Abstract
The p53 tumor suppressor protein plays a key role in the regulation of stress-mediated growth arrest and apoptosis. Stress-induced phosphorylation of p53 tightly regulates its stability and transcriptional activities. Mass spectrometry analysis of p53 phosphorylated in 293T cells by active Jun NH2-terminal kinase (JNK) identified T81 as the JNK phosphorylation site. JNK phosphorylated p53 at T81 in response to DNA damage and stress-inducing agents, as determined by phospho-specific antibodies to T81. Unlike wild-type p53, in response to JNK stimuli p53 mutated on T81 (T81A) did not exhibit increased expression or concomitant activation of transcriptional activity, growth inhibition, and apoptosis. Forced expression of MKP5, a JNK phosphatase, in JNK kinase-expressing cells decreased T81 phosphorylation while reducing p53 transcriptional activity and p53-mediated apoptosis. Similarly transfection of antisense JNK 1 and -2 decreased T81 phosphorylation in response to UV irradiation. More than 180 human tumors have been reported to contain p53 with mutations within the region that encompasses T81 and the JNK binding site (amino acids 81 to 116). Our studies identify an additional mechanism for the regulation of p53 stability and functional activities in response to stress.
The TP53 gene encodes a short-lived transcription factor implicated in the regulation of cell cycle progression, DNA repair, replicative senescence, and programmed cell death (21, 43, 47). The p53 protein is normally expressed at low levels in nonstressed normal growing cells. In response to DNA damage or stress, p53 is modified posttranslationally, with resultant accumulation of transcriptionally active protein (21, 43, 47). Activation of p53 in response to stress leads to temporary growth arrest or apoptosis. Mutations in p53, which are common in human tumors, abrogate its ability to contribute either to growth arrest or to cell death (43, 60). Most mutations found within the DNA-binding domain of p53 impair its binding to target DNA sequences (58). Different p53-responsive genes mediate a variety of cellular functions, including growth arrest by p21WAF1/CIP1 and 14-3-3 (12, 28) and apoptosis by Bax, CD95/Fas, IGF-BP3, and p53-inducible genes (6, 39, 44, 45). However, the mechanisms that enable activation of a selective subset of p53 target genes are poorly understood.
Accumulating evidence suggests that the regulation of p53's transcriptional activities depends on the nature of its phosphorylation (3, 16, 21, 37). Furthermore, a key factor that determines p53's ability to mediate its multiple activities is its stability, which is tightly regulated by Mdm2 and Jun NH2-terminal kinase (JNK) in a phosphorylation-dependent manner (8, 17, 18, 21, 25, 34). In nonstressed cells, Mdm2 targeting of p53 ubiquitination primarily occurs in the G2/M phase of the cell cycle, whereas JNK association and targeting of p53 for ubiquitination take place during the G0 phase (17). In response to stress, p53 phosphorylation coincides with its dissociation from both Mdm2 and JNK (17, 51).
More than a dozen phosphorylation sites have been mapped on p53, but only a few have been characterized with respect to the phosphorylating kinase and the relevance of the phosphorylation to p53 activities. Within the C terminus, human p53 is phosphorylated at amino acids 315, 378, 389, and 392, which enhances the in vitro specific DNA-binding activity of p53 (30, 31, 36, 54). At the p53 N terminus, several potential phosphorylation sites have been identified, including Ser 6, 9, 15, 20, 33, 37, and 46 and Thr 18 and 55 (11, 20, 21, 32, 37). ATM (ataxia telangiectasia mutated) has been implicated in Ser 15 phosphorylation (4, 40, 51, 53), which is induced by DNA damage (40, 53) and which is associated with enhanced p53 transcriptional activities (11, 49, 51). Gamma irradiation-induced ATM-dependent activation of Chk2 results in phosphorylation of Ser 20, which has been implicated in p53 transcriptional activities (10, 29), p53 tetramerization, down-regulation of Mdm2 suppression, p53 stabilization in response to stress (10, 57, 51, 52), and p53-mediated apoptosis (52, 59).
Phosphorylation by p38 kinase on Ser 33 and 46 has been shown to be required for UV-induced p53-mediated apoptosis (7), and inhibition of p38 has been found to attenuate transcriptional activities of p53 and p53-dependent apoptosis induced by chemotherapeutic agents (50). Phosphorylation-driven acetylation of p53 on C-terminus sites activates sequence-specific DNA binding (24, 49) and has also been implicated in transcription-dependent p53-mediated apoptosis (9).
Together, these observations point to a complex array of phosphorylation events occurring in response to stress and DNA damage that involve multiple kinases and multiple phosphoacceptor sites. The regulation of such complex phosphorylation patterns may involve sequential phosphorylation events, as was shown for phosphorylation of Ser 15, which depends on phosphorylation at Ser 46 (7).
In earlier studies we established the contribution of JNK to the regulation of p53 stability. In cells in the G0 and G1 phases of the cell cycle, among other specific conditions, JNK but not Mdm2 is associated with p53 and JNK efficiently targets the ubiquitination and degradation of p53; similarly, JNK serves as a targeting molecule for ubiquitination of nonphosphorylated c-Jun (15), ATF2 (19), and JunB (14). The JNK association with p53 has been mapped to residues 97 to 116 (2), whose deletion prolongs p53's half-life (17). Expression of constitutively active JNK kinase (JNKK) upstream kinase MEKK1 increases p53 stability and transcriptional activity (18). Given the multiple downstream effectors of MEKK1, we further elucidated the possible contribution of JNK to p53's stability and activities. Here we identify and characterize T81 as the phosphoacceptor site for JNK phosphorylation. We show that phosphorylation of T81 in response to stress or DNA damage stabilizes p53 and confers on it transcriptional activities and the ability to elicit apoptosis. In the absence of JNK expression or JNK-mediated phosphorylation, p53 is inactive. The implications of JNK phosphorylation at T81 for p53 activities in the context of the complex phosphorylation of p53 in response to stress are discussed.
MATERIALS AND METHODS
Cells.
293T (ATCC CRL1573) human embryo kidney cells, early passages of normal human fibroblasts (NHF; kind gift from H. Tahara), MCF7 breast cancer cells (ATCC HTB22), and p53 null cells (human lung tumor H1299 and mouse fibroblast 10.1 cell lines; kindly provided by X. Wu, and S. Aaronson, respectively) were grown in Dulbecco modified Eagle medium supplemented with antibiotics and 10% fetal bovine serum.
Oligonucleotide synthesis and treatment.
The oligonucleotides used were synthesized at Isis Pharmaceuticals, Inc. The sequences of the oligonucleotides are as follows: JNK1AS (ISIS12439), 5′-CTC TCT GTA GGC CCG CTT GG-3′; JNK2AS (ISIS18076), 5′-GTC CGG GCC AGG CCA AAG TC-3′; JNK1Scr (ISIS101759), 5′-CTT TCC GTT GGA CCC CTG GG-3′; JNK2Scr (ISIS101769), 5′-GTG CGC GCG AGC CCG AAA TC-3′. All oligonucleotides consisted of 2′-O-methoxyethyl modified sugars (residues 1 to 5 and 16 to 20) and 2-deoxy sugars (residues 6 to 15). The linkages are uniform phosphorothioate. Cells were treated with 0.25 μM oligonucleotides in the presence of 10 μg of Lipofectin reagent (Life Technologies)/ml as described previously (46).
Expression vectors.
Human p53 cDNA was used as a backbone for generating a PCR-based site-directed single-base-pair substitution at amino acid (aa) 81 (Quick Change; Stratagene), generating p53T81A, which was confirmed by sequencing. Rat p53 mutated at aa 101, 105, and 108 or with aa 97 to 116 deleted (ΔP7) and the constitutively active forms of JNKK2 and MEKK1 kinases have been previously described (18). FLAG-tagged MKP5 expression vector was constructed by amplifying the MKP5 open reading frame using reverse transcription-PCR and subcloning the amplification product into a pcDNA3.0 plasmid (Invitrogen). A monoclonal antibody directed against MKP5 was raised against a fragment spanning aa 2 to 154 of MKP5 (A. Bar Shira et al., unpublished data).
