Abstract
Only two complete genomes of the cyanobacterial genus Gloeobacter from two very different regions of the world currently exist. Here, we present the complete genome sequence of a third member of the genus isolated from a waterfall cave in Mexico. Analysis of the average nucleotide identities (ANIs) between published Gloeobacter genomes revealed that the complete genome of this new member is only 92.7% similar to Gloeobacter violaceus and therefore we determined it to be a new species. We propose to name this new species Gloeobacter morelensis after the location in Mexico where it was isolated. The complete genome consists of one circular chromosome (4,921,229 bp), one linear plasmid (172,328 bp), and one circular plasmid (8,839 bp). Its genome is the largest of all completely sequenced genomes of Gloeobacter species. Pangenomic comparisons revealed that G. morelensis encodes 759 genes not shared with other Gloeobacter species. Despite being more closely related to G. violaceus, it features an extremely divergent psbA gene encoding an atypical D1 core subunit of Photosystem II previously only found within the genome of Gloeobacter kilaueensis. In addition, we detected evidence of concerted evolution of psbA genes encoding identical D1 in all three Gloeobacter genomes, a characteristic that seems widespread in cyanobacteria and may therefore be traced back to their last common ancestor.
Keywords: Gloeobacter, oxygenic photosynthesis, cave, waterfall, evolution, photosystem
Significance
Cyanobacteria of the genus Gloeobacter display ancestral features thought to be present during the early stages of the evolution of oxygenic photosynthesis, a process that led to the accumulation of oxygen in the Earth’s atmosphere. Despite their importance to understanding the origin and evolution of cyanobacteria, only two complete genomes of Gloeobacter have been sequenced so far. Additional complete genomes of Gloeobacter are needed to better understand ancestral and derived features of oxygenic photosynthesis in this group of thylakoid-less cyanobacteria. The complete genome of the third member of the cyanobacterial genus Gloeobacter contributes to filling the knowledge gap on how oxygenic cyanobacteria first arose and diversified on the early Earth.
Introduction
Cyanobacteria of the genus Gloeobacter are the earliest-branching members of cyanobacteria capable of oxygenic photosynthesis, a process that led to the accumulation of oxygen gas in the Earth’s atmosphere and paved the way for aerobic organisms to diversify. Members of the genus Gloeobacter lack thylakoid membranes typically associated with cyanobacteria and plastids, a feature conserved in all other oxygenic phototrophs (Rippka et al. 1974). They are widely distributed but poorly represented in terms of genomic sequences and cultivated species. Only two members of the genus Gloeobacter have been cultivated thus far with their complete genomes sequenced (Nakamura et al. 2003; Saw et al. 2013). In 2020, a distantly related sister taxon to Gloeobacter was reported from metagenomes of microbial mats in an Antarctic lake and was named Aurora vandensis (Grettenberger et al. 2020). Recently, a novel strain closely related to A. vandensis that also lacks thylakoids was isolated from a hornwort and named Anthocerotibacter panamensis. Its complete genome was reported recently (Rahmatpour et al. 2021).
Photosystem II is a highly conserved enzymatic complex of cyanobacteria that catalyzes the light-driven oxidation of water to molecular oxygen. Therefore, understanding the evolution of Photosystem II is essential to pinpoint the origin of oxygenic photosynthesis (Rutherford and Faller 2003). Given the phylogenetic position of Gloeobacter in the cyanobacteria tree of life, there is interest in deciphering whether this clade retains ancestral photosystem traits that could provide insight into how oxygenic photosynthesis first emerged.
However, although the genome content of Gloeobacterales appeared to show a slightly decreased number of identifiable accessory photosystem subunits (Nakamura et al. 2003; Saw et al. 2013; Rahmatpour et al. 2021), measurements of oxygen evolution activity in Gloeobacterviolaceus PCC 7421 revealed no functional differences to model organisms (Koyama et al. 2008). The genome of Gloeobacterkilaueensis JS1, on the other hand, revealed an additional Photosystem II subunit with atypical characteristics previously undetected in the closely related G. violaceus genome. This subunit was the least conserved version of any catalytic subunit (D1) of Photosystem II described to date in cyanobacteria and photosynthetic eukaryotes (Cardona et al. 2015). It was denoted Group 0 as it remained a singularity, and its phylogenetic position was interpreted to suggest it originated from an ancient gene duplication event of a Photosystem II D1 subunit (Cardona et al. 2015), antedating the last common ancestor of cyanobacteria by well over half a billion years (Cardona et al. 2019).
