Abstract
The last 18 months, or more, have seen a profound shift in our global experience, with many of us navigating a once-in-100-year pandemic. To date, COVID-19 remains a life-threatening pandemic with little to no targeted therapeutic recourse. The discovery of novel antiviral agents, such as vaccines and drugs, can provide therapeutic solutions to save human beings from severe infections; however, there is no specifically effective antiviral treatment confirmed for now. Thus, great attention has been paid to the use of natural or artificial antimicrobial peptides (AMPs) as these compounds are widely regarded as promising solutions for the treatment of harmful microorganisms. Given the biological significance of AMPs, it was obvious that there was a significant need for a single platform for identifying and engaging with AMP data. This led to the creation of the dbAMP platform that provides comprehensive information about AMPs and facilitates their investigation and analysis. To date, the dbAMP has accumulated 26 447 AMPs and 2262 antimicrobial proteins from 3044 organisms using both database integration and manual curation of >4579 articles. In addition, dbAMP facilitates the evaluation of AMP structures using I-TASSER for automated protein structure prediction and structure-based functional annotation, providing predictive structure information for clinical drug development. Next-generation sequencing (NGS) and third-generation sequencing have been applied to generate large-scale sequencing reads from various environments, enabling greatly improved analysis of genome structure. In this update, we launch an efficient online tool that can effectively identify AMPs from genome/metagenome and proteome data of all species in a short period. In conclusion, these improvements promote the dbAMP as one of the most abundant and comprehensively annotated resources for AMPs. The updated dbAMP is now freely accessible at http://awi.cuhk.edu.cn/dbAMP.
INTRODUCTION
The abuse of traditional antibiotics has resulted in the development of widespread bacterial drug resistance, which can cause serious health problems worldwide (1). It is also becoming increasingly difficult to identify new antibiotics, making the search for alternatives even more important. Antimicrobial peptides (AMPs) are a class of peptides composed of cationic and hydrophobic amino acids with direct antibacterial activity (2). AMPs range in size from <10 to hundreds of amino acids. They are an important part of the innate immune system acting to protect the host from various pathogens and viruses (3,4). These cationic AMPs bind and interact with negatively charged bacterial cell membranes, resulting in changes in their electrochemical potential, which induces cell membrane damage and allows for penetration of larger molecules, such as proteins, destroying cell morphology and ultimately leading to cell death. These AMPs have been proven to have several advantages over traditional antibiotics and exhibit broad-spectrum antimicrobial activities, including antibacterial, antifungal, antiviral and anticancer activities, with some AMPs even able to overcome acquired drug resistance (5). Additionally, the COVID-19 pandemic is unlikely to end until there is a global rollout of treatment that protects against severe disease and drives herd immunity. The COVID-19 yields a severe threat to human health with a high transmission rate, critical symptoms and relatively high mortality rate in some areas. Thus, there is an urgent need to search for effective therapeutic agents targeting the virus. AMPs are widely recognized as promising solutions for harmful microorganisms (2) making them an active target for the development of novel anti-SARS-CoV-2 therapies. Despite this, there have been relatively few descriptions of AMPs or antiviral peptides (AVPs) with any documented antiviral effect. Interestingly, there are a handful of reports describing prophylactic effects for some AVPs used in the treatments of other coronaviruses (6,7). This includes a paper by Zhao et al. (8), which showed that a short peptide, called P9, had robust antiviral effects against a variety of respiratory viruses in vitro and in vivo, including influenza A virus (H1N1, H3N2, H5N1, H7N7 and H7N9), SARS-CoV and MERS-CoV (https://awi.cuhk.edu.cn/dbAMP/information.php?db=dbAMP_19909). In addition to these peptides, various studies have shown that AMPs from amphibian skin, such as caerin (9) and temporin (10–12), have antiviral activities. A recent in silico study by Liscano et al. indicated that two amphibian AMPs, caerin 1.6 and caerin 1.10, had a high affinity for the spike protein of SARS-CoV-2 (13). These results greatly encouraged our expectations that AMPs could be used as alternative drugs in the treatment of COVID-19. Moreover, by failing to address the escalating antimicrobial resistance (AMR) issue, the near-complete beginning of the post-antibiotic era could lead to more infectious deaths and global financial uncertainty by 2050 (14–17). AMPs are a novel class of alternatives that possess potent activity against a wide range of Gram-negative bacteria with little or no capacity to induce AMR (16). This has stimulated the substantial development of new peptide-based antibiotics with improved therapeutic indices (18).
With the fast growing number of AMPs, it becomes challengeable to handle the large quantity of data manually. Therefore, it is of great help to build databases focused on AMPs. Over the last decade, many AMP-related databases have been established to support AMP deposition, query and mining, as a means to develop computational tools for AMP prediction and design. These resources may be separated into two main groups: general and specific databases. For those databases that focus on the collection of general AMPs, the Antimicrobial Peptide Database, established in 2004, is the most popular one, and it acts as a repository for natural AMPs and includes >3200 compounds from a wide variety of organisms (19). Another resource for general AMPs is CAMPR3, which provides AMP family identification based on signature sequences and structural folds, which can help identify key elements during antimicrobial drug design (20). Some other databases for general AMPs, such as DRAMP (21) and LAMP (22), were designed to provide patented peptides and cross-links with other AMP databases, respectively. There was also a recent update to DBAASP, which continues to develop novel prediction tools for the de novo design of peptide-based drugs (23). In addition, many studies have provided experimental data describing the efficacy of various peptide-based antimicrobial agents against Gram-positive or Gram-negative bacteria (24) and some works focused on AMPs with specific functional activities. The databases for antiviral peptides [AVPdb (25)], defensins knowledgebase (26), synthetic peptides [SAPD (27)] and recombinantly produced AMPs [RAPD (28)] were designed to capture these data. There are also other specialized databases, such as CancerPPD, Hemolytik, THPdb, InverPep and AntiTbPdb, which were designed to facilitate the curation of field-specific data. The CancerPPD (29) database describes anticancer peptides and proteins, the Hemolytik (30) database curates data around experimentally confirmed hemolytic and nonhemolytic peptides, THPdb (31) supplies information on FDA-approved peptide and protein therapeutics, InverPep (32) describes the AMPs from invertebrates and the AntiTbPdb (33) includes a description of the experimentally verified antitubercular or antimycobacterial peptides.