In vitro phosphorylation of p53 by purified JNK.
293T cells (108) were calcium transfected with hemagglutinin (HA)-tagged JNK2 (30 μg) and 36 h later treated with UV-C (40 J/m2). Proteins prepared within 45 to 60 min after irradiation were subjected to immunoprecipitation using antibodies to HA. Immunoblotting and silver staining were performed on an aliquot of the purified material to ensure an appropriate degree of purity. Activity of HA-JNK was verified using glutathione S-transferase (GST)–Jun as a substrate in solid-phase kinase reactions. Wild-type (wt) or T81A mutant forms of p53 were in vitro translated using an in vitro translation kit (TNT; Promega). The translated proteins were purified using p53 antibodies (pAb 421) that were covalently bound to Amino link beads (Pierce). Bound p53 was eluted from the antibodies using elution buffer (Pierce) followed by dialysis against kinase buffer (20 mM HEPES [pH 7.6], 1 mM EGTA, 1 mM dithiothreitol, 2 mM MgCl2, 2 mM MnCl2, 5 mM NaF, 1 mM NaVO3, 50 mM NaCl). The amount and purity of p53 were verified by silver staining and immunoblotting before equal aliquots were subjected to an in vitro solid-phase kinase reaction (1). Briefly, bead-bound HA-JNK was incubated with p53 at 37°C for 15 min in the presence of [γ-32P]ATP (50 cpm/fmol) in the presence of kinase buffer. The reaction mixture was then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to Coomassie blue staining to confirm equal loading of p53. The gel was dried, and phosphorylation of p53 was determined by autoradiography. In all cases the buffers used contained a cocktail of protease and phosphatase inhibitors (1).
Stress and DNA damage treatments.
UV irradiation was administered as previously described (1). Briefly, cells were subjected to UV-C irradiation (254 nm; 40 J/m2) in the calibrated area within the tissue culture hood. Medium that had been removed prior to irradiation was returned to minimize serum-induced changes. Treatment with anisomycin (10 μg/ml), hydrogen peroxide (100 μM), sorbitol (0.6 mM), or cis-platinum (0.6 μM) (all purchased from Sigma) was performed by adding them to the medium (time zero) followed by time point analysis as indicated in Results.
Protein stability analysis.
To monitor changes in the stability of p53, pulse-chase analysis was performed using the previously described [35S]methionine-cysteine mixture (17). Briefly, 10.1 p53 null cells (106) were transfected with the wt or T81A mutant form of p53 (2 μg) using Lipofectamine Plus reagent (GIBCO-BRL). Twenty-four hours after transfection the cells were starved (serum without methionine) for 1 h before addition of [35S]methionine-cysteine mixture (1 mCi) to cells for 1 h (time zero followed by chase for the periods of time indicated by the time points in the pertinent figures. Cells were counted and harvested, followed by protein preparation and p53 immunoprecipitation from equal numbers of cells (using 2 μg of DO1 antibodies per 106 cells). Cycloheximide-chase experiments were performed on cells transfected as indicated above, and 24 h later cycloheximide (60 μg/ml) was added to the cultures (time zero). Equal amounts (500 μg) of proteins prepared at various time points were subjected to immunoprecipitation (DO1) followed by immunoblot analysis (polyclonal p53 antibody).
Mass spectrophotometry analysis.
293T cells (108) were transfected (calcium phosphate) with p53 and JNKK2CAA expression vectors. Twenty-four hours after transfection, protein extracts were incubated with p53 antibodies (DO1) immobilized on Aminolink beads (Pierce). Immunoprecipitates were eluted from antibodies reduced and alkylated with iodoacetamide before SDS-PAGE (22). A portion of the gel was electroblotted, followed by immunoblotting to reveal the position of p53. Coomassie blue-stained protein bands were excised from SDS-PAGE gels and digested with trypsin in situ (48). The digests containing the gel bits were extracted in 50% acetonitrile–0.1% trifluoroacetic acid (TFA), concentrated using a SpeedVac to less than 10 μl, and desalted using a Zip-Tip (Millipore, Bedford, Mass.). An aliquot of the digest was mixed with an equal volume of matrix material (α-cyano-4-hydroxy-cinnamic acid) (Sigma) as a saturated solution in 50% acetonitrile–0.05% TFA, and ion masses were determined by matrix-assisted laser desorption–time of flight mass spectroscopy (42) (Kratos; KOMPACT MALDI III). The instrument was calibrated over the range 905.05 to 5,734.59 atomic mass units using peptide mass standards (PerSeptive Biosystems).
Immunoblot analysis.
Polyclonal antibodies to phosphorylated T81 (boldface) were generated (Research Genetics) in rabbits against phosphopeptide PAPAAPTPAAPAP. Serum was precleared on a nonphosphorylated peptide followed by affinity purification on the phosphorylated peptide. Immunoblot analysis with phospho-T81 antibody was performed (using a dilution of 1/100) on proteins that were prenormalized with respect to total p53 levels followed by enhanced chemiluminescence detection (Amersham). In all other cases Western blotting was performed using equal amounts of proteins. Immunoprecipitations were carried out on 1-mg quantities of cellular extracts using the relevant antibodies and protein G beads (GIBCO-BRL).
Confocal immunofluorescence.
Cells grown on 22-mm2 coverslips were fixed in freshly prepared 2.5% para-formaldehyde in phosphate-buffered saline (PBS) (pH 7.4) for 10 min at room temperature. The cells were then washed three times (5 min each) in PBS followed by permeabilization in 0.1% Triton X-100 in PBS (pH 7.4) for 5 min and an additional three 5-min washes in PBS. The cells were incubated on 75-μl drops of anti-p53 antibodies (FL-393; Santa Cruz Biotechnology) diluted (200 ng/ml) in PBS (pH 7.4) containing 1% bovine serum albumin (BSA) for 1 h at room temperature in a humidity chamber. The cells were washed three times in PBS (5 min each) before incubation with 75-μl drops of fluorescein-conjugated anti-rabbit immunoglobulin G (Vector Laboratories) diluted (15 μg/ml) in PBS (pH 7.4) containing 1% BSA for 1 h at room temperature in a humidity chamber in the dark. The cells were washed three times in PBS (5 min each), followed by a brief wash in 0.1% Triton X-100 in PBS, pH 7.4 (5 min), and three additional 5-min washes in PBS, and the coverslips were mounted on glass slides in Vectashield (Vector Laboratories).
Transcriptional activation and growth inhibition assays.
The Mdm2 and p21CIP1/WAF1 minimal promoter-driven luciferase constructs (cotransfected with β-galactosidase [β-Gal] expression vector) were analyzed 24 h after transfection as previously described (18). Growth inhibition assays were performed in six-well plates using H1299 cells transfected with wt or mutant p53T81A. Cells (triplicate wells) were subjected to G418 selection for 3 weeks before clones were stained and counted.
Apoptosis and cell cycle studies.
p53 null cells were exposed to treatments 24 h after transfection. Apoptosis was assessed by annexin-V–fluorescein isothiocyanate staining in the presence of propidium iodide (PI) (Pharmingen). Cell cycle analysis of green fluorescent protein (GFP)-positive cells was carried out using fluorescence-activated cell sorter analysis of PI-stained cells (5 × 104 cells per sample) using a Calibur flow cytometer (Becton Dickinson) under wide-scatter gates. The data were analyzed using CellQuest software.
RESULTS
Mass spectrometry analysis of p53 phosphorylation by JNK.
