Abstract
Background
Preeclampsia (PE) is a major cause of maternal and perinatal morbidity and mortality. Studies on the role of microRNAs (miRNAs), in the pathogenesis of PE through their effects on trophoblast function have been reported, but roles for some miRNAs including miR-513c-5p, have not been identified. We aimed to evaluate potential miRNA candidates that regulate the LRP6 mRNAand to elucidate the possible mechanism in PE. Potential miRNAs were selected by bioinformatics analysis, PCR of placenta tissues and dual luciferase reporter assay of HTR-8/SVneo cells.
Methods
A bioinformatics analysis (Gene Expression Omnibus, GEO; miRWalk) was performed to screen the possible miRNAs that participate in the pathology of PE. Placentas from patients with PE and women with a normal pregnancy were collected to detect the expression of predicted miRNAs by RT-qPCR. A dual luciferase reporter assay was used to test the binding of the potential miRNAs to LRP6. The effects of miR-513c-5p on the biological functions of HTR-8/SVneo cells were further evaluated by performing EdU staining, flow cytometry, wound healing assays and Transwell assays.
Results
GEO and miRWalk predicted 16 miRNAs that might target LRP6. Hsa-miR-371a-5p, hsa-miR-513c-5p, hsa-miR-126-3p, hsa-miR-145-5p, hsa-miR-193b-5p and hsa-miR-296-5p were 6 miRNAs upregulated in the PE placenta. LRP6 was downregulated in patients with PE compared to normal women. miR-513c-5p mimics inhibited LRP6 expression in HTR-8/SVneo cells, and LRP6 is the target gene of miR-513c-5p. miR-513c-5p mimics also inhibited invasion, migration and proliferation of HTR-8/SVneo cells but promoted their apoptosis.
Conclusions
Our study reveals that overexpression of placenta miR-513c-5p is involved in PE by regulating the biological functions of trophoblasts through the inhibition of LRP6.
Keywords: miRNA, Preeclampsia, Trophoblast, miR-513c-5p, Placenta
Background
Preeclampsia (PE) is defined as hypertension (≥140/90 mmHg) developing after 20 weeks’ gestation with one or more of the following: proteinuria, maternal organ dysfunction (including renal, hepatic, hematological, or neurological complications), or fetal growth restriction [1–4]. The main causes of PE remain unclarified, although it is a major cause causing perinatal morbidity and mortality worldwide [5]. Insights into the pathobiology and diagnosis of PE are lacking. Delivery is considered the only cure, indicating that the placenta is of vital importance in the pathogenesis of PE [6]. The search for novel and more effective biomarkers is expected [7].
MicroRNAs (miRNAs) are small noncoding RNAs, usually 22–24 nucleotides long, which bind to messenger RNAs and repress protein expression. More than 2500 mature human miRNAs exist and have important biological functions [8]. Dysregulation of many miRNAs has been reported to play important roles in many diseases, implying that altered circulating miRNA could serve as potential diagnostic and prognostic biomarkers [9, 10].
Low-density lipoprotein receptor-associated protein 6 (LRP6) is an important coreceptor of the Wnt/β-catenin signaling pathway. As shown in our previous study, LRP6 knockdown in the trophoblast cell line HTR-8/SVneo significantly impairs migration, invasion, and tube formation likely mediated by suppressing Wnt/β-catenin signaling pathway [11]. We further revealed that LRP6 regulates Rab7-mediated autophagy through the Wnt/β-catenin pathway to modulate trophoblast cell migration and invasion [12].
According to our previous studies, LRP6 downregulation potentially contributes to PE development. We aimed to evaluate potential miRNA candidates that regulate the LRP6 mRNA and to elucidate the possible mechanism in PE. In vitro experiments were carried out in this study to elucidate the possible mechanism. Potential miRNAs were seleted by bioinformatics analysis (Gene Expression Omnibus database, GEO, RRID:SCR_005012; miRwalk version 2.0, https://zmf.umm.uni-heideberg.de/apps/zmf/mirwalk2, RRID:SCR_016509), RT-qPCR of placenta tissues and dual luciferase reporter assay of HTR-8/SVneo cells to quantify their levels and make sure the regulation of LRP6 and miRNAs.
Materials and methods
Bioinformatics analysis
We selected the GSE15789, GSE69452, GSE84260 and GSE85926 datasets from the GEO, database including patients with PE and normal controls (NCs). Then, an adjusted log2fold change≥1.5 was used as the threshold to identify upregulated miRNAs [13, 14]. miRwalk version 2.0 is a comprehensive database that provides information on predicted miRNAs, as well as the verified binding sites on the target gene. It is currently recognized as miRNA target gene prediction software with a low false-positive rate. “LRP6” was input into the “Gene Targets” frame, select “Human” was selected as the species and 3 ‘UTR as the combined region, and all the prediction software included, this package was used to select miRNAs with frequency ≥ 3/5 [15] and identify miRNAs that may be structurally related to the regulation of LRP6. Next, the GEO and miRWalk results were intersected to identify the candidate miRNAs.
Dual luciferase reporter assay
A dual luciferase reporter assay was utilized to confirm whether the predicted miRNAs bond to the 3’UTR of LRP6. HEK293T cells purchased from American Type Culture Collection (ATCC, RRID:CVCL_0063) were transfected using X-tremegene HP Transfection Reagent (Roche, Basel, Switzerland,), according to the manufacturer’s protocol. Cells (1.0× 105 cells/well) were seeded in 24-well plates and transfection was performed when HEK293T cells reached 60% confluence in a 24-well plate. Plasmids (GeneChem, Shanghai, China) were mixed in Opti-MEM (Gibco, Burlington, Canada) and incubated with the transfection reagent for 20 min at room temperature. Then the cells were cultured in a 5% CO2 incubator at 37 °C for 5 to 6 hours. Twenty-four hours after transfection, the expression of fluorescently labeled genes was observed to determine the transfection efficiency. Forty-eight hours later, luminescence was detected using the Dual-Glo™ Luciferase Assay System (Promega, Madison, USA) according to the manufacturer’s protocol. Data were normalized to Renilla luminescence, and the results are presented relative to the control miRNA transfected group.
Tissue samples
Twelve pregnant women were included in this study, consisting of 6 patients with PE and 6 women with a normal pregnancy. The study was exploratory and no sample size calculation was performed a priori to assess statistical power. The study was approved by the Institutional Research Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong First Medical University (NO.2019–238), and informed consent forms were signed by every donor. Blood pressure (≥140/90 mmHg) and proteinuria occurring after 34-gestationalweeks were the necessary inclusion criteria for patients with PE, all the patients had headaches and only 2 had fetal growth restriction. Patients who had systemic illnesses were excluded. All donors were delivered by cesarean section and were ≤ 35 years old. Villus tissues form placentas were obtained, placed in TRIzol reagent (Invitrogen, Carlsbad, USA, RRID:Addgene_27409), and stored at − 80 °C until RNA extraction and verification of the differentially expressed miRNAs predicted by the bioinformatics analysis.
