Abstract
The transcriptional status of eukaryotic genes is determined by a balance between activation and repression mechanisms. The nuclear hormone receptors represent classical examples of transcription factors that can regulate this balance by recruiting corepressor and coactivator complexes in a ligand-dependent manner. Here, we demonstrate that the equilibrium between activation and repression via a single transcription factor, Elk-1, is altered following activation of the Erk mitogen-activated protein kinase cascade. In addition to its C-terminal transcriptional activation domain, Elk-1 contains an N-terminal transcriptional repression domain that can recruit the mSin3A-histone deacetylase 1 corepressor complex. Recruitment of this corepressor is enhanced in response to activation of the Erk pathway in vivo, and this recruitment correlates kinetically with the shutoff of one of its target promoters, c-fos. Elk-1 therefore undergoes temporal activator-repressor switching and contributes to both the activation and repression of target genes following growth factor stimulation.
The balance between the activation and repression mechanisms that act at a promoter determines the levels of gene transcription. The mechanisms of transcriptional activation have received considerable attention; however, it is becoming clear that transcriptional repression is equally important and can be mediated by several different mechanisms (reviewed in references 9 and 18). One repression mechanism involves the recruitment of corepressor complexes (reviewed in references 35 and 45), many of which contain subunits that possess histone deacetylase activity (HDACs). HDACs act to deacetylate histones and hence convert chromatin into a repressive state (reviewed in references 2, 26, and 28). An example of one such corepressor is the mSin3A-HDAC complex, which contains multiple subunits, including N-CoR (SMRT), mSin3A, HDAC-1, RbAp48, and SAP18/30/46 (45). Within these complexes, N-CoR and mSin3A act as links to transcription factors such as nuclear hormone receptors (via N-CoR [reviewed in reference 45]) and Mad, p53, TEL, and Ikaros (via mSin3A) (13, 27, 29, 33). In the case of the nuclear hormone receptors, ligand binding causes dissociation of these corepressor complexes and replacement with coactivator proteins and associated histone acetylase activities. The net result is a signal-dependent switch from a repressed to an active state (45).
The activation of c-fos following stimulation by a plethora of different extracellular stimuli has been studied intensively (reviewed in reference 7). c-fos exhibits classical immediate-early gene activation kinetics in response to mitogens and growth factors such as serum and epidermal growth factor (EGF), where it is rapidly induced within 15 min of stimulation, followed by a rapid shutoff of transcription back to basal levels within 2 h of stimulation. In the absence of stimulation, c-fos expression is barely detectable. Thus, three phases can be identified: an initial repressed state, activation, and a return to the repressed state. A large number of these stimuli activate c-fos via the serum response element (SRE) (7, 41). In the case of EGF, the signals are primarily transduced via the Erk mitogen-activated protein kinase (MAPK) pathway to the ternary complex factor (TCF) transcription factors that form a complex with the serum response factor (SRF) on the SRE (42). However, serum appears to activate pathways that converge on both the TCF and SRF parts of this complex (19, 20, 25). While it is clear that the TCFs are directly involved in the transcriptional activation process in response to the turning on of the Erk MAPK pathway, it is unclear how c-fos is subsequently turned off and whether the TCFs play a role in this process.
The TCFs are a subfamily of ETS domain transcription factors that currently contains three different proteins, Elk-1, SAP-1, and SAP-2 (Net) (42, 48). These proteins contain four conserved domains (see Fig. 1A); an N-terminal ETS DNA-binding domain; the B box, which binds directly to SRF (39); the D domain, which acts as a docking site for MAPKs (21, 46, 47); and the C domain, which acts as an MAPK-inducible transcriptional activation domain (14, 22, 23, 31, 32, 36). Each TCF appears to respond to a different subset of MAPK cascades, and in the case of Elk-1, evidence has been gathered to implicate the Erk, Jnk, and p38 MAPK pathways in its regulation (43, 48). Both Elk-1 and SAP-1 can act as transcriptional activator proteins, and in the case of Elk-1, both CBP (24) and Sur-2 (4) have been implicated as potential Erk-dependent coactivator proteins. In contrast, SAP-2 appears to be able to act as a transcriptional repressor rather than an activator protein, and in this case, activation of the Erk pathway appears to result in the loss of this repressive activity (15). Two different repression domains have been identified in SAP-2 that are not conserved with Elk-1, the Net inhibitory domain (NID) and the CHBP inhibitory domain (CID) (10, 31).
FIG. 1.
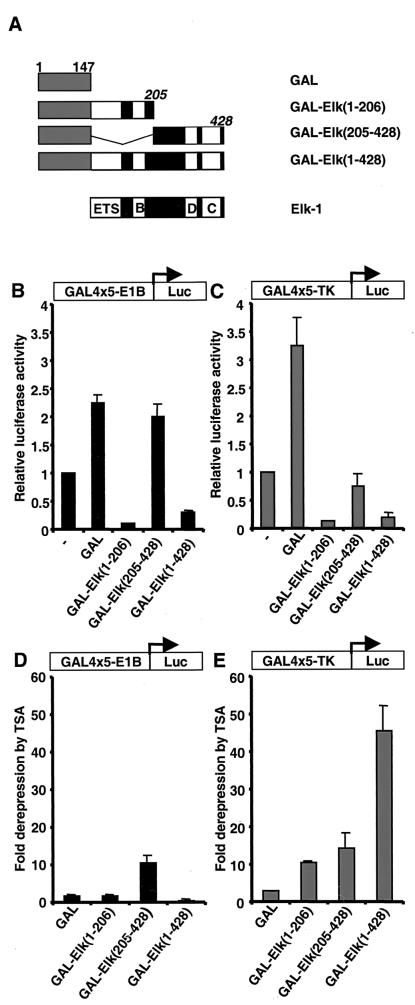
Elk-1 contains a transcriptional repression domain. (A) Diagram illustrating a series of truncated Elk-1 proteins (black boxes, with domains indicated by white boxes) fused to the GAL4 DNA-binding domain (amino acids 1 to 147, grey boxes). Numbers of the C-terminal amino acids in the Elk-1 moiety are indicated (italics). (B and C) GAL-Elk fusion proteins repress GAL4-driven luciferase re- porter genes. 293 cells were cotransfected with 0.1 μg of CMV promoter-driven constructs encoding the indicated GAL4–Elk-1 derivatives and 1 μg of GAL4-driven luciferase reporter plasmids containing the minimal E1B (B) or TK (C) promoter. Cells were maintained in serum-free conditions throughout the experiment. The layout of the reporters is represented as a diagrammatic insert. Luciferase activities relative to the control cells (without transfection of any GAL4 fusions [—]) are presented (means ± standard deviation, n = 2). (D and E) TSA sensitivity of the indicated reporters in the presence of GAL-Elk fusion proteins. Transfection assays were carried out as above. Cells were left in serum-free medium after transfection, treated or not with TSA, and harvested 18 h later. Luciferase activities relative to the untreated cells (fold derepression) are presented (means ± standard deviation, n = 2).
In this study, we have investigated whether Elk-1 might also be able to act as a transcriptional repressor protein and thus play a role in turning off immediate-early genes such as c-fos. We demonstrate that Elk-1 contains an N-terminal transcriptional repression domain and that Elk-1 can recruit the mSin3A–HDAC-1 corepressor complex. Recruitment is enhanced in response to activation of the Erk pathway, and this recruitment correlates kinetically with the shutoff of one of its target promoters, c-fos. It therefore appears that in addition to its role as an activator, Elk-1 also acts as a transcriptional repressor and can undergo activator-repressor switching following activation of the Erk MAPK pathway.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were used for expressing glutathione S-transferase (GST) fusion proteins in Escherichia coli. pAS74 [encoding GST-Elk(1–93); Elk-1 amino acids 1 to 93] (40), pAS77 [encoding GST-Elk(139–168); Elk-1 amino acids 139 to 168] (38), pAS183 [encoding GST–SAP-1(1–92); SAP-1 amino acids 1 to 92] (39), pAS462 [encoding GST-PEA3(341–432); PEA3 amino acids 341 to 432] (6), pAS407 [encoding GST-Elk(205–428); Elk-1 amino acids 205 to 428] (40), and pGNElk [encoding GST-Elk(1–205); Elk-1 amino acids 1 to 205] (14) have been described previously.
pAS278 (encoding hexahistidine-Flag-tagged Elk-1 [amino acids 1 to 428]) was used to express full-length Elk-1 in E coli.
