses and associated factors. Int J Environ Res Public Health 2020; 17(9):3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Milletari F, Navab N, Ahmadi S-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, 2016, 565–71. [Google Scholar]
- 105. Ming W. K., Huang J., and Zhang C. J.. Breaking down of healthcare system: Mathematical modelling for controlling the novel coronavirus (2019-nCoV) outbreak in Wuhan, China, jan 2020.
- 106. Mishra D., Mishra A., Chaturvedi V. K., and Singh M. P.. An overview of COVID-19 with an emphasis on computational approach for its preventive intervention, oct 2020. [DOI] [PMC free article] [PubMed]
- 107. Mitjà O, Corbacho-Monné M, Ubals M, et al. Hydroxychloroquine for Early Treatment of Adults With Mild Coronavirus Disease 2019: A Randomized, Controlled Trial. Clin Infect Dis 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mohamed K, Yazdanpanah N, Saghazadeh A, et al. Computational Drug Discovery and Repurposing for the Treatment of COVID-19: A Systematic Review. SSRN Electron J 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mongelli A. Evidence for biological age acceleration and telomere 2 shortening in covid-19 survivors medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 110. Nabavi S, Ejmalian A, Moghaddam ME, et al. Medical Imaging and Computational Image Analysis in COVID-19 Diagnosis: A Review. 2020. [DOI] [PMC free article] [PubMed]
- 111. Napolitano F, Xu X, Gao X. Impact of computational approaches in the fight against covid-19: an ai guided review of 17,000 studies – the cscov database. Zenodo Jun 2021. 10.5281/zenodo.5495823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Naqvi AAT, Fatima K, Mohammad T, et al. Insights into sars-cov-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2020; 1866(10):165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nguyen A, David JK, Maden SK, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol 2020; 94(13):e00510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ni MY, Yang L, Leung CM, et al. Mental health, risk factors, and social media use during the covid-19 epidemic and cordon sanitaire among the community and health professionals in wuhan, china: cross-sectional survey. JMIR mental health 2020; 7(5):e19009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nour M, Cömert Z, Polat K. A novel medical diagnosis model for covid-19 infection detection based on deep features and bayesian optimization. Appl Soft Comput 2020; 97:106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ojha PK, Kar S, Krishna JG, et al. Therapeutics for COVID-19: from computation to practices-where we are, where we are heading to. Mol Divers 2020; 1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Oli AN, Obialor WO, Ifeanyichukwu MO, et al. Immunoinformatics and Vaccine Development: An Overview. ImmunoTargets and Therapy 2020; 9:13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ong EZ, Chan YFZ, Leong WY, et al. A dynamic immune response shapes covid-19 progression. Cell Host Microbe 2020; 27(6):879–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ozturk T, Talo M, Yildirim EA, et al. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput Biol Med 2020; 121:103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pan X-w, Xu D, Zhang H, et al. Identification of a potential mechanism of acute kidney injury during the covid-19 outbreak: a study based on single-cell transcriptome analysis. Intensive Care Med 2020; 46(6):1114–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Paraskevis D, Kostaki EG, Magiorkinis G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol 2020; 79:104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Paraskevis D, Kostaki EG, Magiorkinis G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event, 2020. [DOI] [PMC free article] [PubMed]
- 123. Park WB, Kwon N-J, Choi S-J, et al. Virus isolation from the first patient with sars-cov-2 in korea. J Korean Med Sci 2020; 35(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Pathak Y, Shukla PK, Tiwari A, et al. Deep transfer learning based classification model for covid-19 disease. Irbm 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu SP, Zhang Q, Wang W, et al. Hyperglycemia is a strong predictor of poor prognosis in COVID-19. Diabetes Res Clin Pract 2020; 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of infectious sars-cov-2 variant b. 1.617. 2 to monoclonal antibodies and sera from convalescent and vaccinated individuals bioRxiv. 2021.
- 127. Rajaraman S, Siegelman J, Alderson PO, et al. Iteratively pruned deep learning ensembles for covid-19 detection in chest x-rays. IEEE Access 2020; 8:115041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus/nCoV-2019 Genomic Epidemiology - Virological, 2020.
- 129. Ranzani OT, Hitchings M, Dorion M, et al. Effectiveness of the coronavac vaccine in the elderly population during a p. 1 variant-associated epidemic of covid-19 in brazil: A test-negative case-control study medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 130. Rao ASS, Vazquez JA. Identification of covid-19 can be quicker through artificial intelligence framework using a mobile phone–based survey when cities and towns are under quarantine. Infection Control & Hospital Epidemiology 2020; 41(7):826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Raw RK, Kelly C, Rees J, et al. Previous covid-19 infection but not long-covid is associated with increased adverse events following bnt162b2/pfizer vaccination medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 132. Redmon J, Farhadi A. Yolo9000: Better, faster, stronger, 2016.
