Abstract
Massive corals of the genus Porites, common, keystone reef builders in the Indo-Pacific Ocean, are distinguished by their relative stress tolerance and longevity. In order to identify genetic bases of these attributes, we sequenced the complete genome of a massive coral, Porites australiensis. We developed a genome assembly and gene models of comparable quality to those of other coral genomes. Proteome analysis identified 60 Porites skeletal matrix protein genes, all of which show significant similarities to genes from other corals and even to those from a sea anemone, which has no skeleton. Nonetheless, 30% of its skeletal matrix proteins were unique to Porites and were not present in the skeletons of other corals. Comparative genomic analyses showed that genes widely conserved among other organisms are selectively expanded in Porites. Specifically, comparisons of transcriptomic responses of P. australiensis and Acropora digitifera, a stress-sensitive coral, reveal significant differences in regard to genes that respond to increased water temperature, and some of the genes expanded exclusively in Porites may account for the different thermal tolerances of these corals. Taken together, widely shared genes may have given rise to unique biological characteristics of Porites, massive skeletons and stress tolerance.
Keywords: scleractinia, Porites, genome sequencing, skeletal matrix protein, gene duplication, gene expression
Significance
The massive stony corals of the genus Porites are common, important reef builders in the Indo-Pacific Ocean. Although massiveness and longevity are their distinguishing characters, molecular mechanisms underlying biological characteristics of Porites are largely unknown. In this study, we sequenced the complete genome of Porites australiensis from Okinawa, Japan. We found that homologs of all genes identified from skeletal matrix protein genes exist in other corals and anemones, although 30% of these were exclusive to Porites skeletons. Conserved genes shared with other animals are selectively expanded in Porites and these include genes that respond to diverse environmental stresses. Taken together, genes conserved among a wide range of organisms explain unique Porites attributes, massive skeletons, and stress tolerance.
Introduction
Coral reefs support the most diverse marine ecosystems on Earth (Wilkinson 2008); however, they face a range of anthropogenic challenges, including ocean acidification, increasing seawater temperatures (Hoegh-Guldberg et al. 2007), and deoxygenation (Hughes et al. 2020). Tropical storms, predation by crown-of-thorns starfish, and coral bleaching, a breakdown of the mutualism between corals and their endosymbiotic photosynthetic algae, are major causes of coral reef decline (De'ath et al. 2012). Bleaching, largely caused by increased seawater temperature, has been observed circumglobally with increasing frequency (Hughes et al. 2017; Nakamura 2017). Because roughly 35% of all marine species depend upon coral reefs at some stage of their life cycles, loss of coral reefs also destroys the habitats of diverse marine species, making extensive loss of reef habitats one of the most pressing environmental issues of our time.
Stony corals of the genus Porites are common, important reef builders in the Indo-Pacific Ocean, and massiveness and longevity are their distinguishing characters. Porites colonies that may have survived more than 400 years have been reported (e.g., Kawakubo et al. 2017). More than 80 named species and numerous unclassified forms have been identified in this genus (Hoeksema and Cairns 2021). Porites species have thicker tissues and appear more robust to thermal stress than other corals, such as Acropora sp., which have thinner tissues (e.g., Loya et al. 2001). Thus, Porites corals have been used for comparative analyses of stress responses (Fitt et al. 2009). Because of their massiveness and longevity, geochemical tracers called “proxies,” such as oxygen isotope ratios, strontium-calcium ratios, and heavy metal concentrations, in growth rings of CaCO3Porites skeleton have been used to monitor changes in sea surface temperature, salinity, and/or marine pollutants (Shen et al. 1987; Beck et al. 1992; Gagan et al. 2000; Inoue and Tanimizu 2008). Seawater acidification reduces Porites calcification (Anthony et al. 2008; Iguchi et al. 2012; Olde et al. 2015). Using coral calcification as an index, Porites taxa have been used to reconstruct past environmental changes so as to better understand tropical climate systems and to predict future climate change (Cobb et al. 2003; Solomon et al. 2007). Recently, detailed geochemical studies on mechanisms of coral calcification have been conducted in order to investigate the validity of proxies and/or to improve the use of proxies (Gagan et al. 2012; McCulloch et al. 2012; Hayashi et al. 2013); however, biological studies on Porites corals have been few in number compared with other coral taxa.
Recent rapid accumulation of coral genomic data enables us to better understand molecular mechanisms underlying coral biology (Shinzato et al. 2011; Prada et al. 2016; Voolstra et al. 2017; Cunning et al. 2018; Ying et al. 2018, 2019; Helmkampf et al. 2019; Shumaker et al. 2019; Shinzato et al. 2021). Reef-building corals are diverse not only morphologically, but also physiologically and ecologically. Susceptibilities to bleaching vary among coral taxa (Marshall and Baird 2000), and in warming sea temperatures, massive Porites became “winners,” increasing relative percent cover after a massive coral bleaching event caused coverage decreases of bleaching-sensitive taxa or “losers” (Loya et al. 2001; van Woesik et al. 2011). Why are Porites corals stress-tolerant and how do they survive longer under adverse conditions? Although a Porites lutea genome was reported previously (Robbins et al. 2019), that study focused only on possible genomic interactions between host corals, symbiotic algae, bacteria, and archaea. Molecular mechanisms underlying biological characteristics of Porites are largely unknown to date.
In this study, we sequenced the complete genome of a common Porites species in Okinawa, Japan, Porites australiensis (fig. 1A), and explored genomic features that might shed light on unique biological characters of Porites, for example, stress tolerance and massive skeletons. Understanding genetic mechanisms underlying unique characters of Porites and comparing them with those of other coral taxa may facilitate predictions about whether and how coral reef ecosystems, which comprise multiple reef-building corals, can survive current global warming.
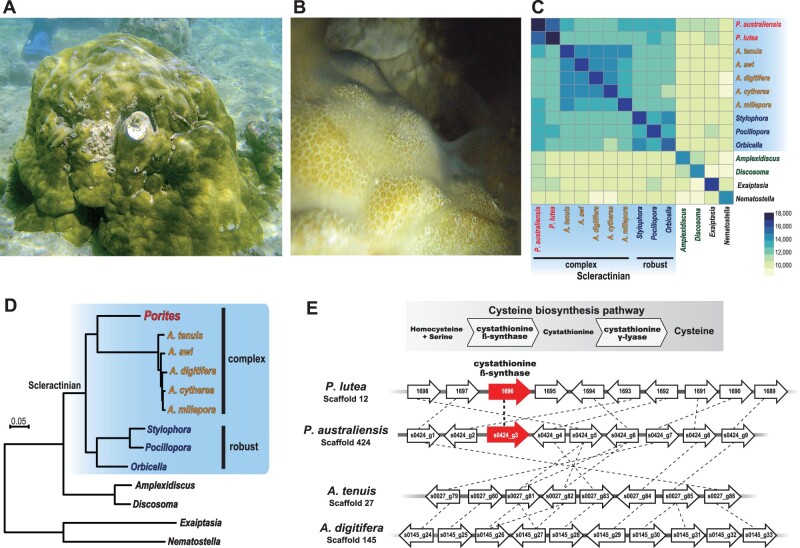
Fig. 1.
Genome sequencing of Porites australiensis. (A) Photo of a P. australiensis colony in Okinawa, Japan. (B) Spawning of a male colony of P. australiensis. Sperm can be observed on the surface of the colony. (C) A heatmap showing numbers of shared HOGs among anthozoans. Scleractinian species are shaded in blue. Porites is in red; Acropora species are in yellow. Robust coral species are in blue and corallimorpharians are in green. (D) Molecular phylogeny of anthozoans using 1,878 single-copy, orthologous genes (429,044 amino acids). All nodes are supported with 100% bootstrap support. (E) The cystathionine β-synthase (CBS) locus in the Porites genomes. The cysteine biosynthesis pathway is shown above, and the syntenic relationship around CBS in the P. australiensis, P. lutea, Acropora tenuis, and A. digitifera genomes is shown below. Genes are shown with arrows (arrow directions correspond to gene directions) and PoritesCBS genes are shown as red arrows. Genes belonging to the same OG are connected by dotted lines.
Results
The Porites australiensis Genome
Because P. australiensis is gonochoric, in order to isolate high-quality P. australiensis genomic DNA, we collected sperm from a male colony (fig. 1B). We obtained about 39 Gigabase pairs (Gbp) of genome sequencing data (supplementary table 1, Supplementary Material online), assembled the data, and then created a P. australiensis draft genome assembly of 576 Mbp with a 554-kb N50 size and 4,983 scaffold sequences (table 1). We confirmed that 99% of the shotgun sequences mapped back to the assembled genome, and among uniquely mapped pairs (mapping quality >10), 99.3% of all pairs were properly mapped on the same scaffold. Estimation of the haploid genome size, based on kmer coverage of the shotgun sequencing data using GenomeScope (Vurture et al. 2017), was close to the assembled genome size (560 Mbp, kmer length 29; supplementary fig. 1, Supplementary Material online). We predicted 30,301 protein coding genes from the assembled genome. Although genome assembly statistics for P. lutea (Robbins et al. 2019) are slightly better than these for P. australiensis, the number of predicted genes was comparable between the two genomes (table 1; Robbins et al. 2019). Benchmarking Universal Single-Copy Orthologs (BUSCO) analyses (Simao et al. 2015; Waterhouse et al. 2017), which assess whether universal single-copy orthologous genes observed in more than 90% of metazoan species (from the OrthoDB database of orthologs) (www.orthodb.org; version 9) are recovered in a genome/transcriptome assembly, yielded completeness scores for the P. australiensis genome assembly and gene models of about 91% and 94% (average of Complete BUSCO %), respectively, indicating that the genome assembly and gene models are of reasonable quality and comparable to those of previously reported coral genomes, including P. lutea (table 1). Among the 30,301 predicted genes, 20,418 exhibited similarities with genes in the Uniprot/Swissprot database, whereas 20,447 were related to entries in the Pfam conserved protein domains database (supplementary table 2, Supplementary Material online). We also obtained the complete sequence of the P. australiensis mitochondrial genome (18,647 bp, accession number: LC605627), which is >99.7% identical to the Porites lobata mitochondrial genome (KU761954.1, 18,604/18,647 bp identity, no gaps).