The following plasmids were constructed and used for mammalian cell transfections. pG5-E1B-Luc contains five GAL4 DNA-binding sites cloned upstream of a minimal E1B promoter element and the firefly luciferase gene; pSRE-Luc contains two copies of the c-fos serum response element (nucleotides −357 to −275) upstream from a minimal thymidine kinase (TK) promoter and the luciferase gene (37). All have been described previously (11). pG5-TK-Luc (pAS1567) contains five GAL4 DNA-binding sites cloned upstream of a minimal TK promoter element and the firefly luciferase gene and was constructed in several steps. The HindIII/BamHI fragment from pG5E4T (kindly provided by Stefan Roberts) was inserted into the same sites in pBLCAT (3) to create pG5-TK-CAT (pAS1565). The HindIII/XhoI fragment from pAS1465 was inserted into the same sites of pBS-SK+ to produce pBG5TK (pAS1566). The XmaI/BglII fragment from pAS1466 (containing the G5-TK promoter fragment) was inserted into the same sites in pT81Luc (kindly provided by E. Oettgen) to produce pG5-TK-Luc (pAS1467). pCMV5-MEK-1 (ΔN S218E-S222D) encodes constitutively active MEK-1; pAS383 (pCMV5-Elk-1, encoding full-length Elk-1 controlled by a cytomegalovirus [CMV] promoter), pAS571 (pCMV-GAL), and pAS572 [pCMV-GAL-Elk(205–428)] were described previously (46). pAS1068 (pSG424-new) encodes the GAL4 DNA-binding domain with a frameshift in the multiple cloning site and was constructed by ligating the BamHI/KpnI-annealed oligonucleotides ADS673 and ADS674 into the same sites of pSG424 (constructed by Alex Galanis). pAS1557 [pCMV-GAL-Elk(1–93)] and pAS1563 [pCMV-GAL-SAP-2(215–281)] were constructed by ligating SalI/XbaI PCR fragments into the same sites of pAS571, pAS1558 [pGAL-Elk(1–206)] was constructed by ligating the BamHI/XbaI PCR fragment into the same sites of pAS1068. pAS1559 [pCMV-GAL-Elk(1–206)] was constructed by ligating the HindIII/XbaI fragment from pAS1458 into the same sites of pCMV5. pAS1561 [pCMV-GAL-Elk(1–428)] was constructed by ligating a StuI/XbaI fragment from pAS728 into the same sites of pAS1458. pAS1654 (encoding Flag-tagged full-length Elk-1[S383A/S389A] mutant) was constructed in several steps. First, a PCR fragment was generated from pAS567, digested with XmaI/XhoI, and ligated into pAS728 to produce pAS1651. The NcoI/XhoI fragment from pAS1651 was then inserted into the same sites in pETnef-PFH to produce pAS1652. The NcoI/HindIII fragment from pAS1652 was then inserted into the same sites in pRSETB (Invitrogen) to produce pAS1653. Finally, the KpnI/HindIII fragment from pAS1653 was ligated into the same sites in pCMV5 to produce pAS1654. pAS1655 (encoding full-length His-Flag-tagged Elk-1 under a ponasterone-inducible promoter) was constructed by inserting the KpnI/BamHI fragment from pAS383 into pIND(SP1) (Invitrogen).
The following plasmids were used for in vitro transcription-translation or transfection purposes: pCS-myc, pCS-myc-mSin3A(1–205), pCS-myc-mSin3A(1–479), pCS-myc-mSin3A(1–680), pCS-myc-mSin3A(1–1015), pCS-myc-mSin3A(1–1275), pcDNA3-HDAC-1, and pCMV5′ 3T-HDAC-1 (encoding N-terminally hemagglutinin [HA]-tagged full-length HDAC-1) (29).
All plasmid constructs made by PCR were verified by automated dideoxy sequencing.
Protein expression and purification.
GST fusion proteins were expressed in E. coli JM101 or X90 and purified as described previously (40). Full-length hexahistidine-tagged polypeptides were expressed in E. coli BL21(DE3)(pLysS) with the pET vector system and quantified as described previously (46).
The synthesis of proteins by in vitro transcription and translation was carried out with the TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's recommendations. Newly synthesized 35S-labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by visualization and quantification by phosphorimager (Fuji and Tina software version 2.08e [Fuji] or Bio-Rad Molecular Imager FX and Quantity One software).
In vitro protein-protein interaction assays.
Interactions between 0.5 to 1 μg each of GST or His-tagged fusion proteins and in vitro-translated proteins were investigated using pull-down assays as described previously (38) with modified buffer conditions (40 mM HEPES [pH 7.9], 100 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 40 mM β-glycerophosphate, 0.5 mM dithiothreitol, 0.1 mM sodium orthovanadate, 0.05% NP-40, 0.5 mM phenylmethylsulfonyl fluoride).
Tissue culture, stable cell line generation, cell transfection, and reporter gene assays.
293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL). Transfection experiments were carried out using Superfect transfection reagent (Qiagen) as described previously (46). The cell lines EcR-293-Elk#1 and EcR-293-Elk#8 inducibly express a Flag-His-tagged version of full-length Elk-1. These clonogenic cell lines were generated according to the manufacturer's instructions (Invitrogen). EcR293 cells were transfected with pAS1655, and clones were isolated by G418 selection. The resulting cell lines were maintained in DMEM with 10% FBS, zeocin (400 μg/ml), and G418 (500 μg/ml). Induction of Elk-1 was achieved by adding ponasterone A (5 μM) for 24 h in serum-free medium. Cells were subsequently stimulated with EGF.
For reporter gene assays, 1 μg of reporter plasmid was cotransfected with various vectors. Cell extracts were prepared, and luciferase and β-galactosidase assays were carried out as described previously (46). Cells were treated with 50 nM EGF (Sigma) or 330 nM trichostatin A (TSA) and left for the times indicated before harvesting.
Kinase reactions.
Kinase reactions were carried out as described previously (46).
Immunoprecipitation and Western blot analysis.
For immunoprecipitations, anti-Flag agarose beads (Sigma) were used for Flag-tagged proteins and for GAL4 fusion proteins. The antibody matrix was prepared by binding the GAL4 antibody (sc-577x; Santa Cruz Biotechnology) to protein G beads. Lysates from 293 cells were prepared from 35-mm dishes in 200 μl of Triton lysis buffer (TLB) containing protease inhibitors as described previously (46). Antibody matrix (20 μl) was incubated with whole-cell extracts with rotation for 2 h at room temperature. Complexes were washed three times with TLB, boiled in sample loading buffer, and subjected to SDS-PAGE followed by Western blot analysis. The M2 anti-Flag (Sigma), anti-phospho-S383 Elk-1 antibody (NEB), anti-Elk-1 (NEB), anti-mSin3A (K-20; Santa Cruz), anti-Myc (Santa Cruz), anti-HA (BDH), anti-HDAC-1 (Upstate Biotechnology) and horseradish peroxidase-conjugated secondary antibodies (Transduction Laboratories) were used in the immunoblot analysis as described previously (46), followed by SuperSignal West Dura extended-duration substrate (Pierce) and visualized by phosphorimager (Bio-Rad Fluor-S MultiImager and Quantity One software).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays using antisera specific to acetylated histone H4, Flag, or mSin3A (Santa Cruz) were performed exactly as specified by the manufacturer (Upstate Biotechnology). PCR of the c-fos promoter was performed on immunoprecipitated chromatin using oligonucleotides ADS859 (5′-AGCAGTTCCCGTCAATCC-3′) and ADS860 (5′-TGAGCATTTCGCAGTTCC-3′). DNAs were amplified for 28 cycles using Biotaq DNA polymerase (Bioline).
Figure generation and data quantification.
Figures were generated electronically from scanned autoradiographic images by using Picture Publisher (Micrografix) or Adobe PhotoDeluxe Business Edition 1.0 and Powerpoint version 7.0 (Microsoft) software. Final images are representative of the original autoradiographic images. Data from Western blots are computer-generated images (Quantity One; Bio-Rad). Phosphorimager data were quantified using either Tina software (version 2.08e; Fuji) or Quantity One (Bio-Rad).
RESULTS
Elk-1 contains an N-terminal transcriptional repression domain.
The transcription factor Elk-1 plays a pivotal role in the induction of c-fos transcription following growth factor stimulation. Elk-1 contains a C-terminal transcriptional activation domain (42, 48). In order to investigate whether Elk-1 also contains a transcriptional repression domain and hence might participate in c-fos downregulation, a series of truncated Elk-1 proteins fused to the GAL4 DNA-binding domain were constructed (Fig. 1A) and tested for their ability to regulate two different GAL4-driven luciferase reporters in 293 cells. These reporters differed only by the basal promoters E1B and TK. Under serum-free conditions, full-length Elk-1 [GAL-Elk(1–428)] and a carboxyl-terminal deletion mutant [(GAL-Elk(1–206)] repressed the activity of both of these luciferase reporter genes. However, in comparison, little repression was observed with the N-terminally truncated protein GAL-Elk(205–428) (Fig. 1B and C). This demonstrates that the amino-terminal part of Elk-1 contains a transcriptional repression domain.
