Abstract
Development of Th2 subset of CD4+ T cells involves the interleukin-4 (IL-4)- and Stat6-dependent increase in GATA-3 expression during primary activation. Recently we reported that the phenotypic stability and factor independence of Th2 cells involves acquisition of an intracellular pathway that maintains GATA-3 expression. Evidence from retroviral expression studies implied that this pathway involved an autoactivation of GATA-3 expression, since Stat6-deficient T cells induced endogenous GATA-3 when infected with GATA-3-expressing retroviruses. That study left unresolved the issue of whether GATA-3 autoactivation was direct or indirect. Several other Th2-specific transcription factors have been described, including c-Maf and JunB. We therefore examined the ability of these other transcription factors to induce GATA-3 expression and promote Th2 development. Neither c-Maf nor JunB induced Th2 development in Stat6-deficient CD4+ T cells, in contrast to GATA-3. Consistent with this indication of a possible direct autoactivation pathway, we also observed that heterologous GATA family proteins GATA-1, GATA-2, and GATA-4 were also capable of inducing GATA-3 expression in developing Stat6-deficient T cells and promote Th2 development. Mutational analysis revealed evidence for two distinct mechanisms of GATA-3 action. IL-4 induction by GATA-3 required each of the functional domains to be present, whereas repression of gamma interferon could occur even when mutants of GATA-3 lacking the second transactivation domain, TA2, were expressed. The GATA-dependent induction of the GATA-3 but not the other GATA genes in T cells suggests that T-cell-specific cis elements within the GATA-3 locus likely cooperate with a general GATA recognition motif to allow GATA-3-dependent autoactivation.
GATA family transcription factors include six known members with a common DNA-binding domain that is highly conserved among vertebrate species (1, 30, 31, 50, 53). Regions outside the DNA-binding domain vary substantially among members of this family but are conserved between species, suggesting conserved functions of homologous GATA factors between species. GATA-1 expression is restricted to hematopoietic lineage cells and plays an important role in erythroid lineage development (58). GATA-2 is less restricted, with expression in hematopoietic, endothelial, and neuronal cells (5, 49). GATA-3 is important for embryonic brain development and T-cell lineage development (8, 33). GATA-4, GATA-5, and GATA-6 are expressed in the endoderm in an overlapping manner; these proteins have been implicated in regulating gut and cardiac tissue formation (1, 9, 16, 19, 24). Thus, GATA transcription factors are significant in lineage specification of many cell types.
While GATA factor expression is lineage and stage specific, they bind a common cis element, WGATAR (21, 29), by a two-C4-zinc finger DNA-binding domain (28). The C-terminal zinc finger may be more important for DNA-target interactions, since its deletion prevents DNA binding and completely eliminates function (28). The N-terminal zinc finger domain may influence DNA binding (28, 38) and interactions between GATA and transcriptional cofactors, such as the FOG-1 and FOG-2 proteins (27, 42, 46, 52). The protein regions surrounding the GATA-1 C-terminal zinc fingers are targets of acetylation (2, 3), which can modify the ability of GATA-1 to interact with CBP/p300 histone acetyltransferases to regulate transcriptional activity (2, 3, 15). These domains, comprising the first 214 amino acids of GATA-3, are required for activation of a GATA-dependent reporter construct (54). The first 119 amino acids of GATA-4 are required for synergistically activating transcription of the atrial natriuretic factor promoter with the homeodomain protein Nkx2.5 (6), suggesting that the amino terminus of GATA-3 may be required for higher-order interactions with additional factors as well.
T-cell-specific GATA-3 activity was initially found by its binding to T-cell receptor (TCR) δα enhancer (10, 13, 17, 22). However, the lethality of GATA-3 targeting (36) prevented conventional analysis for TCR gene expression, which required RAG-1 blastocyst reconstitution (47) and revealed an arrest of thymocyte development at the double negative stage, specifically within the CD44+ CD25− CD4− CD8− stage (11). GATA-3 is also important to later stages of T-cell development, being involved in commitment of CD4+ T cells to the T-helper 2 (Th2) phenotype (34, 35, 57). GATA-3 expression directed by the CD4 promoter in transgenic mice caused the increased production of several Th2 cytokines (57). Furthermore, GATA-3 was found subsequently to inhibit Th1 cytokine expression by a mechanism that was independent of the induction of interleukin-4 (IL-4) (35).
GATA-3 is the predominant GATA member expressed in thymocytes and T cells, with no evidence for expression of any other family members at significant levels (8, 33). In mature CD4 T cells, GATA-3 expression is regulated by cytokines and costimulation during the primary T-cell activation (35, 41). In naive T cells, GATA-3 is expressed at low levels, which is increased by IL-4 in a Stat6-dependent manner and decreased by IL-12 in a Stat4-dependent manner (35). Importantly, GATA-3 may play a role in controlling its own expression through a Stat6-independent autoactivation pathway (34). This pathway represents a potentially important step in the stable commitment of T cells to the Th2 lineage, since GATA-3 expression is maintained in the absence of the Th2-inducing signals initially required for Th2 development (35). Autoactivation of other GATA family members has also been proposed. GATA-1 expression is increased during erythroid development and has been shown to involve autoactivation through WGATAR elements in the GATA-1 promoter enhancer (32, 51). Furthermore, GATA-2 autoactivation may occur during pituitary cell lineage commitment and participate in counterregulation of the transcription factor Pitl (4). Thus, autoactivation may participate generally in lineage commitment programs enacted by GATA family factors.
The present study extends previous studies of GATA-3 regulation during T-cell development by defining the requirements for GATA-3 induction in T cells by GATA factors independent of the c-Jun-activated kinase (JAK)/STAT pathway. In particular, we tested the hierarchy of Th2-specific transcription factors for STAT-independent Th2 development and GATA-3 expression, analyzing potential interactions between GATA-3, c-Maf, and JunB. Furthermore, we examined the structural features of GATA-3 that are required for autoactivation. Our results indicate that activation of the GATA-3 gene can occur in response to forced expression of heterologous GATA factors. This result suggests that T-cell-specific GATA-3 induction results from cooperation between T-cell-specific factors targeting cis-acting elements within the GATA-3 locus, rather than by GATA-3's exerting site-specific actions on targets that are unique within the GATA family. Finally, the GATA-3-dependent activation of Th2 cytokines and the repression of gamma interferon (IFN-γ) may involve structurally distinct mechanisms of transcriptional control.
MATERIALS AND METHODS
Mice, cytokines, and antibodies.
DO11.10 TCR-transgenic Stat6-deficient mice were provided by M. J. Grusby (18). Recombinant cytokines and antibodies have been described previously (39). R. D. Schreiber generously provided monoclonal anti-IFN-γ antibody H22. IL-4, IL-5, and IFN-γ enzyme-linked immunosorbent assay (ELISA) analysis was performed as described (14, 39).
T-cell activation and retroviral infection.
