Abstract
Pneumonia, especially corona virus disease 2019 (COVID-19), can lead to serious acute lung injury, acute respiratory distress syndrome, multiple organ failure and even death. Thus it is an urgent task for developing high-efficiency, low-toxicity and targeted drugs according to pathogenesis of coronavirus. In this paper, a novel disease-related compound identification model–based capsule network (CapsNet) is proposed. According to pneumonia-related keywords, the prescriptions and active components related to the pharmacological mechanism of disease are collected and extracted in order to construct training set. The features of each component are extracted as the input layer of capsule network. CapsNet is trained and utilized to identify the pneumonia-related compounds in Qingre Jiedu injection. The experiment results show that CapsNet can identify disease-related compounds more accurately than SVM, RF, gcForest and forgeNet.
Keywords: capsule network, pharmacological mechanism, pneumonia, drug prescription
Introduction
Pneumonia refers to lung inflammation, which is a frequently occurring and common disease of the respiratory system [1, 2]. Pneumonia can occur in people of any age, but the young, the aged and the people with immune deficiency or poor immune system are high-risk patients [3, 4]. If this disease is not treated in time, septic shock, lung abscess, pleurisy and pleural effusion, lung abscess and empyema may occur. Severe pneumonia could lead to heart failure and insufficient blood supply, which will directly affect the functional operation of major organs of the body, and even lead to asphyxia and death [5]. Especially since the end of 2019, corona virus disease 2019 (COVID-19) caused by new coronavirus (2019-nCoV or SARS-CoV-2) has been wreaking havoc in more than 100 countries. At present, there are more than 195 million confirmed cases in the world, with more than 4180 thousand deaths. The main clinical manifestations of this disease are fever, cough, shortness of breath and dyspnea [6, 7]. In severe cases, it can lead to serious acute lung injury, acute respiratory distress syndrome, multiple organ failure and even death [8, 9]. At present, no specific drug has been developed to treat COVID-19. Therefore, it is an urgent task for the medical community to find high-efficiency, low-toxicity and targeted drugs according to the structure and pathogenesis of coronavirus.
Network pharmacology is a new interdisciplinary in recent years, which integrates biological network and drug function network to analyze the relationships between drugs and nodes or network modules in the network [10, 11]. The main work of network pharmacology is to study pharmacological mechanism of prescription in treating disease through the research methods of system biology, which could use mathematical means such as statistics and complex network to better understand the behaviors of cells and organs at the molecular level, accelerate the identification of drug targets and find new biomarkers [12–14]. In the past two years, a large number of studies on the prevention mechanism of traditional Chinese medicine (TCM) prescriptions treating pneumonia based on network pharmacology methods have been put forward [15–20]. Liu et al. utilized network pharmacology and molecular docking technology to analyze the pharmacological mechanism of matrine in treatment of COVID-19 and liver injury [21]. Peng et al. investigated the pharmacological mechanism of Lianhua Qingwen Prescription for the treatment of COVID-19 and found some compounds in the prescription such as quercetin and kaempferol, which could play an important role in preventing COVID-19 [22]. Cheng et al. investigated Fufang Banlangen Keli (FBK) and found its pharmacological mechanism of treating COVID-19 and severe acute respiratory syndrome (SARS) [23]. Yan et al. constructed and analyzed QingfeiPaidu decoction components–targets–biological function network, and gave further explanation about the pharmacological effect of QingfeiPaidu decoction in the treatment of COVID-19 [24]. Liu et al. analyzed the active components and related targets of Chaihu Guizhi Ganjiang decoction and constructed ‘drug–disease–target’ network [25]. The results of molecular docking showed that this decoction could be utilized to treat COVID-19 with cold dampness depression in the early stage.
Deep learning is a new field in machine learning research [26]. Its motivation is to establish and simulate the neural network of human brain for analytical learning [27, 28]. Because of its strong learning ability, wide coverage and strong adaptability, deep learning has been widely applied to solve the problems of a large number of fields, which promotes the rapid development of artificial intelligence [29–32]. As the most classical deep learning model in the early stage, convolutional neural network (CNN) needs a large number of images for training, cannot deal with ambiguity well and could lose a large amount of information in the pool layer [33, 34]. Therefore, in order to deal with the shortcomings of CNN, Hinton proposed a novel deep learning model namely capsule network (CapsNet) [35]. It is considered that the brain is organized into a module called capsule. These capsules are especially good at dealing with the characteristics of object postures (position, size and direction), deformation, speed, albedo, tone, texture and so on. Compared with other advanced deep learning models, CapsNet performed better in many areas [36–39]. Tao et al. presented wavelet multi-level attention CapsNet to solve texture classification [40]. Afshar et al. utilized CapsNet to identify brain tumor type [41]. Li et al. utilized five-layer CapsNet to identify the rice images in order to monitor the development of rice [42]. Fang et al. proposed inception capsule networks to improve protein gamma-turn identification [43]. Li et al. improved one-dimensional inception capsule network (IICN) to diagnose the bearing pitting errors, which performed better than other deep learning methods [44].
In the research process of network pharmacology, the active components with high degrees are screened according to the structural analysis of the ‘drug–disease–target’ network. Then, the active components with good binding to the target proteins are determined by molecular docking technology, and the pharmacological mechanism of the important components in treating the disease is analyzed further. This process is very cumbersome and difficult to analyze, especially when there are many traditional Chinese medicine components in the prescription. And manual analysis is prone to make errors, waste of time and arbitrariness. In recent years, some machine learning methods have been applied for network pharmacology to improve these problems. In this paper, an identification model of closely related active components in treating diseases based on capsule network is proposed. Taking pneumonia as an example, the prescriptions and active components closely related to the pharmacological mechanism of pneumonia are collected in order to construct training set. The features of each component are extracted as the input layer of capsule network. CapsNet is trained and utilized to identify the active components related to pneumonia in a new drug prescription.