Cell culture and miRNA transfection
The HTR-8/SVneo cell line purchased from ATCC (RRID:CVCL_7162) was maintained in DMEM/F12 (Gibco, Grand Island, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, USA), and 1% penicillin and streptomycin in a 5% CO2 atmosphere at 37 °C. The miR-513c-5p mimics and negative control were obtained from Gemma Pharma and transfected at a concentration of 50 nM. The two established cell lines sequences were named miR-513c-5p (5′- UUCUCAAGGAGGUGUCGUUUAU-3′) and negative control RNA (NC) (5′- UUCUCCGAACGUGUCACGUTT-3′). Briefly, the transfection reagent, Lipofectamine RNAiMAX (Life Technologies, Carlsbad, USA), was diluted in Opti-MEM and added to the medium; 30 min later, cells were seeded in each well. At 6 h after transfection, the culture medium was replaced with fresh medium; 24 h after plating, cells were harvested for detection. Transfection efficiency was determined by analyzing gene expression 24 h later, using “reverse transcription-quantitative polymerase chain reaction” (RT-qPCR).
Literature search for miRNAs of interest
PubMed (https://pm.yuntsg.com/) was searched for the needed literature. The name of one of the miRNAs awaiting for dual luciferase reporter assay verification and “trophoblast” were input respectively to retrieve relevant references.
RNA extraction and RT-qPCR
The quantification and amplification of mRNAs and cDNA quantification in placental tissues and the HTR-8/SVneo cell line were performed using SYBR green-based RT-qPCR, as described previously [16]. TRIzol reagent was used to isolate total RNA from placental tissues or the HTR-8/SVneo cell line. A First Strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, USA) was utilized to synthesize cDNAs from 1 μg of total RNA. RT-qPCR with gene-specific primers was performed on the resulting cDNAs using Fast SYBR green double-stranded DNA binding dye (Applied Biosystems, Foster city, USA) and a Roche LightCycler® 480 II sequence detection system (Roche, Basel, Swizerland). Primer sequences of LRP6, β-actin, miR-513c-5p and U6 are shown in Table 1. The RT-qPCR profile was as follows: amplification at 37 °C for 15 min, 85 °C for 5 s, and 4 °C before removal. This process was followed by 40 cycles of 95 °C for 10 min, 95 °C for 10 s, 57 °C for 30 s and 72 °C for 10 s. U6 and β-actin were used as internal controls for miRNAs and LRP6, respectively. The experiments were performed in triplicate, and the results were analyzed using the 2-ΔΔCt method.
Table 1.
Sequences of RT-qPCR primers
| LRP6 Forward | TATTGTCCCCCGATGGGCTG |
| LRP6 Reverse | AGTACATGAACCCACTTGAAGGA |
| β-actin Forward | TTCCAGCAGATGTGGATCAGC |
| β-actin Reverse | GAAGCATTTGCGGTGGAC |
| miR-513c-5p Forward | AGCAGGTTCTCAAGGAGGTGTC |
| miR-513c-5p Reverse | TAAGGTTCTTCACGACTGGTTCAC |
| U6 Forward | CAGCACATATACTAAAATTGGAACG |
| U6 Reverse | ACGAATTTGCGTGTCATCC |
Cell proliferation analysis
The proliferation of HTR-8/SVneo cells was measured using EdU staining (RuiboBio, Guangzhou, China) according to the manufacturer’s protocol. Briefly, HTR-8/SVneo cells were transfected as mentioned above, and 3000 cells were plated in each well of 96-well plates. Twelve hours later, EdU was added to the medium at a final concentration of 50 μM for 1 h. Immunofluorescence staining was performed using EdU reagent kits according to the manufacturer’s instructions. The stained cells were examined with high content screening (Molecular Devices, Silicon Valley, USA) and photographed with a 10× objective. The experiment was performed in triplicate. The positive incidence was proliferative cells/all cells observed, which was defined as red dots/DAPI counts.
Cell apoptosis analysis
Apoptosis was analyzed using flow cytometry. Twenty-four hours after transfection, HTR-8/SVneo cells were digested with 0.25% trypsin. Apoptotic cells were detected using the Annexin V-FITC/propidium iodide (PI) Apoptosis Detection Kit (Sungene Biotech, Wuhan, China) after treatment. According to the manufacturer’s instructions, the stained cells were assayed using a CytoFLEX flow cytometer (Beckman, Miami, USA) after fixation. The positive cells were calculated and analyzed with CytoExpert 2.0 software (Beckman Coulter, Miami, USA).
Cell migration analysis
The ability of HTR-8/SVneo cells to migrate was tested by performing wound healing assays. The transfected HTR-8/SVneo cells were cultured in a 96-well plate and stained with PKH67 (Sigma, Saint Louis, USA) for 15 min. The necrotic cells were removed, and then the remaining cells were cultured in serum-free DMEM/F12, which was set as 0 h. The area between the scratches was photographed and calculated by high content screening (10x objective) after 12 h, and the migration of the cells was determined by calculating the scratch area at 0 h - scratch area at 12 h.
Cell invasion analysis
Transwell assays were performed to assess the invasive ability of HTR-8/SVneo cells. For this experiment. 50 μl of undiluted Matrigel (BD Biosciences, USA) were added to a 8 μm Transwell plate (Merck Millipore, Billerica, USA). After transfection, 5 × 105 HTR-8/SVneo cells were seeded in a Transwell chamber and incubated for 48 h. Then, 4% paraformaldehyde was used to fix the cells in the Transwell chamber, and 0.1% crystal violet was used to stain the HTR-8/SVneo cells on the underside of the Transwell chamber. The invading cells were examined using a digital microscope at 200 x magnification and counted using ImageJ software. Each experiment was repeated three times.
Statistical analysis
Data is presented as means with standard deviation (Sd). The comparison of demographic characteristics between groups was performed using an unpaired Student’s t test. All experiments were of equal variance. All statistical analyses were performed using GraphPad Prism version 5.01. P values < 0.05 were considered to be significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Potential miRNAs predicted by bioinformatics analysis
We used the online programs GEO and miRWalk to predict candidate miRNAs that might target LRP6 in humans. Placenta tissue miRNAs of PE and normal pregnant women were search by GEO, and 4 related datasets were obtained. Forty-two upregulated miRNAs were identified (Table 2). miRWalk provided 324 miRNAs according to the set frequency (Table 3). Finally, 16 identified miRNAs that were upregulated in PE and that potentially target LRP6 (hsa-miR-129-2-3p, hsa-miR-409-3p, hsa-miR-765, hsa-miR-371a-5p, hsa-miR-296-5p, hsa-miR-874-3p, hsa-miR-605, hsa-miR-513a-5p, hsa-miR-513c-5p, hsa-miR-126-3p, hsa-miR-143-3p, hsa-miR-145-5p, hsa-miR-193b-5p, hsa-miR-27a-5p, hsa-miR-412-5p, and hsa-miR-497-5p) at the junction of the GEO and miRWalk results.