Human p53HA immunopurified from JNKKCAA-expressing 293T cells was separated by SDS-PAGE and digested in situ with trypsin. Analysis of the peptide digests by mass spectrometry identified a peptide mass (m/z) of 3528.5 from the p53/JNKK+ isolate that was not detectable in the p53/JNKK− digest. Conversely, a peptide mass (m/z) of 3446.2 was found in the digest of the p53/JNKK− sample but was not detected in the p53/JNKK+ digest. A theoretical tryptic digest of p53 predicts a unique m/z of 3445.97 corresponding to peptide 66 to 101. Within the 0.1% error of the mass spectrometer, our results are consistent with a single phosphorylation of p53 peptide 66 to 101 (3445.97 + 80 = 3525.97) caused by coexpression of p53 with JNKKCAA. Subsequent analysis was carried out on T81, which is the only residue within this region that is a proline-directed Ser/Thr, a conserved JNK phosphoacceptor site. Analogs of human proline-driven T81 are found in p53 from other species, including mice (Thr 78), rats (Thr 89), and canines (Thr 76).
T81 is phosphorylated by JNK.
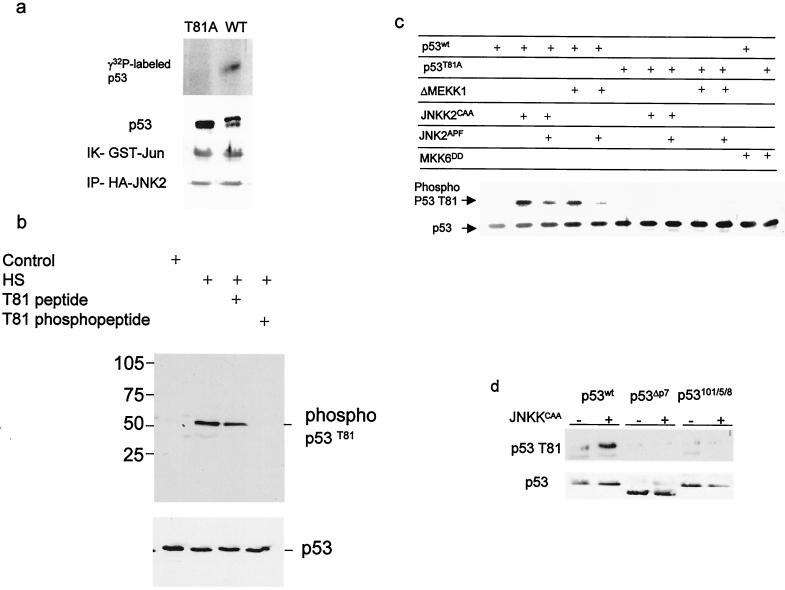
Immunopurified JNK phosphorylated the wt p53 but not the mutant form of p53 in which T81 was replaced with Ala (T81A) (Fig. 1a). To detect T81 phosphorylation in vivo, we raised antibodies to a phosphorylated T81 peptide. Recognition by phospho-T81 antibodies was inhibited by a phosphorylated, but not by a nonphosphorylated, peptide corresponding to this domain (Fig. 1b). Coexpression of the constitutively active forms of MEKK1 (ΔMEKK1) or JNKK (JNKK2CAA), but not MKK6 (MKK6DD), with wt p53 led to efficient phosphorylation of wt p53 detected by phospho-T81 antibodies. Neither kinase could phosphorylate the p53 T81A mutant (Fig. 1c). These data suggest that JNK, but not p38, phosphorylates p53 at T81. In the same vein, forced expression of JNK2APF, a dominant-negative form of JNK decreased MEKK1- as well as JNKK-mediated phosphorylation of p53 on T81 (Fig. 1c). Expression of a 20-aa peptide that corresponds to the JNK association site on p53 (P7) and that disrupts the JNK-p53 complex in vivo and in vitro (16) decreased JNKK-dependent phosphorylation of p53 on T81 (not shown). Similarly, JNKK did not induce phosphorylation of T81 on p53 whose JNK docking site had been deleted or mutated (18), indicating that the association of JNK with p53 is required for its phosphorylation at T81 (Fig. 1d). These experiments establish that JNK phosphorylates p53 on T81.
FIG. 1.
JNK phosphorylates p53 on T81. (a) Wild-type or mutant (T81A) forms of human p53 were in vitro translated, purified, and subjected to phosphorylation using JNK immunopurified from UV-treated 293T cells. Middle, immunoblot analysis of material subjected to autoradiography using DO1 antibodies. The doublet represents the nonphosphorylated (lower band) and phosphorylated (upper band) forms of p53. The control reaction using c-Jun as a substrate (immunokinase [IK] reaction of GST-Jun) and the level of HA-JNK2 that was immunoprecipitated (IP-HA-JNK2) are shown (bottom). (b) Proteins prepared from NHF that were exposed to sham stress (control) or heat shock (HS; 42°C for 1 h) were subjected to immunoblot analysis using the antibodies to phosphorylated T81 in the presence of excess phospho or nonphospho peptides as indicated (top) or to Western blotting using DO1 antibodies (bottom). (c) T81 phosphorylation on wt or T81A forms of p53 was monitored after cotransfection of the respective construct (1 μg) with the indicated upstream kinases (3 μg) into H1299 cells. Analysis of T81 phosphorylation was carried out using the antibodies to phospho-T81. Bottom, p53 levels detected with DO1 antibodies. (d) T81 phosphorylation of p53 impaired in JNK association. Either wt rat p53 or p53 with deletions (ΔP7) or mutations (at aa 101, 105, and 108) within the region, which encompasses the JNK binding site (1, 18) was cotransfected with JNK2CAA into H1299 cells. Proteins were prepared and subjected to Western blot analysis using phospho-T81 antibodies (top) or DO1 antibodies (bottom).
T81 is phosphorylated in vivo following exposure to DNA-damaging agents.
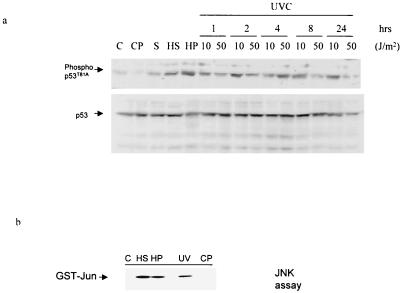
To determine changes in T81 phosphorylation in response to stress and DNA damage, NHF were exposed to heat shock, cis-platinum, anisomycin, H2O2, or UV treatment. Immunoblot analysis using antibodies to phospho-T81 revealed that T81 is phosphorylated in vivo within 1 h after exposure to heat shock, hydrogen peroxide, sorbitol, or UV (Fig. 2a; please note that the level of p53 has been normalized to ensure that changes in phosphorylation are not due to altered expression level). The degree of JNK activation in response to these treatments was monitored via immunokinase reactions using GST-Jun as a substrate (Fig. 2b). Treatment with cis-platinum (Fig. 2a) or X rays (not shown) neither activated JNK nor phosphorylated p53 at T81 in the NHF cells. Time course analysis demonstrated that the degree of T81 phosphorylation depends on the UV dose. Low-dose UV irradiation (10 J/m2), which causes growth arrest, resulted in T81 phosphorylation within as little as 30 min (not shown), with increased phosphorylation after 1 to 2 h. The level of T81 phosphorylation remained above background even 24 h after UV treatment (Fig. 2a). In response to high-dose irradiation (50 J/m2), which causes apoptosis, phosphorylation on T81 was seen within 1 h, peaked within 4 h, and declined to background levels within 24 h (Fig. 2a). The kinetics of T81 phosphorylation was consistent with increased levels of p53 in response to these stress-inducing agents (1, 17).
FIG. 2.
In vivo phosphorylation of T81 in response to stress and DNA damage. (a) NHF cells were subjected to the indicated treatments (heat shock [HS], 42°C for 1 h; UV-C (254 nm), 10 or 50 J/m2 [UV]; H2O2 [HP], 100 μM; cis-platinum [CP], 0.6 μM; sorbitol [S], 0.6 M), and proteins were prepared 1 h later unless indicated otherwise. The quantities of p53 used for the Western blotting were prenormalized to assure equal loading of p53 and thus do not reflect changes in the amount of p53 seen after these treatments. Upper lane, analysis with antibodies to phospho-T81; lower lane, analysis with DO1 antibodies to p53. (b) JNK activity in the protein samples analyzed in panel a, upper lane (1-h time point), was monitored via the solid-phase kinase reaction using GST-Jun1–87 as a substrate.