Wild-type and Stat6-deficient DO11.10 splenocytes were purified by red blood cell lysis (Sigma, St. Louis, Mo.) and activated with ovalbumin (0.5 mg/ml) (Sigma) at 6 × 106/ml in IMDM medium. IL-12 (10 U/ml) and anti-IL-4 (11B11; 10 μg/ml) were added for Th1 development, IL-4 (100 U/ml) was added for Th2 development, and nothing was added for neutral development. Cells were infected at 36 h of activation using 18 × 106 cells, 5 ml of retroviral supernatant, Polybrene (8 μg/ml) (Sigma), and IL-2 (40 U/ml). Cells were harvested 7 days after activation, and murine CD4+ (mCD4+) green fluorescent protein-expressing (GFP+) cells were purified by cell sorting following staining with phycoerythrin-conjugated anti-CD4 (GK1.5; PharMingen). Cells were reactivated on BALB/c splenocytes as antigen-presenting cells (2,000 rad, 2.5 × 106 cells) and expanded for 1 week under the initial conditions. Cells (1.25 × 106) were harvested on day 14 and restimulated on BALB/c splenocytes as antigen-presenting cells (2,000 rad, 2.5 × 106 cells).
Retroviral constructs.
The control retroviral vector GFP-RV and GATA-3RV have been described (39). GATA1-RV was constructed by generating a cDNA using the sense primer 5′GAAGATCTACGCGTCGACCCATGGATTTTCCTGGT3′ and antisense primer 5′CCGCTCGAGGGTCAAGAACTGAGTGGGGCG3′ with the template plasmid pMT2-mGATA-1 (50). GATA2-RV was generated by NotI digestions of plasmid pBSSK GATA-2 (from Naoko Minegishi; unpublished) and treatment with Vent polymerase, to generate a 1.4-kb GATA-2 cDNA with blunt ends. A 1.9-kb GATA-4 cDNA was generated by EcoRI digestion of pMT2-GATA4 (1) and treatment with Vent polymerase. The c-maf cDNA was generated by HindIII-SpeI digest of pBSKS c-maf (23) and treatment with Vent polymerase to generate blunt ends. All fragments were ligated into the blunt vector GFP-RV. JunB-RV was constructed by isolation of the SalI-XhoI fragment of RSV-JunB (26) to produce a full-length cDNA and ligation into the XhoI-digested GFP-RV. GATA-3 deletion mutant constructs were generated by PCR using Pfu polymerase (Stratagene, La Jolla, Calif.) with the following primers: ΔTA1 sense, CGGGTGGTGCGTGTCTGGGTGCTGACCGTT; ΔTA1 antisense, CCTCTGTCCGTTTACCCTCCG; ΔTA2 sense, AACGGACAGAGGCCCTGGAGA; ΔTA2 antisense, GCCCACCACCCCATTACCACCACCTAT;ΔNf sense, TCCGAACCCGGTAGGGGATCC; ΔNf antisense, CTGTCGGCAGCAAGGAGAGCAGGG; ΔCf sense, CAGTCTTCGCTTGGGCTTGATAAGGGGC; and ΔCf antisense, CTCTGGAGGAACGCTAATGGGGACC. The blunt ends of the linear PCR products were ligated using T4 DNA ligase (NEB). Plasmid sequences were confirmed by restriction digestion and sequencing.
RNA, Northern blots, and RNase protection assay.
Total RNA was isolated with the RNeasy kit (Qiagen). Total RNA (10 μg/lane) was separated by electrophoresis at 100 V for 6 h and transferred to a Zeta Probe membrane (Bio-Rad, Hercules, Calif.). Probe (106 cpm/ml) was used for Northern hybridization. The GATA-1 probe was a 1.2-kb EcoRT fragment digested from the vector pMT2-mGATA-1, the GATA2 probe (a full-length, 1.4-kb NotI fragment digested from the vector pBSSK-mGATA-2), the GATA-3 probe (a 1.5-kb cDNA [35]), and the GATA-4 probe (a 327-bp 3′ fragment generated using the sense primer 5′CTAAGCTGTCCCCACAAGGC3′ and the antisense primer 5′CAGAGCTCCACCTGGAAAGG3′ and pMT2-mGATA-4 as the template). The IL-12 receptor beta 2 (IL-12Rβ2) chain and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes have been describes previously (43).
Western blot analysis.
Transduced T cells (107) were stimulated for 16 h with phorbol myristate acetate (PMA) (50 ng/ml) and ionomycin (1 μM), and 293 cells or QT6 cells (54) were transiently transfected for 48 h using Superfect (Qiagen). Cells were harvested and lysed with 5% sodium dodecyl sulfate (SDS)–6.25 mM Tris(pH 6.8)–0.5 mM EDTA (pH 8.0) and centrifuged at 100,000 rpm for 10 min. Lysates were electrophoresed through an SDS–12% polyacrylamide gel electrophoresis (PAGE) gel and transferred to nitrocellulose using semidry transfer of 15 V for 45 min. c-Maf was detected using antibody M173 (Santa Cruz), and JunB was detected using antibody N17 (Santa Cruz) and visualized by ECL (Amersham) using goat anti-rabbit immunoglobulin conjugated to horseradish peroxidase (1:500) (Jackson ImmunoResearch). GATA-3 and Stat1 proteins were detected by Western blotting as previously described (7, 39).
EMSAs.
QT6 cells were transiently transfected with various GATA-3 expression constructs as indicated in the figure legends, and nuclear extracts were prepared after 48 h as described previously (44). For GATA-3 electrophoretic mobility shift assay (EMSA) studies, the amount of nuclear extract used was varied to equalize the amount of mutant GATA-3 protein expression according to Western analysis (Fig. 5B, upper panel), and incubated in 10-μl reactions with 1 μg of poly (dIdC) (Pharmacia) at room temperature and 5 × 104 cpm of the double-stranded probe CAACCCTACGCTGATAAGATTAGTCTGAAAG. After 30 min, complexes were electrophoresed in 6% polyacrylamide at room temperature in 0.4 × Tris-borate-EDTA for 2 h at 150 V.