To determine if associated HDAC activity is required for repression by GAL-Elk fusion proteins, we examined the effect of TSA, a specific inhibitor of HDACs, on repression mediated by GAL-Elk(1–206), GAL-Elk(205–428), and GAL-Elk(1–428). On the E1B-Luc reporter, TSA did not lead to relief of repression by Elk(1–206) and only led to an enhancement of promoter activity in the presence of GAL-Elk(205–428) (Fig. 1D). This suggests the presence of a repression domain in the C-terminal half of Elk-1, which is usually masked by the transcriptional activation domain (TAD) (see Discussion). These results also indicate that the N-terminal end of Elk-1 [Elk(1–206)] represses in an HDAC-independent manner at this promoter. However, on the TK-Luc reporter, TSA led to enhanced promoter activity by all three GAL-Elk fusion proteins (Fig. 1E), indicating a role for HDACs in repressing transcription via these proteins on this promoter. Interestingly, TSA causes more enhancement of promoter activity in the presence of full-length Elk-1 than either of the deleted proteins, suggesting cooperativity between repression domains at the N- and C-terminal ends of Elk-1.
Collectively, these data therefore demonstrate that the Elk-1 repression activity is mediated, at least in part, by histone deacetylation-dependent mechanisms in a promoter-specific manner.
Elk-1 interacts with components of the mSin3A–HDAC-1 complex in vitro.
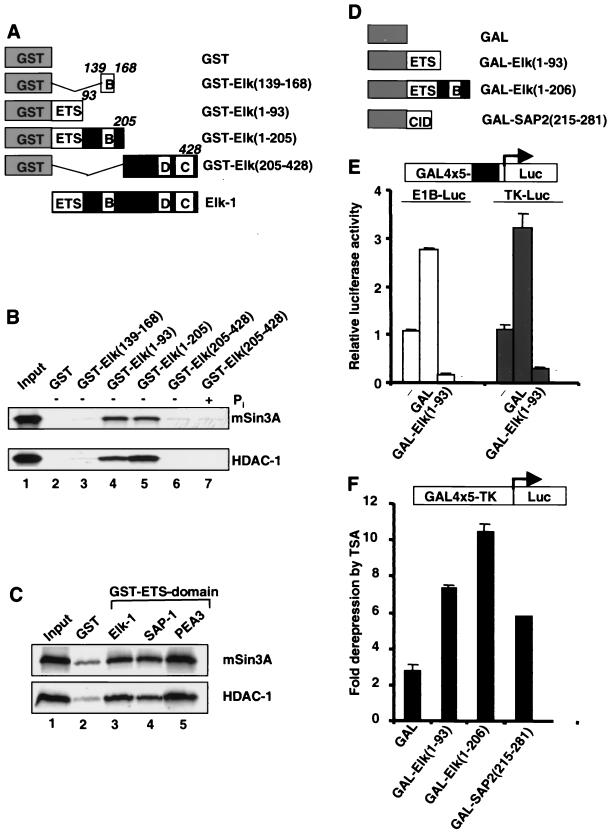
One mechanism for transcriptional repression is via recruitment of HDAC-containing complexes (2, 26, 28). Several corepressor complexes which target HDACs to promoters have been identified (5, 45). One such complex contains mSin3A and HDAC-1. To investigate whether Elk-1 binds to components of this complex, in vitro pull-down experiments were carried out. A series of truncated proteins fused to GST (Fig. 2A) were tested for their ability to interact with either mSin3A (Fig. 2B, top panel) or HDAC-1 (Fig. 2B, bottom panel) in order to map the mSin3A interaction domain in Elk-1. Of the fusion proteins tested, interactions were only obtained with the C-terminally truncated Elk-1 derivatives GST-Elk(1–93) and GST-Elk(1–205) (Fig. 2B, lanes 4 and 5). We were unable to detect interactions with the B box alone [GST-Elk(139–168)] or with either nonphosphorylated or phosphorylated forms of the N-terminally truncated Elk-1 derivative GST-Elk(205–428) (Fig. 2B, lanes 3, 6, and 7). These results indicate that the N-terminal ETS DNA-binding domain contained in Elk(1–93) is sufficient to mediate interactions with mSin3A and HDAC-1.
FIG. 2.
Elk-1 interacts with mSin3A and HDAC-1 in vitro. (A) Schematic illustration of a series of truncated Elk-1 proteins (black boxes) fused to GST (grey boxes). Numbers of the C-terminal amino acids in the Elk-1 moiety are indicated (italics). (B) Mapping the mSin3A and HDAC-1 interaction domain on Elk-1. GST pull-down analysis of 35S-labeled mSin3A (upper panel) and HDAC-1 (bottom panel) with GST (lane 2) and a series of truncated Elk-1 proteins fused to GST (lane 3 to 7). GST-Elk(205–428) was left nonphosphorylated (lane 6) or phosphorylated with Erk2 (lane 7) prior to immobilization on the beads, and 5% of the input proteins are shown in lane 1. (C) Binding of mSin3A–HDAC-1 to different ETS domains. GST pull-down analysis of 35S-labeled mSin3A (upper panel) and HDAC-1 (bottom panel) with GST (lane 2) and a series of GST-ETS domain fusion proteins (lanes 3 to 5); 5% of the input proteins are shown in lane 1. (D) Diagram illustrating a series of protein domains fused to GAL4 DNA-binding domain (amino acids 1 to 147, grey boxes). (E) Either 1 μg of GAL4x5-TK-Luc (open bars) or GAL4x5-E1B-Luc (grey bars) reporter vectors and 0.1 μg of pCMV-GAL or pCMV-GAL-Elk(1–93) were transfected. Cells were left in the serum-free medium for 18 h before harvesting. The luciferase activities relative to GAL are presented (mean ± standard deviation, n = 2). (F) GAL4 reporter gene assays were carried out in 293 cells in the presence of GAL4 fusion proteins; 1 μg of GAL4x5-TK-Luc reporter vector and 0.1 μg of the indicated CMV promoter-driven constructs encoding GAL4 fusion proteins were transfected. Cells were treated and data are presented as in Fig. 1D and E.
The ETS DNA-binding domain is conserved among members of the ETS domain family of transcription factors. To examine if this domain from other family members represents a binding motif for mSin3A, GST fusions to the ETS domain of SAP-1 and PEA3 were tested for their ability to bind to mSin3A and HDAC-1 in GST pull-down experiments (Fig. 2C). The ETS domains of SAP-1 and PEA3 bind to mSin3A (top panel) and HDAC-1 (bottom panel) with an efficiency similar to that observed with Elk-1 (Fig. 2C, lanes 3, 4, and 5). These results demonstrate that the ETS domains from different proteins interact with mSin3A and HDAC-1, and therefore, different ETS proteins may also respond to the mSin3A–HDAC-1 complex.
In order to verify that the ETS DNA-binding domain is sufficient to mediate transcriptional repression by Elk-1, GAL-Elk(1–93) was tested on the E1B-Luc and TK-Luc reporters (Fig. 2E). In both cases, GAL-Elk(1–93) efficiently repressed transcription. Furthermore, GAL-Elk(1–93) can also promote >90% repression of a composite Lex-Gal-driven promoter-reporter construct in the presence of the strong Lex-VP16 activator, demonstrating that it also represents a potent repressor in the presence of highly activated transcription (data not shown). The role of deacetylases in repression mediated by the ETS domain was subsequently determined using TSA (Fig. 2F). TSA causes derepression of the TK-Luc reporter in the presence of GAL-Elk(1–93), indicating that the ETS DNA-binding domain of Elk-1 is sufficient to mediate HDAC-dependent repression in vivo. The level of derepression is similar to that observed with GAL–SAP-2(215–281), which contains an HDAC-dependent repression domain (CID) from SAP-2 (10). The ETS domain therefore represents an HDAC-dependent repression domain that can interact with components of the mSin3A–HDAC-1 complex.
Mapping the Elk-1 interaction determinants in the mSin3A protein.