Substitution of T81 for Ala attenuates UV- and JNK-mediated p53 stabilization.
A prerequisite for p53 ability to elicit its activities is its enhanced stability, which is phosphorylation dependent (18). The association of nonactive JNK with p53, which takes place under nonstressed conditions, targets p53 ubiquitination and degradation in an Mdm2-independent manner (17). Conversely, signaling by upstream JNKK kinase MEKK1 prolongs p53's half-life (16). To monitor the half-life of p53 mutated on T81A, p53 null cells were transfected with the wt or T81A mutant forms of p53 and pulse-chase analysis was carried out under normal growth conditions as well as after UV irradiation or JNKK expression.
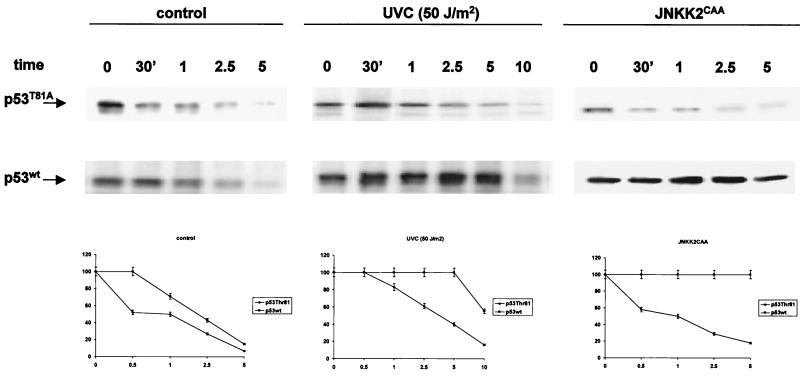
Both wt and T81A forms of p53 exhibited short half-lives in the absence of stress (Fig. 3, left). This finding indicates that both forms are similarly targeted for ubiquitination and degradation. In response to UV treatment the half-life of wt p53 was prolonged, compared with the half-life of the T81A mutant (Fig. 3, middle). These data suggest that mutation in T81 is sufficient to impair the stabilization of p53 in response to UV irradiation. To directly address whether JNK's ability to alter p53 stability is impaired in the T81A mutant, p53 null (10.1) cells were cotransfected with the constitutively active form of JNKK2 (JNK2CAA) and either the wt or the T81A form of p53. Half-life measurements performed on the cells 24 h later revealed that JNKK increased the stability of the wt but not the T81A form of p53 (Fig. 3, right). Differences in the half-life of the T81A form of p53 in response to JNKK were more noticeable than those after UV treatment. This is not surprising in light of the multiple kinases induced by UV treatment, which contribute to p53 phosphorylation and stabilization, including p38 and Chks (7, 10, 21, 40, 51, 52). These observations suggest that T81 phosphorylation is required for JNK-mediated changes in p53 stability. Further support for the role of JNK in stabilizing p53 via T81 was obtained from analysis of p53 stability under cycloheximide-chase conditions (27). Exposure of cells to cycloheximide conditions (60 μg/ml) that activate JNK but block protein synthesis revealed a prolonged half-life for the wt, but not the T81A, form of the protein (data not shown). These data establish the role of T81 in p53 stabilization following UV irradiation or JNK activation.
FIG. 3.
Stability of p53 mutated at T81. Pulse-chase analysis of p53 half-life was performed as indicated in Materials and Methods using 10.1 p53 null cells that had been transfected with either wt or T81A forms of p53. At the indicated time points, p53 was immunoprecipitated with DO1 antibodies followed by autoradiography. The half-lives of p53 under nonstressed conditions and in response to UV irradiation or following JNK activation (forced expression of JNKK2CAA) are shown. Bottom, corresponding densitometry analysis, which was carried out in three independent experiments.
Role of T81 phosphorylation in p53 transcriptional activities, p53-mediated growth inhibition, and growth arrest.
To determine whether T81 phosphorylation is important for activation of p53's biological functions, we first assessed the transactivation capacities of the T81A mutant. Basal transcriptional activities of p21WAF1/CIP1 promoter sequences linked to a luciferase reporter were fivefold higher for wt p53 than for the T81A form, which exhibited activities similar to those seen with the empty expression vector. A substantial increase (16-fold over basal levels) in the level of p21WAF1/CIP1-mediated luciferase activity upon expression of the constitutively active form of MEKK1 (ΔMEKK1) was recorded. In response to the constitutively active JNKK2 (JNKK2CAA) or after UV irradiation, the expression of the wt but not the T81A form led to a sixfold increase in transcriptional activities mediated by p21WAF1/CIP1 (Fig. 4a) or Mdm2 promoter sequences (data not shown). Transcriptional activities of T81A were equal to those of transcriptionally “dead” hot spot p53 mutants (at aa 248 and 273), before as well as after UV irradiation (Fig. 4b). Corresponding to the level of luciferase activity driven by the p21WAF1/CIP1 promoter are expression levels of p21WAF1/CIP1, which were monitored via Western blotting (Fig. 4b, bottom). These data suggest that phosphorylation on T81 contributes to transcriptional activities of p53 in response to stress.
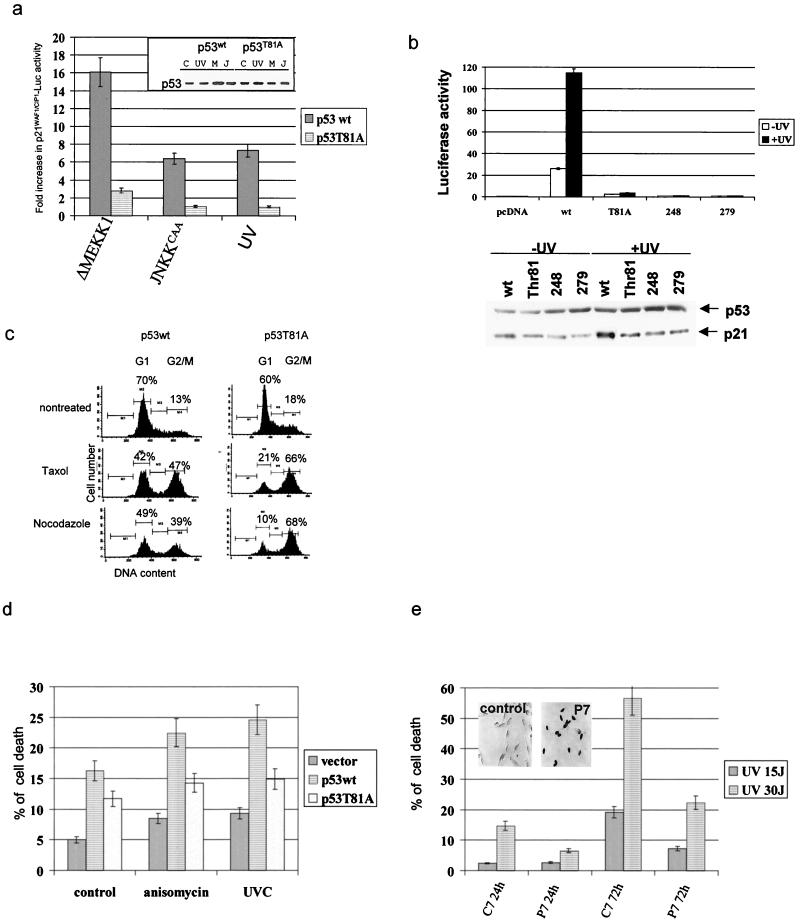
FIG. 4.