Table 1.
Genome Assembly and Gene Prediction Statistics for Porites australiensis and Comparisons with Publicly Available Scleractinian Coral Genomes
| Coral Species | Porites australiensis | Porites lutea | Acropora digitifera | Acropora tenuis | Acropora millepora | Stylophora postillata | Orbicella faveolata | Pocillopora damicornis |
|---|---|---|---|---|---|---|---|---|
| Total assembly size (Mbp) | 576 | 552 | 416 | 403 | 419 | 400 | 486 | 234 |
| Gap rate (%) | 9.7 | 8.7 | 0.4 | 7.4 | 9.7 | 10.5 | 26.7 | 3.7 |
| No. scaffolds | 4,983 | 2,975 | 955 | 1,538 | 3,869 | 5,687 | 1,932 | 4,392 |
| Scaffold N50 (kb) | 555 | 661 | 1,856 | 1,166 | 494 | 457 | 1,162 | 326 |
| No. predicted protein coding genes | 30,301 | 31,126 | 22,221 | 22,802 | 23,710 | 24,833 | 25,916 | 19,935 |
| BUSCO completeness % (upper: genome, lower: gene model) | C: 91.1 [S: 87.0, D: 4.1], F: 1.2, M: 7.7 | C: 92.2 [S: 89.3, D: 2.9], F: 1.3, M: 6.5 | C: 90.2 [S: 88.7, D: 1.5], F: 1.6, M: 8.2 | C: 90.5 [S: 89.4, D: 1.1], F: 1.8, M: 7.7 | C: 91.3 [S: 89.5, D: 1.8], F: 1.5, M: 7.2 | C: 88.3 [S: 86.8, D: 1.5], F: 3.3, M: 8.4 | C: 85.8 [S: 83.2, D: 2.6], F: 4.5, M: 9.7 | C: 89.2 [S: 88.7, D: 0.5], F: 2.4, M: 8.4 |
| C: 94.1 [S: 91.9, D: 2.2], F: 3.2, M: 2.7 | C: 94.8 [S: 93.0, D: 1.8], F: 3.0, M: 2.2 | C: 92.4 [S: 90.6, D: 1.8], F: 3.7, M: 3.9 | C: 94.6 [S: 93.9, D: 0.7], F: 2.8, M: 2.6 | C: 96.0 [S: 94.5, D: 1.5], F: 1.5, M: 2.5 | C: 96.7 [S: 81.3, D: 15.4], F: 1.5, M: 1.8 | C: 90.0 [S: 87.1, D: 2.9], F: 5.0, M: 5.0 | C: 94.0 [S: 93.5, D: 0.5], F: 2.5, M: 3.5 | |
| Reference | This study | Robbins et al. (2019) | Shinzato et al. (2021) | Shinzato et al. (2021) | Ying et al. (2019) a | Voolstra et al. (2017) a | Prada et al. (2016) a | Cunning et al. (2018) a |
Genome data were retrieved from NCBI RefSeq100.
C, complete BUSCOs; S, complete and single-copy BUSCOs; D, complete and duplicated BUSCOs; F, fragmented BUSCOs; M, missing BUSCOs.
In addition to the P. australiensis and P. lutea genomes, we used publicly available gene models of two anemones, Nematostella vectensis (Putnam et al. 2007) and Exaiptasia pallida (Baumgarten et al. 2015), two corallimorpharians, Amplexidiscus fenestrafer and Discosoma spp. (Wang et al. 2017) for Hierarchical Orthogroup (HOG) clustering. Reef-building scleractinians comprise two major clades, “robust” and “complex,” based on molecular phylogenic analyses (Romano and Palumbi 1996; Kitahara et al. 2010). Genomes of three scleractinians from the “robust” clade, Stylophora pistillata (Voolstra et al. 2017), Pocillopora damicornis (Cunning et al. 2018), and Orbicella faveolata (Prada et al. 2016), and five Acropora corals, A. awi, A. cytherea, A. digitifera, A. tenuis (Shinzato et al. 2021), and A. millepora (Ying et al. 2019) from the “complex” clade were also used for HOG clustering (14 anthozoan genomes in total). Examination of HOGs in anthozoan genomes allowed us to identify 33,052 HOGs in all taxa, 25,246 in scleractinians, and 20,786 in Porites (fig. 1C; supplementary table 3, Supplementary Material online). Phylogenomic analysis of these anthozoan genomes using 1,878 single-copy orthologous group (OG) genes yielded robust phylogenetic relationships, with all clades supported by 100% bootstrap values (fig. 1D), clearly indicating that Porites belongs to the “complex” coral clade, as reported by previous molecular phylogenic analyses (e.g., Fukami et al. 2008; Kitahara et al. 2010, 2016).
In previous studies, we reported that cystathionine β-synthase (CBS, fig. 1E), an essential enzyme for cysteine biosynthesis, was probably lost from the A. digitifera (Shinzato et al. 2011) and other acroporid genomes (Shinzato et al. 2021). Because Acropora species are sensitive to bleaching, it is likely that Acropora depends upon symbiotic dinoflagellates to produce cysteine. Genes orthologous with CBS can be found in Porites (P. australiensis and P. lutea), as well as in robust coral, corallimorpharian, and sea anemone genomes (HOG0019269, supplementary table 3, Supplementary Material online). In this study, we compared syntenic relationship around the CBS genomic locus in Porites and Acropora. Although syntenic relationships of genes neighboring the PoritesCBS are well conserved between Porites and Acropora, the CBS locus is missing from the Acropora genomes (fig. 1E), providing additional evidence supporting the notion that this enzyme was lost in acroporids (Shinzato et al. 2021) and that differences in dependency on symbiotic algae could partially explain the high sensitivity of Acropora to bleaching.
Skeletal Proteome of Porites australiensis
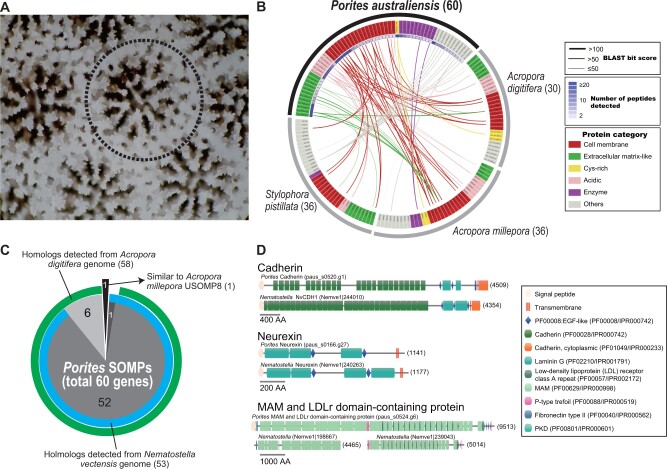
In order to identify genes involved in P. australiensis skeleton formation, we performed a skeletal proteome analysis using liquid chromatography tandem mass spectrometry (LC–MS/MS) (fig. 2). In total, 60 skeletal organic matrix proteins (SOMPs) were detected in the skeleton (fig. 2B; supplementary tables 4 and 5, Supplementary Material online). According to functional domain architecture and predicted subcellular localization, SOMPs were classified into six categories: cell membrane, extracellular matrix-like, cysteine-rich, acidic, enzymes, and others (fig. 2B; supplementary table 4, Supplementary Material online). One-third of SOMPs were categorized as cell membrane proteins, having one or more transmembrane domains (fig. 2B; supplementary table 4, Supplementary Material online). Twelve cell membrane proteins presented high sequence similarity to SOMPs of three other corals (two Acropora and Stylophora) with high BLAST bit score (≥100) (fig. 2B). Among them, orthologs including REJ domain-containing protein (dcp), neurexin, TSP-1 and VWA dcps, MAM and LDLr dcp, and cadherin are found in the other three corals (supplementary tables 4 and 6, Supplementary Material online). Porites galaxin, originally identified from a coral, Galaxea fascicularis (Fukuda et al. 2003), and classified in the cysteine-rich category, has a counterpart in Acropora skeletal proteomes, but not in Stylophora (supplementary tables 4 and 6, Supplementary Material online). Secreted acidic Asp-rich proteins, or SAARPs (Ramos-Silva et al. 2013), are present in all three coral skeletal proteomes (supplementary tables 4 and 6, Supplementary Material online). Three other Porites acidic protein genes (paus_s0019.g24, paus_s0019.g25, and paus_s0019.g26) tandemly located in the P. australiensis genome, also showed weak similarity to other SOMPs, owing to their CUB domains (supplementary table 6, Supplementary Material online).