Methods
Capsule network
Capsule network (CapsNet) is a new neural network structure proposed by Hinton [35]. Its basic structure contains five layers: input layer, convolution layer, primary capsule layer, digital capsule layer and output layer, whose structure is shown in Figure 1. Firstly, in the convolution layer, the convolution layer of traditional CNN is utilized to extract the low-level features. The primary capsule layer is one of the cores of capsule network, which could realize the transformation from scalar to vector. The convolution kernel is utilized for further feature extraction to obtain several capsules. In the next step, the specific process of the output using the capsule principle is given as follows:
Figure 1 .
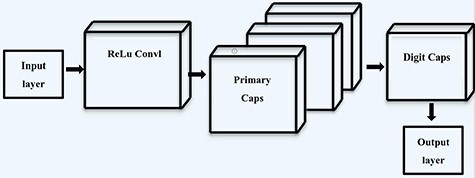
The basic structure of capsule network.
Compared with convolutional neural network, capsule network utilizes vector capsule to replace neurons, dynamic routing to replace pooling operation and squash function to replace ReLu activation function. The schematic diagram of capsule principle is shown in Figure 2, where 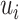 represents the output of the i-th low-level capsule,
represents the output of the i-th low-level capsule, 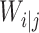 represents the weight matrix between low-level capsule
represents the weight matrix between low-level capsule 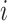 and high-level capsule
and high-level capsule 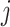 and ûj|i represents the predicted output of low-level capsule
and ûj|i represents the predicted output of low-level capsule 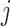 . ûj|i could be calculated as follows:
. ûj|i could be calculated as follows:
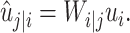 |
(1) |
In the high-level capsule network, 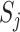 is the input of the high-level capsule network and
is the input of the high-level capsule network and 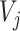 is the output of the high-level capsule network.
is the output of the high-level capsule network. 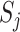 could be calculated as follows:
could be calculated as follows:
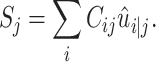 |
(2) |
where 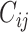 represents the corresponding coupling coefficient of the prediction vector ûj|i of the low-level capsule.
represents the corresponding coupling coefficient of the prediction vector ûj|i of the low-level capsule.
Figure 2 .
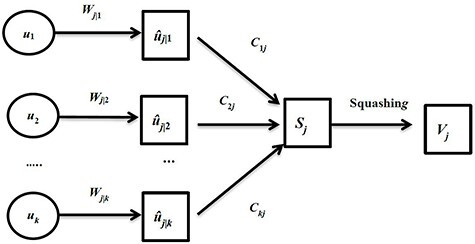
The working principle of capsule.
CapsNet model utilizes activity vector to represent whether an entity appears and the attributes of the entity. The values of the vectors with different dimensions are utilized to represent different attributes, and then the modulus of the vector is utilized to represent the probability of the occurrence of the entity. In order to make the occurrence probability of the entity between 0 and 1, the vector is compressed and standardized through a nonlinear calculation. Squashing nonlinear function can be utilized, which could be calculated as follows:
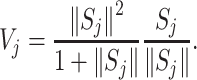 |
(3) |
where 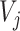 represents the output vector of CapsNet.
represents the output vector of CapsNet.
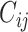 is the coupling coefficient, which could be determined by dynamic routing iteration method. When
is the coupling coefficient, which could be determined by dynamic routing iteration method. When 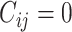 , it means that there is no information transfer between low-level capsule i and high-level capsule j [45]. Coupling coefficient can be calculated as follows:
, it means that there is no information transfer between low-level capsule i and high-level capsule j [45]. Coupling coefficient can be calculated as follows:
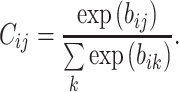 |
(4) |
where 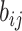 represents the logarithmic probability of coupling between capsule i and capsule j, which is initialized to 0.
represents the logarithmic probability of coupling between capsule i and capsule j, which is initialized to 0. 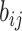 will be dynamically updated in the routing iteration as follows.
will be dynamically updated in the routing iteration as follows.
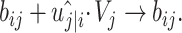 |
(5) |
In capsule network, the vector length of the high-level capsule represents the probability of the category, so in this layer, the category of high-level capsule with the maximum output length of vector is selected as the category predicted by the model. Except that the coupling coefficient C is updated through dynamic routing, other convolution parameters of the whole network and W in CapsNet need to be updated according to the loss function, which is defined as follows:
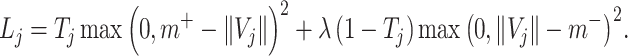 |
(6) |
where 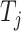 indicates whether the jth class exists;
indicates whether the jth class exists; 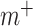 ,
, 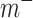 and
and 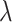 are super parameters that need to be specified in advance.
are super parameters that need to be specified in advance.
Disease-related compound identification algorithm based on capsule network
In this paper, a novel disease-related compound identification model based on capsule network is proposed. The flowchart is shown in Figure 3, which is introduced in detail as follows:
Figure 3 .
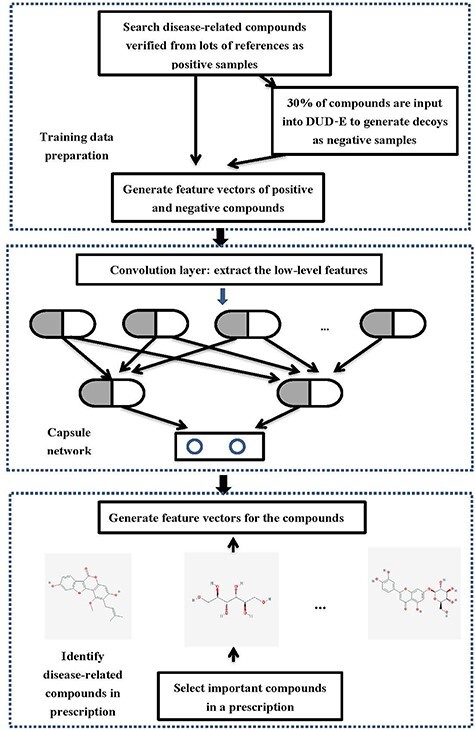
The flowchart of disease-related compound identification model based on capsule network.