Activity of p53 mutated at T81. (a) p53 null cells were transfected with p21WAF1/CIP1-Luc, a constitutively active form of MEKK1 or JNKK2 and wt or mutant forms of p53. (Inset) Expression levels of the p53 forms in extracts used for transcriptional analysis. Basal transcriptional activities of wt p53 were five times higher than those seen with either the T81A mutant or empty vector. The data shown reflect fold increases over basal activity of the construct. C, control; M, ΔMEKK1; J, JNKKCAA. (b) Comparison of T81A transcriptional activities with those of different transcriptionally dead p53 mutants. Mouse 10.1 p53 null cells were transfected with p21WAF1/CIP1-Luc and wt or mutant forms of p53. Cells were treated with UV (15 J/m2) 36 h later, and proteins were prepared after an additional 6 h. Analysis was carried out as indicated in panel a. Bottom, Western blots of proteins prepared from the same experiment using antibodies to p53 and p21WAF1/CIP. (c) p53 vectors and GFP were transfected into 10.1 cells, and 24 h later cells were treated with Taxol or nocodazole (5 μg/ml). PI staining and flow cytometry analyses of GFP+ cells were performed 16 h after treatment as detailed in Materials and Methods. (d) H1299 cells were transfected with the constructs and 24 h later were treated with anisomycin (10 μg/ml) or UV-C irradiation (15 J/m2). Analysis of cell death was performed after an additional 36 h. The fraction of dead cells shown includes annexin-V- and PI-positive cells, thereby including apoptotic and necrotic death. (e) 3T3 mouse fibroblasts were transfected with P7 (corresponding to the JNK binding site on p53) or C7 (P7 sequence in scrambled order) cloned into pcDNA3HA. UV irradiation was administered 24 h later. The degree of apoptosis was calculated 24 or 72 h after irradiation based on fluorescence-activated cell sorter analysis of PI-positive cells. The inset depicts immunocytochemistry analysis of P7 expression.
As p53-dependent transcription of p21WAF1/CIP1 is required for p53-dependent growth arrest in response to stress (21, 43, 47), we next assessed the fraction of 10.1 cells expressing wt or T81A p53 in the G1 phase of the cell cycle upon treatment with Taxol or nocodazole, which were selected to represent the agents known to affect the cell cycle and cell death in a p53-dependent manner (56). Whereas, following exposure to Taxol or nocodazole, 42 and 49% of wt p53-expressing cells, respectively, accumulated in the G1 phase, only 21 and 10% of T81A-expressing cells, respectively, exhibited a G1 arrest (Fig. 4c). The decrease in the fraction of cells found in G1 upon expression of T81A coincided with an elevated fraction of cells within the G2/M phase of the cell cycle, which is characteristic of transcriptionally inactive mutant forms of p53 (66 versus 47% after Taxol treatment and 68 versus 39% after nocodazole treatment; Fig. 4c). Further analysis was carried out to investigate the growth-inhibitory properties of T81A mutant p53. Forced expression of wt or T81A forms in H1299 cells followed by G418 selection revealed that wt p53 expression resulted in 33 ± 6.5 colonies, compared with 82 ± 7.7 colonies upon expression of the T81A mutant and 85 ± 9.3 colonies in cells transfected with the empty vector. These data point to the requirement of T81 phosphorylation as a prerequisite of p53-dependent cell growth inhibition. In agreement with earlier observations that identified aa 65 to 90 as essential for p53 to mediate growth arrest (61), our findings pinpoint the requirement for the single amino acid T81.
Forced expression of wt or T81A forms of p53 in H1299 cells increased the fraction of apoptotic cells, suggesting that T81A p53 is as potent as the wt p53 protein in eliciting this basal degree of apoptosis (Fig. 4d). However, in response to anisomycin or UV irradiation, wt p53 caused a significantly greater extent of cell death than mutant T81A p53 (Fig. 4d). These data suggest that T81 phosphorylation contributes to p53's ability to mediate apoptosis in response to stress and DNA damage. The basal degree of cell death elicited by the T81A form of p53 may be attributed to the ability of p53 to elicit apoptosis even when it is transcriptionally inactive (26).
Forced expression of a peptide (P7) which corresponds to the JNK docking site efficiently blocked JNK association with, and phosphorylation of, p53, whereas it did not affect other JNK substrates (not shown). The ability of p53 to mediate programmed cell death in response to UV irradiation was impaired (15 to 5% after 24 h and 57 to 21% after 72 h) upon expression of P7, but not control peptide C7 (Fig. 4e). These findings further support the role of JNK in regulating the ability of p53 to elicit UV-induced cell death.
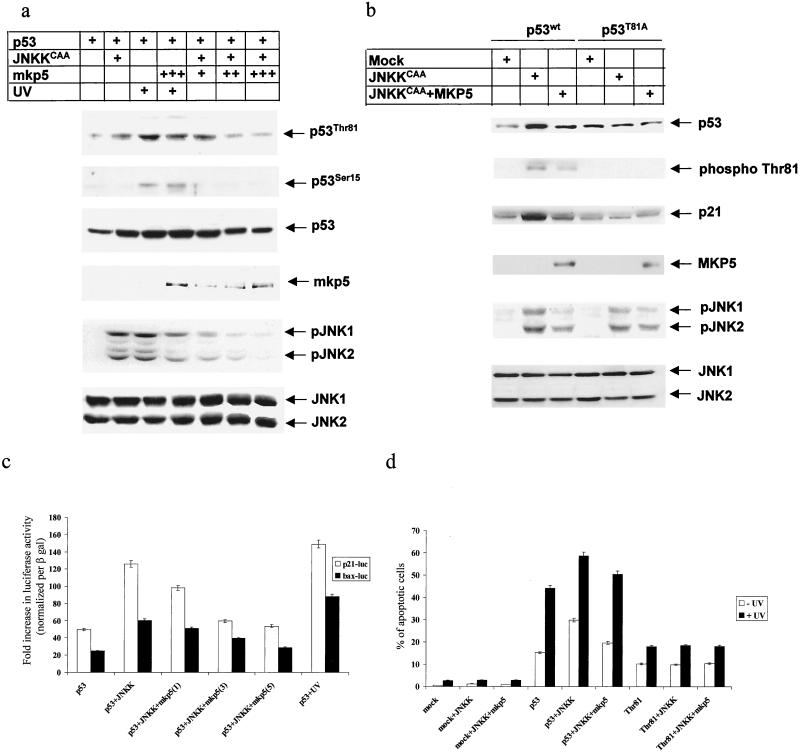
JNK phosphatase MKP5 efficiently attenuates JNK-mediated T81 phosphorylation, p53 transcriptional activities, and p53-mediated apoptosis.
As an independent approach to investigating the role of JNK in T81 phosphorylation and in p53 activities, we used specific JNK/p38 phosphatase MKP5 (55). Forced expression of MKP5 in JNKK-expressing cells reduced the level of T81 phosphorylation in a dose-dependent manner (Fig. 5a). MKP5 expression decreased, albeit less efficiently, T81 phosphorylation in response to UV irradiation (Fig. 5a). Under these conditions, MKP5 did not affect the level of Ser 15 phosphorylation. Differences in MKP5 effects on T81 phosphorylation in JNKK versus UV-treated cells may be attributed to the degree of MKP5 activities. Analysis of JNK phosphorylation revealed that MKP5 was less effective in removing phospho groups from JNK in UV-treated cells than in JNKK-expressing ones. Accordingly, UV may attenuate MKP5 activities, which would also explain why MKP5 was less effective in attenuating T81 phosphorylation in UV-treated cells. Further support for the role of JNK in T81 phosphorylation and activities comes from comparison between wt and T81A mutant forms of p53. Coexpression of JNKK and p53 (wt or T81A) in p53 null mouse fibroblasts in the presence or absence of MKP5 revealed a clear increase in T81 phosphorylation on the wt but not the T81A mutant form of p53, which was attenuated upon MKP5 expression. Decreased T81 phosphorylation coincided with a decrease in p21 expression (Fig. 5b). These results coincide with luciferase assays performed under the same conditions. Expression of JNKK with p53 in p53 null cells caused a threefold increase in p53 transcriptional activities, monitored using Bax and p21 promoter sequences linked to the luciferase reporter gene. However, addition of MKP5 to the transfection mixture decreased p53 transcriptional activities. The degree of decreased p53 transcription depended on the amount of MKP5 transfected (Fig. 5c). MKP5 did not have any effect on Mdm2-luc or Bax-luc activities (data not shown). These observations further support the role of JNK in T81 phosphorylation and transcriptional activities.