Fig. 2.
Skeletal proteome analysis of Porites australiensis. (A) High-magnification photo of a P. australiensis skeleton. The dotted line indicates a polyp. (B) A circos plot showing sequence similarities of PoritesSOMP genes against known SOMPs of other corals, Acropora digitifera, A. millepora, and Stylophora pistillata. Each box along the arc indicates a SOMP gene, and gene names are colored based on protein categories (cell membrane, extracellular matrix-like, cysteine-rich, acidic, enzymes, and others), as shown at the right. SOMPs connected by lines show significant sequence similarity (BLASTP, E-value ≤e−5), and thicknesses of lines reflect BLAST bit scores. Numbers of peptides detected by LC–MS/MS are shown by heatmap inside Porites gene names. (C) A pie chart showing numbers of putative homologs of PoritesSOMP genes (total 60) detected in Acropora digitifera (green) and sea anemone N. vectensis (blue) genomes. One gene (paus_s0004.g93) that was not detected in the two genomes showed similarity to A. milleporaUSOMP8 (Ramos-Silva et al. 2013). (D) Comparison of functional protein domain architectures of Porites cell membrane SOMPs, cadherin, neurexin, and MAM and LDLr dcps (upper) with their putative orthologs in Nematostella (lower). The amino acid length of each gene is shown in brackets.
We searched for putative homologous genes of the 60 P. australiensisSOMPs in the other cnidarian genomes using BLAST searches (fig. 2C). The majority of P. australiensisSOMP genes (59/60) showed significant sequence similarities to genes of Acropora (A. digitifera and A. millepora) and notably, to those of a sea anemone, N.vectensis (fig. 2C). A PoritesSOMP gene (paus_s0004.g93) that does not possess any conserved domains and does not resemble the A. digitifera and N. vectensis genes, shows significant similarity to the A. milleporaUSOMP8 (BLASTP, e-value <e−5, fig. 2B; supplementary table 4, Supplementary Material online). Among the 60 P. australiensisSOMPs, 18 genes were not found among previously reported SOMPs of A. digitifera, A. millepora, and S. pistillata (fig. 2B; supplementary table 6, Supplementary Material online), indicating that these function as SOMPs specifically in Porites but not in the other three corals. Domain architectures of three Porites cell membrane SOMPs (cadherin, neurexin, and MAM and LDLr dcp) are well conserved with their putative orthologs in sea anemones (fig. 2D). These results indicate that all P. australiensisSOMP genes have homologs in other corals and noncoral anthozoans, but some genes do not present in skeletons of other corals (Acropora and Stylophora) are employed as skeletal matrix proteins in P. australiensis.
Porites-Specific Gene Expansions Include Environmental Stress Response Genes
Gene duplication is a major driving force of genome evolution and facilitates acquisition of novel gene functions (Ohno 1970). Therefore, genes supporting unique characters of Porites, such as stress tolerance, may have been duplicated during evolution of the Porites lineage. To eliminate the potential impact of genome assembly and gene prediction errors, we considered HOGs for which gene numbers in both P. australiensis and P. lutea are larger than other coral or anthozoan genomes. We identified 31 HOGs specifically expanded in the Porites lineage (fig. 3A). Among those, 25 HOGs exhibit similarities to proteins in the SwissProt protein database. The 31 expanded HOGs show significantly more Swiss-Prot protein database hits (83%, 257 out of 309 genes) than P. australiensis gene models generally (67%, 20,418 out of 30,301 genes, P < 0.01, Pearson’s Chi-squared test), indicating that conserved genes shared with other organisms may have been selectively expanded in the Porites lineage.
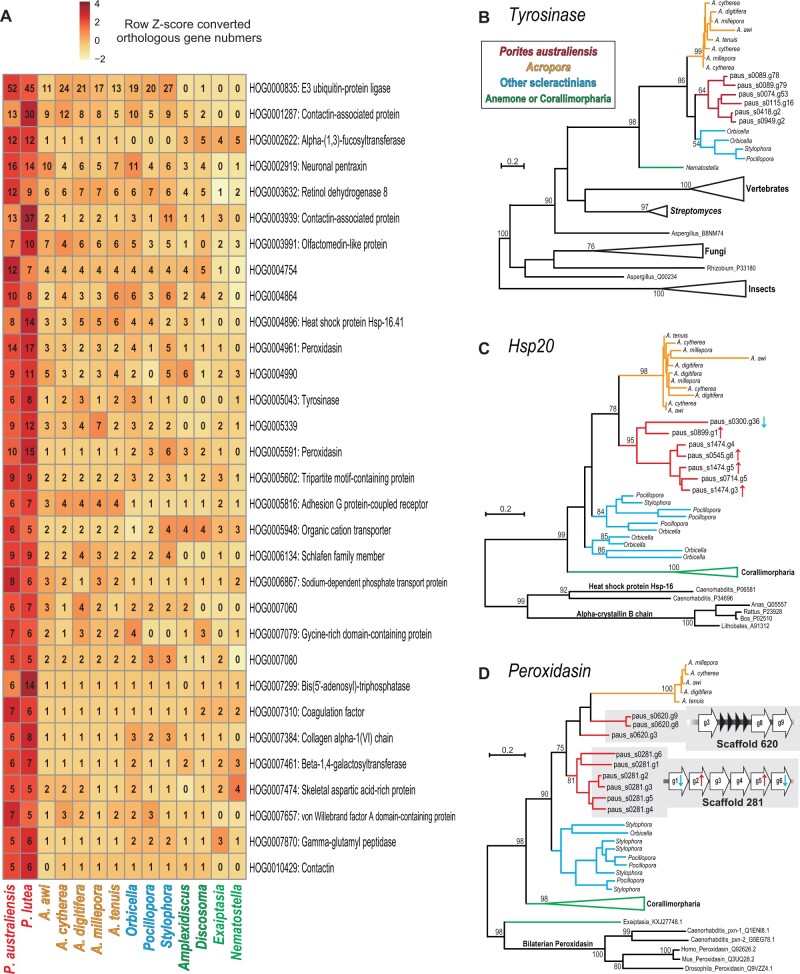
Fig. 3.
Porites-specific gene expansions. (A) Gene families that are expanded in the Porites lineage. Numbers of genes in each gene family, shown in each box, are converted to row Z scores and colored. (B) Maximum likelihood analysis of tyrosinases, or tyrosinase-type phenoloxidases genes. (C) Maximum likelihood analysis of heat shock protein 20 genes. (D) Maximum likelihood analysis of peroxidasin genes. Peroxidasin gene clusters in the P. australiensis genome are shown in gray. Nodes of P. australiensis genes are colored red, Acropora genes are in yellow. Robust coral genes are in blue. Corallimorpharian genes are in green. Upward red arrows and downward blue arrows next to gene names indicate genes significantly upregulated or downregulated by increased seawater temperature (FDR <0.05) reported in figure 4, respectively.
We found that expanded HOGs include genes possibly involved in variety of environmental stress responses, such as tyrosinase (fig. 3B), small heat shock protein (HSP20) (fig. 3C), and peroxidasin (fig. 3D). Molecular phylogenetic analyses of these genes showed that in Porites, most of them are clustered (fig. 3B–D), indicating that gene duplications occurred specifically in the Porites lineage. Peroxidasin genes are tandemly located in the P. australiensis genome with the same gene orientation (three genes in scaffold 681, and six genes in scaffold 281, fig. 3D). Interestingly, five of seven duplicated heat shock protein genes and four of six tandemly duplicated peroxidasin genes on scaffold 281 are differentially expressed when P. australiensis is exposed to increased seawater temperature (fig. 3C and D; for more details, see below). In contrast, none of the expanded tyrosinase genes examined in this study (fig. 3B) exhibited enhanced expression in response to increased seawater temperature or light intensity.
Differential Molecular Responses of Porites and Acropora to Increased Light Intensity and Seawater Temperature
In order to explore differences in transcriptome responses to environmental stressors in stress-tolerant Porites and stress-sensitive Acropora, we performed an experiment subjecting P. australiensis and A. digitifera to increased seawater temperature (25–30 °C) and light intensity (180–200–400 μmol/m2/s). Then transcriptome responses of both corals were investigated and differentially expressed genes (DEGs, P < 0.05) were identified (fig. 4A). In order to compare P. australiensis and A. digitifera DEGs directly, we focused on single-copy HOGs (SC-HOGs) between P. australiensis and A. digitifera (9,078 HOGs; see supplementary table 3, Supplementary Material online). Unexpectedly, few DEGs were induced in either coral by increased light intensity (2 DEGs in P. australiensis and 14 DEGs in A. digitifera, supplementary table 7, Supplementary Material online). Only one SC-HOG, hepatic leukemia factor (HLF), a member of the proline and acidic amino acid-rich basic leucine zipper (PAR) transcription factor family, was upregulated in both corals, in response to increased light (fig. 4A; supplementary table 7, Supplementary Material online).
