Abstract
Understanding the structure-function relationships between diverse cell types in a complex organ environment requires detailed in situ reconstruction of cell-associated molecular properties in the context of 3-dimensional, macro-scale tissue architecture. We recently developed a simple and effective method for tissue clearing, Clearing-enhanced 3D (Ce3D), that achieves excellent transparency, preserves cell morphology, tissue architecture, and reporter molecule fluorescence, while also being robustly compatible with direct immunolabeling. These characteristics in turn permit high-quality multiplex fluorescence microscopy of large tissue volumes, as well as image analysis using advanced platforms such as volumetric Histocytometry, collectively allowing quantitative characterization of cells based on phenotypic and functional markers with respect to their spatial positioning within tissues. The Ce3D clearing is fast, achieving robust transparency of most tissues within 24h, albeit still necessitating additional time for staining, imaging and analysis (1–2wks). Here we provide detailed procedures for tissue clearing using Ce3D, including optimized workflows for tissue processing and staining, as well treatment of difficult-to-clear organs, such as the brain. We also describe a new procedure for RNA detection in Ce3D treated tissues, as well as provide additional details for the volumetric Histocytometry data processing steps. Finally, we discuss the current limitations and work-around strategies for improving antibody-based tissue immunolabeling, fluorophore multiplexing, large-volume microscopy, and computational analysis of large image datasets. Together, these detailed procedures and solutions for high-resolution volumetric microscopy with Ce3D enable quantitative visualization of cells and tissues at a high level of detail, allowing exploration of cellular spatial relationships in a variety of tissue settings.
Keywords: Tissue clearing, volumetric microscopy, immunofluorescent imaging, Histocytometry, image processing, cell segmentation, in situ hybridization, RNAscope, tissue architecture, Ce3D, confocal microscopy, light-sheet imaging
INTRODUCTION:
Recent advances in technologies for high-dimensional cell profiling, such as flow cytometry, CyTOF, and single-cell RNA sequencing, have revealed extensive cellular heterogeneity among what were previously considered homogeneous sets of cells, suggesting the existence of a much greater than anticipated multitude of lineages, populations, and differentiation states among the cellular components of diverse tissues and organs1,2,3. Appropriate localization of these populations within their natural tissue setting is critical for relating this diversity to function, given that positioning within a tissue exposes cells to distinct microenvironments that play a major role in determining their differentiation state and activity4,5. At a higher scale, spatial organization of these diverse cell populations is a fundamental determinant of physiology and disruption of cellular organization can lead to tissue pathology and organ dysfunction.
Acquisition of spatially-resolved datasets has been historically achieved with wide-field or confocal microscopy of relatively thin tissue sections (5–30um) immunolabelled with antibodies against specific cell components, mainly proteins, and conjugated to diverse chromogenic or fluorescent probes. Genetically engineered animal models, in which enzymatic or fluorescent reporter proteins are expressed under the control of cell-type specific promoters, have also been used to identify different cell types within tissues. While earlier generations of microscopes have generally permitted visualization of only a few distinct fluorescent probes, more recent instruments equipped with spectral or tunable detectors and multiple laser sources readily permit visualization of a substantially greater number of fluorophores with unique excitation and/or emission fluorescence characteristics (up to 13 in our hands)6,7. Alternative techniques using iterative staining or use of metal ion-based probes combined with imaging mass cytometry have further extended the multiplexing capabilities of microscopes and allow very extensive cellular profiling within thin tissue sections8,9,10. Such multiplexing is essential for studying phenotypically complex cell populations defined by combinations of different markers. As one example, dendritic cell subsets, complex populations of innate immune cells involved in antigen presentation, require evaluation of at least 5 different surface markers for appropriate classification. Simultaneous visualization of additional cell types or anatomical landmarks within the same tissue sample is required for understanding how the cells are distributed with respect to their surroundings and necessitates further antibody and fluorophore multiplexing11,12
Quantification of cells in tissue sections has also been historically achieved with manual enumeration of specific imaged objects summed across multiple fields of view. As the number of phenotypic markers used for cell identification has increased and automated tiling stages enable collection of larger image datasets, there has also been a need for development of more quantitative, algorithm-driven methods of cellular profiling and enumeration. To this end, we previously established an image analysis pipeline, Histocytometry, which permits phenotypic characterization and quantification of cells in tissue sections in a manner akin to flow cytometry7. With this technology, we have studied the fine-grained spatial organization of diverse immune populations in lymphoid and non-lymphoid organs during homeostatic and disease conditions, and have identified complex spatial relationships between cells that would be challenging to reveal with more conventional image analysis methods11,13–22. As specific examples, our studies on organization of immune cells in lymphoid organs have revealed that the positioning of distinct innate antigen presenting cells in lymph nodes actively sets different thresholds of activation for CD4+ vs. CD8+ T cells in response to vaccination11,13, while localized activity of T regulatory cells promotes the generation of inhibitory microenvironments and limits autoimmunity15. Similarly, in our human studies, quantitative imaging has revealed the precise location of T follicular regulatory (TFR) cells, leading to a redefined model of their mode of action19. Analysis of inflamed human lymph nodes during HIV infection with Histocytometry demonstrated an unexpected localization of effector CD8+ T cells to inflamed germinal centers, and has led to the development of novel therapeutic strategies16.
Nevertheless, even with these quantitative approaches, the use of thin sections to explore cellular relationships within tissues has major limitations. Many cell types are rare and difficult to detect within individual sections in sufficient number for accurate analysis. Non-uniform distribution of cells in organs can also lead to misleading results based on the specific section or region-of-interest chosen for analysis. Thin sections are also problematic for accurately aligning cells to structurally complex anatomical landmarks (e.g., blood or lymphatic vessels), as these may lie just outside of the sampled volume. These issues are further complicated by variability in results obtained even when studying genetically identical tissue donors due to errors introduced by inadequate sampling of tissue space using thin sections. Thus, the ability to accurately study cellular organization in tissues would be best undertaken using methods permitting visualization of larger tissue volumes that maximize tissue sampling while revealing 3-dimensional associations of cells and their surroundings.
This need is amplified by the limitations of current tissue dissociation-based methods, such as flow cytometry or single-cell RNA-seq, which are heavily biased towards preferential analysis of cells that are easily extracted from the tissues. Recent estimates indicate a drastic underestimation (up to 70-fold) of the actual cellularity within tissues even for relatively easy to isolate cells (lymphocytes) as compared to microscopy-based enumeration23. We observed similar results with dendritic cells, which have strong adhesive interactions with various stromal and extracellular matrix elements and are difficult to remove from tissues even with optimized digestion protocols; similar findings have been previously reported for stromal cells7,24. Further, certain activation states of immune cell subsets can also be associated with differential expression of specific adhesion receptors, which can result in non-equivalent dissociation from tissues during isolation and erroneous representation during analysis, as we have observed for recently activated T lymphocytes7,25. For these reasons, techniques exclusively relying on dissociation-based cell profiling (e.g. flow cytometry and RNA-seq) are likely to produce inaccurate information about cellular composition and gene expression within tissues. Therefore, in addition to yielding information on cellular spatial distribution, quantitative volumetric microscopy can in principle provide more accurate information on the composition of various tissue components as compared to dissociation-based methods.
Tissue clarification for volumetric imaging
Imaging of larger tissue volumes has been previously achieved with serial sectioning and post-acquisition image reconstruction. However, such techniques are technically very challenging and time consuming, with the resultant composite images being associated with sectioning or alignment artifacts, which make volumetric visualization and quantitative analysis problematic. More recently, various techniques for tissue clearing have emerged, allowing section-free volumetric microscopy that for some small organs in experimental animals, enables the entire organ to be visualized at a reasonable resolution. These clearing approaches produce tissue transparency by minimizing the refractive index mismatches and subsequent photon scattering that normally occurs when light travels through the lipid and aqueous layers within tissues26,27. Many such clearing methodologies have been described, such as BABB, Clarity, Cubic, iDISCO and several 2nd generation techniques, with each method having distinct advantages with respect to the overall quality of tissue transparency, ability to clear lipid-rich organs, preservation of reporter protein fluorescence, or compatibility with immunolabeling28–43. However, each of these methods has also been associated with specific limitations, such as loss of endogenous protein fluorescence (iDISCO), poor epitope preservation (CLARITY), or distortion of tissue architecture due to substantial volumetric changes (multiple methods). To circumvent these limitations, we recently developed a new tissue clearing method, Clearing-enhanced 3D (Ce3D), which minimizes many of these issues and provides a largely compromise-free methodology for large volumetric microscopy. Important features of Ce3D are that this technique robustly preserves the capacity for direct rather than indirect, amplified antibody-based immunolabeling, maintains fluorescence of endogenous reporter proteins, and minimally impacts overall tissue and cellular morphology, features that in aggregate allow high-quality, multiplex volumetric microscopy of diverse tissues and cell types44.
Overview of the procedure
The Ce3D clarification procedure is simple and inexpensive (Figure 1, Table 1). Clearing is achieved via a single-step incubation of tissues with an easy to make solution (22% weight/volume N-methylacetamide and 86% weight/volume Histodenz; approximate cost $16 per 5ml of reagent). Similar to conventional whole-mount processing techniques, prior to clarification, the tissues are fixed with 1–4% paraformaldehyde or specialized fixation solutions containing paraformaldehyde (Steps 1(A)iii-v, Table 1) and stained with a panel of antibodies against specific molecules of interest (Steps 1(A)vii-ix). Secondary detection with streptavidin or species-specific reagents can also be utilized, although use of directly conjugated antibodies markedly reduces overall incubation times while permitting simultaneous use of several antibodies from the same species such as mouse. This ability to use Ce3D with directly labelled antibodies is a consequence of the method’s high signal-to-noise (S/N) ratio and contrasts with other clearing technologies for which amplification with secondary antibodies is necessary to achieve an adequate signal over background. To date, we have tested >50 different antibodies against distinct protein markers of interest and observed a robust ability to detect these in Ce3D-processed tissues, attaining results similar to conventional immunolabeling of paraformaldehyde-fixed sections. Similarly, major organic fluorophore families, such as Alexa fluorophores, perform extremely well with the Ce3D clearing procedure, thereby permitting robust fluorescent multiplexing. Finally, unlike alcohol-based clarification methods, Ce3D preserves reporter protein fluorescence and to date we have successfully visualized tissues from GFP, YFP, RFP, CFP and DsRed reporter animals without significant loss of fluorescence over time. Once the tissues are stained and cleared (Steps 1(A)xi-xiiii, 1(B)ix-x, 1(C)viii-ix), they can be visualized with confocal microscopy (Steps 2–7), but are also compatible with light-sheet imaging. Using Ce3D, we have achieved large volumetric imaging of various murine tissue types, including lymph node, lung, kidney, liver, mammary gland, small intestine and brain (Figure 3, Table 1), and have also used the method to image bones, heart and muscle. In unpublished studies, we also attained robust staining, Ce3D clarification, and imaging of human biopsy samples of lymphoid tissues, liver, gut, and other tissues, suggesting that this technology can be directly applicable to clinical studies.
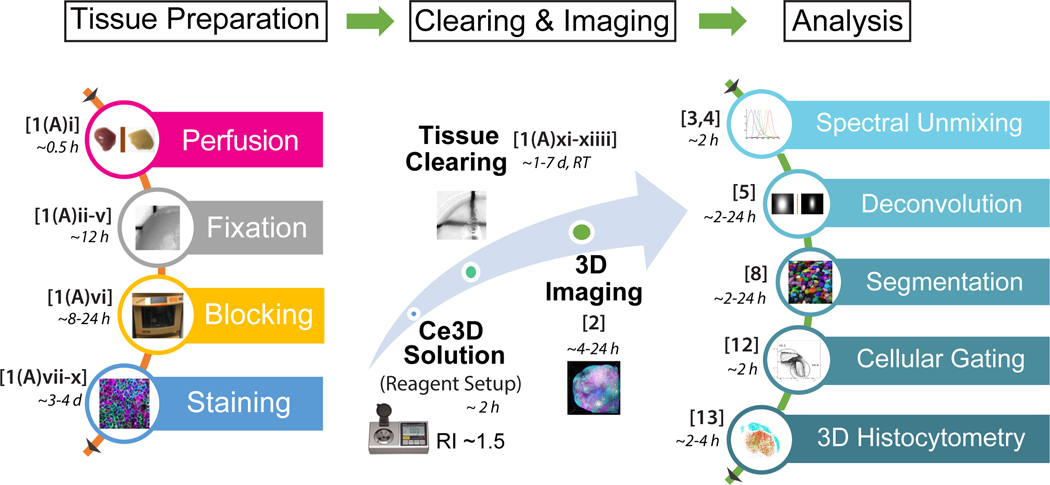
Figure 1. The flowchart of Ce3D clearing, imaging and analysis.