FIG. 5.
JNK phosphatase attenuates T81 phosphorylation and p53 activities. (a) p53 null cells were transfected with p53, a constitutively active form of JNKK, and different concentrations of MKP5. Cells were exposed to 45 J of UV-C/m2, as indicated. Top, specific phosphorylation pattern of T81. Phosphorylation on serine 15 after UV treatment and the expression of p53 are shown. Controls revealing the expression of exogenously transfected MKP5-phosphorylated and nonphosphorylated forms of JNK1 and -2 are also shown. (b) p53 null cells were transfected with wt or T81A mutant forms of p53 together with JNKKCAA and MKP5 as indicated. Analysis for expression of p53 and p53 phosphorylation on T81 and a control analysis for p21WAF1/CIP1 expression and JNK phosphorylation (and nonphosphorylation) levels were carried out 36 h after transfection. (c) p53 null cells were transfected with either p21-luc or Bax-luc constructs, together with the indicated vectors and β-Gal. For transfection efficiency lysates were normalized with respect to β-Gal activity and subjected to a luciferase assay. The diagram reflects changes in luciferase activity in six independent transfections. (d) 10.1 cells were transfected with the indicated constructs together with GFP, and 24 h after transfection cells were treated with 45 J of UV-C PI staining and flow cytometry analysis of GFP+ cells were performed 16 h after UV treatment. The diagram shows changes in apoptotic cell death after six independent transfections.
To further assess the effect of JNK on p53 activities, we monitored the degree of apoptosis in JNKK-transfected cells. Forced expression of p53 in p53 null fibroblasts was sufficient to induce a low level of apoptosis, and cotransfection of p53 and JNKK led to a substantial increase, from 13 to 30%, in the basal degree of apoptosis (Fig. 5d). These results are in accordance with the changes to apoptosis in response to MEKK1 treatment (18). Whereas UV treatment led to a threefold (15 to 45%) increase in the degree of apoptosis, a higher level of apoptosis (58%) was seen in the presence of JNKK (Fig. 5d). Coexpression of MKP5 with JNKK efficiently decreased the basal level of apoptosis (from 30 to 18%, where 13% is the basal level) and, to a lesser extent, the degree of UV-induced apoptosis (from 58 to 50%; Fig. 5d). These findings suggest that JNK efficiently increases p53's ability to elicit apoptosis. In response to UV irradiation, it is the combination of other UV-inducible kinases that contributes, in concert with a UV-mediated decrease in MKP5 activities, to the limited effect of MKP5 on UV-induced apoptosis, compared with the effect on JNKK-expressing cells. Importantly, p53 mutated on T81 was not able to elicit apoptosis in response to JNKK expression, nor was it affected upon MKP5 expression (Fig. 5d). These results further confirm the requirement of T81 phosphorylation for p53's ability to elicit programmed cell death in response to JNK stimuli.
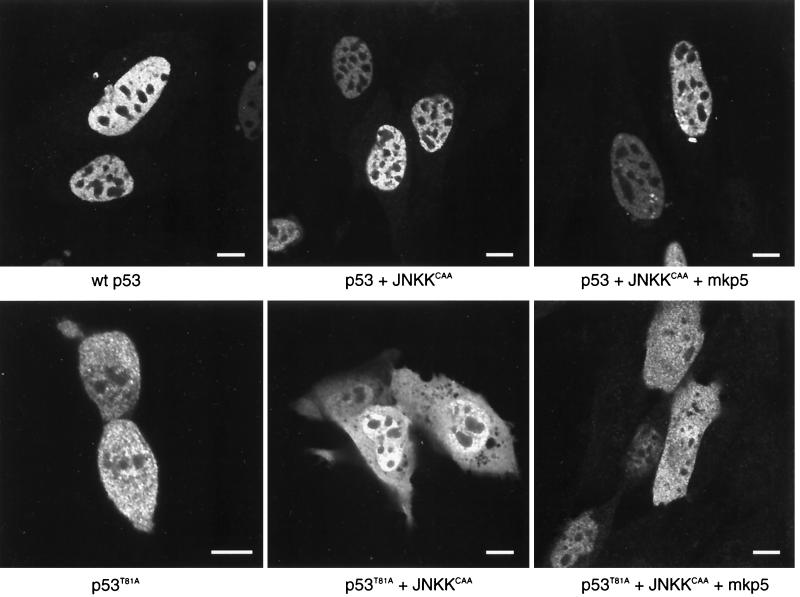
Cellular localization of wt and T81 p53 following JNKK and MKP5 expression.
Given the relationship between cellular localization of p53 and its stability and activities, we monitored possible changes in p53 distribution after JNKK and MKP5 expression. As shown in Fig. 6, the expression of either JNK kinase or its phosphatase did not alter the nuclear localization of p53. These data suggest that JNK phosphorylation does not affect the cellular compartmentalization of p53. A similar analysis performed on the T81A form of p53 revealed that the lack of the JNK phosphorylation site led to equal distribution of this p53 mutant in the cytoplasm and the nucleus, which was not affected by MKP5 or by JNKK expression (Fig. 6, bottom). Cellular distribution of either wt or T81A forms of p53 was not affected by UV irradiation, nor was it changed upon leptomycin B treatment (not shown). Since the phosphoacceptor site studied here lies within the proline-rich domain, which has been implicated in p53 conformation, we also monitored possible changes of p53 conformation due to its mutation on T81. Unlike p53 mutated on aa 248 (13), T81A p53 retained the conformation of wt p53 forms, as determined on the basis of the peptide digest pattern (data not shown).
FIG. 6.
Cellular localization of wt and T81A p53 forms. p53 null 10.1 cells were transfected with wt p53, p53 plus JNKKCAA, p53 plus JNKKCAA plus MKP5, p53T81A, p53T81A plus JNKKCAA, or p53T81A plus JNKKCAA plus MKP5. Cells were immunolabeled with a rabbit polyclonal antibody to p53 (FL-393) (Santa Cruz Biotechnology) (diluted 1:500) followed by detection with fluorescein-conjugated anti-rabbit immunoglobulin G (heavy plus light chains) (Vector Laboratories). Cells were illuminated with the 488-nm line of the argon laser of a Leica TCS-SP (UV) confocal laser scanning microscope and examined using a 100×, 1.4 -numerical-aperture objective lens. For wt p53 (top three panels), labeling is restricted primarily to the nucleus (excluding the nucleoli) and nuclear bodies. Little or no cytoplasmic labeling is seen. For p53T81A (bottom three panels), labeling is divided between the nucleus and the cytoplasm. The lower left panel shows a pair of cells that are completing division. In the lower right panel, p53T81A is divided evenly between the nucleus and the cytoplasm in cells that express the protein at higher levels; at lower expression levels, it is found primarily in the nucleus. In all cases, p53 labeling is restricted from the nucleolus. Bars, 10 nm
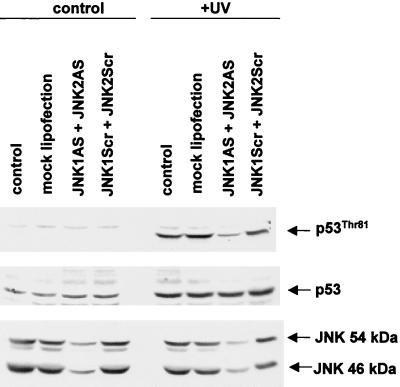
T81 phosphorylation is attenuated in cells treated with antisense JNK.