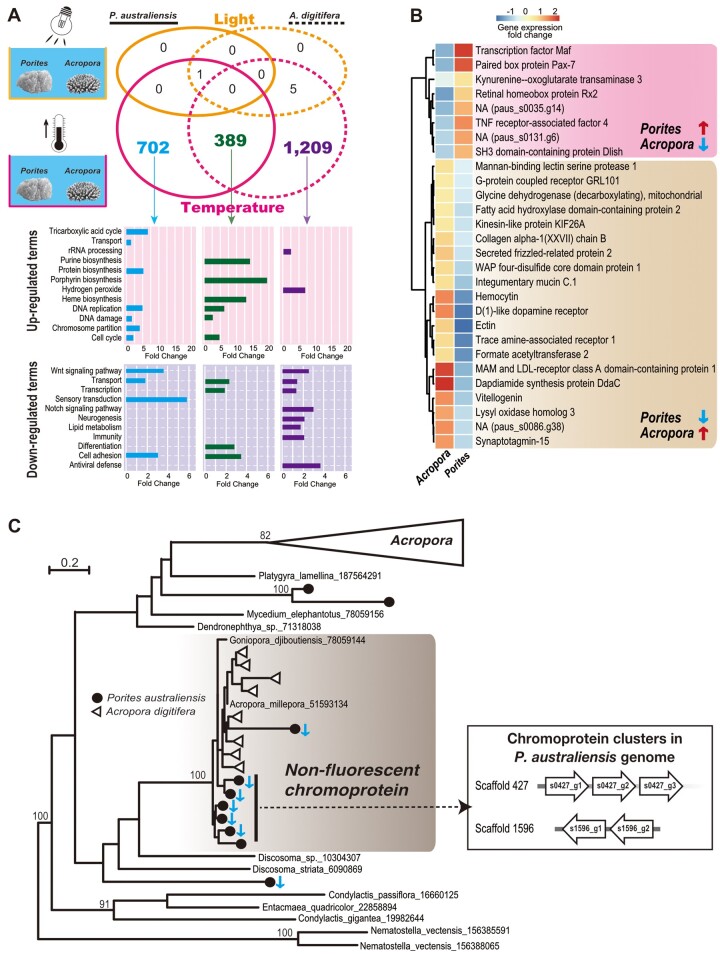
Fig. 4.
Comparison of transcriptomic responses of Porites australiensis and Acropora digitifera to increased seawater temperature and light intensity. (A) Upper: Venn diagram showing numbers of DEGs included in single-copy Hierarchical Orthogroups (SC-HOGs) between P. australiensis and A. digitifera. SC-HOGs differentially expressed under increased light intensity are colored yellow, and SC-HOGs differentially expressed under increased seawater temperature are colored pink. Lower: significantly upregulated or downregulated UniProt keywords (biological process) detected from DEGs in SC-HOGs under increased seawater temperature. Blue indicates Porites-specific SC-HOGs that were differentially expressed. Green indicates SC-HOGs differentially expressed in both Porites and Acropora. Purple indicates Acropora-specific SC-HOGs that were differentially expressed. (B) Gene expression changes of SC-HOGs differentially expressed in both Porites and Acropora, but in which expression patterns were opposite. Putative gene names are based on the Uniprot/Swissprot database. Pink shading denotes SC-HOG gene pairs that were significantly upregulated in Porites and downregulated in Acropora. Brown shading indicates SC-HOGs that were significantly downregulated in Porites and upregulated in Acropora. Gene expression changes compared with control conditions are shown in a colored heatmap. (C) Maximum likelihood analysis of anthozoan fluorescent protein (FP)-like genes using 215 aligned translated amino acids. Bootstrap values more than 80% are shown. Porites australiensis genes are shown with circles and Acropora digitifera genes are shown with triangles. The Acropora-specific FP clade is collapsed and nonfluorescent chromoproteins are shaded in gray. Downward blue arrows indicate P. australiensischromoprotein genes significantly downregulated by increased seawater temperature (FDR <0.05). Chromoprotein gene clusters in the P. australiensis genome, all of which were downregulated by increased seawater temperature, are shown in the box to the right.
Despite the small number of genes affected by increased light intensity, large numbers of genes in P. australiensis (2,749) and A. digitifera (3,800) were differentially expressed in response to increased seawater temperature (supplementary tables 8 and 9, Supplementary Material online). A total of 389 SC-HOGs were differentially expressed in both Porites and Acropora. Seven hundred and two SC-HOGs were differentially expressed solely in P. australiensis (418 upregulated and 284 downregulated genes), and 1,209 SC-HOGs were differentially expressed only in Acropora (659 upregulated and 550 downregulated genes) (fig. 4A). Functional enrichment analysis based on UniProt keywords revealed different responses between Porites and Acropora to increased seawater temperature (fig. 4A). Genes involved in purine biosynthesis, porphyrin biosynthesis, and heme biosynthesis were upregulated, and genes involved in transcription and differentiation were downregulated in both Porites and Acropora. DNA replication, DNA damage, and cell cycle genes were upregulated, whereas sensory transduction genes participating in cellular conversion of extracellular stimuli into electric signals, were downregulated in P. australiensis, although genes encoding proteins that process hydrogen peroxide were specifically upregulated, whereas notch signaling, lipid metabolism, and immunity were specifically downregulated in A. digitifera (fig. 4A). Interestingly, among the 389 SC-HOGs that were differentially expressed in both P. australiensis and A. digitifera (fig. 4A), 28 SC-HOGs, including transcription factors (Maf and homeobox) and receptors, showed opposing gene expression patterns, for example, upregulated in Porites, but downregulated in Acropora, or vice versa (fig. 4B), suggesting that these promote different transcriptome responses in Porites and Acropora.
We found that gene expression responses of fluorescent protein (FP)-like genes to increased seawater temperature differed significantly between Porites and Acropora. We identified ten FP-like genes in the P. australiensis genome, and molecular phylogenetic analysis of FP-like genes revealed that nonfluorescent chromoproteins clustered together with 100% bootstrap support (fig. 4C). Interestingly, all FP-like genes differentially expressed in response to increased seawater temperature in Porites (six downregulated genes, of which five are tandemly located in the P. australiensis genome) are chromoproteins; however, no A. digitiferachromoproteins displayed altered gene expression patterns (fig. 4C). In contrast, seven Acropora-specific FP-like genes in A. digitifera were differentially expressed in response to increased seawater temperature (one upregulated and six downregulated; supplementary fig. 2, Supplementary Material online).
Discussion
Conserved Genes May Contribute to Unique Characters of Massive Porites
Massive Porites are regarded as stress-tolerant and long-lived, and a recent study showed that these corals have higher thermal tolerance and that they may acclimatize fast enough to keep pace with current global warming (DeCarlo et al. 2019). We found that some conserved genes shared with other organisms are underwent duplication in the Porites lineage (fig. 3). Interestingly, these expanded genes include those involved in environmental stress responses in a variety of animals, and involvement of these genes in environmental stress responses in corals has been suggested and investigated. Consistent with our finding that numbers of HSP20 genes in Porites genomes are larger than those of other corals, corallimorpharians, and sea anemones (fig. 3A), Ying et al. (2018) reported that numbers of HSP20 genes in P. lutea, as well as in a Goniastrea genomes, are larger than those of Acropora corals and that different numbers of HSP20 genes may correlate with coral stress tolerance. Tyrosinases, or tyrosinase-type phenoloxidases are responsible for the immune response of the phenoloxidase pathway in invertebrates via melanin synthesis, and coral tyrosinase-type phenoloxidases respond to various environmental stressors, including heat stress, disease, pathogens, sedimentation, nutrient loading, and damage (Mydlarz et al. 2008, 2009; Palmer et al. 2011, 2012; Sheridan et al. 2014; van de Water et al. 2015, 2018; Kelly et al. 2016; Wall et al. 2018; Dougan et al. 2020). Peroxidasin genes are involved in oxidative stress responses and are differentially expressed in corals under heat stress (Voolstra et al. 2009; Barshis et al. 2013; Louis et al. 2017). Taken together, conserved environmental stress−response genes may contribute to the stress tolerance of massive Porites. Although gene expression of Porites-specific expanded tyrosinase genes was not affected by increased seawater temperature or increased light intensity, four out of seven expanded HSP20 genes and two out of six tandem-duplicated peroxidasin genes were upregulated by increased seawater temperature (four upregulated and one downregulated; fig. 3), suggesting that some Porites-specific expanded HSP20 and peroxidasin genes may be involved in its high-temperature response, but that tyrosinase genes serve other functions.