Here, we present the complete genome sequence of an additional member of the Gloeobacter. Comparisons of complete genomes and pangenomes revealed that this member is a different species closely related to G.violaceus. Surprisingly, the genome of this new Gloeobacter has a gene encoding the atypical Group 0 D1 that is not found in the genome of the more closely related G. violaceus but is found in the genome of the distantly related G. kilaueensis. Very few complete genomes of Gloeobacter and related species exist and the addition of this novel Gloeobacter species is therefore crucial to further bridge the knowledge gap that exists in our understanding of how oxygenic photosynthesis arose on Earth and how cyanobacteria first acquired this important biological function.
Results and Discussion
Genome Assembly, Annotation, and Comparison with Other Gloeobacter spp
A combined total of 2,157,121,864 clean and trimmed high-quality bases were obtained from both Illumina and Nanopore sequence reads. A hybrid assembly using both short-read Illumina and long-read Nanopore sequences produced a genome assembly without any gaps (table 1). The genome assembly performed using Unicycler (Wick et al. 2017) resulted in three contigs and the analysis of Unicycler genome assembly paths using the Bandage tool (Wick et al. 2015) indicated a circular chromosome of 4,921,229 bp, a linear plasmid of 172,328 bp, and a circular plasmid of 8,839 bp (supplementary fig. S1, Supplementary Material online). CheckM and micomplete analyses revealed high genome completeness (>99% and 100% complete, respectively) and low level of contamination (<1%) (supplementary table S1, Supplementary Material online). The level of completeness reported by CheckM and micomplete tools may not accurately reflect the actual completeness due to the use of so-called universal single-copy genetic markers. These marker gene sets vary between divergent organisms and low completeness scores may be reported even for highly complete genomes. To obtain an independent estimate of the quality of genome assembly, we used BUSCO to assess genome completeness and marker gene contents of closely related Gloeobacter and associated lineages. This tool was originally designed for eukaryotic genomes but has been updated to assess marker genes in prokaryotes as well (Manni et al. 2021). BUSCO analysis using universal and cyanobacteria marker gene sets revealed similar levels of genome completeness and marker gene presence in the genome sequences of three Gloeobacter species (supplementary fig. S3, Supplementary Material online).
Table 1.
General Features of the Genome
| Feature | Chromosome (Circular) | Plasmid 1 (Linear) | Plasmid 2 (Circular) |
|---|---|---|---|
| Length (bp) | 4,921,229 | 172,328 | 8,839 |
| GC content (%) | 61.86 | 58.85 | 59.07 |
| CDS | 4,569 | 177 | 9 |
| Pseudogenes | 107 | 1 | 1 |
| CRISPR arrays | 3 | 0 | 0 |
| rRNA (5S, 16S, 23S) | 1, 1, 1 | 0, 0, 0 | 0, 0, 0 |
| tRNA | 44 | 0 | 0 |
A total of 4,755 coding sequences (CDS) were identified by the NCBI PGAP automatic annotation pipeline (v4.6) (table 1). Three CRISPR (Clustered Regularly Interspaced Palindromic Repeats) regions were identified in the circular chromosome. The genome of this newly sequenced Gloeobacter is the largest among the three Gloeobacter with complete genomic sequences: 4.92 Mbp for the circular chromosome compared with 4.72 Mbp for G. kilaueensis and 4.66 Mbp for G. violaceus PCC 7421. PyANI calculation using ANIb (Pritchard et al. 2016) revealed that Gloeobacter violaceus PCC 7421 and the newly sequenced Gloeobacter share only 92.6% average nucleotide identity (ANI) despite having 99.93% 16S rRNA gene sequence identity. This value is below the threshold value for delineating species (Jain et al. 2018). Genome-to-genome distance calculator (Meier-Kolthoff et al. 2013) revealed a value of 49.4% between G. violaceus PCC 7421 and the new Gloeobacter genome, which is well below the 70% cutoff value for delineating species based on in silico DNA–DNA hybridization (Wayne 1988; Stackebrandt and Goebel 1994). The new Gloeobacter genome has 98.7% 16S rRNA gene sequence identity and ANI of 75.73% to G. kilaueensis. Based on these metrics, this newly sequenced genome of Gloeobacter likely belongs to a new species and we propose to name it Gloeobacter morelensis based on the origin of the biofilm materials (Morelos, Mexico) from which this cyanobacterium was isolated. The genus and species names of G.violaceus, the first cultivated member of this cyanobacterial lineage, was first published in 1974 (Rippka et al. 1974) and validated in 2013 (Mareš et al. 2013). Here, we have decided to preserve the genus name of the lineage. Formal description of the species is beyond the scope of this study as we have not yet deposited the pure culture to a culture collection agency, but we plan to further characterize this novel species in greater detail in the near future.