Data preparation
Firstly, according to four search terms, pneumonia, new coronavirus pneumonia, COVID-19 and lung injury, the related literatures are searched. Deleted duplicate literatures and 86 important literatures are obtained. The literatures involve many prescriptions, such as Scutellaria, honeysuckle, Jinzhen oral liquid, Yinqiao Jiedu Soft Capsules, Huanglian Jiedu Decoction, Huopo Xialing Decoction, etc. Among these prescriptions, 88 compounds closely related to the pathogenesis of pneumonia are obtained, which are confirmed by experimental verification or molecular docking, such as luteolin, baicalein, wogonin, oroxylina, Alpinetin, quercetin, etc.
The 88 compounds collected are utilized as positive samples. 30% of the 88 compounds are randomly selected and put into the DUD-E website (http://dude.docking.org/) [46]. The decoys corresponding to the input compounds are generated as the control group. A certain number of decoys are randomly selected from the control group as negative samples, so that the ratio of positive samples and negative samples reaches 1:3.
CapsNet training
In order to train the CapsNet with positive and negative compounds, the molecular descriptor of each compound is obtained as the feature set of the compound. The collected data are input into the input layer of CapsNet model for training. This is a two-class classification problem. The super parameters 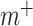 ,
, 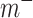 and
and 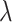 are set 0.9, 0.1 and 0.5, respectively. By the iterations, the optimal CapsNet model is obtained.
are set 0.9, 0.1 and 0.5, respectively. By the iterations, the optimal CapsNet model is obtained.
Disease-related compound identification
In order to study whether a new prescription or medicament can prevent pneumonia-related diseases, the compounds are extracted from the prescription or medicament. And according to the properties, the important compounds are selected. The molecular descriptors of important compounds are also extracted as feature sets, which are input into the optimal capsule network model trained in the previous step in order to give the scores of each important compound. If the score of a compound is higher than 0.5, this compound can be identified to be related to diseases and can be used for the further analysis of network pharmacology. On the contrary, compounds with scores lower than 0.5 cannot be considered.
Experiment Results and Discussion
Relevant and non-relevant compounds about pneumonia are collected to test the effectiveness of capsule network. The data contain 88 positive samples and 264 negative samples. The molecular descriptor is utilized to represent each compound. In order to reflect the effectiveness of CapsNet, SVM [47], RF [48], gcForest [49] and forgeNet [50] are also used to screen the effective compounds in traditional Chinese medicine prescriptions for treating diseases. ROC curve and AUC value are utilized to evaluate the performance of the classifiers. SN, SP, ACC, MCC and F1 are also utilized, which are defined as follows:
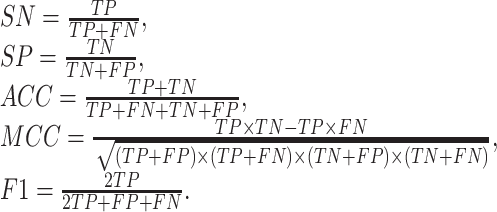 |
(7) |
where TP is the number of true related compounds identified, FN is the number of true related compounds identified as non-related ones, TN is the number of true non-related compounds identified and FN is the number of true non-related compounds identified as related ones.
Model testing
Using 4-cross validation, 6-cross validation, 8-cross validation and 10-cross validation methods, the ROC curves and AUC values identified by the five methods are shown in Figures 4–7, respectively. From Figures 4–6, it could be seen clearly that the ROC curves obtained by CapsNet and RF are very close, which are better than ones obtained by SVM, gcForest and forgeNet. From Figure 7, we also see that the ROC curves of CapsNet, forgeNet, gcForest and RF are very close, which perform better than SVM. The ROC results could show that that CapsNet and RF have the higher accuracy and generalization ability. In order to compare the performances of these five methods more accurately, the AUC values of five methods with 4-cross validation, 6-cross validation, 8-cross validation and 10-cross validation methods could be also obtained. The AUC performances show that the AUC values obtained by the five methods are also very similar, but CapsNet performs best.
Figure 4 .
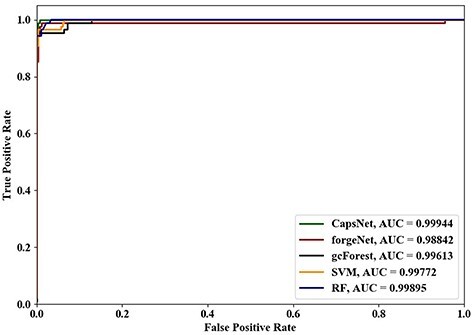
ROC curves and AUC values of five methods using 4-cross validation.
Figure 7 .
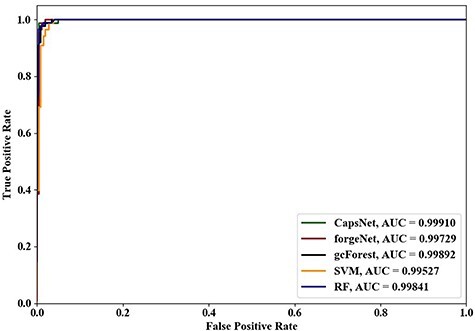
ROC curves and AUC values of five methods using 10-cross validation.
Figure 6 .