Massive skeletons are unique to Porites corals. Sequence similarity searches revealed that a set of orthologs encoding nine SOMPs (cadherin, neurexin, MAM and LDLr dcp, TSP1 and VWA dcp, SAARPs, and USOMP8) is shared among Porites, Acropora, and Stylophora (fig. 2B; supplementary table 6, Supplementary Material online), suggesting that these functioned as basic SOMPs in the common ancestor of extant scleractinians. An acidic protein family (SAARPs) is commonly found in coral skeletons (fig. 2B), indicating its essential role in coral biomineralization. Negatively charged side chains of aspartic acid residues may interact with calcium ions to regulate nucleation, inhibition, and orientation of calcium carbonate crystal growth (Addadi and Weiner 1985; Albeck et al. 1993; Marin and Luquet 2007). In P. australiensisSOMPs, we identified novel acidic SOMPs that have CUB domains (Pau-SAARP, supplementary table 4, Supplementary Material online), which may also act as calcium binding sites (Blanc et al. 2007) and/or may interact with other proteins to construct the skeletal organic matrix. Other acidic SOMPs, skeletal acidic proteins, or SAPs that are present in Acropora skeletons (Shinzato et al. 2011; Ramos-Silva et al. 2013) were not detected in Porites. This supports previous reports that SAPs are unique to Acropora species (Shinzato et al. 2011; Takeuchi et al. 2016). In summary, our comparative study of Porites and other coral SOMPs highlights essential proteomic components for coral skeletal formation, such as cell membrane proteins and acidic proteins. Notably, all PoritesSOMP genes have putative homologs in other corals and even in a sea anemone, which has no skeleton (fig. 2C; supplementary table 3, Supplementary Material online). In particular, domain architectures of three cell membrane SOMPs (cadherin, neurexin, and MAM and LDLr dcp) are well conserved between Porites and Nematostella (fig. 2D), suggesting that these proteins originally functioned in cell adhesion, attaching ancestors of these organisms to the substrate. Taken together, most SOMP genes existed in the ancestor of scleractinians and anemones, dating back about 500 Ma (Shinzato et al. 2011), and were co-opted for coral skeleton formation in the scleractinian lineage (Takeuchi et al. 2016).
Different Molecular Responses of Acropora and Porites to Increased Temperature May Reflect Their Divergent Stress Tolerance
Because of their algal endosymbionts, light is important to maintain coral health. Light-enhanced calcification of reef-building corals is a well-known phenomenon (Cohen et al. 2016). However, in this study, expression of very few genes was affected by increased light intensity in either Porites or Acropora (fig. 4A; supplementary table 7, Supplementary Material online), indicating that host corals show no universal molecular response to increased irradiation and that increased light intensity has a limited impact on either coral host. The small number of DEGs that responded to increased irradiation, which did not include SOMPs, is consistent with a previous experiment using symbiotic and apo-symbiotic coral polyps, showing that light-enhanced coral calcification could be caused by increased pH of the calcifying fluid induced by algal photosynthesis, rather than by the increase of symbiont photosynthetic products supplied to host corals (Inoue et al. 2018). Nevertheless, HLF transcription factors are the only genes that were commonly upregulated in both Porites and Acropora (fig. 4A; supplementary table 7, Supplementary Material online). Although the function of HLF genes in corals is unclear, HLF transcription factors regulate expression of apoptotic and circadian clock genes (Waters et al. 2013; Takahashi 2017), suggesting that increased light may affect circadian rhythm of Porites and Acropora, although no expression changes of apoptotic and circadian clock genes were detected in this study.
Among the 2,300 SC-HOGs differentially expressed in response to increased seawater temperature, 16% (389) were differentially expressed in both Porites and Acropora, and 29% (702) and 53% (1,209) were exclusive to Porites and Acropora, respectively (fig. 4A). Upregulation of protein biosynthesis, DNA replication, DNA damage, and cell cycle genes were observed in Porites, but not in SC-HOGs solely in Acropora, indicating that Porites actively reacts to higher temperatures at the cellular level and that these mechanisms may contribute to its stress tolerance. Another interesting point is that expression patterns of some transcription factors, controlling expression of numerous downstream genes and affecting various biological processes, were reversed in Porites and Acropora under increased seawater temperature (fig. 4B). Maf transcription factors are involved in cellular responses against oxidants and heavy metals (Suzuki et al. 2001; Bensellam et al. 2015). These transcription factor genes may reveal the diversity of molecular responses of corals to environmental changes.
FPs are thought to serve multiple functions in corals, including maintenance of obligate symbioses with dinoflagellates, quenching of oxygen radicals, and stress responses (e.g., Bou-Abdallah et al. 2006; Dove et al. 2001; Kawaguti 1969; Matz et al. 2002; Rodriguez-Lanetty et al. 2009; Salih et al. 2000; Seneca et al. 2010). Corals possess three FPs [cyan (CFP), green (GFP), and red (RFP)], and nonfluorescent blue/purple chromoprotein (Kelmanson and Matz 2003; Field et al. 2006). Expression of coral FP genes is affected by external stimuli, such as light, heat, and injury (D’Angelo et al. 2008, 2012; Rodriguez-Lanetty et al. 2009; Roth et al. 2010; Seneca et al. 2010; DeSalvo et al. 2012; Roth and Deheyn 2013). Chromoproteins exhibit higher absorption and lower emission (Matz et al. 2002; Bou-Abdallah et al. 2006) and may have higher antioxidant activity than FPs (Palmer et al. 2009). Although it has been reported that expression of a chromoprotein in Porites astreoides was upregulated by heat stress (Kenkel et al. 2011), in this study, all differentially expressed chromoprotein genes were downregulated under increased temperature (fig. 4C). Although it could be that seawater temperature in this study was not increased enough to induce upregulation of chromoprotein genes, their functions in Porites may be more diverse, for example, maintenance of homeostasis. Studying functions of chromoproteins and FPs in corals should enable greater understanding of coral biology.
Conclusions
We sequenced the complete genome of P.australiensis with reasonable quality. We showed that homologs of all Porites skeletal matrix protein genes exist in other corals and anemones. Moreover, we revealed that conserved genes shared with other taxa may be selectively expanded in the Porites lineage and that these expanded genes include those that respond to diverse environmental stresses. This suggests that genes conserved among many organisms may have contributed significantly to distinctive Porites biological characters, their massive skeletons and high stress tolerance. Comparison of transcriptomic responses to changing environments using Porites and Acropora highlighted dynamic differences of their responses to environmental changes. Some genes expanded exclusively in Porites, including HSP20 and peroxidasin, respond to increased seawater temperature, possibly accounting for the different stress tolerances of these corals. The present genomic resources, together with other coral genomic data, will provide a powerful resource to understand the divergent ecology of reef-building corals as well as mechanisms of coral calcification.
Materials and Methods
Sample Collection, Genome Sequencing, and Assembly
The coral sample used in this study was collected at Sesoko Island, Okinawa, Japan, under Okinawa prefectural permit number 20-69. Small P.australiensis colonies had been maintained at the Sesoko Research Station of the University of the Ryukyus for 5 years. Because P. australiensis is gonochoric, gonad development for each colony was checked and male colonies were identified. Sperm was isolated from a male colony when spawning occurred (June 11, 2012). Unfertilized eggs were collected from a female colony (June 30, 2013) and snap-frozen until needed.
DNA was isolated from sperm using the phenol-chloroform method and fragmented into approximately 600-bp lengths. Two hundred nanograms of DNA were used for PCR-free shotgun library preparation. For mate-pair libraries, DNA of different lengths (approximately 3, 7, 10, and 15 kb) was separated using SageELF (Sage Science). Nextera Mate Pair Library Prep Kits (Illumina) were used for library preparation following manufacturer instructions. Prepared libraries were sequenced using MiSeq and HiSeq sequencers (Illumina) according to the manufacturer’s protocols (Illumina).
Sequence data (supplementary table 1, Supplementary Material online) were assembled using Newbler version 2.8 (Roche) with “-het” and “-large” options. Possible diploid scaffolds were merged with HaploMerger2 (Huang et al. 2017) and further scaffolding was performed with SSPACE (ver. 3) with “-k 3” option (Boetzer et al. 2011). Gaps inside scaffolds were closed using GapCloser (ver. 1.10) with default settings (Huang et al. 2017). Possible errors in genome assembly were corrected with Pilon version 1.22 (Walker et al. 2014) using Illumina shotgun data with default settings. In the end, we identified scaffold sequences with high or low coverage or those that may have originated from one of the two allelic copies of heterozygous regions, using Purge Haplotigs with Illumina shotgun data to calculate coverages for each scaffold (Roach et al. 2018), and these were excluded from subsequent analyses. To check genome assembly correctness, we mapped back 10% of randomly selected paired-end shotgun sequencing data to the assembled genome using Bowtie 2 version 2.4.4 with the very-sensitive-local option (Langmead and Salzberg 2012). Mapped reads and properly paired reads, both forward and reverse reads mapped to the same scaffold, were counted using SAMtools version 0.1.19 (Li et al. 2009). Estimation of the haploid genome size using shotgun sequencing data was performed using GenomeScope (Vurture et al. 2017) with a kmer length of 29 and max kmer coverage of 10,000. We assessed genome assembly completeness with BUSCO ver. 3.0.2 (Simao et al. 2015; Waterhouse et al. 2017) using the Metazoa set (978 genes). To reconstruct the complete P. australiensis mitochondrial genome, we BLAST-searched raw Newbler assembled scaffolds using Porites mitochondrial genomes as queries and identified the best candidate sequence. Then the sequence was annotated using GeSeq (Tillich et al. 2017).