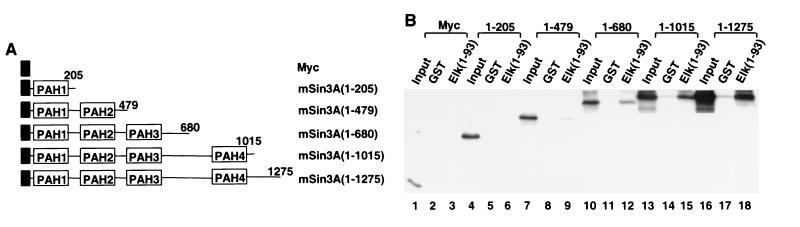
In order to determine the region in mSin3A that interact with Elk-1, a series of 35S-labeled C-terminally truncated mSin3A proteins that contain one or more paired amphipathic helix (PAH) domains were generated (Fig. 3A): PAH1 (amino acids 1 to 205), PAH1→2 (amino acids 1 to 479), PAH1→3 (amino acids 1 to 680), PAH1→4 (amino acids 1 to 1015), and full-length (amino acids 1 to 1275). The proteins were used in GST pull-down assays with GST-Elk(1–93) (Fig. 3B). mSin3A(1–680), mSin3A(1–1015), and mSin3A(1–1275) interact strongly with GST-Elk(1–93) (Fig. 3B, lanes 12, 15, and 18). However, little interaction is seen with either mSin3A(1–479) or mSin3A(1–205) (Fig. 3B, lanes 6 and 9). This analysis therefore demonstrates that the region containing amino acids 479 to 680 of mSin3A that encompasses PAH3 is required to mediate interactions with the ETS domain of Elk-1.
FIG. 3.
Mapping the Elk-1 interaction domain in mSin3A. (A) Schematic representation of mSin3A indicating the locations of the four PAH domains (white boxes). The numbers of the C-terminal amino acids in each truncated mSin3A construct are indicated. N-terminal Myc epitope tags are shown by black boxes. (B) Interaction of the indicated 35S-labeled Myc-tagged mSin3A deletion mutants or full-length protein with GST (lanes 2, 5, 8, 11, 14, and 17) or GST-Elk(1–93) (lanes 3, 6, 9, 12, 15, and 18) was investigated by GST pull-down analysis. Equal molar amounts of 35S-labeled proteins were used in each reaction, and 5% of the input proteins are shown (lanes 1, 4, 7, 10, 13, and 16).
mSin3A interacts with Elk-1 in vivo.
To examine whether the interactions between Elk-1 and mSin3A observed in vitro could be recapitulated in mammalian cells, either native full-length Elk-1 or truncated GAL4–Elk-1 fusions were coexpressed with mSin3A or HDAC-1 in 293 cells, and immunoprecipitation experiments were performed.
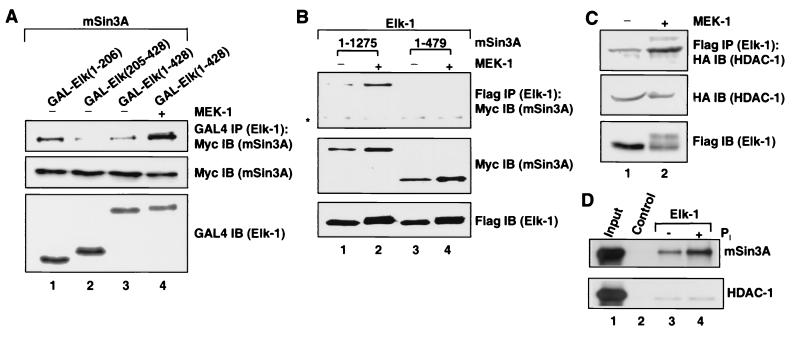
First, coimmunoprecipitation experiments were carried out with a series of GAL–Elk-1 fusion proteins (Fig. 1A and 2E) and Myc-tagged full-length mSin3A to identify the interaction surface on Elk-1 (Fig. 4A). The GAL fusion protein containing the N-terminal end of Elk-1 [GAL4-Elk(1–206)] interacted with mSin3A (Fig. 4A, lane 1, top panel); however, in comparison, weaker interactions were observed in N-terminally truncated GAL-Elk(205–428) and full-length GAL4-Elk(1–428) (Fig. 4A, lanes 2 and 3, top panel). Transfection of constitutively activated MEK-1 leads to enhanced activation of Erk (data not shown). Under these conditions, interactions were strongly enhanced between mSin3A and GAL-Elk(1–428) (Fig. 4A, lane 4, top panel). In all cases, mSin3A and GAL fusion proteins are expressed at comparable levels (Fig. 4A, middle and bottom panels). These data are fully consistent with the in vitro mapping experiments (Fig. 2B) and demonstrate that the N-terminal end of Elk-1, containing the ETS domain, is sufficient for binding to mSin3A in vivo. In the context of full-length Elk-1, activation of the Erk pathway stimulates this interaction in vivo.
FIG. 4.
Activation of the Erk pathway enhances Elk-1 interactions with mSin3A and HDAC-1 in vivo. (A) Mapping the mSin3A binding domain on Elk-1. Coimmunoprecipitation (IP) of mSin3A with GAL–Elk-1 truncations from 293 cells. Cells were cotransfected with 2 μg of pCS2-mSin3A (Myc tagged), 2 μg of the indicated CMV promoter-driven GAL-Elk fusion proteins, and 1 μg of pCMV5-MEK-1 (to trigger Elk-1 phosphorylation) (lane 4). GAL fusion proteins were immunoprecipitiated with an anti-GAL4 antibody, and coprecipitated mSin3A was subsequently detected with a Myc antibody (top panel). The expression level of each protein was detected by immunoblotting (IB) with the indicated antibodies (middle and bottom panels). (B) The Elk-1 binding motif in mSin3A is required for the interaction of mSin3A with full-length Elk-1 in vivo. Cells were cotransfected with 2 μg of the indicated Myc-tagged mSin3A derivatives and 2 μg of pCMV5-Elk-1 (Flag tagged) with or without 1 μg of pCMV5-MEK-1. Elk-1 was immunoprecipitated with anti-Flag-agarose. Immunocomplexes were subsequently subjected to immunoblotting with a Myc antibody to detect mSin3A (top panel). Total cell extracts were also analyzed by immunoblot with the antibodies indicated on the right to detect total levels of epitope-tagged proteins (middle and bottom panels). Asterisk represents a cross-reacting nonspecific band. The input levels of mSin3A increase by ∼2-fold in the presence of MEK, whereas the levels of the Elk-1–mSin3A complex increase by ∼12-fold. (C) HDAC-1 and Elk-1 coexist in a complex in vivo. 293 cells were transfected with pCMV5-Elk-1 (Flag tagged), pCMV5-HDAC-1 (HA tagged), and pCMV5-MEK-1 (where indicated). Coimmunoprecipitation assays were carried out with anti-Flag-agarose and then immunoblotting with anti-HA antibody to detect HDAC-1 coprecipitates (top panel). The expression levels of each protein in total cell lysates were monitored by immunoblot with the indicated antibodies (middle and bottom panels). (D) Elk-1 interaction with mSin3A in vitro is stimulated by phosphorylation of Elk-1 by Erk2. Pull-down analysis of full-length mSin3A (upper panel) and HDAC-1 (bottom panel) with nonphosphorylated (lane 3) and phosphorylated (lane 4) full-length Elk-1 immobilized onto Ni-nitrilotriacetic acid-agarose beads. The beads alone (lane 2) were used as a control. 35S-labeled mSin3A and HDAC-1 were generated by in vitro translation, whereas Elk-1 was prepared from bacteria, and 5% of the input proteins are shown in lane 1.
To confirm these interactions and determine the interaction surface on mSin3A, Flag-tagged full-length Elk-1 was coexpressed with Myc-tagged mSin3A derivatives. Parallel experiments were also carried out in the presence of cotransfected constitutively active MEK-1. Elk-1 and associated proteins were initially precipitated with Flag-agarose beads, followed by detection of coprecipitating mSin3A with an anti-Myc antibody. In the absence of cotransfected MEK-1, mSin3A could be weakly detected in immunocomplexes from Elk-1. However, the efficiency of complex formation was enhanced in the presence of cotransfected MEK-1 (Fig. 4B, lanes 1 and 2, top panel). No detectable coprecipitation was found when the truncated mSin3A(1–479) was used in either the presence or absence of MEK-1 (Fig. 4B, lanes 3 and 4, top panel). This is consistent with the in vitro interaction data, showing that full-length mSin3A binds to Elk-1 but the truncated protein mSin3A(1–479) does not (Fig. 3B). Together, these results demonstrate that Elk-1 and mSin3A interact in vivo and this interaction is enhanced upon activation of the Erk pathway.
To examine if HDAC-1 exists as part of Elk-1-associated protein complexes, similar coimmunoprecipitation experiments were performed. HDAC-1 can be found in Flag (Elk-1) immunoprecipitates (Fig. 4C, lane 1) and, in reciprocal experiments, Elk-1 is also found in HDAC-1 immunoprecipitates (data not shown). Activation of the Erk pathway leads to enhanced HDAC-1 interaction in vivo (Fig. 4C, lane 2, top panel). In contrast, no Erk-dependent recruitment of a different HDAC, HDAC-4, to Elk-1 was observed (data not shown).
