Abstract
We analyzed the relationship between histone acetylation and transcriptional regulation at 40 Saccharomyces cerevisiae promoters that respond to specific activators and repressors. In accord with the general correlation between histone acetylation and transcriptional activity, Gcn4 and the general stress activators (Msn2 and Msn4) cause increased acetylation of histones H3 and H4. Surprisingly, Gal4-dependent activation is associated with a dramatic decrease in histone H4 acetylation, whereas acetylation of histone H3 is unaffected. A specific decrease in H4 acetylation is also observed, to a lesser extent, at promoters activated by Hap4, Adr1, Met4, and Ace1. Activation by heat shock factor has multiple effects; H4 acetylation increases at some promoters, whereas other promoters show an apparent decrease in H3 and H4 acetylation that probably reflects nucleosome loss or gross alteration of chromatin structure. Repression by targeted recruitment of the Sin3-Rpd3 histone deacetylase is associated with decreased H3 and H4 acetylation, whereas repression by Cyc8-Tup1 is associated with decreased H3 acetylation but variable effects on H4 acetylation; this suggests that Cyc8-Tup1 uses multiple mechanisms to reduce histone acetylation at promoters. Thus, individual activators confer distinct patterns of histone acetylation on target promoters, and transcriptional activation is not necessarily associated with increased acetylation. We speculate that the activator-specific decrease in histone H4 acetylation is due to blocking the access or function of an H4-specific histone acetylase such as Esa1.
Transcription in eukaryotes occurs in the context of DNA packaged into chromatin. The basic unit of chromatin is the nucleosome, in which DNA is wrapped around the core histones H2A, H2B, H3, and H4. Nucleosome remodeling complexes such as Swi-Snf can facilitate opening of repressive chromatin structures in promoter regions to provide access for DNA-binding activator proteins or general transcription factors (32). In addition, reversible chromatin modifications such as acetylation, phosphorylation, and methylation of N-terminal histone tails can modulate accessibility of DNA within chromatin (56). Acetylation of lysines in the histone tails neutralizes their positive charge, thereby weakening electrostatic interactions with DNA (25) and interactions between neighboring nucleosomes (42). The tails of histones H3 and H4 are important for transcriptional regulation of numerous genes, because mutations in these histone tails result in both derepression and diminished activation (15, 44). Furthermore, histone acetylases and deacetylases can be recruited to specific promoters, whereupon they serve as transcriptional regulators (33, 58).
It is generally believed that transcriptional activity is correlated with histone acetylation (22, 58), and this relationship was first described nearly 40 years ago (3, 51). Silenced domains in Saccharomyces cerevisiae such as telomeres and the silent mating type loci form heterochromatin-like structures, and they are deacetylated relative to surrounding regions (4, 5). This silencing is dependent on the Sir proteins, notably the NAD-dependent histone deacetylase Sir2 (27). Similarly, transcriptional inactivation of one of the two female X chromosomes in mammals is associated with a lack of H4 acetylation (29). In contrast, dosage compensation in flies occurs by increasing transcription at the single male X chromosome (41), which is accompanied by increased acetylation at lysine 16 of histone H4 (60) and recruitment of MOF histone acetylase (23). Hyperacetylation is also associated with large domains of potentially and transcriptionally active chromatin such as the human β-globin locus (24).
Histone acetylation is also involved in transcriptional repression and activation at the single-gene level. Genes repressed by targeted recruitment of the Sin3-Rpd3 histone deacetylase complex contain deacetylated histones in their promoter region (31, 54), and the histone deacetylase activity of Rpd3 is essential for repression (30). Histone acetylation has also been suggested to be involved in repression by the Cyc8-Tup1 corepressor, which is recruited to promoters by pathway-specific DNA-binding repressors (17, 55). The Tup1 repression domain interacts with underacetylated histone H3 and H4 tails in vitro (16, 45), histone tail mutations partially alleviate repression of Tup1-regulated genes (17, 67), and Cyc8-Tup1 can interact with Rpd3 and Hos2 histone deacetylases in vitro (66). Transcriptional activation by Gcn4 is augmented by Gcn5 histone acetylase activity (37, 65), and it is associated with a localized increased histone acetylation at the promoter (36, 37). Gcn4 interacts with the Gcn5-containing SAGA complex in vitro (13, 47), and it presumably increases histone acetylation in vivo by recruiting SAGA to target promoters. Similarly, the Swi5 activator is required for recruitment of SAGA (10) and for increased histone acetylation (34, 35) at the HO promoter. In mammalian cells, histone hyperacetylation occurs at promoters induced by hormones or interferon, presumably due to recruitment of the p300 (also known as CREB binding protein) or ACTR histone acetylases (8, 50).
As the above studies involve a limited number of individual promoters, it is difficult to assess whether activator-dependent acetylation or repressor-dependent deacetylation is a general phenomenon. Although there are some cases in which histone acetylation appears unchanged upon transcriptional induction (11, 49), these experiments generally involved analysis of entire mRNA coding regions and would be unable to detect promoter-localized changes in histone acetylation, such as those observed in targeted recruitment of SAGA or Sin3-Rpd3 complexes. In the case of the mouse mammary tumor virus (MMTV) promoter, transcriptional activation is unexpectedly blocked when histone acetylation is globally increased by treatment with sodium butyrate or trichostatin A (6, 7, 46). However, histone acetylation at the MMTV promoter was not examined in these experiments, and it is unknown whether the observed effects on MMTV transcription are an indirect consequence of the drug treatments. Finally, previous studies analyzed a very limited number of promoters affected by a particular activator or repressor. Hence, it is unknown whether the histone acetylation status is specifically directed by the activators and repressors, is related to transcriptional activity per se, or is determined individually by the underlying chromatin structure of each promoter.
In this study, we analyze acetylation of histone H3 and H4 tails at a variety of native yeast promoters that are regulated by well-defined activators and repressors. For each transcriptional regulator, we analyze multiple promoters that either are responsive or nonresponsive to the regulator. We show that individual activators direct specific histone acetylation patterns at responsive promoters, that some activators cause a dramatic decrease in acetylation of histone H4, and that the Cyc8-Tup1 corepressor inhibits histone acetylation by multiple mechanisms. More generally, our results indicate that transcriptional activation is not necessarily associated with increased histone acetylation.
