Abstract
Due to bursts in the expression of thousands of germline-specific genes, the testis has the most diverse and complex transcriptome of all organs. By analyzing the male germline of mice, we demonstrate that the genome-wide reorganization of super-enhancers (SEs) drives bursts in germline gene expression after the mitosis-to-meiosis transition. SE reorganization is regulated by two molecular events: the establishment of meiosis-specific SEs via A-MYB (MYBL1), a key transcription factor for germline genes, and the resolution of SEs in mitotically proliferating cells via SCML2, a germline-specific Polycomb protein required for spermatogenesis-specific gene expression. Prior to entry into meiosis, meiotic SEs are preprogrammed in mitotic spermatogonia to ensure the unidirectional differentiation of spermatogenesis. We identify key regulatory factors for both mitotic and meiotic enhancers, revealing a molecular logic for the concurrent activation of mitotic enhancers and suppression of meiotic enhancers in the somatic and/or mitotic proliferation phases.
Introduction
Meiosis is an essential step in the preparation of haploid gametes, and the transition from mitotic proliferation to meiosis is a fundamental event in the maturation of germ cells. In the male germline, this mitosis-to-meiosis transition coincides with a fundamental alteration to the transcriptome: a dynamic and massive change in genome-wide gene expression1–5. Because of bursts in the expression of thousands of germline genes, the testis has the most diverse, complex, and rapidly evolved transcriptome of all organs3,6,7. During spermatogenesis, the mitosis-to-meiosis transition accompanies the dynamic reorganization of epigenetic modifications, accessible chromatin, and 3D chromatin conformation—all in preparation for the next generation of life5,8–13. However, it remains largely unknown how DNA regulatory elements underlie massive, dynamic transcriptional changes in the mammalian male germline.
Enhancers play key roles in the control of cell type-specific gene expression programs through the binding of transcription factors (TFs) and interactions with promoters14–16. Some of these enhancers aggregate to regulate the expression of genes important for establishing cellular identity17–20; such aggregates have been termed “super-enhancers” (SEs)17. SEs are prevalent in various cell and tissue types and are also found in cancer cells, where they direct the expression of key tumor pathogenesis genes20. However, for the most part, the characterization of SEs has been limited to somatic and/or mitotically proliferating cells. Given the massive scale of the mitosis-to-meiosis transcriptome alteration, there are compelling questions as to the detailed profiles of active enhancers in spermatogenesis as well as the existence of meiosis-specific SEs.
Here, we determine the profiles of active enhancers in representative stages of mouse spermatogenesis. We identify a meiotic type of SE, and we demonstrate that the dynamic transcriptome at the mitosis-to-meiosis transition is driven by a switch from mitotic to meiotic SEs. Through functional and systematic analyses, we identified key regulatory factors for both mitotic and meiotic enhancers. Our analyses expose the molecular logic for SE switching at the mitosis-to-meiosis transition, revealing activation mechanisms for mitotic enhancers that are concurrent with suppression mechanisms for meiotic enhancers in the somatic and/or mitotic proliferation phases.
Results
The landscape of active enhancers during spermatogenesis
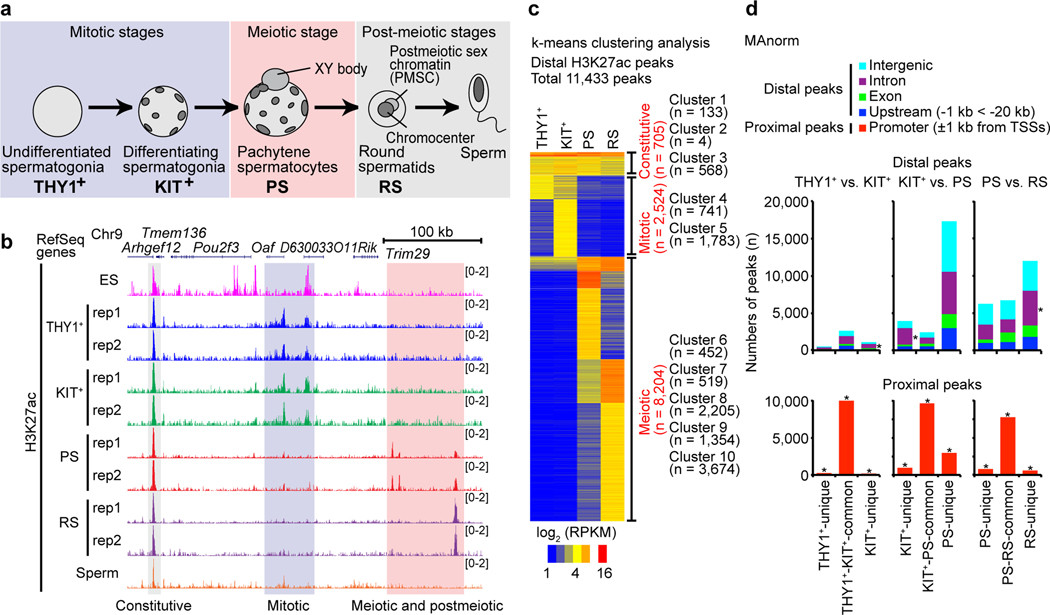
To determine the landscape of active enhancers in spermatogenesis, we performed chromatin immunoprecipitation with sequencing (ChIP-seq) for the histone modification H3K27ac, a marker of active enhancers21. We analyzed four representative stages of wild-type spermatogenesis: THY1+ undifferentiated spermatogonia, a population that contains both spermatogonial stem cells and progenitor cells; KIT+ differentiating spermatogonia; pachytene spermatocytes (PS) in meiotic prophase; and postmeiotic round spermatids (RS) (Fig. 1a). We carried out H3K27ac cross-linking ChIP-seq with ChIPmentation22 for two independent biological replicates and obtained reproducible, detailed profiles of putative active enhancers during spermatogenesis (see Methods, Fig. 1b, Extended Data Fig. 1a). Consistent with the massive, dynamic transcriptional changes that occur at the mitosis-to-meiosis transition, we observed three types of H3K27ac peaks: (1) those in mitotically proliferating spermatogonia (i.e., the “mitotic phase:” blue shadow); (2) those that appeared in meiotic spermatocytes and continued to grow in postmeiotic spermatids (red shadow); and (3) constitutive peaks (gray shadow: Fig. 1b).
Figure 1. The landscape of active enhancers during spermatogenesis.
(a) Schematic of mouse spermatogenesis and the four representative stages analyzed in this study: THY1+, undifferentiated spermatogonia; KIT+, differentiating spermatogonia; PS, pachytene spermatocytes; and RS, round spermatids. (b) Track views of H3K27ac ChIP-seq enrichment with biological replicates for representative stages of spermatogenesis. ES: embryonic stem cells. (c) k-means clustered heatmap of distal H3K27ac peaks (detected by MACS2) in spermatogenesis. (d) MAnorm analysis for H3K27ac peaks at each transition of spermatogenesis. The genomic distribution for each peak is shown with colored bars. H3K27ac peak-enrichment for each genomic entity (intergenic, intronic, etc.) was compared to randomly selected genomic regions. *P < 0.00001, chi-square test with Yates’s correction. Data for panels in c and d are available as source data.
For the quantitative comparison of putative active enhancers during spermatogenesis, we analyzed H3K27ac ChIP-seq peaks ±1 kb outside transcription start sites (TSSs: “distal peaks”, see Methods). We detected 11,433 distal H3K27ac ChIP-seq peaks that were present in at least one stage of spermatogenesis (Supplementary Data Set 1). Through k-means clustering, the distal peaks were categorized into three classes (10 clusters) as follows (Fig. 1c): (1) The first class (705 peaks) represents constitutive active enhancers, i.e., those observed throughout spermatogenesis; (2) the second class (2,524 peaks) represents enhancers that are active in the mitotic phase but are inactive in meiotic and postmeiotic phases; and (3) the third class (8,204 peaks) consists of enhancers that are largely inactive in the mitotic phase yet are highly active in meiotic and postmeiotic stages. In contrast to a previous view that enhancer activation may not be involved in mouse spermatogenesis9, these results demonstrate that large numbers of putative enhancers are activated de novo in spermatogenesis.
To elucidate how active enhancers change in the course of spermatogenesis, we examined the dynamics of H3K27ac peaks at various transitions from one stage to another. H3K27ac-proximal peaks (within ±1 kb of TSSs: promoters) were largely common at each transition, and THY1+ and KIT+ spermatogonia share a common active-enhancer profile (distal H3K27ac peaks) (Fig. 1d). In contrast, there is a dynamic change in the distribution of active enhancers at the mitosis-to-meiosis transition (i.e., between KIT+ spermatogonia and PS). These results suggest that a majority of active enhancers in KIT+ spermatogonia disappear prior to meiosis, and an extensive de novo formation of active enhancers takes place in meiotic prophase. Additionally, the level of H3K27ac is augmented at the promoters in the KIT+ to PS transition (Extended Data Fig. 1b). The continued alteration of active enhancers occurred from meiotic PS to postmeiotic RS, and we observed an additional de novo activation of enhancers in RS (Fig. 1d).