To investigate whether direct phosphorylation of Elk-1 by Erk can promote enhanced recruitment of the mSin3A–HDAC-1 complex, in vitro pull-down experiments were carried out using either phosphorylated or nonphosphorylated full-length recombinant Elk-1 and in vitro-translated mSin3A and HDAC-1 (Fig. 4D). Weak interactions between Elk-1 and mSin3A or HDAC-1 were obtained (Fig. 4D, lane 3). However, upon phosphorylation of Elk-1, the efficiency of this interaction with mSin3A was enhanced, whereas binding to HDAC-1 was unaffected (Fig. 4D, lane 4).
Taken together, these data demonstrate that Elk-1 and mSin3A interact in vivo and that the N-terminal end of Elk-1 is necessary for these interactions. HDAC-1 is also found in complexes with Elk-1. Activation of the Erk pathway results in the phosphorylation of Elk-1 and enhancement of interactions with mSin3A and HDAC-1.
Regulation of the mSin3A–Elk-1 interaction by the Erk pathway.
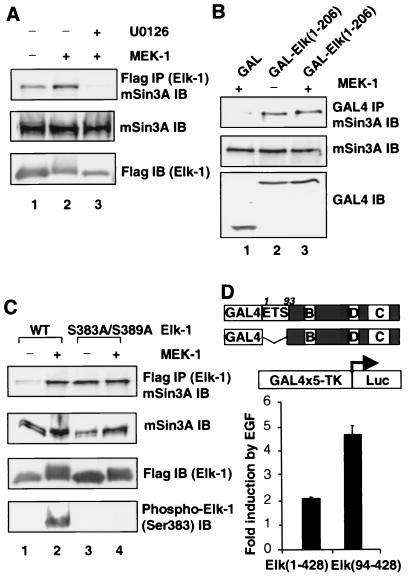
To further probe the links between the activation of the Erk pathway and the recruitment of the mSin3A–HDAC-1 complex, an additional series of experiments were performed. First, we confirmed that the catalytic activity of the overexpressed MEK-1 was required to promote enhanced Elk-1–mSin3A complex formation. In the absence of the MEK inhibitor U0126, MEK-1 promoted increased Elk-1–mSin3A complex formation (Fig. 5A, lanes 1 and 2). However, in the presence of U0126, the levels of complex formation were reduced to below basal levels (Fig. 5A, lane 3), demonstrating the importance of downstream signaling events in promoting complex formation.
FIG. 5.
Regulation of the mSin3A–Elk-1 interaction by the Erk pathway. (A) The MEK inhibitor U0126 blocks interactions between Elk-1 and mSin3A. 293 cells were transfected with expression vectors for Elk-1, mSin3A, and a constitutively active form of MEK-1 (where indicated), in the presence or absence of U0126. Elk-1 was immunoprecipitated with Flag-agarose beads, and coprecipitated mSin3A was detected using an anti-Sin3A antibody (top panel). Input levels of mSin3A and Elk-1 were determined by immunoblot (IB) (bottom two panels). (B) Deletion of the Elk-1 C terminus stops MEK-inducible mSin3A recruitment. 293 cells were transfected with expression vectors for the indicated GAL fusion proteins and a constitutively active form of MEK-1 (where indicated). GAL fusion proteins were immunoprecipitated, and coprecipitated mSin3A was detected using an anti-Sin3A antibody (top panel). Input levels of mSin3A and Elk-1 were determined by immunoblot (bottom two panels). (C) Ser383 and Ser389 are required to allow MEK-inducible mSin3A recruitment. 293 cells were transfected with expression vectors for the indicated wild-type (WT) and mutant Elk-1 proteins, mSin3A, and a constitutively active form of MEK-1 (where indicated). Elk-1 proteins were immunoprecipitated, and coprecipitated endogenous mSin3A was detected using an anti-Sin3A antibody (top panel). Input levels of mSin3A and Elk-1 were determined by immunoblot (middle two panels), and the phosphorylation status of Elk-1 was determined by using a phospho-Ser383 antibody (bottom panel). (D) Deletion of the ETS domain leads to enhancement of EGF-mediated Elk-1 activation. 293 cells were transfected with expression vectors for the indicated GAL–Elk-1 fusion proteins (shown schematically) and a TK-Luc reporter construct. Cells were serum starved for 12 h, followed by EGF stimulation for 12 h before harvesting. Results show the average fold induction by EGF of the reporter construct (means ± standard deviation, n = 2).
To investigate the requirement for Elk-1 phosphorylation in this recruitment process in vivo, we first investigated the effect of MEK-1 on the interactions between mSin3A and GAL-Elk(1–206). This Elk-1 derivative retains the mSin3A binding domain but lacks the C-terminal Erk phosphoacceptor sites. Constitutive binding of mSin3A is observed which is not further augmented by the presence of MEK-1 (Fig. 5B, lanes 2 and 3). This is in contrast to full-length Elk-1, where MEK-inducible binding of mSin3A is clearly observed (Fig. 4A). Second, we investigated MEK-inducible binding of mSin3A to a mutant Elk-1 protein that lacks two important C-terminal phosphoacceptor motifs (S383A and S389A). While mSin3A binding to wild-type Elk-1 is clearly inducible (Fig. 5C, lanes 1 and 2), binding to Elk-1(S383A/S389A) is no longer stimulated by MEK-1 (Fig. 5C, lanes 3 and 4). The basal level of mSin3A binding to this mutant protein is also elevated, suggesting a positive role for these serine residues in blocking mSin3A interactions which is lost upon mutation or phosphorylation (see Discussion). Similarly, no increase in mSin3A binding to Elk-1(S383A/S389A) was observed in response to EGF stimulation (data not shown). Together, these data demonstrate that the Erk phosphoacceptor sites in Elk-1 play a critical role in the EGF-MEK-inducible recruitment of mSin3A.
Finally, one prediction of these results is that activation of the Erk pathway by EGF will promote both activation and repression of transcription via Elk-1. Therefore, in the absence of the repression-mSin3A recruitment domain, enhanced activation in response to EGF stimulation should be observed due to the loss of mSin3A-mediated attenuation. We therefore compared the ability of GAL fusions of full-length Elk-1 and an N-terminally truncated version of Elk-1 to activate transcription in response to EGF stimulation (Fig. 5D). While EGF promotes a twofold increase in the ability of GAL-Elk(1–428) to activate transcription, deletion of the mSin3A binding domain in GAL-Elk(94–428) led to a 4.5-fold increase in activity. Thus, the mSin3A-binding domain in Elk-1 plays an important role in attenuating its response to Erk pathway activation.
Kinetic studies of interactions between mSin3A–HDAC-1 and Elk-1 in vivo following EGF stimulation.
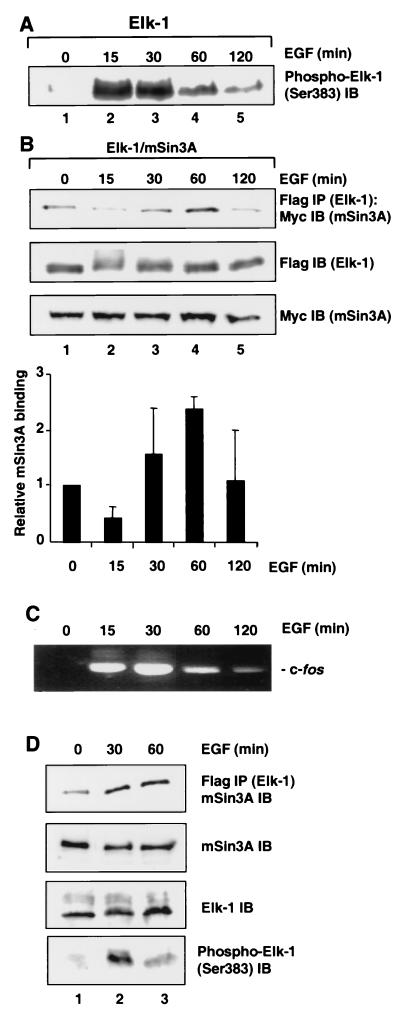
EGF leads to the rapid activation of the Erk MAPK pathway and subsequent activation of transcription factors such as Elk-1 and target immediate-early genes such as c-fos (reviewed in reference 42). This rapid activation is followed by a rapid shutoff of these genes. The previous series of experiments suggested that the phosphorylation status of Elk-1 plays an important role in recruiting both mSin3A and HDAC-1. In order to test whether recruitment of the corepressor complex correlates kinetically with EGF-mediated Elk-1 activation and c-fos downregulation in vivo, we investigated the ability of Elk-1 and the mSin3A–HDAC-1 complex to associate following EGF stimulation of 293 cells. First, the phosphorylation status of Elk-1 was monitored following EGF treatment of 293 cells using an antibody directed against phosphoserine 383. Phosphorylated Elk-1 was strongly detected after 15 and 30 min but at much lower levels preceding and following this time period (Fig. 6A). Interactions between mSin3A and Elk-1 were then investigated, and weak interactions could be observed during the first 15 min of EGF stimulation. However, these interactions were strongly enhanced after 30 min, maintained at a higher level after 60 min, and subsequently reduced to their original levels 2 h after stimulation (Fig. 6B, top panel and graphic representation). All proteins are expressed at comparable levels within each experiment (Fig. 6B, middle and bottom panels).