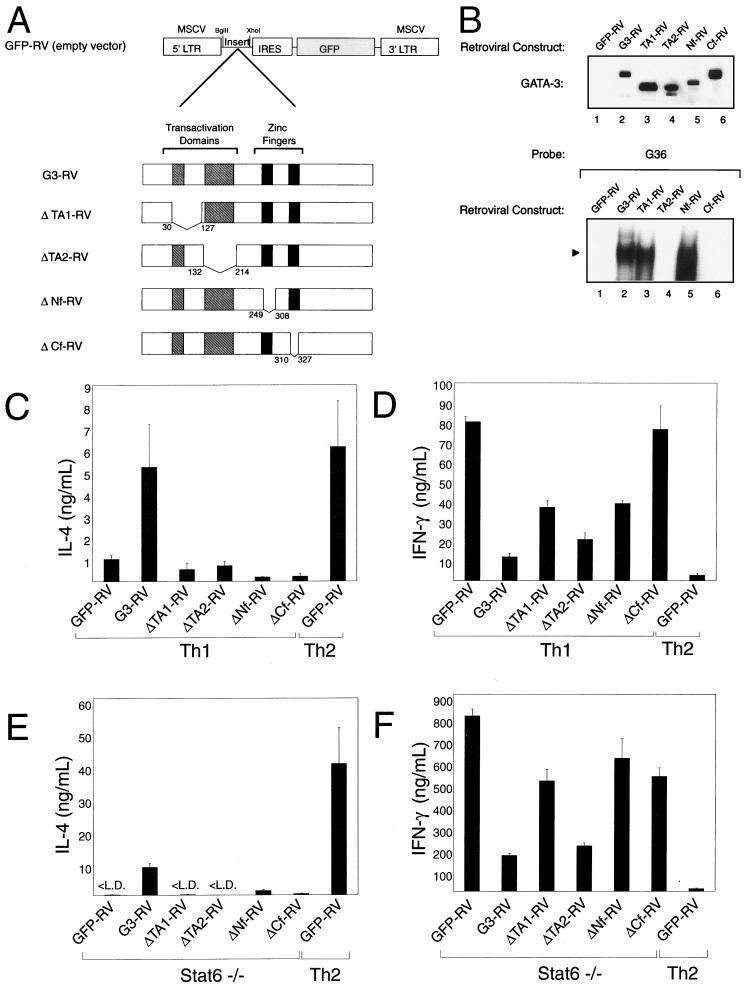
FIG. 5.
Distinct domains couple GATA-3 to IL-4 activation and IFN-γ inhibition. (A) Schematic of GATA-3 mutations, illustrating the four described functional domains of GATA-3 and the amino acids (aa) deleted from each specific expression construct: ΔTA1-RV, deletion of GATA-3 residues 29 to 128; ΔTA2-RV, deletion of GATA-3 132 to 214; ΔNf, deletion of GATA-3 249 to 308, encompassing the entire N-terminal zinc finger; and ΔCf, deletion of GATA-3 309 to 328, a portion of the C-terminal zinc finger region. (B) QT6 cells were transiently transfected with the indicated GATA-3 expression constructs, and nuclear extracts were prepared after 48 h. Nuclear extracts were electrophoresed by SDS–12% PAGE, transferred to nitrocellulose, and probed for GATA-3 expression (upper panel). Nuclear extracts were incubated in 10-μl reactions with the GATA-3 probe for 30 min and resolved by 6% polyacrylamide electrophoresis at room temperature. The solid triangle indicates the probe-bound GATA complex. (C and E) Mutant GATA proteins fail to activate IL-4 in developing Th1 and Stat6-deficient DO11.10 cells. Data are presented as in Fig. 1B. (D and F) Mutant GATA proteins differentially repress IFN-γ production in developing Th1 (D) and Stat6-deficient (F) T cells. Data are presented as in Fig. 1B. L.D., limit of detection.
RESULTS
GATA-3 but not c-Maf or JunB induces IL-4 production in Stat6-deficient T cells.
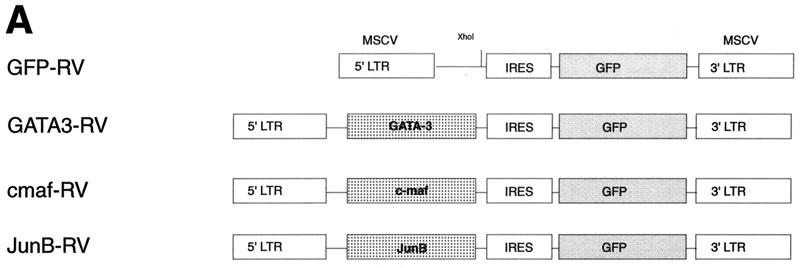
Recently, GATA-3 expression in Th2 cells was suggested to involve a process of Stat6-independent autoactivation (34). However, that study did not determine whether autoactivation involved direct actions of GATA-3 on the GATA-3 gene or promoter or whether an intermediate GATA-3-induced factor was involved. At present, three candidate transcription factors could participate in the Th2-specific activation of GATA-3, including GATA-3 itself (34, 57), c-Maf (12), and JunB (40). GATA-3 and c-Maf are selectively transcribed in Th2 cells, whereas JunB expression involves translational or posttranslational control (25, 40). Forced expression of GATA-3 in Stat6-deficient T cells induced expression of c-Maf, suggesting that c-Maf may potentially mediate an indirect and reciprocal induction of GATA-3 (34).
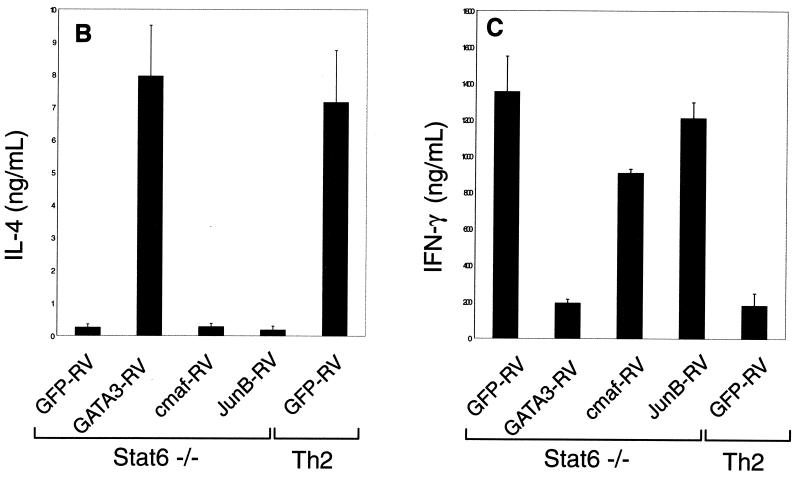
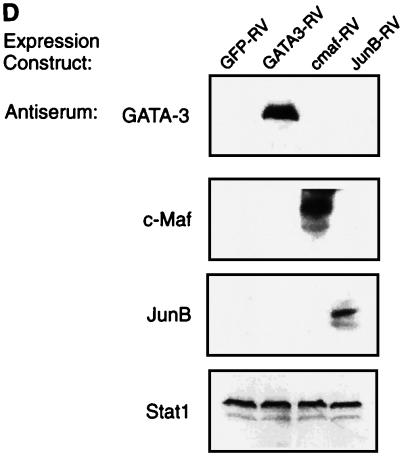
In an attempt to distinguish whether GATA-3-induced GATA-3 expression is direct or indirect, we expressed GATA-3, c-Maf, and JunB by retroviral infection of naive Stat6-deficient T cells (Fig. 1A), purified virus-infected cells by cell sorting, and analyzed the effects on Th2 development (Fig. 1A). Infection by control retrovirus did not enhance IL-4 production or decrease IFN-γ production relative to uninfected cells (Fig. 1B and C). In contrast, infection by GATA-3-expressing retrovirus increased IL-4 and repressed IFN-γ production (Fig. 1B and C) as previously described (35). However, c-Maf- and JunB-expressing viruses did not induce IL-4 or inhibit IFN-γ (Fig. 1B and C), but were comparable to the control retrovirus. T cells infected by c-Maf and JunB also did not induce expression of the endogenous GATA-3 gene, although protein expression of c-Maf and JunB constructs was confirmed by Western analysis (Fig. 1D). This inability of c-Maf or JunB expression to induce IL-4 suggests that they are not sufficient to induce GATA-3 expression. In addition, direct analysis of these cells for GATA-3 expression confirms that c-Maf and JunB do not induce GATA-3 expression (data not shown), implying either that GATA-3 directly autoactivates or that some unidentified Th2-specific factor acts to induce GATA-3 expression.