- 133. Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019-nCoV, 2020. [DOI] [PMC free article] [PubMed]
- 134. Riou J, Althaus CL. Pattern of early human-to-human transmission of wuhan 2019 novel coronavirus (2019-ncov), december 2019 to january 2020. Eurosurveillance 2020; 25(4):2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Roberts K, Alam T, Bedrick S, et al. Trec-covid: rationale and structure of an information retrieval shared task for covid-19. J Am Med Inform Assoc 2020; 27(9):1431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, 234–41. [Google Scholar]
- 137. Rossi R, Socci V, Pacitti F, et al. Mental health outcomes among frontline and second-line health care workers during the coronavirus disease 2019 (covid-19) pandemic in italy. JAMA Netw Open 2020; 3(5):e2010185–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sauvat A, Ciccosanti F, Colavita F, et al. On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2. Cell Death and Disease 2020; 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. SE G, CC DS, DB O, et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med Exp 2020; 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sempowski GD, Saunders KO, Acharya P, et al. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19, 2020. [DOI] [PMC free article] [PubMed]
- 141. Shan F, Gao Y, Wang J, et al. Lung infection quantification of covid-19 in ct images with deep learning arXiv preprint arXiv:2003.04655. 2020.
- 142. Shao P, Shan Y. Beware of asymptomatic transmission: Study on 2019-nCoV prevention and control measures based on extended SEIR model, 2020.
- 143. Shen Z, Xiao Y, Kang L, et al. Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis 2020; 71(15):713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Shinde V, Bhikha S, Hossain Z, et al. Preliminary efficacy of the nvx-cov2373 covid-19 vaccine against the b. 1.351 variant MedRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 145. Shoukat A, Wells CR, Langley JM, et al. Projecting demand for critical care beds during covid-19 outbreaks in canada. Cmaj 2020; 192(19):E489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Shrock E, Fujimura E, Kula T, et al. Viral epitope profiling of covid-19 patients reveals cross-reactivity and correlates of severity. Science 2020; 370(6520). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shu T, Ning W, Wu D, et al. Plasma proteomics identify biomarkers and pathogenesis of covid-19. Immunity 2020; 53(5):1108–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Siemieniuk RA, Bartoszko JJ, Ge L, et al. Drug treatments for covid-19: Living systematic review and network meta-Analysis. The BMJ 2020; 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Simons D, Shahab L, Brown J, et al. The association of smoking status with SARS-CoV-2 infection, hospitalization and mortality from COVID-19: a living rapid evidence review with Bayesian meta-analyses (version 7), 2020. [DOI] [PMC free article] [PubMed]
- 150. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition arXiv preprint arXiv:1409.1556. 2014.
- 151. SINGH AK, Phatak SR, SINGH R, et al. Antibody response after second-dose of chadox1-ncov (covishieldtm) and bbv-152 (covaxintm) among health care workers in india: Final results of cross-sectional coronavirus vaccine-induced antibody titre (covat) study medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 152. Sliwoski G, Kothiwale S, Meiler J, et al. Computational methods in drug discovery, 2014. [DOI] [PMC free article] [PubMed]
- 153. Soderberg CK, Errington TM, Nosek BA. Credibility of preprints: an interdisciplinary survey of researchers. R Soc Open Sci 2020; 7(10):201520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tahamtan I, Safipour Afshar A, Ahamdzadeh K. Factors affecting number of citations: a comprehensive review of the literature. Scientometrics 2016; 107(3):1195–225. [Google Scholar]
- 155. Taylor S, Landry CA, Paluszek MM, et al. Covid stress syndrome: Concept, structure, and correlates. Depress Anxiety 2020; 37(8):706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (sars-cov-2) lineage with multiple spike mutations in south africa medRxiv. 2020.
- 157. Tu W, Tang H, Chen F, et al. Epidemic update and risk assessment of 2019 novel coronavirus-china, january 28, 2020. China CDC Weekly 2020; 2(6):83–6. [PMC free article] [PubMed] [Google Scholar]
- 158. Vale RD. Accelerating scientific publication in biology. Proc Natl Acad Sci U S A 2015; 112(44):13439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Van Der Made CI, Simons A, Schuurs-Hoeijmakers J, et al. Presence of Genetic Variants among Young Men with Severe COVID-19. JAMA - Journal of the American Medical Association 2020; 324(7):663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Virhammar J, Kumlien E, Fällmar D, et al. Acute necrotizing encephalopathy with SARS-CoV-2 RNA confirmed in cerebrospinal fluid. Neurology 2020; 95(10):445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Voorhees E, Alam T, Bedrick S, et al. Trec-covid: constructing a pandemic information retrieval test collection. In: ACM SIGIR Forum, Vol. 54. NY, USA: ACM New York, 2021, 1–12. [Google Scholar]
- 162. Wang C, Li X, Ning W, et al. Multi-omic profiling of plasma identify biomarkers and pathogenesis of covid-19 in children medRxiv. 2021.