MATERIALS AND METHODS
Plasmids and strains.
The plasmid YIP-His3A5 used to create modified HIS3 alleles has been described previously (28). All upstream activating sequences (UASs) of this HIS3 allele are deleted and replaced with different activator binding sites. The HIS3 reporter genes were introduced into FT5 by two-step gene replacement (α ura3-53 trp1-Δ63 his3-Δ200 leu2::PET56). Strain JDY4251 carries a Gcn4 site, JDY7482 carries a Gal4 site, and JDY8702 carries two Ace1 sites as the sole UAS in the HIS3 promoters. The rpd3, sin3, and ume6 mutant strains were derived from FT5 by deleting the respective open reading frame using hisG-based constructs (1). The isogenic tup1 mutant strain has been described previously (61). The yeast strains used for the methionine and ethanol induction are based on W303-1A (39). All yeast strains were grown in yeast-peptone-dextrose (YPD) unless indicated otherwise. For the galactose induction strain JDY7482 was grown in YPD and shifted to yeast-peptone containing 2% galactose for 8 h. To induce Gcn4 activated genes, strain JDY4251 was grown in glucose minimal medium supplemented with all essential amino acids to mid-log phase. Half of the culture was then shifted to medium lacking histidine and containing 10 mM 3-aminotriazole for 4 h. Respiratory genes were induced by growing cells in synthetic complete medium containing 4% glucose, washing in medium lacking glucose, and transferring to medium containing 3% ethanol as a nonfermentable carbon source for 6 h. Methionine-regulated genes were induced by growth in glucose minimal medium lacking methionine. As a control, noninduced cultures were grown in the presence of 1 mg of methionine per ml. Copper response genes were induced by growing strain JDY8702 in glucose minimal medium containing 0.5 mM CuSO4 for 15 min. To induce the heat shock response, strain JDY7462 was grown at 25°C to mid-log phase and shifted to 39°C for 20 min.
Chromatin IPs.
Formaldehyde-cross-linked chromatin was immunoprecipitated essentially as described previously (39) with the following modifications. Many of the samples used in these experiments were previously analyzed for TATA binding protein (TBP) occupancy (39). For the TBP occupancy experiments performed here, the chromatin solution was subject to immunoprecipitations with 10 μl of polyclonal TBP antibody (obtained from Laurie Stargell). Approximately 1/100 of the material recovered after the IP and 1/10,000 of the input DNA was used as a template for PCR containing 0.1 mCi of [α-32P]dATP per ml. The PCR profile used was 90 s at 94°C; which was followed by 26 cycles of 30 s at 94°C, 45 s at 53°C, and 1 min at 72°C; and a final 5-min extension at 72°C. To measure histone acetylation levels, chromatin was immunoprecipitated with 2 μl of antibodies raised against acetylated forms of H3 and H4 N-terminal tails (Upstate Biotech). Approximately 1/100 of the precipitated chromatin and 1/10,000 of the input DNA was used as a template in a 24-cycle PCR. Alternatively, 8 μl of an antibody against unacetylated H4 tails (Serotec) was used, followed by a 26-cycle PCR with 1/100 of the immunoprecipitated chromatin and 1/100,000 of the input DNA. All PCR products were separated on 8% polyacrylamide gels and quantified using a PhosphorImager. The relative acetylation level of a given gene was calculated as the ratio between the amount of PCR product obtained with the immunoprecipitated chromatin and with the input DNA. The value obtained for the PGK1 control promoter was arbitrarily set to 10 and all other values are presented relative to this standard. Similarly, for the heat shock experiment the ADH1 promoter was used as a standard. The relative H3 and H4 deacetylation caused by Rpd3 or Tup1 was calculated by dividing the acetylation level of a mutant strain by that of a wild-type strain. All quantitative values of histone acetylation status represent the average of at least three independent assays.
RESULTS
General approach.
Previous studies demonstrating gene-specific increases in histone acetylation associated with increased transcription have focused on a small number of genes and specific transcriptional regulators. As a more general evaluation of the relationship between histone acetylation and transcriptional activation, we used chromatin immunoprecipitation (57) to analyze the level of H3 and H4 acetylation at a large number of promoters under conditions under which transcription was activated or repressed by well-defined regulators. Formaldehyde-cross-linked chromatin from living yeast cells was immunoprecipitated with antibodies directed against acetylated forms of histones. The H3 antibody was raised against an H3 N-terminal tail peptide acetylated at lysines 9 and 14, while the H4 antibody was raised against a peptide acetylated at lysines 5, 8, 12, and 16 of H4. The amount of immunoprecipitated DNA was assayed by quantitative PCR using primers spanning the region of interest and compared to the amount of input DNA prior to immunoprecipitation. The resulting IP efficiency is a measure of H3 and H4 acetylation of this region. Due to the nature of the antibodies, our experiments measure an averaged acetylation status of H3 and H4, and they do not address the possibility of differential acetylation of distinct lysine residues within the H3 and H4 tails.
Gal4 causes H4-specific deacetylation during transcriptional activation.
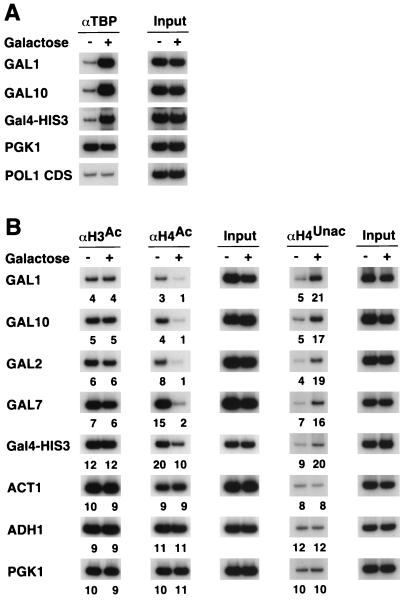
We first analyzed histone acetylation at promoters regulated by the Gal4 activator protein in cells grown under repressing (glucose) or activating (galactose) conditions. Efficient and specific activation of the GAL promoters was verified by monitoring TBP occupancy in the same samples used to measure histone acetylation (Fig. 1A). As expected (39, 40), growth in galactose is accompanied by increased TBP recruitment at the GAL1 and GAL10 promoter, while TBP occupancy is unchanged at the constitutively active PGK1 promoter and negligible at the POL1 coding region.