FIG. 1.
GATA-3 but not c-Maf or JunB induces IL-4 expression in Stat6-deficient T cells. (A) Schematic of GFP-RV-based retroviral expression constructs containing GATA-3 (GATA3-RV), c-Maf (cmaf-RV), or JunB (JunB-RV). (B) Induction of IL-4 by GATA-3 but not by c-Maf or JunB in Stat6-deficient T cells. Activated Stat6-deficient or wild-type DO11.10 splenocytes developed under Th2 conditions were transduced with the indicated retrovirus as described (39). Cells were sorted 7 days after the first activation, restimulated with ovalbumin (0.5 mg/ml) and irradiated BALB/c splenocytes (2.5 × 106 cells, 2,000 rad) for 1 week under the initial conditions, then harvested (day 14), and restimulated. Supernatants were collected after 48 h (day 16) for IL-4 ELISA analysis. Data bars represent the average of four independent cytokine readings for each point, and error bars show the standard deviation. (C) c-Maf and JunB fail to suppress production of IFN-γ in Stat6-deficient T cells. Supernatants described in for panel B were subjected to quantitation of IFN-γ levels by ELISA as described (14). Data are presented as in panel B. (D) Expression of murine stem cell virus (MSCV) long terminal repeat (LTR)-driven GATA-3, c-Maf, and JunB. 293 cells were transiently transfected with 20 μg of GATA3-RV, cmaf-RV, and JunB-RV using Superfect (Quiagen). After 48 h, cells lysates were electrophoresed by SDS–12% PAGE, transferred to nitrocellulose, and analyzed by Western analysis. GATA-3 was detected using antibody HG3-31 (Santa Cruz), c-Maf was detected using M173 (Santa Cruz), JunB was detected using N17 (Santa Cruz), and Stat1 was detected using E23 (Santa Cruz).
Heterologous GATA factors induce Th2 development.
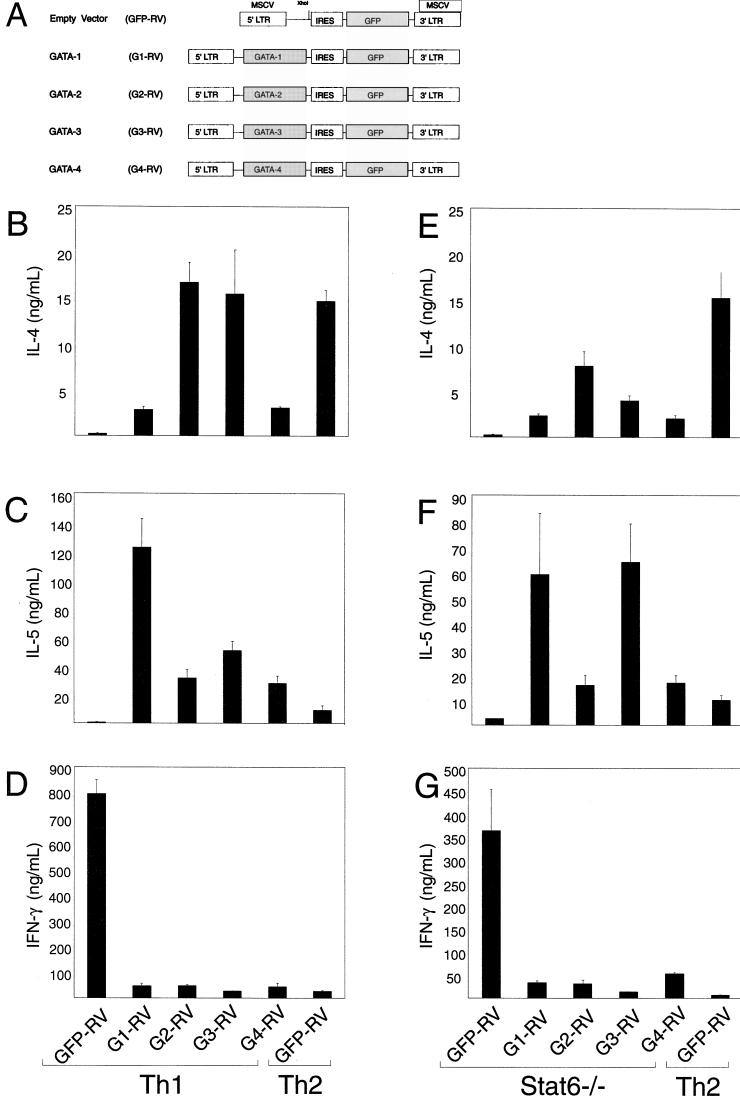
Recent studies have suggested that distinct members of the GATA family may be able to exert some overlapping functions, since targeting GATA-3 to the GATA-1 locus allowed a partial rescue of the erythroid defect seen in GATA-1-deficient embryos (48). Furthermore, an early study of GATA-3 gene regulation (8) identified a potential GATA-binding site within the first GATA-3 gene intron, a region shown to be required for high GATA-3 promoter activity. This site could be a target for direct GATA-3 gene autoactivation, although that study did not directly examine this issue. Addressing this issue experimentally requires either direct promoter mapping in transgenic mice or the development of an efficient reporter system in primary T cells, an approach beyond current capabilities. To address this issue, we asked if GATA factors besides GATA-3 could affect Th2 commitment. Thus, full-length cDNAs for murine GATA-1, GATA-2, GATA-3, and GATA-4 were expressed by retrovirus in naive Stat6-deficient T cells. Retrovirally infected cells were purified by cell sorting and analyzed for Th2 development. As expected, wild-type T cells activated in Th1 conditions and infected by the empty retroviral control vector showed minimal IL-4 and IL-5 production (Fig. 2B and C). By contrast, expression of each GATA protein substantially increased IL-4 and IL-5 production (Fig. 2B and C). Likewise, each GATA protein increased IL-4 and IL-5 production by Stat6-deficient T cells activated under neutral conditions relative to the control retrovirus (Fig. 2E and F). In both the wild-type and Stat6-deficient T cells, GATA-2 and GATA-3 caused the highest levels of IL-4 production. Interestingly, IL-5 appeared to be more strongly augmented by GATA-1 and GATA-3, although the basis for these subtle differences between GATA factors is not clear.