Phylogenomic Analysis
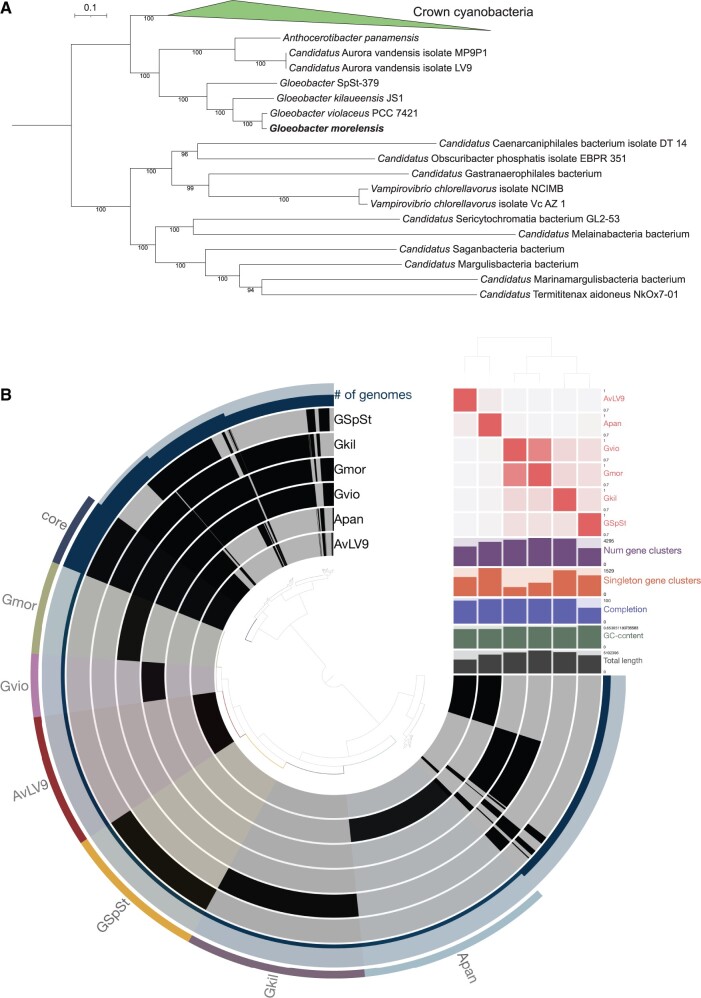
PhyloPhlAn (v3.0.51) (Asnicar et al. 2020) identified 395 unique single-copy genes in 159 cyanobacterial genomes (including the genome of G. morelensis) with species designations from the NCBI database. This workflow aligned, trimmed, and concatenated the single-copy genes and the concatenated alignment was used as an input to run IQ-TREE (v2.0.3) (Minh et al. 2020) with the best-fit model predicted by the tool. The resulting maximum likelihood phylogenomic tree was rooted with sequences from Melainabacteria, Sericytochromatia, and Margulisbacteria (Soo et al. 2019). The phylogenomic tree shows two deeply branching and distinct sister clades of oxyphotobacteria, one consisting of G. violaceus, G. kilaueensis, Gloeobacter SpSt-379 (from a metagenome bin), and G. morelensis and the other consisting of A.panamensis and A.vandensis isolates LV9 and MP9P1 with 100% ultrarapid bootstrap support (fig. 1A; supplementary fig. S2, Supplementary Material online). Gloeobactermorelensis is the sister lineage of G. violaceus PCC 7421 in the phylogenomic tree. The topology of the tree is similar to the tree obtained by Rahmatpour et al. (2021).