Robust expression of key spermatogenesis genes is facilitated by de novo establishment of super-enhancers after the mitosis-to-meiosis transition
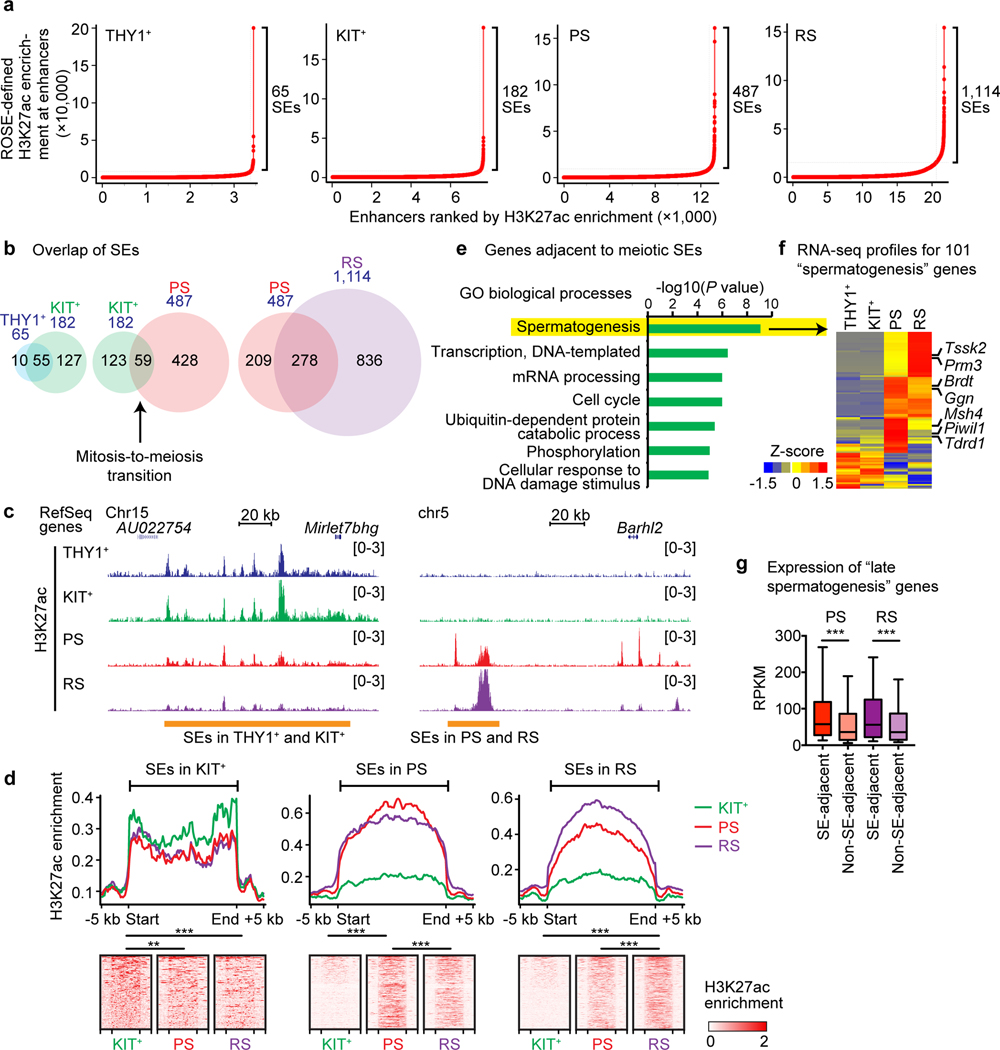
To elucidate how massive, dynamic transcriptional change is stimulated at the mitosis-to-meiosis transition, we sought to test the following hypothesis: The transcriptional change of the mitosis-to-meiosis transition is associated with the establishment of SEs. SEs have been defined as large chromatin domains enriched with H3K27ac and/or other active enhancer marks23; drawing on this definition, we identified SEs based on elevated H3K27ac enrichment in spermatogenesis using the same criteria and algorithm as previously described17,19. We found that SEs are established in the course of spermatogenesis, and we also found they increase in number as germ cells mature: We identified 65 SEs in THY1+ spermatogonia, 182 SEs in KIT+ spermatogonia, 487 SEs in PS, and 1,114 SEs in RS (Fig. 2a, Supplementary Data Set 2). Among the 65 SEs in THY1+ spermatogonia, 85% (55/65) are common to SEs identified in KIT+ spermatogonia (Fig. 2b), indicating a common SE profile in mitotically dividing THY1+ and KIT+ spermatogonia. However, among the 182 SEs in KIT+ spermatogonia, only 32% (59/182) are common to SEs in PS (Fig. 2b). These data reveal the dynamic, de novo formation of SEs at the mitosis-to-meiosis transition. After the mitosis-to-meiosis transition, 57% (278/487) of SEs in PS were common to RS; we observed the establishment of 836 new SEs in RS (Fig. 2b).
Figure 2. The identification of super-enhancers during spermatogenesis.
(a) Identification of super-enhancers (SEs) in representative stages of spermatogenesis. H3K27ac signal (as defined by ROSE) enrichment represents, for each enhancer region, density × length. (b) SE overlap at the stage transitions of spermatogenesis. (c) Track views of H3K27ac ChIP-seq enrichment for representative SEs in spermatogenesis. (d) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment at SEs. H3K27ac enrichment values: RPM. (e) Gene ontology analysis of genes adjacent to meiotic SEs (i.e., genes within 20 kb upstream-to-50 kb downstream of SEs). (f) RNA-seq profiles for 101 “spermatogenesis” genes out of SE adjacent genes. (g) Box-and-whisker plots show the distributions of RNA-seq RPKM values for late spermatogenesis genes. Among 2,623 “late spermatogenesis” genes (i.e., genes that are not highly expressed in spermatogonia but are highly expressed in PS and/or RS; a gene list is included in Supplementary Data Set 4), 652 genes are located adjacent to meiotic SEs (genes within 20 kb upstream-to-50 kb downstream of SEs). Central bars represent medians, the boxes encompass 50% of data points, and the whiskers indicate 90% of the data points. ***P < 0.0001, Mann-Whitney U test. Data for panels in f and g are available as source data.
We identified distinct characteristics for SEs common to the mitotic stages (i.e., between THY1+ and KIT+ spermatogonia) as well as distinct characteristics for SEs common to the meiotic (i.e., PS) and postmeiotic (i.e., RS) stages. First, with respect to SEs common to mitotic stages (THY1+ and KIT+ spermatogonia: termed “mitotic SEs”), H3K27ac was decreased in PS but tended to persist throughout spermatogenesis into at least as late as the RS stage (Fig. 2c, d). On the other hand, with respect to PS and RS SEs (termed “meiotic SEs”), H3K27ac was largely absent from corresponding genomic loci in THY1+ and KIT+ spermatogonia, and was robustly established during the mitosis-to-meiosis transition (Fig. 2c, d). Intriguingly, meiotic SEs tend to consist of large and broad H3K27ac peaks, while mitotic SEs tend to comprise clusters of distinct, narrow H3K27ac peaks (Fig. 2c, d); SEs with this distinct, narrow conformation have been reported to be a general feature of tissue-specific SEs in mitotically proliferating cells17,18,20.
Gene ontology analysis revealed that genes adjacent to meiotic SEs are enriched for roles in “spermatogenesis” (Fig. 2e). We identified 101 genes that were categorized for “spermatogenesis,” and this gene group is highly expressed after the mitosis-to-meiosis transition (Fig. 2f, Supplementary Data Set 3). This group includes key regulators of spermatogenesis such as Brdt, a key spermatogenic chromatin regulator24; Trdr1 and Piwil1 (also known as Miwi), components of the PIWI-interacting RNA pathway25,26; Msh4, an essential gene for meiotic recombination27; and other spermatogenesis genes, including Ggn, Prm3, and Tssk2 (Fig. 2f). As well, among 2,623 late spermatogenesis genes (Supplementary Data Set 4), we found that genes adjacent to meiotic SEs are subject to higher expression than genes that are not (Fig. 2g). These results suggest that the robust expression of key spermatogenesis genes is facilitated by the de novo establishment of meiotic SEs.
A-MYB establishes meiotic SEs for the targeted activation of germline genes
Enhancers contain transcription factor (TF)-binding sites that regulate the expression of target genes28. To identify TF-binding sites indicative of the mitotic phase versus the meiotic phase, we compared H3K27ac ChIP-seq peaks between KIT+ spermatogonia and PS. Among the H3K27ac peaks unique to KIT+, the most significant TF-binding motif was one common to STAT family transcription factors (STAT1, STAT3, and STAT5A, Fig. 3a). This observation is in line with the function of STAT3 in spermatogonial differentiation29. The second-most-significant motif was one common to FOX family transcription factors (FOXK1, FOXJ3, and FOXG1, Fig. 3a).
Figure 3. A-MYB-binding sites occupy a central location within the H3K27ac peaks of meiotic SEs.
(a) Representative TF-binding sites detected via MEME-ChIP of in H3K27ac enrichment in KIT+ vs. PS. E values: expected values output by the MEME expectation maximization algorithm. (b) Track views of H3K27ac ChIP-seq enrichment at meiotic SEs in KIT+ vs. PS. (c) MAnorm analysis for whole testis tissue A-MYB peaks and PS H3K27ac peaks. (d) Pie charts represent the genomic-entity distributions of A-MYB-H3K27ac-common distal peaks as detected by MAnorm. Meiotic SEs are shown apart from all other enhancers. (e) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment around the summits of A-MYB-binding peaks at SEs (left) and all other enhancers (right). H3K27ac enrichment values: RPM. (f) Overlap between SEs and piRNA clusters, and the numbers of A-MYB-binding peaks within piRNA cluster-meiotic SE intersections. Data for panels in d and f are available as source data.