Table 2.
42 upregulated miRNAs filtrated by 4 datasets
| Name | logFC |
|---|---|
| hsa-miR-497-5p | 2.506 |
| hsa-miR-199b-5p | 2.316 |
| hsa-miR-615-3p | 2.301 |
| PREDICTED_MIR192 | 2.280 |
| hsa-miR-1247-5p | 2.193 |
| hsa-miR-455-3p | 2.110 |
| PREDICTED_MIR206 | 2.097 |
| hsa-miR-145-5p | 2.066 |
| hsa-miR-513-5p | 2.062 |
| hsa-miR-3178 | 2.042 |
| hsa-miR-455-5p | 2.024 |
| hsa-miR-409-3p | 1.968 |
| hsa-miR-371-5p | 1.950 |
| hsa-miR-199a-5p | 1.918 |
| PREDICTED_MIR145 | 1.888 |
| PREDICTED_MIR172 | 1.879 |
| PREDICTED_MIR143 | 1.876 |
| hsa-miR-765 | 1.871 |
| PREDICTED_MIR112 | 1.869 |
| hsa-miR-675-5p | 1.852 |
| hsa-miR-129-3p | 1.831 |
| hsa-miR-27a-5p | 1.827 |
| PREDICTED_MIR88 | 1.814 |
| hsa-miR-1248 | 1.744 |
| hsa-miR-143-3p | 1.731 |
| PREDICTED_MIR160 | 1.686 |
| hsa-miR-125a-3p | 1.665 |
| hsa-miR-199a-3p | 1.639 |
| hsa-miR-126-3p | 1.637 |
| hsa-miR-668-3p | 1.628 |
| hsa-miR-888 | 1.603 |
| hsa-miR-412-5p | 1.596 |
| hsa-miR-6821-5p | 1.586 |
| hsa-miR-296-5p | 1.582 |
| has-PreMIR-194-2 | 1.563 |
| hsa-miR-526b-5p | 1.557 |
| hsa-miR-10b-5p | 1.550 |
| hsa-miR-199b-3p | 1.538 |
| hsa-miR-874 | 1.531 |
| hsa-miR-193b-5p | 1.521 |
| hsa-miR-605 | 1.512 |
| hsa-miR-10a-5p | 1.506 |
A total of 42 upregulated miRNAs(log2 Fold Change≥1.5)obtained by GEO when the 4 sets of miRNAs mentioned were analyzed
Table 3.
324 miRNAs predicted by miRWalk
| Gene | miRNA | StemLoopID | miRanda | miRDB | miRWalk | RNA22 | Targetscan | SUM |
|---|---|---|---|---|---|---|---|---|
| LRP6 | hsa-miR-183 | hsa-mir-183 | 1 | 1 | 1 | 1 | 1 | 5 |
| LRP6 | hsa-miR-641 | hsa-mir-641 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-30d | hsa-mir-30d | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-195 | hsa-mir-195 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-381 | hsa-mir-381 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-424 | hsa-mir-424 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-501-5p | hsa-mir-501 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-29a | hsa-mir-29a | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548i | hsa-mir-548i-4 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-545 | hsa-mir-545 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-497 | hsa-mir-497 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-450b-5p | hsa-mir-450b | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-30a | hsa-mir-30a | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-603 | hsa-mir-603 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-570 | hsa-mir-570 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-30b | hsa-mir-30b | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-448 | hsa-mir-448 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-624 | hsa-mir-624 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-21 | hsa-mir-21 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548 h | hsa-mir-548 h-4 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-454 | hsa-mir-454 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-651 | hsa-mir-651 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548b-5p | hsa-mir-548b | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-204 | hsa-mir-204 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-942 | hsa-mir-942 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-559 | hsa-mir-559 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-577 | hsa-mir-577 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-590-5p | hsa-mir-590 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-205 | hsa-mir-205 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548c-5p | hsa-mir-548c | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-15a | hsa-mir-15a | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-224 | hsa-mir-224 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-579 | hsa-mir-579 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-16 | hsa-mir-16-2 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548d-5p | hsa-mir-548d-2 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548p | hsa-mir-548p | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-590-3p | hsa-mir-590 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-211 | hsa-mir-211 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-409-3p | hsa-mir-409 | 1 | 0 | 1 | 1 | 1 | 4 |
| LRP6 | hsa-miR-548c-3p | hsa-mir-548c | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-1270 | hsa-mir-1270 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-580 | hsa-mir-580 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-29c | hsa-mir-29c | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-29b | hsa-mir-29b-2 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-15b | hsa-mir-15b | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-495 | hsa-mir-495 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-300 | hsa-mir-300 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-30c | hsa-mir-30c-1 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548a-5p | hsa-mir-548a-3 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-620 | hsa-mir-620 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-548j | hsa-mir-548j | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-582-5p | hsa-mir-582 | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-30e | hsa-mir-30e | 1 | 1 | 1 | 0 | 1 | 4 |
| LRP6 | hsa-miR-518a-5p | hsa-mir-518a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1285 | hsa-mir-1285-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-17 | hsa-mir-17 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-23b | hsa-mir-23b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 h | hsa-mir-548 h-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-193b | hsa-mir-193b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-582-3p | hsa-mir-582 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-509-5p | hsa-mir-509-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-200a | hsa-mir-200a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548f | hsa-mir-548f-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-654-3p | hsa-mir-654 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-361-3p | hsa-mir-361 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520f | hsa-mir-520f | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-600 | hsa-mir-600 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-543 | hsa-mir-543 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-623 | hsa-mir-623 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-214 | hsa-mir-214 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1261 | hsa-mir-1261 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-488 | hsa-mir-488 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-569 | hsa-mir-569 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320c | hsa-mir-320c-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-105 | hsa-mir-105-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-126 | hsa-mir-126 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320d | hsa-mir-320d-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520c-3p | hsa-mir-520c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518a-3p | hsa-mir-518a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-645 | hsa-mir-645 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1285 | hsa-mir-1285-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-20a | hsa-mir-20a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-27b | hsa-mir-27b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 h | hsa-mir-548 h-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-584 | hsa-mir-584 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-7 | hsa-mir-7-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-346 | hsa-mir-346 | 1 | 1 | 0 | 0 | 1 | 3 |
| LRP6 | hsa-miR-340 | hsa-mir-340 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-182 | hsa-mir-182 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548f | hsa-mir-548f-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-20b | hsa-mir-20b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-502-5p | hsa-mir-502 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-655 | hsa-mir-655 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-141 | hsa-mir-141 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1279 | hsa-mir-1279 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-362-5p | hsa-mir-362 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-515-5p | hsa-mir-515-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-217 | hsa-mir-217 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1262 | hsa-mir-1262 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-491-3p | hsa-mir-491 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-487b | hsa-mir-487b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-34b | hsa-mir-34b | 1 | 0 | 0 | 1 | 1 | 3 |
| LRP6 | hsa-miR-1323 | hsa-mir-1323 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-129-3p | hsa-mir-129-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320c | hsa-mir-320c-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-369-3p | hsa-mir-369 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-330-3p | hsa-mir-330 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520 h | hsa-mir-520 h | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-513a-3p | hsa-mir-513a-2 | 1 | 1 | 0 | 0 | 1 | 3 |