Gene Prediction and Annotation
A training set for gene prediction was prepared from all open reading frames in the P. australiensis adult transcriptome assembly (Shinzato et al. 2014) using PASA, following instructions (Haas et al. 2003), then we trained Augustus version 3.2.3 (Stanke et al. 2006) to prepare parameters specific to Porites. We also used Augustus for gene prediction from repeat-masked genomes produced by RepeatMasker (Smit et al. 1996–2010) with “–UTR=on” option using RNA-seq data from eggs and adults as hints (mapping both raw RNA-Seq reads and transcriptome assembly data). We named the P. australiensis gene models using the pattern, paus.s[scaffold number].g[number of gene in the scaffold]. For example, “paus_s0019.g26,” is located in scaffold 19 and is the 26th gene in the scaffold. In order to remove gene models that sometimes originate from different haplotypes, the predicted proteome was clustered using CDHIT (98% sequence identity) (Li and Godzik 2006), and proteins shorter than 30 amino acids were excluded from subsequent analyses. Predicted gene models were BLASTed against the Uniprot/Swissprot (UniProt Consortium 2018) database and were analyzed with InterProScan 5 with e-value cutoff of 1e−5 (Jones et al. 2014).
Clustering of Orthologous Genes and Molecular Phylogenetic Analysis
We used publicly available gene models of the two anemones, N.vectensis (Putnam et al. 2007) and E.pallida (Baumgarten et al. 2015), two corallimorpharians, A.fenestrafer and Discosoma spp. (Wang et al. 2017), and three scleractinians from the “robust” clade, S.pistillata (Voolstra et al. 2017), P.damicornis (Cunning et al. 2018), and O.faveolata (Prada et al. 2016). Five Acropora corals, A. awi, A. cytherea, A. digitifera, A. tenuis (Shinzato et al. 2021), and A. millepora (Ying et al. 2019), representing the “complex” clade, were also included in our analyses. For the A. millepora, S. pistillata, O. faveolata, and E. pallida genomes, we downloaded data from the NCBI RefSeq database. Then, using OrthoFinder version 2.4.0 (Emms and Kelly 2015), we performed clustering of orthogroups (OGs). Genes descended from a single gene in the last common ancestor of a group of species, Hierarchical Orthogroups (HOGs) were used for subsequent analyses.
For phylogenomic analysis of anthozoan genomes, we used 1,878 genes that were assigned by OrthoFinder as single-copy in all of the above anthozoan genomes. All amino acid sequences belonging to same OG were aligned with MAFFT (ver. 7.310. with –auto option) (Katoh and Standley 2013) and all gaps in the alignment were removed with TrimAL (Capella-Gutierrez et al. 2009) with the –nogaps option. Then all sequences from the same species were concatenated, and finally, a maximum likelihood analysis was performed using concatenated sequences (429,044 amino acids in length) from RAxML with 100 bootstraps replicates and “protgammaauto” option.
In order to identify HOGs specifically expanded in the Porites lineage, we selected HOGs as follows: 1) gene numbers of P. australiensis and P. lutea should be more than four; 2) gene numbers of both Porites species should be larger than those of the other 12 anthozoan species used in OG clustering; 3) genes from at least 11 of 14 species were included; 4) z-score converted gene numbers of P. australiensis or P. lutea must be over 1.92 (P < 0.05).
For phylogenetic analysis of each gene, amino acid sequences were aligned using MAFFT (ver. 7.310. with –auto option) (Katoh and Standley 2013), and gaps in aligned sequences were trimmed using TrimAL (Capella-Gutierrez et al. 2009) with the –gappyout option. After that, poorly aligned sequences were removed (-resoverlap 0.75 -seqoverlap 80). Then we performed molecular phylogenetic analysis of the selected alignments using RAxML (maximum likelihood method) with 100 bootstrap replicates and “protgammaauto” option (Stamatakis 2014).
Skeletal Proteome Analysis of P. australiensis
A colony of P. australiensis kept at the Sesoko Station of the University of the Ryukyus, was used for skeletal proteomic analysis. Methods for sample cleaning, skeletal organic matrix extraction, and identification of SOMPs have been described by Takeuchi et al. (2018). Briefly, the coral sample was put in 3 L of 10× diluted household bleach solution overnight, and then washed with water. This procedure continued until animal tissue and other organisms on the surface were removed (initial bleaching). The coral skeleton was rinsed, air-dried and crushed into ∼2-mm fragments with a Jaw-crusher (Retsch BB200). Fragments were immersed in a 10× dilution of sodium hypochlorite 10–15% (SIGMA) for 50 h (second bleaching or 2bl), and then they were washed with purified water, dried, and powderized using a rotary mill (Frisch Pulverisette 2). The powder was sieved (pore size <200µm) and separated into two batches. One batch was subsequently decalcified, whereas the other was bleached in NaOCl solution again, washed, and air-dried (third bleaching or 3bl) before decalcification. Cleaned powder samples were suspended in cold water and decalcified by adding acetic acid (10% v/v) with an electric burette (Titronic Universal, Schott, Mainz, Germany) at 4 °C overnight. The solution was centrifuged, after which the pellet (acid insoluble matrix or AIM) and supernatant (acid soluble matrix or ASM) were treated separately. The AIM pellet was washed with MilliQ water and freeze-dried. The ASM solution was concentrated by ultrafiltration (Amicon Ultracel 10 kDa), dialyzed for 4 days, and freeze-dried. Proteomic analyses were conducted on bulk ASM and AIM matrices after in-gel digestion with trypsin. For MS and MS/MS, analyses were performed using an Ultimate 3000 Rapid Separation Liquid Chromatographic (RSLC) system (Thermo Fisher Scientific) online with a hybrid LTQ-Orbitrap-Velos mass spectrometer (Thermo Fisher Scientific). All technical details are provided in Immel et al. (2016). Database searches were carried out using Mascot version 2.4 and 2.5 (MatrixScience, London, UK) on gene models of P. australiensis. The false discovery rate (FDR) was set to 0.05. Proteins were treated as identified SOMPs and further analyzed when more than two unique peptides or more than ten total peptides were detected (supplementary table 5, Supplementary Material online).
In order to find functional domains in SOMPs, amino acid sequences were analyzed using InterProScan 5.3 (Jones et al. 2014). Signal peptides were predicted with SignalP 4.1 (Petersen et al. 2011). Transmembrane domains were assessed with TMHMM 2.0 software (Krogh et al. 2001). Theoretical molecular weights and isoelectric points of SOMP sequences without signal peptides were calculated using IPC 1.0 (Kozlowski 2016). All programs were run using default settings and thresholds. Sequence similarities between SOMPs of P. australiensis and other corals, including A. digitifera (Takeuchi et al. 2016), A. millepora (Ramos-Silva et al. 2013), and S.pistillata (Drake et al. 2013), were evaluated using BLASTP with a threshold of ≤e−5. We also BLASTP-searched all protein models encoded in the A. digitifera genome (Shinzato et al. 2011) to find homologous gene candidates of P. australiensisSOMPs. A. digitifera protein-coding gene(s) significantly (cutoff ≤e−5) similar to P. australiensisSOMPs were identified from the A. digitifera genome and were used for pairwise alignments of amino acid sequences using MAFFT with the “einsi” command (Katoh and Standley 2013). Then poorly aligned regions were trimmed using TrimAL (Capella-Gutierrez et al. 2009). If lengths of a high-quality alignment region between a P. australiensisSOMP gene were longer than 40% of full-length, we assumed that the A. digitifera gene is a putative homolog of the P. australiensisSOMP gene, regardless of whether gene products were detected in the A. digitifera skeleton. The homologous gene candidate search was also conducted against the genome of a sea anemone, N.vectensis (Putnam et al. 2007).
Transcriptomic Responses of P. australiensis and A. digitifera to Increased Seawater Temperature and Light Intensity
Three colonies of P. australiensis (2 cm diameter) and A. digitifera (2 cm branch length) were collected and three fragments from each colony were isolated and maintained in a tank with running seawater under natural light conditions at Sesoko Station, Tropical Biosphere Research Center, University of the Ryukyus, Okinawa, Japan, before starting the experiment. Three fragments from each colony were isolated and maintained in each aquarium as reported in Iguchi et al. (2012). Seawater temperature in each aquarium was gradually increased from 21 °C (ambient, March 11, 2016) to 25 °C in 4 days and was maintained at approximately 25 °C for 3 days with a 12:12 light:dark photoperiod (180–200 μmol/m2/s) under metal-halide lamps (Funnel2 150 W, Kamihata, Japan). Then two aquaria subjected to increased seawater temperature were set to 30 °C (180–200 μmol/m2/s) and two aquaria subjected to increased light intensity were set to 400 μmol/m2/s (25 °C) for 3 days, because coral calcification rates are reportedly saturated at about 400 μmol/m2/s (Suggett et al. 2013). Three fragments each from three colonies (nine fragments total) were used for each condition (control: 25 °C and 180–200 μmol/m2/s; increased temperature: 30 °C and 180–200 μmol/m2/s; intensity light: 25 °C and 400 μmol/m2/s). As three fragments from the same colony of A. digitifera maintained at increased seawater temperature died when the experiment finished, these were excluded from subsequent analyses. Coral fragments were snap frozen in liquid nitrogen and stored at −80 °C until use.