Fig. 1.
(A) Phylogenomic tree displaying placement of Gloeobacter morelensis with closely related lineages. Only the names of Gloeobacter-associated lineages and members of the outgroup are shown. Full tree with names of all lineages can be found in supplementary figure S1, Supplementary Material online. (B) Pangenomic comparison of three complete genomes and a draft genome of Gloeobacter species using the Anvi’o tool. Core gene clusters shared by all species are highlighted and labeled as “core” and gene clusters present in each species are highlighted and labeled as “Gmor,” “Gvio,” “AvLV9,” “GSpSt,” “Gkil,” and “Apan” to represent G. morelensis, G. violaceus, A. vandensis LV9, G. SpSt-379, G. kilaueensis, and A. panamensis, respectively. ANIs between them are shown as a heatmap in red near the top right quadrant of the figure. Placement of the genomes is based on ANI similarities as predicted by the PyANI tool.
Pangenomic Analysis
To identify unique and shared genes among the four Gloeobacter species and two sister taxa, we used Anvi’o (Eren et al. 2021) to perform a pangenomic comparison of their genomes. The analysis revealed that the genome of G. morelensis encodes 729 unique gene clusters (759 unique genes) not shared with other genomes (fig. 1B; supplementary table S2, Supplementary Material online). Aurora vandensis LV9 and A.panamensis form a distinct clade diverged from the Gloeobacter clade based on ANI values. They share very low ANI values with Gloeobacter species: between 69.93% and 70.42% (fig. 1). Core gene clusters shared between genomes included in this analysis is 580. Gloeobactermorelensis and G. violaceus PCC 7421 share 798 unique gene clusters (1,717 genes total) between them (supplementary table S2, Supplementary Material online).
Photosystem II D1 Evolution
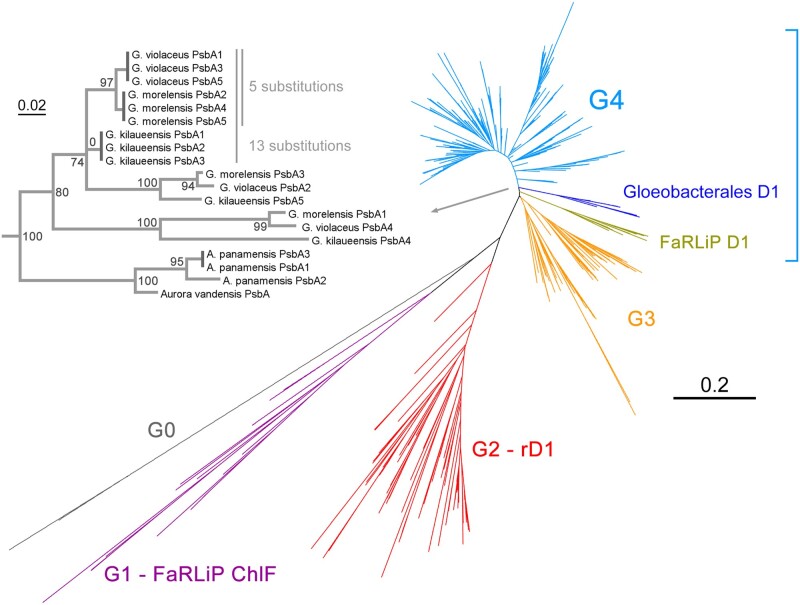
The unrooted phylogeny of D1 is shown in figure 2 and is similar to those presented by previous authors (Sheridan et al. 2020). Photosystem II adapts to changing environments by swapping D1 subunits to variant forms better tuned to new conditions. These sequence variants can range from just a few amino acid changes to forms exhibiting >40% amino acid substitutions when compared with the well characterized standard D1 (Group 4, G4, blue). The more divergent forms, like for example, “ChlF” (Group 1, G1, purple), enables Photosystem II to acquire new functions beyond oxygen evolution, such as chlorophyll f synthesis in the far-red light photoacclimation response (FaRLiP) (Ho et al. 2016; Trinugroho et al. 2020). In the same response, another variant D1 (olive), uses chlorophyll f in a Photosystem II complex to enable oxygen evolution under far-red light conditions (Gan et al. 2014; Nürnberg et al. 2018). Nonetheless, all known cyanobacteria capable of oxygenic photosynthesis retain at least one, but often several standard D1, which is the main catalytic form of D1 used in oxygen evolution activity. The branches designated G0 (gray) represents the most divergent D1 amino acid sequences known as reported here. These sequences are of unknown function and have accumulated many amino acid substitutions likely to disrupt Photosystem II function if assembled into a complex (Cardona et al. 2015).