| LRP6 | hsa-miR-646 | hsa-mir-646 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1287 | hsa-mir-1287 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 h | hsa-mir-548 h-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548a-3p | hsa-mir-548a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-874 | hsa-mir-874 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320a | hsa-mir-320a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302a | hsa-mir-302a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548f | hsa-mir-548f-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-503 | hsa-mir-503 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-656 | hsa-mir-656 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-31 | hsa-mir-31 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-143 | hsa-mir-143 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1274b | hsa-mir-1274b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-519c-3p | hsa-mir-519c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-605 | hsa-mir-605 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-301b | hsa-mir-301b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1236 | hsa-mir-1236 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-219-5p | hsa-mir-219-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548n | hsa-mir-548n | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-146b-3p | hsa-mir-146b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-554 | hsa-mir-554 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-573 | hsa-mir-573 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1271 | hsa-mir-1271 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-105 | hsa-mir-105-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320d | hsa-mir-320d-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-371-5p | hsa-mir-371 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518c | hsa-mir-518c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-328 | hsa-mir-328 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-522 | hsa-mir-522 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-650 | hsa-mir-650 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1290 | hsa-mir-1290 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-124 | hsa-mir-124-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181d | hsa-mir-181d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-587 | hsa-mir-587 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-7 | hsa-mir-7-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-219-5p | hsa-mir-219-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548f | hsa-mir-548f-4 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-505 | hsa-mir-505 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-659 | hsa-mir-659 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-4 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-145 | hsa-mir-145 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1288 | hsa-mir-1288 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-607 | hsa-mir-607 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-509-3-5p | hsa-mir-509-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1237 | hsa-mir-1237 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-let-7d | hsa-let-7d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-221 | hsa-mir-221 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 m | hsa-mir-548 m | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-555 | hsa-mir-555 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-576-5p | hsa-mir-576 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-149 | hsa-mir-149 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1826 | hsa-mir-1826 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-372 | hsa-mir-372 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-519d | hsa-mir-519d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-629 | hsa-mir-629 | 1 | 1 | 0 | 0 | 1 | 3 |
| LRP6 | hsa-miR-323-3p | hsa-mir-323 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-519a | hsa-mir-519a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1291 | hsa-mir-1291 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-124 | hsa-mir-124-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302e | hsa-mir-302e | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-512-5p | hsa-mir-512-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-889 | hsa-mir-889 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181b | hsa-mir-181b-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548f | hsa-mir-548f-5 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-513a-5p | hsa-mir-513a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-660 | hsa-mir-660 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-5 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520a-3p | hsa-mir-520a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-612 | hsa-mir-612 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-935 | hsa-mir-935 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-628-5p | hsa-mir-628 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1238 | hsa-mir-1238 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-222 | hsa-mir-222 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548o | hsa-mir-548o | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-202 | hsa-mir-202 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-557 | hsa-mir-557 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-576-3p | hsa-mir-576 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320b | hsa-mir-320b-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-106a | hsa-mir-106a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-186 | hsa-mir-186 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1827 | hsa-mir-1827 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520d-3p | hsa-mir-520d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-148b | hsa-mir-148b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-527 | hsa-mir-527 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548d-5p | hsa-mir-548d-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 k | hsa-mir-548 k | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-23a | hsa-mir-23a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-124 | hsa-mir-124-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302f | hsa-mir-302f | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548a-3p | hsa-mir-548a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-875-5p | hsa-mir-875 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-7 | hsa-mir-7-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-128 | hsa-mir-128-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-452 | hsa-mir-452 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-513a-5p | hsa-mir-513a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-758 | hsa-mir-758 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-93 | hsa-mir-93 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-152 | hsa-mir-152 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-664 | hsa-mir-664 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302b | hsa-mir-302b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-630 | hsa-mir-630 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1201 | hsa-mir-1201 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1266 | hsa-mir-1266 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-802 | hsa-mir-802 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-373 | hsa-mir-373 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520 g | hsa-mir-520 g | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-194 | hsa-mir-194-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-331-5p | hsa-mir-331 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-519a | hsa-mir-519a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548d-3p | hsa-mir-548d-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-128 | hsa-mir-128-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1277 | hsa-mir-1277 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-512-5p | hsa-mir-512-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-190b | hsa-mir-190b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-506 | hsa-mir-506 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1264 | hsa-mir-1264 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-153 | hsa-mir-153-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-519b-3p | hsa-mir-519b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-633 | hsa-mir-633 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1206 | hsa-mir-1206 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1269 | hsa-mir-1269 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-432 | hsa-mir-432 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-561 | hsa-mir-561 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-765 | hsa-mir-765 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-188-5p | hsa-mir-188 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-374a | hsa-mir-374a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-106b | hsa-mir-106b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-338-3p | hsa-mir-338 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-499-5p | hsa-mir-499 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 