Next, we sought to understand how and to what extent the motif enrichment data for KIT+ spermatogonia differ from motif enrichment data for KIT+ spermatogonia reported in our previous study of accessible chromatin (detected via ATAC-seq; like H3K27ac ChIP-seq peaks, ATAC-seq peaks are indicators of cis-regulatory elements)10. Notably, in KIT+ spermatogonia, consensus motifs for DMRT1, a key TF that regulates the mitosis-to-meiosis transition30, were enriched only in ATAC-positive/H3K27ac-negative peaks, suggesting that DMRT1 functions outside of active enhancers (Extended Data Table 1, Supplementary Data Set 5). A similar feature was also found in PS: The binding motifs for the POU/OCT family of TFs (POU2F, POU3F, and POU1F1) were found only in ATAC-positive/H3K27ac-negative peaks, suggesting that the POU/OCT family of TFs functions outside of active enhancers in PS (Extended Data Table 1). Among the TFs that have binding motifs in PS H3K27ac peaks, nine TFs are highly expressed in PS, suggesting their involvement in the regulation of meiotic enhancers; the nine candidate TFs are A-MYB, a key TF for germline genes31; CREM, another key TF in spermatogenesis32; ELF2; OVOL1; OVOL2; TCFL5; RFX1; RFX4; and RFX8 (Extended Data Fig. 2).
Among the candidate TFs, binding motifs of A-MYB (MYBL1: GGCAGTT), a key transcription factor for germline genes31,33, exhibit the most significant enrichment relative to PS-unique motifs found within H3K27ac peaks (Fig. 3a). Of note, we identified A-MYB ChIP-seq peaks at the center of SE-associated H3K27ac peaks in PS (Fig. 3b). Genome-wide, most A-MYB peaks overlap H3K27ac peaks (Fig. 3c). Consistent with the reported function of A-MYB on promoters33, the majority of peaks common to A-MYB and H3K27ac are located at promoters (Fig. 3c). However, among the peaks common to A-MYB and H3K27ac, 518 are located at meiotic SEs and 2,005 are located at other meiotic enhancers (Fig. 3d). Many of these peaks are located in intergenic regions (Fig. 3d), and H3K27ac peaks in PS can be found around the summits of A-MYB peaks (Fig. 3e), suggesting a function for A-MYB in the activation of meiotic enhancers and SEs.
Importantly, we found that meiotic SEs overlap pachytene piRNA clusters, which produce pachytene piRNAs (Fig. 3b, right panel; and Fig. 3f). Among pachytene piRNA clusters, we found that BTBD18-dependent piRNA loci are highly likely to overlap meiotic SEs (Fig. 3f); BTBD18 is an essential factor for pachytene piRNA production by way of transcriptional elongation34. A-MYB is essential for the production of pachytene piRNA33, and the numbers of A-MYB peaks are in proportion to the numbers of meiotic SEs on piRNA clusters (Fig. 3f). Thus, A-MYB and meiotic SEs may comprise, in part or whole, a potential mechanism for the production of pachytene piRNA.
To understand the functions of A-MYB in spermatogenesis, we examined A-myb mutant (Mybl1repro9) mice, which exhibit complete meiotic arrest in the mid pachytene stage and subsequent cell death (Extended Data Fig. 3)31. In normal meiosis, H3K27ac was present throughout the nucleus with enrichment on the X chromosome domain in the pachytene stage (Fig. 4a)35. Yet in A-myb mutants, meiotic nuclei were largely devoid of H3K27ac and, when present, H3K27ac was clustered in large, ectopic puncta (arrowheads in Fig. 4a) and along the axis of chromosome X (magnified panel in Fig. 4a). These observations indicate a global alteration of H3K27ac deposition in A-myb mutants.
Figure 4. A-MYB establishes meiotic SEs for the targeted activation of germline genes.
(a) Chromosome spreads of wild-type (WT) and A-myb mutant (A-myb Mut) pachytene spermatocytes (PS) immunostained with antibodies raised against SYCP3 and H3K27ac. Dashed squares: sex chromosomes, which are magnified in panels at bottom. White arrowheads indicate ectopic clusters of H3K27ac signal. Immunostaining intensity in arbitrary units (a.u.) is quantified by densitometry across the indicated path (‘ to ”). Scale bars, top panels: 10 μm; bottom panels: 5 μm. (b) Track views of H3K27ac ChIP-seq enrichment for representative meiotic SEs in WT and A-myb Mut PS. (c) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment at SEs in WT and A-myb Mut PS. H3K27ac enrichment values: RPM. (d) RNA-seq analysis of A-myb Mut vs. A-myb heterozygous control testes at postnatal day 14 (P14). The 1,705 genes exhibiting significant changes in expression P adj < 0.01, binomial test with Benjamini-Hochberg correction) in A-myb mutants are represented by blue circles. ***P = 1.0 × 10−79, hypergeometric probability test. 211 dysregulated meiotic SE-adjacent genes (red circles) ÷ 652 meiotic SE-adjacent genes; 1,705 dysregulated differentially expressed genes (blue circles) ÷ 22,661 NCBI RefSeq genes. (e) Activation of a representative meiotic SE via CRISPR activation (CRISPRa). Top: Schematic for CRISPRa experiments. Four guide RNAs (gRNAs) were designed to target adjacent regions (less than 1kb) from two A-MYB binding sites (Two gRNAs for each A-MYB binding site). Bottom: CRISPRa-dependent expression of the gene Zfp37 as measured by qRT-PCR. Error bars represent as mean ± s.e.m.: *P < 0.05, **P < 0.01, unpaired t tests. Three biological replicates were examined. (f) A model for the A-MYB-dependent establishment of meiotic SEs. Data for panels in a, d, and e are available as source data.
To evaluate the role of A-MYB in the establishment of meiotic SEs, we performed ultra-low-input native ChIP-seq36 for H3K27ac in A-myb mutant PS due to their limited availability. H3K27ac deposition was significantly reduced at sites of meiotic SEs in A-myb mutant PS compared to wild-type controls (Fig. 4b, c). Consistent with this result, we observed a significant overlap of meiotic SE adjacent genes and genes differentially expressed in A-myb mutants at postnatal day 14 (P14)35, and many of the differentially expressed genes were found to be downregulated in A-myb mutants. Meanwhile, there is no significant correlation between A-MYB enrichment on the promoters and the degree of gene deregulation of these SE target genes in the A-myb mutants (Extended Data Fig. 3d), further supporting the function of meiotic SEs in the regulation of adjacent genes. To confirm the activation of meiotic genes by adjacent meiotic SEs, we performed CRISPR activation (CRISPRa) experiments using embryonic stem (ES) cells, in which meiotic SEs and germline genes are not active. Using doxycycline (Dox)-inducible CRISPRa ES cells (J1 ES cells harboring a Dox-inducible dCas9-VPR transgene), the germline gene Zfp37, which is adjacent to a meiotic SE, was activated upon introduction of four guide RNAs (gRNAs) targeting its adjacent meiotic SE (Fig. 4e). Induction of A-MYB expression further stimulated CRISPRa-mediated activation (Fig. 4e). Taken together, we conclude that A-MYB establishes meiotic SEs that activate adjacent germline genes (Fig. 4f).
SCML2 facilitates the resolution of mitotic SEs during meiosis
Next, we sought to determine a mechanism underlying the resolution of mitotic SEs at the mitosis-to-meiosis transition. We focused our investigation on the function of SCML2, a germline specific Polycomb protein that is responsible for suppression of somatic/progenitor genes on autosomes after the mitosis-to-meiosis transition4. Since Scml2-knockout (KO) spermatogenesis does not exhibit meiotic arrest4, we were able to obtain sufficient numbers of mutant cells to perform a cross-linking-with-ChIPmentation method for H3K27ac22. Although H3K27ac peaks were comparable between Scml2-KO mice and wild-type littermate controls in THY1+ and KIT+ spermatogonia (Extended Data Fig. 4a), a large number of intergenic and intronic H3K27ac peaks were found in Scml2-KO PS compared to wild-type PS (Fig. 5a). Intriguingly, the increased numbers of H3K27ac peaks in Scml2-KO PS appeared to result from the retention of mitotic enhancers after the mitosis-to-meiosis transition: H3K27ac peaks at mitotic SEs, which were resolved in wild-type PS, were, for the most part, retained in Scml2-KO PS and RS (Fig. 5b, Extended Data Fig. 4b). These results suggest that the SCML2-mediated resolution of mitotic SEs constitutes a potential mechanism for the suppression of somatic/progenitor genes.
Figure 5. SCML2 is required for the resolution of mitotic SEs during meiosis.
(a) MAnorm analysis for H3K27ac peaks in wild-type (WT) and Scml2-knockout (Scml2-KO) PS and RS. (b) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment at KIT+ SEs in representative stages of spermatogenesis (KIT+, PS, RS). H3K27ac enrichment values: RPM. (c) A model for the SCML2-dependent resolution of mitotic SEs. (d) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment at meiotic SEs in PS and RS. (e) RNA-seq analysis of Scml2-KO vs. wild-type PS. The 4,812 genes exhibiting significant changes in expression (P adj < 0.01, binomial test with Benjamini-Hochberg correction) in Scml2-KO are represented by blue circles. **P = 0.00422, hypergeometric probability test. 6 differentially expressed genes (represented by red circles) ÷ 9 meiotic SE-associated TFs; 4,812 differentially expressed genes ÷ 22,661 NCBI RefSeq genes. Data for panels in a and e are available as source data.