The dbAMP launched its first manually annotated AMP data storage in 2018 (34), focusing on collecting natural and synthetic AMPs and providing general, structural and >20 types of functional activities linked to published works. In addition, given the wide application of next-generation sequencing (NGS) and third-generation sequencing, the dbAMP was also designed to provide a platform for AMP exploration and functional prediction supported by in silico determination of critical physicochemical properties from high-throughput data. Here, we describe the updates and new features in the dbAMP platform, which may serve as a helpful resource for AMP study and design. dbAMP 2.0 provides a homology-based gene prediction program, an integrated tool stream that combines open reading frame (ORF) prediction and AMP classification to identify probably AMPs directly from genome or proteome sequencing data. Considering the rapid development of computational tools, it is expected that highly accurate prediction models could help researchers improve scoring functions for the design and prediction of AMP sequences while reducing their development costs. These updates move us toward a more harmonized system for AMP production and provide a powerful unified source for initial AMP investigation.
SYSTEM OVERVIEW AND DATABASE UPDATES
Updated database content and data statistics
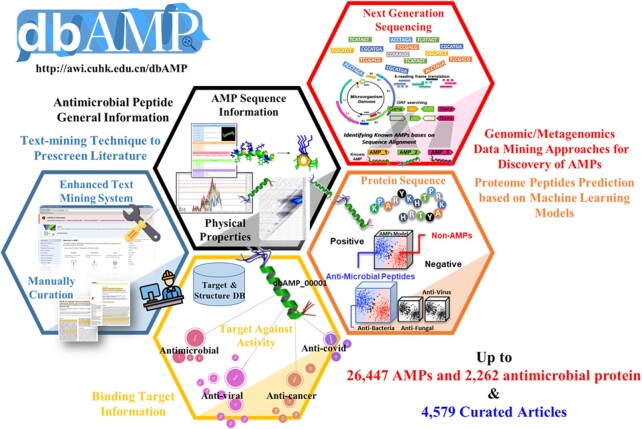
Since the first version of dbAMP (34) was released in 2018, the number of natural and artificial AMPs has increased drastically, along with the attention of global research groups to drug resistance issues. Table 1 describes the improvements and updated content in dbAMP 2.0. These improvements include an increase in the number of curated articles, AMP sequences and source organisms. Additionally, to extract useful information from the curated articles, a new text mining system was built to enhance the automated recognition of AMP-related articles through a scoring system. Specifically, natural language processing was adopted to extract needed information from articles. The extracted information was then integrated into the updated database. As of June 2021, this update had significantly increased the number of AMPs, >2-fold when compared with dbAMPv1, and included a total of 9454 AMP–target interactions between 28 709 entries (including 26 447 AMPs and 2262 antimicrobial proteins) and 5531 target organisms as described in 4579 research articles (Figure 1). The distribution of the AMP source organisms is shown in Supplementary Figure S1 with the most common source organisms being the amphibia (28.1%), mammals (22.1%), arthropods (11.1%) and Viridiplantae (10.1%). In addition, literature and related database records allowed for the functional characterization of these AMPs within the dbAMP categorizing the AMP data into eight major functional classes with 53 functional activities. The most populated classes were the antibacterial peptides (68.33%), followed by new functional peptides (30.88%), antifungal peptides (19.29%), disease-associated peptides (11.48%), antiviral peptides (6.2%), antiparasitic peptides (1.43%), toxic peptides (0.78%) and new mechanism-associated peptides (0.63%) (Table 2). It is of interest to note that this update included over 180 anticoronavirus peptides, with experimental validation.
Table 1.
Comparison between this update and existing AMP databases
| Features | dbAMP | LAMPv2 | DBAASPv3 | DRAMPv2 | dbAMP 2.0 |
|---|---|---|---|---|---|
| Release date (latest) | January 2019 | March 2020 | November 2020 | May 2021 | June 2021 |
| Number of AMPs | 12 389 | 23 253 | >15 700 | 22 151 | 26 447 AMPs and 2,262 antimicrobial proteins |
| Organisms | 2048 | – | – | – | 3044 |
| Tertiary structures (PDB structure) | 1169 | – | >3600 (molecular dynamic trajectory >3200) | 283 | 3444 |
| Tertiary structures (predicted structure) | – | – | – | 263 | 458 |
| Number of biological activities | 26 | 38 | – | 11 | 53 |
| Number of target organisms | 1737 | – | 6560 | – | 5531 |
| Curated AMP–target interactions | 6338 | – | – | – | 9454 |
| Text mining technique to prescreen literature | Text extraction system | – | – | – | Enhanced NLP system |
| Download dataset | Yes | Yes | Yes | Yes | Yes |
| Benchmark datasets for prediction | Yes | – | – | – | Yes |
| Application utilities | |||||
| Antimicrobial potency analysis | Yes | – | – | – | Yes |
| Detection of cryptic region in AMPs | Yes | – | – | – | Yes |
| AMP prediction | AMP prediction models based on multiple species | – | Prediction of general antimicrobial activity/against activity | – | AMPpredictor: enhance AMP prediction models |
| NGS data analysis | AMP sequence alignment based on Bowtie2 | – | – | – | AMPfinder: genomic/proteomic data mining approaches for the discovery of AMPs |
The terms that could not be identified or missing are recorded as ‘–’.
Figure 1.
Highlighted improvements in dbAMP 2.0. dbAMP is the most comprehensive resource for AMPs with this update bringing the total values for the AMP sequences and curated articles to >28 000 and >4500, respectively.
Table 2.