FIG. 6.
Kinetic studies of the interaction of mSin3A and HDAC-1 with Elk-1 following EGF stimulation. (A) Phosphorylation of Elk-1 at Ser383 following stimulation by EGF was detected by immunoblot (IB) with an anti-phospho-S383 antibody. (B) Kinetics of mSin3A binding. 293 cells were cotransfected with CMV promoter-driven expression vectors encoding mSin3A together with Elk-1. Cells were serum starved for 24 h following transfection, and total-cell extracts were taken at the indicated times after EGF stimulation. Coimmunoprecipitation assays were carried out with Flag-agarose beads, and coprecipitated mSin3A (Myc tagged) was detected with anti-Myc antibody. Total-cell extracts were analyzed by immunoblot with the indicated antibodies for total expression levels of epitope-tagged proteins (middle and bottom panels). Quantification of mSin3A binding to Elk-1 relative to the zero time point is shown graphically. Data are averages and standard deviations from four independent experiments. (C) RT-PCR analysis of c-fos transcription following the indicated times of EGF stimulation. (D) Inducible binding of endogenous mSin3A to Elk-1 following EGF induction. The expression of Flag-tagged Elk-1 was induced in the EcR-293-Elk#8 cell line, and endogenous mSin3A was coprecipitated using Flag-agarose beads at the indicated times after EGF stimulation (top panel). The total levels of mSin3A, Elk-1, and phosphorylated Elk-1 (Ser383) are shown in the bottom three panels.
Reverse transcription (RT)-PCR analysis was performed in order to confirm that the kinetics of Elk-1 phosphorylation and mSin3A–HDAC recruitment correlate with the kinetics of c-fos induction and subsequent repression following EGF stimulation of 293 cells (Fig. 6C). c-fos expression is rapidly induced after 15 min, maintained at 30 min, and then reduced back to near basal levels 2 h after stimulation. Thus, induction correlates with Ser383 phosphorylation kinetics, whereas repression correlates with the kinetics of mSin3A recruitment.
We were unable to detect endogenous Elk-1 using the currently available antisera. Therefore, in order to investigate whether endogenous mSin3A can interact with Elk-1 in the absence of transient-transfection analysis, we constructed cell lines which inducibly express a Flag-tagged Elk-1 protein (EcR-293-Elk#1 and -Elk#8). Immunoprecipitation analysis was carried out on the EcR-293-Elk#8 cell line following EGF induction and demonstrated that, as observed with transiently transfected constructs (Fig. 6B), Elk-1 interactions with endogenous mSin3A were enhanced after 30 min and maintained at a high level after 60 min (Fig. 6D, top panel). The levels of input proteins were similar (Fig. 6D, middle panels), and phosphorylation of Elk-1 occurred with the expected kinetics (Fig. 6D, bottom panel). Similar results were observed with an independent cell line (EcR-293-Elk#1) (data not shown).
Collectively, these data demonstrate that Elk-1 is rapidly activated by phosphorylation at Ser383 following 15 to 30 min of EGF treatment and subsequently dephosphorylated by 60 min. In contrast, enhanced recruitment of mSin3A corepressor complex is first observed after 30 min, maintained at 60 min, and reduced to basal levels thereafter. Thus, kinetically, recruitment of the mSin3A complex overlaps the peak of Elk-1 phosphorylation, with enhanced recruitment occurring with a temporal delay. The recruitment of the corepressor complex occurs immediately before c-fos expression is turned off.
HDACs are involved in the regulation of Elk-1-controlled promoters.
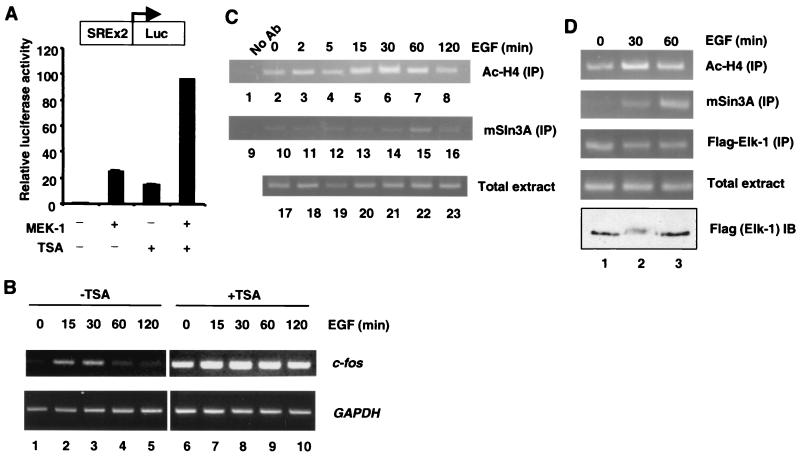
The TCFs play a pivotal role in regulating c-fos induction following growth factor and mitogen stimulation. The results presented here suggest that the TCFs might also be involved in repression of c-fos by recruitment of HDAC-containing complexes. In order to determine whether HDACs are involved in regulating c-fos promoter activity, we first examined the effect of the HDAC inhibitor TSA on the activity of a c-fos SRE-driven luciferase reporter. Transfection of 293 cells with constitutively active MEK-1 leads to stimulation of the SRE reporter. When cells were treated with TSA alone, stimulation of the SRE reporter was also observed, indicating a role for HDACs in maintaining a low basal level of promoter activity. Moreover, synergistic activation was obtained when MEK-1-transfected cells were also treated with TSA (Fig. 7A). These results therefore indicate that HDAC is involved in the regulation of the SRE from the c-fos promoter.
FIG. 7.
Histone deacetylation plays a role in regulating Elk-1-dependent promoters. (A) Effect of TSA on the activation of the Elk-1-regulated reporter gene SRE-Luc. The layout of the reporter is represented as a diagrammatic insert. 293 cells were transfected with 0.25 μg of SREx2-TK-Luc reporter vector together with 0.25 μg of pCMV5-MEK-1, where indicated. Serum-starved cells (18 to 24 h of starvation) were left untreated or treated with TSA for 18 h. The luciferase activities relative to the control cells (without MEK-1 cotransfection and TSA treatment) are presented (mean ± standard deviation, n = 2). (B) Inhibition of HDACs results in loss of basal repression and downregulation of the c-fos gene. RT-PCR analysis of c-fos (top panels) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (bottom panels) transcription following the indicated times of EGF stimulation, in the absence (lanes 1 to 5) and presence (lanes 6 to 10) of TSA treatment. (C) EGF stimulation results in mSin3A recruitment and alterations in the histone acetylation status of the endogenous c-fos SRE. ChIP analysis of the c-fos SRE promoter in 293 cells using antisera specific for acetylated histone H4 (top panel) and mSin3A (middle panel). Total-cell extracts were taken at the indicated times after EGF stimulation. Following immunoprecipitation (IP) of formaldehyde-cross-linked lysates, PCR of eluted DNA using oligonucleotides specific for the c-fos SRE promoter was performed. Total chromatin extracts were used for input controls (bottom panel). As negative controls, lysis buffer alone (data not shown) and protein A-precipitated lysate (15 min of EGF stimulation) were used (lanes 1 to 9). (D) ChIP assays were carried out in the EcR-293-Elk cell lines with antibodies for acetylated histone H4 (Ac-H4), mSin3A, and Flag-tagged Elk-1. Coprecipitated c-fos promoter DNA was detected by PCR. The total levels of Elk-1 in the extracts are shown by Western blotting (bottom panel).
In order to examine a potential role for histone deacetylation in regulating the endogenous c-fos gene in its natural chromatin context, the role of acetylation and deacetylation processes in regulation c-fos promoter induction was investigated. First, the effect of TSA on the expression of c-fos was examined. In the absence of TSA, c-fos is expressed at a low basal level, is induced rapidly by EGF, and returns to basal levels by 120 min (Fig. 7B, lanes 1 to 5). In contrast, in the presence of TSA, the basal level of c-fos expression is elevated, the induction is less pronounced, and the expression does not return to basal levels (Fig. 7B, lanes 6 to 10). Thus, HDACs are important in maintaining the low basal levels of c-fos expression and in reestablishing this basal level following growth factor activation.
Next, we used ChIP analysis to monitor the acetylation status of the endogenous c-fos gene following EGF stimulation. Strikingly, the acetylation status of the c-fos promoter closely mirrored the activation of Elk-1, with high levels of acetylation observed between 15 and 60 min after EGF stimulation. A return to basal acetylation levels was observed between 60 and 120 min (Fig. 7C, top panel). Thus, acetylation and subsequent deacetylation play an important role in the activation and shut-off of c-fos transcription.