FIG. 1.
Gal4-dependent activation is associated with deacetylation of histone H4. Cross-linked chromatin preparations from strain JDY7482 grown in the presence (+) or absence (−) of galactose were immunoprecipitated with antibodies against TBP (A) and acetylated (superscript Ac) H3 and H4 tails or unacetylated (superscript Unac) H4 tails (B). Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
All four galactose-inducible genes tested (GAL1, GAL10, GAL2, and GAL7) show similar level, of H3 acetylation under repressing or activating conditions (Fig. 1B). Surprisingly, all four GAL genes exhibit a four- to sixfold decrease in H4 acetylation during growth in galactose. Such H4-specific deacetylation was also observed, albeit to a lesser degree, at an artificial GAL-HIS3 promoter containing a single Gal4 binding site upstream of the HIS3 core promoter region. The stronger effects at the natural Gal4-dependent promoters might be due to multiple Gal4 binding sites at these promoters. This H4-specific decrease is not observed at the ADH1, PGK1, and ACT1 promoters, indicating that it is specific to Gal4-dependent, galactose-induced genes and is not a general effect of growth conditions or transcriptional activity.
These observations suggest that Gal4-dependent activation is associated with a striking decrease in H4 acetylation and no effect on H3 acetylation. However, it was formally possible that the pattern of histone acetylation in response to galactose induction was in fact due to a loss of nucleosomes at the GAL promoters accompanied by an increase of H3 acetylation at the remaining nucleosomes. To address this issue we assayed the same samples using antibodies raised against a peptide corresponding to the unacetylated H4 tail. The amounts of DNA associated with unacetylated H4 would increase if Gal4-dependent induction resulted in actual deacetylation of H4, whereas it would decrease upon nucleosome loss. As shown in Fig. 1B, the GAL promoters show a three- to sixfold increase in the amount of unacetylated H4, while no effect is observed at the control promoters. Thus, the Gal4-dependent changes are not the result of nucleosome loss but rather arise from an actual decrease in H4 acetylation.
Gcn4 activation increases H3 and H4 acetylation.
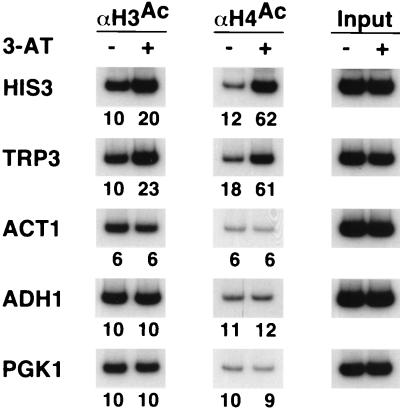
Gcn4 activation of the HIS3 promoter is associated with a localized increase in acetylation of H3 (36, 37). H3 hyperacetylation requires Gcn5 histone acetylase and presumably reflects recruitment of the SAGA complex by Gcn4 (12, 47). In agreement with these results, we observe a two- to threefold increase in acetylated H3 at two Gcn4-dependent promoters (HIS3 and TRP3) under conditions of Gcn4 activation (Fig. 2). In addition, the levels of H4 acetylation at the HIS3 and TRP3 promoters also increase by a factor of 3. Increased H3 and H4 acetylation is specific to Gcn4-regulated promoters, because histone acetylation is unaffected at the PGK1, ACT1, and ADH1 promoters. As the histone acetylase activity of Gcn5 in the context of SAGA or ADA complexes is directed towards H3 (20), these observations suggest that Gcn4 recruits an H4-specific acetylase to promoters. Esa1, the catalytic subunit of the NuA4 complex (2, 20), is a likely candidate; it is the major H4 acetylase in vivo (52), and it interacts with Gcn4 and stimulates Gcn4-dependent transcription in vitro (63).
FIG. 2.
Gcn4-dependent activation results in H3 and H4 hyperacetylation. Cross-linked chromatin preparations from strain JDY4251 grown in glucose minimal medium in the presence (+) or absence (−) of 10 mM aminotriazole were immunoprecipitated with antibodies against the acetylated tails of H3 and H4. Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
H4-specific deacetylation is associated with activation by Hap4, Adr1, and Met4.
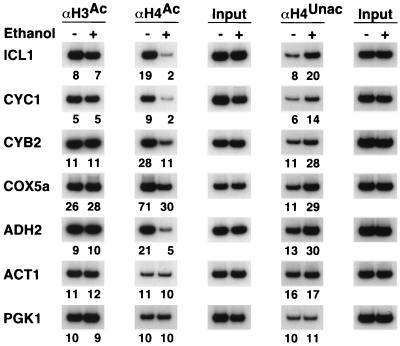
In the presence of a nonfermentable carbon source, transcription of many respiratory genes is induced by the Hap activator complex, in which the transcriptional activation domain is provided by Hap4 (48). We analyzed histone acetylation at Hap4-regulated promoters in cells grown in glucose or ethanol (Fig. 3). For all four Hap4-regulated promoters tested, H3 acetylation is unaffected by carbon source, whereas H4 acetylation decreases in ethanol medium, conditions of Hap-dependent activation. Reduced acetylation of H4 is more pronounced at the ICL1 (eightfold) and CYC1 (fivefold) promoters than at the COX5a and CYB2 promoters (two- to threefold). Decreased H4 acetylation is specific to Hap4-regulated genes, as it not observed at the unregulated PGK1 and ACT1 promoters. In accord with the idea that Hap4-dependent activation is associated with actual deacetylation of H4, we observe a two- to threefold increase in the amount of DNA immunoprecipitated by antibodies against the unacetylated tail. A fourfold decrease in H4, but not H3, acetylation is observed at the ADH2 promoter in ethanol medium, conditions under which transcription of this gene is activated by Adr1 (18). Thus, activation by Hap4 and probably Adr1 is associated with reduced acetylation of H4.