Comparison of the data statistics from this update and the previous version in terms of functional activities
| Function classes | Against activity | dbAMP | dbAMP 2.0a (%) |
|---|---|---|---|
| Antibacterial | Antibacterial | 3006 | 4837 (18.29) |
| Anti-Gram-positive | 2726 | 11 652 (44.06) | |
| Anti-Gram-negative | 2323 | 12 405 (46.91) | |
| Antimicrobial | 4816 | 8654 (32.72) | |
| Antibiofilm | 40 | 40 (0.14) | |
| Mollicute | – | 36 (0.14) | |
| Antiyeast | 4 | 5 (0.02) | |
| Antilisterial | – | 2 (0.01) | |
| Antifungal | Antifungal | 1623 | 5454 (20.62) |
| Antiviral | Antiviral | 300 | 1745 (6.60) |
| Anti-SARS/CoV | – | 186 (0.70) | |
| Antiparasitic | Antiparasitic | 123 | 186 (0.70) |
| New function peptides | Mammalian cells | 308 | 402 (1.52) |
| Anuran defense | – | 7256 (27.44) | |
| Insecticidal | 35 | 1791 (6.77) | |
| Antiprotozoal | 6 | 195 (0.74) | |
| Chemotactic | 59 | 61 (0.23) | |
| Antimalarial | 26 | 46 (0.17) | |
| Antinematode | – | 46 (0.17) | |
| Antiplasmodial | – | 35 (0.13) | |
| Cell penetrating | – | 29 (0.11) | |
| Enzyme inhibitor | 26 | 26 (0.10) | |
| Wound healing | 19 | 21 (0.07) | |
| Antibiotic | – | 19 (0.07) | |
| Immunomodulant | – | 17 (0.06) | |
| Spermicidal | 13 | 13 (0.05) | |
| Edema inducer | – | 11 (0.04) | |
| Disease-associated peptides | Anticancer | 227 | 2290 (7.98) |
| Anti-HIV | 109 | 2286 (8.64) | |
| Antitumor | 9 | 1018 (3.85) | |
| Anti-HCV | – | 67 (0.25) | |
| Antiangiogenesis | – | 13 (0.05) | |
| Anti-HSV | – | 10 (0.04) | |
| Antiallodynic | – | 1 (0.01) | |
| New mechanism-associated peptides | Antihypertensive | – | 1 (0.01) |
| Anti-MRSA | – | 874 (3.30) | |
| Antidiabetic | – | 113 (0.43) | |
| Antioxidant | 22 | 31 (0.12) | |
| Surface immobilized | 19 | 27 (0.10) | |
| Mast cell degranulating | – | 18 (0.07) | |
| Uterotonic | – | 6 (0.02) | |
| Anti-inflammatory | – | 6 (0.02) | |
| Antineurotensive | – | 4 (0.02) | |
| Plasma anticlotting | – | 3 (0.01) | |
| Proteolytic | – | 2 (0.01) | |
| Antinociceptive | – | 2 (0.01) | |
| Hypotensive | – | 1 (0.01) | |
| Sodium channel blocker | 2 | 2 (0.01) | |
| Toxic | Cytotoxin | – | 1 (0.01) |
| Hemolytica | – | 115 (0.43) | |
| Cytolytic | – | 98 (0.37) | |
| Ichthyotoxic | – | 14 (0.05) | |
The numbers in parentheses are displayed as theproportion of entries in the dbAMP.
3D structure visualization of AMPs
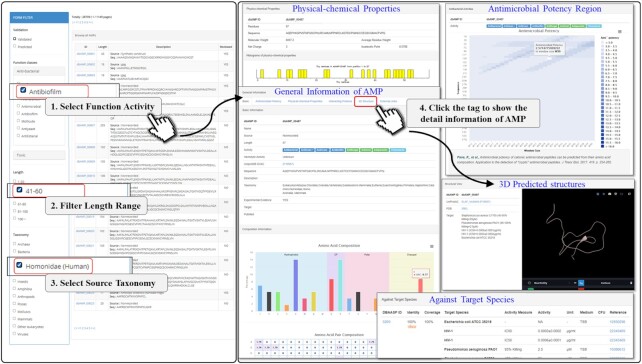
AMPs have experienced a resurgence in interest resulting from the increasingly serious problem of antibiotic resistance. These peptides have attracted significant attention as potential therapeutic agents because they combine the high selectivity, potency and advantages of biological agents with the low toxicity, conformational limitation and cost reductions (35–38). Natural AMPs have been applied in traditional medicine and appear to be reasonable choices for clinical trials and practical applications (35,39). However, the structural characteristics of these peptides are often unstable, and their pharmacokinetic characteristics are poorly described, which seriously hinders their further application as drugs (40). Scientists hope to find an alternative to antibiotics as soon as possible. So far, we have collected 2442 validated AMP structures with 100% sequence identities by alignment from PDB. Meanwhile, 1002 validated structures of AMPs are matched with the criteria of sequence identities ≥90% and E-values ≤10–5. After the sequence alignment, totally 1059 AMPs can be mapped onto the 3444 entries of PDB (Table 1). Moreover, certain AMPs [e.g. segments from most plants (41) or histidine-rich human histatin (42)] are nested on their parent proteins. These AMPs are consequently extracted and isolated on demand (43). As a matter of fact, the isolated segments of the parent proteins possess different structures from the crystallization of their sources. Therefore, to provide complete AMP structure information and accelerate the development of these therapeutic drugs, there is an urgent need to combine computational methods with classical functional evaluation to provide a streamlined approach to novel antimicrobial development. The use of simulated structural evaluations would allow for high-throughput screening and a more robust hit ratio for downstream development. Due to a lack of validated 3D structures for these stapled peptides, structural prediction may be an alternative way to realize structure visualization (44). In this update, the 3D structure for each AMP without experimentally confirmed PDB entries was predicted using I-TASSER allowing for automated protein structure prediction and structure-based functional annotation (45). The I-TASSER server is an online platform for protein structure and function predictions that can produce novel structural predictions using known structures or ab initio using sequence data alone. Thus, we retrieved the relevant structural templates from PDB using the multithread splitting method and then constructed a novel structure prediction model using segment assembly simulation. We then matched the predicted structural model to known proteins in the functional database and added the relevant functional information. Until now, there are 458 3D structures for the current entries of the database (Table 1) that have been developed by this tool and are available for the further utilizations. Figure 2 reveals that dbAMP 2.0 can provide comprehensive functional analysis and predicted structures for each peptide in the ‘3D structure’ information pages.