SCML2 is required for the establishment of H3K27me3 during meiosis, forming two major classes of bivalent promoters comprised of H3K27me3 and H3K4me2/3: Class I domains, which are associated with developmental regulator genes; and Class II domains, which are associated with somatic/progenitor genes37. We observed an increase in H3K27ac signal intensity at promoters of both classes of bivalent genes in Scml2-KO mice (Extended Data Fig. 4c). We presume that this is the consequence—at least in part—of an antagonistic relationship between H3K27me3 and H3K27ac at promoters, since both post-translational modifications occupy the same amino acid residue (K27) of the histone H3 tail.
Next, we sought to evaluate the binding of SCML2 to mitotic SEs and the establishment of H3K27me3 upon the resolution of mitotic SEs. We found that SCML2 peaks overlap with H3K27ac peaks at mitotic SEs (Extended Data Fig. 5a). Although the majority of SCML2 peaks appeared on promoters, SCML2 peaks are present on mitotic SEs and other enhancers in spermatogonia (Extended Data Fig. 5b, c). H3K27me3 was established at SCML2-containing mitotic SEs after the resolution of mitotic SEs (Extended Data Fig. 5d). These data suggest SCML2 may bind a portion of mitotic SEs to establish H3K27me3 (Fig. 5c).
We also observed a decrease in H3K27ac intensity at meiotic SEs in Scml2-KO PS and RS (Fig. 5d)—observations that are consistent with the down-regulation of late-spermatogenesis genes in Scml2-KO PS and RS4. Since SCML2 is a suppressor, we suspected that meiotic SEs were indirectly impaired in Scml2-KO PS and RS. Of note, six out of nine of the candidate regulators of meiotic enhancers (identified in Extended Data Fig. 2b and including A-MYB) were downregulated in Scml2-KO PS (Fig. 5e). Therefore, our results suggest meiotic SEs are indirectly downregulated in Scml2-KO PS and RS.
SCML2 is required for the formation of SEs on the X chromosome during meiosis
Switching our focus to meiosis, we performed analyses to elucidate mechanisms governing the activation of enhancers on the male sex chromosomes. During male meiosis, the sex chromosomes undergo regulation distinct from autosomes due to meiotic sex chromosomes inactivation (MSCI)38,39. MSCI engages a DNA damage response (DDR) pathway to regulate gene silencing in PS 40. RNF8, an E3 ligase and key DDR factor, is responsible for the establishment of ubiquitination and active histone modifications—including H3K27ac—on the sex chromosomes during meiosis, thereby regulating the activation of a subset of sex chromosome genes that escape post-meiotic silencing35,41. While we observed a paucity of distal H3K27ac peaks on the sex chromosomes of THY1+ and KIT+ spermatogonia, we identified the de novo establishment of H3K27ac peaks in the mitosis-to-meiosis transition (Fig. 6a).
Figure 6. The distinct regulation of meiotic SEs on autosomes versus the sex chromosomes.
(a) MAnorm analysis for H3K27ac peaks on the sex chromosomes in THY1+ and KIT+ spermatogonia, and in KIT+ spermatogonia and PS. (b) MAnorm analysis for H3K27ac peaks on the sex chromosomes in PS and RS in wild-type and Scml2-KO. (c) Numbers of SEs on the X chromosome in each stage of spermatogenesis. (d) Numbers of SEs on the X chromosome and autosomes in PS. (e) Track views of H3K27ac ChIP-seq enrichment at a representative meiotic SE in spermatogenesis. (f, g) Box-and-whisker plots show the distributions of ChIP-seq enrichment around autosomal meiotic SEs (f) and X-linked meiotic SEs (g). Central bars represent medians, the boxes encompass 50% of the data points, and the whiskers indicate 90% of the data points. ***P < 0.0001, Mann-Whitney U test. (h) Model showing poised meiotic SEs in THY1+ spermatogonia facilitate the establishment of active meiotic SEs in pachytene spermatocytes. Data for panels in a, b, d, f, and g are available as source data.
To dissect the regulatory mechanisms underlying this process, we focused on SCML2, which has a critical regulatory function on the sex chromosomes independent of its functions on autosomes4. SCML2 functions downstream of the DDR pathway that initiates MSCI, where it cooperates with RNF8 to establish H3K27ac35. On the sex chromosomes of PS and RS, a large portion of distal H3K27ac peaks (particularly those in intergenic and intronic regions) depend on SCML2 (Fig. 6b). Interestingly, ATAC-seq peaks appeared specifically on the sex chromosomes of PS in an SCML2-dependent fashion too10. Thus, SCML2 is a key regulatory factor for chromatin accessibility and H3K27ac deposition on the meiotic sex chromosomes. Accordingly, 26 SEs are established on the X chromosomes in meiosis (Fig. 6c), and these largely depend on SCML2 (Fig. 6d). This is unlike SCML2’s function to resolve mitotic SEs (Fig. 5); as such, we observed increased numbers of SEs on the autosomes of Scml2-KO PS (Fig. 6d). These results demonstrate autosome- and sex chromosome-specific functions for SCML2 in the regulation of enhancers in spermatogenesis.
Meiotic super-enhancers on autosomes are poised in undifferentiated spermatogonia
Since SEs on the sex chromosomes are established downstream of the DDR pathway in meiosis, we suspected SEs on the autosomes are regulated by a distinct mechanism. Thus, to determine the mechanism by which autosomal SEs are established, we examined the epigenetic status of meiotic SEs in progenitor cells, specifically the active marks H3K4me2 and H3K4me3, which were previously reported to be associated with poised gene promoters during spermatogenesis5. Notably, prior to the establishment of H3K27ac, H3K4me2 was present on autosomal meiotic SEs in THY1+ and KIT+ spermatogonia (Fig. 6e, f). Additionally, H3K4me3 is also enriched on autosomal meiotic SEs in THY1+ spermatogonia (Fig. 6f). These features were unique to meiotic SEs; other meiotic enhancers detected through analyses of distal H3K27ac peaks did not exhibit these features (Extended Data Fig. 6a). These results suggest that meiotic SEs are poised as early as the THY1+ spermatogonia phase to prepare for the expression of key spermatogenesis genes after the mitosis-to-meiosis transition. Such features were not observed on meiotic SEs associated with the X chromosome (Fig. 6g), lending further support for the distinct regulation of meiotic SEs between autosomes and chromosome X. We also found that the TSSs of late spermatogenesis genes are broadly poised for activation in spermatogonia (Extended Data Fig. 6b)5 and that the TSSs of genes adjacent to these SEs retain similar epigenetic patterns in each stage (Extended Data Fig. 7). Together, these data indicate that SE-associated late spermatogenesis genes on autosomes are poised for activation in two layers: SEs and TSSs (Fig. 6h). We propose that this form of epigenomic programming ensures the unidirectional differentiation of spermatogenesis.
Next, we sought to identify mechanisms for the expression of postmeiotic spermatid-specific genes. On autosomes, a proportion of distal H3K27ac peaks around RS-specific genes are poised with H3K4me2 in THY1+ spermatogonia (Extended Data Fig. 6c). On the PS X chromosome, at distal H3K27ac peaks around RS-specific genes, H3K27ac and H3K4me2 became temporarily enriched (Extended Data Fig. 6d)35. These results underscore distinct gene activation mechanisms for autosomes versus the sex chromosomes in haploid RS.
Identification of key regulatory factors for both mitotic and meiotic enhancers
Finally, we took advantage of our new data sets to infer general mechanisms underlying the regulation of mitotic and meiotic enhancers. Since the meiotic gene program is largely repressed in cell types that undergo mitotic divisions, we sought to identify putative TFs that meet one of two counteractive conditions: (1) Those that can operate on and/or promote the activity of mitotic enhancers, and (2) those that can suppress meiotic enhancers. To this end, we used our recently published Regulatory Element Locus Intersection (RELI) algorithm42 to compare the genomic locations of our H3K27ac peak data with a large collection of publicly available ChIP-seq data. Taking the genomic location information for H3K27ac peaks detected to be mitotic enhancers in KIT+ spermatogonia, we analyzed the intersections between these data and publicly available ChIP-seq data sets for many TFs in many contexts. Since the overwhelming majority of public ChIP-seq data are from somatic cells that undergo mitotic divisions in between cell cycles, this informs (at least) one interpretation for such an experiment: The enrichment of intersections between mitotic enhancers and TFs could be indicative of general mechanisms that operate on mitotic enhancers. In addition to TFs that were previously associated with spermatogonia such as STAT3, TCF3, MAZ, and ETS129,43,44, we identified additional factors with enriched ChIP-seq peaks at distal H3K27ac peaks in KIT+ spermatogonia: SRF, TCF12, GATA4, BCL6, CEBPB, and MAX (Fig. 7a, Supplementary Data Set 6). When we applied the same analysis to mitotic SEs, we identified UBTF, RBP1, CHD1, ZFX, and KLF4 as specific factors that may be involved in their regulation (Fig. 7b, Supplementary Data Set 7). Among them, ZFX was previously implicated in spermatogenesis45. Next, we applied this strategy to identify factors that suppress meiotic enhancers in the mitotic phase. Compellingly, at sites of meiotic enhancers as determined by the loci of distal H3K27ac peaks in PS, we revealed high enrichment for factors that comprise, in part, transcriptional silencing machinery, including REST, TRIM28, RCOR2, SIN3A, and YY1 (Fig. 7c, Supplementary Data Set 8). Of note, when we applied this analysis to meiotic SEs in the mitotic phase, we identified KDM5A, a histone demethylase that acts on H3K4me3, as the factor having the highest enrichment at meiotic SEs (Fig. 7d, Supplementary Data Set 9). Together, these analyses systematically identify putative regulators of mitotic and meiotic enhancers, providing insightful clues as to their underlying molecular mechanisms.