The flowchart demonstrates the individual steps (brackets) and their approximate timing requirements for the Ce3D clearing, imaging and Histocytometry analysis protocol, as denoted by numbers in parentheses and the time guidelines below. Tissues are first perfused [Step 1(A)i] and fixed by either PLP or BD fixative at 4°C [Steps 1(A)ii-v]. Tissues are next blocked and permeabilized at 37°C to enhance antibody penetration and minimize nonspecific antibody binding [Step 1(A)vi]. Tissues are then stained with directly conjugated antibodies with the total incubation time depending on the size of the tissue [Steps 1(A)vii-x]. The Ce3D solution is made during the blocking/staining steps and the RI is checked at room temperature (RT). Tissues are next cleared at RT [Step 1(A)xi-xiiii] and imaged using confocal [Step 2], two-photon or light-sheet microscopy. After image acquisition, spectral unmixing (compensation) is conducted to remove spillover [Steps 3, 4]. Deconvolution is also used to enhance the spatial allocation of imaged signal [Step 5]. Segmentation of individual cells is finally performed [Step 8] to extract data for Cellular gating [Step 12] and 3D Histocytometry [Step 13] for quantitative analysis of cellular position within tissues.
Table 1.
Recommended tissue processing workflow
| Tissue Type | Tissue processing | Volume (mm3) | Preferred Fixative | Blocking/Staining Buffer | Temp | Staining Time | Washing Time | Clearing Time |
|---|---|---|---|---|---|---|---|---|
| Lymph node | whole-mount | 10 | PLP/BD Cytofix | Conventional | 37°C | 4 days | 3 days | 1–2 days |
| Lung | whole-mount | 1000 | PLP/BD Cytofix | Conventional | 37°C | 4 days | 2 days | 1–2 days |
| Small Intestine | whole-mount | 170 | PLP/BD Cytofix | Conventional | 4°C | 12 hours | 1 day | 1 day |
| Thymus | whole-mount | 60 | PLP/BD Cytofix | Conventional | 37°C | 3 days | 2 days | 1–2 days |
| Bone | whole-mount (split longitudonally) | 70 | PLP/BD Cytofix | Conventional | 37°C | 1 day | 1 day | 1–2 days |
| Kidney | whole-mount | 250 | 4% PFA Perfusion | Conventional | 37°C | 2–4 days | 2–3 days | 2–3 days |
| Liver | slice (500 um) | 150 | BD Cytofix/Cy toperm | Alternative | 4°C | 4 days | 2–3 days | 2–3 days |
| Brain | slice (up to 2mm) | 200 | BD Cytofix/Cy toperm | Alternative | 37°C | 2 days | 2–3 days | 1 day |
This table provides general guidelines for effective tissue staining based on the indicated organ type and sample volume, and these can be modified further based on empirical testing. The preferred fixative, blocking buffer, staining time and temperature, as well as the approximate washing and clearing time requirements are indicated for various tested tissues. While most organs can be processed using the thick-slice methodology, certain tissues, indicated here, are more permissible for whole-mount antibody labeling. Optimal bone staining and clearing can be achieved by splitting the organ into two separate halves longitudinally after fixation but prior to blocking. Empirical testing also indicates that 4% PFA perfusion fixation is preferred for kidney processing. Similarly, rapid staining of the vascular endothelium can be achieved with perfusion-based labeling for various organ types.
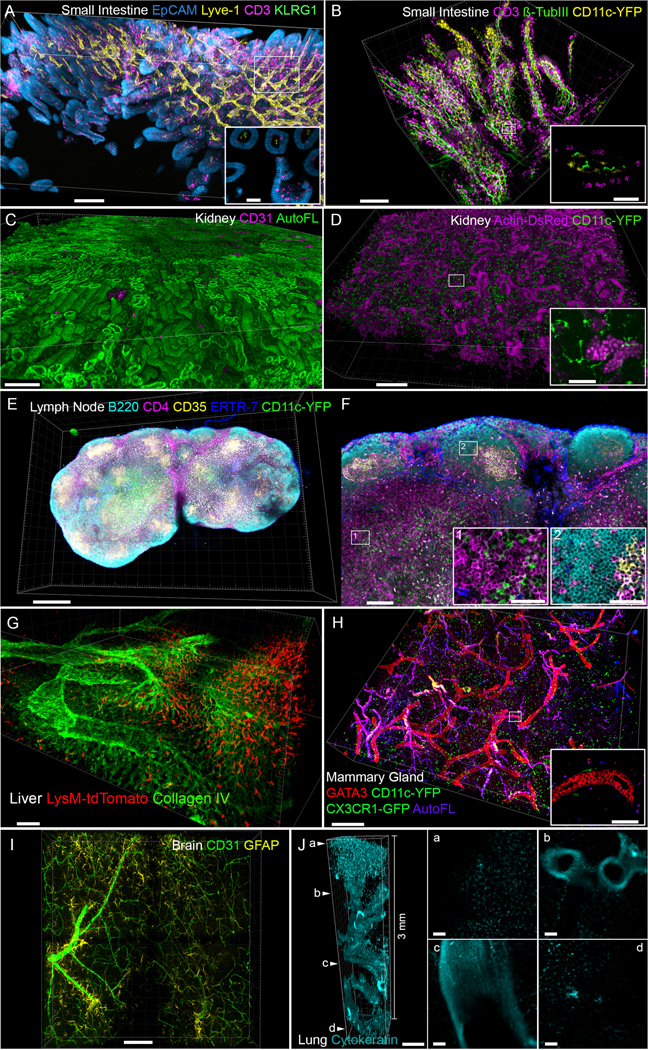
Figure 3. Volumetric Ce3D microscopy of diverse organs.
A-B) Small intestine: the spatial relations of lymphatic vessels, nerves and immune cells inside villi and basal part of small intestine were revealed by their corresponding markers: EpCAM (blue), Lyve-1 (yellow), Beta-tubulin III (green), CD3 (magenta), KLRG-1 (green) and CD11c-YFP (yellow). Quality of individual cell imaging is shown in a zoom-in inset. Scale bars in A: main − 200 μm; inset − 40 μm; Scale bars in B: main − 100 μm; inset − 30 μm. C) The tubular and glomerular structures of kidneys were visualized with CD31 (magenta) staining and autofluorescence (green). Scale bar − 400 μm. D) Actin-DsRed and CD11c-YFP were used to visualize kidney tubular structure and the localization of immune cells. Individual cells are shown in a zoom-in inset. Scale bars: main − 200 μm; inset − 50 μm. E) 3D reconstruction of the whole lymph node labeled with B220 (cyan), CD4 (magenta), CD35 (yellow), ERTR-7 (blue) and a genetically-encoded fluorescent protein, CD11c-YFP (green). Scale bar − 300 μm. F) A single virtual z-slice view of the lymph node (from E). Individual cells were revealed in inset 1 (B cell zone) and inset 2 (T cell zone). Scale bars: main − 100 μm; inset − 30 μm. G) Liver macrophages were visualized using a genetically-encoded fluorescent protein reporter, LysM-tdTomato (red), with respect to the vascular bed revealed with Collagen IV (green) staining. Scale bar − 60 μm. H) The 3D organization of ducts in the mammary gland was revealed by GATA3 antibody (red). Immune cells were revealed by genetically-encoded fluorescent proteins CD11c-YFP and CX3CR1-GFP in the same channel (green), while blood vessels were revealed using autofluorescence signal (blue). Scale bars: main − 400 μm; inset − 60 μm. I) The spatial relationships between astrocytes (GFAP, yellow) and blood vessels (CD31, green) were revealed in the 3D brain slice. Scale bar − 100 μm. J) 3mm deep image stack of a lung was acquired by using two-photon microscope and the motCORR dipping objective to reveal the bronchial airways with Cytokeratin (cyan) staining. Scale bar − 300 μm. Individual virtual Z-slices demonstrate the quality of the obtained signal throughout the Z stack. Scale bars − 50 μm in a, b, c, d. All tissues were isolated from mice maintained at an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility at NIAID. All procedures were approved by the NIAID Animal Care and Use Committee (NIH).
In more recent work, we have found that Ce3D is also compatible with detection of RNA transcripts in large tissue volumes (Figure 4). Such staining can be essential for detection of analytes that are not readily revealed with conventional antibody probes, such as expression of certain cytokines and chemokines, while also providing some insight into how cell positioning within tissues influences gene expression on a single-cell level. Our RNA detection approach is based on a recently described signal amplification technology, RNAscope, which allows robust detection of RNA transcripts at single-molecule resolution directly in tissue sections and is compatible with Ce3D45. The use of this RNA detection method together with Ce3D antibody staining allows robust and high-quality detection of RNA transcripts in thick clarified tissue volumes in association with phenotypic analysis of cells expressing the RNA. The workflow for combining RNA detection with Ce3D tissue clearing is presented in detail in Box 1. When combined with more conventional antibody-based staining approaches, this now collectively allows for robust visualization of diverse analytes of interest and detection of highly complex cellular states in large tissue volumes. The combined method is likely to be especially valuable in leveraging the information from single cell RNA sequencing that provides RNA marker sets for newly described cell subsets.
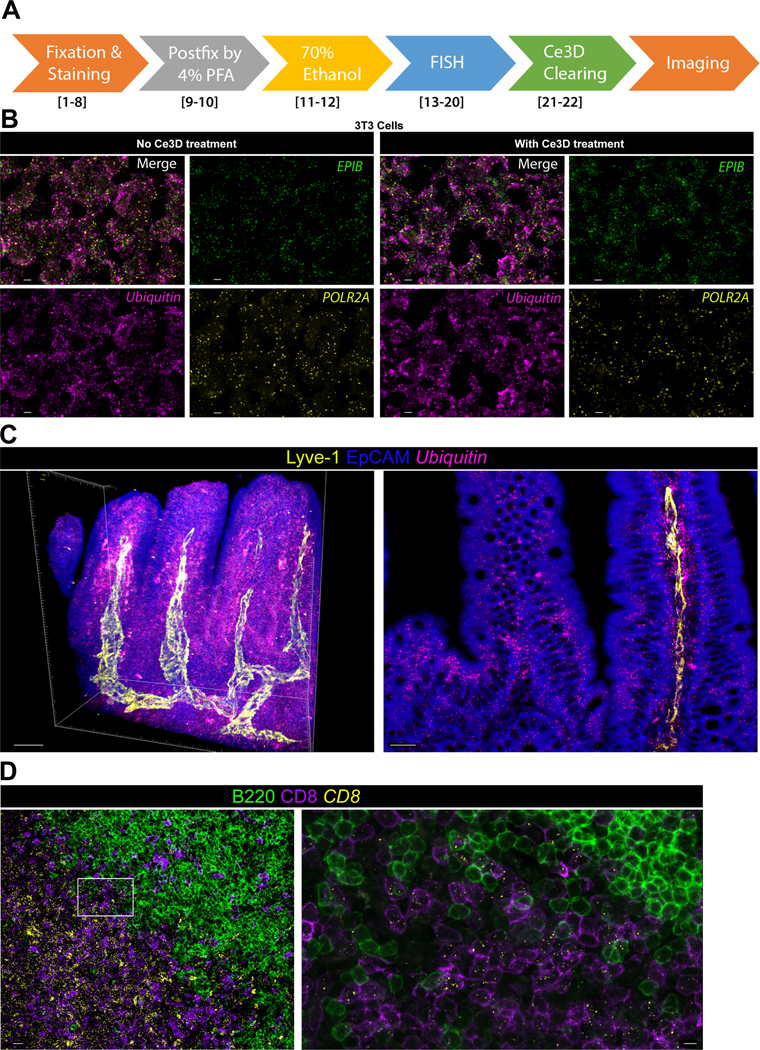
Figure 4. 3D in situ hybridization with immunostaining.