Figure 2.
The predicted structure viewer was integrated into the platform during this update. A case study describing the production of AMP, elafin (dbAMP_00487), which is the major antiviral protein in cervicovaginal lavage fluid, using human γδ T cells.
A systematic pipeline for the discovery of AMPs on genomic and transcriptomic data
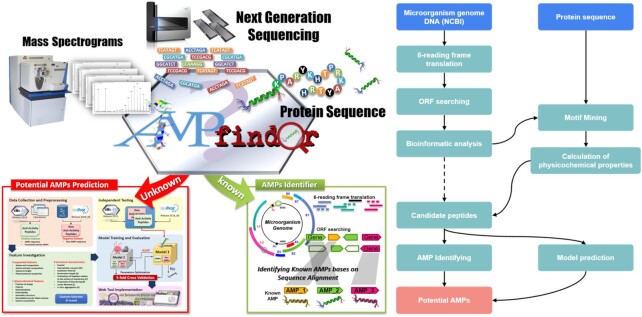
AMPfinder is a simple, yet accurate, computational pipeline that processes either whole genome/metagenome or proteome sequences, and combines ORF prediction with accurate AMP classification to facilitate AMP prediction from protein or nucleotide data. The search for AMPs is based on alignment searching the existing AMP databases and predicting the feature model from the amino acid sequences obtained from the translation of the original transcriptome sequence data (Figure 3). AMPfinder provides a powerful alignment tool for both DNA and protein sequences using the data available in dbAMP. Prodigal is a free, open-source bioinformatics-based algorithm that efficiently predicts protein-coding genes (46). AMPfinder predicts ORFs using Prodigal when queried using a DNA sequence, which translates the input transcriptome data and selects short sequences containing ORFs and signal peptide cleavage sites. If protein sequences are submitted, AMPfinder will skip the prediction of ORFs and directly use these protein sequences. Then, BLAST (47) (for the command-line tool) or Diamond (48) (for dbAMP website) was used for homology detection and machine learning prediction model for the search of potential AMPs, in which case all known or potential motifs will be revealed and classified. AMPfinder used the AMP prediction module built in the first release of dbAMP. The training set was adapted from the dataset proposed by Wang, Hu and coworkers (49,50). After removing redundant sequences, the training set (containing 2399 AMPs and 26 850 non-AMPs with a ratio of ∼1:10) was classified into seven common species according to their source organisms. As reported by Chung et al. (51), the random forest was the best classifier for predicting AMPs in these seven categories of organisms. The accuracies of all the predictive models were >93% (Supplementary Table S1). With the promising performance in predicting AMPs on microorganisms, AMPfinder can effectively identify AMPs throughout large-scale genome sequences of all species, whereas current general-purpose gene prediction programs mainly focus on specific species (52,53). Therefore, AMPfinder could be an efficient and effective tool for the rapid screening of potential AMPs.
Figure 3.
Main pipeline workflow for AMPfinder. AMPfinder is an efficient online tool, which can accurately identify AMPs within genome/transcriptome and proteome data in a short period of time.
Enhanced prediction of AMPs using proteomic data
A previous iteration of this database used an alternative prediction tool to facilitate computer-aided AMP identification based on different species (51). It only focused on identifying general antimicrobial activity. However, the mechanisms of AMP targeting different microbes need to be emphasized. Thus, we proposed an enhanced prediction scheme for this version of dbAMP that uses a machine learning-based prediction model to identify specific targets based on the collected annotations of related AMP functional activities from the dbAMP. A schematic framework for this enhanced prediction is illustrated in Figure 4. We adopted a two-stage classification scheme (38,54) in which the first stage distinguished AMPs from regular peptides and the second stage was responsible for characterizing the specific function that targets different microbes, including bacteria (Gram-positive and Gram-negative, separately), viruses, fungi, and cancer and mammalian cells. This prediction scheme combines several peptide descriptors (55), which can encrypt the combinatorial and physicochemical properties of specific amino acids. The gradient boosting decision tree (GBDT) algorithm (56) was used at each of the tasks to establish the classifiers and the imbalanced learning strategies (57) were applied to improve the classifier’s performance by reducing the curse of insufficient positive labels within specific tasks. The prediction results are confidence values (ranging from 0 to 1) for each of the input peptides, which indicate the putative activities against different targets. Statistics about the predicted confidence value of training/test datasets are summarized in Supplementary Figure S2 and Supplementary Table S2. Users can conduct their screening process arbitrarily with the predicted confidence values, such as choosing the peptides with the largest confidences or directly discriminating with the default cutoff value (0.5). This prediction scheme achieved considerable performance in assisting with AMP design (Table 3). We also introduce our previously developed machine learning-based prediction scheme, AVPIden, for antivirus peptide target prediction (58). This method can characterize the specific targets of AVPs, including six different virus families and eight specific viruses, such as coronavirus. These prediction tools are provided on the ‘Analyze’ page.
Figure 4.