Total RNA was isolated from each fragment using an RNeasy plant kit (QIAGEN) and sequenced on an Illumina HiSeq2000 platform. Illumina adaptor sequences and low-quality reads (Quality score <20, length <25 bp) were trimmed with CUTADAPT v1.16 (Martin 2011). Cleaned reads of both P. australiensis and A. digitifera were mapped to each gene model using MiniMap v2.9 (Li 2018) with default settings. Statistical tests were performed using EdgeR v3.28.1 (Robinson et al. 2010; McCarthy et al. 2012) in R v3.6.3 (R Core Team 2015), because EdgeR permits the estimation of gene-specific biological variation, even for experiments with minimal levels of biological replication. As implemented in EdgeR, prior to statistical testing, we removed genes with expression levels below 1 count per million in at least half the RNA-Seq samples. Then we performed normalization with the trimmed mean of M-values method, and exact tests between control and treated groups. P-values were adjusted using the Benjamini–Hochberg method, and genes exhibiting an FDR <0.05 were considered differentially expressed genes (DEGs). In order to compare stress responses between P. australiensis and A. digitifera, single-copy HOGs (SC-HOGs) between P. australiensis and A. digitifera were identified. Functional enrichment analysis using DEGs was performed on the web platform DAVID v6.8 (Huang da et al. 2009) with default settings. UniProt IDs from all SC-HOGs of P. australiensis and A. digitifera were used as the background data set of the enrichment analysis, and UniProt IDs assigned to SC-HOGs in both P. australiensis and A. digitifera, or to P. australiensis- or A. digitifera-specific DEGs, were analyzed. Because 99.7% of UniProtIDs assigned to SC-HOGs had UniProt keywords, we focused on UniProt keywords for the functional enrichment analysis. UniProt keywords (biological process) representing P < 0.05 were considered significantly enriched terms.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by The Canon Foundation and Interdisciplinary Collaborative Research Program of the Atmosphere and Ocean Research Institute, the University of Tokyo, and in part by KAKENHI (17KT0027, 20H03235, and 20K21860 to CS). We thank Ms Mariia Khalturina (OIST) for performing genome sequencing of P. australiensis and Dr Steven D. Aird for editing the manuscript. We also thank Prof. Takashi Nakamura (University of the Ryukyus) for sharing coral specimens and Ms Hiromi Kinjo for her help and support with the preparation and performance of the tank experiments at Sesoko Station. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Author Contributions
C.S. and M.I. conceptualized and C.S., Ma.K., N.S., and M.I. designed the project. A.I., Y.Y., and C.S. performed specimen collection, species identification, sample collection, coral culturing, and tank experiments. Mi.K. performed RNA sequencing. C.S. and I.T. assembled genome, performed gene prediction, and analyzed genomic data. T.T., C.B., and F.M. performed LC–MS/MS analysis and T.T. analyzed proteomic data. Y.Y. and C.S. analyzed transcriptomic data. T.T. and Y.Y. participated in writing and C.S. wrote the main manuscript. All authors reviewed and approved the final version of the manuscript.
Data Availability
Data regarding the Porites australiensis genome assembly have been registered at GenBank under the BioProject accession PRJDB4187. The genome assembly has been deposited in the DNA DataBank of Japan/European Nucleotide Archive/GenBank under accession numbers BOPM01000001–BOPM01004983. The P. australiensis genome assembly and gene prediction data are also available from https://drive.google.com/drive/folders/1xZCIAHLmuCcdIQlvDHbN7GGJzxp6VIs0. RNA-Seq sequencing data are available from DRA submission number DRA011516.
Literature Cited
- Addadi L, Weiner S.. 1985. Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc Natl Acad Sci U S A. 82(12):4110–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck S, Aizenberg J, Addadi L, Weiner S.. 1993. Interactions of various skeletal intracrystalline components with calcite crystals. J Am Chem Soc. 115(25):11691–11697. [Google Scholar]
- Anthony KRN, Kline DI, Diaz-Pulido G, Dove S, Hoegh-Guldberg O.. 2008. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc Natl Acad Sci U S A. 105(45):17442–17446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis DJ, et al. 2013. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci U S A. 110(4):1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112(38):11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JW, et al. 1992. Sea-surface temperature from coral skeletal strontium calcium ratios. Science 257(5070):644–647. [DOI] [PubMed] [Google Scholar]
- Bensellam M, Montgomery MK, Luzuriaga J, Chan JY, Laybutt DR.. 2015. Inhibitor of differentiation proteins protect against oxidative stress by regulating the antioxidant-mitochondrial response in mouse beta cells. Diabetologia 58(4):758–770. [DOI] [PubMed] [Google Scholar]
- Blanc G, et al. 2007. Insights into how CUB domains can exert specific functions while sharing a common fold: conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J Biol Chem. 282(23):16924–16933. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Bou-Abdallah F, Chasteen ND, Lesser MP.. 2006. Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta 1760(11):1690–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb KM, Charles CD, Cheng H, Edwards RL.. 2003. El Nino/Southern oscillation and tropical Pacific climate during the last millennium. Nature 424(6946):271–276. [DOI] [PubMed] [Google Scholar]
- Cohen I, Dubinsky Z, Erez J.. 2016. Light enhanced calcification in hermatypic corals: new insights from light spectral responses. Front Mar Sci. 2:122. [Google Scholar]
- Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N.. 2018. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep. 8(1):16134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo C, et al. 2008. Blue light regulation of host pigment in reef-building corals. Mar Ecol Prog Ser. 364:97–106. [Google Scholar]
- D’Angelo C, et al. 2012. Locally accelerated growth is part of the innate immune response and repair mechanisms in reef-building corals as detected by green fluorescent protein (GFP)-like pigments. Coral Reefs. 31(4):1045–1056. [Google Scholar]
- De'ath G, Fabricius KE, Sweatman H, Puotinen M.. 2012. The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci U S A. 109(44):17995–17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarlo TM, et al. 2019. Acclimatization of massive reef-building corals to consecutive heatwaves. Proc Biol Sci. 286(1898):20190235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalvo MK, Estrada A, Sunagawa S, Medina M.. 2012. Transcriptomic responses to darkness stress point to common coral bleaching mechanisms. Coral Reefs. 31(1):215–228. [Google Scholar]
- Dougan KE, et al. 2020. Nutrient pollution and predation differentially affect innate immune pathways in the Coral Porites porites. Front Mar Sci. 7:563865. [Google Scholar]
- Dove S, Hoegh-Guldberg O, Ranganathan S.. 2001. Major colour patterns of reef building corals are due to a family of GFP like proteins. Coral Reefs 19:197–204. [Google Scholar]
- Drake JL, et al. 2013. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A. 110(10):3788–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SF, Bulina MY, Kelmanson IV, Bielawski JP, Matz MV.. 2006. Adaptive evolution of multicolored fluorescent proteins in reef-building corals. J Mol Evol. 62(3):332–339. [DOI] [PubMed] [Google Scholar]
- Fitt WK, et al. 2009. Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to short-term thermal stress: the host does matter in determining the tolerance of corals to bleaching. J Exp Mar Biol Ecol. 373(2):102–110. [Google Scholar]
- Fukami H, et al. 2008. Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PLoS One 3(9):e3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, et al. 2003. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem Biophys Res Commun. 304(1):11–17. [DOI] [PubMed] [Google Scholar]
- Gagan MK, et al. 2000. New views of tropical paleoclimates from corals. Quat Sci Rev. 19(1–5):45–64. [Google Scholar]
- Gagan MK, Dunbar GB, Suzuki A.. 2012. The effect of skeletal mass accumulation in Porites on coral Sr/Ca and δ18O paleothermometry. Paleoceanography. 27(1):n/a. [Google Scholar]
- Haas BJ, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi E, et al. 2013. Growth-rate influences on coral climate proxies tested by a multiple colony culture experiment. Earth Planet Sci Lett. 362:198–206. [Google Scholar]
- Helmkampf M, Bellinger MR, Geib SM, Sim SB, Takabayashi M.. 2019. Draft genome of the rice Coral Montipora capitata obtained from linked-read sequencing. Genome Biol Evol. 11(7):2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, et al. 2007. Coral reefs under rapid climate change and ocean acidification. Science 318(5857):1737–1742. [DOI] [PubMed] [Google Scholar]
- Hoeksema BW, Cairns S.. 2021. World List of Scleractinia. Available from: http://www.marinespecies.org/scleractinia. Accessed February 20, 2021.