l | hsa-mir-548 l | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-130a | hsa-mir-130a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-301a | hsa-mir-301a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-885-5p | hsa-mir-885 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1256 | hsa-mir-1256 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-507 | hsa-mir-507 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-671-5p | hsa-mir-671 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-29b | hsa-mir-29b-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-153 | hsa-mir-153-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-513b | hsa-mir-513b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302c | hsa-mir-302c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-297 | hsa-mir-297 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-635 | hsa-mir-635 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1208 | hsa-mir-1208 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-494 | hsa-mir-494 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-770-5p | hsa-mir-770 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-190 | hsa-mir-190 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-377 | hsa-mir-377 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518a-5p | hsa-mir-518a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181a | hsa-mir-181a-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-6 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-339-5p | hsa-mir-339 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-499-3p | hsa-mir-499 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548d-3p | hsa-mir-548d-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-132 | hsa-mir-132 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548i | hsa-mir-548i-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-296-5p | hsa-mir-296 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520e | hsa-mir-520e | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-593 | hsa-mir-593 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-212 | hsa-mir-212 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1257 | hsa-mir-1257 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-412 | hsa-mir-412 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-509-5p | hsa-mir-509-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-767-5p | hsa-mir-767 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-513c | hsa-mir-513c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518f | hsa-mir-518f | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-618 | hsa-mir-618 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1184 | hsa-mir-1184 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548e | hsa-mir-548e | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-16 | hsa-mir-16-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1272 | hsa-mir-1272 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-567 | hsa-mir-567 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-581 | hsa-mir-581 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-193a-3p | hsa-mir-193a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518a-3p | hsa-mir-518a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181b | hsa-mir-181b-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-7 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-500 | hsa-mir-500 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-27a | hsa-mir-27a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548i | hsa-mir-548i-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-130b | hsa-mir-130b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-515-5p | hsa-mir-515-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1184 | hsa-mir-1184 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181a | hsa-mir-181a-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1259 | hsa-mir-1259 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-485-5p | hsa-mir-485 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-510 | hsa-mir-510 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-767–3p | hsa-mir-767 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1197 | hsa-mir-1197 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-302d | hsa-mir-302d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-520b | hsa-mir-520b | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1274a | hsa-mir-1274a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-892a | hsa-mir-892a | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-30c | hsa-mir-30c-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-194 | hsa-mir-194-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-380 | hsa-mir-380 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518d-3p | hsa-mir-518d | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-181c | hsa-mir-181c | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-8 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-384 | hsa-mir-384 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1302 | hsa-mir-1302-2 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-137 | hsa-mir-137 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548i | hsa-mir-548i-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548a-3p | hsa-mir-548a-3 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-873 | hsa-mir-873 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-622 | hsa-mir-622 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-1184 | hsa-mir-1184 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-548 g | hsa-mir-548 g | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-532-5p | hsa-mir-532 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-320b | hsa-mir-320b-1 | 1 | 0 | 1 | 0 | 1 | 3 |
| LRP6 | hsa-miR-518b | hsa-mir-518b | 1 | 0 | 1 | 0 | 1 | 3 |
324 miRNAs were predicted by miRWalk that might involve in LRP6 regulation (frequency ≥ 3/5)
MiR-513c-5p was selected as the target miRNA
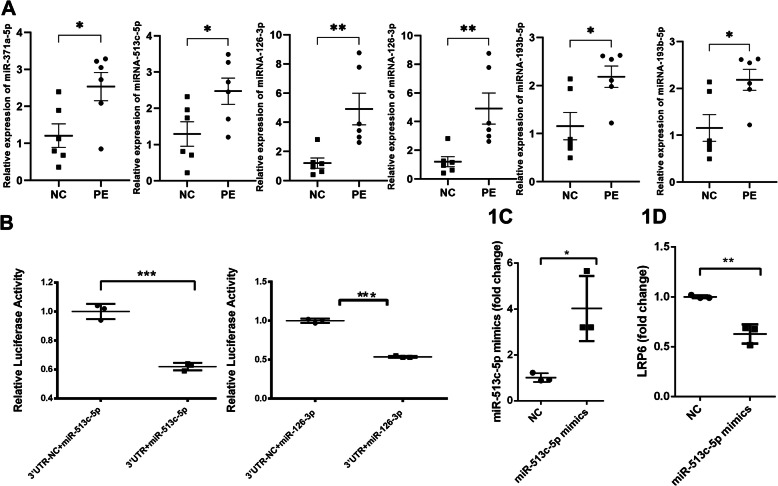
RT-qPCR of placental tissue from 6 patients with PE and 6 women with normal pregnancies was performed to validate whether the levels of the 16 miRNAs in the placenta were higher in pregnant women with PE than in normal pregnant women (the demographics of the patients are shown in Table 4). Our data revealed that 6 miRNAs (hsa-miR-371a-5p, hsa-miR-513c-5p, hsa-miR-126-3p, hsa-miR-145-5p, hsa-miR-193b-5p and hsa-miR-296-5p) were upregulated and showed a trend toward significance in association with PE (Fig.1a, miR-371a: P < 0.05, 1.210 ± 0.714, 2.538 ± 0.853; miR-513c-5p: P < 0.05, 1.292 ± 0.754, 2.474 ± 0.809; miR-126-3p: P < 0.01, 1.204 ± 0.780, 4.915 ± 2.424; miR-145-5p: P < 0.05, 1.116 ± 0.523, 2.060 ± 0.670; miR-193b-5p: P < 0.05, 1.170 ± 0.637; miR-296-5p: P < 0.05, 1.870 ± 0.602).
Table 4.
Patient demographics
| PE patients | Normal pregnant women | P value | |
|---|---|---|---|
| Gestational weeks (minimum-maximum) | 35–37 | 38–40 | < 0.000 |
| Systolic blood pressure (mm Hg) | 159.2 ± 2.5 | 108.7 ± 2.1 | < 0.000 |
| Diastolic blood pressure (mm Hg) | 105.8 ± 3.7 | 75.3 ± 1.7 | < 0.000 |
| Proteinuria(mean ± Sd) | 0.771 ± 0.113 | 0.000 ± 0.000 | <0.000 |
| Maternal age | 31.830 ± 1.014 | 29.830 ± 0.792 | 0.151 |
Fig. 1.
MiR-513c-5p and miR-126-3p targeted the 3’UTR of LRP6. A RT-qPCR detected the expression of 16 miRNAs in placental tissues, and 6 miRNAs with increased expression were identified. B The 3’UTR of LRP6 harbors one complementary binding site for miR-513c-5p and the miR-126-3p sequences. C MiR-513c-5p mimics increased the expression of miR-513c-5p. D MiR-513c-5p mimics inhibited LRP6 expression
Meanwhile, a literature search was needed to better understand the relationship of 6 candidate miRNAs, LPP6 and gestation-related diseases. After a literature search in PubMed, 2 studies at most for each miRNA were acquired. A previous study reported that miR-371a-5p regulates an X-linked inhibitor of apoptosis protein in the pathogenesis of recurrent pregnancy loss [17], and miR-371a-5p promoted the proliferation, migration, and invasion of choriocarcinoma cells [18]. A trend toward the downregulation of miR-126-3p was observed in women with relevant pregnancy-related complications (PE, gestational hypertension and intrauterine growth restriction) compared with women with a normal pregnancy [19, 20]. Tumor necrosis factor-alpha suppresses the invasion of HTR-8/SVneo trophoblast cells through microRNA-145-5p-mediated downregulation of Cyr61 [21]. Higher miR-193b-5p expression in placentae from patients with early-onset pregnancy complications might be involved in the pathogenesis of PE and intrauterine growth restriction [22]. Exosomes containing miR-296-5P have been successfully delivered to recipient cells and might play a biological role in conceptus-endometrial cross-talk crucial for a successful pregnancy [23]. Although the roles of these six miRNAs in the relationship between LRP6 and gestation-related diseases were not explored, five of the six (except miR-513c-5p) mentioned miRNAs were reported to be involved in gestational-related diseases. From a different perspective, the aforementioned literature retrieval process provided evidence for the credibility of GEO and miRWalk analyses, and we wondered whether the candidate miRNAs described above bind to LRP6 and affect trophoblast function.