Figure 7. The identification of key regulatory factors for mitotic and meiotic enhancers, and SEs.
(a, b) Identification of putative regulators for mitotic enhancers and SEs. The y-axis indicates the -log of the P value for the overlap between publicly available ChIP-seq datasets for various TFs and mitotic enhancers (a) or SEs (b) based on the RELI algorithm (see Methods). TFs of interest are highlighted. (c, d) Comparisons between TFs enriched in meiotic PS and KIT+ enhancers (c) or SEs (d). The y-axis indicates the ratio of the -log of the P-value for the overlap between publicly available ChIP-seq datasets for various TFs and enhancers (c) or SEs (d) based on the RELI algorithm.
Discussion
In this study, we determined the profiles of putative active enhancers in representative stages of spermatogenesis and demonstrated that SE-switching underlies the dynamic transcriptome change of the mitosis-to-meiosis transition. Using an improved H3K27ac ChIP-seq method, we were able to profile active enhancers at high resolution and with high signal-to-noise ratios. Since SEs regulate gene expression that underlies cellular identity17,18, the A-MYB-dependent regulation of meiotic SEs becomes a conceivable mechanism for gene expression that defines the cellular identity of germ cells in late spermatogenesis. Our analyses revealed SE-adjacent genes that are critical for late spermatogenesis. In particular, Piwil1 and Tdrd1 are involved in the regulation of piRNA25,26. Thus, meiotic SEs may be central to the robust production of pachytene piRNA that, in turn, determines cellular identity. Interestingly, the A-MYB-dependent activation of germline genes is an ancient mechanism also found in rooster testes33. Given the robust and evolutionarily conserved nature of germline gene activation via SEs, such a mechanism stands in stark contrast to a concomitant mechanism whereby rapidly evolved enhancers, driven by endogenous retroviruses, activate species-specific germline genes (Sakashita et al.,)46.
In tumor cells, SEs are regulated by BRD4, a member of the bromodomain and extraterminal (BET) subfamily of proteins19. In spermatogenesis, a testis-specific member of the BET family, BRDT, is required for the meiotic gene expression program24,47. Given the molecular similarities between BRD4 and BRDT, it is possible that BRDT could be a binding protein for meiotic SEs, and loss of function of BRDT could represent loss of function of meiotic SEs. Curiously, another protein containing bromodomains, BRWD1, can also recognize acetylated lysine residues and is required for postmeiotic transcription in spermatids48. Likewise, BRD4 is also associated with gene expression in spermatids49. Thus, BRWD1 and BRD4 may be involved in the increased numbers of active enhancers established in the transition from PS to RS. Of note, many of the highly expressed genes in RS are initially transcribed in PS5, and SEs that have been identified in RS were initially enriched with K3K27ac in PS (Fig. 2d). Hence, it is likely that, in PS, A-MYB may also initiate RS SE formation to facilitate RS gene expression.
Our study has shown that SCML2 mediates distinct forms of regulation for active enhancers on autosomes versus active enhancers on sex chromosomes. On the autosomes, SCML2, a highly expressed protein in undifferentiated spermatogonia4, is involved in the resolution of mitotic SEs after the mitosis-to-meiosis transition (Fig. 5), while meiotic SEs are already poised with H3K4me2 in undifferentiated spermatogonia (Fig. 6). Therefore, it is conceivable that dual mechanisms preprogram meiotic gene expression in progenitor cells, defining unidirectional differentiation of spermatogenesis. On the other hand, our results indicate that active enhancers and postmeiotic gene expression are directly downstream of a DDR pathway specific to the sex chromosomes.
Finally, through genome-wide analyses, we revealed transcription factors that might bind mitotic and meiotic enhancers, as well as mitotic and meiotic SEs (Fig. 7). Among the factors we identified, the transcriptional repressor KDM5A (also known as RBP2 or JARID1A), which evinced the highest enrichment value for meiotic SEs, is of particular interest because KDM5A was originally implicated in tumorigenesis50. Since many germline genes are expressed in many cancer types—so-called cancer/testis genes51—it is interesting to consider that the regulation of meiotic SEs could, in turn, drive or otherwise regulate germline gene expression in various cancers.
In summary, our current study provides a framework to understand the regulation of gene expression during spermatogenesis. Because our study focuses on representative stages, it will be important to further dissect the complex and well-coordinated nature of spermatogenesis. Recent studies using single-cell analyses have revealed new details for the transcriptomes of progressive cell types in human and mouse spermatogenesis52–57. Such dynamism is achievable through the functional interplay of TFs and enhancers, as well as other regulatory elements. Indeed, more than a thousand TFs are differentially expressed in spermatogenesis10. Of note, the testis has the largest number of specifically expressed TFs of all organs58. The systematic determination of germline cis-regulatory elements makes for a compelling future research direction. And given the evolutionary divergent nature of piRNA loci in modern humans59, investigation of the evolutionary aspect of meiotic SEs makes for a promising research direction too.
Methods
Animals.
Mice were maintained and used according to the guidelines of the Institutional Animal Care and Use Committee (protocol no. IACUC2018–0040) at Cincinnati Children’s Hospital Medical Center. A-mybmut/mut (Mybl1repro9) mice, which were ENU-induced on the C57BL/6J background, have been previously reported31,33. Scml2-knockout (KO) mice on the C57BL/6J background have been previously reported4.
Cell lines.
Wild-type J1 male embryonic stem cells (henceforth “ES cells”) derived from male agouti 129S4/SvJae embryos have been described previously60. CRISPRa ES cell line has been generated in Sakashita et al.46. Since these cells were easily distinguished based on colony morphologies, cell lines have been authenticated by microscopic inspection. None of the cell lines has been tested for mycoplasma contamination.
Cell culture.
ES cells were cultured in ESC media (15% FBS, 25 mM HEPES, 1× GlutaMAX, 1× MEM Non-essential Amino Acids Solution, 1× Penicillin/Streptomycin, and 0.055 mM β-Mercaptoethanol in DMEM High Glucose (4.5 g/L)) containing 2i (1 μM PD0325901, LC Laboratories; and 3μM CHIR99021, LC Laboratories) and LIF (1300 U/mL, in-house) on cell culture plates coated with 0.2% gelatin under feeder-free conditions. The expanded ES colonies were dissociated using 0.25% trypsin-EDTA solution for passaging.
Germ cell fractionation.
Wild-type and Scml2-KO male mice on the C57BL/6J background (at least 12 independent mice at 90–120 days of age or at least 30 independent mice at 7 days of age) were used for isolation of germ cells for ChIP-seq experiments.
Pachytene spermatocytes and round spermatids were isolated via BSA gravity sedimentation as previously described61. Purity was confirmed by nuclear staining with Hoechst 33342 using fluorescence microscopy. In keeping with previous studies from the Namekawa lab4,10,12,37, only fractions with a mean purity of ≥90% were used to generate Next Generation Sequencing data sets.
Spermatogonia were isolated as described previously9 and collected from C57BL/6J wild-type or Scml2-KO mice aged 6–8 days. Testes were collected in a 24-well plate in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with GlutaMax (Thermo Fisher Scientific), non-essential amino acids (NEAA) (Thermo Fisher Scientific), and penicillin and streptomycin (Thermo Fisher Scientific). After removing the tunica albuginea membrane, testes were digested with collagenase (1 mg/ml) at 34°C for 20 min to remove interstitial cells, then centrifuged at 188×g for 5 min. Tubules were washed with the medium and then digested with trypsin (2.5 mg/ml) at 34°C for 20 min to obtain a single cell suspension. Cells were filtered with a 40-μm strainer to remove Sertoli cells, and the cell suspension was plated in a 24-well plate for 1 h in the medium supplemented with 10% fetal bovine serum, which promotes adhesion of remaining somatic cells. Cells were washed with magnetic cell-sorting (MACS) buffer (PBS supplemented with 0.5% BSA and 5 mM EDTA) and incubated with CD117 (KIT) MicroBeads (Miltenyi Biotec) on ice for 20 min. Cells were washed and resuspended with MACS buffer and filtered with a 40-μm strainer. Cells were separated by autoMACS Pro Separator (Miltenyi Biotec) with the program “possel.” Cells in the flow-through fraction were washed with MACS buffer and incubated with CD90.2 (THY1) MicroBeads (Miltenyi Biotec) on ice for 20 min. Cells were washed and resuspended with MACS buffer and filtered with a 40-μm strainer. Cells were separated by autoMACS Pro Separator (Miltenyi Biotec) with the program “posseld.” Purity was confirmed by immunostaining.
Histology and immunofluorescence analyses.
Wild-type and A-myb mutant male mice on the C57BL/6J background (three independent mice, at 90–120 days of age) were used for histological analysis. For preparation of testicular paraffin blocks, testes were fixed with 4% paraformaldehyde (PFA) overnight at 4°C with gentle inverting. Testes were dehydrated and embedded in paraffin. For histological analysis, 5 μm-thick paraffin sections were deparaffinized and stained with hematoxylin and eosin. For immunofluorescence analysis of testicular sections, antigen retrieval was performed by boiling the slides in target retrieval solution (DAKO) for 10 min and letting the solution cool for 30 min. Sections were blocked with Blocking One Histo (Nacalai) for 1 h at room temperature and then incubated with anti-H1T antibody (1/1000 dilution, gift from Dr. Mary Ann Handel) overnight at 4°C. The resulting signals were detected by incubation with secondary antibodies conjugated to fluorophores (Thermo Fisher Scientific). Sections were counterstained with DAPI. Images were obtained via TiE fluorescence microscopy (Nikon) and processed with NIS-Elements (Nikon) and ImageJ (National Institutes of Health)62.