A key prediction of our results is that Elk-1-mediated recruitment of mSin3A to the c-fos promoter would occur maximally around 30 to 60 min after EGF stimulation as acetylation levels drop (Fig. 7C) and c-fos promoter activity is abolished (Fig. 7B). ChIP analysis was used to test this directly, using antibodies against mSin3A. Strikingly, recruitment of endogenous mSin3A to the c-fos promoter can be observed, with a peak around 60 min following EGF stimulation (Fig. 7C, middle panel, lane 17). This recruitment occurs with virtually identical kinetics to maximal Elk-1 interactions (Fig. 5B) and correlates well with the loss of acetylation of the c-fos promoter (Fig. 7C, lanes 7 and 8).
In order to demonstrate that Elk-1 occupies the promoter at the same time as mSin3A, we used the EcR-293-Elk cell lines. The enhancement and reduction of histone H4 acetylation and induction of Elk-1 phosphorylation (shown by a shift of the Elk-1 band) were observed with the expected kinetics in both cell lines (Fig. 7D, top and bottom panels, respectively, and data not shown). Moreover, the kinetics of mSin3A recruitment were similar to that seen in 293 cells (Fig. 7D). Elk-1 was present on the c-fos promoter before induction and was maintained up to 1 h after EGF induction (Fig. 7D). Thus, Elk-1 is present on the c-fos promoter as mSin3A is recruited.
These results therefore demonstrate that HDACs, and in particular the mSin3A complex, play a role in regulating Elk-1-dependent promoters and are consistent with our observation that Elk-1 is able to recruit the mSin3A-HDAC corepressor complex.
DISCUSSION
Transcriptional repression is mediated, at least in part, by corepressor complexes containing HDACs. In this study, we demonstrate that the mSin3A-HDAC complex is recruited by the Elk-1 transcription factor following growth factor stimulation and activation of the Erk pathway. This complex contributes to the repression of Elk-1 target promoters such as c-fos.
Determinants of corepressor complex binding.
Elk-1 contains a C-terminal transcriptional activation domain. However, recruitment of the Sin3A-HDAC-1 corepressor complex by Elk-1 is mediated by the N-terminal part of the protein. Indeed, the ETS DNA-binding domain appears to be sufficient to repress transcription (Fig. 1) and to recruit this corepressor complex (Fig. 2 and 4). Furthermore, deletion of the ETS domain also leads to an enhanced response to EGF stimulation (Fig. 5). The ETS domains of other ETS domain transcription factors are also able to bind to the mSin3A-HDAC complex (Fig. 2), raising the possibility that this might be a general property of ETS domain transcription factors. It is, however, possible that additional domains will regulate (either positively or negatively) the ability of other family members to bind to this corepressor complex in a similar way to Elk-1, which uses its C-terminal domain to regulate these interactions (see below). In this regard, it is interesting that the ETS domain protein TEL is a repressor protein (30) and can recruit mSin3A-HDAC complexes (13), and at least in the case of the full-length protein, the ETS domain plays an important role in this repressive process. In vitro pull-down assays (Fig. 2) suggest that either the mSin3A component or the HDAC-1 part of this complex can interact directly with Elk-1, although a role for an adapter protein contained in the reticulocyte lysate cannot be discounted. However, by analogy with other transcription factors, including Mad1 (29), p53 (33), TEL (13), and Ikaros (27), mSin3A is an attractive candidate for the component that binds to Elk-1. It cannot, however, be ruled out that further mSin3A complex components play a role in augmenting interactions in vivo. Mapping of the interaction domain on mSin3A indicates that Elk-1 binding requires sequences in or close to the PAH3 domain, as observed previously for Ikaros (27). This differs significantly from Mad1, which binds to mSin3A via PAH2 (12), and p53, which binds to the linker between PAH2 and PAH3. This indicates that mSin3A has multiple interaction surfaces that can bind to different classes of transcription factors, consistent with a role as a scaffold protein.
As the other TCFs have a domain structure similar to that of Elk-1, it is likely that they may also recruit the mSin3A–HDAC-1 complex following growth factor stimulation. In the case of SAP-2, this would represent a third type of repression domain, as this also possesses two additional repression domains (NID and CID) (10, 31). Thus, by acquiring or evolving different domains, members of the TCF subfamily have evolved into either efficient repressors, such as SAP-2, or proteins of a bipotential nature, such as Elk-1, which can either activate or repress transcription.
MAPK pathway-dependent recruitment of the mSin3A–HDAC-1 corepressor complex.
Signal-mediated switching of transcription factors from a repressive to an active mode has been observed previously for the nuclear hormone receptor family, in which ligand binding triggers the displacement of corepressors and recruitment of coactivator proteins (reviewed in reference 45). Recently, Cdk-mediated phosphorylation has been shown to promote the sequential dissociation of HDACs from retinoblastoma protein (Rb) and Rb from the activator protein E2F (17). In this study, we demonstrate that activation of MAPK pathways can also lead to regulation of the recruitment of corepressor complexes. Activation of the Erk pathway in vivo or phosphorylation of Elk-1 in vitro by Erk-2 results in enhanced binding of the mSin3A-HDAC complex. Phosphorylation of Ser383 is essential for the enhancement of mSin3A recruitment (Fig. 5), although the kinetics of recruitment are slightly delayed in comparison to phosphorylation of Elk-1 at Ser383 (Fig. 6), indicating a temporal delay in corepressor binding. It is currently unclear why this delay occurs, but this suggests that Ser383 phosphorylation acts as a trigger for binding rather than mediating the binding event itself. Interestingly, the replacement of Ser383 and Ser389 with Ala residues results in constitutively higher but noninducible mSin3A binding in vivo (Fig. 5C), indicating that the presence of the Ser residues is actually required to prevent mSin3A binding, presumably by stabilizing a refractory protein conformation. The temporal delay might also reflect that although Ser383 phosphorylation is detectable after 15 min, stoichiometric phosphorylation at multiple sites might not occur until later, and this could be required for corepressor recruitment. Consistent with this hypothesis is the observation that phosphorylation-dependent conformational changes and the resulting allosteric stimulation of DNA binding by Elk-1 only occur at maximal phosphorylation levels (49). It is likely that this conformational change also regulates the interaction of Elk-1 with mSin3A-HDAC as well as the DNA, as it is the same domain (ETS domain) that is directly involved in these two intramolecular binding processes. Indeed, truncated Elk-1 proteins that lack the C-terminal regulatory domain can constitutively interact with the corepressor complex (Fig. 2 and 4). This implies that the C-terminal end either masks the interaction site or promotes an inhibitory conformation, which is subsequently reversed upon phosphorylation. As Elk-1 is initially stimulated to activate transcription by Erk-mediated phosphorylation, a second reason for this temporal delay in corepressor binding might be occlusion by coactivator complexes. Although Erk-mediated Elk-1 phosphorylation promotes the binding of mSin3A-HDAC to Elk-1 in vitro and this is mirrored upon stimulation of the Erk pathway in vivo, it is also possible that this pathway might also modify components of the corepressor complex to potentiate the repressive effect. However, HDAC-1 does not appear to be an Erk substrate (data not shown), and to date there is no other evidence to support this hypothesis. Finally, it is possible that additional Elk-1 modifications are required in addition to Ser383 phosphorylation to promote mSin3A recruitment in vivo. These modifications might be triggered downstream of the Erk MAPK pathway. However, it is clear that activation of the Erk pathway leads to temporal recruitment of the mSin3A complex to Elk-1 in vivo.
Role of Elk-1-mediated corepressor recruitment.
Elk-1 plays a pivotal role in the upregulation of immediate-early genes such as c-fos following growth factor stimulation (reviewed in references 7 and 42). Here we demonstrate that it is also involved in the downregulation of these promoters by recruiting corepressor complexes. The kinetics of corepressor complex recruitment closely follows the kinetics of c-fos shutdown (Fig. 6). Furthermore, the recruitment of the corepressor complex also correlates kinetically with deacetylation occurring at the c-fos promoter (Fig. 7). Thus, Elk-1 is ideally placed to act as both an activator and repressor of c-fos transcription following growth factor stimulation. It is likely that a balance between coactivator and corepressor complex recruitment is achieved following phosphorylation of Elk-1, which initially favors activation but ultimately dictates repression. Thus, one additional role of corepressor recruitment might be to set a ceiling on the degree and duration of activation mediated by Elk-1 in response to growth factor signaling. A similar role has been proposed for the TGIF-HDAC complex that is recruited by Smad2 (44). Our observation that HDAC-containing corepressor complexes are important in c-fos regulation is consistent with previous studies that have indicated that HDACs and the histone acetylation status of the c-fos promoter are important determinants of its activity (1, 8, 34).