Targeting relationship verified by luciferase report assay
Bioinformatics analysis and RT-qPCR selected 6 miRNAs as candidate miRNAs. The miRNA-target relationship was verified by performing a luciferase reporter assay. All 6 miRNAs were identified, but only the binding of miR-513c-5p and miR-126-3p to the targeting sites of LRP6 reduced luciferase expression. The incubation of the 3’UTR plasmids with miR-513c-5p and miR-126-3p resulted in decreased luciferase activity compared to the 3’UTR-NC groups, indicating that miR-513c-5p and miR-126-3p targeted LRP6 (Fig. 1b, miR-513c-5p: P < 0.001,1.000 ± 0.053, 0.620 ± 0.027; miR-126-3p:P < 0.001, 1.000 ± 0.026,0.537 ± 0.012). Based on these results, miR-513c-5p and miR-126-3p directly target LRP6 and downregulate its expression. In subsequent experiments, miR-513c-5p was randomly selected from the two miRNAs. Further assays of miR-126-3p are still needed.
MiR-513c-5p regulated the expression of LRP6 in HTR-8/SVneo cells in vitro
The miR-513c-5p mimics and their negative control (NC) were synthesized to further determine the roles of miR-513c-5p in the development of PE. Then, the transfection efficiency of miR-513c-5p mimics in HTR-8/SVneo cells was tested by performing RT-qPCR assay. As presented in Fig. 1c, the introduction of miR-513c-5p mimics detected was higher than the negative control in HTR-8/SVneo cells (P < 0.05, 1.011 ± 0.189, 4.022 ± 1.416), suggesting that miR-513c-5p mimics would be useful for subsequent gain-of-function experiments. The LRP6 levels in the miR-513c-5p group and NC group were detected, and the expression of the LRP6 mRNA was reduced in the miR-513c-5p group (Fig. 1d, P < 0.01, 1.000 ± 0.0161, 0.627 ± 0.097).
Knockdown of miR-513c-5p inhibited proliferation, invasion migration and promoted apoptosis in HTR-8/SVneo cells.
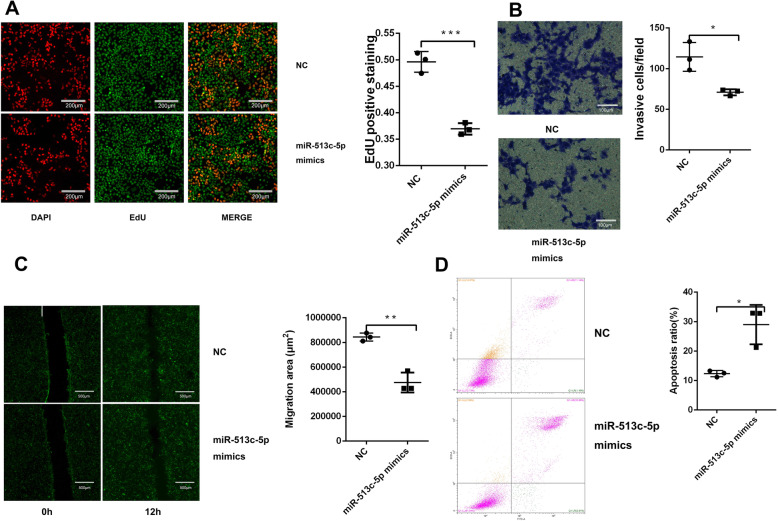
We determined the effect of miR-513c-5p on HTR-8/SVneo cell proliferation using an EdU assay. HTR-8/SVneo cell proliferation was significantly decreased in the miR-513c-5p group compared to the NC group (Fig.2a, P < 0.01, 0.496 ± 0.0120, 0.369 ± 0.011). These data indicated that the proliferative ability of trophoblast cells was obviously affected by miR-513c-5p overexpression. Invasion assay depending on Matrigel demonstrated that invasion activity of HTR-8/SVneo cells was remarkably suppressed with excessive miR-513c-5p expression (Fig.2b, P < 0.01, 114 ± 18, 71 ± 4). The growth rate of miR-513c-5p overexpressing cells was significantly decreased compared to that of NC cells. Wound healing assays showed that the miR-513c-5p mimic treatment markedly inhibited the motility of HTR-8/SVneo cells, as determined by the migration area (Fig.2c, P < 0.01, 843,815 ± 32,623, 474,689 ± 81,106). The cells were analyzed using flow cytometry to determine late apoptotic activity. As shown in Fig.2d, the mimics induced the apoptosis of 28.97 ± 3.87% of HTR-8/SVneo cells, while NC induced the apoptosis of only 12.31 ± 0.62% of cells. Overexpression of miR-513c-5p significantly increased HTR-8/SVneo cells apoptosis (P < 0.05, 12 ± 1, 29 ± 7).
Fig. 2.
MiR-513c-5p was selected as a miRNA of interest based on the results of the bioinformatic analysis and RT-qPCR. The miR-513c-5p mimic suppressed cell proliferation, invasion and migration but promoted cell apoptosis in HTR-8/SVneo cells. The cell proliferative ability and cell apoptotic rate were determined at 24 h after transfection. A The EdU assay showed that the proliferation of HTR-8/SVneo cells was reduced by the miR-513c-5p mimic (n = 3). B Invasion assays revealed that the invasive ability was inhibited by the miR-513c-5p mimic. The Transwell chamber assay detected the changes in the invasive capabilities of HTR-8/SVneo cells. Compared to NC, trophoblast cells transfected with the miR-513c-5p mimic were significantly less invasive (P < 0.05). C The miR-513c-5p mimic inhibited the migration of HTR-8/SVneo cells, as analyzed using the wound healing assay. Quantitative analysis of the migration area was assessed. D The flow cytometry analysis revealed an increase in the percentage of apoptotic cells after transfection with miR-513c-5p mimics compared with NC
Discussion
Due to the severity of PE, its pathology has always been a research focus. Increasing evidence reveals that miRNAs may participate in the pathology of PE [9, 24, 25]. Accumulating evidence highlights the role of LRP6 in PE. Low LRP6 expression might be responsible for lower trophoblast migration and invasion and subsequent PE, and the mechanisms showed a strong association with Wnt/β-catenin pathway [26]. The overexpression of miR-95-5p regulates the expression of matrix metalloproteinase-2, matrix metalloproteinase-9 and tissue inhibitors of metalloproteinase-1 in trophoblast cells by targeting LRP6, thereby participating in the metastasis of trophoblast cells and causing the occurrence and progression of PE [27]. LRP6 was reported to be involved in the proliferation, migration and invasion of trophoblast cells via miR-346 [28]. Notably, miR-590-3p might inhibit trophoblast-dependent maternal spiral artery remodeling by regulating both trophoblast invasion and endovascular formation through the repression of LRP6 [29]. Based on the results, LRP6 might be regulated by different miRNAs that changing trophoblast function.