Preparation and immunofluorescence of meiotic chromosome spreads.
Meiotic chromosome spreads from testes were prepared as described63. Briefly, testes were excised from mice and detunicated in 1× PBS. Seminiferous tubules were dissociated from whole testes, and approximately one-quarter of the tubules were transferred into each well of 4-well plate containing 1 ml hypotonic extraction buffer (HEB: 30 mM Tris base, 17 mM trisodium citrate, 5 mM ethylenediaminetetraacetic acid (EDTA), 50 mM sucrose, 5 mM dithiothreitol (DTT), 1× cOmplete Protease Inhibitor Cocktail (Sigma, 11836145001)). The seminiferous tubules were incubated in HEB on ice for 2 h with gentle stirring every 15 min. After incubation, a small clump of seminiferous tubules was gently pulled and mashed between tweezer tips in 30 μL of sucrose (100 mM) on an uncharged microscope slide (Gold Seal: ThermoFisher Scientific, 3010–002). Testicular cell suspension was applied to positively charged slides; prior to application of the suspension, the slides were incubating in a chilled fixation solution (2% paraformaldehyde, 0.1% Triton X-100, 0.02% sodium monododecyl sulfate, adjusted to pH 9.2 with sodium borate buffer) for a minimum of 2 min. After applying the cell suspension/sucrose mixture, the slides were placed in humid chambers and incubated overnight at RT. The following day, slides were washed twice with 0.4% Photo-Flo 200 (Kodak, 146– 4510), at RT, 2 min per wash. Slides were dried completely at RT before staining or storage in slide boxes at −80°C.
For immunofluorescence analysis of meiotic chromosome spreads, slides were washed once with PBS containing 0.1% Tween 20 (PBST) and blocked with antibody dilution buffer (PBST containing 0.15% BSA) for 30 min at RT. Then, slides were incubated with primary antibodies overnight at 4°C. This study made use of the following primary antibodies: anti-H3K27ac (1/500 dilution, ab4729, Abcam) and anti-SYCP3 (1/1000, ab97672, Abcam). The resulting signals were detected via incubation with secondary antibodies conjugated to fluorophores (Thermo Fisher Scientific). Slides were counterstained with DAPI. Images were obtained with a TiE fluorescence microscope (Nikon) and processed with NIS-Elements (Nikon) and ImageJ (National Institutes of Health)62.
Cross-linking ChIP-seq library preparation with the ChIPmentation method and sequencing.
We used ~2–4 × 106 THY1+ and KIT+ spermatogonia, ~1–2 × 106 PS, ~5–10 × 106 RS isolated from wild-type and Scml2-KO testes. Cells were suspended in chilled 1× PBS. One-eleventh volume of crosslinking solution (50 mM HEPES-NaOH pH 7.9, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 8.8% formaldehyde) was added to the cell suspension and incubated on ice for 8 min. One-twentieth volume of 2 M glycine was added to the cell suspension and incubated at room temperature for 5 min to stop the reaction. Cells were washed twice with PBS, frozen at −80°C, and lysed at 4°C for 10 min each in ChIP lysis buffer 1 (50 mM HEPES pH 7.9, 140 mM NaCl, 10% glycerol, 0.5% IGEPAL-630, 0.25% Triton X-100). After centrifugation at 2,000×g for 10 min at 4°C, pellets were resuspended with ChIP lysis buffer 2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) and incubated at 4°C for 10 min. After centrifugation at 2,000×g for 10 min at 4°C, pellets were washed with TE containing 0.1% SDS and protease inhibitors (Sigma; 11836145001) and resuspended with the same buffer. Chromatin was sheared to approximately 200–500 bp by sonication using a Covaris sonicator at 10% duty cycle, 105 pulse intensity, 200 burst for 2 min. Sheared chromatin was cleared by centrifugation at 20,000×g for 20 min, followed by pre-incubation with Dynabeads Protein G (Thermo Fisher Scientific). Chromatin immunoprecipitation was carried out on an SX-8X IP-STAR compact automated system (Diagenode). Briefly, Dynabeads Protein G were pre-incubated with 0.1% BSA for 2 h. Then, the cleared chromatin was incubated with beads conjugated to antibodies against H3K27ac (Active Motif, 39133) at 4°C for 8 h, washed sequentially with wash buffer 1 (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% NaDOC, and 1% Triton X-100), wash buffer 2 (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% NaDOC, and 1% Triton X-100), wash buffer 3 (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NaDOC, and 0.5% NP-40), wash buffer 4 (10 mM Tris-HCl pH 8.0, 1 mM EDTA, and 0.2% Triton X-100), and wash buffer 5 (10 mM Tris-HCl).
DNA libraries were prepared through the ChIPmentation method22. Briefly, beads were resuspended in 30 μl of the tagmentation reaction buffer (10 mM Tris-HCl pH 8.0 and 5 mM MgCl2) containing 1 μl Tagment DNA Enzyme from the Nextera DNA Sample Prep Kit (Illumina) and incubated at 37°C for 10 min in a thermal cycler. The beads were washed twice with 150 μl cold wash buffer 1, incubated with elution buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 250 mM NaCl, 0.3% SDS, 0.1 μg/μl Proteinase K) at 42°C for 30 min, and then incubated at 65°C for another 5 h to reverse cross-linking. DNA was purified with the MinElute Reaction Cleanup Kit (Qiagen) and amplified with NEBNext High-Fidelity 2× PCR Master Mix (NEB). Amplified DNA was purified by Agencourt AMPure XP (Beckman Coulter). Afterwards, DNA fragments in the 250- to 500-bp size range were prepared by agarose gel size selection. DNA libraries were adjusted to 5 nM in 10 mM Tris-HCl pH 8.0 and sequenced with an Illumina HiSeq 2500.
Please note: while generated for and analyzed in this study, our H3K27ac ChIP-seq data for wild-type PS and RS were initially introduced in another study that analyzed active enhancers on the sex chromosomes35.
Ultra-low-input native ChIP-seq library preparation and sequencing.
Ultra-low-input native ChIP-seq36 library preparation and sequencing were performed using small numbers of A-myb mutant PS and littermate wild-type PS and described in Sakashita et al.46.
CRISPR activation.
We performed functional evaluations of a representative SE locus adjacent to the Zfp37 gene locus using the CRISPRa ES cells that was established in Sakashita et al., co-submitted. We chose the Zfp37 gene locus because A-MYB binding was not observed in the Zfp37 promoter. To that end, we used CRISPOR tools64 to design four gRNAs for the locus at flanking regions of two A-MYB binding sites at the SE locus. Four gRNA sequences are as follows: Zfp37-se_sgRNA_1: GAU CGG UCG GGU CGG UUG UC; Zfp37-se_sgRNA_2: GCC GAC CCA GUC GAU GUG UC; Zfp37-se_sgRNA_3: UCC ACC UUU GGC UUA GUC CC; Zfp37-se_sgRNA_4: ACU AAC UUU AGU GGA GCU GA. The gRNAs were synthesized as TrueGuide Modified Synthetic sgRNA (Thermo Fisher). One day before transduction, 2×105 CRISPRa ES cells were seeded into each well of a 24-well plate coated with 0.2% gelatin. At day 1, transient transfections were performed with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher) following the manufacturer’s instructions. 240 ng of equimolar pooled sgRNA was used. At day 2, 500 ng of A-MYB expression vector (PGK-A-MYB plasmid: established in Sakashita et al., co-submitted) was transfected with Lipofectamine 3000 Transfection Reagent (Thermo Fisher) following the manufacturer’s instruction. The cell culturing media was changed with ESC media without 2i and LIF, containing hygromycin B Gold with or without 1 μg/mL doxycycline every day after one wash with PBS. At day 4, the adherent cells in each well were lysed for RNA extraction.
RNA extraction and RT-qPCR.
Total RNA was isolated using an RNeasy Plus Mini Kit (Qiagen). First-strand cDNA synthesis was performed using 200 ng of total RNA with the SuperScript IV Reverse Transcriptase and oligo-dT (20) primer (Thermo Fisher) according to the manufacturer’s instructions. Real-time PCR was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems) with Fast SYBR Green Master Mix (Thermo Fisher) and specific primer sets (Zfp37-RT_Fw: ATACGAAGCTAGAGAACTGCAG; Zfp37-RT_Rv: TCAGACTACTTGGGCTGGCT.). Relative gene expression was quantified with the ΔΔCT method and normalized to Hprt expression.
ChIP-seq and RNA-seq data analysis.
Data analysis for ChIP-seq was performed in the BioWardrobe Experiment Management System (https://biowardrobe.com/landing)65. Briefly, reads were aligned to mouse genome mm10 with Bowtie (version 1.2.0)66, assigned to NCBI RefSeq genes or isoforms, and coverage was displayed on a local mirror of the UCSC genome browser.