A) The flowchart of steps involved in 3D in situ hybridization and Ce3D tissue clearing. Step numbers referring to the procedure steps as detailed in Box 1 are indicated in brackets. B) The standard control slide from ACD was used to compare the signal density with (right) and without (left) Ce3D treatment using the probes of three housekeeping gene (EPIB, green; FOLR2A, yellow; Ubiquitin, magenta). Data demonstrate that Ce3D tissue treatment does not lead to noticeable loss of mRNA signal. Scale bar, 5μm. C) Volumetric in situ hybridization for the Ubiquitin mRNA (magenta) was combined with antibody staining to detect lymphatic channels (Lyve-1, yellow) and intestinal epithelium (EpCAM, blue). Total volumetric reconstruction of the imaged slice (Z depth = 350μm, scale bar − 50μm, left), as well as a single virtual Z-slice view (right, scale bar = 15 μm) is shown. D) Volumetric imaging of a LN stained by the combination of in situ hybridization (CD8, yellow) and immunofluorescence (CD8 and B220, magenta and green, respectively). Scale bars: main − 15 μm; inset − 5 μm. All tissues were isolated from mice maintained at an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility at NIAID. All procedures were approved by the NIAID Animal Care and Use Committee (NIH).
Box 1: Detection of RNA with Ce3D FISH. Timing: ~6–7d.
This box describes how to combine RNA FISH and immunostaining with Ce3D clearing. See Figure 4a for a schematic overview of the procedure.
Procedure:
!CRITICAL: Only use RNase-free Eppendorf tubes and DEPC-treated water throughout this Procedure.
Obtain fixed whole tissues as described in Step 1(A)i-iii and wash with PBS plus RNase Inhibitor (1:20 dilution, vol/vol) for 20min a total of 3 times
Incubate the tissue in 30% (wt/vol) sucrose diluted in PBS plus RNase inhibitor (1:20 dilution, (vol:vol) at 4°C overnight.
Embed the sample in OCT and freeze on dry ice.
Use a cryostat to section the sample into 100–300um thick slices (as in Step 1(C)ii-iii). Make sure to use RNase-free slides.
Prepare a humidified chamber for RNA detection by spraying the inside of the chamber with RNaseZAP.
Prepare RNase Blocking and Staining Buffers. Use the same recipe as in the Reagent Setup, but use PBS plus RNase inhibitor (1:20 dilution, vol/vol) as base for adding other reagents
Block tissue slices for 1h at RT
Stain tissues with antibodies overnight at RT (as in Step 1(A)vii).
Wash tissues with PBS plus RNase inhibitor (1:20 dilution, vol:vol) at RT for 1.5h a total of 3 times.
-
Fix the section at RT for 1h with 4% PFA in PBS (vol/vol) plus RNase inhibitor (1:20 dilution, vol/vol)
!CRITICAL STEP: This second PFA incubation step fixes the antibodies to the tissue
Wash the section with PBS plus RNase Inhibitor (1:20 dilution, vol/vol) as buffer for 20min, 3 times at RT
-
Place the slides into pre-chilled 70% ethanol (vol/vol) at 4°C overnight.
!CRITICAL STEP: 70% Ethanol (vol/vol) should be made using a previously unopened 100% ethanol bottle and RNase-free DEPC-treated water
Wash tissues with RNase-free water for 20min, 3 times at RT
Hybridization (Steps 13–20): Use RNAscope manufacturer’s manual for hybridization. In short: Start by setting the temperature of the hybridization oven to 40°C.
Place a SecureSeal spacer around the tissue.
Add mRNA probes to the tissue and incubate overnight inside the oven.
Remove the spacer and wash tissues with 1x RNAscope Washing buffer for 20min, 2 times at RT.
Add Amp1 and incubate at 40°C for 1h. Then wash with 1x Washing buffer for 20min, 2 times at RT.
Add Amp2, incubate at 40°C for 30min. Then wash with 1x Washing buffer for 20min, 2 times at RT.
Add Amp3, incubate at 40°C for 1h. Then wash with 1x Washing buffer for 20min, 2 times at RT.
Add Amp4, incubate at 40°C for 30min. Then wash with 1x Washing buffer for 20min, 2 times at RT.
-
Gently dab the samples on a Kimwipe to get rid of excess Washing Buffer. Add Ce3D solution to the tissue, incubate for 20min at RT. Replace with new Ce3D solution and incubate for an additional 30min.
!CRITICAL STEP: Perform this step as quickly as possible to prevent tissue dehydration
!CRITICAL STEP: Presence of excess Washing Buffer can dilute the Ce3D clearing solution and change the refractive index (e.g., liquid transferred with forceps). The clearing reagent may need replacement.
-
Mount the cleared sample as described in Step 1(A)xii-xiii.
PAUSE POINT: The mounted samples can be stored in the dark at 4°C for up to 1 week.
Practical limitations of tissue clearing
Certain limitations do remain with respect to several aspects of large tissue volume immunolabeling, clearing, and high-resolution microscopy. Optimal transparency (low scattering) is difficult to achieve with some tissues using Ce3D. In particular, brains treated with conventional fixatives and Ce3D reagents require extended clarification times, presumably due to a high degree of tissue lipidation and marked reduction in the penetration and equilibration speed of the Ce3D clearing reagent. Inclusion of saponin-containing detergents in the Ce3D protocol and incubation of brain slices at 33–37°C for 1–2 days prior to Ce3D clearing markedly reduces the overall clearing time (1–3mm brain slices cleared within 24hr), and the inclusion of this detergent into the blocking and antibody staining mixtures also minimizes the total tissue processing time (3–6 days total, Table 1). In addition, certain tissues, such as the spleen or liver, have large quantities of heme-containing red blood cells (RBCs), which are not readily cleared with Ce3D, and perfusion of animals prior to tissue isolation helps minimize this issue. We have also noted that the Ce3D reagent can generate minor tissue discoloration during clearing, and this may lead to somewhat reduced laser penetration deep into the tissue. Such discoloration has been observed with other clearing methodologies, and as previously reported by other groups, addition of 0.5% thioglycerol to the Ce3D clearing reagent can help minimize this issue.
One major challenge with large volume tissue immunolabeling in general is that antibodies are relatively large in molecular weight (>150kDa) and do not rapidly diffuse into the tissues. It may take several days for antibodies to penetrate uniformly into the deeper tissue regions and each organ type necessitates empirical testing of the concentration and staining duration for specific probes and fluorophores. Extended time for washing the stained tissues is also important for removal of residual unbound antibody that may generate non-specific background signals during imaging. In our hands, optimal antibody labeling is achieved by incubating smaller mouse organs for 2–3 days on a (rocker/shaker) at 32–37°C using an antibody staining buffer containing a mild detergent (0.3% Triton-X100), while the washing step involved further tissue incubation at 24–33°C for at least one additional day with mild agitation (rocker/shaker). Additional incubation time may be required for staining larger tissue volumes. In future iterations, it may be possible to improve the speed of antibody tissue penetration with microwave technology, as has been previously demonstrated for thin sections46. Alternatively, Fab antibody fragments or single-domain nanobodies are of markedly lower molecular weight (50kDa and 12–15kDa, respectively) and can achieve a similar degree of target specificity as conventional antibodies, thus potentially leading to increased speed of tissue labeling due to their more rapid permeation of the tissue volume47.
In addition to antibody penetration, substantial care must be paid to fluorophore selection for tissue volume labeling. Probe penetration is markedly reduced when using bulky fluorophores and this can lead to non-homogeneous tissue labeling. Certain dyes are in particular problematic, such as the Brilliant Violet non-tandem polymers (BV421, BV480 and BV510) and certain large MW fluorescent proteins (phycoerythrin, allophycocyanin). Additional issues may also arise from use of highly charged dyes, with the presence of multiple charged residues on certain fluorophores preventing probe penetration deep inside the tissue. If using such fluorophores, non-uniform labeling of cells may be observed even after extended labeling periods, with centrally localized cells being stained with substantially lower quantities of antibody as compared cells in the tissue periphery. Similar non-homogeneous labeling is also observed for nuclear DNA-intercalating dyes (e.g. DAPI, Hoechst), presumably due to concentration gradients of the dye generated during diffusion from the incubation solution towards the tissue interior. We were unable to eliminate such gradient effects in whole-mount tissues by varying the dye concentration and duration of labeling, making nuclear staining with such probes problematic. It may be possible to achieve homogeneous nuclear staining using antibodies against certain anti-nuclear proteins (e.g. histone, laminin, etc…), especially in thick tissue slices (see below), although in pilot whole-mount trials with several anti-histone antibodies, we were unable to attain sufficiently homogeneous labeling for accurate cell segmentation. In addition, volumetric imaging necessitates that special consideration be paid to the fluorophore photostability, as certain dyes may not be sufficiently stable for repeated illumination during volumetric confocal imaging (e.g., Pacific Blue, PE, Alexa Fluor 700). In confocal microscopy molecules above and below the focal plane are actively excited with the laser light and will photobleach during imaging. It may be possible to reduce fluorophore photobleaching by increasing the raster scan speed, while also increasing the scan averaging for improved image quality. Alternatively, light-sheet microscopes (LSM) can completely alleviate such depth-dependent photobleaching, as these instruments selectively illuminate a single plane of interest within the tissue without eliciting out of focus fluorophore excitation. However, most LSM instruments use low NA objectives to provide a large field of view and this prevents collection of adequately resolved data for proper segmentation of closely packed cells in many tissues, so this technology is not yet optimized for the type of quantitative imaging at high resolution we discuss here. One additional point of consideration is that residual light absorbance and scattering can persist even after tissue clarification, and this can differentially affect distinct excitation or emission light wavelengths of different fluorophores. These various factors mean that substantial effort is required with respect to the design and testing of multiplexed antibody panels if optimized tissue immunolabeling and volumetric microscopy is to be accomplished with high quality data output.
EXPERIMENTAL DESIGN
Considerations for imaging instrumentation.
The instrumentation and optics used for volumetric imaging of cleared tissues are of critical importance. Confocal microscopes equipped with an array of lasers and filterless detectors, as well as with motorized tiling stages, are commonly available in most research institutions and are capable of acquiring large tissue volumes. Objectives with high numerical apertures (NA) are essential for obtaining high-resolution images, albeit at the cost of limiting working distance and hence, imaging depth. In our hands, a 20× 0.7NA objective generates relatively high-quality image datasets of Ce3D clarified organs with a maximal working distance of ~600um. In certain instances, the resolving power of this objective can still be insufficient for discriminating signals between closely adjoining cells using Histocytometry, with signal overlap being particularly problematic in the axial plane of imaging44. More accurate signal allocation can be achieved with 40X 1.3NA optics, complemented with image deconvolution, although this further restricts the working distance to ~200–250um and limits the ability to reconstruct large tissue volumes. Much larger imaging depths can be attained using conventional 5–10X objectives (>2mm working distance), although the low NA of such optics results in reduced signal detail and quality and is most appropriate for reconstruction of general tissue features and not for detailed cellular phenotyping. Recently developed motorized correction collar objectives designed specifically for imaging cleared tissues combine high NA optics with long working distances, allowing high-resolution microscopy at extended depths (up to 6mm for Leica Microsystems microscopes). However, these objectives are currently costly, pre-specified to the refractive index of specific clearing immersion media, and available only for certain microscope configurations.
The time required to acquire large volumetric datasets with confocal instruments must also be taken into consideration. Even the use of a 20X objective to visualize relatively small murine lymph nodes (1mm^3) can take several (4–12) hours for acquisition, with the total time being dependent on the voxel size and scan speed, as well as on the number of sequential laser scans for fluorescence multiplexing. Thus, volumetric reconstruction of larger tissue samples becomes highly time-demanding with conventional confocal systems. While LSM can allow very rapid microscopy of larger organs, as noted above, current commercial instruments generate fairly low-resolution images and are associated with various light penetration artifacts. The Betzig group has recently combined lattice light-sheet microscopy with adaptive optics to achieve distortion-free, high-resolution imaging of relatively large tissue volumes48. Although such instruments are not yet widely available, they hold promise for allowing fast and detailed volumetric microscopy of large tissue volumes.