FIG. 3.
Transcriptional activation by Hap4 and Adr1 correlates with histone H4-specific deacetylation. Cross-linked chromatin preparations from cells grown in medium containing either glucose or ethanol as the sole carbon source were immunoprecipitated with antibodies against the acetylated (superscript Ac) tails of H3 and H4 or unacetylated (superscript Unac) H4 tails. Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
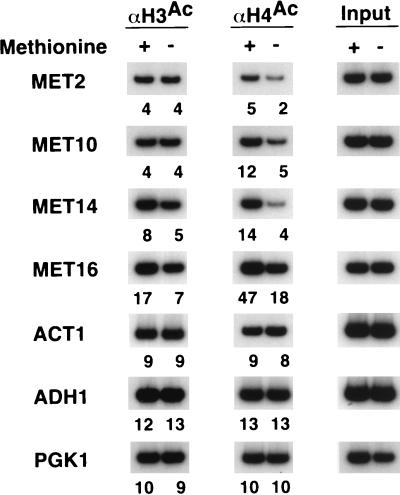
The MET genes are coordinately induced in the absence of methionine by a heteromeric DNA-binding complex containing the Met4 activator (38). As is the case with genes activated by Gal4, Hap4, and Adr1, H3 acetylation was unaffected by Met4-dependent activation (except perhaps for MET16), whereas H4 acetylation decreased two- to threefold at the MET10, MET14, and MET16 promoters; the decrease was minor at the MET2 promoter (Fig. 4). Again, the unregulated PGK1, ADH1, and ACT1 promoters showed no change in either H3 or H4 acetylation under these conditions, suggesting that Met4 activation is associated with decreased H4 acetylation.
FIG. 4.
Activation by Met4 causes histone H4 deacetylation. Cross-linked chromatin preparations from cells grown in the presence (+) or absence (−) of methionine were immunoprecipitated with antibodies against the acetylated tails of H3 and H4. Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
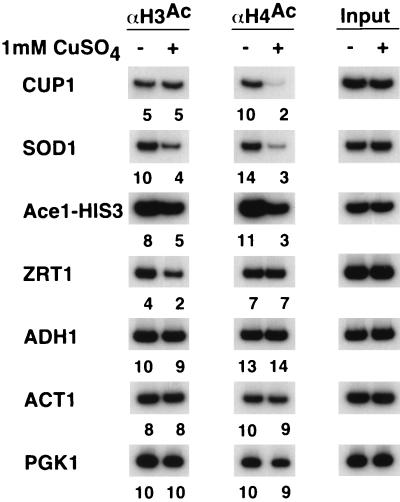
Ace1 and Zap1 differentially affect histone acetylation in response to copper.
Several copper-responsive genes are activated by Ace1, a protein whose specific DNA binding activity depends on the concentration of copper in the medium (19). For both Ace1-dependent promoters tested, CUP1 and SOD1, we observe decreased H4 acetylation upon copper induction (Fig. 5). The SOD1 also displays a mild (twofold) decrease in H3 acetylation, whereas the CUP1 promoter is unaffected. In accord with these observations, a minimal HIS3 promoter whose expression is controlled by two Ace1 sites displays a decrease in H4 acetylation. Thus, Ace1-dependent activation is associated with decreased H4 acetylation. In contrast, the ZRT1 promoter, which is activated in response to copper addition by Zap1 (70), shows a slight decrease in H3 acetylation and is unaffected for H4 acetylation. The ADH1, PGK1, and ACT1 promoters, whose activities are independent of copper, do not show a change in histone acetylation. These observations suggest that Ace1 and Zap1 differentially affect histone acetylation.
FIG. 5.
Activation by Ace1 and Zap1 results in decreased histone acetylation. Cross-linked chromatin preparations from cells grown in the presence (+) or absence (−) of copper were immunoprecipitated with antibodies against the acetylated tails of H3 and H4. Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
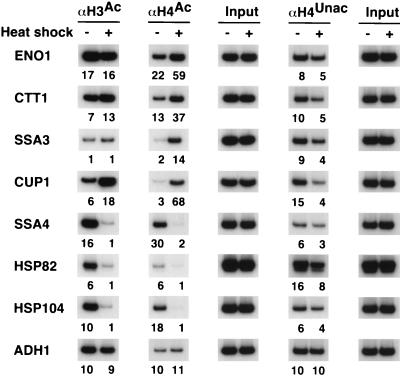
Histone acetylation and nucleosome perturbation upon heat shock induction.
When yeast cells are subjected to a brief heat shock, transcription of a large number of genes is rapidly induced. The classic heat shock genes are activated by heat shock factor (Hsf1), whereas the general stress-inducible genes are activated by Msn2 and Msn4 (59). For two promoters activated by Msn2 and Msn4, ENO1 and CTT1, heat shock treatment results in increased H4 acetylation, but no effect on H3 acetylation (Fig. 6). Increased H4 acetylation at these promoters was confirmed by the decreased association with unacetylated H4. As expected, histone acetylation is unaffected at the ADH1 promoter, which does not respond to heat shock. Thus, Msn2 and Msn4 activation is associated with increased H4 acetylation at target promoters.
FIG. 6.
Histone acetylation in response to heat shock. Cross-linked chromatin preparations from cells that were (+) or were not (−) subjected to a 20-min heat shock were immunoprecipitated with antibodies against the acetylated (superscript Ac) tails of H3 and H4 or unacetylated (superscript Unac) H4 tails. Immunoprecipitated and input material was analyzed by PCR with primers corresponding to the indicated promoters. Relative acetylation levels are indicated below each gel lane and were calculated as described in Materials and Methods.