ChIP-seq peaks for H3K27ac, H3K4me2, H3K4me3, and A-MYB were identified using MACS2 (version 2.1.1.20160309)67 with default parameter setting for narrow peak detection in BioWardrobe. For the MACS2 analysis of H3K27ac, we permitted only peaks ±1 kb outside transcription start sites, i.e., ‘distal peaks,’ with a normalized enrichment value of ≥4. In this study, we refer to peaks ±1 kb outside TSSs as ‘distal peaks,’ and we refer to peaks within ±1 kb of TSSs as ‘proximal peaks.
Pearson correlations for the genome-wide enrichment of H3K27ac peaks among ChIP-seq library replicates were analyzed using SeqMonk (Babraham Institute).
MAnorm, software designed for quantitative comparisons of ChIP-seq data sets68, was used to compare the genome-wide ChIP-seq peaks among stages in spermatogenesis. Unique peaks were defined using the following criteria: (1) defined as “unique” by the MAnorm algorithm; (2) P-value <0.01; (3) raw counts of unique reads >10. Peaks common to two stages were defined using the following criteria: (1) defined as “common” by MAnorm algorithm; (2) raw read counts of both stages >10.
SEs were identified based on elevated H3K27ac enrichment in spermatogenesis using the exactly same criteria and algorism as previously described17,19. The reads were normalized for each dataset before differential peak calling. We used normalized H3K27ac ChIP-seq data and MACS2 peak calls to rank all enhancers in spermatogenic cell type categories by increasing the total background (i.e., input)-subtracted ChIP-seq occupancy of H3K27ac; this was done by calling the ROSE_main.py program18,27 with default parameters. We plotted the total background (i.e., input)-subtracted ChIP-seq occupancy of H3K27ac. Enhancers within 12.5 kb of each other were consolidated into a single entity. The visualized plot allowed us to define a clear inflection point to separate SEs in each cell type from all other enhancers. Enhancer entities enriched with an H3K27ac signal above the inflection point were defined as SEs.
Average tag density profiles were calculated around transcription start sites for gene sets of somatic/progenitor genes, late spermatogenesis genes, constitutive active genes, and constitutive inactive genes as described previously5. Resulting graphs were smoothed in 200-bp windows. Enrichment levels for ChIP-seq experiments were calculated for 4-kb windows, promoter regions of genes (±2 kb surrounding TSSs) and enhancer regions. To normalize tag value, read counts were multiplied by 1,000,000 and then divided by the total number of reads in each nucleotide position. The total amount of tag values in promoter or enhancer regions were calculated as enrichment. The program ngs.plot was used to draw heatmaps for ChIP-seq read enrichment.
The k-means clustering of differential enhancer peaks were analyzed using Cluster 3.0 software. The results were further analyzed using JavaTreeview software69 to visualize as heatmaps.
MEME-ChIP70 was used for motif discovery as described in the text. For all motif analyses, we used only peak regions (±250 bp from the peak summit) outside of ±1 kb from TSSs; we chose a maximum of 3,000 peak regions from the lowest P-values (P <0.01) via MAnorm analysis, and we extracted those sequences using the Table Browser71.
HOMER motif analyses72 were performed using an expanded TF-binding motif library taken from the Cis-BP database73). The HOMER software package72 was used for motif enrichment analyses using a customized version of HOMER that employs a log2 scoring system and motifs contained in the Cis-BP motif database73.
RELI (Regulatory Element Locus Intersection) analysis was performed as described42. In brief, genomic regions of interest (e.g., ChIP-seq peaks) were systematically aligned with a large collection of publicly available ChIP-seq data for various TFs in various cellular contexts largely taken from mouse ENCODE, and the significance of the intersection of each data set was calculated using RELI.
Analyses of RNA-seq data were performed in the BioWardrobe Experiment Management System65. Briefly, reads were aligned by STAR (version STAR_2.5.3a)74 with default arguments except --outFilterMultimapNmax 1 and --outFilterMismatchNmax 2. The --outFilterMultimapNmax parameter was used to allow unique alignments only, and the --outFilterMismatchNmax parameter was used to allow a maximum of 2 errors. NCBI RefSeq annotation from the mm10 UCSC genome browser 75 was used, and canonical TSSs (1 TSS per gene) were analyzed. All reads from the resulting .bam files were split for related isoforms with respect to RefSeq annotation. Then, the EM algorithm was used to estimate the number of reads for each isoform. To detect differentially expressed genes between two biological samples, a read count output file was input to the DESeq2 package (version 1.16.1); then, the program functions DESeqDataSetFromMatrix and DESeq were used to compare each gene’s expression level between two biological samples. Differentially expressed genes were identified through binominal tests, thresholding Benjamini-Hochberg-adjusted P values to <0.01. To perform gene ontology analyses, the functional annotation clustering tool in DAVID (version 6.8) was used and a background of all mouse genes was applied. Biological process term groups with a significance of P < 0.05 (modified Fisher’s exact test) were considered significant.
Statistics.
Statistical methods and p values for each plot are listed in the figure legend and/or in the corresponding Methods section. In brief, all grouped data are represented as mean ± s.e.m. All box-and-whisker plots are represented as: center lines, median; box limits, interquartile range (25 and 75 percentiles); whiskers, >90% of the data points. Unless stated otherwise. Statistical significances for pairwise comparisons were determined using the two-sided Mann-Whitney U tests and unpaired t-tests. All quantitative analyses are represented as the mean ± s.e.m. of three biological replicates. Fisher exact test and hypergeometric test were used for detection of significantly enriched GO terms, genes and loci compared with backgrounds. Differentially expressed genes were determined in DESeq2 package. NGS data (RNA-seq and ChIP-seq) are based on two independent replicates. For all experiments, no statistical methods were used to predetermine sample size. Experiments were not randomized and investigator were not blinded to allocation during experiments and outcome assessments.
Extended Data
Extended Data Fig. 1. Biological replicates for H3K27ac ChIP-seq data.
(a) Scatter plots show the reproducibility of H3K27ac ChIP-seq enrichment at individual peaks between biological replicates. Each peak was identified using MACS (P < 1×10−5). H3K27ac ChIP-seq enrichment levels are shown in log2 RPKM values. The color scale indicates H3K27ac ChIP-seq peak density. Pearson correlation values (R) are shown. While generated for and analyzed in this study, our H3K27ac ChIP-seq data for wild-type PS and RS were initially introduced in another study that analyzed active enhancers on the male sex chromosomes (these data are adapted from Adams et al., PLOS Genet 2018)35. (b) Average tag density and heatmaps for H3K27ac ChIP-seq at proximal peak regions in KIT+ and PS. Proximal peaks identified in Fig.1d were used for this analysis.
Extended Data Fig. 2. Identification of candidate TFs whose binding sites are enriched in meiotic SEs.
Identification of candidate TFs whose binding sites are enriched in meiotic SEs. HOMER analyses identify 72 TFs that have binding motifs in meiotic SEs. Among the 72 TFs, 9 TFs are highly expressed in PS and RS (≥4-fold in comparison to their expression levels in spermatogonia).
Extended Data Fig. 3. Spermatogenic phenotypes of A-myb mutant mice.
(a) Testis and epididymis sections from wild-type (WT) littermate control (left panels) and A-myb mutants (right panels) at 8 weeks of age. The sections were stained with hematoxylin and eosin. Scale bars: 200 μm. (b) Testis sections from WT and A-myb mutant mice immunostained with antibodies raised against H1T. Scale bars: 200 μm. Numbers of H1T+ cells per seminiferous tubule as mean and whiskers, which indicate 25% (bottom) and 75% (top) of the data points from two independent littermate pairs (right panel). ***P < 0.001, unpaired t test. (c) Chromosome spreads of wild-type and A-myb mutant PS immunostained with antibodies raised against SYCP3 and H3K27ac. Scale bars: 10 μm. Late pachytene spermatocytes were not detected (N.D.) in A-myb mutant samples. (d) Scatter plot depicts relationship between A-MYB enrichments at promoters (TSS±1 kb) and dysregulation of meiotic SE-adjacent genes that were also differentially expressed in A-myb mutant testis (211 genes identified in Figure 4D). Red line represents a regression line. R: Pearson correlation coefficient.
Extended Data Fig. 4. Comparison of H3K27ac ChIP-seq enrichment between wild-type and Scml2-KO cells.
(a) MAnorm analysis for H3K27ac peaks in THY1+ and KIT+ spermatogonia between wild-type and Scml2-KO. (b) Track views of H3K27ac ChIP-seq enrichment on representative mitotic SEs in spermatogenesis. (c) Average tag densities and heatmaps for H3K27ac ChIP-seq enrichment at genomic bivalent domains in PS and in RS.
Extended Data Fig. 5. SCML2 binds to and regulates the resolution of mitotic SEs.
(a) Track views of SCML2 enrichment in GS cells and H3K27ac and H3K27me3 ChIP-seq enrichment at a representative mitotic SE in spermatogenesis. Light blue bars represent SCML2-binding sites with the peaks of H3K27ac in spermatogonia. After the mitosis-to-meiosis transition, SCML2 establishes H3K27me3 at these sites. (b) MAnorm analysis for SCML2 peaks in GS and THY1+-H3K27ac peaks. (c) Pie charts represent the genomic-entity distributions of SCML2-H3K27ac-common distal peaks detected by MAnorm. Meiotic SEs are shown apart from all other enhancers. (d) Average tag densities and heatmaps for H3K27ac and H3K27me3 ChIP-seq signal at mitotic SEs that intersect with SCML2 peaks (n = 84). H3K27ac enrichment values: RPM.
Extended Data Fig. 6. ChIP-seq enrichment at various genomic loci.