Image processing and data analysis.
Visualization and analysis of volumetric images can be achieved with various software. In our hands, Imaris (Bitplane) excels at intuitive visualization of multiparameter volumetric datasets, but we have not exhaustively compared its performance with other platforms (Figures 3 and 4). Further quantitative and phenotypic analysis of imaged cells (Histocytometry) can be achieved by segmenting cells into individual 3D objects. To this end, sparsely distributed cells can be segmented using the Imaris Surface creation module (Step 8(A)ii), which utilizes watershed-based algorithms for object splitting. This segmentation becomes more challenging if multiple cells are physically touching one another or are clustered, especially if these are visualized using only cell membrane markers. The signals from adjoining cells are not sufficiently resolved with conventional confocal imaging, and watershed-based algorithms identify the brightest-stained regions (i.e., adjoining area between the clustered cells) as the object center, leading to inaccurate cell segmentation. Although we previously resolved this issue in thin sections using nuclear-based segmentation, this was not applicable to whole-organ segmentation due to lack of homogeneous nuclear staining (see above). While the Imaris Cell module can in principle perform 3D segmentation of clustered cells based on membrane signals alone, in our hands the current software iteration (Imaris 9) does not permit processing of large image datasets due to software instability.
Instead, we overcame these issues by implementing a relatively straightforward signal inversion technique (Figure 5). In this method, all visualized membrane stain signals are intensity-normalized and summed together to generate a new Composite Sum Channel representing all imaged cells (Step 8(B)i). This Sum Channel is next inverted to make the nuclear and cytoplasmic areas bright and the stained membrane regions dark, thus generating discrete boundaries between cells. The Inverted Channel is further processed with Gaussian pixel smoothing, as well as adjusted for contrast and gamma-correction to enhance the signal separation between individual cells (Steps 8(B)ii-v). We were able to further enhance the cell boundaries using the 2D-Skeletonization plugin in ImageJ, generating discrete pixel-wide boundaries between cells that are further added to the Inverted Channel (Steps 8(C)i-xi). Finally, the enhanced Inverted Channel is segmented using the Imaris Surface creation module, generating objects for all imaged cells, as well as the unstained extracellular regions (Step 8(C)xii). Even though many of the stains of interest are localized on the plasma membranes, which are actively excluded from the generated cell objects with this signal inversion technique, sufficient residual membrane signals within the cell objects remain for phenotyping. Further, exclusion of the outermost cell membrane voxels also minimizes the artifactual incorporation of signals from neighboring cells, thus helping improve cellular analysis in denser tissues. While the Membrane Sum technique works well in our hands for cell segmentation, it may also be possible to achieve similar inversion-based segmentation using dedicated ubiquitous cell membrane marker staining. However, the use of such a dedicated membrane marker would also eliminate one channel from an already limited panel for cellular phenotypic analysis, diminishing the utility of the technique for multiplex analysis.
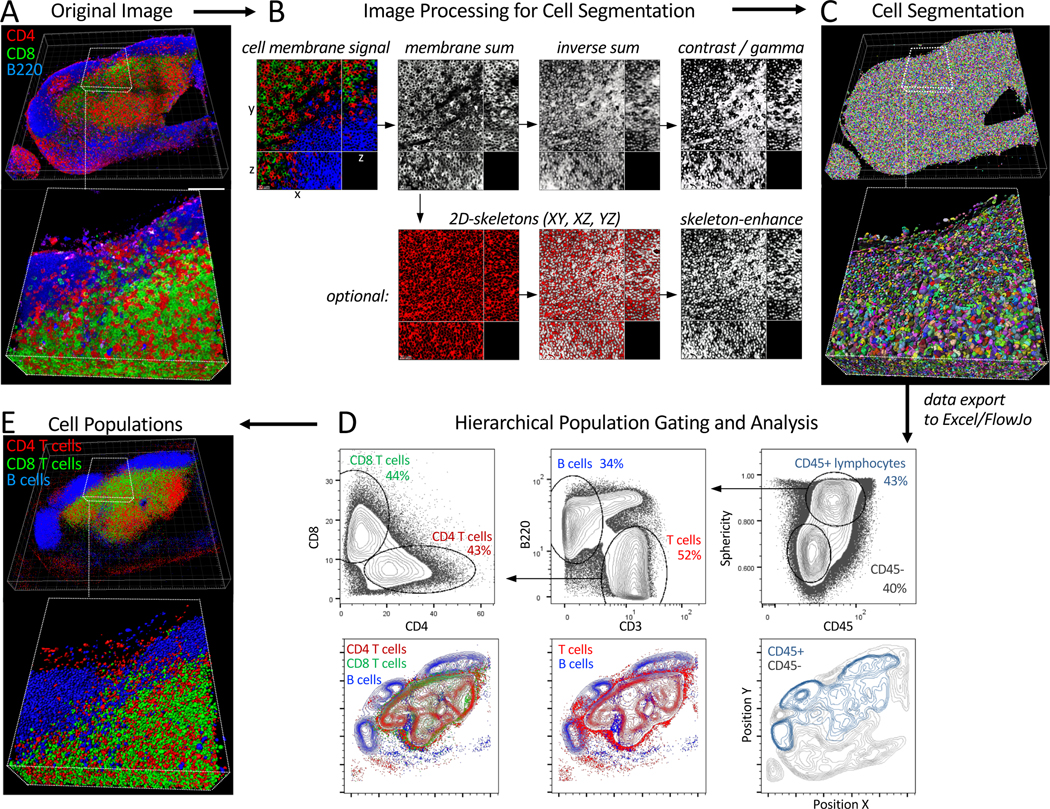
Figure 5. Volumetric Histocytometry example workflow.
250um fixed sections of steady-state murine lymph nodes were stained with antibodies against various lymphocyte populations (CD3, CD4, CD8, B220, CD45), Ce3D cleared, imaged with confocal microscopy (40x objective), deconvolved, and corrected for depth attenuation. A) Volumetric reconstruction of the 3D image in Imaris. Scale bar – 200 μm. B) Image processing steps for cell segmentation. All cell membrane signals were normalized with respect to each other and summed together using Imaris Channel Arithmetics XTension. The Sum channel was next inverted, smoothed and corrected for contrast and gamma to enhance separation between cells. An optional enhancement step (bottom panels) was also performed by generating 2D skeletons on the Sum channel in ImageJ. The skeletons were next used for Boolean gating of the Inverse Sum channel outside of the skeleton signal using Channel Arithmetics. C) The enhanced Inverse Sum channel was used to generate cell surface objects in Imaris. D) Data on all cell objects was exported into Excel, concatenated into a single CSV file, and imported into FlowJo for hierarchical population gating and analysis (top panels). Positional visualization of the gated cells, presented as density distributions, was also performed in FlowJo (bottom panels). E) Cell object gating and visualization was also performed in Imaris using Object Filters and gating thresholds from (D). All tissues were isolated from mice maintained at an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility at NIAID. All procedures were approved by the NIAID Animal Care and Use Committee (NIH).
Once the cell surface objects are generated, the relevant object statistics (mean channel fluorescence, object volume, sphericity, XYZ position) are either analyzed utilizing built-in tools in Imaris (Vantage) or passed for further analysis using additional software (e.g., Flowjo), (Steps 9–13). The extracellular regions or other artifacts generated during the inversion-based segmentation can be excluded from analysis using morphologic and phenotypic gating (cells are more spherical and homogeneous in size than artifactual regions). Finally, hierarchical population gating is performed on the remaining cell objects and the gated populations are visualized using positional plotting directly in Flowjo. Positional gating of cells within specific regions of interest can also be performed to study the composition of different cell populations within a given region.
It is important to note that many of the processing steps described above require substantial hands-on expertise and processing time. Cell segmentation often requires empirical testing of different object settings during surface creation. Further, current segmentation software can have trouble with very large datasets, necessitating dedicated workstations with large amounts of RAM (128GB minimum) and splitting of image files into multiple adjoining regions for processing. In addition, phenotypic analysis software such as FlowJo is not optimized for positional visualization of cells, nor for studying the spatial relationships between different cell types. Thus, there is a need for continued development of dedicated software platforms for quantitative cellular visualization and spatial distribution analysis. In addition, it must be noted that the costs for dedicated workstations equipped with the necessary analysis software can be extensive (roughly $10K for a capable workstation, plus $12–25K for dedicated software). Many facilities have dedicated workstations for such image analysis, which can help minimize expenses associated with large volume image analysis.
Recommendations for productive imaging of clarified tissues.
Given the limitations with respect to antibody tissue penetration, restricted working distances of conventional objectives, low speed of high-resolution imaging, as well as in the available software platforms (see “Practical limitations of tissue clearing”), comprehensive analysis of cells in large tissue volumes is challenging and can be impractical for studies requiring analysis of many samples and timepoints. Thus, we recommend a modified workflow to make tissue clearing more applicable for conventional studies. Specifically, we advise use of thick tissue slices varying from 250–600um in thickness instead of whole isolated organs or multi-mm samples. The slices can be generated using either a vibratome with agarose-embedded fixed tissues (Step 1 Option B) or with a cryostat for OCT (Optimal cutting temperature) -embedded frozen tissues (Step 1 Option C). The duration of antibody staining is minimized with thick slices and allows use of large MW dyes, while the reduced depth of imaging also permits use of less photostable fluorophores, thus collectively maximizing probe multiplexing. Such thick tissue slices can also be easily imaged in their entirety using conventional high-NA objectives at high resolution, and the image visualization and data analysis also becomes more practical using existing software. Use of OCT-embedded tissues for generating thick slices also allows side-by-side comparison of volumetric vs. thin section images, as both can be readily generated during sectioning. While these thick slices lack the ability to provide information on structures spanning several mm or across entire organs of small animals, they still generate 50–100X more (volumetric) data than conventional sections, while demanding minimal alterations to the staining panels and processing workflows already established in labs using conventional section-based immunofluorescence microscopy. Finally, unless laboratories already own dedicated microscopes and workstations, the use of thick slices can help reduce the overall costs associated with large volume imaging. While the Ce3D clearing on its own is inexpensive, substantial additional costs can be incurred for extended time necessary to image larger tissue volumes at high resolution using confocal microscopes at core facilities (typically $30–60 per hour). In the future, access to properly configured light-sheet microscopes that can collect data at 10–100x the rate of standard confocal instruments may address this time-of-use cost factor.
MATERIALS
Biological materials
-
Mice. Reporter mouse lines used for the examples provided in this manuscript include: CD11c-YFP [B6.Cg-Tg(Itgax-Venus)1Mnz/J], actin-DsRed [B6.Cg-Tg(CAG-DsRed*MST)1Nagy], Cxcl12-DsRed (Cxcl12tm2.1Sjm/J), Cx3cr1-GFP (B6.129P-Cx3cr1tm1Litt/J) and were obtained from The Jackson Laboratory. Two- to 6-month-old male or female mice were used for all experiments and were randomly allocated into treatment groups. The investigators were not blinded to allocation during experiments and outcome assessment. All mice were maintained in specific pathogen-free conditions at an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility at the National Institute of Allergy and Infectious Diseases (NIAID). All procedures described in this Protocol were approved by the NIAID Animal Care and Use Committee (NIH).
!CAUTION: Any experiments involving live animals must conform to relevant Institutional and National regulations.
Reagents
Tissue Fixation and Blocking
PBS (Catalog # 10010023, pH=7.4, Gibco)
Heparin sodium, (Catalog # 27602, Fresenius Kabi)
Optimum Cutting Temperature (O.C.T.) Compound, Tissue-Tek (Catalog # 62550, Electron Microscopy Sciences)
0.1 M Tris Buffer (Catalog # RGF3340, KD Medical)
- Paraformaldehyde (PFA) Solution, 16% (vol/vol) (Catalog # 15710, Electron Microscopy Sciences)
- !CAUTION: PFA is toxic. Use a chemical hood and appropriate personal protective equipment (gloves and lab coat) when using PFA.