For promoters activated by Hsf1, the results are somewhat more complicated. Two promoters, SSA3 and CUP1, show a significant increase in H4 acetylation upon heat shock. The Hsf1-dependent increase in H4 acetylation at CUP1 is noteworthy, because this promoter shows decreased H4 acetylation during copper induction via Ace1. This discordant behavior at CUP1 driven by Hsf1 or Ace1 provides further evidence that changes in histone acetylation are activator-specific. The Hsf1-dependent increase in H4 acetylation is consistent with the Hsf1-dependent recruitment of Esa1, the catalytic subunit of the major H4 acetylase (52). H3 acetylation is unaffected at SSA3 but is increased at CUP1.
In contrast, three other Hsf1-activated promoters (SSA4, HSP104, and HSP82) show a dramatic decrease in the association of acetylated H3 and H4 in response to heat shock. In addition, these promoters show a significant decrease in the amount of unacetylated H4. These results indicate that heat shock causes a dramatic change in chromatin structure that decreases the amount of histones H3 and H4 cross-linked to the promoters. We suspect that this change in chromatin structure reflects a loss of nucleosomes, which is consistent with previous studies (21), although other perturbations cannot be excluded. In any event, this Hsf1-dependent alteration in chromatin structure makes it impossible to assess the effect of Hsf1 on histone acetylation at the SSA4, HSP104, and HSP82 promoters. However, Esa1 is recruited to the SSA4 and HSP104 promoters in response to heat shock (52).
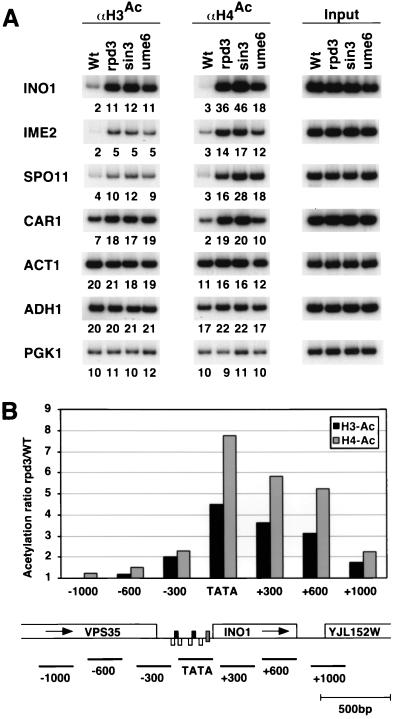
Histone deacetylation in response to the Cyc8-Tup1 and Sin3-Rpd3 corepressors.
Previous work indicated that Ume6-dependent recruitment of the Sin3-Rpd3 histone deacetylase complex generated a local domain of histone deacetylation centered at the site of recruitment (31, 54). Consistent with these results, we observe a strong (four- to eightfold) Sin3- and Rpd3-dependent decrease in H3 and H4 acetylation at all four repressed genes tested (INO1, IME2, SPO11, and CAR1) (Fig. 7A), and mapping experiments on INO1 show that deacetylation is most pronounced at the Rpd3 recruitment site (Fig. 7B). Analysis of control promoters not regulated by the Sin3-Rpd3 corepressor reveals that sin3 and rpd3 mutations have no effect on H3 acetylation and result in only a very slight increase in H4 acetylation. This slight increase at unregulated promoters is likely due to untargeted histone deacetylation by the Sin3-Rpd3 complex that occurs throughout the genome (53).
FIG. 7.
Genes repressed by Sin3-Rpd3 contain deacetylated histones H3 and H4 at their upstream regions. Chromatin extracted from formaldehyde-cross-linked cultures of strain JDY7841 and isogenic rpd3, sin3, and ume6 deletion strains was immunoprecipitated using antibodies against acetylated H3 and H4. (A) PCR products corresponding to the indicated promoters were generated from immunoprecipitated and input DNAs. Wt, wild type. (B) Rpd3-mediated deacetylation was mapped across the INO1 genomic locus using PCR primers centered around the indicated distances from the TATA region. The relative histone deacetylation caused by Rpd3 at each amplified region was calculated as described in Materials and Methods. A map of the INO1 locus indicates the position of the URS elements (black boxes), the UAS sequences (white boxes), and the TATA region (gray box).
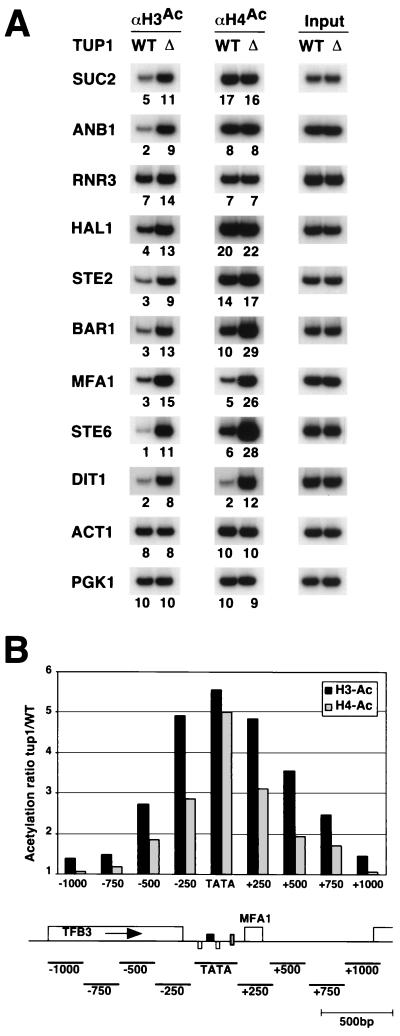
To analyze the effect of the Cyc8-Tup1 corepressor on histone acetylation, we analyzed several repressed promoters in wild-type and tup1 mutant strains. We chose representative genes of different regulons: ANB1, an oxygen-regulated gene; SUC2, a glucose-repressed gene; RNR3, a DNA damage-inducible gene; HAL1, an osmotic-stress-inducible gene; STE6, STE2, BAR1, and MFA1, a-specific genes; and DIT1, a sporulation-specific gene (reviewed in reference 55). As shown in Fig. 8A, Tup1 repression is associated with a decrease in H3 acetylation at all promoters tested, although the magnitude of the decrease (2- to 10-fold) varies depending on the individual promoter. In addition, Tup1 causes a 5- to 10-fold decrease in H4 acetylation at the a-specific promoters—MFA1, BAR1, and STE6—and the sporulation-specific DIT1 promoter but surprisingly does not affect H4 acetylation at five other Tup1-regulated promoters tested. Histone acetylation was unaffected at several control promoters not repressed by Tup1, indicating that Tup1-dependent histone deacetylation is limited to promoters to which Cyc8-Tup1 is recruited. Thus, Tup1 repression results in histone deacetylation, but unlike the case for repression by Sin3-Rpd3, the pattern of deacetylated histones depends on the promoter.