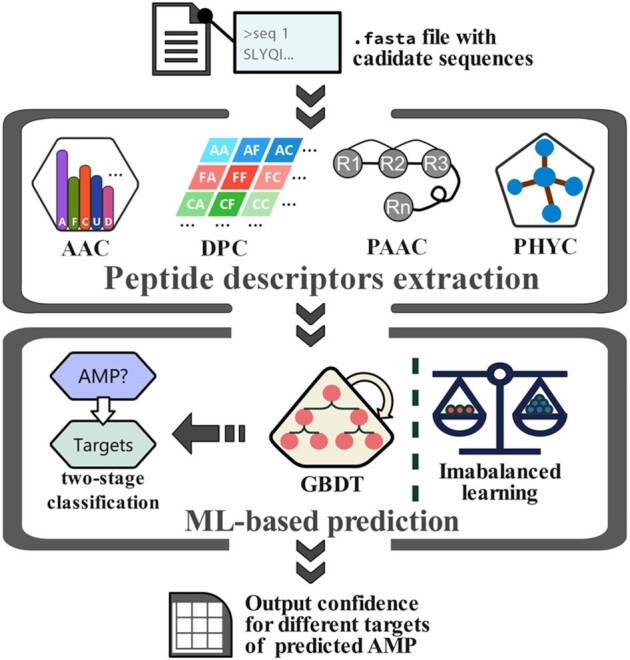
The schematic framework underlying AMP target prediction. First, the user-input sequences are transformed into one of four different groups of peptide descriptors including amino acid composition (AAC), dipeptide composition (DPC), pseudo-amino acid composition (PAAC) and physiochemical properties (PHYC). These descriptors are then used as the feature vector for processing during the two-stage classification process that relies on GBDT and imbalanced learning. This evaluation will then produce a confidence value (ranging from 0 to 1 as the potency level for targeting different microbes) for each of the predicted AMPs.
Table 3.
Prediction performance for the test dataset for each of the tasks in two-stage AMP prediction
| Stage | Prediction task | SEN (%) | SPEC (%) | MeanACC (%) |
|---|---|---|---|---|
| First | AMP | 93.34 | 91.37 | 92.36 |
| Second | Anti-Gram-positive | 91.14 | 88.42 | 89.78 |
| Anti-Gram-negative | 89.58 | 88.79 | 89.19 | |
| Antivirus | 88.87 | 81.05 | 84.96 | |
| Antifungal | 93.86 | 57.92 | 75.89 | |
| Anticancer | 84.33 | 81.24 | 82.79 | |
| Mammalian inhibition | 78.49 | 79.08 | 78.79 |
TP, TN, FP and FN denote true positive, true negative, false positive and false negative, respectively. The sensitivity (SEN), specificity (SPEC) and mean accuracy (MeanACC) are defined as follows: SEN = TP/(TP + FN); SPEC = TN/(TN + FP); and MeanACC = 0.5(SEN + SPEC). The value 0.5 is a default cutoff of confidence values used to determine the positive or negative predictions.
DISCUSSION
New tools for identifying AMPs in large genomes
In this update, we launched a new integrated online tool designed to improve the prediction accuracy of small peptides (AMPfinder). This system uses the integrated AMP information in dbAMP to search for potential AMPs using genome/metagenome or proteome data and applied either via the dbAMP website or as a command-line tool. AMPfinder provides a preliminary annotation of the submitted DNA sequences based on the data available in dbAMP. AMPfinder can accept GenBank accession or GI numbers, pasted sequences or uploaded nucleotide sequence files in FASTA format. These files can contain more than one FASTA formatted sequence, such as whole genome sequencing assembly contigs or multiple proteins. AMPfinder analyzes these sequences and provides a detailed output of the predicted AMPs and source organism class. The dbAMP website also includes a new AMPfinder visualization tool for short peptide predictions in environmental samples using known AMP detection (Supplementary Figure S3A) and unknown AMP prediction and includes their source category (Supplementary Figure S3B). AMPfinder then provides preliminary annotations of AMP sequences based on the data available in dbAMP.
AMPfinder is the first package specifically designed for the identification of AMPs in large genomes. Previous software packages have also included tools for the prediction of AMPs. However, their main purpose was limited to plant species (52,53). Given this, we evaluate the accuracy of AMPfinder classifications using the representative Periplaneta americana (American cockroach) genome (ASM293952v1) as a test case. AMPfinder identified 11 highly homologous AMPs (Supplementary Table S3) in this genome when using the default search E-value threshold and model prediction values, which was consistent with previous studies (59). In addition, our prediction identified an additional 16 potential AMPs (Supplementary Table S4), highlighting the value of these tools.
Collection of the coronavirus targeting peptides
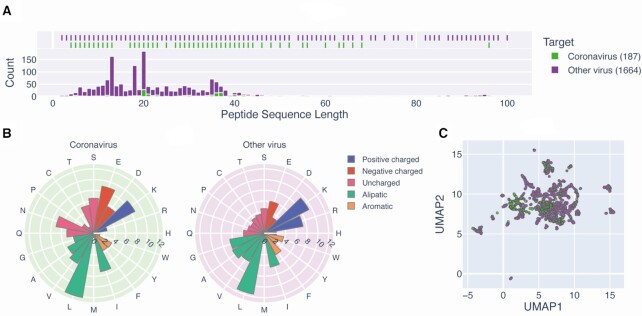
It has been reported that several AVPs have shown some functional activity against coronavirus (6,60). Due to the current pandemic, the development of new therapeutic agents relies on prior knowledge of existing data, including existing computer-assisted methods to date (61). We collected a variety of anticoronavirus peptides from various resources and summarized their basic properties compared to normal AVPs. The length distribution (Figure 5A) of the regular antivirus peptides is slightly concentrated at <20 amino acids, but some of these AVPs can be longer than 100 residues. The mean value and actual distribution of these amino acids are shown in Figure 5B and Supplementary Figure S4, and the high frequency of positively charged and aliphatic amino acids in both categories of peptides may be related to their penetration of the viral membrane. Moreover, we computed the latent sequence encodings of these peptides using tape (62) and performed dimension reduction using UMAP (63) to inspect the differences between the anticoronavirus peptides and regular AVPs (Figure 5C). Although there is a clear distinction, the distance between the anticoronavirus peptides and regular AVPs remains small. This collection of anticoronavirus peptides may offer valuable information for the development of novel therapeutic agents against related pathogens.
Figure 5.
Summary of the properties of the peptides shown to target coronavirus and other viruses. (A) Length distribution of the anticoronavirus peptides (n = 187) and regular AVPs (n = 1664). To distinguish between AMPs and antimicrobial proteins, the entries with sequence length >100 amino acids (n = 1 for anticoronavirus peptides and n = 57 for other regular AVPs, respectively) are not shown in the histogram. (B) Average amino acid composition. Amino acids are categorized according to their physicochemical properties. (C) Dimension reduction of peptide sequences as extracted by tape, which reveals the differences between these peptides and where each point represents a peptide sequence with anticoronavirus (green) or regular antivirus (purple) activity.