- !CRITICAL: Methanol is included in some PFA solutions as a stabilizer, but presence of methanol can cause less ideal fixation.
BD Perm/Wash (Catalog # 51–2091K, BD Biosciences)
BD Cytofix/Cytoperm (Catalog # 51–52090KZ, BD Biosciences)
Na2HPO4 (Catalog # S0751, Sigma-Aldrich)
NaH2PO4 (Catalog # S5136, Sigma-Aldrich)
L-Lysine (Catalog # L5501, Sigma-Aldrich)
NaIO4 (Catalog # 311448, Sigma-Aldrich)
Triton X-100 (Catalog # T-9284, Sigma-Aldrich)
Albumin from bovine serum (Catalog # A7906, Sigma-Aldrich)
- Mouse serum (Catalog # 10410, ThermoFisher Scientific)
- !CRITICAL: - Mouse serum is used for blocking mouse tissues
- NaN3 (Catalog # S2002, Sigma-Aldrich)
- !CAUTION: NaN3 is toxic. Use a chemical hood and appropriate personal protective equipment (gloves and lab coat) when using NaN3.
Ce3D Tissue Clearing
N-methylacetamide (Catalog # M26305–500G, Sigma-Aldrich)
Histodenz (Catalog # D-2158, Sigma-Aldrich)
1-Thioglycerol (Catalog # M1753, Sigma-Aldrich)
- Slygard-184 Elastomer Kit (Catalog # 634165S, VWR)
- !CAUTION: Direct skin contact with Slygard-184 can cause skin cancer! Use appropriate personal protective equipment (gloves and lab coat) when using Slygard-184.
High Vacuum grease (Catalog # DOWC1597418, VMR)
Agarose (Catalog # 16500500, ThermoFisher Scientific)
Ce3D FISH
RNAscope Fluorescent Multiplex Detection Reagents (Catalog # 2001989, Advanced Cell Diagnostics)
HybEZ™ Hybridization System (Catalog # 310010, Advanced Cell Diagnostics)
100% ethanol (Catalog # 64–17-5, Warner-Graham Company)
RNAse inhibitor: Ribonucleoside Vanadyl Complex (Catalog # S1402S, NEB)
RNaseZAP (Catalog # AM9780, Thermofisher Scientific)
DEPC-treated water (Catalog # 221–036-10, Crystalgen)
4% PFA (vol/vol, diluted from 16% PFA with DEPC-treated dH20)
Control Slide-Mouse 3T3 Cell Pellet (Catalog # 310023, Advanced Cell Diagnostics)
Probes: 3-plex Positive Control Probe- Mm (Catalog # 320881, Advanced Cell Diagnostics); Positive Control Probe- Mm-Ubc (Catalog # 310771, Advanced Cell Diagnostics)
Microscopy
UltraComp eBeads (Catalog # 01–2222-41, Fisher Scientific)
Equipment
Tissue Fixation and Blocking
Dissection microscope with fiberoptic illumination
0.45μm Sterile Disposable Vacuum Filter (Catalog # SE1M003M00, Fisher Scientific)
Ce3D Tissue Clearing
Refractometer (Model 30034, Sper Scientific or similar)
Parafilm (Catalog # P7793, Sigma-Aldrich)
Glass bottom culture dishes (Catalog # P35G-0–10-C, MatTek Corporation)
-
Coverglass, 22×22mm, No. 0 (Catalog # 3206, Ted Pella, Inc.)
!CRITICAL: No. 0 coverglass maximizes working distance of the objective
Ce3D FISH
RNase free slides (Catalog # 71869–10, Electron Microscopy Sciences)
SecureSeal™ Hybridization Chambers (Catalog # 621211, GraceBio-Labs)
Microscopy and image processing
Confocal microscope equipped with a motorized tiling stage (e.g., Lecia TCS SP8 or similar)
Microscope Objectives (20x oil, NA=0.75, working distance=680μm; 40x oil, NA=1.3, working distance=240 μm or similar).
Computer Workstation with high RAM (e.g., HP Z840, Graphic Card: nVIDIA M6000, RAM 256GB or similar)
Offline LASX software (Leica Microsystems) for tile stitching and spectral unmixing (or similar microscope manufacturer’s software for offline image processing)
Huygens deconvolution software (Scientific Volume Imaging, http://svi.nl/)
Imaris 9.2 (Bitplane, http://www.bitplane.com) or higher for 3D image rendering, surface object construction and data analysis
ImageJ/FIJI (National Institutes of Health, https://fiji.sc) for additional image processing
Microsoft Office – Excel (https://products.office.com/en-us/excel)
Additional Common Supplies and Equipment
Eppendorf tubes (1ml, 5ml)
Conical tubes: 15ml, 50ml (Catalog # 14–432-22, Fisher Scientific)
Multi-well plates for tissue harvest / processing (Greiner or similar)
Pipettes (5ml, 10ml, 25ml)
Kimwipes (Catalog # 06–666, Fisher Scientific)
Uncharged glass slides (Catalog # 4445, Thermo Scientific - or similar)
PAP pen (Catalog # H-4000, Vector Laboratories)
Dark, humidified slide staining tray
Forceps (#5 Dumont, Fine Science Tools or similar)
Water bath
Vortex (Catalog # SI-0236, Scientific Industries, Inc. or similar)
pH meter
Rotator
Incubation Shaker (e.g., Multitron standard, Infors HT or similar)
Vibratome (Catalog # VT1000s, Leica or similar)
Certified Chemical Hood
Reagent Setup
!CRITICAL: PLP buffer is the optimal fixative for preserving membrane glycoproteins and carbohydrates. If using BD Cytofix or BD Cytofix/Cytoperm Buffers for fixation and the PBS-based buffer for washing, the preparation of P-buffer, L-Lysine Buffer and PLP Buffer is not necessary. Refer to Table 1 for specific tissue recommendations.
- Fixation Reagents: P-Buffer
- Prepare stock 0.2M Na2HPO4 solution - store at room temperature (RT: 20–25°C), sterile
- Prepare stock 0.2M NaH2PO4 solution (store RT, sterile)
-
Prepare 4% (vol/vol) PFA by diluting 16% PFA (vol/vol) in PBS. Aliquot 2.5ml in 5ml tubes and store at −20˚C!CAUTION: PFA is toxic. Use chemical hood, gloves and lab coat throughout.
-
For preparing 200ml P-buffer, add together the following and mix well:
- 81ml of 0.2M Na2HPO4
- 19ml of 0.2M NaH2PO4
- 100ml of H20
!CRITICAL: The buffer can be scaled accordingly. - Check the pH – this should be 7.4.
- Store at 4˚C for up to 6 months.
-
Fixation Reagents: L-Lysine buffer
Add L-Lysine to P-buffer to make final L-Lysine concentration 0.2M. Aliquot into 50ml conical tubes and store at −20˚C for up to 6 months.
-
Fixation Reagents: PLP buffer
The recipe below is for 10ml but it can be scaled accordingly. Add together the following and mix well:- 2.5 ml 4% PFA (vol/vol)
- 0.0212 g of NaIO4 (sodium periodate)
- 3.75 ml L-Lysine buffer
- 3.75 P-buffer
!CRITICAL: PLP buffer should be freshly prepared as PFA can degrade over time.
-
Alternative Fixation Reagents: BD Cytofix
BD Cytofix is a commercial fixative that can be used instead of the PLP fixation buffer. Dilute 1-part BD Cytofix with 3-parts PBS.
!CRITICAL: BD fixation buffers should be freshly diluted before use. Make sure reagents are not expired as PFA can degrade over time. Prepare sufficient fixation buffer based on utilized tissues: 1–2ml for small organs (1–2 mm3); ~10ml for larger tissues (e.g., liver, lung, kidney, etc.); ~5ml of BD Cytofix/Cytoperm buffer per 2–3mm brain sections.
-
Alternative Fixation Reagents: BD Cytofix/Cytoperm
BD Cytofix/Cytoperm is optimal fixative for brain and other lipid-rich tissues (Table 1). Dilute 1-part BD Cytofix/Cytoperm with 3-parts PBS (1:4 final).
!CRITICAL: BD fixation buffers should be freshly diluted before use. Make sure reagents are not expired as PFA can degrade over time. Prepare sufficient fixation buffer based on utilized tissues: 1–2ml for small organs (1–2 mm3); ~10ml for larger tissues (e.g., liver, lung, kidney, etc.); ~5ml of BD Cytofix/Cytoperm buffer per 2–3mm brain sections.
-
Conventional Blocking Buffer
Prepare 1x PBS (or P-buffer) supplemented with 0.3% Triton-X100 (vol/vol), 1% Bovine Serum Albumin (vol/vol), and 1% Normal Mouse Serum (vol/vol).
!CRITICAL: Blocking buffer should be freshly prepared. Filter the Blocking Buffer using a 0.45um filter to minimize bacterial contamination. Prepare sufficient Blocking Buffer based on utilized tissues: ~1ml for small organs (1–2 mm3); ~10ml for larger tissues (e.g., liver, lung, kidney, etc.).
!CRITICAL: Normal mouse serum is used with mouse tissues. Adjust specific blocking sera according to specific tissue and secondary detection reagent needs.
-
Alternative Blocking Buffer
Prepare 1x BD Perm Buffer (generated by diluting 10x stock buffer to 1x with PBS) supplemented with 0.3% Triton-X100 (vol/vol), 1% Bovine Serum Albumin (vol/vol), 1% Normal Mouse Serum (vol/vol).
!CRITICAL: Blocking buffer should be freshly prepared. Filter the Blocking Buffer using a 0.45um filter to minimize bacterial contamination. Prepare sufficient Blocking Buffer based on utilized tissues: ~1ml for small organs (1–2 mm3); ~10ml for larger tissues (e.g., liver, lung, kidney, etc.).
!CRITICAL: Normal mouse serum is used with mouse tissues. Adjust specific blocking sera according to specific tissue and secondary detection reagent needs.
-
Conventional Washing Buffer
Prepare 1x PBS supplemented with 0.3% Triton-X100 (vol/vol), and 0.5% 1-Thioglycerol (vol/vol). This buffer can be stored at 4°C for up to 1 week.
!CRITICAL This buffer can be used based on convenience. We have observed no differences between PBS (Conventional)- vs. P-Buffer (Alternative)-based Washing buffers.
-
Alternative Washing Buffer
Prepare 1X P-buffer supplemented with 0.3% Triton-X100 (vol/vol), and 0.5% 1-Thioglycerol (vol/vol). This buffer can be stored at 4°C for up to 1 week.
!CRITICAL: This buffer can be used based on convenience. We have observed no differences between PBS (Conventional) vs. P-Buffer (Alternative)-based Washing buffers.
-
40% N-methylacetimide (vol/vol)
!CAUTION: N-methylacetamide is a presumed human reproductive and fetal toxicant. Use a chemical hood and appropriate personal protective equipment (gloves and lab coat) throughout preparation of the N-methylacetamide. Collect waste in appropriate container for EH&S disposal.- Place the entire 100% N-methylacetamide bottle into a 37˚C incubator for >1 hour until fully dissolved. N-methylacetamide is solid at RT.
-
Pre-warm a 25ml pipet and use it to transfer 20ml of 100% N-methylacetamide (vol/vol) into a 50ml conical tube.!CRITICAL STEP: N-methylacetamide will stick to the pipette wall if pipette is not prewarmed.
-
Add 30ml of PBS at RT to the 20ml N-methylacetamide and mix to make 40% (vol/vol) N-methylacetamide stock solution!CRITICAL STEP: The 40% N-methylacetamide (vol/vol) can be stored tightly capped at RT for 2 months.
-
Ce3D clearing solution
!CAUTION: N-methylacetamide is a presumed human reproductive and fetal toxicant. Use a chemical hood and appropriate personal protective equipment (gloves and lab coat) throughout preparation of the Ce3D clearing solution. Collect waste in appropriate container for EH&S disposal.
!CRITICAL: It takes 2h to make the Ce3D clearing solution – it is usually prepared during the tissue staining/washing steps.