FIG. 2.
Heterologous GATA factors induce Th2 development. (A) Schematic of constructs driving the expression of control vector (GFP-RV), GATA-1 (G1-RV), GATA2 (G2-RV), GATA-3 (G3-RV), and GATA-4 (G4-RV). IL-4 (B and E) and IL-5 (C and F) induction by GATA-1, GATA-2, GATA-3, and GATA-4 in developing DO11.10 Th1 cells and Stat6-deficient T cells. Activated splenocytes were developed under the indicated conditions and transduced with the indicated retroviral expression constructs 36 h postactivation. Cells were sorted 7 days after the first activation for expression of GFP and murine CD4 to greater than 95% puarity, restimulated with ovalbumin protein (0.5 mg/ml) and irradiated BALB/c splenocytes (2.5 × 106 cells, 2,000 rad) for 1 week under the initial conditions, harvested on day 14, and restimulated for cytokine analysis. Supernatants were collected after 48 h (day 16) and subjected to IL-4 and IL-5 ELISA analysis. Data are presented as in Fig. 1B. (D and G) GATA-1, GATA-2, GATA-3, and GATA-4 repress IFN-γ production by developing wild-type Th1 cells (D) and Stat6-deficient cells (G). The supernatants collected from the cells described above were quantified for their levels of IFN-γ by ELISA. Data are presented as in Fig. 1B.
Repression of Th1-associated genes by heterologous GATA factors.
GATA-3 also inhibits IFN-γ production through an IL-4-independent mechanism (35). We examined repression of IFN-γ by heterologous GATA factors in retrovirally infected and sort-purified cells as described above. Each GATA protein produced a nearly complete repression of IFN-γ in both wild-type T cells (Fig. 2D) and Stat6-deficient T cells (Fig. 2G) relative to high levels of IFN-γ produced by cells infected with the control retrovirus. Thus, heterologous GATA factors GATA-1, GATA-2, and GATA-4 were capable of mimicking the developmental effects of GATA-3 in T cells even though they are normally not expressed in T cells (see Fig. 4A).
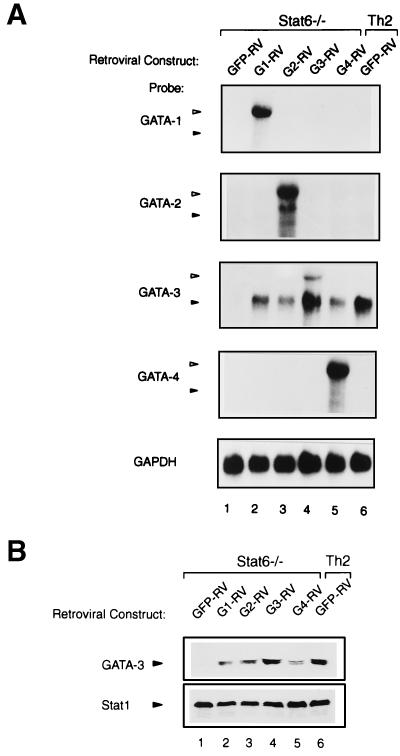
FIG. 4.
Heterologous GATA family proteins induce endogenous GATA-3 expression. (A) Northern analysis of retroviral and endogenous mRNA in cells transduced with GATA-1, GATA-2, GATA-3, and GATA-4. The cells described in the legend to Fig. 2E were analyzed for expression of GATA-1, GATA-2, GATA-3, and GATA-4 as described for Fig. 3. Open triangles indicate the predicted retroviral GATA mRNA, and solid triangles indicate the predicted endogenous GATA mRNA. (B) Western analysis of GATA-3 protein levels. Cells were harvested on day 28 and stimulated with PMA (50 ng/ml) and ionomycin (1 μM) for 16 h. Cytoplasmic and nuclear proteins were extracted as described in Materials and Methods, then electrophoresed through an SDS–12% PAGE gel, and transferred to nitrocellulose. GATA-3 was detected using antibody HG3-31, and Stat1 was detected using antibody E23 (Santa Cruz) as a normalization control.
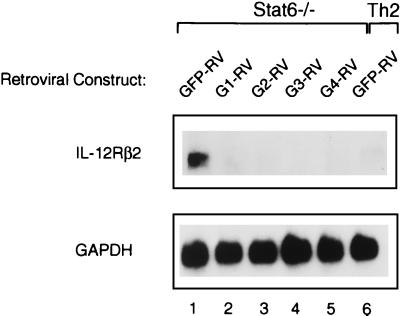
Th1 cells but not Th2 cells acquire IL-12Rβ2 subunit expression (43), consistent with functional loss of IL-12 signaling in Th2 cells (45). Furthermore, forced expression of GATA-3 represses IL-12Rβ2 in T cells activated under Th1-inducing conditions (35). Thus, we analyzed IL-2Rβ2 expression in Stat6-deficient T cells retrovirally transduced with each heterologous GATA factor (Fig. 3). Similar to the previously described effect of GATA-3, each of the other GATA factors produced a similar, nearly complete repression of IL-12Rβ2 mRNA compared to control vector-infected cells (Fig. 3). In summary, the forced expression of GATA-1, GATA-2, GATA-3, and GATA-4 leads in each case both to nearly complete repression of IFN-γ production and IL-12Rβ2 mRNA expression.
FIG. 3.
Repression of IL-12Rβ2 expression by heterologous GATA factors. Northern analysis of IL-12Rβ2 mRNA in cells transduced with GATA-1, GATA-2, GATA-3, and GATA-4. Stat6-deficient lymphocytes transduced with GATA proteins (as described in the legend to Fig. 2E) were restimulated and expanded for subsequent Northern analysis. Cells were harvested on day 28 and restimulated with PMA (50 ng/ml) and ionomycin (1 M) for 6 h prior to RNA isolation; 10 μg of total RNA was subjected to electrophoresis on a 1.1% agarose gel, transferred to Zeta-probe membrane, and probed sequentially with IL-12Rβ2 and GAPDH probes as described (43).
Heterologous GATA factors induce endogenous GATA-3 expression.