FIG. 8.
Tupl-mediated repression is associated with local deacetylation of histone tails. Cross-linked chromatin preparations from strain FT5 and the isogenic tup1 deletion strain were immunoprecipitated with antibodies against the acetylated tails of H3 and H4. (A) The promoter region of the indicated genes was PCR amplified from immunoprecipitated and input DNAs. WT, wild type; Δ, mutant. (B) PCR was performed with primer pairs spanning the MFA1 locus and centered approximately at the distance indicated from the α2-Mcm1 binding sites that serve to recruit the Cyc8-Tup1 corepressor. A map of the MFAl locus indicates the position of the α2-Mcml operator (black box), the pheromone response elements bound by Ste 12 (white boxes), and the TATA element (grey box). The relative H3 and H4 deacetylation associated with Tupl was calculated as detailed in Materials and Methods.
Tup1-dependent histone deacetylation could either be confined to the regulatory region of repressed genes or spread over a larger chromosomal domain, and it has been reported that Tup1 is associated with the entire reading frame of the repressed STE6 gene (14). At the MFA1 locus (Fig. 8B), Tup1-dependent deacetylation is strongest around the region immediately downstream of the α2-Mcm1 operator, which serves as the recruitment site of the Cyc8-Tup1 complex, although reduced acetylation is still observed with primer pairs centered 750 bp upstream or downstream from the operator. Taking into account the size of the fragmented chromatin and PCR primers (31), we estimate the domain of histone deacetylation extends approximately 500 bp in either direction from the site of recruitment. The extent and magnitude of the deacetylated domain following recruitment of Cyc8-Tup1 are roughly comparable to that following recruitment of Sin3-Rpd3 histone deacetylase, although the Tup1-dependent domain appears to extend further upstream.
DISCUSSION
Transcriptional activation is often not associated with increased histone acetylation.
To address the correlation between histone acetylation and transcriptional activity, we analyzed acetylation of histone H3 and H4 tails at 40 yeast promoters that are regulated by well-defined activators and repressors. In accord with the expected correlation, transcriptional repression by the Cyc8-Tup1 and Sin3-Rpd3 corepressors is always associated with histone deacetylation at the target promoters. In contrast, transcriptional enhancement by DNA-binding activators does not necessarily result in increased histone acetylation. Most unexpectedly, the level of H4 acetylation decreases in response to certain activators, and this decrease can be as dramatic as that observed at promoters repressed by targeted recruitment of Sin3-Rpd3 histone deacetylase. Thus, increased histone acetylation at promoters is often not associated with, and hence is not a prerequisite for, enhanced transcription by activators.
There are several explanations for why efficient transcriptional activation can occur in the absence of increased histone acetylation. First, analysis of bulk histones in yeast cells indicates that histones H3 and H4 contain an average of approximately two acetylated lysines per histone tail (66). This average level of histone acetylation may be sufficient for transcriptional activation for many promoters, such that activator-dependent hyperacetylation is not required. Second, increased histone acetylation might not be important for transcriptional activation of promoters whose chromatin structures are not inherently inhibiting. In this regard, yeast promoter regions are often preferentially accessible to nuclear proteins (43), and the majority of yeast genes are transcriptionally unaffected upon loss of histone H4 (68). Third, activators that function primarily by directly recruiting the polymerase II transcription machinery might not cause increased histone acetylation, particularly if they are unable to recruit histone acetylase complexes to promoters. In any event, the level of histone acetylation at yeast promoters can often be a poor indicator for the level of gene expression.
Individual activators confer distinct patterns of histone acetylation or deacetylation at target promoters.
Our results strongly suggest that individual activators are the primary determinants of the histone acetylation patterns that arise upon transcriptional induction. In general, natural promoters affected by a particular activator show a similar pattern of histone acetylation. For example, activation by Gcn4 and the stress-inducible activators (Msn2 and Msn4) show increased histone acetylation, whereas activation by Gal4, Hap4, Ace1, and Met4 is associated with decreased H4 acetylation. Conversely, HIS3 promoter derivatives that differ solely by the activator binding sites show different patterns of histone acetylation, and the observed patterns resemble those that occur on natural promoters that respond to the same activator. Finally, Ace1- and Hsf1-dependent activation of the CUP1 promoter results in distinct patterns of histone acetylation that are in accord with those mediated by the activator. Our conclusion for activator-directed patterns of histone acetylation is based on the analysis of 27 promoters and nine activators and hence is likely to apply to most activators and promoters in yeast.
Although activators are the primary determinant of histone acetylation patterns, our results also provide evidence for promoter-specific effects that are independent of the activator. The magnitude of the activator-dependent effect on histone acetylation can vary depending on the individual promoter. In part, this variability in fold effect is due to the fact that individual promoters can have different absolute levels of histone acetylation (as measured by immunoprecipitation efficiency of promoter fragments) prior to transcriptional induction. In addition, there are a few examples in which H3 acetylation differs at natural promoters responding to a common activator (e.g., CUP1 and SOD1 in response to Ace1 and SSA3 and CUP1 in response to Hsf1). The clearest example of promoter-specific effects is provided by the two classes of Hsf1-activated promoters. One class (SSA3 and CUP1) shows increased histone acetylation, whereas the other class (SSA4, HSP104, and HSP82) shows nucleosome loss or some other major change in chromatin structure that is manifested as an apparent decrease in acetylated H3 and H4 as well as nonacetylated H4. Promoter-specific effects on histone acetylation are likely to reflect differences in (i) the proteins bound to the promoter, (ii) inherent nucleosome positioning, density, or stability, and (iii) accessibility of the promoter to the untargeted actions of the various histone acetylases and deacetylases.