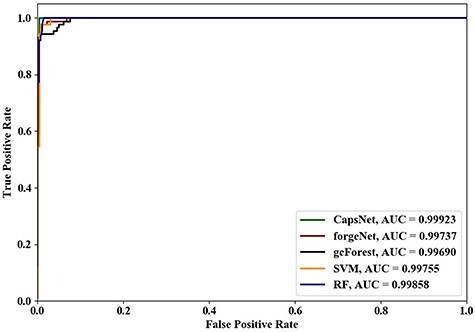
ROC curves and AUC values of five methods using 8-cross validation.
Figure 5 .
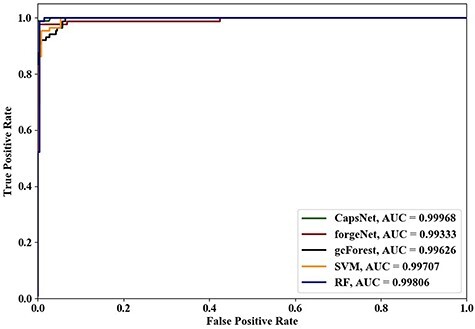
ROC curves and AUC values of five methods using 6-cross validation.
SN, SP, ACC, MCC and F1 performances of disease-related compound identification by five methods with the above four kinds of cross-validation methods are depicted in Figures 8–11, respectively. From the results obtained by 4-cross validation, CapsNet and gcForest could obtain the same SN performance, which is 2.4% higher than SVM, 7.4% higher than RF and 1.2% higher than forgeNet. The SN performance shows that CapsNet and gcForest could identify more true disease-related compounds than SVM, RF and forgeNet. In terms of SP, RF could obtain the best performance, which is 1.0. CapsNet and SVM could obtain the second best performances, which is 0.996 and very close to the performance of RF. In terms of ACC, CapsNet is 0.6% higher than SVM, 1.4% higher than RF, 5.7% higher than gcForest and 0.6% higher than forgeNet, which reveal that CapsNet could obtain the best accuracy among five methods. In terms of MCC, CapsNet is 1.6% higher than SVM, 4% higher than RF, 14.6% higher than gcForest and 1.6% higher than forgeNet. In terms of F1, CapsNet is 1.2% higher than SVM, 3.1% higher than RF, 10.7% higher than gcForest and 1.2% higher than forgeNet, which show that CapsNet performs best for disease-related compound inference overall.
Figure 8 .
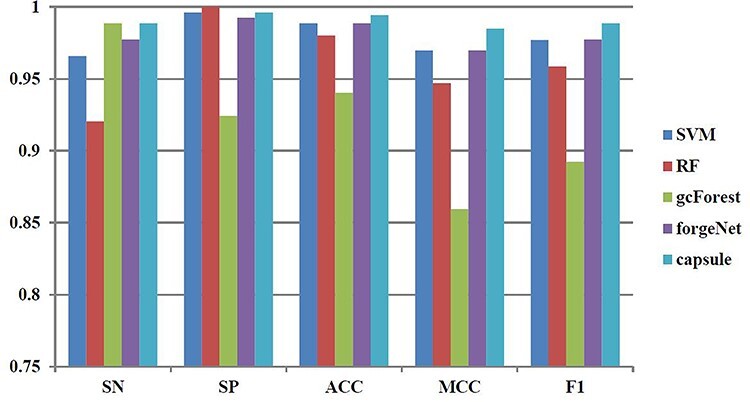
SN, SP, ACC, MCC and F1 performances of disease-related compound identification by five methods and 4-cross validation.
Figure 11 .
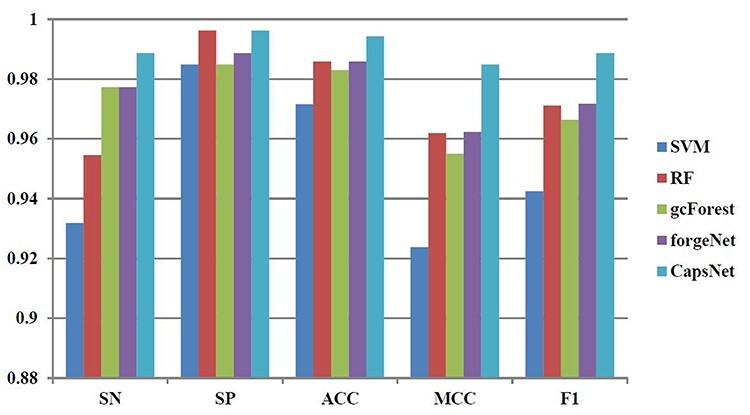
SN, SP, ACC, MCC and F1 performances of disease-related compound identification by five methods and 10-cross validation.
With 6-cross validation method, Figure 9 shows that it could be seen that CapsNet performs best, which is 3.5% higher than SVM, 4.8% higher than RF, 1.2% higher than gcForest and 1.2% higher than forgeNet in terms of SN. RF also obtains the best SP performance, and CapsNet could obtain the second best value, which is 0.996212. In terms of ACC, CapsNet is 2.3% higher than SVM, 0.9% higher than RF, 4.5% higher than gcForest and 1.2% higher than forgeNet. In terms of MCC, CapsNet is 6.5% higher than SVM, 2.4% higher than RF, 11.8% higher than gcForest and 3.1% higher than forgeNet. In terms of F1, CapsNet is 4.7% higher than SVM, 1.8% higher than RF, 8.6% higher than gcForest and 2.3% higher than forgeNet.
Figure 9 .