Each of the heterologous GATA factors induced IL-4 and inhibited IFN-γ expression in Stat6-deficient T cells, suggesting that they share developmental effects with GATA-3 (Fig. 2). As a control, we wished to compare the relative levels of each retrovirus-derived GATA factor. Thus, we carried out Northern analysis in Stat6-deficient T cells infected by GATA-1, GATA-2, GATA-3, and GATA-4 retroviruses and purified them by cell sorting as described for the experiment in Fig. 2 (Fig. 4A). We used hybridization probes specific to each GATA factor to measure the relative level of mRNA derived from each retroviral construct. As expected, we observed expression of the appropriate GATA factor in each case at the size appropriate for the retroviral construct (Fig. 4A, open triangles). In the case of GATA-3-expressing retrovirus, endogenous GATA-3 mRNA was also induced to levels similar to that in the Th2 control (Fig. 4A, lanes 4 and 6, solid triangles), consistent with our previous report (34). Unexpectedly, endogenous GATA-3 mRNA was induced by the other GATA factors as well, but not by the empty retrovirus. To confirm this induction of GATA-3, we examined the level of GATA-3 by Western analysis, finding levels of GATA-3 induced by the heterologous GATA factors generally similar to that induced by GATA-3 and present in the Th2 control (Fig. 4B). Interestingly, the actions of these heterologous GATA factors were specifically restricted to the activation of the GATA-3 locus, since we did not observe activation of other GATA genes in these T cells (Fig. 4A). Thus, it appears that the GATA-3 locus is the only one of the tested GATA loci that is inducible in T cells, but that any GATA factor is capable of mediating this activation of GATA-3.
Distinct domains couple GATA-3 to IL-4 activation and to IFN-γ inhibition.
To determine the domains of GATA-3 that are important for its various activities in Th2 development, we generated a number of the previously described mutations used to define the functional domains of GATA-3 (Fig. 5A) (54). The ΔTA1 GATA-3 mutant lacks amino acids 29 to 128, encompassing the first transactivation domain, while ΔTA2 deletes amino acids 132 to 214. The ΔNf GATA-3 mutant has a deletion of the entire N-terminal zinc finger (amino acids 249 to 308), and the ΔCf mutant has a deletion of amino acids 309 to 328 in the C-terminal finger. First, we verified the protein expression and expected molecular weights of these mutant GATA proteins (Fig. 5B, upper panel). As expected, ΔTA1 and ΔTA2 migrated with the lowest molecular sizes, with ΔNf migrating at a larger size and ΔCf migrating at a size smaller than wild-type GATA-3. Next we determined the effects of these mutations in GATA-3 on its ability to bind to a consensus GATA-binding motif (Fig. 5B, lower panel). Deletion of amino acids 29 to 128 did not reduce DNA binding as measured by EMSA. Likewise, deletion of the N-terminal zinc finger region left DNA binding intact. As expected, in contrast, deletion of the C-terminal zinc finger abrogated DNA binding, as did deletion of the region between amino acids 132 and 214. Thus, this panel contains GATA mutants that both retain and lose DNA-binding capacity.
These GATA-3 mutants were expressed by retrovirus in naive wild-type and Stat6-deficient T cells undergoing primary activation, and the infected cells were purified by cell sorting for cytokine analysis as before (Fig. 5C to F). All four GATA-3 deletion mutants failed to induce IL-4 in both wild-type Th1 cells (Fig. 5C) and Stat6-deficient T cells (Fig. 5E). This result indicates a strong requirement for fully intact GATA-3 for inducing IL-4 gene expression, as deletion of any of these parts of GATA-3 protein completely abrogated the induction of IL-4. However, variable results were found for the ability of these mutants to mediate the inhibition of IFN-γ. ΔTA2 was able to repress IFN-γ nearly as well as wild-type GATA-3, both in wild-type (Fig.D) and Stat6-deficient (Fig.F) T cells, despite its apparent loss of DNA binding, suggesting that it may exert these effects by indirect interactions with other protein factors. In contrast, less inhibition of IFN-γ was observed for the ΔCf, ΔTA1, and ΔNf mutants, which were also somewhat variable between the wild-type and Stat6-deficient T cells, making interpretation of their effects on inhibiting IFN-γ difficult. The inability of the ΔCf mutant to activate IL-4 and to inhibit IFN-γ production is consistent with a requirement for GATA-3 to bind DNA in mediating both of these activities, since the C-terminal zinc finger mediates GATA-3 DNA interactions.
Structural requirements for GATA-3 autoactivation.
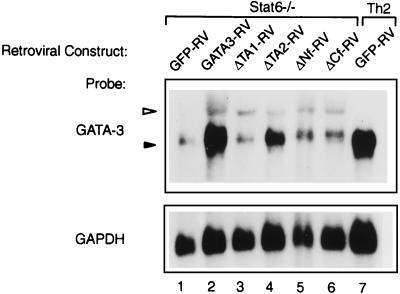
To determine how these mutations influence the ability of GATA-3 to activate the endogenous GATA-3 locus, Northern analysis was performed in Stat6-deficient T cells infected with control vector (GFP-RV), GATA3-RV, and the mutants ΔTA1-RV, ΔTA2-RV, ΔNf-RV and ΔCf-RV in the previous experiment. As expected, retroviral GATA-3 expression strongly induced the expression of the endogenous GATA-3 gene (Fig. 6, lane, 2) relative to T cells infected with only the control vector (GFP-RV). In contrast, retroviral expression of the GATA-3 mutants ΔTA1, ΔNf, and ΔCf failed to activate expression of the endogenous GATA-3 gene above the low level of background evident in the empty retroviral control lane (Fig. 6, compare lanes 1 to 6). The ΔTA2 GATA-3 mutant unexpectedly showed some weak activation of the endogenous GATA-3 gene, consistent with its greater inhibition of IFN-γ production (Fig. 5D and F), although it is not clear whether this is a direct or an indirect effect mediated by squelching interactions with other protein factors.
FIG. 6.
Structural requirements for GATA-3 autoactivation. Cells described in the legends to Fig. 5D and 5E were harvested on day 28, and total RNA was isolated; 10 μg of total RNA was subjected to electrophoresis on a 1.1% agarose gel, transferred to Zeta-probe membrane, and probed sequentially with GATA-3 (35) and GAPDH (43) probes as described. The open triangle indicates the predicted retroviral GATA-3 mRNA, and the solid triangle indicates the predicted endogenous GATA-3 mRNA.
DISCUSSION
At present there are three transcription factors that are selectively expressed in Th2 cells. GATA-3 augments several Th2 cytokines, including IL-4, IL-5, and IL-10, (55, 57). c-Maf is reported to exert a selective action that is restricted to the IL-4 promoter (12, 20). JunB selectively accumulates in Th2 cells, based on translational rather than transcriptional control (40), and may augment IL-4 promoter activity in synergy with c-Maf (25). Since the commitment step in Th2 development may involve an intracellular feedback pathway of GATA-3 autoactivation, we became interested in characterizing GATA-3 regulation, specifically to address whether GATA-3 expression was dependent on other Th2-specific transcription factors or was direct in nature. By comparison, for example, erythroid cell development appears to involve GATA-1 autoactivation by a direct mechanism that relies on a GATA motif in the distal GATA-1 enhancer (32, 51). Furthermore, since replacement of GATA-1 by GATA-3 partially restored erythroid development (48), we wished to directly examine functional overlap of heterologous GATA proteins in directing Th2 development. Finally, we sought to determine the structural requirements underlying GATA-3 autoactivation and Th2 development.