To further establish the role of JNK in the phosphorylation of T81, we used antisense oligonucleotides to both JNK1 and JNK2 (JNK1AS and JNK2AS). Previous studies established that treatment of cells with JNK-AS efficiently blocks JNK expression at the RNA and protein levels (46), thereby creating a transient nearly null environment for JNK. Using MCF7 cells (which contain the wt form of p53), we monitored changes in endogenous p53 phosphorylation on T81 following UV irradiation of cells pretreated with JNK1AS and JNK2AS. Transfection of JNK1AS and JNK2AS led to a significant decrease in the level of JNK1 and JNK2 RNA (not shown) and in protein levels (Fig. 7). Within 2 h after UV irradiation, T81 was similarly phosphorylated in mock-, pcDNA3-, and control antisense-oligonucleotide-transfected cells. However, a substantially lower degree of phosphorylation was evident in the JNK1AS- and JNK2AS-transfected cells (Fig. 7). These results establish that in vivo phosphorylation of T81 in response to UV treatment is mediated by JNK.
FIG. 7.
JNK antisense oligonucleotides attenuate T81 phosphorylation in vivo. MCF7 cells were transfected with either specific antisense oligonucleotides directed against JNK1 and JNK2 or nonspecific antisense oligonucleotides (250 nM each). Forty-eight hours after transfection, cells were either sham treated or UV treated and harvested 2 h after treatment. Top, specific T81 phosphorylation; middle, expression level of p53 in response to indicated treatments; bottom, expression of JNK1 and -2 in the presence or absence of transfected oligonucleotides. Control lane, no treatment; mock lipofection, cells maintained in concentrations of Lipofectin equal to those for the oligonucleotide-transfected samples; JNK1AS and JNK2AS, JNK antisense oligonucleotides that were transfected; JNKScr, control “scrambled” oligonucleotides.
T81 and the JNK docking site on p53 are often mutated in human cancer.
A further indication of the importance of JNK phosphorylation on T81 for p53 functions comes from the finding that human tumors contain p53 with mutations within the JNK binding and phosphorylation sites. A search of the p53 mutant database for mutations within T81 identified two cases of breast and skin tumors with p53 mutated at this site. More than 180 additional tumors were found to harbor p53 with mutations on residues within, or adjacent to, the JNK docking/phosphorylation sites (aa 81 to 117), indicating that mutations occur frequently in this area (5). This finding provides additional evidence of the importance of JNK association and phosphorylation of p53.
DISCUSSION
Increasing evidence points to the physiological importance of posttranslational modifications for p53 stability and activities. Those modifications include phosphorylation and sumoylation, which have been associated with the transcriptionally active form of p53 (21, 23, 37). Whereas p53 modification by conjugation to SUMO-1 on a single lysine residue (K386) is thought to enhance the transactivation activity of p53, multiple phosphorylation sites appear to be required for p53 stability and transcriptional activities.
The present study examines the role of in vivo phosphorylation of the newly recognized T81 site in response to various stress stimuli and shows that JNK is the phosphorylating kinase. Support for T81 as the JNK phosphoacceptor site has been provided by seven independent approaches, including (i) in vivo phosphorylation of p53 by JNKK as revealed by mass-spectrometric analysis of immunopurified p53, (ii) in vitro phosphorylation of bacterially expressed p53 by a purified form of active JNK, (iii) lack of phosphorylation by p38, (iv) inhibition of T81 phosphorylation by the dominant-negative form of JNK, (v) lack of T81 phosphorylation on p53 that lacks the JNK binding site, (vi) inhibition of JNK phosphorylation in vivo in cells that exhibit the nearly null JNK environment generated by JNK antisense oligonucleotides, and (vii) inhibition of T81 phosphorylation in vivo by JNK phosphatase MKP5.
Phosphorylation at this site is of crucial importance in relation to p53's ability to elicit transcriptional activation under conditions where JNK is activated (i.e., UV) and was abolished in p53 mutated at T81. Abrogated transactivation of p53 impaired its ability to elicit growth arrest, growth inhibition, and programmed cell death, all of which are key regulatory events in p53-mediated responses of cells to DNA damage. A lower degree of nuclear accumulation, as revealed by our confocally based immunohistochemistry analysis, may account for some, but not all, of the observed decrease in T81A p53's ability to elicit transcriptional activities of p21, Mdm2, or Bax. Whereas 50% of the T81A form of p53 is localized within the nucleus, 90% of p53 transcriptional activities were lost. Furthermore, T81A p53's loss of function also cannot be attributed to altered conformation, since peptide digestion revealed a pattern similar to that of the wt but not that of the conformationally altered 248 mutant. Lack of a significant difference in the half-life of p53 under nonstressed conditions also supports the notion that the T81A protein is under regulation similar to that of the wt counterpart.
Independent confirmation of the role of JNK phosphorylation of T81 on p53 was provided by the use of JNK1 and -2 antisense oligonucleotides and JNK phosphatase MKP5. Expression of antisense JNK1 and -2 efficiently attenuated UV-mediated T81 phosphorylation in cells. Along those lines, forced expression of MKP5 effectively reduced T81 phosphorylation following JNKK expression. A less-pronounced effect was seen in UV-treated cells, probably as result of the activation of additional p53 kinases that also contribute to p53 stability as well as to the attenuation of MKP5's activities. MKP5 was also efficient in attenuating both JNKK-mediated p53 transcriptional activity and JNKK-dependent p53-elicited apoptosis. Taken together, our data provide direct support for the finding that JNK phosphorylation of p53 takes place at T81 and that this phosphorylation is important for p53 stability and activities.
It is important to note that p53 phosphorylation by JNK is conformation sensitive; attempts to phosphorylate in vitro the GST-p53 fusion protein with JNK failed unless the GST portion of the protein was first removed (2). In this connection it is notable that an earlier study suggested that JNK phosphorylates p53, but at a residue different from that reported in the present study (38). This discrepancy may also relate to the use of murine p53; the site in question (Ser 34) has no human counterpart.
It is necessary to understand the relationship between phosphorylation on T81 and that on other sites that are phosphorylated in response to DNA damage, including Ser 15, 20, 33, 46, and 389. Growing evidence suggests that the type and dose of stress or damage incurred are important factors in determining the type of kinases induced and the pattern of p53 phosphorylation (8). It is likely that phosphorylation on certain residues may precede that on others. Accordingly, phosphorylation at one site may be required to acquire a certain conformation, enabling subsequent phosphorylation events to take place on other domains of p53. Such a model is supported by the finding that Ser 15 phosphorylation is affected by mutation of Ser 33 and Ser 46, which are p38 phosphorylation sites (7), which may explain the different kinetics that are often observed in an examination of p53 phosphorylation on different residues. Along these lines, although phosphorylation of p53 by JNK at T81 is important for p53 transactivation, it is likely to be augmented by other stress- and DNA damage-responsive kinases (e.g., ATM, p38, Chk2) that phosphorylate p53 on other residues. The requirement for coordinated phosphorylation of p53 by different kinases implies that JNK phosphorylation on T81 may not be sufficient for p53 stabilization and transcriptional activities, thereby explaining why certain JNK activators (e.g., interleukin 1) do not stabilize or activate p53 (41). Lack of JNK phosphorylation under such conditions may be overruled by other stress-inducible kinases that can phosphorylate p53 on other residues.
An important indication of the importance of T81 in p53 functions comes from the observation that close to 200 human tumors contain p53 with mutations within the domain that encompasses JNK binding and JNK phosphorylation sites. Those mutants are expected to exhibit impaired JNK association and phosphorylation and, as a result, to lack transcriptional activities.
In establishing the role of JNK and T81 in the stability and activity of p53, our study identifies an additional mechanism underlying the p53-dependent cellular response to stress and damage.