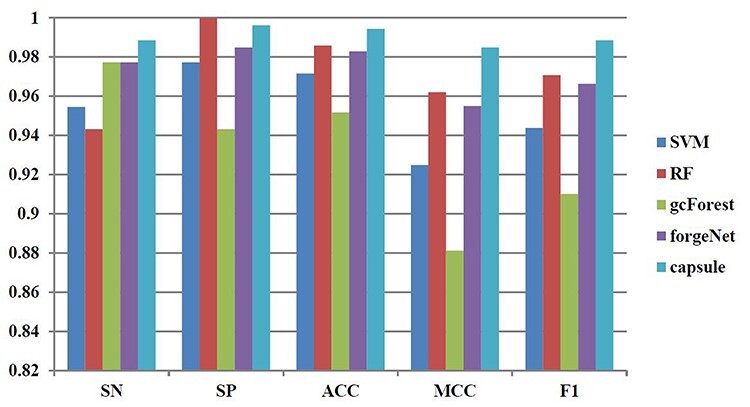
SN, SP, ACC, MCC and F1 performances of disease-related compound identification by five methods and 6-cross validation.
From Figure 10, it could be seen that CapsNet performs best, which is 2.3% higher than SVM, 8.6% higher than RF, 1.1% higher than gcForest and 2.3% higher than forgeNet in terms of SN. In terms of SP, CapsNet, forgeNet and RF could obtain the same performance, which is 0.4% higher than SVM and 6.1% higher than gcForest. In terms of ACC, CapsNet performs best, which is 0.8% higher than SVM, 2% higher than RF, 4.8% higher than gcForest and 0.6% higher than forgeNet. CapsNet has the best MCC performance, which is 2.4% higher than SVM, 5.8% higher than RF, 12.3% higher than gcForest and 1.6% higher than forgeNet. In terms of F1, CapsNet is 1.8% higher than SVM, 4.4% higher than RF, 9.1% higher than gcForest and 1.2% higher than forgeNet.
Figure 10 .
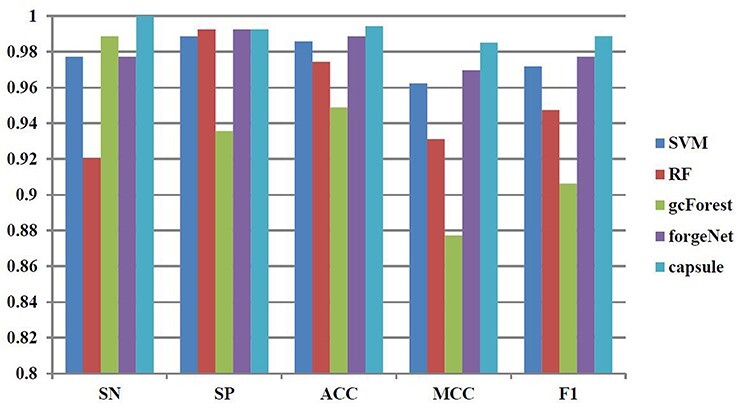
SN, SP, ACC, MCC and F1 performances of disease-related compound identification by five methods and 8-cross validation.
From the results obtained by 10-cross validation, it could be seen that CapsNet performs best, which is 6.1% higher than SVM, 3.6% higher than RF, 1.2% higher than gcForest and 0.8% higher than forgeNet in terms of SN. The SN performance shows that CapsNet could identify more true disease-related compounds than SVM, RF, gcForest and forgeNet. In terms of SP, CapsNet and RF could obtain the same performance, which is 0.996212. These two methods are 1.12% higher than SVM and gcForest, and 0.7% higher than forgeNet. By comparison, it could be seen that CapsNet and RF could identify more true unrelated compounds. CapsNet has the best ACC performance, which is 2.3% higher than SVM, 0.9% higher than RF, 1.2% higher than gcForest and 0.9% higher than forgeNet. In terms of MCC, CapsNet is 6.6% higher than SVM, 2.4% higher than RF, 3.1% higher than gcForest and 2.3% higher than forgeNet. In terms of F1, CapsNet is 4.9% higher than SVM, 1.8% higher than RF, 2.3% higher than gcForest and 1.7% higher than forgeNet, which show that CapsNet performs best overall.
Compound identification
In order to test the effectiveness of data collection and capsule network further, our proposed method is utilized to identify the pneumonia-related compounds in Qingre Jiedu injection. Qingre Jiedu injection contains 11 Traditional Chinese medicines, which are Anemarrhena asphodeloides, Radix Scrophulariae, Rehmannia glutinosa, Radix Isatidis, gardenia, gentian, forsythia, Scutellaria baicalensis, honeysuckle, Ophiopogon japonicus and viola. From TCSMP website (https://old.tcmsp-e.com/tcmsp.php) [51], according to the drug likeness (DL) and oral bioavailability (OB) criteria, 151 compounds are screened from the 11 medicines in Qingre Jiedu injection. The molecular descriptor of each compound is also obtained as the feature set of the compound. The collected data are input into the optimal CapsNet model and 59 compounds are identified to be closely related disease. By analyzing 59 compounds, six compounds quercetin, luteolin, kaempferol, baicalein, wogonin and stigmasterol have been confirmed in a large number of literatures to have clear antiviral, anti-inflammatory and lung, liver and cardiovascular protection and have potential therapeutic effects on lung injury, liver injury, cardiovascular disease and inflammatory response induced by COVID-19. Our method could also identify ent-Epicatechin, Chinoinin, flavanone, Baicalin, acteoside and Forsythiaside, which are the effective components of treating COVID-19. Ent-Epicatechin can regulate multiple signaling pathways by inhibiting the binding of SARS-CoV-2 protein and angiotensin-converting enzyme II (ACE2) in order to prevent COVID-19 [52]. Chinoinin can inhibit NF-κB signaling pathway to improve pneumonia induced by Staphylococcus aureus in mice [53]. Eriodyctiol (flavanone) can play an antiviral role by inhibiting virus replication and reducing cytokine production, which may play an important role in anti-SARS-COV-2 [54]. Acteoside can prevent lung injury by regulating COX-2 and 5-LOX in the lung and has a certain protective effect on the lung [55]. Forsythiaside can combine with MPRO to effectively inhibit the cleavage of virus precursor protein, so as to block virus replication and play the role of anti-COVID-19 [56]. The past researches have proved that many flavonoids have the characteristics of multi-target, multi-channel and multi-system regulation of diseases and can regulate viral inflammatory pathway and respiratory tract infection pathway [57]. Our method also identifies a variety of other flavonoids, such as 5,7,4′-Trihydroxy-8-methoxyflavone, 5,8,2′-Trihydroxy-7-methoxyflavone and 5,7,2,5-tetrahydroxy-8,6-dimethoxyflavone, etc.