The following is a 5ml Ce3D clearing solution preparation – it can be scaled according to experimental needs:-
Add together in order into a 5ml Eppendorf tube:
- 2ml 40% N-methylacetamide (vol/vol)
- 4g Histodenz
- An additional 750ul 40% N-methylacetamide (vol/vol)
- 5ul of Triton X-100
!CRITICAL STEP: Histodenz is layered between 40% N-methylacetamide (vol/vol) to promote faster dissolution. - Seal the tube with parafilm.
-
Place the tube with Ce3D mix into a 37˚C shaking incubator set to 150–225RPM for > 1h until fully dissolved.!CRITICAL STEP: Ce3D clearing solution will go from agglomerated whitish liquid to fully mixed clear liquid.
-
Use a disposable transfer pipette or other mixing tool to ensure that the Ce3D solution is fully mixed (Ce3D density gradients can occur inside the tube due to the high viscosity of the reagent).!CRITICAL STEP: Ensure complete Ce3D clearing solution mixing, as incomplete mixing will change the Ce3D clearing properties.
- Add 25ul 1-Thioglycerol to the mixed Ce3D solution. This reagent helps reduce tissue discoloration during clearing.
-
Check the refractive index (RI) of Ce3D solution by using a refractometer. Expected RI = 1.495~1.505!CRITICAL STEP: Its critical to check the RI of the final Ce3D clearing solution as improper RI can cause less than ideal clearing!CRITICAL STEP: This buffer can be stored at RT for up to 4 weeks in the Eppendorf tube sealed by parafilm.
-
PROCEDURE
Tissue Preparation
-
1
For preparation of whole-mount tissues, follow Option A. Whole-mount tissue processing is time intensive and can be challenging for obtaining homogeneous tissue labeling (see Introduction). Use of 100–600um thick tissue slices reduces the processing time requirements, while also permitting use of larger MW fluorophores. For vibratome slice generation from non-frozen tissues, follow Option B. For cryostat slice generation from OCT-embedded frozen tissues, follow Option C.
Option A: Whole-mount.
Timing: ~3–5d for tissue processing/staining; ~1–7d for tissue clearing.
! CRITICAL: Whole-mount tissue staining can also be performed in combination with RNA FISH. In this case, follow the procedure described in Box 1 instead.
-
For large organs like lung, kidney, heart and liver, perfuse mice with PBS with heparin (10 unit/ml) to eliminate vascular blood content. The optimal perfusion sites are:
- liver: portal vein
- kidney: abdominal aorta
- lung and heart: left atrium
!CRITICAL STEP: Perfusion for lungs is optional as most erythrocytes will be washed away during the staining and washing procedure.
!CRITICAL STEP: Perfusion fixation with 4% PFA (vol/vol) can be used to maximally retain epitopes.
? TROUBLESHOOTING
-
Harvest tissues into a tissue culture plate with PBS on ice (e.g., 6-well plate with 500ul PBS per well for lymph nodes) and use a dissection microscope and forceps to carefully remove attached fat or connective tissue.
!CRITICAL STEP: Samples can overheat if kept too close to the light.
-
Fix tissues overnight at 4°C using one of the following fixation buffers based on the tissue type (Table 1) and target molecule of interest (see Reagent Setup). Tissues can be placed on a shaker to enhance the speed of fixative penetration.
- PLP fixation buffer: optimal fixative for preserving membrane glycoproteins and carbohydrates
- BD Cytofix: off the shelf commercial fixative that can be used instead of the PLP fixation buffer
- BD Cytofix/Cytoperm: optimal fixative for brain and other lipid-rich tissues
!CRITICAL STEP: Use an approximate volume of 1ml for small organs (1–2 mm3), larger volumes for larger tissues (e.g., 10ml for liver, lung, kidney, etc.), and approximately 5ml of BD Cytofix/Cytoperm buffer per 2–3mm thick brain section.
? TROUBLESHOOTING
-
Wash fixed tissues for 30–60min with either PBS-based Conventional Washing Buffer or P-Buffer based Alternative Washing Buffer (see Reagent Setup).
!CRITICAL STEP: Washing buffers can be used based on convenience. We have observed no differences between PBS vs. P-Buffer-based Washing buffers.
!CRITICAL STEP: Use an approximate volume of 1ml for small organs (1–2 mm3). Use larger volumes for larger tissues (e.g., 10ml for liver, lung, kidney, etc.)
Repeat Washing (Step iv) 2x more times for a total of 3 washes.
-
Block and permeabilize tissues by incubating at 37°C for 8–24h on a shaker in either Conventional Blocking Buffer (most tissues), or in Alternative Blocking Buffer (use with brain or other lipid-rich tissues, Table 1).
!CRITICAL STEP: Larger organs (liver, lung, kidney etc.) may require additional blocking time.
-
Stain tissues by incubating with a panel of antibodies diluted in appropriate blocking buffer. Incubate using a shaking incubator set to 34–37°C, 150–220rpm for 3–4 days (e.g. volume: 500ul-1ml of staining buffer is used to stain 2–3 mouse lymph nodes).
!CRITICAL STEP: Antibodies are usually diluted at 1:100 – 1:200 (vol:vol) from commercial supplier stocks, but the precise dilution depends on original antibody concentration, manufacturer recommendation and empirical testing.
!CRITICAL STEP: Minimize use of antibodies conjugated to large-MW fluorophores (e.g. APC, PE, Brilliant Violet 421) as these do not readily penetrate thicker tissues (see the “Practical limitations of tissue clearing” section of the Introduction). These dyes do however work for 200–500um thick sections generated in Option B and C.
!CRITICAL STEP: Prevent buffer evaporation by sealing the tube using parafilm. For incubating multi-well plates at 37°C, make sure to cover the wells with parafilm to minimize evaporation, block from light exposure, and do not use a rotator.
!CRITICAL STEP: Alternatively, samples can be incubated for 5–6 days at RT, or 7–10 days at 4°C. Different temperature and incubation times influence antibody penetration and can affect the quality of staining. Optimal staining conditions may depend on tissue type and probe target, and should be determined experimentally (Table 1).
? TROUBLESHOOTING
-
Wash tissues with Washing Buffer at 37°C degree for 8–14h. (e.g. volumes: 1ml Wash Buffer per lymph node; 10ml for liver, lung, kidney etc.)
!CRITICAL STEP: extending the washing time at 37°C can destabilize tissue staining and lead to loss of signal
Replace with new Washing Buffer and incubate at RT for 1–4 days (based on tissue size, Table 1), changing the buffer every 12–24h.
OPTIONAL: If using primary / secondary antibody detection, repeat staining (Steps vii-ix) using the appropriate secondary detection reagents (e.g., anti-rabbit Alexa-488) at an empirically determined dilution (typically 1:400 vol:vol) from commercial supplier stocks.
-
Gently dab the samples on a Kimwipe to get rid of excess Washing Buffer and place samples directly into the Ce3D clearing solution in a 1–5ml Eppendorf tube. We suggest using 1ml Ce3D clearing solution for lymph nodes, and 10ml for liver, lung, kidney etc.
!CRITICAL STEP: Perform this step as quickly as possible to prevent tissue dehydration
!CRITICAL STEP: Presence of excess Washing Buffer can dilute the Ce3D clearing solution and change the refractive index (e.g., liquid transferred with forceps). The clearing reagent may need replacement.
!CRITICAL STEP: For difficult to penetrate tissues, follow the gradient clearing procedure described in Box 2.
-
Cover the sample tube with tin foil and attach to a rotor. Rotate samples at RT until clear. Most samples will clear in 12–24h, but larger organs may take longer (Table 1).
!CRITICAL STEP: Clearing at 37°C is not recommended due to discoloration.
? TROUBLESHOOTING
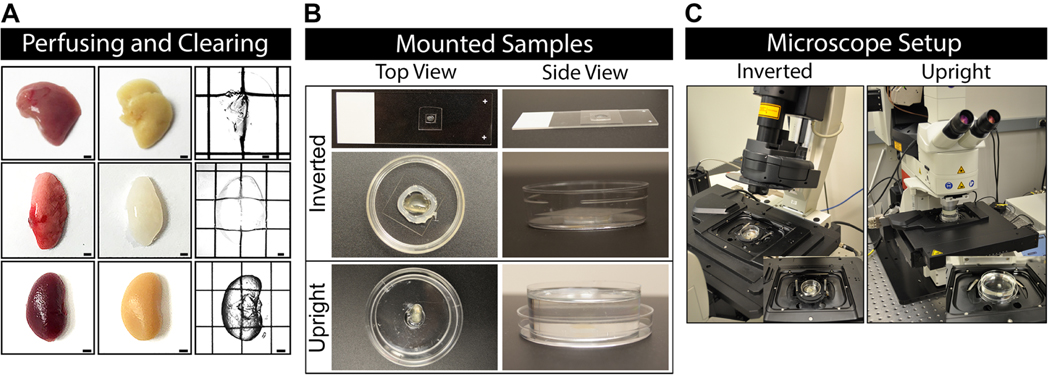
Mount the cleared sample in a No. 0 glass-bottom dish filled with Ce3D clearing solution. This is ideal for imaging on an inverted microscope.
-
To minimize use of Ce3D reagent, make a spacer out of Slyguard 184 inside the No. 0 glass-bottom dish, creating an inner chamber. The spacer thickness depends on tissue size. Spacers of varying thicknesses can be made by solidifying Slyguard 184 in 10cm culture plates at RT. Alternatively, vacuum grease can be used as an alternative to Slygard 184 to make a chamber for smaller tissues. Fill the inner chamber with Ce3D solution and place tissues inside.
PAUSE POINT: The mounted samples can be stored in the dark at RT for up to 1 week.
Box 2: Gradient Clearing. Timing: ~15h.
This clearing variant is optional. For difficult to penetrate tissues (e.g., brain slices), gradient clearing can enhance the penetration of Ce3D clearing reagent (due to reduced viscosity) and reduce tissue discoloration.
Perform a series of step-wise Ce3D gradient switches, each time increasing the concentration of the Ce3D solution (see Reagent Setup) diluted with PBS, supplemented with 5% 1-Thioglycerol (vol/vol).
30% Ce3D (vol/vol) for 1h at RT
50% Ce3D (vol/vol) for 1h at RT
70% Ce3D (vol/vol) for 1h at RT
80% Ce3D (vol/vol) for 2h at RT
90% Ce3D (vol/vol) for 2h at RT
100% Ce3D for >8h at RT until clear
!CRITICAL STEP: The above gradient has been optimized for 1mm thick samples. It is suggested to scale up time for thicker samples.
Option B: Thick tissue slices: vibratome slice generation from non-frozen tissues.
Timing: ~1–5d for tissue processing/staining; ~3h-1d for tissue clearing.
Obtain fixed and washed tissues, as described in Steps 1(A)i-v.
Melt 2% agarose in PBS (wt/vol) using a microwave and pour liquid into wells of a multi-well plastic plate or into plastic tissue molds.
Allow liquid to cool until no longer too hot to touch, but before agarose solidifies. iv. Gently embed the tissues into the melted agarose using forceps.
Once solidified, use a pointed end of a pipette tip or forceps to dislodge the agar block containing tissue and place into chilled PBS on ice.
Use a vibratome to slice the agar tissue blocks to desired thickness (200–600um)
Transfer slices to 12–48 multi-well plates filled with PBS making sure not to touch the tissue with forceps.
-
Proceed with blocking, staining and washing of tissues as described in Steps 1(A)vi-x.
!CRITICAL STEP: Incubation time for blocking, staining and washing will decrease as compared to whole mount tissues (e.g., for 250um slices, homogeneous probe labeling can be attained with ~12h blocking at 37°C, 2 days of staining at 37°C, and ~12h of washing at RT)
? TROUBLESHOOTING
-
Gently dab the samples on a Kimwipe to get rid of excess Washing Buffer. Transfer the agar-embedded washed sections to a new well filled with the Ce3D clearing reagent. Incubate in the dark at RT with mild rocking, replacing the Ce3D clearing reagent at least once, until slices become transparent (usually within 1–2h).
!CRITICAL STEP: Perform the transfer step quickly to prevent tissue dehydration.
!CRITICAL STEP: For difficult to penetrate tissues, such as brain sections, follow the gradient clearing procedure described in Box 2.