- 163. Wang G, Liu X, Li C, et al. A noise-robust framework for automatic segmentation of covid-19 pneumonia lesions from ct images. IEEE Trans Med Imaging 2020; 39(8):2653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wang LL, Lo K, Chandrasekhar Y, et al. Cord-19: The covid-19 open research dataset ArXiv, 2020.
- 165. Wang Q, Li M, Wang X, et al. Covid-19 literature knowledge graph construction and drug repurposing report generation arXiv preprint arXiv:2007.00576. 2020.
- 166. Wang S-H, Govindaraj VV, Górriz JM, et al. Covid-19 classification by fgcnet with deep feature fusion from graph convolutional network and convolutional neural network. Information Fusion 2021; 67:208–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang X, Guan Y. COVID-19 drug repurposing: A review of computational screening methods, clinical trials, and protein interaction assays. Med Res Rev page med.217282020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Wang Z, Xu X. scrna-seq profiling of human testes reveals the presence of the ace2 receptor, a target for sars-cov-2 infection in spermatogonia, leydig and sertoli cells. Cell 2020; 9(4):920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020; 584(7821):430–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Wise C, Ioannidis VN, Calvo MR, et al. Covid-19 knowledge graph: accelerating information retrieval and discovery for scientific literature arXiv preprint arXiv:2007.12731. 2020.
- 171. World Health Organization . Epidemic intelligence from open sources (EIOS).
- 172. World Health Organization . Tracking sars-cov-2 variants.
- 173. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 579(7798):265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. The BMJ 2020; 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in covid-19 patients. Emerging microbes & infections 2020; 9(1):761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Xu Z, Cao Y, Jin C, et al. Gasnet: Weakly-supervised framework for covid-19 lesion segmentation arXiv preprint arXiv:2010.09456. 2020.
- 177. Yelin I, Katz R, Herzel E, et al. Associations of the bnt162b2 covid-19 vaccine effectiveness with patient age and comorbidities at daily resolution medRxiv. 2021.
- 178. Yu Q, Wang Y, Huang S, et al. Multicenter cohort study demonstrates more consolidation in upper lungs on initial CT increases the risk of adverse clinical outcome in COVID-19 patients. Theranostics 2020; 10(12):5641–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020; 368(6491):630–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zehender G, Lai A, Bergna A, et al. Genomic characterization and phylogenetic analysis of sars-cov-2 in italy. J Med Virol 2020; 92(9):1637–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019-ncov infection: a bioinformatics analysis based on single-cell transcriptomes BioRxiv, 2020.
- 182. Zhang H, Rostami MR, Leopold PL, et al. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med 2020; 202(2):219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Zhang J, Litvinova M, Wang W, et al. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside hubei province, china: a descriptive and modelling study. Lancet Infect Dis 2020; 20(7):793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Zhang L, Zhou R. Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase. Journal of Physical Chemistry B 2020; 124(32):6955–62. [DOI] [PubMed] [Google Scholar]
- 185. Zhang X, Yang Q, Albaradei S, et al. Rise and fall of the global conversation and shifting sentiments during the COVID-19 pandemic. Humanities and Social Sciences Communications 2021; 8(1):120. [Google Scholar]
- 186. Zhang XJ, Qin JJ, Cheng X, et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab 2020; 32(2): 176–187.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet 2020; 395(10229):1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhou H, Chen X, Hu T, et al. A novel bat coronavirus closely related to sars-cov-2 contains natural insertions at the s1/s2 cleavage site of the spike protein. Curr Biol 2020; 30(11):2196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Zhou L, Li Z, Zhou J, et al. A Rapid, Accurate and Machine-Agnostic Segmentation and Quantification Method for CT-Based COVID-19 Diagnosis. IEEE Trans Med Imaging 2020; 39(8):2638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579(7798):270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-ncov/sars-cov-2. Cell discovery 2020; 6(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Zhou Y, Wang F, Tang J, et al. Artificial intelligence in COVID-19 drug repurposing, 2020. [DOI] [PMC free article] [PubMed]
- 193. Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of covid-19 patients. Cell Host Microbe 2020; 27(6): 883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zhou Z, Siddiquee MMR, Tajbakhsh N, et al. Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, 2018, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Zhu Z, Xu S, Wang H, et al. Covid-19 in wuhan: immediate psychological impact on 5062 health workers MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 196. Feng Shi, Jun Wang, Jun Shi, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE reviews in biomedical engineering 2020; 14: 4–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CSCoV database is publicly available from Zenodo repository: https://zenodo.org/record/5495823 [111]. The code used in the study is publicly available from the GitHub repository: https://github.com/SFB-KAUST/covid-review.