Mechanisms of activator-dependent acetylation or deacetylation at target promoters.
The simplest mechanism for activator-dependent increases in H3 and/or H4 acetylation is that activators recruit histone-specific acetylases to target promoters, whereupon they locally acetylate histones. In yeast cells, H3 is acetylated primarily by Gcn5, whereas H4 is acetylated primarily by Esa1. Cells lacking Gcn5 or Esa1 show, respectively, decreased H3 or H4 acetylation of bulk histones (9, 69) and numerous genomic regions (36, 52, 64). Gcn5 is the catalytic subunit of the SAGA and ADA complexes (20), and Esa1 is the catalytic subunit of the NuA4 complex (2). Thus, activator-specific recruitment of these Gcn5-containing and/or Esa1-containing complexes would result in increased H3 and/or H4 acetylation at target promoters.
Gcn4 is likely to increase H3 and H4 acetylation by recruiting both Gcn5- and Esa1-containing complexes to target promoters. Gcn4 interacts in vitro with SAGA (13, 47) and NuA4 (26, 63), and Gcn4-dependent hyperacetylation of H3 depends on Gcn5 but not on transcriptional activity of the target promoter (36, 37). We suspect that the Msn2 and Msn4 activators cause increased acetylation of stress-inducible promoters by recruiting Esa1 and perhaps Gcn5, although there is no additional evidence beyond the histone acetylation patterns. In the case of Hsf1-dependent promoters, heat shock results in increased occupancy by Esa1, thereby providing direct evidence for targeted recruitment (52). Hsf1-dependent recruitment of Esa1 occurs at all promoters tested, even those (SSA3 and HSP104) that appear to undergo Hsf1-dependent nucleosome loss and hence show no apparent increase in histone acetylation.
There are two potential mechanisms to account for the unexpected observation that certain activators (particularly Gal4 and Hap4) are associated with a specific decrease in H4 acetylation. In one model, these activators recruit an H4-specific histone deacetylase to target promoters. This model seems unlikely because there is no evidence for an H4-specific histone deacetylase in yeast and because it requires that a variety of distinct activation domains share a common feature that permits recruitment of a specific deacetylase(s). Thus, we favor the second model, in which these activators restrict the access or inhibit the activity of an H4-specific acetylase in the vicinity of the promoter. This model is supported by the facts that Esa1 is the catalytic subunit of NuA4, an H4-specific histone acetylase (2), and that Esa1 is responsible for the vast majority of genome-wide H4 acetylation in vivo (52). Activator-dependent H4 deacetylation could be accomplished through active masking of a critical functional domain(s) in the NuA4 complex or through the generation of an active transcription complex that passively blocks the association of NuA4 with the promoter region. The latter scenario seems more plausible, as it provides a common mechanism by which diverse activators mediate a very similar pattern of H4 acetylation changes. However, activator-dependent blocking of Esa1 would not apply at promoters at which activators actually recruit Esa1 (52).
Implications for the mechanism of repression by Cyc8-Tup1.
In principle, a given activator or repressor should confer the same effect on histone acetylation at target promoters. This prediction is generally observed for a variety of activators discussed above and for the Sin3-Rpd3 corepressor, a histone deacetylase complex that represses transcription upon recruitment to target promoters. Specifically, repression by targeted recruitment of Sin3-Rpd3 is associated with deacetylation of both H3 and H4 at all promoters tested. Thus, our observation that all nine Cyc8-Tup1-repressed promoters tested show decreased H3 acetylation strongly suggests a mechanistic connection between repression by Cyc8-Tup1 and deacetylation of H3.
Cyc8-Tup1 could mediate H3 deacetylation either by recruiting a histone deacetylase or by blocking the access or activity of a histone acetylase. Hda1 is a candidate for a histone deacetylase recruited by Cyc8-Tup1 because it deacetylates H3 much more efficiently than H4 in vitro (53). However, if Tup1-dependent recruitment of Hda1 occurs, it is unlikely to be the sole mechanism for repression, because hda1 mutants efficiently mediate repression by Cyc8-Tupl (17). Although Cyc8-Tupl interacts with Rpd3 in vitro (67), our results do not support the model in which Rpd3 is specifically recruited to promoters repressed by Cyc8-Tup1, because the pattern of histone deacetylation differs from the situation when Rpd3 is recruited to promoters. In this regard, the in vitro interaction of Cyc8-Tup1 with Rpd3 is mediated by the TPR domains of Cyc8 (67), which are dispensable for repression in vivo (61, 62). In the alternative model in which Cyc8-Tup1 blocks a histone acetylase, Gcn5 (in the context of the SAGA or ADA complex) is the likely candidate given its specificity for H3 over H4. Cyc8-Tup1 could block activator-dependent recruitment or untargeted action of a Gcn5 complex. Models invoking recruitment of a histone deacetylase or blocking of an acetylase are consistent with previous observations that mutations in histone tails or histone deacetylases weaken Cyc8-Tup1 repression in vivo (16, 17) and that Tup1 preferentially binds hypoacetylated histone tails in vitro (16).
It is important to note that, while Tup1-dependent deacetylation is limited to H3 in the case of promoters regulated by glucose, oxygen, osmotic stress, and DNA damage, both H3 and H4 are deacetylated in the case of three a-specific promoters and a sporulation-specific promoter. This difference in acetylation specificity could be due to the fact that distinct DNA-binding repressors interact with different surfaces of the Cyc8-Tup1 complex (62); hence, there might be some flexibility in the structure of Cyc8-Tup1 that permits differential recruitment of histone deacetylases at different promoters. Alternatively, Cyc8-Tup1 might repress transcription, in part, by inhibiting the function of activators. As individual promoters repressed by Cyc8-Tup1 respond to different activators and individual activators can direct distinct patterns of histone acetylation, this activator-inhibition mechanism can easily explain the differential effect of Cyc8-Tup1 on histone acetylation.