SUMMARY AND PERSPECTIVES
AMPs are promising candidates for resolving post-antibiotic effects, with an increasing number of studies suggesting that AMPs may act as potential therapeutic agents against various pathogens (64). Some studies have also suggested that some AMPs may inhibit COVID-19 (65). In silico peptide design can assist and accelerate the development process for novel AMPs (61,66), but relies on the related data stored in large databases and computer-aided analysis tools. Thus, dbAMP was established to provide a single platform combining any and all information on and computational analyses of novel AMPs. This revision of the dbAMP platform incorporates >28 000 unique AMP sequence entries from the literature and related databases with detailed annotations and computation-based physiochemical properties. We have improved the summary statistics of the entire database in an effort to provide more valuable perspective for researchers and established a novel 3D structure viewer for validated peptides and computationally derived putative structures for those without experimentally validated structures. This tool was designed to allow users to investigate the crucial mechanisms of AMP interaction with different pathogens. In addition, we integrated AMPfinder and AMP functional target prediction to produce a single platform solution for AMP development. The combination of these two analytical tools allows for the application of both proteome and genome data in the screening and identification of potential AMP sequences and provides their putative activity scores for a wide variety of pathogens. Eventually, we hope to maintain the dbAMP platform in real time to include cutting-edge developments and studies of novel therapeutic candidates and will try to establish a comprehensive encyclopedia of AMPs for scientific research.
DATA AVAILABILITY
The dbAMP data content will be maintained and updated quarterly via the continuous survey of public resources and research articles. The database assistant system is now freely accessed online at http://awi.cuhk.edu.cn/dbAMP and all manually confirmed AMP resources can be accessed via the download page (https://awi.cuhk.edu.cn/dbAMP/download.php), allowing researchers to independently analyze our data. We also provide all previous versions of the database, which can be accessed through the ‘Previous Release’ page. Finally, the source code for AMPfinder can be accessed via https://github.com/BiOmicsLab/AMPfinder.
Supplementary Material
Contributor Information
Jhih-Hua Jhong, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Lantian Yao, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Science and Engineering, The Chinese University of Hong Kong, Shenzhen 518172, China.
Yuxuan Pang, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Science and Engineering, The Chinese University of Hong Kong, Shenzhen 518172, China.
Zhongyan Li, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Chia-Ru Chung, Department of Computer Science and Information Engineering, National Central University, Taoyuan 32001, Taiwan.
Rulan Wang, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Science and Engineering, The Chinese University of Hong Kong, Shenzhen 518172, China.
Shangfu Li, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Wenshuo Li, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Science and Engineering, The Chinese University of Hong Kong, Shenzhen 518172, China.
Mengqi Luo, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Renfei Ma, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Yuqi Huang, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Xiaoning Zhu, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Jiahong Zhang, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Hexiang Feng, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Qifan Cheng, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Chunxuan Wang, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Kun Xi, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Li-Ching Wu, Department of Biomedical Sciences and Engineering, National Central University, Taoyuan 32001, Taiwan.
Tzu-Hao Chang, Graduate Institute of Biomedical Informatics, Taipei Medical University, Taipei 10675, Taiwan.
Jorng-Tzong Horng, Department of Computer Science and Information Engineering, National Central University, Taoyuan 32001, Taiwan.
Lizhe Zhu, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Ying-Chih Chiang, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
Zhuo Wang, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China.
Tzong-Yi Lee, Warshel Institute for Computational Biology, The Chinese University of Hong Kong, Shenzhen 518172, China; School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen 518172, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [32070659]; Guangdong Province Basic and Applied Basic Research Fund [2021A1515012447]; Ganghong Young Scholar Development Fund [2021E007]; Science, Technology and Innovation Commission of Shenzhen Municipality [JCYJ20200109150003938]; Warshel Institute for Computational Biology. Funding for open access charge: The Chinese University of Hong Kong, Shenzhen Futian Biomedical Innovation R&D Center [P2-2021-ZYL-001-A].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hansen M.P., Hoffmann T.C., McCullough A.R., van Driel M.L., Del Mar C.B.. Antibiotic resistance: what are the opportunities for primary care in alleviating the crisis?. Front. Public Health. 2015; 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huan Y., Kong Q., Mou H., Yi H.. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 2020; 11:582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li W., Separovic F., O’Brien-Simpson N.M., Wade J.D.. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021; 50:4932–4973. [DOI] [PubMed] [Google Scholar]
- 4. Zakaryan H., Chilingaryan G., Arabyan E., Serobian A., Wang G.. Natural antimicrobial peptides as a source of new antiviral agents. J. Gen. Virol. 2021; 102: 10.1099/jgv.0.001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung C.R., Kuo T.R., Wu L.C., Lee T.Y., Horng J.T.. Characterization and identification of antimicrobial peptides with different functional activities. Brief. Bioinform. 2019; 21:1098–1114. [DOI] [PubMed] [Google Scholar]