? TROUBLESHOOTING
-
Mount the sample by transferring cleared agar slices to either a microscope slide (for 100–250um thick slices) or to a No. 0 glass-bottom dish (for 200–500um thick slices). Add additional Ce3D solution on top of the slice as mounting medium and gently place coverglass (No. 0 preferable) on top of the tissue. For slide-mounted tissue slices, seal the coverslip edges with nail polish.
PAUSE POINT: The mounted samples can be stored in the dark at RT for up to 1 week.
Option C: Thick tissue slices - Cryostat slice generation from OCT-embedded frozen tissues.
Timing: ~1–5d for tissue processing/staining; ~3h-1d for tissue clearing.
Obtain frozen OCT tissue blocks (previously PFA fixed, embedded in OCT and stored at − 80°C).
-
Equilibrate tissue blocks to 1–2°C above normal cryostat cutting temperature (e.g., −17°C for lymph nodes).
!CRITICAL STEP: Cutting at slightly warmer temperature minimizes the cracking of OCT and tissues. Cutting of thicker slices promotes cracking and should be avoided.
Use the cryostat to section tissues at 100–250um thickness (may need to use the Trim option, based on the cryostat model).
-
Use forceps to transfer OCT-embedded slices to uncharged glass slides.
!CRITICAL STEP: Uncharged slides allow tissues to float during blocking and staining, thus allowing antibody penetration from both sides of the tissue.
Circle tissues with PAP pen and further surround the tissues with a thick layer of vacuum grease to create a well-like enclosure.
-
Proceed with blocking, staining and washing of tissues (as in Steps 1(A)vi-x) by adding the Blocking / Staining / Washing Buffer to the enclosure in order to fully immerse the tissues.
!CRITICAL STEP: Gently cover and seal the enclosure with parafilm to minimize evaporation during incubation at 37°C.
!CRITICAL STEP: Place slide inside a humidifying dark chamber and place inside a 34–37°C incubator to block or stain tissues. Do not use shaker or rotator.
!CRITICAL STEP: Incubation time for blocking, staining and washing will decrease as compared to whole mount tissues (e.g., for 250um slices, homogeneous probe labeling can be attained with ~12h blocking at 37°C, 2 days of staining at 37°C, and ~12h of washing at RT).
!CRITICAL STEP: To remove or replace liquids at various steps during incubation, gently lift off the parafilm with forceps and use a 200ul pipette to remove liquids from the enclosure, exercising caution not to touch the tissues with the pipette tip.
? TROUBLESHOOTING
-
Remove Washing buffer using a pipette and carefully lift off the vacuum grease layer with a pipette tip.
!CRITICAL STEP: Proceed to next step as quickly as possible to prevent tissue dehydration
-
Clear the tissue slice by adding the Ce3D clearing reagent onto the tissue slice. Incubate in the dark without rotation, replacing the Ce3D clearing reagent at least once, until slices become transparent (usually within 1–2h).
!CRITICAL STEP: Presence of excess Washing Buffer can dilute the Ce3D clearing solution. It is important to replace the Ce3D clearing solution at least once during incubation.
? TROUBLESHOOTING
-
Mount the sample by removing excess Ce3D solution, placing a thin vacuum grease spacer (~1mm) around the cleared thick tissue slices, and adding back additional Ce3D solution as mounting medium. Gently place coverglass (No. 0 preferable) on top of the tissue and pressing evenly around the edges to make a seal with the spacer. Seal the edges using nail polish.
PAUSE POINT: The mounted samples can be stored in the dark at RT for up to 1 week.
Imaging and Image Pre-Processing.
Timing: ~4–24h for tissue imaging; ~2–24h for unmixing/deconvolution.
!CRITICAL: The following section (Steps 6–7) has been worked out for specific software (LASX, Huygens Essential, Imaris) and have not been rigorously compared to or validated using alternative software platforms.
-
2
Acquire high-resolution, multiparameter confocal image stacks of cleared tissues
!CRITICAL STEP: The suggested minimum voxel density is 500–600nm in X,Y; 1000nm in Z, with the images acquired using high NA objectives. Quality of the image and the capacity to perform accurate Histocytometry analysis will improve with higher voxel density (~300nm in X, Y) and use of higher NA objectives (~1.3NA), albeit at the cost of the objective’s working distance and total image volume.
? TROUBLESHOOTING
-
3
Acquire individual images of single-stained control samples using the same microscope settings as for the original sample. We typically utilize UltraComp eBeads stained with the individual antibodies/fluorophores, with each single-stained control sample mounted separately on a microscope slide.
-
4
Transfer the data to a workstation and use LASX software (or similar software based on microscope manufacturer) to perform image tile stitching and Channel Dye Separation (also known as spectral unmixing or compensation) of the multi-parameter image.
-
5
Deconvolve the image using Huygens Essential software.
!CRITICAL STEP: Deconvolution is optional for image analysis. It minimizes the contribution of artifactual external neighboring signals into the segmented cell objects, helping improve the accuracy of cell profiling with Histocytometry.
-
6
Import the images into Imaris for visualization and further processing.
!CRITICAL STEP: While Imaris version 9 has improved 3D surface object processing for Histocytometry, in our hands Imaris 8 performed better at basic image rendering.
-
7
Correct for signal depth attenuation in all channels displaying depth attenuation using the Normalize Layers function in Imaris.
!CRITICAL STEP: The Attenuation Correction XTension plugin can be used instead of Normalize Layers and may lead to improved results, but will take substantially longer to complete.
? TROUBLESHOOTING
Cell Segmentation for Volumetric Histocytometry.
-
8
For Histocytometry analysis, use one the following two pipeline options to generate individual surface objects around the imaged cells. These options are based on the local density of cells and signals of interest and the capacity to accurately segment cells using Imaris object creation module for downstream analysis (see Experimental Design for details). For cell segmentation of sparsely distributed cells, follow Option A. For densely packed membrane-stained cells, follow Option B.
!CRITICAL: The following step has been worked out for specific software (Imaris) and have not been rigorously compared to or validated using alternative software platforms.
Option A: Cell segmentation of sparsely distributed cells.
Timing: ~30min-4h
!CRITICAL: This is the simplest method of segmentation that applies to non-clustered cells and for studying cell markers not directly overlapping with highly proximal cells (see Experimental Design for details).
-
Identify the channel of interest that represents a specific cell type for analysis. If multiple cell types can be defined in different channels, these channels can be summed together to generate a Composite Channel using the Channel Arithmetics XTension in Imaris.
!CRITICAL STEP: When summing channels, use a multiplier factor to normalize the channels to one another. Boolean operators can also be used to restrict the summation of voxels meeting specific criteria (e.g., greater than a specific intensity value, not overlapping with signal in other channels).
-
Use Surface Object Creation module in Imaris to generate cell objects on the identified channel of interest. Specify the following parameters:
- Smoothness: usually set to width of two voxels
- Background subtraction: allows segmentation of data with varying levels of background signal based on tissue location, as well as for cells with low signal to noise ratios.
- Threshold: should be set to a level that clearly demarcates the desired signal on cells, but does not make the surface objects so inclusive that they incorporate non-specific background
- Splitting seed size: should be set to the average size of the cells of interest
- Quality: use a quality threshold to exclude the majority of low signal events
- Number of Voxels: use at least 100 voxels (typically >200) to exclude small irrelevant artifacts
!CRITICIAL STEP: Imaris 9 is recommended for building cell objects in large 3D datasets.
!CRITICAL STEP: Nuclear restricted signals (e.gs., transcription factors, Ki67) can be similarly used for cell segmentation. Sufficient plasma membrane signal is usually incorporated into the nuclear-based cell objects for accurate phenotypic analysis.
Option B: Cell segmentation of densely packed membrane-stained cells.
Timing: ~1–24h, based on dataset size
!CRITICAL: This method allows segmentation of densely packed cells defined solely by membrane staining. This is also done when nuclear-based staining and segmentation is problematic (see Experimental Design for details).
-
Sum the cell membrane channels representing the cell types of interest using the Channel Arithmetics XTension in Imaris to generate a Composite Sum channel.
!CRITICAL STEP: When summing, use a multiplier factor to normalize the channels to one another. Boolean operators can also be used to restrict the summation of voxels meeting specific criteria (e.g., greater than a specific intensity value)
Use Gaussian Blur tool in Imaris to smooth out the cell boundaries
Invert the blurred Sum channel so that inner cell areas appear light (high signal) and cell membranes appear dark (low signal)
Use the Gamma Correction and Linear Stretch tools in Imaris to optimally enhance the contrast and signal-to-noise ratios of the Inverted Sum Channel
Use Surface Object Creation module in Imaris to generate surfaces on the processed Inverted Sum Channel (as above).
Option C: Cell Skeleton Enhanced Segmentation.
Timing: ~2–24h, based on dataset size
!CRITICAL: If poor segmentation of individual cells is observed in Step.8(B)v above (i.e. frequent objects representing multiple cells fused together), try this cell skeleton enhanced segmentation pipeline. These steps can enhance cell segmentation in low signal-to-noise areas of the image.
!CRITICAL: Once imported into ImageJ/FIJI (step.vi below), it is helpful to duplicate the images throughout the processing steps described below to easily recover progress if errors are made.
Import the Inverted Sum Channel (from Step 8(B)ii above) into ImageJ/FIJI
Binarize the image stack using the Auto Local Threshold plugin. With our image data, mean auto-thresholding performed the best
-
Perform 2D Skeletonization using the Skeletonize plugin to generate single pixel-width reconstructions of the plasma membranes
!CRITICAL STEP: You may need to invert the image (plugin works on black signal)
On the original imported Inverted Sum Channel, use the Stack/Reslice plugin to generate both XZ and YZ resliced images
Binarize both resliced images using Auto Local Threshold tool as described in Step 10.
Skeletonize both resliced images using the Skeletonize plugin as described in Step 11.
Use the Stack/Reslice plugin again to reconvert these skeletonized resliced images to the original XYZ orientation. You may need to perform additional image transformations using the Rotate or Flip tools to get the images back into the original orientation.
Invert all three of the Skeletonized images (XY, XZ, YZ planes) to make sure that the membrane signal is white and background is dark
Sum all three Skeletonized images together to generate a Composite Skeleton image
Save and Import the Composite Skeleton into the original file open in Imaris as a separate channel.
Use Boolean operations with Channel Arithmetics in Imaris to exclude all signal within the Inverted Sum Channel (from step.iv) overlapping with the Composite Skeleton Channel, or simply Subtract the Composite Skeleton Channel from the Inverted Sum Channel
-
Use Surface Object Creation module in Imaris to generate surfaces on the processed Inverted Sum Channel (as above)
!CRITICIAL STEP: Imaris 9 is recommended for building cell objects in large 3D datasets.
Data Export and Histocytometry. Timing: ~2–4h
-
9
Once the surface objects representing the cells of interest are generated (from Steps 8(A)ii, 8(B)v, or 8(C)xii), export relevant statistics for all cell objects into CSV or XLS files. In our hands, mean channel intensity, sphericity, volume, and position are the most relevant for analysis.
-
10
Combine all the parameters of interest into a composite statistics matrix file by copying/pasting the individual statistic columns side-by-side next to one another in a new Excel file. Use the channel/stain names as column headers (single row only).
!CRITICAL STEP: While statistics data can be combined manually, Excel plugins (e.g., RDB Merge, http://www.rondebruin.nl/win/addins/rdbmerge.htm) can be used to automate this process.
-
11
Save as a new CSV file and import the data into FlowJo by simply dragging the CSV file into the workspace. This will generate a new FCS file in the same folder as the CSV file.
-
12
Exclude segmentation artifacts by using Sphericity and Volume gating to identify single cells. Data can now be used for phenotypic gating, as typically performed in flow cytometry.
!CRITICAL STEP: All channels will require rescaling for better cell visualization. Linear and ArcSinH (maximum must be a multiple of 256) transformations are most useful.
? TROUBLESHOOTING
-
13
Visualize the location of gated cell populations using X- vs. Y-position 2-dimensional plots. Cells can also be gated directly on X- vs. Y-positional plots for positional gating (e.g. cells located within a germinal center). Multiple populations can be overlayed together in Layout Editor to generate a visual reconstruction of the entire tissue.