Fig. 2.
Unrooted ML tree of D1 amino acid sequences. Different types of D1 are denoted G0 to G4, representing Group 0 to Group 4 and based on the categorization by Cardona et al. (2015). FaRLiP D1 denotes the standard D1 used in the far-red light acclimation response, whereas FaRLiP ChlF denotes the divergent D1 known to confer “chlorophyll f synthase” activity to Photosystem II. Group 2 or rD1 denotes the “rogue” D1, a widespread but atypical form of D1 of unresolved function. The subtree in light gray zooms in on the Group 4 D1 of the Gloeobacterales. The dark gray vertical branches represent intraspecific identical D1 forms (branches of no length). Scale bar denotes amino acid substitutions per site.
The genome of G. morelensis encodes six distinct D1 proteins: Three of these were identical to each other, following the same pattern of D1 diversification as the other two Gloeobacter. Five of these, the standard D1 (G4) subunits, branched as sister to G. violaceus with the sixth being the second atypical Group 0 sequence (fig. 2). The genome of A. panamensis encodes three D1 subunits, two of these identical, whereas only one D1 is known for A. vandensis. The Aurora/Anthocerotibacter sequences clustered as sister to all other Gloeobacter standard D1, and thus diverged before the duplication of the main D1 subunits found in the genus Gloeobacter. In addition, the tree revealed that the three identical D1 sequences in each of the Gloeobacter genomes and the two identical sequences in A. panamensis are in fact distantly related to each other. The three identical sequences in G. morelensis compared with the three identical ones in G. violaceus differ by five amino acid substitutions, whereas differing by 13 substitutions when compared with that from G. kilaueensis and by 52 substitutions when compared with the two identical ones in A. panamensis. These patterns suggest not only frequent gene duplications and losses, but also a degree of concerted gene evolution.
Concerted evolution of duplicated genes, in which identical copies evolve as a unit via intragenomic homologous recombination, is known to serve as a molecular mechanism maintaining sequence homogeneity through time of highly similar or identical paralogous genes in both prokaryotes and eukaryotes (Santoyo and Romero 2005). It is plausible that concerted evolution of psbA genes (encoding D1), among other factors (Shi et al. 2005), contributed to Photosystem II maintaining one of the slowest rates of protein evolution since the origin of photosynthesis (Oliver et al. 2021). It is worth noting that this pattern of D1 evolution is not unique to Gloeobacterales and similar arrays of identical and variant D1 are found often in other strains of cyanobacteria, including the relatively late emerging heterocystous clade (Cardona et al. 2015). For example, both Nostoc sp. PCC 7120 and Nostoc punctiforme PCC 73102 each have five standard D1, three of which are also identical in each species, but differing by 25 amino acid substitutions when compared between species. These patterns indicate that this mechanism of photosystem evolution is a feature present since at least the last common ancestor of cyanobacteria.