- 6. Elnagdy S., AlKhazindar M.. The potential of antimicrobial peptides as an antiviral therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020; 3:780–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laneri S., Brancaccio M., Mennitti C., De Biasi M.G., Pero M.E., Pisanelli G., Scudiero O., Pero R.. Antimicrobial peptides and physical activity: a great hope against COVID 19. Microorganisms. 2021; 9:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K., Chan C.C., Leung H.C., Fai N., Lin Y.P.et al.. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016; 6:22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mousavi Maleki M.S., Rostamian M., Madanchi H.. Antimicrobial peptides and other peptide-like therapeutics as promising candidates to combat SARS-CoV-2. Expert Rev. Anti-infect. Ther. 2021; 19:1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marimuthu S.K., Nagarajan K., Perumal S.K., Palanisamy S., Subbiah L.. In silico alpha-helical structural recognition of temporin antimicrobial peptides and its interactions with middle east respiratory syndrome-coronavirus. Int. J. Pept. Res. Ther. 2019; 26:1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Angelis M., Casciaro B., Genovese A., Brancaccio D., Marcocci M.E., Novellino E., Carotenuto A., Palamara A.T., Mangoni M.L., Nencioni L.. Temporin G, an amphibian antimicrobial peptide against influenza and parainfluenza respiratory viruses: insights into biological activity and mechanism of action. FASEB J. 2021; 35:e21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcocci M.E., Amatore D., Villa S., Casciaro B., Aimola P., Franci G., Grieco P., Galdiero M., Palamara A.T., Mangoni M.L.et al.. The amphibian antimicrobial peptide temporin B inhibits in vitro herpes simplex virus 1 infection. Antimicrob. Agents Chemother. 2018; 62:e02367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liscano Y., Onate-Garzon J., Ocampo-Ibanez I.D.. In silico discovery of antimicrobial peptides as an alternative to control SARS-CoV-2. Molecules. 2020; 25:5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith R., Coast J.. The true cost of antimicrobial resistance. BMJ. 2013; 346:f1493. [DOI] [PubMed] [Google Scholar]
- 15. Laxminarayan R., Duse A., Wattal C., Zaidi A.K., Wertheim H.F., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H.et al.. Antibiotic resistance: the need for global solutions. Lancet Infect. Dis. 2013; 13:1057–1098. [DOI] [PubMed] [Google Scholar]
- 16. Spohn R., Daruka L., Lazar V., Martins A., Vidovics F., Grezal G., Mehi O., Kintses B., Szamel M., Jangir P.K.et al.. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019; 10:4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Wang H.-Y., Chung C.-R., Horng J.-T., Lu J.-J., Lee T.-Y.. Large-scale mass spectrometry data combined with demographics analysis rapidly predicts methicillin resistance in Staphylococcus aureus. Brief. Bioinform. 2020; 22:bbaa293. [DOI] [PubMed] [Google Scholar]
- 18. Jiang Z., Vasil A.I., Vasil M.L., Hodges R.S.. “Specificity determinants” improve therapeutic indices of two antimicrobial peptides piscidin 1 and dermaseptin S4 against the Gram-negative pathogens Acinetobacter baumannii and Pseudomonas aeruginosa. Pharmaceuticals (Basel). 2014; 7:366–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 2020; 29:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waghu F.H., Barai R.S., Gurung P., Idicula-Thomas S.. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016; 44:D1094–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang X., Dong F., Shi C., Liu S., Sun J., Chen J., Li H., Xu H., Lao X., Zheng H.. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data. 2019; 6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye G., Wu H., Huang J., Wang W., Ge K., Li G., Zhong J., Huang Q.. LAMP2: a major update of the database linking antimicrobial peptides. Database (Oxford). 2020; 2020:baaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pirtskhalava M., Amstrong A.A., Grigolava M., Chubinidze M., Alimbarashvili E., Vishnepolsky B., Gabrielian A., Rosenthal A., Hurt D.E., Tartakovsky M.. DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021; 49:D288–D297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vishnepolsky B., Zaalishvili G., Karapetian M., Nasrashvili T., Kuljanishvili N., Gabrielian A., Rosenthal A., Hurt D.E., Tartakovsky M., Grigolava M.et al.. De novo design and in vitro testing of antimicrobial peptides against Gram-negative bacteria. Pharmaceuticals (Basel). 2019; 12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wi C.I., Sohn S., Rolfes M.C., Seabright A., Ryu E., Voge G., Bachman K.A., Park M.A., Kita H., Croghan I.T.et al.. Application of a natural language processing algorithm to asthma ascertainment. An automated chart review. Am. J. Respir. Crit. Care Med. 2017; 196:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seebah S., Suresh A., Zhuo S., Choong Y.H., Chua H., Chuon D., Beuerman R., Verma C.. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2007; 35:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baltzer S.A., Brown M.H.. Antimicrobial peptides: promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 2011; 20:228–235. [DOI] [PubMed] [Google Scholar]
- 28. Chu H.L., Yip B.S., Chen K.H., Yu H.Y., Chih Y.H., Cheng H.T., Chou Y.T., Cheng J.W.. Novel antimicrobial peptides with high anticancer activity and selectivity. PLoS One. 2015; 10:e0126390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tyagi A., Tuknait A., Anand P., Gupta S., Sharma M., Mathur D., Joshi A., Singh S., Gautam A., Raghava G.P.. CancerPPD: a database of anticancer peptides and proteins. Nucleic Acids Res. 2015; 43:D837–D843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gautam A., Chaudhary K., Singh S., Joshi A., Anand P., Tuknait A., Mathur D., Varshney G.C., Raghava G.P.. Hemolytik: a database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res. 2014; 42:D444–D449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Usmani S.S., Bedi G., Samuel J.S., Singh S., Kalra S., Kumar P., Ahuja A.A., Sharma M., Gautam A., Raghava G.P.S.. THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One. 2017; 12:e0181748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gomez E.A., Giraldo P., Orduz S.. InverPep: a database of invertebrate antimicrobial peptides. J. Glob. Antimicrob. Resist. 2017; 8:13–17. [DOI] [PubMed] [Google Scholar]
- 33. Usmani S.S., Kumar R., Kumar V., Singh S., Raghava G.P.S.. AntiTbPdb: a knowledgebase of anti-tubercular peptides. Database (Oxford). 2018; 2018:bay025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jhong J.H., Chi Y.H., Li W.C., Lin T.H., Huang K.Y., Lee T.Y.. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019; 47:D285–D297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Craik D.J., Fairlie D.P., Liras S., Price D.. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013; 81:136–147. [DOI] [PubMed] [Google Scholar]