ACKNOWLEDGMENTS
We thank Victor Adler, Karen Quadrini, and Ekaterina Matusevich for technical assistance, Roger Davis, Michael Karin, and Audrey Minden for stress kinase constructs, Xei Wu and Toru Ouchi for p53 constructs, and Hidetoshi Tahara for NHF. We also thank James Manfredi for advice on conformation analysis.
This study was supported by grant CA78419 from the National Cancer Institute (to Z.R.) and by an NIH shared instrumentation grant (1 S10 RR0 9145-01) and NSF Major Research Instrumentation grant (DBI-9 724504).
REFERENCES
- 1.Adler V, Schaffer A, Kim J, Dolan L, Ronai Z. UV-irradiation and heat shock mediate JNK activation via alternate pathways. J Biol Chem. 1995;270:26071–26077. doi: 10.1074/jbc.270.44.26071. [DOI] [PubMed] [Google Scholar]
- 2.Adler V, Pincus M R, Minamoto T, Fuchs S Y, Bluth M J, Brandt-Rauf P W, Friedman F K, Robinson R C, Chen J M, Wang X W, Harris C C, Ronai Z. Conformation-dependent phosphorylation of p53. Proc Natl Acad Sci USA. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashcroft M, Taya Y, Vousden K H. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 5.Beroud C, Soussi T. p53 gene mutation: software and database. Nucleic Acids Res. 1998;26:200–204. doi: 10.1093/nar/26.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 7.Bulavin D V, Saito S, Hollander M C, Sakaguchi K, Anderson C W, Appella E, Fornace A J., Jr Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18:6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buschmann T, Adler V, Matusevich E, Fuchs S Y, Ronai Z. p53 phosphorylation and association with murine double minute 2, c-Jun NH2-terminal kinase, p14ARF, and p300/CBP during the cell cycle and after exposure to ultraviolet irradiation. Cancer Res. 2000;60:896–900. [PubMed] [Google Scholar]
- 9.Chao C, Saito S, Kang J, Anderson C W, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chehab N H, Malikzay A, Stavridi E S, Halazonetis T. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumaz N, Meek D W. Serine 15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Diery W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J N, Lin D, Mercer W E, Kinzier K W, Vogelstien B. Waf1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander P, Legros Y, Soussl T, Prives C. Regulation of mutant p53 temperature-sensitive DNA binding. J Biol Chem. 1996;271:25468–25478. doi: 10.1074/jbc.271.41.25468. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs S Y, Xie B, Adler V A, Fried V A, Davis R J, Ronai Z. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J Biol Chem. 1997;272:32163–32168. doi: 10.1074/jbc.272.51.32163. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs S Y, Dolan L R, Davis R, Ronai Z. Phosphorylation dependent targeting of c-jun ubiquitination by JNK. Oncogene. 1996;13:1529–1533. [PubMed] [Google Scholar]
- 16.Fuchs S Y, Fried V A, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs S Y, Adler V, Buschmann T, Yin Z, Wu X, Jones S N, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs S Y, Adler V, Pincus M R, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs S Y, Ronai Z. Ubiquitination and degradation of ATF2 are dimerization dependent. Mol Cell Biol. 1999;19:3289–3298. doi: 10.1128/mcb.19.5.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatti A, Li H H, Traugh J A, Liu X. Phosphorylation of human p53 on Thr-55. Biochemistry. 2000;39:9837–9842. doi: 10.1021/bi992454i. [DOI] [PubMed] [Google Scholar]
- 21.Giaccia A, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 22.Glickman M H, Rubin D M, Fried V A, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;95:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 25.Haupt Y, Maya R, Kazaz A, Oren M. MDM2 promotes the rapid degradation of p53. Nature. 1999;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 26.Haupt Y, Rowan S, Shaullan E, Kazaz A, Vousden K, Oren M. p53 mediated apoptosis in HeLa cells: transcription dependent and independent mechanisms. Leukemia. 1997;3:339. [PubMed] [Google Scholar]
- 27.Hengstermann A, Whitaker N J, Zimmer D, Zentgraf H, Scheffner M. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene. 1998;17:2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 28.Hermeking H, Lengauer C, Polyak K, He T, Zhang L, Thiagalingan S, Kinzier K W, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 29.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Ma W, Maxiner A, Sun Y, Dong Z. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J Biol Chem. 1999;274:12229–12235. doi: 10.1074/jbc.274.18.12229. [DOI] [PubMed] [Google Scholar]
- 31.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 32.Khanna K, Keating K, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, Lavin M F. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 33.Kim A L, Raffo A J, Brandt-Rauf P W, Pincus M R, Monaco R, Abarzua P, Fine R L. Conformation and molecular basis for induction of apoptosis by a p53 c-terminal peptide in human cancer cells. J Biol Chem. 1999;274:34924–34931. doi: 10.1074/jbc.274.49.34924. [DOI] [PubMed] [Google Scholar]
- 34.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 35.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H, Fisher R P, Bailey P, Levine A J. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53 enhancing its sequence-specific DNA binding activity in vitro. Mol Cell Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meek D W. Multiple phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 38.Milne D M, Campbell L E, Campbell D G, Meek D W. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 39.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa A, Taya Y, Tamal K, Yamaizumi M. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol Cell Biol. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naica A, Rangnekar V M. The G1-phase growth-arresting action of interleukin-1 is independent of p53 and p21/WAF1 function. J Biol Chem. 1998;273:30517–30523. doi: 10.1074/jbc.273.46.30517. [DOI] [PubMed] [Google Scholar]
- 42.Ndubuisi M I, Guo G G, Fried V A, Etllnger J D, Sehgal P B. Cellular physiology of STAT3; where's the cytoplasmic monomer? J Biol Chem. 1999;274:25499–25509. doi: 10.1074/jbc.274.36.25499. [DOI] [PubMed] [Google Scholar]
- 43.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 44.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L, Santee S, Fujiwara T, Roth J, Deisseroth A B, Zhang W-W, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polyak K, Xia Y, Zweler J, Kinzler K, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 46.Potapova O, Gorospe M, Dougherty R H, Dean N M, Gaarde W A, Holbrook N J. Inhibition of c-Jun N-terminal kinase 2 expression suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner. Mol Cell Biol. 2000;20:1713–1722. doi: 10.1128/mcb.20.5.1713-1722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prives C, Hall P A. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Rosenfeld J, Capdeville J, Guillernot J C, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 49.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez-Prieto R, Rojas J M, Taya Y, Gutkind J S. A role for the p38 mitogen-activated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res. 2000;60:2464–2472. [PubMed] [Google Scholar]
- 51.Shieh S-Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphoryiate p53 at multiple DNA damage inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 52.Shieh S-Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siliciano J, Canman C, Taya Y, Sakaguchi K, Apella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takenaka I, Morin F, Seizinger B, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 55.Theodosiou A, Smith A, Gillieron C, Arkinstall S, Ashworth A. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene. 1999;18:6981–6988. doi: 10.1038/sj.onc.1203185. [DOI] [PubMed] [Google Scholar]
- 56.Tishler R B, Lamppu D M, Park S, Price B D. Microtubule-active drugs taxol, vinblastine, and nocodazole increase the levels of transcriptionally active p53. Cancer Res. 1995;55:6021–6025. [PubMed] [Google Scholar]
- 57.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unger T, Mietz J A, Scheffner M, Yee C, Howley P M. Functional domains of wild-type and mutant p53 proteins involved in transcriptional regulation, transdominant inhibition, and transformation suppression. Mol Cell Biol. 1993;13:5186–5194. doi: 10.1128/mcb.13.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Unger T, Sionov R, Moallem E, Yee C, Howley P M, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 60.Velculescu V E, El-Deiry W S. Biological and clinical importance of the p53 tumor suppressor gene. Clin Chem. 1996;42:858–868. [PubMed] [Google Scholar]
- 61.Walker K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]