- Huang da W, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Huang S, Kang M, Xu A.. 2017. HaploMerger2: rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly. Bioinformatics 33(16):2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, et al. 2020. Coral reef survival under accelerating ocean deoxygenation. Nat Clim Chang. 10(4):296–307. [Google Scholar]
- Hughes TP, et al. 2017. Global warming and recurrent mass bleaching of corals. Nature 543(7645):373–377. [DOI] [PubMed] [Google Scholar]
- Iguchi A, et al. 2012. Effects of acidified seawater on coral calcification and symbiotic algae on the massive coral Porites australiensis. Mar Environ Res. 73:32–36. [DOI] [PubMed] [Google Scholar]
- Immel F, et al. 2016. The shell of the invasive bivalve species Dreissena polymorpha: biochemical, elemental and textural investigations. PLoS One 11(5):e0154264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, et al. 2018. A simple role of coral-algal symbiosis in coral calcification based on multiple geochemical tracers. Geochim Cosmochim Acta 235:76–88. [Google Scholar]
- Inoue M, Tanimizu M.. 2008. Anthropogenic lead inputs to the western Pacific during the 20th century. Sci Total Environ. 406(1-2):123–130. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguti S. 1969. Effect of the green fluorescent pigment on the productivity of reef corals. Micronesica 5:313. [Google Scholar]
- Kawakubo Y, Alibert C, Yokoyama Y.. 2017. A reconstruction of subtropical Western North Pacific SST variability back to 1578, based on a Porites Coral Sr/Ca record from the Northern Ryukyus, Japan. Paleoceanography 32(12):1352–1370. [Google Scholar]
- Kelly LA, Heintz T, Lamb JB, Ainsworth TD, Willis BL.. 2016. Ecology and pathology of novel plaque-like growth anomalies affecting a reef-building coral on the Great Barrier Reef. Front Mar Sci. 3:151. [Google Scholar]
- Kelmanson IV, Matz MV.. 2003. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: faviida). Mol Biol Evol. 20(7):1125–1133. [DOI] [PubMed] [Google Scholar]
- Kenkel CD, et al. 2011. Development of gene expression markers of acute heat-light stress in reef-building corals of the genus Porites. PLoS One 6(10):e26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ.. 2010. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One 5(7):e11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, Fukami H, Benzoni F, Huang D.. 2016. The new systematics of scleractinia: integrating molecular and morphological evidence. In: Goffredo S, Dubinsky Z, editors. The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters. Cham (Switzerland): Springer International Publishing. p. 41–59. [Google Scholar]
- Kozlowski LP. 2016. IPC—isoelectric point calculator. Biol Direct. 11(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Louis YD, Bhagooli R, Kenkel CD, Baker AC, Dyall SD.. 2017. Gene expression biomarkers of heat stress in scleractinian corals: promises and limitations. Comp Biochem Physiol C Toxicol Pharmacol. 191:63–77. [DOI] [PubMed] [Google Scholar]
- Loya Y, et al. 2001. Coral bleaching: the winners and the losers. Ecol Lett. 4(2):122–131. [Google Scholar]
- Marin F, Luquet G.. 2007. Unusually acidic proteins in biomineralization. In: Bäuerlein E, editor. Handbook of biomineralization. Weinheim, Germany: WILEY-VCH Verlag GmbH & Co. KGaA. p. 273–290. [Google Scholar]
- Marshall PA, Baird AH.. 2000. Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19(2):155–163. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10. [Google Scholar]
- Matz MV, Lukyanov KA, Lukyanov SA.. 2002. Family of the green fluorescent protein: journey to the end of the rainbow. Bioessays 24(10):953–959. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch M, Falter J, Trotter J, Montagna P.. 2012. Coral resilience to ocean acidification and global warming through pH up-regulation. Nat Clim Change 2(8):623–627. [Google Scholar]
- Mydlarz LD, Couch CS, Weil E, Smith G, Harvell CD.. 2009. Immune defenses of healthy, bleached and diseased Montastraea faveolata during a natural bleaching event. Dis Aquat Organ. 87(1–2):67–78. [DOI] [PubMed] [Google Scholar]
- Mydlarz LD, Holthouse SF, Peters EC, Harvell CD.. 2008. Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS One 3(3):e1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. 2017. Mass coral bleaching event in Sekisei lagoon observed in the summer of 2016. J Jpn Coral Reef Soc. 19(1):29–40. [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. London: George Allen & Unwin.
- Olde K, et al. 2015. Geochemical and palynological sea-level proxies in hemipelagic sediments: a critical assessment from the Upper Cretaceous of the Czech Republic. Palaeogeogr Palaeoclimatol Palaeoecol. 435:222–243. [Google Scholar]
- Palmer CV, Bythell JC, Willis BL.. 2011. A comparative study of phenoloxidase activity in diseased and bleached colonies of the coral Acropora millepora. Dev Comp Immunol. 35(10):1098–1101. [DOI] [PubMed] [Google Scholar]
- Palmer CV, Bythell JC, Willis BL.. 2012. Enzyme activity demonstrates multiple pathways of innate immunity in Indo-Pacific anthozoans. Proc Biol Sci. 279(1743):3879–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CV, Modi CK, Mydlarz LD.. 2009. Coral fluorescent proteins as antioxidants. PLoS One. 4(10):e7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786. [DOI] [PubMed] [Google Scholar]
- Prada C, et al. 2016. Empty Niches after extinctions increase population sizes of modern corals. Curr Biol. 26(23):3190–3194. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramos-Silva P, et al. 2013. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 30(9):2099–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR.. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins SJ, et al. ; ReFuGe2020 Consortium. 2019. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat Microbiol. 4(12):2090–2100. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lanetty M, Harii S, Hoegh-Guldberg O.. 2009. Early molecular responses of coral larvae to hyperthermal stress. Mol Ecol. 18(24):5101–5114. [DOI] [PubMed] [Google Scholar]
- Romano SL, Palumbi SR.. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271(5249):640–642. [Google Scholar]
- Roth MS, Deheyn DD.. 2013. Effects of cold stress and heat stress on coral fluorescence in reef-building corals. Sci Rep. 3:1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Latz MI, Goericke R, Deheyn DD.. 2010. Green fluorescent protein regulation in the coral Acropora yongei during photoacclimation. J Exp Biol. 213(Pt 21):3644–3655. [DOI] [PubMed] [Google Scholar]
- Salih A, Larkum A, Cox G, Kühl M, Hoegh-Guldberg O.. 2000. Fluorescent pigments in corals are photoprotective. Nature 408(6814):850–853. [DOI] [PubMed] [Google Scholar]
- Seneca FO, et al. 2010. Patterns of gene expression in a scleractinian coral undergoing natural bleaching. Mar Biotechnol (NY) 12(5):594–604. [DOI] [PubMed] [Google Scholar]
- Shen GT, Boyle EA, Lea DW.. 1987. Cadmium in corals as a tracer of historical upwelling and industrial fallout. Nature 328(6133):794–796. [Google Scholar]
- Sheridan C, et al. 2014. Sedimentation rapidly induces an immune response and depletes energy stores in a hard coral. Coral Reefs 33(4):1067–1076. [Google Scholar]
- Shinzato C, Inoue M, Kusakabe M.. 2014. A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One 9(1):e85182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2021. Eighteen coral genomes reveal the evolutionary origin of Acropora strategies to accommodate environmental changes. Mol Biol Evol. 38(1):16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323. [DOI] [PubMed] [Google Scholar]
- Shumaker A, et al. 2019. Genome analysis of the rice coral Montipora capitata. Sci Rep. 9(1):2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P.. 1996. –2010. RepeatMasker Open-3.0. Available from: http://www.repeatmasker.org. Accessed December 15, 2015.
- Solomon S; Intergovernmental Panel on Climate Change Working Group I. 2007. Climate change 2007: The physical Science Basis: Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, New York: Cambridge University Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server Issue):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggett DJ, et al. 2013. Light availability determines susceptibility of reef building corals to ocean acidification. Coral Reefs 32(2):327–337. [Google Scholar]
- Suzuki T, Blank V, Sesay JS, Crawford DR.. 2001. Maf genes are involved in multiple stress response in human. Biochem Biophys Res Commun. 280(1):4–8. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 18(3):164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, et al. 2018. Biochemical characterization of the skeletal matrix of the massive coral, Porites australiensis—the saccharide moieties and their localization. J Struct Biol. 203(3):219–229. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N.. 2016. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS One 11(6):e0156424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, et al. 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46(5):2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Water J, et al. 2018. Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef-building coral. Mol Ecol. 27(4):1065–1080. [DOI] [PubMed] [Google Scholar]
- van de Water JA, et al. 2015. The coral immune response facilitates protection against microbes during tissue regeneration. Mol Ecol. 24(13):3390–3404. [DOI] [PubMed] [Google Scholar]
- van Woesik R, Sakai K, Ganase A, Loya Y.. 2011. Revisiting the winners and the losers a decade after coral bleaching. Mar Ecol Prog Ser. 434:67–76. [Google Scholar]
- Voolstra CR, et al. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 7(1):17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra CR, et al. 2009. Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genomics 10:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurture GW, et al. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33(14):2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall CB, et al. 2018. The effects of environmental history and thermal stress on coral physiology and immunity. Mar Biol. 165(3):56. [Google Scholar]
- Wang X, et al. 2017. Draft genomes of the corallimorpharians Amplexidiscus fenestrafer and Discosoma sp. Mol Ecol Resour. 17(6):e187–e195. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2017. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters KM, Sontag RL, Weber TJ.. 2013. Hepatic leukemia factor promotes resistance to cell death: implications for therapeutics and chronotherapy. Toxicol Appl Pharmacol. 268(2):141–148. [DOI] [PubMed] [Google Scholar]
- Wilkinson C. 2008. Status of coral reefs of the world. Townsville (Australia): Global Coral Reef Monitoring Network and Reef and Rainforest Research Center.
- Ying H, et al. 2018. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 19(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, et al. 2019. The whole-genome sequence of the Coral Acropora millepora. Genome Biol Evol. 11(5):1374–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data regarding the Porites australiensis genome assembly have been registered at GenBank under the BioProject accession PRJDB4187. The genome assembly has been deposited in the DNA DataBank of Japan/European Nucleotide Archive/GenBank under accession numbers BOPM01000001–BOPM01004983. The P. australiensis genome assembly and gene prediction data are also available from https://drive.google.com/drive/folders/1xZCIAHLmuCcdIQlvDHbN7GGJzxp6VIs0. RNA-Seq sequencing data are available from DRA submission number DRA011516.