Conclusion
Our study reports the complete genome sequence of the third Gloeobacter species; only two complete genomes had been published thus far in the past 18 years. Gloeobacter are considered conditionally rare microbial species that are important and significant to our understanding of the origin and evolution of photosystems in diverse lineages of microbes (Saw 2021). Additional work will be required to further characterize the genes encoded in the plasmids to determine their roles in the ecology of G. morelensis. We also found putative molecular evidence for concerted evolution of psbA genes (D1) in this lineage, which may have implications for the mechanisms of molecular evolution required for the early diversification of photosystem core proteins before the last common ancestor of cyanobacteria. This study improves our knowledge of Gloeobacter species and their genetic diversity, but more genomes are needed to reconstruct a high-resolution view of the origin and evolution of oxygenic photosynthesis.
Materials and Methods
Sample Collection and Cultivation
Samples were collected from a waterfall cave in Morelos, Mexico and brought back to the laboratory of G.M. Samples were grown on BG11 medium as previously described (Montejano et al. 2018). Biofilm sample collection and cultivation were reported in a previous study and pure culture of G. morelensis obtained from the biofilm was sent to the laboratory of J.S. at The George Washington University. To obtain enough cells to extract DNA from, the culture obtained from Mexico was restreaked on a BG11 agar plate and the plates were incubated in a light incubator at 28 °C for several weeks until large quantities of cells were visible on the agar plates. DNA was extracted from a single large colony.
Genome Sequencing, Assembly, and Annotation
Genomic DNA of G.morelensis was extracted from a single large colony on a quadrant-streaked BG11 agar plate using Zymo Microbiomics DNA extraction kit and quantified with Qubit 4 fluorometer. Purified genomic DNA was sent to Microbial Genomes Sequencing Center (MiGS) in Pittsburgh, PA. Two separate Illumina samples and one Nanopore sample were obtained from MiGS. Raw sequences were trimmed for quality and contamination using the following tools: bbduk for Illumina reads and filtlong for Nanopore reads. Trimming parameters for bbduk were as follows: qtrim=rl trimq=20. Trimming parameters for filtlong were as follows: –min_length 1000 –keep_percent 90 –target_bases 1000000000. Quality-trimmed Illumina and Nanopore sequences were coassembled using Unicycler (v0.4.8) with default parameters. Polishing of the final assembly was done with Pilon, which is integrated in the Unicycler assembler. Circularity of assembled contigs were determined by examining Unicycler assembly paths using the Bandage tool (v0.8.1). Genome completeness and contamination was estimated using CheckM (v1.1.0) (Parks et al. 2015), miComplete (v1.1.1) (Hugoson et al. 2020), and BUSCO (Manni et al. 2021).
Phylogenetic Analysis of D1 Subunits
A total of 1,198 D1 amino acid sequences were retrieved from the NCBI database using BLAST in February 18, 2020. A redundancy cutoff of 98% sequence identity reduced the number of sequences to 540. However, all sequences from all Gloeobacter strains, including the identical ones, were kept in the data set. An additional D1 from the metagenome-assembled genome of A.vandensis (Grettenberger et al. 2020) and the three amino acid sequences from the genome of A. panamensis were later added to the data set (Rahmatpour et al. 2021). Alignments were produced using Clustal Omega (Sievers et al. 2011) using ten combined guided-tree and HMM iterations. Maximum likelihood phylogenetic trees were executed using IQ-TREE 2.0 (Minh et al. 2020), with automatic best model selection, applying ultrafast bootstrap for branch support and run until the bootstrap correlation coefficient of split occurrence frequencies reached a value of 0.99.