(a–d) Box-and-whisker plots show the distributions of enrichment for ChIP-seq enrichment for the indicated genomic loci. Central bars represent medians, the boxes encompass 50% of the data points, and the whiskers indicate 90% of the data points.
Extended Data Fig. 7. Enrichment of H3K4me2, H3K4me3, and H3K27me3 at the promoters of genes adjacent to SEs.
Box-and-whisker plots show the distributions of ChIP-seq enrichment at TSSs ±2 kb for genes adjacent to SEs in spermatogenesis. Central bars represent medians, the boxes encompass 50% of the data points, and the whiskers indicate 90% of the data points.
Supplementary Material
Acknowledgements
We thank members of the Namekawa and Maezawa laboratories for discussion and helpful comments regarding the manuscript. We also thank the CCHMC Research Flow Cytometry Core for sharing MACS equipment, X. Li at the University of Rochester Medical Center for sharing A-myb mutant mice, and M.A. Handel at the Jackson Laboratory for sharing the H1T antibody. Funding sources: The research project grant by the Azabu University Research Services Division, Ministry of Education, Culture, Sports, Science and Technology (MEXT)-Supported Program for the Private University Research Branding Project (2016–2019), Grant-in-Aid for Research Activity Start-up (19K21196), the Takeda Science Foundation (2019), and the Uehara Memorial Foundation Research Incentive Grant (2018) to S.M.; Lalor Foundation Postdoctoral Fellowship and JSPS Overseas Research Fellowships to A.S.; Albert J. Ryan Fellowship to K.G.A.; CCHMC Endowed Scholar and CpG grant awards to M.T.W.; National Institute of Health (NIH) DP2 GM119134 to A.B.; NIH R01 GM122776 and GM098605 to S.H.N.
Footnotes
Code availability. Source code for all software and tools used in this study with documentation, examples and additional information, is available at following URLs:
・https://github.com/alexdobin/STAR (STARRNA-seq aligner)
・http://younglab.wi.mit.edu/super_enhancer_code.html (ROSE)
・http://crispor.tefor.net (CRISPOR)
・https://pypi.org/project/MACS2 (MACS2)
・https://bedtools.readthedocs.io/en/latest/content/installation.html (BEDTools)
・https://bioconductor.org/packages/release/bioc/html/DESeq2.html (DESeq2)
・https://david.ncifcrf.gov/summary.jsp (DAVID)
・https://www.bioinformatics.babraham.ac.uk/projects/seqmonk (SeqMonk)
・https://github.com/shenlab-sinai/ngsplot (ngsplot)
・https://github.com/tidyverse/ggplot2 (ggplot2)
・http://homer.ucsd.edu/homer (HOMER)
・http://great.stanford.edu/public/html (GREAT)
・https://imagej.net/Fiji/Downloads (Fiji—ImageJ)
Information for the BioWardrobe Experiment Management Platform, which is commercial software, is available at https://biowardrobe.comandhttps://github.com/Barski-lab/biowardrobe.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Cross-linking H3K27ac ChIP-seq data reported in this study have been deposited to the Gene Expression Omnibus (GEO) under the accession number GSE130652. H3K27ac native ChIP-seq data in WT and A-myb mutant PS reported in this study are described in the accompanying study (Sakashita et al.)46 and deposited under the accession number GSE142173. All other next-generation sequencing datasets used in this study are publicly available. RNA-seq data from THY1+ spermatogonia, PS, and RS were downloaded from the Gene Expression Omnibus (accession number: GSE55060)4. ATAC-seq data from KIT+ spermatogonia and PS were downloaded from the Gene Expression Omnibus (accession number: GSE102954)10. ChIP-seq data for A-MYB and RNA-seq data from A-MYB mutant and control testes were downloaded from the Gene Expression Omnibus (accession number: GSE44690)33. ChIP-seq data for H3K4me3, and H3K4me2, and RNA-seq data from KIT+ spermatogonia, were downloaded from the Gene Expression Omnibus (GSE89502)37. While generated for and analyzed in this study, our H3K27ac ChIP-seq data for wild-type PS and RS were initially introduced in another study that analyzed active enhancers on the sex chromosomes35; ChIP-seq data for H3K27ac in wild-type PS and RS were downloaded from the Gene Expression Omnibus (GSE107398)35. ChIP-seq data for H3K27ac from embryonic stem cells was downloaded from Gene Expression Omnibus (GSE29184)76. ChIP-seq data for H3K27ac from sperm were downloaded from Gene Expression Omnibus (accession number: GSE79230)77.
The source data for Figs. 1c,d; 2f,g,3;d,f; 4a,d,e; 5a,e; 6a,b,d,f,g are available with the paper online.
Competing Interest Statement
A.B. is a cofounder of Datirium, LLC.
References
- 1.Shima JE, McLean DJ, McCarrey JR & Griswold MD The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71, 319–30 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Namekawa SH et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16, 660–7 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Soumillon M. et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3, 2179–90 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa K. et al. SCML2 Establishes the Male Germline Epigenome through Regulation of Histone H2A Ubiquitination. Dev Cell 32, 574–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sin HS, Kartashov AV, Hasegawa K, Barski A. & Namekawa SH Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline. BMC Biol 13, 53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramskold D, Wang ET, Burge CB & Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brawand D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–8 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Kimmins S. & Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature 434, 583–9 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Hammoud SS et al. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239–53 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Maezawa S, Yukawa M, Alavattam KG, Barski A. & Namekawa SH Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res 46, 593–608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y. et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol Cell 73, 547–561.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Alavattam KG et al. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol 26, 175–184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel L. et al. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat Struct Mol Biol 26, 164–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulger M. & Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong CT & Corces VG Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12, 283–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calo E. & Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker SC et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A 110, 17921–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loven J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creyghton MP et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidl C, Rendeiro AF, Sheffield NC & Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 12, 963–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adam RC et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaucher J. et al. Bromodomain-dependent stage-specific male genome programming by Brdt. Embo j 31, 3809–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuma S. et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A 103, 15894–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W. & Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2, 819–30 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Kneitz B. et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14, 1085–97 (2000). [PMC free article] [PubMed] [Google Scholar]
- 28.Spitz F. & Furlong EE Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13, 613–26 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Oatley JM, Kaucher AV, Avarbock MR & Brinster RL Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol Reprod 83, 427–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matson CK et al. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19, 612–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolcun-Filas E. et al. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foulkes NS, Mellstrom B, Benusiglio E. & Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature 355, 80–4 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Li XZ et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50, 67–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou L. et al. BTBD18 Regulates a Subset of piRNA-Generating Loci through Transcription Elongation in Mice. Dev Cell 40, 453–466.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Adams SR et al. RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes. PLoS Genet 14, e1007233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brind’Amour J. et al. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6, 6033 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Maezawa S. et al. Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc Natl Acad Sci U S A 115, 4957–4962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichijima Y, Sin HS & Namekawa SH Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci 69, 2559–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner JM Meiotic Silencing in Mammals. Annu Rev Genet 49, 395–412 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Ichijima Y. et al. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25, 959–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sin HS et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev 26, 2737–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harley JB et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet 50, 699–707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z. et al. Dynamics of the Transcriptome during Human Spermatogenesis: Predicting the Potential Key Genes Regulating Male Gametes Generation. Sci Rep 6, 19069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryser S. et al. Gene expression profiling of rat spermatogonia and Sertoli cells reveals signaling pathways from stem cells to niche and testicular cancer cells to surrounding stroma. BMC Genomics 12, 29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luoh SW et al. Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development 124, 2275–84 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Sakashita A. et al. Endogenous retroviruses drive species-specific germline transcriptomes in mammals. Nat Struct Mol Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matzuk MM et al. Small-molecule inhibition of BRDT for male contraception. Cell 150, 673–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pattabiraman S. et al. Mouse BRWD1 is critical for spermatid postmeiotic transcription and female meiotic chromosome stability. J Cell Biol 208, 53–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant JM et al. Characterization of BRD4 during mammalian postmeiotic sperm development. Mol Cell Biol 35, 1433–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Defeo-Jones D. et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 352, 251–4 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Scanlan MJ, Simpson AJ & Old LJ The cancer/testis genes: review, standardization, and commentary. Cancer Immun 4, 1 (2004). [PubMed] [Google Scholar]
- 52.Sohni A. et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep 26, 1501–1517.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo J. et al. The adult human testis transcriptional cell atlas. Cell Res 28, 1141–1157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green CD et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev Cell 46, 651–667.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hermann BP et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep 25, 1650–1667.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y. et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res 28, 879–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M. et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 23, 599–614.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Lambert SA et al. The Human Transcription Factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Ozata DM et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol 4, 156–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li E, Bestor TH & Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–26 (1992). [DOI] [PubMed] [Google Scholar]
- 61.Bellve AR Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 225, 84–113 (1993). [DOI] [PubMed] [Google Scholar]
- 62.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alavattam KG, Abe H, Sakashita A. & Namekawa SH Chromosome Spread Analyses of Meiotic Sex Chromosome Inactivation. Methods Mol Biol 1861, 113–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Concordet JP & Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46, W242–w245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kartashov AV & Barski A. BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. Genome Biol 16, 158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M. & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao Z, Zhang Y, Yuan GC, Orkin SH & Waxman DJ MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13, R16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saldanha AJ Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–8 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Machanick P. & Bailey TL MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karolchik D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32, D493–6 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weirauch MT et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer LR et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41, D64–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jung YH et al. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep 18, 1366–1382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.