Relationship to higher organisms.
In yeast, nucleosomes are moderately acetylated, with each histone tail containing an average of approximately two acetylated lysines (66). This average level probably reflects the balance between genome-wide (i.e., untargeted) action of histone acetylases and deacetylases (36, 52, 64). Such moderately acetylated chromatin might be generally permissive for molecular events on DNA, thereby explaining why histone acetylation is often not correlated with transcriptional activity in yeast. In multicellular organisms, the average level of histone acetylation is considerably lower, and a greater proportion of the genome is present in heterochromatin or other kinds of large chromosomal domains that are transcriptionally inert. We suggest that an important component of the classical relationship between histone acetylation and transcriptional activity is the relief of repressive chromatin domains that contain deacetylated histones. However, at the level of individual genes, we suggest that many activators stimulate transcription by mechanisms that do not involve increased histone acetylation. In this view, yeast and multicellular eukaryotes utilize similar molecular mechanisms to connect histone acetylation and transcriptional regulation, but they differ in the proportions of the genome that contain permissive or restrictive chromatin.
ACKNOWLEDGMENTS
We thank Laurent Kuras for cross-linked chromatin samples and advice on chromatin immunoprecipitation, Laurie Stargell for TBP antibodies, and Juliet Reid and Elmar vom Baur for discussing unpublished observations on histone acetylation.
This work was supported by grants to K.S. from the National Institutes of Health (GM30186 and GM53720).
ADDENDUM IN PROOF
Since the submission of this paper, J. Wu et al. (Mol. Cell 7:117–126, 2001) showed that Tup1 repression is associated with deacetylation of histones H3 and H2B and that Tup1 interacts in vitro with an isolated subunit of the HDAI histone deacetylase complex.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard S, Utley R T, Savard J, Clarke A, Grant P, Brandl C J, Pillus L, Workman J L, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esalp and the ATF-related cofactor Tralp. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allfrey V, Faulkner R M, Mirsky A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnick E H, John S, Berard D S, LeFebvre P, Hager G L. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the MMTV promoter is prevented by sodium butyrate. Proc Natl Acad Sci USA. 1990;87:3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresnick E H, John S, Hager G L. Histone hyperacetylation does not alter the position or stability of phased nucleosomes on the MMTV LTR. Biochemistry. 1991;30:3490–3497. doi: 10.1021/bi00228a020. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 9.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Esalp is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 11.Crane-Robinson C, Hebbes T R, Clayton A L, Thorne A W. Chromosomal mapping of core histone acetylation by immunoselection. Methods. 1997;12:48–56. doi: 10.1006/meth.1997.0446. [DOI] [PubMed] [Google Scholar]
- 12.Drysdale C M, Duenas E, Jackson B M, Reusser U, Braus G H, Hinnebusch A G. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drysdale C M, Jackson B M, Klebanow E R, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil A, Hinnebusch A G. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducker C E, Simpson R T. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durrin L K, Mann R K, Kayne P S, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 16.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor TUP1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 17.Edmondson D G, Zhang W, Watson A, Xu W, Bone J R, Yu Y, Stillman D, Roth S Y. In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gen5p activation. Cold Spring Harbor Symp Quant Biol. 1998;63:459–468. doi: 10.1101/sqb.1998.63.459. [DOI] [PubMed] [Google Scholar]
- 18.Eisen A, Taylor W E, Blumberg H, Young E T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988;8:4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furst P, Hu S, Hackett R, Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- 20.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gen5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 21.Gross D S, Adams C C, Lee S, Stentz B. A critical role for the heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Szauter P, Lucchesi J C. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong L, Schroth G P, Matthews H R, Yau P, Bradbury E M. Studies of the DNA binding properties of histone H4 amino terminus: thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 26.Ikeda K, Steger D J, Eberharter A, Workman J L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai S-I, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 28.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeppesen P, Turner B M. The inactive X chromsosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 30.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 33.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 34.Krebs J E, Fry C J, Samuels M L, Peterson C L. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 35.Krebs J E, Kuo M-H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo M-H, vom Baur E, Struhl K, Allis C D. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 37.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gen5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 39.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;389:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 40.Li X-L, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;389:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 41.Lucchesi J C. Dosage compensation in files and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev. 1998;8:179–184. doi: 10.1016/s0959-437x(98)80139-1. [DOI] [PubMed] [Google Scholar]
- 42.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 43.Mai X, Chou S, Struhl K. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol Cell Biol. 2000;20:6668–6676. doi: 10.1128/mcb.20.18.6668-6676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann R K, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukai Y, Matsuo E, Roth S Y, Harashima S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1 homolog. Mol Cell Biol. 1999;19:8461–8468. doi: 10.1128/mcb.19.12.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers C A, Schmidhauser C, Fragoso G, Mellentin-Michelotti J, Casperson G F, Pujuguet P, Hager G L, Bissell M J. Characterization of BCE-1: a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natarajan K, Jackson B M, Zhou H, Winston F, Hinnebusch A G. Transcriptional activation by Gcn4 involves independent interactions with the Swi/Snf complex and the Srb/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 48.Olesen J T, Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- 49.O'Neill L P, Turner B M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-b promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 51.Pogo B G T, Allfrey V G, Mirsky A E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci USA. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid J L, Iyer V R, Brown P O, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 53.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 55.Smith R L, Johnson A D. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 56.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 57.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 59.Treger J M, Schmitt A P, Simon J R, McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:26875–26879. doi: 10.1074/jbc.273.41.26875. [DOI] [PubMed] [Google Scholar]
- 60.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 61.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 62.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 co-repressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 63.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 64.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 65.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gen5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waterborg J H. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]
- 67.Watson A D, Edmondson D G, Bone J R, Mukai Y, Yu Y, Du W, Stillman D J, Roth S Y. Ssn6-Tupl interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyrick J J, Holstege F C P, Jennings E G, Causton H C, Shore D, Grunstein M, Lander E S, Young R A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Bone J R, Edmondson D G, Turner B M, Roth S Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao H, Eide D J. Zaplp, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]