Phylogenomic Analysis
PhyloPhlAn version 3.0.51 was used to identify conserved single-copy genes with the following commands: “phylophlan_write_config_file -o custom_phylophlan.ctg -d a –db_aa diamond –map_aa diamond –msa mafft –trim trimal –tree1 fasttree,” “phylophlan -f custom_phylophlan.ctg -i faas –proteome_extension .faa -d phylophlan –diversity low –fast –nproc 24 -o phylophlan -t a –verbose 2>&1 | tee log.phylophlan.txt.” The resulting concatenated multiple sequence alignment was then used to construct a maximum likelihood phylogenomic tree using IQ-TREE (v2.0.3). The best-fitting model was identified using the command “iqtree -s faas_concatenated.aln -m TESTONLY -T 24 -pre modeltest,” and the final tree was constructed using the following command “iqtree -s faas_concatenated.aln -m LG+F+G4 -alrt 1000 -bb 1000 -T 24.” Resulting phylogenomic tree was visualized with a custom Python script utilizing the ETE3 Python library (Huerta-Cepas et al. 2016). The script is accessible at the following Github repository: https://github.com/SawLabGW/Gmor.
Pangenomic and Comparative Genomic Analyses
Four genomes of Gloeobacter (G. violaceus, G. kilaueensis, G. morelensis, G. SpSt-379) and two genomes of sister taxa (A.vandensis LV9 and A.panamensis) were used in this analysis. The accession numbers of the genomes used were: CP062698.1 (A.panamensis), JAAXLT010000001.1 (A.vandensis LV9), DSQC00000000.1 (Gloeobacter SpSt-379), CP003587.1 (G. kilaueensis JS1), and BA000045 (G. violaceus PCC 7421). Gloeobacter SpSt-379 is an incomplete metagenomic bin obtained from a previous study (Zhou et al. 2020). Aurora vandensis LV9 and A.panamensis genomes were first annotated with Prokka version 1.13 to identify coding regions. These four annotated genomes were analyzed using Anvi’o version 7.0 (Eren et al. 2021) to identify unique and shared gene clusters between them. To perform pangenomic comparisons, “anvi-pan-genome” command was used with default parameters. ANIs between the strains were calculated with the “anvi-compute-genome-similarity” command with “–program pyANI” option. Complete list of commands and workflow used in this analysis are documented in a Markdown file deposited to a Github repository (https://github.com/SawLabGW/Gmor).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Karina Osorio Santos for technical support with the isolation of Gloeobacter morelensis. J.S. is supported by the startup funds and University Facilitating Fund from The George Washington University. T.C. is supported by a UKRI Future Leaders Fellowship (MR/T017546/1). We gratefully acknowledge the computing resources provided on the High-Performance Computing Cluster operated by Research Technology Services at The George Washington University and at Imperial College London. We thank Alex Pyron for helpful comments to improve the manuscript.
Data Availability
The complete genome sequence of Gloeobacter morelensis has been deposited to GenBank under accession numbers CP063845–CP063847. Raw Illumina and Nanopore sequences have been deposited to NCBI Sequence Reads Archive (SRA) under accession numbers SRR12931218 and SRR12931219, respectively.
Data deposition: The complete genome sequence of Gloeobacter morelensis has been deposited at GenBank under accessions accession numbers CP063845–CP063847. This project has been deposited at NCBI under the Bioproject accession number PRJNA671611.