- 36. Bui V.M., Weng S.L., Lu C.T., Chang T.H., Weng J.T., Lee T.Y.. SOHSite: incorporating evolutionary information and physicochemical properties to identify protein S-sulfenylation sites. BMC Genomics. 2016; 17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bui V.M., Lu C.T., Ho T.T., Lee T.Y.. MDD-SOH: exploiting maximal dependence decomposition to identify S-sulfenylation sites with substrate motifs. Bioinformatics. 2016; 32:165–172. [DOI] [PubMed] [Google Scholar]
- 38. Kao H.J., Huang C.H., Bretana N.A., Lu C.T., Huang K.Y., Weng S.L., Lee T.Y.. A two-layered machine learning method to identify protein O-GlcNAcylation sites with O-GlcNAc transferase substrate motifs. BMC Bioinformatics. 2015; 16:S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramakers B.E., van Hest J.C., Lowik D.W.. Molecular tools for the construction of peptide-based materials. Chem. Soc. Rev. 2014; 43:2743–2756. [DOI] [PubMed] [Google Scholar]
- 40. Mendive-Tapia L., Preciado S., Garcia J., Ramon R., Kielland N., Albericio F., Lavilla R.. New peptide architectures through C–H activation stapling between tryptophan–phenylalanine/tyrosine residues. Nat. Commun. 2015; 6:7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barashkova A.S., Rogozhin E.A.. Isolation of antimicrobial peptides from different plant sources: does a general extraction method exist?. Plant Methods. 2020; 16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oppenheim F., Xu T., McMillian F., Levitz S., Diamond R., Offner G., Troxler R.. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988; 263:7472–7477. [PubMed] [Google Scholar]
- 43. Lee H.-T., Lee C.-C., Yang J.-R., Lai J.Z., Chang K.Y.. A large-scale structural classification of antimicrobial peptides. Biomed. Res. Int. 2015; 2015:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang K.Y., Lee T.Y., Kao H.J., Ma C.T., Lee C.C., Lin T.H., Chang W.C., Huang H.D.. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019; 47:D298–D308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su M.G., Huang K.Y., Lu C.T., Kao H.J., Chang Y.H., Lee T.Y.. topPTM: a new module of dbPTM for identifying functional post-translational modifications in transmembrane proteins. Nucleic Acids Res. 2014; 42:D537–D545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 49. Wang P., Hu L., Liu G., Jiang N., Chen X., Xu J., Zheng W., Li L., Tan M., Chen Z.et al.. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS One. 2011; 6:e18476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiao X., Wang P., Lin W.Z., Jia J.H., Chou K.C.. iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013; 436:168–177. [DOI] [PubMed] [Google Scholar]
- 51. Chung C.R., Jhong J.H., Wang Z., Chen S., Wan Y., Horng J.T., Lee T.Y.. Characterization and identification of natural antimicrobial peptides on different organisms. Int. J. Mol. Sci. 2020; 21:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shelenkov A.A., Slavokhotova A.A., Odintsova T.I.. Cysmotif searcher pipeline for antimicrobial peptide identification in plant transcriptomes. Biochemistry (Moscow). 2018; 83:1424–1432. [DOI] [PubMed] [Google Scholar]
- 53. Zhou P., Silverstein K.A., Gao L., Walton J.D., Nallu S., Guhlin J., Young N.D.. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinformatics. 2013; 14:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang C.H., Su M.G., Kao H.J., Jhong J.H., Weng S.L., Lee T.Y.. UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst. Biol. 2016; 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pang Y., Wang Z., Jhong J.-H., Lee T.-Y.. Identifying anti-coronavirus peptides by incorporating different negative datasets and imbalanced learning strategies. Brief. Bioinform. 2021; 22:1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Friedman J.H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 2001; 29:1189–1232. [Google Scholar]
- 57. Krawczyk B. Learning from imbalanced data: open challenges and future directions. Prog. Artif. Intell. 2016; 5:221–232. [Google Scholar]
- 58. Pang Y., Yao L., Jhong J.-H., Wang Z., Lee T.-Y.. AVPIden: a new scheme for identification and functional prediction of antiviral peptides based on machine learning approaches. Brief. Bioinform. 2021; 22:bbab263. [DOI] [PubMed] [Google Scholar]
- 59. Kim I.W., Lee J.H., Subramaniyam S., Yun E.Y., Kim I., Park J., Hwang J.S.. De novo transcriptome analysis and detection of antimicrobial peptides of the American cockroach Periplaneta americana (Linnaeus). PLoS One. 2016; 11:e0155304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahendran A.S.K., Lim Y.S., Fang C.-M., Loh H.-S., Le C.F.. The potential of antiviral peptides as COVID-19 therapeutics. Front. Pharmacol. 2020; 11:575444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barreto-Santamaría A., Patarroyo M.E., Curtidor H.. Designing and optimizing new antimicrobial peptides: all targets are not the same. Crit. Rev. Clin. Lab. Sci. 2019; 56:351–373. [DOI] [PubMed] [Google Scholar]
- 62. Rao R., Bhattacharya N., Thomas N., Duan Y., Chen X., Canny J., Abbeel P., Song Y.S.. Evaluating protein transfer learning with TAPE. Adv. Neural Inform. Process. Syst. 2019; 32:9689. [PMC free article] [PubMed] [Google Scholar]
- 63. McInnes L., Healy J., Melville J.. Umap: uniform manifold approximation and projection for dimension reduction. 2018; arXiv doi:09 February 2018, preprint: not peer reviewedhttps://arxiv.org/abs/1802.03426.
- 64. Lei J., Sun L., Huang S., Zhu C., Li P., He J., Mackey V., Coy D.H., He Q.. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019; 11:3919–3931. [PMC free article] [PubMed] [Google Scholar]
- 65. Tonk M., Růžek D., Vilcinskas A.. Compelling evidence for the activity of antiviral peptides against SARS-CoV-2. Viruses. 2021; 13:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Melo M.C.R., Maasch J.R.M.A., de la Fuente-Nunez C.. Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 2021; 4:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dbAMP data content will be maintained and updated quarterly via the continuous survey of public resources and research articles. The database assistant system is now freely accessed online at http://awi.cuhk.edu.cn/dbAMP and all manually confirmed AMP resources can be accessed via the download page (https://awi.cuhk.edu.cn/dbAMP/download.php), allowing researchers to independently analyze our data. We also provide all previous versions of the database, which can be accessed through the ‘Previous Release’ page. Finally, the source code for AMPfinder can be accessed via https://github.com/BiOmicsLab/AMPfinder.