It is likely that other mechanisms in addition to HDAC recruitment might also contribute to repression of the c-fos promoter. For example, exchange of Elk-1 for other negatively acting TCFs such as SAP-2 or Net-b might also occur (16). The observation that Id helix-loop-helix proteins (Ids) can promote dissociation of the TCFs suggests a possible mechanism for the exchange of promoter-bound factors (50). However, as the Ids are expressed outside the time window in which the initial promoter shutdown occurs, they are unlikely to contribute to this phase of promoter regulation. Furthermore, we also demonstrate that the repression domain in Elk-1 can repress transcription by different mechanisms at different promoters (Fig. 1). The mechanistic basis for this is unknown, but Elk-1 might repress different targets by either HDAC-dependent or HDAC-independent mechanisms or a combination of both.
Finally, it is interesting that there appears to be a cryptic repression domain embedded within the C-terminal end of the protein that acts via HDACs (Fig. 1). One attractive possibility is that this domain serves to repress Elk-1 activity in the absence of MAPK cascade activation. Future experiments will address this possibility.
Conclusions.
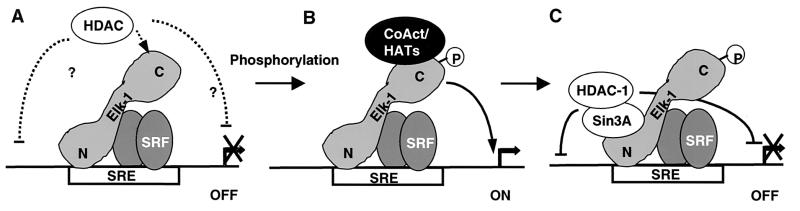
In this study, we demonstrate that Elk-1 can act as both a transcriptional activator and transcriptional repressor protein. Both of these activities are promoted by activation of the Erk MAPK pathway. A model for how this bipotential regulatory role affects the regulation of Elk-1-controlled promoters such as the c-fos SRE is shown in Fig. 8. Upon activation of the Erk pathway, both coactivator and the mSin3A-HDAC corepressor complexes are recruited, with corepressor recruitment incurring a temporal delay (Fig. 8B and C). Removal of the coactivators will then lead to a shutdown of the promoter by the corepressor complex. It is currently unclear what maintains the promoter in a repressive state, although an attractive hypothesis is that a second corepressor complex binds to Elk-1 in its nonphosphorylated state (Fig. 8A). Thus, Elk-1 plays a pivotal role in both the upregulation of immediate-early gene transcription and their subsequent rapid shutdown. It will be interesting to determine whether it also contributes to the maintenance of promoters in their basal repressive states.
FIG. 8.
Role(s) of Elk-1 in up- and downregulating the c-fos SRE. (A) In the absence of Elk-1 phosphorylation, the SRE is inactive. A putative HDAC-dependent pathway is indicated by dotted lines, where HDACs are recruited in an mSin3A-independent manner to the C-terminal end of Elk-1 to repress transcription. (B) Phosphorylation (P) results in activation of Elk-1 and stimulation of the promoter via coactivators (CoAct) and/or histone acetylases (HATs). (C) Following phosphorylation, the HDAC-mSin3A complex is recruited to the N-terminal end of Elk-1 via mSin3A in a phosphorylation-regulatable manner to repress transcription.
ACKNOWLEDGMENTS
We thank Margaret Bell and for excellent technical assistance; Paul Shore, Adam West, and members of our laboratories for comments on the manuscript and stimulating discussions; and Alan Whitmarsh, Roger Davis, Stefan Roberts, and Erik Jansen for reagents.
This work was supported by grants from the AICR (T.K.), Cancer Research Campaign (CRC) (T.K. and A.D.S.), and the Wellcome Trust (A.D.S.) and a Lister Institute of Preventative Medicine Research Fellowship to A.D.S.
REFERENCES
- 1.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 3.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the CAT gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 4.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 6.Brown L A, Amores A, Schilling T F, Jowett T, Baert J L, de Launoit Y, Sharrocks A D. Molecular characterization of the zebrafish PEA3 ETS-domain transcription factor. Oncogene. 1998;17:93–104. doi: 10.1038/sj.onc.1201911. [DOI] [PubMed] [Google Scholar]
- 7.Cahill M A, Janknecht R, Nordheim A. Signal uptake by the c-fos serum response element. In: Bauerle P A, editor. Inducible gene expression. Boston, Mass: Birkhauser; 1995. pp. 39–72. [Google Scholar]
- 8.Cheung P, Tanner K G, Cheung W L, Sassone-Corsi P, Denu J M, Allis C D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 9.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 10.Criqui-Filipe P, Ducret C, Maira S M, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalgleish P, Sharrocks A D. The mechanism of complex formation between Fli-1 and SRF transcription factors. Nucleic Acids Res. 2000;28:560–569. doi: 10.1093/nar/28.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers A L, Billin A N, Liu J, Ayer D E. A 13-amino-acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem. 1999;274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- 13.Fenrick R, Amann J M, Lutterbach B, Wang L, Westendorf J J, Downing J R, Hiebert S W. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovane A, Pintzas A, Maira S M, Sobieszczuk P, Wasylyk B. Net, a new ets transcription factor that is activated by Ras. Genes Dev. 1994;8:1502–1513. doi: 10.1101/gad.8.13.1502. [DOI] [PubMed] [Google Scholar]
- 16.Giovane A, Sobieszczuk P, Ayadi A, Maira S M, Wasylyk B. Net-b, a Ras-insensitive factor that forms ternary complexes with serum response factor on the serum response element of the fos promoter. Mol Cell Biol. 1997;17:5667–5678. doi: 10.1128/mcb.17.10.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbour J W, Luo R X, Dei Santi A, Postigo A A, Dean D C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 18.Herschbach B M, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 19.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 20.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs D, Glossip D, Xing H, Muslin A J, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 22.Janknecht R, Zinck R, Ernst W H, Nordheim A. Functional dissection of the transcription factor Elk-1. Oncogene. 1994;9:1273–1278. [PubMed] [Google Scholar]
- 23.Janknecht R, Ernst W H, Nordheim A. SAP1a is a nuclear target of signaling cascades involving ERKs. Oncogene. 1995;10:1209–1216. [PubMed] [Google Scholar]
- 24.Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 25.Johansen F E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson C A, Turner B M. Histone deacetylases: complex transducers of nuclear signals. Semin Cell Dev Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 27.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 29.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 30.Lopez R G, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. J Biol Chem. 1999;274:30132–30138. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 31.Maira S-M, Wurtz J-M, Wasylyk B. Net (ERP/SAP2), one of the Ras-inducible TCFs, has a novel inhibitory domain with resemblance to the helix-loop-helix motif. EMBO J. 1996;15:5849–5865. [PMC free article] [PubMed] [Google Scholar]
- 32.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 33.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naranjo J R, Mellstrom B, Auwerx J, Mollinedo F, Sassone-Corsi P. Unusual c-fos induction upon chromaffin PC12 differentiation by sodium butyrate: loss of fos autoregulatory function. Nucleic Acids Res. 1990;18:3605–3610. doi: 10.1093/nar/18.12.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 36.Price M A, Rogers A E, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seth A, Gonzalez F A, Gupta S, Raden D L, Davis R J. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 38.Shore P, Sharrocks A D. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shore P, Sharrocks A D. The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 1995;23:4698–4706. doi: 10.1093/nar/23.22.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shore P, Bisset L, Lakey J, Waltho J P, Sharrocks A D. Characterization of the Elk-1 ETS DNA-binding domain. J Biol Chem. 1995;270:5805–5811. doi: 10.1074/jbc.270.11.5805. [DOI] [PubMed] [Google Scholar]
- 41.Treisman R. The serum response element. Trends Biochem Sci. 1992;17:423–426. doi: 10.1016/0968-0004(92)90013-y. [DOI] [PubMed] [Google Scholar]
- 42.Treisman R. Ternary complex factors: growth regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 43.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 44.Wotton D, Lo R S, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 45.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 46.Yang S-H, Yates P R, Whitmarsh A J, Davis R J, Sharrocks A D. The Elk-1 ETS-domain transcription factor contains a MAP kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang S-H, Whitmarsh A J, Davis R J, Sharrocks A D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang S-H, Yates P R, Sharrocks A D. The ETS-domain transcription factors: lessons from the TCF subfamily. Gene Ther Mol Biol. 1999;3:355–371. [Google Scholar]
- 49.Yang S-H, Shore P, Willingham N, Lakey J H, Sharrocks A D. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999;18:5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates P R, Atherton G T, Deed R W, Norton J D, Sharrocks A D. Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J. 1999;18:968–976. doi: 10.1093/emboj/18.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]