Literature Cited
- Asnicar F, et al. 2020. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun. 11(1):2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona T, Murray JW, Rutherford AW.. 2015. Origin and evolution of water oxidation before the last common ancestor of the cyanobacteria. Mol Biol Evol. 32(5):1310–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona T, Sánchez-Baracaldo P, Rutherford AW, Larkum AW.. 2019. Early Archean origin of Photosystem II. Geobiology 17(2):127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, et al. 2021. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 6(1):3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan F, et al. 2014. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345(6202):1312–1327. [DOI] [PubMed] [Google Scholar]
- Grettenberger CL, et al. 2020. A phylogenetically novel cyanobacterium most closely related to Gloeobacter. ISME J. 14(8):2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M-Y, Shen G, Canniffe DP, Zhao C, Bryant DA.. 2016. Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 353(6302):aaf9178. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P.. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoson E, Lam WT, Guy L.. 2020. miComplete: weighted quality evaluation of assembled microbial genomes. Bioinformatics 36(3):936–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S.. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 9(1):5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama K, et al. 2008. Oxygen evolution in the thylakoid-lacking cyanobacterium Gloeobacter violaceus PCC 7421. Biochim Biophys Acta 1777(4):369–378. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM.. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareš J, Komárek J, Compère P, Oren A.. 2013. Validation of the generic name Gloeobacter Rippka et al. 1974, Cyanophyceae. Cryptogam Algol. 34(3):255–262. [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M.. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montejano G, Becerra-Absalón I, Gold-Morgan M, Osorio-Santos K.. 2018. Gloeobacter violaceus: primitive reproductive scheme and its significance. Plant Syst Evol. 304(10):1221–1229. [Google Scholar]
- Nakamura Y, et al. 2003. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 10(4):137–145. [DOI] [PubMed] [Google Scholar]
- Nürnberg DJ, et al. 2018. Photochemistry beyond the red limit in chlorophyll f-containing photosystems. Science 360(6394):1210–1213. [DOI] [PubMed] [Google Scholar]
- Oliver T, Sánchez-Baracaldo P, Larkum AW, Rutherford AW, Cardona T.. 2021. Time-resolved comparative molecular evolution of oxygenic photosynthesis. Biochim Biophys Acta Bioenerg. 1862(6):148400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW.. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25(7):1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK.. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 8(1):12–24. [Google Scholar]
- Rahmatpour N, et al. 2021. A novel thylakoid-less isolate fills a billion-year gap in the evolution of cyanobacteria. Curr Biol. 31(13):2857–2867.e4. [DOI] [PubMed] [Google Scholar]
- Rippka R, Waterbury J, Cohen-Bazire G.. 1974. A cyanobacterium which lacks thylakoids. Arch Microbiol. 100(1):419–436. [Google Scholar]
- Rutherford AW, Faller P.. 2003. Photosystem II: evolutionary perspectives. Philos Trans R Soc Lond B Biol Sci. 358(1429):245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G, Romero D.. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev. 29(2):169–183. [DOI] [PubMed] [Google Scholar]
- Saw JHW. 2021. Characterizing the uncultivated microbial minority: towards understanding the roles of the rare biosphere in microbial communities. mSystems 6(4):e0077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw JHW, et al. 2013. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kīlauea Caldera, Hawai’i. PLoS One 8(10):e76376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan KJ, Duncan EJ, Eaton-Rye JJ, Summerfield TC.. 2020. The diversity and distribution of D1 proteins in cyanobacteria. Photosynth Res. 145(2):111–128. [DOI] [PubMed] [Google Scholar]
- Shi T, Bibby TS, Jiang L, Irwin AJ, Falkowski PG.. 2005. Protein interactions limit the rate of evolution of photosynthetic genes in cyanobacteria. Mol Biol Evol. 22(11):2179–2189. [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo RM, Hemp J, Hugenholtz P.. 2019. Evolution of photosynthesis and aerobic respiration in the cyanobacteria. Free Radic Biol Med. 140:200–205. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM.. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 44(4):846–849. [Google Scholar]
- Trinugroho JP, et al. 2020. Chlorophyll f synthesis by a super-rogue photosystem II complex. Nat Plants. 6(3):238–244. [DOI] [PubMed] [Google Scholar]
- Wayne LG. 1988. International committee on systematic bacteriology: announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Zentralbl Bakteriol Mikrobiol Hyg A 268(4):433–434. [DOI] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE.. 2015. Bandage: interactive visualization of de novo genome assemblies: fig. 1. Bioinformatics 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE.. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 13(6):e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. 2020. Genome- and community-level interaction insights into carbon utilization and element cycling functions of Hydrothermarchaeota in hydrothermal sediment. mSystems 5(1):e00795–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequence of Gloeobacter morelensis has been deposited to GenBank under accession numbers CP063845–CP063847. Raw Illumina and Nanopore sequences have been deposited to NCBI Sequence Reads Archive (SRA) under accession numbers SRR12931218 and SRR12931219, respectively.