In recent years, an increasing number of investigations have regarded miRNAs as participants in the pathogenesis of PE. MiR-513c-5p may have potential value as a cancer marker and has implications for further understanding the molecular basis of different tumor types. MiR-513c-5p is upregulated in breast cancer [30] and is more abundant in sex cord stromal tumors than in ovarian germ cell tumors [31].
In addition, this study also showed that miR-513c-5p expression was negatively associated with the expression of LRP6. A dual luciferase reporter assay indicated that LRP6 was a direct target of miR-513c-5p and that the expression of miR-513c-5p negatively regulated LRP6 expression. Subsequent functional studies showed that miR-513c-5p promoted apoptosis but inhibited the proliferation, invasion and migration of HTR-8/SVneo cells. These results suggested that miR-513c-5p was closely related to the development of PE. Our study partially clarified the role of miR-513c-5p in the development of preeclampsia by regulating LRP6 and provided new suggestions for its specific diagnosis and treatment.
Taken together, miR-513c-5p inhibits trophoblast function by downregulating LRP6. The identification of the inhibitory effects of miR-513c-5p overexpression on trophoblasts may provide insights into potential miRNA-targeted strategies for PE associated with trophoblast dysfunction. According to a recent study, infectious agents, such as human herpesvirus 6, may modulate miRNA expression associated with trophoblast behaviors [32]. Considering the complicated modulatory connections, further studies will be needed to confirm the relationships of miRNAs, trophoblast function and related diseases. Pathological pregnancies were elucidated by trophoblast cell research, and large numbers of studies on the roles of miRNAs in the development of PE have been performed using the HTR8/SVneo cell line due to its efficiency. Because of the drawbacks of cell-based research, for example, the HTR8/SVneo cell line is a mix of trophoblast and stromal/mesenchymal cells [33], further in vivo experiments are needed. In addition, due to the limited sample size of the patients included and differences in gestational age between women with PE and women with normal pregnancies, in vivo experiments are also necessary.
Conclusions
Our study reveals that overexpression of miR-513c-5p is involved in PE by regulating the biological functions of trophoblasts via inhibition of LRP6.
Acknowledgements
We would like to thank our patients for cooperating with placenta collecting.
Abbreviations
- PE
Preeclampsia
- miRNA
microRNA
- LRP6
Low-density lipoprotein receptor-associated protein 6
- GEO
Expression Omnibus
- NC
Normal controls
- RT-qPCR
Reverse transcription-quantitative polymerase chain reaction
Authors’ contributions
Q Z was responsible for conceiving the work and performing all the procedures. HY L wrote and Y Z substantively revised the manuscript. The first three authors contributed equally to the manuscript. W P drafted and revised the manuscript, and MQ G analysed and interpreted data for the work. FY Z and HY H analysed the data and processed the figures. XT W, X G and LL designed the work; they also gave the final approval of the version to be published. All authors have read and approved the manuscript.
Funding
This research received the fund of National Key Research and Development Programme of China (2018YFC1002900) and National Natural Science Foundation of China (81801473, 81971409), and the programme supported the cost of reagent and consumable materials.
Availability of data and materials
All data included in this study are available upon request by contact with the corresponding author.
Declarations
Ethics approval and consent to participate
The ethical committee of Shandong Provincial Hospital affiliated to Shandong First Medical University approved this study and all patients who agreed to participate in the study signed written informed consent.
Consent for publication
Written informed consent was obtained from the patients for publication of this study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qian Zhou, Hongyan Li and Yan Zhang contributed equally to this work.
Contributor Information
Xietong Wang, Email: wxt65@vip.163.com.
Xiao Gu, Email: 15153158512@163.com.
Lei Li, Email: sdobstetricsli@126.com.
References
- 1.WHO . WHO Recommendations for Prevention and Treatment of Pre-Eclampsia and Eclampsia. Geneva: WHO; 2011. [PubMed] [Google Scholar]
- 2.Duhig K, Vandermolen B, Shennan A. Recent advances in the diagnosis and management of pre-eclampsia. F1000Res. 2018;7:242. doi: 10.12688/f1000research.12249.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phipps E, Prasanna D, Brima W, Jim B. Preeclampsia: updates in pathogenesis, definitions, and guidelines. Clin J Am Soc Nephrol. 2016;11(6):1102–1113. doi: 10.2215/CJN.12081115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mol BWJ, Roberts CT, Thangaratinam S, Magee LA, de Groot CJM, Hofmeyr GJ. Pre-eclampsia. Lancet. 2016;387(10022):999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 5.Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semin Perinatol. 2012;36(1):56–59. doi: 10.1053/j.semperi.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Jena MK, Sharma NR, Petitt M, Maulik D, Nayak NR. Pathogenesis of Preeclampsia and Therapeutic approaches targeting the placenta. Biomolecules. 2020;10(6):953. doi: 10.3390/biom10060953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Park M, Kim JY, Kim T, Hwang JY, Ha KS, Won MH, Ryoo S, Kwon YG, Kim YM. Circulating miRNAs associated with dysregulated vascular and Trophoblast function as target-based diagnostic biomarkers for Preeclampsia. Cells. 2020;9(9):2003. doi: 10.3390/cells9092003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed]
- 9.Lv Y, Lu C, Ji X, Miao Z, Long W, Ding H, Lv M. Roles of microRNAs in preeclampsia. J Cell Physiol. 2019;234(2):1052–1061. doi: 10.1002/jcp.27291. [DOI] [PubMed] [Google Scholar]