SVM, RF, gcForest and forgeNet are also used to screen important compounds in Qingre Jiedu injection. SVM could identify 22 compounds, but many important compounds were not identified, such as wogonin, baicalin, acteoside, stigmasterol, baicalein, chlorogenic acid, some other important flavonoids, etc. RF predicts that 151 compounds are not related with COVID-19. By sorting the predicted scores of 151 compounds, it is found that some important compounds such as forsythiaside, acteoside and luteolin, rank top. But some important compounds such as acacetin, baicalin and wogonin have lower ranks, which reveal that these important compounds are not closely related to COVID-19 disease. gcforest identified 40 compounds, but some important compounds such as stigmasterol, wogonin, acacetin and sitosterol were not identified. forgeNet identifies 126 compounds, and the most important compounds are identified, but a large number of false-positive compounds are also identified to be related to diseases, resulting in the identification results with low accuracy. By comparing the identification results of disease-related compounds obtained by five methods, it can be clearly seen that capsule network can identify disease-related compounds more accurately than the other four methods.
In order to further compare the performance of the five methods, the ROC curves and AUC values of the prediction results are also shown in Figure 12. Through the results, it can be seen that CapsNet can get the better ROC curve than other four methods. For AUC values, CapsNet is 12.9% higher than RF, 6.2% higher than SVM, 2.1% higher than forgeNet and 1.7% higher than gcForest.
Figure 12 .
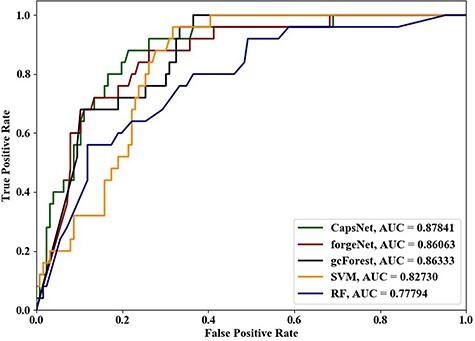
ROC curve and AUC value of the prediction results by five methods.
Conclusions
In order to identify disease-related compounds more accurately, this paper proposes a novel disease-related compound identification model–based capsule network. In this model, five-layer capsule network is utilized. According to pneumonia-related keywords, the prescriptions and active components related to the pharmacological mechanism of disease are collected and utilized to constitute training data. The compounds in Qingre Jiedu injection are collected and utilized as testing data. SVM, RF, gcForest and forgeNet are also utilized for compound identification. ROC curves and AUC performances show that CapsNet can get the better ROC curves than other four methods, which could also make the 1.7–12.9% improvement in terms of AUC compared with SVM, RF, gcForest and forgeNet. By analysis of the identification results, we could see that CapsNet performs best, which could identify disease-related compounds more accurately than the other four methods.
Key Points
A novel disease-related compound identification model–based capsule network (CapsNet) is proposed.
According to pneumonia-related keywords, the prescriptions and active components related to the pharmacological mechanism of disease are collected and extracted in order to construct training set.
In the process of network pharmacology research, the active components with high degree are screened according to the structural analysis of the ‘drug–disease–target’ network.
Bin Yang is a professor in School of Information Science and Engineering at Zaozhuang University. His research interests include bioinformatics, network pharmacology and intelligent computing. His affiliation is with School of Information Science and Engineering, Zaozhuang University, Zaozhuang 277160, P. R. China.
Wenzheng Bao is a lecturer in School of Information Engineering at Xuzhou University of Technology. His research interests include protein-protein interactions network and deep learning. His affiliation is with School of Information Engineering, Xuzhou University of Technology, Xuzhou 221018, P. R. China.
Jinglong Wang achieved his Ph.D. degree from Shandong University of traditional Chinese Medicine, and is now working in college of food science and pharmaceutical engineering, Zaozhuang University. He is engaged in the processing of Chinese Materia Medica and the analysis of pharmacodynamic substance. His affiliation is college of food science and pharmaceutical engineering, Zaozhuang University, Zaozhuang 277160, P.R. China.
Contributor Information
Bin Yang, School of Information science and Engineering, Zaozhuang University, Zaozhuang, China 277160.
Wenzheng Bao, School of Information and Electrical Engineering, Xuzhou University of Technology, Xuzhou, China 221018.
Jinglong Wang, College of Food Science and Pharmaceutical Engineering, Zaozhuang University, Zaozhuang 277160, China.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Author Contribution Statement
W.B. conceived the method. B.Y. designed the method. B.Y. and J.W. conducted the experiments and W.B. wrote the main manuscript text. All authors reviewed the manuscript.
Funding
The talent project of ‘Qingtan scholar’ of Zaozhuang University, Jiangsu Provincial Natural Science Foundation, China (SBK2019040953), Youth Innovation Team of Scientific Research Foundation of the Higher Education Institutions of Shandong Province, China (2019KJM006), the Key Research Program of the Science Foundation of Shandong Province (ZR2020KE001), the PhD research startup foundation of Zaozhuang University (2014BS13) and Zaozhuang University Foundation (2015YY02), the Natural Science Foundation of China (61902337), Natural Science Fund for Colleges and Universities in Jiangsu Province (19KJB520016), Xuzhou Natural Science Foundation (KC21047) and Young talents of science and technology in Jiangsu.