We first directly tested the hypothesis that c-Maf or JunB could induce the Th2-specific expression of GATA-3. We reasoned that if either of these factors was responsible for the Th2-specific expression of GATA-3, forcing c-Maf or JunB expression by retrovirus would induce the endogenous GATA-3 gene, which leads to Th2 development (34). We expressed c-Maf, JunB, and GATA-3 by retrovirus in Stat6-deficient CD4+ T cells and followed T-cell development. Neither c-Maf nor JunB induced IL-4 production or inhibited IFN-γ production (Fig. 1), nor was endogenous GATA-3 expression altered (data not shown). The simplest interpretation is that neither c-Maf nor JunB is sufficient for inducing Stat6-independent Th2 development or GATA3 expression. Rather, GATA-3 autoactivation may depend on other unrecognized Th2-specific transcription factors, or alternatively, GATA-3 autoactivation may be dependent on GATA-3 alone.
Previously, evidence has been presented for a functional overlap among the various members of the GATA family. GATA-3 can partially restore the erythroid development that is normally dependent on expression of GATA-1 (48). To ask if functional overlap extends to GATA-3-dependent Th2 development, we tested the abilities of several GATA proteins to induce Th2 development using retroviral infection of wild-type and Stat6-deficient T cells. GATA-1, GATA-2, GATA-3, and GATA-4 each enhanced IL-4 and IL-5 production and inhibited IFN-γ production (Fig. 2). GATA-2 and GATA-3 are 55% identical at the amino acid level, whereas GATA-1 and GATA-4 have only between 20 and 25% identity to GATA-3. The greatest identity among all four of these GATA family members is within the N- and C-terminal zinc fingers, which contain a domain found to interact with FOG-1 (52) and the DNA-binding domain (54), where the GATA factors are all approximately 90% identical. It is possible, therefore, that these GATA factors significantly overlap in their ability to bind to common GATA target sites within DNA, although perhaps with different affinities, and to bind cofactors, such as FOG-1, that interact with the GATA N-terminal zinc finger.
Unexpectedly, the forced expression of each GATA factor induced the expression of the endogenous GATA-3 gene in T cells (Fig. 4). Notably, while each retroviral GATA factor increased the expression of the endogenous GATA-3 gene, none of these factors induced expression of the other endogenous GATA genes. This may suggest that only the GATA-3 locus possesses an enhancer element that can confer expression in T cells. Second, expression of GATA-3 but not other GATA genes appears to be inducible by the activity of GATA factors, not limited to autoactivation by GATA-3, but also responsive to GATA-1, GATA-2, and GATA-4. Indeed, GATA-3 gene expression may actually rely upon induction by other GATA factors in some tissues (37). This conclusion was reached from studies in which the expression of GATA-2, under control of the HOXb1 gene, was found to be required for the normal expression of GATA-3 in the ventral rhombomere 4. This result suggests that the induction of GATA-3 in T cells caused by GATA-2 expression in this study may have a normal physiologic role during development. In Jurkat T cells, GATA-3 gene regulation was reported to involve sequences between −308 and +1004 relative to its transcriptional start site (8), containing a double GATA-binding consensus within the first intron. In addition, a T-cell-specific DNase-hypersensitive region was also reported to reside 10 kb upstream of the transcriptional start site (8). Testing the role of these regions in the Th2-specific expression of GATA-3 will require more direct analysis in an appropriate T-cell developmental system.
Autoactivation appears to be a common paradigm in stabilizing developmental programs involving transcription factors. Positive autoactivation by GATA transcription factors was previously established in the GATA-1 system. In erythroid lineage cells, GATA-1 gene autoactivation is thought to provide a mechanism for progressive accumulation of GATA-1 protein and to promote erythroid differentiation (51). Analysis of the GATA-1 regulatory elements has identified GATA sites in both the promoter and the enhancer that are functionally involved in GATA-1 gene activation (32, 51). Using a transgenic reporter analysis, the enhancer and promoter were found to cooperate in driving reporter expression in both primitive and definitive erythroid populations (32). The GATA-1 enhancer contains a GATA-binding consensus that is critical for reporter activity, implying a model of direct GATA-1 autoactivation and providing a paradigm for direct autoactivation for GATA-3 in the T-cell lineage.
There are additional mechanisms besides a direct GATA-3-dependent autoactivation to explain its Th2-selective expression. GATA-1 directly antagonizes the actions of the Ets family transcription factor PU.1 in the myeloid lineage, which may prevent the positive regulation of myeloid genes promoting erythroid development (56). Thus, direct interference with a repressor of GATA-3 expression could also explain apparent GATA-3-dependent induction in developing Th2 cells. Furthermore, GATA proteins associate with FOG family coactivators (52), which can act as activators or repressors of GATA-mediated transcription, in a context-dependent manner (27, 42, 46, 52). In summary, there are several possible mechanisms that could underlie the Th2-specific expression of GATA-3 and that are consistent with promiscuous activation of GATA-3 by heterologous family members.
Structural analysis of GATA-3 in Th2 development suggested the possibility of distinct requirements for IL-4 induction and IFN-γ repression. The GATA-3 mutant lacking the C-terminal zinc finger completely failed to inhibit IFN-γ expression or activate IL-4. The GATA-3 mutants lacking the first transactivation domain (TA1) or the N-terminal zinc finger (Nf) showed only partial inhibition of IFN-γ, which varied between wild-type and Stat6-deficient T cells, but also completely failed to activate IL-4 production (Fig. 5). Interestingly, the TA2 mutant completely failed to induce IL-4 production, as expected, but did retain some ability to repress IFN-γ production in both wild-type and Stat6-deficient T cells and to activate endogenous GATA-3. Since this mutant lacked apparent DNA binding, at least as measured by EMSA, these effects could potentially be mediated by indirect interactions, for example, by binding and sequestering GATA-interacting factors. For example, this mutant conceivably could bind factors such as FOG-1, thereby limiting the ability of FOG-1 to influence the transcriptional activity of the native GATA-3 protein. Thus, it is difficult at present to determine whether the inhibition of IFN-γ observed with this TA2 mutant is a direct effect of the TA2 protein on IFN-γ expression or is mediated by the partial induction of the endogenous GATA-3, which is known to be able to inhibit IFN-γ expression (35). Distinguishing between these possibilities will require analysis in a system where activation of the endogenous GATA-3 gene is blocked, for example, by using ES cells in which both GATA-3 alleles have been targeted by homologous recombination (11, 47).