- 10.Qin W, Tang Y, Yang N, Wei X, Wu J. Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertil Steril. 2016;105(5):1247–1254. doi: 10.1016/j.fertnstert.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Wan J, Xue J, Li H, Wang S, Zhou Q, Zhou Y, Wang X. Low-density lipoprotein receptor-related protein 6 is essential for trophoblast survival and invasion. Reprod Sci. 2019;26(1):35–43. doi: 10.1177/1933719118756773. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Peng W, Zhou Q, Wan JP, Wang XT, Qi HB. LRP6 regulates Rab7-mediated autophagy through the Wnt/beta-catenin pathway to modulate trophoblast cell migration and invasion. J Cell Biochem. 2020;121(2):1599–1609. doi: 10.1002/jcb.29394. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Liu SH, Liu QY, Zou QQ, Lv L, Liu GL, et al. Analysis of the role and regulatory mechanism of hsa-miR-504 in cervical Cancer based on the Cancer genome atlas database. Cancer Biother Radiopharm. 2021;36(6):511–20. [DOI] [PubMed]
- 14.Bo LJ, Wei B, Li ZH, Wang ZF, Gao Z, Miao Z. Bioinformatics analysis of miRNA expression profile between primary and recurrent glioblastoma. Eur Rev Med Pharmacol Sci. 2015;19(19):3579–3586. [PubMed] [Google Scholar]
- 15.Gao H, Jin H, Li G. Predicting miRNA targets for head and neck squamous cell carcinoma using an ensemble method. Int J Biol Markers. 2018;33(1):87–93. doi: 10.5301/ijbm.5000285. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, Lian Y, Zhang Y, Li L, Li H, Shen D, Zhou Y, Zhang M, Lu Y, Liu J, Xia Y, Wang X. Platelet-derived microparticles from recurrent miscarriage associated with antiphospholipid antibody syndrome influence behaviours of trophoblast and endothelial cells. Mol Hum Reprod. 2019;25(8):483–494. doi: 10.1093/molehr/gaz019. [DOI] [PubMed] [Google Scholar]
- 17.Du E, Cao Y, Feng C, Lu J, Yang H, Zhang Y. The possible involvement of miR-371a-5p regulating XIAP in the pathogenesis of recurrent pregnancy loss. Reprod Sci. 2019;26(11):1468–1475. doi: 10.1177/1933719119828051. [DOI] [PubMed] [Google Scholar]
- 18.Zhao JR, Cheng WW, Wang YX, Cai M, Wu WB, Zhang HJ. Identification of microRNA signature in the progression of gestational trophoblastic disease. Cell Death Dis. 2018;9(2):94. doi: 10.1038/s41419-017-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hromadnikova I, Kotlabova K, Hympanova L, Krofta L. Gestational hypertension, preeclampsia and intrauterine growth restriction induce dysregulation of cardiovascular and cerebrovascular disease associated microRNAs in maternal whole peripheral blood. Thromb Res. 2016;137:126–140. doi: 10.1016/j.thromres.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Gu Y, Lewis DF, Cooper DB, McCathran CE, Wang Y. Downregulation of vitamin D receptor and miR-126-3p expression contributes to increased endothelial inflammatory response in preeclampsia. Am J Reprod Immunol. 2019;82(4):e13172. doi: 10.1111/aji.13172. [DOI] [PubMed] [Google Scholar]
- 21.Wen Z, Chen Y, Long Y, Yu J, Li M. Tumor necrosis factor-alpha suppresses the invasion of HTR-8/SVneo trophoblast cells through microRNA-145-5p-mediated downregulation of Cyr61. Life Sci. 2018;209:132–139. doi: 10.1016/j.lfs.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Awamleh Z, Han VKM. Potential pathophysiological role of microRNA 193b-5p in human placentae from pregnancies complicated by preeclampsia and intrauterine growth restriction. Mol Biol Rep. 2020;47(9):6531–6544. doi: 10.1007/s11033-020-05705-y. [DOI] [PubMed] [Google Scholar]
- 23.Bidarimath M, Khalaj K, Kridli RT, Kan FW, Koti M, Tayade C. Extracellular vesicle mediated intercellular communication at the porcine maternal-fetal interface: a new paradigm for conceptus-endometrial cross-talk. Sci Rep. 2017;7(1):40476. doi: 10.1038/srep40476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong K, Zhang X, Ma L, Gao N, Tang H, Jian F, Ma Y. Downregulations of circulating miR-31 and miR-21 are associated with preeclampsia. Pregnancy Hypertens. 2019;17:59–63. doi: 10.1016/j.preghy.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Niu ZR, Han T, Sun XL, Luan LX, Gou WL, Zhu XM. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am J Obstet Gynecol. 2018;218(2):249 e241–249 e212. doi: 10.1016/j.ajog.2017.11.568. [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Zhang Y, Yang P, Gao X, Wang Y. Downregulated low-density lipoprotein receptor-related protein 6 induces the maldevelopment of extravillous trophoblast via Wnt/beta-catenin signaling pathway. Mol Cell Probes. 2019;44:21–28. doi: 10.1016/j.mcp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Ni H, Wang X, Qu H, Gao X, Yu X. MiR-95-5p involves in the migration and invasion of trophoblast cells by targeting low density lipoprotein receptor-related protein 6. J Obstet Gynaecol Res. 2021;47(1):184–197. doi: 10.1111/jog.14451. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Li H, Li M, Zhang W, Yang Z, Zhang S. LRP6 is involved in the proliferation, migration and invasion of trophoblast cells via miR346. Int J Mol Med. 2020;46(1):211–223. doi: 10.3892/ijmm.2020.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Pan X, Yu X, Li L, Qu H, Li S. MicroRNA-590-3p inhibits trophoblast-dependent maternal spiral artery remodeling by repressing low-density lipoprotein receptor-related protein 6. Mol Genet Genomic Med. 2018;6(6):1124–1133. doi: 10.1002/mgg3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muti P, Donzelli S, Sacconi A, Hossain A, Ganci F, Frixa T, Sieri S, Krogh V, Berrino F, Biagioni F, Strano S, Beyene J, Yarden Y, Blandino G. MiRNA-513a-5p inhibits progesterone receptor expression and constitutes a risk factor for breast cancer: the hOrmone and diet in the ETiology of breast cancer prospective study. Carcinogenesis. 2018;39(2):98–108. doi: 10.1093/carcin/bgx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang RK, Li X, Mu N, Hrydziuszko O, Garcia-Majano B, Larsson C, Lui WO. MicroRNA expression profiles in nonepithelial ovarian tumors. Int J Oncol. 2018;52(1):55–66. doi: 10.3892/ijo.2017.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bortolotti D, Soffritti I, D'Accolti M, Gentili V, Di Luca D, Rizzo R, Caselli E. HHV-6A infection of endometrial epithelial cells affects miRNA expression and trophoblast cell attachment. Reprod Sci. 2020;27(3):779–786. doi: 10.1007/s43032-019-00102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abou-Kheir W, Barrak J, Hadadeh O, Daoud G. HTR-8/SVneo cell line contains a mixed population of cells. Placenta. 2017;50:1–7. doi: 10.1016/j.placenta.2016.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.