References
- [1]. Kumar V, Chibber S. A comparison of acute lung inflammation in Klebsiella pneumoniae B5055-induced pneumonia and sepsis in BALB/c mice. Crit Care 2009;13:P2. [DOI] [PubMed] [Google Scholar]
- [2]. Tuvim MJ, Evans SE, Clement CG, et al. Augmented lung inflammation protects against influenza a pneumonia. PLoS One 2009;4(1):e4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Nunes S, Sa-Leao R, Carrico J, et al. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care Centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J Clin Microbiol 2005;43(3):1285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Ruiz M, Ewig S, Marcos M, et al. Etiology of community-acquired pneumonia: impact of age, comorbidity, and severity. Am J Respir Crit Care Med 1999;160(2):397–405. [DOI] [PubMed] [Google Scholar]
- [5]. Avila WS, Kirschbaum M, Devido MS, et al. Covid-19, congenital heart disease and pregnancy: dramatic conjunction. Case Rep Dermatol 2021;ytab291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Zakary O, Bidah S, Rachik M, et al. Mathematical model to estimate and predict the COVID-19 infections in Morocco: optimal control strategy. J Appl Math 2020;2020:1–13. [Google Scholar]
- [7]. Juan J, Gil MM, Rong Z, et al. Effects of coronavirus disease 2019 (COVID) on maternal, perinatal and neonatal outcomes: a systematic review. Ultrasound Obstet Gynecol 2020;56:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Chen Q, Wang Y, Jiao F, et al. The mechanism and intervention strategies of inflammatory cytokine storm in corona virus disease 2019. Chin J Infect Diseases 2020;38(00):E013–3. [Google Scholar]
- [9]. Bandt J, Monin C. Obesity, nutrients and the immune system in the era of COVID-19. Nutrients 2021;13(2):610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Shao LA, Qyda B. New progress of interdisciplinary research between network toxicology, quality markers and TCM network pharmacology. Chin Herb Med 2019;11(4):347–8. [Google Scholar]
- [11]. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682–90. [DOI] [PubMed] [Google Scholar]
- [12]. Bezhentsev V, Ivanov S, Kumar S, et al. Identification of potential drug targets for treatment of refractory epilepsy using network pharmacology. J Bioinforma Comput Biol 2018;16(1):1840002. [DOI] [PubMed] [Google Scholar]
- [13]. Kumar A. Sharma D, Aggarwal ML, et al. Cancer/testis antigens as molecular drug targets using network pharmacology. Tumor Biol 2016;37(12):1–9. [DOI] [PubMed] [Google Scholar]
- [14]. Aguda BD. Network pharmacology of glioblastoma. Curr Drug Discov Technol 2013;10(2):125–38. [DOI] [PubMed] [Google Scholar]
- [15]. Zhang Y, Yao Y, Yang Y, et al. Investigation of anti-SARS, MERS, and COVID-19 effect of Jinhua Qinggan granules based on a network pharmacology and molecular docking approach. Nat Prod Commun 2021;16(5):1934578X2110206. [Google Scholar]
- [16]. Gu YY, Zhang M, Cen H, et al. Quercetin as a potential treatment for COVID-19-induced acute kidney injury: based on network pharmacology and molecular docking study. PLoS One 2021;16(1):e0245209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Zhao Y, Hu J, Song J, et al. Exploration on Shufeng Jiedu capsule for treatment of COVID-19 based on network pharmacology and molecular docking. Chin Med 2020;11(1):9–18. [Google Scholar]
- [18]. Cai Y, Zeng M, Chen YZ. The pharmacological mechanism of Huashi Baidu formula for the treatment of COVID-19 by combined network pharmacology and molecular docking. Ann Palliat Med 2021;10(3):16. [DOI] [PubMed] [Google Scholar]
- [19]. Wang Y, Zhao J, Yao Y, et al. To explore the potential mechanism of Yiqing capsule in the treatment of COVID-19 based on network pharmacology. Chin Med 2020;11(2):96–104. [Google Scholar]
- [20]. Wang Y, Ru Y, Zhuo G, et al. Investigation of the potential mechanism governing the effect of the Shen Zhu san on COVID-19 by network pharmacology. Evid Based Complement Alternat Med 2020;2020(13):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Liu F, Li Y, Yang Y, et al. Study on mechanism of matrine in treatment of COVID-19 combined with liver injury by network pharmacology and molecular docking technology. Drug Deliv 2021;28(1):325–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Peng W, Han D, Xu Y, et al. To explore the material basis and mechanism of Lianhua Qingwen prescription against COVID-19 based on network pharmacology. Integrat Respirat Med 2020;1:3. [Google Scholar]
- [23]. Cheng L, Wang F, Zhang SB, et al. Network pharmacology integrated molecular docking reveals the anti-COVID-19 and SARS mechanism of Fufang Banlangen Keli. Nat Prod Commun 2021;16(1):1934578X2098842. [Google Scholar]
- [24]. Yan H, Zou Y, Zou C. Mechanism of QingfeiPaidu decoction for treatment of COVID-19: analysis based on network pharmacology and molecular docking technology. Nan fang yi ke da xue xue bao = J South Med Univ 2020;40(5):616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Liu J, Fan MY, Sun KB, et al. Exploring active ingredients and function mechanism of Chaihu Guizhi Ganjiang decoction against coronavirus disease 2019 based on molecular docking technology. Chin Tradit Herb Drug 2020;51(7):1704–12. [Google Scholar]
- [26]. Dargan S, Kumar M, Ayyagari MR, et al. A survey of deep learning and its applications: a new paradigm to machine learning. Arch Computat Methods Eng 2020;27:1071–92. [Google Scholar]
- [27]. Liang M. Optimization of quantitative financial data analysis system based on deep learning. Complexity 2021;2021:5527615. [Google Scholar]
- [28]. Hassan YF. Deep learning architecture using rough sets and rough neural networks. Kybernetes 2017;46(4):693–705. [Google Scholar]
- [29]. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42(9):60–88. [DOI] [PubMed] [Google Scholar]
- [30]. Alipanahi B, Delong A, Weirauch MT, et al. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 2015;33:831–8. [DOI] [PubMed] [Google Scholar]
- [31]. Jian Z, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 2015;12(10):931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Chen Y, Lin Z, Xing Z, et al. Deep learning-based classification of hyperspectral data. IEEE J Sel Top Appl Earth Obs Remote Sens 2017;7(6):2094–107. [Google Scholar]
- [33]. Tan Y, Tang P, Zhou Y, et al. Photograph aesthetical evaluation and classification with deep convolutional neural networks. Neurocomputing 2016;228:165–75. [Google Scholar]
- [34]. Tachibana Y, Obata T, Kershaw J, et al. The utility of applying various image preprocessing strategies to reduce the ambiguity in deep learning-based clinical image diagnosis. Magn Reson Med Sci 2020;19(2):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Sabour S, Frosst N,Hinton GE. Dynamic routing between capsules. In: Proceedings of the 31st Conference on Neural Information Processing Systems. Curran Associates Inc., 2017:3859-69.