ACKNOWLEDGMENTS
This work was supported by NIH grants AI31328 and AI/DK39676. K.M.M. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Arceci R J, King A A, Simon M C, Orkin S H, Wilson D B. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 4.Dasen J S, O'Connell S M, Flynn S E, Treier M, Gleiberman A S, Szeto D P, Hooshmand F, Aggarwal A K, Rosenfeld M G. Reciprocal interactions of Pit1 and GATA2 mediate signaling gradient-induced determination of pituitary cell types. Cell. 1999;97:587–598. doi: 10.1016/s0092-8674(00)80770-9. [DOI] [PubMed] [Google Scholar]
- 5.Dorfman D M, Wilson D B, Bruns G A, Orkin S H. Human transcription factor GATA-2: evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 6.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrar J D, Smith, Murphy T L, Murphy K M. Recruitment of stat4 to the human interferon-alpha/beta receptor requires activated stat2. J Biol Chem. 2000;275:2693–2697. doi: 10.1074/jbc.275.4.2693. [DOI] [PubMed] [Google Scholar]
- 8.George K M, Leonard M W, Roth M E, Lieuw K H, Kioussis D, Grosveld F, Engel J D. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 9.Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson A J, McDougall S, Leiden J, Calame K L. GATA elements are necessary for the activity and tissue specificity of the T-cell receptor beta-chain transcriptional enhancer. Mol Cell Biol. 1994;14:4286–4294. doi: 10.1128/mcb.14.6.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendriks R W, Nawijn M C, Engel J D, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Ho I C, Hodge M R, Rooney J W, Glimcher L H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 13.Ho I C, Vorhees P, Marin N, Oakley B K, Tsai S F, Orkin S H, Leiden J M. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh C S, Macatonia S E, O'Garra A, Murphy K M. Pathogen-induced Th1 phenotype development in CD4+ alpha beta-TCR transgenic T cells is macrophage dependent. Int Immunol. 1993;5:371–382. doi: 10.1093/intimm/5.4.371. [DOI] [PubMed] [Google Scholar]
- 15.Hung H L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Evans T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev Biol (Orlando) 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- 17.Joulin V, Boris D, Eliot J F, Laborite M C, Chretien S, Mattei M G, Romeo P H. A T-cell specific TCR delta DNA binding protein is a member of the human GATA family. EMBO J. 1991;10:1809–1816. doi: 10.1002/j.1460-2075.1991.tb07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 19.Kelley C, Blumberg H, Zon L I, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- 20.Kim J I, Li T, Ho I C, Grusby M J, Glimcher L H. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko L J, Yamamoto M, Leonard M W, George K M, Ting P, Engel J D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991;11:2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurschner C, Morgan J I. The maf proto-oncogene stimulates transcription from multiple sites in a promoter that directs Purkinje neuron-specific gene expression. Mol Cell Biol. 1995;15:246–254. doi: 10.1128/mcb.15.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 25.Li B, Tournier C, Davis R J, Flavell R A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Chambard J C, Karin M, Olson E N. Fos and Jun repress transcriptional activation by myogenin and MyoD: the amino terminus of Jun can mediate repression. Genes Dev. 1992;6:676–689. doi: 10.1101/gad.6.4.676. [DOI] [PubMed] [Google Scholar]
- 27.Lu J R, McKinsey T A, Xu H, Wang D Z, Richardson J A, Olson E N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin D I, Orkin S H. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 29.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrisey E E, Ip H S, Lu M M, Parmacek M S. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol (Orlando) 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 31.Morrisey E E, Ip H S, Tang Z, Lu M M, Parmacek M S. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol (Orlando) 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura S, Takahashi S, Kuroha T, Suwabe N, Nagasawa T, Trainor C, Yamamoto M. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol Cell Biol. 2000;20:713–723. doi: 10.1128/mcb.20.2.713-723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oosterwegel M, Timmerman J, Leiden J, Clevers H. Expression of GATA-3 during lymphocyte differentiation and mouse embryogenesis. Dev Immunol. 1992;3:1–11. doi: 10.1155/1992/27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy K M. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang W, Ranganath S H, Weindel K, Bhattacharya D, Murphy T L, Sha W C, Murphy K M. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 36.Pandolfi P P, Roth M E, Karis A, Leonard M W, Dzierzak E, Grosveld F G, Engel J D, Lindenbaum M H. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 37.Pata I, Studer M, van Doorninck J H, Briscoe J, Kuuse S, Engel J D, Grosveld F, Karis A. The transcription factor GATA3 is a downstream effector of Hoxb1 specification in rhombomere 4. Development. 1999;126:5523–5531. doi: 10.1242/dev.126.23.5523. [DOI] [PubMed] [Google Scholar]
- 38.Pedone P V, Omichinski J G, Nony P, Trainor C, Gronenborn A M, Clore G M, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranganath S, Ouyang W, Bhattarcharya D, Sha W C, Grupe A, Peltz G, Murphy K M. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 40.Rincon M, Derijard B, Chow C W, Davis R J, Flavell R A. Reprogramming the signalling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T-cells into effector Th1 and Th2 cells. Genes Function. 1997;1:51–68. doi: 10.1046/j.1365-4624.1997.00007.x. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Palmero M, Hara T, Thumbs A, Hunig T. Triggering of T cell proliferation through CD28 induces GATA-3 and promotes T helper type 2 differentiation in vitro and in vivo. Eur J Immunol. 1999;29:3914–3924. doi: 10.1002/(SICI)1521-4141(199912)29:12<3914::AID-IMMU3914>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 42.Svensson E C, Tufts R L, Polk C E, Leiden J M. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szabo S J, Dighe A S, Gubler U, Murphy K M. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo S J, Gold J S, Murphy T L, Murphy K M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol Cell Biol. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. . (Erratum, 13:5928.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szabo S J, Jacobson N G, Dighe A S, Gubler U, Murphy K M. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity. 1995;2:665–675. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 46.Tevosian S G, Deconinck A E, Cantor A B, Rieff H I, Fujiwara Y, Corfas G, Orkin S H. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ting C N, Olson M C, Barton K P, Leiden J M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 48.Tsai F Y, Browne C P, Orkin S H. Knock-in mutation of transcription factor GATA-3 into the GATA-1 locus: partial rescue of GATA-1 loss of function in erythroid cells. Dev Biol (Orlando) 1998;196:218–227. doi: 10.1006/dbio.1997.8842. [DOI] [PubMed] [Google Scholar]
- 49.Tsai F Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 50.Tsai S F, Martin D I, Zon L I, D'Andrea A D, Wong G G, Orkin S H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 51.Tsai S F, Strauss E, Orkin S H. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 52.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto M, Ko L J, Leonard M W, Beug H, Orkin S H, Engel J D. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Gu L, Romeo P H, Boris D, Motohashi H, Yamamoto M, Engel J D. Human GATA-3 trans-activation, DNA-binding, and nuclear localization activities are organized into distinct structural domains. Mol Cell Biol. 1994;14:2201–2212. doi: 10.1128/mcb.14.3.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang D H, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 56.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska H S, Auron P E, Tenen D G, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng W, Flavell R A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 58.Zon L I, Tsai S F, Burgess S, Matsudaira P, Bruns G A, Orkin S H. The major human erythroid DNA-binding protein (GF-1): primary sequence and localization of the gene to the X chromosome. Proc Natl Acad Sci USA. 1990;87:668–672. doi: 10.1073/pnas.87.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]