!CRITICAL STEP: FlowJo does not easily plot 3D data. Initial gates around specific Z-positions can help visualize complex 3D datasets using simple XY positional plots. This can be achieved by plotting cells along the Z-dimension on a histogram or 2-dimensional plot and next creating a gate around a Z-region of interest (virtual Z slice). Apply the same gating tree as created in Step 12 to cells within the virtual Z-slice and visualize cell populations on a X- vs. Y-positional plot. You can change the virtual Z-slice position within the image dataset by changing the Z gate.
TIMING
Step 1, Tissue Preparation
Option A, Whole-mount: ~3–5d for tissue processing/staining; ~1–7d for tissue clearing
Option B, Thick tissue slices: vibratome slice generation from non-frozen tissues: ~1–5d for tissue processing/staining; ~3h-1d for tissue clearing.
Option C: Thick tissue slices - Cryostat slice generation from OCT-embedded frozen tissues: ~1–5d for tissue processing/staining; ~3h-1d for tissue clearing.
Steps 2–7, Image Pre-Processing. Timing: ~4–24h for tissue imaging; ~2–24h for unmixing/deconvolution
Step 8, Cell Segmentation for Volumetric Histocytometry.
Option A, Cell segmentation of sparsely distributed cells: Timing: ~30min-4h
Option B, Cell segmentation of densely packed membrane-stained cells: Timing: ~1–24h, based on dataset size
Option C: Cell Skeleton Enhanced Segmentation: Timing: ~2–24h, based on dataset size
Steps 9–13, Data Export and Histocytometry: ~2–4h
Box 1, Whole-mount in combination with RNA FISH: ~6–7d
Box 2, Gradient Clearing: ~15h
ANTICIPATED RESULTS
As shown in Figure 2A, Ce3D can achieve a high degree of transparency in multiple different tissue types. While some tissue discoloration (browning) is frequently observed after clarification, in our hands this minimally affects laser light tissue penetration and the subsequent capacity to achieve useful volumetric confocal imaging. If problematic, such discoloration can potentially be minimized by clearing tissues at 4°C. The ability of Ce3D to preserve cellular morphology and reporter fluorescence, coupled with its robust compatibility with antibody-based labeling or RNA detection in turn allows high-resolution, multiplexed and multimodal imaging and can generate datasets with very high signal-to-noise image quality (Figures 3 and 4). We include examples here of Ce3D-enabled imaging of the small intestine (Figure 3a/b), kidney (Figure 3 c/d), lymph node (Figure 3e/f), liver (Figure 3g), mammary gland (Figure 4h), brain (Figure 4i) and lung (Figure 4j). Although tissue penetration of antibody-based probes can be challenging, especially for whole-mount samples, the workflows included here provide guidelines for the steps and timelines necessary for optimized volumetric labeling. The ability to generate high-quality multiplexed image datasets not only allows direct visualization of tissue structure and general cellular positioning, but can be coupled with Histocytometry to achieve a quantitative understanding of cellular phenotypic states directly within 3D tissues (Figure 5). Ultimately, this can lead to a better understanding of how tissue microenvironments shape cellular states and conversely how cellular positioning can influence the function of the examined tissue in both healthy and diseased states.
Figure 2. Clearing, Mounting and Microscope Setup.
A) To achieve tissue clearing, the samples are perfused (left vs. middle) and cleared using the Ce3D solution (right). Scale bar − 1 mm. B) Samples are next mounted on either a slide or in a petri dish based on sample volume and the type of utilized objective and microscope. C) Inverted or upright microscope setups can be used for sample imaging.
TROUBLESHOOTING
Troubleshooting guidance can be found in Table 2.
Table 2.
Troubleshooting Table
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| Tissue preparation Step 1(A)i | Remaining blood in the tissue is observed | Too little PBS with heparin was used; too much pressure was employed during perfusion | Perfuse the organ at a slower rate with the needle in the optimal perfusion site |
| Tissue preparation Step 1(A)ii | Dark areas observed in the tissue / imaging stack | Cleared fat residue attached to the surface of cleared sample may obscure light penetration. | Remove of as much fat residue as possible during tissue isolation. Use a dissection microscope to assist. |
| Tissue preparation Step 1(A)iii | Poor fixation - Lack of detectable signal for multiple epitopes observed during imaging. | Fixative was not fresh and the PFA has degraded. | Prepare fixative right before experiment; Dilute the 16% PFA into 4% using 1X PBS and store aliquots for use at −20°C (see Reagent Setup). |
| Tissue preparation Steps 1(A)vi-vii | Bacterial contamination | Re-filter the blocking buffer, use sterile Eppendorf tubes and forceps. | |
| Tissue preparation Step 1(A)vii | Poor or non-homogeneous labeling of tissues for specific probes observed during imaging | Insufficient staining | Increase the concentration of antibody; increase the staining time. |
| Tissue preparation Step 1(A)vii | Poor tissue penetration for certain probes observed during imaging. Usually seen as good staining at tissue periphery and lack of staining in the deeper regions. | The size or charge of fluorophore conjugated to antibody is not permissive to antibody tissue penetration | Change panel to reagents involving smaller sized or less charged fluorophores (see “Practical limitations of tissue clearing” In the Introduction) |
| Tissue preparation Steps 1(A)viii | Poor overall signal for multiple probes observed during imaging | Washing too long at 37°C may cause excess antibody elution | Wash tissues at RT |
| Tissue preparation Steps 1(A)xi-xii, 1(B)ix, 1(C)viii-ix | Poor clearing – as detected by lack of tissue transparency after incubation with the Ce3D reagent | Incorrect refractive index of the Ce3D clearing buffer | Double-check refractive index of the Ce3D reagent before use (see Reagent Setup) |
| Washing buffer was transferred together with sample into clearing solution (e.g., on forceps used to transfer tissues) | Minimize transfer of washing buffer by gently wiping off tools. | ||
| Sample was attached to slide, blocking the clearing solution from getting into tissue. | Make sure the samples are floating in clearing solution to ensure penetration from all sides. | ||
| Clearing solution became too concentrated due to evaporation | Incubate samples in a tightly-sealed, small container. Seal plates with parafilm. Seal mounted slides with nail polish. | ||
| Imaging and Image Pre-processing Step 2 | Lack of image clarity obtained during acquisition | The refractive index right above the tissue surface is not homogeneous when the tissue is mounted for upright microscope; the temperature between the objective and clearing solution is different. | Let the sample sit on the stage of microscope for 0.5 – 1 hour before image acquisition. |
| The refractive index optimal for the objective lens does not match the refractive index of clearing solution | Useing a lens with refractive index with adjustable range including 1.49–1.50 | ||
| Bubbles in clearing solution or mounted tissue | Air trapped in clearing solution or within the tissue (e.g., lung) during sample clearing on the rotor | Let the tube with the clearing solution and samples sit on a stable / non-moving rack for 0.5 – 1 hours before mounting | |
| Bubbles underneath the objective lens | Air trapped under objective lens when it is being submerged into clearing solution | Adding a little clearing solution on the objective lens | |
| Histocytometry Steps 8–13 | Software or computer crashes during visualization, segmentation or analysis | RAM is not large enough | Increase RAM; upgrade graphics card; split the image stack into several segments along the Z direction and analyze individually. Merge cell object statistics data from the segments afterwards in Flowjo or Excel. |
| Poor ability to phenotypically characterize cells | Z-attenuation distorts the signal in the Zdimension, limiting cell subset discrimination | Ensure proper Z-attenuation correction is applied in Imaris, or alternatively apply additional Z-correction in FlowJo or Excel; | |
| Poor splitting of individual cells is leading to fusion of fluorescence signals between neighboring cell objects. | Use more aggressive contrast/gamma corrections and thresholding to delineate the borders of individual cells in all 3 dimensions. Reduce the object creation threshold to reduce cell object fusion. If these fail, ensure that sufficient Z-resolution is used to segment cell objects in 3D. This may require use of a higher NA objective. |
Acknowledgements
We would like to thank Dr. Kairui Mao for helping with gut and mammary gland tissue preparation. In addition, we thank all members of the Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases (NIAID) at the NIH for many helpful comments during the course of these experiments. This research was supported by the Intramural Research Program, NIAID, NIH, as well as by NIH R01AI134713 MYG.
Footnotes
Competing financial interests
The patent for the methodology described in this paper was filed with the US Patent Office, PCT Patent Application—PCT/US2017/049133, HHS Reference No. E–168–2016, Title: “Enhanced Tissue Clearing Solution, Clearing-Enhanced 3D (Ce3D), Compatible With Advanced Fluorescence Microscopy Imaging”
Data availability
The image datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
TWEET Multiplexed fluorescent microscopy and histocytometry data analysis of tissues using Ce3D tissue clearing #Ce3D
COVER TEASER Multiplexed imaging of large tissue volumes
References
- 1.Spitzer MH & Nolan GP Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcantara-Hernandez M et al. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 47, 1037–1050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papalexi E & Satija R Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Altschuler SJ & Wu LF Cellular heterogeneity: do differences make a difference? Cell 141, 559–563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Junttila MR & de Sauvage FJ Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354, doi: 10.1038/nature12626 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Cutrale F et al. Hyperspectral phasor analysis enables multiplexed 5D in vivo imaging. Nat Methods 14, 149–152 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Gerner MY, Kastenmuller W, Ifrim I, Kabat J & Germain RN Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JR, Fallahi-Sichani M & Sorger PK Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun 6, 8390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goltsev Y et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerner MY, Torabi-Parizi P & Germain RN Strategically localized dendritic cells promote rapid T cell responses to lymph-borne particulate antigens. Immunity 42, 172–185 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Vu Manh TP, Bertho N, Hosmalin A, Schwartz-Cornil I & Dalod M Investigating Evolutionary Conservation of Dendritic Cell Subset Identity and Functions. Front Immunol 6, 260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerner MY, Casey KA, Kastenmuller W & Germain RN Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J Exp Med 214, 3105–3122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im SJ et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovas C et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci Transl Med 9, aag2285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrovas C et al. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest 122, 3281–3294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preite S et al. Hyperactivated PI3Kdelta promotes self and commensal reactivity at the expense of optimal humoral immunity. Nat Immunol 19, 986–1000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayin I et al. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J Exp Med 215, 1531–1542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao K et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fonseca DM et al. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell 163, 354–366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radtke AJ et al. Lymph-node resident CD8alpha+ dendritic cells capture antigens from migratory malaria sporozoites and induce CD8+ T cell responses. PLoS Pathog 11, e1004637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinert EM et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda A et al. An improved method for isolation of epithelial and stromal cells from the human endometrium. J Reprod Dev 62, 213–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabbari A, Legge KL & Harty JT T cell conditioning explains early disappearance of the memory CD8 T cell response to infection. J Immunol 177, 3012–3018 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Richardson DS & Lichtman JW Clarifying Tissue Clearing. Cell 162, 246–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tainaka K, Kuno A, Kubota SI, Murakami T & Ueda HR Chemical Principles in Tissue Clearing and Staining Protocols for Whole-Body Cell Profiling. Annu Rev Cell Dev Biol 32, 713–741 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Erturk A et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Renier N et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Hama H et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Hama H et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14, 1481–1488 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Tainaka K et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray E et al. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell 163, 1500–1514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodt HU et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Kuwajima T et al. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke MT, Fujimoto S & Imai T SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154–1161 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Ke MT et al. Super-Resolution Mapping of Neuronal Circuitry With an Index-Optimized Clearing Agent. Cell Rep 14, 2718–2732 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Pan C et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 13, 859–867 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Murakami TC et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat Neurosci 21, 625–637 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Tainaka K et al. Chemical Landscape for Tissue Clearing Based on Hydrophilic Reagents. Cell Rep 24, 2196–2210 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Kubota SI et al. Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution. Cell Rep 20, 236–250, doi: 10.1016/j.celrep.2017.06.010 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Li W, Germain RN & Gerner MY Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci U S A 114, E7321–E7330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K Microwave-Assisted Tissue Preparation for Rapid Fixation, Decalcification, Antigen Retrieval, Cryosectioning, and Immunostaining. Int J Cell Biol 2016, 7076910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beghein E & Gettemans J Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein-Protein Interaction Analysis, and Protein Function Exploration. Front Immunol 8, 771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TL et al. Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]