- [36]. Acharya HR, Bhat AD, Avinash K, et al. LegoNet-classification and extractive summarization of Indian legal judgments with capsule networks and sentence embeddings. J Intell Fuzzy Syst 2020;39(2):2037–46. [Google Scholar]
- [37]. Al-Nuaimi DH, Akbar MF, Salman LB, et al. AMC2N: automatic modulation classification using feature clustering-based two-lane capsule networks. Electronics 2021;10(1):76. [Google Scholar]
- [38]. Afshar P, Oikonomou A, Naderkhani F, et al. 3D-MCN: a 3D multi-scale capsule network for lung nodule malignancy prediction. Sci Rep 2020;10:7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Guo Z, Liu H, Zheng Z, et al. Accurate extraction of mountain grassland from remote sensing image using a capsule network. IEEE Geosci Remote Sens Lett 2021;18(6):964–8. [Google Scholar]
- [40]. Tao Z, Wei T, Li J. Wavelet multi-level attention capsule network for texture classification. IEEE Signal Process Lett 2021;28:1215–9. [Google Scholar]
- [41]. Afshar P, Mohammadi A, Plataniotis K N. Brain tumor type classification via capsule networks. In: 25th IEEE International Conference on Image Processing (ICIP), IEEE. 2018, 3129-33.
- [42]. Li Y, Qian M, Liu P, et al. The recognition of rice images by UAV based on capsule network. Clust Comput 2019;22:9515–24. [Google Scholar]
- [43]. Fang C, Shang Y, Dong X. Improving protein gamma-turn prediction using inception capsule networks. Sci Rep 2018;8(1):15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Li X, Kong X, Zhang J, et al. A study on fault diagnosis of bearing pitting under different speed condition based on an improved inception capsule network. Measurement 2021;181:109656. [Google Scholar]
- [45]. Liu N, Ju S, Xiong X, et al. Drug-Drug Relationship Extraction Based on Capsule Networks. Journal of Chinese Information Processing 2020;34(1):80–86+96. [Google Scholar]
- [46]. Mysinger MM, Carchia M, Irwin JJ, et al. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 2012;55(14):6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Furey TS, Cristianini N, Duffy N, et al. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 2000;16(10):906–14. [DOI] [PubMed] [Google Scholar]
- [48]. Breiman L. Random forest. Mach Learn 2001;45:5–32. [Google Scholar]
- [49]. Zhou ZH, Feng J. Deep forest: Towards an alternative to deep neural networks. In: Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence, IJCAI. 2017;2017:3553–59. [Google Scholar]
- [50]. Kong Y, Yu T. forgeNet: a graph deep neural network model using tree-based ensemble classifiers for feature graph construction. Bioinformatics 2020;36(11):3507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Chem 2014;6(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Zhan Q, Huang Y, Lin S, et al. Study on active compounds of Yupingfeng San for prevention of coronavirus disease 2019 (COVID-19) based on network pharmacology and molecular docking. Chin Tradit Herb Drug 2020;51(07):1731–40. [Google Scholar]
- [53]. Qiu F, Song C, Qin F, et al. Mangiferin relieves Staphylococcus aureus-induced pneumonia by inhibiting the NF-κB signalling pathway. Chin J Gen Pract 2021;19(4):558–60. [Google Scholar]
- [54]. Xie L, Lin X, He P, et al. Research on Shuanghuanglian oral liquid for treatment of COVID-19 based on network pharmacology and molecular docking technology. J Hunan Univ Chin Med 2020;40(9):1123–31. [Google Scholar]
- [55]. Zhang H, Yu P, Zhang N, et al. Acteoside attenuates acute lung injury induced by lipopolysaccharide in mice. Pharmacol Clin Chin Mater Med 2015;31(3):41–3. [Google Scholar]
- [56]. Su Z, Zhang X, Ke Z, et al. Network pharmacology study of Yinqiao Jiedu soft capsules in treatment of COVID-19. Chin Tradit Herb Drug 2020;51(9):2354–60. [Google Scholar]
- [57]. Qu Y, Xu F, Wang Y, et al. Mechanism of flavonoids in the treatment of coronavirus disease – 19 (COVID – 19) based on network pharmacology and molecular docking. J Baotou Medical College 2020;36(03):74–78+86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


